A 10-kD cytoplasmic protein, Ssl3829, plays an important role in NDH-1 assembly by accumulating subunit maturation factor and assembly intermediate in a cyanobacterium.
Abstract
Despite significant progress in clarifying the subunit compositions and functions of the multiple NDH-1 complexes in cyanobacteria, the assembly factors and their roles in assembling these NDH-1 complexes remain elusive. Two mutants sensitive to high light for growth and impaired in NDH-1-dependent cyclic electron transport around photosystem I were isolated from Synechocystis sp. strain PCC 6803 transformed with a transposon-tagged library. Both mutants were tagged in the ssl3829 gene encoding an unknown protein, which shares significant similarity with Arabidopsis (Arabidopsis thaliana) CHLORORESPIRATORY REDUCTION7. The ssl3829 product was localized in the cytoplasm and associates with an NDH-1 hydrophilic arm assembly intermediate (NAI) of about 300 kD (NAI300) and an NdhI maturation factor, Slr1097. Upon deletion of Ssl3829, the NAI300 complex was no longer visible on gels, thereby impeding the assembly of the NDH-1 hydrophilic arm. The deletion also abolished Slr1097 and consequently reduced the amount of mature NdhI in the cytoplasm, which repressed the dynamic assembly process of the NDH-1 hydrophilic arm because mature NdhI was essential to stabilize all functional NAIs. Therefore, Ssl3829 plays an important role in the assembly of the NDH-1 hydrophilic arm by accumulating the NAI300 complex and Slr1097 protein in the cytoplasm.
Cyanobacterial NDH-1 complexes participate in a variety of bioenergetic reactions, including respiration, cyclic electron transport around PSI, and CO2 acquisition (Ogawa, 1991; Mi et al., 1992; Ohkawa et al., 2000). They are predominantly, if not totally, located in the thylakoid membrane (Ohkawa et al., 2001, 2002; Zhang et al., 2004; Xu et al., 2008; Battchikova et al., 2011a). Structurally, the cyanobacterial NDH-1 complexes closely resemble energy-converting complex I in eubacteria and the mitochondrial respiratory chain, regardless of the absence of homologs of three subunits in cyanobacterial genomes that constitute the catalytically active core of complex I (Friedrich et al., 1995; Friedrich and Scheide, 2000; Arteni et al., 2006). Over the past few years, significant achievements have been made in resolving the subunit compositions and functions of the multiple NDH-1 complexes in several cyanobacterial strains (for review, see Battchikova and Aro, 2007; Ogawa and Mi, 2007; Ma, 2009; Battchikova et al., 2011b; Ma and Ogawa, 2015). Four types of NDH-1, NDH-1L, NDH-1L′, NDH-1MS, and NDH-1MS′, have been predicted in the cyanobacterium Synechocystis sp. strain PCC 6803 (hereafter Synechocystis 6803) based on results of reverse genetics experiments (Ohkawa et al., 2000; Shibata et al., 2001; Battchikova and Aro, 2007). Proteomics studies have confirmed the presence of NDH-1L, NDH-1MS, and NDH-1MS′ in either Synechocystis 6803 or Thermosynechococcus elongatus (Herranen et al., 2004; Zhang et al., 2005; Xu et al., 2008; Wulfhorst et al., 2014), although NDH-1L′ is not yet identified in cyanobacteria. Furthermore, Bernát et al. (2011) demonstrated that all four types are involved in NDH-1-dependent cyclic electron transport around PSI (NDH-CET). NDH-CET plays an important role in coping with various environmental stresses regardless of its elusive mechanism. For example, this function can greatly alleviate high light-sensitive growth phenotypes (Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015). Therefore, a high-light strategy can help in identifying the proteins that affect NDH-CET activity.
Proteomics and electron microscopy analyses revealed the presence of two major NDH-1 complexes in cyanobacteria: an L-shaped NDH-1L complex and a U-shaped NDH-1MS complex, with molecular masses of about 460 and 490 kD, respectively (Herranen et al., 2004; Zhang et al., 2004, 2005; Arteni et al., 2006). Each of them consists of three parts: a membrane-embedded arm, a hydrophilic connecting arm, and a mysterious catalytic domain (Prommeenate et al., 2004; Ma and Ogawa, 2015). Recently, NdhS was suggested to be included in this mysterious catalytic domain because it may be a ferredoxin-binding subunit (Battchikova et al., 2011a; Ma and Ogawa, 2015). Although the membrane arm included in NDH-1L and NDH-1MS shares five common hydrophobic subunits (NdhA to NdhC, NdhE, and NdhG) and may be involved in proton pumping and plastoquinone binding, other subunit compositions are significantly different. In addition to these five common subunits, the membrane arm of NDH-1L contains NdhD1, NdhF1, NdhP, and NdhQ (Zhang et al., 2005, 2014; Nowaczyk et al., 2011; Wulfhorst et al., 2014; Ma and Ogawa, 2015; Zhao et al., 2015), and that of NDH-1MS includes NdhD3, NdhF3, CupA, and CupS (Herranen et al., 2004; Zhang et al., 2005). In spite of this difference, NDH-1L and NDH-1MS contain an identical hydrophilic arm that consists of eight subunits (NdhH to NdhO; Prommeenate et al., 2004; Battchikova et al., 2005; Zhao et al., 2014; He et al., 2016) carrying redox centers, FMN, and Fe-S clusters, which is extrapolated from bacterial complex I (Sazanov and Hinchliffe, 2006; Efremov et al., 2010; Baradaran et al., 2013). The subunit compositions of the hydrophilic arm are also identical to that from the chloroplast NDH-1 complex (for review, see Battchikova et al., 2011b; Ifuku et al., 2011; Peng et al., 2011b; Ma and Ogawa, 2015).
Despite the progress made in revealing the structure of multiple NDH-1 complexes in several cyanobacterial strains, few reports have focused on the assembly of these complexes. Because of the low abundance and fragile nature of these NDH-1 complexes, it is hard to use biochemical approaches, such as pulse-chase labeling, to investigate the NDH-1 assembly process. In a complementary approach to biochemistry, forward or reverse genetics has identified a large number of assembly factors specific to an individual photosynthetic complex, including PSII and PSI (Nixon et al., 2010; Rochaix, 2011), resulting in the accumulation of knowledge on the assembly machinery and on the dynamics that regulate the process. Over the last decade, forward or reverse genetics studies have identified many nucleus-encoded assembly factors that are required for the assembly of the chloroplast NDH-1 hydrophilic arm, such as Arabidopsis (Arabidopsis thaliana) CHLORORESPIRATORY REDUCTION6 (CRR6) and CRR7 (for review, see Suorsa et al., 2009; Ifuku et al., 2011; Peng et al., 2011b). With the assistance of these assembly factors, a dynamic assembly process of the chloroplast NDH-1 hydrophilic arm has been suggested (Peng et al., 2012). Recently, a cyanobacterial CRR6 protein, Slr1097, also has been identified in Synechocystis 6803 and is found to be required for efficient assembly of the NDH-1 hydrophilic arm involving NdhI maturation (Dai et al., 2013). Except for Slr1097, however, no other assembly factors for the cyanobacterial NDH-1 complexes have been identified and characterized, even though homologous sequences of certain assembly factors for the chloroplast NDH-1 complex, such as CRR7 (Munshi et al., 2005), have been found in cyanobacterial genomes.
Here, we report the discovery of a cyanobacterial CRR7 protein, Ssl3829, in Synechocystis 6803 using a forward genetics strategy. The assembly factor was found to interact with an NDH-1 hydrophilic arm assembly intermediate (NAI) complex with molecular mass of about 300 kD (NAI300) and an NdhI maturation factor, Slr1097 (Dai et al., 2013), and its absence reduced the abundance of the NAI300 complex and Slr1097 protein to levels no longer detectable on gels. We further demonstrate an important role of Ssl3829 in the assembly process of the NDH-1 hydrophilic arm by accumulating NAI300 complex and Slr1097 protein in the cytoplasm of Synechocystis 6803.
RESULTS
Isolation of NDH-CET-Defective Mutants
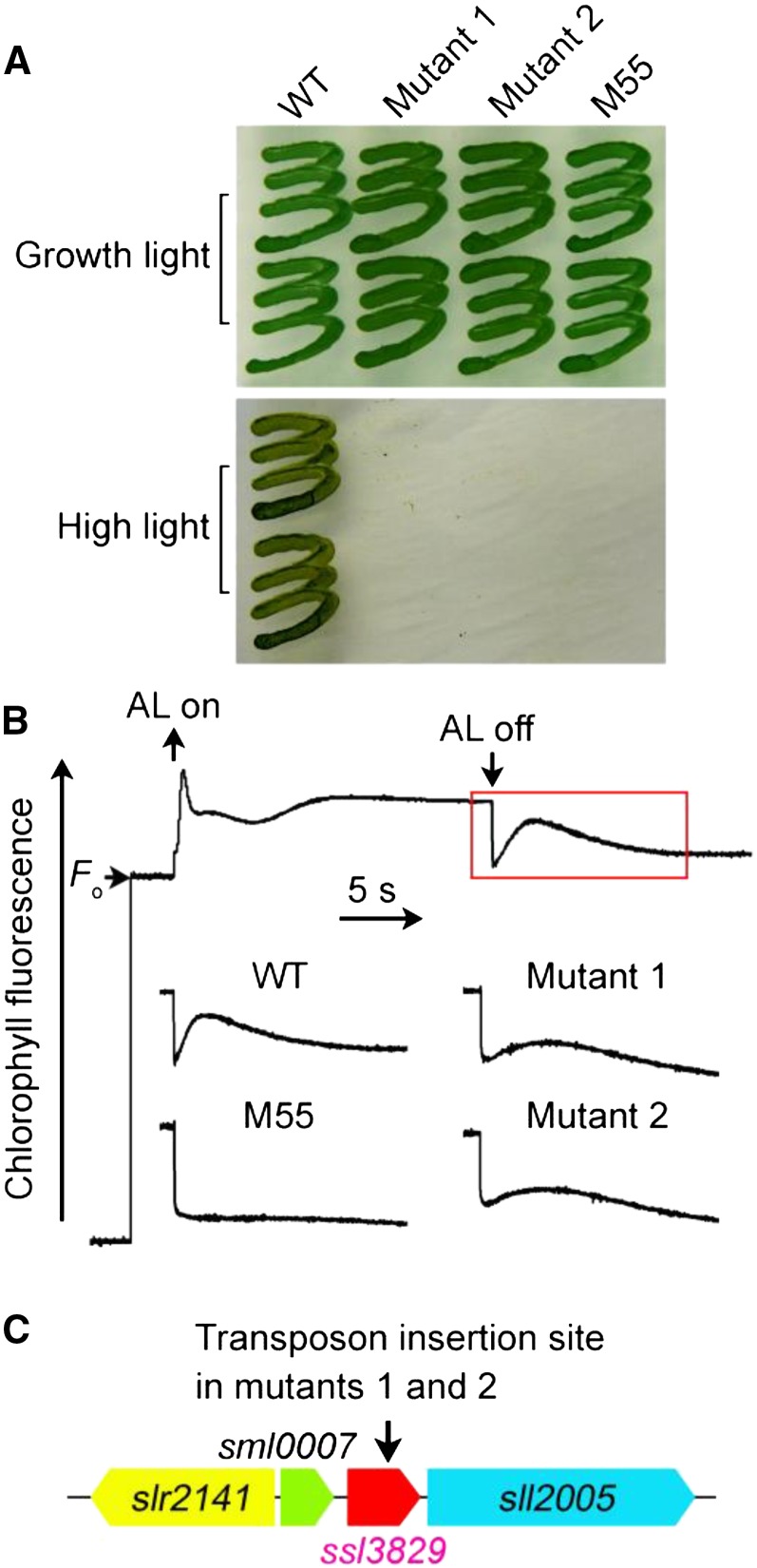
NDH-CET plays an important role in alleviating high-light stress in cyanobacteria (Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015). Therefore, upon exposure of cells to high light (about 200 µmol photons m−2 s−1), the growth of NDH-CET-defective mutants, such as the NdhB-deficient mutant (M55; Fig. 1A), is retarded significantly when compared with the wild type, regardless of similar growth under normal growth light irradiation. To screen the NDH-CET-defective mutants, we transformed wild-type cells with a transposon-bearing library, thus tagging and inactivating many genes randomly, and then cultured the mutant cells under high-light conditions. After analyzing the growth phenotypes of about 650 transposon-tagged mutant clones under conditions of normal light and high light, we successfully obtained 11 high-light-sensitive mutants. Among them, we isolated two NDH-1-less mutant strains, which were unable to grow on plates under high light but grew similar to the wild type under growth light conditions (Fig. 1A).
Figure 1.
Growth, NDH-CET activity, and transposon insertion site of high-light-sensitive mutants of Synechocystis 6803. A, Growth of the wild type (WT) and mutants under normal light (40 µmol photons m−2 s−1) and high light (200 µmol photons m−2 s−1). B, Analysis of NDH-CET activity using Chl fluorescence. The top curve shows a typical trace of Chl fluorescence in wild-type Synechocystis 6803. The Chl a concentration was adjusted to 10 µg mL−1 before measurement. Cells were exposed to actinic light (AL; 620 nm, 45 µmol photons m−2 s−1) for 30 s. AL was turned off, and the subsequent change in the Chl fluorescence level was recoded. The activity of NDH-CET in the wild type and mutants was compared from the height and the relative rate of postillumination increase in Chl fluorescence. C, Schematic with the arrow indicating the transposon insertion site in mutants 1 and 2 probed by PCR analysis using the primers listed in Supplemental Table S1.
To test whether the high-light-dependent phenotype of the two mutants resulted from defective NDH-CET, we monitored the postillumination rise in chlorophyll (Chl) fluorescence. This technique has been used extensively to monitor the NDH-CET activity in vivo in cyanobacteria (Mi et al., 1995; Deng et al., 2003; Ma and Mi, 2005; Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015; Gao et al., 2016) and higher plants (Burrows et al., 1998; Shikanai et al., 1998; Hashimoto et al., 2003; Wang et al., 2006; Peng et al., 2009, 2011a, 2012; Sirpiö et al., 2009; Yamamoto et al., 2011; Armbruster et al., 2013). Figure 1B shows that the activity of NDH-CET in both mutants was lower than that in the wild type as assessed by the height and the relative rate of postillumination rise in Chl fluorescence. These results suggested that NDH-CET was affected in mutants 1 and 2.
To identify the genes inactivated by transposon tagging, we analyzed the sites of transposon insertion in the two mutants. Sequencing of the PCR products revealed that both mutants were tagged in the same gene of unknown function (ssl3829; Fig. 1C). The transposon insertion occurred at position 1,267,815 of the Synechocystis 6803 genome (National Center for Biotechnology Information no. 16330313; Kaneko et al., 1996). This implies that deletion of ssl3829 affects NDH-CET activity.
Deletion of ssl3829 Impairs NDH-CET Activity
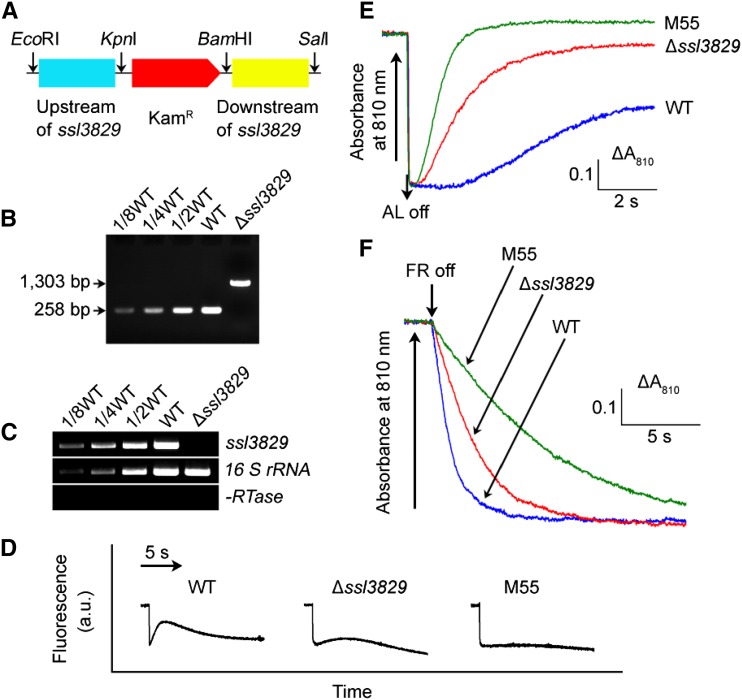
To verify that deletion of ssl3829 affects NDH-CET activity, we replaced the ssl3829 coding region using a kanamycin resistance marker (KamR; Fig. 2A). PCR analysis of the ssl3829 locus confirmed the complete segregation of the ∆ssl3829 mutant allele (Fig. 2B). Reverse transcription (RT)-PCR analysis of the ssl3829 gene in the ∆ssl3829 mutant demonstrated the absence of its transcript in the mutant (Fig. 2C). Furthermore, deletion of ssl3829 had no effect on the expression levels of its neighboring genes, slr2141, sml0007, and sll2005 (Fig. 1C), as deduced from the results of RT-PCR analysis (Supplemental Fig. S1).
Figure 2.
Deletion of ssl3829 and its effect on NDH-CET. A, Construction of the plasmid used to generate the ssl3829 deletion mutant (∆ssl3829). B, PCR segregation analysis of the ∆ssl3829 mutant using the ssl3829-G and ssl3829-H primer sequences (Supplemental Table S1). C, Transcript levels of ssl3829 in the wild-type (WT) and ∆ssl3829 strains. The transcript level of 16 S rRNA in each sample is shown as a control. The absence of contamination of DNA was confirmed by PCR without reverse transcriptase. D, Analysis of NDH-CET activity by Chl fluorescence. The experimental procedure was as in Figure 1B. M55 is an ndhB deletion mutant (ΔndhB); a.u., arbitrary units. E, Redox kinetics of P700 after termination of AL illumination (800 µmol photons m−2 s−1 for 30 s) under a background of FR light. The cells were illuminated by AL supplemented with FR light to store electrons in the cytoplasmic pool. After termination of AL illumination, P700+ was reduced transiently by electrons from the plastoquinone pool; thereafter, P700 was reoxidized by background FR light. The redox kinetics of P700 was recorded. The P700+ levels were standardized by their maximum levels attained by exposure to FR light. The reoxidation level of P700 was evaluated by the height and the relative rate of redox kinetics of P700. F, Kinetics of the P700+ rereduction in darkness after turning off FR light with a maximum at 720 nm in the presence of 10 µm 3-(3,4-dichlorophenyl)-1,1-dimethylurea in wild-type, ∆ssl3829, and M55 strains. The Chl a concentration was adjusted to 20 µg mL−1 before measurement, and curves are normalized to the maximal signal. The rereduction level of P700+ was assessed by the initial reduction rate of P700+.
As expected, the activity of NDH-CET, as measured by the postillumination rise in Chl fluorescence, was lower in ∆ssl3829 than in the wild type. The activity, however, remained relatively high when compared with that in M55 (Fig. 2D). A similar result was obtained by measuring the oxidation of P700 by far-red (FR) light after AL illumination. When AL was turned off after 30 s of illumination by AL (800 µmol photons m−2 s−1) supplemented with FR light, P700+ was transiently reduced by electrons from the plastoquinone pool and subsequently reoxidized by background FR light. The operation of the NDH-1 complexes, which transfer electrons from the reduced cytoplasmic pool to plastoquinone, hinders the reoxidation of P700 (Shikanai et al., 1998; Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015; Gao et al., 2016). The reoxidation of P700 in ∆ssl3829 was about 4 times faster than that in the wild type, as deduced from the half-time for reaching the maximum P700+ rise levels, although it was still relatively slow compared with the M55 mutant (Fig. 2E). We also measured NDH-CET by monitoring the reduction rate of P700+ in darkness after illumination of cells with FR light. The rereduction of P700+ was remarkably slower in ∆ssl3829 when compared with that in the wild type, although it was still relatively fast compared with the rate in the M55 mutant (Fig. 2F). Under high light intensities, ∆ssl3829 grew slower than the wild type despite similar growth under growth light (Supplemental Fig. S2). Based on the above results, we conclude that deletion of a cyanobacterial gene, ssl3829, impairs NDH-CET activity.
Ssl3829 Is Required for the Efficient Assembly of NDH-1 Complexes in the Thylakoid Membranes
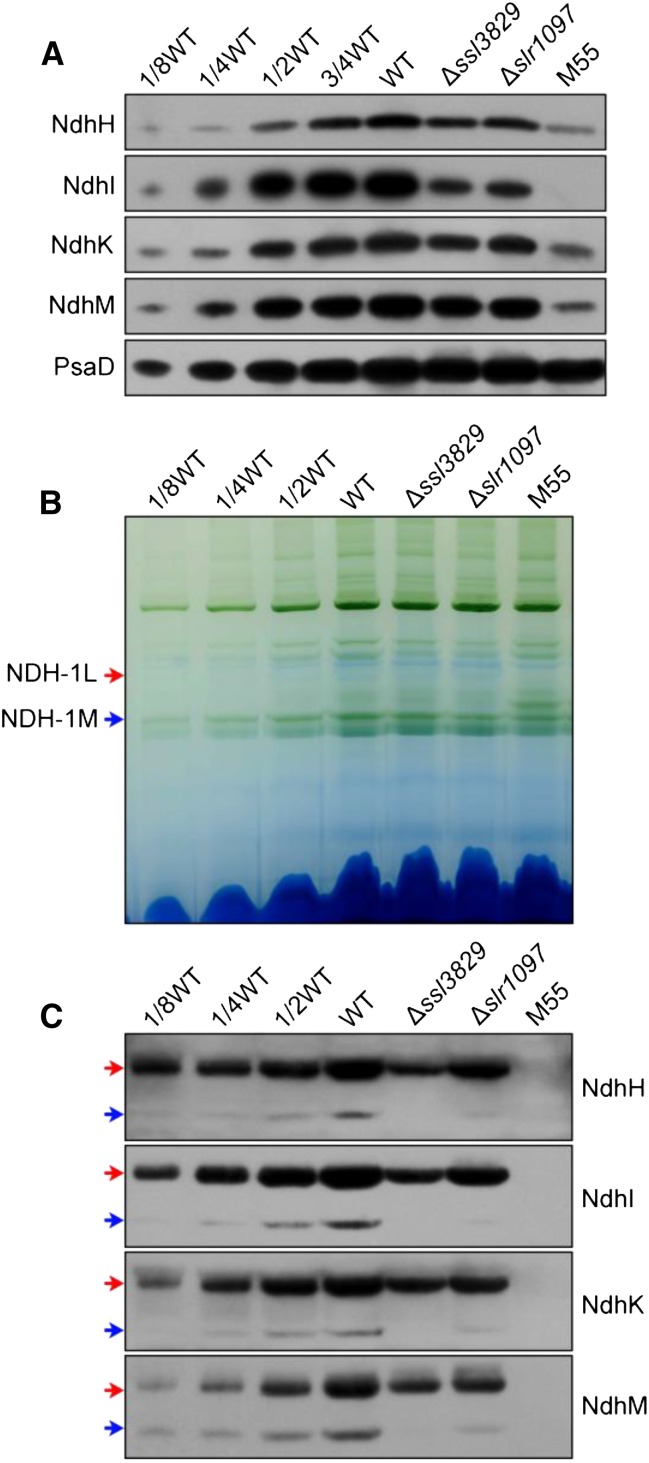
In Synechocystis 6803, NDH-1L and NDH-1M are two typical NDH-1 complexes observed from the blue native (BN)-PAGE gel of wild-type membranes (Herranen et al., 2004; Prommeenate et al., 2004), and the functional complex NDH-1L is involved in NDH-CET (Bernát et al., 2011). To uncover how the deletion of ssl3829 impairs NDH-CET activity, we separated NDH-1L and NDH-1M complexes from thylakoid membranes of the wild type, ∆ssl3829, and M55 strains and deduced the amount of these complexes from the density of NdhH, NdhI, NdhK, and NdhM bands visualized by the western analyses. Deletion of ssl3829 evidently decreased the protein abundance of total NDH-1 in the thylakoid membranes (Fig. 3A), and the deletion reduced components associated with NDH-1L by 50% to 75% and those associated with NDH-1M by at least 90% (Fig. 3, B and C). This indicates that ssl3829 deletion significantly impeded the accumulation of NDH-1L and NDH-1M in the thylakoid membranes, thereby impairing NDH-CET activity.
Figure 3.
Accumulation and assembly of NDH-1L and NDH-1M complexes from the thylakoid membrane of wild-type (WT), ∆ssl3829, ∆slr1097, and M55 strains. A, Immunodetection of Ndh subunits in thylakoid membranes from the wild type (including the indicated serial dilutions) and ∆ssl3829, ∆slr1097, and M55 mutants. Immunoblotting was performed with antibodies against hydrophilic Ndh subunits (NdhH, NdhI, NdhK, and NdhM). Lanes were loaded with thylakoid membrane proteins corresponding to 1 µg of Chl a (100%). At bottom, PsaD was used as a loading control. B, Thylakoid protein complexes isolated from the wild type and mutants were separated by BN-PAGE. Thylakoid membrane extract corresponding to 9 µg of Chl a was loaded onto each lane. The positions of NDH-1L and NDH-1M complexes are indicated by red and blue arrows, respectively. C, Protein complexes were electroblotted to a polyvinylidene difluoride membrane, and the membrane was cross-reacted with anti-NdhH, anti-NdhI, anti-NdhK, and anti-NdhM to probe the assembly of NDH-1L and NDH-1M complexes.
Ssl3829 Is a Cytoplasmic Protein Conserved in Phototrophs
To obtain insights into how ssl3829 deletion affected the assembly of NDH-1 complexes, we investigated the localization of the Ssl3829 protein. To perform this investigation, we constructed a mutant, WT-Ssl3829-YFP-His6 (hereafter referred as to WT-Ssl3829-YH), which has a yellow fluorescent protein (YFP)-His6 tag on the C terminus of Ssl3829 (Supplemental Fig. S3A). PCR analysis indicated a complete segregation of the tagged gene (Supplemental Fig. S3B). Western analysis of the total protein from the tagged strain and confocal analysis of the tagged cells confirmed the expression of Ssl3829-YH (Supplemental Fig. S3, C and D). The tagging did not influence the activity of NDH-1 complexes (Supplemental Fig. S3, E–G). The soluble and total membrane fractions were isolated from the tagged cells, and their purity was confirmed by western analysis using antibodies against Rubisco large subunit (RbcL) and D1, which cross-reacted with soluble and total membrane fractions, respectively (Zhang et al., 2009, 2012; Dai et al., 2013). The Ssl3829 protein as assessed by the protein abundance of GFP and His was detected only in the soluble fraction but not in the total membrane fraction, in contrast to the localization of the NDH-1 hydrophilic arm subunits, NdhH and NdhK, mainly in the total membrane fraction (Fig. 4). It is worthy of note that we did not observe the cleavage band of the fusion protein Ssl3829-YH on gels. Therefore, our results suggest that Ssl3829 protein is confined to the cytoplasm.
Figure 4.
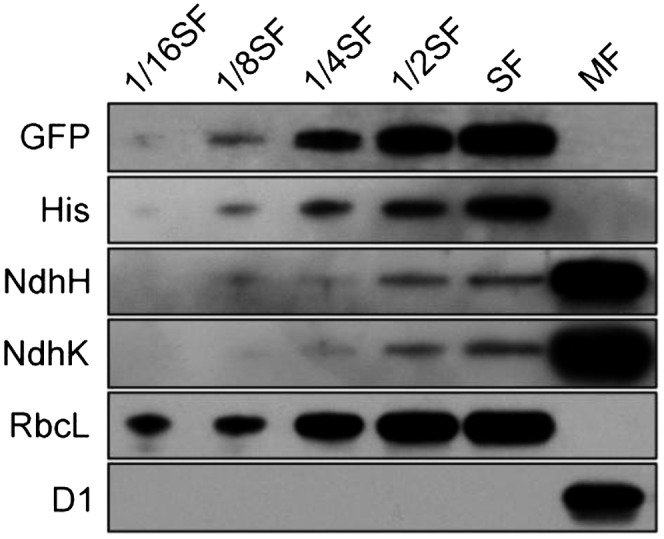
Localization of Ssl3829. The proteins of soluble cytoplasm and total membrane were isolated from the WT-Ssl3829-YH strain, and immunoblot analyses were performed with the indicated antibodies. Cytoplasmic and membrane proteins (10 µg) were loaded on each well (100%). Serial dilutions of cytoplasmic proteins are indicated, and detection of RbcL was used as the loading control. SF and MF represent soluble and total membrane fractions, respectively.
As shown by Munshi et al. (2005), Ssl3829 is a homolog of CRR7 (Supplemental Fig. S4), and the homologs are present in phototrophic organisms but not in Chlamydomonas reinhardtii, which lacks the photosynthetic ndh genes (Maul et al., 2002). This fact, and the localization of Ssl3829 and CRR7 in the cytoplasm (Fig. 4) and stroma (Peng et al., 2010), respectively, suggest that Ssl3829 might play a similar role to CRR7 as a factor essential to the assembly of the NDH-1 hydrophilic arm, thereby influencing the assembly of NDH-1 complexes in the thylakoid membranes.
Ssl3829 Associates with an NAI300 Complex
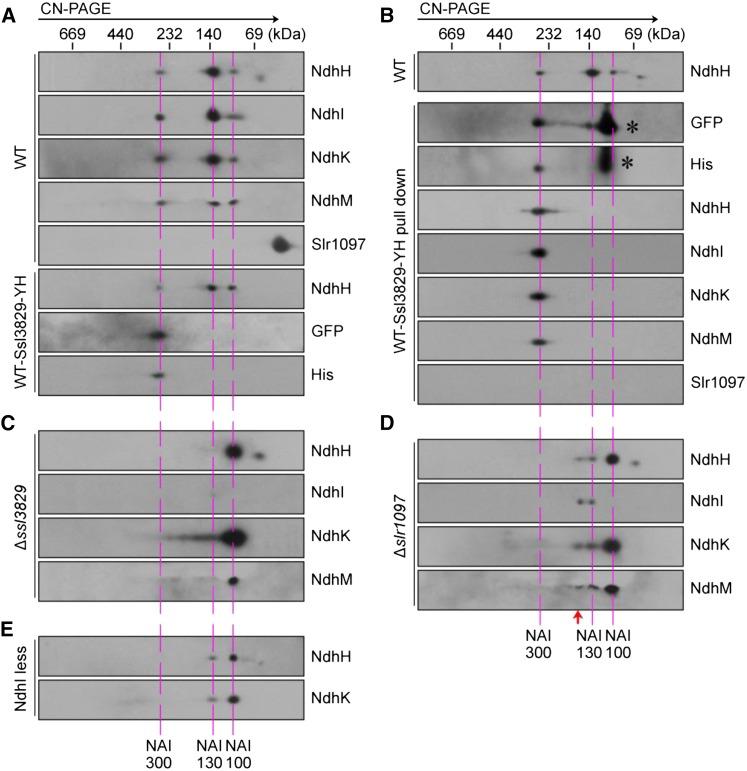
In higher plants, the assembly process of the NDH-1 hydrophilic arm is mainly involved in a dynamic transition of several NAIs with the aid of various assembly factors (Peng et al., 2012). To test whether these NAIs also were present in cyanobacteria, we used clear native (CN)-PAGE to separate the protein complexes from the cytoplasm of the wild-type and WT-Ssl3829-YH strains. Subsequent second-dimension SDS-PAGE and immunoblot analysis detected several complexes that included NdhH, NdhI, NdhK, and NdhM and had molecular masses of about 300, 130, and 100 kD, respectively (Fig. 5A). Recently, He et al. (2016) also detected these three complexes from the cytoplasm of the wild-type strain, although there are differences in the subunit compositions identified in their complexes from those in our complexes. Based on their apparent molecular masses, we designated them as NAI300, NAI130, and NAI100, respectively (Fig. 5A). We further found that the Ssl3829 protein was present only in the NAI300 complex, as judged by the trace of GFP and His proteins (Fig. 5A). This suggests that Ssl3829 may associate with the NAI300 complex.
Figure 5.
Formation of NAIs in the cytoplasm of various mutants. Cytoplasmic protein complexes isolated from wild-type (WT) and WT-Ssl3829-YH (A), ∆ssl3829 (C), ∆slr1097 (D), and NdhI-less (E) strains and purified from the WT-Ssl3829-YH using an Ni2+ column (B) were separated by CN-PAGE followed by two-dimensional SDS-PAGE. Proteins were immunodetected with two antibodies against NdhH and NdhK or the specific antibodies indicated. Dashed lines indicate the positions of NAIs, and the red arrow indicates the position of a new complex of about 150 kD produced by Slr1097 deletion. Asterisks represent free Ssl3829-YH proteins.
To consolidate this possibility, we purified the NAI300 complex from the solubilized cytoplasm of the WT-Ssl3829-YH strain using Ni2+ affinity chromatography. As expected, the NAI300 complex but not the NAI130 and NAI100 complexes was isolated successfully (Fig. 5B). Liquid chromatography-tandem mass spectrometry analysis of the purified NAI300 complex revealed the presence of Ssl3829 protein and several NDH-1 hydrophilic arm subunits, NdhH to NdhK and NdhM, in this complex (Table I; Supplemental Data Set S1). Taking all these results together, we conclude that Ssl3829 protein associates with the NAI300 complex.
Table I. Summary of the NDH-1 subunits and assembly factor identified from Q-Exactive mass analysis of the purified NAI300 complex samples.
The complete list of proteins identified in the purified NAI300 complex samples can be found in Supplemental Data Set S1.
| Open Reading Frame | Protein Name | Mascot Score | Protein Match | Coverage (%) |
|---|---|---|---|---|
| slr0261 | NdhH | 82.7 | 4 | 14.4 |
| sll0520 | NdhI | 84.3 | 3 | 19.7 |
| slr1281 | NdhJ | 34.8 | 1 | 6.7 |
| slr1280 | NdhK | 43.7 | 2 | 13.3 |
| slr1623 | NdhM | 49.9 | 2 | 14.9 |
| ssl3829 | Ssl3829 | 52.0 | 2 | 35.3 |
Ssl3829 Is Required to Accumulate the NAI300 Complex
To examine the effect of Ssl3829 on the accumulation of the NAI300 complex, we analyzed the cytoplasmic protein complexes isolated from wild-type and ∆ssl3829 strains. The results showed that deletion of ssl3829 decreased the abundance of the NAI300 complex to levels no longer detectable on a gel (Fig. 5C). We then compared the amount of several NDH-1 hydrophilic arm subunits in the cytoplasm of wild-type and ∆ssl3829 strains. As expected, deletion of ssl3829 significantly increased the components of the NDH-1 hydrophilic arm in the cytoplasm when compared with the wild type, as deduced from the abundance of NdhH, NdhK, and NdhM proteins (Fig. 6). These results indicate that Ssl3829 is required to accumulate the NAI300 complex in the cytoplasm. Thus, Ssl3829 is involved in the assembly of the NDH-1 hydrophilic arm in the cytoplasm via accumulation of the NAI300 complex.
Figure 6.
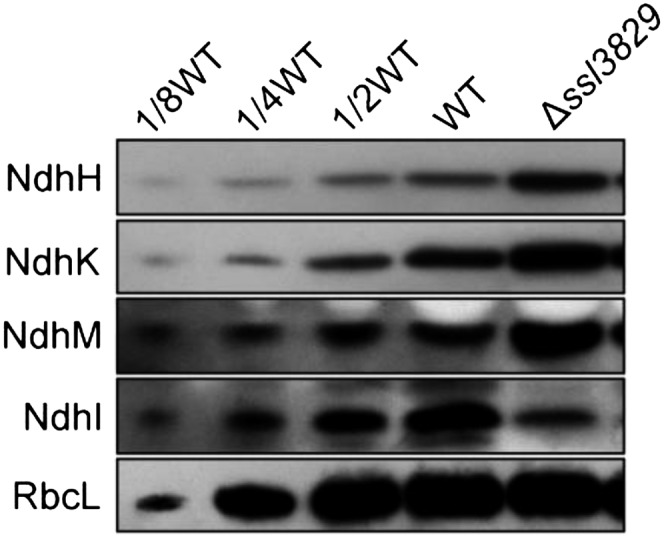
Abundance of several NDH-1 hydrophilic arm subunits in the soluble fractions. Cytoplasmic proteins were isolated from the wild-type (WT) and ∆ssl3829 strains, and immunoblot analysis was performed with the indicated antibodies. Cytoplasmic proteins (50 µg) were loaded on each well (100%), and RbcL was detected as the loading control.
Ssl3829 Interacts with Slr1097
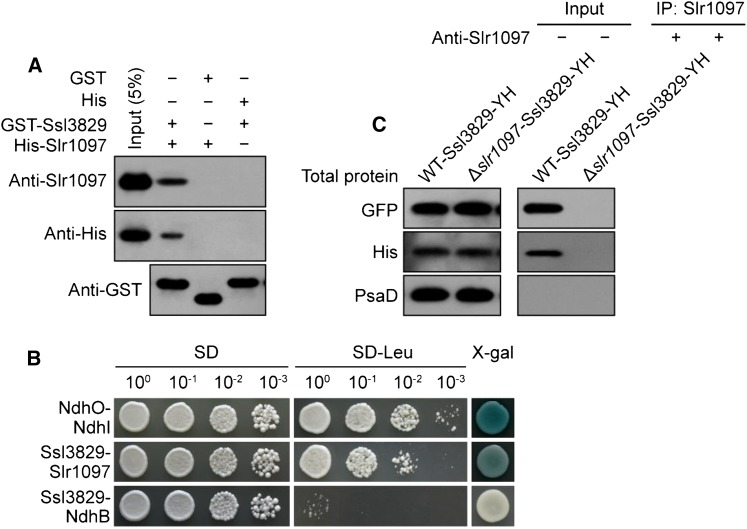
Deletion of ssl3829 decreased the amount of NdhI in the cytoplasm significantly, although the deletion increased the amount of NdhH, NdhK, and NdhM considerably (Fig. 6). To clarify why the deletion of ssl3829 exclusively decreased the amount of NdhI in the cytoplasm, we determined the interactions of Ssl3829 with NdhI and its maturation factor, Slr1097 (Dai et al., 2013), using a glutathione S-transferase (GST) pull-down strategy. A protein-protein interaction was found only between Ssl3829 and Slr1097 (Fig. 7A) but not between Ssl3829 and NdhI (Supplemental Fig. S5). The interaction was reinforced by the results of yeast two-hybrid (Fig. 7B) and coimmunoprecipitation (Fig. 7C) assays. All these results support the view that Ssl3829 interacts with Slr1097.
Figure 7.
Interaction of Ssl3829 with Slr1097. A, GST pull-down assay shows the interaction of Ssl3829 with Slr1097. The expressed proteins were mixed and incubated with Glutathione Sepharose 4B beads on a rotating shaker at 4°C overnight. After washing, the interaction of GST-Ssl3829 with His-Slr1097 was detected using the antibodies against Slr1097 and His. The interactions of GST (without Ssl3829) with His-Slr1097 and of His (without Slr1097) with GST-Ssl3829 were used as negative controls. The left lane represents an input (5%) control. B, Ssl3829 and Slr1097 were separately constructed into bait and prey vectors and transformed into the yeast (Saccharomyces cerevisiae) strain EGY48. Transformed yeast was diluted and dropped onto synthetic dropout (SD) medium, Leu-negative (−Leu) SD medium, or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid (X-gal) medium. The interactions of NdhO-NdhI and Ssl3829-NdhB were assayed as positive and negative controls, respectively. C, Coimmunoprecipitation assay of the interaction of Ssl3829 with Slr1097. Total proteins from the WT-Ssl3829-YH and ∆slr1097-Ssl3829-YH strains were incubated with Protein A Sepharose-coupled anti-Slr1097 antiserum. The immunoprecipitates were probed with specific antibodies, as indicated at left.
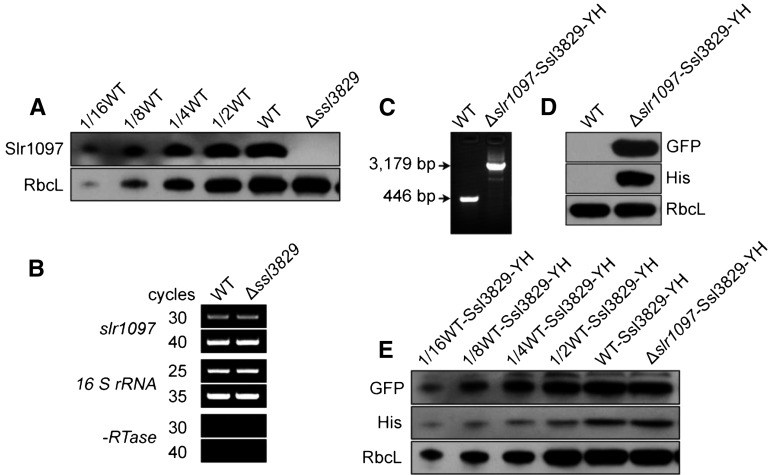
We assume that the interaction of Ssl3829 with Slr1097 may be required to stabilize the NdhI maturation factor, Slr1097, thereby influencing the amount of NdhI in the cytoplasm. To test this hypothesis, we analyzed the amount of Slr1097 protein accumulated in the cytoplasm from the wild-type and ∆ssl3829 strains. Deletion of ssl3829 resulted in a complete absence of Slr1097 in the cytoplasm (Fig. 8A), but the deletion did not influence the transcript levels of slr1097 (Fig. 8B). Conversely, deletion of slr1097 did not influence the accumulated levels of Ssl3829 in the cytoplasm, as judged by the amount of GFP and His proteins (Fig. 8, C–E). This indicates that Ssl3829 is required to stabilize Slr1097, an NdhI maturation factor, in the cytoplasm. Thus, deletion of ssl3829 destabilizes Slr1097 and consequently decreases the amount of mature NdhI in the cytoplasm.
Figure 8.
Relationship between Ssl3829 and Slr1097. A, Western analysis of Slr1097 from the wild-type (WT; including indicated serial dilutions) and ∆ssl3829 strains. Lanes were loaded with the cytoplasmic proteins corresponding to 50 µg (100%). RbcL was used as a loading control. B, RT-PCR analysis of slr1097 from the wild-type and ∆ssl3829 strains. The transcript level of 16 S rRNA in each sample is shown as a control. The absence of contamination of DNA was confirmed by PCR without reverse transcriptase. C, PCR segregation analysis of ssl3829-yfp-his6 from the wild-type and Δslr1097-Ssl3829-YH strains using the ssl3829-yfp-his6-E and -F primer sequences (Supplemental Table S1). D, Western analysis of proteins from the wild-type and Δslr1097-Ssl3829-YH strains using GFP and His antibodies. Total protein corresponding to 1 µg of Chl a was loaded onto each lane, and RbcL was detected as a loading control. E, Western analysis of Ssl3829 from the wild-type and Δslr1097 strains having Ssl3829-YH. Lanes were loaded with the cytoplasmic proteins corresponding to 50 µg (100%). RbcL was used as a loading control.
Slr1097 Stabilizes the NAIs via NdhI
Although Slr1097 was not detected in any NAIs, NdhI was present in the NAI300 and NAI130 complexes (Fig. 5A). Therefore, the absence of Slr1097 caused by ssl3829 deletion may influence the rate of formation of these two NAI complexes by decreasing the amount of mature NdhI in the cytoplasm. To explore this possibility, we analyzed the cytoplasmic protein complexes isolated from the wild-type and ∆slr1097 strains. Deletion of slr1097 reduced the abundance of the NAI300 complex to levels no longer detectable on a gel and significantly decreased the accumulation of the NAI130 complex (Fig. 5D). We then analyzed the cytoplasmic protein complexes isolated from the wild type and NdhI-less mutant (Supplemental Fig. S6, A–C). The allele segregation experiment of the NdhI-less mutant has been made for at least 2 years, but we were unable to achieve its complete segregation (Supplemental Fig. S6, B and C), just like the ndhI deletion mutant of He et al. (2016), thus suggesting that the ndhI gene might be necessary for the survival of Synechocystis 6803. Although the exact reason why the ndhI deletion mutant was not completely segregated remains elusive, the amount of NdhI was reduced to less than half the wild-type value in the NdhI-less mutant, similar to the amount in the Δslr1097 mutant (Fig. 3A; Supplemental Fig. S6C). The partial deletion of ndhI in the NdhI-less mutant caused the NAI300 complex to be no longer visible on a gel and significantly reduced the amount of the NAI130 complex (Fig. 5E), in line with the result with the ∆slr1097 mutant (Fig. 5D). Taking all these results together, we conclude that Slr1097 stabilizes the NAI300 and NAI130 complexes through involvement in the maturation process of NdhI in the cytoplasm. This indicates that Ssl3829 is involved in the assembly of the NDH-1 hydrophilic arm in the cytoplasm also through stabilizing Slr1097.
DISCUSSION
Distinct Role of Ssl3829 and Its Counterpart CRR7 in NDH-1 Hydrophilic Arm Assembly
Over the past few years, numerous assembly factors that are mainly involved in assembling the NDH-1 hydrophilic arm have been discovered and characterized in higher plants (Suorsa et al., 2009; Peng et al., 2011b, 2012). However, such assembly factors are still poorly understood in cyanobacteria, although we recently identified an assembly factor, Slr1097, in Synechocystis 6803 (Dai et al., 2013). Slr1097 plays a similar role to CRR6 in stabilizing NdhI in the cytoplasm, which possibly assists the assembly of the NDH-1 hydrophilic arm (Peng et al., 2012; Dai et al., 2013). Here, we successfully identified another assembly factor, Ssl3829 (Figs. 1–4), a counterpart of CRR7 in cyanobacteria (Supplemental Fig. S4), and found that it plays an important role in assembling the NDH-1 hydrophilic arm by accumulating the NAI300 complex (Figs. 5 and 6) and Slr1097 protein in the cytoplasm (Figs. 5, 7, and 8).
In higher plants, CRR7 does not interact with any NAIs, and its absence did not influence the stability of any NAIs in the stroma (Peng et al., 2012). Therefore, CRR7 is suggested to be involved in the final step of NDH-1 hydrophilic arm assembly, in which the fully assembled NAI, including NdhN, is inserted into thylakoids (Peng et al., 2012). By contrast, in cyanobacteria, Ssl3829 interacts with the NAI300 complex, and its absence caused the complex to be no longer visible on gels (Fig. 5; Table I; Supplemental Data Set S1). This suggests the involvement of Ssl3829 in an earlier step of NDH-1 hydrophilic arm assembly. Therefore, Ssl3829 and its counterpart CRR7 may be involved in different steps of NDH-1 hydrophilic arm assembly.
Furthermore, CRR7 does not interact with CRR6 (Peng et al., 2012), and its deletion did not influence the stability of CRR6 (Peng et al., 2010, 2012), which is required for the biogenesis of NdhI (Peng et al., 2012). Therefore, the deletion does not influence the amount of NdhI accumulated in the stroma (Peng et al., 2012). By contrast, Ssl3829 interacts with Slr1097, a counterpart of CRR6 in cyanobacteria (Dai et al., 2013), and its deletion completely abolished the accumulation of Slr1097 in the cytoplasm (Fig. 6). Consequently, the deletion almost completely abolished the accumulation of NdhI in the cytoplasm (Fig. 6). The decrease in mature NdhI caused by ssl3829 deletion destabilized the NAIs (Fig. 5), thereby influencing the assembly of the NDH-1 hydrophilic arm in the cytoplasm. Based on the above results, we suggest that there is a distinct role of Ssl3829 and its counterpart CRR7 during the assembly process of the NDH-1 hydrophilic arm subcomplex.
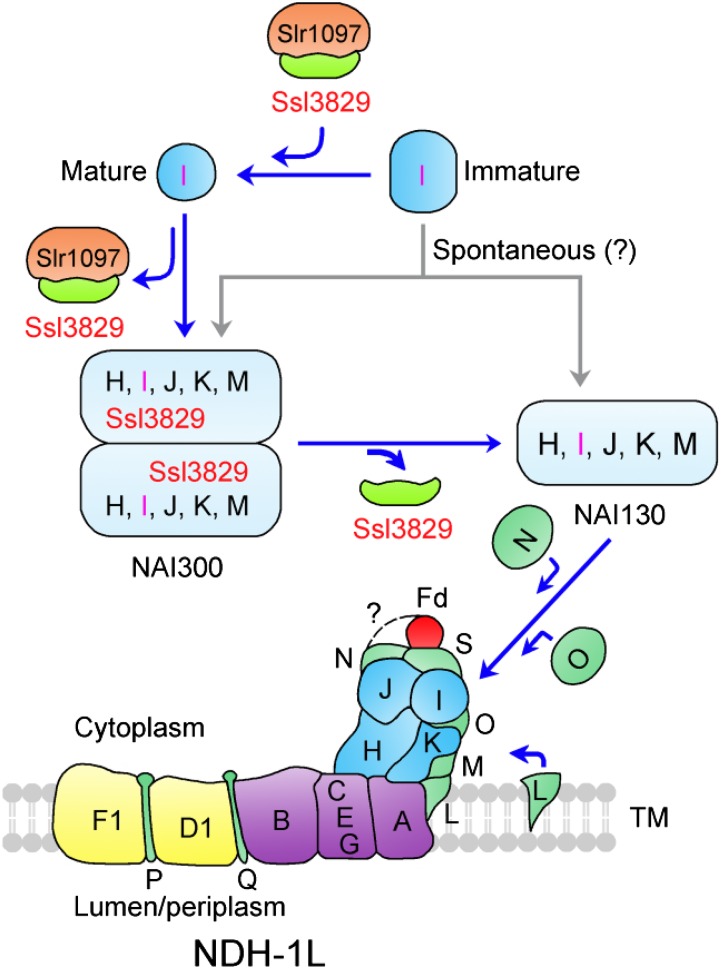
A Model for the Assembly of the NDH-1 Hydrophilic Arm
Ssl3829 interacts with NAI300 but not with NAI130, and its absence resulted in a significant abolishment of these two NAIs, although the residual amount of NAI130 may be slightly more than that of NAI300 (Fig. 5; Table I; Supplemental Data Set S1). It seems likely that the significant abolishment of NAI300 caused by the deletion of ssl3829 impeded the formation of NAI130. This possibility and their molecular masses suggest that the formation of NAI300 occurs before NAI130 formation and that NAI130 may be closer to the fully assembled form of the NDH-1 hydrophilic arm, as represented schematically in Figure 9 (see blue arrows). In addition, the formation of NAI100 increased significantly in ∆ssl3829, ∆slr1097, and NdhI-less mutants with the abolishment of NAI300 and NAI130 complexes (Fig. 5). This implies that NAI100 was either a degradation product of the NAI300 and NAI130 complexes or a precursor of the NAI300 complex. If the latter is true, Ssl3829 also may be involved in the transition from NAI100 to NAI300.
Figure 9.
Model schematically representing an important role of Ssl3829 in assembling the NDH-1 hydrophilic arm. NAI300 and NAI130 may be involved in the assembly process of the NDH-1 hydrophilic arm in Synechocystis 6803, and Slr1097 has been suggested to participate in NdhI maturation. Ssl3829 plays an important role in assembling the NDH-1 hydrophilic arm by accumulating Slr1097 protein and the NAI300 complex. TM, Thylakoid membrane.
Ssl3829 localizes in NAI300, and Slr1097 is not detected in any NAIs (Fig. 5). Besides Ssl3829, NAI300 contains NdhH, NdhI, NdhJ, NdhK, and NdhM (Fig. 5; Table I; Supplemental Data Set S1). Since the formation of NAI130 occurs after NAI300 formation, these five Ndh hydrophilic subunits also may be included in NAI130. In addition to these five subunits, the NDH-1 hydrophilic arm subcomplex also may contain NdhL, NdhN, and NdhO, based on biochemical evidence and extrapolation analysis (Prommeenate et al., 2004; Peng et al., 2009; Zhao et al., 2014), although single-particle electron microscopy analysis suggested that they are localized in the hydrophobic arm (Birungi et al., 2010). However, these three Ndh subunits did not associate with any NAIs (Supplemental Fig. S7) and were even not detected in the cytoplasm (Supplemental Fig. S8). Therefore, if these three subunits were true members of the NDH-1 hydrophilic arm, they were assembled into the hydrophilic arm after NAI130, as represented schematically in Figure 9.
Based on the above analyses, we propose a model for the assembly of the NDH-1 hydrophilic arm in cyanobacteria, as illustrated in Figure 9. NdhH to NdhK, NdhM, and Ssl3829 form NAI300. After the release of Ssl3829, NAI300 produces NAI130 (Fig. 9), but its absence causes NAI300 and NAI130 to be highly unstable on gels (Fig. 5C). Deduced from the molecular masses of NAI300 and NAI130, NAI300 appears to be an NAI130 dimer with multiple copies of Ssl3829 (Fig. 9) and/or other assembly factors, which are present mostly in proteins copurified with Ssl3829 (Supplemental Data Set S1). In addition, deletion of Slr1097 produces a new complex of about 150 kD (Fig. 5D, red arrow). Thus, it appears that Slr1097 is involved in forming the NAI130 dimer with assembly factors including Ssl3829, although it is not detected in any NAIs (Fig. 5). Finally, NAI130 accepts NdhL, NdhN, and NdhO subunits to form an integral NDH-1 hydrophilic arm and is incorporated into the thylakoids.
Role of Ssl3829 in Assembling the NDH-1 Hydrophilic Arm
We demonstrated in this study that Ssl3829-YH associates with NAI300 and that Ssl3829-YH or Ssl3829 interacts with Slr1097 (Figs. 5 and 7; Table I; Supplemental Data Set S1). The absence of Ssl3829 reduced the abundance of the NAI300 complex to levels no longer detectable on a gel (Fig. 5), thereby impeding the assembly process of the NDH-1 hydrophilic arm (Fig. 9, blue arrows). In addition, the absence of Ssl3829 completely abolished Slr1097 (Fig. 8A) and significantly decreased the amount of mature NdhI (Fig. 6) and the accumulation of the NAI300 and NAI130 complexes in the cytoplasm because of the presence of NdhI in these two NAIs (Fig. 5), thereby influencing the assembly efficiency of the NDH-1 hydrophilic arm (Fig. 9, blue arrows). Thus, Ssl3829 plays an important role in assembling the NDH-1 hydrophilic arm by accumulating the NAI300 complex and Slr1097 protein in the cytoplasm. Repression of the NDH-1 hydrophilic arm assembly caused by the deletion of ssl3829 hindered the assembly of NDH-1L and NDH-1M in the thylakoid membrane (Fig. 3), thereby impairing NDH-CET activity (Figs. 1 and 2) and generating a high-light-sensitive growth phenotype (Fig. 1; Supplemental Fig. S2).
MATERIALS AND METHODS
Culture Conditions
Synechocystis sp. strain PCC 6803 Glc-tolerant strain (the wild type) and its mutants ∆ssl3829, ∆slr1097 (Dai et al., 2013), NdhI-less, M55 (Ogawa, 1991), WT-Ssl3829-YH, WT-NdhL-YH (Birungi et al., 2010), and ∆slr1097-Ssl3829-YH were cultured at 30°C in BG-11 medium (Allen, 1968) buffered with Tris-HCl (5 mm, pH 8) and bubbled with 2% (v/v) CO2 in air. Solid medium was the same BG-11 supplemented with 1.5% agar. Continuous illumination was provided by fluorescent lamps at 40 or 200 µmol photons m–2 s–1. The mutant strains were grown in the presence of appropriate antibiotics.
Isolation and Construction of Mutants
A cosmid library of the Synechocystis 6803 genome was constructed. The library contained 105 clones with inserts of 35 to 38.5 kb and was subjected to in vitro transposon mutagenesis using the EZ-Tn5 < KAN-2 > Insertion Kit (Epicentre Biotechnologies) and then used to transform the wild-type cells of Synechocystis 6803. Following transformation, cells were spread on 1.5% BG-11 agar plates (5 µg kanamycin mL−1), and KamR mutants that grew slowly on the plates under conditions of continuous illumination with high light (200 µmol photons m–2 s–1) but normally under conditions with growth light (40 µmol photons m–2 s–1) were isolated. Genomic DNA isolated from each mutant was digested with HhaI and, after self-ligation, was used as a template for inverse PCR with primers (Supplemental Table S1) complementary to the N- and C-terminal regions of the KamR cassette. The exact position of the cassette in the mutant genome was determined by sequencing the PCR product.
The Δssl3829 mutant was constructed as follows. The upstream and downstream regions of ssl3829 were amplified by PCR, creating appropriate restriction sites. A DNA fragment encoding a KamR cassette also was amplified by PCR, creating KpnI and BamHI sites using appropriate PCR primers, ssl3829-C and ssl3829-D (Supplemental Table S1). These three PCR products were ligated into the MCS of pUC19 (Fig. 2A) and used to transform the wild-type cells of Synechocystis 6803 to generate the Δssl3829 mutant. The transformants were spread on agar plates containing BG-11 medium and kanamycin (10 µg mL−1) buffered at pH 8, and the plates were incubated in 2% (v/v) CO2 in air under illumination by fluorescent lamps at 40 µmol photons m–2 s–1. The mutated ssl3829 in the transformants was segregated to homogeneity (by successive streak purification) as determined by PCR amplification and RT-PCR analysis (Fig. 2, B and C).
A DNA fragment containing ssl3829 and its upstream region was amplified by PCR, creating SalI and KpnI sites on both ends, and was ligated to the SalI and KpnI sites in the MCS of the pEYFP-His6-SpR plasmid (Birungi et al., 2010). A fragment containing the downstream region of ssl3829 also was amplified by PCR, creating EcoRI and SpeI sites, and was ligated to the downstream region of the SpR gene (Supplemental Fig. S3A). The vector thus constructed was used to transform the wild-type and ∆slr1097 cells of Synechocystis 6803 to generate the WT-Ssl3829-YH and ∆slr1097-Ssl3829-YH mutant strains. The transformation was performed as described previously (Williams and Szalay, 1983; Long et al., 2011). The yfp and his6 regions in the transformants were segregated to homogeneity (by successive streak purification) as determined by PCR amplification (Supplemental Fig. S3B and Fig. 8C).
RNA Extraction and RT-PCR Analysis
Total RNA was isolated and analyzed as described previously (McGinn et al., 2003). RT-PCR was performed using the Access RT-PCR system (Promega) to generate products corresponding to ssl3829, slr2141, sml0007, sll2005, slr1097, and 16 S rRNA, with 0.5 µg of DNase-treated total RNA as starting material. RT-PCR conditions were 95°C for 5 min followed by cycles of 95°C, 62°C, and 72°C for 30 s each. The reactions were stopped after 25 or 35 cycles for 16 S rRNA and after 30 or 40 cycles for ssl3829, slr2141, sml0007, sll2005, and slr1097. The primers used are summarized in Supplemental Table S1.
Chl Fluorescence and P700 Analysis
The transient increase in Chl fluorescence after AL had been turned off was monitored as described (Ma and Mi, 2005). The redox kinetics of P700 was measured according to previously described methods (Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015; Gao et al., 2016). The rereduction of P700+ in darkness was measured with a Dual-PAM-100 (Walz) with an emitter-detector unit ED-101US/MD by monitoring absorbance changes at 830 nm and using 875 nm as a reference. Cells were kept in the dark for 2 min, and 10 µm 3-(3,4-dichlorophenyl)-1,1-dimethylurea was added to the cultures prior to the measurement. The P700 was oxidized by far-red light with a maximum at 720 nm from a light-emitting diode lamp for 30 s, and the subsequent rereduction of P700+ in the dark was monitored.
Isolation of Crude Thylakoid Membranes
The cell cultures (800 mL) were harvested at the logarithmic phase and washed twice by suspending in 50 mL of fresh BG-11 medium, and the thylakoid membranes were isolated according to Gombos et al. (1994) with some modifications as follows. The cells suspended in 5 mL of disruption buffer (10 mm HEPES-NaOH, 5 mm sodium phosphate, pH 7.5, 10 mm MgCl2, 10 mm NaCl, and 25% [v/v] glycerol) were supplemented by zirconia/silica beads and broken by vortexing 15 times at the highest speed for 20 s at 4°C with 5 min of cooling on ice between the runs. The crude extract was centrifuged at 5,000g for 5 min to remove the glass beads and unbroken cells. By further centrifugation at 20,000g for 30 min, we obtained crude thylakoid membranes from the precipitation.
Isolation of Membrane and Soluble Cell Fractions
The total membrane and soluble fractions of Synechocystis 6803 cells were isolated as described previously (Zhang et al., 2009, 2012; Dai et al., 2013) with slight modifications. In brief, the cells were pelleted from batch cultures and broken with glass beads by vortexing at 4°C in disruption buffer. The cell debris and glass beads were removed by 5 min of centrifugation at 5,000g. The total membrane and cytoplasmic soluble fractions were separated by centrifugation at 110,000g for 30 min.
Electrophoresis and Immunoblotting
BN-PAGE of Synechocystis 6803 membranes was performed as described previously (Kügler et al., 1997) with slight modifications (Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015; Gao et al., 2016). Isolated membranes were prepared for BN-PAGE as follows. Membranes were washed with 330 mm sorbitol, 50 mm Bis-Tris, pH 7, and 0.5 mm phenylmethylsulfonyl fluoride (Sigma) and resuspended in 20% (w/v) glycerol, 25 mm Bis-Tris, pH 7, 10 mm MgCl2, 0.1 units of RNase-free DNase RQ1 (Promega) at a Chl a concentration of 0.3 mg mL−1, and 0.5 mm phenylmethylsulfonyl fluoride. The samples were incubated on ice for 10 min, and an equal volume of 3% n-dodecyl β-d-maltoside (DM) was added. Solubilization was performed for 40 min on ice. Insoluble components were removed by centrifugation at 18,000g for 15 min. The collected supernatant was mixed with a one-tenth volume of sample buffer, 5% Serva Blue G, 100 mm Bis-Tris, pH 7, 30% (w/v) Suc, 500 mm ε-amino-n-caproic acid, and 10 mm EDTA. Solubilized membranes were then applied to a 0.75-mm-thick, 5% to 12.5% acrylamide gradient gel (Hoefer Mighty Small mini-vertical unit). Samples were loaded on an equal Chl a basis per lane. Electrophoresis was performed at 4°C by increasing the voltage gradually from 50 to 200 V during the 5.5-h run.
CN-PAGE was performed with 0.01% DM and 0.025% deoxycholate additives to the cathode buffer as described by Wittig et al. (2007). Cytoplasmic soluble fractions were applied to a 0.75-mm-thick, 5% to 12.5% acrylamide gradient gel (Hoefer Mighty Small mini-vertical unit). Samples were loaded on an equal protein basis of 80 µg per lane. Electrophoresis was performed at 4°C by increasing the voltage gradually from 50 to 200 V during the 5.5-h run.
For electrophoresis in the second dimension, several lanes of the CN gel were cut out and incubated in Laemmli SDS sample buffer containing 5% β-mercaptoethanol and 6 m urea for 1 h at 25°C. The lanes were then laid onto a 1-mm-thick 12% polyacrylamide gel with 6 m urea (Laemmli, 1970).
For immunoblotting, the proteins were electrotransferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore) and detected by protein-specific antibodies using an ECL assay kit (Amersham Pharmacia) according to the manufacturer’s protocol. Antibody against NdhN protein of Synechocystis 6803 was raised in our laboratory. To amplify the ndhN gene, primer sequences were designed and appear in Supplemental Table S1. The PCR products were ligated into a vector, pET32a, and the construct was amplified in Escherichia coli DH-5α. The plasmid was used to transform E. coli strain BL21 (DE3) pLysS for expression. The gene expression products from E. coli were purified and used as antigens to immunize rabbits to produce polyclonal antibodies. Antibodies against NdhH, NdhI, NdhK, NdhM, NdhO, and Slr1097 were raised previously in our laboratory (Ma and Mi, 2005; Dai et al., 2013; Zhao et al., 2014). Antibody against His was purchased from Shanghai Immune Biotech, and antibodies against GFP, GST, a PSII subunit (D1) and a PSI subunit (PsaD), and RbcL were purchased from Agrisera.
Affinity Chromatography
Purification of proteins containing the His6 tag was performed using a nickel-nitrilotriacetic acid His-Bind Resin column (Novagen, Merck). Chromatography buffers contained 20 mm HEPES, pH 7.5, 10% (w/v) glycerol, 100 mm NaCl, and 0.015% DM supplemented with 5, 10, and 200 mm imidazole for binding, washing, and elution of the proteins, respectively. After Ni2+ affinity chromatography and prior to CN-PAGE and tandem mass spectrometry analysis, the eluted material was concentrated using Microcon YM-100 (Millipore) centrifuge filter devices.
Peptide Preparation for Tandem Mass Spectrometry Analysis
Peptide preparation and liquid chromatography-electrospray ionization-tandem mass spectrometry analyses were performed as described previously (Battchikova et al., 2011a). The purified NAI300 complex samples were treated twice with 50 mm ammonium bicarbonate in 30% (v/v) acetonitrile for 10 min and 100% (v/v) acetonitrile for 15 min and then dried in a vacuum concentrator. The dried gel pieces were treated with 0.01 mg mL−1 trypsin (sequence grade; Promega)/50 mm ammonium bicarbonate and incubated at 37°C for 20 h. The digested peptides on the gel pieces were recovered twice with 20 µL of 5% (v/v) formic acid/50% (v/v) acetonitrile. The extracted peptides were combined and then dried in a vacuum concentrator.
Mass Spectrometry Analysis and Database Searching
Liquid chromatography-tandem mass spectrometry analyses were performed on a Q-Exactive mass spectrometer (Thermo Finnigan) coupled with an Easy-nLC1000 HPLC system (Thermo Fisher Scientific). Trypsin-digested peptides were dissolved in 12 µL of 2% formic acid, loaded onto a C18 reverse-phase column (10 cm × 75 μm i.d.) in buffer A (2% acetonitrile and 0.1% formic acid), and separated by a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) at a flow rate of 250 nL min−1 controlled by InterlliFlow technology over 90 min. Mass spectrometry data were acquired using a data-dependent top-10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1,800 mass-to-charge ratio [m/z]) for higher-energy collision dissociation fragmentation. Survey scans were acquired at a resolution of 70,000 at m/z 200, and resolution for higher-energy collision dissociation spectra was set to 17,500 at m/z 200. The raw data were processed with Protein Discoverer software, version 1.1 (Thermo Fisher Scientific), using default parameters to generate concatenated Mascot generic files. Database searches were performed using an in-house Mascot server (version 2.2) against a UniProt database of Synechocystis 6803 proteins. The search criteria allowed for one missed cleavage of trypsin, oxidation of Met, and 6-ppm and 0.1-D mass accuracies for mass spectrometry and tandem mass spectrometry modes, respectively.
Expression and Purification of Fusion Proteins
The target proteins Slr1097, Ssl3829, and NdhI were fused to various tags as follows. The fragments containing slr1097 and ndhI genes were amplified by PCR and inserted between the BamHI and SalI sites and the BamHI and SacI sites of pET32a, respectively, to form the His-tagged fusion protein constructs (primers are shown in Supplemental Table S1). The PCR-amplified ssl3829 gene was cloned in frame into EcoRI and XhoI sites of pGEX-5X-1 to form the GST-tagged fusion protein constructs (primers are shown in Supplemental Table S1). Subsequently, these constructs were transformed into E. coli strain BL21 (DE3) pLysS and induced by 1 mm isopropyl-β-d-thiogalactoside for 16 h at 16°C to express His- and GST-tagged fusion proteins. These fusion proteins were purified at 4°C using a nickel column (GE Healthcare) and Glutathione Sepharose 4B (GE Healthcare), respectively, according to the manufacturers’ instructions.
GST Pull-Down Assay
The GST pull-down assay was performed as described previously (Zhao et al., 2014) with some modifications. To test the interaction of Ssl3829 with Slr1097, an equal amount of GST and GST-Ssl3829 proteins was separately incubated with 20 µL of Glutathione Sepharose 4B beads (GE Healthcare) in 300 µL of binding buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5 mm EDTA, 0.5% [v/v] Triton X-100, 1 mm phenylmethanesulfonyl fluoride, and 1 mm dithiothreitol) at 4°C for 1 h. The beads were washed three times with 1 mL of binding buffer, and then equal amounts of His and His-Slr1097 were separately added and incubated in a 300-µL reaction system on a rotating shaker at 4°C overnight. After the overnight incubation, the beads were pelleted by centrifugation and washed four times with the ice-cold binding buffer described above. The washed pellet were resuspended in 30 µL of 1× SDS (2%) loading buffer and then boiled for 5 min. After centrifugation, the supernatant was collected and subjected to immunoblot analysis.
Coimmunoprecipitation
The coimmunoprecipitation assay was performed as described previously (Komenda et al., 2005) with some modifications. Total protein at 300 µg isolated from strain WT-Ssl3829-YH or Δslr1097-Ssl3829-YH in 500 µL of TNE buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 2 mm EDTA) was solubilized with 0.05% (w/w) DM. After centrifugation at 110,000g for 30 min at 4°C, the supernatant was incubated with 20 µL of anti-Slr1097 serum overnight at 4°C with gentle rotation. The solution was then incubated with an equal amount of Protein A Sepharose (GE Healthcare) for 1 h at 4°C. The Protein A Sepharose 4B with bound protein immunoglobulin complexes were subsequently sedimented and washed twice with 1 mL of TNE buffer containing 0.05% (w/w) DM and twice with TNE buffer. The beads were then suspended in 30 µL of 1× SDS (2%) loading buffer and boiled for 5 min. After centrifugation, the supernatant was collected and subjected to immunoblot analysis.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed using the LexA system (Clontech). The PCR-amplified ssl3829- and ndhO-encoding gene fragments were cloned in frame into EcoRI and XhoI sites of pLexA to form the bait construct (primers are shown in Supplemental Table S1). The fragments containing slr1097, ndhB, and ndhI genes were amplified by PCR and inserted into the XhoI sites of pJG4-5 to form the prey construct (primers are shown in Supplemental Table S1). The bait and prey constructs together with the reporter vector pSH18-34 were cotransformed into yeast (Saccharomyces cerevisiae) strain EGY48 according to the manufacturer’s instructions for the Matchmaker LexA two-hybrid system (Clontech). Transformed yeast was diluted and dropped onto synthetic dropout medium, Leu-negative synthetic dropout medium, or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid medium and then grown at 30°C in darkness as described earlier (Sun et al., 2009; Dai et al., 2013).
Growth Curve
The cell density of wild-type and ∆ssl3829 cells cultured at growth light and high light was determined every 12 h by measuring the A730 using a spectrophotometer (UV3000; Shimadzu).
Wild-type and ∆ssl3829 cells (1.5 mL) were collected by centrifugation, and pigments in the pellet were extracted by 1.5 mL of methyl alcohol. Chl a in the extract was determined using a spectrophotometer (UV3000; Shimadzu) according to the formula Chl a (µg mL−1) = 13.9 × A665 (Arnon, 1949).
Confocal Laser Scanning Microscopy
Cell imaging was observed using a laser scanning confocal microscope (FV1000; Olympus) with a 100×/1.4 plan apochromat oil-immersion objective and with an 80-µm confocal pinhole. The argon laser line, 515 nm, and the helium laser line, 543 nm, were used for the excitation of YFP and autofluorescence, respectively. YFP fluorescence emission was selected with a 515-nm dichroic mirror and a band-pass filter of 535 to 590 nm. Autofluorescence was collected through a 545-nm dichroic mirror and a long-pass filter of 560 nm.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Ssl3829 (NP_441041.1), Slr1097 (NP_440076.1), and CRR7 (At5g39210).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-PCR analysis of ssl3829 and its neighboring genes in the wild-type and ∆ssl3829 strains.
Supplemental Figure S2. Growth of wild-type and ∆ssl3829 cells under different light intensities.
Supplemental Figure S3. Construction and characterization of the WT-Ssl3829-YH strain.
Supplemental Figure S4. Sequence comparison between Ssl3829 and CRR7 (At5g39210).
Supplemental Figure S5. GST pull-down assay shows no interaction between Ssl3829 and NdhI.
Supplemental Figure S6. Deletion mutation and molecular identification of the ndhI gene.
Supplemental Figure S7. Analysis of NdhL, NdhN, and NdhO in NAIs in the cytoplasm.
Supplemental Figure S8. Comparison of NdhL, NdhN, and NdhO protein abundances between soluble and membrane fractions.
Supplemental Table S1. Primers used in this study.
Supplemental Data Set S1. Total proteins identified in the purified NAI300 complex samples.
Supplementary Material
Acknowledgments
We thank Hualing Mi (Shanghai Institutes for Biological Sciences) and Hongquan Yang (Fudan University) for the NdhI antibody of Synechocystis 6803 and the yeast two-hybrid system, respectively; mass spectrometry was carried out at the Shanghai Applied Protein Technology and confocal microscopy was performed at the Shanghai Institutes for Biological Sciences, and Dr. Xiaoshu Gao of the Shanghai Institutes is thanked for help in cell imaging.
Glossary
- Chl
chlorophyll
- RT
reverse transcription
- AL
actinic light
- FR
far-red
- BN
blue native
- NAI
NDH-1 assembly intermediate
- CN
clear native
- DM
n-dodecyl β-d-maltoside
- m/z
mass-to-charge ratio
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31570235 and 31370270) and the Shanghai Natural Science Foundation (grant no. 14ZR1430000).
References
- Allen MM. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Rühle T, Kreller R, Strotbek C, Zühlke J, Tadini L, Blunder T, Hertle AP, Qi Y, Rengstl B, et al. (2013) The photosynthesis affected mutant68-like protein evolved from a PSII assembly factor to mediate assembly of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell 25: 3926–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteni AA, Zhang P, Battchikova N, Ogawa T, Aro EM, Boekema EJ (2006) Structural characterization of NDH-1 complexes of Thermosynechococcus elongatus by single particle electron microscopy. Biochim Biophys Acta 1757: 1469–1475 [DOI] [PubMed] [Google Scholar]
- Baradaran R, Berrisford JM, Minhas GS, Sazanov LA (2013) Crystal structure of the entire respiratory complex I. Nature 494: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battchikova N, Aro EM (2007) Cyanobacterial NDH-1 complexes: multiplicity in function and subunit composition. Physiol Plant 131: 22–32 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Eisenhut M, Aro EM (2011b) Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim Biophys Acta 1807: 935–944 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Wei L, Du L, Bersanini L, Aro EM, Ma W (2011a) Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J Biol Chem 286: 36992–37001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battchikova N, Zhang P, Rudd S, Ogawa T, Aro EM (2005) Identification of NdhL and Ssl1690 (NdhO) in NDH-1L and NDH-1M complexes of Synechocystis sp. PCC 6803. J Biol Chem 280: 2587–2595 [DOI] [PubMed] [Google Scholar]
- Bernát G, Appel J, Ogawa T, Rögner M (2011) Distinct roles of multiple NDH-1 complexes in the cyanobacterial electron transport network as revealed by kinetic analysis of P700+ reduction in various Ndh-deficient mutants of Synechocystis sp. strain PCC6803. J Bacteriol 193: 292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birungi M, Folea M, Battchikova N, Xu M, Mi H, Ogawa T, Aro EM, Boekema EJ (2010) Possibilities of subunit localization with fluorescent protein tags and electron microscopy exemplified by a cyanobacterial NDH-1 study. Biochim Biophys Acta 1797: 1681–1686 [DOI] [PubMed] [Google Scholar]
- Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J 17: 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Zhang L, Zhang J, Mi H, Ogawa T, Ma W (2013) Identification of a cyanobacterial CRR6 protein, Slr1097, required for efficient assembly of NDH-1 complexes in Synechocystis sp. PCC 6803. Plant J 75: 858–866 [DOI] [PubMed] [Google Scholar]
- Deng Y, Ye J, Mi H (2003) Effects of low CO2 on NAD(P)H dehydrogenase, a mediator of cyclic electron transport around photosystem I in the cyanobacterium Synechocystis PCC6803. Plant Cell Physiol 44: 534–540 [DOI] [PubMed] [Google Scholar]
- Efremov RG, Baradaran R, Sazanov LA (2010) The architecture of respiratory complex I. Nature 465: 441–445 [DOI] [PubMed] [Google Scholar]
- Friedrich T, Scheide D (2000) The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett 479: 1–5 [DOI] [PubMed] [Google Scholar]
- Friedrich T, Steinmüller K, Weiss H (1995) The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett 367: 107–111 [DOI] [PubMed] [Google Scholar]
- Gao F, Zhao J, Wang X, Qin S, Wei L, Ma W (2016) NdhV is a subunit of NADPH dehydrogenase essential for cyclic electron transport in Synechocystis sp. strain PCC 6803. Plant Physiol 170: 752–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombos Z, Wada H, Murata N (1994) The recovery of photosynthesis from low-temperature photoinhibition is accelerated by the unsaturation of membrane lipids: a mechanism of chilling tolerance. Proc Natl Acad Sci USA 91: 8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36: 541–549 [DOI] [PubMed] [Google Scholar]
- He Z, Xu M, Wu Y, Lv J, Fu P, Mi H (2016) NdhM subunit is required for the stability and the function of NAD(P)H dehydrogenase complexes involved in CO2 uptake in Synechocystis sp. strain PCC 6803. J Biol Chem 291: 5902–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranen M, Battchikova N, Zhang P, Graf A, Sirpiö S, Paakkarinen V, Aro EM (2004) Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol 134: 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K, Endo T, Shikanai T, Aro EM (2011) Structure of the chloroplast NADH dehydrogenase-like complex: nomenclature for nuclear-encoded subunits. Plant Cell Physiol 52: 1560–1568 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3: 109–136 [DOI] [PubMed] [Google Scholar]
- Komenda J, Tichý M, Eichacker LA (2005) The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol 46: 1477–1483 [DOI] [PubMed] [Google Scholar]
- Kügler M, Jänsch L, Kruft V, Schmitz UK, Braun HP (1997) Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE). Photosynth Res 53: 35–44 [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Long Z, Zhao J, Zhang J, Wei L, Wang Q, Ma W (2011) Effects of different light treatments on the natural transformation of Synechocystis sp. strain PCC 6803. Afr J Microbiol Res 5: 3603–3610 [Google Scholar]
- Ma W. (2009) Identification, regulation and physiological functions of multiple NADPH dehydrogenase complexes in cyanobacteria. Front Biol China 4: 137–142 [Google Scholar]
- Ma W, Mi H (2005) Expression and activity of type 1 NAD(P)H dehydrogenase at different growth phases of the cyanobacterium, Synechocystis PCC6803. Physiol Plant 125: 135–140 [Google Scholar]
- Ma W, Ogawa T (2015) Oxygenic photosynthesis-specific subunits of cyanobacterial NADPH dehydrogenases. IUBMB Life 67: 3–8 [DOI] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14: 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn PJ, Price GD, Maleszka R, Badger MR (2003) Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC6803. Plant Physiol 132: 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Endo T, Ogawa T, Asada K (1995) Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediated cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 36: 661–668 [Google Scholar]
- Mi H, Endo T, Schreiber U, Ogawa T, Asada K (1992) Electron donation from cyclic and respiratory flows to the photosynthetic intersystem chain is mediated by pyridine nucleotide dehydrogenase in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol 33: 1233–1237 [Google Scholar]
- Munshi MK, Kobayashi Y, Shikanai T (2005) Identification of a novel protein, CRR7, required for the stabilization of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant J 44: 1036–1044 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot (Lond) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyk MM, Wulfhorst H, Ryan CM, Souda P, Zhang H, Cramer WA, Whitelegge JP (2011) NdhP and NdhQ: two novel small subunits of the cyanobacterial NDH-1 complex. Biochemistry 50: 1121–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T. (1991) A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC6803. Proc Natl Acad Sci USA 88: 4275–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Mi H (2007) Cyanobacterial NADPH dehydrogenase complexes. Photosynth Res 93: 69–77 [DOI] [PubMed] [Google Scholar]
- Ohkawa H, Pakrasi HB, Ogawa T (2000) Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J Biol Chem 275: 31630–31634 [DOI] [PubMed] [Google Scholar]
- Ohkawa H, Sonoda M, Hagino N, Shibata M, Pakrasi HB, Ogawa T (2002) Functionally distinct NAD(P)H dehydrogenases and their membrane localization in Synechocystis sp. PCC6803. Funct Plant Biol 29: 195–200 [DOI] [PubMed] [Google Scholar]
- Ohkawa H, Sonoda M, Shibata M, Ogawa T (2001) Localization of NAD(P)H dehydrogenase in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 183: 4938–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Cai W, Shikanai T (2010) Chloroplast stromal proteins, CRR6 and CRR7, are required for assembly of the NAD(P)H dehydrogenase subcomplex A in Arabidopsis. Plant J 63: 203–211 [DOI] [PubMed] [Google Scholar]
- Peng L, Fukao Y, Fujiwara M, Shikanai T (2012) Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 24: 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Fukao Y, Fujiwara M, Takami T, Shikanai T (2009) Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21: 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Fukao Y, Myouga F, Motohashi R, Shinozaki K, Shikanai T (2011a) A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biol 9: e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng L, Yamamoto H, Shikanai T (2011b) Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim Biophys Acta 1807: 945–953 [DOI] [PubMed] [Google Scholar]
- Prommeenate P, Lennon AM, Markert C, Hippler M, Nixon PJ (2004) Subunit composition of NDH-1 complexes of Synechocystis sp. PCC 6803: identification of two new ndh gene products with nuclear-encoded homologues in the chloroplast Ndh complex. J Biol Chem 279: 28165–28173 [DOI] [PubMed] [Google Scholar]
- Rochaix JD. (2011) Assembly of the photosynthetic apparatus. Plant Physiol 155: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov LA, Hinchliffe P (2006) Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311: 1430–1436 [DOI] [PubMed] [Google Scholar]
- Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A, Ogawa T (2001) Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc Natl Acad Sci USA 98: 11789–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S, Allahverdiyeva Y, Holmström M, Khrouchtchova A, Haldrup A, Battchikova N, Aro EM (2009) Novel nuclear-encoded subunits of the chloroplast NAD(P)H dehydrogenase complex. J Biol Chem 284: 905–912 [DOI] [PubMed] [Google Scholar]
- Sun SY, Chao DY, Li XM, Shi M, Gao JP, Zhu MZ, Yang HQ, Luan S, Lin HX (2009) OsHAL3 mediates a new pathway in the light-regulated growth of rice. Nat Cell Biol 11: 845–851 [DOI] [PubMed] [Google Scholar]
- Suorsa M, Sirpiö S, Aro EM (2009) Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol Plant 2: 1127–1140 [DOI] [PubMed] [Google Scholar]
- Wang P, Duan W, Takabayashi A, Endo T, Shikanai T, Ye JY, Mi H (2006) Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol 141: 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JGK, Szalay AA (1983) Stable integration of foreign DNA into the chromosome of the cyanobacterium Synechococcus R2. Gene 24: 37–51 [DOI] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schägger H (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6: 1215–1225 [DOI] [PubMed] [Google Scholar]
- Wulfhorst H, Franken LE, Wessinghage T, Boekema EJ, Nowaczyk MM (2014) The 5 kDa protein NdhP is essential for stable NDH-1L assembly in Thermosynechococcus elongatus. PLoS ONE 9: e103584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Ogawa T, Pakrasi HB, Mi H (2008) Identification and localization of the CupB protein involved in constitutive CO2 uptake in the cyanobacterium, Synechocystis sp. strain PCC 6803. Plant Cell Physiol 49: 994–997 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Peng L, Fukao Y, Shikanai T (2011) An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gao F, Zhao J, Ogawa T, Wang Q, Ma W (2014) NdhP is an exclusive subunit of large complex of NADPH dehydrogenase essential to stabilize the complex in Synechocystis sp. strain PCC 6803. J Biol Chem 289: 18770–18781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Allahverdiyeva Y, Eisenhut M, Aro EM (2009) Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS ONE 4: e5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Battchikova N, Jansen T, Appel J, Ogawa T, Aro EM (2004) Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16: 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Battchikova N, Paakkarinen V, Katoh H, Iwai M, Ikeuchi M, Pakrasi HB, Ogawa T, Aro EM (2005) Isolation, subunit composition and interaction of the NDH-1 complexes from Thermosynechococcus elongatus BP-1. Biochem J 390: 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Eisenhut M, Brandt AM, Carmel D, Silén HM, Vass I, Allahverdiyeva Y, Salminen TA, Aro EM (2012) Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24: 1952–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Gao F, Zhang J, Ogawa T, Ma W (2014) NdhO, a subunit of NADPH dehydrogenase, destabilizes medium size complex of the enzyme in Synechocystis sp. strain PCC 6803. J Biol Chem 289: 26669–26676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Rong W, Gao F, Ogawa T, Ma W (2015) Subunit Q is required to stabilize the large complex of NADPH dehydrogenase in Synechocystis sp. strain PCC 6803. Plant Physiol 168: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.