miR858a targets MYB transcription factors involved in flavonoid biosynthesis and plant growth and development.
Abstract
MicroRNAs (miRNAs) are endogenous, noncoding small RNAs that function as critical regulators of gene expression. In plants, miRNAs have shown their potential as regulators of growth, development, signal transduction, and stress tolerance. Although the miRNA-mediated regulation of several processes is known, the involvement of miRNAs in regulating secondary plant product biosynthesis is poorly understood. In this study, we functionally characterized Arabidopsis (Arabidopsis thaliana) miR858a, which putatively targets R2R3-MYB transcription factors involved in flavonoid biosynthesis. Overexpression of miR858a in Arabidopsis led to the down-regulation of several MYB transcription factors regulating flavonoid biosynthesis. In contrast to the robust growth and early flowering of miR858OX plants, reduction of plant growth and delayed flowering were observed in Arabidopsis transgenic lines expressing an artificial miRNA target mimic (MIM858). Genome-wide expression analysis using transgenic lines suggested that miR858a targets a number of regulatory factors that modulate the expression of downstream genes involved in plant development and hormonal and stress responses. Furthermore, higher expression of MYBs in MIM858 lines leads to redirection of the metabolic flux towards the synthesis of flavonoids at the cost of lignin synthesis. Altogether, our study has established the potential role of light-regulated miR858a in flavonoid biosynthesis and plant growth and development.
MicroRNAs (miRNAs) are small, mostly noncoding (22–24 nucleotides) RNAs that modulate gene expression by pairing to corresponding target mRNA and promote mRNA degradation and/or translation inhibition (Llave et al., 2002; Brodersen et al., 2008; Filipowicz et al., 2008). miRNAs are important regulators of many physiological and developmental processes (Voinnet, 2009), and their misexpression leads to a variety of defects in plants (Wu, 2013). Various studies have demonstrated that miRNAs regulate flowering, leaf, seed, and root morphology, as well as stress and hormonal responses in plants (Leung and Sharp, 2010; Liu and Chen, 2010; Stauffer and Maizel, 2014; Hong and Jackson, 2015; Zhang, 2015). Recent developments in genome-wide experimental approaches involving miRNA overexpression (Wu, 2013), use of miRNA target mimics (Franco-Zorrilla et al., 2007; Eamens and Wang, 2011; Meng et al., 2012), and detailed analysis of miRNA promoters (Baek et al., 2013; Sharma et al., 2015) were helpful to understand the physical interactions between miRNAs and their target mRNAs as well as their biological significance in plant growth and development (de Lima et al., 2012; Wu, 2013). The majority of miRNAs are known to target transcription factors, which regulate development, hormonal regulation, and stress responses in plants (Sunkar et al., 2012; Wu, 2013; Zhang, 2015; Zhang and Wang, 2015).
Flavonoids are low-Mr polyphenolic compounds and encompass a diverse class of phenolic compounds, including flavonols, flavones, isoflavones, anthocyanins, proanthocyanidins, and phlobaphene pigments (Taylor and Grotewold, 2005; Lepiniec et al., 2006; Buer et al., 2010). The biological functions of flavonoids are as pigments, signaling molecules, UV protectants, phytoalexins, and regulators of auxin transport in plants (Winkel-Shirley, 2001; Buer and Muday, 2004; Santelia et al., 2008). The phenylpropanoid pathway is the main route for flavonoid biosynthesis, and the transcriptional regulation of this pathway is one of the most studied pathways in plants (Quattrocchio et al., 2006; Stracke et al., 2007). The synthesis of most flavonoid compounds shares common enzymes, and sometimes the enzymes of the specific branch are controlled by distinct regulators. MYB11, MYB12, and MYB111 have been demonstrated to regulate the biosynthesis of flavonols by targeting genes encoding early pathway enzymes such as chalcone synthase (CHS), chalcone isomerase (CHI), flavonoid 3′ hydroxylase (F3′H), and flavonol synthase (FLS) recruited in the phenylpropanoid pathway (Mehrtens et al., 2005; Stracke et al., 2007, 2010). MYB123 (TT2) controls the biosynthesis of proanthocyanidins in the seed coat of Arabidopsis (Arabidopsis thaliana; Xu et al., 2014). Similarly, members of the R2R3-MYB protein family, such as MYB58, MYB63, and MYB85, regulate lignin biosynthesis in fibers and vessels, whereas MYB46 and MYB83 act as switch regulators during secondary wall biosynthesis (Zhong et al., 2008; McCarthy et al., 2009; Zhou et al., 2009; Zhong and Ye, 2012). Flavonol biosynthesis is a highly complex, tissue and developmentally regulated process in Arabidopsis that is orchestrated by the group of R2R3-MYB transcription factors, including MYB11, MYB12, and MYB111. These MYBs differentially affect the spatial accumulation of specific flavonols in Arabidopsis (Stracke et al., 2007).
In the past few years, studies have demonstrated that miRNAs may act as master regulators of flavonoid biosynthesis in Arabidopsis. It has been shown that the miR156-SPL9 network directly influences anthocyanin production through targeting genes encoding PAP1 and dihydroflavonol 4-reductase (Gou et al., 2011; Cui et al., 2014). The miR163 targets S-adenosyl-Met-dependent methyltransferases, which methylate secondary metabolites and signaling molecules (Ng et al., 2011). Studies also have revealed that miR397 regulates lignin biosynthesis by altering laccase expression in Arabidopsis and Populus spp. (Lu et al., 2013; Wang et al., 2014). It has been reported that the miR159, miR319, and miR828 families deregulate MYB transcription factors in apple (Malus domestica), tomato (Solanum lycopersicum), grape (Vitis vinifera), and Salvia miltiorrhiza (Xia et al., 2012; Rock, 2013; Li et al., 2014; Jia et al., 2015). Similarly, a study demonstrated that miR828 regulates MYB2, which is involved in Arabidopsis trichome and cotton (Gossypium hirsutum) fiber development (Guan et al., 2014).
In this study, we investigated the role of miR858a in the regulation of the phenylpropanoid pathway and development in Arabidopsis. The genetic, molecular, cellular, and biochemical experiments using transgenic lines expressing miR858a (miR858OX) and artificial miR858 target mimic (MIM858) show that miR858a targets flavonol-specific MYBs involved in regulating early steps of the flavonoid biosynthetic pathway. Genome-wide expression and detailed metabolite analyses of transgenic lines suggest that miR858a plays an important role in plant growth and development. We conclude that the miR858a-MYB network is an additional regulatory sector of the module that coordinates flavonoid and lignin biosynthesis in plants.
RESULTS
Identification of miRNAs Regulating Flavonoid Biosynthesis
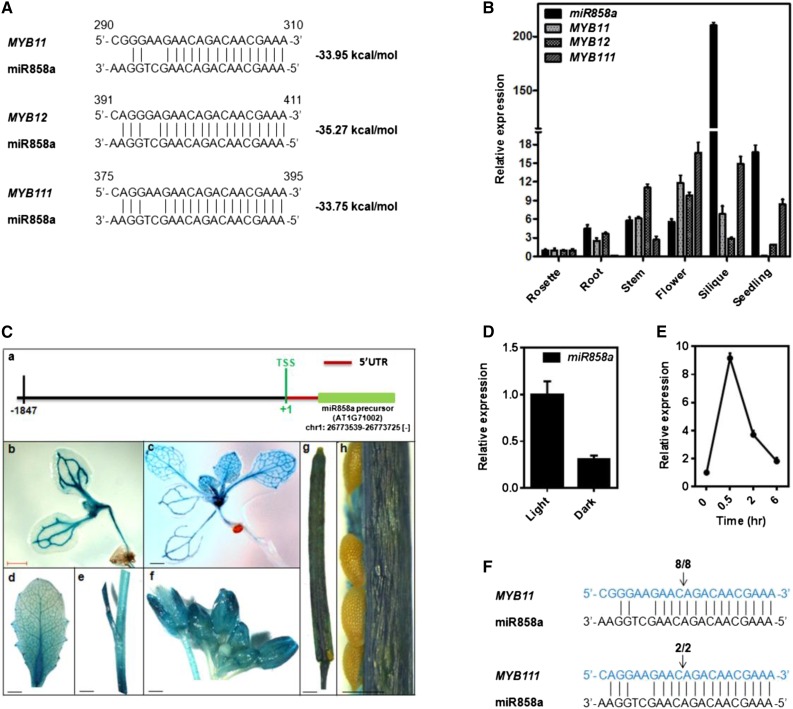
As a first step to identify the potential miRNAs regulating MYB11, MYB12, and MYB111 transcription factors, nucleotide sequences of the mRNAs were used for BLAST searches against the Sanger miRNA database (http://microrna.sanger.ac.uk). Ath-miR858 showed significant sequence homology with these MYB transcription factors. Besides, analysis using the WMD3 miRNA target identification tool (Schwab et al., 2006) identified a number of putative targets of miR858. The result suggested that MYB11, MYB12, and MYB111 could be the best potential targets of miR858, with hybridization energy of the duplex structure less than or equal to −35 kcal mol−1 and average homology of 80% or greater (Fig. 1A). A detailed bioinformatic analysis revealed that miR858 sequences are evolutionarily conserved in dicot genera such as Arabidopsis, Gossypium, Solanum, and Cucumis (Supplemental Table S1). Similar to our observation, Xia et al., (2012) reported that miR858 and its targets are well conserved in diverse dicot species. There are two members of the miR858 family (Ath-miR858a/b) present in the Arabidopsis genome, as listed in miRBase release 21. The mature sequence of miR858a differs from that of miR858b by a single nucleotide (Supplemental Table S1). However, miR858a and miR858b are transcribed from different loci but target similar sets of MYB transcription factors. Therefore, we focused on studying the role of miR858a as a putative regulator of MYB transcription factors in this study.
Figure 1.
Identification and characterization of miR858a in Arabidopsis. A, Sequences of MYB11, MYB12, and MYB111 mRNAs (5′ to 3′) aligned with the mature sequence of miR858a (3′ to 5′). Dashed lines and spaces represent perfect match and G:U wobble pairing, respectively. The free energy of binding is indicated at right. B, Expression of miR858a, MYB11, MYB12, and MYB111 in rosette, root, stem, flower, and silique from 5-week-old plant and 10-d-old seedling of Arabidopsis Columbia-0 (Col-0). For miR858a, SnoR41Y RNA was used, and for MYB11, MYB12, and MYB111, TUBULIN was used as the endogenous control to normalize the relative expression levels. Bars represent means ± se (n = 3). C, Histochemical staining showing GUS activity in different developmental organs in the ProMIR858:GUS transgenic lines: a, schematic representation of the miR858a promoter upstream of the precursor; b, 5-d-old seedling; c, 12-d-old seedling; d, rosette leaf; e, stem; f, inflorescence; g, silique; and h, seed. UTR, Untranslated region. Bars = 1 cm for b to f and 500 µm for g. D, qRT-PCR analysis of the mRNA level of miR858a in wild-type (Col-0) seedlings grown in light and dark conditions for 5 d. Bars represent means ± se (n = 3) from three biological replicates. E, qRT-PCR analysis of the mRNA levels of miR858a in wild-type upon exposure to light (150 µmol m−2 s−1) at the indicated times. Bars represent means ± se (n = 3) from three biological replicates. F, Target validation for miR858a by RLM-RACE. Cleavage products of MYB11 and MYB111 mRNA are shown. The fractions of clones with the expected 5′ end among all sequenced products are indicated above the target site. Arrows indicate the cleavage sites detected by RLM-RACE.
Widespread Expression of miR858a in Arabidopsis
To investigate the spatiotemporal expression of endogenous miR858a, quantitative real-time (qRT)-PCR analysis of mature miR858a was carried out at different stages of Arabidopsis development. The analysis suggested that miR858a is expressed in rosette, root, stem, flower, silique, and seedling (Fig. 1B). Comparative expression analysis revealed that miR858a is found predominantly in the silique, at least 1 order of magnitude higher compared with seedling, root, stem, rosette leaves, and flower of the mature plant. These results revealed that miR858a is expressed extensively in all the studied tissues and indicated that it may have an important role in plant growth and development.
To study the tissue-specific expression of miR858a, a promoter-reporter (ProMIR858a:GUS) expressing transgenic lines of Arabidopsis was developed. Histochemical GUS analysis in tissues of various developmental stages revealed that ProMIR858a is active in seedling, rosette leaf, stem, flower buds, and mature silique (Fig. 1C). It is important to mention that an intense GUS staining was observed in the vascular tissues of hypocotyl, root, and leaf veins of the seedling (Fig. 1C, b and c) as well as in rosette leaf, stem, flower, and silique of mature plants (Fig. 1C, d–h). Although GUS staining patterns (Fig. 1C) are, in general, consistent with the miR858a transcript abundance (Fig. 1B), differences in relative GUS staining between rosette, flower, and silique tissues were observed. These differences might be due to the limitation of the promoter:GUS approach in studying the spatial expression of genes. Altogether, these results suggested that miR858a is expressed in all developmental stages and might be one of the key regulators of plant growth and development.
The search for cis-regulatory motifs in the promoter region of pre-miR858a using the Plant-CARE database revealed an assortment of putative light-responsive elements (LREs; Supplemental Table S2). Light is one the most important factors affecting MYB transcription factors, and thereby flavonoid biosynthesis, in Arabidopsis (Stracke et al., 2010). Based on the abundance of LREs in the miR858a promoter, we assumed that light may be an important factor for the transcriptional activation of miR858a. To verify this, we measured the expression of miR858a and the ProMIR858a:GUS reporter gene under light and dark cycles. In the light-to-dark transition assay, the miR858a precursor transcript level was reduced considerably in the wild-type (Col-0) background (Fig. 1D). We further confirmed the transcriptional activation of the miR858a precursor gene in response to light (Fig. 1E). A marked decrease in GUS staining and GUS gene expression was observed when seedlings were grown under continuous dark for 5 d as compared with standard growth conditions (Supplemental Fig. S1, A, a and b, and B). Interestingly, GUS staining and GUS gene expression increased after the transfer of dark-grown seedlings to light for different time periods (Supplemental Fig. S1, A, c–e, and C). The pattern of GUS staining was comparable to GUS gene expression under a light/dark cycle (Supplemental Fig. S1). Together, our results demonstrate that miR858a is expressed extensively in Arabidopsis and that its transcript levels are highly dependent on light.
miR858a Targets R2R3-MYB Transcription Factors
To predict possible targets of miR858a in Arabidopsis, we used the online prediction algorithm WMD3 and psRNAtarget. The analysis identified more than 40 putative targets of miR858a, including MYB11, MYB12, and MYB111 (Supplemental Table S3). Most of the identified targets were members of the Arabidopsis R2R3 domain-containing MYB family, including MYB3, MYB4, MYB6, MYB11, MYB12, MYB32, MYB63, MYB85, MYB111, MYB123, MYB305, and MYBL2. Our analysis suggests that miR858a may target these transcription factors, as it has a sequence complementary to the mRNA sequences encoding a conserved part of the R2R3-MYB DNA-binding domain. To correlate the expression of miR858a and their putative targets, the abundance of the MYB11, MYB12, and MYB111 genes in different tissues of Arabidopsis was analyzed through qRT-PCR. The results demonstrated differential expression of miR858a as well as MYB11, MYB12, and MYB111 in rosette, stem, flower, silique, and seedling. The maximum expression of miR858a was observed in siliques and the least in rosettes (Fig. 1B). Interestingly, the expression of MYB11, MYB12, and MYB111 overlapped with the abundance of miR858a in the stem and flower (Fig. 1B). These results correlate with previous reports where both miRNA and target mRNAs were detected predominantly in the same tissues in Arabidopsis (Llave et al., 2002; Zhang and Li, 2013). The highest expression of miR858a was observed in the seedling, with least expression of MYB11 and MYB12. The observations clearly suggest that miR858a may target flavonol-specific MYBs in Arabidopsis.
Moreover, to experimentally confirm the target of miR858a, we analyzed cleavage products of RISCmiR858a action through RNA ligase-mediated 5′ amplification of cDNA ends (RLM-RACE). MYB11 and MYB111 were confirmed as targets of miR858a (Fig. 1F). Although, during RLM-RACE analysis, we were unable to get cleavage product of MYB12, an earlier study had shown MYB12 to be a direct target of miR858 (Fahlgren et al., 2007). In addition, cleavage products for MYB6, MYB13, MYB20, MYB42, MYB48, MYB83, TT2, and MYBL2 also were identified through RLM-RACE (Supplemental Fig. S2). The analysis confirmed MYB13 as a direct target of miR858a (Supplemental Fig. S2B). A majority of the cleavage products for MYB6, MYB20, MYB42, MYB48, MYB83, TT2, and MYBL2 were detected outside of the canonical miRNA cleavage sites (Supplemental Fig. S2). It is likely that such cleavage might be mediated by variants of the mature miRNA, abundant miRNA* sequences, or miRNA/miRNA* duplexes from the same hairpin, as suggested by Addo-Quaye et al. (2008). Overall, the results showed that flavonol-specific MYBs, namely MYB11, MYB12, and MYB111, are direct targets of miR858a.
miR858a Modulates the Phenylpropanoid Pathway in Arabidopsis
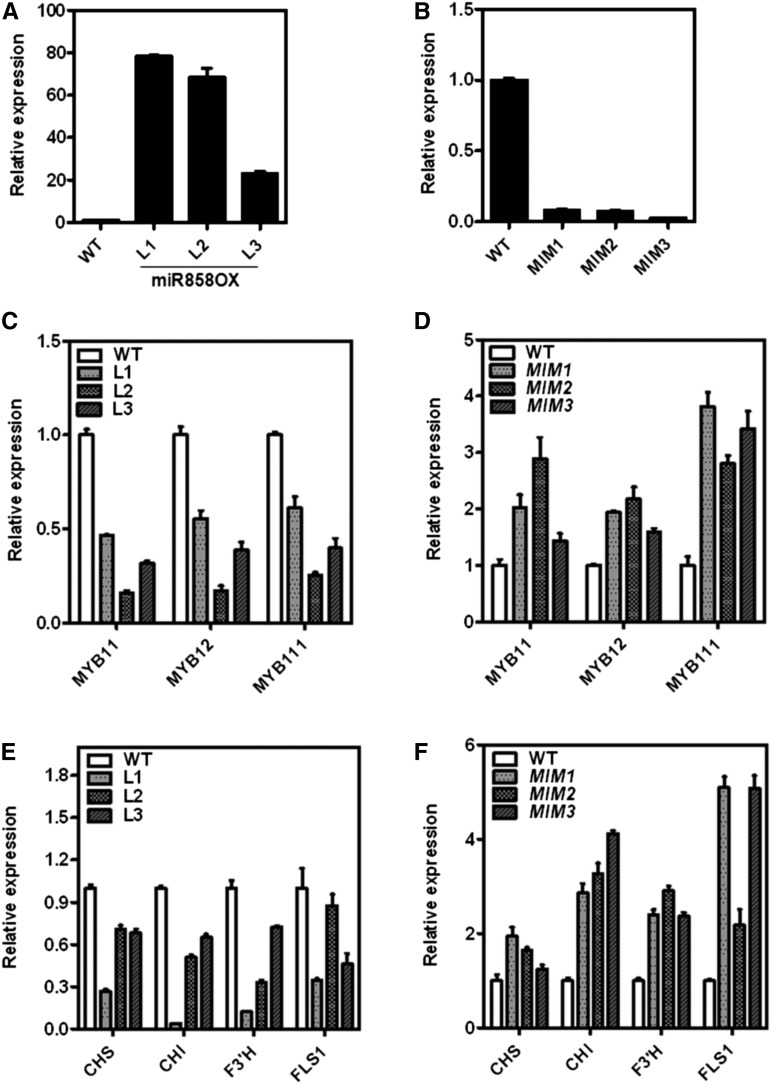
To investigate the function of miR858a, we developed miR858a-overexpressing transgenic lines (miR858OX) under the control of the cauliflower mosaic virus 35S promoter in Arabidopsis. To further strengthen the study, we generated the artificial target mimic (MIM858)-expressing transgenic lines that are supposed to nullify the effect of the endogenous miR858a activity. Homozygous transgenic lines for both miR858OX and MIM858 were selected for further analysis. Significant modulation in the expression of miR858a was observed in miR858OX and MIM858 transgenic lines (Fig. 2, A and B). As expected, the expression of miR858a was significantly higher in miR858OX lines (Fig. 2A). Surprisingly, the expression of miR858a was compromised significantly in transgenic lines expressing MIM858 compared with wild-type plants (Fig. 2B). Previous studies in Arabidopsis and bacteria also have described decreased levels of the targeted miRNA after MIM expression. Those studies suggested that the unproductive interaction of RISCmiRNA with MIM affects miRNA stability (Overgaard et al., 2009; Todesco et al., 2010). We suggest that a similar mechanism might be the reason for the decreased levels of miR858a in MIM858 transgenic lines. Mature miR858a accumulated in miR858OX lines at significantly higher levels in all tissues compared with the wild-type (Supplemental Fig. S3), indicating the successful biogenesis of ectopic miR858a precursors.
Figure 2.
Characterization of miR858OX and MIM858 transgenic lines. A, qRT-PCR analysis showing the expression of mature miR858a in rosette tissue of different miR858OX transgenic lines. B, qRT-PCR analysis showing the expression of mature miR858a in rosette tissue of different MIM858 transgenic lines. C, Expression of MYB11, MYB12, and MYB111 in miR858OX transgenic lines. D, Expression of MYB11, MYB12, and MYB111 in MIM858 transgenic lines. E, Expression of CHS, CHI, F3′H, and FLS1 in miR858OX transgenic lines. F, Expression of CHS, CHI, F3′H, and FLS1 in MIM858 transgenic lines. For miR858a, snoR41Y RNA was used, and for MYBs and pathway genes, TUBULIN was used as the endogenous control to normalize the relative expression levels. WT, wild-type. Bars represent means ± se (n = 3).
To study whether overexpression and sequestration of miR858a affected the expression of MYB11, MYB12, and MYB111 genes, the transcript levels of these MYBs were analyzed in the rosette tissue. The expression of MYB11, MYB12, and MYB111 was reduced significantly in miR858OX lines (Fig. 2C), which suggests a negative correlation between miR858a and target gene expression. On the other hand, as a result of the sequestration of endogenous miR858a by the expression of MIM858, elevated levels of the MYB11, MYB12, and MYB111 transcripts were observed (Fig. 2D). Detailed study of the expression patterns of miR858a targets revealed that most of these genes were down-regulated in different tissues in miR858OX lines (Supplemental Fig. S4).
To study whether the modulation of miR858a levels affects flavonoid pathway genes, the expression of CHS, CHI, F3′H, and FLS1 was studied in transgenic lines. Analysis revealed that the expression of these genes was reduced in miR858OX lines in comparison with the wild type (Fig. 2E). Contrasting results were obtained in MIM858 transgenic plants, with significantly enhanced expression of CHS, CHI, F3′H, and FLS1 as compared with the wild-type (Fig. 2F). This suggests that miR858a expression negatively affects the flavonoid biosynthetic pathway by repressing the activities of the MYB11, MYB12, and MYB111 transcription factors. Moreover, we also performed comparative expression analysis between the wild-type and miR858OX lines using the Affymetrix microarray platform. Analysis revealed that the expression of the putative targets (MYB40, MYB65, MYB83, MYB86, and MYB111) and the flavonoid biosynthesis genes (CHS, CHI, and FLS1) was negatively correlated with the miR858a expression levels (Supplemental Table S4; Supplemental Fig. S5, A and B). Altogether, the data imply that miR858a controls the phenylpropanoid pathway by regulating the expression of the flavonol-specific MYB transcription factors in Arabidopsis.
miR858a Is Required for Plant Growth and Development
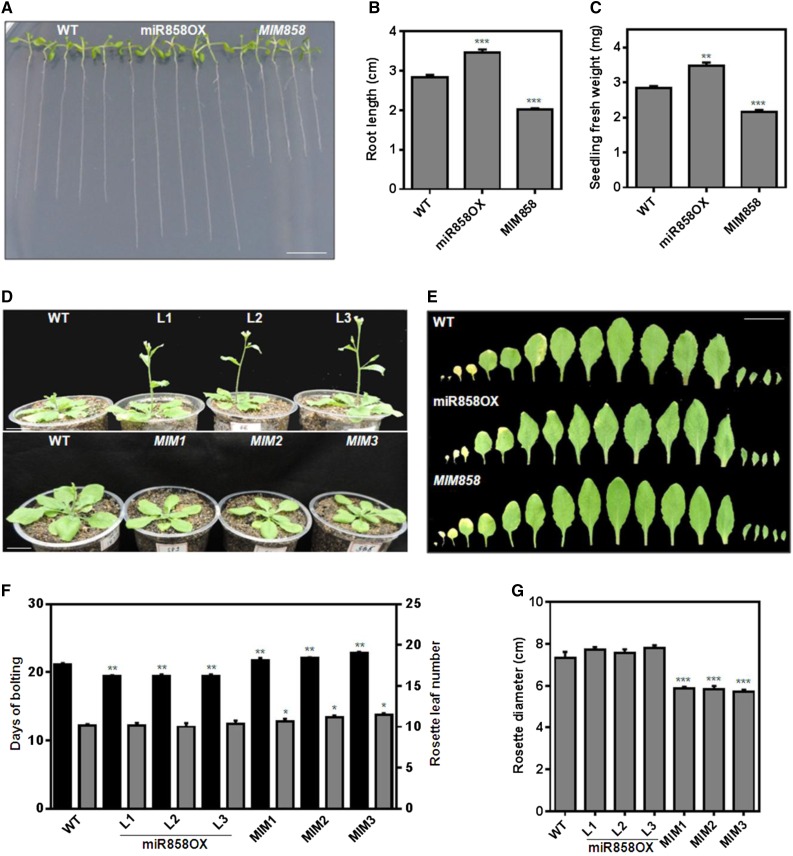
Our genome-wide expression analysis and in silico prediction along with the RLM-RACE analysis confirmed MYB20, MYB42, and MYB83 as targets of miR858a (Supplemental Data Set S1; Supplemental Fig. S2). These MYBs are known to be associated with plant growth and development (Zhong et al., 2008; McCarthy et al., 2009; Dubos et al., 2010; Zhong and Ye, 2012). Therefore, we studied growth parameters of the transgenic and wild-type plants throughout the life cycle. The results suggested that ectopic expression of miR858a and MIM858 changes the various morphological characteristics, with contrasting phenotypes between miR858OX and MIM858 transgenic lines. Transgenic plants overexpressing miR858a showed an elevated vegetative growth phenotype, while MIM858 exhibited reduced vegetative growth compared with wild-type plants just after germination. Significantly longer root length, cotyledonary growth, and seedling fresh weight were observed in miR858OX lines compared with the wild type. In addition, shorter primary roots with smaller cotyledonary growth and lower seedling fresh weight were observed in MIM858 lines compared with miR858OX lines and the wild type (Fig. 3, A–C; Supplemental Table S5; Supplemental Fig. S6A).
Figure 3.
Alteration of morphology in the miR858a transgenic lines. A, Seedling phenotypes of the wild type (WT) and miR858OX, and MIM858 transgenic lines grown on one-half-strength Murashige and Skoog (MS) medium for 10 d. B, Root length of wild-type, miR858OX, and MIM858 seedlings after growth for 10 d on one-half-strength MS medium. C, Seedling fresh weight (mg) of the wild type, miR858OX, and MIM858 after growth for 10 d on one-half-strength MS medium. Phenotype (A) and other data included in B and C for other transgenic lines are shown in Supplemental Figure S6. D, Rosette phenotypes of wild-type, miR858OX (L1–L3), and MIM858 (MIM1–MIM3) transgenic plants. E, Leaf size and pattern of each rosette leaf beginning from the oldest one of wild-type, miR858OX, and MIM858 transgenic plants. F, Number of rosette leaves (black bars) and days of bolting (gray bars) of wild-type, miR858OX, and MIM858 transgenic plants grown under long-day conditions (16-h-light/8-h-dark cycle). G, Rosette diameters of transgenic lines and the wild type. Plants were grown for 3 weeks under a 16-h-light/8-h-dark photoperiod before photography. Bars represent means ± se of eight to 10 individual plants. The level of significance was evaluated by one-way ANOVA with the Newman-Keuls test as a posthoc test. Asterisks indicate significant differences from the wild type: *, P < 0.01; **, P < 0.001; and ***, P < 0.0001. Bars in A, D, and E = 1 cm.
Rapid growth and more serrate margins of rosette leaves were observed in mature miR858OX transgenic lines compared with the wild type (Fig. 3, D and E). However, no significant difference was seen in the leaf number, rosette diameter, rosette fresh weight, and plant height in mature miR858OX transgenic plants (Fig. 3, E–G; Supplemental Tables S5 and S6; Supplemental Fig. S6B). Concomitantly, MIM858 lines displayed a smaller rosette phenotype during the early developmental stage. Even at the time of bolting, the rosette diameter was reduced significantly compared with the wild type (Fig. 3D; Supplemental Table S5). The rosette leaves of mature MIM858 plants were elongated with sinuate margins compared with wild-type and miR858OX rosette leaves (Supplemental Fig. S6B). Marked differences in rosette fresh weight and plant height also were observed (Supplemental Table S6; Supplemental Fig. S6). These results suggest that miR858a is one of the crucial regulators of plant growth and development.
Altered Reproductive Phase Parameters in Transgenic Lines
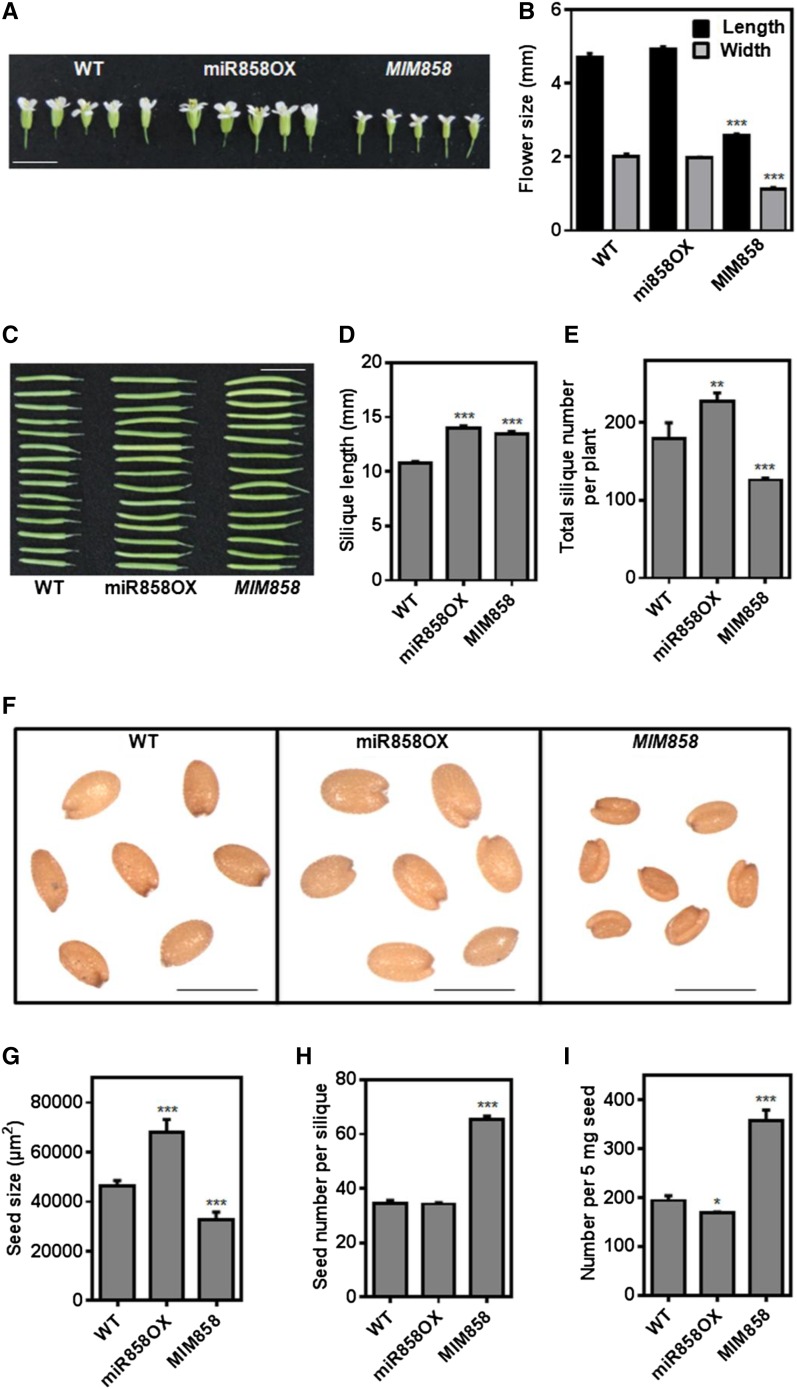
Variations in the flowering time and flower size of miR858OX and MIM858 transgenic lines were observed compared with wild-type plants (Figs. 3 and 4). Under standard growth conditions, early bolt formation and flowering were observed in the miR858OX plants in comparison with the wild type, whereas bolt formation and flowering time were delayed significantly in the MIM858 plants relative to the wild type (Fig. 3, D and F). More rosette leaves compared with wild-type leaves were observed in the MIM858 transgenic lines, although no significant change was observed in the miR858OX lines. Unlike miR858OX lines, a significantly reduced rosette diameter was observed in the MIM858 transgenic lines compared with the wild type (Fig. 3, D and G). Aside from flowering time, significant reductions in the length and width of flowers were observed in MIM858 transgenic lines compared with the wild type (Fig. 4, A and B; Supplemental Table S7). However, no such difference was observed in the miR858OX lines. These results suggested that modulation in the expression of miR858a affected flowering time and flower size, which might be controlled coordinately by miR858 in flowering plants.
Figure 4.
Variation of flower, silique, and seed parameters in the transgenic lines. A, Flower size, both length and width, was reduced in MIM858 transgenic plants, while no significant change was observed in miR858OX flowers. Photographs of flowers were taken after 5 weeks of growth under standard conditions. Bar = 1 cm. B, Mean length and width of independent transgenic lines, with variation represented as se (n = 8–12). C, Phenotypes of transgenic lines producing longer siliques than the wild type (WT). Siliques from 6-week-old plants are shown. Bar = 1 cm. D, The transgenic lines produced longer siliques than the wild type. E, The transgenic lines produced more siliques per plant than the wild type. F, Representative photographs of the seed size of transgenic and wild-type plants. The seed size of miR858OX was bigger, and that of MIM858 was significantly smaller, compared with the wild type. Bars = 500 µm. G, Seed size of different transgenic lines and wild-type plants. H, Seed number per silique was higher in the MIM858 transgenic line, while no difference was observed in the miR858OX line. I, Number of seeds per 5 mg was lower in miR858OX but higher in MIM858 transgenic lines. Data are presented as means ± se (n = 8–12). Data were recorded from 15 siliques per plant and 8 to 12 plants for each line and the wild type. Significance was determined by one-way ANOVA with the Newman-Keuls test as a posthoc test. Asterisks indicate significant differences from the wild type: *, P < 0.01; **, P < 0.001; and ***, P < 0.0001.
Furthermore, observations also suggest that miR858OX lines have increased number and length of siliques per plant (Fig. 4, C–E; Supplemental Table S8; Supplemental Fig. S6C) as well as enlarged seed size (Fig. 4, F and G) compared with the wild type. In contrast, MIM858 lines produced a lower number but longer siliques per plant, with seeds significantly smaller in size (Fig. 4, C–F; Supplemental Table S8; Supplemental Fig. S6C). Additionally, miR858OX lines had a smaller number of seeds per silique while MIM858 produced more seeds per silique compared with the wild type (Fig. 4H; Supplemental Table S8). We also counted the number of seeds per unit (5 mg) weight and observed a significant increase in seed count in MIM858 compared with miR858OX and wild-type plants (Fig. 4I; Supplemental Table S8), indicating larger seed size in miR858OX and smaller seed size in MIM858 transgenic lines. Enhanced vegetative growth with increased seed size and weight in miR858OX suggested an important role of miR858a during seed development and plant yield.
To identify differentially expressed genes in miR858OX lines compared with the wild type, microarray analysis was carried out. Analysis revealed differential expression of genes involved in jasmonate, ethylene, and gibberellin signaling pathways and regulatory mechanisms. In addition, the expression of many development- and stress-responsive genes was modulated in the transgenic lines (Supplemental Data Set S1). The expression of significantly up-regulated genes identified through microarray analysis was validated using qRT-PCR (Supplemental Fig. S7) and showed a similar expression trend. Altogether, significant changes in the phenotypes and microarray expression analysis suggested that miR858a plays a versatile role in plant growth and development of Arabidopsis.
Metabolite Profiling in miR858OX and MIM858 Transgenic Plants
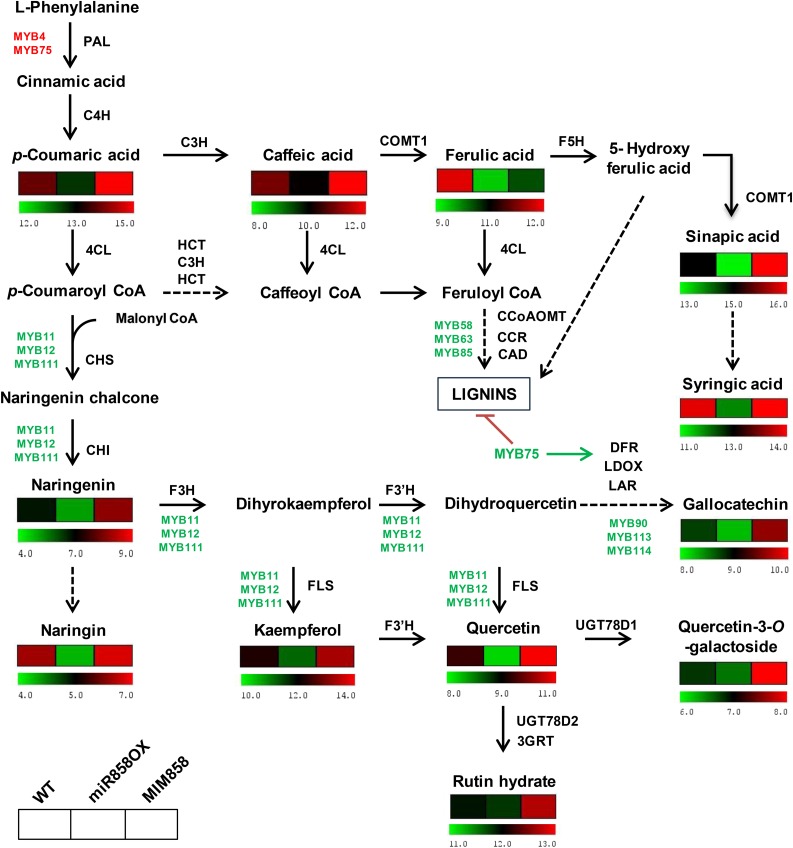
To investigate the impact of miR858a on the synthesis of metabolites via the phenylpropanoid pathway, Q-Trap mass spectrometric analysis of methanolic extracts of rosette and stem of 5-week-old wild-type, miR858OX, and MIM858 plants was carried out (Table I). The modulation in contents of molecules of various steps of the phenylpropanoid pathway in miR858OX and MIM858 transgenic plants compared with the wild type is summarized in Figure 5. The analysis revealed higher accumulation of the major flavonoids, such as kaempferol, quercetin, and rutin hydrate, in MIM858 compared with wild-type plants. In contrast, a significant decrease in these flavonoids was observed in the miR858OX rosette tissue (Table I; Fig. 5). Interestingly, these compounds are among the most abundant flavonol derivatives in wild-type plants (Stracke et al., 2007). Kaempferol was significantly higher (10,199.6 ± 161.0 µg per 100 g dry weight) in rosette tissues of MIM858 compared with the wild type (4,808.3 ± 93.3 µg per 100 g dry weight) and miR858OX (2,320.4 ± 36.6 µg per 100 g dry weight). In addition, other flavonols, such as quercetin-3-O-galactoside, and metabolites belonging to different classes of flavonoids, such as flavanones (naringenin and naringin) and flavan-3-ols (gallocatechin), showed similar accumulation patterns (Table I; Fig. 5).
Table I. Metabolite content in rosette tissues of wild-type, miR858OX, and MIM858 transgenic plants.
Rosette tissue samples from plants were taken and metabolite contents analyzed by liquid chromatography-tandem mass spectrometry. Values presented are means ± se (n = 6; µg per 100 g dry weight) of measurements from at least five biological replicates per genotype. Statistically significant values from the wild type are based on one-way ANOVA: *, P < 0.01; **, P < 0.001; and ***, P < 0.0001.
| Molecule | Wild Type | miR858OX | MIM858 |
|---|---|---|---|
| Naringenin | 108.6 ± 1.4 | 34.91 ± 0.4*** | 276.89 ± 3.7*** |
| Naringin | 77.4 ± 1.2 | 19.5 ± 0.2** | 104.73 ± 1.4** |
| Kaempferol | 4,861.65 ± 85.6 | 2,331.8 ± 38.9** | 10,249.41 ± 171.2*** |
| Quercetin | 701.59 ± 13.6 | 285.84 ± 3.9*** | 1,995.2 ± 33.3*** |
| Quercetin-3-O-galactoside | 110.71 ± 2.7 | 92.8 ± 1.5* | 430.84 ± 7.1*** |
| Rutin hydrate | 3,898.99 ± 65.1 | 3,540.58 ± 59.1* | 6,683.54 ± 111.6** |
| Gallocatechin | 431.4 ± 7.2 | 306.93 ± 4.0** | 763.9 ± 10.2*** |
| p-Coumaric acid | 14,434.29 ± 194.1 | 7,120.55 ± 118.9** | 32,506.44 ± 444.3*** |
| Caffeic acid | 1,961 ± 34.2 | 1,062.07 ± 17.7* | 4,516.59 ± 75.4*** |
| Ferulic acid | 3,793.51 ± 66.8 | 679.3 ± 11.3*** | 1,333.91 ± 18.2** |
| Sinapic acid | 32,940.7 ± 550.3 | 8,701.96 ± 145.3** | 66,980.51 ± 915.5*** |
| Syringic acid | 15,325.29 ± 269.9 | 3,901.08 ± 36.7** | 19,290.53 ± 252.7* |
Figure 5.
Metabolic profiling of miR858OX and MIM858 transgenic lines. The schematic diagram shows the phenylpropanoid pathway. Heat maps indicate log2-transformed values of the concentrations of metabolites in rosette tissue of wild-type (WT), miR858OX, and MIM858 transgenic plants (for values, see Table I). Liquid chromatography-tandem mass spectrometry analysis was performed with the methanolic extracts of dried rosette tissues. Arrows indicate the order of steps in the pathway; dashed arrows represent multiple enzymatic steps. The main enzymes are as follows: PAL, Phe ammonia lyase; C4H cinnamate 4-hydroxylase; 4CL, 4-coumaroyl-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′ hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; LDOX, leucoanthocyanidin dioxygenase; LAR, leucoanthocyanidin reductase; UGT78D1, flavonol 3-O-rhamnosyltransferase; UGT78D2, flavonoid 3-O-glucosyltransferase; 3GRT, flavonol-3-O-glucoside l-rhamnosyltransferase; HCT, p-hydroxycinnamoyl-CoA:quinate shikimate p-hydroxycinnamoyltransferase; C3H, p-coumaroyl ester 3-hydroxylase; COMT I, caffeic acid O-methyltransferase of class I; CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; F5H, ferulic acid 5-hydroxylase. MYB transcription factors are shown in green and red (green for activation and red for inhibition). Likewise, the green arrow is for activation and the red arrow is for inhibition.
Other phenylpropanoids, such as p-coumaric acid, caffeic acid, ferulic acid, sinapic acid, and syringic acid, were increased significantly in MIM858, while their accumulation decreased in miR858OX, in comparison with the wild type (Table I; Fig. 5). The accumulation of metabolites in stem varied significantly compared with rosette samples in the transgenic lines. Surprisingly, higher metabolite accumulation was detected in stem tissue of miR858OX as well as MIM858 in contrast to the wild type (Supplemental Table S9). These differences were indicative of the existence of a differential regulation of metabolic pathways by miR858a in different tissues. Collectively, the higher accumulation of flavonoids in MIM858 and their reduced levels in miR858OX in rosette tissue of transgenic lines indicate that miR858a regulates the biosynthesis of flavonoids through the transcriptional regulation of upstream transcription factors that control the expression of genes involved in the flavonoid biosynthesis pathway.
miR858a Affects Lignin Biosynthesis in Transgenic Plants
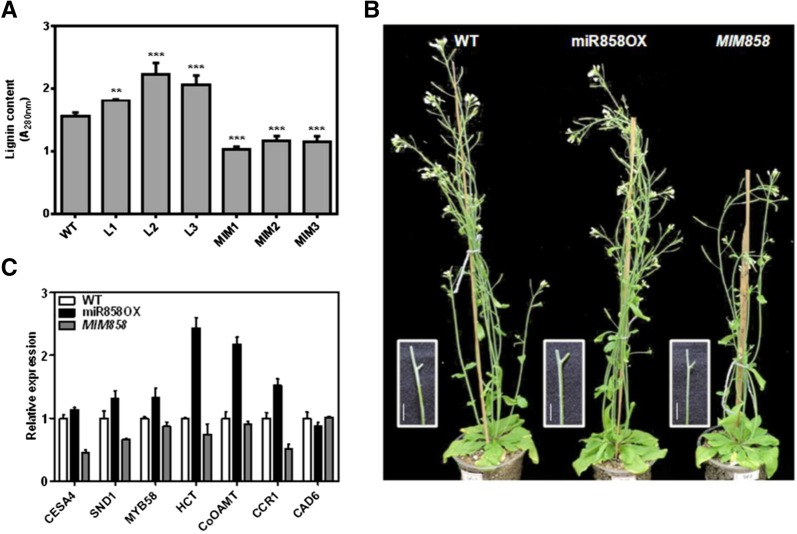
Lignin accumulation analysis suggested modulated lignin content in the vascular and interfascicular tissues of transgenic lines compared with the wild type. Lignin staining and quantification of stem cross sections of the wild type and miR858OX, and MIM858 lines revealed enhanced lignification in the vascular and interfascicular tissues of the miR858OX lines (Fig. 6A; Supplemental Fig. S8). Although miR858a overexpression caused ectopic deposition of lignin, no prominent secondary wall thickening was observed in the epidermal cells. These results are distinct from the observations of the MIM858 lines, which showed less lignification of cells (Supplemental Fig. S8). The examination of vascular bundles and epidermal cells showed that the secondary wall thickness of both vessels and fibers was reduced in the MIM858 line compared with the wild type. Surprisingly, these results are inconsistent with phenotypic observations showing weaker inflorescence stems of MIM858 lines than the wild-type plants (Fig. 6B). Consistent with the histological data, enhanced expression of several secondary wall-promoting and lignin biosynthetic genes, including CESA4, SND1, MYB58, HCT, CCoAOMT1, CCR1, and CAD6, was observed in the stem tissue of miR858OX transgenic lines (Fig. 6C). At the same time, reduced expression of these genes was observed in the MIM858 lines. These observations explicitly suggest that the miR858a-MYB interaction maintains the metabolic flux balance between flavonoid and lignin biosynthesis by activating a subset of the secondary wall and lignin biosynthetic genes.
Figure 6.
miR858a expression affects lignin deposition in Arabidopsis. A, Quantification of total lignin content in the wild-type and the transgenic lines. Mature stems of wild-type, miR858OX, and MIM858 transgenic plants were equally weighed and used for quantification. Bars represent means ± se of three biological replicates at each sampling time point. Significance was determined by one-way ANOVA with the Newman-Keuls test as a posthoc test. B, Growth phenotypes of 5-week-old wild-type, miR858OX, and MIM858 plants. Bars = 1 cm. C, Expression profiles of CESA4, SND1, MYB58, HCT, CCoAOMT1, CCR1, and CAD6 in the stem of wild-type, miR858X, and MIM858 plants. The transcript abundance of each gene was normalized to the level of the TUBULIN gene. Bars represent means ± se (n = 3).
DISCUSSION
The regulation of gene expression in specific cells and tissues is essential for normal growth of the plant. Over the past few years, a large number of experiments have shown that a subgroup of R2R3-MYBs, such as MYB11, MYB12, and MYB111, apparently influence the spatial accumulation of flavonols by activating a branch of the phenylpropanoid pathway (Mehrtens et al., 2005; Czemmel et al., 2009; Misra et al., 2010; Stracke et al., 2010; Liu et al., 2015). These MYBs participate in fine-tuning the transcriptional activation of a set of genes engaged in flavonol biosynthesis and maintain a subtle balance with other branches of the phenylpropanoid pathway (Xu et al., 2015). Studies suggest that the organ and temporal distributions of flavonols are the consequences of the differential expression of these MYBs and environmental factors in Arabidopsis (Stracke et al., 2007). The fundamental question arises about whether only cis-acting elements present in the promoters of these MYBs are responsible for differential expression or whether any other trans-factors also determine their regulatory mechanisms. Numerous past studies have suggested that miR858 can target R2R3-MYBs in apple, grape, and S. miltiorrhiza (Xia et al., 2012; Rock, 2013; Li et al., 2014). In a quest for such factors, we have shown that miR858a controls the expression of flavonol-specific MYBs, such as MYB11, MYB12, and MYB111 in addition to other members of the MYB family, and studied its detailed role further in Arabidopsis.
miR858 is transcribed from two genomic loci in Arabidopsis and targets similar genes. The additional targets of miR858a identified in this study might be involved in regulating diverse developmental aspects in the plant, which would confirm the significance of miR858a in specific developmental programs in Arabidopsis. The earlier function of miR858 has been recognized in trichome fiber development by regulating MYB2 in Arabidopsis (Guan et al., 2014) and anthocyanin biosynthesis in tomato (Jia et al., 2015). These reports are in agreement with the idea that miR858 might target a number of R2R3-MYB genes and may be involved in the regulatory responses leading to secondary metabolite production and specific developmental programs. The expression of miR858a was found to be widespread in Arabidopsis, as is evident from qRT-PCR results and promoter activity (Fig. 1). The GUS staining observed in the different developmental stages throughout the life cycle of Arabidopsis suggests an active participation of miR858a-mediated regulation in plant growth and development. Despite the in planta expression, miR858a was specifically active in vascular tissues of the seedling.
Light serves as a critical factor in the transcriptional activation of the miR858a precursor, which indicates that, similar to that of MYBs, this aspect of flavonol biosynthesis regulation also is under the tight control of light. In silico analysis of the promoter of miR858a revealed the presence of many LREs, specifically HY5-binding sites, indicating the putative light-mediated regulation of miR858a expression (Supplemental Table S2). HY5 is a member of the bZIP/bHLH transcription factor family that plays a pivotal role in light signaling and photomorphogenesis processes by interacting with various pathways (Lee et al., 2007; Delker et al., 2014). It has been shown that the HY5-mediated transcriptional activation of MYB12 and MYB111 genes is under the control of UV-B and visible light (Stracke et al., 2010). Our light-to-dark transition experiment revealed that miR858a is reduced substantially under the dark condition in Col-0 (Fig. 1D). The expression of miR858a is induced so rapidly that it reached the maximum within 0.5 h of exposure to light, showing the relevance of light in its regulation (Fig. 1E). Therefore, it could be concluded that light is the major factor that controls the expression of miR858a and various genes of the phenylpropanoid pathway.
miR858a cleaves flavonol-specific MYBs such as MYB11, MYB12, and MYB111, which are predicted to be potential targets using online target prediction tools (Supplemental Table S3). MYB11 and MYB111 have been verified as targets through RLM-RACE analysis; however, we were unable to detect the cleavage product of MYB12 in our experimental conditions, although MYB12 was confirmed to be an actual target of miR858 in an earlier study (Fahlgren et al., 2007). MYB transcription factors are considered potential targets of miR858 in several plant species (Addo-Quaye et al., 2008; Xia et al., 2012; Jeong et al., 2013; Guan et al., 2014; Jia et al., 2015). In addition, our in silico prediction and cleavage product analysis for another set of MYBs (Supplemental Fig. S2) suggest that miR858a offers complex regulation in Arabidopsis and is not confined only to flavonol-specific MYBs.
miR858a Expression Is Important for Arabidopsis Growth and Development
miR858a caused a decrease in the expression of target MYBs and affected the growth and morphology in Arabidopsis in miR858OX and MIM858 lines. To nullify the effect of endogenous miR858a, we employed the sponge decoy strategy and developed MIM858 in such a way that it contains two repeats of the exact complementary sequence of miR858a, and three repeats have a three-nucleotide bulge between positions 9 and 11 (Ebert et al., 2007). As expected, MIM858 lines successfully delivered the knockdown phenotype in Arabidopsis. A significant reduction in miR858a and an increase in target gene expression were observed in MIM858 lines (Fig. 2, A–D). Phenotypic examination of both types of transgenic lines revealed that miR858OX and MIM858 lines produced contrasting features of various growth parameters (Figs. 3 and 4; Supplemental Fig. S6; Supplemental Tables S5 and S6). Although the expression of miR858a was least in rosette tissue (Fig. 1B), transgenic lines overexpressing miR858a significantly changed rosette size, flowering time, and metabolite content. This indicates that even a slight variation of miR858a expression in rosette tissue could lead to a much bigger impact on the morphology of the plant where this miRNA is lowly abundant. It is well known that MYB transcription factors play an important role in many developmental processes (Ishida et al., 2007; Reyes and Chua, 2007; McCarthy et al., 2009; Dubos et al., 2010; Cui et al., 2013; Chaonan et al., 2014). Previous studies have found that AtMYB11 modulates overall growth in plants by reducing the proliferation activity of the meristematic cells and delaying plant development, whereas the mutant showed faster development of the inflorescence and initiated more lateral inflorescences and fruits (Petroni et al., 2008). Similarly, contrasting phenotypes were obtained in miR858OX and MIM858 transgenic lines, in which the expression of MYB11 is altered significantly (Figs. 3 and 4; Supplemental Fig. S6; Supplemental Tables S5 and S6).
miRNAs such as miR156, miR172, and miR159 regulate key genes involved in flowering-related processes (Hong and Jackson, 2015; Teotia and Tang, 2015). The contrasting observations of bolt formation, flowering, and flower size between miR858OX and MIM858 lines might be due to the perturbation of flowering-related MYBs. MYBs and flavonoids play a very crucial role in Arabidopsis seed development (Routaboul et al., 2006; Doughty et al., 2014; Xu et al., 2014); therefore, a significant change in seed size in transgenic lines may be an outcome of varied flavonoid content brought on by miR858a. On the other hand, the validation of several other MYBs participating in various developmental processes as targets of miR858a (Supplemental Table S3) could be another reason for such developmental alteration in the transgenic lines. The relation of these transcription factors with miR858a and the downstream cascade could be a subject of future study.
miR858a-Mediated Regulation Affects the Phenylpropanoid Pathway
The regulation of flavonol biosynthesis mainly takes place at the transcriptional level and by feedback inhibition of the end products. The flavonol-specific MYBs induced the expression of structural genes such as CHS, CHI, F3H, and FLS1 of the phenylpropanoid pathway (Stracke et al., 2007; Dubos et al., 2010; Xu et al., 2015). Previously, a plethora of reports suggested that the expression of such MYBs in tobacco (Nicotiana tabacum) and tomato enhances the expression of structural genes of the pathway leading to enhanced flavonoid content, nutritional quality, and insect resistance (Luo et al., 2008; Misra et al., 2010; Pandey et al., 2012, 2014a, 2014b, 2015a, 2015b; Zhang et al., 2015; Choudhary et al., 2016). Here, we describe the participation of miR858a in the transcription factor regulon responsible for the biosynthesis of flavonoids and development in Arabidopsis. Our analysis suggested that the expression of CHS, CHI, F3H, and FLS1 was down-regulated in miR858OX but induced in MIM858 lines (Fig. 2, E and F). These results indicate that MIM858 titrates the endogenous miR858a activity in vivo and enhances the expression of genes encoding enzymes of flavonoid biosynthesis.
Metabolic profiling demonstrated a significant rise in major flavonoids, such as kaempferol and rutin (Table I), and other phenylpropanoids in the MIM858 transgenic lines. Previous studies suggested that flavonoids influence plant architecture (Taylor and Grotewold, 2005; Buer and Djordjevic, 2009; Buer et al., 2010) mainly through their effects on auxin transport. Flavonoid-defective mutants display a wide range of alterations in myriad developmental processes, such as root and shoot development, flower organ number, overall architecture and stature, and seed organ density (Buer and Djordjevic, 2009). Modulation in major flavonoids, namely kaempferol and quercetin, in our metabolic profiles (Table I; Supplemental Table S9) clearly provides the justification for the diverse alteration of phenotypes in the transgenic lines. This notion also was supported by the microarray and qRT-PCR analyses, which revealed the down-regulation of flavonoid-specific MYBs and early pathway genes (Supplemental Data Set S1; Supplemental Table S4; Supplemental Fig. S5).
CHS and HCT share a common substrate (p-coumaroyl-CoA) leading to the biosynthesis of flavonoids and lignin in the phenylpropanoid pathway (Besseau et al., 2007). The repression of lignin biosynthesis by HCT silencing impacts flavonoid accumulation significantly in Arabidopsis (Hoffmann et al., 2004; Besseau et al., 2007; Li et al., 2010). We quantified lignin in the stem of the wild-type and transgenic lines and measured the expression of genes such as CESA4, SND1, MYB58, HCT, CoOAMT, CCR1, and CAD6. The results clearly showed a high lignin content in the stem of miR858OX in contrast to a lower lignin content in MIM858 transgenic lines (Fig. 6). This might be due to the atypical lignification of the mechanical tissues in the stem of MIM858 lines. These results indicate that the miR858a-mediated perturbation of the flavonoid branch of the phenylpropanoid pathway can affect the lignin biosynthesis branch of the pathway. The same interpretation could be applied to explain the rosette diameter and fresh weight phenotype of MIM858 transgenic plants (Supplemental Table S6). Together, flavonoid profiling and transcriptome data from a combinatorial analysis of miR858OX and MIM858 plants suggest that miR858a responses affect the flavonoid biosynthetic pathway. Our results provide new insights into the regulation of the phenylpropanoid pathway by miR858a besides various transcription factors.
MATERIALS AND METHODS
Bioinformatic Analyses
To identify miRNAs targeting Arabidopsis (Arabidopsis thaliana) MYB11, MYB12, and MYB111 genes, BLASTn searches were performed on the Sanger database for miRNAs (http://microrna.sanger.ac.uk; release 15) and for available genomic sequences from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). Sequences of the miR858a precursor and mature isoforms in Arabidopsis and other plant species were retrieved from the miRBase database. To evaluate the hybridization energies and percentage homology between miR858a and their putative targets, alignments of 21 bp between the mature miRNA and the target mRNAs were carried out using psRNAtarget server (http://plantgrn.noble.org/psRNATarget; Dai and Zhao, 2011) and WMD3 (http://wmd3.weigelworld.org; Schwab et al., 2006).
Plant Material and Growth Conditions
Arabidopsis ecotype Col-0 was used as the wild-type plant in this study. For germination, seeds were surface sterilized and placed on one-half-strength MS medium (Sigma-Aldrich) containing 1% Suc. After stratification for 2 to 3 d at 4°C in the dark, seeds were transferred to a growth chamber (Conviron) under controlled conditions of 16-h-light/8-h-dark photoperiod cycle, 22°C temperature, 150 to 180 µmol m−2 s−1 light intensity, and 60% relative humidity for 10 d unless mentioned otherwise. Measurements of the root length and fresh weight were performed on 10-d-old seedlings. Rosette fresh weight and diameter were determined for 20-d-old and 1-month-old plants. Plant height and silique data were taken from the mature plants. For the light-to-dark transition assay, wild-type (Col-0) and ProMIR858a:GUS transgenic seeds were transferred to standard light and complete dark conditions for 5 d. After dark treatment, seedlings were exposed to light for different time periods (0.5, 2, and 6 h) and used for further analysis.
Construct Preparation and Transformation
To overexpress miR858a, the miR858a precursor (187 bp; AT1G71002) was amplified from the single-stranded complementary DNA (cDNA) library prepared from Arabidopsis rosette leaves through PCR using specific primers (Supplemental Table S10). Amplicon was cloned into pT/Z subcloning vector (Fermentas) and then transferred into plant expression vector pBI121 containing the cauliflower mosaic virus 35S promoter. To reduce miR858a activity, a target-mimic approach (Franco-Zorrilla et al., 2007) was used. To synthesize and clone the miR858a target mimic, overlapping forward and reverse primers were designed (Supplemental Table S10). The amplified PCR product was cloned into pT/Z subcloning vector (Fermentas) and then transferred into plant expression vector pBI121. All the constructs were transformed into Agrobacterium tumefaciens (strain GV3101) and used to transform Arabidopsis (Col-0) using the floral dip method (Clough and Bent, 1998). Transformants were screened on one-half-strength MS agar plates containing kanamycin (50 µg mL−1). For promoter analysis, the 1,847-bp upstream region from the transcription start site of miR858a precursor (Fig. 1Ca) was PCR amplified using high-fidelity enzyme mix (Fermentas) from genomic DNA. Amplicon was cloned into pCAMBIA1303 plant binary vector, resulting in the construct ProMIR858:GUS. For identification of the transcription start site of miR858a, a plant promoter analysis navigator (PlantPAN; http://PlantPAN2.itps.ncku.edu.tw) resource was used. Transgenic plants were generated and screened on hygromycin (10 mg mL−1). Out of a minimum of six transgenic lines generated using different constructs, three independent lines each for miR858OX, MIM858, and ProMIR858:GUS were selected for further analysis. All the studies were performed in T4 homozygous lines.
Expression Analysis
For expression analysis, microRNA858a TaqMan PCR assays were used following the recommendations of the manufacturer (Applied Biosystems). A TaqMan PCR assay for the small nuclear RNA snoR41Y was used as a normalization control. For qRT-PCR, total RNA was treated with TURBO DNAfree (Ambion), and 2 µg was reverse transcribed using the RevertAid H minus First-Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer’s instructions. The cDNA was diluted 20 times with nuclease-free water, and 1 µL was used as a template for quantitative PCR performed using Fast SYBR Green mix (Applied Biosystems) in a Fast 7500 Thermal Cycler instrument (Applied Biosystems). Expression was normalized using TUBULIN and analyzed through the comparative ∆∆CT method (Livak and Schmittgen, 2001). Primer sequences used for expression analysis are listed in Supplemental Table S10.
Microarray Analysis
Genome-wide expression analysis was performed using GeneChip Arabidopsis Genome ATH1 microarrays (Affymetrix) and according to Minimum Information About a Microarray Experiment guidelines (Brazma et al., 2001). Total RNA was extracted from 3-week-old rosette leaves of Col-0 and miR858OX plants using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). Briefly, total RNA (250 ng) of each sample was taken as starting material and strictly followed the Affymetrix protocol during RNA amplification, hybridization, washing-staining, and scanning of the arrays. After hybridization, raw data from CEL files were initially analyzed using the Affymetrix Expression Console with library files supplied by Affymetrix. Analyses were performed using robust multiarray average for gene levels without normalization. The log2-transformed data were exported for analysis using Affymetrix Transcriptome Analysis Console 2.0 software. qRT-PCR was carried out to validate microarray data.
RLM-RACE
To map cleavage sites in the candidate targets of miR858a in planta, modified RLM-RACE was performed using the First Choice RLM-RACE Kit (Ambion). Total RNA was extracted from wild-type and miR858OX rosette leaves and ligated directly to RNA oligonucleotide adapter without CIP and TAP treatments. cDNA synthesis was performed using oligo(dT) primer using the RevertAid H minus First-Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer’s instructions. RACE and nested-RACE PCR were carried out subsequently with adaptor-specific forward primers and gene-specific reverse primers for the detection of cleavage products of the target mRNAs. PCR products were cloned into the pT/Z subcloning vector (Fermentas) and sequenced to locate miRNA cleavage sites.
Histochemical Assays
GUS staining was performed using a standard method (Jefferson, 1989). Different tissues of ProMIR858:GUS transgenic lines were incubated in a solution containing 100 mm sodium phosphate buffer, pH 7.2, 10 mm EDTA, 0.1% Triton X-100, 2 mm potassium ferricyanide, 2 mm potassium ferrocyanide, and 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide at 37°C for 4 to 6 h. The chlorophyll was removed by incubation and multiple washes using 70% ethanol.
Flavonoid Accumulation Analysis
Dried tissues (100–500 mg of rosette and stem from mature plants) were ground into the fine powder in liquid N2 and extracted in 80% methanol using an Ultra-Turrax T-25 Digital Homogenizer (IKA) at 16,000 rpm for 2 min. The homogenate was centrifuged at 20,000g for 10 min at 4°C, and the supernatant was filtered through a 0.22-µm polytetrafluoroethylene syringe filter (Millipore). The filtrate was collected in a clear-glass HPLC vial (Agilent Technologies) and injected into the HPLC-mass spectrometry device. All samples were analyzed on the 1260 Infinity series HPLC system (Agilent Technologies) equipped with 1260 Infinity series pump, autosampler, column compartment, and thermostat using a Zorbax Eclipse Plus column (4.6 × 100 mm, 3.5 µm) maintained at 40°C. The mobile phase consisted of an aqueous solution of 0.1% liquid chromatography-mass spectrometry-grade formic acid (solution A) and 0.1% formic acid in liquid chromatography-mass spectrometry-grade acetonitrile (solution B). The gradient was programmed as follows: 0 to 2 min, solution B (2%); 2 to 22 min, solution B (2%–98%); and 22.01 to 25 min, solution B (98%–2%). Other chromatographic parameters included a constant flow of 600 µL min−1 (injection volume, 5 µL), autosampler temperature of 10°C, and run time of 25 min including equilibration. Before analysis, all of the samples were filtered through 0.22-µm syringe membrane filters (Millipore).
The detection of different molecules was performed using an HPLC system (Agilent Technologies 1260 Infinity series) coupled to a triple quadrupole system (QTRAP5500; ABSciex) using an electrospray ionization probe in both modes of ionization. The voltage was set at 5,500 V for positive ionization and −4,500 V for negative ionization. The values of gas 1 and gas 2 (50 psi), curtain gas (30 psi), collision-assisted dissociation (medium), and temperature of the source (550°C) were used. The mass spectrometer was used in multiple reaction monitoring mode for identification and quantification analysis. Analytical standards of phenolics were purchased from Sigma-Aldrich. Identification and quantitative analysis were carried out using Analyst software (version 1.5.2) and MultiQuant software (version 2.0.2). The Signal finder algorithm was used in Multiquant software for quantitative analysis.
Lignin Analysis
To visualize lignified cells in stems, hand-cut sections were stained for 2 min using Toluidine Blue O (Sigma-Aldrich) and visualized on a Leica DM2500 microscope (Leica Microsystems). Lignin quantification was carried out using the method described by Bruce and West (1989). Briefly, samples (600 mg) were pulverized in liquid N2 and suspended in 2 mL of ethanol. Following centrifugation at 12,000g (30 min at 4°C), the pellet was dried at 25°C for 12 h. Dried pellet was resuspended in 5 mL of 2 n HCl and 0.5 mL of thioglycolic acid and incubated at 95°C for 8 h followed by cooling to room temperature. The suspension was centrifuged at 12,000g for 30 min, and the pellet was washed with 2.5 mL of double distilled water. After recentrifuging at 12,000g and 4°C for 5 min, the resulting pellet was suspended in 5 mL of 1 n NaOH and agitated gently at 25°C for 18 h. After centrifugation at 12,000g and 4°C for 30 min, 1 mL of HCl was added to the supernatant and the solution was allowed to precipitate at 4°C overnight. Following centrifugation at 12,000g and 4°C for 30 min, the pellet was dissolved in 3 mL of 1 n NaOH and the A280 was recorded as lignin content.
Statistical Analysis
The experiments were arranged in a completely randomized design. Each sample time point, for each treatment, was composed of three independent biological replicates. Data are plotted as means ± se in the figures. Phenotypic observations were monitored for two to three generations for consistency. One-way ANOVA followed by posthoc Newman-Keuls test was used for multiple comparisons of significance using GraphPad Prism 6.05 software.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of the miR858a is regulated by light.
Supplemental Figure S2. miR858a-mediated cleavage of MYB genes.
Supplemental Figure S3. Expression analysis of miR858a in Arabidopsis.
Supplemental Figure S4. Relative expression of MYB transcription factors in miR858OX transgenic lines.
Supplemental Figure S5. Modulation of gene expression inmiR858OX transgenic lines.
Supplemental Figure S6. Phenotypic variations in miR858OX transgenic lines.
Supplemental Figure S7. Expression analysis of significantly up-regulated genes in miR858OX transgenic lines.
Supplemental Figure S8. miR858a affects lignin depositions in Arabidopsis stem.
Supplemental Table S1. Alignment of miR858a mature sequence among different plant species.
Supplemental Table S2. Putative cis-acting elements in the miR858a promoter.
Supplemental Table S3. Putative targets of Arabidopsis miR858a.
Supplemental Table S4. Differentially expressed MYB transcription factors and flavonoid structural genes.
Supplemental Table S5. Comparisons of seedling root length, fresh weight, and rosette diameter in the wild type and miR858a transgenic lines.
Supplemental Table S6. Comparisons of plant height, rosette fresh weight, and rosette diameter in mature wild-type plants and miR858a transgenic lines.
Supplemental Table S7. Comparisons of silique, seed number, and morphology in the wild type and miR858a transgenic lines.
Supplemental Table S8. Comparisons of flower size in the wild type and miR858a transgenic lines.
Supplemental Table S9. Metabolite content in stem tissues of wild-type plants and miR858OX, and MIM858 transgenic lines.
Supplemental Table S10. Primers used.
Supplemental Data Set S1. List of differentially expressed genes in miR858OX identified through microarray analysis.
Supplementary Material
Glossary
- miRNA
microRNA
- qRT
quantitative real-time
- LREs
light-responsive elements
- Col-0
Columbia-0
- RLM-RACE
RNA ligase-mediated 5′ amplification of cDNA ends
- MS
Murashige and Skoog
- cDNA
complementary DNA
Footnotes
This work was supported by the Council of Scientific and Industrial Research (grant no. BSC–0107 and senior research fellowships to D.S., A.P., and C.B.) and the Indian Council of Medical Research (research fellowship to M.T.).
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Kim MC, Chun HJ, Kang S, Park HC, Shin G, Park J, Shen M, Hong H, Kim WY, et al. (2013) Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol 161: 362–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. (2001) Minimum Information About a Microarray Experiment (MIAME): toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Bruce RJ, West CA (1989) Elicitation of lignin biosynthesis and isoperoxidase activity by pectic fragments in suspension cultures of castor bean. Plant Physiol 91: 889–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Djordjevic MA (2009) Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot 60: 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaonan L, Ng CKY, Liu-Min F (2014) MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot 114: 80–91 [Google Scholar]
- Choudhary D, Pandey A, Adhikary S, Ahmad N, Bhatia C, Bhambhani S, Trivedi PK, Trivedi R (2016) Genetically engineered flavonol enriched tomato fruit modulates chondrogenesis to increase bone length in growing animals. Sci Rep 6: 21668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2014) The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80: 1108–1117 [DOI] [PubMed] [Google Scholar]
- Cui MH, Yoo KS, Hyoung S, Nguyen HT, Kim YY, Kim HJ, Ok SH, Yoo SD, Shin JS (2013) An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett 587: 1773–1778 [DOI] [PubMed] [Google Scholar]
- Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, Robinson SP, Bogs J (2009) The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol 151: 1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima JC, Loss-Morais G, Margis R (2012) MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genet Mol Biol (Suppl) 35: 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J, et al. (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep 9: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Doughty J, Aljabri M, Scott RJ (2014) Flavonoids and the regulation of seed size in Arabidopsis. Biochem Soc Trans 42: 364–369 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Eamens AL, Wang MB (2011) Alternate approaches to repress endogenous microRNA activity in Arabidopsis thaliana. Plant Signal Behav 6: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, Chen ZJ (2014) miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5: 3050. [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Jackson S (2015) Floral induction and flower formation: the role and potential applications of miRNAs. Plant Biotechnol J 13: 282–292 [DOI] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, et al. (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19: 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Jeong DH, Thatcher SR, Brown RS, Zhai J, Park S, Rymarquis LA, Meyers BC, Green PJ (2013) Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiol 162: 1225–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Shen J, Liu H, Li F, Ding N, Gao C, Pattanaik S, Patra B, Li R, Yuan L (2015) Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242: 283–293 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Reichel M, Li Y, Millar AA (2014) The functional scope of plant microRNA-mediated silencing. Trends Plant Sci 19: 750–756 [DOI] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8: 689–708 [DOI] [PubMed] [Google Scholar]
- Liu Q, Chen YQ (2010) A new mechanism in plant engineering: the potential roles of microRNAs in molecular breeding for crop improvement. Biotechnol Adv 28: 301–307 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Li Q, Wei H, Chang MJ, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun YH, Yuan L, et al. (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci USA 110: 10848–10853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Butelli E, Hill L, Parr A, Niggeweg R, Bailey P, Weisshaar B, Martin C (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J 56: 316–326 [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50: 1950–1964 [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Shao C, Wang H, Jin Y (2012) Target mimics: an embedded layer of microRNA-involved gene regulatory networks in plants. BMC Genomics 13: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra P, Pandey A, Tiwari M, Chandrashekar K, Sidhu OP, Asif MH, Chakrabarty D, Singh PK, Trivedi PK, Nath P, et al. (2010) Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol 152: 2258–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Zhang C, Miller M, Palmer G, Whiteley M, Tholl D, Chen ZJ (2011) Cis- and trans-regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23: 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P (2009) Switching off small RNA regulation with trap-mRNA. Mol Microbiol 73: 790–800 [DOI] [PubMed] [Google Scholar]
- Pandey A, Misra P, Bhambhani S, Bhatia C, Trivedi PK (2014a) Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci Rep 4: 5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Misra P, Chandrashekar K, Trivedi PK (2012) Development of AtMYB12-expressing transgenic tobacco callus culture for production of rutin with biopesticidal potential. Plant Cell Rep 31: 1867–1876 [DOI] [PubMed] [Google Scholar]
- Pandey A, Misra P, Choudhary D, Yadav R, Goel R, Bhambhani S, Sanyal I, Trivedi R, Trivedi PK (2015a) AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues. Sci Rep 5: 12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Misra P, Khan MP, Swarnkar G, Tewari MC, Bhambhani S, Trivedi R, Chattopadhyay N, Trivedi PK (2014b) Co-expression of Arabidopsis transcription factor, AtMYB12, and soybean isoflavone synthase, GmIFS1, genes in tobacco leads to enhanced biosynthesis of isoflavones and flavonols resulting in osteoprotective activity. Plant Biotechnol J 12: 69–80 [DOI] [PubMed] [Google Scholar]
- Pandey A, Misra P, Trivedi PK (2015b) Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep 34: 1515–1528 [DOI] [PubMed] [Google Scholar]
- Petroni K, Falasca G, Calvenzani V, Allegra D, Stolfi C, Fabrizi L, Altamura MM, Tonelli C (2008) The AtMYB11 gene from Arabidopsis is expressed in meristematic cells and modulates growth in planta and organogenesis in vitro. J Exp Bot 59: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R (2006) PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18: 1274–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49: 592–606 [DOI] [PubMed] [Google Scholar]
- Rock CD. (2013) Trans-acting small interfering RNA4: key to nutraceutical synthesis in grape development? Trends Plant Sci 18: 601–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224: 96–107 [DOI] [PubMed] [Google Scholar]
- Santelia D, Henrichs S, Vincenzetti V, Sauer M, Bigler L, Klein M, Bailly A, Lee Y, Friml J, Geisler M, et al. (2008) Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J Biol Chem 283: 31218–31226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Tiwari M, Lakhwani D, Tripathi RD, Trivedi PK (2015) Differential expression of microRNAs by arsenate and arsenite stress in natural accessions of rice. Metallomics 7: 174–187 [DOI] [PubMed] [Google Scholar]
- Stauffer E, Maizel A (2014) Post-transcriptional regulation in root development. Wiley Interdiscip Rev RNA 5: 679–696 [DOI] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R (2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203 [DOI] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8: 317–323 [DOI] [PubMed] [Google Scholar]
- Teotia S, Tang G (2015) To bloom or not to bloom: role of microRNAs in plant flowering. Mol Plant 8: 359–377 [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang CY, Zhang S, Yu Y, Luo YC, Liu Q, Ju C, Zhang YC, Qu LH, Lucas WJ, Wang X, et al. (2014) MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol J 12: 1132–1142 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. (2013) Plant microRNAs and development. J Genet Genomics 40: 217–230 [DOI] [PubMed] [Google Scholar]
- Xia R, Zhu H, An YQ, Beers EP, Liu Z (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20: 176–185 [DOI] [PubMed] [Google Scholar]
- Xu W, Lepiniec L, Dubos C (2014) New insights toward the transcriptional engineering of proanthocyanidin biosynthesis. Plant Signal Behav 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. (2015) MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot 66: 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Q (2015) MicroRNA-based biotechnology for plant improvement. J Cell Physiol 230: 1–15 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li L (2013) SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J 74: 98–109 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Alseekh S, Tohge T, Rallapalli G, Luo J, Kawar PG, Hill L, Santino A, Fernie AR, et al. (2015) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2012) MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53: 368–380 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.