In the green alga C. reinhardtii, reversible S-nitrosylation of a cytosolic translation repressor acts as a redox switch that fine-tunes light harvesting in response to a fluctuating light supply.
Abstract
Photosynthetic eukaryotes are challenged by a fluctuating light supply, demanding for a modulated expression of nucleus-encoded light-harvesting proteins associated with photosystem II (LHCII) to adjust light-harvesting capacity to the prevailing light conditions. Here, we provide clear evidence for a regulatory circuit that controls cytosolic LHCII translation in response to light quantity changes. In the green unicellular alga Chlamydomonas reinhardtii, the cytosolic RNA-binding protein NAB1 represses translation of certain LHCII isoform mRNAs. Specific nitrosylation of Cys-226 decreases NAB1 activity and could be demonstrated in vitro and in vivo. The less active, nitrosylated form of NAB1 is found in cells acclimated to limiting light supply, which permits accumulation of light-harvesting proteins and efficient light capture. In contrast, elevated light supply causes its denitrosylation, thereby activating the repression of light-harvesting protein synthesis, which is needed to control excitation pressure at photosystem II. Denitrosylation of recombinant NAB1 is efficiently performed by the cytosolic thioredoxin system in vitro. To our knowledge, NAB1 is the first example of stimulus-induced denitrosylation in the context of photosynthetic acclimation. By identifying this novel redox cross-talk pathway between chloroplast and cytosol, we add a new key element required for drawing a precise blue print of the regulatory network of light harvesting.
Photosynthetic organisms constantly adapt their light-harvesting machinery to a changing environment to ensure optimal photosynthetic efficiency (Anderson et al., 1995). Expression control of the major light-harvesting proteins at photosystem II (LHCII) provides an efficient means to regulate the initial step of photosynthesis. In unicellular green algae, including the model organism Chlamydomonas reinhardtii, exposure to high light causes a precipitous drop in LHCII transcript levels, while a decreased light availability has an opposite effect (Escoubas et al., 1995; Teramoto et al., 2002; Durnford et al., 2003; Chen et al., 2004). This strong modulation of transcript levels is mainly achieved by controlling nuclear transcription initiation rates (Escoubas et al., 1995), which is in agreement with an unchanged stability of LHCII transcripts in high-light-grown C. reinhardtii cells (Durnford et al., 2003). In this alga, translation of LHCII mRNAs also is controlled by light, and its repression occurs after exposure to elevated light intensity (Durnford et al., 2003; Mussgnug et al., 2005).
C. reinhardtii contains the cytosolic RNA-binding protein NAB1, which acts as a translation repressor by sequestrating LHCBM mRNAs into translationally silent mRNP (messenger ribonucleoprotein) complexes, thus preventing their translation (Mussgnug et al., 2005). NAB1 shows a preference for certain LHCBM isoform transcripts, and in accordance with its function as a LHCII translation repressor, the NAB1 knockout mutant overaccumulates light-harvesting proteins and displays a dark-green phenotype (Mussgnug et al., 2005). The extent of NAB1-mediated translation repression is controlled at multiple levels, which perfectly reflects the vital role that light harvesting plays in a phototrophic organism (Wobbe et al., 2009; Blifernez et al., 2011; Berger et al., 2014).
For instance, carbon dioxide limiting conditions trigger the accumulation of NAB1 via nuclear promoter activation, which in turn reduces the synthesis of LHCII proteins when antenna size reduction is required to maintain a normal photosystem II (PSII) excitation pressure, while the reductant sink capacity of the Calvin-Benson cycle is restricted (Berger et al., 2014). Promoter activation can be abolished by blocking photosynthetic electron transport, indicating that NAB1 expression is modulated by chloroplast retrograde signals. In fact, several lines of evidence suggest that NAB1 can be regarded as a regulatory hub connecting short- and long-term photoacclimation mechanisms.
The RNA-binding activity of NAB1 is regulated by two types of posttranslational modifications with Arg methylation acting as a master switch that adjusts activity to the prevailing metabolic situation (Blifernez et al., 2011). In addition, NAB1 activity is redox controlled via modification of Cys residues. The protein contains two cysteines, Cys-181 and Cys-226, both located in the RNA recognition motif (RRM; Wobbe et al., 2009). Oxidative modification of NAB1 decreases both RNA-binding and translation repressor activity of the enzyme. Replacement of either amino acid with Ser impairs NAB1 deactivation, causing a pale-green, small antenna phenotype (Beckmann et al., 2009; Wobbe et al., 2009), but single mutation of Cys-226 causes a much stronger phenotype than the replacement of Cys-181 (Wobbe et al., 2009). Together with biochemical data, the phenotypic difference between the two Cys single mutants demonstrated that single Cys modification and not the formation of intramolecular disulfide bonds must be the mechanism of NAB1 deactivation (Wobbe et al., 2009).
Two redox posttranslational modifications of cysteines, namely, nitrosylation and glutathionylation, recently emerged as key mechanisms of cell regulation and signaling that can affect the function of hundreds of proteins and are implicated in a broad spectrum of human diseases, including cancer, diabetes, and several neurodegenerative, cardiovascular, muscular, or pulmonary diseases (Martínez-Ruiz and Lamas, 2007; Mieyal et al., 2008; Foster et al., 2009). Protein glutathionylation consists of the formation of a mixed disulfide between glutathione (GSH; γ-l-glutamyl-l-cysteinyl-Gly) and a protein thiol. This modification can protect protein thiols from overoxidation but can also have regulatory and signaling functions (Zaffagnini et al., 2012). Protein S-nitrosylation results from the covalent binding of nitric oxide (NO) to a Cys thiol moiety and plays a major role in numerous cellular processes in mammals (Benhar et al., 2009). NO production has also been reported in plants where it has been detected in different tissues or subcellular compartments, such as plastids, peroxisomes, or mitochondria (Baudouin, 2011; Fröhlich and Durner, 2011). In animals, the main source of NO is a reaction catalyzed by NO synthases, but the relevance of this synthesis pathway for plant NO metabolism is still a matter of debate (Fröhlich and Durner, 2011). Although a NO synthase homolog could be characterized in the microalga Ostreococcus tauri (Foresi et al., 2010), nitrite and nitrate reductase seem to be the main NO-producing systems in the plant kingdom (Baudouin, 2011; Fröhlich and Durner, 2011), where a multitude of physiological processes have been shown to rely on NO signaling (Astier et al., 2011). Nitric oxide can not only react with protein thiols directly to form protein S-nitrosothiols, but can also form adducts with the most abundant intracellular Cys-containing tripeptide glutathione. S-nitrosoglutathione (GSNO) is regarded as the main mobile NO reservoir of the cell and a major transnitrosylating agent. Its concentration is controlled by GSNO reductase, which catalyzes the reduction of GSNO to oxidized glutathione (GSSG) and ammonia, thereby modulating indirectly the level of nitrosylated proteins (Liu et al., 2001). Recently, a proteomic study conducted with C. reinhardtii demonstrated that S-nitrosylation could constitute a major regulatory mechanism, when this organism is exposed to nitrosative stress and numerous identified S-nitrosylation targets are implicated in photosynthesis (Morisse et al., 2014). In this study, we set out to determine the physiological role and type of single Cys modification occurring on NAB1 in vivo. Overall our data clearly demonstrate that reversible nitrosylation of a specific Cys residue in the RRM domain of NAB1 modulates translation of light-harvesting protein-encoding mRNAs in the cytosol of C. reinhardtii cells. The fact that denitrosylation of NAB1 is catalyzed by a cytosolic thioredoxin (TRX) system in vitro provides important insights into the cross talk between chloroplast and cytosol, which is needed for a stoichiometric fine-tuning of photosynthetic apparatus components encoded by the nuclear genome.
RESULTS
Cys-226 of the LHCII Translation Regulator NAB1 Is a Potential Site of Nitrosylation
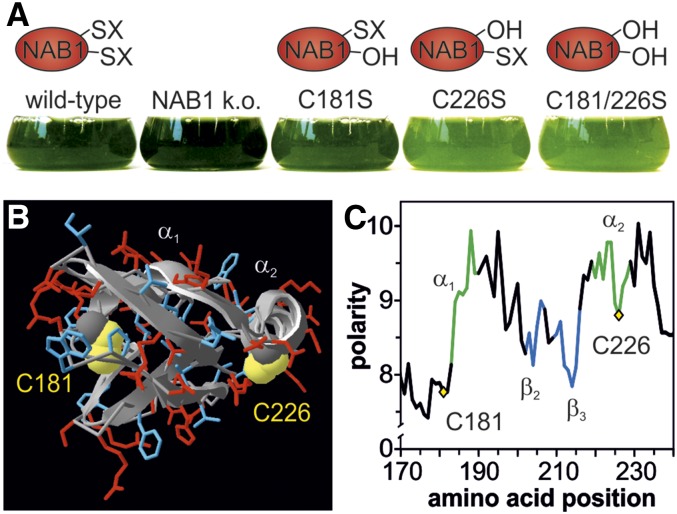
The cytosolic protein NAB1 has a central role as a control hub in the complex regulatory network responsible for adjusting the light-harvesting antenna system to changing environmental conditions (Mussgnug et al., 2005; Wobbe et al., 2009; Blifernez et al., 2011; Berger et al., 2014). In particular, the redox state of two cysteines at amino acid positions 181 and 226 was shown to be critical for the LHCII translation repressor activity of NAB1 (Wobbe et al., 2009). Expression of three distinct NAB1 Cys mutants C181S, C226S, and C181/226S in a knockout strain lacking endogenous NAB1 revealed a pale-green phenotype in case of the double mutant and single mutant C226S (Fig. 1A). The Ser residues replacing cysteines in these NAB1 variants mimic the unmodified form of the protein, which was shown to be the more active state in RNA-binding studies (Wobbe et al., 2009). The low chlorophyll content displayed by mutants C226S and C181/226S is the result of a decreased accumulation of light-harvesting proteins (LHCBM), which can in turn be explained by the disruption of a crucial deactivation mechanism and translation repression being locked in the “on” state (Wobbe et al., 2009). The phenotype of the single Cys mutant C226S clearly shows that this residue alone is sufficient for a redox control of NAB1 activity.
Figure 1.
Essentiality of Cys-226 for NAB1 redox control and in silico indication for its nitrosylation. A, Pale-green phenotype displayed by cell lines expressing NAB1 variants that lack Cys-226, which is essential for Cys-based redox control of its repressor activity. In these variants, cysteines amenable to redox-based modifications (-SX) are replaced with nonreactive serines (-OH) that mimic the free thiol state (-SH). B, In silico model of the NAB1-RRM domain. Polar and charged amino acids in the surrounding of cysteines 181 and 226 are shown in red, and uncharged/nonpolar amino acids are depicted in blue. The protein backbone is shown in a ribbon presentation. C, Polarity plot of the RRM domain. The relative polarity is shown on the y axis, while the amino acid position is given on the x axis. α-helices, β-sheets, and loop regions are indicated by a green, blue, and black color, respectively.
Previous in silico analyses based on homology modeling of the C-terminal RRM domain of NAB1, which harbors both cysteines, demonstrated a distinct surface exposure of Cys-181 and Cys-226. In line with its more crucial function for NAB1 redox control, Cys-226 turned out to be the more accessible Cys (Wobbe et al., 2009). Moreover, the great spatial distance between Cys-181 and Cys-226 suggested that intramolecular disulfide formation was unlikely, and this view was further supported by in vitro experiments conducted with recombinant NAB1 (Wobbe et al., 2009). Glutathionylation of NAB1 could be demonstrated after treatment of recombinant NAB1 with glutathione disulfide, but if this type of modification is relevant for NAB1 redox control in vivo remained unclear (Wobbe et al., 2009). As a starting point for further investigations regarding the precise chemical nature of NAB1 Cys modifications, we extended previous in silico analyses by inspecting the microenvironment of both cysteines more closely (Fig. 1B). Cys-181 is surrounded by nonpolar amino acids (Fig. 1C; blue residues in Fig. 1B) in an environment with a low electrostatic potential, whereas the environment of Cys-226 is characterized by a high electrostatic potential (Fig. 1C). Out of 14 neighboring residues (−7 to +7), seven are charged and two are polar (red residues in Fig. 1B) in the case of Cys-226. This Cys is part of an α-helix (α2; Figure 1B) in a rather surface-exposed area, overall an environment with characteristics of S-nitrosylation sites (Gould et al., 2013). Furthermore, lysines are found overrepresented in proximity to cysteines amenable to nitrosylation (Lindermayr et al., 2006; Morisse et al., 2014), and relative to Cys-226 in NAB1, two lysines are located at positions −7 (K219) and +2 (K228), respectively. These position-specific characteristics support that Cys-226 rather than Cys-181 is the main target for thiol-based NAB1 activity control, as assumed before (Wobbe et al., 2009) and that Cys-226 is potentially amenable to nitrosylation.
NAB1 Is Deactivated under Nitrosative Stress Conditions by Nitrosylation of Cys-226
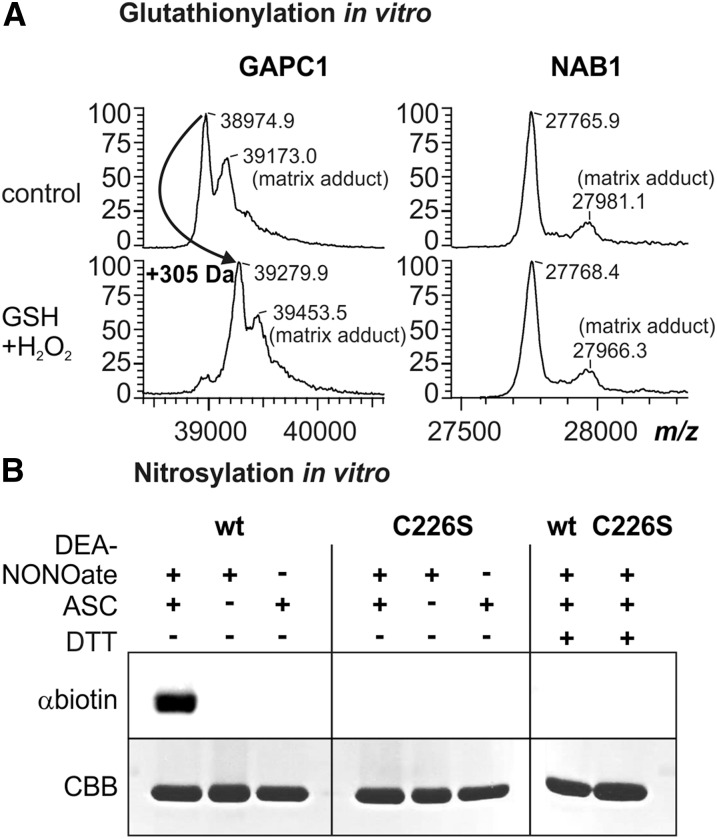
Earlier results indicated that NAB1 could be glutathionylated in vitro in the presence of 5 mm GSSG (Wobbe et al., 2009). While such a treatment is classically used to trigger protein glutathionylation in vitro, it is not physiologically relevant as the in vivo GSSG concentration is much lower, even under severe oxidative stress conditions (Noctor et al., 2012). The predominant mechanisms of glutathionylation in vivo are considered to rely on an initial oxidation of the target Cys, e.g. to sulfenic acid in the presence of H2O2, followed by reaction with reduced glutathione (Mieyal et al., 2008; Dalle-Donne et al., 2009; Zaffagnini et al., 2012). We therefore purified recombinant wild-type NAB1 (Mussgnug et al., 2005) and analyzed if it could undergo glutathionylation in the presence of 0.1 mm H2O2 and 0.5 mm GSH (Fig. 2A; GSH+H2O2). The recombinant Arabidopsis (Arabidopsis thaliana) cytosolic glyceraldehyde-3-phosphate dehydrogenase GAPC1 was used as control since this enzyme is a well established model protein undergoing both glutathionylation (Bedhomme et al., 2012) and nitrosylation (Zaffagnini et al., 2013b). Mass spectrometry showed that GAPC1 is shifted by 305 D after glutathionylation treatment, while this is not the case for NAB1 (Fig. 2A). This shift is consistent with the presence of one glutathione adduct per GAPC1 monomer as previously described (Bedhomme et al., 2012). Therefore, these data demonstrate that NAB1 is not amenable to glutathionylation in vitro under physiologically relevant conditions.
Figure 2.
NAB1 can be nitrosylated in vitro. A, Analysis of NAB1 and GAPC1 in vitro glutathionylation following treatment with hydrogen peroxide (0.1 mm) and glutathione (0.5 mm) via MALDI-TOF mass spectrometry. Left panel: Mass spectrum of GAPC1 obtained after treatment with GSH+H2O2 and subsequent reduction using DTT (2.5 mm; used as a control). A mass increase of 305 D corresponds to one glutathione molecule covalently bound per protein monomer. The peaks labeled “matrix adducts” correspond to proteins with a sinapinic acid adduct. Differences between mass peaks of unmodified NAB1 and GAPC1 are within the experimental error of the instrument. Right panel: Mass spectrometric analysis of recombinant NAB1 under identical conditions. B, Treatment of recombinant NAB1 (wt) and NAB1C226S (C226S) with the NO-donor DEA-NONOate followed by the biotin switch technique. Addition (+) or omission (−) of the reaction components DEA-NONOate (1 mm), ascorbate (ASC; 40 mm), and DTT (20 mm) during the assay is indicated in the upper part. NAB1-biotinylation as an indicator for prior nitrosylation was detected by immunoblotting with a biotin-specific antiserum (αbiotin), and NAB1 protein amounts were assessed by Coomassie staining (CBB) after SDS-PAGE separation.
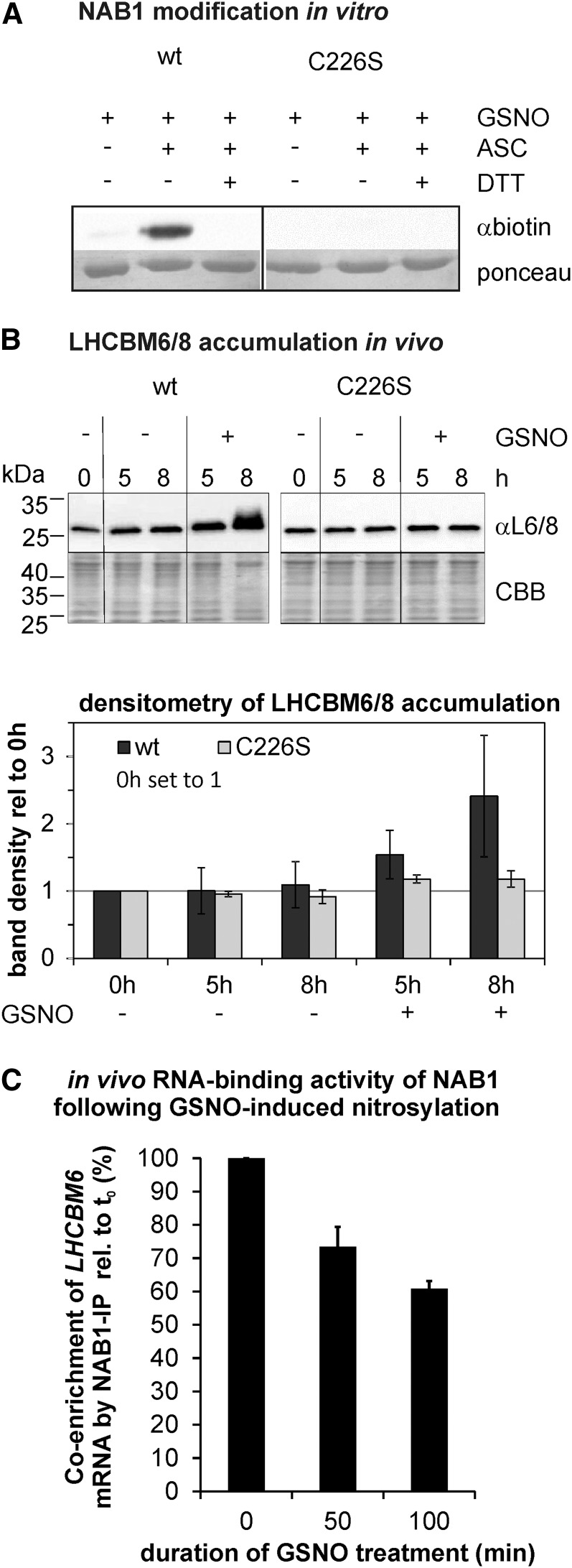
To investigate whether NAB1 can be nitrosylated in vitro, we applied the NO donor DEA-NONOate and monitored nitrosylation using the biotin switch technique (BST; Jaffrey and Snyder, 2001; Fig. 2B). The principle of this method is a three-step reaction starting with blocking of all free thiols (-SH) with alkylating agents, followed by specific reduction of S-nitrosylated cysteines with ascorbate. The final step is the biotin labeling of the previously S-nitrosylated Cys residues with the thiol-specific biotinylating agent HPDP-Biotin via intermolecular disulfide bond formation (-S-S-biotin). The biotin-labeled proteins can then be detected with antibiotin antibodies or can be purified by avidin affinity chromatography and DTT elution. Application of the BST subsequent to incubation of recombinant NAB1 with DEA-NONOate revealed that wt-NAB1 is nitrosylated (Fig. 2B, left panel, wt, +DEA-NONOate, +ASC, and αbiotin). As previous results (Wobbe et al., 2009) and in silico analyses (Fig. 1, B and C) suggested that Cys-226 is the main target for redox-based NAB1 activity regulation, a recombinant protein variant, in which Cys-226 is replaced by a nonreactive Ser (C226S; Wobbe et al., 2009), was also analyzed. Absence of detectable nitrosylation in the NAB1C226S variant following DEA-NONOate treatment clearly demonstrated that Cys-226 is the main target of nitrosylation in vitro (Fig. 2B, middle panel; C226S). Omission of either NO donor or ascorbate during the BST prevented tagging of NAB1 (Fig. 2B, wt, +DEA-NONOate/−ASC, and –DEA-NONOate/+ASC), demonstrating the specificity of the assay. In addition, disulfide reduction with DTT was performed to show reversibility of labeling, as expected for a nitrosothiol-dependent signal (Fig. 2B, right panel, wt, Cys-226, and +DTT).
To further confirm that Cys-226 is amenable to nitrosylation, we applied the BST to recombinant wt-NAB1 and NAB1C226S following treatment with the transnitrosylating agent GSNO (Fig. 3A). Only in the case of wt-NAB1 did exposure to GSNO result in a detectable nitrosylation (Fig. 3A, wt, C226S, and +GSNO). Moreover, since GSNO can trigger both nitrosylation and glutathionylation, MALDI-TOF mass spectrometry analyses performed as in Figure 2A indicated that no glutathionylation of NAB1 could be detected after GSNO treatment (data not shown). Together, the data demonstrate that Cys-226 of NAB1 can be nitrosylated in vitro.
Figure 3.
Nitrosylation of NAB1 at Cys-226 decreases its translation repressor activity. A, In vitro nitrosylation of recombinant NAB1 with GSNO (2 mm). The wild-type version of NAB1 was used along with NAB1C226S. The biotin switch technique was applied to detect S-nitrosylation via immunodetection of the biotin label (αbiotin). Omission of GSNO (−) and addition of DTT (+DTT) served as a control to assess stringency of the assay. Ponceau staining served as loading control. B, Effects of Cys-226 nitrosylation on the accumulation of major light-harvesting proteins LHCBM6/8. The cellular amount of LHCBM6/8 was determined by immunodetection (αL6/8; upper panel) 5 and 8 h following GSNO addition (+) to cultures expressing either wt-NAB1 (wt) or NAB1C226S (C226S) under control of the PSAD promoter. Negative controls (−GSNO) were included to exclude effects unrelated to nitrosative stress. Changes in the level of LHCBM6/8 relative to the t0 level of the wild type (black bars) or C226S (gray bars) were quantified by densitometric scanning (lower panel). Mean values from two biological replicates including two technical replicates are given along with standard deviations (n = 4). C, Effects of Cys-226 nitrosylation on the RNA-binding activity of NAB1 in vivo. Cys nitrosylation was induced by addition of GSNO to liquid cultures expressing wt-NAB1 under control of the PSAD promoter, and samples for the coimmunoprecipitation of LHCBM6-mRNA via NAB1 were taken at indicated time points (x axis). Enrichment factors for LHCBM6-mRNA were determined by comparing the relative LHCBM6 amount in input and coprecipitated samples for each time point by applying quantitative RT-PCR. The GSNO-induced change in LHCBM6-mRNA enrichment during NAB1 immunoprecipitation was calculated by setting the enrichment factor obtained before (t0min) GSNO addition to 100%. Error bars represent standard errors derived from four technical replicates (n = 4).
We further investigated whether nitrosylation of NAB1 in vivo has an impact on its translation repressor activity. Changes in the accumulation of the PSII-associated major light-harvesting protein LHCBM6, whose mRNA represents the prime target of NAB1 (Mussgnug et al., 2005), were chosen as a proxy for alterations in NAB1 activity induced by nitrosylation (Fig. 3B). GSNO was added to liquid cultures of cells overexpressing either wt-NAB1 or the NAB1C226S variant to induce nitrosylation of the protein. In the time range of 5 to 8 h, a strong accumulation of LHCBM6/8 was detected in cultures expressing wt-NAB1 after adding GSNO (Fig. 3B, wt, +GSNO, 5 h, 1.54 ± 0.35 [sd; n = 4] with wt t0 set to 1; 8 h, 2.41 ± 0.90), whereas GSNO addition had no effect on LHCBM6/8 accumulation in cultures of NAB1C226S (Fig. 3B, C226S, +GSNO; 5 h, 1.17 ± 0.06 [sd; n = 4] with C226S set to 1; 8 h, 1.17 ± 0.12).
In order to analyze if the accumulation of LHCBM6, seen in Wt-NAB1-expressing cells following GSNO addition, is caused by an altered translation repressor activity, we determined the impact of nitrosative stress on the RNA-binding activity of NAB1 (Fig. 3C). The in vivo binding capacity of NAB1 for LHCBM6-mRNA was assessed by coimmunoprecipitating this transcript with an antiserum directed against NAB1 following GSNO addition to liquid cultures of a strain overexpressing wt-NAB1. Enrichment factors were calculated before (t0) and after GSNO addition (t50min; t100min) by comparing the relative amount of LHCBM6-mRNA in cells subjected to NAB1 immunoprecipitation and the amount present in coprecipitated RNA. NAB1-mediated enrichment of LHCBM6-mRNA declined from 100% (t0, before GSNO addition) to 73.4% ± 6% (se; n = 4) within 50 min of GSNO treatment and prolongation of GSNO exposure to 100 min brought about a further decrease to 60.8% ± 4.5%.
These results indicate that artificial in vivo nitrosylation of NAB1 at Cys-226 leads to a decreased repressor activity followed by an accumulation of light-harvesting proteins.
Light Intensity Modulates the Degree of NAB1 Nitrosylation
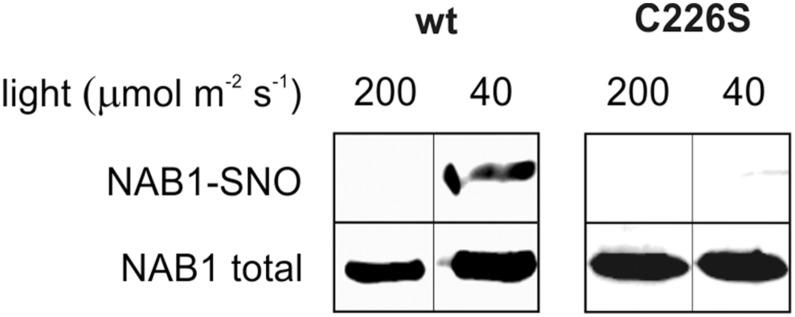
The results presented in Figures 2 and 3 demonstrate that NAB1 can be nitrosylated artificially in vitro using nitrosylating agents and that nitrosative stress triggers the deactivation of NAB1 in vivo. To investigate if NAB1 is nitrosylated in vivo under physiological conditions, the degree of nitrosylation was analyzed under different light conditions using the BST coupled to immunoblot detection of NAB1 after avidin affinity chromatography (Fig. 4). Cultivation of cells with wt-NAB1 under elevated light conditions (200 µmol m−2 s−1) was not accompanied by a detectable nitrosylation of NAB1 (Fig. 4, wt, 200, and NAB1-SNO). However, a decrease of the light intensity to 40 µmol m−2 s−1 during cultivation caused nitrosylation of wt-NAB1 (Fig. 4, wt, 40, and NAB1-SNO). Absence of nitrosylation in NAB1C226S under these conditions again underscored the relevance of Cys-226 for NAB1 redox control by Cys nitrosylation (Fig. 4, C226, 40, and NAB1-SNO).
Figure 4.
NAB1 is nitrosylated in vivo at Cys-226 under low-light conditions. C. reinhardtii cells overexpressing wt-NAB1 or NAB1C226S were grown under low light (40 μmol m−2 s−1) or elevated light (200 μmol m−2 s−1) for 8 h and then subjected to BST. After BST, biotinylated proteins were purified by streptavidin affinity chromatography and eluted with DTT. Eluted proteins were analyzed by western blot using αNAB1 antibodies (NAB1-SNO). Immunoblot detection of NAB1 before chromatography (NAB1 total) served as control. Bands shown in one panel derive from the same blot and were rearranged for clarity.
Low-Light-Induced Deactivation of NAB1 Requires the Intracellular Production of Nitric Oxide
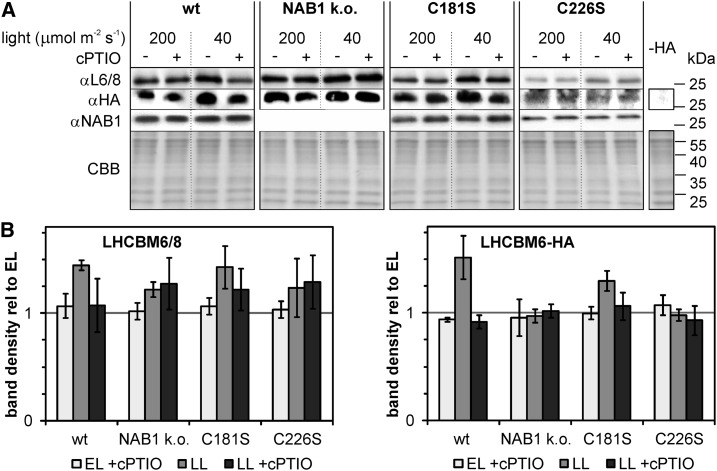
The LHCBM6 translation repressor NAB1 is clearly nitrosylated under low light (Fig. 4), primarily at Cys-226 (Figs. 1–4), and as an oxidative modification, this lowers its RNA binding activity (Wobbe et al., 2009; Fig. 3B). It is well known that C. reinhardtii cells accumulate LHCBM proteins in response to light limitation, and a higher transcription rate of LHCBM genes contributes to the observed accumulation (Escoubas et al., 1995; Teramoto et al., 2002). We next wanted to answer the question whether a low-light-induced deactivation of translation control via NAB1 nitrosylation is needed for antenna size enlargement in addition to LHCBM promoter activation. To this end, algal strains devoid of NAB1 (NAB1 k.o.) or carrying the wild-type protein or mutated versions (C181S and C226S) were subjected to a shift from elevated to low light (Fig. 5). For all strains, a HA-tagged LHCBM6 expressed under control of the constitutive PSAD promoter was introduced to allow differentiation between LHC transcription and translation control. Algal cultures were grown under elevated light conditions (200 µmol m−2 s−1) before transferring them to low light intensities (40 µmol m−2 s−1) or continuing elevated light cultivation for 5 h. The nitric oxide scavenger cPTIO was applied to inhibit nitrosylation of NAB1 (Fig. 5). When cultures expressing wt-NAB1 were subjected to the shift from elevated to low light, LHCBM6/8 levels rose as expected (Fig. 5, wt, αL6/8, 200 versus 40; −cPTIO, 1.44 ± 0.05 [±sd; n = 4] with EL/−cPTIO set to 1). A strong accumulation of the HA-tagged LHCBM6 protein under these conditions (wt, αHA, 1.51 ± 0.20), whose expression is decoupled from endogenous LHCBM promoter control, indicated that a derepression of translation could significantly contribute to the observed LHCBM6 accumulation. As was recently shown, cellular NAB1 levels determine the extent of LHCBM translation repression under certain conditions (Berger et al., 2014). However, the shift to low light did not result in a changed NAB1 level (αNAB1) and indicated that an altered repressor activity is the main trigger for LHCBM6 accumulation. This could be confirmed by supplementing cultures with the NO scavenger cPTIO to inhibit NAB1 nitrosylation during low-light acclimation, which prevented accumulation of LHCBM6 (αL6/8, 40, +cPTIO, 1.07 ± 0.25) and HA-LHCBM6 (αHA, 40, +cPTIO, 0.91 ± 0.06 [±sd; n = 4]). A similar effect of cPTIO was noted for the strain expressing a NAB1C181S variant (αHA, C181S, 40; +cPTIO [1.06 ± 0.13] versus –cPTIO [1.29 ± 0.09]) but could not be observed in the case of NAB1C226S (C226S, 40, +cPTIO [0.93 ± 0.13] versus –cPTIO [0.97 ± 0.05]). Further supporting the NO-dependent specific nitrosylation of NAB1 at Cys-226, nitric oxide scavenging had no effect on LHCBM6 accumulation in the NAB1 knockout mutant (αHA, NAB1 k.o., 40, +cPTIO [1.02 ± 0.06] versus –cPTIO [0.97 ± 0.06]). We therefore conclude that under the conditions tested, the RNA-binding activity of NAB1 and, thus, translation control on LHCBM6 is inhibited by nitrosylation of Cys-226.
Figure 5.
LHCBM6 accumulation under low light is triggered by NAB1 nitrosylation. Algal strains without NAB1 (NAB1 k.o.), expressing the wild-type (wt) protein or mutated versions (C181S and C226S), were used. A wild-type strain expressing NAB1 from the endogenous promoter was used here to determine light-dependent NAB1 expression. All strains additionally contain a HA-tagged LHCBM6 under control of the constitutive PSAD promoter. Strains were precultivated under elevated light (EL; 200 μmol m−2 s−1) and either remained in this light or were shifted to low-light intensities (LL; 40 μmol m−2 s−1) for 5 h, with (+) or without (−) the addition of 0.1 mm cPTIO. A, Immunoblot detection of LHCBM6/8 (αL6/8), LHCBM6-HA (αHA), and NAB1 (αNAB1) in whole-cell lysates is shown, and a Coomassie stain (CBB) served as a loading control. To probe specificity of HA-tag immunodetection, a strain without LHCBM6-HA was loaded as negative control (-HA). B, Densitometric quantification of changes in the LHCBM6/8 (left panel) and HA-LHCBM6 (right panel) protein levels relative to the level found in elevated light conditions (set to 1). Mean values are derived from two biological replicates, each including two technical replicates (n = 4).
NAB1 Can Be Efficiently Denitrosylated by Thioredoxin h1 in Vitro
Results presented in Figure 5 clearly indicate that NAB1 undergoes nitrosylation, when C. reinhardtii cultures are transferred from higher to lower light intensities. The resulting decrease in NAB1 repressor activity ensures that translation repression of light-harvesting protein encoding mRNAs is relieved and that LHCBM proteins accumulate in C. reinhardtii cells exposed to limiting light supply. We next wanted to address the molecular mechanisms behind the reverse process that reactivates NAB1 by denitrosylation in response to an increased supply of light (Fig. 4). The main denitrosylation mechanism occurring in eukaryotic cells operates via reduced GSH in conjunction with the enzyme GSNO reductase (Paige et al., 2008). Although most nitrosothiols can be denitrosylated by GSH, a small subset appears to be specifically denitrosylated by the thioredoxin/thioredoxin reductase system (Benhar et al., 2009).
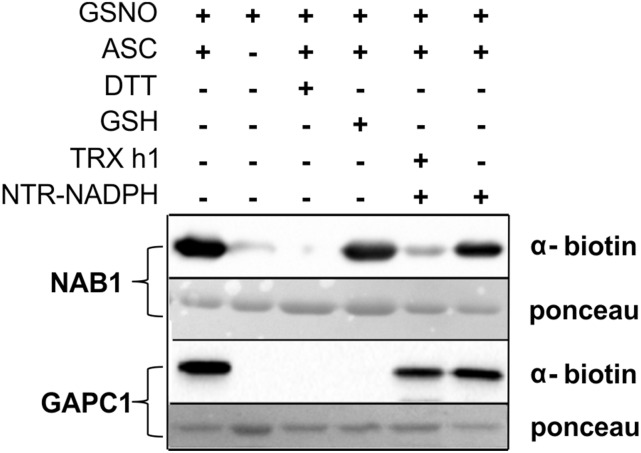
To gain insight into the mechanisms activating NAB1 in cells exposed to elevated light quantities, we analyzed if GSH or a TRX system are capable of denitrosylating NAB1 in vitro (Fig. 6). Recombinant NAB1 was nitrosylated at Cys-226 in the presence of GSNO and directly subjected to the BST (Fig. 6, lanes 1–3) or treated with either glutathione (lane 4, +GSH) or cytosolic TRX h1 in combination with its reduction system (lane 5, +TRX h1, + NTR-NADPH) prior to the detection of protein nitrosylation.
Figure 6.
Thioredoxin h1 denitrosylates NAB1 in vitro. The purified recombinant proteins NAB1 and GAPC1 (cytosolic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis) were nitrosylated with 2 mm GSNO in vitro (+GSNO; lanes 1–6) and subjected to the biotin switch technique (+GSNO; +ASC) for tagging of S-nitrosylation sites with biotin prior to immunodetection (α-biotin). Specificity of the assay was confirmed by omitting ascorbate (−ASC; lane 2) or including DTT (+DTT; lane 3), and protein amounts on the blotting membrane were visualized by Ponceau staining. GSH (5 mm) and recombinant thioredoxin reducing system (1 µm NTR + 2 mm NADPH), alone or in combination with thioredoxin h1 (TRX h1; 20 µm), were tested for their denitrosylation capacity with NAB1-SNO and GAPC1-SNO prior to application of BST (lanes 4–6).
Nitrosylated NAB1 could be efficiently denitrosylated only when TRX h1 was provided together with a NADPH-dependent TRX reductase (NTR) and NADPH (Fig. 6, lane 5). In contrast, addition of glutathione had no significant effect on NAB1 nitrosylation, whereas nitrosylation of a cytosolic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis (Fig. 6, GAPC1) was reduced to undetectable levels only following GSH treatment (lane 4) but not when the TRX system was used (lane 5). The specific denitrosylation of GAPC1 by glutathione has been demonstrated before (Zaffagnini et al., 2013b) and confirms reliability of the in vitro assay. As additional specificity controls for the assay, the absence of biotin label on NAB1 or GAPC1 cysteines was demonstrated after omitting ascorbate (Fig. 6, lane 2, −ASC) or adding DTT (lane 3, −DTT). NAB1 denitrosylation could only be observed when both TRX h1 and a reduction system were present (Fig. 6, lane 5 versus lane 6). The finding that NAB1 is efficiently denitrosylated by the cytosolic TRX h1 in vitro provides new insights into the chloroplast-to-cytosol cross talk that fine-tunes cytosolic translation of nucleus-encoded light-harvesting proteins.
DISCUSSION
NAB1-Mediated Control of Light Harvesting Protein Synthesis Is Regulated at Multiple Levels
In photosynthetic organisms, long-term acclimation to changes in the availability of light or other external factors having a strong impact on photosynthetic activity requires a modulated expression of genes encoding components of the photosynthetic apparatus (Woodson and Chory, 2008). The resulting stoichiometric adjustments ensure sufficient photosynthetic performance under adverse conditions and help prevent damage to the photosynthetic machinery induced by the formation of reactive oxygen species (Anderson et al., 1995). During evolution, many endosymbiont genes encoding essential photosynthetic proteins have been transferred to the nucleus. A tight regulation of their expression in response to environmental changes requires a cross talk between different cellular compartments, including the nucleus, chloroplast, cytosol, and mitochondria (Woodson and Chory, 2008). The amount of PSII-associated major light-harvesting proteins determines the light capture capacity of algal and leaf cells, and the adjustment of cellular LHCBM levels represents a prime example for the molecular complexity of long-term acclimation processes (Erickson et al., 2015). In C. reinhardtii, NAB1 was determined to be a central hub within the network controlling light-harvesting protein availability in the thylakoid membrane (Mussgnug et al., 2005; Wobbe et al., 2009; Blifernez et al., 2011; Berger et al., 2014). In accordance with its crucial function, NAB1-mediated translation control of LHCBM proteins was shown to be itself regulated on multiple levels (Wobbe et al., 2009; Blifernez et al., 2011; Berger et al., 2014). Apart from a modulation of cellular NAB1 levels during the acclimation to an altered carbon availability (Berger et al., 2014), the activity state of NAB1, regulated via posttranslational modifications, was demonstrated to determine the translational status of LHCBM mRNAs (Wobbe et al., 2009; Blifernez et al., 2011). Two types of posttranslational modifications with distinct physiological functions were identified. Arg methylation is a costly posttranslational modification, as measured by the ATP input required, and has a rather low turnover rate (Fackelmayer, 2005). Methylation and demethylation of NAB1 Arg residues can therefore be considered a less dynamic process, which supports the view that this type of modification acts as a master control switch (Blifernez et al., 2011). Indeed, the degree of NAB1 methylation was found to be low when C. reinhardtii cells were cultivated under strictly heterotrophic conditions, whereas photoautotrophic growth was accompanied by a high extent of Arg methylation (Blifernez et al., 2011). High methylation levels under photoautotrophic conditions ensure that the slowly responding activity switch is in the “on” state, when a fine-tuning of light harvesting plays a vital role for the algal cell.
Nitrosylation of Cys-226 Acts as a Redox Switch for Fine-Tuning of Light Harvesting
Unperturbed photoautotrophic growth in a natural environment requires a continuous and fast readjustment of LHCBM translation rates in the cytosol to perfectly meet the current demand for light-harvesting proteins in the chloroplast, which is defined by factors such as light availability. Redox control via Cys modification is a typical example for a dynamic type of protein activity control (Spadaro et al., 2010). In addition to Arg methylation, a second activity control switch has been identified in NAB1 and is based on the reversible modification of Cys residues present in its RRM domain, as was shown in a previous study (Wobbe et al., 2009). In this study, the general significance of Cys modification for NAB1 activity control and the more critical role of Cys-226 could be revealed, but the precise chemical nature of Cys modifications occurring on NAB1 in vivo and their function in a physiological context remained to be elucidated (Wobbe et al., 2009). Inspection of a structural model of the NAB1-RRM domain generated in silico together with an analysis of the protein microenvironment surrounding Cys-226 suggested that S-nitrosylation could potentially occur at this site (Fig. 1). This was confirmed by demonstrating specific nitrosylation of Cys-226 in vitro using nitrosylating agents (Figs. 2B and 3A). Nitrosative stress resulted in a strong accumulation of LHCBM6 proteins, whose RNA is the prime target of NAB1-mediated translation control (Fig. 3B). RNA coimmunoprecipitation experiments clearly demonstrated that nitrosylation of NAB1 decreases its affinity toward LHCBM6 transcripts and, hence, its translation repressor activity (Fig. 3C). This is in line with previous results (Wobbe et al., 2009) demonstrating that Cys modification of recombinant NAB1 either by oxidation or by alkylation in vitro was accompanied by a decrease in RNA-binding affinity for the target mRNA sequence. Importantly, NAB1 nitrosylation could also be detected under physiological conditions, in the absence of nitrosylating agents, and the degree of Cys-226 nitrosylation was found to be negatively correlated with the light intensity used for cultivation (Fig. 4). This could be further confirmed by analyzing the effect of an elevated to low-light shift on the accumulation of a LHCBM6 protein, whose expression is driven by a constitutive promoter instead of the low-light-inducible endogenous promoter (Fig. 5). Moreover, LHCBM6 accumulation following a shift to low light could be suppressed by adding a nitric oxide scavenger.
The Activation of NAB1 by the Cytosolic Thioredoxin System Is the First Example of Stimulus-Induced Denitrosylation as Part of Photosynthetic Light Acclimation Mechanisms
In the cell, a vast majority of nitrosylated proteins are in the thiol state (-SH) under basal conditions and become nitrosylated (-SNO) under stress conditions associated with increased NO production. Once the stress is relieved, the protein returns to the thiol state due to decreased NO production and active denitrosylation mediated most generally by GSH. Indeed, most protein nitrosothiols (75 to 80%) were shown to be susceptible to reduction by GSH. For example, denitrosylation of Arabidopsis GAPC1 was found to be specifically performed by GSH and to be directly controlled by the [GSH]/[GSNO] ratio but independent from the [GSH]/[GSSG] ratio (Zaffagnini et al., 2013b). The nitrosothiols that are resistant to GSH are reduced by specific denitrosylases, the most prominent being cytosolic and mitochondrial thioredoxins, which were shown to denitrosylate numerous proteins (Benhar et al., 2010; Sengupta and Holmgren, 2013). The regulation of NAB1 activity by nitrosylation is atypical because the enzyme is nitrosylated under basal low-light conditions and becomes denitrosylated under stress conditions, i.e. higher light intensities (Fig. 4). Hence, by contrast with GAPC1, NAB1-SNO is resistant to GSH but is efficiently denitrosylated by cytosolic TRX h1 (Fig. 6). This mechanism of stimulus induced denitrosylation is reminiscent of the activation of caspase 3 by mitochondrial TRX during apoptotic signaling in mammalian cells (Benhar et al., 2008) or the activation of Arabidopsis NPR1 by salicylic acid-induced plant immune responses (Kneeshaw et al., 2014). To our knowledge, NAB1 is the first example of stimulus-induced denitrosylation within photosynthetic light acclimation processes.
The Redox Control Mechanism Fine-Tuning NAB1 Activity Provides Novel Insights into the Cross Talk between Chloroplast and Cytosol Required for Long-Term Light Acclimation
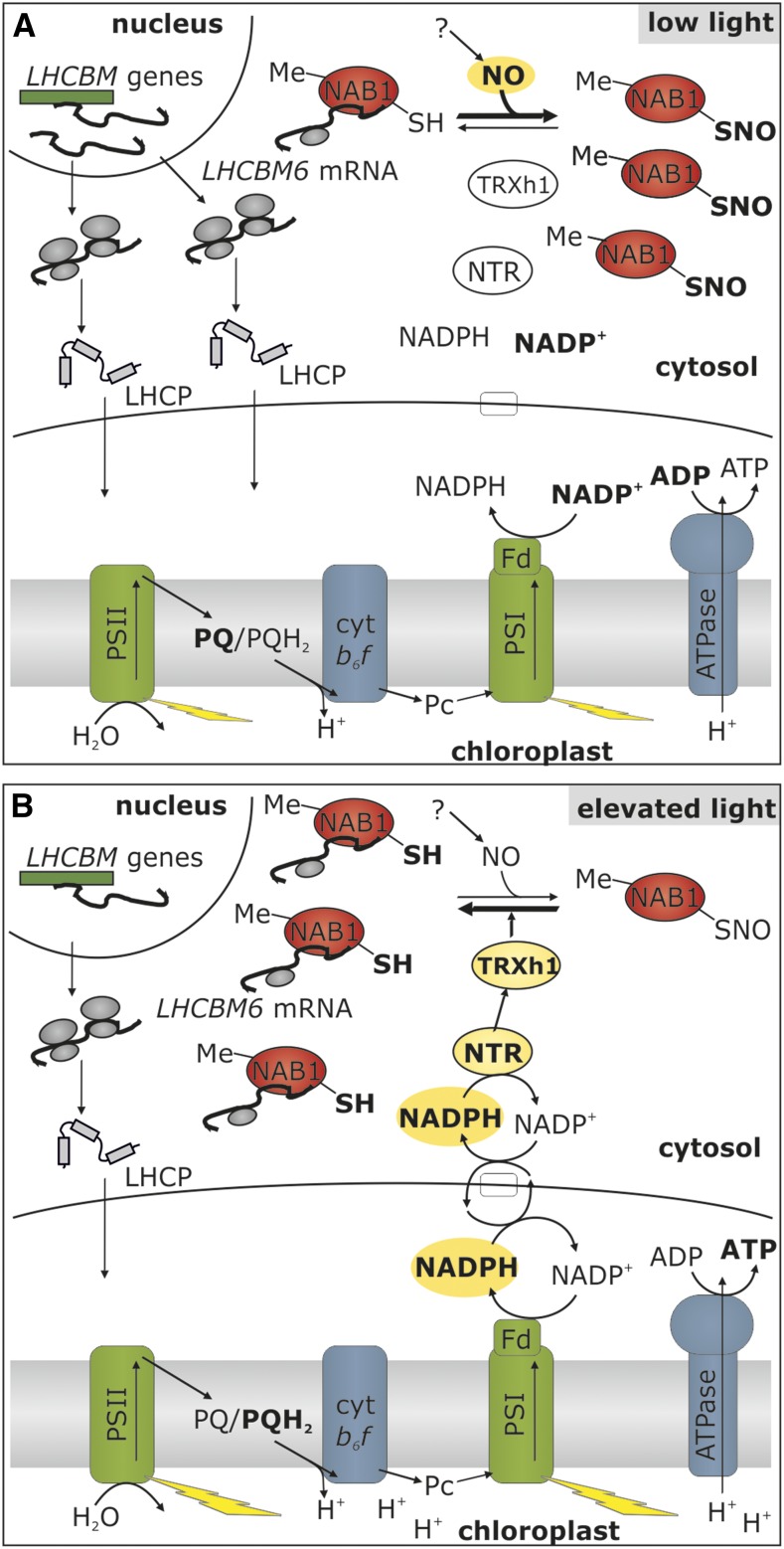
Taken together, our results enable us to propose a new view on the regulatory network of light-harvesting control in a photosynthetic eukaryote by introducing the mechanism of nitrosylation-based activity regulation of NAB1 in response to light quantity changes (Fig. 7). Under low-light conditions (Fig. 7A), C. reinhardtii cells accumulate LHCBM proteins (Bonente et al., 2012), which is achieved by an induction of the nuclear LHCBM promoters (Teramoto et al., 2002; Durnford et al., 2003). NAB1 is expressed under these conditions and a significant part of the NAB1 population is methylated (Blifernez et al., 2011), meaning that the “master switch” is set to the “on” position. Nitrosylation of Cys-226, which inhibits the activity of NAB1 in low-light-acclimated cells, ensures that induction of the LHCBM promoter is not counteracted by translation repression, thus permitting the required increase in cellular LHCBM levels. Although the requirement of intracellular NO formation for NAB1 nitrosylation under low-light conditions could be demonstrated in this study (Fig. 5), it is currently difficult to depict the whole nitric oxide metabolism in C. reinhardtii (Wei et al., 2014). For the NO-dependent nitrosylation in response to light limitation (Fig. 5), we can only rule out an implication of the reactions catalyzed by nitrate reductase as a source of NO (Sakihama et al., 2002), since the strain used in this study lacks, like most laboratory strains, a functional nitrate reductase (Hyams and Davies, 1972; Harris, 2009). Nevertheless, nitrate reductase-independent NO production is still possible and several enzymatic systems have been proposed as alternative NO sources (Fröhlich and Durner, 2011). This possible nitrate reductase-independent production of NO was recently suggested to occur in C. reinhardtii. Indeed, several strains deficient for nitrate reductase were shown to produce NO under diverse conditions, including dark anoxia (Hemschemeier et al., 2013a, 2013b) and nitrogen starvation (Wei et al., 2014). These data suggest the existence of alternative mechanisms of NO production in algal cells, which could account for NAB1 nitrosylation in vivo.
Figure 7.
Light modulation of light-harvesting protein synthesis by nitrosylation and thioredoxin-dependent denitrosylation. A, Under low light, the demand for light-harvesting proteins is high, which is met by high rates of nuclear LHCBM transcription (Teramoto et al., 2002). NAB1 is Arg methylated (Me), but a high nitrosylation level (SNO) results in a low LHCBM RNA binding activity, allowing accumulation of light-harvesting apoproteins (LHCP). B, Elevation of light intensity leads to the accumulation of reducing power (NADPH), and shuttle systems (e.g. malate valve or triose phosphate shuttle; white box) export reducing equivalents to the cytosol. Via a system of NADPH-dependent thioredoxin reductase (NTR) and thioredoxin h1 (TRX h1), this reducing power is used to denitrosylate NAB1, which activates cytosolic LHCBM translation repression. Together with a low LHCBM transcription (Teramoto et al., 2002), the concerted cytosolic and nuclear LHCII expression control ensures a low abundance of light-harvesting proteins when light is in excess.
When C. reinhardtii cells are exposed to elevated light supply (Fig. 7B), a bulk part of the NAB1 population is in its active state, due to the low level of nitrosylation (Figs. 4 and 5). In this situation, nuclear LHCBM promoter control and translation control work in concert to efficiently decrease the rate of LHCBM protein synthesis in the cytosol. Efficient denitrosylation of NAB1 by the cytosolic TRX system in vitro (Fig. 6) gives an idea of how the NAB1 activity state could be quickly adjusted to the prevailing light situation in vivo, in relation with the intracellular redox state.
Light activation of photosynthetic gene translation via Cys-based redox control has been previously described in the chloroplast of C. reinhardtii. An involvement of distinct TRX systems, coupled either directly to the photosynthetic electron transport chain through the reduction of TRX by ferredoxin-dependent enzymes (Trebitsh et al., 2000) or enzymes using NADPH as electron donor (Schwarz et al., 2012), has been proposed. Our data highlight a novel type of redox control mechanism regulating the abundance of nucleus-encoded components constituting the photosynthetic apparatus in the chloroplast. A light-dependent modulation of nuclear photosynthetic gene translation in the cytosol of higher plants has been demonstrated for PSI subunits (Sherameti et al., 2002), ferredoxin (Petracek et al., 1997, 1998), and PSII-associated light-harvesting proteins (Petracek et al., 1997) along with a requirement of photosynthetic electron transport for translational activation. However, these studies could not provide insights into the underlying mechanisms enabling the redox cross talk between chloroplast and cytosol.
Efficient denitrosylation of NAB1 by TRX h1 is an important finding because it exemplifies how translation of plastid-targeted photosynthetic proteins could be coupled to the cytosolic redox state via NADPH-dependent TRX reductase (Huppe et al., 1991; Fig. 7B, NTR). TRX h1 is one of the two isoforms residing in the cytosol of C. reinhardtii cells (Lemaire and Miginiac-Maslow, 2004) and was shown to play an essential role within the alkylation-induced response to DNA damage (Sarkar et al., 2005). The question arises as to how alterations in the cytosolic NADPH/NADP+ ratio might be connected to photosynthetic activity in the chloroplast, which is itself intimately linked to the light capture capacity at PSII. The malate-oxaloacetate shuttle seems to be a reasonable candidate for such an interorganellar cross talk (Scheibe, 2004). Under elevated light conditions, part of the NADPH produced by an increased linear electron transport activity could be used to convert oxaloacetate into malate, a form of reducing power that can shuttle between chloroplast and cytosol. In addition, triose phosphate metabolites exported from the chloroplast and produced in a light-dependent manner by the Calvin-Benson cycle could also contribute to the interorganellar cross talk. Indeed, the cytosol contains a nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase that oxidizes G3P directly to 3-phosphoglycerate and NADPH (Rius et al., 2006; Zaffagnini et al., 2013a). These shuttles could alter the redox state of the cytosol by increasing both the NADH/NAD+ and NADPH/NADP+ ratio, which would result in increased availability of reduced thioredoxin h1 required for the denitrosylation of NAB1. Overall the proposed mechanism could quickly adjust the translation rate of LHCBM mRNAs to the chloroplast redox state. Perturbations of photosynthesis leading to the accumulation of reducing equivalents would transiently activate the translation repressor NAB1, while in the absence of bottlenecks downstream of photosystem I, translation of light-harvesting proteins would be permitted. NAB1 represents a key regulatory hub for the long-term adjustment of photosynthetic light-harvesting capacities in the cytosol of C. reinhardtii cells. Details regarding the precise adjustment of NAB1-mediated translation control on various levels and in response to distinct physiological cues have been elucidated (Wobbe et al., 2009; Blifernez et al., 2011; Berger et al., 2014). Our new finding, that this protein is regulated via reversible nitrosylation/denitrosylation in the cytosol, provides essential insights in the molecular mechanisms underlying the intense interorganellar communication required for long-term photoacclimation in a phototrophic eukaryotic cell.
MATERIALS AND METHODS
Chemicals and Enzymes
GSH, DTT, ascorbate (ASC), NADP (NADPH), 1,1-diethyl-2-hydroxy-2-nitroso-hydrazine sodium salt (DEA-NONOate), 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO), and GSNO were purchased from Sigma-Aldrich. GSNO and DEA-NONOate concentrations were determined spectrophotometrically using molar extinction coefficients of 920 m−1 cm−1 at 335 nm and 6700 m−1 cm−1 at 250 nm, respectively. N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (HPDP-biotin) was purchased from Pierce. Recombinant proteins were prepared as previously described for TRX h1 from Chlamydomonas reinhardtii (Goyer et al., 1999), NADPH-dependent thioredoxin reductase (NTR) from Saccharomyces cerevisiae (Pérez-Pérez et al., 2014), and cytoplasmic glyceraldehyde-3-phosphate dehydrogenase (GAPC1) from Arabidopsis (Arabidopsis thaliana; Bedhomme et al., 2012).
Strains and Culture Conditions
The wild-type C. reinhardtii strain CC849 (cw10, mt-; Chlamydomonas Resource Center, St. Paul, MN) is the parental strain of the NAB1 knockout mutant stm3 (Mussgnug et al., 2005). The Cys mutants, carrying NAB1 with either Cys-181 or Cys-226 replaced by Ser, as well as the respective control strain with a wild-type NAB1, were created by introducing the NAB1 gene under the control of the PSAD promoter into stm3 (Wobbe et al., 2009), thereby obviating endogenous expression regulation. To distinguish between LHC transcription and translation control, HA-tagged LHCBM6 under the control of the PSAD promoter was introduced into the wild-type strain, NAB1 knockout, and Cys mutants (Mussgnug et al., 2005; Wobbe et al., 2009).
The strains were cultured in Tris-acetate-phosphate medium under low (40 µmol photons m−2 s−1) or elevated (200 µmol photons m−2 s−1) fluorescent white light. Cell density was monitored by determining the optical density at 750 nm as well as cell count (Beckmann Coulter hemocytometer). For artificial nitrosative stress, cultures were supplemented with 2 mm GSNO or 1 mm DEA-NONOate. To prevent nitrosylation, 0.1 mm cPTIO was added as NO scavenger.
In Vitro Glutathionylation of NAB1 and GAPC1
Reduced recombinant proteins (40 μm; Mussgnug et al., 2005; Bedhomme et al., 2012) were incubated in 30 mm Tris-HCl, pH 7.9, and 1 mm EDTA in the presence of 0.1 mm H2O2 and 0.5 mm GSH for 30 min at 25°C. For each protein, a control was performed by replacing H2O2 and GSH with Tris-HCl buffer, and the reversibility of the induced glutathionylation was verified after incubation with 2.5 mm DTT for 15 min at 25°C. After each treatment, proteins were analyzed by MALDI-TOF mass spectrometry.
MALDI-TOF Mass Spectrometry
One microliter of protein solution was mixed with 4 µL of a saturated solution of sinapinic acid in 30% acetonitrile containing 0.3% trifluoroacetic acid. Then, 1.5 µL of this premix was deposited onto the sample plate and allowed to dry under a gentle air stream at room temperature. Mass determination of NAB1 and GAPC1 was carried out after calibration on monocharged and dicharged yeast enolase ions (m/z: 23,336 and 46,671 D, respectively) in positive linear mode on a Performance Axima MALDI-TOF mass spectrometer (Shimadzu) with pulse extraction fixed at 28,000.
Nitrosylation/Denitrosylation Assays
Purified recombinant NAB1 (Mussgnug et al., 2005) and GAPC1 (Bedhomme et al., 2012) were nitrosylated in sample buffer (50 mm Tris-HCl, pH 7.9, 1 mm EDTA, and 100 mm NaCl) in the presence of 2 mm GSNO for 20 min in the dark at 25°C. Denitrosylation of NAB1-SNO and GAPC1-SNO was performed for 30 min in sample buffer supplemented with GSH (5 mm) or a mixture containing 1 μm NTR and 2 mm NADPH in the presence or absence of 20 μm TRX h1. Addition of DTT (20 mm) served as a positive control for this denitrosylation assay. Cys nitrosylation should be abolished after DTT treatment.
Biotin Switch Technique
The extent of protein nitrosylation was assessed by the adapted BST (Jaffrey and Snyder, 2001). After nitrosylation/denitrosylation treatments, proteins (0.8–1 mg/mL) were precipitated with 4 volumes of 80% cold acetone at −20°C during 20 min and pelleted by centrifugation at 4°C for 10 min at 15,000g. The pellet was resuspended in TENS buffer (50 mm Tris-HCl, pH 7.9, 1 mm EDTA, 100 mm NaCl, and 1% SDS) supplemented with alkylating reagents (20 mm iodoacetamide and 20 mm N-ethylmaleimide), to allow blocking of free thiols. After 60 min incubation at 25°C under shaking, the samples were acetone precipitated, as described above, to remove unreacted alkylating reagents. After resuspension in TENS buffer, proteins were incubated in the presence of 40 mm ascorbate and 0.2 mm HPDP-biotin for 30 min. This step allows reduction of S-nitrosylated cysteines and their derivatization with biotin. Proteins were then acetone precipitated to remove unreacted labeling compounds, pelleted by centrifugation as above, and resuspended in TENS buffer. All steps were performed in the dark. All BST assays included a negative control where ascorbate was omitted to prevent reduction of S-nitrosothiols and subsequent biotinylation. This control without ascorbate allows assessing the efficiency of the initial thiol-blocking step. After the final precipitation, proteins were analyzed by gel electrophoresis and immunoblotting or purified by avidin affinity chromatography.
Purification of S-Nitrosylated Proteins by Affinity Chromatography
Biotinylated proteins were resuspended in TENS buffer, and 200 µg was loaded onto a 1-mL streptavidin-agarose column preequilibrated with TENS buffer and incubated for 30 min at room temperature. The column was extensively washed with 10 volumes of TENS buffer supplemented with NaCl up to 600 mm and then with 10 volumes of TENS buffer. Finally, the proteins retained on the column were eluted with 2 mL of 10 mm DTT dissolved in TEN buffer (TENS buffer without SDS). The eluted proteins were concentrated using a vacuum or filter concentrator (Amicon Ultra; Millipore) and analyzed by gel electrophoresis and immunoblotting with anti-NAB1 antibodies as described below.
Coimmunoprecipitation of LHCBM6-mRNA via NAB1
Liquid cultures of the strain overexpressing wt-NAB1 were grown in Tris-acetate-phosphate medium under continuous white light (200 µmol m−2 s−1) until the late exponential phase was reached. Samples representing t0 (before GSNO addition) were taken, and Cys nitrosylation was induced by adding 2 mm GSNO, which was followed by incubation for 2 h at room temperature under constant orbital shaking (110 rpm) and 200 µmol m−2 s−1 white light illumination. The coimmunoprecipitation of NAB1-mRNA complexes was carried out as described (Wobbe et al., 2009).
Gel Electrophoresis and Immunoblotting
Whole cell lysates were subjected to immunoblotting as described (Wobbe et al., 2009; Blifernez et al., 2011). Tris-Gly polyacrylamide gels (Laemmli, 1970) were used for Coomassie staining and blots against NAB1 and LHCBM6/8. Tris-tricine polyacrylamide gels (Schägger and von Jagow, 1987) were used for detection of HA-tagged LHCBM6. Antiserum raised against NAB1 was generated as described (Mussgnug et al., 2005), and anti-LHCBM6/8 antibody was a gift of M. Hippler (Münster, Germany). The antibody detecting the HA-tag was purchased (Roche).
For nitrosylation/denitrosylation assays, proteins were quantified using the bicinchoninic acid assay, separated by nonreducing SDS-PAGE, and transferred onto nitrocellulose membranes. Protein loading and transfer were assessed by Ponceau staining of the membrane. Proteins were then analyzed by western blotting using a primary antibiotin antibody (Sigma-Aldrich) and an anti-mouse secondary antibody coupled to peroxidase (Sigma-Aldrich). Signals were visualized by ECL, as described previously (Bedhomme et al., 2012).
In Silico Protein Analysis
ExPASy tools were used for protein in silico analysis. The predicted structure of the NAB1 RRM domain was visualized with Swiss pdb viewer (Guex and Peitsch, 1997) available at http://www.expasy.org/spdbv/. Grantham polarity plot was obtained from http://web.expasy.org/protscale/ with a window size of nine amino acids (Gasteiger et al., 2005).
Acknowledgments
We acknowledge M. Zaffagnini for providing AtGAPC1 protein and for critical reading of the manuscript. We also thank M. Hippler for providing the antibody against LHCBM6/8. We are grateful to the Center for Biotechnology (CeBiTec) at Bielefeld University for access to the Technology Platforms. No conflict of interest declared.
Glossary
- RRM
RNA recognition motif
- GSH
glutathione
- GSNO
S-nitrosoglutathione
- GSSG
oxidized glutathione
- TRX
thioredoxin
- BST
biotin switch technique
References
- Anderson JM, Chow WS, Park Y-I (1995) The grand design of photosynthesis: Acclimation of the photosynthetic apparatus to environmental cues. Photosynth Res 46: 129–139 [DOI] [PubMed] [Google Scholar]
- Astier J, Rasul S, Koen E, Manzoor H, Besson-Bard A, Lamotte O, Jeandroz S, Durner J, Lindermayr C, Wendehenne D (2011) S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci 181: 527–533 [DOI] [PubMed] [Google Scholar]
- Baudouin E. (2011) The language of nitric oxide signalling. Plant Biol (Stuttg) 13: 233–242 [DOI] [PubMed] [Google Scholar]
- Beckmann J, Lehr F, Finazzi G, Hankamer B, Posten C, Wobbe L, Kruse O (2009) Improvement of light to biomass conversion by de-regulation of light-harvesting protein translation in Chlamydomonas reinhardtii. J Biotechnol 142: 70–77 [DOI] [PubMed] [Google Scholar]
- Bedhomme M, Adamo M, Marchand CH, Couturier J, Rouhier N, Lemaire SD, Zaffagnini M, Trost P (2012) Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem J 445: 337–347 [DOI] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Hess DT, Stamler JS (2008) Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320: 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Stamler JS (2009) Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol 10: 721–732 [DOI] [PubMed] [Google Scholar]
- Benhar M, Thompson JW, Moseley MA, Stamler JS (2010) Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry 49: 6963–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger H, Blifernez-Klassen O, Ballottari M, Bassi R, Wobbe L, Kruse O (2014) Integration of carbon assimilation modes with photosynthetic light capture in the green alga Chlamydomonas reinhardtii. Mol Plant 7: 1545–1559 [DOI] [PubMed] [Google Scholar]
- Blifernez O, Wobbe L, Niehaus K, Kruse O (2011) Protein arginine methylation modulates light-harvesting antenna translation in Chlamydomonas reinhardtii. Plant J 65: 119–130 [DOI] [PubMed] [Google Scholar]
- Bonente G, Pippa S, Castellano S, Bassi R, Ballottari M (2012) Acclimation of Chlamydomonas reinhardtii to different growth irradiances. J Biol Chem 287: 5833–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Durnford DG, Koblizek M, Falkowski PG (2004) Plastid regulation of Lhcb1 transcription in the chlorophyte alga Dunaliella tertiolecta. Plant Physiol 136: 3737–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A (2009) Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci 34: 85–96 [DOI] [PubMed] [Google Scholar]
- Durnford DG, Price JA, McKim SM, Sarchfield ML (2003) Light-harvesting complex gene expression is controlled by both transcriptional and post-transcriptional mechanisms during photoacclimation in Chlamydomonas reinhardtii. Physiol Plant 118: 193–205 [Google Scholar]
- Erickson E, Wakao S, Niyogi KK (2015) Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J 82: 449–465 [DOI] [PubMed] [Google Scholar]
- Escoubas JM, Lomas M, LaRoche J, Falkowski PG (1995) Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92: 10237–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackelmayer FO. (2005) Protein arginine methyltransferases: guardians of the Arg? Trends Biochem Sci 30: 666–671 [DOI] [PubMed] [Google Scholar]
- Foresi N, Correa-Aragunde N, Parisi G, Caló G, Salerno G, Lamattina L (2010) Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 22: 3816–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Hess DT, Stamler JS (2009) Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med 15: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich A, Durner J (2011) The hunt for plant nitric oxide synthase (NOS): is one really needed? Plant Sci 181: 401–404 [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In JM Walker, ed, The Proteomics Protocols Handbook. Humana Press, Totowa, NJ, pp 571–607 [Google Scholar]
- Gould N, Doulias P-T, Tenopoulou M, Raju K, Ischiropoulos H (2013) Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J Biol Chem 288: 26473–26479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer A, Decottignies P, Lemaire S, Ruelland E, Issakidis-Bourguet E, Jacquot J-P, Miginiac-Maslow M (1999) The internal Cys-207 of sorghum leaf NADP-malate dehydrogenase can form mixed disulphides with thioredoxin. FEBS Lett 444: 165–169 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Harris EH. (2009) The genus Chlamydomonas In EH Harris, ed, The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use, Ed 2, Vol 1. Academic Press, San Diego, CA, pp 1–18 [Google Scholar]
- Hemschemeier A, Casero D, Liu B, Benning C, Pellegrini M, Happe T, Merchant SS (2013a) Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell 25: 3186–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A, Düner M, Casero D, Merchant SS, Winkler M, Happe T (2013b) Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc Natl Acad Sci USA 110: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppe HC, Picaud A, Buchanan BB, Miginiac-Maslow M (1991) Identification of an NADP/thioredoxin system in Chlamydomonas reinhardtii. Planta 186: 115–121 [PubMed] [Google Scholar]
- Hyams J, Davies DR (1972) The induction and characterisation of cell wall mutants of Chlamydomonas reinhardi. Mutat Res 14: 381–389 [Google Scholar]
- Jaffrey SR, Snyder SH (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: pl1. [DOI] [PubMed] [Google Scholar]
- Kneeshaw S, Gelineau S, Tada Y, Loake GJ, Spoel SH (2014) Selective protein denitrosylation activity of Thioredoxin-h5 modulates plant Immunity. Mol Cell 56: 153–162 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Miginiac-Maslow M (2004) The thioredoxin superfamily in Chlamydomonas reinhardtii. Photosynth Res 82: 203–220 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Bahnweg G, Durner J (2006) Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J Biol Chem 281: 4285–4291 [DOI] [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410: 490–494 [DOI] [PubMed] [Google Scholar]
- Martínez-Ruiz A, Lamas S (2007) Signalling by NO-induced protein S-nitrosylation and S-glutathionylation: convergences and divergences. Cardiovasc Res 75: 220–228 [DOI] [PubMed] [Google Scholar]
- Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD (2008) Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal 10: 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisse S, Zaffagnini M, Gao X-H, Lemaire SD, Marchand CH (2014) Insight into protein S-nitrosylation in Chlamydomonas reinhardtii. Antioxid Redox Signal 21: 1271–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussgnug JH, Wobbe L, Elles I, Claus C, Hamilton M, Fink A, Kahmann U, Kapazoglou A, Mullineaux CW, Hippler M, et al. (2005) NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. Plant Cell 17: 3409–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH (2012) Glutathione in plants: an integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Paige JS, Xu G, Stancevic B, Jaffrey SR (2008) Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol 15: 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Zaffagnini M, Marchand CH, Crespo JL, Lemaire SD (2014) The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 10: 1953–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Huber SC, Thompson WF (1997) Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 9: 2291–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Nguyen TT, Gatz C, Sowinski DA, Allen GC, Thompson WF (1998) Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc Natl Acad Sci USA 95: 9009–9013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF (2006) Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol Biol 61: 945–957 [DOI] [PubMed] [Google Scholar]
- Sakihama Y, Nakamura S, Yamasaki H (2002) Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: an alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol 43: 290–297 [DOI] [PubMed] [Google Scholar]
- Sarkar N, Lemaire S, Wu-Scharf D, Issakidis-Bourguet E, Cerutti H (2005) Functional specialization of Chlamydomonas reinhardtii cytosolic thioredoxin h1 in the response to alkylation-induced DNA damage. Eukaryot Cell 4: 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Scheibe R. (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Schwarz C, Bohne AV, Wang F, Cejudo FJ, Nickelsen J (2012) An intermolecular disulfide-based light switch for chloroplast psbD gene expression in Chlamydomonas reinhardtii. Plant J 72: 378–389 [DOI] [PubMed] [Google Scholar]
- Sengupta R, Holmgren A (2013) Thioredoxin and thioredoxin reductase in relation to reversible S-nitrosylation. Antioxid Redox Signal 18: 259–269 [DOI] [PubMed] [Google Scholar]
- Sherameti I, Nakamura M, Yamamoto YY, Pfannschmidt T, Obokata J, Oelmüller R (2002) Polyribosome loading of spinach mRNAs for photosystem I subunits is controlled by photosynthetic electron transport. Plant J 32: 631–639 [DOI] [PubMed] [Google Scholar]
- Spadaro D, Yun BW, Spoel SH, Chu C, Wang YQ, Loake GJ (2010) The redox switch: dynamic regulation of protein function by cysteine modifications. Physiol Plant 138: 360–371 [DOI] [PubMed] [Google Scholar]
- Teramoto H, Nakamori A, Minagawa J, Ono T-A (2002) Light-intensity-dependent expression of Lhc gene family encoding light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Physiol 130: 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Levitan A, Sofer A, Danon A (2000) Translation of chloroplast psbA mRNA is modulated in the light by counteracting oxidizing and reducing activities. Mol Cell Biol 20: 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Derrien B, Gautier A, Houille-Vernes L, Boulouis A, Saint-Marcoux D, Malnoë A, Rappaport F, de Vitry C, Vallon O, Choquet Y, Wollman FA (2014) Nitric oxide-triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell 26: 353–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe L, Blifernez O, Schwarz C, Mussgnug JH, Nickelsen J, Kruse O (2009) Cysteine modification of a specific repressor protein controls the translational status of nucleus-encoded LHCII mRNAs in Chlamydomonas. Proc Natl Acad Sci USA 106: 13290–13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Bedhomme M, Marchand CH, Morisse S, Trost P, Lemaire SD (2012) Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxid Redox Signal 16: 567–586 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P (2013a) Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci 4: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Morisse S, Bedhomme M, Marchand CH, Festa M, Rouhier N, Lemaire SD, Trost P (2013b) Mechanisms of nitrosylation and denitrosylation of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. J Biol Chem 288: 22777–22789 [DOI] [PMC free article] [PubMed] [Google Scholar]