Defects in a NAD salvage pathway trigger the accumulation of nicotinamide and increase histone H3K9 acetylation and expression of senescence-related genes in rice.
Abstract
Premature leaf senescence affects plant growth and yield in rice. NAD plays critical roles in cellular redox reactions and remains at a sufficient level in the cell to prevent cell death. Although numerous factors affecting leaf senescence have been identified, few involving NAD biosynthetic pathways have been described for plants. Here, we report the cloning and characterization of Leaf Tip Senescence 1 (LTS1) in rice (Oryza sativa), a recessive mutation in the gene encoding O. sativa nicotinate phosphoribosyltransferase (OsNaPRT1) in the NAD salvage pathway. A point mutation in OsNaPRT1 leads to dwarfism and the withered leaf tip phenotype, and the lts1 mutant displays early leaf senescence compared to the wild type. Leaf nicotinate and nicotinamide contents are elevated in lts1, while NAD levels are reduced. Leaf tissue of lts1 exhibited significant DNA fragmentation and H2O2 accumulation, along with up-regulation of genes associated with senescence. The lts1 mutant also showed reduced expression of SIR2-like genes (OsSRT1 and OsSRT2) and increased acetylation of histone H3K9. Down-regulation of OsSRTs induced histone H3K9 acetylation of senescence-related genes. These results suggest that deficiency in the NAD salvage pathway can trigger premature leaf senescence due to transcriptional activation of senescence-related genes.
Leaf senescence is the final stage of leaf development wherein the expression levels of numerous genes change radically (Lim et al., 2007). Cellular components and metabolism are altered during senescence. For example, macromolecules such as proteins, nucleic acids, and carbohydrates are hydrolyzed (Woo et al., 2001; Kim et al., 2006). Leaf senescence is beneficial for nutrient recycling from source to sink tissues and contribute to reproductive success (Yang et al., 2011; Guo et al., 2015). This process is developmentally regulated during plant growth. Despite numerous studies investigating senescence in leaves, the underlying molecular mechanism remains largely unknown due to its complexity.
NAD and its derivative NADP are crucial metabolites for redox reactions in living organisms and form a basis for almost every metabolic pathway in the cell (Berger et al., 2004; Hunt et al., 2004; Noctor et al., 2006). There is increasing evidence that the biosynthesis, degradation, and recycling of NAD(P)(H) are associated with signal transduction, developmental regulation, and stress resistance (Amor et al., 1998; Imai et al., 2000; North et al., 2003; Mittler et al., 2004; Sánchez et al., 2004; Chai et al., 2005, 2006; De Block et al., 2005). The amount of NAD(P)(H) should be sufficient for preventing cell death due to NAD depletion in vivo. Therefore, every organism must continuously resynthesize NAD(P)(H) to maintain balance of the internal environment (Hashida et al., 2009).
Like other organisms, Arabidopsis (Arabidopsis thaliana) contains two NAD(P)(H) biosynthetic pathways: the de novo and salvage pathways (Katoh et al., 2006; Wang and Pichersky, 2007). In the salvage pathway, nicotinate mononucleotide (NaMN; an intermediate in the de novo pathway) is formed from nicotinamide (a product of NAD degradation) through two enzymatic steps (Katoh et al., 2006; Wang and Pichersky, 2007). Specifically, nicotinamide is hydrolyzed to nicotinate by nicotinamidase and is converted to NaMN by nicotinate phosphoribosyltransferase (NaPRTase; Wang and Pichersky, 2007; Hashida et al., 2009). NaPRTase is believed to be the rate-limiting enzyme in the NAD salvage pathway (Magni et al., 1999). Homologs of NaPRTase are found in plants like Arabidopsis and rice (Oryza sativa; Katoh and Hashimoto, 2004). However, very little has been learned about their physiological functions and how the activity of NaPRTase is regulated in plants.
On the other hand, new functions of NAD and its derivatives in the salvage pathway have been found involving the regulation of complex cellular processes, including transcriptional regulation via NAD-dependent deacetylation and poly(ADP-ribosylation) (Imai et al., 2000; North et al., 2003; Sánchez et al., 2004; De Block et al., 2005). For instance, NAD can be broken down into nicotinamide by silent information regulator 2 (SIR2) and poly(ADP-ribose)polymerase. SIR2 family (or sirtuin) proteins are NAD-dependent histone deacetylases involved in chromatin silencing at the mating-type loci in yeasts (Frye, 2000; Moazed, 2001).
From archaea to humans, SIR2 family proteins can generally be divided into four classes, I to IV (Brachmann et al., 1995; Frye, 2000). Yeast SIR2 belongs to class I of sirtuin genes and plays important roles in transcriptional silencing, genome stability, and longevity (Blander and Guarente, 2004). Histone H3 and H4 hyperacetylation of the mating-type loci is induced when yeast SIR2 is deleted (Robyr et al., 2002). Hyperacetylation of histones leads to active transcription, while hypoacetylation of histones results in repressed expression of genes (Carrozza et al., 2003). SIR2 family members in mammalian cells, designated SIRT1 to SIRT7, are categorized into four classes (Frye, 2000). The nearest human homolog, SIRT1, a negative regulator of p53, inhibits apoptosis through deacetylation of this transcription factor. Furthermore, SIRT6 deacetylates H3K9 and plays an important role in repressing aging-related gene expression and preventing genome instability (Luo et al., 2001; Vaziri et al., 2001; Michishita et al., 2008; Dang et al., 2009; Tennen and Chua, 2011).
Plant genomes contain fewer SIR2 homologs than animal genomes (Hu et al., 2009). Arabidopsis contains two SIR2 family genes (designated AtSRT1 and AtSRT2) belonging to two of the four classes of this family (Pandey et al., 2002). Similarly, rice contains two SIR2 family genes: the class IV gene OsSRT1 and the class II gene OsSRT2 (Pandey et al., 2002). Additionally, OsSRT1 is directly responsible for histone H3 Lys 9 (H3K9) deacetylation, a process required for transcriptional repression of transposable elements, metabolism, and apoptosis-related genes (Huang et al., 2007; Zhong et al., 2013).
Since deacetylation of SIR2 is NAD dependent, the NAD de novo and salvage pathways play crucial roles in regulating the activity of SIR2 enzymes. Regulation of SIR2 activity can be achieved by intermediates in the NAD salvage pathway and NAD metabolites/derivatives (Schmidt et al., 2004). A nuclear NAD salvage pathway in yeast has been implicated as a regulatory control point for SIR2 (Smith et al., 2000; Anderson et al., 2002; Bitterman et al., 2002; Sandmeier et al., 2002). Increasing doses of several salvage pathway genes or NAD flux in yeast results in increased gene silencing and life span extension via the activation of SIR2 (Anderson et al., 2002). Moreover, altering the level of cellular nicotinamide in yeast cells, either genetically or by exogenous addition, strongly inhibits gene silencing, increases rDNA recombination, and shortens the replicative life span of the yeast to that of the sir2 mutant (Bitterman et al., 2002). SIRT1-mediated deacetylation of p53 can be inhibited by nicotinamide both in vitro and in vivo (Luo et al., 2001). Changes in both free nicotinamide and NAD levels provide the greatest contribution to the cellular activity of SIR2, but nicotinamide has a more dramatic effect in the presence of smaller fluctuations in concentration (Schmidt et al., 2004).
In this study, we cloned and characterized a new gene encoding OsNaPRT1 (O. sativa NaPRTase 1), designated Leaf Tip Senescence 1 (LTS1), in rice. A mutation in LTS1 resulted in dwarfism and withered leaf tips. We describe the characterization of the physiological function of OsNaPRT1 in rice and how this protein dominates other intermediates and genes in the NAD salvage pathway.
RESULTS
The lts1 Mutant Exhibits Dwarfism and Withered Leaf Tips
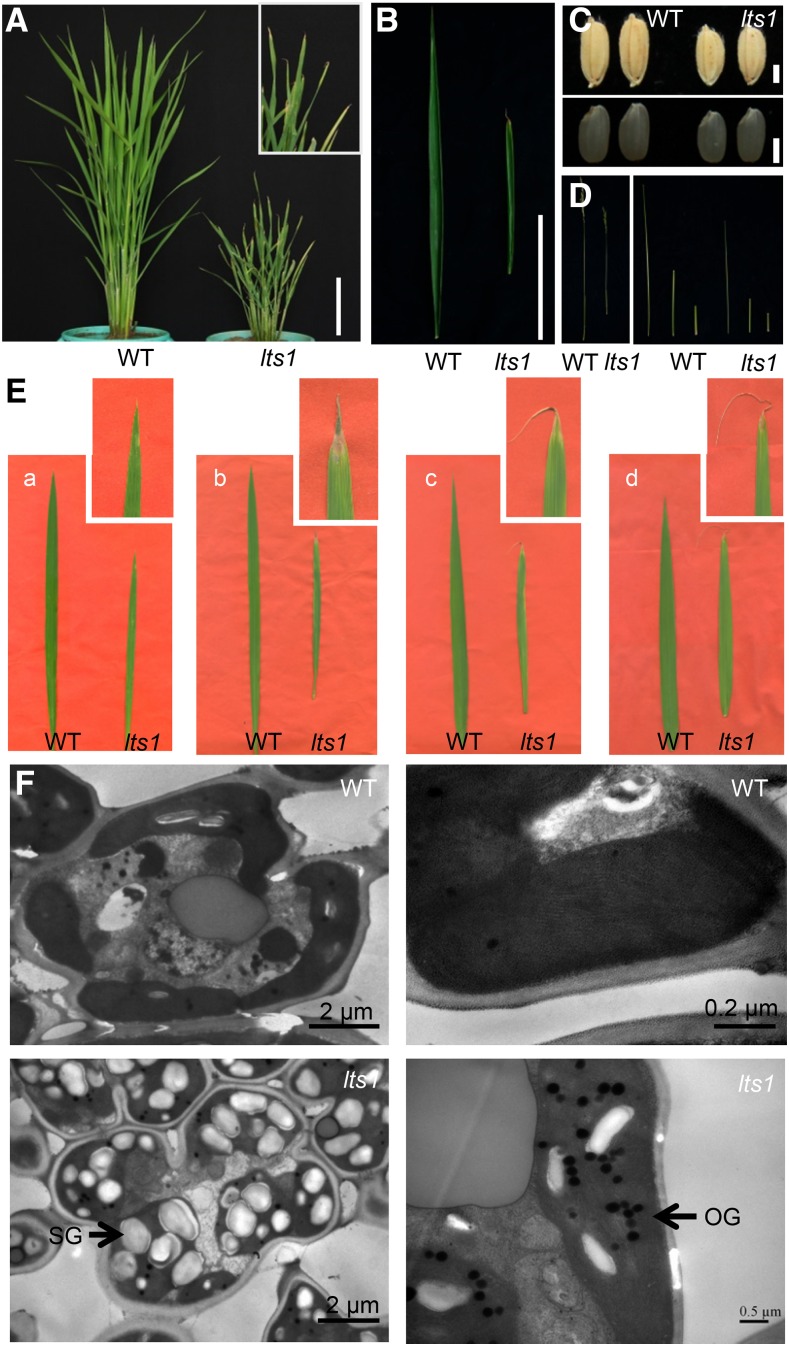
The lts1 mutant, with dwarf stems and withered leaf tips, was isolated from a mutant population of japonica rice variety Nipponbare. Each internode of lts1 was almost uniformly shortened, resulting in an elongation pattern similar to that of the wild type. However, the leaves and grains of lts1 were smaller than those of the wild type (Fig. 1, A–D).
Figure 1.
Phenotypic characteristics of the wild type and lts1. A, Wild-type and lts1 plants at the tillering stage. Inset shows a magnified view of a leaf tip of lts1. B, Wild-type and lts1 leaves at the tillering stage. C, Wild-type and lts1 grains (top); wild-type and lts1 brown rice (bottom). D, Wild-type and lts1 stems. E, Kinetic phenotypes of wild-type and lts1 leaves. Insets show magnified views of leaf tip in lts1: a to d, at 30, 50, 70, and 90 d postgermination, respectively. F, TEM analysis of cells close to leaf tips of the wild type and lts1 at the tillering stage. SG, starch grain; OG, osmiophilic globule. Bars = 10 cm in A and B and 2 mm in C.
Leaf lesions of lts1 appeared at 4 weeks postgermination under natural field conditions (25–35°C). Initially, a few yellow spots emerged at the tip of each leaf approximately 30 d postgermination. As the plants grew, the spotted areas withered at the tillering stage (50 d postgermination), and the leaves shriveled in the area of senescence progressively over time (Fig. 1E).
Transmission electron microscopy (TEM) analysis revealed that numerous starch grains accumulated in mesophyll cells in the spotted leaf area of lts1, whereas few were detected in the wild type. This observation suggests that starch degradation is reduced in the mutant compared to the wild type. Additionally, the number of osmiophilic globules was obviously elevated in the spotted leaf area of lts1, representing a characteristic of ongoing cell senescence (Fig. 1F).
LTS1 Encodes a NaPRTase
To isolate LTS1, we crossed the lts1 mutant with indica cultivar Taichung Native 1 to generate a segregation population for gene mapping. The F1 plants from the cross were normal, and segregation occurred in the F2 generation. Among the F2 individuals, 556 showed the wild-type phenotype and 177 showed the mutant phenotype. This result is in accordance with a segregation ratio of 3:1 (χ2 = 0.28 < χ2 0.05 = 3.84), indicating that the phenotype shown by lts1 is caused by a single recessive mutation.
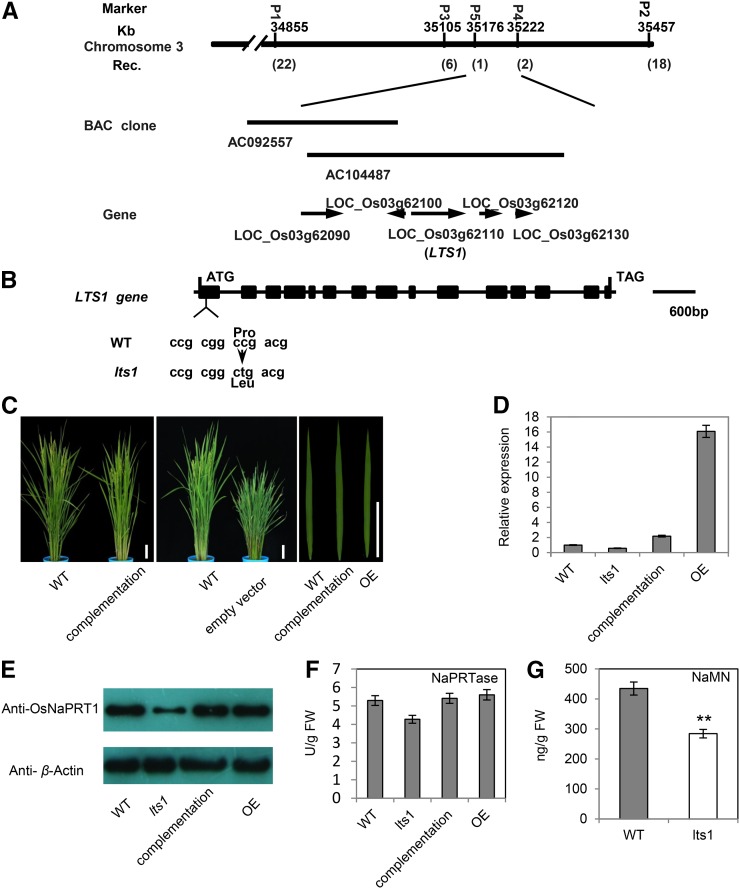
The LTS1 locus was roughly mapped on chromosome 3. Subsequent fine mapping delimited the LTS1 locus to a 46-kb region between markers P4 and P5 (Fig. 2A). DNA sequencing analysis of the entire region in lts1 revealed a single C-to-T nucleotide substitution in the first exon of LOC_Os03g62110 (MSU Rice Genome Annotation Project; http://rice.plantbiology.msu.edu/). This point mutation resulted in a Pro-to-Leu substitution at the 21th residue, causing the expression of the gene at the transcriptional level to decline by ∼40% in fully expanded leaves of lts1 compared to the wild type (Fig. 2, B and D).
Figure 2.
Molecular characteristics of LTS1. A, Fine mapping of LTS1. Markers used for mapping are indicated. Numbers in parentheses indicate the number of recombinants. AC092557 and AC104487 are accession numbers of BACs. B, Schematic representation of LTS1. Black rectangles represent exons. C, Complementation of lts1 by genomic DNA of OsNaPRT1/LTS1. Bars = 10 cm. D, Quantitative RT-PCR analysis of OsNaPRT1/LTS1 expression levels in fully expanded leaves of wild-type, lts1, complementation, and overexpression lines at 80 d postgermination. Data are the mean values. Error bars refer to sd of three biological repeats. E, Western-blot analysis of OsNaPRT1 in fully expanded leaves of wild-type, lts1, complementation, and overexpression lines at 80 d postgermination. F, Enzyme activity of NaPRTase in wild-type, lts1, complementation, and overexpression lines determined in leaves of 50 d postgermination. Data are the mean values. Error bars refer to sd of three biological repeats. FW, fresh weight. G, Leaf contents of NaMN in 50-d-old wild-type and lts1 plants. Data are the mean values. Error bars refer to sd of three biological repeats. **P ≤ 0.01; Student’s t test.
To investigate the linkage of the lts1 mutation and LOC_Os03g62110, we transformed the mutant with a genomic fragment encompassing the wild-type gene including its native promoter. Forty-three T0-independent transformants were obtained, all of which resembled the wild type (Fig. 2C; Supplemental Fig. S2). None of the progeny plants (T1 and T2) containing the transgene exhibited the dwarfism or withered leaf tip phenotypes, and the expression of LTS1 in these transgenic plants was approximately 2-fold higher than that of the wild type (Fig. 2D).
We then performed western-blot analysis using specific antibodies against LTS1. The expression of LTS1 was obviously reduced in lts1 but returned to normal wild-type levels in the complementation and overexpression lines (Fig. 2E). These results confirm that LTS1 is LOC_Os03g62110, whose repression causes premature leaf senescence in lts1.
The open reading frame of LTS1 is predicted to encode a protein of 560 amino acids that is a rice NaPRTase (OsNaPRT). To confirm that NaPRTase activity is affected by the mutation of LTS1, we measured the NaPRTase activity in leaf tissues of wild-type, lts1, complementation, and overexpression plants. The results show that NaPRTase activity was lower in lts1 than in the other lines (Fig. 2F). We then measured the content of NaMN, which is produced by the enzymatic reaction involving NaPRTase, finding that lts1 had significantly lower NaMN levels at 50 d postgermination compared to the wild type (Fig. 2G).
Sequence analysis revealed the presence of two copies of OsNaPRT in the rice genome (designated OsNaPRT1/LTS1 and OsNaPRT2). Protein sequence alignment showed that almost 95% of amino acids are conserved in the two encoded proteins (data not shown). Compared to homologous proteins from other species (sorghum [Sorghum bicolor], maize [Zea mays], barley [Hordeum vulgare], turnip [Brassica rapa], and Arabidopsis), almost 90% of amino acids throughout the protein are conserved in OsNaPRT1, as revealed by ClustalW alignment (Supplemental Fig. S1).
Expression of OsNaPRT1 in Different Rice Tissues
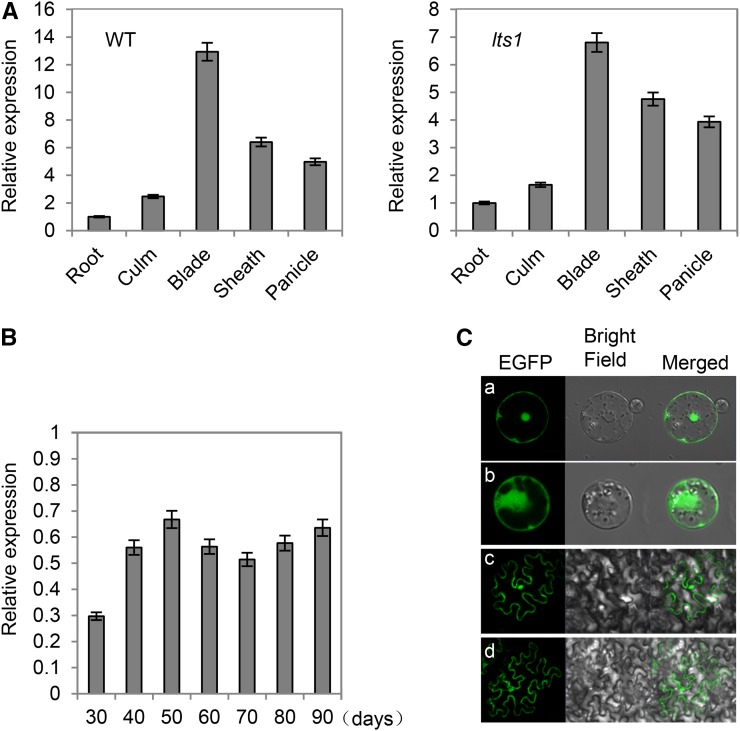
Real-time PCR experiments revealed that OsNaPRT1 is widely expressed in different rice tissues. The highest level of OsNaPRT1 transcript was detected in leaves, followed by leaf sheaths and panicles; low levels of OsNaPRT1 were detected in stems and roots. The same expression pattern was observed in lts1 and the wild type (Fig. 3A). We then examined OsNaPRT1 expression in fully expanded leaves at different developmental stages, finding that OsNaPRT1 transcript levels exhibited similar reductions at 50 d postgermination and later stages, when compared with those in the wild type (Fig. 3B).
Figure 3.
Expression profiles of OsNaPRT1. A, Expression of OsNaPRT1 in various organs, including root, culm, leaf blade, leaf sheath, and panicle at the heading stage in the wild type and lts1. Data are the mean values. Error bars refer to sd of three biological repeats. B, Change over time in OsNaPRT1 transcription levels of fully expanded leaves at different days postgermination in lts1. OsNaPRT1 transcription level in the wild type was set at 1.0. Data are the mean values. Error bars refer to sd of three biological repeats. C, Subcellular localization of OsNaPRT1-EGFP fusion. Transient expression of OsNaPRT1-EGFP fusion in rice protoplast (a) and epidermal cell of N. benthamiana leaves (c) and expression of EGFP in rice protoplast (b) and epidermal cell of N. benthamiana leaves (d) were photographed under a confocal microscope at 488 nm.
To detect the subcellular localization of OsNaPRT1, the coding region of the cDNA was fused to an eGFP-coding sequence under the control of two 35S promoters for transient expression analysis in rice protoplasts. The fusion protein was localized to the nucleus, cytoplasm, and cell membrane. Similar results were obtained from tobacco (Nicotiana benthamiana) leaf epidermal cells that contained the same constructs (Fig. 3C).
Down-Regulation of OsNaPRT1 Leads to Leaf Senescence
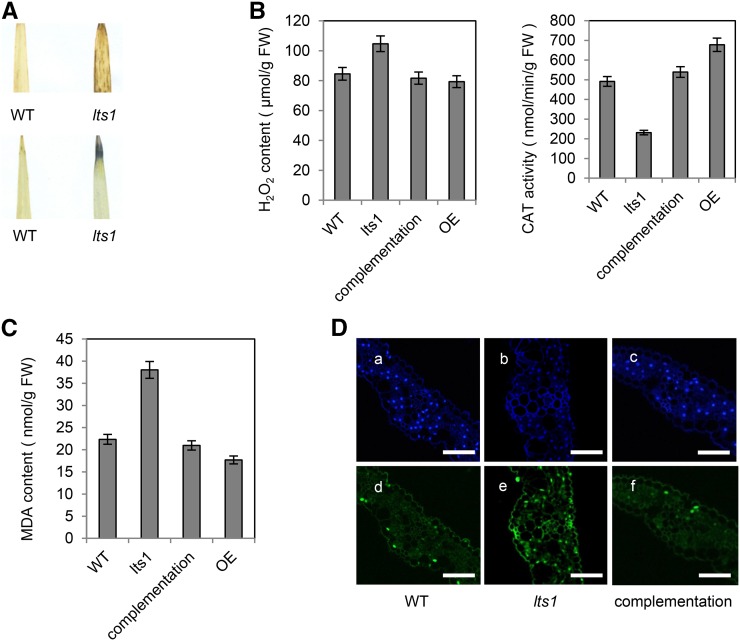
To investigate the role of OsNaPRT1 in leaf senescence, we incubated young leaf tips of the wild type and lts1 in 3,3′-diaminobenzidine (DAB) and nitrotetrazolium blue chloride (NBT), revealing H2O2 production in numerous leaf tip cells in lts1 at 50 d postgermination (Fig. 4A). We then measured the concentrations of senescence-related substances, including H2O2 and malondialdehyde (MDA), as well as catalase (CAT) activity. The results showed that concentrations of both H2O2 and MDA were higher in lts1 leaves than in the wild-type leaves and the activity of CAT was lower in the mutant (Fig. 4, B and C). Therefore, the withered leaf tip phenotype in lts1 plants suggests that down-regulation of OsNaPRT1 could initiate cell senescence in rice.
Figure 4.
lts1-induced H2O2 production and genomic DNA fragmentation. A, DAB assay of H2O2 in leaf tip tissue (top) of 50-d-old wild type (left) and lts1 (right) plants. NBT assay of superoxide in leaf tip tissue (bottom) of 50-d-old wild-type (left) and lts1 (right) plants. B, H2O2 contents and CAT activity in 50-d-old plants. Data are the mean values. Error bars refer to sd of three biological repeats. FW, fresh weight. C, MDA contents in 50-d-old plants. Data are the mean values. Error bars refer to sd of three biological repeats. D, TUNEL assay of leaves. Blue signal is 4′,6-diamino-phenylindole staining; green color represents positive result. a to c are negative controls for d to f, respectively. a and d are wild-type leaves; b and e are lts1 leaves; c and (f) are complementation leaves. Bars = 50 µm.
To confirm that down-regulation of OsNaPRT1 induces cell death, leaf tissues of wild-type, lts1, and complementation plants at 50 d postgermination were subjected to a terminal deoxyribonucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay. The same leaf sections were simultaneously stained with 4′,6-diamino-phenylindole to reveal the nuclei (blue) in each section. Few of the nuclei in leaf sections of the wild-type and complementation plants were TUNEL positive, whereas numerous nuclei in leaf sections of lts1 were TUNEL positive (green; Fig. 4D). These results indicate that down-regulation of OsNaPRT1 generally induces DNA damage and accelerates cell senescence in lts1leaf tissues.
Inhibition of OsSRTs Expression in lts1
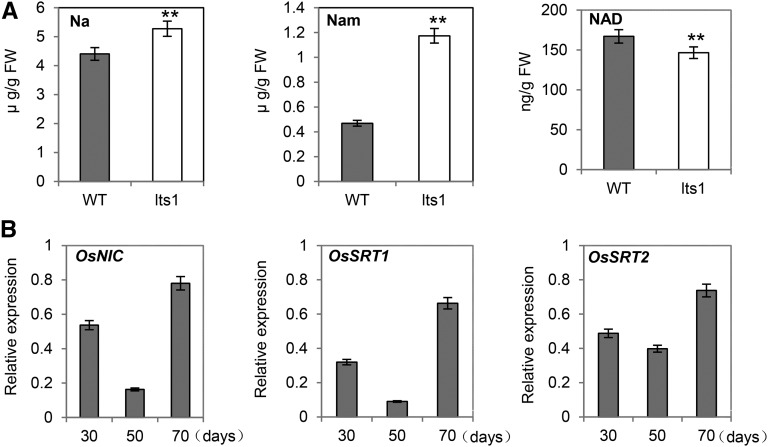
Because NaPRTase is the rate-limiting enzyme in the NAD salvage pathway, we investigated whether down-regulation of OsNaPRT1 would affect the contents of specific intermediates in this pathway. Measurement of leaf tissue contents revealed that lts1 had significantly higher nicotinate and nicotinamide levels but lower NAD levels at 40 d postgermination compared to the wild type (Fig. 5A). Additionally, we examined the expression levels of three critical genes in the salvage pathway, OsNIC and OsSRTs (OsSRT1 and OsSRT2). These genes were obviously down-regulated in lts1 compared to the wild type (Fig. 5B). These results suggest that the mutant form of OsNaPRT1 disrupts the NAD salvage pathway.
Figure 5.
Leaf contents of intermediates and expression levels of key genes in the salvage pathway. A, Leaf contents of nicotinate (Na), nicotinamide (Nam), and NAD in 40-d-old wild-type and lts1 plants. Data are the mean values. Error bars refer to sd of three biological repeats. **P ≤ 0.01; Student’s t test. FW, fresh weight. B, Leaf expressional levels of OsNIC, OsSRT1, and OsSRT2 in lts1 plants at 30, 50, and 70 d postgermination. The corresponding transcription levels of genes in the wild-type plants were set at 1.0. Data are the mean values. Error bars refer to sd of three biological repeats.
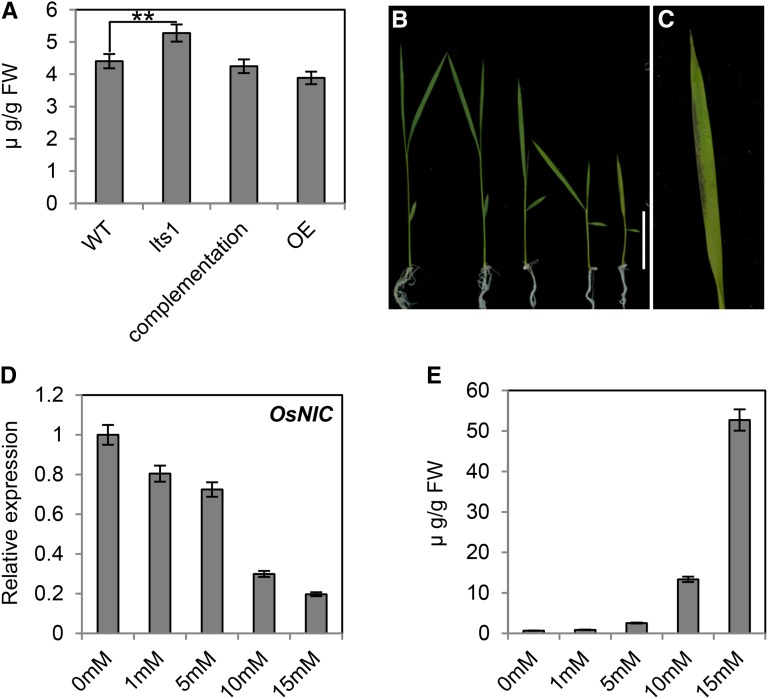
As nicotinate is the substrate of OsNaPRT1, it is understandable that the level of nicotinate increased because of the weakening of OsNaPRT1 in lts1. Thereafter, we measured nicotinate levels in leaf tissue, finding that the complementation and overexpression lines had lower nicotinate levels than lts1 (Fig. 6A). To explore the effects of abnormal nicotinate levels in the mutant on the intermediates and key genes in the NAD salvage pathway, we germinated wild-type seedlings on half-strength Murashige and Skoog (MS) medium for 10 d, followed by growth on medium containing different concentrations of nicotinate for 1 week. The results show that with increasing nicotinate concentration, the expression of OsNIC gradually decreased (Fig. 6D). The results indicated that higher concentrations of nicotinate can inhibit the expression of OsNIC. Additionally, as the concentration of nicotinate increased, plants growth was inhibited, but no obvious leaf senescence was observed except for the plants grown under the maximum concentration of nicotinate (Fig. 6, B and C). These results indicate that abundant nicotinate can negatively regulate the height of plants but does not have an obvious effect on senescence of leaf in rice.
Figure 6.
Inhibition of seedling growth and expression of OsNIC in wild-type plants grown on nicotinate. A, Leaf contents of nicotinate in 40-d-old wild-type, lts1, complementation, and overexpression lines plants. Data are the mean values. Error bars refer to sd of three biological repeats. **P ≤ 0.01; Student’s t test. FW, fresh weight. B and C, Ten-day-old seedlings of wild-type plants were transferred from half-strength MS medium to medium supplemented with different concentrations of nicotinate and grown for an additional 7 d (the concentration of nicotinate for the seedling was 0, 1, 5, 10, and 15 mm from left to right). Close-up of the leaf of the seedling grown on 15 mm nicotinate is shown in C. Bar = 5 cm. D, The expression of OsNIC in the leaves of seedlings treated with different concentrations of nicotinate. Data are the mean values. Error bars refer to sd of three biological repeats. E, The contents of nicotinamide in the leaves of seedling treated with different concentrations of nicotinate. Data are the mean values. Error bars refer to sd of three biological repeats. FW, fresh weight.
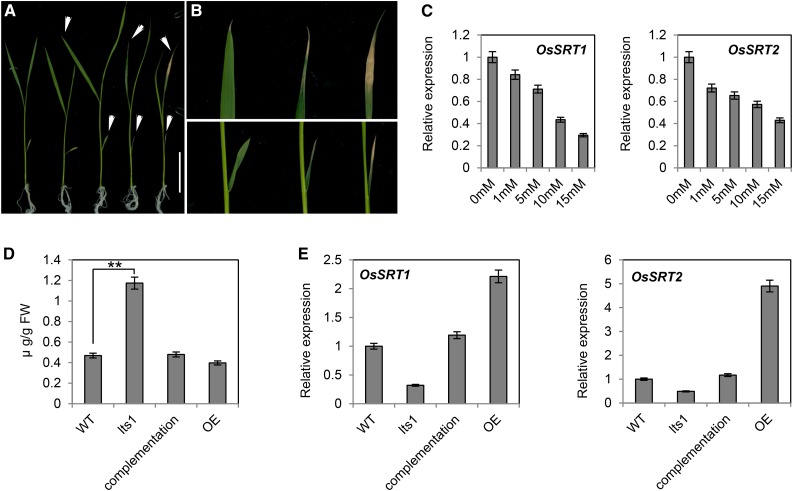
Afterward, we measured the content of nicotinamide in the plants that were treated with different concentrations of nicotinate, which revealed increasing nicotinamide levels in response to treatment with increasing concentrations of nicotinate (Fig. 6E). Thus, the rise of nicotinamide, the substrate of OsNIC, was likely due to the repression of OsNIC expression by high concentrations of nicotinate. In yeast and mammalian cells, high concentrations of nicotinamide can inhibit SIR2 activity; to test whether this is also the case in rice, we placed wild-type seedlings in media with increasing concentrations of nicotinamide and examined the levels of OsSRT expression a week later using real-time PCR. The results showed that, with the increased nicotinamide concentration, the level of OsSRTs decreased (Fig. 7C). The degree of leaf tip senescence became more severe with the elevation of nicotinamide concentration but the height of plant wasn’t impacted (Fig. 7, A and B). These results suggest that leaf senescence in the mutant is caused by the accumulation of nicotinamide and the down-regulated OsSRTs indirectly. Furthermore, we measured the nicotinamide contents in wild-type, lts1, complementation, and overexpression lines. The nicotinamide level was lower in complementation and overexpression lines than in the lts1 mutant, but the expression of OsSRTs was higher (Fig. 7, D and E). All of these results indicated that the accumulation of nicotinamide inhibits the expression of OsSRTs and leads to senescence in rice leaves.
Figure 7.
Phenotype of leaf tip and expression of OsSRTs in wild-type plants grown on nicotinamide. A and B, Ten-day-old seedlings of wild-type plants were transferred from half-strength MS medium to medium supplemented with different concentrations of nicotinamide and grown for an additional 7 d (the concentration of nicotinamide for the seedling was 0, 1, 5, 10, and 15 mm from left to right). Close-up of the senescent leaf tips of the seedling indicated by the white arrowheads is shown in order in B. Bar = 5 cm. C, The expression of OsSRTs in the leaves of seedling treated with different concentrations of nicotinamide. Data are the mean values. Error bars refer to sd of three biological repeats. D, Leaf contents of nicotinamide in 40-d-old wild-type, lts1, complementation, and overexpression lines plants. Data are the mean values. Error bars refer to sd of three biological repeats. **P ≤ 0.01; Student’s t test. FW, fresh weight. E, Leaf expressional levels of OsSRTs in wild-type, lts1, complementation, and overexpression lines at 40 d postgermination. Data are the mean values. Error bars refer to sd of three biological repeats.
Activation of Senescence-Associated Genes Revealed by Transcriptomic Analysis
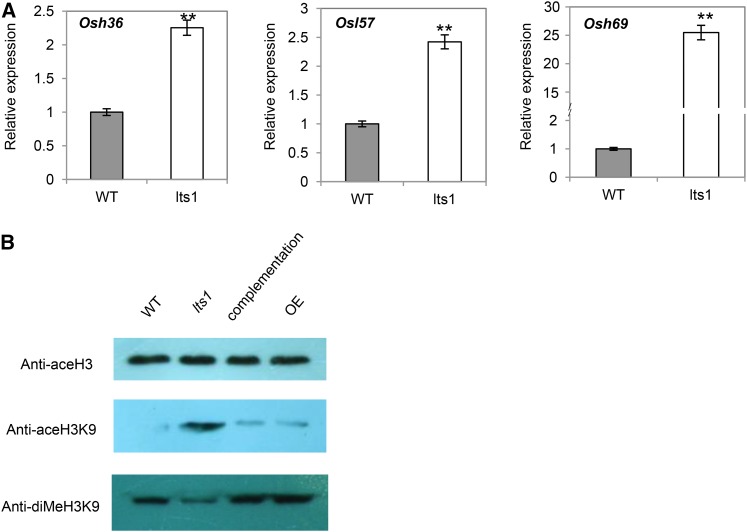
To further characterize senescence in lts1, we investigated the expression of three established senescence marker genes by quantitative real-time PCR. The senescence-associated genes (SAGs) Osh36, Osl57, and Osh69 were expressed at higher levels in fully expanded leaves of the lts1 mutant than in those of the wild type (Fig. 8A). This result indicates that the transcriptional levels of the three SAGs were higher and the leaf senescence process was significantly accelerated in lts1 compared to the wild type.
Figure 8.
Expression of three SAGs in lts1 and acetylation and dimethylation of total H3K9 in lts1. A, Relative transcript levels of Osh36, Osl57, and Osh69 in wild-type plants were set at 1.0. Data are the mean values. Error bars refer to sd of three biological repeats. **P ≤ 0.01; Student’s t test. B, Western-blot analysis of histone extracted from wild-type, lts1, complementation, and overexpression lines with antibodies against acetylated histone H3, acetylated H3K9, and dimethylated H3K9.
To investigate the effects of down-regulation of OsNaPRT1 on gene expression, we performed a genome-wide analysis of lts1 using an Affymetrix whole-genome microarray. In total, 625 differentially expressed genes (DEGs) were up-regulated and 256 DEGs were down-regulated in lts1 compared to the wild type (Supplemental Table S1). We conducted functional classification of these DEGs using the biological process category of the Rice Gene Ontology. We found that many of the DEGs are related to oxidation reduction and transcriptional regulation (Supplemental Table S1). This result suggests that the function of oxidation reduction is disturbed in the mutant due to the reduced transcription of OsNaPRT1.
Additionally, various DEGs were categorized to the metabolism of proteins, carbohydrates, macromolecules, and amino acid derivatives, and a number of genes involved in ion transport and transmembrane transport were notably up- or down-regulated in lts1 (Supplemental Table S1). These results indicate that the mutation of OsNaPRT1 contributes to the metabolism and remobilization of nutrients during the senescence process. Importantly, according to the results of microarray analysis, some of the DEGs are related to cell senescence (i.e. response to oxidative stress, defense response, and cell death), including more up-regulated than down-regulated genes (Supplemental Tables S1 and S2). The results indicate that the degree of leaf senescence is higher in lts1 than in the wild type, which is in agreement with the expression patterns of the three SAGs (Osh36, Osl57, and Osh69).
Histone H3K9 Acetylation of SAGs in lts1
In light of the down-regulation of OsSRTs, we wanted to investigate whether the H3K9 deacetylation activity of OsSRTs is reduced in lts1. Thus, we performed western-blot analysis using antibodies raised specifically against acetylated histone H3 and acetylated H3K9, using dimethylated H3K9 as a supplement. The results show that total histone H3 acetylation was not significantly affected in lts1 compared to the wild type. However, acetylation of H3K9 was induced, while its dimethylation was clearly decreased in the mutant (Fig. 8B), suggesting that the ability of OsSRTs to deacetylate H3K9 is impaired in the mutant.
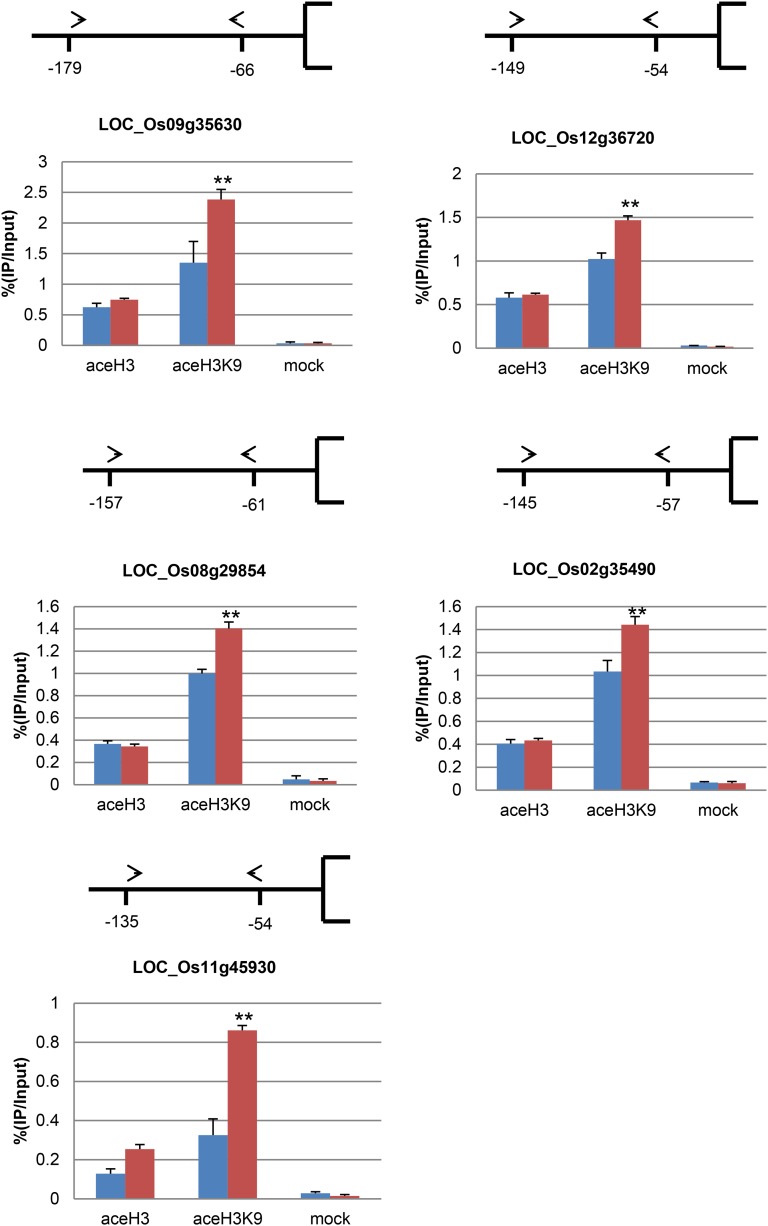
To determine whether the activation of senescence-associated genes in the mutant is due to increased H3K9 acetylation via down-regulation of OsSRTs, we performed chromatin immunoprecipitation (ChIP) assays. Chromatin fragments were isolated from 3-week-old leaves of wild-type and lts1 plants and immunoprecipitated with antibodies against acetylated histone H3 or acetylated H3K9. We analyzed precipitated chromatin DNA by real-time PCR to test for enrichment relative to nonprecipitated (input) genomic DNA. H3K9 acetylation was clearly induced in the selected senescence-related genes of lts1 (Fig. 9), which is in agreement with the expression data from Affymetrix whole-genome microarray analysis. These results suggest that down-regulation of OsSRTs leads to up-regulated expression of SAGs in lts1.
Figure 9.
Histone H3 and H3K9 acetylation of senescence-related genes in lts1. DNA was immunoprecipitated with antibodies specific to acetylated histone H3 (aceH3), acetylated H3K9 (aceH3K9), or without antibody (mock). Positions of primers relative to the initiation ATG codon are indicated by arrowheads and numbers. LOC numbers of selected genes are indicated below. Relative amounts of PCR products compared to input chromatin from wild-type extracts (arbitrarily set at 100) are shown in the graphs. Blue bars, the wild type; red bars, lts1. Data are the mean values. Error bars refer to sd of three biological repeats. **P ≤ 0.01; Student’s t test.
DISCUSSION
Mutant OsNaPRT1 Disturbs the NAD Salvage Pathway and Leads to Multiple Abnormal Phenotypes in Rice
Many organisms, including plants, have evolved an NAD salvage pathway that reuses nicotinamide released from NAD to maintain NAD(P)(H) balance (Ashihara et al., 2005). Previous studies have investigated the biological significance of some genes associated with NAD(P)(H) biosynthesis in plants, including NMNAT, AtNIC1, NADK2, and so on (Hashida et al., 2007, 2009; Wang and Pichersky, 2007). Here, we demonstrate that a mutation in OsNaPRT1, which encodes a NaPRTase capable of converting nicotinate to NaMN in the salvage pathway of NAD biosynthesis, accelerates the progress of leaf senescence in rice.
Sequence analysis revealed the presence of two genes encoding NaPRTase in the rice genome, i.e. OsNaPRT1 (LOC_Os03g62110) and OsNaPRT2 (LOC_Os04g35060). Protein sequence alignment showed that amino acids are highly conserved in OsNaPRT1. Similar results were obtained from the alignment of homologs in sorghum, maize, barley, turnip, and Arabidopsis (Supplemental Fig. S1). Relative RNA expression analysis demonstrated that the expression level of OsNaPRT1 was reduced by approximately one-third in lts1 compared to the wild type, while the level of OsNaPRT1 protein was even more reduced (Fig. 2, D and E; Supplemental Fig. S1). Meanwhile, the expression of OsNaPRT2 exhibited no obvious difference between lts1 and the wild type (data not shown). Compared to the wild type, the abnormal phenotypes of dwarf stems and withered leaf tips in the lts1 mutant are only due to the change of a single conserved amino acid at the beginning of the OsNaPRT1 sequence. These results suggest that OsNaPRT1 plays an indispensable role in the development of rice plants.
TEM observation, histochemical analysis, TUNEL assays, and senescence marker gene expression analysis showed that cell senescence was induced in lts1, resulting in withered leaf tips (Fig. 1F, 4, and 8A). Moreover, lts1 showed multiple phenotypes different from those of the wild type, such as dwarf stems and small grains (Fig. 1, C and D). We hypothesize that the deficiency of OsNaPRT1 leads to a disturbance of the NAD salvage pathway in leaf tissues of lts1 and affects redox reactions in vivo leading to pleiotropic phenotypes.
Subsequent assays of intermediates and examination of the expression of pivotal genes in the NAD salvage pathway demonstrated that these parameters are significantly altered in lts1 compared to the wild type (Fig. 5A). Furthermore, microarray analysis showed that a number of DEGs encoding oxidation-reduction proteins were up- or down-regulated in lts1 (Supplemental Table S1). These results suggest that numerous oxidation-reduction reactions are indeed altered in the mutant due to the deficient NAD salvage pathway. Considering the important contribution of this pathway to metabolism, the decrease in NAD levels should alter metabolic processes. The up- and down-regulation of numerous genes involved in various metabolic processes confirms that the metabolism in lts1 is disturbed. These disorders in redox reactions and metabolism are likely the main reasons for the abnormal phenotypes of this mutant.
Our TEM data show that abundant starch grains accumulated in mesophyll cells of lts1 (Fig. 1F), indicating that the carbohydrate metabolic process is blocked in this mutant. Since carbohydrates are the major source of energy for the plant, a carbohydrate deficiency would inevitably affect plant development. Consequently, a lack of essential nutrients is likely one of the reasons for the multiple defective phenotypes of lts1 compared to the wild type.
OsNaPRT1 Deficiency Initiates Leaf Cell Senescence by Altering Nicotinamide Contents
Nicotinamide is thought to function as an initial signal in the defensive response to DNA strand breakage caused by oxidative stress and/or specific chemicals (Berglund et al., 1996). In this study, histochemical staining showed that H2O2 production in leaf cells occurred at higher level in the lts1 mutant than in the wild type. H2O2 production is an indicator of hypersensitive senescence in plant cells during incompatible plant-pathogen interactions (Brodersen et al., 2002). The TUNEL-positive signals detected in nuclei indicate that DNA fragmentation generally occurred in leaf cells of lts1 (Fig. 4). Microarray analysis also showed that many more genes involved in the response to oxidative stress and defense were up-regulated than down-regulated (Supplemental Table S1).
Additionally, nicotinamide promotes apoptosis in mammalian cells by inhibiting the SIR2 homolog SIRT1 (Luo et al., 2001; Vaziri et al., 2001; Bitterman et al., 2002). Evidence indicates that depleting the intracellular inhibitor nicotinamide is sufficient to activate SIR2, which is the mechanism by which PNC1 regulates longevity in yeast (Bitterman et al., 2002). Moreover, PNC1 levels are elevated under every condition known to extend the life span of cells, and these cells have increased rates of nicotinamide hydrolysis (Anderson et al., 2003). Because leaf cell senescence is similar to apoptosis in mammalian cells, we speculate that the accumulation of nicotinamide is another key factor that contributes to the initiation of cell senescence in rice.
The levels of two important intermediates in the NAD salvage pathway, i.e. nicotinate and nicotinamide, obviously increased in lts1 leaves (Fig. 5A). However, it was unclear whether the accumulation of nicotinamide causes leaf cell senescence. Feeding experiments using different concentrations of nicotinate and nicotinamide showed that nicotinamide indeed plays a role in leaf cell senescence, but nicotinate mainly influences the height of rice plants. The detailed mechanism underlying how nicotinate regulates plant height in rice requires further study.
As the key enzyme contacts nicotinate with nicotinamide, the nicotinamidase in Arabidopsis has been designated AtNIC1 as the homolog of PNC1 (Wang and Pichersky, 2007). The level of OsNIC can be controlled by excessive nicotinate via feedback regulation. The content of the substrate nicotinamide increases because OsNIC is repressed by the redundant amount of nicotinate. Therefore, high concentrations of nicotinate can also cause the leaf cell senescence. Thus, our findings suggest that mutant OsNaPRT1 in lts1 affects nicotinamide content in the leaves, thereby indirectly accelerating leaf cell senescence. We also aimed to discover the crucial downstream factors causing cell senescence in lts1.
Repressed Expression of OsSRTs Activates Senescence-Related Genes
SIR2 belongs to the NAD salvage pathway and plays a role in converting NAD to nicotinamide. Rice contains two SIR2-related genes of different classes, i.e. OsSRT1 (LOC_Os04g20270) and OsSRT2 (LOC_Os12g07950) (Pandey et al., 2002). In this study, the down-regulation of the salvage pathway led to increased nicotinamide levels and reduced total NAD levels. According to previous reports, nicotinamide and NAD have a strong impact on SIR2 activity in yeast and mammals: The latter plays a positive role, whereas the former plays a negative role (Schmidt et al., 2004). In this study, real-time PCR revealed that the expression levels of OsSRT1 and OsSRT2 were reduced in the lts1 mutant (Fig. 5B). To demonstrate that nicotinamide negatively regulates the expression of OsSRTs in rice, we treated plants with various concentrations of nicotinamide. We also monitored the expression of OsSRTs in response to different concentrations of nicotinate treatment because the nicotinamide contents also increased in response to increasing nicotinate levels. We found that excessive nicotinamide levels inhibited the expression of OsSRTs in a dose-dependent manner. By measuring the content of nicotinamide and detecting the level of OsSRTs in wild-type, lts1, complementation, and overexpression lines, we confirmed that it is effectively to promote the expression of OsSRTs with reduced amount of nicotinamide.
As OsSRT1 has been reported to be mainly involved in histone H3K9 deacetylation in rice (Huang et al., 2007), we investigated whether this activity is impaired in lts1. Molecular analysis showed that H3K9 acetylation increased while H3K9 dimethylation decreased in lts1 (Fig. 8B). Since H3K9 acetylation is associated with active chromatin (Carrozza et al., 2003), we examined the expression of selected senescence-related genes using microarray analysis. The senescence-related genes were up-regulated in lts1 (Supplemental Tables S1 and S2), indicating that the down-regulation of OsSRTs affects H3K9 deacetylation and senescence-related gene expression in rice. ChIP assays revealed a clear increase in H3K9 acetylation of the tested senescence-related genes in lts1 (Fig. 9), which is in agreement with the transcriptional activation of these genes. These results provide strong evidence that the up-regulation of senescence-related genes is a consequence of the down-regulation of OsSRTs.
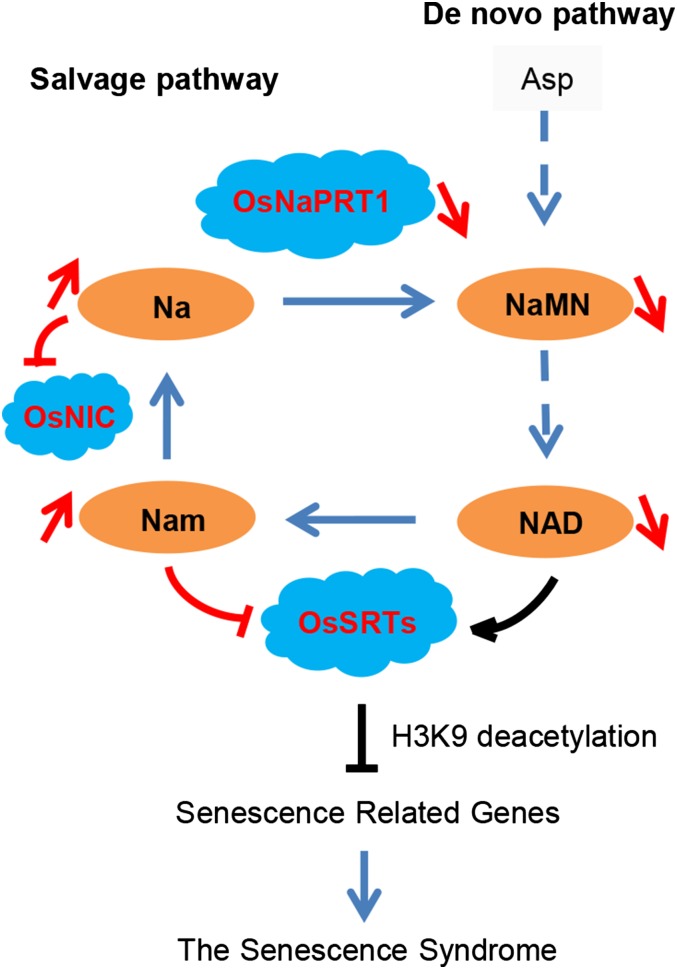
Acetylation of p53 induced by DNA damage leads to its activation and either growth arrest or cell death in mammalian cells (Vaziri et al., 2001; Chowdhury et al., 2006). SIR2 protein binds and deacetylates p53 via a specific modification, which has been implicated in the activation of p53 as a transcription factor (Vaziri et al., 2001). In this study, OsSRTs functioned in a similar manner to SIR2, while the behavior of senescence-related plant genes was analogous to that of p53. In conclusion, we used lts1 to demonstrate the role of OsNaPRT1 in cell senescence in rice. We found that deficient OsNaPRT1 triggers the accumulation of nicotinamide and thus represses the expression of OsSRTs. Meanwhile, deficient OsNaPRT1 is associated with a decrease in NAD levels. As an NAD-dependent deacetylase, decreased NAD would result in a lower deacetylation ability of OsSRTs. This leads to increased H3K9 acetylation and transcriptional activation of senescence-related genes, thereby initiating the onset of leaf senescence (Fig. 10).
Figure 10.
Proposed model for the function of OsNaPRT1 in leaf senescence in rice. Na, nicotinate; Nam, nicotinamide; OsNIC, O. sativa nicotinamidase; OsSRTs, O. sativa sirtuins (NAD-dependent protein deacetylases). The red marks show changes in the salvage pathway in lts1.
MATERIALS AND METHODS
Plant Materials and Culture Conditions
The lts1 mutant was isolated from japonica rice (Oryza sativa) variety Nipponbare by mutagenesis with ethyl methanesulfonate (Sigma-Aldrich). Mutant plants were grown in the paddy field of the China National Rice Research Institute in Hangzhou from May to October of each year to observe the development of withered leaf tips and other phenotypes. To determine the fine expression pattern of OsNaPRT1, various organs of wild-type and mutant plants were collected starting from the heading stage. For kinetic analysis of OsNaPRT1 expression, flag leaves of wild-type and mutant plants were harvested every 10 d postgermination. All experiments were repeated three times independently.
Gene Cloning and Complementation
In total, ∼1,800 F2 mutant plants were generated from the cross of lts1 with indica rice variety Taichung Native 1 for map-based cloning. LTS1 was ultimately mapped to a 46-kb region on chromosome 3. The predicted LTS1 locus was PCR amplified and sequenced to detect the mutation. The DNA fragment encompassed a wild-type copy of OsNaPRT1/LTS1 under the control of its own promoter. The vector pCambia1300 was used to produce the plasmid 1300-OsNaPRT1, which was then introduced into lts1 mutant plants by Agrobacterium tumefaciens-mediated transformation for genetic complementation. Forty-three T0-independent transformants restored to the wild-type phenotypes were generated. For the OsNaPRT1 overexpression construct, a 1,683-bp fragment of full-length cDNA was amplified by PCR from the cDNA library of Nipponbare and inserted into the overexpression vector pCambia1300S (pCambia1300 containing the 35S promoter). The resulting plasmid (35S-OsNaPRT1) was transformed into japonica cultivar Nipponbare to generate 17 T0-independent OsNaPRT1 overexpression plants. The primer sequences used in this study are listed in Supplemental Table S3.
Gene Expression Analysis
Total RNA was extracted from leaves and various organs of plants using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The extracted RNA was reverse transcribed using a ReverTra Ace quantitative PCR RT Master Mix Kit with gDNA remover (Toyobo). Quantitative real-time PCR experiments were performed using a Power SYBR Green PCR Master Mix kit (Applied Biosystems). The rice Actin1 gene was used as an internal control. Considering that OsNaPRT2 is highly similar to OsNaPRT1, sequences specific to OsNaPRT1 were chosen for the real-time PCR experiments to avoid amplifying OsNaPRT2. Primer sequences can be found in Supplemental Table S3. Values are the means ± sd of three biological repeats. The Student’s t test was used for statistical analysis.
Transmission Electron Microscopy
Tissue sections close to the withered leaf tips of lts1 and sections from the same region in Nipponbare were selected at the tillering stage. The sections were cut into 0.5- to 1-cm segments, immediately placed in precooled 2.5% glutaraldehyde in phosphate buffer (0.1 m, pH 7.0), and fixed for at least 4 h. After three washes with phosphate buffer (0.1 m, pH 7.0), the fixed segments were treated overnight with 1% OsO4. Then the segments were processed as previously described (Li et al., 2011), embedded in paraffin wax, mounted, and observed under an H-7650 transmission electron microscope (Hitachi).
Western-Blot Analysis
Total protein extraction from leaf tissue was performed as previously described (Rubio et al., 2005). Two to three leaves were ground into powder in liquid nitrogen and transferred into a 1.5-mL centrifuge tube. One-half to one milliliter of plant extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% NP-40, and 1 mm PMSF) was added into the tube. The leaf tissue was grinded at once and the extract was centrifuged at 12,000 rpm for 5 min. The supernatant fraction was transferred into a new vial.
Histone protein extraction from leaf tissue was performed according to Tariq et al. (2003). Three to five leaves were ground into powder in liquid nitrogen and transferred into a 1.5-mL centrifuge tube. One-half to one milliliter of HE buffer (10 mm Tris-HCl, pH 7.5, 2 mm EDTA, 0.25 m HCl, 5 mm DTT, and 0.2 mm PMSF) was added into the tube. The leaf tissue was immediately disaggregated and the extract was centrifuged at 12,000 rpm for 10 min. The supernatant fraction was transferred into a new vial.
Trichloroacetic acid was added to the vial to a concentration of 25% and the mixture was centrifuged at 15,000 rpm for 30 min. After being washed with acetone and dried, the leaf protein was resuspended in Laemmli sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, 25% glycerol, 0.01% bromophenol blue, and 10% β-mercaptoethanol). Total protein was separated by 12% SDS-PAGE and histone protein was separated by 15% SDS-PAGE. The following steps were performed according to a previous report (Huang et al., 2007).
NaPRTase Activity Assay
Enzyme extracts were prepared from frozen leaf tissue and all extraction procedures were performed at 4°C. The assay was performed by following the conversion of nicotinate to nicotinate mononucleotide. The reaction mixture contained 0.15 mm nicotinate, 1.5 mm 5-phosphoribosyl-1-pyrophosphate, 4.5 mm ATP, 50 mm potassium phosphate, 50 mm Tris-HCl, pH 7.8, and 15 mm MgCl2. Appropriate concentration of samples were added to 120 μL of reaction mixture and incubated at room temperature (30°C) for 5 min. One unit of enzyme activity is defined as the number of nanomoles of product formed per minute. Specific activity is expressed as unit/gram of fresh leaves. The detailed experimental procedure was described by Wubbolts et al. (1990).
TUNEL Assays
The TUNEL assays were performed using a Fluorescein In Situ Cell Death Detection Kit (Roche) following the manufacturer’s instructions. The methods used for sectioning and fluorescence labeling were described elsewhere (Huang et al., 2007).
Histochemical Analysis, H2O2 and MDA Determination, and Detection of CAT Activity
The DAB assay was carried out according to a previous study (Thordal-Christensen et al., 1997). Young leaves of 50-d-old plants were placed in DAB dissolved in 10 mm PBS (1 mg/mL) for 8 h. The leaves were then boiled in 80% ethanol for 10 min and stored in 100% ethanol. The NBT solution was prepared following the same procedure used for DAB, except that the concentration was 0.5 mg/mL. Leaves were transferred into the NBT solution for 3 h, boiled in 80% ethanol for 10 min, and stored in 100% ethanol. The H2O2 and MDA contents and CAT activity were determined as described by Moradi and Ismail (2007).
Metabolite Determination
Leaves of a specific age were collected and ground in liquid nitrogen. The homogenate was immediately added to the appropriate extraction buffer. The relative levels of metabolites were determined using an API 4000 system (Applied Biosystems). Data analysis was performed as described previously (Schippers et al., 2008). Values are means ± sd of three biological repeats. The Student’s t test was used for statistical analysis.
Microarray Hybridization and Data Analysis
RNA samples from fully expanded flag leaves of wild-type and lts1 plants were used for microarray hybridization in CapitalBio Corporation. Probe labeling and chip hybridization were carried out according to a standard protocol through the Affymetrix custom service. To compare the samples for each set of experiments, normalization was performed following the standard Affymetrix protocol. DEGs in wild-type and lts1 plants were classified functionally using the biological process category of Rice Gene Ontology (www.geneontology.com/). Three independent biological replicates were conducted for wild-type and lts1 plants. DEGs with log2 ratios ≥ 1.00 or ≤ –1.00 were analyzed.
ChIP Assays
ChIP assays were performed using 3-week-old leaves of wild-type and lts1 plants (Benhamed et al., 2006). The antibodies for histone acetylation tests were purchased from Upstate Biotech. Immunoprecipitated DNA was analyzed by real-time PCR using a 7500 Fast Real-PCR System (Applied Biosystems). Fragments (2,000 bp) of sequences upstream of ATG were chosen as the promoter regions for primers design. All primers amplified 80- to 120-bp products and were annealed at 58°C. Three independent biological repeats were performed. The primer sequences are listed in Supplemental Table S3.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: OsNaPRT1 (Os03g0837300), OsNaPRT2 (Os04g0429800), OsSRT1 (Os04g0271000), OsSRT2 (Os12g0179800), OsNIC (Os02g0606800), Osh36 (Os05g0475400), Osl57 (Os02g0817700), and Osh69 (Os08g0495800).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein sequence alignment of OsNaPRT1 and homologs.
Supplemental Figure S2. The phenotypes of wild type, complementation, and overexpression grains (top) and brown rice (bottom).
Supplemental Table S1. Functional classification of differentially expressed genes in lts1 mutant compared to wild-type plants.
Supplemental Table S2. Senescence-related genes identified by Affymetrix microarray analysis as being differentially expressed in lts1 mutant plants.
Supplemental Table S3. PCR primers used in this study.
Supplementary Material
Glossary
- NaMN
nicotinate mononucleotide
- NaPRTase
nicotinate phosphoribosyltransferase
- TEM
transmission electron microscopy
- DAB
3,3′-diaminobenzidine
- NBT
nitrotetrazolium blue chloride
- MDA
malondialdehyde TUNEL: terminal deoxyribonucleotidyl transferase-mediated dUTP nick-end labeling
- MS
Murashige and Skoog
- SAG
senescence-associated gene
- DEG
differentially expressed gene
- ChIP
chromatin immunoprecipitation
Footnotes
Articles can be viewed without a subscription.
References
- Amor Y, Babiychuk E, Inzé D, Levine A (1998) The involvement of poly(ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett 440: 1–7 [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA (2003) Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA (2002) Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem 277: 18881–18890 [DOI] [PubMed] [Google Scholar]
- Ashihara H, Stasolla C, Yin YL, Loukanina N, Thorpe TA (2005) De novo and salvage biosynthetic pathways of pyridine nucleotides and nicotinic acid conjugates in cultured plant cells. Plant Sci 169: 107–114 [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou DX (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Ramírez-Hernández MH, Ziegler M (2004) The new life of a centenarian: signalling functions of NAD(P). Trends Biochem Sci 29: 111–118 [DOI] [PubMed] [Google Scholar]
- Berglund T, Kalbin G, Strid A, Rydström J, Ohlsson AB (1996) UV-B- and oxidative stress-induced increase in nicotinamide and trigonelline and inhibition of defensive metabolism induction by poly(ADP-ribose)polymerase inhibitor in plant tissue. FEBS Lett 380: 188–193 [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA (2002) Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem 277: 45099–45107 [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L (2004) The Sir2 family of protein deacetylases. Annu Rev Biochem 73: 417–435 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9: 2888–2902 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jørgensen LB, Brown RE, Mundy J (2002) Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev 16: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, Côté J (2003) The diverse functions of histone acetyltransferase complexes. Trends Genet 19: 321–329 [DOI] [PubMed] [Google Scholar]
- Chai MF, Chen QJ, An R, Chen YM, Chen J, Wang XC (2005) NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol Biol 59: 553–564 [DOI] [PubMed] [Google Scholar]
- Chai MF, Wei PC, Chen QJ, An R, Chen J, Yang S, Wang XC (2006) NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant J 47: 665–674 [DOI] [PubMed] [Google Scholar]
- Chowdhury I, Tharakan B, Bhat GK (2006) Current concepts in apoptosis: the physiological suicide program revisited. Cell Mol Biol Lett 11: 506–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL (2009) Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block M, Verduyn C, De Brouwer D, Cornelissen M (2005) Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J 41: 95–106 [DOI] [PubMed] [Google Scholar]
- Frye RA. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273: 793–798 [DOI] [PubMed] [Google Scholar]
- Guo M, Li RD, Yao J, Zhu J, Fan XY, Wang W, Tang SZ, Gu MH, Yan CJ (2015) RL3(t), responsible for leaf shape formation, delimited to a 46-kb DNA fragment in rice. Rice Sci 22: 44–48 [Google Scholar]
- Hashida SN, Takahashi H, Uchimiya H (2009) The role of NAD biosynthesis in plant development and stress responses. Ann Bot (Lond) 103: 819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida SN, Takahashi H, Kawai-Yamada M, Uchimiya H (2007) Arabidopsis thaliana nicotinate/nicotinamide mononucleotide adenyltransferase (AtNMNAT) is required for pollen tube growth. Plant J 49: 694–703 [DOI] [PubMed] [Google Scholar]
- Hu Y, Qin F, Huang L, Sun Q, Li C, Zhao Y, Zhou DX (2009) Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem Biophys Res Commun 388: 266–271 [DOI] [PubMed] [Google Scholar]
- Huang L, Sun Q, Qin F, Li C, Zhao Y, Zhou DX (2007) Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol 144: 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Lerner F, Ziegler M (2004) NAD - new roles in signalling and gene regulation in plants. New Phytol 163: 31–44 [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Katoh A, Hashimoto T (2004) Molecular biology of pyridine nucleotide and nicotine biosynthesis. Front Biosci 9: 1577–1586 [DOI] [PubMed] [Google Scholar]
- Katoh A, Uenohara K, Akita M, Hashimoto T (2006) Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol 141: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhong S, Li G, Li Q, Mao B, Deng Y, Zhang H, Zeng L, Song F, He Z (2011) Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res 21: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W (2001) Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107: 137–148 [DOI] [PubMed] [Google Scholar]
- Magni G, Amici A, Emanuelli M, Raffaelli N, Ruggieri S (1999). Enzymology of NAD+ synthesis. In DL Purich, ed, Advances in Enzymology. John Wiley & Sons, New York, pp 135–182. [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TLA, Barrett JC, et al. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Moazed D. (2001) Enzymatic activities of Sir2 and chromatin silencing. Curr Opin Cell Biol 13: 232–238 [DOI] [PubMed] [Google Scholar]
- Moradi F, Ismail AM (2007) Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot (Lond) 99: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Queval G, Gakière B (2006) NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J Exp Bot 57: 1603–1620 [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437–444 [DOI] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M (2002) Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446 [DOI] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J 41: 767–778 [DOI] [PubMed] [Google Scholar]
- Sánchez JP, Duque P, Chua NH (2004) ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J 38: 381–395 [DOI] [PubMed] [Google Scholar]
- Sandmeier JJ, Celic I, Boeke JD, Smith JS (2002) Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD(+) salvage pathway. Genetics 160: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers JHM, Nunes-Nesi A, Apetrei R, Hille J, Fernie AR, Dijkwel PP (2008) The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20: 2909–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MT, Smith BC, Jackson MD, Denu JM (2004) Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem 279: 40122–40129 [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD (2000) A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA 97: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennen RI, Chua KF (2011) Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem Sci 36: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149–159 [DOI] [PubMed] [Google Scholar]
- Wang G, Pichersky E (2007) Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J 49: 1020–1029 [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubbolts MG, Terpstra P, van Beilen JB, Kingma J, Meesters HA, Witholt B (1990) Variation of cofactor levels in Escherichia coli. Sequence analysis and expression of the pncB gene encoding nicotinic acid phosphoribosyltransferase. J Biol Chem 265: 17665–17672 [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Zhang H, Zhao Y, Sun Q, Hu Y, Peng H, Zhou DX (2013) The rice NAD(+)-dependent histone deacetylase OsSRT1 targets preferentially to stress- and metabolism-related genes and transposable elements. PLoS One 8: e66807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.