Dicotyledonous castor seeds with persistent endosperms exhibit novel genomic DNA methylation profiles mediated by 24-nt siRNAs.
Abstract
Investigations of genomic DNA methylation in seeds have been restricted to a few model plants. The endosperm genomic DNA hypomethylation has been identified in angiosperm, but it is difficult to dissect the mechanism of how this hypomethylation is established and maintained because endosperm is ephemeral and disappears with seed development in most dicots. Castor bean (Ricinus communis), unlike Arabidopsis (Arabidopsis thaliana), endosperm is persistent throughout seed development, providing an excellent model in which to dissect the mechanism of endosperm genomic hypomethylation in dicots. We characterized the DNA methylation-related genes encoding DNA methyltransferases and demethylases and analyzed their expression profiles in different tissues. We examined genomic methylation including CG, CHG, and CHH contexts in endosperm and embryo tissues using bisulfite sequencing and revealed that the CHH methylation extent in endosperm and embryo was, unexpectedly, substantially higher than in previously studied plants, irrespective of the CHH percentage in their genomes. In particular, we found that the endosperm exhibited a global reduction in CG and CHG methylation extents relative to the embryo, markedly switching global gene expression. However, CHH methylation occurring in endosperm did not exhibit a significant reduction. Combining with the expression of 24-nucleotide small interfering RNAs (siRNAs) mapped within transposable element (TE) regions and genes involved in the RNA-directed DNA methylation pathway, we demonstrate that the 24-nucleotide siRNAs played a critical role in maintaining CHH methylation and repressing the activation of TEs in persistent endosperm development. This study discovered a novel genomic DNA methylation pattern and proposes the potential mechanism occurring in dicot seeds with persistent endosperm.
Methylated cytosine residue, also called the fifth nucleotide, is one of the most well-studied epigenetic marks found extensively in eukaryotes (Feng et al., 2010; Zemach et al., 2010b), in spite of the fact that the levels and patterns of cytosine DNA methylation appear to exhibit drastic changes among different eukaryotic organisms (Zhu, 2009; Zemach et al., 2010b). DNA methylation has shown broad-ranging functions, including the regulation of gene expression, involvement in chromatin organization, and protection of the genome from invading and mobile DNA elements (Heard and Disteche, 2006; Klose and Bird, 2006; Feinberg, 2007). Usually, DNA methylation can be maintained through mitotic cell division; thus, it is a relatively stable and heritable epigenetic marker (Henderson and Jacobsen, 2007). In plants, DNA methylation generally refers to three different nucleotide sequence contexts: symmetric CG and CHG methylation and asymmetric CHH methylation (where H = C, T, or A), which seems to be distinct from that in mammals, where they are mostly limited to the CG context.
Studies have been made to elucidate the mechanisms that result in DNA methylation levels and patterns at the genome level in plants. Generally, CG methylation is generated by the conserved DNA methyltransferase METHYLTRANSFERASE1 (MET1; Vongs et al., 1993; Genger et al., 1999; Kankel et al., 2003; Law and Jacobsen, 2010) and CHG methylation is produced by the plant-specific DNA methyltransferase CHROMOMETHYLASE3 (CMT3; Cao and Jacobsen, 2002a; Law and Jacobsen, 2010; Zemach et al., 2010b), whereas CHH de novo methylation is established by a 24-nucleotide small interfering RNA (siRNA) directed DNA methylation (RdDM) pathway to guide the DNA methyltransferases DOMAINS REARRANGED METHYLTRANSFERASE1 (DRM1) and DRM2 in plants (Cao and Jacobsen, 2002b; Law and Jacobsen, 2010; Mosher and Melnyk, 2010). Also, several studies have demonstrated that de novo CHH methylation could be established by the plant-specific DNA methyltransferase CMT2 in an independent RdDM pathway (Zemach et al., 2013; Stroud et al., 2014). In addition, the DNA methyltransferase-like Dnmt2, first identified from bacteria, also was detected in plants, although its function in regulating DNA methylation remained largely unknown (Hermann et al., 2003; Mund et al., 2004; Ponger and Li, 2005). According to observations of genomic DNA methylation, there is generally a substantial amount of CG methylation and a small amount of non-CG methylation, although the genomic DNA methylation level varies broadly in the plants tested (Zemach et al., 2010b). Additionally, it is known that active DNA demethylation in Arabidopsis (Arabidopsis thaliana) depends mainly upon the activity of the DNA glycosylase DEMETER (DME; Kinoshita et al., 2004; Gehring et al., 2006), which is able to recognize and remove methylated cytosines by a base excision-repair pathway (Law and Jacobsen, 2010). In particular, studies have revealed that DME is expressed specifically in the central cell of the female gametophyte (Choi et al., 2002), leading to a tissue-specific removal of DNA methylation marks on the maternal genome and the establishment of genomic imprinting in Arabidopsis (Gehring et al., 2009). DNA glycosylases such as REPRESSOR OF SILENCING1 (ROS1), DEMETER-LIKE2 (DML2), and DML3 also are involved in the removal of 5-methylated cytosines in Arabidopsis (Gong et al., 2002), preventing the hypermethylation of specific genome regions (Penterman et al., 2007). Loss-of-function mutants of ROS1, DML2, and DML3 would result in significant changes of DNA methylation levels in all sequence contexts, particularly the CG dinucleotide in Arabidopsis (Penterman et al., 2007). These studies clearly suggest that the extents and patterns of genomic DNA methylation are maintained by the interaction of DNA methyltransferases and demethylases.
Increasing evidence demonstrates that DNA methylation plays crucial roles in the control of seed development and seed size (Xiao et al., 2006a, 2006b; Hu et al., 2014; Xing et al., 2015). In particular, DNA methylation was considered as a major regulator of gene imprinting that mainly manifests itself in the endosperm of angiosperm plants, closely associated with endosperm and seed development, such as the imprinted genes MEA (Grossniklaus et al., 1998; Köhler et al., 2003), FWA (Kinoshita et al., 2004), and FIS2 (Jullien et al., 2006). Investigations of genomic DNA methylation in the seeds of Arabidopsis (Gehring et al., 2009; Hsieh et al., 2009), rice (Oryza sativa; Zemach et al., 2010a), and maize (Zea mays; Zhang et al., 2014; Lu et al., 2015) revealed extensive hypomethylation in the endosperm genome. However, the mechanism of how the extent and pattern of DNA methylation are established and whether the extent and pattern of DNA methylation are conserved in plant seeds is largely unclear. Specifically, in most dicots, including Arabidopsis, their endosperms are ephemeral and gradually consumed by the embryo tissue during seed development (Sreenivasulu and Wobus, 2013). For this reason, it appears difficult to dissect the potential mechanism of genomic DNA methylation occurring in a dicot’s seeds. In contrast to Arabidopsis, castor bean (Ricinus communis), a member of Euphorbiaceae family, has relatively large and persistent endosperm throughout seed development (Greenwood and Bewley, 1982). It has been considered as a model plant for studies on endosperm and seed development among dicots (Houston et al., 2009; Nogueira et al., 2012). Also, castor bean is one of the most important nonedible oilseed crops, and its seed oil stored in endosperm is used broadly in industry (Ogunniyi, 2006). Importantly, the castor bean genome data have been completed (Chan et al., 2010), which facilitates the genome-wide investigation of DNA methylation in castor bean seed. In this study, we investigated the genomic DNA methylome occurring in endosperm and embryo of castor bean by deep bisulfite sequencing and revealed a conserved and distinct profile. Specifically, the extensive hypomethylation of CG and CHG in the endosperm genome could significantly influence the expression of flanking genes. However, the methylation of CHH did not exhibit a significant reduction in endosperm relative to the embryo. With the integration of the expression of the small RNAs (mediating DNA methylation) and DNA methylation-related genes, we propose a potential model to dissect how DNA methylation (particularly for the rich CHH methylation identified) was established and maintained in embryo and endosperm of castor bean.
RESULTS
Identification and Characterization of DNA Methylation-Related Genes
Genomic DNA methylation may be maintained by the interaction of DNA methylation and DNA demethylation processes in a given tissue. The main components involved in DNA methylation and demethylation processes are well studied in Arabidopsis. To dissect the mechanism of DNA methylation in castor bean, we globally inspected the methylation-related genes encoding DNA methyltransferase and demethylases at the genome scale. In total, we identified and characterized the sequence structures of eight genes encoding DNA methyltransferase and three genes encoding demethylase (Table I). Similar to the amino acid sequences in Arabidopsis, identified DNA methyltransferases (including MET1, DRM, CMT, and Dnmt2) and demethylases (including DME, ROS1, DML2, and DML3) from castor bean shared the conserved domains (Supplemental Figs. S1–S5), as reported previously in other plants (Finnegan and Dennis, 1993; Mok et al., 2010; Qian et al., 2014). Based on the structural features of these genes, in particular the presence and distribution of domains and motifs (Supplemental Fig. S6, A and B), the DNA methyltransferase genes could be identified easily as RcMET1-1 and RcMET1-2 in the MET group, RcCMT1 and RcCMT2 in the CMT group, RcDRM1, RcDRM2, and RcDRM3 in the DRM group, and RcDnmt2 in the Dnmt2 group (Supplemental Fig. S6C). By analogy, three DNA demethylase genes were identified as RcDME, RcROS1, and RcDML3 (Supplemental Fig. S6D).
Table I. Identification of genes encoding DNA methyltransferases and demethylases in castor bean.
| Gene Locus | Location | Coding Sequence Length | Protein Length | Homologs | Annotation |
|---|---|---|---|---|---|
| bp | amino acids | ||||
| DNA methyltransferases | |||||
| 29983.m003308 | 29983:1075551-1081789 | 4,629 | 1,542 | AT5G49160 | MET1-1 |
| 29609.m000606 | 29609:246194-255135 | 4,755 | 1,584 | AT5G49160 | MET1-2 |
| 28582.m000332 | 28582:146814-156888 | 2,538 | 845 | AT1G80740 | CMT1 |
| 29827.m002677 | 29827:858418-869538 | 2,205 | 734 | AT4G19020 | CMT2 |
| 29848.m004665 | 29848:1192243-1195163 | 1,215 | 404 | AT5G25480 | Dnmt2 |
| 29631.m001043 | 29631:330007-332680 | 1,440 | 479 | AT5G14620 | DRM1 |
| 29889.m003366 | 29889:694336-699871 | 2,037 | 678 | AT3G17310 | DRM2 |
| 29917.m001982 | 29917:300487-305138 | 2,061 | 686 | AT5G15380 | DRM3 |
| DNA demethylases | |||||
| 29428.m000327 | 29428:105887-115737 | 5,621 | 1,876 | AT5G04560 | DME |
| 29092.m000452 | 29092:87494-96521 | 4,905 | 1,634 | AT2G36490 | ROS1 |
| 29991.m000647 | 29991:222724-231238 | 5,139 | 1,712 | AT4G34060 | DML3 |
As shown in Supplemental Figure S6C, the phylogenetic analysis of DNA methyltransferase genes generated four distinct clades corresponding to the MET, CMT, DRM, and Dnmt2 groups with well-supported bootstrap values. It seemed that each group displayed a monophyly with an evolutionary lineage from dicots to monocots consistent with previous studies (Zemach et al., 2010b; Jurkowski and Jeltsch, 2011). Within the DNA demethylase genes, phylogenetic analysis generated six clades (I–VI). The DME homologs were restricted to dicots and absent in monocots, confirming previous observations (Zemach et al., 2010a; Zhang et al., 2011). The ROS1 genes were clustered into clades II (nested by the homologs from dicots), III, and IV (both nested by the homologs from monocots). However, both clade I and clade II were rooted by monkey flower (Mimulus guttatus), a basal dicotyledonous plant (Zemach et al., 2010a), suggesting that the DME may be phylogenetically monophyletic in dicots after gene duplication arising in the early angiosperm. In addition, the DML3 genes were clustered into clades V and VI nested by the homologs from dicots and monocots, respectively.
Expression Profiles of DNA Methylation-Related Genes
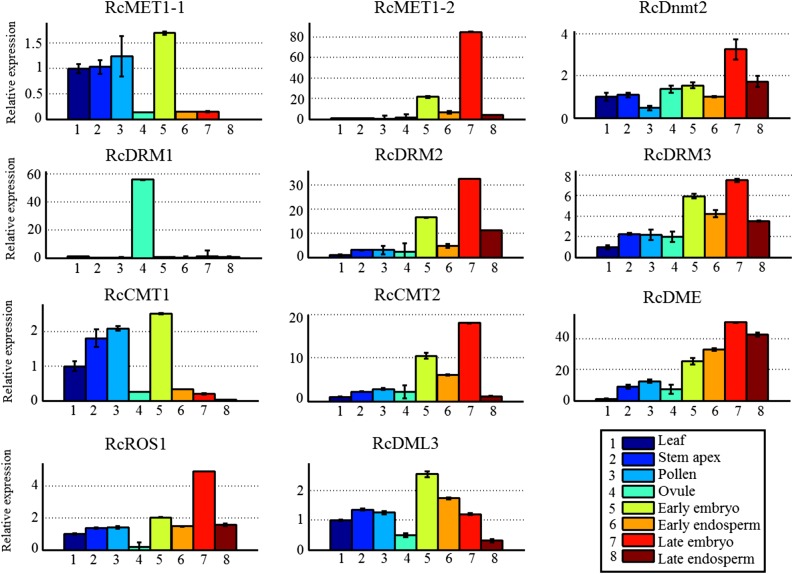
To understand how DNA methylation is established and maintained, we assayed the expression profiles of DNA methylation-related genes (including DNA methyltransferase and demethylase genes) in different tissues and developing seeds at the early and late stages. As shown in Figure 1, among the MET1 genes, RcMET1-1 was highly expressed in leaf, stem apex, pollen, and early embryo but lowly expressed in ovule and developing endosperms (including the early and late stages), while RcMET1-2 was expressed only in developing embryo tissue (particularly at the late stage). Similarly, among the CMT genes, responsible for mediating CHG methylation, RcCMT1 was highly expressed in leaf, stem apex, pollen, and early embryo but lowly expressed in ovule and developing endosperms (including the early and late stages), while RcCMT2 was expressed mainly in the developing embryo and early endosperm tissues. The substantial reduction of transcript levels of both RcMET1 and RcCMT genes in castor bean endosperm compared with embryo was consistent with a report on Arabidopsis (Jullien et al., 2012). Within the DRM genes, responsible for guiding CHH methylation, RcDRM1 was expressed specifically in ovule, similar to DRM1 expression in Arabidopsis (Jullien et al., 2012), while RcDRM2 and RcDRM3 were expressed mainly in developing embryo and endosperm tissues, distinctly different from their expression in Arabidopsis. In Arabidopsis, DRM2 was strongly transcribed in the embryo tissue but not detected in endosperm (Jullien et al., 2012). In addition, the RcDnmt2 gene, in spite of its functional uncertainty, was broadly expressed in diverse tissues.
Figure 1.
Expression profiles of DNA methyltransferase and demethylase genes in different tissues of castor bean. The castor bean ACTIN2 gene was used as an internal reference for quantitative reverse transcription-PCR analysis. The expression level of each gene in different tissues was normalized to the leaf. Error bars indicate the se with five biological replicates.
Among the DNA demethylase genes, intriguingly, RcDME was broadly expressed in diverse tissues, particularly in both embryo and endosperm tissues. This observation is distinct from the expression patterns of the DME gene in Arabidopsis, which was expressed specifically in the central cell of the female gametophyte or the vegetative cell of the male gametophyte (Choi et al., 2002; Schoft et al., 2011). Both RcROS1 and RcDML3, which may be involved in mediating DNA demethylation, were expressed extensively in diverse tissues. Also, using our previous RNA sequencing data (Xu et al., 2014), we reassayed the expression profiles of DNA methylation-related genes in endosperm and embryo tissues and obtained similar results (Supplemental Fig. S7). In addition, we noted that the expression levels of most genes were distinct between the early and late stages of endosperm and embryo development, implying that the genomic DNA methylation level within endosperm and embryo might be dynamic with seed development.
Genomic DNA Methylation in Castor Bean Seed
To obtain the genomic DNA methylation profiles at single nucleotides, we examined the DNA methylation extent and pattern in endosperm and embryo tissues by high-throughput bisulfite-treated DNA sequencing techniques, which had been broadly applied in genomic DNA methylation studies (Feng et al., 2010; Zemach et al., 2010b). Our bisulfite sequencing yielded about 120 million paired-end reads, and 88% of reads were mapped into the castor bean genome with an effective depth of 31× coverage (Supplemental Table S1). Moreover, about 83% of cytosines were covered by at least two reads in the castor bean genome. As the sequencing depth reached 30×, the number of detected cytosines reached saturation (Supplemental Fig. S8), meaning that our sequencing data were sufficient for further analysis.
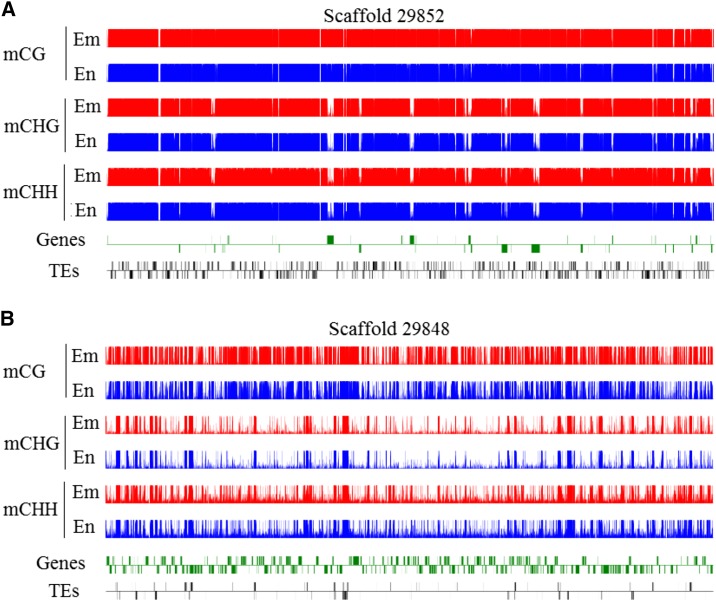
After sequencing data were mapped into the castor bean genome, we identified, in total, 19,444,574 and 21,157,129 methylated cytosine residues from endosperm and embryo, respectively. Global DNA methylation profiles demonstrated that all three sequence contexts, CG, CHG, and CHH, were, in the example shown in Figure 2, heavily methylated in the transposable element (TE)-rich regions (Fig. 2A), whereas the methylation patterns of the three sequence contexts appeared to be quite different in the gene-rich regions (Fig. 2B). The density peaks of CG methylation were found mainly in the gene body regions, but the peaks of both CHG and CHH methylation were found mainly in the TE regions and distributed sparely in the gene body regions. Clearly, these observations showed that DNA methylation occurred mainly in the TE-rich regions, consistent with previous reports from other plants (Gehring et al., 2009; Zemach et al., 2010a), and the global methylation patterns between CG and non-CG contexts were differentiated in castor bean.
Figure 2.
DNA methylation profiles in castor bean seeds. A, Global distribution of the methylation of three sequences in scaffold 29852 (TE-rich scaffold). B, Global distribution of the methylation of three sequences in scaffold 29848 (gene-rich scaffold). Red and blue tracks represent the cytosine methylation in embryo (Em) and endosperm (En), respectively. Genes and TEs oriented 5′ to 3′ and 3′ to 5′ are shown above and below the lines, respectively.
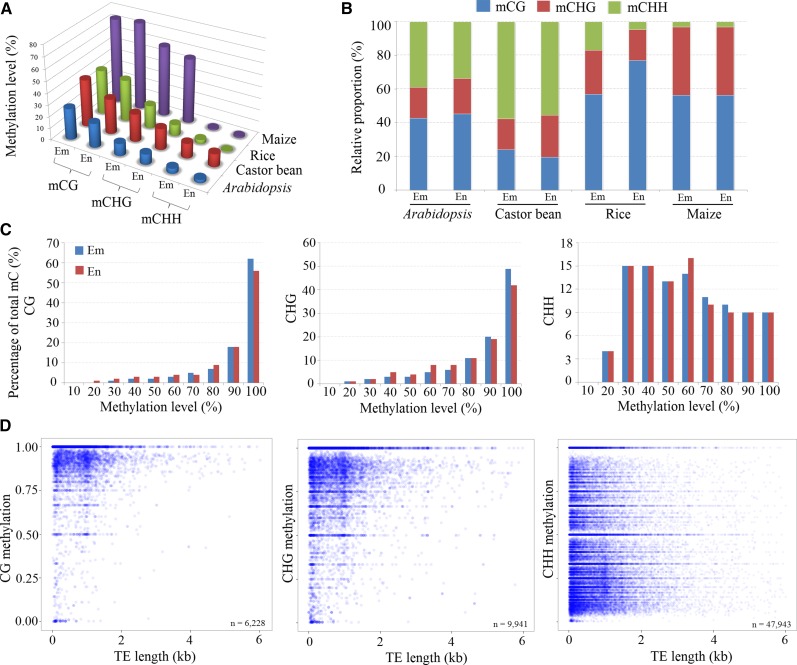
While investigating the genomic methylation percentage of CG, CHG, and CHH in embryo and endosperm tissues, we found that the methylation of CG, CHG, and CHH was, respectively, 30.3%, 18.3%, and 11.2% within the endosperm and 40.7%, 24%, and 12.7% within the embryo tissues. Obviously, the endosperm exhibited a hypomethylation, with a reduction of 10.4% CG methylation and 5.7% CHG methylation, and a nearly equal CHH methylation, compared with the embryo (Fig. 3A). Compared with the genomic DNA methylation levels in Arabidopsis (Hsieh et al., 2009), rice (Zemach et al., 2010a), and maize (Zhang et al., 2014; Lu et al., 2015), we found that the genomic DNA hypomethylation of endosperm tissues relative to embryo tissues was extensive in angiosperms, particularly for dicotyledons (Fig. 3A), although the biological roles of endosperm hypomethylation remain unknown. Interestingly, the global CHH methylation of endosperms was not reduced substantially compared with the embryo in castor bean, distinct from that in Arabidopsis (Hsieh et al., 2009) and rice (Zemach et al., 2010a). In contrast to other plants tested, in which CG methylation is most abundant, castor bean showed rich CHH methylation (68%) in both endosperm and embryo (Fig. 3B). When inspecting the proportions of CG, CHG, and CHH in the genomes of Arabidopsis, rice, maize, and castor bean, we found that they did not display a significant difference (Supplemental Table S2). This meant that the higher proportion of CHH methylation occurring in castor bean endosperm and embryo did not result from a higher CHH content in its genome compared with Arabidopsis, rice, and maize.
Figure 3.
DNA methylation levels and distribution in castor bean seed. A, Genomic methylation levels of CG, CHG, and CHH in endosperm (En) and embryo (Em) among different species. B, Relative proportions of CG, CHG, and CHH methylation contexts in endosperm and embryo among different species. C, Histogram of methylation frequencies on the y axis for different levels (%) of CG, CHG, and CHH methylation (x axis) in endosperm and embryo tissues. D, Point scatter of the correlation between TE lengths and average methylation levels in CG, CHG, and CHH contexts.
In addition, it is a remarkable fact that the methylated level of the CHH context apparently exhibited a more uniform distribution between 30% and 100% compared with the CG and CHG contexts, although the CHH methylation is predominant in number (Fig. 3C). While inspecting the methylated ratios of the CG and CHG sites, we found that the majority of CG sites (greater than 80%) were highly methylated in both embryo and endosperm. The methylated ratios of the major CHG sites displayed a similar pattern to the CG sites (Fig. 3C), consistent with the observation from cassava (Manihot esculenta) leaves (Wang et al., 2015). This observation seems to be distinct from the previous report in Arabidopsis (Greaves et al., 2012), where major CHG sites are not highly methylated. Additionally, we noted that the methylated ratios of the major CG and CHG sites were reduced slightly in endosperm compared with the embryo. Furthermore, we examined the average methylation levels of CG, CHG, and CHH in TEs and found that about 6,228 and 9,941 TEs contained CG and CHG methylation, respectively, whereas about 47,943 TEs harbored CHH methylation. In particular, more than 80% of TEs with very short sequence length (less than 1,000 bp) were heavily methylated (about 80%) in the CG and CHG contexts (Fig. 3D). Most TEs with CHH methylation (about 51.9%) were methylated in the medium range. Moreover, a large number of TEs were more densely methylated in CHH than in CG and CHG. The CHH context in long TEs (greater than 1,000 bp) was more heavily methylated (Fig. 3D). These observations show that CG and CHG methylation are more effectively maintained in short TEs than CHH methylation during DNA replication, while CHH methylation is more randomly distributed across short and long TEs, with a relatively low level of methylation compared with CG and CHG in both embryo and endosperm. Compared with previously studied plants such as Arabidopsis (Hsieh et al., 2009), rice (Zemach et al., 2010a), maize (Zhang et al., 2014; Lu et al., 2015), and soybean (Glycine max; Song et al., 2013), the genomic methylation profiles in castor bean seem to be distinct.
Taken together, although there are general features in the methylation patterns among different plant species, castor bean exhibits some specific patterns such as a very high proportion of CHH methylation, more robust maintenance of CHG methylation at a high level in TE regions, and a very similar level of CHH methylation arising between endosperm and embryo.
DNA Methylation Profiles in Gene and TE Regions
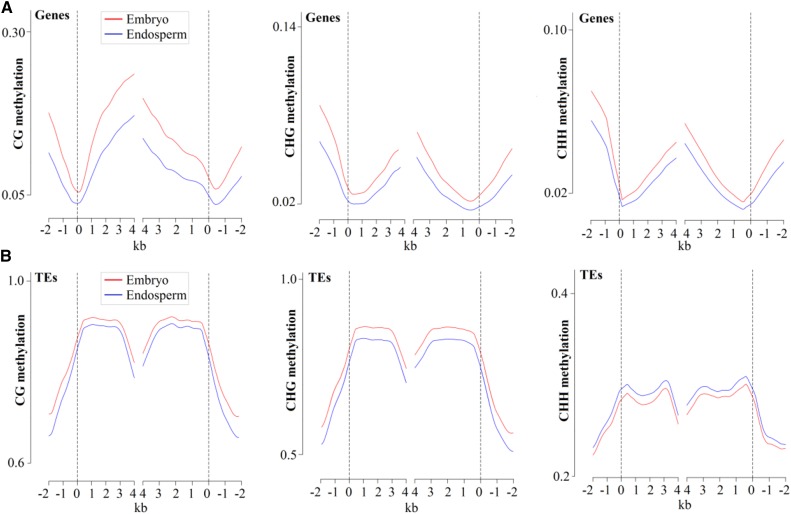
While inspecting the distribution of CG, CHG, and CHH methylations in gene and TE regions, we observed that the CG methylation occurred preferentially in the gene body regions relative to the flanking regions, similar to previous reports in other plants (Hsieh et al., 2009; Zemach et al., 2010a; Song et al., 2013; Lu et al., 2015; Wang et al., 2015), whereas the extents of CHG and CHH methylation were low in gene body regions and relatively higher in flanking regions (Fig. 4A), consistent with their global DNA methylation patterns (Fig. 2). The DNA methylation extents of the CG, CHG, and CHH contexts were very low near transcriptional start sites and transcriptional end sites but increased gradually when departing from these sites (Fig. 4A). In contrast to gene body regions, the TEs were highly methylated in all CG, CHG, and CHH sequence contexts (Fig. 4B), similar to previous findings in Arabidopsis (Hsieh et al., 2009), rice (Zemach et al., 2010a), and soybean (Song et al., 2013), suggesting that DNA methylation in transposon silencing might be conserved in plants. Furthermore, we noted that most of the methylated TEs belonged to class I (retrotransposons), especially for LTR/Gypsy and LTR/Copia, consistent with their abundances in the castor bean genome. Among class II (DNA transposons), the TE-type DNA/MuDR was more frequently methylated than others (Supplemental Fig. S9A).
Figure 4.
Genomic DNA methylation profiles in endosperm (blue lines) and embryo (red lines) tissues. A, DNA methylation patterns across genes. B, DNA methylation patterns across TEs. Castor bean annotated genes and TEs were aligned at the 5′ end (left) or the 3′ end (right). The average methylation level for each 100-bp interval is plotted. The dashed lines represent the point of alignment.
Unsurprisingly, the endosperm clearly exhibited the CG, CHG, and CHH hypomethylation in both gene body and flanking regions compared with the embryo. Moreover, the endosperm hypomethylation of CG and CHG was shown in TE regions. The endosperm exhibited a higher CHH methylation level in TE regions than in the embryo (Fig. 4B). Consistent with the reduced methylation extent of TEs in endosperm, the average methylation extents of both class I and II TEs were slightly lower in CG and CHG contexts relative to the embryo. The TEs within class I exhibited a higher methylation extent than those within class II. The CHH methylation extent of the class I TEs in endosperm, however, appeared to be slightly higher compared with the embryo, and the CHH methylation extents of the class II TEs appeared to be equal in endosperm and embryo (Supplemental Fig. S9B).
Characterization of Endosperm Differential Methylation Regions
To further characterize the genomic hypomethylation regions in endosperm, we analyzed the differential methylation regions (DMRs) within a sliding window by subtracting the endosperm methylation loci from the embryo methylation loci. Initially, we identified 64,733 CG, 70,074 CHG, and 3,298 CHH discreet DMRs between endosperm and embryo. To sort out the substantial and significant differences of DNA methylation levels at DMRs, we applied a stringent standard: at least a 40% difference (Fisher’s exact test, P < 0.05) with at least either methylation level greater than 60% for the CG and CHG contexts, and at least a 20% difference (Fisher’s exact test, P < 0.05) with at least either methylation level greater than 40% for the CHH context, between endosperm and embryo. In total, 20,464 CG and 25,940 CHG DMRs were identified, of which 99.9% of loci were significantly hypomethylated in endosperm (Table II; Supplemental Table S3). However, only 2,727 CHH DMRs were identified, of which 55.9% of loci exhibited a hypomethylation in endosperm and 44.1% of loci exhibited a hypomethylation in embryo (Table II; Supplemental Table S3).
Table II. DMRs between embryo and endosperm in castor bean.
| DNA Context | Total No. of DMRs | Hypermethylation in Embryo | Hypermethylation in Endosperm |
|---|---|---|---|
| CG | 20,464 | 20,443 (99.9%) | 21 (0.01%) |
| CHG | 25,940 | 25,903 (99.9%) | 37 (0.01%) |
| CHH | 2,727 | 1,526 (55.9%) | 1,201 (44.1%) |
Among these CG DMRs, approximately 30.8% of DMRs were located in the gene body, 27.9% and 25.6% of DMRs were located in 2-kb upstream and downstream regions of the gene, respectively, while only 15.7% of DMRs were located in TE regions. On the contrary, most CHG DMRs (about 42%) were found in TE regions, just 10.1% of DMRs were found in the gene body regions, and 27.4% and 20.6% of DMRs were found in upstream and downstream regions, respectively. For the CHH DMRs, most of them were located in the upstream and downstream regions of the gene (47.3% and 30.1%, respectively), while only 6.9% of DMRs were located in the gene body and 15.7% were located in TE regions (Supplemental Fig. S10). These analyses provided a clear landscape of genomic methylation differentiation between embryo and endosperm (Fig. 4): almost all DMRs were linked to the genomic hypomethylation of endosperm and resulted mainly from the reduction of CG and CHG methylation. The CG DMRs were located mainly in the gene body regions, while most CHG DMRs were found in the TE regions.
Effects of DNA Methylation on Gene Expression
To determine whether the genomic DNA methylation level might affect global gene expression in castor bean seeds, we initially investigated the methylation changes of CG, CHG, and CHH contexts for all transcripts obtained from previous studies (Xu et al., 2014) in embryo tissues. Based on their expression levels, genes were divided into four classes: no expression (reads per kilobase per million reads mapped [RPKM] ≤ 1), low expression (1 < RPKM ≤ 10), middle expression (10 < RPKM ≤ 100), and high expression (RPKM > 100), according to the criteria of Lu et al. (2015). As shown in Supplemental Figure S11, it seemed that the CG methylation levels occurring in the upstream and downstream regions of genes were negatively correlated with their gene expression, but in the gene body regions, the moderately expressed genes showed the highest CG methylation level, although it might be difficult to predict the interaction between the methylation extents (occurring in the upstream, downstream, or gene body region) and gene expression levels. However, it appeared that the nonexpressed (or rarely expressed) genes exhibited the highest CHG and CHH methylation levels in the upstream, downstream, and gene body regions, although the changes of methylation levels were slight across the different expression levels. These observations implied that DNA methylation, in a way, repressed gene expression in castor bean embryo tissues.
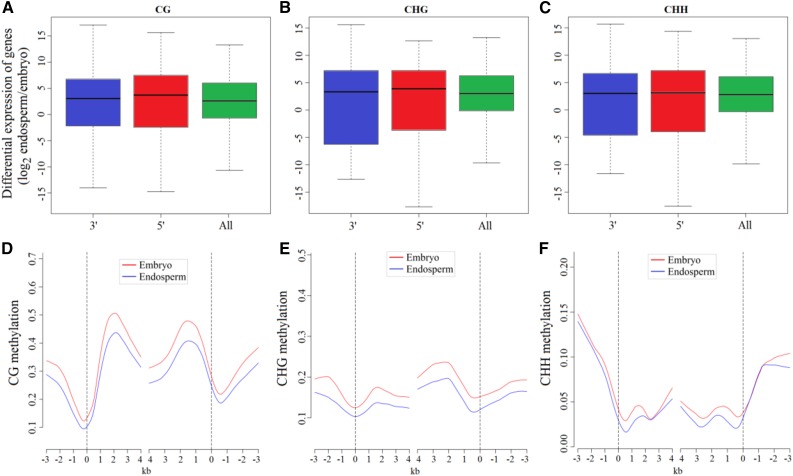
To characterize the effect of hypomethylation regions in endosperm on gene expression, we collected the genes with substantially reduced DNA methylation within 2 kb of either the 5′ or 3′ end and examined their expression between endosperm and embryo based on our previous mRNA sequencing data from embryo and endosperm (Xu et al., 2014). The results showed that genes with reduced CG and CHG methylation upstream of the start of transcription were slightly, but not significantly, up-regulated in their expression in the endosperm relative to the embryo, whereas genes with hypomethylation occurring in the 3′ end did not alter their expression levels (Fig. 5, A and B). In particular, the expression levels of genes exhibiting reduced CHH methylation in the 5′ and 3′ ends were not affected (Fig. 5C). However, we investigated the transcriptome of endosperm and embryo and identified subsets of the genes that were expressed preferentially (at least 4-fold higher) in endosperm than in embryo (Supplemental Table S4). By calculating the average methylation levels in the gene body and the 5′ and 3′ ends, we found that these genes exhibiting endosperm-preferential expression showed significantly low levels of CG and CHG methylation (Fisher’s exact test, P < 0.05), but not in CHH loci of endosperm compared with embryo. It was clear that these genes, which were preferentially up-expressed in endosperm relative to embryo, exhibited reduced CG and CHG methylation in endosperm throughout the 5′ end, gene body, and 3′ end (Fig. 5, D and E) and a tiny CHH methylation reduction, if any, around these genes as well (Fig. 5F).
Figure 5.
Effect of hypo-DMR regions in endosperm on gene expression. A to C, Box plots indicate the differences of expression levels between endosperm and embryo for genes with either 5′ hypomethylation or 3′ hypomethylation in endosperm, compared with the average of all genes. D to F, Average methylation levels for endosperm-preferred genes with 4-fold expression levels relative to embryo are plotted for each 100-bp interval.
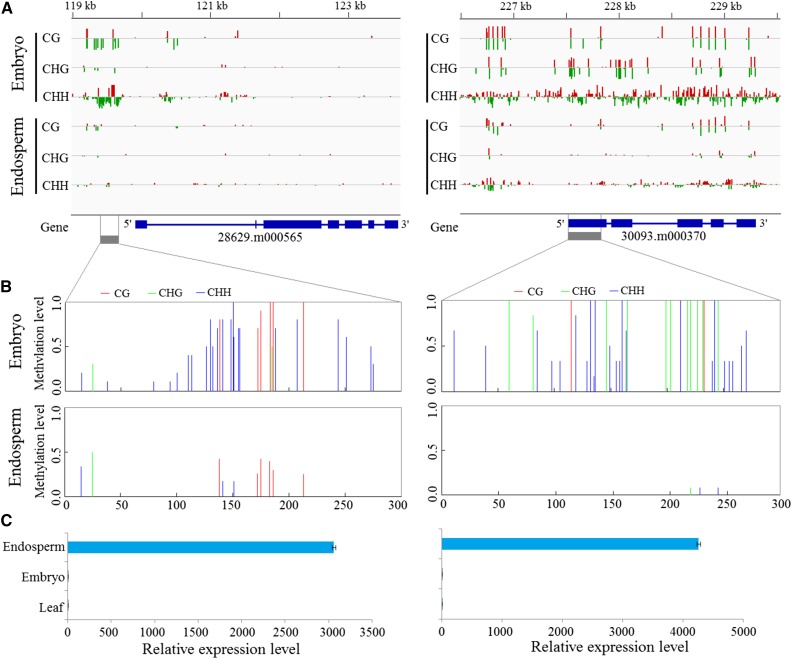
Indeed, several studies also showed that DNA hypomethylation could alter gene-preferential expression in endosperm (Hsieh et al., 2009; Zemach et al., 2010a). As illustrated in Figure 6, endosperm hypomethylation in some specific loci markedly promoted gene expression, whereas embryo hypermethylation near the promoter regions repressed gene transcription. For example, gene 28629.m000565 encoding a β-fructofuranosidase involved in carbohydrate metabolic processes was strongly expressed in endosperm. Gene 30093.m000370, homologous to AtYUCCA10 (AT1G48910) involved in regulating seed development in Arabidopsis (Cheng et al., 2007), was expressed strongly in endosperm. The two genes exhibited substantially lower DNA methylation extents and higher expression levels in endosperm than in embryo. Furthermore, the DNA methylation extents and gene expression levels of the two genes were validated experimentally using the combination method of bisulfite treatment and clone sequencing (Fig. 6, B and C). In addition, Gene Ontology analysis showed significant evidence for the functional enrichment of genes affected by endosperm hypomethylation DMRs (Supplemental Fig. S12). The Gene Ontology terms including binding, catalytic, biological regulation, cellular, and metabolic processes were enriched substantially (χ2 test, P < 0.05). These findings strongly imply that the endosperm hypomethylation is likely involved in the regulation of specific endospermogenesis and storage material biosynthesis in endosperm, although its potential mechanism is still unclear. In short, our data indicate that the CG and CHG hypomethylations might be the potential mechanism for altering genes preferentially expressed in castor bean endosperm, particularly when the CG and CHG hypomethylations occur near or within a gene.
Figure 6.
Effects of two hypomethylation regions in endosperm on gene expression. A, DNA methylation levels of all three sequence contexts in endosperm and embryo. The gray boxes indicate the regions that were experimentally confirmed. B, Validation of DNA methylation levels by bisulfite PCR sequencing. C, Relative expression levels of two genes (28629.m000565 and 30093.m000370) overlapping the hypomethylation regions by quantitative PCR (qPCR) analysis.
Association Analyses of Small RNA Expression and DNA Methylation
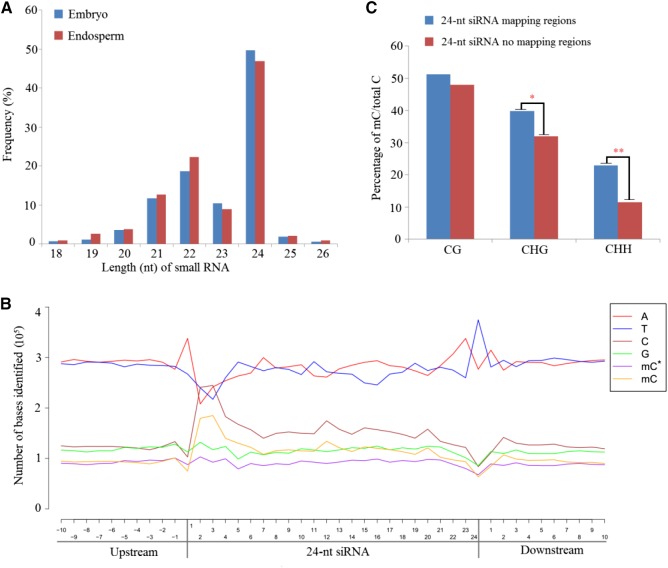
Increasing evidence has indicated that small RNAs can direct de novo DNA methylation at their target loci through RdDM (Law and Jacobsen, 2010; Mosher and Melnyk, 2010). Here, we investigated the small RNA expression profiles in endosperm and embryo tissues by high-throughput deep sequencing. A total of 10,323,788 and 12,382,489 clean reads were obtained from embryo and endosperm libraries, respectively. After filtration of raw data (see “Materials and Methods”), we found that the 24-nucleotide class was the most abundant group of small RNAs in castor bean seeds based on their distribution in length (Fig. 7A), consistent with our previous study in castor bean (Xu et al., 2013).
Figure 7.
Length distribution of small RNAs and relationship between 24-nucleotide siRNA and DNA methylation. A, Length size distribution of small RNAs (from 18 to 26 nucleotides) in castor bean endosperm and embryo. B, Nucleotide frequency and distribution of the 24-nucleotide siRNA mapping region and within 10-nucleotide flanking regions. mC represents the methylcytosine on the sense strand; mC* represents the methylcytosine on the antisense strand. C, Comparison of DNA methylation levels between a 24-nucleotide (nt) siRNA uniquely mapping region and without that region in each sequence context.
Considering that the functions of different small RNAs in mediating DNA methylation were unclear in castor bean seeds, the base composition and strand-specific DNA methylation state of bisulfite-converted genomic DNA sequences were analyzed within the regions homologous to all 21- to 24-nucleotide unique small RNAs sequenced from castor bean seeds. Theoretically, the cytosine distribution pattern of a small RNA sequence could influence the methylated cytosine distribution of the DNA strand identical to the small RNA sequence (termed the sense strand) and guanine in small RNAs could influence the methylated cytosine distribution of the DNA strand complementary to the small RNA sequence (termed the antisense strand). We investigated all uniquely mapping small RNA loci and counted the number of methylated cytosines on either DNA strand of the nuclear genome. As a result, we found that the changes of methylated cytosines on the sense and antisense strands were strongly associated with the altered cytosine and guanine residues in 24-nucleotide siRNAs (Fig. 7B), whereas this tendency was not obvious in mapping regions of 21- to 23-nucleotide- small RNAs (Supplemental Fig. S13, A–C), suggesting that DNA methylation might be closely linked to the expression of 24-nucleotide siRNAs in castor bean seed. Therefore, we focused on the 24-nucleotide siRNA mapping regions for subsequent analyses.
We next sought to investigate the average methylation levels between 24-nucleotide siRNA mapping regions and no mapping regions at each methylation sequence context. Interestingly, the methylation levels of both CHG and CHH contexts were significantly higher in siRNA-covered regions than the regions without siRNA mapping (Fisher’s exact test, P < 0.05), whereas the CG methylation level of siRNA regions was not increased substantially compared with the regions without siRNA (Fig. 7C). Potentially, the increase in CHH methylation level in 24-nucleotide siRNA mapping regions might be caused by activation of the RdDM pathway. To test this, we identified five genes, RcPoI IV (30190.m011113), RcPoI V (30131.m007197), RcRDR2 (30147.m013981), RcDCL3 (29739.m003642), and RcAGO4 (29684.m000322), involved in the RdDM pathway from castor bean based on genome data and examined their expression levels in different tissues, including leaf, embryo, and endosperm. Compared with leaf tissue, these five genes were up-regulated in endosperm and embryo tissues. Further comparing their expression levels between endosperm and embryo, we found that these five genes were markedly down-regulated in endosperm relative to embryo (Supplemental Fig. S13D). These findings suggest that genes involved in the RdDM pathway participate in altering the non-CG methylation in castor bean endosperm and embryo.
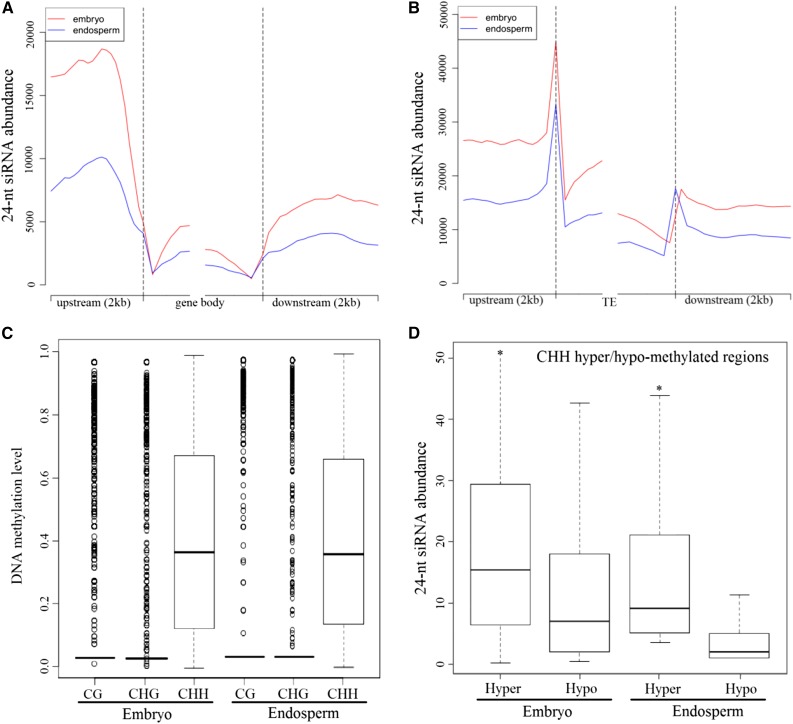
In addition, we found that the 24-nucleotide siRNA abundance of the gene body was smaller than that in the upstream and downstream flanking sequence regions. More siRNAs were identified in the upstream sequence rather than in the downstream sequence (Fig. 8A). In particular, the abundance of siRNAs in the flanking regions in endosperm was substantially lower than that in embryo, consistent with the methylation pattern of non-CG contexts in the gene region (Fig. 4). Although the distribution pattern of siRNAs in the TE and its flanking regions was similar to that in gene regions, the abundance of siRNAs in TE regions was significantly higher than that in gene regions, consistent with the denser and heavier methylation level occurring in TE regions (Fig. 8B). Interestingly, a large number of TEs with siRNA coverage (85.2%) were methylated at a moderate level in the CHH context, whereas only 7.8% and 15% of TEs were methylated in CG and CHG contexts, respectively (Fig. 8C). Also, we found significant evidence of the enrichment of these TEs around the genes within the 4-kb regions (Fisher’s exact test, P < 0.01). In particular, we found that there were more 24-nucleotide siRNAs enriched in CHH hypermethylated regions but fewer siRNAs in the CHH hypomethylated regions in endosperm and embryo tissues (Wilcoxon rank-sum test, P < 0.01; Fig. 8D). These results clearly indicated that 24-nucleotide siRNAs could significantly induce the increase of DNA methylation level in non-CG sequence contexts, especially for CHH methylation, which might be guided by the canonical RdDM pathway. Also, we noted that the number of CHH methylations guided by the RdDM pathway made up about 27% of the total CHH methylation loci in castor bean endosperm.
Figure 8.
Abundance of 24-nucleotide siRNAs in castor bean embryo and endosperm tissues. A, Number distribution of siRNAs in the gene body from the 5′ end (left) to the 3′ end (right) and flanking 2-kb regions. B, Number distribution of siRNAs in the TE and flanking 2-kb regions. C, Methylation levels of all three sequence contexts in TEs within siRNA coverage. D, Abundance of 24-nucleotide siRNAs located to the CHH hypermethylated and hypomethylated regions.
DISCUSSION
In this study, we globally identified the DNA methylation-related genes, comprehensively investigated DNA methylation levels and patterns in castor bean endosperm and embryo integrated with the expression profiles of small RNA and mRNA, and found a new landscape of genomic DNA methylation from a dicotyledonous seed with persistent endosperm throughout seed development. As reported above, our observations showed that key methyltransferases and demethylases were independently evolved in monocotyledons and dicotyledons, although these methylation-related genes were conserved among species. However, this study still leaves open the question of whether global methylation or demethylation has a common evolutionary origin among angiosperms, although the overall process of extensive DNA methylation or demethylation is likely conserved between monocots and dicots.
Genome-wide bisulfite sequencing reveals that rich CHH methylation occurred in castor bean endosperm and embryo tissues, which appears to be distinct from previous reports from other plants. The higher proportion of CHH methylation arose in both embryo and endosperm of castor bean, consistent, in a way, with the high proportion of CHH methylation identified from the leaves of cassava (about 58%; Wang et al., 2015). In spite of the global CG and CHG DNA hypomethylation that arose in endosperm relative to embryo, CHH methylation did not exhibit a significant reduction in endosperm. Comparing the genomic DNA methylation in seeds of angiosperms such as Arabidopsis (Hsieh et al., 2009), rice (Zemach et al., 2010a), maize (Lu et al., 2015), and soybean (Song et al., 2013), our observation of the genomic DNA methylation of castor bean endosperm and embryo exhibits a novel pattern, implying that a distinct mechanism maintaining DNA methylation occurs in dicot castor bean seeds with persistent endosperm. However, we do not rule out whether the nonreduction of CHH methylation that arose in endosperm relative to embryo in castor bean resulted from the different stages of seed development in this study, compared with Arabidopsis (in which the endosperm exists only at the early stage of seed development).
Generally, the low methylation level in the upstream genes could promote gene expression and the presence of DNA methylation inhibits transcriptional initiation. In this study, we noted that the relationship between DNA methylation and global gene expression is not strong in castor bean seed tissues, consistent with previous reports in Arabidopsis (Hsieh et al., 2009) and rice (He et al., 2010). Our investigations appeared to demonstrate that DNA methylation, in a way, mainly repressed gene expression in castor bean seeds, although the associations were variable, depending on the methylated regions (such as upstream, downstream, and gene body) and different methylation sequence contexts. For the genes with preferential expression in endosperm, however, there is a significant decrease of CG and CHG methylation level in the promoter region of genes relative to embryo tissues. These genes affected by hypomethylation in endosperm are involved in some specific development processes.
It is well known that the final levels and patterns of DNA methylation are determined by the activities of both DNA methyltransferase and demethylases. While inspecting the expression profiles of these DNA methylation-related genes, we found a distinct expression pattern of the RcDME and RcDRM genes in castor bean seeds. In Arabidopsis, the DME gene was expressed specifically in the central cell of the female gametophyte (Choi et al., 2002), which may play a critical role in maintaining genomic DNA methylation extent and result in global genomic DNA hypomethylation in the developing endosperm (Hsieh et al., 2009). Our results demonstrated that, as shown in Figure 1, RcDME was broadly expressed in various tissues of castor bean, strongly suggesting that the mechanism of maintaining genomic DNA methylation might be different in castor bean and Arabidopsis. Additionally, the expression of RcMET1 and RcCMT, encoding CG and CHG methyltransferase, was substantially lower in the ovule or the developing endosperm relative to the embryo, implying that the activities of CG and CHG methyltransferase might be insufficient in endosperm, which could result in a CG and CHG hypomethylation in some regions. As shown in Supplemental Figure S7, however, the expression level of RcDRM3 was higher than that of RcMET1 and RcCMT in endosperm and was not repressed (unlike in Arabidopsis, where this gene was repressed in endosperm; Jullien et al., 2012), suggesting the potential roles of RcDRM3 in castor bean seed development with persistent endosperm. Indeed, there is evidence that the loss of function of DRM could result in abnormal reproductive growth and seed morphology (Garcia-Aguilar et al., 2010; Moritoh et al., 2012).
In the RdDM pathway, the 24-nucleotide siRNAs can direct DRM2 to methylate the CHH sequence context (Law and Jacobsen, 2010; Mosher and Melnyk, 2010). This study revealed that a large number of 24-nucleotide siRNAs were transcribed in endosperm and embryo and were mapped with TE regions where CHH methylation arose more densely and heavily. In addition, the expression levels of genes (RcPoI IV, RcPoI V, RcRDR2, RcDCL3, RcAGO4, and RcDRM3) involved in activating the RdDM pathway were significantly up-regulated in endosperm and embryo tissues than those in leaf tissue, clearly suggesting that the RdDM pathway might be activated and play a critical role in directing CHH methylation in endosperm and embryo. Although the biological interests of the rich CHH methylation in castor bean endosperm remain unclear, it is possible that when CG and CHG demethylation occurs extensively in endosperm, the maintenance of CHH methylation would help to repress the movement of many TEs, safeguarding global gene expression and persistent endosperm development. However, it should be noted that CHH methylation directed by the RdDM pathway made up only about 27% of total CHH methylation loci in castor bean endosperm. This implies that the rich CHH methylation in endosperm might not only be established and maintained by the RdDM pathway but that other uncertain factors or pathways probably are involved in mediating CHH methylation in castor bean seeds.
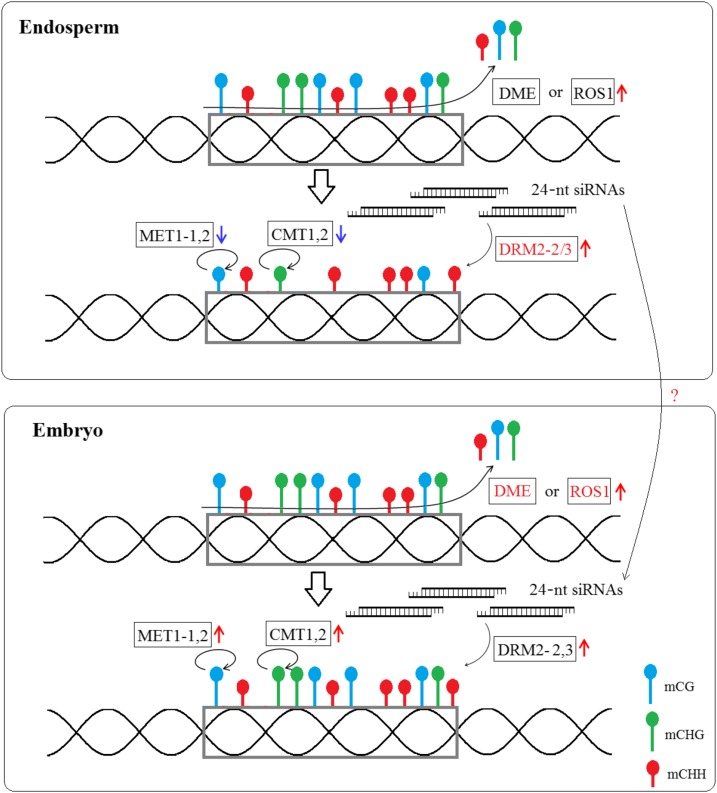
Taken together, as shown in Figure 9, we propose a potential scenario to explain how DNA methylation was maintained in castor bean seed. The repression of RcMET1 and RcCMT expression and the activity of RcDME in endosperm result in genomic hypomethylation of CG and CHG at some discrete loci, which may be a conserved mechanism for the establishment of endosperm hypomethylation among flowering plants, at least in Arabidopsis, rice, and castor bean. In contrast with RcMET1 and RcCMT, the expression of RcDRM3 and the abundant 24-nucleotide siRNAs in endosperm might result in an effective maintenance of CHH methylation within siRNA mapping regions during endosperm development in castor bean. Similarly, the relatively higher expression of RcMET1, RcCMT, and RcDRM genes maintains the genomic CG, CHG, and CHH methylation extents in embryo development. Although this scenario provides just a preliminary hypothesis to explain the potential mechanism of genomic DNA methylation in castor bean seed, this study adds new insights into our understanding of the molecular mechanism of genomic DNA methylation in embryo and endosperm of angiosperms.
Figure 9.
Proposed model to explain the DNA methylation pattern in castor bean endosperm and embryo. RcDME or RcROS1 removes the methylated cytosine at the genome scale (blue lollipops for CG, green lollipops for CHG, and red lollipops for CHH methylation), and demethylation leads to the generation of 24-nucleotide siRNAs that could guide de novo CHH DNA methylation by RcDRM2 in the endosperm and embryo. The insufficient expression of RcMET1 and RcCMT in the endosperm tissue, which maintain the CG and CHG methylation levels (denoted by looping arrows), results in reduced CG and CHG methylation (denoted by short-handled lollipops) compared with embryo (denoted by long-handled lollipops).
CONCLUSION
We globally characterized the DNA methylation-related genes encoding DNA methyltransferases and demethylases and comprehensively dissected the association of genomic DNA methylation and global gene expression in castor bean seeds. Our findings revealed that the CHH methylation extent in endosperm and embryo was, unexpectedly, substantially higher than in previously studied plants, irrespective of the CHH percentage in their genomes. In particular, we found that the global hypomethylation of CG and CHG in endosperm markedly switched the global gene expression and was closely associated with endosperm development. Curiously, within the siRNA coverage regions, the CHH methylation occurring in endosperm associated tightly with the expression of 24-nucleotide siRNAs. The rich CHH methylation occurring in castor bean seeds and the nonreduction of CHH methylation in endosperm relative to embryo result, at least partially, from the RdDM pathway, where the 24-nucleotide siRNAs play a critical role in repressing the activation of TEs around the genes by guiding CHH methylation in TE regions. These results reveal a novel genomic DNA methylation pattern in dicot seeds with persistent endosperms.
MATERIALS AND METHODS
Identification of DNA Methylation-Related Genes
An extensive search was carried out to detect all DNA methylation-related genes encoding DNA methyltransferases and demethylases based on the castor bean (Ricinus communis) genome database (downloaded from http://castorbean.jcvi.org/index.php). The sequences of Arabidopsis (Arabidopsis thaliana) DNA methylation-related genes were obtained from https://www.arabidopsis.org. The amino acid sequences obtained from Arabidopsis were designed as queries against the castor bean protein database by running the NCBI-blast-2.2.24+ software. Subsequently, all hit amino acid sequences were extracted using a custom Perl script based on their identifier numbers, and then these sequences were confirmed by analyzing the characterized domains in the candidate sequences by SMART tools (http://smart.embl-heidelberg.de/) and InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/). Multiple alignments of amino acid sequences were carried out using ClustalW (Larkin et al., 2007), and conserved motifs and domains of each group were identified. Additionally, we constructed an unrooted phylogenetic tree using the neighbor-joining criteria in MEGA 5.0 (Tamura et al., 2011) with 10,000 bootstrap replicates.
Expression Profile Analysis of DNA Methylation-Related Genes by qPCR
In order to investigate the expression profiles of DNA methylation-related genes, we collected diverse tissues, including tender leaf, stem apex, pollen, ovule, early embryo and endosperm (25 d after pollination [DAP]), and late embryo and endosperm (40 DAP), from the inbred line ZB107 (kindly provided by the Zibo Academy of Agricultural Sciences). Total mRNA was isolated from the various tissues using the RNAprep Pure Plant Kit (Tiangen). mRNA with DNA enzyme digestion was used for first-strand complementary DNA synthesis using the PrimeScript RT Reagent Kit (TaKaRa), and then these complementary DNAs were subjected to real-time qPCR using SYBR Green Master Mix (TaKaRa). The qPCR for each tissue was performed with five independent biological replications in the Bio-Rad CFX Manager system as follows: precycling steps at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 58°C for 20 s, and 72°C for 30 s. The RcACTIN2 gene was used as an internal reference to normalize the relative amount of mRNAs for all samples. Also, the expression of genes involved into the RdDM pathway was examined by qPCR, as described above. All primers used in this study are listed in Supplemental Table S5.
Bisulfite Sequencing
Based on previous morphological descriptions (Greenwood and Bewley, 1982), developing seeds at the early stage of cotyledon formation (35 DAP; inbred line ZB107) were collected and dissected. Bisulfite sequencing from five replicates of endosperm and embryo tissues was performed as described in our previous study (Xu et al., 2014). First, high-quality genomic DNA (about 5 μg) was isolated using the Plant Genomic DNA Kit (Tiangen). Second, genomic DNA was fragmented between 300 and 500 bp and end repaired. Third, custom Illumina adapters were ligated following the manufacturer’s protocols, and then fragments with adapters were bisulfite converted using EpiTech Bisulfite Kits (Qiagen). Finally, the converted DNA fragments were PCR amplified with Pfu DNA polymerase (TaKaRa) and sequenced at BGI, generating paired-end 90-bp reads from each library.
Small RNA Library Construction and Sequencing Data
Small RNA sequencing with three replicates was performed as described in our previous study (Xu et al., 2013) using 35-DAP endosperm and embryo tissues. Briefly, after total RNA isolation and PAGE, the small RNAs (16–30 nucleotides) were isolated and purified using the sRNA Gel Extraction Kit (TaKaRa). Then, adapters were ligated to the 5′ and 3′ termini of small RNAs, and reserve transcription was performed followed by low-cycle PCR. Finally, small RNA libraries were constructed and sequenced using an Illumina Genome Analyzer.
After sequencing, we clipped the adapter sequences and filtered out the low-quality tags. The reads with identical sequences, including ribosomal RNA, tRNA, small nuclear RNA, and small nucleolar RNA, were removed from the raw data. The clean small RNA sequences that ranged from 16 to 30 nucleotides were collected and mapped to the castor bean genome using the SOAP 2.0 program (Li et al., 2009). The unique sequences were subjected to subsequent analysis.
Analysis of DMRs
The clean data we obtained were mapped to the castor bean Hale reference genome using BSMAP software (Xi and Li, 2009), allowing up to two mismatches. Then we used custom Perl scripts to call methylated cytosine and calculate its methylation level based on cytosine percentage in a given position. The methylation profile at flanking 2-kb regions and gene body (or TE) is plotted based on the average methylation level for each 100-bp interval. Additionally, we identified DMRs for each sequence context (CG, CHG, and CHH) between endosperm and embryo tissue using the tDMR package (Rakyan et al., 2008) with the following stringent criteria: (1) at least five methylated cytosine sites; (2) more than 10 reads coverage on average; (3) the distance between adjacent methylated sites is less than 200 bp; (4) greater than 20% methylation level in at least one sample; and (5) Fisher’s exact test P < 0.05 and false discovery rate < 0.05. The putative DMRs overlapping at adjacent 2-kb (upstream or downstream) or body regions of genes or TEs were sorted for further study.
Effect of DNA Methylation on Gene Expression
We used publicly available global gene expression data (downloaded from our previous study under accession no. SRX485027) for endosperm and embryo at the same stage. The gene’s expression levels were normalized, and differentially expressed genes also were identified between endosperm and embryo using the TopHat software package (Trapnell et al., 2012). Expression scores for genes with DMRs in the endosperm within 2 kb of either the 5′ or 3′ end were used in the next analysis.
Validation of the DMRs and DMR-Related Gene Expression
To experimentally confirm whether the differential DNA methylation alters gene expression, we picked two hypomethylated DMRs identified in endosperm and tested the expression of adjacent genes in endosperm and embryo tissues. About 500 ng of genomic DNA was extracted from 35-DAP endosperm and embryo and was subjected to bisulfite treatment using the EZ DNA Methylation-Lightning Kit (ZYMO Research) according to the manufacturer’s instructions. The bisulfite-converted DNA was PCR amplified with AceTaq DNA Polymerase (Vazyme). For the DMR identified at the upstream region of gene 28629.m000565, primers 5′-TGGTTGGTTTTGGTGYAATTAAATAYTGGTGG-3′ and 5′-TTACATTATTTRACATATRTCAAACTTTCATC-3′ were used; for the DMR identified within the first exon of gene 30093.m000370, primers 5′-TAGGGTTTATTTTGAAAAATGAAATGTAGT-3′ and 5′-AAATACACACACACTCTCTCTCTCATA-3′ were used (primers were designed by Kismeth software [http://katahdin.mssm.edu/kismeth]; Gruntman et al., 2008). The purified PCR products were cloned into the pGEM-T vector, and 15 clones were randomly selected for sequencing. The final sequencing results were summarized and visualized with MATLAB software.
In addition, the relative expression levels of these two genes tested in endosperm and embryo tissue were carried out by qPCR as described above. For the genes 28629.m000565 and 30093.m000370, primer pairs 5′-AGCCCAAAGACAATCCGGTA-3′/5′-ATACCAGTTTCCGCTTTCGC-3′ and 5′-ATCGTCACAGCAATGGATCG-3′/5′-CCAGAGCCAACAACCAAGAC-3′ were used for qPCR amplification.
The raw data used for this study have been submitted to the National Center for Biotechnology Information Sequence Read Archive under accession number SRX1267331.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of amino acid sequences of DNA methyltransferases MET1 between Arabidopsis thaliana and castor bean.
Supplemental Figure S2. Comparison of amino acid sequences of DNA methyltransferases DRM between Arabidopsis thaliana and castor bean.
Supplemental Figure S3. Comparison of amino acid sequences of DNA methyltransferases CMT between Arabidopsis thaliana and castor bean.
Supplemental Figure S4. Comparison of amino acid sequences of DNA methyltransferases DNMT2 between Arabidopsis thaliana and castor bean.
Supplemental Figure S5. Comparison of amino acid sequences of the domains of DNA demethylases between Arabidopsis thaliana and castor bean.
Supplemental Figure S6. Schematic structures of DNA methyltransferases and demethylases (A and B) and phylogenetic analysis (C and D).
Supplemental Figure S7. Heatmaps representing the expression profiles of methyltransferase and demethylase genes in RNA-seq data in castor bean endosperm and embryo tissues.
Supplemental Figure S8. Sequencing depth and saturation analysis of the embryo and endosperm tissues in castor bean.
Supplemental Figure S9. DNA methylation and TEs.
Supplemental Figure S10. The percentage of DMRs distribution in TE, gene and 2kb flanking gene regions.
Supplemental Figure S11. Effect of DNA methylation on global gene expression in castor bean embryo tissues.
Supplemental Figure S12. GO enrichment analysis of genes affected by endosperm DMRs.
Supplemental Figure S13. Relationship between small RNAs and methylated cytosines, and genes expression involved in RdDM pathway.
Supplemental Table S1. Information on bisulfite sequencing data.
Supplemental Table S2. The percentages of CG, CHG and CHH in genomes of Arabidopsis (At), rice (Os), maize (Zm) and castor bean (Rc).
Supplemental Table S3. Substantial differentially methylated regions in CG, CHG and CHH sequence contexts between embryo and endosperm.
Supplemental Table S4. Endosperm-preferred genes and the differentially methylated regions in each sequence context.
Supplemental Table S5. Primers used in this study for gene expression profiles based on qPCR.
Supplementary Material
Acknowledgments
We thank the Guangzhou Gene Denovo Biotechnology Co., Ltd., for assisting in bisulfite sequencing analysis. This study was facilitated by the Germplasm Bank of Wild Species at the Kunming Institute of Botany.
Glossary
- siRNA
small interfering RNA
- RdDM
RNA-directed DNA methylation
- TE
transposable element
- DMR
differential methylation region
- RPKM
reads per kilobase of transcript per million reads mapped
- qPCR
quantitative PCR
- DAP
days after pollination
References
- Cao X, Jacobsen SE (2002a) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA (Suppl 4) 99: 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002b) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1138–1144 [DOI] [PubMed] [Google Scholar]
- Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G, et al. (2010) Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol 28: 951–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42 [DOI] [PubMed] [Google Scholar]
- Feinberg AP. (2007) Phenotypic plasticity and the epigenetics of human disease. Nature 447: 433–440 [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES (1993) Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res 21: 2383–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D (2010) Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22: 3249–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genger RK, Kovac KA, Dennis ES, Peacock WJ, Finnegan EJ (1999) Multiple DNA methyltransferase genes in Arabidopsis thaliana. Plant Mol Biol 41: 269–278 [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Ying H, Taylor JM, Peacock WJ, Dennis ES (2012) Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA 109: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood JS, Bewley JD (1982) Seed development in Ricinus communis castor bean. I. Descriptive morphology. Can J Bot 60: 1751–1760 [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE (2007) Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Hermann A, Schmitt S, Jeltsch A (2003) The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J Biol Chem 278: 31717–31721 [DOI] [PubMed] [Google Scholar]
- Houston NL, Hajduch M, Thelen JJ (2009) Quantitative proteomics of seed filling in castor: comparison with soybean and rapeseed reveals differences between photosynthetic and nonphotosynthetic seed metabolism. Plant Physiol 151: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li N, Xu C, Zhong S, Lin X, Yang J, Zhou T, Yuliang A, Wu Y, Chen YR, et al. (2014) Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc Natl Acad Sci USA 111: 10642–10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F (2006) Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22: 1825–1830 [DOI] [PubMed] [Google Scholar]
- Jurkowski TP, Jeltsch A (2011) On the evolutionary origin of eukaryotic DNA methyltransferases and Dnmt2. PLoS ONE 6: e28104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W (2003) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Lu X, Wang W, Ren W, Chai Z, Guo W, Chen R, Wang L, Zhao J, Lang Z, Fan Y, et al. (2015) Genome-wide epigenetic regulation of gene transcription in maize seeds. PLoS ONE 10: e0139582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok YG, Uzawa R, Lee J, Weiner GM, Eichman BF, Fischer RL, Huh JH (2010) Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc Natl Acad Sci USA 107: 19225–19230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritoh S, Eun CH, Ono A, Asao H, Okano Y, Yamaguchi K, Shimatani Z, Koizumi A, Terada R (2012) Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J 71: 85–98 [DOI] [PubMed] [Google Scholar]
- Mosher RA, Melnyk CW (2010) siRNAs and DNA methylation: seedy epigenetics. Trends Plant Sci 15: 204–210 [DOI] [PubMed] [Google Scholar]
- Mund C, Musch T, Strödicke M, Assmann B, Li E, Lyko F (2004) Comparative analysis of DNA methylation patterns in transgenic Drosophila overexpressing mouse DNA methyltransferases. Biochem J 378: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FCS, Palmisano G, Schwämmle V, Campos FAP, Larsen MR, Domont GB, Roepstorff P (2012) Performance of isobaric and isotopic labeling in quantitative plant proteomics. J Proteome Res 11: 3046–3052 [DOI] [PubMed] [Google Scholar]
- Ogunniyi DS. (2006) Castor oil: a vital industrial raw material. Bioresour Technol 97: 1086–1091 [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL (2007) DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA 104: 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponger L, Li WH (2005) Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol Biol Evol 22: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Qian Y, Xi Y, Cheng B, Zhu S (2014) Genome-wide identification and expression profiling of DNA methyltransferase gene family in maize. Plant Cell Rep 33: 1661–1672 [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S, Tomazou EM, Bäckdahl L, Johnson N, Herberth M, et al. (2008) An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res 18: 1518–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, Slusarz L, Mosiolek M, Park JS, Park GT, et al. (2011) Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci USA 108: 8042–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Lu X, Li QT, Chen H, Hu XY, Ma B, Zhang WK, Chen SY, Zhang JS (2013) Genome-wide analysis of DNA methylation in soybean. Mol Plant 6: 1961–1974 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Wobus U (2013) Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol 64: 189–217 [DOI] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Wang H, Beyene G, Zhai J, Feng S, Fahlgren N, Taylor NJ, Bart R, Carrington JC, Jacobsen SE, Ausin I (2015) CG gene body DNA methylation changes and evolution of duplicated genes in cassava. Proc Natl Acad Sci USA 112: 13729–13734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Li W (2009) BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL (2006a) Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol 142: 1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL (2006b) DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing MQ, Zhang YJ, Zhou SR, Hu WY, Wu XT, Ye YJ, Wu XX, Xiao YP, Li X, Xue HW (2015) Global analysis reveals the crucial roles of DNA methylation during rice seed development. Plant Physiol 168: 1417–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Cui Q, Li F, Liu A (2013) Transcriptome-wide identification and characterization of microRNAs from castor bean (Ricinus communis L.). PLoS ONE 8: e69995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dai M, Li F, Liu A (2014) Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res 42: 6987–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D (2010a) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D (2010b) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zhang M, Xie S, Dong X, Zhao X, Zeng B, Chen J, Li H, Yang W, Zhao H, Wang G, et al. (2014) Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res 24: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhao H, Xie S, Chen J, Xu Y, Wang K, Zhao H, Guan H, Hu X, Jiao Y, et al. (2011) Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci USA 108: 20042–20047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2009) Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 43: 143–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.