OsDET1 modulates ABA signaling pathway and ABA biosynthesis, leading to contradictory phenotypes related to ABA in OsDET1 deficiency transgenic plants.
Abstract
DEETIOLATED1 (DET1) plays a critical role in developmental and environmental responses in many plants. To date, the functions of OsDET1 in rice (Oryza sativa) have been largely unknown. OsDET1 is an ortholog of Arabidopsis (Arabidopsis thaliana) DET1. Here, we found that OsDET1 is essential for maintaining normal rice development. The repression of OsDET1 had detrimental effects on plant development, and leaded to contradictory phenotypes related to abscisic acid (ABA) in OsDET1 interference (RNAi) plants. We found that OsDET1 is involved in modulating ABA signaling in rice. OsDET1 RNAi plants exhibited an ABA hypersensitivity phenotype. Using yeast two-hybrid (Y2H) and bimolecular fluorescence complementation assays, we determined that OsDET1 interacts physically with DAMAGED-SPECIFIC DNA-BINDING PROTEIN1 (OsDDB1) and CONSTITUTIVE PHOTOMORPHOGENIC10 (COP10); DET1- and DDB1-ASSOCIATED1 binds to the ABA receptors OsPYL5 and OsDDB1. We found that the degradation of OsPYL5 was delayed in OsDET1 RNAi plants. These findings suggest that OsDET1 deficiency disturbs the COP10-DET1-DDB1 complex, which is responsible for ABA receptor (OsPYL) degradation, eventually leading to ABA sensitivity in rice. Additionally, OsDET1 also modulated ABA biosynthesis, as ABA biosynthesis was inhibited in OsDET1 RNAi plants and promoted in OsDET1-overexpressing transgenic plants. In conclusion, our data suggest that OsDET1 plays an important role in maintaining normal development in rice and mediates the cross talk between ABA biosynthesis and ABA signaling pathways in rice.
Leaf senescence is the final stage of leaf development. The most obvious phenotypic change during senescence is leaf yellowing caused by chlorophyll degradation (Bleecker and Patterson, 1997; Lim et al., 2007). Leaf structure, cellular metabolism, and gene expression also undergo dramatic changes during this process. Valuable nutrient components in the cell are remobilized to young tissues, while senescent leaves die slowly (Hörtensteiner and Feller, 2002; Buchanan-Wollaston et al., 2005; Lim et al., 2007). Leaf senescence is controlled by a complex regulatory network that is governed by developmental age. Leaf senescence also can be affected by other endogenous factors (such as phytohormones, the levels of some metabolites, and the state of photosystem complexes) and external factors (biotic and abiotic stresses). Among these, darkness is one of the most effective known external stimuli that cause leaf senescence, and it is frequently used to simulate synchronous senescence (Kim et al., 2006; Kong et al., 2006; Liang et al., 2014). Abscisic acid (ABA) plays a central role in regulating leaf senescence governed by developmental age (Breeze et al., 2011; Lee et al., 2011). The ABA content increases during leaf senescence in many plants, such as oat (Avena sativa; Gepstein and Thimann, 1980), rice (Oryza sativa; Philosoph-Hadas et al., 1993), and Arabidopsis (Arabidopsis thaliana; Zhao et al., 2010; Breeze et al., 2011). Simultaneously, ABA biosynthesis and signaling genes are up-regulated during age-dependent leaf senescence (Tan et al., 2003; Buchanan-Wollaston et al., 2005). Moreover, numerous biotic and abiotic stresses usually induce ABA biosynthesis and increase ABA levels, activate ABA signaling pathways, and lead to leaf senescence. Exogenously applied ABA increases the expression of chlorophyll degradation-related genes (CDGs) and senescence-associated genes (SAGs) and promotes leaf senescence (Liang et al., 2014). Furthermore, ABA plays multiple roles in plant developmental processes in addition to leaf senescence, such as seed formation, dormancy, and germination (Yoshida et al., 2006; Chinnusamy et al., 2008; Zhao et al., 2010; Dong et al., 2014).
ABA signaling is mediated by the pyrabactin resistance/pyrabactin resistance-like/regulatory components of the ABA receptor (PYR/PYL/RCAR; Cutler et al., 2010; Hubbard et al., 2010). In the presence of ABA, PYR/RCARs bind with ABA and clade A phosphatases type 2C (PP2Cs; these include ABA INSENSITIVE1, HYPERSENSITIVE TO ABA1 [HAB1], and HAB2) to form a PP2C-ABA-PYL ternary complex (Park et al., 2009; Cutler et al., 2010; Hubbard et al., 2010), inhibiting the phosphatase activity of PP2Cs, allowing SUCROSE NONFERMENTING1-related subfamily 2 kinase (SnRK2) activation and phosphorylation of the ABA-responsive element binding factor family transcription factors, ultimately regulating the transcriptional response to ABA (Cutler et al., 2010; Hubbard et al., 2010). Simultaneously, the PYL ABA receptors are recognized and ubiquitinated by DET1- AND DDB1-ASSOCIATED1 (DDA1) and the COP10-DET1-DDB1 (CDD) complex to alleviate the adverse effects of continuous ABA responses. CDD components cooperate to regulate PYL stability. DEETIOLATED1 (DET1), part of the CDD complex, also modulates ABA responses (Irigoyen et al., 2014).
DET1 is a well-known repressor of photomorphogenesis (Pepper et al., 1994). This nucleus-localized protein interacts with DAMAGED-SPECIFIC DNA-BINDING PROTEIN1 (DDB1) to regulate plant photomorphogenesis (Schroeder et al., 2002). DET1 and DDB1 interact with CONSTITUTIVE PHOTOMORPHOGENIC10 (COP10) to form the CDD complex in living plant cells (Yanagawa et al., 2004). The CDD complex cooperates with CULLIN4 (CUL4) in response to light and mediates plant development (Chen et al., 2006, 2010). DET1 also was shown recently to interact directly with phytochrome-interacting factors and to modulate their stability, thereby further repressing photomorphogenesis (Dong et al., 2014). A mutation in DET1 damages the normal functioning of the CDD complex in Arabidopsis. Furthermore, adding a C-terminal GFP tag interferes with the formation of the CCD complex by DET1, DDB1, and COP10 and influences the normal functioning of DET1 in Arabidopsis (Schroeder et al., 2002; Irigoyen et al., 2014). In addition to regulating photomorphogenesis, DET1 also plays a key role in many biological processes in plants. DET1 functions in some stress responses, and it joins with CUL4, DDB1, and DDB2 to form a complex to maintain genome integrity in Arabidopsis during UV light stress (Castells et al., 2011). DET1 also influences plant responses to ABA by modulating the functioning of CDD in Arabidopsis. The CDD complex interacts with DDA1 to form the COP10-DET1-DDB1-DDA1 (CDDD) complex, which provides substrate specificity for CUL4-RING E3 ubiquitin ligase (CRL4) ubiquitination of the PYR/PYL/RCAR family of ABA receptors. Reduced CDD function causes ABA hypersensitivity in Arabidopsis. As part of the CDD substrate adaptor module, DET1 indirectly affects the recognition and ubiquitination of PYLs by DDA1. The Arabidopsis det1-1 mutant exhibits an increased response to the ABA-mediated inhibition of germination and seedling establishment compared with wild-type plants (Irigoyen et al., 2014). However, interestingly, the detached leaves of the Arabidopsis det1-1 mutant exhibit significantly delayed dark-induced leaf senescence compared with the wild type (Chory et al., 1994).
Although DET1 plays an important role in plant development, the roles of OsDET1 in rice remain unclear, except for its role in chlorophyll biosynthesis (Huang et al., 2013). Here, we report that OsDET1 influences the sensitivity of rice plants to ABA. As OsDET1 is an ortholog of Arabidopsis DET1, we found that OsDET1 deficiency caused pleiotropic phenotypes related to ABA hypersensitivity in OsDET1 RNA interference transgenic plants (OsDET1 RNAi plants). We also observed similar ABA hypersensitivity phenotypes in OsDET1-GFP plants. yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays showed that OsDET1 interacts physically with OsDDB1 and OsCOP10 in vivo. OsDDA1 also binds to the ABA receptors OsPYL5 and OsDDB1. Further experiments demonstrated that the degradation of OsPYL5 was delayed in OsDET1 RNAi and OsDET1-GFP plants. These results suggest that deficiency or structural changes to OsDET1 may disturb the functioning of CDD and cause ABA hypersensitivity in rice. Surprisingly, OsDET1 also influenced ABA biosynthesis and inactivation. The level of endogenous ABA was reduced in OsDET1 RNAi plants, while ABA biosynthesis was promoted in OsDET1-overexpressing transgenic plants (OE-OsDET1 plants), suggesting that OsDET1 also modulates ABA biosynthesis in rice. Overall, our results suggest that OsDET1 mediates the cross talk between ABA biosynthesis and ABA signaling pathways in rice.
RESULTS
Stress-Induced and Spatial Expression Profiles of OsDET1 in Rice
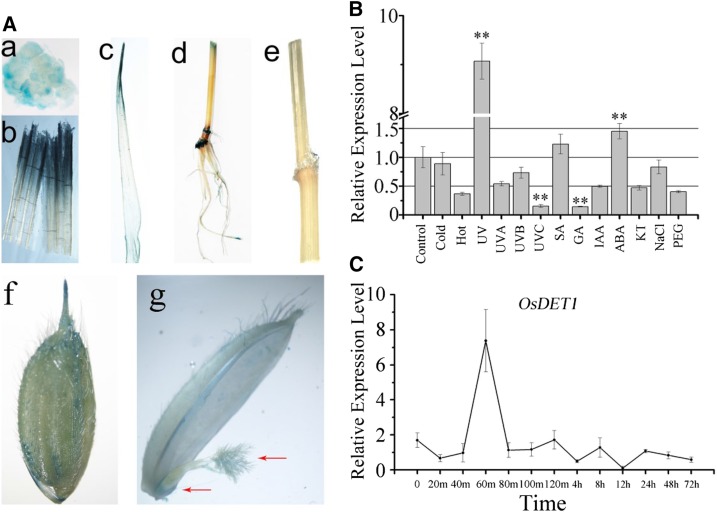
DET1 plays an important role in regulating plant development. To investigate the role of OsDET1 in rice development, we constructed the vector POsDET1::GUS and introduced it into cv Nipponbare rice. Consistent with our previous results (Huang et al., 2013), GUS activity was detected in almost all tissues and organs, such as young callus, roots, stems, nodes, internodes, leaf sheaths, leaves, and panicles (Fig. 1A). GUS activity was stronger in leaves, leaf sheaths, and internodes than in roots and panicles. Surprisingly, GUS staining was observed distinctly in the stigma and ovary, suggesting that OsDET1 affects the fertility of rice spikelets (Fig. 1A). We examined the expression pattern of OsDET1 in plants under abiotic stresses (cold, hot, UV light, UV-C light, polyethylene glycol, and NaCl) and phytohormone treatments (gibberellin, auxin, salicylic acid, 6-furfurylaminopurine, and ABA) by quantitative reverse transcription (qRT)-PCR, finding that OsDET1 was induced by ABA (Fig. 1B). Kinetic expression analysis showed that the expression of OsDET1 was up-regulated after 60 min of ABA treatment, followed by rapid down-regulation (Fig. 1C), suggesting that OsDET1 might be involved in the ABA signaling pathway. OsDET1 expression also was affected by other abiotic stresses and phytohormones; for example, OsDET1 was up-regulated by UV light treatment and down-regulated by UV-C light treatment (Fig. 1B). These results suggest that OsDET1 plays an important role in developmental and environmental responses in rice.
Figure 1.
Analysis of OsDET1 expression. A, Histochemical staining of POsDET1::GUS transgenic plants. Transgenic plants were tested during protoplast culture and the flowering stage. Images are as follows: callus (a), leaf sheaths (b), leaves (c), roots, stems, and internodes (d), branches (e), panicle spikelets (f), and pistils and ovaries (g). B, Effects of phytohormones and abiotic stresses on OsDET1 expression. Two-week-old cv Nipponbare plants were used for the experiment. GA, Gibberellic acid; IAA, indole-3-acetic acid; KT, 6-furfurylaminopurine; PEG, polyethylene glycol; SA, salicylic acid. **, P ≤ 0.01 (Student’s t test). C, Kinetic analysis by qRT-PCR of OsDET1 expression after treatment with 50 μm ABA.
OsDET1 Deficiency Accelerates Dark-Induced Leaf Senescence
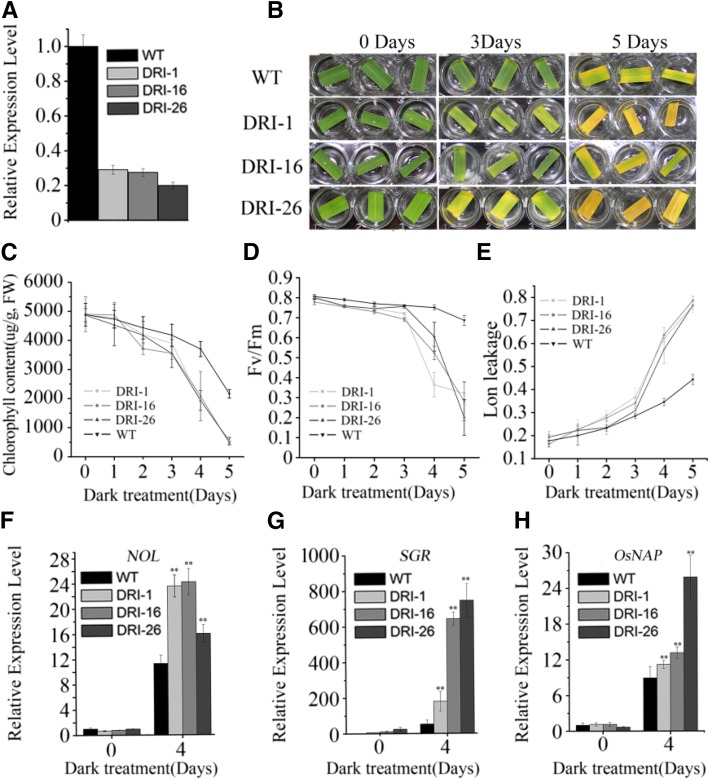
Detached leaves of the det1-1 mutant exhibit delayed dark-induced leaf senescence compared with the wild type in Arabidopsis (Chory et al., 1994). Therefore, we investigated the role of OsDET1 in leaf senescence in rice by generating transgenic OsDET1 RNAi plants. The expression of OsDET1 was reduced markedly in these plants (Fig. 2A). Therefore, we utilized the penultimate leaves of these lines at the tillering stage for further dark-induced leaf senescence experiments. OsDET1 RNAi plants exhibited significantly accelerated leaf yellowing and reduced chlorophyll contents compared with the wild type during dark-induced leaf senescence (Fig. 2, B and C). We also examined the ratio of variable fluorescence to maximum fluorescence (Fv/Fm) and the membrane ion leakage values in these plants. Fv/Fm values reflect PSII activity and membrane ion leakage reflects cell death, and they are frequently used as parameters of leaf senescence (Morita et al., 2009). As shown in Figure 2D, the Fv/Fm values decreased rapidly with treatment time in OsDET1 RNAi plants, indicating that chloroplast damage was accelerated in the leaves of these plants. Similarly, the membrane ion leakage values increased sharply in transgenic plant leaves (Fig. 2E), indicating that the process of programmed cell death was accelerated in OsDET1 RNAi plant leaves. Furthermore, the expression of major key CDGs (including PAO, SGR, NYC1, NYC3, RCCR1, PPH, and NOL) and SAGs (including OsNAP, OsI57, and OsI85) was markedly higher in leaves of OsDET1 RNAi plants than in wild-type leaves during 4 d of dark induction (Fig. 2, F–H; Supplemental Fig. S1, A and B; Morita et al., 2009; Rong et al., 2013; Yamatani et al., 2013; Liang et al., 2014).
Figure 2.
OsDET1 deficiency accelerates dark-induced leaf senescence in OsDET1 RNAi plants. A, qRT-PCR analysis of the expression of OsDET1 in OsDET1 RNAi lines and the wild type (WT). B, Suppression of OsDET1 promoted dark-induced leaf senescence. C, Changes with time of chlorophyll content in OsDET1 RNAi lines and wild-type plants during dark incubation. Values are means ± sd of nine measurements. The penultimate leaves were detached and incubated with water in darkness at the tillering stage. FW, Fresh weight. D, Changes with time of Fv/Fm values in OsDET1 RNAi lines and wild-type plants during dark incubation. Values are means ± sd of nine measurements. E, Changes with time of membrane ion leakage in OsDET1 RNAi lines and wild-type plants during dark incubation. Values are means ± sd of nine measurements. F to H, Expression of NOL, SGR, and OsNAP in the wild type and OsDET1 RNAi lines during dark incubation. **, P ≤ 0.01 (Student’s t test).
Given that OsDET1 deficiency accelerated the expression of CDGs and chlorophyll degradation during dark-induced leaf senescence, we performed western-blot analysis to investigate the stability of photosynthetic proteins in OsDET1 RNAi plants and the wild type during dark-induced leaf senescence. After 4 d of dark treatment, the PSII proteins (Lhcb1, Lhcb4, Lhcb6, and D1) and PSI proteins (Lhca1 and PSa) were quickly degraded in OsDET1 RNAi plants compared with the wild type, whereas Lhcb2 and RbcL were retained in OsDET1 RNAi plants and wild-type plants (Supplemental Fig. S2). These results suggest that OsDET1 deficiency impairs the stability of photosynthetic proteins during dark-induced leaf senescence in rice.
We observed the chloroplast ultrastructure in the plants using transmission electron microscopy. Interestingly, the grana stacks were thicker in OsDET1 RNAi plants than in wild-type plants at the tillering stage (Supplemental Fig. S3, A, B, F, G, K, L, P, and Q), suggesting that OsDET1 also modulates chloroplast development in rice. During dark incubation, the grana stacks and stroma membranes were rapidly degraded in OsDET1 RNAi plants compared with wild-type plants. After 4 d of dark treatment, the grana stack arrays and intergranal lamellae were only slightly disordered in the wild type. By contrast, the volume of the thylakoid membrane was reduced significantly in OsDET1 RNAi plants, and the grana and intergranal lamellae were fused and disordered (Supplemental Fig. S3, C, D, H, I, M, N, R, and S). After 6 d of dark treatment, the chloroplast components were almost decomposed in OsDET1 RNAi plants (Supplemental Fig. S3, J, Q, and T), whereas unbroken chloroplasts with fused grana stacks were observed in the wild type (Supplemental Fig. S3E). These observations suggest that OsDET1 deficiency accelerates the degradation of the thylakoid membrane system during dark-induced leaf senescence. In the dark, the Arabidopsis det1-1 mutant displays impaired chloroplast development and the accumulation of mRNA for several chloroplast genes (Chory et al., 1989). The detached leaves of this mutant exhibit delayed dark-induced leaf senescence compared with the wild type (Chory et al., 1994). However, along with the obvious disruption of chloroplasts, the degradation of photosynthetic proteins was accelerated in OsDET1 RNAi plants compared with the wild type during dark-induced leaf senescence, implying that DET1 performs different roles in rice and Arabidopsis.
Overall, unlike the phenotype of the Arabidopsis det1-1 mutant, our results demonstrate that OsDET1 deficiency accelerates leaf senescence during dark treatment in rice. Furthermore, OsNAP, an important link between ABA signaling and leaf senescence, is induced specifically by ABA, which directly activates the expression of CDGs and other SAGs (Liang et al., 2014). Thus, it appears that ABA signaling is intensified in OsDET1 RNAi plants during dark-induced leaf senescence. However, while the increased dark-induced leaf senescence phenotype is usually closely linked with premature leaf senescence in rice (Morita et al., 2009; Rong et al., 2013; Yamatani et al., 2013; Liang et al., 2014), in this study, OsDET1 deficiency accelerated dark-induced leaf senescence, but no visible difference in leaf senescence was observed in OsDET1 RNAi plants compared with wild-type plants at the vegetative and late filling stages (Fig. 2; Supplemental Fig. S4, A and B).
Inhibited OsDET1 Expression Accelerates ABA-Induced leaf Senescence
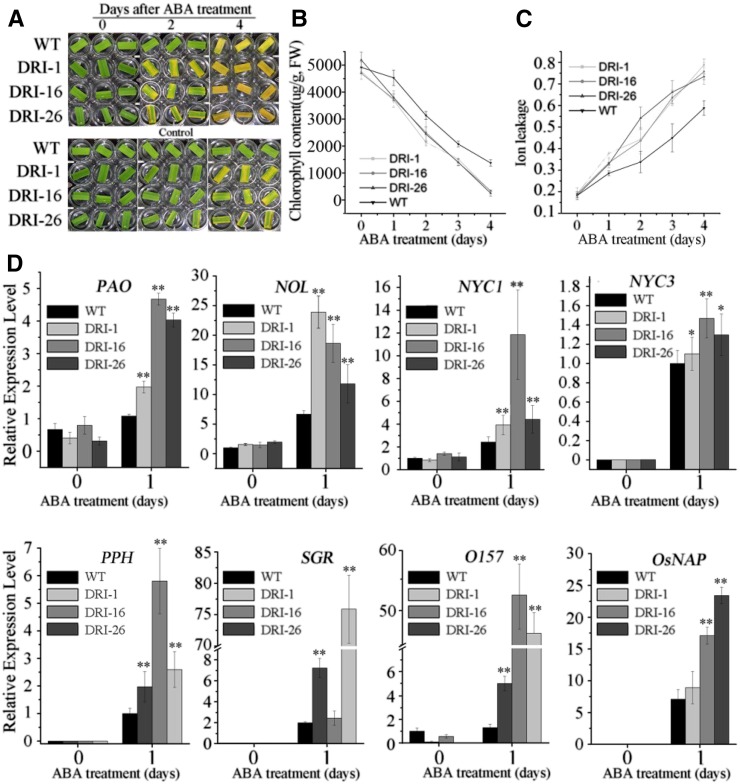
ABA plays a central role in modulating leaf senescence (Lim et al., 2007). To further investigate the intrinsic functions of OsDET1 in leaf senescence, we explored leaf senescence phenotypes in response to ABA treatment. Detached leaves from the tillering stage were treated with ABA in the light. As expected, the detached leaves of OsDET1 RNAi plants exhibited obviously accelerated senescence. The chlorophyll contents decreased rapidly and membrane ion leakage increased quickly in OsDET1 RNAi plants (Fig. 3, A–C). Simultaneously, the expression of CDGs and SAGs was up-regulated significantly in OsDET1 RNAi plants compared with the wild type during treatment (Fig. 3D). Taken together, these results demonstrate that OsDET1 deficiency accelerates ABA-induced leaf senescence, implying that ABA signaling is enhanced in OsDET1 RNAi plants.
Figure 3.
OsDET1 deficiency effects on ABA-induced leaf senescence. A, OsDET1 deficiency accelerates ABA-induced leaf senescence. The penultimate leaves were detached and incubated with water containing 50 μm ABA in light, with no added ABA as a control. B, Changes with time of chlorophyll contents in OsDET1 RNAi lines and wild-type (WT) plants after treatment with 50 μm ABA. Values are means ± sd of nine measurements. FW, Fresh weight. C, Changes with time of membrane ion leakage in OsDET1 RNAi lines and wild-type plants after treatment with 50 μm ABA. Values are means ± sd of nine measurements. D, Expression of CDGs (PAO, SGR, NYC1, NYC3, RCCR, PPH, and NOL) and SAGs (OsNAP and OsI57) in wild-type plants and OsDET1 RNAi plants after treatment with 50 μm ABA. *, P ≤ 0.05 and **, P ≤ 0.01 (Student’s t test).
OsDET1 Influences ABA Biosynthesis in Rice
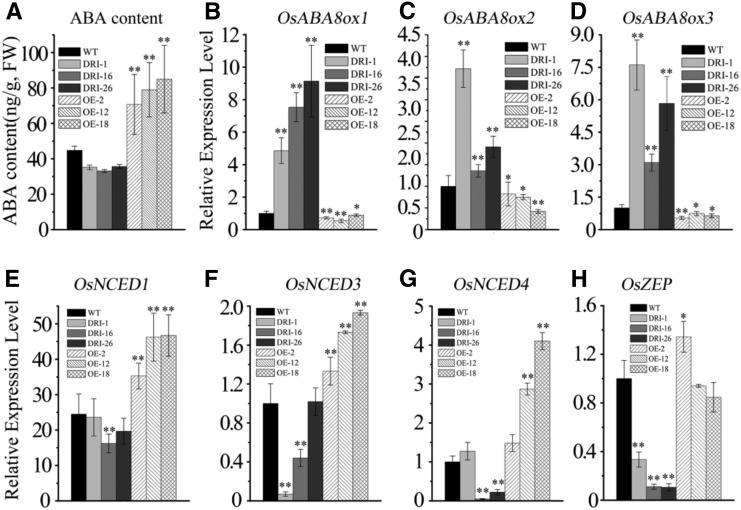
To help explain the discrepancy between the leaf senescence phenotype during dark and ABA stress and the normal growth of OsDET1 RNAi plants, we investigated the ABA contents in OsDET1 RNAi plants and wild-type plants at the vegetative and late filling stages; T2 OE-OsDET1 plants served as a positive control (Supplemental Fig. S5A). Surprisingly, compared with the wild type, the ABA contents were reduced in OsDET1 RNAi plants and increased in OE-OsDET1 plants (Fig. 4A; Supplemental Fig. S6A). We also analyzed the expression of key ABA biosynthesis genes (OsNCED1, OsNCED3, OsNCED4 [for 9-cis-epoxycarotenoid dioxygenase], and OsZEP [for zeaxanthin epoxidase]) and ABA inactivation genes (OsABA8ox1, OsABA8ox2, and OsABA8ox3) in these plants. As expected, the transcript levels of ABA biosynthesis genes declined in OsDET1 RNAi plants compared with the wild type (Fig. 4, E and H). Conversely, OsNCED1, OsNCED3, and OsNCED4 were up-regulated in OE-OsDET1 plants (Fig. 4, E–G). In addition, the ABA inactivation genes OsABA8ox1, OsABA8ox2, and OsABA8ox3 were markedly up-regulated in OsDET1 RNAi plants but down-regulated in OE-OsDET1 plants (Fig. 4, B–D). Taken together, these results suggest that OsDET1 acts as a positive regulator of ABA biosynthesis by modulating the expression of ABA biosynthesis and inactivation genes. Therefore, although OsDET1 deficiency enhances ABA signaling and the ABA contents were reduced in OsDET1 RNAi plants, this new balance maintained the leaf in normal functioning at the vegetative and filling stages in OsDET1 RNAi plants.
Figure 4.
OsDET1 influences rice ABA synthesis and inactivation. A, ABA content in wild-type (WT) and transgenic plants. The penultimate leaves were detected at the tillering stage. Values are means ± sd of six measurements. **, P ≤ 0.01 (Student’s t test). FW, Fresh weight. B to D, Expression of ABA inactivation genes (OsABA8ox1, OsABA8ox2, and OsABA8ox3) in the wild type and transgenic lines. *, P ≤ 0.05 and **, P ≤ 0.01 (Student’s t test). E to H, Expression of ABA biosynthesis genes (OsNCED1, OsNCED3, OsNCED4, and OsZEP) in the wild type and transgenic lines. *, P ≤ 0.05 and **, P ≤ 0.01 (Student’s t test).
Darkness forcefully stimulates ABA biosynthesis (Gepstein and Thimann, 1980) and causes the rapid senescence of leaves in OsDET1 RNAi plants. Senescence also is a powerful factor for promoting ABA biosynthesis. The contents of ABA are increased tremendously with the leaves in senescence in many plants (Zhao et al., 2010; Breeze et al., 2011). By contrast, the repression of ABA biosynthesis by OsDET1 deficiency was gentle in OsDET1 RNAi plants (Fig. 4; Supplemental Fig. S6A). Therefore, in dark treatment, the balance was shifted by darkness. Consistent with the premature leaf senescence, the ABA contents were much higher in OsDET1 RNAi plants than in the wild type after a 4-d dark treatment (Supplemental Fig. S7A). Moreover, qRT-PCR demonstrated that ABA biosynthesis was promoted and ABA inactivation was repressed in OsDET1 RNAi plants compared with the wild type (Supplemental Fig. S7, B–H). These results suggest that the factors of darkness and senescence are more powerful than OsDET1 in regulating ABA biosynthesis during dark-induced leaf senescence and clearly indicate that the enhanced ABA signaling is the main reason for the rapid leaf senescence of OsDET1 RNAi plants in dark incubation.
Since ABA promotes leaf senescence in plants, activating endogenous ABA biosynthesis causes ABA hypersensitivity in many plants (Suzuki et al., 2002; Oliver et al., 2007; Zhu et al., 2009). To further assess the function of OsDET1 in ABA biosynthesis, we examined the leaf senescence phenotype of OE-OsDET1 plants during dark incubation. As expected, OE-OsDET1 plants also exhibited an accelerated leaf senescence phenotype (Supplemental Fig. S5). These results suggest that the positive regulation of ABA biosynthesis by OsDET1 also causes ABA hypersensitivity in OE-OsDET1 plants during dark treatment.
OsDET1 Influences Seed Germination and Seedling Growth
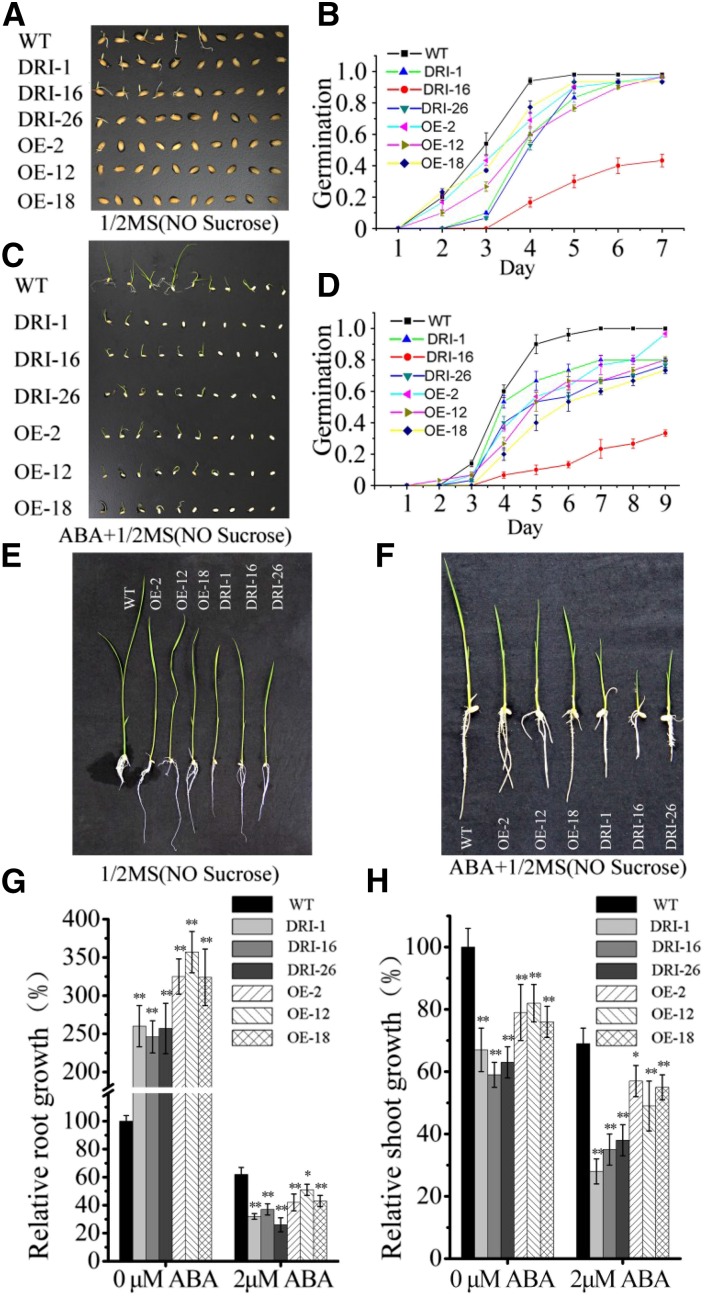
To further explore the role of OsDET1 in linking ABA signaling and ABA biosynthesis, we performed germination and postgermination assays to investigate the ABA-related phenotypes of the transgenic lines. Compared with the wild type, OsDET1 RNAi plants showed delayed germination on one-half-strength Murashige and Skoog (MS) medium (Fig. 5, A and B). Moreover, ABA treatment significantly delayed the germination of OsDET1 RNAi seeds (Fig. 5, C and D). After germination, wild-type and transgenic seedlings of similar size were transferred to one-half-strength MS medium containing 2 µm ABA. In the absence of ABA, shoot growth was inhibited markedly. Conversely, root growth was promoted in the OsDET1 RNAi lines compared with the wild type (Fig. 5, E, G, and H). Application of ABA strongly inhibited root and shoot growth in the OsDET1 RNAi lines compared with the wild type (Fig. 5, F–H). Overall, these finding indicate that down-regulating OsDET1 confers ABA hypersensitivity in OsDET1 RNAi plants during seed germination and seedling growth.
Figure 5.
OsDET1 influences seed germination and seedling growth. A, Overexpression or suppression of OsDET1 inhibited seed germination in transgenic plants. B, Germination time courses on one-half-strength MS medium. C, Overexpression or suppression of OsDET1 inhibited seed germination in transgenic plants during ABA treatment. D, Germination time courses on one-half-strength MS medium containing 2 µm ABA. E, Overexpression or suppression of OsDET1 inhibited shoot growth and promoted root growth. F, Overexpression or suppression of OsDET1 inhibited the growth of shoot and root during ABA treatment. G and H, Root and shoot growth rates of the wild type (WT) and OsDET1 RNAi and OE-OsDET1 plants. Three independent experiments were carried out with similar results. Representative graphs are shown (n = 30 seeds in each experiment). *, P ≤ 0.05 and **, P ≤ 0.01 (Student’s t test).
Some phenotypes of the OE-OsDET1 plants were similar to those of the OsDET1 RNAi lines. For example, on one-half-strength MS medium, seed germination was obviously delayed, shoot growth was inhibited, and root growth was promoted (Fig. 5). These results imply that higher levels of endogenous ABA also cause ABA hypersensitivity in OE-OsDET1 plants during seed germination and seedling growth. However, unlike the OsDET1 RNAi lines, the inhibition of root and shoot growth was slight in the OE-OsDET1 lines compared with the wild type during ABA treatment (Fig. 5, F–H).
OsDDA1 Interacts with OsPYL5 and the CDD Complex in Vivo
To further unravel the mechanism by which OsDET1 deficiency causes ABA hypersensitivity in rice, we performed yeast two-hybrid (Y2H) assays to identify proteins that interact physically with OsDET1. The OsDET1 coding sequence was inserted into PGBKT7 as bait, and a cv Nipponbare complementary DNA (cDNA) library previously generated in our laboratory was used for yeast two-hybrid (Y2H) screening. Nine candidate interacting proteins were identified in these assays (Supplemental Fig. S8). OsDDB1 (Os05g0592400), an ortholog of Arabidopsis DDB1 (Kimura et al., 2004), was identified as an interacting protein of OsDET1.
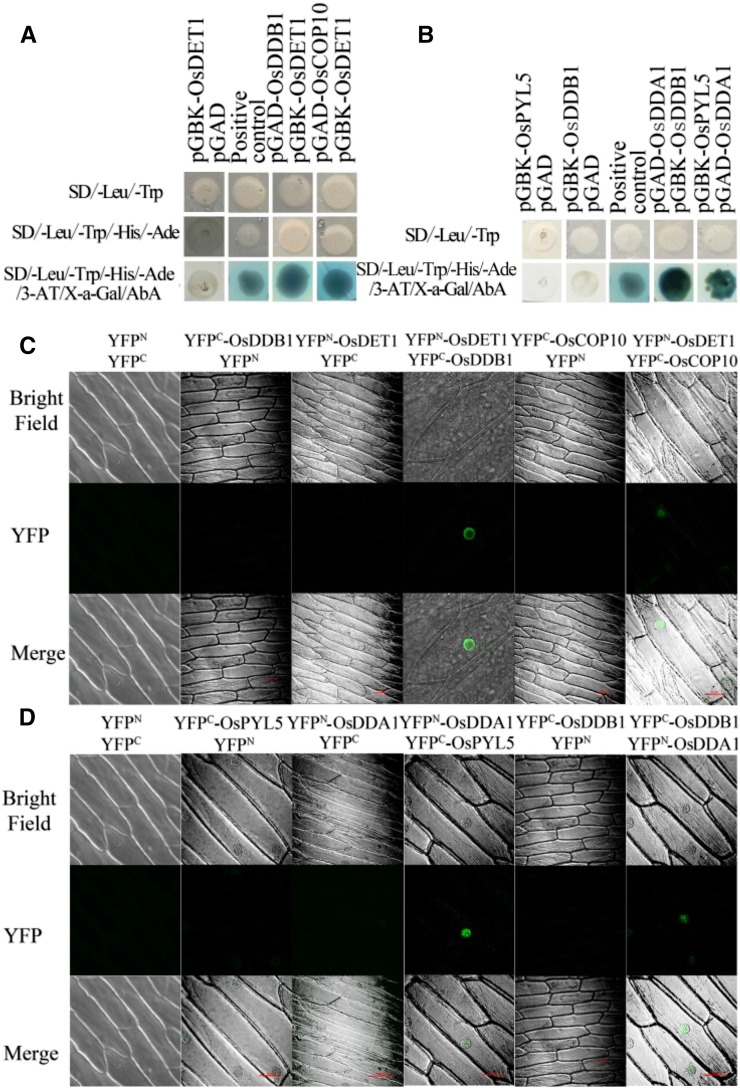
Indeed, a series of studies within the last decade demonstrated that DET1 interacts with COP10 and DDB1 to form the CDD complex in Arabidopsis and tomato (Solanum lycopersicum; Kimura et al., 2004; Lau and Deng, 2012). The CDD complex interacts with DDA1 to form the CDDD complex, which mediates the ubiquitination of the PYR/PYL/RCAR family of ABA receptors in Arabidopsis (Irigoyen et al., 2014). Since our results suggest that OsDET1 deficiency increases ABA sensitivity in rice, we hypothesized that a similar mechanism might occur in rice. Therefore, we cloned the orthologous genes of DDA1, COP10, and PYL8-related from cv Nipponbare. The full-length coding sequences of OsDDB1, OsCOP10, and OsDDA1 and truncated versions of the ABA receptor gene OsPYL5 (encoding amino acids 90–209) were fused separately to pGADT7 or PGBKT7, and a yeast two-hybrid (Y2H) assay was performed to determine the interaction of OsDET1 with OsDDB1, OsCOP10, OsDDA1, and OsPYL5 (Ishibashi et al., 2003; Kim et al., 2012, 2014; Irigoyen et al., 2014). We found that OsDET1 interacts physically with OsDDB1 and OsCOP10 (Fig. 6A) and that OsDDA1 binds to OsPYL5 and OsDDB1 in yeast (Y2H) cells (Fig. 6B).
Figure 6.
Interaction of OsDET1, OsDDB1, OsCOP10, OsDDA1, and OsPYL5 in vivo. A, OsDET1 interacts with OsDDB1 and OsCOP10 in yeast cells. The coding range of OsDET1 was inserted into pGBKT7 as bait, and the coding ranges of OsDDB1 and OsCOP10 were inserted into pGADT7 as prey. B, OsDDA1 interacts with OsDDB1 and OsPYL5 in yeast cells. The coding range of OsDDA1was inserted into pGBKT7 as bait, and the full-length coding sequence of OsDDB1 and a truncated version of the ABA receptor OsPYL5 (amino acids 90–209) were fused separately to pGADT7 as prey. AbA, Aureobasidin A; 3-AT, 3-amino-1,2,4-triazole; SD, synthetic dextrose. C, OsDET1 interacts with OsDDB1 and OsCOP10 in vivo. YFPN-OsDET1/YFPC-OsDDB1 and YFPN-OsDET1/YFPC-OsCOP10 were transformed into onion epidermal cells by Agrobacterium tumefaciens-mediated transformation to test their interactions. After 48 h of incubation in the dark, YFP signal was detected by confocal microscopy. Bright field was used to indicate the localization of nuclei. Empty vectors were used as negative controls. D, OsDDA1 interacts with OsDDB1 and OsPYL5 in vivo. YFPN-OsDDA1/YFPC-OsDDB1 and YFPN-OsDDA1/YFPC-OsPYL5 were transformed into onion epidermal cells by A. tumefaciens-mediated transformation to test their interactions.
To confirm the results of the yeast two-hybrid (Y2H) assay, we performed BiFC assays using agroinfiltrated onion (Allium cepa) epidermal cells. The full-length coding sequences of OsDET1, Os-DDB1, OsCOP10, OsDDA1, and OsPYL5 were fused separately with the N- or C-terminal portions of yellow fluorescent protein (YFP). OsDET1 interacted with OsDDB1 and OsCOP10 in the BIFC assays (Fig. 6C), further indicating that the CDD complex may be present in rice. In addition, OsDDA1 interacted with OsPYL5 and OsDDB1 (Fig. 6D), suggesting that OsDDA1 functions in combining the CDD complex with OsPYL5 in rice. Taken together, these results support the notion that OsDET1 interacts with OsCOP10 and OsDDB1 and modulates OsPYL degradation in rice.
OsDET1 Influences OsPYL5 Degradation in Rice
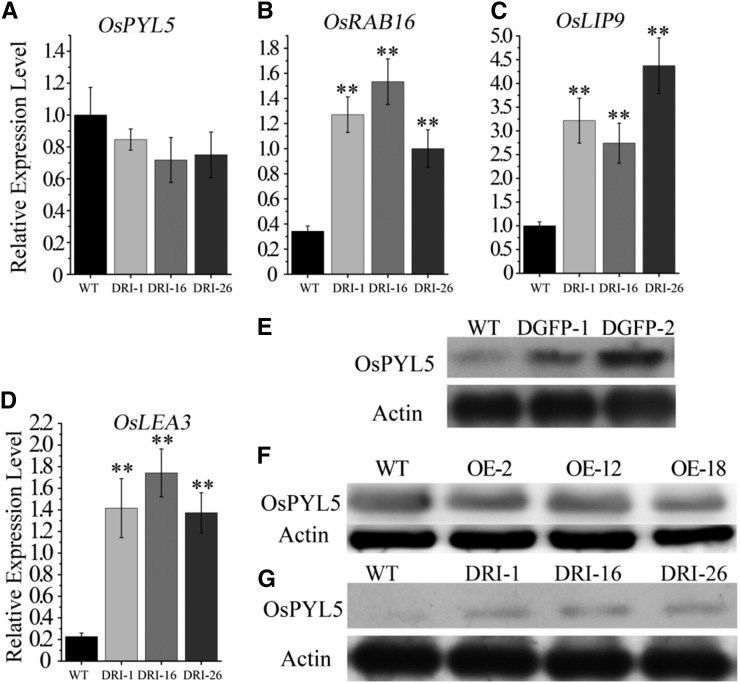
To determine whether OsDET1 takes part in the regulation of OsPYL degradation in rice, we monitored the expression of OsPYL5 by qRT-PCR. The expression of OsPYL5 was reduced slightly in OsDET1 RNAi plants after 12 h of ABA treatment compared with the wild type (Fig. 7A). Conversely, western blotting showed that OsPYL5 levels were markedly higher in OsDET1 RNAi plants than in wild-type plants (Fig. 7G), implying that the degradation of OsPYL5 was delayed in OsDET1 RNAi plants. In addition, we observed no significant differences of OsPYL5 level in OE-OsDET1 plants and the wild type (Fig. 7F), suggesting that the overexpression of OsDET1 was unable to promote the degradation of OsPYL5 in OE-OsDET1 plants. The expression of ABA-inducible marker genes, including OsRAB16A, OsLEA, and OsLIP9, is promoted by OsPYL5 in plants under ABA treatment (Kim et al., 2012). Therefore, we investigated the transcription of these genes in the transgenic lines. Consistent with the higher level of OsPYL5, the expression of OsRAB16A, OsLEA, and OsLIP9 was significantly higher in OsDET1 RNAi plants than in wild-type plants (Fig. 7, B–D). Taken together, these results suggest that the degradation of OsPYL5 is impaired in OsDET1 RNAi plants, leading to ABA hypersensitivity.
Figure 7.
OsDET1 influences the degradation of OsPYL5 and activates the expression of ABA mark genes. Fourteen-day-old seedlings were treated with ABA solution (one-half-strength MS medium with 5 μm ABA) for 12 h. Shoots were detected by qRT-PCR. A to D, Expression of OsPYL5, OsRAB16, OsLIP9, and OsLEA3 in the wild type (WT) and OsDET1 RNAi lines during ABA treatment. **, P ≤ 0.01 (Student’s t test). E, Western-blot analysis of OsPYL5 from detached OsDET1-GFP leaves at the tillering stage after 12 h of ABA treatment. F, Western-blot analysis of OsPYL5 from detached OE-OsDET1 leaves at the tillering stage after 12 h of ABA treatment. G, Western-blot analysis of OsPYL5 from detached OsDET1 RNAi leaves after 12 h of ABA treatment.
Pleiotropic Function of OsDET1 in Rice
OsDET1 deficiency caused pleiotropic phenotypes in addition to accelerated dark- and ABA-induced leaf senescence. Excessive RNA interference treatment led to plant death in T0 transgenic plants at the vegetative growth stage or a failure of T1 transgenic seeds to germinate (Supplemental Fig. S9, A–C). In addition, OsDET1 deficiency caused an overall reduction in biomass. The number of tillers, panicles, and spikelets as well as fertility were reduced significantly in OsDET1 RNAi plants (Fig. 8A; Supplemental Fig. S9, D and E; Supplemental Table S1). Meanwhile, the plant height and leaf length were reduced in OsDET1 RNAi plants compared with the wild type (Supplemental Fig. S9, E and F). Conversely, OE-OsDET1 plants were vigorous and fertile (Supplemental Figs. S10A and S11). The leaf length was increased in transgenic plants compared with wild-type plants, which caused drooping leaves in OE-OsDET1 plants (Supplemental Fig. S10, A and B).
Figure 8.
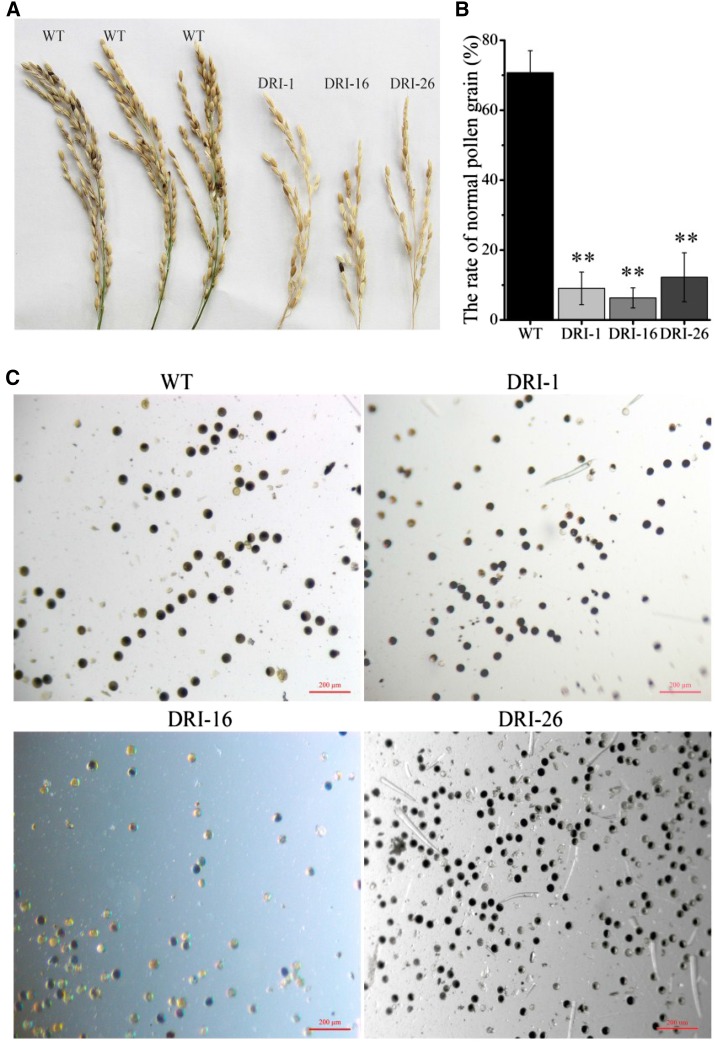
OsDET1 deficiency affects the pollination and fertilization processes in flower. A, Spikelets from the wild type (WT) and OsDET1 RNAi lines showed reductions in the number of filled seeds in the transgenic lines. B and C, Pollen grain morphology analysis showing the pollen grains of OsDET1 RNAi plants under ABA stress. Pollen grains were isolated from the wild type and OsDET1 RNAi plants and tested with a microscope. Three photographs were used for statistical analysis. Representative graphs are shown. **, P ≤ 0.01 (Student’s t test). Bars = 200 μm.
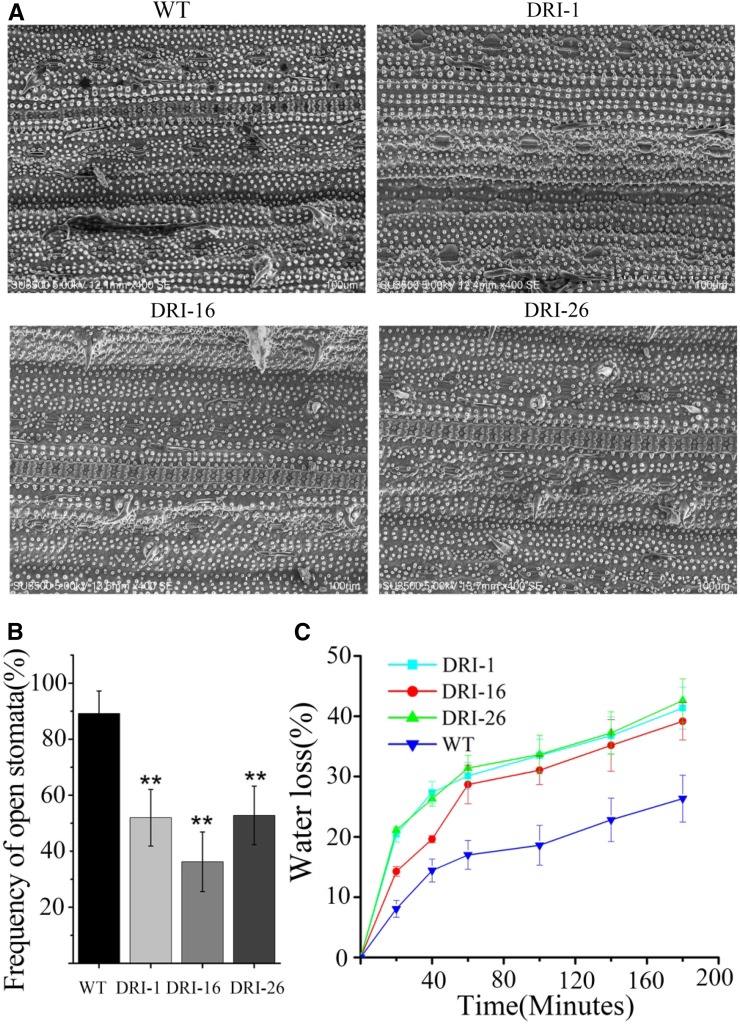
We observed some phenotypes related to ABA hypersensitivity in OsDET1 RNAi plants. The transgenic plants had fewer filled spikelets than wild-type plants (Fig. 8A) as well as slightly smaller pollen grains and a higher proportion of irregularly shaped pollen grains (Fig. 8, B and C). Similarly, more stomatal pores were completely closed in OsDET1 RNAi plants compared with the wild type, as revealed by scanning electron microscopy (SEM: Fig. 9, A and B). These results further support the notion that OsDET1 deficiency increases ABA sensitivity in rice. Interestingly, less wax crystallization was observed on the OsDET1 RNAi leaf surface compared with the wild type (Fig. 9A), and more rapid water loss occurred in OsDET1 RNAi plants as well (Fig. 9C). Wax crystallization functions as the outermost barrier against nonstomatal water loss (Zhou et al., 2015). Therefore, the accelerated water loss may have been due to the deficient wax crystallization in OsDET1 RNAi plants. ABA promotes cuticular wax biosynthesis in many plants. Thus, these phenotypes are consistent with the reduced ABA levels in OsDET1 RNAi plants, but they appear to contradict the ABA sensitivity of these plants.
Figure 9.
OsDET1 deficiency leads to contradictory ABA-related phenotypes in leaf. A, Stomatal aperture and wax crystallization of OsDET1 RNAi plants and wild-type plants (WT) observed by SEM. B, Frequency of open stomata. Values are means ± sd (n = 6). **, P ≤ 0.01 (Student’s t test). C, Water loss assay of leaves from OsDET1 RNAi plants and wild-type plants.
In this study, ABA biosynthesis was accelerated in OE-OsDET1 plants. We noticed that more wax crystallization existed on the OE-OsDET1 leaf surface than on wild-type leaves (Supplemental Fig. S10C), which was consistent with the increase of endogenous ABA in OE-OsDET1 plants. But the gentle increase of ABA was unable to change the morphology of pollen in OE-OsDET1 plants. The ratio of normal pollen in OE-OsDET1 plants showed only a slight decrease compared with that in the wild type (Supplemental Fig. S11, B and C). In addition, the spikelet fertility of OE-OsDET1 plants was normal (Supplemental Fig. S11A). Taken together, these pleiotropic phenotypes of OE-OsDET1 and OsDET1 RNAi plants suggest the diverse role of OsDET1 in rice development.
Overexpressing OsDET1-GFP Enhances ABA Sensitivity in Rice
Adding a C-terminal GFP tag to DET1 disrupts the functioning of the CDD complex in Arabidopsis (Schroeder et al., 2002). We hypothesized that a C-terminal GFP tag also would disrupt the functioning of OsDET1 in rice. Therefore, we introduced the pCOMBIA1301-OsDET1-GFP vector into cv Nipponbare rice. OsDET1-GFP transcript levels were significantly higher than endogenous OsDET1 levels in OsDET1-GFP transgenic plants (OsDET1-GFP plants), whereas there was no obvious change in endogenous OsDET1 expression relative to the wild type (Supplemental Fig. S12A). Different from OE-OsDET1 plants, the phenotype of OsDET1-GFP plants was similar to that of OsDET1 RNAi plants. The transgenic plants showed growth retardation, dwarf, and some ABA sensitivity phenotypes (Supplemental Fig. S12B). The morphology of OsDET1-GFP pollen grains also was changed, and the spikelets were almost completely infertile in OsDET1-GFP plants (Supplemental Fig. S11). Moreover, OsDET1-GFP plants exhibited an accelerated leaf senescence phenotype during dark or ABA treatment (Supplemental Figs. S13 and S14). These results demonstrate that overexpressing OsDET1-GFP enhances ABA sensitivity in rice. Meanwhile, unlike OE-OsDET1 plants, the ABA content was reduced in OsDET1-GFP leaves in plants at the tillering stage (Supplemental Fig. S6B), and OsPYL5 degradation was delayed in OsDET1-GFP plants compared with the wild type after ABA treatment (Fig. 7E). These results suggest that the ABA hypersensitivity phenotypes of OsDET1-GFP plants are not caused by posttranscriptional gene silencing or overexpression of OsDET1. Instead, the misshapen OsDET1-GFP protein might disturb the normal functioning of OsDET1. Overall, these findings suggest that structural changes in OsDET1 may impair the normal functioning of the CDD complex and further influence ABA receptor stability in rice.
Transcriptome Analysis of OsDET1 RNAi Plants
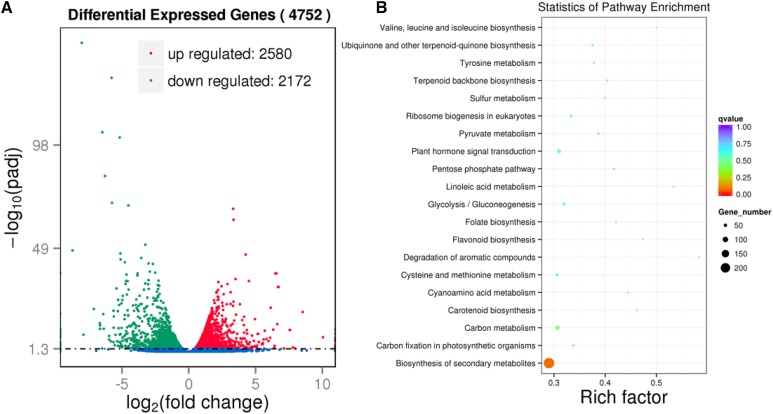
To obtain clues about the pleiotropic roles of OsDET1 in rice, we performed mRNA sequencing (RNA-SEQ) to examine the genome-wide effects of the OsDET1 gene in OsDET1 RNAi plants (Supplemental Table S2). A total of 4,752 differentially expressed genes (DEGs) were identified after comparing the transcriptome profiles of DRI-16 versus wild-type plants under normal growth conditions based on the criteria of DESeq (Anders et al., 2015). There were more up-regulated genes (2,580) than down-regulated genes (2,172; Fig. 10A; Supplemental Table S3). The DEGs were classified into different functional categories based on Gene Ontology (GO) annotation analysis. A wide range of categories were affected in OsDET1 RNAi plants, including biological process, molecular function, and cellular component GO terms (Supplemental Table S4). A number of GO terms, including chlorophyll catabolic process and chlorophyllase activity, were assigned to the DEGs (Supplemental Table S4). In addition, OsDET1 deficiency strongly affected the expression of photosynthesis- and thylakoid-related genes (Supplemental Table S5). DET1 modulates photomorphogenesis by regulating large numbers of light-mediated genes in Arabidopsis, as determined by microarray-based expression profiling analyses (Ma et al., 2003; Dong et al., 2014). Together, these findings suggest that OsDET1 modulates chlorophyll metabolic processes and chloroplast development and that the role of DET1 in photomorphogenesis is conserved in rice.
Figure 10.
Transcriptome comparison between leaves of the wild type and DRI-16 using RNA-SEQ. A, Volcano plot showing DEGs of the wild type and DRI-16. Biological significance (log2 fold change) is depicted on the x axis and statistical significance (log10) is depicted on the y axis. Statistical significance was corrected at P < 0.05. B, Scatterplot of KEGG pathway enrichment statistics from wild-type and DRI-16 leaves. Rich factor is the ratio of the number of DEGs to the number of background genes in a KEGG pathway. The top 20 enriched KEGG pathways are listed.
Furthermore, 15 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were enriched in OsDET1 RNAi plants based on corrected P < 0.05 values (Fig. 10B; Supplemental Table S6). This result is consistent with the multiple phenotypes of OsDET1 RNAi plants and further supports the pleiotropic functions of OsDET1 in rice. In addition, plant hormone signal transduction pathways were markedly affected by OsDET1 deficiency. In particular, 16 differentially expressed ABA signal transduction genes were detected, most of which were significantly up-regulated in OsDET1 RNAi plants; eight genes encoding PP2C family proteins were markedly up-regulated in these plants (Table I). ABA increases the expression levels of all OsPP2C genes from subfamily A (Xue et al., 2008). In this study, three basic Leu zipper transcription factor (bZIP) family genes were markedly up-regulated (Table I); bZIPs play important roles in the ABA signaling pathway and were induced by ABA in previous studies (Nijhawan et al., 2008; Amir Hossain et al., 2010). Moreover, OsPYL1 was down-regulated in OsDET1 RNAi plants, whereas OsPYL10 was up-regulated (Table I). These results are consistent with previously reported OsPYL expression patterns after ABA treatment (Tian et al., 2015). Overall, these results indicate that the OsDET1 RNAi plants suffered from ABA stress, further supporting the notion that OsDET1 deficiency leads to ABA hypersensitivity.
Table I. Examples of the DEGs in DRI-16 transgenic lines that are predicted to be involved in the ABA signaling pathway.
| Gene Identifier | Log2 Fold Change | Associated Gene Name | Description |
|---|---|---|---|
| Protein phosphatase 2C | |||
| OS03G0268600 | 1.6772 | Protein phosphatase 2C | |
| OS09G0325700 | 1.3708 | Protein phosphatase 2C | |
| OS01G0583100 | 2.0733 | Protein phosphatase 2C | |
| OS05G0592800 | 0.65232 | Protein phosphatase 2C | |
| OS01G0846300 | 1.7045 | Protein phosphatase 2C | |
| OS05G0537400 | 0.54579 | Protein phosphatase 2C | |
| OS04G0167900 | 1.8707 | Protein phosphatase 2C | |
| OS04G0167875 | 2.1824 | Protein phosphatase 2C | |
| Ser/Thr-protein kinase SRK2 | |||
| OS10G0564500 | −1.2467 | SAPK3 | |
| OS03G0390200 | 0.61378 | SAPK1 | |
| OS04G0691100 | −0.62471 | SAPK5 | |
| ABA receptor PYR/PYL family | |||
| OS02G0255500 | 0.5072 | OSJNBA0052K15.19 | Polyketide cyclase/dehydrase||START-like domain |
| OS01G0827800 | Infinitesimal | B1088C09.11 | Polyketide cyclase/dehydrase||START-like domain |
| OS03G0297600 | −2.3167 | OS03G0297600 | Polyketide cyclase/dehydrase||START-like domain |
| ABA-responsive element-binding factor | |||
| OS01G0867300 | −1.2092 | P0677H08.2-1 | Basic-Leu zipper domain |
| OS01G0813100 | 1.3435 | P0432B10.10 | Basic-Leu zipper domain |
| OS09G0456200 | 1.2221 | B1342C04.26 | Basic-Leu zipper domain |
DISCUSSION
DET1 encodes an evolutionarily conserved protein. DET1, a central repressor of plant photomorphogenesis, indirectly regulates the expression of a vast number of light-regulated genes by modulating the stability of some key transcription factors involved in light signaling. The mutation of DET1 causes pleiotropic phenotypes in Arabidopsis and tomato (Chory et al., 1994; Pepper et al., 1994; Mustilli et al., 1999; Huang et al., 2014). In Arabidopsis, DET1 represses light-induced seed germination. The det1 mutants display light-grown phenotypes in the dark. The mutation of DET1 also leads to chlorophyll accumulation in Arabidopsis and tomato leaves during the vegetative stage. In addition, DET1 play a negative role in controlling flowering time in Arabidopsis (Kang et al., 2015). Therefore, DET1 plays a prominent role in modulating plant development (Lau and Deng, 2012). Despite the rapid progress in dissecting the roles of DET1 in many plants several decades ago, little is known about the role of OsDET1 in rice. This study demonstrates that changes in OsDET1 expression cause pleiotropic phenotypes in transgenic rice. Specifically, OsDET1 deficiency had strong detrimental effects on basic plant development, indicating that OsDET1 is essential for maintaining normal development in rice and is required throughout the plant life cycle.
Notably, the pollen grain morphology was altered in OsDET1 RNAi plants (Fig. 8, B and C). Similar phenotypes were observed in rice treated with exogenous ABA or with increased endogenous ABA content (i.e. transgenic plants overexpressing OsAP2-39; Oliver et al., 2007; Yaish et al., 2010). In addition, more stomatal pores were closed in OsDET1 RNAi plants than in wild-type plants (Fig. 9, A and B). ABA, which regulates stomatal movement, is biosynthesized mainly in vascular parenchyma cells and is transported to guard cells by ABA transporters (such as AtBCG25 and AtABCG40; Dörffling and Tietz, 1985; Cheng et al., 2002; Koiwai et al., 2004; Endo et al., 2008; Pandey et al., 2009; Kanno et al., 2010). Subsequently, ABA is recognized by the PYR/PYL/PCAR ABA receptor and further associates with PP2Cs to form the PP2C-ABA-PYL ternary complex, which actives the ABA response. The accumulation of ABA in guard cells leads to stomatal closure (Cutler et al., 2010; Kim et al., 2010; Gonzalez-Guzman et al., 2012). Therefore, these phenotypes imply that OsDET1 RNAi plants suffered from ABA stress. However, the ABA content was reduced in OsDET1 RNAi leaves (Fig. 4A; Supplemental Fig. S6). These seemingly contradictory results indicate that OsDET1 deficiency most likely increases ABA sensitivity in OsDET1 RNAi plants. ABA mediates plant germination and early seedling growth (Kim et al., 2012; Irigoyen et al., 2014). In this study, down-regulation of OsDET1 produced ABA-hypersensitive phenotypes during seed germination and early seedling growth in OsDET1 RNAi plants (Fig. 5), which strongly supports the role of OsDET1 in ABA sensitivity. This notion was further confirmed by our investigation of dark- and ABA-induced leaf senescence (Figs. 2 and 3; Supplemental Figs. S1–S3), where leaf senescence was accelerated under dark and ABA treatment in OsDET1 RNAi plants, indicating that OsDET1 deficiency increases the sensitivity of ABA signaling in rice.
ABA signaling is mediated by the PYR/PYL/RCAR family of ABA receptors (Gonzalez-Guzman et al., 2012; Wang et al., 2013; Rodriguez et al., 2014). DET1 negatively regulates ABA signaling by modulating the ubiquitination of the PYL ABA receptor in Arabidopsis. DET1 interacts with COP10 and DDB1 to form the CDD complex, which further associates with DDA1 to form the CDDD complex. This complex provides substrate specificity for CRL4 by the recognition of DDA1 for the ubiquitination targets. Although the CDD complex does not interact with PYLs in vivo, it influences the recognition of DDA1 by PYLs (Irigoyen et al., 2014). In Arabidopsis, damage to the CDD complex obstructs the ubiquitination of PYLs, leading to their accumulation. The det1-1 mutants exhibit increased responses to ABA-mediated inhibition of germination and seedling establishment compared with the wild type (Irigoyen et al., 2014). Given that OsDET1 deficiency also leads to ABA hypersensitivity in OsDET1 RNAi plants and that the orthologs of DDA1, COP10, and PYL8 were discovered in rice, we speculate that a similar ABA signal regulatory mechanism also exists in rice (Fig. 11). The results of protein interaction analysis and the abundance of PYR/PYL/RCAR ABA receptors strongly support this model. First, we found that OsDET1 interacts with OsDDB1 and OsCOP10 in vivo (Fig. 6). Next, we demonstrated that OsDDA1 interacts with OsPYL5 and OsDDB1 (Fig. 6). Moreover, the degradation of OsPYL5 was impaired in OsDET1 RNAi plants. In addition, OsPYL5 was shown previously to promote the expression of ABA-inducible marker genes (including OsRAB16A, OsLEA, and OsLIP9) during ABA treatment and to play an important role in mediating ABA signaling during seed germination and early seedling growth (Kim et al., 2012, 2014). In this study, these ABA-inducible marker genes were up-regulated significantly in OsDET1 RNAi plants during ABA treatment compared with wild-type plants (Fig. 7). These results are consistent with the finding that OsPYL5 accumulated in OsDET1 RNAi plants, which further supports our hypothesis. The correct structure of DET1 is necessary to maintain the function of the CDD complex: a C-terminal GFP tag on DET1 hinders the formation of the CDD complex by DET1, COP10, and DDB1 in Arabidopsis (Schroeder et al., 2002). We found that overexpressing OsDET1-GFP also increased plant sensitivity to ABA signaling (Supplemental Figs. S11–S14) and delayed the degradation of OsPYL5 in OsDET1-GFP plants (Fig. 7E). These findings suggest that structural changes in OsDET1 impair the normal functioning of CDD and may affect ABA receptor stability in rice, further supporting the role of OsDET1 in modulating ABA signaling.
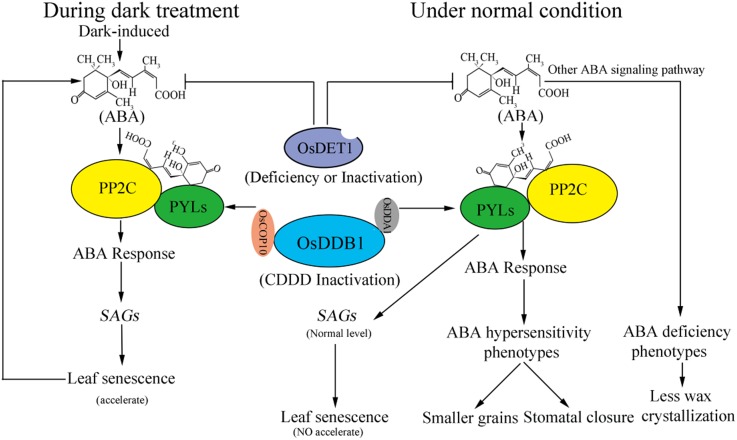
Figure 11.
Model of how OsDET1 deficiency leads to contradictory phenotypes related to ABA in OsDET1 RNAi plants. OsDDB1 interacts with OsCOP10, OsDET1, and OsDDA1 to form the CDDD complex. The reduction of CDDD complex function hinders the ubiquitination of OsPYLs and causes the accumulation of OsPYLs. The accumulation of PYLs and ABA promotes the stability of the PP2C-ABA-PYL ternary complex (Irigoyen et al., 2014). However, OsDET1 deficiency also leads to a decline in the content of ABA in normal conditions. Thus, due to these opposing factors, OsDET1 deficiency only partly causes the ABA hypersensitivity phenotype, such as closed stomatal pores and changes of pollen grain morphology, although it is unable to induce leaf senescence in normal development. The modulation of cuticular wax biosynthesis by the ABA signaling pathway is distinct from the governing stomatal regulation, and the accumulation of cuticular waxes is reduced in OsDET1 RNAi plants. During dark treatment, ABA is induced by continuous dark, which further enhances the ABA response and finally leads to increased leaf senescence. The leaf senescence in turn promotes the synthesis of ABA. Finally, OsDET1 RNAi plants exhibit a significantly accelerated leaf senescence phenotype.
Interestingly, OsDET1 also influences ABA biosynthesis in rice. Endogenous ABA levels are controlled by the balance of ABA biosynthesis and inactivation. ZEP and NCED are rate-limiting enzymes in ABA biosynthesis. ABA 8′-hydroxylase is the major enzyme for ABA inactivation (Nambara and Marion-Poll, 2005). Our results show that the ABA contents decreased in OsDET1 RNAi and OsDET1-GFP plants and increased in OE-OsDET1 plants compared with wild-type plants (Fig. 4A; Supplemental Fig. S6). Further analysis suggested that OsDET1 regulates ABA biosynthesis by fine-tuning the expression of ABA biosynthesis and inactivation genes (Fig. 4). ABA mediates plant germination and early seedling growth (Mulkey et al., 1983; Cornish and Zeevaart, 1985). Stimulating ABA biosynthesis inhibits seed germination as well as root and shoot growth in rice (Ikeda et al., 2002; Zhu et al., 2009; Cai et al., 2015). We found that seed germination was delayed in OE-OsDET1 plants and that root and shoot growth was inhibited during ABA treatment (Fig. 5). Similarly, mutation of the SENESCENCE-ASSOCIATED E3 UBIQUITIN LIGASE1 gene (SAUL1) increases ABA biosynthesis and causes early leaf senescence under low light conditions in the saul1 mutants (Raab et al., 2009). OE-OsDET1 plants also showed premature leaf senescence during dark treatment (Supplemental Fig. S5). These phenotypes are in accordance with the higher ABA levels in OE-OsDET1 plants, and they also support our conclusion that OsDET1 plays a positive role in ABA biosynthesis. In addition, ABA promotes cuticular wax biosynthesis in many plants (Fricke et al., 2006; Seo et al., 2011; Lee and Suh, 2015; Zhou et al., 2015). Interestingly, this study found that there was less wax crystallization on the leaf surface of OsDET1 RNAi plants than in wild-type plants (Fig. 9A). On the contrary, more wax crystallization existed on the OE-OsDET1 plant leaf surface than in the wild type (Supplemental Fig. S10C). These results are consistent with the change of ABA contents in transgenic plants, further supporting the notion that OsDET1 modulates ABA biosynthesis.
Therefore, we propose a model explaining the contradictory ABA-related phenotypes of OsDET1 RNAi plants (Fig. 11). The PP2C-ABA-PYL ABA signaling pathway regulates leaf senescence. OsDET1 deficiency damages the functioning of the CDD complex and causes the accumulation of PYLs. Meanwhile, OsDET1 deficiency also reduces ABA contents in OsDET1 RNAi plants. PYLs and ABA positively regulate the stability of the PP2C-ABA-PYL ternary complex, leading to ABA responses (Irigoyen et al., 2014; Zhao et al., 2016). Therefore, the reduced ABA content alleviates the damage caused by ABA hypersensitivity in OsDET1 RNAi plants, thereby preventing early leaf senescence in these plants. Nonetheless, OsDET1 RNAi plants also partially exhibit ABA hypersensitivity phenotypes, including stomatal closure and changes in pollen grain morphology. Furthermore, ABA promotes cuticular wax biosynthesis, but not via the PP2C-ABA-PYL ABA signaling pathway (Seo et al., 2011; Gonzalez-Guzman et al., 2012). Thus, wax crystallization and accumulation are weakened in OsDET1 RNAi plants. ABA signaling is induced by dark treatment, which increases dark-induced leaf senescence, in turn promoting ABA biosynthesis. Indeed, the OsDET1 RNAi plants exhibited rapidly accelerated leaf senescence.
In conclusion, the disruption of OsDET1 expression leads to a range of altered phenotypes. OsDET1 deficiency or structural changes impair the degradation of OsPYL5, stimulating ABA signaling, thereby causing ABA hypersensitivity in rice. OsDET1 also plays a positive role in ABA biosynthesis. Due to these two effects, OsDET1 RNAi plants exhibited some seemingly contradictory phenotypes related to ABA. In addition, overexpression of OsDET1 also caused ABA hypersensitivity in OE-OsDET1 plants. Overall, our work demonstrates that OsDET1 not only is involved in regulating the ABA signaling pathway but also regulates the ABA biosynthesis pathway in rice, implying that this protein has diverse effects on the ABA signaling pathway.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa japonica ‘Nipponbare’) plants were used in this study. Recombinant plasmids were transformed into rice callus by Agrobacterium tumefaciens (EHA105) medium transformation as described previously (Huang et al., 2013). The plants were grown in a greenhouse under a 12-h-light/12-h-dark cycle (300 mmol photons m−2 s−1) at 30°C and 60% relative humidity.
For OsDET1 induction analysis, cv Nipponbare seeds were germinated and cultured in one-half-strength MS liquid medium in a greenhouse for 2 weeks at 30°C under a 12-h-light/12-h-dark cycle. Different types of phytohormones were added to the nutrition solution, including gibberellin (50 μm), auxin (50 μm), 6-benzylaminopurine (50 μm), salicylic acid (50 μm), and ABA (50 μm). In addition, the plants were treated with NaCl (200 mm) or polyethylene glycol 6000 (20%) for 8 h for various lengths of time. For UV light treatment, the seedlings were treated with UV light for 40 min. Five plants were collected per treatment for RNA isolation. All experiments were repeated three times independently.
Physiological Measurements
The penultimate leaves at the tillering stage were used for dark- and ABA-induced leaf senescence tests. Photosynthetic pigments were extracted with 80% (v/v) acetone from dark-induced senescent and fresh leaves. Chlorophyll contents were determined as described (Lichtenthaler, 1987). Fv/Fm was measured with a PAM 2000 fluorometer (Heinz Walz) as described previously (Sakuraba et al., 2014). To measure membrane ion leakage, freshly excised leaves were washed several times with Milli-Q water and incubated in Milli-Q water in the dark at 30°C. Conductivity was measured with a DDSJ-318 conductivity meter (INESA). To measure water-loss rates, the penultimate leaves were detached from rice plants and weighed over time at room temperature at the tillering stage. To determine leaf stomatal closure status, the penultimate leaves were detached from rice plants of tillering stage and monitored by SEM (S-3400; Hitachi; Sang et al., 2012). To determine pollen fertility, pollen iodine staining was performed as described previously (Yaish et al., 2010). To explore leaf cell death and chlorophyll degradation, leaves were examined by transmission electron microscopy (H-7650; Hitachi) using a voltage of 80 kV as described previously (Morita et al., 2009). Leaf ABA contents were measured using a Phytodetek competitive ELISA kit (Agdia). ABA was extracted as described previously and measured following the manufacturer’s instructions (Fukumoto et al., 2013). Three independent biological repeats were performed.
Histochemical Staining for GUS Activity
The vector pCAMBIA-1381Z was used to construct the fusion plasmid POsDET1::GUS, containing the 2-kb OsDET1 promoter region and the GUS reporter gene. The construct was stably transformed into cv Nipponbare rice. GUS staining and observation were carried out as described previously (Liang et al., 2014).
Dark- and ABA-Induced Leaf Senescence
To analyze dark- and ABA-induced senescence, the penultimate leaves were excised from the plants at the tillering stage. The detached leaves were incubated in water at 30°C in the dark to analyze dark-induced leaf senescence. Detached leaves were incubated in 50 μm ABA at 30°C in the light to analyze ABA-induced leaf senescence.
Germination and Postgermination Assays
After surface sterilization, the seeds were planted on one-half-strength MS medium supplemented with 0 and 2 μm ABA. Seeds that formed green shoots were scored as germinated. Seed germination was record every day until day 10. For postgermination assays, seedlings were grown on one-half-strength MS medium for 4 to 5 d. Similarly sized transgenic plants were transferred to one-half-strength MS medium supplemented with 0 and 2 μm ABA. Seedling growth was measured at 7 d after transfer. The plants were grown in a greenhouse under a 12-h-light/12-h-dark cycle (300 mmol photons m−2 s−1) at 20°C and 60% relative humidity.
Expression Analysis
Total RNA was extracted using RNAiso Plus (Takara; http://www.takara.com.cn), and the RNA was reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara) according to the manufacturer’s instructions. Transcript levels of each gene were measured by qRT-PCR using CFX1000 (Bio-Rad) with the SYBR Premix ExTaq II Kit (Takara). Primer sets used for qRT-PCR are listed in Supplemental Table S7. The relative expression levels of each gene were normalized to the expression of OsACTIN1. Values are means ± sd of three biological repeats.
Yeast Two-Hybrid and BiFC Assays
yeast two-hybrid (Y2H) analysis was performed using the Matchmaker Gold Yeast Two-Hybrid System as described in the Yeastmaker Yeast Transformation System 2 User Manual (Clontech). First, the coding sequence of OsDET1 was amplified and inserted into PGBKT7 as bait using the primers listed in Supplemental Table S7. Then, the PGBK-OsDET1 plasmid was transformed into Y2H Gold yeast cells. After autoactivation and toxicity experiments were performed, the cv Nipponbare cDNA library (generated previously in our laboratory) was screened with PGBK-OsDET1 as bait in the Y2H Gold strain (Clontech) on SDO/X-α-gal/aureobasidin A and synthetic dextrose/−Trp media.
The plasmids were purified from positive clones and transformed into Escherichia coli DH5α. Plasmids with an open reading frame fused in frame to PGADT7 were identified by nucleotide sequencing, followed by BLAST analysis against the National Center for Biotechnology Information database. To confirm the positive interactions, bait and prey plasmids were cotransformed into Y2H Gold yeast cells, followed by incubation on synthetic dropout (SD) medium lacking tryptophan (-Trp), leucine (-Leu), histidine (-His) and adenine (-Ade) and containing the X-α-gal, Aureobasidin A and 3-amino-1, 2, 4-triazole (SD/-Trp/-Leu/-His/-Ade/X/AbA/3-AT).
To confirm protein interactions, full-length OsDDA1, OsDDB1, and OsCOP10 cDNA were cloned into pGBKT7. The full-length OsDET1 and truncated versions of OsPYL5 (amino acids 84–207) were fused to pGADT7, and the plasmids were cotransformed into Y2H Gold yeast cells, followed by incubation on the SD/-Trp/-Leu/-His/-Ade/X/AbA/3-AT medium.
The vectors (pFGC-N-YFP and pFGC-C-YFP) used in the BiFC assay were a kind gift from Chen Zhixiang. For BiFC analysis, the full-length coding sequences of OsDET1 and OsDDA1 without terminators were subcloned adjacent to pFGC-N-YFP to form the pFGC-OsDET1-YFP and pFGC-OsDDA1-YFP constructs, respectively. The full-length coding sequences of OsCOP10, OsPYL5, and OsDDB1, without terminators, were subcloned adjacent to pFGC-N-YFP to form the pFGC-OsCOP10-YFP, pFGC-OsPYL5-YFP, and pFGC-OsDDB1-YFP constructs, respectively. The primers used are listed in Supplemental Table S7. Pairs of plasmids were introduced into onion (Allium cepa) epidermal cells by A. tumefaciens-mediated transformation as described previously. The onion cells were then incubated at 28°C for 48 h. YFP fluorescence was examined and photographed by confocal microscopy (A1+R; Nikon) at 48 to 72 h after infiltration.
Antibody Production
Two specific polypeptides of OsPYL5 were synthesized (HMRRLHSHAPGE and AVQSPTSPLEQ) and injected separately into rabbits. Rabbit preimmune serum was used to check for anti-OsPYL5 specificity.
Immunoblot Analysis
Total leaf protein was prepared essentially as described (Sangon Biotech; http://www.sangon.com/). SDS-PAGE and subsequent immunoblot analysis were performed as described previously (Morita et al., 2009). Protein content was detected by the Bradford method (Bio-Rad). The following antibodies were used: polyclonal antibodies against LHCa1, LHCb1, LHCb2, LHCb4, LHCb6, Rubisco large subunit (Agrisera; http://www.agrisera.com/), and OsPYL5. The antibodies were detected using an enhanced chemiluminescence detection system (Qiagen).
RNA-SEQ Analysis
The penultimate leaves were dissected from 8-week-old wild-type and DRI-16 plants. Samples from five siblings grown in the same bucket were pooled as one biological repeat; three biological repeats were performed per genotype. The RNA-SEQ experiments were performed by Novogene. Total RNA was extracted with TRIzol reagent (Invitrogen). Library construction was performed according to Illumina instructions and sequenced on a HiSeq 2000 sequencer. All paired-end reads were mapped to the rice cv Nipponbare genome using TopHat2 (Langmead et al., 2009; Trapnell et al., 2009; Kim and Salzberg, 2011). Expression levels were calculated using the reads per kb per million reads method (Mortazavi et al., 2008). The DESeq R package (1.10.1) was used for analysis the differential expression genes (DEGs) of wild-type and DRI-16 plant. The DEGs were filtered for a corrected P-value threshold of 0.005 (P values were adjusted using the Benjamini & Hochberg method).
The clustered genes were assigned to biological process categories based on GO analysis using the Web tool DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/home.jsp; Huang et al., 2008, 2009). Significantly enriched GO terms for the DEGs compared with the genomic background (corrected P < 0.05) were identified using a hypergeometric test. KEGG pathway-based analysis was performed using the blastall program against the KEGG database (http://www.genome.jp/kegg). Significantly enriched metabolic pathways or signal transduction pathways for the DEGs were identified by pathway enrichment analysis (corrected P < 0.05; Kanehisa et al., 2008).
Sequence data from this article can be found in the National Center for Biotechnology Information Short Read Archive sequence database under accession number SRR2989983. Sequence data also can be found in GenBank libraries with the following accession numbers: OsDET1 (AK100613), OsDDB1 (AK065508), Osh36 (AF251070), OsI57 (AF251076), OsNCED1 (LOC_Os02g47510), OsNCED2 (LOC_Os12g24800), OsNCED3 (LOC_Os03g44380), OsNCED4 (LOC_Os07g05940), AK101547, AK070447, AK102951, AK111970, AK106213, and AK060647. Genes and their associated accession codes from the Rice Annotation Project Database are as follows: OsNAP (LOC_Os03g21060), SGR (LOC_Os09g36200), NYC1 (LOC_Os01g12710), NYC3 (LOC_Os06g24730), RCCR1 (LOC_Os10g25030), OsZEP (LOC_Os04g37619), OsABA8ox1 (LOC_Os02g47470), OsABA8ox2 (LOC_Os08g36860), OsABA8ox3 (LOC_Os09g28390), LOC_Os02g0104700, LOC_Os05g0468600, RAB16A (LOC_Os11g26790), LEA3 (LOC_Os05g46480), LIP9 (LOC_Os02g44870), and OsPYL5 (LOC_Os05g12260).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. OsDET1 deficiency accelerates the expression of CDGs and SAGs during dark-induced leaf senescence.
Supplemental Figure S2. Photosynthetic protein degradation during dark-induced leaf senescence.
Supplemental Figure S3. Ultrastructural changes of chloroplasts in OsDET1 RNAi lines and the wild type during dark incubation.
Supplemental Figure S4. Characterization of OsDET1 RNAi plants at the late filling stage.
Supplemental Figure S5. Overexpression of OsDET1 accelerates dark-induced leaf senescence.
Supplemental Figure S6. OsDET1 deficiency and overexpressing OsDET1-GFP decrease the ABA content in leaves.
Supplemental Figure S7. ABA biosynthesis of OsDET1 RNAi lines during dark treatment.
Supplemental Figure S8. OsDET1 interacts with both candidate proteins in vivo.
Supplemental Figure S9. OsDET1 deficiency leads to pleiotropic phenotypes in OsDET1 RNAi lines.
Supplemental Figure S10. Overexpressing OsDET1 causes pleiotropic phenotypes in OE-OsDET1 plants.
Supplemental Figure S11. Different functions of overexpressing OsDET1-GFP or OsDET1 in pollination and fertilization processes in the flower.
Supplemental Figure S12. Phenotypes of OsDET1-GFP plants.
Supplemental Figure S13. Overexpressing OsDET1-GFP accelerates ABA-induced leaf senescence.
Supplemental Figure S14. Overexpressing OsDET1-GFP accelerates dark-induced leaf senescence in rice.
Supplemental Table S1. Agronomic traits of OsDET1 RNAi lines.
Supplemental Table S2. Summary of the mRNA sequencing data mapping.
Supplemental Table S3. Transcriptome analysis identified 4,752 DEGs in DRI-16 transgenic plants.
Supplemental Table S4. GO biological process assignments and their enrichment.
Supplemental Table S5. Examples of the DEGs in OsDET1 RNAi plants that are predicted to be involved in photosynthesis and thylakoid.
Supplemental Table S6. KEGG pathway enrichment results for DEGs.
Supplemental Table S7. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Bing Yang (Iowa State University) for advice on the RNAi vector, Zhixiang Chen (Purdue University) for providing the vectors pFGC-N-YFP and pFGC-C-YFP, and Guanghua He (Southwest University, China) for SEM analyses.
Glossary
- ABA
abscisic acid
- CDGs
chlorophyll degradation-related genes
- SAGs
senescence-associated genes
- BiFC
bimolecular fluorescence complementation
- qRT
quantitative reverse transcription
- Fv/Fm
ratio of variable fluorescence to maximum fluorescence
- MS
Murashige and Skoog
- cDNA
complementary DNA
- RNA-SEQ
mRNA sequencing
- DEGs
differentially expressed genes
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- SEM
scanning electron microscopy
Footnotes
This work was supported by the National Genetically Modified Organisms Breeding Major Projects (grant no. 2009ZX08009–103B), the National Key Technology R&D Program of China (grant no. 2011BAD35B 02–05), the Shuzhou Rice Research Institute, and the Public Experiment Center of the State Bioindustrial Base.
References
- Amir Hossain M, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB (2010) The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72: 557–566 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Cai S, Jiang G, Ye N, Chu Z, Xu X, Zhang J, Zhu G (2015) A key ABA catabolic gene, OsABA8ox3, is involved in drought stress resistance in rice. PLoS One 10: e0116646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells E, Molinier J, Benvenuto G, Bourbousse C, Zabulon G, Zalc A, Cazzaniga S, Genschik P, Barneche F, Bowler C (2011) The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress. EMBO J 30: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al. (2006) Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Gong Z, Zhu JK (2008) Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol 50: 1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999 [DOI] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M (1994) A role for cytokinins in de-etiolation in Arabidopsis: det mutants have an altered response to cytokinins. Plant Physiol 104: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish K, Zeevaart JA (1985) Movement of abscisic acid into the apoplast in response to water stress in Xanthium strumarium L. Plant Physiol 78: 623–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, Chen H (2014) Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26: 3630–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörffling K, Tietz D (1985) Abscisic acid in leaf epidermis of Commelina communis L.: distribution and correlation with stomatal closure. J Plant Physiol 117: 297–305 [DOI] [PubMed] [Google Scholar]
- Endo A, Sawada Y, Takahashi H, Okamoto M, Ikegami K, Koiwai H, Seo M, Toyomasu T, Mitsuhashi W, Shinozaki K, et al. (2008) Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol 147: 1984–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W, Akhiyarova G, Wei W, Alexandersson E, Miller A, Kjellbom PO, Richardson A, Wojciechowski T, Schreiber L, Veselov D, et al. (2006) The short-term growth response to salt of the developing barley leaf. J Exp Bot 57: 1079–1095 [DOI] [PubMed] [Google Scholar]
- Fukumoto T, Kano A, Ohtani K, Inoue M, Yoshihara A, Izumori K, Tajima S, Shigematsu Y, Tanaka K, Ohkouchi T, et al. (2013) Phosphorylation of D-allose by hexokinase involved in regulation of OsABF1 expression for growth inhibition in Oryza sativa L. Planta 237: 1379–1391 [DOI] [PubMed] [Google Scholar]
- Gepstein S, Thimann KV (1980) Changes in the abscisic acid content of oat leaves during senescence. Proc Natl Acad Sci USA 77: 2050–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53: 927–937 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Qin F, Zang G, Kang Z, Zou H, Hu F, Yue C, Li X, Wang G (2013) Mutation of OsDET1 increases chlorophyll content in rice. Plant Sci 210: 241–249 [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Deng XW (2014) Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr Opin Plant Biol 21: 96–103 [DOI] [PubMed] [Google Scholar]
- Huang WL, Tung CW, Ho SW, Hwang SF, Ho SY (2008) ProLoc-GO: utilizing informative Gene Ontology terms for sequence-based prediction of protein subcellular localization. BMC Bioinformatics 9: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Sonoda Y, Vernieri P, Perata P, Hirochika H, Yamaguchi J (2002) The slender rice mutant, with constitutively activated gibberellin signal transduction, has enhanced capacity for abscisic acid level. Plant Cell Physiol 43: 974–979 [DOI] [PubMed] [Google Scholar]
- Irigoyen ML, Iniesto E, Rodriguez L, Puga MI, Yanagawa Y, Pick E, Strickland E, Paz-Ares J, Wei N, De Jaeger G, et al. (2014) Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26: 712–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Kimura S, Yamamoto T, Furukawa T, Takata K, Uchiyama Y, Hashimoto J, Sakaguchi K (2003) Rice UV-damaged DNA binding protein homologues are most abundant in proliferating tissues. Gene 308: 79–87 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MY, Yoo SC, Kwon HY, Lee BD, Cho JN, Noh YS, Paek NC (2015) Negative regulatory roles of DE-ETIOLATED1 in flowering time in Arabidopsis. Sci Rep 5: 9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, Seo M (2010) Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol 51: 1988–2001 [DOI] [PubMed] [Google Scholar]
- Kim D, Salzberg SL (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG (2012) A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot 63: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Kim H, Lee K, Hwang H, Bhatnagar N, Kim DY, Yoon IS, Byun MO, Kim ST, Jung KH, Kim BG (2014) Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J Exp Bot 65: 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Tahira Y, Ishibashi T, Mori Y, Mori T, Hashimoto J, Sakaguchi K (2004) DNA repair in higher plants: photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells. Nucleic Acids Res 32: 2760–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai H, Nakaminami K, Seo M, Mitsuhashi W, Toyomasu T, Koshiba T (2004) Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol 134: 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z, Li M, Yang W, Xu W, Xue Y (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141: 1376–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lee IC, Hong SW, Whang SS, Lim PO, Nam HG, Koo JC (2011) Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol 52: 651–662 [DOI] [PubMed] [Google Scholar]
- Lee SB, Suh MC (2015) Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol 56: 48–60 [DOI] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, et al. (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111: 10013–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler FW. (1987) Karl Freudenberg, Burckhardt Helferich, Hermann O. L. Fischer: a centennial tribute. Carbohydr Res 164: 1–22 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Ma L, Zhao H, Deng XW (2003) Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130: 969–981 [DOI] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2009) Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59: 940–952 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mulkey TJ, Evans ML, Kuzmanoff KM (1983) The kinetics of abscisic acid action on root growth and gravitropism. Planta 157: 150–157 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C (1999) Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Dennis ES, Dolferus R (2007) ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol 48: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Oliver SN, Dennis ES, Dolferus R (2007b) ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol 48: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J (1994) DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78: 109–116 [DOI] [PubMed] [Google Scholar]
- Philosoph-Hadas S, Hadas E, Aharoni N (1993) Characterization and use in ELISA of a new monoclonal antibody for quantitation of abscisic acid in senescing rice leaves. Plant Growth Regul 12: 71–78 [Google Scholar]
- Raab S, Drechsel G, Zarepour M, Hartung W, Koshiba T, Bittner F, Hoth S (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J 59: 39–51 [DOI] [PubMed] [Google Scholar]
- Rodriguez L, Gonzalez-Guzman M, Diaz M, Rodrigues A, Izquierdo-Garcia AC, Peirats-Llobet M, Fernandez MA, Antoni R, Fernandez D, Marquez JA, et al. (2014) C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. Plant Cell 26: 4802–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong H, Tang Y, Zhang H, Wu P, Chen Y, Li M, Wu G, Jiang H (2013) The Stay-Green Rice Like (SGRL) gene regulates chlorophyll degradation in rice. J Plant Physiol 170: 1367–1373 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Park SY, Kim YS, Wang SH, Yoo SC, Hörtensteiner S, Paek NC (2014) Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Mol Plant 7: 1288–1302 [DOI] [PubMed] [Google Scholar]
- Sang X, Li Y, Luo Z, Ren D, Fang L, Wang N, Zhao F, Ling Y, Yang Z, Liu Y, et al. (2012) CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol 160: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J (2002) De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol 12: 1462–1472 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng XW (2002) Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev 16: 554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tian X, Wang Z, Li X, Lv T, Liu H, Wang L, Niu H, Bu Q (2015) Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice (N Y) 8: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen ZH, Zhang B, Hills A, Blatt MR (2013) PYR/PYL/RCAR abscisic acid receptors regulate K+ and Cl− channels through reactive oxygen species-mediated activation of Ca2+ channels at the plasma membrane of intact Arabidopsis guard cells. Plant Physiol 163: 566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y (2008) Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish MW, El-Kereamy A, Zhu T, Beatty PH, Good AG, Bi YM, Rothstein SJ (2010) The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet 6: e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamatani H, Sato Y, Masuda Y, Kato Y, Morita R, Fukunaga K, Nagamura Y, Nishimura M, Sakamoto W, Tanaka A, et al. (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence. Plant J 74: 652–662 [DOI] [PubMed] [Google Scholar]
- Yanagawa Y, Sullivan JA, Komatsu S, Gusmaroli G, Suzuki G, Yin J, Ishibashi T, Saijo Y, Rubio V, Kimura S, et al. (2004) Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HF, Qiu K, Ren GD, Zhu Y, Kuai BK (2010) A pleiotropic phenotype is associated with altered endogenous hormone balance in the developmentally stunted mutant (dsm1). J Plant Biol 53: 79–87 [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li L, Xiang J, Gao G, Xu F, Liu A, Zhang X, Peng Y, Chen X, Wan X (2015) OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS ONE 10: e116676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Ye N, Zhang J (2009) Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol 50: 644–651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.