Sulfide, but not other sulfur-containing molecules, represses autophagy irrespective of the redox conditions.
Abstract
Accumulating experimental evidence in mammalian, and recently plant, systems has led to a change in our understanding of the role played by hydrogen sulfide in life processes. In plants, hydrogen sulfide mitigates stress and regulates important plant processes such as photosynthesis, stomatal movement, and autophagy, although the underlying mechanism is not well known. In this study, we provide new experimental evidence that, together with our previous findings, demonstrates the role of hydrogen sulfide in regulating autophagy. We used green fluorescent protein fluorescence associated with autophagic bodies and immunoblot analysis of the ATG8 protein to show that sulfide (and no other molecules such as sulfur-containing molecules or ammonium) was able to inhibit the autophagy induced in Arabidopsis (Arabidopsis thaliana) roots under nitrogen deprivation. Our results showed that sulfide was unable to scavenge reactive oxygen species generated by nitrogen limitation, in contrast to well-established reducers. In addition, reducers were unable to inhibit the accumulation of autophagic bodies and ATG8 protein forms to the same extent as sulfide. Therefore, we conclude that sulfide represses autophagy via a mechanism that is independent of redox conditions.
Historically, sulfide has been considered to be a toxic molecule that is hazardous to life and the environment. However, hydrogen sulfide is currently recognized as an important signaling molecule that functions as a physiological gasotransmitter and is of comparable importance to nitric oxide (NO) and carbon monoxide in mammalian systems. The number of biological roles of sulfide has expanded rapidly in recent times and is the subject of many reviews that have emphasized the physiological importance of sulfide, because most mammalian cells produce and metabolize sulfide in a precise and regulated manner (Szabó, 2007; Li and Moore, 2008; Gadalla and Snyder, 2010; Kimura, 2011; Wang, 2012). Accumulating evidence from numerous studies in plant biology also have shown that hydrogen sulfide is a signaling molecule that can be as important as NO and hydrogen peroxide (H2O2); thus, hydrogen sulfide has also been the subject of several recent reviews (García-Mata and Lamattina, 2013; Lisjak et al., 2013). Sulfide has been found to mediate increases in tolerance and protection against certain plant stresses, primarily through the increased performance of antioxidant defenses. For example, sulfide alleviates the inhibitory effects of copper and aluminum stress on wheat (Triticum aestivum) germination and barley (Hordeum vulgare) seedlings (Zhang et al., 2008, 2010; Dawood et al., 2012), the effect of boron on cucumber (Cucumis sativus) root elongation (Wang et al., 2010), and the toxicity of cadmium in Populus euphratica (Sun et al., 2013) and alfalfa (Medicago sativa) seedlings (Li et al., 2012a). Sulfide also improves drought and hypoxia resistance and heat and salinity tolerance (Jin et al., 2011; Li et al., 2012b; Cheng et al., 2013; Christou et al., 2013). Furthermore, sulfide has been suggested to play a role in regulating photosynthesis and flower senescence and in prolonging the postharvest shelf life of fruits (Chen et al., 2011; Zhang et al., 2011; Hu et al., 2012). Interestingly, hydrogen sulfide also has been identified as a component of the abscisic acid signaling pathway in guard cells (García-Mata and Lamattina, 2010; Lisjak et al., 2010). Recently, it was demonstrated that sulfide acts upstream of NO to modulate abscisic acid-dependent stomatal closure (Scuffi et al., 2014). The cross talk between NO and sulfide also has been suggested based on the induced alleviation of cadmium toxicity in alfalfa seedlings (Li et al., 2012a), the improved heat tolerance of maize (Zea mays) seedlings (Li et al., 2013), and the enhanced salinity tolerance of alfalfa seeds during germination (Wang et al., 2012b). All of these findings clearly demonstrate the importance of sulfide as a signaling molecule that is involved in regulating numerous essential processes in plants.
The endogenous production of hydrogen sulfide in mammalian tissues occurs through the enzymatic reactions of l-Cys (Szabó, 2007; Li and Moore, 2008; Gadalla and Snyder, 2010; Kimura, 2011; Wang, 2012). In plants, we identified an enzyme, DES1, with l-Cys desulfhydrase activity that is located in the cytosol of Arabidopsis (Arabidopsis thaliana; Álvarez et al., 2010). This is the only enzyme that has been unequivocally established to catalyze the desulfuration of l-Cys to sulfide plus ammonia and pyruvate; therefore, it is responsible for the release of hydrogen sulfide in the plant cytosol. Mutations in the DES1 gene impede the formation of sulfide and strongly affect plant metabolism and stress responses. Thus, the detailed characterization of des1 null mutants reveals premature leaf senescence, a dramatically altered plant transcriptional profile at the mature developmental stage, and the induction of autophagy (Álvarez et al., 2012b). The des1 mutants also present increased tolerance to conditions that promote oxidative stress, a high resistance to biotrophic and necrotrophic pathogens, salicylic acid accumulation, and WRKY DOMAIN CONTAINING TRASCRIPTION FACTOR54 and PATHOGENESIS RELATED1 induction; therefore, des1 mutants resemble constitutive systemic acquired resistance mutants (Álvarez et al., 2010, 2012a). Moreover, in des1 mutants, stomata do not close in response to abscisic acid. This effect is restored by the application of exogenous sulfide or genetic complementation, demonstrating the involvement of DES1 in abscisic acid signaling in guard cells. Further studies have shown that DES1 is required for abscisic acid-dependent NO production (Scuffi et al., 2014). Thus, in plant cells, DES1 could be responsible for modulating the generation of sulfide for important signaling processes (Romero et al., 2013; Gotor et al., 2015) such as autophagy.
Autophagy is a universal mechanism with a prosurvival role in eukaryotic cells and involves the digestion of cell contents to recycle the necessary nutrients or to degrade damaged or toxic components. The most important feature of autophagy (we refer to the macroautophagy) is the de novo synthesis of double membrane-bound structures called autophagosomes, which engulf and deliver materials to the vacuole to be broken down. Proteins involved in autophagy (ATG proteins) have been used to monitor autophagic activity in plants; the most commonly used protein is ATG8, which is tethered to autophagosomes by lipidation (Thompson and Vierstra, 2005; Bassham et al., 2006; 2007; Yoshimoto et al., 2010; Li and Vierstra, 2012; Yoshimoto, 2012). DES1 deficiency promotes the accumulation and lipidation of ATG8 isoforms in Arabidopsis leaves. Because mutation of the DES1 gene impedes sulfide generation in the cytosol, ATG8 protein accumulation and lipidation are prevented in des1 mutants when sulfide is generated by genetic complementation or exogenous application. Interestingly, exogenous sulfide also rescues the activation of autophagy that results from dark-induced carbon starvation in wild-type Arabidopsis leaf tissues (Álvarez et al., 2012b).
The underlying mechanism for transforming the sulfide signal into a biological response is largely unknown. Two mechanisms of action have been proposed based on the chemical properties of hydrogen sulfide. The nucleophilic properties of sulfide and its capacity to react with different oxygen species and nitrogen oxides suggest that it can act as an antioxidant to reduce oxidative stress (Fukuto et al., 2012). The second mechanism consists of the posttranslational modification of the –SH groups of protein Cys residues to generate the persulfide group –SSH and, in this way, alter protein activities or functions (Mustafa et al., 2009; Paul and Snyder, 2012; Aroca et al., 2015).
The aim of this work was to determine the mechanism underlying the regulation of autophagy by sulfide. For this purpose, we have investigated the role of sulfide in the induction of autophagy under nitrogen deprivation.
RESULTS
Sulfide Represses Autophagy When It Is Induced under Nitrogen Deprivation in Arabidopsis Roots
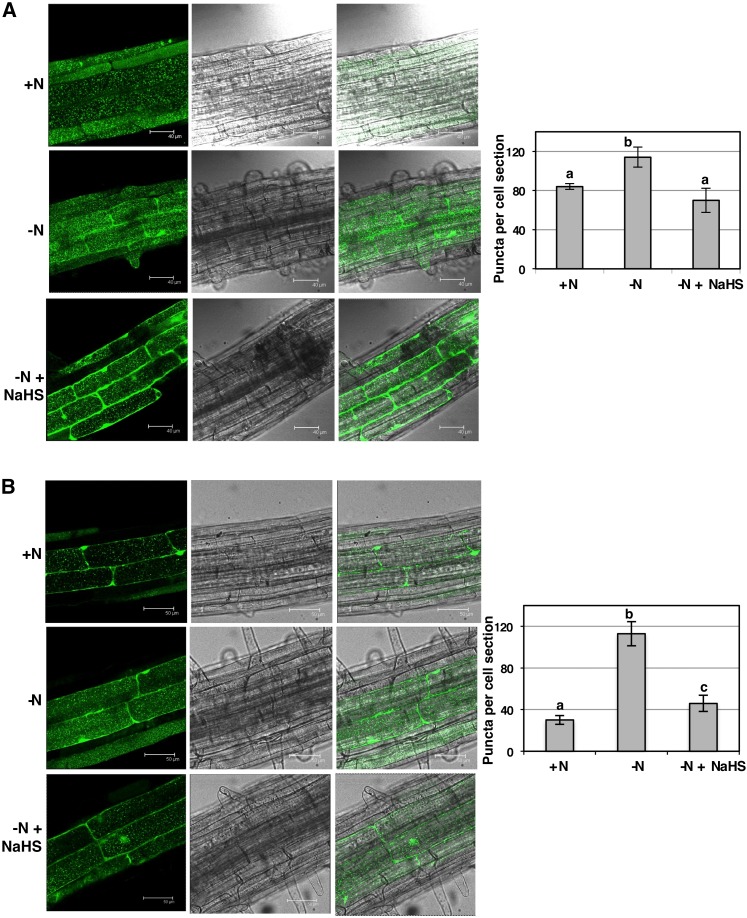
Previously, we showed that the application of exogenous sulfide rescues the induction of autophagy in Arabidopsis leaves, both in the des1 mutant and in carbon-starved wild-type plants, by analyzing the accumulation of ATG8 protein isoforms (Álvarez et al., 2012b). These results prompted us to suggest that sulfide generated in the cytosol by DES1 behaves as a repressor of autophagy (Gotor et al., 2013, 2015; Romero et al., 2013). To complement our previous studies and decipher the mechanism underlying the role of sulfide in autophagy, we established a new experimental system in which autophagy was induced by a different condition, in a different tissue, and using a different molecular tool. Thus, wild-type Arabidopsis plants expressing the GFP-ATG8a fusion protein were subjected to nitrogen limitation, and the effects of exogenous sulfide on seedling roots were analyzed by confocal microscopy of the GFP fluorescence. Seedlings were grown for 1 week on nitrogen-sufficient medium, and a portion of the seedlings was transferred to fresh nitrogen-rich medium. The other seedlings were transferred to medium lacking nitrogen, and a third set of seedlings was placed on the same nitrogen-deficient medium supplemented with 200 μm sodium hydrosulfide (NaHS). After the transfer, the seedlings were grown for an additional 2 or 4 d and then removed and treated with concanamycin A prior to observing them by confocal microscopy. In the roots of plants grown under nitrogen-rich conditions, we observed fluorescent punctate structures that were identified previously as GFP-ATG8-tagged autophagosomes and autophagic bodies (Yoshimoto et al., 2004; Contento et al., 2005; Thompson et al., 2005; Merkulova et al., 2014; Bassham, 2015). However, when the seedlings were subjected to nitrogen deprivation, independently of the time period, there was an increase in the number of autophagic bodies (Fig. 1), as demonstrated previously (Thompson et al., 2005; Phillips et al., 2008). Therefore, our results indicated that our nitrogen-limited conditions successfully induced autophagy. Interestingly, the presence of NaHS in the medium significantly inhibited the accumulation of fluorescent vesicles observed during nitrogen deprivation (Fig. 1). Without concanamycin A treatment, we failed to detect dotted structures; in contrast, we observed diffuse fluorescent staining of only the cytoplasm, with no change in response to nitrogen limitation (Supplemental Fig. S1).
Figure 1.
Effect of exogenous sulfide on autophagy induced by nitrogen deprivation in Arabidopsis roots. Wild-type seedlings expressing the GFP-ATG8a fusion were grown for 7 d on nitrogen-rich medium and then transferred to the same medium (+N), to a nitrogen-deficient medium (−N), or to a nitrogen-deficient medium containing 200 μm NaHS (−N + NaHS) for an additional 2 d (A) or 4 d (B). Root cells were visualized by confocal fluorescence microscopy and exposed to concanamycin A prior to observation, as described in “Materials and Methods.” Representative single optical sections of fluorescence, visible, and overlaid images are shown. The number of GFP-ATG8 puncta per cell section was determined using PDQuest software as described in “Materials and Methods.” Values are average numbers ± sd of fluorescent vesicles within the central vacuoles per cell of 25 to 35 cells in each of three independent experiments. Different letters indicate significant differences (P < 0.05).
To determine whether the changes in the number of autophagic bodies observed by confocal microscopy were significant, these structures were quantified using the images of 25 to 35 different cells (Fig. 1). We quantified a large number of fluorescent vesicles per root cell in nitrogen-deprived seedlings, both when they were grown for an additional 2 or 4 d, compared with those grown under nitrogen-rich conditions. The number of autophagic bodies was more than 3-fold higher in nitrogen-deficient medium than in rich medium after 4 d of growth. Moreover, the presence of exogenous sulfide produced an important decrease in the number of vesicles that was more significant during a treatment period of 4 d, reaching levels close to those observed in nitrogen-rich medium. Both the increase in autophagic bodies under nitrogen-limited conditions and their reduction by exogenous sulfide were statistically significant.
It is important to note that the nitrogen-deficient medium used in this study contained all of the sulfate salts at the same concentrations that were present in the Murashige and Skoog (MS) medium and, therefore, was a sulfate-rich medium. These results confirmed that sulfide represses autophagy in Arabidopsis independently of sulfur limitation.
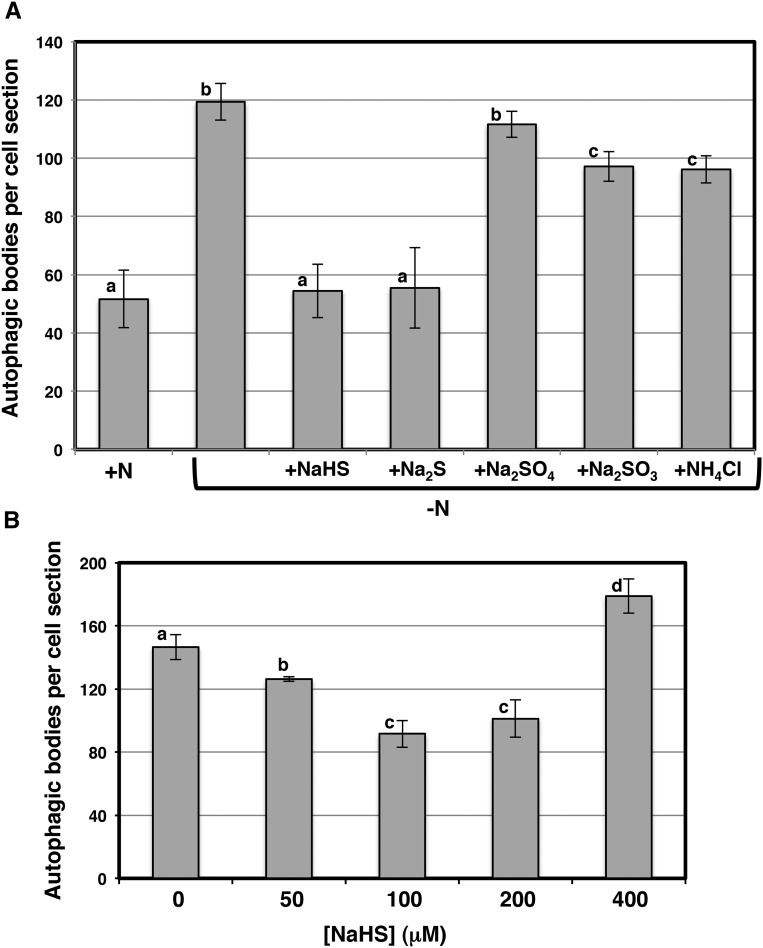
To establish that the role of sulfide in regulating autophagy was specific to this molecule and, therefore, different from that of other inorganic sulfur-containing compounds, we performed a similar nitrogen limitation experiment in the presence of different sodium salts for 4 d (Fig. 2A). As expected, the sulfate and sulfite sodium salts that did not release sulfide were unable to reverse the accumulation of autophagic bodies under nitrogen deprivation to the same extent as sulfide. However, both the Na2S and NaHS treatments produced a similar reduction in the number of autophagic bodies under nitrogen limitation. We concluded that only sulfide donor molecules are responsible for the inhibition of autophagic body accumulation induced by nitrogen deprivation in Arabidopsis roots. In addition, we included a treatment with the same concentration of ammonium and under the same nitrogen limitation conditions, and we did not observe a strong inhibition of fluorescent vesicle accumulation similar to that produced by sulfide (Fig. 2A). This result again confirmed that sulfide acts as a signaling molecule during the repression of autophagy. Furthermore, sulfide acted in a dose-dependent manner (Fig. 2B). The effects of sulfide were optimal at 100 to 200 μm NaHS; higher concentrations of the donor were less effective and even induced an increase in the accumulation of autophagic bodies. Thus, at 400 μm NaHS and nitrogen deprivation, we observed an increase of approximately 20% in the number of vesicles relative to roots under nitrogen limitation stress alone. This pattern may stem from sulfide toxicity, the generation of reactive sulfur and oxygen species after a certain concentration threshold that results in major oxidative damage, another condition that is known to induce autophagy (Xiong et al., 2007). We selected concentrations of 100 or 200 μm for further experiments to avoid any potential toxic effects on seedling growth that were not been observed in previous experiments.
Figure 2.
Effects of different chemicals (A) and concentrations (B) on the autophagy induced by nitrogen starvation in Arabidopsis roots. Wild-type seedlings expressing the GFP-ATG8a fusion were grown for 7 d on nitrogen-rich medium and then transferred for an additional 4 d to the same medium (+N), to a nitrogen-deficient medium (−N), or to a nitrogen-deficient medium containing 200 μm different salts as indicated (A) or, alternatively, to a nitrogen-deficient medium containing different concentrations of NaHS as indicated (B). Root cells were visualized by confocal fluorescence microscopy and, prior to observation, were exposed to concanamycin A as described in “Materials and Methods.” The number of autophagic bodies per cell section was determined using PDQuest software as described in “Materials and Methods.” Values are average numbers ± sd of fluorescent vesicles within the central vacuoles per cell of 25 to 35 cells in each of three independent experiments. Different letters indicate significant differences (P < 0.05). NaHS, sodium hydrosulfide. Na2S, sodium sulfide. Na2SO4, sodium sulfate. Na2SO3, sodium sulfite. NH4Cl, ammonium chloride.
Sulfide Repression of Autophagy Is Also Observed at the ATG8 Protein Level
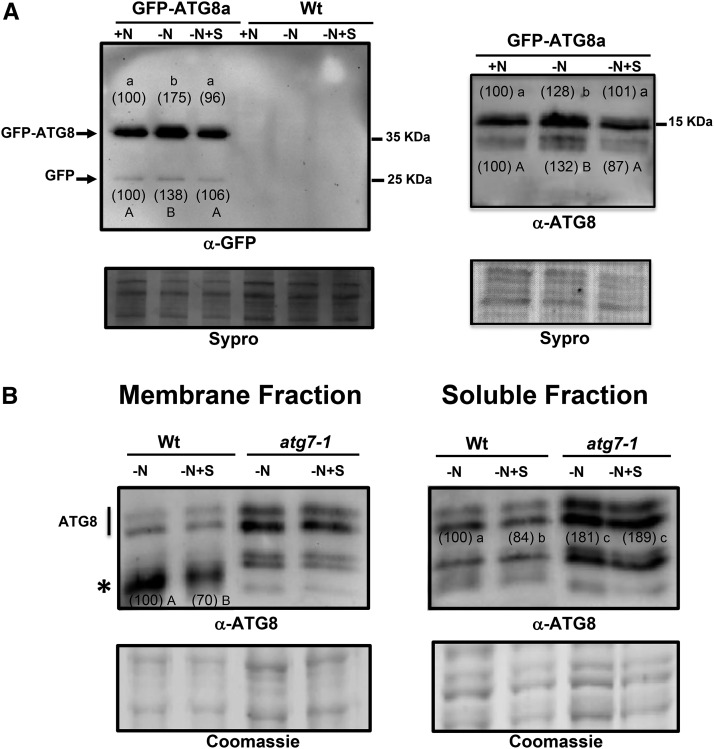
We also studied the protein profiles of ATG8 in root protein extracts prepared from seedlings that had been subjected to nitrogen limitation in the presence or absence of sulfide, using the same methods as presented above. We used polyclonal antibodies raised against the recombinant ATG8 protein from Chlamydomonas reinhardtii, a method that has been useful for the detection of ATG8 proteins in Arabidopsis (Pérez-Pérez et al., 2010; Álvarez et al., 2012b), to detect the GFP-ATG8a fusion protein as well as endogenous ATG8 proteins and used commercial anti-GFP antibodies to detect the fusion protein and the free GFP to show the autophagic flux. Total protein extracts were electrophoresed on 10% (w/v) acrylamide gels to detect the GFP-ATG8a fusion protein or, alternatively, on 15% (w/v) acrylamide gels to detect endogenous ATG8 proteins (Fig. 3). The immunoblot analysis revealed that the anti-GFP antibodies detected an intense protein band that corresponded to the fusion protein. Consistent with previous data, the ATG8 fusion protein level increased in seedling roots under nitrogen deprivation compared with those grown under nitrogen-replete conditions. The presence of sulfide in the nitrogen-deficient medium reduced the accumulation of the ATG8 fusion protein (Fig. 3A, left). This observation was reproducible in different experiments, considering the intensity of the bands relative to the total protein-loading control. The amount of each immunodetected protein band relative to the loading control was quantified in the three replicates of the experiment and the average is shown in parentheses, with a value of 100% assigned to the band corresponding to the +N sample. Multivariate ANOVA statistical analysis of the data was performed, demonstrating that the effect of sulfide on ATG8 fusion protein accumulation under nitrogen-deficient medium was statistically significant. Interestingly, the anti-GFP antibodies also detected the accumulation of free GFP, as observed previously when the same GFP-ATG8a seedlings were subjected to nitrogen starvation conditions (Chung et al., 2010; Suttangkakul et al., 2011; Li et al., 2014). This free GFP released from the fusion protein after autophagic transport into the vacuole correlates with the autophagic flux and represents another marker for the autophagy process. We have to take into account that the GFP-ATG8a transgene is under the control of the strong cauliflower mosaic virus 35S promoter and that, therefore, the ratio of the fusion protein band intensity to that of the free GFP is higher than the one expected (Shin et al., 2014). We also found that the level of free GFP accumulation decreased in the presence of sulfide in the nitrogen-deficient medium (Fig. 3A, left), reflecting a repression of the autophagic flux by this molecule. To corroborate that the free GFP bands were specific and therefore correlated with the autophagic fluxes, immunoblot analysis of wild-type seedling roots subjected to the same conditions was performed in parallel. No band was detected using the anti-GFP antibodies, excluding the possibility of nonspecific bands (Fig. 3A, left).
Figure 3.
Immunoblot analysis of GFP-ATG8a fusion and endogenous ATG8 protein accumulation in Arabidopsis roots. A, Wild-type seedlings (Wt) and wild-type seedlings expressing the GFP-ATG8a fusion were grown for 7 d on nitrogen-rich medium and then transferred to the same medium (+N), to a nitrogen-deficient medium (−N), or to a nitrogen-deficient medium containing 200 μm NaHS (−N+S) for an additional 4 d. Total protein extracts were prepared from roots as described in “Materials and Methods,” and 3 μg of each extract was resolved by 10% (w/v) SDS-PAGE and subjected to immunoblot analysis with anti-GFP antibodies (left) or 20 μg of each extract was resolved by 15% (w/v) SDS-PAGE and subjected to immunoblot analysis with anti-ATG8 (right). As the protein-loading control, before immunodetection, the membrane was stained with SYPRO Ruby. The experiment was repeated three times, and representative images are shown. The amount of each immunodetected protein band relative to the loading control was quantified in the three replicates of the experiment, and the average is shown in parentheses, with a value of 100% assigned to the band corresponding to the +N sample. B, Wild-type and atg7-1 mutant seedlings were grown for 7 d on nitrogen-rich medium and then transferred to a nitrogen-deficient medium (−N), or to a nitrogen-deficient medium containing 200 μm NaHS (−N+S) for an additional 4 d. Total protein extracts were prepared from roots and membrane, soluble fractions were separated by centrifugation as described in “Materials and Methods,” and 15 μL of each fraction was resolved by 15% (w/v) SDS-PAGE and subjected to immunoblot analysis with anti-ATG8. As the protein-loading control, a gel was run in parallel and stained with Coomassie Brilliant Blue. In the membrane fraction, the amount of the lipidated ATG8 protein band (asterisk) was quantified relative to the loading control, with a value of 100% assigned to the band corresponding to the −N sample of the wild type; in the soluble fraction, all immunodetected protein bands were quantified relative to the loading control, with a value of 100% assigned to the −N sample of the wild type. Different letters indicate significant differences (P < 0.05).
Our results indicated that the GFP-ATG8a fusion protein accumulated in nitrogen-deficient conditions and that the presence of sulfide reduced its accumulation. However, we might expect that the GFP-ATG8a protein would be degraded via autophagy and, therefore, observe a decrease in the amount of the fusion protein. The reason for this apparent contradiction is due to our experimental design in the following way. To perform SDS-PAGE with the best loading control possible, protein samples were diluted to the same concentration to ensure equal loading of the samples. However, a detailed observation of the protein concentration quantified in each extract showed that, systematically, the protein concentration of total protein extracts prepared from roots grown on nitrogen-deficient medium was lower than that of the extracts in nitrogen-rich conditions and of those in nitrogen-deficient conditions containing sulfide (Supplemental Table S1). Differences in protein concentrations also were observed when the protein extracts from the aerial part of the same seedlings were subjected to SDS-PAGE, mainly in the Rubisco large subunit protein band intensity (Supplemental Fig. S2). The net degradation of cellular proteins was already observed in tobacco (Nicotiana tabacum) cells cultured in Suc starvation and was concluded to be due to autophagy (Moriyasu and Ohsumi, 1996; Takatsuka et al., 2004). Our data again reinforce the conclusion that sulfide represses autophagy.
When immunoblot analysis was performed to specifically detect endogenous ATG8 proteins in the root protein extracts from seedlings expressing GFP-ATG8a, nearly identical results were obtained (Fig. 3A, right). Two groups of ATG8 proteins were detected using the anti-Cr-ATG8 antibodies that should correspond to the unmodified ATG8 protein forms and the conjugated ATG8-Phosphatidylethanolamine (PE) proteins, as observed previously in Arabidopsis (Phillips et al., 2008; Chung et al., 2010; Álvarez et al., 2012b). To confirm that the bands with faster mobility represented ATG8-PE adducts, we performed membrane fractionation on both the wild-type and atg7-1 backgrounds. In the atg7-2 mutant, ATG8 lipidation is blocked and, thus, the ATG8-PE adducts are absent (Chung et al., 2010; Suttangkakul et al., 2011). By centrifugal separation of protein extracts prepared from wild-type and atg7-1 seedlings that had been subjected to nitrogen limitation in the presence or absence of sulfide, we obtained membrane and soluble protein fractions that were subjected to immunoblot analysis (Fig. 3B). When the protein profiles were compared, we observed protein bands with faster mobility enriched in the membrane fraction of the wild type that were absent in the atg7-1 background (Fig. 3B, asterisk). Moreover, consistent with these proteins representing the ATG8-PE adducts, they were absent in both the wild-type and the atg7-1 soluble protein fractions. Other protein bands observed in the membrane fractions correspond to contamination of the soluble fractions. The SDS-PAGE profiles also showed the reversal of the accumulation of both types of ATG8 protein, in the soluble fraction and in the membrane fraction of the wild type, in nitrogen-limited root seedlings when exogenous sulfide was present in the medium. Curiously, the sulfide reduction of ATG8 protein accumulation was not observed in atg7-1 fractions. Collectively, these results provide evidence that sulfide regulates autophagy also under conditions of nitrogen deprivation. Our results confirmed this role at the cellular and protein levels.
The Mechanism of Autophagy Reversion via Sulfide Is Not Dependent on Its Antioxidant Activity
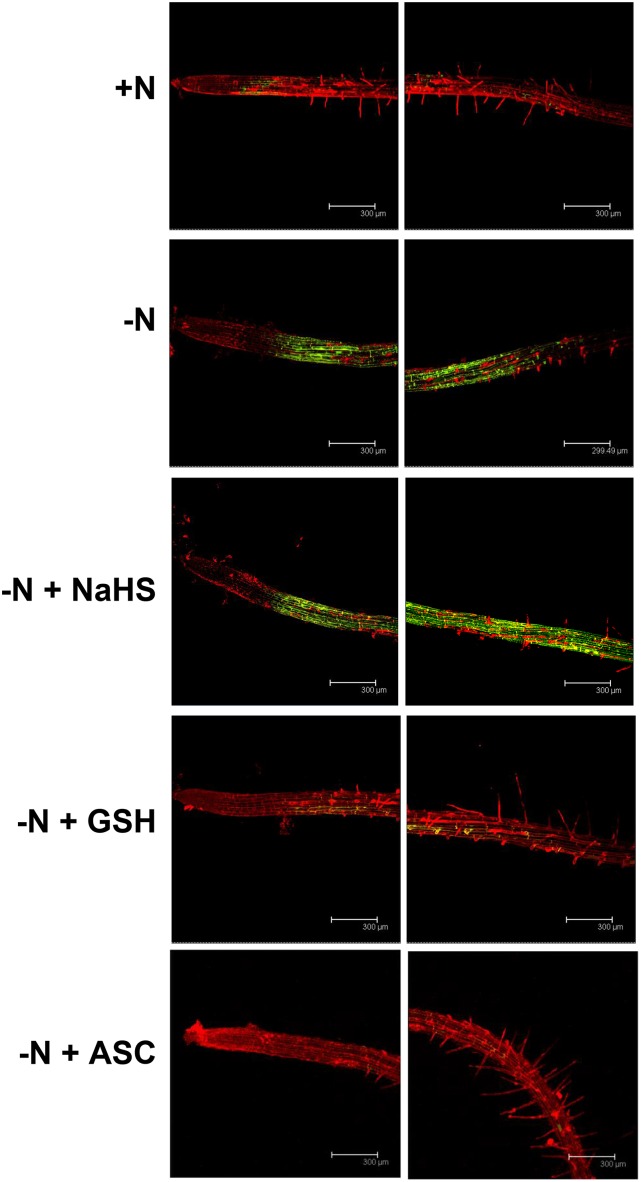
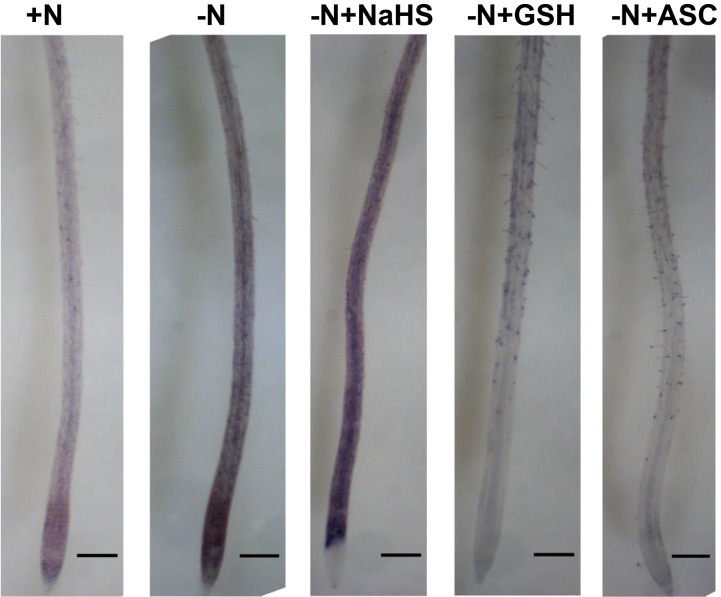
Several reports have suggested that exogenous hydrogen sulfide increases the antioxidant capability of plant cells, thereby alleviating oxidative damage induced by plant stresses. This finding has led researchers to question whether sulfide functions as a signaling molecule (Hancock and Whiteman, 2014). Oxidative stress has been shown to induce autophagy (Xiong et al., 2007; Pérez-Pérez et al., 2012a; Pérez-Martín et al., 2014), especially under conditions of nutrient limitation, in which the induction of autophagy has been suggested to involve reactive oxygen species (ROS; Liu et al., 2009). To determine whether the effect of sulfide on the induction of autophagy was mediated by H2O2 scavenging, we performed the same nitrogen deprivation experiment in the presence of sulfide or one of the established antioxidants glutathione and ascorbate (Foyer and Noctor, 2011), except that the roots were specifically stained for H2O2 (Fig. 4). After 4 d of growth on nitrogen-deficient medium, we clearly observed an increase in fluorescence emission (pseudocolored in green) in the roots of the seedlings that resulted from oxidation of the nonfluorescent H2DCFDA to the fluorescent product. This result suggested that H2O2 was being produced at high rates. In contrast, the fluorescence emission was nearly undetectable in roots from plants grown under nitrogen-rich conditions. Interestingly, the presence of 200 μm NaHS in the nitrogen-deficient growth medium did not have any effect on the intensity of the fluorescence emission. A similar signal was observed compared with roots grown under conditions of nitrogen starvation. Furthermore, when the nitrogen-deficient medium was supplemented with either 200 μm reduced glutathione or ascorbate, a decrease in the fluorescent signal was observed, reaching the same signal intensity as observed under nitrogen-sufficient conditions, as expected for well-known antioxidants. These findings showed that hydrogen sulfide does not behave as an H2O2 scavenger in our conditions. We cannot exclude that other ROS different from H2O2 might mediate the repressive effect of sulfide. Such is the case of the superoxide radical anion, whose function in root development is well established (Foreman et al., 2003). Consequently, superoxide production was visualized by nitroblue tetrazolium (NBT) staining of the same root tissues, and basically the same results were obtained (Fig. 5). The dark-blue stain due to the superoxide was observed most intensively and extensively distributed in roots after 4 d of growth on nitrogen-deficient medium alone and in the presence of sulfide by comparison with the other studied conditions, mainly in the root differentiation zone used for monitoring GFP-ATG8.
Figure 4.
Detection of H2O2 in Arabidopsis roots. Wild-type seedlings were grown for 7 d on nitrogen-rich medium and then transferred to the same medium (+N), to a nitrogen-deficient medium (−N), or to a nitrogen-deficient medium containing 200 μm NaHS (−N + NaHS), glutathione (−N + GSH), or ascorbate (−N + ASC) for an additional 4 d. The roots were loaded with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) for 5 min to detect H2O2 (pseudocolored in green) in the presence of propidium iodide to visualize cell walls (pseudocolored in red). Representative images of two different zones of the root for each sample are shown.
Figure 5.
Detection of superoxide in Arabidopsis roots. Wild-type seedlings were grown for 7 d on nitrogen-rich medium and then transferred to the same medium (+N), to a nitrogen-deficient medium (−N), or to a nitrogen-deficient medium containing 200 μm NaHS (−N+NaHS), glutathione (−N+GSH), or ascorbate (−N+ASC) for an additional 4 d. The roots were stained with NBT as described in “Materials and Methods.” Representative images are shown. Bars = 250 μm.
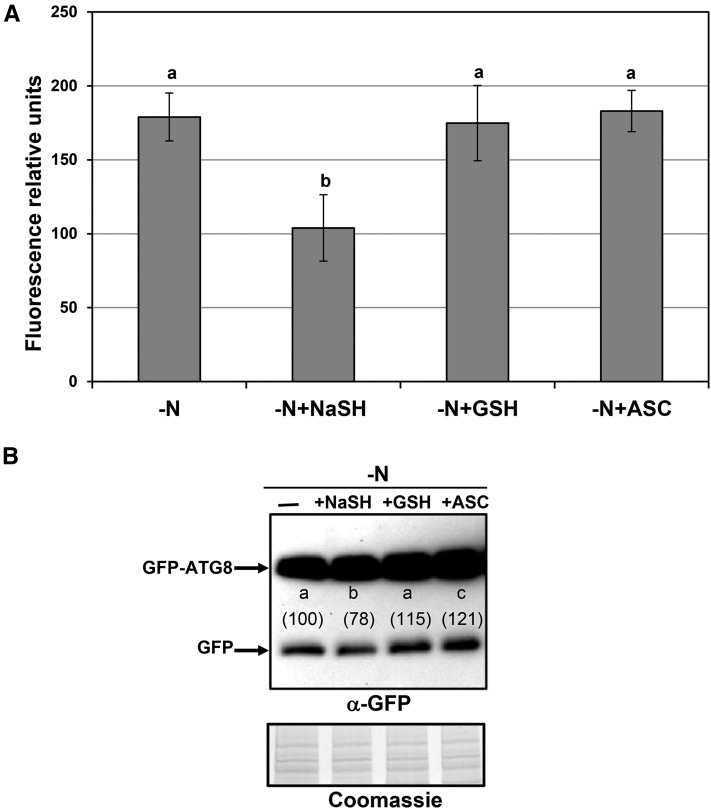
We further analyzed the effects of both glutathione and ascorbate on the increased accumulation of autophagic bodies in response to nitrogen limitation in plants expressing the GFP-ATG8a fusion protein (Fig. 6). Only the presence of NaHS in the medium significantly inhibited the accumulation of fluorescent vesicles, while glutathione and ascorbate were unable to produce any effect (Fig. 6A). Representative single optical sections of the images visualized by confocal microscopy are shown in Supplemental Figure S3. Similar results also were obtained when immunoblot analysis was performed. The presence of sulfide and no glutathione and ascorbate significantly decreased the accumulation of the free GFP level, indicative of a repression of the autophagic flux by sulfide (Fig. 6B). These data clearly suggested that the effect of sulfide on the progression of autophagy is independent of ROS. In a previous work, however, a reduction of punctate structures was observed in the presence of antioxidants (Liu et al., 2009). Differences in methodological procedures, antioxidant concentrations, and timing of the treatments make both investigations incomparable. In this work, our antioxidant treatment was performed in solid medium at 200 μm for 4 d in nitrogen-deficient conditions, exactly the same conditions used to check the effect of sulfide as a possible antioxidant. On the contrary, in the previous work (Liu et al., 2009), the treatment was performed in liquid nitrogen-rich medium with the antioxidant at 20 mm in one case (imidazole) and 2 mm in other case (ascorbate) for 4 h.
Figure 6.
Effects of exogenous sulfide, reduced glutathione, and ascorbate on the autophagy induced by nitrogen deprivation in Arabidopsis roots. Wild-type seedlings expressing the GFP-ATG8a fusion were grown for 7 d on nitrogen-rich medium and then transferred to a nitrogen-deficient medium (−N), or to a nitrogen-deficient medium containing 200 μm NaHS (−N+NaHS), glutathione (−N+GSH), or ascorbate (−N+ASC) for an additional 4 d. A, Root cells were visualized by confocal fluorescence microscopy and, prior to observation, were exposed to concanamycin A as described in “Materials and Methods.” The mean fluorescence inside the vacuoles was determined using ImageJ software. Values are expressed as relative units ± sd of mean fluorescence within the central vacuoles of three independent experiments. Representative single optical section of fluorescence, visible, and overlaid images are shown in Supplemental Figure S3. B, Immunoblot analysis of the GFP-ATG8a fusion protein. Total protein extracts were prepared from roots as described in “Materials and Methods,” and 8 μg of each extract was resolved by 10% SDS-PAGE and subjected to immunoblot analysis with anti-GFP antibodies. As the protein-loading control, a gel was run in parallel and stained with Coomassie Brilliant Blue. The experiment was repeated three times, and representative images are shown. The amount of the free GFP protein band relative to the loading control was quantified, and average values are shown in parentheses, with a value of 100% assigned to the band corresponding to the −N sample. Different letters indicate significant differences (P < 0.05).
Correlation between Autophagy and DES1
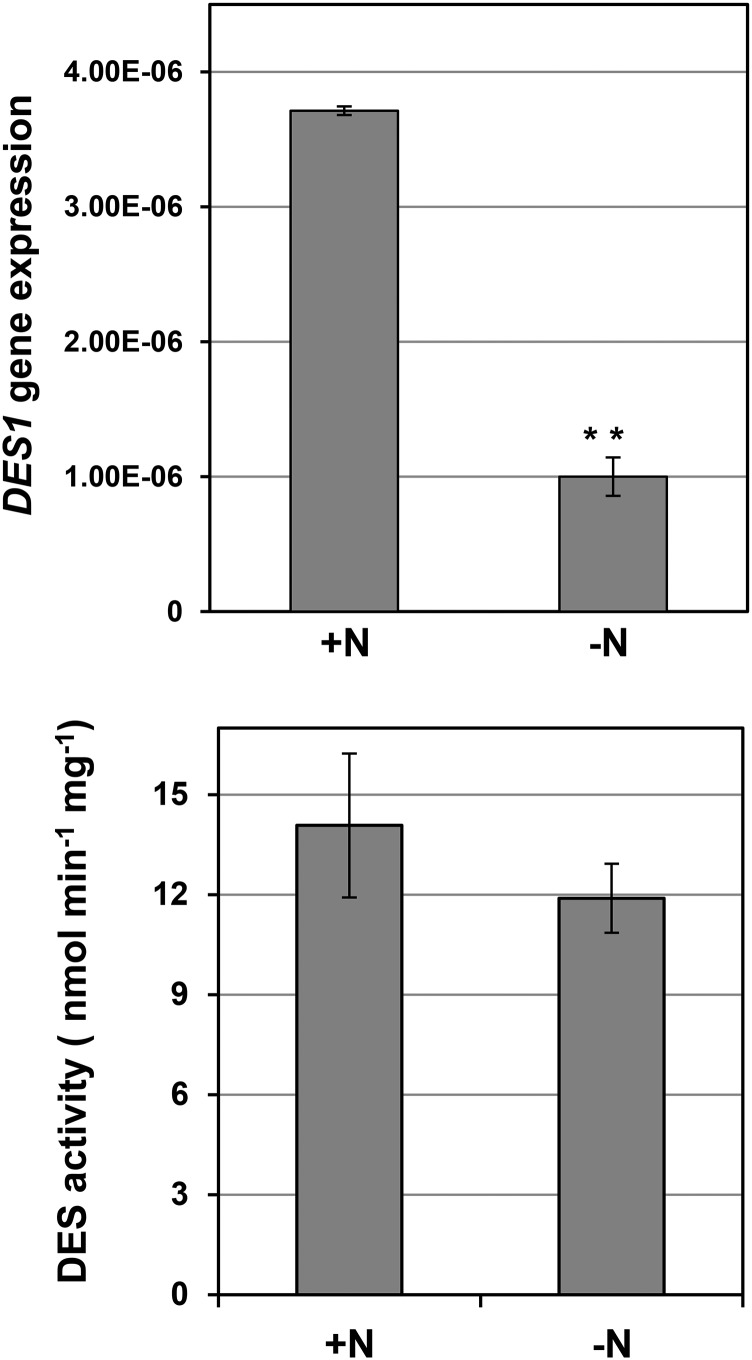
Previously, we demonstrated that DES1 deficiency leads to the accumulation and lipidation of ATG8 proteins. Through genetic complementation or exogenous application of sulfide, we were able to rescue autophagy induction in des1 null mutants (Álvarez et al., 2012b). Consequently, we suggested that DES1 should modulate the generation of sulfide in the plant cytosol for autophagy signaling (Gotor et al., 2013; Romero et al., 2013). In addition, we wanted to determine whether stressful conditions that induce autophagy, such as nutrient limitation, had any effect on DES1. To test this hypothesis, we performed the same experiment as described above and compared seedlings grown under nitrogen-rich and nitrogen-deficient conditions in terms of DES1 gene expression and DES enzyme activity (Fig. 7). A strong reduction (73%) in DES1 gene expression was observed in the roots of seedlings grown in nitrogen-deficient medium compared with those grown in nitrogen-replete medium. The total DES activity was also decreased slightly when the seedlings were transferred to nitrogen-deprived conditions, but this reduction was not statistically significant. Other enzymes with hydrogen sulfide-releasing activity, which is the method used to measure DES activity, are present in plants; therefore, the contribution of cytosolic DES1 protein to total DES activity is expected to be low.
Figure 7.
Effects of nitrogen starvation on DES1 gene expression and DES activity in Arabidopsis roots. Wild-type seedlings were grown for 7 d on nitrogen-rich medium and then transferred to the same medium (+N) or to a nitrogen-deficient medium (−N) for an additional 4 d. Root samples were collected and used for quantitative real-time reverse transcription (RT)-PCR to analyze the expression of the DES1 gene and to measure DES activity as described in “Materials and Methods.” Values are means ± sd of three independent experiments. **, P < 0.01.
The Effect of Sulfide Is Observable at the Phenotypic Level
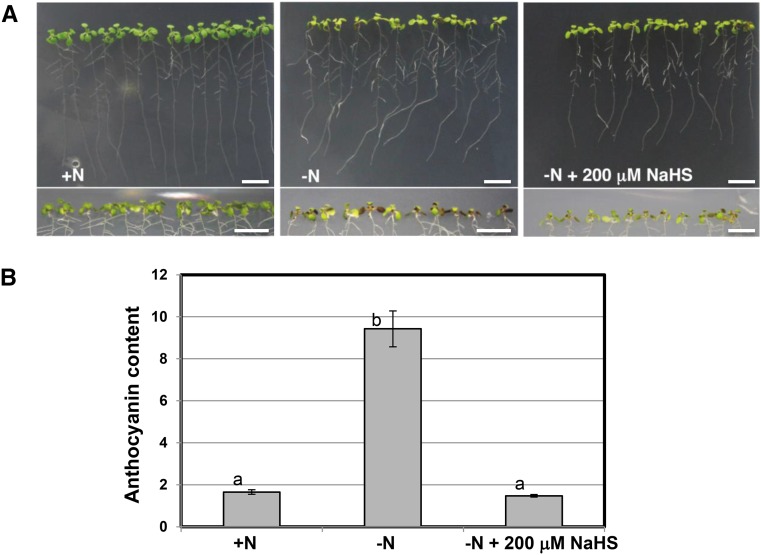
To establish that particular concentrations of sulfide molecule donors did not have a toxic effect on seedling growth, we studied the phenotypic characteristics of seedlings expressing GFP-ATG8a and grown in the different conditions described above. When the seedlings were transferred from nitrogen-rich to nitrogen-deficient medium, a decrease in shoot and root biomass was evident at the beginning of the transfer period. After 4 d, a change in leaf color was appreciable in the nitrogen-deficient medium and reflected the repression of chlorophyll synthesis and the induction of anthocyanin synthesis, which are characteristic plant responses to nitrogen deprivation (Scheible et al., 2004; Peng et al., 2007). In contrast, the presence of NaHS apparently reduced the extent of browning, which was observable primarily in the abaxial portion of the leaves, resulting in a healthier phenotype (Fig. 8A). The level of anthocyanin was approximately 6-fold higher in the nitrogen-starved seedlings than in the nitrogen-sufficient seedlings. The presence of NaHS resulted in identical (or even slightly lower) anthocyanin content compared with that in the nitrogen-sufficient seedlings (Fig. 8B). Furthermore, the presence of sulfide in the nitrogen-limited medium was effective in alleviating the stress induced by the absence of a nitrogen source during the longer treatment periods (6 and 8 d). During these long periods of nitrogen deprivation, the application of lower concentrations of sulfide and the use of NaHS as a sulfide donor were more effective for reducing stress (Supplemental Figs. S4 and S5). Therefore, we conclude that hydrogen sulfide can play important roles in plants, with phenotypically observable outcomes.
Figure 8.
Phenotypes of wild-type seedlings expressing GFP-ATG8a under different conditions. Wild-type seedlings expressing the GFP-ATG8a fusion were grown for 7 d on nitrogen-rich medium and then transferred to the same medium (+N), to a nitrogen-deficient medium (−N), or to a nitrogen-deficient medium containing 200 μm NaHS for an additional 4 d. A, Representative bright-field images of whole seedlings. The bottom images show the undersides of the leaves (abaxial part of the leaves). Bars = 1 cm. B, Anthocyanin content per gram fresh weight was measured as described in “Materials and Methods.” Values are means ± sd of three independent experiments. Different letters indicate significant differences (P < 0.01).
DISCUSSION
The conceptualization of the role of hydrogen sulfide in both animal and plant systems has changed dramatically in recent years. Currently, hydrogen sulfide is viewed not as a toxic molecule to life but, rather, as a regulator of essential life processes. In mammals, hydrogen sulfide has been considered to be a gasotransmitter with major physiological functions in different body systems (Gadalla and Snyder, 2010). Alterations of hydrogen sulfide metabolism have important pathological consequences, establishing the clinical relevance of this molecule (Łowicka and Bełtowski, 2007; Szabó, 2007; Wang, 2012). In addition to the protective effects of hydrogen sulfide against a wide array of different stresses that have been demonstrated in plants (Zhang et al., 2008, 2010; Wang et al., 2010; Jin et al., 2011; Dawood et al., 2012; Li et al., 2012a, 2012b; Cheng et al., 2013; Christou et al., 2013; Sun et al., 2013), this molecule has also been shown to regulate plant processes that are critical for plant performance, such as photosynthesis (Chen et al., 2011), stomatal movement (García-Mata and Lamattina, 2010; Lisjak et al., 2010; Scuffi et al., 2014), and autophagy (Álvarez et al., 2012b; Gotor et al., 2013). In this study, we provide new experimental evidence that, together with our previous findings, demonstrates the role of hydrogen sulfide in regulating the progression of autophagy independently of the condition in which the autophagy originates. Most importantly, our findings contribute to our understanding of the mechanism underlying the regulation of autophagy by sulfide in plant systems by confirming that its role is not dependent on the redox state.
As in other organisms, autophagy is constitutively active in plant cells under favorable growth conditions (Sláviková et al., 2005). However, when plants are exposed to adverse environmental conditions, one of their responses to cope with stress and survive is to increase the induction of autophagy. These stresses include nitrogen and carbon deprivation. Several studies have reported that autophagy can be induced by carbon and nitrogen deprivation, which is detected in many cases through a substantial increase in ATG8 protein levels (Doelling et al., 2002; Hanaoka et al., 2002; Thompson et al., 2005; Xiong et al., 2005; Chung et al., 2009; Guiboileau et al., 2013). In this study, we observed basal levels of autophagy under nitrogen-sufficient conditions. However, under stressful growth conditions, increased levels of autophagy were observed under nitrogen deprivation. Interestingly, the presence of sulfide during stress prevented an increase in the activation of autophagy, revealing the same level of basal autophagy as observed in the favorable growth conditions. The observed effect of sulfide was unrelated to its availability as a nutrient, as the growth medium contained sulfate salts as a source of sulfur nutrients. Furthermore, the presence of ammonium at the same concentration did not alter the activation of autophagy.
This study was performed in root cells from young seedlings overexpressing GFP-ATG8a under nitrogen deprivation using GFP fluorescence detection to monitor autophagy. Identical effects of sulfide on autophagy have been observed previously in different tissue (leaves), at different plant developmental stages (mature plants), under different stress conditions (carbon starvation), and in different genetic backgrounds (des1 mutant; Álvarez et al., 2012b). Therefore, the evidence indicates that sulfide regulates the progression of autophagy by acting as a repressor in plant systems. In mammals, the effect of sulfide on autophagy is unclear, because recent findings have indicated that sulfide can both suppress and induce autophagy (Wang et al., 2012a; Kundu et al., 2014). Recently, the sulfur amino acids Cys and Met have been shown to regulate the translational capacity and metabolic homeostasis in eukaryotic cells and to inhibit autophagy and promote cell growth (Laxman et al., 2013; Sutter et al., 2013). In the latter report, the importance was placed on Met as a precursor of the methyl donor S-adenosyl-Met, which has been reported to inhibit autophagy through the action of methyltransferase enzymes, although Cys also has been shown to suppress autophagy.
Deciphering the mechanism underlying the transformation of the sulfide signal into a biological response and identifying the molecular targets of sulfide remain challenging. It has been shown previously that the hydrogen sulfide anion can generate Cys and glutathione persulfides with antioxidant properties that are superior to glutathione (Francoleon et al., 2011; Ida et al., 2014). The persulfides have been shown to act as reactive reductants that are capable of reacting rapidly with the electrophilic H2O2; therefore, persulfides exhibit H2O2-scavenging activity. Moreover, several studies in photosynthetic organisms have demonstrated the activation of autophagy in response to several conditions that increase ROS generation, suggesting the presence of a strong link between autophagy and ROS (Pérez-Pérez et al., 2012b). Thus, we sought to determine whether the mechanism underlying the repression of autophagy by sulfide involves some ROS, such as H2O2 and superoxide radical, the latter with well-established functions in roots. Our results showed that sulfide was unable to scavenge the ROS generated by nitrogen limitation, as observed for reduced glutathione and ascorbate. Moreover, upon nitrogen limitation, we clearly differentiated the inhibitory effect of sulfide, both on the accumulation of autophagic bodies visualized by GFP fluorescence and on the accumulation of ATG8 protein forms detected by immunoblotting, from the effect produced by reduced glutathione and ascorbate, which were unable to reverse to any extent the progression of autophagy. Therefore, our results suggest that the effect of sulfide on the progress of autophagy is independent of its capacity to react with H2O2 or with superoxide radical anion.
The second mechanism proposed in animal systems to explain the physiological effects of hydrogen sulfide is the posttranslational modification of proteins known as S-sulfhydration (Paul and Snyder, 2012). Previous studies have indicated that this modification can be regulated by competition between the nitrosylation and sulfhydration of the same Cys residues. S-Sulfhydration changes an –SH to an –SSH, modifying the chemical reactivity of enzymes and possibly their access to their respective targets, as observed for glyceraldehyde-3-phosphate dehydrogenase (Gadalla and Snyder, 2010; Kabil and Banerjee, 2010; Paul and Snyder, 2012). Very recently, S-sulfhydration of proteins was confirmed to occur in Arabidopsis leaf tissues under physiological conditions, and the presence of an S-sulfhydrated Cys residue in a plant protein was demonstrated recently for the first time (Aroca et al., 2015). Therefore, two questions arise: whether S-sulfhydration regulates autophagy, and what are the specific targets of sulfide. The thiol redox state can profoundly influence autophagy, especially during the initiation and completion of autophagosomes, due to the ability of several proteins that are involved in autophagy to sense alterations in the cellular redox state by means of reactive Cys residues (Filomeni et al., 2010). Examples of this are the ubiquitin-like systems ATG7-ATG10 and ATG7-ATG3, the functions of which rely on the Cys-based transfer of ATG12 and ATG8, respectively, which are essential proteins for autophagosome membrane elongation (Filomeni et al., 2010). Moreover, redox regulation of the Cys protease ATG4 has been characterized in detail in mammalian systems, and a Cys residue located near the catalytic site has been found to be critical for this regulation (Scherz-Shouval et al., 2007). The ATG4 protein cleaves ATG8 near the C terminus, downstream of a conserved Gly, allowing the conjugation to PE to proceed through the exposed Gly. ATG4 also plays a role in the deconjugation of ATG8-PE to release ATG8 from the autophagosome membrane. Because of its role as both a conjugating and deconjugating enzyme, the activity of ATG4 is expected to be tightly regulated. A detailed study of the ATG8-processing activity by two ATG4 isoforms from Arabidopsis was reported recently (Woo et al., 2014). That study demonstrated a dose-dependent inhibition of Arabidopsis ATG4 Cys protease activity under oxidative stress that was restored by dithiothreitol, indicating that this plant protein is also under redox regulation similar to the mammalian protein. In addition, a study of yeast ATG4 demonstrated that this protein is regulated by the oxidoreduction of a single disulfide bond that involves thioredoxin (Pérez-Pérez et al., 2014). Thus, both ubiquitin-like system proteins and the ATG4 protein could be specific targets of sulfide, and further studies are currently being developed.
In this work, we provide more data that, together with previous studies, highlight the role of the DES1 protein in autophagy. Not only does the deficiency in DES1 promote the induction of autophagy (Álvarez et al., 2012b), but a condition that induces autophagy also promotes the repression of DES1. Further research will be required to determine whether this reverse correlation can be generalized to other situations in which autophagy is activated and to ascertain the underlying regulatory mechanisms. In addition, this study, like previous ones, demonstrates that hydrogen sulfide plays an important signaling role in plants. This effect is observable phenotypically and is measurable by anthocyanin content, because the presence of sulfide alleviated nitrogen limitation stress. When hydrogen sulfide is present below a toxic threshold, it plays a general role in improving plant performance. This finding is corroborated by a recent report that found that exposing plants to extremely low concentrations of sulfide results in significant increases in biomass (Dooley et al., 2013).
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Treatments
The Arabidopsis (Arabidopsis thaliana) wild-type plants expressing GFP-ATG8a used in this work have been described previously (Thompson et al., 2005), and the atg7-1 mutant was obtained from the Nottingham Arabidopsis Stock Centre. Arabidopsis seeds were sown on MS solid medium containing 0.8% (w/v) agar and synchronized at 4°C for 2 d. The plates were then incubated vertically in a growth chamber under a regime of 16 h of light at 22°C/8 h of dark at 20°C. For exposure to nitrogen-starvation conditions, nitrogen-deficient MS medium was prepared by replacing the nitrate salts with chloride salts, without altering the sulfate salts. One-week-old seedlings were transferred to nitrogen-deficient MS solid medium for an additional 2 or 4 d of growth. Alternatively, the 1-week-old seedlings were transferred to the same nitrogen-deficient MS medium containing 200 or 100 μm Na2S or NaHS as the source of exogenous sulfide. For concanamycin A treatment, seedlings were incubated in liquid nitrogen-deficient MS medium containing 0.5 μm concanamycin A (Santa Cruz Biotechnology) for 16 h at room temperature under agitation at 80 rpm. After the treatment, the roots were washed in water, and the differentiation zone was observed using a confocal microscope. Concanamycin A was prepared as a 100 μm stock solution in dimethyl sulfoxide. To ensure that dimethyl sulfoxide did not produce any effect on the results of the experiments, the seedlings also were incubated in liquid nitrogen-deficient MS medium in the presence of dimethyl sulfoxide alone as a control.
Visualization and Quantification of GFP-Tagged Autophagic Bodies
Root cells were viewed using a TCS SP2 spectral confocal microscope (Leica Microsystems). GFP was excited using the 488-nm line of an argon ion laser, and emission was detected between 510 and 580 nm. Single optical images were processed with PDQuest software (Bio-Rad) as follows. First, a black-and-white image was automatically generated from the original image, and a minimal intensity and size of the spot were defined. Second, a new image was generated with the selected spots circled in red, a manual correction was performed to adjust the selected spots to the cells to be counted, and a file with automatic counting was obtained (Supplemental Fig. S6). The number of fluorescent vesicles within the central vacuoles was counted in 25 to 35 different cells. The data represent average numbers ± sd from three independent experiments. Images were also processed using ImageJ software. In the latter, the mean fluorescence within the vacuoles was counted within a 400-μm2 section from the same cells. The results, when expressed as a percentage relative to the value obtained in the nitrogen-rich medium, were the same using both software programs.
Immunoblot Analysis
Plant root material (20–100 mg) was ground in liquid nitrogen with 100 to 400 μL of extraction buffer (100 mm Tris-HCl, pH 7.5, 400 mm Suc, 1 mm EDTA, 10 mg mL−1 sodium deoxycholate, 0.1 mm phenylmethylsulfonyl fluoride, 10 mg mL−1 pepstatin A, and 4% [v/v] protease inhibitor cocktail [Roche]) using a pestle and mortar and centrifuged at 500g for 10 min to obtain the supernatant fraction as described previously (Yoshimoto et al., 2004; Álvarez et al., 2012b). The total amount of protein in the resulting supernatant was determined using a previously described method (Bradford, 1976). For subcellular fractionation, the supernatant was centrifuged at 13,000g for 15 min to generate the membrane fraction and the supernatant (Yoshimoto et al., 2004). For the immunoblot analyses, 3 μg of root protein extracts was electrophoresed on 10% (w/v) acrylamide gels or, alternatively, 20 μg of root protein extracts was electrophoresed on 15% (w/v) acrylamide gels before transfer to polyvinylidene fluoride membranes (Bio-Rad) according to the manufacturer’s instructions. Anti-Cr-ATG8 (Pérez-Pérez et al., 2010; Álvarez et al., 2012b), anti-GFP (eBioscience), and secondary antibodies were diluted 1:2,000, 1:1,000, and 1:50,000, respectively, in phosphate-buffered saline containing 0.1% (v/v) Tween 20 (Sigma-Aldrich) and 5% (w/v) milk powder. The ECL Select Western Blotting Detection Reaction (GE Healthcare) was used to detect the proteins with horseradish peroxidase-conjugated anti-rabbit secondary antibodies. For a protein-loading control, an identical gel was run in parallel and stained with Coomassie Brilliant Blue (Sigma) or, alternatively, the membrane before immunodetection was stained with SYPRO Ruby (Life Technologies) to detect all protein bands. The immunodetected protein bands were quantified relative to the Coomassie Brilliant Blue-stained gel or the SYPRO Ruby-stained membrane using Quantity One software (Bio-Rad).
Detection of ROS
For the fluorimetric detection of H2O2, roots were incubated for 5 min with 10 mm H2DCFDA (Life Technologies) in the presence of 10 mm propidium iodide (Life Technologies) to visualize the cell walls. The samples were observed using a TCS SP2 spectral confocal microscope (Leica Microsystems) with the following settings: excitation, 488 nm; emission, 500 to 550 nm for fluorescein detection and 600 to 650 nm for propidium iodide detection. For detection of the superoxide radical anion, roots were stained with NBT chloride (Sigma-Aldrich) as described previously (García et al., 2010). The seedlings were incubated in 0.1 m Tris-HCl, 0.1 m NaCl, 0.05 m MgCl2, and 0.5 mg mL−1 NBT (pH 9.5) for 2 h at room temperature in the dark. After rinsing, the roots were observed with an Olympus SZ-PT stereoscope equipped with a DFC300FX Leica camera.
Real-Time RT-PCR
Quantitative real-time RT-PCR was used to analyze the expression of the DES1 gene as described previously (Laureano-Marín et al., 2014). The primer sequences used were as follows: qDES1-F, 5′-TCGAGTCAGTCAGATATGAAGCT-3′, and qDES1-R, 5′-TGTAACCTTGGTACCAACATCTCT-3′, for the DES1 gene; qUBQ-F, 5′-GGCCTTGTATAATCCCTGATGAATAAG-3′, and qUBQ-R, 5′-AAAGAGATAACAGGAACGGAAACATAGT-3′, for the constitutive UBIQUITIN10 (UBQ10) gene. The DES1 expression levels were normalized to that of the constitutive UBQ10 gene by subtracting the cycle threshold value of UBQ10 from the cycle threshold value of the DES1 gene. The results shown are means ± sd of at least three independent RNA samples.
Determination of DES Activity
Plant root material was ground in 20 mm Tris-HCl (pH 8) using a mortar and pestle with liquid nitrogen. After centrifugation at 15,000g for 15 min at 4°C, the resulting supernatant was used as a plant soluble extract for DES activity. The total amount of protein in the extracts was determined by the Bradford method using the Bio-Rad protein assay. DES activity was measured by the release of sulfide from l-Cys as described previously (Álvarez et al., 2010).
Determination of Anthocyanin Content
Approximately 40 to 70 mg of aerial tissues from seedlings was homogenized in 1 mL of propanol:HCl:water (18:1:81) and further extracted in a boiling-water bath for 3 min. The mixture was centrifuged at 5,000g for 40 min. The absorbance of the supernatant was measured at 535 and 650 nm, and the total anthocyanin content per gram fresh weight was determined using previously described methods (Lange et al., 1971).
Statistical Analysis
Multivariate ANOVA statistical analysis of the data was performed using the program Statgraphics Centurion.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At5g28030.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Autophagy induced by nitrogen deprivation in Arabidopsis roots.
Supplemental Figure S2. Representative SDS-PAGE of protein extracts.
Supplemental Figure S3. Representative single optical simages corresponding to the experiment described in Figure 5.
Supplemental Figure S4. Phenotypes of wild type seedlings expressing GFP-ATG8a under different conditions grown for 6 d.
Supplemental Figure S5. Phenotypes of wild type seedlings expressing GFP-ATG8a under different conditions grown for 8 d.
Supplemental Figure S6. Image processing with PDQuest software (Bio-Rad).
Supplemental Table S1. Protein concentration of root extracts.
Supplementary Material
Acknowledgments
We thank Dr. Irene García for critical reading of the article, Dr. Alicia Orea for confocal microscopy service, Dr. José Luis Crespo for providing the anti-ATG8 antibodies, and Dr. Richard Vierstra for providing seeds of wild-type Arabidopsis expressing GFP-ATG8a.
Glossary
- NO
nitric oxide
- H2O2
hydrogen peroxide
- MS
Murashige and Skoog
- ROS
reactive oxygen species
- H2DCFDA
2′,7′-dichlorodihydrofluorescein diacetate
- NBT
nitroblue tetrazolium
- RT
reverse transcription
Footnotes
This work was supported in part by the European Regional Development Fund through the Ministerio de Economia y Competitividad (grant no. BIO2013–44648–P and fellowship support to A.M.L.-M. through the Formación de Personal Investigador program).
References
- Álvarez C, Bermúdez MA, Romero LC, Gotor C, García I (2012a) Cysteine homeostasis plays an essential role in plant immunity. New Phytol 193: 165–177 [DOI] [PubMed] [Google Scholar]
- Álvarez C, Calo L, Romero LC, García I, Gotor C (2010) An O-acetylserine(thiol)lyase homolog with l-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol 152: 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, García I, Moreno I, Pérez-Pérez ME, Crespo JL, Romero LC, Gotor C (2012b) Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 24: 4621–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca Á, Serna A, Gotor C, Romero LC (2015) S-Sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol 168: 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC. (2007) Plant autophagy: more than a starvation response. Curr Opin Plant Biol 10: 587–593 [DOI] [PubMed] [Google Scholar]
- Bassham DC. (2015) Methods for analysis of autophagy in plants. Methods 75: 181–188 [DOI] [PubMed] [Google Scholar]
- Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K (2006) Autophagy in development and stress responses of plants. Autophagy 2: 2–11 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chen J, Wu FH, Wang WH, Zheng CJ, Lin GH, Dong XJ, He JX, Pei ZM, Zheng HL (2011) Hydrogen sulphide enhances photosynthesis through promoting chloroplast biogenesis, photosynthetic enzyme expression, and thiol redox modification in Spinacia oleracea seedlings. J Exp Bot 62: 4481–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Zhang L, Jiao C, Su M, Yang T, Zhou L, Peng R, Wang R, Wang C (2013) Hydrogen sulfide alleviates hypoxia-induced root tip death in Pisum sativum. Plant Physiol Biochem 70: 278–286 [DOI] [PubMed] [Google Scholar]
- Christou A, Manganaris GA, Papadopoulos I, Fotopoulos V (2013) Hydrogen sulfide induces systemic tolerance to salinity and non-ionic osmotic stress in strawberry plants through modification of reactive species biosynthesis and transcriptional regulation of multiple defence pathways. J Exp Bot 64: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung T, Suttangkakul A, Vierstra RD (2009) The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC (2005) Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Dawood M, Cao F, Jahangir MM, Zhang G, Wu F (2012) Alleviation of aluminum toxicity by hydrogen sulfide is related to elevated ATPase, and suppressed aluminum uptake and oxidative stress in barley. J Hazard Mater 209-210: 121–128 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Dooley FD, Nair SP, Ward PD (2013) Increased growth and germination success in plants following hydrogen sulfide administration. PLoS ONE 8: e62048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomeni G, Desideri E, Cardaci S, Rotilio G, Ciriolo MR (2010) Under the ROS…thiol network is the principal suspect for autophagy commitment. Autophagy 6: 999–1005 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoleon NE, Carrington SJ, Fukuto JM (2011) The reaction of H2S with oxidized thiols: generation of persulfides and implications to H2S biology. Arch Biochem Biophys 516: 146–153 [DOI] [PubMed] [Google Scholar]
- Fukuto JM, Carrington SJ, Tantillo DJ, Harrison JG, Ignarro LJ, Freeman BA, Chen A, Wink DA (2012) Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem Res Toxicol 25: 769–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla MM, Snyder SH (2010) Hydrogen sulfide as a gasotransmitter. J Neurochem 113: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García I, Castellano JM, Vioque B, Solano R, Gotor C, Romero LC (2010) Mitochondrial β-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana. Plant Cell 22: 3268–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2010) Hydrogen sulphide, a novel gasotransmitter involved in guard cell signalling. New Phytol 188: 977–984 [DOI] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2013) Gasotransmitters are emerging as new guard cell signaling molecules and regulators of leaf gas exchange. Plant Sci 201-202: 66–73 [DOI] [PubMed] [Google Scholar]
- Gotor C, García I, Crespo JL, Romero LC (2013) Sulfide as a signaling molecule in autophagy. Autophagy 9: 609–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotor C, Laureano-Marin AM, Moreno I, Aroca A, Garcia I, Romero LC (2015) Signaling in the plant cytosol: cysteine or sulfide? Amino Acids 47: 2155–2164 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Avila-Ospina L, Yoshimoto K, Soulay F, Azzopardi M, Marmagne A, Lothier J, Masclaux-Daubresse C (2013) Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol 199: 683–694 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JT, Whiteman M (2014) Hydrogen sulfide and cell signaling: team player or referee? Plant Physiol Biochem 78: 37–42 [DOI] [PubMed] [Google Scholar]
- Hu LY, Hu SL, Wu J, Li YH, Zheng JL, Wei ZJ, Liu J, Wang HL, Liu YS, Zhang H (2012) Hydrogen sulfide prolongs postharvest shelf life of strawberry and plays an antioxidative role in fruits. J Agric Food Chem 60: 8684–8693 [DOI] [PubMed] [Google Scholar]
- Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, et al. (2014) Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci USA 111: 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Shen J, Qiao Z, Yang G, Wang R, Pei Y (2011) Hydrogen sulfide improves drought resistance in Arabidopsis thaliana. Biochem Biophys Res Commun 414: 481–486 [DOI] [PubMed] [Google Scholar]
- Kabil O, Banerjee R (2010) Redox biochemistry of hydrogen sulfide. J Biol Chem 285: 21903–21907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. (2011) Hydrogen sulfide: its production and functions. Exp Physiol 96: 833–835 [DOI] [PubMed] [Google Scholar]
- Kundu S, Pushpakumar S, Khundmiri SJ, Sen U (2014) Hydrogen sulfide mitigates hyperglycemic remodeling via liver kinase B1-adenosine monophosphate-activated protein kinase signaling. Biochim Biophys Acta 1843: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Shropshire W, Mohr H (1971) An analysis of phytochrome-mediated anthocyanin synthesis. Plant Physiol 47: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureano-Marín AM, García I, Romero LC, Gotor C (2014) Assessing the transcriptional regulation of L-cysteine desulfhydrase 1 in Arabidopsis thaliana. Front Plant Sci 5: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP (2013) Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154: 416–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD (2014) AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Li L, Moore PK (2008) Putative biological roles of hydrogen sulfide in health and disease: a breath of not so fresh air? Trends Pharmacol Sci 29: 84–90 [DOI] [PubMed] [Google Scholar]
- Li L, Wang Y, Shen W (2012a) Roles of hydrogen sulfide and nitric oxide in the alleviation of cadmium-induced oxidative damage in alfalfa seedling roots. Biometals 25: 617–631 [DOI] [PubMed] [Google Scholar]
- Li ZG, Gong M, Xie H, Yang L, Li J (2012b) Hydrogen sulfide donor sodium hydrosulfide-induced heat tolerance in tobacco (Nicotiana tabacum L) suspension cultured cells and involvement of Ca2+ and calmodulin. Plant Sci 185-186: 185–189 [DOI] [PubMed] [Google Scholar]
- Li ZG, Yang SZ, Long WB, Yang GX, Shen ZZ (2013) Hydrogen sulphide may be a novel downstream signal molecule in nitric oxide-induced heat tolerance of maize (Zea mays L.) seedlings. Plant Cell Environ 36: 1564–1572 [DOI] [PubMed] [Google Scholar]
- Lisjak M, Srivastava N, Teklic T, Civale L, Lewandowski K, Wilson I, Wood ME, Whiteman M, Hancock JT (2010) A novel hydrogen sulfide donor causes stomatal opening and reduces nitric oxide accumulation. Plant Physiol Biochem 48: 931–935 [DOI] [PubMed] [Google Scholar]
- Lisjak M, Teklic T, Wilson ID, Whiteman M, Hancock JT (2013) Hydrogen sulfide: environmental factor or signalling molecule? Plant Cell Environ 36: 1607–1616 [DOI] [PubMed] [Google Scholar]
- Liu Y, Xiong Y, Bassham DC (2009) Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963 [DOI] [PubMed] [Google Scholar]
- Łowicka E, Bełtowski J (2007) Hydrogen sulfide (H2S): the third gas of interest for pharmacologists. Pharmacol Rep 59: 4–24 [PubMed] [Google Scholar]
- Merkulova EA, Guiboileau A, Naya L, Masclaux-Daubresse C, Yoshimoto K (2014) Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles. Plant Cell Physiol 55: 715–726 [DOI] [PubMed] [Google Scholar]
- Moriyasu Y, Ohsumi Y (1996) Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol 111: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH (2009) H2S signals through protein S-sulfhydration. Sci Signal 2: ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Snyder SH (2012) H2S signalling through protein sulfhydration and beyond. Nat Rev Mol Cell Biol 13: 499–507 [DOI] [PubMed] [Google Scholar]
- Peng M, Bi YM, Zhu T, Rothstein SJ (2007) Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol 65: 775–797 [DOI] [PubMed] [Google Scholar]
- Pérez-Martín M, Pérez-Pérez ME, Lemaire SD, Crespo JL (2014) Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol 166: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Couso I, Crespo JL (2012a) Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 8: 376–388 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Florencio FJ, Crespo JL (2010) Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 152: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Lemaire SD, Crespo JL (2012b) Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Zaffagnini M, Marchand CH, Crespo JL, Lemaire SD (2014) The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 10: 1953–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero LC, García I, Gotor C (2013) L-Cysteine Desulfhydrase 1 modulates the generation of the signaling molecule sulfide in plant cytosol. Plant Signal Behav 8: e24007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuffi D, Álvarez C, Laspina N, Gotor C, Lamattina L, García-Mata C (2014) Hydrogen sulfide generated by l-cysteine desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiol 166: 2065–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KD, Lee HN, Chung T (2014) A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells 37: 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sláviková S, Shy G, Yao Y, Glozman R, Levanony H, Pietrokovski S, Elazar Z, Galili G (2005) The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot 56: 2839–2849 [DOI] [PubMed] [Google Scholar]
- Sun J, Wang R, Zhang X, Yu Y, Zhao R, Li Z, Chen S (2013) Hydrogen sulfide alleviates cadmium toxicity through regulations of cadmium transport across the plasma and vacuolar membranes in Populus euphratica cells. Plant Physiol Biochem 65: 67–74 [DOI] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter BM, Wu X, Laxman S, Tu BP (2013) Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 154: 403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó C. (2007) Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov 6: 917–935 [DOI] [PubMed] [Google Scholar]
- Takatsuka C, Inoue Y, Matsuoka K, Moriyasu Y (2004) 3-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol 45: 265–274 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Wang BL, Shi L, Li YX, Zhang WH (2010) Boron toxicity is alleviated by hydrogen sulfide in cucumber (Cucumis sativus L.) seedlings. Planta 231: 1301–1309 [DOI] [PubMed] [Google Scholar]
- Wang D, Ma Y, Li Z, Kang K, Sun X, Pan S, Wang J, Pan H, Liu L, Liang D, et al. (2012a) The role of AKT1 and autophagy in the protective effect of hydrogen sulphide against hepatic ischemia/reperfusion injury in mice. Autophagy 8: 954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. (2012) Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev 92: 791–896 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li L, Cui W, Xu S, Shen W, Wang R (2012b) Hydrogen sulfide enhances alfalfa (Medicago sativa) tolerance against salinity during seed germination by nitric oxide pathway. Plant Soil 351: 107–119 [Google Scholar]
- Woo J, Park E, Dinesh-Kumar SP (2014) Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci USA 111: 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K. (2012) Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53: 1355–1365 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Takano Y, Sakai Y (2010) Autophagy in plants and phytopathogens. FEBS Lett 584: 1350–1358 [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu LY, Hu KD, He YD, Wang SH, Luo JP (2008) Hydrogen sulfide promotes wheat seed germination and alleviates oxidative damage against copper stress. J Integr Plant Biol 50: 1518–1529 [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu SL, Zhang ZJ, Hu LY, Jiang CX, Wei ZJ, Liu J, Wang HL, Jiang ST (2011) Hydrogen sulfide acts as a regulator of flower senescence in plants. Postharvest Biol Technol 60: 251–257 [Google Scholar]
- Zhang H, Tan ZQ, Hu LY, Wang SH, Luo JP, Jones RL (2010) Hydrogen sulfide alleviates aluminum toxicity in germinating wheat seedlings. J Integr Plant Biol 52: 556–567 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.