Redundant key roles of ERN1/ERN2 during root hair endosymbiotic infection.
Abstract
Legumes improve their mineral nutrition through nitrogen-fixing root nodule symbioses with soil rhizobia. Rhizobial infection of legumes is regulated by a number of transcription factors, including ERF Required for Nodulation1 (ERN1). Medicago truncatula plants defective in ERN1 are unable to nodulate, but still exhibit early symbiotic responses including rhizobial infection. ERN1 has a close homolog, ERN2, which shows partially overlapping expression patterns. Here we show that ern2 mutants exhibit a later nodulation phenotype than ern1, being able to form nodules but with signs of premature senescence. Molecular characterization of the ern2-1 mutation reveals a key role for a conserved threonine for both DNA binding and transcriptional activity. In contrast to either single mutant, the double ern1-1 ern2-1 line is completely unable to initiate infection or nodule development. The strong ern1-1 ern2-1 phenotype demonstrates functional redundancy between these two transcriptional regulators and reveals the essential role of ERN1/ERN2 to coordinately induce rhizobial infection and nodule organogenesis. While ERN1/ERN2 act in concert in the root epidermis, only ERN1 can efficiently allow the development of mature nodules in the cortex, probably through an independent pathway. Together, these findings reveal the key roles that ERN1/ERN2 play at the very earliest stages of root nodule development.
Legumes form mutualistic associations with soil microorganisms including rhizobial bacteria, giving rise to nitrogen-fixing root nodules. The development of a functional nitrogen-fixing nodule in legumes depends on a successful molecular dialogue between the plant and respective bacterial partner before the bacteria can enter the host root to colonize the developing nodules. As part of the preinfection dialogue, rhizobia secrete lipo-chitooligosaccharide signaling molecules known as Nod (or nodulation) factors (NFs; Dénarié et al., 1996) that are required for subsequent steps of rhizobial infection and nodule organogenesis. Surface bacterial polysaccharides are also important bacterial components required for the development of infected root nodules (Kawaharada et al., 2015). In many legumes, rhizobia penetrate the host root intracellularly through epidermal root hairs (RHs; reviewed by Murray, 2011). After entrapment within a curled RH, rhizobia multiply to form a microcolony during an initial phase of infection chamber remodeling (Fournier et al., 2015). Subsequently, an infection thread (IT) initiated from the infection chamber progresses through underlying root cortical cells to reach the nodule primordium that has formed via cell division of specific root cortical cells (Xiao et al., 2014). Bacteria within ITs are then released into specialized compartments of host cells and differentiate into bacteroids that fix nitrogen within the newly developed nodule (reviewed by Popp and Ott, 2011; Oldroyd, 2013).
NFs are perceived within the host root epidermis, leading to the rapid activation of symbiosis-associated genes including the Medicago truncatula ENOD11 gene, encoding a repetitive proline-rich protein (Journet et al., 2001; Charron et al., 2004). Genetic studies conducted in model legumes have led to the identification of host NF signaling components including plasma membrane-localized LysM or LRR domain-containing receptor kinases implicated in NF perception (Broghammer et al., 2012; Oldroyd, 2013; Antolín-Llovera et al., 2014). Sustained oscillations in nuclear calcium (Ca2+) levels occur downstream of NF perception, and this requires both nuclear membrane-localized cation channels and nuclear pore-associated proteins (Ehrhardt et al., 1996; Sieberer et al., 2009; Capoen et al., 2011; Oldroyd, 2013). Symbiotic calcium oscillations are decoded within the nucleus by a calcium- and calmodulin-dependent protein kinase (CCaMK; Singh and Parniske, 2012) that, by regulating the phosphorylation status of the transcription factor CYCLOPS/IPD3, allows the transduction of the calcium signaling to symbiotic gene expression (Messinese et al., 2007; Yano et al., 2008; Singh et al., 2014). Many of the early NF signaling genes are also required during root endosymbiotic interactions with arbuscular mycorrhizal fungi and are together considered as part of the common symbiotic signaling pathway (Gutjahr and Parniske, 2013).
A number of transcription factors (TFs) are specifically associated with nodulation (Libault et al., 2009; Soyano and Hayashi, 2014), and in most cases are genetically positioned downstream of CCaMK. A few of them have been shown to be directly involved in early NF signaling and these include the GRAS-type NSP1/NSP2 (Smit et al., 2005; Kaló et al., 2005; Heckmann et al., 2006; Murakami et al., 2006), the CCAAT-box binding NF-YA1/A2 (Laloum et al., 2014), Nodule Inception (NIN; Marsh et al., 2007; Yoro et al., 2014; Vernié et al., 2015), and the ERF Required for Nodulation1 (ERN1; Andriankaja et al., 2007; Middleton et al., 2007; Cerri et al., 2012). Gene expression and functional studies indicate that these TFs are involved in both NF signaling and rhizobial infection and can participate in overlapping transcription networks. For example, this is the case for NF-YA and NSP1/NSP2, directly involved in the transcriptional regulation of ERN1 (Hirsch et al., 2009; Madsen et al., 2010; Cerri et al., 2012; Laloum et al., 2014).
The M. truncatula ERN1 TF has been identified as a direct regulator of ENOD11 in root epidermal cells responding to NFs (Andriankaja et al., 2007). Knocking out ERN1 abolishes both ENOD11 transcription and the formation of functional infected nodules (Middleton et al., 2007; Pislariu et al., 2012). However, in contrast to most of the early NF signaling genes, limited host responses are still observed in the ern1-1 mutant, including residual RH infection and cortical cell divisions leading to arrested noninfected nodules (Middleton et al., 2007; Cerri et al., 2012). This finding raised the question of whether partial functional redundancy could exist with the closely related ERN2, which can functionally replace ERN1 when expressed under the control of the ERN1 promoter in the ern1-1 mutant (Cerri et al., 2012). Furthermore, both ERN1 and ERN2 are able to activate the transcription of ENOD11 via the NF-box, a 30-bp regulatory unit sufficient for conferring NF-elicited gene expression of ENOD11 within the nodulation competence root zone (Andriankaja et al., 2007). Although expressed at lower levels compared to ERN1, ERN2 displays partially overlapping expression profiles in RHs in response to NFs as well as during subsequent sequential stages of rhizobial infection (Cerri et al., 2012). However, the relative importance of ERN2 during nodulation has not yet been addressed. Here we describe the identification and characterization of two mutant alleles for ERN2 and the generation of a double mutant defective for both ERN1 and ERN2. We show that unlike ern1-1, loss-of function ern2 mutant alleles are still able to form nitrogen-fixing nodules, although these nodules are partially defective in rhizobial root colonization and can show premature senescence. Significantly, the ern1-1 ern2-1 double mutant is blocked very early during the symbiotic interaction prior to infection, thus demonstrating the essential roles of these TFs for initiating RH infection and nodule development.
RESULTS
Identification of Two ern2 Mutant Alleles
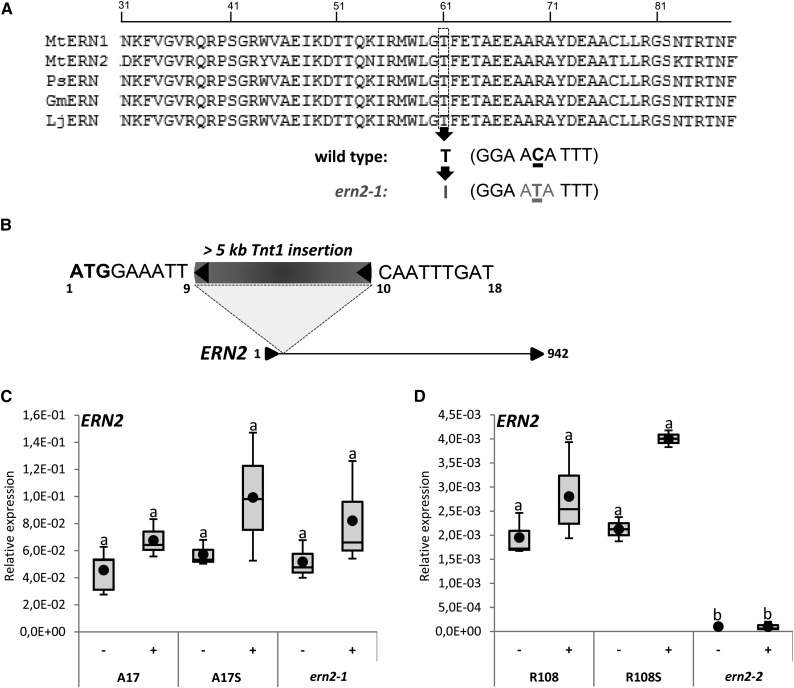
High-throughput reverse genetic strategies were used to identify mutations in the ERN2 locus. Firstly, a TILLING approach was employed to identify mutations within a 1 kb region of the ERN2 coding sequence by screening an ethyl methanesulfonate (EMS)-mutagenized collection (Le Signor et al., 2009). A single-nucleotide substitution mutant, designated ern2-1, was shown to carry a threonine (Thr, T) to isoleucine (Ile, I) transition at position 61 within the ERN2 DNA binding domain (Fig. 1A). This mutation is particularly interesting since T61 is a highly conserved residue within the AP2/ERF DNA binding domain. Secondly, by screening a Tnt1 transposon-tagged M. truncatula R108 collection (Pislariu et al., 2012), we were able to identify the ern2-2 mutant that carries a transposon insertion at the beginning of the ERN2 coding sequence (Fig. 1B). Homozygous ern2-1 and ern2-2 mutant lines and the respective nonmutated sibling plants, referred to as A17 Sibling (A17S) and R108 Sibling (R108S), were used for subsequent phenotypic analyses. Quantitative (Q) RT-PCR analysis demonstrated that ERN2 transcript levels can be detected at comparable levels in wild-type and ern2-1 mutant roots (Fig. 1C), indicating that the ern2-1 point mutation does not affect the accumulation of ERN2 transcript levels. As previously demonstrated, ERN2 transcript levels do not significantly change upon NF-treatment (Cerri et al., 2012). Finally, as expected, ERN2 expression is totally abolished in the ern2-2 mutant (Fig. 1D), confirming that this is a null mutation.
Figure 1.
Identification of ERN2 mutant alleles. A, Alignment of M. truncatula ERN1 (accession: EU038802) and ERN2 (accession: EU038803) DNA binding domains with closest Pisum sativum (PsERN, EF396329), Glycine max (GmERN, XM_006604176), and L. japonicus (LjERN, AP006677.1) ortholog sequences using Clustal W (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The conserved Thr (residue T61 in MtERN2) is replaced by an Ile (I) in the ern2-1 EMS mutant. B, The ern2-2 line is a knockout mutant with an approximately 5.3-kb Tnt1 retrotransposon inserted between nucleotides 9 and 10 downstream of the ERN2 start codon (position 1). Nucleotide sequences flanking the Tnt-1 insertion are indicated. The stop codon is at position 942. C and D, QRT-PCR analyses of ERN2 transcript levels in total RNA samples from M. truncatula roots treated for 6 h with water (−) or 10−9 m NF solution (+). RNA was extracted from wild type (A17 and R108), wild-type sibling (A17S and R108S), and ern2-1 (A17 background) and ern2-2 (R108 background). Box plots represent relative expression levels of two to four independent biological experiments after normalization against reference transcript levels. Box plots represent first and third quartile (horizontal box sides), minimum and maximum (outside whiskers). Black circles depict mean values. One-way ANOVA followed by a Tukey honest significant difference (HSD) test of the values was performed (in C, P > 0.05; in D, P < 0.001). Classes sharing the same letter are not significantly different.
The ern2-1 Mutation Affects the Ability of ERN2 to Bind to the NF-box and Activate ENOD11 Transcription
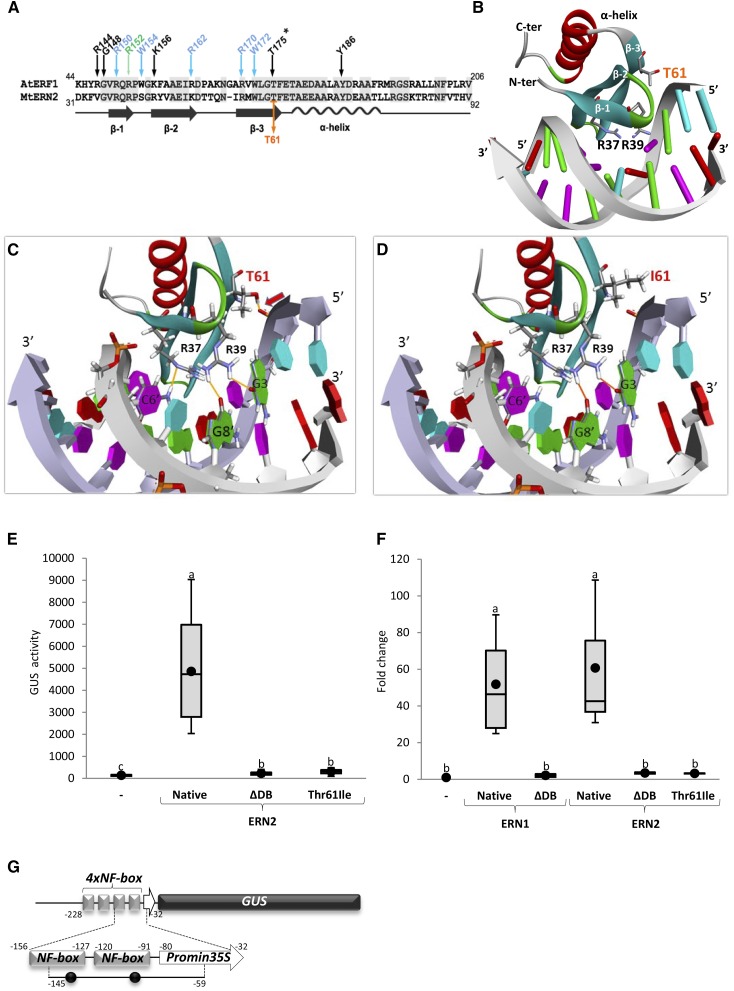
As stated above, the ern2-1 point mutation targets a highly conserved Thr located at the C-terminal extremity of the predicted third β-strand within the AP2/ERF DNA binding domain (Fig. 2, A and B). Previous solution NMR studies of the complex between the AtERF1 DNA binding domain and its GCC-box revealed that this conserved Thr residue forms a strong hydrogen bond with the sugar-phosphate (P) backbone, alongside other sugar-P backbone interactions and DNA base recognitions by neighboring conserved amino acids (Allen et al., 1998). To evaluate to what extent the T61I transition in ern2-1 affects the DNA binding capacity of ERN2, we first made use of a molecular modeling strategy.
Figure 2.
Mutation of threonine 61 affects the ability of ERN2 to bind to the NF-box and to activate transcription. A, Alignment of the AP2/ERF DNA binding domains of AtERF1 and MtERN2, comprising three associated beta strands (β-1, β-2, and β-3) and a single α-helix. Important residues involved in base interactions (green arrow), base and sugar-P backbone interactions (blue arrows), or sugar-P backbone interactions (black arrows) are depicted according to Allen et al. (1998). Conserved Arg (R), Gly (G), Trp (W), Lys (K), Thr (T), and Tyr (Y) residues are numbered according to the ATG start codon of AtERF1. The T61 residue mutated in ern2-1 is indicated in orange and corresponds to the conserved T175 residue in AtERF1 (asterisk). B and C, 3D modeling of the DNA binding domain of wild-type MtERN2 in the presence of its GCC-like target sequence based on the known 3D structure of the DNA binding domain of AtERF1 in the presence of its target sequence, the canonical GCC-box (Allen et al., 1998). In the more detailed view shown in C, three critical DNA binding interactions are highlighted: the T61 located at the extremity of β-3 (arrow) and two base-contacted Arg residues located at either ends of β-1. D, The T61I replacement in the ern2-1 mutant allele abolishes the T61 backbone interaction and modeling/docking predicts the additional loss of the H-bond interaction between R37 and the C6′ base. Bases are labeled as follows: thymidine in blue, guanine in green, cytosine in pink, and adenine in red. Note that the complex illustrated in D is predicted to be unstable, and hence that the mutated ERN2 is unable to bind its GCC-like DNA target. E to G, Transcription activation assays and in vivo ChIP analyses were performed in N. benthamiana leaves to study the ability of the mutated Thr61 to Ile (T61I) ERN2 to either activate or bind to the ENOD11 NF-box target sequence. E and F, N. benthamiana cells were transformed with the 4xNF-box-GUS reporter alone (−), or with 3xHA-tagged ERN1 and ERN2 TFs (Native), respective DNA binding domain deleted versions (ΔDB), and the T61I ERN2 point-mutation. E, Transcription activation of the 4xNF-box-GUS reporter was measured by quantifying the fluorimetric GUS activity (pmol/MU/min/mg protein) of individual N. benthamiana leaf discs. Box plots represent GUS activity measurements of 16 to 18 independent N. benthamiana plants obtained from three independent experiments. F, ChIP followed by QPCR showed association of native 3xHA-ERN1 and ERN2 to an 86-bp amplicon encompassing two GCC-like ERN binding motifs (black dots) within the NF-box (G), as confirmed by DNA sequencing. ChIP data are presented as fold change relative to the negative 4xNF-box-GUS reporter alone (−) after normalization to Input DNA. Box plots represent data from two to four technical replicates from three to four independent biological experiments (n = 12 or 16). G, Schematic view of the 4xNF-box-GUS reporter construct used in E and F, which is composed of four copies of the approximately 30-bp NF-box unit placed upstream the minimum −47 bp 35S promoter. The QPCR-amplified region, including two GCC motifs (black circles), is between −145 and −59 positions, as indicated. Box plots depict mean value (black circles), first and third quartile (horizontal box sides), and minimum and maximum (outside whiskers). One-way ANOVA followed by a Tukey HSD test of the values were performed (in E and F, P < 0.001). Classes sharing the same letter are not significantly different.
The solution structures of the AP2/ERF domain of AtERF1 either alone or bound to its DNA motif “GCC-box” (5′-AGCCGCC-3′) have previously shown that three antiparallel β-strands, stabilized by an α-helix, are responsible for the major groove contacts (Allen et al., 1998; Fig. 2A). The majority of the DNA contacts are made by Arg and Trp residues located within the β-strands. Based on the structure of the AtERF1 DNA binding domain/GCC-box complex, we used homology modeling and molecular docking to construct a three-dimensional (3D) structural model for the native ERN2 DNA binding domain in the presence of its target GCC-like DNA sequence (Fig. 2B). It should be noted that despite sequence differences, the key interactions between the ERN2 DNA binding domain and its GCC-like target are essentially conserved (Supplemental Fig. S1). According to this model, the ERN2 T61 residue is similarly involved in a key sugar-P backbone interaction and this crucial hydrogen bond is therefore lost in the case of the T61I ern2-1 mutation (compare Fig. 2, C and D, and Supplemental Fig. S1). Modeling also predicts that the replacement of the T61 side chain by the hydrophobic Ile (I) side chain results in the loss of the R37 hydrogen-bonding to the corresponding 6′ cytosine.
To verify the predicted loss of target DNA binding for the mutated ERN2, we then examined the impact of the T61 to I mutation on the ability of ERN2 to recognize its target DNA sequence and activate transcription. ERN2 was previously shown to recognize the NF-box regulatory sequence, essential for NF-elicited expression of ENOD genes in M. truncatula RHs (Andriankaja et al., 2007). By constructing a T61I mutated ERN2 that mimics the ern2-1 mutant allele, we tested the ability of this mutated protein to activate transcription of the 4xNF-box-GUS reporter by employing a transient expression assay in Nicotiana benthamiana cells (Andriankaja et al., 2007; Cerri et al., 2012). While the native ERN2 protein successfully activated the 4xNF-box-GUS reporter, the mutated T61I protein was totally unable to transactivate the reporter construct, similarly to the truncated form of ERN2 devoid of its DNA binding domain (referred to as ΔDB; Fig. 2E; Supplemental Fig. S2A). Western-blot analysis and subcellular localization studies of YFP-fused protein versions confirmed that the three tested proteins were correctly expressed and targeted to the nuclear compartment (Supplemental Fig. S2, B and C). These results therefore show that the ern2-1 point mutation does not affect either protein stability or subcellular nuclear localization but severely impairs the transcriptional capacity of the mutated ERN2 protein. Chromatin immunoprecipitation (ChIP) followed by QPCR was then performed to examine whether the mutated ERN2 was also impaired in DNA binding to the NF-box target sequence. Primer pairs spanning the NF-box sequence were used in ChIP experiments from N. benthamiana cells expressing native or modified ERN2 versions in the presence of the 4xNF-box-GUS reporter (Fig. 2, F and G). The closely related ERN1 TF and an inactive DNA binding domain-deleted version (referred as ERN1ΔDB) were used in these experiments as positive and negative controls (Fig. 2F). In line with the transcriptional activation experiments described above, a significant enrichment of the NF-box sequence was only observed in samples expressing native ERN1 or ERN2 (Fig. 2F), thus demonstrating that the mutated ERN2 is unable to associate with the NF-box. Taken together, and consistent with the predictions of the molecular modeling experiments, these results indicate that the T61I mutation of ERN2 in the ern2-1 mutant allele drastically affects the ability of ERN2 to bind to NF-box-containing promoters and activate transcription of target gene sequences.
ern2 Mutants Show Reduced Colonization But Are Able to Form Nitrogen-Fixing Nodules That Senesce Prematurely
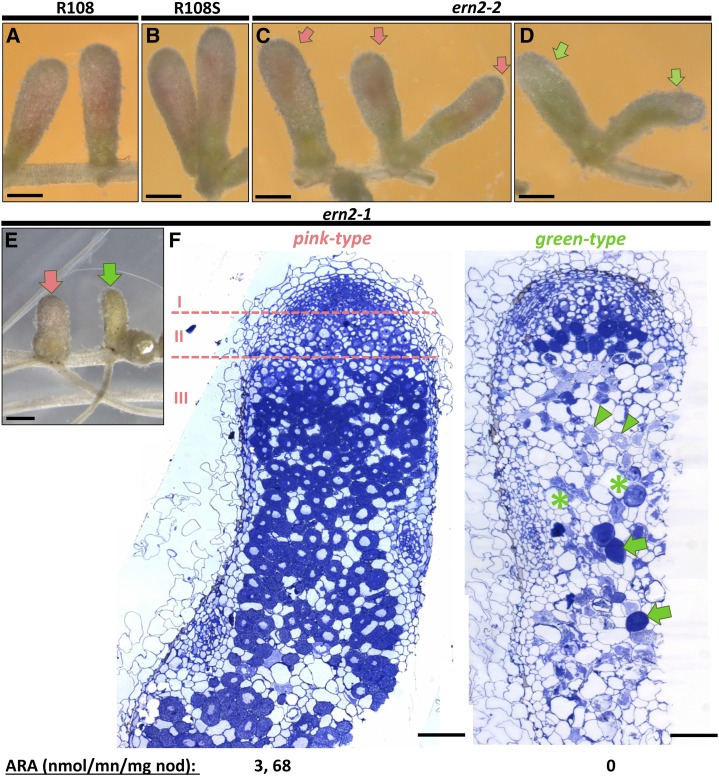
While ern1 mutants are impaired in the formation of mature infected nodules (Middleton et al., 2007; Pislariu et al., 2012), both ern2-1 and ern2-2 mutant lines can form functional pink-colored nodules (compare Fig. 3, A–C). However, even if ern2-1 and ern2-2 nodules appeared similar to those of wild-type plants, approximately 25% of them became elongated and green-colored about 4 weeks postinoculation (wpi; Fig. 3, D and E). Acetylene reduction assays of the ern2 nodules revealed that the pink-colored nodules were able to fix nitrogen as efficiently as wild-type nodules, whereas the green-colored ones were devoid of nitrogen-fixation capacity (Fig. 3). A closer inspection of ern2 nodule sections revealed that whereas the pink-colored nodules possessed the characteristic differentiation zones of functional nodules, the green nodules were abnormally empty and either lacked or possessed a reduced meristematic zone (Fig. 3F). These nonfunctional nodules contained collapsed cells, characteristic of advanced senescence. To examine whether a senescence-like process was already underway in the pink ern2 nodules, we compared the expression of senescence-associated marker genes (van de Velde et al., 2006) in pink nodules from both wild-type and ern2 roots. QRT-PCR analysis indeed revealed increased transcript levels of senescence-associated cysteine protease and vacuolar processing enzyme-encoding genes in both ern2-1 and ern2-2 nodules by comparison with wild-type counterparts (Supplemental Fig. S3, A and B).
Figure 3.
The ern2 mutant lines form nitrogen-fixing nodules that prematurely senesce. Phenotypic analyses of wild-type R108 (A), wild- type R108S (B), ern2-2 (C and D), and ern2-1 (E and F) nodules, collected 4 wpi with S. meliloti. At this stage, nodules from R108 and R108S have a major pink-colored zone indicative of nitrogen-fixation activity. The majority of nodules formed on ern2 (C and E, pink arrows) also have pink-colored zones, although often reduced in size by comparison to the wild-type nodules. Approximately 25% of mutant nodules are green-colored (green arrows in D and E), indicative of nodule senescence. F, Toluidine blue-stained 1-μm longitudinal sections of both pink and green-type ern2-1 nodules. Pink-type nodules exhibit the characteristic zonation of wild-type nodules with meristematic (I), infection (II), and nitrogen-fixation zones (III) while green-type nodules lack a clear nodule zone organization. The meristematic zone is hardly seen and cells in the central nodule region are either undergoing advanced degradation (green arrowheads), are collapsed (green asterisks), or are invaded by saprophytic bacteria (green arrows). Nitrogen-fixation measurements of isolated nodules by the acetylene reduction assay confirmed the absence of nitrogen fixation of green-type nodules. Bars in A to E = 1 mm and in F = 0.2 mm.
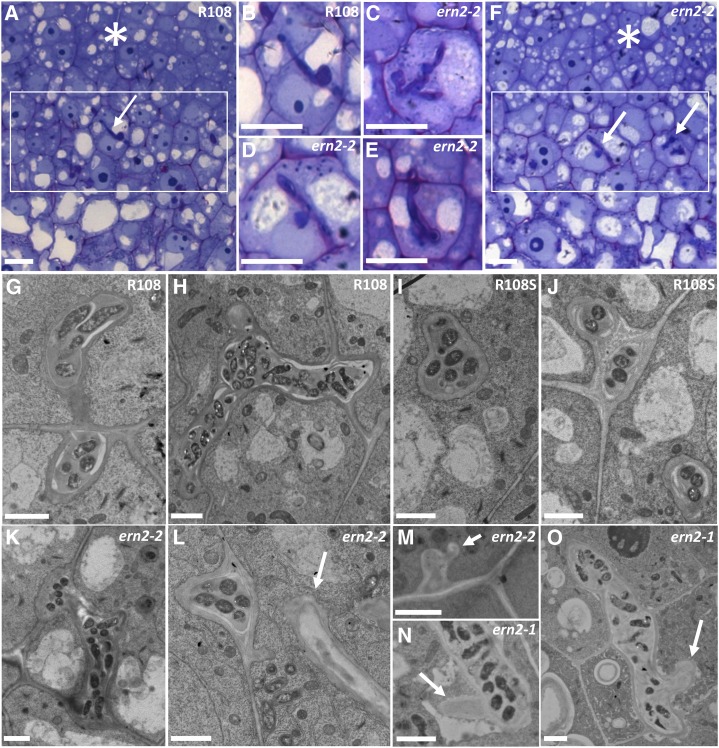
Because ERN2 is mainly expressed in the nodule infection zone II (Cerri et al., 2012), we decided to examine this zone in more detail in the pink ern2 nodules. Optical and electron microscopy analyses did not reveal major changes in ern2 nodule differentiation except for a number of unusual ITs with zones devoid of bacteria in six out of 16 nodule sections analyzed (Fig. 4). Such IT structures were not observed in the 10 analyzed wild-type nodule sections. Taken together, these findings demonstrate that ERN2 is not essential for the formation of mature nitrogen-fixing nodules, but it is required for optimal nodulation. The absence of ERN2 in nodule zone II results in the formation of abnormal ITs, and we propose that this then leads to accelerated nodule senescence.
Figure 4.
The ern2 nodules are partially defective in IT development. Detailed views of infection zones (rectangles) of both wild-type R108 (A) and pink ern2-2 nodules (F) from toluidine-blue/basic fuchsin-stained 1-μm longitudinal sections. ITs within these regions (arrows) are highlighted in B (R108) and in C to E (ern2-2). ITs in ern2-2 are either similar to wild-type ITs or can exhibit a thickened appearance associated with less staining (C–E). Electron microscopy analysis has shown that some ITs of ern2-2 or ern2-1 can have zones devoid of bacteria (arrows in L–O) while others resemble wild-type ITs (compare R108 in G and H; R108S in I and J; ern2-2 in K–M; and ern2-1 in N and O). Nodule meristematic regions are highlighted by asterisks in A and F. Bars in A to F = 100 μm, G to O = 2 μm.
The ern1-1 ern2-1 Double Mutant Is Totally Impaired in RH Rhizobial Infection But Not in Arbuscular Mycorrhizal Colonization
Knowing that ERN2 is also expressed during early stages of RH infection (Cerri et al., 2012), we decided to examine whether ern2 mutant lines would also be altered during these early root colonization steps. Although major phenotypic differences were not observed between wild-type and mutant lines, ern2 mutant roots exhibited overall reduced root colonization rates, leading to the formation of less infected nodule primordia (Supplemental Fig. S4, A and B). Close inspection of mutant roots revealed root cortical colonization sites with unusual IT ramifications, but were not always sufficiently obvious to validate a clear infection phenotype (Supplemental Fig. S4C). To investigate whether this mild phenotype was due to functional redundancy with the close homolog ERN1, known to be coexpressed with ERN2 (and at higher levels) during these early stages of root colonization (Cerri et al., 2012), we decided to generate a double mutant by crossing the ern1-1 (Middleton et al., 2007) and ern2-1 (this work) mutant lines. Progeny from this cross carrying the ern1-1 allele were individually analyzed for the presence of the ERN2 mutation to obtain homozygous double mutants for further phenotypic analysis. Different wild-type and mutant genotypes were inoculated with Sinorhizobium meliloti and analyzed at various time points postinoculation. At 3 dpi, rhizobial infection had already reached the root cortex in both wild-type and ern2 lines and some infected primordia/nodules were also observed (Fig. 5A; Supplemental Fig. S5, A, C, and E). As expected, rhizobial infection in ern1-1 roots remained blocked at the root surface (Fig. 5B; Supplemental Fig. S5G), with a large number of RHs arrested at the microcolony stage (Fig. 5C) and several growth-arrested ITs within RHs (Fig. 5D). In contrast, ern1-1 ern2-1 double mutant roots were totally devoid of rhizobial RH infection (Fig. 5, E and F; Supplemental Fig. S5I). At 4 wpi, normal nodule development was observed in both wild-type and ern2 roots (Supplemental Fig. S5, B, D, and F) while no colonized nodules were observed in either ern1-1 or ern1-1 ern2-1 roots (Supplemental Fig. 5, H and J). As described previously in Middleton et al. (2007), typical uninfected and under-developed nodules with arrested ITs on the upper surface could be observed on ern1-1 roots, but these structures were not observed in the double mutant roots (Supplemental Figs. S5, H–J, and S7A). Despite the strong nodulation phenotype, ern1-1 ern2-1 roots were not affected during root colonization by Rhizophagus irregularis (Supplemental Fig. S6).
Figure 5.
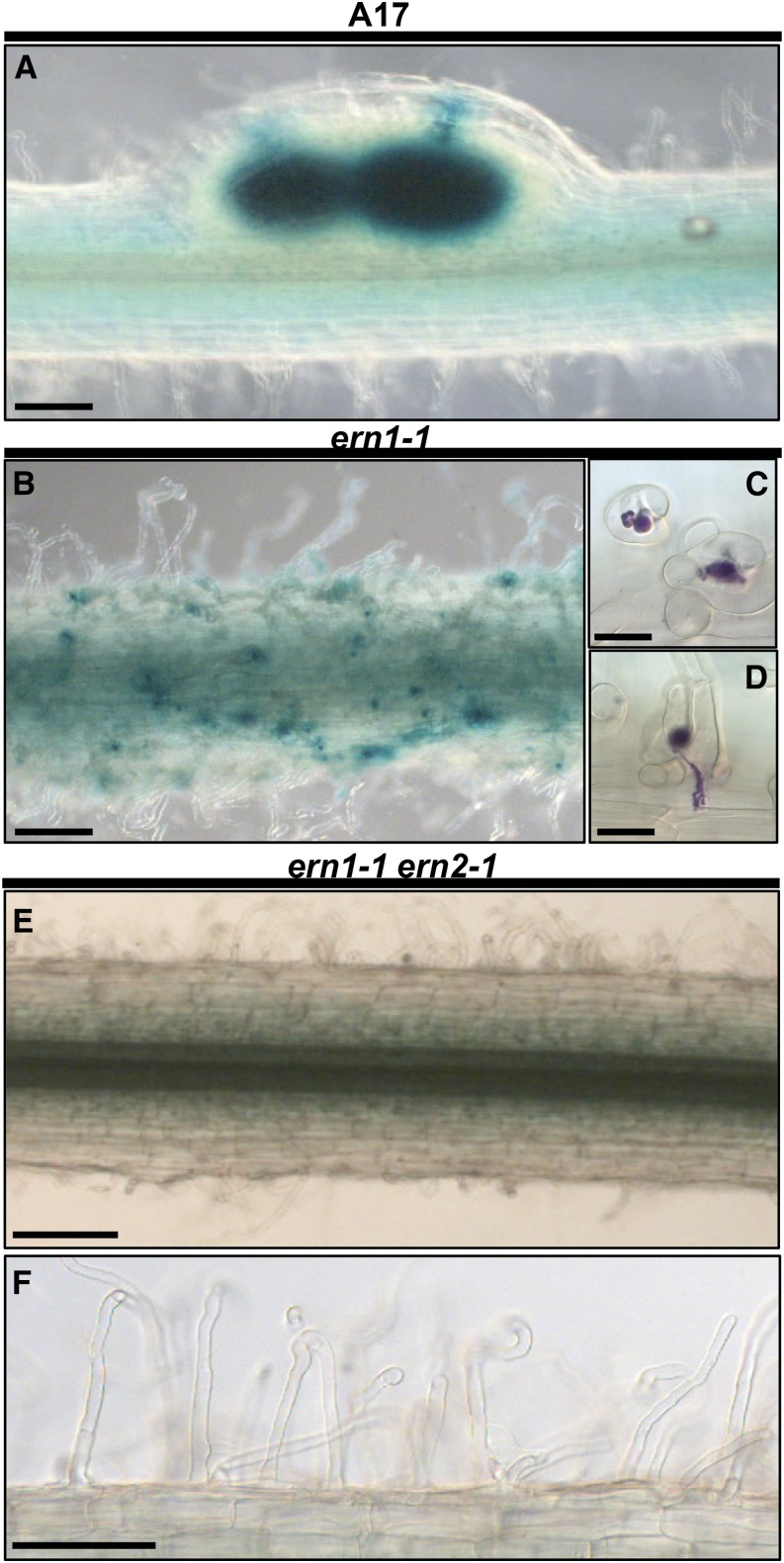
The double ern1-1 ern2-1 mutant is totally defective for rhizobial infection. S. meliloti-inoculated aeroponic-grown plants were analyzed 3 d postinoculation (3 dpi) in wild-type A17 (A), ern1-1 (B–D) and the ern1-1 ern2-1 double mutant (E and F). Histochemical staining using either X-Gal (blue in A, B, E, and F) or Magenta-Gal (purple in C and D) substrates allows the visualization of the constitutive bacterial β-galactosidase activity. At this stage, wild-type A17 (A; see also Supplemental Fig. S5), wild-type A17S, and ern2-2 (see Supplemental Fig. S5) exhibit infected young nodules or nodule primordia. In contrast, rhizobial infection is arrested after successful initiation in ern1-1 (B), as illustrated by the numerous root hair curlings (RHCs) with entrapped bacteria (C) or RH ITs that do not reach the inner root cortical tissues (D) as previously described. Strikingly, the roots of the ern1-1 ern2-1 double mutant do not display any RHC with entrapped bacteria or RH ITs (E and F). Bars = 250 μm (A, B, and E); 20 μm (C and D); 100 μm (F).
A detailed analysis of ern1-1 ern2-1 roots revealed that they were impaired very early during the symbiotic interaction, and were unable to form typical RH curls with entrapped rhizobia. On the rare occasions where RHs initiated curling in ern1-1 ern2-1 (Fig. 6F), the curls appeared defective, since bacteria were not entrapped as observed in wild-type RH curls (Fig. 6C). Instead, the double mutant displayed predominantly spatula-like RHs (Fig. 6E, arrows) that become even more pronounced over time as illustrated at 4 dpi (Fig. 6G, arrows). This RH phenotype is only observed in S. meliloti-inoculated roots, indicating that it is dependent on symbiotic signaling (Fig. 6H).
Figure 6.
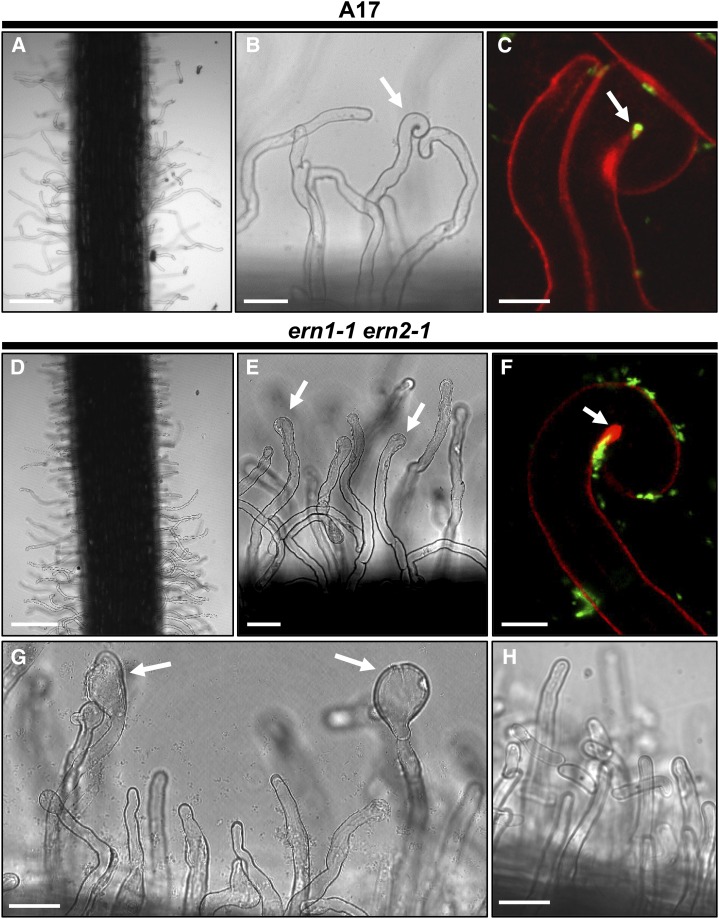
RHC with enclosed rhizobia is absent in the ern1-1 ern2-1 double mutant. Wild-type A17 and ern1-1 ern2-1 RHs were imaged after inoculation with S. meliloti (visualized in green). At 2 dpi, RHs are usually deformed in both A17 (A and B) and ern1-1 ern2-1 (D and E) lines. At this stage, RHCs are frequent in A17 (arrow in B) while in the double mutant most RHs display RH tip swelling (arrows in E). This RH swelling becomes more pronounced over time as illustrated at 4 dpi (arrows in G) and is not observed in noninoculated conditions (H). In some rare cases, loose curls can be observed in ern1-1 ern2-1 roots (F). But in this case, curling is not complete and although numerous rhizobia are associated with these RHs, they are not entrapped within the curl, in contrast to the wild-type situation shown in C. Bars = 200 μm (A and D); 40 μm (B, E, G, and H); 10 μm (C and F).
ERN1 and ERN2 Are Both Required for Accurate Expression of NF Signaling Genes
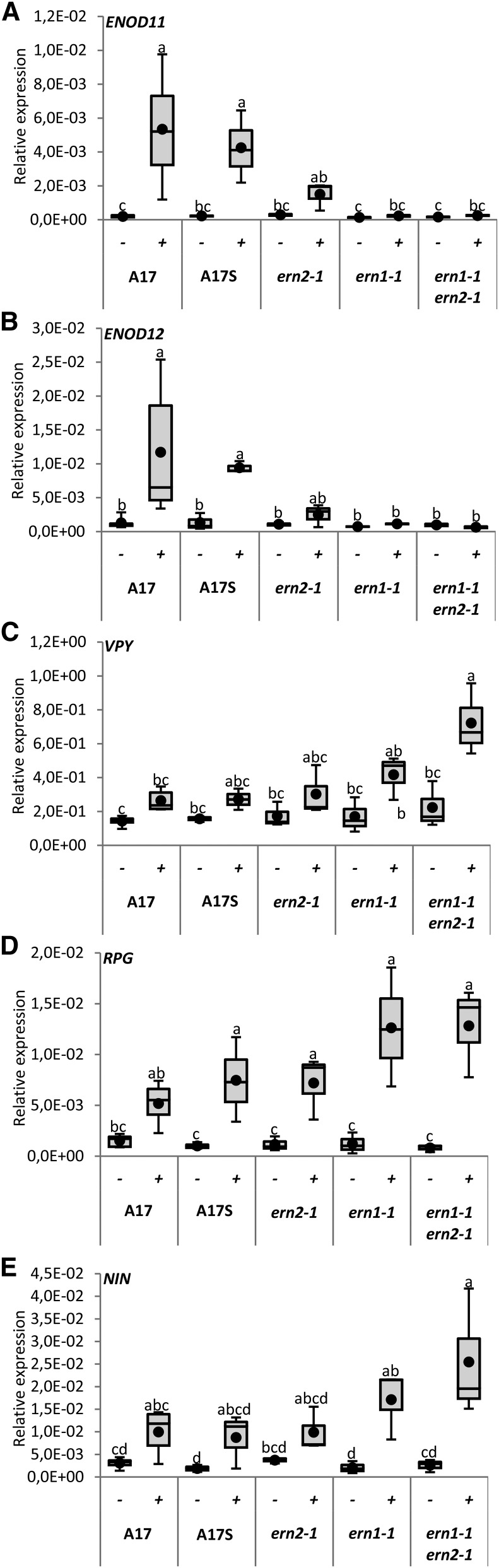
To evaluate the effect of inactivating either ERN1, ERN2 or both TFs on host gene expression, we analyzed the expression of a number of symbiotic genes by QRT-PCR in the various single and double mutant genotypes during NF signaling. As shown in Figure 7, A and B, NF-elicited expression of the NF-box containing target genes ENOD11 and ENOD12 is totally abolished in the double mutant line (Fig. 7, A and B). A less drastic but significant reduction of ENOD12 and ENOD11 transcript levels is also observed in NF-treated samples of the single ern2-1 mutant (Fig. 7, A and B). This indicates that ERN1 and ERN2 are both required to assure optimal expression levels of these genes. In contrast, transcript levels of the infection-related genes NIN (Marsh et al., 2007), RPG (Arrighi et al., 2008), and VPY (Murray et al., 2011) are either at higher (ern1-1 and ern1-1 ern2-1) or comparable levels (ern2-1) to wild-type plants (Fig. 7, C–E). This suggests that these genes are likely to act upstream ERN1/ERN2 or in parallel pathways that are up-regulated in the absence of functional ERN1/ERN2.
Figure 7.
Symbiotic gene expression is either down- or up-regulated in the ern1-1 ern2-1 double mutant. QRT-PCR analyses of ENOD11, ENOD12, VPY, RPG, and NIN transcripts was performed using total RNA samples extracted from wild type A17S, ern2-1, ern1-1, and the double ern1-1 ern2-1 mutant after 6 h treatment with either water control (−) or 10−9 m NF solution (+). Values represent the average from RNA samples from pooled plants (n = 26 A17S, n = 24 ern2-1, n = 35 ern1-1, and n = 35 ern1-1 ern2-1) from three to five independent biological experiments after normalization against reference transcript levels. Box plots represent first and third quartile (horizontal box sides), and minimum and maximum (outside whiskers). Black circles depict mean values. One-way ANOVA followed by a Tukey HSD test of the values was performed (in A, B, D, and E, P < 0.001; in C, P < 0.01). Classes sharing the same letter are not significantly different.
Expression of ERN1 or ERN2 under Native Promoters Is Sufficient to Restore Nodule Organogenesis
Complementation of the double mutant with either ProERN1:YFP-ERN1 or ProERN2:YFP-ERN2 fully restored RH curling and IT formation (Supplemental Fig. S7), demonstrating that ERN1 or ERN2 alone are sufficient for rhizobial infection. ProERN2:YFP-ERN2 also fully restored the formation of arrested ern1-1-like noninfected nodules in the double mutant, confirming that endogenous expression of ERN2 is sufficient to induce nodule initiation. To assess whether the defects in nodule organogenesis are simply a reflection of defects in rhizobial infection or instead reflect a direct role for ERN1 and ERN2 during nodule initiation, we expressed ERN1 and ERN2 from the epidermal-specific Expansin7 promoter (Vernié et al., 2015) and tested for complementation of the double mutant. Epidermal expression of either ERN1 or ERN2 could only restore RH curling and IT formation, and not nodule organogenesis (Fig. 8), thus indicating that ERN1 and ERN2 expression by endogenous promoters is required to activate the nodule organogenesis-signaling pathway. Furthermore, only ERN1 expressed from its native promoter can efficiently restore the formation of infected nodules (Supplemental Fig. S7). Taken together, our phenotypic analyses and complementation studies demonstrate the essential roles of ERN1 and ERN2 in regulating the earliest steps of rhizobial infection and nodule development (Fig. 9). During early infection, ERN1 or ERN2 present in the root epidermis control the initiation of RH curling, microcolony proliferation, and formation of ITs. These TFs are also essential for inducing at a distance nodule organogenesis in the inner root cortex, but this requires expression under the control of their native promoters. The later steps of root cortical cell infection and nodule formation are exclusively dependent on ERN1.
Figure 8.
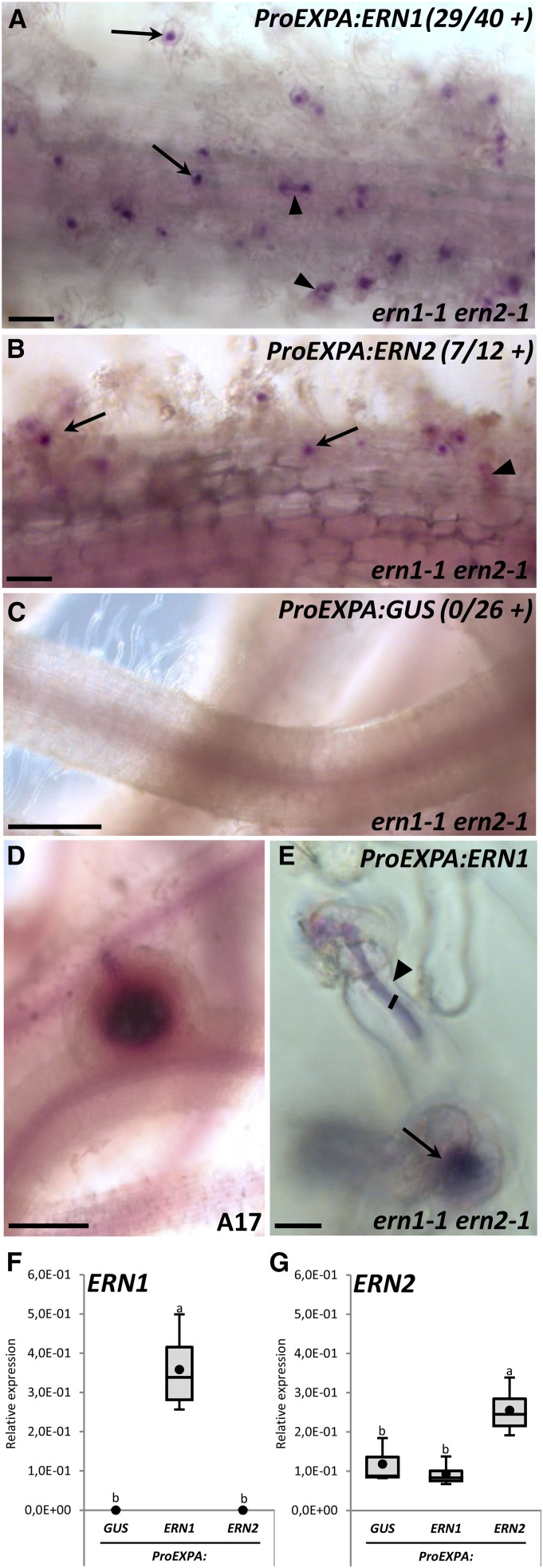
Epidermal expression of ERN1 and ERN2 can only restore RH infection. The ern1-1 ern2-1 double mutant was transformed by A. rhizogenes with a ProEXPA:ERN1 (A and E), ProEXPA:ERN2 (B), and ProEXPA:GUS negative control (C) constructs. Transformation of A17 with the ProEXPA:GUS construct was used as a positive control to validate nodule formation in the same experimental conditions (D). Transformed root systems were analyzed 3 to 4 wpi with S. meliloti-lacZ after staining for β-galactosidase bacterial activity (magenta staining). Typical “ern1-like” noninfected arrested nodules or infected nodules were not observed in any of the ern1-1 ern2-1 transgenic roots analyzed. Only RHCs or RH ITs (arrows and arrowheads in A, B, and E) were observed in complemented roots. Numbers in A to C represent the number of independent composite plants exhibiting restored RHC or RH infection from three (for ProEXPA:ERN1 and ProEXPA:GUS) and two (for ProEXPA:ERN2) independent experiments. F and G, QRT-PCR analyses in total root RNA samples from individual composite plants confirmed the expression of ERN1 or ERN2 in complemented roots. Values represent average of three to four independent plants after normalization against reference transcript levels. Black circles depict mean values. One-way ANOVA followed by a Tukey HSD test of the values were performed (in F, P < 0.001; in G, P < 0.01). Classes sharing the same letter are not significantly different. Bars = 100 μm (A and B); 500 μm (C and D); 10 μm (E).
Figure 9.
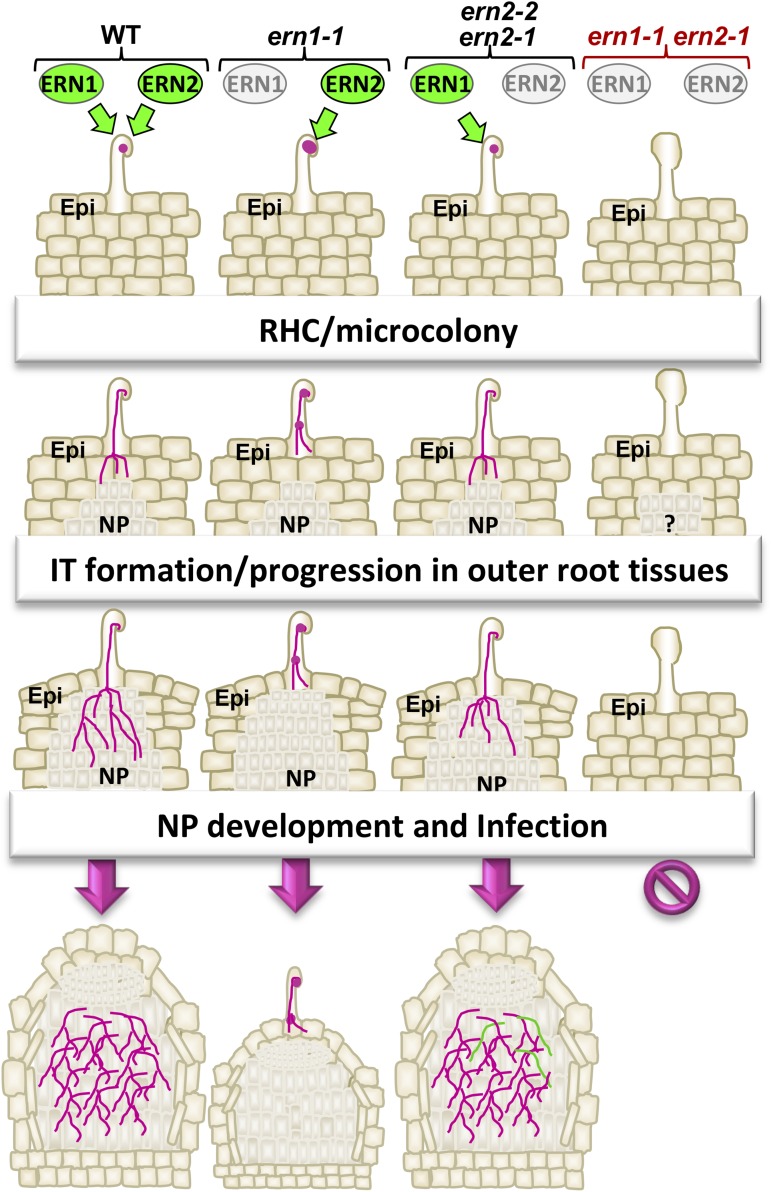
Overview of the different rhizobial-infection steps regulated by ERN1 and ERN2 TFs. Schematic representation of the different nodulation steps in wild type, ern1-1, ern2 (ern2-1 and ern2-2), and ern1-1 ern2-1 mutants. The ern1-1 ern2-1 double mutant is completely unable to initiate infection or nodule development. ERN1 alone can assure all the early steps of root colonization (RHC, IT initiation/progression) and nodule organogenesis as shown by the phenotype of ern2 and ern1-1 ern2-1 mutants complemented by ProERN1-ERN1. Nevertheless, ern2 mutants are less efficient in root colonization and develop nodules that can exhibit abnormal ITs (green lines) and premature senescence. In the absence of ERN1 (ern1-1), ERN2 alone can assure all the early steps of colonization (RHC, IT initiation/progression) of outer root tissues and nodule organogenesis. This leads to the formation of small underdeveloped nodules with infection sites on the top. Proliferation of cell division in the root cortex is abolished in the double mutant and can only be induced when ERN1 or ERN2 are expressed under the control of their native promoters. ERN1 (and not ERN2) is essential for rhizobial colonization of dividing nodule primordia in the root cortex to assure the formation of mature infected nodules.
DISCUSSION
The establishment of the nitrogen-fixing root endosymbiosis in legumes involves a number of TFs required for orchestrating transcriptional changes during host reprogramming for rhizobial infection and nodule development. Forward and reverse genetic approaches have helped to demonstrate their central roles in the development of nitrogen-fixing nodules. However, despite clear-cut nodulation phenotypes, many TF mutants are still able to partially initiate early symbiotic signaling or rhizobial infection (Middleton et al., 2007; Yano et al., 2008; Horváth et al., 2011; Ovchinnikova et al., 2011; Laporte et al., 2014). The recent discovery of gene duplications for some of these genes (Young et al., 2011) thus raises the question of the extent to which genetic redundancy is operating during these early symbiotic stages, as recently shown for NF-Y TFs (Laloum et al., 2014). This article focuses on the phylogenetically related M. truncatula ERF transcription factors ERN1 and ERN2 (Andriankaja et al., 2007). Previous knockout mutant analysis had shown that ERN1 was essential for nodule development (Middleton et al., 2007), yet the ern1-1 (bit1-1) knockout displayed an intermediate phenotype that could be the result of potential redundancy with ERN2, a gene with a partially overlapping expression pattern (Cerri et al., 2012; Chen et al., 2015). Here we have used a genetic approach to test for functional redundancy between these two genes during root nodulation. The analysis of ern2 single mutants and a double ern1-1 ern2-1 mutant reveal a more extreme symbiotic phenotype for the double mutant compared to either single mutant. As discussed in detail below, while ern1-1 can still initiate RH infection and root cortical cell proliferation, the double mutant is totally impaired in rhizobial RH infection and nodule development.
Mild Infection Defects May Contribute to the Accelerated Senescence of ern2 Nodules
We have isolated and characterized single point mutation (ern2-1) and knockout (ern2-2) lines that are both still able to form mature nitrogen-fixing nodules. Despite the fact that the two ern2 lines were isolated from A17 and R108 mutant collections (Le Signor et al., 2009; Pislariu et al., 2012), they display similar phenotypes. Although forming functional nitrogen-fixing nodules, ern2 mutant lines nevertheless exhibit some limited nodulation defects since we observe an overall 2-fold decrease in the number of infected nodules and nodule primordia as well as certain nodules containing a limited number of abnormal ITs (see Fig. 4 and Supplemental Fig. S3). On the other hand, no strong differences were observed during early stages of RH infection of ern2 mutant lines, most likely due to the presence of ERN1. Our data indicate that ERN1 is the predominant TF required for infection and nodule formation. Although ERN2 is dispensable for organogenesis, the subtle infection abnormalities observed suggest that ERN2 has limited but unique functions that cannot be replaced by ERN1 and this is most likely explained by the different expression patterns of the two genes (Cerri et al., 2012). In particular, ern2 nodules exhibit certain ITs devoid of bacteria (Fig. 4) in the zone where ERN2 is known to be preferentially expressed (Cerri et al., 2012; Roux et al., 2014), suggesting that ERN2 is required for appropriate bacterial development and/or maintenance within the ITs, a function that apparently cannot be totally fulfilled by ERN1.
Molecular and cytological analyses have also revealed a precocious senescence program in ern2 nodules that can lead to elongated nodules that no longer fix nitrogen (see Fig. 3F). Nodule senescence involves modifications of the redox cellular status and proteolytic activities that ultimately lead to massive degradation of cellular contents (van de Velde et al., 2006; Dupont et al., 2012). This process occurs naturally in the proximal zone as the nodule ages, but can be accelerated or prematurely orchestrated in a number of Lotus japonicus and M. truncatula symbiotic mutants, often leading to either highly reduced or completely abolished nitrogen fixation capacity as observed for a proportion of ern2 nodules (Hossain et al., 2006; Kumagai et al., 2007; Pislariu et al., 2012; Bourcy et al., 2013; Berrabah et al., 2014). It has also been suggested that nodule senescence can be triggered by the host to avoid the energetic cost of maintaining ineffective nodules (Suganuma et al., 2003; Magori and Kawaguchi, 2009). This reasoning could apply in the case of ern2, where partially defective colonization might lead to early nodule senescence. In this scenario, senescence would not be a direct consequence of the mutated gene, but rather a consequence of a host-triggered response to ineffective nodulation.
Mutation of a Conserved Thr Residue Impacts ERF DNA Binding Activity
The ern2-1 mutant allele exhibits a point mutation leading to the replacement of a highly conserved Thr (position 61) by an Ile residue within the DNA binding domain. The phenotypic and molecular characterization of this mutation has revealed that T61 has a crucial role in the DNA binding activity of ERN2. ChIP and transactivation assays have demonstrated that the mutated ERN2 protein, although correctly targeted to the nucleus, does not associate in vivo to its target promoter sequence and as a result is unable to properly activate transcription. The negative effect of this mutation on DNA binding can be adequately explained by 3D homology modeling, based on the earlier solution NMR structure determined for the AP2/ERF domain of AtERF1 interacting with the GCC-box DNA motif (Allen et al., 1998; Yamasaki et al., 2012). ERF TFs exhibit an unusual mode of DNA interaction via three β-strands contacting the DNA backbone and specific bases within the major groove, a global structure that is conserved even in the distantly related apicomplexan ERF-like proteins (Lindner et al., 2010). The T61I replacement abolishes a key hydrogen-bond interaction with the sugar-P backbone, in addition to negatively affecting the DNA base interaction of R37 (Fig. 2). The homology modeling predicts that these modifications drastically affect the overall stability of the DNA-protein complex, in line with the loss of DNA binding and transcriptional activity of the mutated T61I ERN2 (Fig. 2, E and F). Consistent with these findings, T61 has recently been shown to play an important role in stabilizing the binding complex of a barley ERF TF (Pandey et al., 2015) and that R37 is important for DNA binding by group IXa Orca3 ERF TFs (Shoji et al., 2013).
ERN1 and ERN2 Together Regulate NF Signaling Genes
While individual ERN1 or ERN2 TFs are sufficient to activate ENOD gene expression in transient assays, they are both required in the native M. truncatula context to assure optimal levels of ENOD11 and ENOD12 expression in RHs responding to NF treatments. Although these TFs are coexpressed at this key early symbiotic stage, there are nevertheless slight differences in both levels and spatio-temporal expression patterns (Cerri et al., 2012). ERN1, which is up-regulated within 1 to 3 hpi in roots treated with NFs or following inoculation with S. meliloti (Cerri et al., 2012; Larrainzar et al., 2015), probably plays a predominant role in the early activation of target genes, while ERN2, whose constitutive level is increased slightly later (approximately 6 hpi), is likely to assure optimal expression levels at later time points or in cooperation with ERN1. To date there is no evidence that ERN1 and ERN2 can function directly in cooperation, although heterodimerization has been reported to occur with a number of ERF TFs (Licausi et al., 2013; Tripathi et al., 2015; Zhao et al., 2015). The comparison of the transcriptome from single versus double mutants should help in identifying downstream genes under the control of these two complementary TFs.
While ENOD gene expression is strongly down-regulated in the ern mutant backgrounds, the expression of a number of other symbiosis-associated genes increased in ern1-1 or ern1-1 ern2-1 backgrounds (Fig. 7). ERN1 has recently been shown to be under the negative control of an ethylene signaling pathway proposed to attenuate NF signaling responses via a negative feedback loop (Larrainzar et al., 2015). In this scenario, ERN1/ERN2 could contribute to this negative feedback-loop by modulating the expression of certain NF signaling genes. Among these genes NIN has recently been shown to be coexpressed and interconnected with ERN1 and also to participate in this negative feedback loop (Yoro et al., 2014; Larrainzar et al., 2015; Soyano et al., 2015). The interconnection between these TFs has been suggested to occur via the NIN target NF-YA1, which is able to directly regulate the expression of ERN1 (Laloum et al., 2014; Larrainzar et al., 2015). However, NIN can also negatively affect ERN TF function by competitively inhibiting the induction of ERN NF-responsive target genes (Marsh et al., 2007; Vernié et al., 2015). The antagonistic function between them may allow the fine-tuning of gene expression at particular symbiotic stages. Future genetic analyses and a spatio-temporal analysis of NIN in relation to ERN TFs should help to elucidate these functional relationships.
ERN1 and ERN2 Coordinately Regulate Rhizobial Infection and Nodule Organogenesis
The ern1-1 ern2-1 double mutant exhibits a very severe symbiotic phenotype, being unable to form normal curled RHs with entrapped bacteria. Nevertheless, RHs still respond to rhizobial inoculation, with the formation of spatula-like tip swellings, reminiscent of the so-called “hair swelling” phenotype reported for other early NF signaling mutants such as dmi (Catoira et al., 2000, and reviewed by Charpentier and Oldroyd, 2010). This pinpoints a new role for these factors at a very early stage preceding bacterial entrapment, before complete RH curling and infection chamber development (Fournier et al., 2015). NIN was also recently shown to be required during this early stage of infection chamber remodeling (Fournier et al., 2015). This suggests that ERN1/ERN2 and NIN are all associated with RH infection via similar or parallel pathways. It is possible that combinatorial networks of TFs are required to allow proper RH remodeling for the initiation of ITs. This RH developmental program is probably unique to rhizobial nodulation and not part of the common symbiotic pathway, as both nin and the double ern1-1 ern2-1 mutant are not defective for arbuscular mycorrhizal colonization (Supplemental Fig. S6).
While ERN1 and ERN2 have redundant roles during rhizobial infection, this is not the case during root cortical entry that appears to be dependent only on ERN1. We presume that this is due to subtle differences in tissue-specific expression levels rather than functional differences between the two symbiotic TFs, since ERN2 can fully replace ERN1 when expressed under the ERN1 promoter (Cerri et al., 2012). In contrast with initial root infection stages, ERN1 plays a predominant role during nodulation, which is in line with the fact that ERN1 is highly expressed throughout root nodulation as compared to ERN2. Previous phylogenetic and transcriptomic studies have suggested that ERN1, originally derived from ERN2 via gene duplication, might have a more specialized function during nodulation, whereas ERN2 would be specialized for arbuscular mycorrhization (Young et al., 2011). However, although ERN2 is indeed expressed in arbuscule-containing cells (Czaja et al., 2012), the ern1-1 ern2-1 double mutant does not have a pronounced arbuscular mycorrhizal colonization phenotype (Supplemental Fig. S6). Further detailed analysis will now be needed to examine whether mutations in either of the ERN TF genes modify the efficiency of arbuscular mycorrhizal colonization, as shown for NSP1 and NSP2 (Maillet et al., 2011; Delaux et al., 2013).
Several lines of evidence point to a major role of ERN1 during the development of mature nitrogen-fixing nodules. The ern1-1 mutant is unable to form mature nodules but can initiate root cortical cell divisions that lead to small noninfected underdeveloped nodules that can be visualized within weeks after bacterial inoculation (Middleton et al., 2007). ERN1 was thus suggested to be required at a differentiation stage allowing the transition from proliferating cells to the development of mature infected nodules (Cerri et al., 2012). In contrast, the double mutant line, which is blocked prior to rhizobial infection, is also completely blocked for nodule organogenesis. This is consistent with previous work demonstrating that ern1-1 roots over-expressing an autoactive form of CCaMK are unable to support spontaneous nodule formation (Middleton et al., 2007). Epidermal-specific expression of ERN1 or ERN2 can restore rhizobial infection in the double mutant but not nodule organogenesis that can be promoted only by the expression of ERN1 or ERN2 under the control of their native promoters (Fig. 8). Thus, plants expressing only ERN1 (either ern2 lines or the double mutant complemented with ERN1) can form mature infected nodules whereas plants expressing only ERN2 (either ern1 or the double mutant complemented with ERN2) develop preferentially noninfected underdeveloped nodules. This suggests that the presence of either ERN1 or ERN2 can independently trigger nodule organogenesis upon rhizobial inoculation. However, this occurs despite the fact that ERN2 is not expressed locally in developing nodule primordia (Cerri et al., 2012). We therefore suggest that the expression of ERN1/ERN2 in their native environments (most likely in the epidermis and outer cortex during infection) is required for the induction of the nodule organogenesis signaling pathway in inner cortical cells. Xiao et al. (2014) have recently established a M. truncatula nodule fate map, describing the involvement of different root tissues in nodule formation. Applying this fate map to the single ern1 mutant and the double mutant should help to identify the precise developmental steps that are impaired in these mutants. While root cortical cell proliferation is likely dependent on ERN1/ERN2 function during early infection, the subsequent development of nodules requires a unique ERN1 function, probably through another signaling program involving the local expression of ERN1 (Cerri et al., 2012).
MATERIALS AND METHODS
Plant Material, Bacterial Strains, and Fungal Spores
Both of the Medicago truncatula genotypes A17 and R108 were used in this study. The ern2-1 mutant was obtained following reverse screening of a chemically EMS mutagenesis M. truncatula A17 population (Le Signor et al., 2009) and contains a point mutation in the ERN2 coding sequence leading to the replacement of Thr-61 by an Ile residue. The mutant was back-crossed twice with the A17 wild type, and phenotypic analysis was subsequently performed on the homozygous mutant and the wild-type line derived from the same mutagenized population and referred to as A17S. The insertion of a Tnt1 retrotransposon in NF5892 (R108 genotype; Tadege et al., 2008; Pislariu et al., 2012) was identified via the transposon display PCR protocol (Ratet et al., 2010) using primers ERN2-219-Fw, ERN2-220-Rev, LTR6-Rev, and ERN2-222-Rev (Supplemental Table S1), and the R1 population from which we subsequently obtained the segregating homozygous ern2-2 mutant and a sibling wild-type line, referred to as R108S. The double mutant ern1-1 ern2-1 was generated by manual crossing between ern1-1 (carrying the stably integrated ProENOD11:GUS fusion; Middleton et al., 2007) as female parent and ern2-1 (this study). The progeny of two independent crosses was screened for the presence of the GUS gene through PCR using the primers ProENOD11-935-Fw and GUS-1-Rev, as described in Cerri et al. (2012). Nodulation and GUS-minus F2 progeny plants were selected for genomic DNA extraction, PCR amplifications (using primer pairs ProERN1-654-Fw/ERN1-Rev1 and ERN2-11-Fw/ERN2-1060-Rev listed in Supplemental Table S1), and sequencing to select F2 plants harboring both the ern1-1 deletion and the ern2-1 point mutation. Selected F2 plants were then used for seed production. Phenotyping was performed on F3-F4 homozygous plants for both mutations. Plant seeds were scarified and surface-sterilized prior to germination on inverted soft Campbell agar plates, as described in Cerri et al. (2012). Seedlings were transferred to aeroponic growth conditions or used for Agrobacterium rhizogenes transformation as described in the Medicago Handbook (http://www.noble.org/medicagohandbook). Plasmid DNAs were used to transform Escherichia coli DH5, A. rhizogenes ARquA1 (Quandt et al., 1993), or Agrobacterium tumefaciens GV3101 and GV3103 bacterial strains. Sinorhizobium meliloti Sm 2011-lacZ (constitutively expressing a hemA-lacZ fusion; Ardourel et al., 1994) or Sm 2011-CFP (constitutively expressing CFP, kindly provided by P. Smit) were propagated on tryptone yeast medium supplemented with 6 mm CaCl2 in the presence of the appropriate antibiotic, and used for M. truncatula nodulation assays. Spores of Rhizophagus irregularis were purchased at Agronutrition (Carbonne, France).
DNA Constructs
To generate the ERN2 construct mimicking the ern2-1 mutation for transactivation and DNA binding studies in Nicotiana benthamiana, a 537-bp amplicon was generated by PCR amplification using the ern2-1 genomic DNA as template and the ERN2-11-Fw and ERN2-Rev1 primers (Supplemental Table S1). This DNA fragment was further subcloned into pGEM-T (Promega, Madison, WI), and then digested by HpaI/KpnI restriction enzymes, thus generating a 288-bp fragment containing the ern2-1 mutation (Fragment A). Starting from the pDONOR-207-ERN2 plasmid (Andriankaja et al., 2007), a 325-bp DNA fragment was isolated by ApaI/HpaI restriction (Fragment B) containing both vector and 5′ ERN2 sequences. Fragments A and B were then ligated into pDONOR-207-ERN2 digested with ApaI/KpnI, thus generating a new pDONOR-207-ERN2* plasmid where the ERN2 sequence carries the ern2-1 T61I mutation. pDONOR-207-ERN2* was then used for LR recombination in the destination vectors PAMPAT-35S-3HA and PAMPAT-35S-YFP, subsequently used for transactivation assays, subcellular location, and chromatin immunoprecipitation (ChIP) in N. benthamiana. PAMPAT-35S vectors expressing native HA or YFP-tagged ERN1, ERN2, and DNA-binding deleted versions ΔDB and pLP100 binary vectors expressing the GUS reporter, YFP-ERN1 and YFP-ERN2 fusions under the control of ProERN1 or ProERN2 were generated before (Andriankaja et al., 2007; Cerri et al., 2012), while ProEXPA:GUS, ProEXPA:ERN1, and ProEXPA:ERN2 constructs were obtained as follows: the coding sequences of GUS, ERN1, and ERN2 in the pDON207 vector were used for recombination into the gateway destination vector pK7WG2-R-ProEXPA comprising 401 bp of promoter sequences of the EXPA gene (Vernié et al., 2015).
Modeling the ERN2 DNA Complex
Homology models were generated using Accelrys Discovery Studio v. 3.1 (DS 3.1, v. 2011; Accelrys, San Diego, CA). The NMR-derived structure of the AtERF1 DBD (PDB: 1GCC; chain A) was used as a structural template to model MtERN2 DBD (62% sequence identity). Structure-guided sequence alignment was carried out using the Align 123 program (from InsightII, Accelrys) and served as an input for the automated homology modeling program MODELLER v. 9.8 (https://salilab.org/modeller/). Out of 50 possible models, we selected the one with the lowest total energy as well as the best Profiles-3D score, reflecting an optimal folding consistency. The structure of the mutated T61I ERN2 DBD was derived from the native ERN2 DBD model within MODELLER (https://salilab.org/modeller/). DNA building and molecular docking were performed using the Biopolymer, Discover, and Docking modules (from InsightII v. 2005, Accelrys). The 11-bp GCC-like promoter DNA fragment 5′-TTGCAGGCCTA-3′ was modeled within the Biopolymer module using the inter-base-pair structural parameters (rise, twist, tilt, and roll) inferred from the homologous NMR-derived DNA structure (PDB: 1GCC; chains B-C). The resulting structure was minimized using the Discover consistent valence force field (from InsightII v. 2005, Accelrys). Note that the nucleotides of the coding and complementary strands in the 5′-3′ direction are numbered 1 to 11 and 1′ to 11′, respectively (Supplemental Fig. S1; Fig. 2, B–D). The solution structure of the AtERF1 DBD/GCC-box complex (PDB: 1GCC) was used as a framework to pre-position the ERN2 DBD 3D model relative to its GCC-like DNA target. The structure of the complex was then processed by the Affinity program (https://affinity.serif.com/en-us/), an automatic docking refinement procedure within the Docking module (from InsightII, Accelrys) that follows a Monte Carlo interaction energy minimization protocol and allows flexibility to predefined atoms of both the ligand-DBD and receptor-DNA. The criterion used for identifying hydrogen bonds was based on a donor-acceptor distance ≤ 3 Å and a minimum donor-proton-acceptor angle of 120°.
ChIP and Quantitative PCR/RT-PCR Analyses
For ChIP quantitative (Q) PCR experiments, N. benthamiana leaves were harvested 36 h postinfiltration with A. tumefaciens suspensions and cross linked in 1% formaldehyde for 20 min, at room temperature, under vacuum infiltration. After quenching with 125 mm glycine, leaves were rinsed twice in water, then dried and ground in liquid nitrogen. Powdered tissue (3–4 g) was resuspended in Extraction Buffer 1 (EB1), filtered through a 100-μm nylon filter (Millipore, Billerica, MA), and centrifuged (125g, 4°C, 20 min). The pellet was resuspended in EB2, centrifuged (397g, 4°C, 10 min), and the pellet resuspended in EB3. After an additional centrifugation (18,400g, 4°C, 1h), the resulting nuclei pellet was resuspended in Nuclei Lysis buffer prior to sonication (5 cycles of 10 min with 30 s high-intensity pulse, with 30 s interval without pulse; Bioruptor, Diagenode, Liège, Belgium). Composition of EB1, EB2, and EB3 buffers is described in Singh et al. (2014). Chromatin shearing was monitored on 1% agarose gel prior to immunoprecipitation. Input aliquots (10% of the total volume) were removed and immunoprecipitation was performed on the remaining purified chromatin for 2 to 3 h with μMACS anti-HA magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany) and washed with low (150 mm NaCl) and high salt (500 mm NaCl) buffers according to Yamaguchi et al. (2014). Input and anti-HA coupled beads were resuspended in NaCl solution (0.2 m) and chromatin was de-cross-linked at 65°C for 2 h, before incubating for 30 min at room temperature with RNAseA 100 μg/μL (2–5 μL/sample) and for 45 min at 45°C with proteinase K 10 μg/μL (1 μL/1 sample).
After phenol/chloroform extraction, DNA was precipitated (0.3 m sodium acetate, 1:3 v/v 100% ethanol, 30 μg glycogen) and dissolved in water. The presence of NF-box specific sequences in the chromatin immunoprecipitate was detected by QPCR (cycling conditions: 95°C, 5 min followed by 45 cycles of denaturation at 95°C, 15 s, annealing at 62°C, 20 s, and elongation at 72°C, 20 s) using primer pairs Ch-NF-box-Fw and Ch-NF-box-Rev (see Supplemental Table S1). The % of input was calculated for each sample using the equation 100 × 2(Adjust Ct Input − Ct ChIP), whereby Ct = Cycle threshold; Adjust Ct Input = Raw Ct Input − log2 (dilution factor). Data represents fold enrichment for each sample calculated as follows: (% Input TF + % Input 4xNF-box)/% Input 4xNF-box. Values were obtained from three to four independent biological replicates (n = 12–16 leaves) with two to four technical replicates. QRT-PCR analysis was performed as described previously in Cerri et al. (2012). The primer pairs used in this study to amplify TC100437, TC106338, NIN, VPY, RPG, Mt0089_00067, Mt0085_00020, and ENOD12 transcripts are listed in Supplemental Table S1. Those used to amplify ERN1, ERN2, ENOD11, and Ubiquitin transcripts can be found in Cerri et al. (2012). The data represent mean values of three to five independent biological experiments, with at least two technical replicates after normalization with the three reference genes, Ubiquitin, Mt0089_00067, and Mt0085_00020, shown before to exhibit invariable expression levels (Cerri et al., 2012; Roux et al., 2014). Primer pairs to amplify PT4 and Mtr3213.1S1_at (Hogekamp and Küster, 2013) are listed in Supplemental Table S1. PT4 transcript levels represent mean values obtained after normalization with the three reference genes and Mtr.3213.1.S1_at levels.
Transient Expression in N. benthamiana Leaves
A. tumefaciens GV3101 and GV31303 strains containing PAMPAT-35S binary vectors harboring either 3-HA tagged ERN1 and ERN2 proteins (native, ΔDB, and T61I) or the gain-of-function 4xNFbox:GUS fusion (Andriankaja et al., 2007 and this study) were grown on Luria-Bertani medium supplemented with the appropriate antibiotics at 28°C, harvested and resuspended in Agro Mix before infiltration of N. benthamiana leaves as described previously in Andriankaja et al. (2007) and Cerri et al. (2012). Leaf discs were harvested 36 h postinfiltration for direct histochemical GUS assays or frozen in liquid nitrogen prior to quantitative enzymatic GUS fluorimetric assays or western-blot analyses using anti-HA HRP coupled rat monoclonal antibodies (catalog no. 3F10; Roche, Basel, Switzerland).
Bacterial Inoculation and Nodulation Factor Treatment of M. truncatula roots
M. truncatula plants grown in aeroponic conditions were treated with S. meliloti, nodulation factor (NF), or water control treatments as follows. Germinated seedlings were transferred to aeroponic conditions (nitrogen-free Farhaeus media) and nitrogen-starved for 3 d before being inoculated with a S. meliloti bacterial suspension (OD600 = 0.1). Root or nodule samples were then collected at different days or weeks post-bacterial inoculation (1–3 d, 1–6 weeks). For NF treatments, 3-d nitrogen-starved plantlets were transferred to 50-ml Falcon tubes (Becton Dickinson, Franklin Lakes, NJ) wrapped with a black plastic bag and filled with either water (control) or a solution of 10−9 m purified NFs. Samples were then harvested for RNA extraction after 6 h for NF/control treatments. For complementation studies, plants were grown in attapulgite (Oil Dri US Special; http://www.oildri.com/)- containing pots. Briefly, transformed composite plants carrying the appropriate binary vectors were transferred to attapulgite pots two-three weeks after A. rhizogenes transformation (Cerri et al., 2012). Then three or four plants/pots were placed in mini heated greenhouses in the growth chamber for 3 d before inoculation with the S. meliloti 2011-lacZ rhizobial strain (4 mL/pot, OD600 = 0.5). Pots were watered every day, and supplied with liquid Fahraeus medium once a week. Roots were collected 2 to 4 weeks after inoculation for microscopic analysis or RNA extractions.
Inoculation of M. truncatula Plants with R. irregularis
M. truncatula A17 and ern1-1 ern2-2 germinated seedlings were transferred to 50-ml Falcon tubes (Becton-Dickinson) with autoclaved attapulgite (Oil Dri) substrate mixed with 20 mL of low phosphate Long Ashton medium (LALP; Maillet et al., 2011). Inoculation with R. irregularis was performed by mixing 50 spores per plant, by adding the spore suspension to the humidified substrate. Falcon tubes (Becton-Dickinson) were then placed into water-containing pots wrapped in black plastic bags, in a 25°C growth chamber under a plastic dome to maintain high humidity. Three to four days later, plantlets were placed under direct light (400 μmol m−2 s−1 intensity). The entire root system of individual plants contained in the Falcon tube (Becton-Dickinson) was collected after 2 to 3 weeks postinoculation by rinsing in water, and either frozen in liquid nitrogen for subsequent RNA extractions or ink-stained for later microscopic observations. To obtain blue-colored fungal structures, roots were cleared by boiling (95°C) in a 10% KOH solution for 3 to 5 min, rinsed in water, and stained by boiling roots in a 5% black ink solution (Sheaffer Pen, Shelton, CT) with 5% acetic acid at 95°C for 5 min (Vierheilig et al., 1998).
GUS and β-Galactosidase Assays
Histochemical (blue) staining for GUS activity was performed using the substrate X-Gluc (5-bromo-4-chloro-3-indoxyl-β-d-GlcA, cyclohexylammonium salt, B7300; Biosynth, Staad, Switzerland) as described in Cerri et al. (2012). For enzymatic GUS assays, leaf tissues were ground in liquid nitrogen and homogenized in the GUS extraction buffer as described in Andriankaja et al. (2007). The Magenta-Gal substrate (5-bromo-6-chloro-3-indoxyl-β-d-galactopyranoside; B7200; Biosynth) or the blue X-Gal substrate (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, W5376C; Thermo Fisher Scientific, Guilford, CT) were used for histochemical staining of the constitutive β-galactosidase activity within S. meliloti-containing infection threads as described before in Cerri et al. (2012).
Acetylene Reduction Assay
Nitrogenase activity of A17, A17S, and ern2-1 plants was determined using the acetylene reduction assay on either whole-root systems or isolated nodules from plants grown in aeroponic conditions 4 or 6 weeks postinoculation with S. meliloti 2011-lacZ, respectively. Individual root systems or isolated nodules were incubated at 25°C in 60-ml sealed vials containing 0.5 mL of Fahraeus liquid media in the presence of 10% (vol/vol) acetylene. After a 30 min reaction, a time series of three 200 μL aliquots per vial was sampled and the ethylene production was quantified by gas chromatography (model no. GC 7280A; Agilent Technologies, Lexington, MA) by comparison with ethylene standard samples (100 mg/liter).
Microscopy Methods
Microscopic observations with a stereomicroscope (Leica Microsystems, Wetzlar, Germany), a light microscope (AxioPlan 2 Imaging; Carl Zeiss, Oberkochen, Germany), and/or a CCD camera (AxioCam MRc; Carl Zeiss) were done with root or nodule tissues before or after staining for β-galactosidase activity. Nodulated roots mounted in slides were also imaged using a digital slide nanoscanner (Hamamatsu Photonics, Hamamatsu City, Japan). Nodules from A17 wild type, R108 wild type, ern2-1, and ern2-2 mutants grown in aeroponic conditions were isolated and embedded in Epon 812 resin (Electron Microscopy Sciences, Hatfield, PA) to obtain either thin (1 μm) or ultra-thin sections (70–90 nm), respectively, using a Reichert-Jung Ultracut E Ultramicrotome (Ametek Reichert Technologies, Depew, NY). Briefly, the nodules were fixed in 2.5% glutaraldehyde diluted in 0.2 m cacodylate buffer pH = 7.2 for 1 h, including 30 min under vacuum. Then, the nodules were rinsed in the same buffer and postfixed in 2% osmium tetroxyde (diluted in 0.2 m sodium cacodylate), then rinsed again, and progressively dehydrated in ethanol (25, 50, 70, and 90% for 1 h and pure for 2 h), propylene oxide (2 × 1 h) and finally embedded in Epon 812 resin following the manufacturer’s instructions. Ultra-thin sections were subsequently prepared for electron microscopy observations (1200 EX; JEOL USA, Peabody, MA). For optical microscopy, nodule sections were stained with methylene blue (0.2%) and toluidine blue (1%) in borax (1%) and basic fuchsin (0.07%) in water before observations with optic microscope (Axioplan Imaging; Carl Zeiss).
For in vivo observations of double mutant and A17 wild-type root hairs after rhizobial inoculation, plants were grown with their roots covered with a gas-permeable and transparent Lumox plastic film (Sarstedt, Numbrecht, Germany) before inoculation with GFP-expressing rhizobia (Cerri et al., 2012). Inoculated roots were imaged using a light microscope (Axiophot 2, Carl Zeiss) and root infection sites were imaged using a TCS SP2 AOBS confocal laser-scanning microscope (Leica Microsystems) equipped with a long-distance 40× water-immersion objective (HCX Apo L 0.80; Leica Microsystems). The 488-nm argon laser was used to excite GFP and a 561-nm diode to excite cell-wall autofluorescence. Specific emission windows of 500 to 525 nm and 620 to 720 nm were used for GFP and autofluorescence signals, which were colored, respectively, in green and red. Images were processed using the confocal laser-scanning microscope (Leica Microsystems) and the Fiji (fiji.sc; Schindelin et al., 2012; Schneider et al., 2012) and Velocity v. 6.0.1 (Perkin-Elmer, Waltham, MA) softwares. The fluorescence images shown are maximal projections of a Z-stack.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers M. truncatula ERN1 (EU038802) and ERN2 (EU038803); Pisum sativum PsERN (EF396329); Glycine max GmERN (XM_006604176); and Lotus japonicus LjERN (AP006677.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of DNA binding interactions of AtERF1 and MtERN2 AP2/ERF domains.
Supplemental Figure S2. The mutated T61I ERN2 is targeted to the nuclear compartment but is no longer able to activate transcription.
Supplemental Figure S3. ern2 mutant lines exhibit a nodule senescence-like phenotype.
Supplemental Figure S4. ern2 mutants display a slightly reduced nodulation phenotype.
Supplemental Figure S5. The double ern1ern2 mutant is totally defective in root nodulation.
Supplemental Figure S6. Colonization of A17 wild-type and ern1-1 ern2-1 mutant roots by R. irregularis.
Supplemental Figure S7. Complementation of the double ern1-1 ern2-1 mutant with either ERN1 or ERN2 driven by their respective promoters.
Supplemental Table S1. List of the primers used in this study.
Supplementary Material
Acknowledgments
Thanks to M. Beck for critical reading of the manuscript. We also thank F. Maillet and C. Gough for providing S. meliloti NFs and R. irregularis spore suspension; E.-P. Journet, C. Chervin, and W. Bian for advice and technical assistance to perform gas chromatography; A. Niebel for advices with ChIP experiments; Y. Martinez from the FR AIB Imagery platform for helping with slide scans; and T. Vernié for providing the pEXPA sequence and C. Satgé and P. Gamas for providing reference Mt0089_00067 and Mt0085_00020 primers. Thanks to L. Salaoui for technical assistance for ProEXPA complementation and M. Castaingts for helping in ern2 mutant inoculation experiments.
Glossary
- ChIP
chromatin immunoprecipitation
- EMS
ethyl methanesulfonate
- IT
infection thread
- ITs
infection threads
- NF
Nod factor
- NFs
Nod factors
- NIN
Nodule Inception
- NP
nodule primordia
- RH
root hair
- RHs
root hairs
- RHC
root hair curling
- RHCs
root hair curlings
- TF
transcription factor
- TFs
transcription factors
Footnotes
Articles can be viewed without a subscription.
References
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M (1998) A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J 17: 5484–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F (2007) AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolín-Llovera M, Ried MK, Parniske M (2014) Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr Biol 24: 422–427 [DOI] [PubMed] [Google Scholar]
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Godfroy O, de Billy F, Saurat O, Jauneau A, Gough C (2008) The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc Natl Acad Sci USA 105: 9817–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B, et al. (2014) A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol 203: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Bourcy M, Brocard L, Pislariu CI, Cosson V, Mergaert P, Tadege M, Mysore KS, Udvardi MK, Gourion B, Ratet P (2013) Medicago truncatula DNF2 is a PI-PLC-XD-containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol 197: 1250–1261 [DOI] [PubMed] [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109: 13859–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Sun J, Wysham D, Otegui MS, Venkateshwaran M, Hirsch S, Miwa H, Downie JA, Morris RJ, Ané JM, et al. (2011) Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA 108: 14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GE, Barker DG, Fournier J, de Carvalho-Niebel F (2012) Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol 160: 2155–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Oldroyd G (2010) How close are we to nitrogen-fixing cereals? Curr Opin Plant Biol 13: 556–564 [DOI] [PubMed] [Google Scholar]
- Charron D, Pingret JL, Chabaud M, Journet EP, Barker DG (2004) Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol 136: 3582–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DS, Liu CW, Roy S, Cousins D, Stacey N, Murray JD (2015) Identification of a core set of rhizobial infection genes using data from single cell-types. Front Plant Sci 6: 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja LF, Hogekamp C, Lamm P, Maillet F, Martinez EA, Samain E, Dénarié J, Küster H, Hohnjec N (2012) Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipochitooligosaccharides. Plant Physiol 159: 1671–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux P-M, Bécard G, Combier J-P (2013) NSP1 is a component of the Myc signaling pathway. New Phytol 199: 59–65 [DOI] [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Promé JC (1996) Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65: 503–535 [DOI] [PubMed] [Google Scholar]
- Dupont L, Alloing G, Pierre O, El Msehli S, Hopkins J, Hérouart D, Frendo P (2012) The legume root nodule: from symbiotic nitrogen fixation to senescence. In Senescence, Ed. Tetsuji Nagata, Intech Publisher doi: 10.5772/34438: 137–157 [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Fournier J, Teillet A, Chabaud M, Ivanov S, Genre A, Limpens E, de Carvalho-Niebel F, Barker DG (2015) Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol 167: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29: 593–617 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GE (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogekamp C, Küster H (2013) A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 14: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B, Yeun LH, Domonkos A, Halász G, Gobbato E, Ayaydin F, Miró K, Hirsch S, Sun J, Tadege M, et al. (2011) Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol Plant Microbe Interact 24: 1345–1358 [DOI] [PubMed] [Google Scholar]
- Hossain MS, Umehara Y, Kouchi H (2006) A novel fix- symbiotic mutant of Lotus japonicus, Ljsym105, shows impaired development and premature deterioration of nodule infected cells and symbiosomes. Mol Plant Microbe Interact 19: 780–788 [DOI] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312 [DOI] [PubMed] [Google Scholar]
- Kumagai H, Hakoyama T, Umehara Y, Sato S, Kaneko T, Tabata S, Kouchi H (2007) A novel ankyrin-repeat membrane protein, IGN1, is required for persistence of nitrogen-fixing symbiosis in root nodules of Lotus japonicus. Plant Physiol 143: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, Baudin M, Frances L, Lepage A, Billault-Penneteau B, Cerri MR, Ariel F, Jardinaud MF, Gamas P, de Carvalho-Niebel F, et al. (2014) Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J 79: 757–768 [DOI] [PubMed] [Google Scholar]
- Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud MF, Mun JH, Larrainzar E, Cook DR, Gamas P, et al. (2014) The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J Exp Bot 65: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E, Riely BK, Kim SC, Carrasquilla-Garcia N, Yu H-J, Hwang H-J, Oh M, Kim GB, Surendrarao AK, Chasman D, et al. (2015) Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol 169: 233–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Signor C, Savois V, Aubert G, Verdier J, Nicolas M, Pagny G, Moussy F, Sanchez M, Baker D, Clarke J, et al. (2009) Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol J 7: 430–441 [DOI] [PubMed] [Google Scholar]
- Libault M, Joshi T, Benedito VA, Xu D, Udvardi MK, Stacey G (2009) Legume transcription factor genes: what makes legumes so special? Plant Physiol 151: 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Lindner SE, De Silva EK, Keck JL, Llinás M (2010) Structural determinants of DNA binding by a P. falciparum ApiAP2 transcriptional regulator. J Mol Biol 395: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magori S, Kawaguchi M (2009) Long-distance control of nodulation: molecules and models. Mol Cells 27: 129–134 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S (2006) Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res 13: 255–265 [DOI] [PubMed] [Google Scholar]
- Murray JD. (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Murray JD, Muni RR, Torres-Jerez I, Tang Y, Allen S, Andriankaja M, Li G, Laxmi A, Cheng X, Wen J, et al. (2011) Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J 65: 244–252 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Ovchinnikova E, Journet EP, Chabaud M, Cosson V, Ratet P, Duc G, Fedorova E, Liu W, den Camp RO, Zhukov V, et al. (2011) IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago Spp. Mol Plant Microbe Interact 24: 1333–1344 [DOI] [PubMed] [Google Scholar]
- Pandey B, Sharma P, Tyagi C, Goyal S, Grover A, Sharma I (2015) Structural modeling and molecular simulation analysis of HvAP2/EREBP from barley. J Biomol Struct Dyn 19: 1–17 [DOI] [PubMed] [Google Scholar]
- Pislariu CI, Murray JD, Wen J, Cosson V, Muni RR, Wang M, Benedito VA, Andriankaja A, Cheng X, Jerez IT, et al. (2012) A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiol 159: 1686–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Ott T (2011) Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr Opin Plant Biol 14: 458–467 [DOI] [PubMed] [Google Scholar]
- Quandt HJ, Puhler A, Boer I (1993) Transgenic root-nodules of Vicia hirsuta, a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Ratet P, Porcedu A, Tadege M, Myosore K (2010). Insertional mutagenesis in Medicago truncatula using Tnt1 retrotransposon. In Medicago truncatula Handbook. http://www.noble.org/Global/medicagohandbook/pdf/InsertionalMutagenesis.pdf
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Mishima M, Hashimoto T (2013) Divergent DNA-binding specificities of a group of ETHYLENE RESPONSE FACTOR transcription factors involved in plant defense. Plant Physiol 162: 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG (2009) A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol 151: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152 [DOI] [PubMed] [Google Scholar]
- Singh S, Parniske M (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15: 444–453 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Soyano T, Hayashi M (2014) Transcriptional networks leading to symbiotic nodule organogenesis. Curr Opin Plant Biol 20: 146–154 [DOI] [PubMed] [Google Scholar]
- Soyano T, Shimoda Y, Hayashi M (2015) NODULE INCEPTION antagonistically regulates gene expression with nitrate in Lotus japonicus. Plant Cell Physiol 56: 368–376 [DOI] [PubMed] [Google Scholar]
- Suganuma N, Nakamura Y, Yamamoto M, Ohta T, Koiwa H, Akao S, Kawaguchi M (2003) The Lotus japonicus Sen1 gene controls rhizobial differentiation into nitrogen-fixing bacteroids in nodules. Mol Genet Genomics 269: 312–320 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tripathi P, Rabara RC, Choudhary MK, Miller MA, Huang YS, Shen QJ, Blachon S, Rushton PJ (2015) The interactome of soybean GmWRKY53 using yeast 2-hybrid library screening to saturation. Plant Signal Behav 10: e1028705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Velde W, Guerra JC, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S (2006) Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernié T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford M, de Carvalho-Niebel F, Oldroyd GED (2015) The NIN transcription factor coordinates diverse nodulation programmes in different tissues of the root. Plant Cell 27: 3410–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierheilig H, Coughlan AP, Wyss U, Piche Y (1998) Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64: 5004–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T (2014) Fate map of Medicago truncatula root nodules. Development 141: 3517–3528 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kwon CS, William DA, Wagner D (2014) PROTOCOLS: chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S (2012) DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci 18: 267–276 [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoro E, Suzaki T, Toyokura K, Miyazawa H, Fukaki H, Kawaguchi M (2014) A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus. Plant Physiol 165: 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Cheng S, Song Y, Huang Y, Zhou S, Liu X, Zhou DX (2015) The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27: 2469–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.