A repression domain-fused form of VIP1 represses the expression of a subset of touch-responsive genes and enhances touch-induced root waving, suggesting that VIP1 regulates root touch responses.
Abstract
VIP1 is a bZIP transcription factor in Arabidopsis (Arabidopsis thaliana). VIP1 transiently accumulates in the nucleus when cells are exposed to hypoosmotic conditions, but its physiological relevance is unclear. This is possibly because Arabidopsis has approximately 10 close homologs of VIP1 and they function redundantly. To examine their physiological roles, transgenic plants overexpressing a repression domain-fused form of VIP1 (VIP1-SRDXox plants), in which the gene activation mediated by VIP1 is expected to be repressed, were generated. Because hypoosmotic stress can mimic mechanical stimuli (e.g. touch), the touch-induced root-waving phenotypes and gene expression patterns in those transgenic plants were examined. VIP1-SRDXox plants exhibited more severe root waving and lower expression of putative VIP1 target genes. The expression of the VIP1-green fluorescent protein (GFP) fusion protein partially suppressed the VIP1-SRDX-induced increase in root waving when expressed in the VIP1-SRDXox plants. These results suggest that VIP1 can suppress the touch-induced root waving. The VIP1-SRDX-induced increase in root waving was also suppressed when the synthetic auxin 2,4-dichlorophenoxy acetic acid or the ethylene precursor 1-aminocyclopropane-1-carboxylic acid, which is known to activate auxin biosynthesis, was present in the growth medium. Root cap cells with the auxin marker DR5rev::GFP were more abundant in the VIP1-SRDXox background than in the wild-type background. Auxin is transported via the root cap, and the conditions of outermost root cap layers were abnormal in VIP1-SRDXox plants. These results raise the possibility that VIP1 influences structures of the root cap and thereby regulates the local auxin responses in roots.
VIP1 is a bZIP transcription factor in Arabidopsis (Arabidopsis thaliana). Arabidopsis bZIP proteins have been classified into 11 groups (groups A to I and S as well as one unnamed group) based on the similarities in amino acid sequences and features. VIP1 and 11 other bZIP proteins belong to group I (Jakoby et al., 2002). VIP1 and its close homologs function in plant development and responses to various stimuli. VIP1 is thought to help regulate responses to Agrobacterium tumefaciens and flg22, which is a microbe-associated molecular pattern (Tzfira et al., 2001, 2002, 2004; Lacroix et al., 2005; Li et al., 2005; Djamei et al., 2007; Pitzschke et al., 2009), although this role has been questioned (Shi et al., 2014). VIP1 also is involved in regulating responses to sulfur deficiency and hypoosmotic stress (Wu et al., 2010; Tsugama et al., 2012a). VSF-1, a close VIP1 homolog in tomato (Solanum lycopersicum), is expressed specifically in vascular tissues and regulates vascular gene expression (Torres-Schumann et al., 1996; Ringli and Keller, 1998). RF2a and RF2b, two close VIP1 homologs in rice (Oryza sativa), also are involved in regulating vascular gene expression and in regulating responses to the Rice tungro virus (Yin et al., 1997; Petruccelli et al., 2001; Dai et al., 2004, 2006, 2008). RSG, a close VIP1 homolog in tobacco (Nicotiana tabacum), regulates the biosynthesis of a phytohormone, GA (Fukazawa et al., 2000, 2010, 2011; Igarashi et al., 2001; Ishida et al., 2004, 2008; Ito et al., 2010, 2014).
The 12 members of the Arabidopsis group I bZIP proteins could be further classified into subgroups 1 and 2, as well as others on the basis of amino acid sequence similarities, and the genes encoding the subgroup 1 proteins (VIP1, PosF21, bZIP29, bZIP52, bZIP69, and bZIP30) are more highly expressed than those encoding the other group I bZIP proteins. Many of the Arabidopsis group I bZIP proteins show transcriptional activation potential in a yeast one-hybrid system and can interact physically with each other (Tsugama et al., 2014). The functional similarities among the group I bZIP proteins may explain why the phenotype of vip1-1, in which transfer DNA (T-DNA) is present in the 3′ region in the coding sequence (CDS) of VIP1, is similar to that of the wild-type plant under various conditions (Tsugama et al., 2012a, 2014). No mutants of group I bZIP proteins have been characterized other than vip1-1.
To further characterize the physiological roles of VIP1, we used the SRDX transcriptional repression domain. Overexpression of a transcription factor-SRDX fusion protein can dominantly repress the expression of genes normally activated by the transcription factor and its homologs. This may result in significant changes in plant phenotypes, enabling the evaluation of the physiological functions of these transcription factors (Mitsuda et al., 2006). Using the transgenic plants overexpressing the VIP1-SRDX fusion protein, we show that VIP1 is involved in regulating touch-induced root waving, which has implications for responses to hypoosmotic stress. Because auxin influences root-bending responses (Luschnig et al., 1998), the responses of the VIP1-SRDX-overexpressing (VIP1-SRDXox) plants to auxin also were examined.
RESULTS
Similarities in Subcellular Localization and DNA-Binding Ability of Group I bZIP Proteins
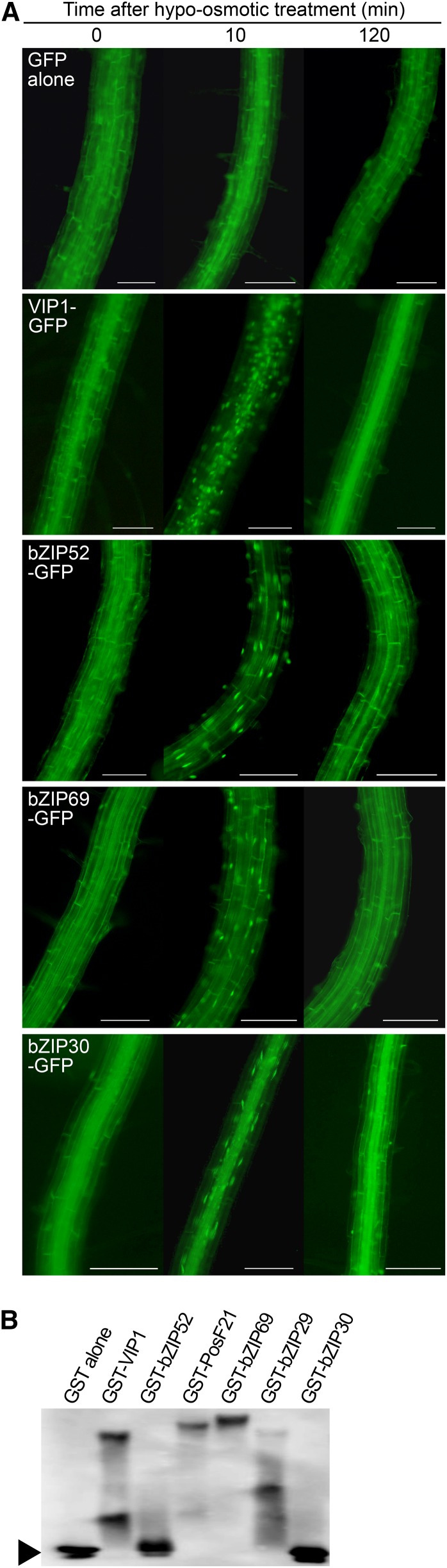
In a previous study, VIP1 and two other Arabidopsis group I subgroup 1 bZIP proteins, PosF21 and bZIP29, exhibited a similar nuclear-cytoplasmic shuttling pattern under a hypoosmotic condition (Tsugama et al., 2014). Three more group I subgroup 1 bZIP proteins (i.e. bZIP52, bZIP69, and bZIP30) were expressed as GFP-fused proteins in Arabidopsis to study their subcellular localization. The bZIP52-GFP, bZIP69-GFP, and bZIP30-GFP signals, as well as the VIP1-GFP signal, were detected mainly in the cytosol 0 and 120 min after the plants were submerged in 20 mm Tris-HCl, pH 6.8. The signals were observed in the nucleus 10 min after the plants were submerged in the same solution (Fig. 1A). Similar results were obtained when a 0.5× Murashige and Skoog (MS) liquid medium was used instead of 20 mm Tris-HCl, pH 6.8 (Supplemental Fig. S1). These proteins also were transiently expressed in lettuce (Lactuca sativa) epidermal cells, and the intensity of their signals in the nucleus increased after the cells were treated with 20 mm Tris-HCl, pH 6.8 (Supplemental Fig. S2). These results further support the idea that the nuclear localizations of the group I bZIP proteins is similarly enhanced by hypoosmotic stress.
Figure 1.
Functional similarities between the Arabidopsis group I bZIP proteins. A, Signal patterns of the group I bZIP proteins fused to GFP. One-week-old transgenic plants expressing GFP alone or the VIP1-GFP, bZIP52-GFP, bZIP69-GFP, or bZIP30-GFP fusion protein were submerged in a hypotonic solution of 20 mm Tris-HCl, pH 6.8, and analyzed by fluorescence microscopy at the indicated time points to detect GFP signals in roots. For each line and time point, more than 10 plants were used, and representative images are shown. Bars = 100 μm. B, Gel-shift assay using the group I bZIP proteins. The GST-VIP1, GST-bZIP52, GST-PosF21, GST-bZIP69, GST-bZIP29, and GST-bZIP30 fusion proteins and GST alone were expressed in E. coli, purified, and used for a gel-shift assay with the digoxigenin-labeled CYP707A1 promoter fragment as the probe. The arrowhead indicates the position of the free probe. The experiments were performed three times, and a representative result is shown.
CYP707A1 is one of the putative VIP1 target genes, and its promoter is bound by VIP1 (Tsugama et al., 2012a). The glutathione S-transferase (GST)-fused forms of bZIP52 PosF21, bZIP69, bZIP29, and bZIP30, as well as VIP1, were expressed in Escherichia coli, purified, and subjected to a gel-shift assay with the CYP707A1 promoter fragment as the probe. The GST-bZIP52 and GST-bZIP30 proteins were not expressed in E. coli, but the others were expressed at detectable levels (Supplemental Fig. S3) and decreased the electrophoretic mobility of the probe in the gel-shift assay (Fig. 1B). This result suggests that these proteins can bind to the CYP707A1 promoter in vitro and raises the possibility that they share somewhat similar DNA-binding abilities.
Overexpression of VIP1-SRDX Enhances Root Waving
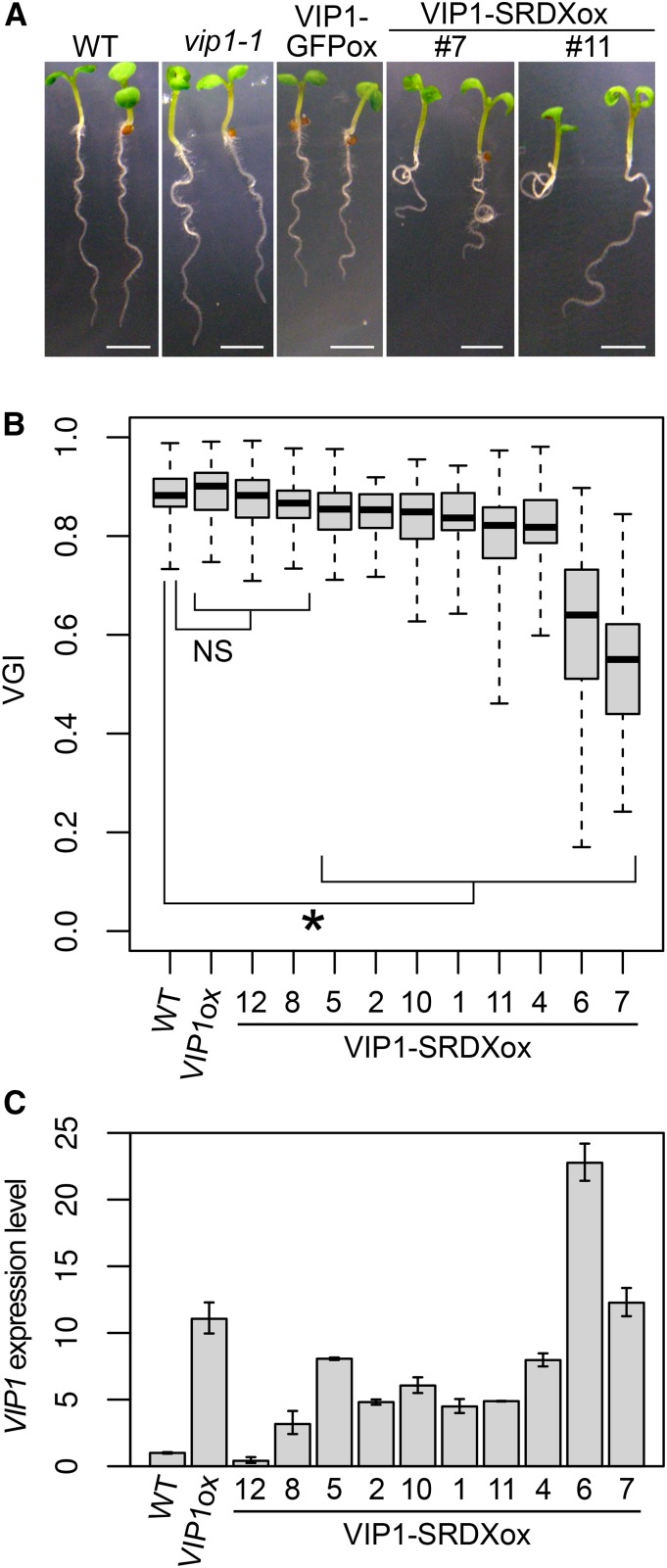
Hypoosmotic stress can induce responses similar to those induced by mechanical stimuli such as touch (Shih et al., 2014). This led us to examine the roles of VIP1 and other group I bZIP proteins in regulating root waving, which is a touch-induced response. When plants are grown on tilted impenetrable agar medium, their root tips are in continuous contact with the surface of the medium, which induces root bending. On the other hand, roots influenced by gravity (i.e. gravitropism) tend to grow in the direction of the gravitational force. The touch-induced bending and the gravitropism cause the roots to have a wavy appearance (for review, see Oliva and Dunand, 2007). The extent of root waving can be determined using the vertical growth index (VGI), which is calculated by dividing the length of the vertical projection of the primary root by the length of the primary root (Grabov et al., 2005). The VGIs of the vip1-1, bzip29-1, and vip1-1/bzip29-1 mutants were similar to those of the wild-type plants (Supplemental Fig. S4). This might be because the group I bZIP proteins have functional redundancies, which mask the effects of the T-DNA insertions in VIP1 and bZIP29. However, some of the VIP1-SRDXox lines, in which gene expression regulated by VIP1 and other group I bZIP proteins is expected to be disturbed, exhibited greater root waving than the wild-type plants (Fig. 2A). Eight (of 10) VIP1-SRDXox lines had lower root VGIs than the wild-type plants (Fig. 2B). The VIP1 expression level, which reflects the VIP1-SRDX expression level in a VIP1-SRDXox line, was highest in VIP1-SRDXox #6, which had the second lowest VGI, followed by #7, which had the lowest VGI (Fig. 2C). This result supports the idea that the root VGI and the VIP1-SRDX expression level are negatively correlated. The overexpression of VIP1 alone did not result in a decrease in the VGI (Fig. 2, B and C). No clear differences were observed between wild-type and VIP1-SRDXox plants regarding responses to either the stress-related phytohormone abscisic acid (ABA) or the hyperosmotic stress induced by mannitol (Supplemental Fig. S5).
Figure 2.
Effects of the overexpression of VIP1-SRDX on root waving and the root VGI. A, Wavy root phenotype. Wild-type (WT), vip1-1, VIP1-GFP-overexpressing (VIP1-GFPox), and VIP1-SRDX-overexpressing (VIP1-SRDXox #4 and #7) seedlings were grown for 10 d on an agar medium tilted at a 45° angle. Representative images are presented for each genotype. Bars = 2.5 mm. B, Root VGIs. Wild-type plants along with one line overexpressing VIP1 alone (VIP1ox) and VIP1-SRDXox lines (line numbers are indicated) were grown for 1 week on agar medium tilted at a 45° angle, and the root VGIs were calculated. The top and bottom edges and the line in the middle of each box indicate the quartiles, and the bar corresponds to the data range (n = 60 for the wild type, 65 for VIP1ox, 40 for VIP1-SRDXox #12, 56 for #8, 57 for #5, 45 for #2, 40 for #10, 40 for #1, 57 for #11, 44 for #4, 56 for #6, and 46 for #7). NS, P > 0.05; and *, P < 0.05 according to the Games-Howell test. Only results of the comparisons with the wild-type sample are shown for simplicity. The mean ± sd for each sample is presented in Supplemental Figure S12B. C, Quantitative reverse transcription (qRT)-PCR analysis of VIP1 expression. Total RNA was extracted from the roots of 10-d-old plants and used to synthesize complementary DNA (cDNA). The relative VIP1 expression level was calculated using the comparative cycle threshold (CT) method with UBIQUITIN5 (UBQ5) as the internal control. Values are presented as means ± sd of three biological replicates.
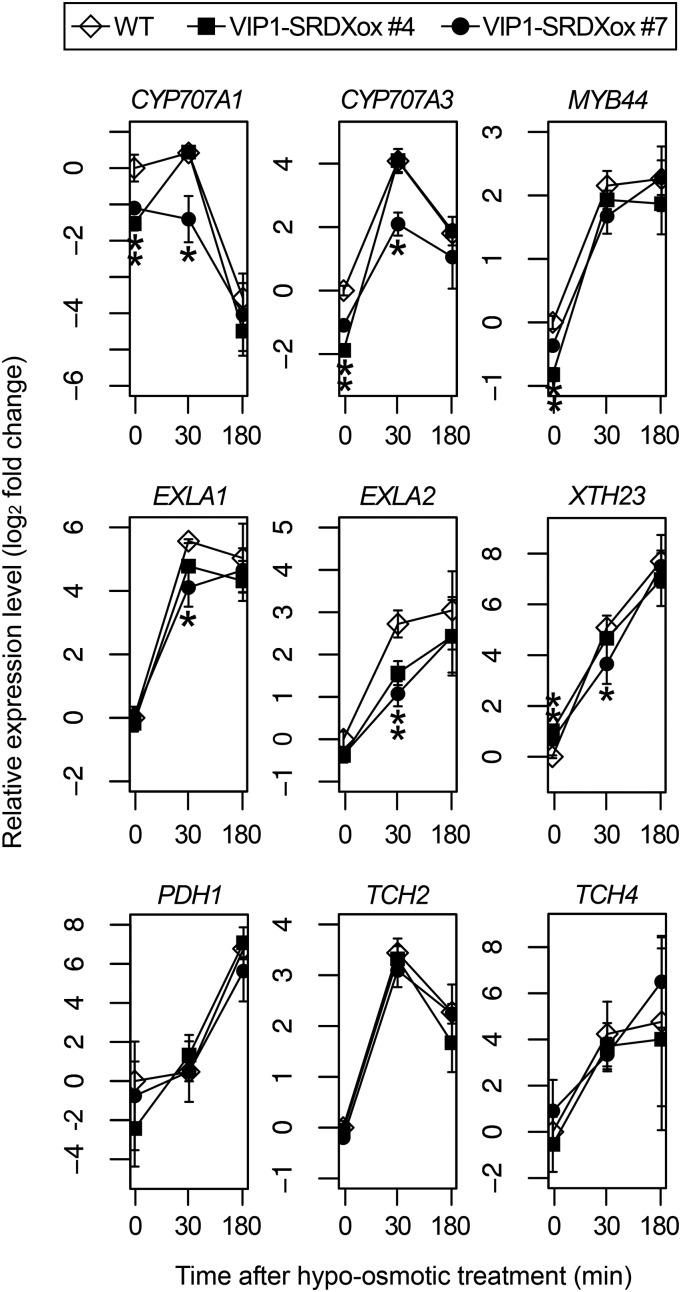
The expression levels of CYP707A1 (a putative VIP1 target gene), CYP707A3, MYB44, EXPANSIN-LIKE A1 (EXLA1), EXLA2, and XTH23 (putative VIP1 target genes up-regulated by hypoosmotic stress; Pitzschke et al., 2009; Tsugama et al., 2014), TCH2 and TCH4 (touch-responsive genes; Lee et al., 2005), and PDH1 (a hypoosmotic stress-responsive gene; Nakashima et al., 1998) were examined in VIP1-SRDXox #7 and #4, the latter of which has a lower expression level of VIP1-SRDX (Fig. 2C; Supplemental Fig. S4) and a higher root VGI than #7 (Fig. 2B). Under unstressed conditions, the CYP707A1, CYP707A3, and EXLA2 expression levels were lower in both VIP1-SRDXox #4 and #7 than in wild-type plants, and the EXLA1 expression level was lower in VIP1-SRDXox #7 than in wild-type plants. Following incubation in 20 mm Tris-HCl, pH 6.8, for 30 min, the EXLA2 expression level was lower in both VIP1-SRDXox #4 and #7 than in wild-type plants, and the CYP707A1, CYP707A3, and EXLA1 expression levels were lower in VIP1-SRDXox #7 than in wild-type plants (Fig. 3). These results support the idea that VIP1-SRDX represses the expression of VIP1 target genes.
Figure 3.
qRT-PCR analysis of the expression of hypoosmotic stress-responsive genes. Wild-type (WT) and VIP1-SRDX-overexpressing (VIP1-SRDXox #4 and #7) seedlings were grown for 1 week. The samples were then incubated in 20 mm Tris-HCl, pH 6.8, for 0, 30, or 180 min (x axes) and used for RNA extraction and cDNA synthesis. The relative gene expression levels were calculated using the comparative CT method with UBQ5 as the internal control and are presented as log2 fold changes (y axes). Values are presented as means ± sd of three biological replicates. *, P < 0.05 versus the wild type in the Tukey-Kramer test conducted using data for the wild type and VIP1-SRDXox #4 and #7 for each time point.
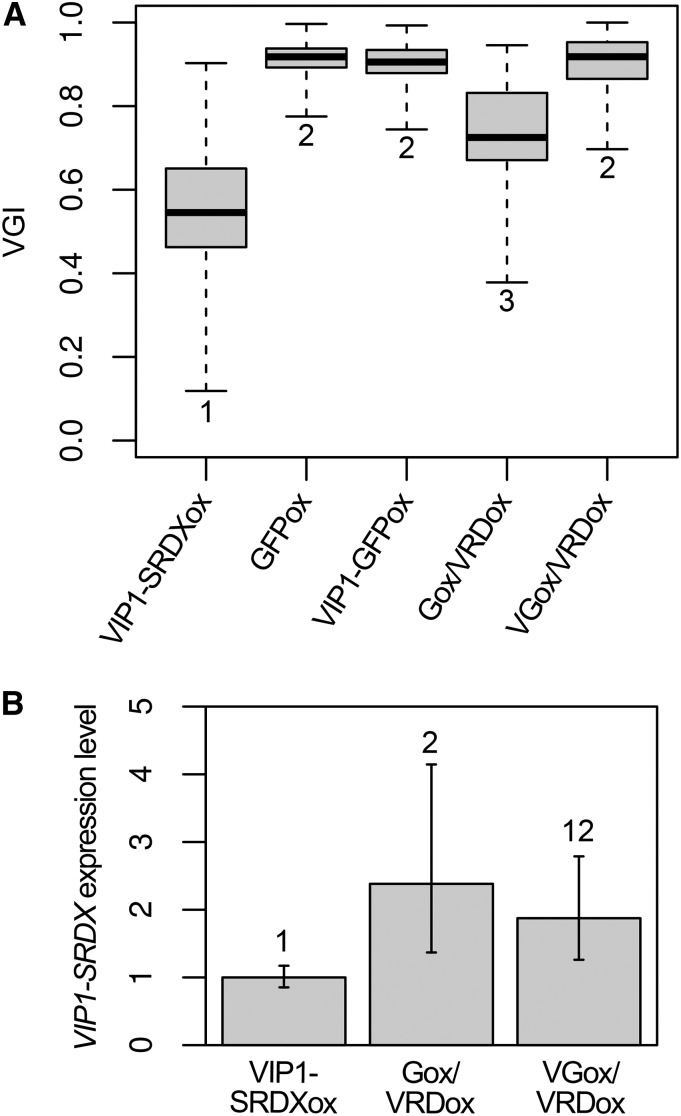
To further characterize the VIP1-SRDX-mediated decrease in the root VGI, the lines Gox/VRDox and VGox/VRDox, which express GFP and VIP1-GFP, respectively, in the VIP1-SRDXox #7 background, were generated by crossing VIP1-SRDXox #7 with either a line containing a 35S promoter-driven GFP (GFPox) or a line containing a 35S promoter-driven VIP1-GFP (VIP1-GFPox). The VGI of Gox/VRDox was higher than that of VIP1-SRDXox #7 but lower than that of GFPox. On the other hand, the VGI of VGox/VRDox was higher than that of VIP1-SRDXox #7 and as high as those of GFPox and VIP1-GFPox (Fig. 4A). The expression level of VIP1-SRDX was not lower in either Gox/VRDox or VGox/VRDox than in VIP1-SRDXox #7 (Fig. 4B). Similar results were obtained when the line expressing the auxin marker DR5rev::GFP (Friml et al., 2003) in the VIP1-SRDXox #7 background was used instead of Gox/VRDox (Supplemental Fig. S6). These results suggest that VIP1-GFP suppresses the VIP1-SRDX-mediated decrease in the root VGI. The VIP1-SRDX expression level was higher in Gox/VRDox than in VIP1-SRDXox #7, but the VGI of Gox/VRDox was higher than that of VIP1-SRDXox #7 (Fig. 4). GFP and/or some transgenes might somewhat inhibit VIP1-SRDX transcription or the functions of VIP1-SRDX. The GFP expression level was lower in Gox/VRDox than in GFPox, and the VIP1-GFP expression level was lower in VGox/VRDox than in VIP1-GFPox (Supplemental Fig. S7A). This may have been due to transgene silencing. Consistent with this result, western blotting revealed that the VIP1-GFP level was lower in VGox/VRDox than in VIP1-GFPox (Supplemental Fig. S7B). In the absence of hypoosmotic stress, the VIP1-GFP signals were detected in the cytosol but not in the nucleus of cells in wavy roots in either VIP1-GFPox or VGox/VRDox (Supplemental Fig. S7C, left), suggesting that the touch stress was insufficient to induce a detectable level of VIP1 nuclear localization. No differences were observed between VIP1-GFPox and VGox/VRDox in the nuclear-cytoplasmic shuttling pattern of VIP1-GFP under hypoosmotic conditions (Supplemental Fig. S7C), suggesting that VIP1-SRDX does not affect the subcellular localization of VIP1.
Figure 4.
Effects of expressing the VIP1-GFP fusion protein in the VIP1-SRDXox background. A, Root VGIs. Plants of each line were grown for 1 week on an agar medium tilted at a 45° angle, and the root VGIs were calculated. The following lines were used: VIP1-SRDXox, which overexpresses the VIP1-SRDX fusion protein (VIP1-SRDXox #7); GFPox, which overexpresses GFP alone; VIP1-GFPox, which overexpresses VIP1-GFP; Gox/VRDox, which is derived from crossing GFPox and VIP1-SRDXox #7; and VGox/VRDox, which is derived from crossing VIP1-GFPox and VIP1-SRDXox #7. The top and bottom edges and the line in the middle of each box indicate the quartiles, and the bar corresponds to the data range (n = 55 for VIP1-SRDXox, 51 for GFPox, 72 for VIP1-GFPox, 53 for Gox/VRDox, and 50 for VGox/VRDox). Data for the boxes indicated by the same numbers are not significantly different (P > 0.05) according to the Games-Howell test. The mean ± sd for each sample is presented in Supplemental Figure S12C. B, qRT-PCR analysis of VIP1-SRDX expression. Total RNA was extracted from roots of 10-d-old plants and used to synthesize cDNA. The relative VIP1-SRDX expression level was calculated using the comparative CT method with UBQ5 as the internal control. Values are presented as means ± sd of three biological replicates. Data for the bars indicated by the same numbers are not significantly different (P > 0.05) according to the Tukey-Kramer test.
Auxin Responses in the VIP1-SRDXox Plants
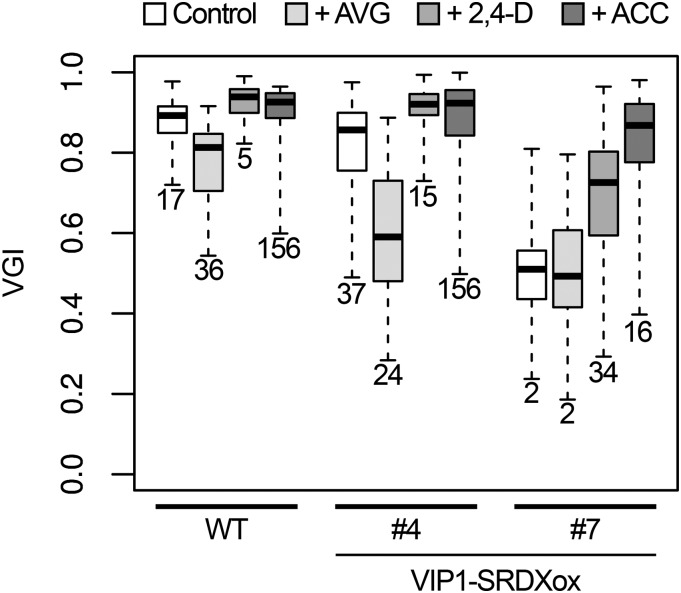
Ethylene can stimulate auxin biosynthesis and basipetal auxin transport in roots, both of which help regulate root tropisms (Růzicka et al., 2007). The ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG) can decrease the endogenous auxin level and auxin responses (Soeno et al., 2010). Consistent with these earlier findings, the root VGIs of wild-type and VIP1-SRDXox #4 plants were lower in the presence of AVG than in its absence, but the extent of the decrease in VGI was greater in VIP1-SRDXox #4 than in wild-type plants. The VGI of VIP1-SRDXox #7 was unaffected by AVG, but it was lower than that of wild-type or VIP1-SRDXox #4 plants regardless of the presence of AVG. This might be because the decrease in the VGI is saturated in VIP1-SRDXox #7, even in the absence of AVG. The VGIs of VIP1-SRDXox #4 and #7 were higher in the presence of either 2,4-dichlorophenoxy acetic acid (2,4-D; synthetic auxin) or 1-aminocyclopropane-1-carboxylic acid (ACC; ethylene precursor) than their absence (Fig. 5). The responses of VIP1-SRDXox #10 and #11 to AVG, 2,4-D, and ACC were similar to those of VIP1-SRDXox #4 (Supplemental Fig. S8). Under these conditions, no clear difference was observed in root length between the wild-type and VIP-SRDXox plants (Supplemental Fig. S9). These results support the idea that auxin is involved in the VIP1-SRDX-dependent decrease in the root VGI.
Figure 5.
Effects of the ethylene biosynthesis inhibitor AVG, the synthetic auxin 2,4-D, and the ethylene precursor ACC on the root VGI. Seedlings of the wild type (WT) and transgenic lines overexpressing the VIP1-SRDX fusion protein (VIP1-SRDX #4 and #7) were grown on a tilted agar medium (Control) or a medium supplemented with 0.5 μm AVG (+AVG), 0.2 μm 2,4-D (+2,4-D), 0.5 μm ACC (+ACC), and the root VGIs were calculated. The top and bottom edges and the line in the middle of each box indicate the quartiles, and the bar corresponds to the data range (n = 46 for the wild-type control, 39 for the wild type + AVG, 33 for the wild type + 2,4-D, 24 for the wild type + ACC, 45 for VIP1-SRDXox #4 control, 43 for #4 + AVG, 48 for #4 + 2,4-D, 40 for #4 + ACC, 41 for #7 control, 37 for #7 + AVG, 44 for #7 + 2,4-D, and 47 for #7 + ACC). Data for the boxes indicated by the same numbers are not significantly different (P > 0.05) according to the Games-Howell test. The mean ± sd for each sample is presented in Supplemental Figure S12E.
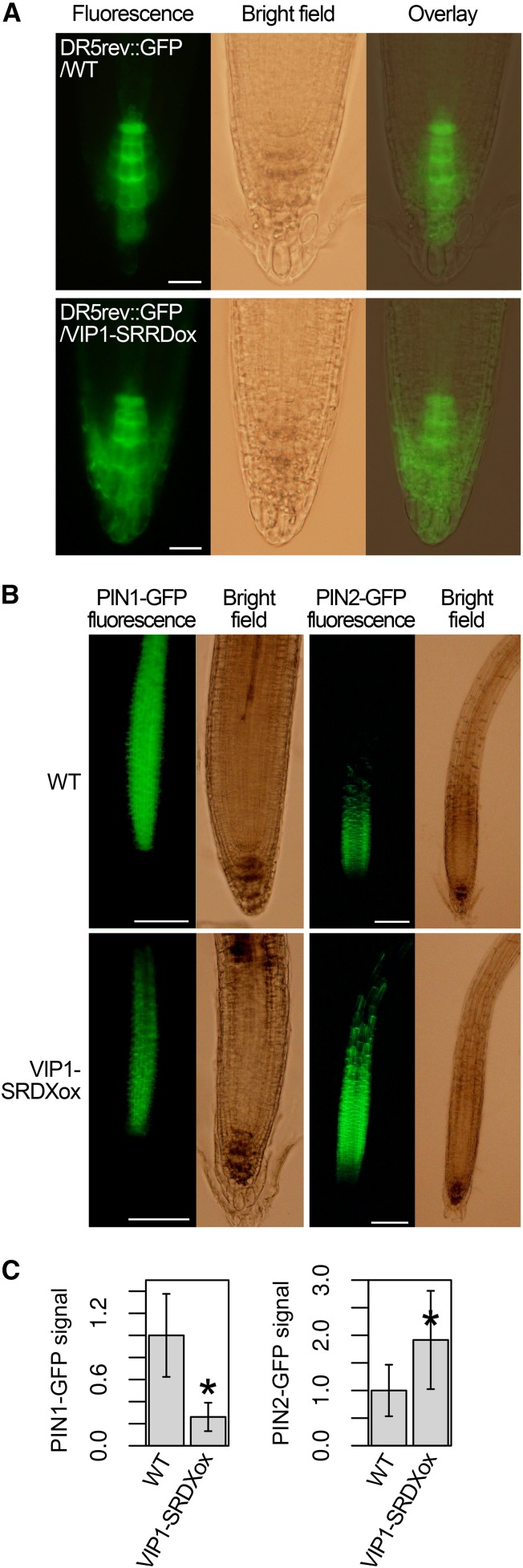
To visualize the local auxin responses in VIP1-SRDXox roots, DR5rev::GFP was introduced into the VIP1-SRDXox #7 background. DR5rev::GFP labels cells exhibiting strong auxin responses (Friml et al., 2003). The DR5rev::GFP signals were limited to the columella cells in the root cap in the wild-type background, but in the VIP1-SRDXox background, they were also detected in the lateral root cap cells (Fig. 6A). To visualize the PIN auxin transporters, PIN1::PIN1-GFP (Benková et al., 2003), PIN2::PIN2-GFP (Xu and Scheres, 2005), PIN3::PIN3-GFP (Zádníková et al., 2010), PIN4::PIN4-GFP (Vieten et al., 2005), and PIN7::PIN7-GFP (Blilou et al., 2005), which express the PIN proteins fused to GFP using their own promoters, were introduced into VIP1-SRDXox #7. No differences were observed between the wild-type background and the VIP1-SRDXox background regarding either the subcellular localization or the tissue specificity of the expression of the PIN-GFP fusion proteins. No differences were observed in the intensities of the PIN3-GFP, PIN4-GFP, and PIN7-GFP signals, either (Supplemental Fig. S10). However, the intensities of the PIN1-GFP and PIN2-GFP signals were lower and higher, respectively, in the VIP1-SRDXox background than in the wild-type background (Fig. 6, B and C). In qRT-PCR, no large differences were observed between wild-type and VIP1-SRDXox plants regarding the expression of either ethylene biosynthesis genes or auxin-related genes, including PIN1 and PIN2. No differences were observed in the expression of the genes encoding the GOLVEN (GLV) secretory peptides, which induce the accumulation of PIN2 and inhibit gravitropic root growth (Whitford et al., 2012; Fernandez et al., 2013), either (Supplemental Table S1). These results suggest that those genes are not VIP1 target genes.
Figure 6.
Local auxin responses and PIN1-GFP and PIN2-GFP levels in the VIP1-SRDXox background. A, Signal patterns of the auxin marker DR5rev::GFP. The transgenic line with DR5rev::GFP in either the wild-type (WT) background (DR5rev::GFP/WT) or the VIP1-SRDXox line #7 background (DR5rev::GFP/VIP1-SRDXox) was analyzed by fluorescence microscopy to detect the GFP signals in roots. For each line, more than 20 plants were used, and representative images are presented. Bars = 20 μm. B, Signal patterns of PIN1::PIN1-GFP and PIN2::PIN2-GFP. A transgenic line with PIN1::PIN1-GFP or PIN2::PIN2-GFP in the wild-type background was crossed with VIP1-SRDXox #7 to obtain a line with PIN1::PIN1-GFP or PIN2::PIN2-GFP in the VIP1-SRDXox background. These lines were grown for 1 week on an agar medium tilted at a 45° angle and then analyzed by fluorescence microscopy. Representative images are presented for each line. Bars = 100 μm. C, Quantification of the PIN1-GFP and PIN2-GFP signals. PIN1-GFP and PIN2-GFP signals were extracted from the images in B. The total areas of the extracted signals were used as the PIN1-GFP and PIN2-GFP levels. Values are presented as means ± sd (n = 12 for PIN1-GFP, wild-type background; 7 for PIN1-GFP, VIP1-SRDXox background; 26 for PIN2-GFP, wild-type background; and 20 for PIN2-GFP, VIP1-SRDXox background). *, P < 0.05 versus the wild type according to the Mann-Whitney U test.
Characterization of the Cells around the Root Tips of VIP1-SRDXox Plants
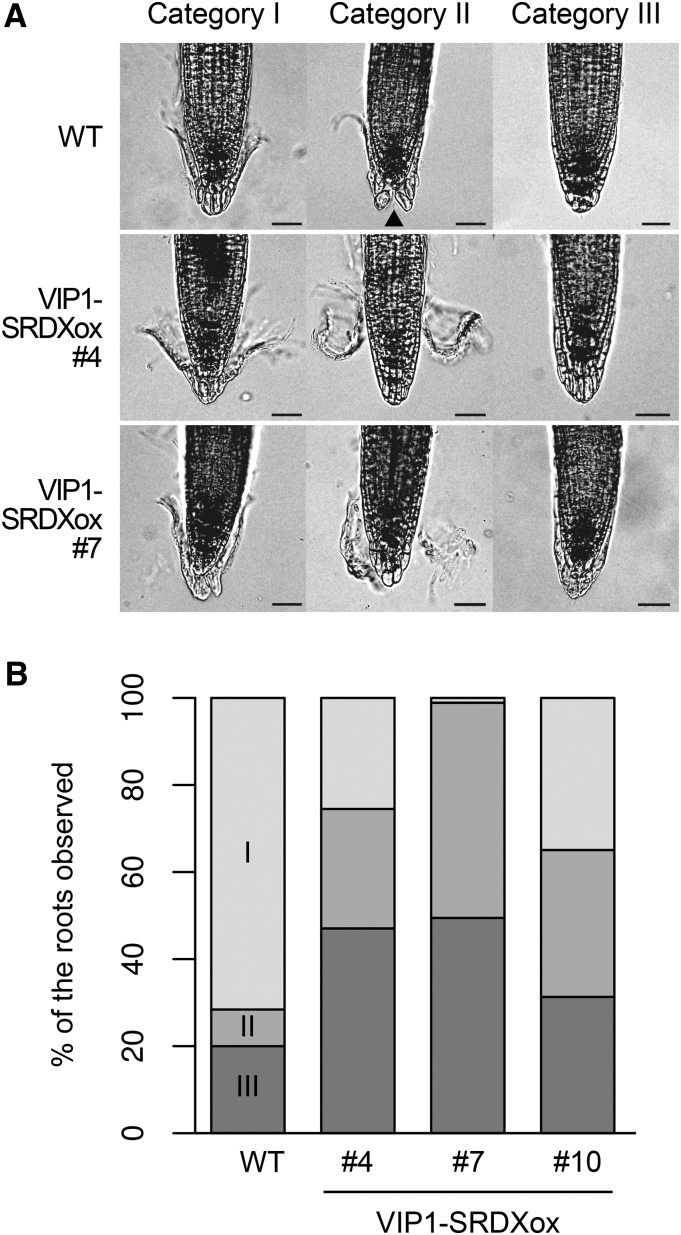
Starch grain accumulation in columella cells is necessary for full gravitropic responses of roots (Kiss et al., 1989), but no differences were observed between wild-type and VIP1-SRDXox plants regarding starch grain accumulation in columella cells (Supplemental Fig. S11A). The number of meristematic cells in the root cortex reflects the activity of the root apical meristem (Willemsen et al., 1998), but no differences were observed in the abundance of such meristematic cells between wild-type and VIP1-SRDXox plants (Supplemental Fig. S11, B and C). These results suggest that VIP1-SRDX affects neither starch accumulation nor the meristem activity in roots. On the other hand, a closer examination of VIP1-SRDXox lines revealed that their outermost root cap layers were abnormal. In the wavy roots, the conditions of the outermost root cap layers were classified into the following three categories: I, the edges of the layer are being removed from the other root cap parts; II, the layer has ruptured in the middle; and III, the layer is completely attached to the inner root cap parts (Fig. 7A). Most of the outermost root cap layers in wild-type plants belonged to categories I and III, whereas most of the layers in VIP1-SRDXox plants belonged to categories II and III (Fig. 7B; Table I). This result supports the idea that VIP1-SRDX affects the adhesion of the cell layers and/or structures of the root cap.
Figure 7.
Effects of the overexpression of VIP1-SRDX on the outermost root cap layers. A, Categorization of the conditions of the outermost root cap layers. The wild type (WT) and two transgenic lines overexpressing the VIP1-SRDX fusion protein (VIP1-SRDXox #4 and #7) were grown for 1 week on an agar medium tilted at a 45° angle. Representative images of the root tips are presented for each line and category for the conditions of the outermost root cap layers. B, Proportions of the root cap layers classified into each category. The images in A were used to classify the conditions of the outermost root cap layers. VIP1-SRDXox #10 plants also were grown and photographed as described in A. The number of plants in each category is provided in Table I. The indicated percentages were calculated based on these numbers.
Table I. Numbers of plants in each genotype and category according to the conditions of the outermost root cap layers.
#4, #7, and #10 are VIP1-SRDXox lines. Categories and genotypes were not independent, according to the χ2 test (P < 10−7).
| Category | Genotype |
Total | |||
|---|---|---|---|---|---|
| Wild Type | #4 | #7 | #10 | ||
| Category I | 68 | 26 | 1 | 26 | 121 |
| Category II | 8 | 28 | 45 | 28 | 109 |
| Category III | 19 | 48 | 45 | 29 | 141 |
| Total | 95 | 102 | 91 | 83 | 371 |
DISCUSSION
VIP1-SRDX caused decreases in the root VGI (Fig. 2) and the expression of the putative VIP1 target genes (Fig. 3). The VIP1-SRDX-mediated decrease in the VGI was suppressed by VIP1-GFP (Fig. 4). These results provide evidence that VIP can be involved in regulating responses to mechanical stimuli such as touch. This idea is relevant to the finding that the nuclear accumulation of VIP1 is induced by hypoosmotic stress, which can mimic such mechanical stimuli. On the other hand, although VIP1-mediated gene expression should occur in the nucleus,VIP1 is likely present in the cytosol but not in the nucleus in the absence of hypoosmotic stress, even in wavy roots responding to touch (Fig. 1; Supplemental Fig. S7C). Even under such conditions, a low level of VIP1 might be present in the nucleus and might be sufficient to properly regulate the expression of their target genes and root growth. However, it is also possible that they have some functions in the cytosol as well as in the nucleus.
VIP1-SRDXox plants exhibited abnormal outermost root cap layers (Fig. 7). This could be attributed to changes in the cell wall properties in root cap cells. In a previous study, antisense-mediated repression of a pectin methylesterase gene, which modifies cell wall properties, inhibited the separation of root cap cells in pea (Pisum sativum; Wen et al., 1999). In other studies, knockouts of the glycosyltransferase gene QUASIMODO1 and the pectin methyltransferases gene QUASIMODO2, both of which also modify cell wall properties, caused defects in the adhesion of root cap cells in Arabidopsis (Bouton et al., 2002; Mouille et al., 2007; Durand et al., 2009). Expansins also regulate cell wall properties (for review, see Lee et al., 2001), and two expansin genes, EXLA1 and EXLA2, are repressed in VIP1-SRDXox plants (Fig. 3). Touch-responsive genes include many other genes that affect cell wall properties (Lee et al., 2005). It is possible that VIP1-SRDX represses the expression of those genes as well. The receptor-like kinase FERONIA regulates the hypoosmotic stress-induced up-regulation of TCH2 and TCH4 (Shih et al., 2014), but the expression of TCH2 and TCH4 is not affected by VIP1-SRDX (Fig. 3), suggesting that VIP1-mediated signaling is distinct from FERONIA-mediated signaling. Because auxin is transported via the root cap, where many of the PIN members are expressed (for review, see Feraru and Friml, 2008), changes in the root cap structures can affect the auxin distribution and/or the local auxin responses around the root tip. Both PIN1 and PIN2 levels can be affected by local auxin levels (Abas et al., 2006; Omelyanchuk et al., 2016). Thus, the differences in the patterns of the DR5rev::GFP, PIN1-GFP, and PIN2-GFP signals between the wild-type and VIP1-SRDX backgrounds (Fig. 6) might be due to differences in their root cap structures. Putative roles of VIP1 and other group I bZIP proteins in responses to mechanical stimuli are proposed in Figure 8, although further studies are required to elucidate the cause-and-effect relationship between the VIP1-SRDX-induced changes in the root cap structures and those in the local auxin responses.
Figure 8.
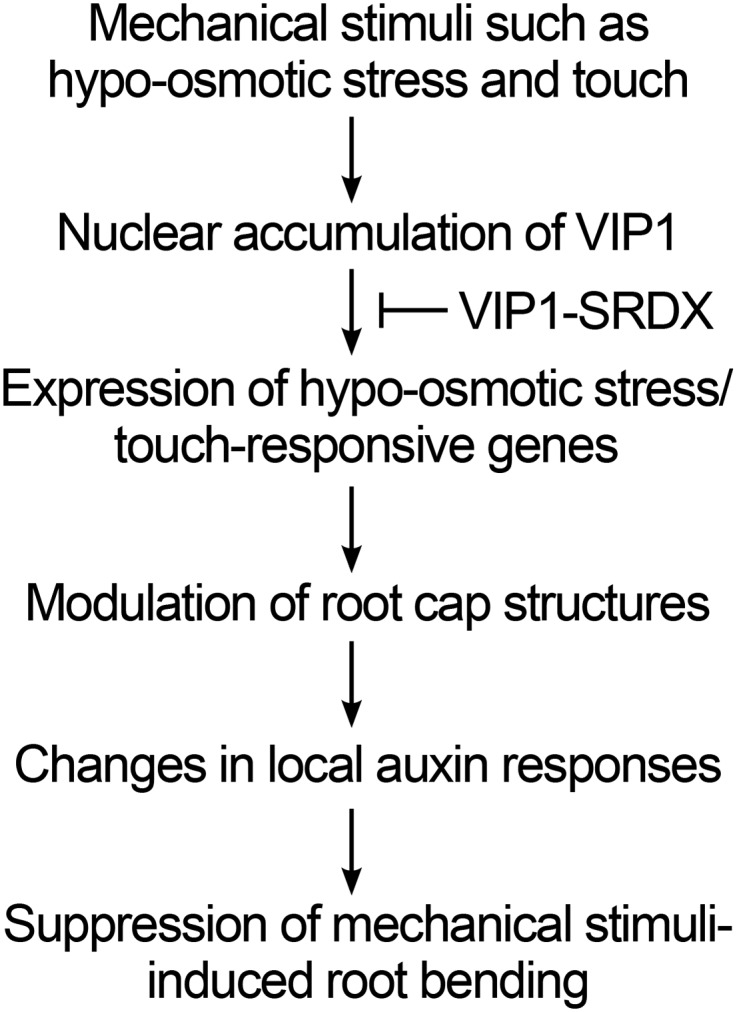
VIP1-mediated responses to mechanical stimuli.
CONCLUSION
VIP1-SRDX represses the expression of touch-responsive and/or hypoosmotic stress-responsive genes, changes root cap structures and local auxin responses, and decreases the root VGI in Arabidopsis. These findings support the idea that VIP1 is involved in regulating the root touch responses. This idea is relevant to the previous finding and our finding here that VIP1 accumulates in the nucleus when cells are exposed to hypoosmotic stress. VIP1-SRDXox plants would be useful to further characterize the roles of the group I bZIP proteins not only in touch responses but also in plant growth and other stress responses.
MATERIALS AND METHODS
Plasmid Construction
To overexpress VIP1 and the VIP1-SRDX fusion protein in Arabidopsis (Arabidopsis thaliana), the oligonucleotides 5′-CTAGACTGGTACCCGGGTCGACTGACTAGTACGAGCT-3′ and 5′-CGTACTAGTCAGTCGACCCGGGTACCAGT-3′ (complementary sequences are underlined) were mixed. The resulting double-stranded DNA was cloned into the XbaI and SacI sites of pBI121-35SMCS-GFP (Tsugama et al., 2012b), which removed the GFP CDS to generate pBI121-35SMCS. The full-length VIP1 CDS was amplified by PCR using VIP1-containing pBS-35SMCS-GFP (Tsugama et al., 2014) as the template and the following primer pair: 5′-CCCCACTAGTCATATGGAAGGAGGAGGAAGAGGACC-3′ and 5′-CCCCGTCGACAGCCTCTCTTGGTGAAATCC-3′ (SpeI, NdeI, and SalI sites are underlined). The PCR products were digested with SpeI and SalI and cloned into the XbaI and SalI sites of pBI121-35SMCS to generate pBI121-35S-VIP1. The oligonucleotides 5′-TCGACTCGACCTGGATCTTGAGCTAAGGTTGGGCTTCGCGA-3′ and 5′-CTAGTCGCGAAGCCCAACCTTAGCTCAAGATCCAGGTCGAG-3′ (complementary sequences are underlined) were mixed. The resulting double-stranded DNA was cloned into the SalI and SpeI sites of pBI121-35S-VIP1 to generate pBI121-35S-VIP1-SRDX.
To express the bZIP52-GFP fusion protein, the bZIP52 CDS was amplified by PCR using bZIP52-containing pGADT7-Rec (Tsugama et al., 2014) as the template and the following primer pair: 5′-CCCTCTAGAGAATTCCATGGAGAAATCAGATCCTCCACC-3′ and 5′-CCCGTCGACCATAGGCAGAGCTACTCTCACTAGC-3′ (XbaI, EcoRI, and SalI sites are underlined). The PCR products were digested with XbaI and SalI and cloned into the XbaI and SalI sites of pBI121-35SMCS-GFP to generate pBI121-35S-bZIP52-GFP. To express the bZIP69-GFP fusion protein, the bZIP69 CDS was amplified by PCR using bZIP69-containing pGADT7-Rec (Tsugama et al., 2014) as the template and the following primer pair: 5′-CCCTCTAGAGGATCCATGGATAAGGAGAAATCTCCTGC-3′ and 5′-GAGGTCGACCGTCTTTTGTCAAGGGAGTTTCACG-3′ (XbaI, BamHI, and SalI sites are underlined). The PCR products were digested with XbaI and SalI and cloned into the XbaI and SalI sites of pBI121-35SMCS-GFP to generate pBI121-35S-bZIP69-GFP. To express the bZIP30-GFP fusion, the bZIP30 CDS was amplified by PCR using bZIP30-containing pGADT7-Rec (Tsugama et al., 2014) as the template and the following primer pair: 5′-CCCTCTAGAGGATCCATGGGTGGTGGTGGTGATACAAC-3′ and 5′-CCCGTCGACCGTCGTTTGAAGTGTGCTTTGC-3′ (XbaI, BamHI, and SalI sites are underlined). The PCR products were digested with XbaI and SalI and cloned into the XbaI and SalI sites of pBI121-35SMCS-GFP to generate pBI121-35S-bZIP69-GFP.
To express the GST-bZIP52 fusion protein in Escherichia coli, the bZIP52 CDS was obtained by digesting pBI121-35S-bZIP52-GFP with EcoRI and SalI. The purified fragment was cloned into the EcoRI and SalI sites of pGEX-6P-3 (GE Healthcare) to generate pGEX-6P-bZIP52. To express the GST-bZIP69 and GST-bZIP30 fusion proteins in E. coli, the bZIP69 and bZIP30 CDSs were obtained by digesting pBI121-35S-bZIP69-GFP and pBI121-35S-bZIP30-GFP with BamHI and SalI. The purified fragments were cloned into the BamHI and SalI sites of pGEX-6P-3 to generate pGEX-6P-bZIP69 and pGEX-6P-bZIP30. To express the GST-PosF21 fusion protein in E. coli, the PosF21 CDS was amplified by PCR using PosF21-containing pGADT7-Rec (Tsugama et al., 2014) as the template and the following primer pair: 5′-CCCGTCGACATGGATAAGGAGAAATCTCCAGC-3′ and 5′-GAGGTCGACGTTCTCTTTCTGGGCTTGTG-3′ (SalI sites are underlined). The PCR products were digested with SalI and cloned into the SalI site of pGEX-6P-3 to generate pGEX-6P-PosF21. To express the GST-bZIP29 fusion protein in E. coli, the bZIP29 CDS was amplified by PCR using bZIP29-containing pGADT7-Rec (Tsugama et al., 2014) as the template and the following primer pair: 5′-GAGGTCGACATGGGTGATACAGAGAAGTGT-3′ and 5′-GGGGTCGACCTTCATTTGATTCAGATTTTGTTGCC-3′ (SalI sites are underlined). The PCR products were digested with SalI and cloned into the SalI site of pGEX-6P-3 to generate pGEX-6P-bZIP29.
Plant Materials
Arabidopsis ecotype Columbia-0 was used as the wild-type control for all experiments. Seeds of vip1-1 (SALK_001014C; Li et al., 2005) and bzip29-1 (SALK_065254C) mutants were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). The double mutant vip1-1/bzip29-1 was generated by crossing vip1-1 and bzip29-1 plants as the pollen and pod parents, respectively. The F2 plants generated from the cross were analyzed by genomic PCR (see “Genomic PCR” below), and homozygous lines with T-DNA insertions in VIP1 and bZIP29 were selected. The F3 seeds produced from the homozygous lines were used for subsequent analyses. Transgenic plants overexpressing VIP1 alone, VIP1-SRDX, or a GFP-fused form of bZIP52, bZIP69, or bZIP30 were generated with pBI121-35S-VIP1, pBI121-35S-VIP1-SRDX, pBI121-35S-bZIP52-GFP, pBI121-35S-bZIP69-GFP, or pBI121-35S-bZIP30-GFP, respectively, using the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). Transgenic lines with DR5rev::GFP (Friml et al., 2003), PIN1::PIN1-GFP (Benková et al., 2003), PIN2::PIN2-GFP (Xu and Scheres, 2005), and PIN7::PIN7-GFP (Blilou et al., 2005) were provided by Dr. Jiří Friml (Institute of Science and Technology). A transgenic line with PIN3::PIN3-GFP (Zádníková et al., 2010) was provided by Dr. Miyo Terao Morita (Nagoya University). A transgenic line containing PIN4::PIN4-GFP (Vieten et al., 2005) was obtained from the Nottingham Arabidopsis Stock Centre (stock no. N9576; http://arabidopsis.info/). Transgenic plants overexpressing GFP and VIP1-GFP (Tsugama et al., 2012a) and transgenic plants with DR5rev::GFP and PIN::PIN-GFP (pod parents) were crossed with VIP1-SRDXox #7 (pollen parent) to introduce these transgenes into the VIP1-SRDXox background. The F2 generation plants were used for subsequent analyses. Genomic PCR analysis was performed to ensure that only data from individuals with the VIP1-SRDX transgene were used.
Growth Tests and Characterization of Root Cells
Seeds were surface sterilized and sown on 1.5% (w/v) agar (Wako Pure Chemical Industries) containing 0.5× MS salts (Wako), 1% (w/v) Suc (Wako), and 0.5 g L−1 MES (Dojindo Laboratories), pH 5.8. Seeds were chilled at 4°C in darkness for 48 h and then germinated at 22°C. Plants were grown at 22°C under a 16-h/8-h light/dark cycle (light intensity of 120 µmol m−2 s−1). To examine plant responses to either ABA or osmotic stress, the plants of those lines were grown for 3 weeks on the same MS agar medium supplemented with 0.5 μm ABA (Sigma-Aldrich) or 200 mm mannitol (Wako).
To evaluate root waving and the removal of outermost layers of root caps, plants were grown for 1 week on MS agar medium that was tilted at a 45° angle. The plants were then photographed, and the lengths of the primary roots and the vertical projections from those roots were measured using ImageJ software (Schneider et al., 2012) to calculate the VGIs as described previously (Grabov et al., 2005). AVG (0.5 μm final concentration; Wako), 2,4-D (0.2 μm; Wako), and ACC (0.5 μm; Wako) also were added to the MS medium to assess their effects on VGIs. Plants were grown on the prepared media until their primary roots grew to approximately 1 to 1.5 cm. Box and whisker plots for VGIs are presented in Figures 2B, 4A, and 5 and Supplemental Figures S4C, S6A, and S8. Bar graphs for them are presented in Supplemental Figure S12.
Starch grains in the root cap were observed and the number of meristematic cortex cells was counted essentially as described previously (Willemsen et al., 1998). Briefly, to visualize starch granules in the root cap cells, 10-d-old plants grown on MS agar medium tilted at a 45° angle were incubated for 3 min at room temperature in Lugol solution (Sigma-Aldrich). The plants were then washed with distilled water and incubated for 10 min at room temperature in transparentizing solution (8 g of chloral hydrate dissolved in a mixture of 3 mL of distilled water and 1 mL of glycerol). Roots from the transparentized samples were then analyzed using a microscope (BX51; Olympus). To count the numbers of meristematic cortex cells, 10-d-old plants grown on MS agar medium tilted at a 45° angle were incubated in a 10 μm propidium iodide (Wako) solution for 5 min at room temperature. Root cells were then observed using an epifluorescence microscope (BX51 equipped with the mercury-vapor lamp USH-102D [Ushio], the fluorescence filter/mirror cube sets U-MNIBA3, U-MWIB3, and U-MWIG [Olympus], and the CCD camera DP73 [Olympus]). Microscope images were obtained using the cellSens software (Olympus). The region in the root tip where the length of a cell was greater than its width was regarded as the starting point of the elongation zone. The number of cells in a cortical layer between the quiescent center and the starting point of the root elongation zone was then counted. The numbers of plants used in these experiments are indicated in the legends for the relevant figures (Figs. 2B, 4A, and 5; Supplemental Figs. S3C, S5, S6A, S8, and S10) and Table I.
Detection and Quantification of GFP-Fused Proteins
Transgenic plants with DR5rev::GFP, PIN1::PIN1-GFP, PIN2::PIN2-GFP, PIN3::PIN3-GFP, PIN4::PIN4-GFP, and PIN7::PIN7-GFP were grown for 1 week on tilted MS agar medium, and the GFP signals in their roots were detected by epifluorescence microscopy. To assess the effects of hypoosmotic stress on the distribution of signals for GFP, VIP1-GFP, bZIP52-GFP, bZIP69-GFP, and bZIP30-GFP, approximately 1-week-old transgenic plants expressing these proteins were submerged in either 20 mm Tris-HCl, pH 6.8, or a liquid medium containing 0.5× MS salts, 1% (w/v) sucrose, and 0.5 g L−1 MES, pH 5.8 (i.e. hypotonic solution), and analyzed by epifluorescence microscopy at the time points indicated in the relevant figures (Fig. 1A; Supplemental Figs. S1 and S7C).
To transiently express GFP, VIP1-GFP, bZIP52-GFP, PosF21-GFP, bZIP69-GFP, and bZIP29-GFP in lettuce (Lactuca sativa), romaine lettuce leaf epidermal cells were transformed using an agroinfiltration method as described previously (Wroblewski et al., 2005). The transformed lettuce cells were incubated in darkness at room temperature for 48 h, submerged in 20 mm Tris-HCl, pH 6.8, incubated at room temperature for 0 or 20 min, and then analyzed by epifluorescence microscopy. The resulting images were processed with the GIMP (http://www.gimp.org/), Inkscape (http://www.inkscape.org), and ImageJ programs.
To quantify the PIN1-GFP and PIN2-GFP signals, the corresponding microscopy images were converted to grayscale using ImageJ. The threshold value was then set to 30 for PIN1-GFP or 27 for PIN2-GFP, and the GFP signals were extracted using the Analyze Particles tool. The areas from which signals were extracted were totaled, and the resulting values were used as the PIN1-GFP and PIN2-GFP levels.
For the western-blot analysis of GFP and VIP1-GFP, proteins were extracted from approximately 10-d-old GFP-overexpressing and VIP1-GFP-overexpressing plants (see “Plant Materials” above) using a chemical cell lysis method (Tsugama et al., 2011). GFP in the proteins was then analyzed by SDS-PAGE followed by western blotting using an anti-GFP antibody (Medical & Biological Laboratories) as described previously (Tsugama et al., 2012a). The numbers of plants and the numbers of replications used for these experiments are indicated in the legends for the relevant figures (Figs. 1A and 6; Supplemental Figs. S1, S7, B and C, and S9).
Genomic PCR
To select the vip1-1/bzip29-1 double mutant, mature leaves (approximately 0.5 cm × 0.5 cm) of F2 plants generated by the cross between vip1-1 and bzip29-1 (see “Plant Materials” above) were ground with a pestle in sample tubes containing 200 μL of extraction solution (200 mm Tris-HCl, pH 7.5, 250 mm NaCl [Wako], 25 mm EDTA [Wako], and 0.5% [w/v] SDS [Wako]). The solutions were centrifuged at 12,000g for 2 min to precipitate the plant debris. The supernatants were mixed with 150 μL of 2-propanol (Wako) and centrifuged at 12,000g for 5 min. The supernatants were discarded, and the precipitates were washed with 70% ethanol (Wako). The precipitates were then dried, dissolved in distilled water, and used as templates for genomic PCR. To examine whether the VIP1-SRDX CDS was present in transgenic plants, genomic DNA was extracted from the plants using a chemical cell lysis method (Tsugama et al., 2011). The PCR was completed using the KOD FX Neo DNA polymerase (Toyobo) and the primers listed in Supplemental Table S2. Images were processed using Inkscape.
Gel-Shift Assay
The gel-shift assay was completed essentially as described previously (Tsugama et al., 2014). Briefly, the pGEX constructs (see “Plasmid Construction” above) were transformed into E. coli strain BL21 (DE3) pLysS cells (Thermo Fisher Scientific). Production of the GST-fused proteins was induced by culturing the transformed E. coli cells for 2 h at 28°C in the presence of 0.2 mm isopropyl β-d-1-thiogalactopyranoside (Wako). The proteins were then purified by affinity chromatography using Glutathione Sepharose 4 Fast Flow (GE Healthcare). The digoxigenin-labeled DNA probe containing the CYP707A1 promoter sequence (approximately 250 bp) was generated as described previously (Tsugama et al., 2014), mixed with the purified proteins, incubated for 20 min at room temperature, and subjected to agarose gel electrophoresis. The DNA probe was transferred to a Hybond-N+ membrane (GE Healthcare), reacted with an alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche Diagnostics), and detected using CDP-Star (Roche) as the chemiluminescent substrate for alkaline phosphatase. The experiments were completed three times, and similar results were obtained. Images were processed using Inkscape.
qRT-PCR
To examine the expression of VIP1, VIP1-SRDX, and auxin-related genes, approximately 20 roots of 10-d-old plants were harvested for each genotype. To examine the expression of hypoosmotic stress-responsive genes, approximately 20 1-week-old plants of each genotype were submerged in 20 mm Tris-HCl, pH 6.8, and incubated for 0, 30, or 60 min at room temperature. Samples were frozen in liquid nitrogen and ground to a fine powder with a mortar and pestle. Total RNA was extracted from the ground plant material using the Favor Prep Plant Total RNA Purification Mini Kit (Favorgen Biotech). cDNA was synthesized from approximately 1 μg of total RNA using the oligo(dT) primer and PrimeScript Reverse Transcriptase (Takara Bio-PCR), which was completed using the StepOne Real-Time PCR System (Thermo Fisher Scientific) with SYBR Premix Ex Taq II (Tli RNaseH Plus; Takara Bio). Relative expression levels were calculated using the comparative CT method with UBQ5 as the internal control. Each experiment was conducted with three biological replicates. The sequences of the primers used for qRT-PCR are provided in Supplemental Table S3.
Details regarding the sequences of the genes used in this study can be obtained with the following Arabidopsis Genome Initiative accession numbers: AT1G06070 (bZIP69), AT1G06850 (bZIP52), AT1G43700 (VIP1), AT2G21230 (bZIP30), AT2G31370 (PosF21), AT4G38900 (bZIP29), AT4G19230 (CYP707A1), AT5G45340 (CYP707A3), AT5G67300 (MYB44), AT3G45970 (EXLA1), AT4G38400 (EXLA2), AT4G25810 (XTH23), AT3G30775 (PDH1), AT5G37770 (TCH2), AT5G57560 (TCH4), AT1G73590 (PIN1), AT5G57090 (PIN2), AT1G70940 (PIN3), AT2G01420 (PIN4), AT1G23080 (PIN7), AT2G38120 (AUX1), AT1G28130 (GH3.17), AT1G70560 (TAA1), AT2G19590 (ACO1), AT1G62380 (ACO2), AT1G03400 (ACOL1), AT1G12010 (ACOL2), AT1G01480 (ACS2), AT5G65800 (ACS5), AT4G11280 (ACS6), AT3G30350 (GLV3), AT3G02240 (GLV4), AT2G03830 (GLV6), AT2G04025 (GLV7), AT3G02242 (GLV8), AT5G51451 (GLV10), and AT5G60810 (GLV11).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Signal patterns for the Arabidopsis group I bZIP proteins fused to GFP in transgenic Arabidopsis plants.
Supplemental Figure S2. Signal patterns for the Arabidopsis group I bZIP proteins fused to GFP in a transient protein expression system.
Supplemental Figure S3. Western-blot analysis of GST alone and the GST-VIP1, GST-bZIP52, GST-PosF21, GST-bZIP69, GST-bZIP29, and GST-bZIP30 fusion proteins.
Supplemental Figure S4. Characterization of the Arabidopsis vip1/bzip29 double mutant.
Supplemental Figure S5. Responses of the Arabidopsis transgenic VIP1-SRDXox lines to ABA and mannitol stress.
Supplemental Figure S6. Effects of expressing the VIP1-GFP fusion protein in the Arabidopsis VIP1-SRDXox background.
Supplemental Figure S7. Signal patterns for the VIP1-GFP fusion protein in the Arabidopsis VIP1-SRDXox background.
Supplemental Figure S8. Effects of the ethylene biosynthesis inhibitor AVG, the synthetic auxin 2,4-D, and the ethylene precursor ACC on the root VGI in Arabidopsis.
Supplemental Figure S9. Relative root length of the Arabidopsis transgenic VIP1-SRDXox lines.
Supplemental Figure S10. PIN::PIN-GFP signals in the Arabidopsis VIP1-SRDXox background.
Supplemental Figure S11. Characterization of cells in and around the root cap of the Arabidopsis VIP1-SRDXox plants.
Supplemental Figure S12. Bar graphs for the VGIs presented in this study.
Supplemental Table S1. Summary of the qRT-PCR analysis of the expression of auxin-related genes in Arabidopsis.
Supplemental Table S2. Primers used for the genomic PCR.
Supplemental Table S3. Primers used for the qRT-PCR.
Supplementary Material
Acknowledgments
We thank Momoka Kobayashi for assisting in experiments; Drs. Nobuhiro Tsutsumi (University of Tokyo), Kiyoshi Masuda, and Kaien Fujino (Hokkaido University) for facilitating this work; Dr. Stanton B. Gelvin (Purdue University) for giving advice on the article; and Drs. Jiří Friml and Miyo Terao Morita, the Arabidopsis Biological Resource Center, and the Nottingham Arabidopsis Stock Centre for providing plant materials.
Glossary
- T-DNA
transfer DNA
- MS
Murashige and Skoog
- VGI
vertical growth index
- ABA
abscisic acid
- AVG
aminoethoxyvinylglycine
- 2,4-D
2,4-dichlorophenoxy acetic acid
- ACC
1-aminocyclopropane-1-carboxylic acid
- qRT
quantitative reverse transcription
- CDS
coding sequence
- cDNA
complementary DNA
- CT
cycle threshold
Footnotes
This work was supported by a Grant-in-Aid for Japan Society for the Promotion of Science fellows (grant no. 25–7247 to D.T.).
Articles can be viewed without a subscription.
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wiśniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C (2006) Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker MT, Talbotec J, Granier F, Lahaye M, Höfte H, Truong HN (2002) QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14: 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dai S, Wei X, Alfonso AA, Pei L, Duque UG, Zhang Z, Babb GM, Beachy RN (2008) Transgenic rice plants that overexpress transcription factors RF2a and RF2b are tolerant to rice tungro virus replication and disease. Proc Natl Acad Sci USA 105: 21012–21016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Zhang Z, Bick J, Beachy RN (2006) Essential role of the box II cis element and cognate host factors in regulating the promoter of Rice tungro bacilliform virus. J Gen Virol 87: 715–722 [DOI] [PubMed] [Google Scholar]
- Dai S, Zhang Z, Chen S, Beachy RN (2004) RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc Natl Acad Sci USA 101: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H (2007) Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318: 453–456 [DOI] [PubMed] [Google Scholar]
- Durand C, Vicré-Gibouin M, Follet-Gueye ML, Duponchel L, Moreau M, Lerouge P, Driouich A (2009) The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol 150: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E, Friml J (2008) PIN polar targeting. Plant Physiol 147: 1553–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Nguyen A, Beeckman T, Madder A, Hilson P (2013) Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol 161: 954–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Nakata M, Ito T, Matsushita A, Yamaguchi S, Takahashi Y (2011) bZIP transcription factor RSG controls the feedback regulation of NtGA20ox1 via intracellular localization and epigenetic mechanism. Plant Signal Behav 6: 26–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Nakata M, Ito T, Yamaguchi S, Takahashi Y (2010) The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J 62: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Ashley MK, Rigas S, Hatzopoulos P, Dolan L, Vicente-Agullo F (2005) Morphometric analysis of root shape. New Phytol 165: 641–651 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Fukazawa J, Yuasa T, Takahashi Y (2004) Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell 16: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Yuasa T, Nakata M, Takahashi Y (2008) A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20: 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y (2010) Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell 22: 1592–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y (2014) Scaffold function of Ca2+-dependent protein kinase: tobacco Ca2+-DEPENDENT PROTEIN KINASE1 transfers 14-3-3 to the substrate REPRESSION OF SHOOT GROWTH after phosphorylation. Plant Physiol 165: 1737–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Kiss JZ, Hertel R, Sack FD (1989) Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177: 198–206 [PubMed] [Google Scholar]
- Lacroix B, Vaidya M, Tzfira T, Citovsky V (2005) The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J 24: 428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Polisensky DH, Braam J (2005) Genome-wide identification of touch- and darkness-regulated Arabidopsis genes: a focus on calmodulin-like and XTH genes. New Phytol 165: 429–444 [DOI] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H (2001) Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 4: 527–532 [DOI] [PubMed] [Google Scholar]
- Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V (2005) Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA 102: 5733–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Hiratsu K, Todaka D, Nakashima K, Yamaguchi-Shinozaki K, Ohme-Takagi M (2006) Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol J 4: 325–332 [DOI] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hématy K, McCartney L, Truong HN, Gaudon V, Thibault JF, et al. (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50: 605–614 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol 118: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva M, Dunand C (2007) Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytol 176: 37–43 [DOI] [PubMed] [Google Scholar]
- Omelyanchuk NA, Kovrizhnykh VV, Oshchepkova EA, Pasternak T, Palme K, Mironova VV (2016) A detailed expression map of the PIN1 auxin transporter in Arabidopsis thaliana root. BMC Plant Biol (Suppl 1) 16: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruccelli S, Dai S, Carcamo R, Yin Y, Chen S, Beachy RN (2001) Transcription factor RF2a alters expression of the rice tungro bacilliform virus promoter in transgenic tobacco plants. Proc Natl Acad Sci USA 98: 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Djamei A, Teige M, Hirt H (2009) VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Natl Acad Sci USA 106: 18414–18419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C, Keller B (1998) Specific interaction of the tomato bZIP transcription factor VSF-1 with a non-palindromic DNA sequence that controls vascular gene expression. Plant Mol Biol 37: 977–988 [DOI] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lee LY, Gelvin SB (2014) Is VIP1 important for Agrobacterium-mediated transformation? Plant J 79: 848–860 [DOI] [PubMed] [Google Scholar]
- Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB (2014) The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol 24: 1887–1892 [DOI] [PubMed] [Google Scholar]
- Soeno K, Goda H, Ishii T, Ogura T, Tachikawa T, Sasaki E, Yoshida S, Fujioka S, Asami T, Shimada Y (2010) Auxin biosynthesis inhibitors, identified by a genomics-based approach, provide insights into auxin biosynthesis. Plant Cell Physiol 51: 524–536 [DOI] [PubMed] [Google Scholar]
- Torres-Schumann S, Ringli C, Heierli D, Amrhein N, Keller B (1996) In vitro binding of the tomato bZIP transcriptional activator VSF-1 to a regulatory element that controls xylem-specific gene expression. Plant J 9: 283–296 [DOI] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T (2011) A rapid chemical method for lysing Arabidopsis cells for protein analysis. Plant Methods 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T (2012a) A bZIP protein, VIP1, is a regulator of osmosensory signaling in Arabidopsis. Plant Physiol 159: 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T (2012b) Drought-induced activation and rehydration-induced inactivation of MPK6 in Arabidopsis. Biochem Biophys Res Commun 426: 626–629 [DOI] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T (2014) Analysis of functions of VIP1 and its close homologs in osmosensory responses of Arabidopsis thaliana. PLoS ONE 9: e103930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V (2001) VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 20: 3596–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V (2002) Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc Natl Acad Sci USA 99: 10435–10440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V (2004) Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431: 87–92 [DOI] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Wen F, Zhu Y, Hawes MC (1999) Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, Pérez AC, Kleine-Vehn J, Vanneste S, Drozdzecki A, Leitner J, Abas L, Aerts M, et al. (2012) GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell 22: 678–685 [DOI] [PubMed] [Google Scholar]
- Willemsen V, Wolkenfelt H, de Vrieze G, Weisbeek P, Scheres B (1998) The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125: 521–531 [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore R (2005) Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3: 259–273 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhao Q, Gao L, Yu XM, Fang P, Oliver DJ, Xiang CB (2010) Isolation and characterization of low-sulphur-tolerant mutants of Arabidopsis. J Exp Bot 61: 3407–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Scheres B (2005) Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Zhu Q, Dai S, Lamb C, Beachy RN (1997) RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J 16: 5247–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, et al. (2010) Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.