Label-free quantitative proteomics analysis of Arabidopsis phloem exudates collected during the induction of systemic acquired resistance (SAR) identifies novel components of the SAR response.
Abstract
Systemic acquired resistance (SAR) is a plant defense response that provides long-lasting, broad-spectrum pathogen resistance to uninfected systemic leaves following an initial localized infection. In Arabidopsis (Arabidopsis thaliana), local infection with virulent or avirulent strains of Pseudomonas syringae pv tomato generates long-distance SAR signals that travel from locally infected to distant leaves through the phloem to establish SAR. In this study, a proteomics approach was used to identify proteins that accumulate in phloem exudates in response to the induction of SAR. To accomplish this, phloem exudates collected from mock-inoculated or SAR-induced leaves of wild-type Columbia-0 plants were subjected to label-free quantitative liquid chromatography-tandem mass spectrometry proteomics. Comparing mock- and SAR-induced phloem exudate proteomes, 16 proteins were enriched in phloem exudates collected from SAR-induced plants, while 46 proteins were suppressed. SAR-related proteins THIOREDOXIN h3, ACYL-COENZYME A-BINDING PROTEIN6, and PATHOGENESIS-RELATED1 were enriched in phloem exudates of SAR-induced plants, demonstrating the strength of this approach and suggesting a role for these proteins in the phloem during SAR. To identify novel components of SAR, transfer DNA mutants of differentially abundant phloem proteins were assayed for SAR competence. This analysis identified a number of new proteins (m-type thioredoxins, major latex protein-like protein, ULTRAVIOLET-B RESISTANCE8 photoreceptor) that contribute to the SAR response. The Arabidopsis SAR phloem proteome is a valuable resource for understanding SAR long-distance signaling and the dynamic nature of the phloem during plant-pathogen interactions.
Plants responding to their environment must communicate over short and long distances to optimize growth and development. At short distances, growth- and stress-related signals move cell to cell through plasmodesmata (symplastically) or diffuse through the apoplast for communication with neighboring cells. At greater distances, macromolecules must access the plant vasculature for long-distance movement from one organ to another. A large body of evidence demonstrates the importance of the xylem and phloem as conduits for the long-distance movement of a diverse set of signals/macromolecules, such as micronutrients/macronutrients, small molecules, phytohormones, lipids, peptides/proteins, and coding/noncoding RNA (for review, see Lucas et al., 2013). These molecules are involved in a number of interorgan signaling responses, ranging from processes governing growth and development to stress-related responses to abiotic and biotic stimuli. Not surprisingly, some pathogens have coopted the plant vasculature to better exploit their hosts. Classic examples of this strategy include the systemic movement of plant viruses through the phloem (Hipper et al., 2013), vasculature-infecting microbes (Yadeta and Thomma, 2013), and phloem-feeding herbivores (Kaloshian and Walling, 2005; Howe and Jander, 2008). In response, plants have developed sophisticated interorgan resistance responses to limit the spread of infecting pathogens as well as to prevent and/or limit the effectiveness of future infection(s). Such responses include virus-induced RNA interference (Yoo et al., 2004), induced systemic resistance caused by beneficial microbes (Pieterse et al., 2014), and systemic acquired resistance (SAR; Champigny and Cameron, 2009).
SAR is classically described as a plant defense response that provides long-lasting, broad-spectrum pathogen resistance to uninfected systemic leaves following an initial localized infection. In Arabidopsis (Arabidopsis thaliana), SAR is induced after a localized infection with compatible or incompatible strains of the hemibiotrophic bacterial phytopathogen Pseudomonas syringae (Cameron et al., 1994). During the compatible interaction with virulent P. syringae, Arabidopsis pattern recognition receptors recognize conserved microbial motifs known as pathogen-associated molecular patterns (PAMPs) to induce PAMP-triggered immunity. However, virulence effector proteins secreted into plant cells by P. syringae suppress this response and promote susceptibility in locally infected tissue (for review, see Xin and He, 2013). Incompatible or avirulent P. syringae strains carry effector proteins that are recognized in plant cells by cognate resistance receptors to induce a robust local defense response termed effector-triggered immunity, which is usually associated with programmed cell death in the form of the hypersensitive response (for review, see Cui et al., 2015). Classic SAR studies suggested that a necrotizing infection was important for SAR induction (for review, see Sticher et al., 1997); however, recent studies demonstrate that the induction of PAMP-triggered immunity is sufficient to induce SAR in Arabidopsis (Mishina and Zeier, 2007). Nevertheless, local infection with virulent or avirulent P. syringae strains leads to the generation of mobile SAR signals that travel from locally infected to distant leaves to initiate SAR.
SAR studies in non-Arabidopsis model systems first suggested that SAR signals move via the phloem. Early grafting experiments in cucumber (Cucumis sativus) determined that SAR signals traveled from induced rootstocks to distant scions to induce SAR (Jenns and Kuc, 1979). A specific role for the phloem in the long-distance transport of SAR signals was identified in cucumber, where restricting vascular connections of induced leaf petioles using a wool/hot-water girdling technique prevented the manifestation of SAR in distant leaves (Guedes et al., 1980). Experiments performed in tobacco (Nicotiana tabacum) demonstrated that the removal of stem sheath also resulted in a loss of systemic immunity (Tuzun and Kuc, 1985), further supporting a role for the plant vasculature in long-distance immune signaling. In Arabidopsis, the transport of SAR signals from locally infected to distant leaves also occurs via the phloem, as demonstrated by overlapping translocation patterns for radiolabeled photosynthate and SAR signals (Kiefer and Slusarenko, 2003). Interestingly, the results did not preclude additional mechanisms of transport, as SAR signal movement was not strictly limited to the orthostichy (vascular bundle) of the induced leaf, suggesting that SAR signals move cell to cell from one orthostichy to another to better disseminate the signal. This idea was supported recently by the observation that plant lines with reduced cell-to-cell movement through plasmodesmata are defective in SAR and the long-distance movement of DEFECTIVE IN INDUCED RESISTANCE1 (DIR1; Carella et al., 2015). Taken together, these studies demonstrate that long-distance SAR signaling is dependent on the phloem for efficient interorgan communication.
The identification of long-distance SAR signals remains an active area of research, as they may represent novel bioprotective agents suitable for use in agriculture (Conrath et al., 2015). Both genetic and analytical biochemical screens have been performed to isolate genes and metabolites important for SAR. A common approach for identifying SAR-activating small molecules is to perform biochemical screens with phloem exudates collected from SAR-induced Arabidopsis leaves. Activity-guided analytical screening of SAR-induced phloem exudates was used to identify the SAR activators azelaic acid and dehydroabietinal (Jung et al., 2009; Chaturvedi et al., 2012) and to analyze amino acid levels during SAR, leading to the identification of pipecolic acid (Návarová et al., 2012). Together, these studies demonstrate that phloem exudates are a rich source of SAR-activating small molecules that may work in concert to induce SAR in distant tissues.
In comparison, our knowledge of protein composition within the phloem during SAR is extremely limited. The lipid transfer protein (LTP) DIR1 is currently the only protein demonstrated to move from SAR-induced to distant tissues via the phloem (Champigny et al., 2013). Recent studies demonstrate that DIR1 interacts with other SAR-related LTPs in untreated tobacco leaves (Yu et al., 2013; Cecchini et al., 2015) and is associated with a dehydroabietinal-containing, trypsin-sensitive, high-molecular-weight fraction of phloem exudates collected from SAR-induced leaves (Shah et al., 2014). This suggests that DIR1 is a member of a large proteinaceous complex that travels to distant leaves in the phloem during SAR. Additionally, total protein levels are typically higher in phloem exudates collected from SAR-induced versus mock-inoculated leaves (Champigny et al., 2013; Carella et al., 2015), supporting the notion that numerous proteins are loaded into the phloem during SAR.
In this study, a proteomics approach was taken to identify proteins that accumulate in phloem exudates during the induction of SAR and, therefore, could be involved in the long-distance signaling stage of SAR. Label-free quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomics was used to identify and quantify proteins present in phloem exudates collected from leaves that were mock inoculated or induced for SAR with virulent or avirulent Pseudomonas syringae pv tomato (Pst). By comparing mock- and SAR-induced exudate proteomes, 16 proteins accumulated and 46 proteins decreased in abundance in phloem exudates during SAR. The functional relevance of these proteins to SAR was explored by performing SAR assays on the corresponding transfer DNA (T-DNA) mutants. This analysis identified a role in SAR for m-type thioredoxins, a putative major latex protein, and the UV-B photoreceptor ULTRAVIOLET-B RESISTANCE8 (UVR8). Further investigation of the UVR8 UV-B signaling pathway revealed a role for the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) and the bZIP transcription factor ELONGATED HYPOCOTYL5 (HY5) in the development of SAR. The Arabidopsis SAR phloem proteome provides new insights into the dynamic nature of the phloem during biotic stress and reveals that a number of previously unknown proteins accumulate in the phloem during SAR.
RESULTS
Quantitative Proteomics of Phloem Exudates during SAR
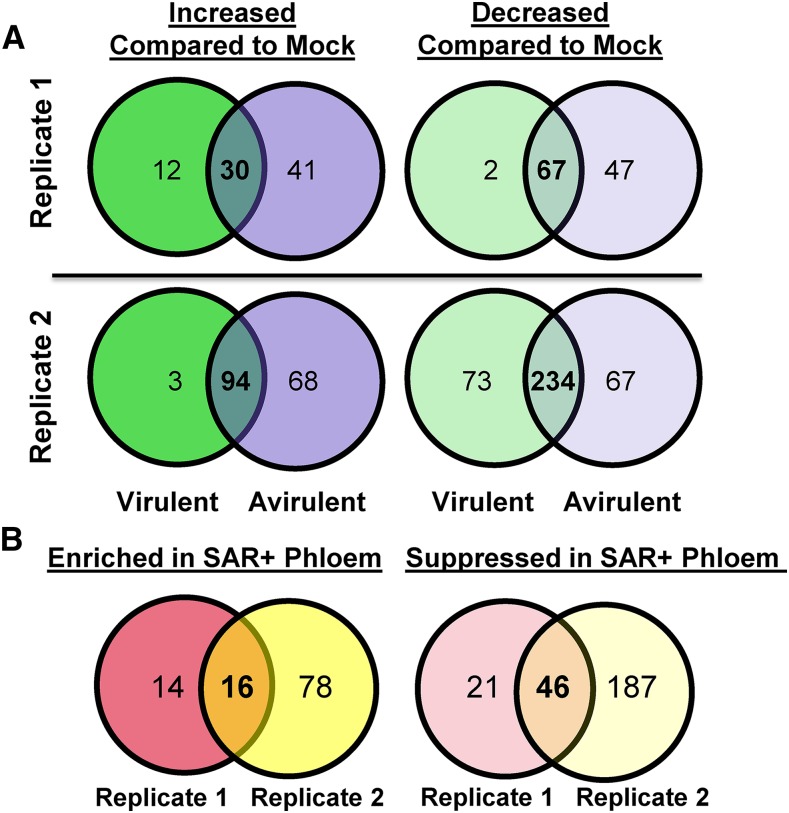
To identify proteins that accumulate in the phloem in response to the induction of SAR, we subjected phloem exudates collected from mock- and SAR-induced Arabidopsis leaves to quantitative label-free LC-MS/MS. Phloem exudates were collected from 24 to 48 h post inoculation (hpi) because the SAR-mobile DIR1 protein accumulates to high levels during this interval (Champigny et al., 2013). Phloem exudates were collected from leaves that were mock inoculated (10 mm MgCl2) or induced for SAR by inoculation with Pst strains that are virulent (Pst DC3000) or avirulent (Pst DC3000/avrRpt2) on Columbia-0 (Col-0) plants. SAR assays performed alongside exudate collection experiments confirmed that SAR was induced by both strains (Supplemental Fig. S1A). This was further supported by observing DIR1 antibody signals in immunoblots of phloem exudates collected from SAR-induced, but not mock-inoculated, leaves (data not shown). To obtain protein levels suitable for LC-MS/MS, exudates from more than 90 plants per treatment were collected and concentrated using centrifugal concentrators (3-kD cutoff) followed by lyophilization. Similar to previous reports (Champigny et al., 2013; Carella et al., 2015), phloem exudates collected from SAR-induced leaves contained higher total protein levels than exudates collected from mock-induced leaves (Supplemental Fig S1B). Concentrated phloem exudates from two independent experimental replicates were subjected to quantitative LC-MS/MS (Supplemental Data S1). Venn diagrams in Figure 1A show the number of proteins that were significantly enriched or suppressed in SAR-induced exudates relative to mock-inoculated controls. Not surprisingly, the exudate proteomes of leaves treated with virulent or avirulent Pst were not identical, as several proteins displayed strain-specific differences in abundance (Supplemental Tables S1 and S2). Since infection with either strain induces SAR to the same extent in Col-0 (Supplemental Fig. S1A), we reasoned that key proteins involved in SAR should accumulate to a similar degree after either treatment. Therefore, we compiled a list of proteins that were differentially abundant in phloem exudates collected from leaves induced for SAR by both Pst strains relative to mock-inoculated phloem exudates (Fig. 1B). A total of 16 proteins were enriched in phloem exudates collected from SAR-induced (virulent and avirulent Pst) leaves compared with mock-inoculated controls (Table I). In contrast, 46 proteins displayed decreased abundance in exudates collected from SAR-induced versus mock-inoculated leaves (Table II; Supplemental Table S3).
Figure 1.
Comparative proteomics analysis of phloem exudates collected during the induction of SAR. Quantitative proteomics data of phloem exudates were collected from mock-inoculated (10 mm MgCl2) and SAR-induced (virulent, Pst DC3000; and avirulent, Pst DC3000/avrRpt2) leaves of two experimental replicates. Values inside Venn diagrams represent the number of unique proteins (at least two peptides) that were differentially abundant (Student’s t test, P < 0.05) between treatments. A, Proteins with increased or decreased abundance in phloem exudates of SAR-induced (virulent or avirulent) leaves compared with mock-inoculated controls in each experimental replicate. B, Proteins that are similarly enriched or suppressed in phloem exudates collected from SAR-induced (virulent and avirulent) compared with mock-inoculated leaves. Venn diagrams generated in Venny 2.0 (Oliveros, 2015; http://bioinfogp.cnb.csic.es/tools/venny/index.html) were remade using Microsoft Office Powerpoint.
Table I. Proteins enriched in the phloem during SAR.
| Locus | Gene Symbol | Description | Relative Abundance (Virulent/Mock) |
Relative Abundance (Avirulent/Mock) |
Peptides Used for Quantitation |
|||
|---|---|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 1 | Replicate 2 | Replicate 1 | Replicate 2 | |||
| AT3G52960 | PrxIIE | Peroxiredoxin | 8.7 | 8.6 | 12.6 | 19.70 | 8 | 10 |
| AT1G03680 | TRXm1 | Thioredoxin | 7.5 | 2 | 174.3 | 9.5 | 6 | 8 |
| AT1G06680 | PsbP1 | PSII subunit | 2.9 | 5.3 | 10.7 | 15.1 | 6 | 6 |
| AT5G42980 | TRXh3 | Thioredoxin | 4.2 | 4.3 | 3.5 | 7.2 | 5 | 4 |
| AT4G03520 | TRXm2 | Thioredoxin | 16.5 | 3.7 | 79.7 | 14.9 | 4 | 5 |
| AT2G43570 | CHI/AED15 | Chitinase | 3.9 | 1.5 | 2.8 | 1.7 | 4 | 3 |
| AT2G44920 | – | Tetratricopeptide-like | 13 | 13.9 | 18.6 | 41.4 | 3 | 4 |
| AT1G20340 | PETE2 | Plastocyanin | 2.9 | 15.6 | 16.7 | 23.9 | 3 | 8 |
| AT5G40370 | GRXC2 | Glutaredoxin | 10.9 | 11.1 | 8.4 | 17.6 | 3 | 4 |
| AT3G50820 | PsbO2 | PSII subunit | 13.7 | 6.8 | 37.4 | 57.3 | 3 | 3 |
| AT2G14610 | PR1 | Pathogenesis-related | 4.8 | 3.8 | 12.2 | 14.6 | 3 | 2 |
| AT4G34050 | CCoAOMT1 | S-Adenosyl-l-Met methyltransferase | 4.3 | 2.4 | 3.3 | 1.8 | 2 | 2 |
| AT2G19760 | PFN1 | Profilin | 5.6 | 1.5 | 21.6 | 1.8 | 2 | 4 |
| AT4G02450 | – | HEAT SHOCK PROTEIN20 (HSP20)-like | 7.5 | 4.7 | 7.2 | 8.2 | 2 | 3 |
| AT2G29450 | GSTU5 | Glutathione S-transferase | 3.3 | 1.8 | 6.2 | 3.5 | 2 | 2 |
| AT1G55260 | LTPG6 | Lipid transfer protein | 4.3 | 4.8 | 9.2 | 9 | 2 | 2 |
| AT1G31812 | ACBP6 | Acyl-CoA-binding protein | 119.4 | 4.4 | 111.1 | 7.9 | 1a | 4 |
| AT3G15360b | TRXm4 | Thioredoxin | 5.5 | 0.6 | 9.3 | 1.9 | 4 | 5 |
| AT4G23670b | MLP | Major latex protein-like | 5.7 | 1.1c | 30.2 | 6 | 2 | 4 |
Only one peptide was available for quantitation.
Peptides with significant enrichment in SAR plus phloem in one of two replicates.
Not statistically significant.
Table II. Selected proteins suppressed in the phloem during SAR.
| Locus | Gene Symbol | Description | Relative Abundance (Virulent/Mock) |
Relative Abundance (Avirulent/Mock) |
Peptides Used for Quantitation |
|||
|---|---|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 1 | Replicate 2 | Replicate 1 | Replicate 2 | |||
| AT3G05900 | – | Neurofilament protein-related | 0.07 | 0.03 | 0.03 | 0.03 | 10 | 15 |
| AT5G66190 | FNR1 | Ferrodoxin oxidoreductase | 0.37 | 0.14 | 0.20 | 0.21 | 4 | 17 |
| AT2G04030 | HSP90.5 | Chaperone protein | 0.51 | 0.13 | 0.26 | 0.18 | 6 | 6 |
| AT5G26000 | TGG1 | Thioglucoside glucohydrolase | 0.29 | 0.40 | 0.28 | 0.75 | 6 | 10 |
| AT3G16470 | JR1 | Man-binding lectin | 0.20 | 0.26 | 0.04 | 0.20 | 5 | 8 |
| AT1G55490 | CPN60B | Chaperonin | 0.24 | 0.15 | 0.14 | 0.19 | 5 | 7 |
| AT3G16400 | NSP1 | Nitrile specifier protein | 0.16 | 0.11 | 0.03 | 0.08 | 4 | 5 |
| AT1G09210 | CRT1b | Calreticulin | 0.09 | 0.08 | 0.03 | 0.08 | 4 | 2 |
| AT1G56340 | CRT1a | Calreticulin | 0.15 | 0.09 | 0.04 | 0.09 | 4 | 4 |
| AT5G54770 | THI1 | Thiazole biosynthetic enzyme | 0.08 | 0.09 | 0.02 | 0.03 | 3 | 2 |
| AT5G28540 | BiP1 | HSP70 | 0.38 | 0.35 | 0.36 | 0.47 | 3 | 7 |
| AT2G28000 | CPN60A | Chaperonin | 0.20 | 0.29 | 0.12 | 0.27 | 3 | 4 |
| AT1G72150 | PATL1 | Patellin | 0.06 | 0.09 | 0.06 | 0.10 | 2 | 3 |
| AT1G76180 | ERD14 | Dehydrin | 0.06 | 0.05 | 0.03 | 0.04 | 2 | 6 |
| AT1G35720 | ANNAT1 | Annexin | 0.17 | 0.08 | 0.06 | 0.06 | 2 | 6 |
| AT4G22670 | HIP1 | HSP70-interacting | 0.03 | 0.01 | 0.00 | 0.04 | 2 | 2 |
| AT2G21660 | GRP7 | Gly-rich protein | 0.19 | 0.10 | 0.06 | 0.06 | 2 | 5 |
| AT5G63860 | UVR8 | UVB photoreceptor | 0.27 | 0.05 | 0.18 | 0.06 | 2 | 1a |
Only one peptide was used for quantitation.
Comparison with Published Phloem Exudate Proteomes
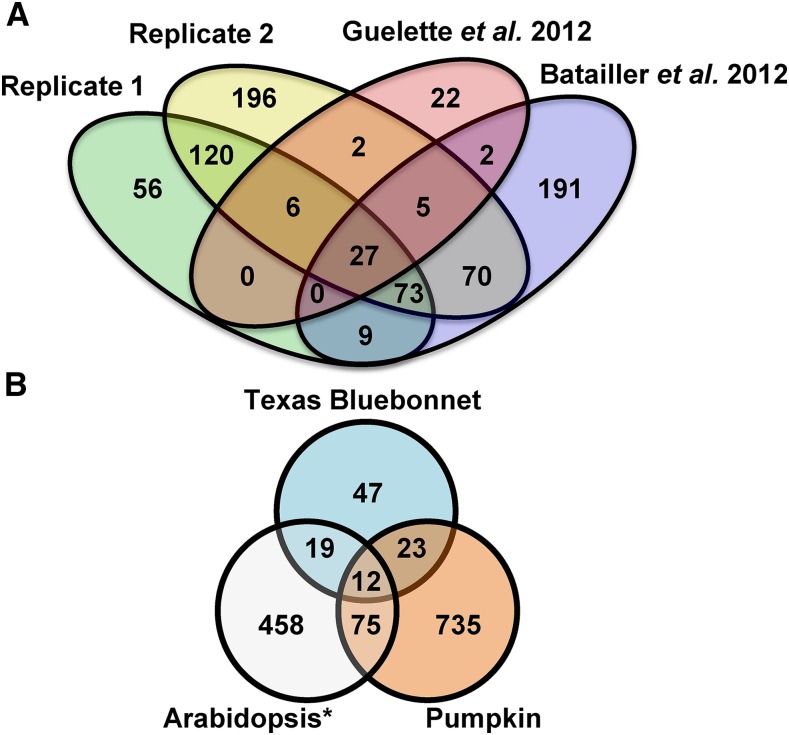
To assess the quality of our proteomes, we compared our data set (all proteins, regardless of treatment) with previously published phloem exudate proteomes. Comparisons were performed with two studies that used LC-MS/MS-based proteomics to identify proteins in phloem exudates collected from untreated Arabidopsis leaves (Batailler et al., 2012; Guelette et al., 2012). A total of 27 common phloem proteins were identified in all three proteomes (Fig. 2A; Supplemental Table S4). Our combined phloem proteome (replicates 1 and 2) overlapped with 49% of the proteins identified by Batailler et al. (2012) and 63% of those described by Guelette et al. (2012). By comparison, the Batailler et al. (2012) data set overlapped with 47% of proteins identified by Guelette et al. (2012). Furthermore, we compared our proteome with phloem proteomes obtained from pumpkin (Cucurbita maxima; Lin et al., 2009) and Texas bluebonnet (Lupinus texensis; Lattanzio et al., 2013; Fig. 2B). Only 12 proteins were present in the proteomes of all three species (Supplemental Table S5). Our Arabidopsis phloem proteome overlapped with 10% of proteins identified in pumpkin exudates and 31% of proteins identified in Texas bluebonnet exudates. In comparison, the Batailler et al. (2012) proteome overlapped with 8% of pumpkin and 22% of Texas bluebonnet phloem proteins. This demonstrates that although there is variation in the protein profiles of phloem exudates within and between species, the phloem proteome generated in this study shares similarity with previously published phloem proteomes.
Figure 2.
Comparing phloem exudate proteomes. A, Venn diagram comparing all proteins identified in replicates 1 and 2 of this study with the Arabidopsis phloem exudate proteomes described by Guelette et al. (2012) and Batailler et al. (2012). B, Comparison of all Arabidopsis proteins identified in this study (Arabidopsis*) with phloem exudate proteomes of pumpkin (Lin et al., 2009) and Texas bluebonnet (Lattanzio et al., 2013). Venn diagrams obtained from Venny 2.0 (Oliveros, 2015; http://bioinfogp.cnb.csic.es/tools/venny/index.html) were remade in Microsoft Office Powerpoint.
GO Slim Analysis of SAR-Enriched Versus SAR-Suppressed Phloem Proteins
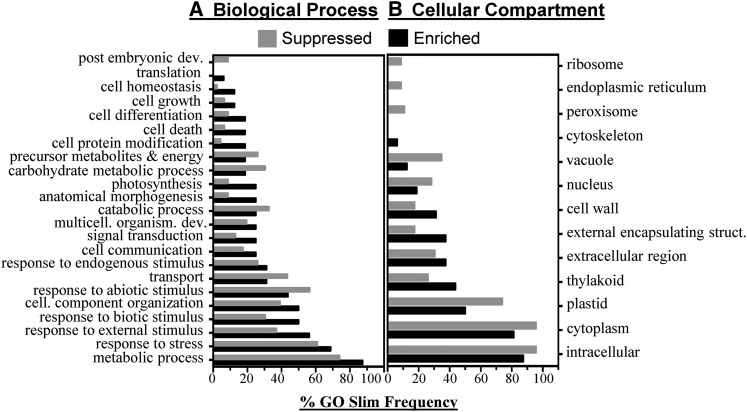
To gain insight into the nature of SAR-enriched and SAR-suppressed phloem proteins, comparative GO Slim analysis was performed (Supplemental Fig. S2). GO Slim terms with a difference of 5% or greater between SAR-enriched and SAR-suppressed phloem proteins were included in Figure 3. SAR-enriched phloem proteins were associated with the Gene Ontology (GO) terms response to stress, response to biotic stimulus, cell death, and response to external stimulus; however, the metabolic process, anatomical morphology, and photosynthesis terms also were more frequent in SAR-enriched compared with SAR-suppressed phloem proteins. In contrast, SAR-suppressed phloem proteins were associated with the GO terms response to abiotic stress, transport, catabolic process, carbohydrate metabolic process, and metabolite precursor and energy (Fig. 3A). In comparing cellular compartment GO terms, it was evident that SAR-enriched phloem proteins were frequently associated with terms representing extracellular (cell wall, external encapsulating structure, and extracellular) and thylakoid localization, while SAR-suppressed phloem proteins were associated with intracellular terms (ribosome, endoplasmic reticulum, vacuole, nucleus, plastid, cytosol, and intracellular; Fig. 3B). The molecular function GO terms catalytic activity, nucleotide binding, RNA binding, transferase activity, and enzyme regulator activity were more frequent in SAR-enriched phloem proteins, whereas binding, protein binding, transporter, carbohydrate binding, and hydrolase were more frequent in suppressed phloem proteins. Although qualitative, the GO Slim analysis demonstrates that the induction of SAR leads to the accumulation and suppression of two distinct sets of proteins.
Figure 3.
GO Slim analysis of proteins enriched or suppressed in SAR-induced phloem exudates. GO Slim terms are given pertaining to biological process (A) and cellular compartment (B) of SAR-enriched (Enriched; n = 16) compared with SAR-suppressed (Suppressed; n = 46) proteins. Only GO Slim terms with a difference in frequency of at least 5% between the enriched and suppressed groups are shown. The full GO analysis can be found in Supplemental Fig. S2.
SAR Phloem Proteome Validation
Among the 16 SAR-enriched phloem proteins, two known regulators of SAR were present. The cytosolic THIOREDOXIN h3 (TRXh3) and ACYL-COENZYME A-BINDING PROTEIN6 (ACBP6) were significantly enriched in phloem exudates collected from SAR-induced compared with mock-inoculated leaves (Table I). TRXh3 regulates the oligomeric status of the master SAR signaling protein NPR1 along with TRXh5 to control the induction of SAR (Tada et al., 2008). Single mutants trxh3 and trxh5 are modestly impacted in SAR; however, loss of the NADPH-DEPENDENT THIOREDOXIN REDUCTASE A protein that regulates their activity results in a full loss of SAR, suggesting that TRXs are important components of the SAR response (Tada et al., 2008). ACBPs including ACBP6 also have been implicated in SAR, such that acbp6 mutants are defective in the generation and/or translocation of SAR signals (Xia et al., 2012). Unexpectedly, DIR1 was not identified in our proteomes despite being readily observed via immunoblot analysis (Champigny et al., 2013). This may be explained by the demonstrated resistance of LTPs to proteolytic degradation (Lindorff-Larsen and Winther, 2001; Scheurer et al., 2004), preventing DIR1 detection during quantitative proteomics analysis of phloem exudates. In support of this idea, recombinant DIR1 protein was not detected using LC-MS/MS. Lastly, the accumulation of the SAR molecular marker PATHOGENESIS-RELATED1 (PR1) was detected in SAR-induced phloem exudates, which together with finding TRXh3 and ACBP6 indicates that the phloem proteomes from pathogen-inoculated leaves represent SAR-activated phloem sap.
To further assess the validity of our SAR proteome, immunoblot experiments were performed to confirm PR1 protein accumulation in phloem exudates during SAR. PR1 was selected because it is an important SAR molecular marker and a reliable antibody was available (Wang et al., 2005). Phloem exudates from mock-inoculated (10 mm MgCl2) and SAR-induced (Pst DC3000/avrRpt2) Col-0 leaves were collected from 25 to 48 hpi, concentrated, and probed with a polyclonal PR1 antibody. As a positive control, exudates also were probed for DIR1, a protein with demonstrated phloem accumulation during SAR (Champigny et al., 2013). As an additional control, total protein extracts from mock- and Pst DC3000/avrRpt2-inoculated leaf tissue (48 hpi) were assayed for PR1 and DIR1 accumulation. As expected, DIR1 antibody signals (7 and 14 kD) were detected in phloem exudates collected from SAR-induced but not mock-inoculated leaves and were undetectable in leaf extracts (Fig. 4). In comparison, PR1 was detected in total protein extracts of Pst DC3000/avrRpt2- but not mock-inoculated leaves. Importantly, PR1 was detected in phloem exudates collected from SAR-induced but not mock-inoculated leaves, confirming that PR1 protein accumulates in the phloem during SAR. This observation further validates the proteomics data set and identifies PR1 as a marker for SAR-activated phloem sap.
Figure 4.
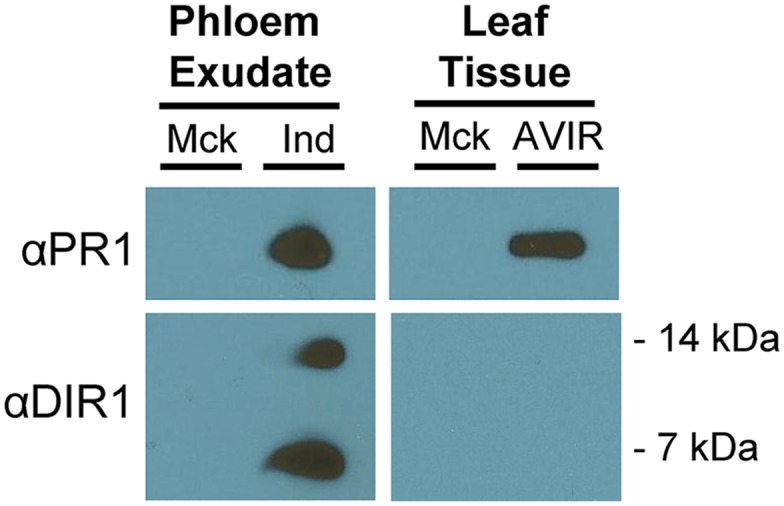
PR1 accumulates in phloem exudates of SAR-induced leaves. Immunoblots are from phloem exudates and leaf tissue collected from 4-week-old Col-0 plants that were mock inoculated (Mck; 10 mm MgCl2) or induced (Ind) for SAR (106 colony-forming units [cfu] mL−1 Pst DC3000/avrRpt2). Phloem exudates were collected from 24 to 48 hpi, and leaf tissue was harvested at 48 hpi. Immunoblotting was performed using PR1 (1:3,000) and DIR1 (1:10,000) antibodies. Similar results were obtained in three independent experiments. AVIR, Avirulent.
Functional Characterization of SAR-Enriched Phloem Proteins
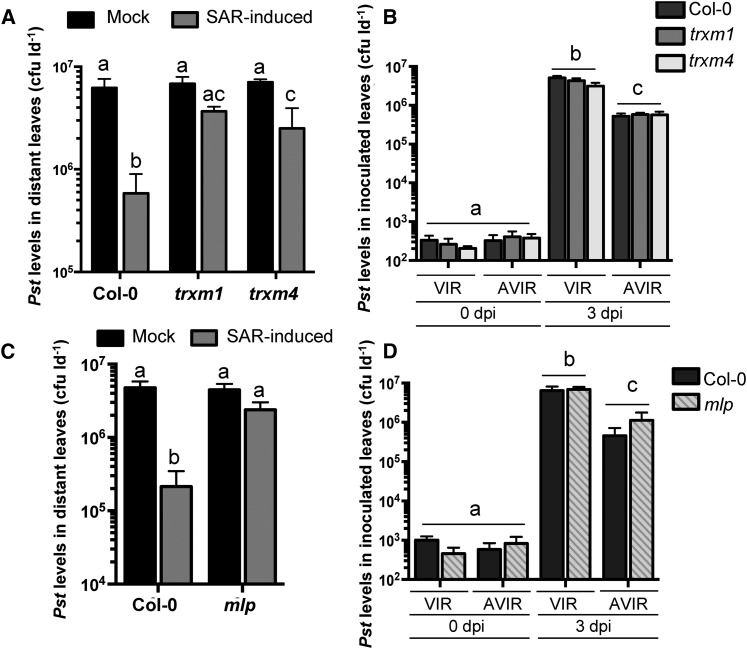
SAR assays were performed on a number of T-DNA insertion mutants corresponding to SAR-enriched phloem exudate proteins to determine if they contribute to SAR. TRXh3 and ACBP6 mutants were not tested because these proteins have been shown to be required for SAR (Tada et al., 2008; Xia et al., 2012). Three members of the TRXm family (TRXm1, TRXm2, and TRXm4) were identified in the proteomics analysis. Both TRXm1 and TRXm2 were enriched in exudates from SAR-induced leaves, while TRXm4 was enriched in exudates collected from leaves induced with avirulent Pst. To determine if this protein family is important for SAR, we compared the SAR phenotypes of the trxm1, trxm2, and trxm4 mutants with that of wild-type Col-0. Distant leaves of SAR-induced Col-0 plants supported 29-fold less bacterial growth than mock-inoculated controls, indicative of a strong SAR response. In comparison, both trxm1 and trxm4 displayed partial defects in the SAR response compared with wild-type Col-0, such that trxm1 and trxm4 plants were 2.5- and 3.5-fold more resistant to Pst in distant leaves of induced versus mock-inoculated plants (Fig. 5A). The SAR phenotype of the trxm2 mutant ranged from partially SAR defective to fully competent in three independent experiments (Supplemental Fig. S3). The partial SAR-defective phenotypes of trxm1 and trxm4 and the variable phenotype of trxm2 may be due to genetic redundancy in the TRXm family. This idea is supported by the observation that TRXm1, TRXm2, and TRXm4 all share high amino acid sequence similarity (greater than 74%) to one another (Supplemental Table S6). To ensure that the partial SAR defects observed in the trxm1 and trxm4 mutants were not caused by a defect in local immune responses, we performed disease resistance assays to assess local responses to virulent and avirulent Pst. In planta bacterial levels of virulent and avirulent Pst in trxm1 and trxm4 were similar to those in wild-type Col-0 at both 0 and 3 dpi (Fig. 5B), demonstrating that trxm1 and trxm4 are not impaired in local immune responses to Pst.
Figure 5.
The SAR-enriched phloem proteins TRXm1, TRXm4, and MLP are involved in SAR. A and D, Standard SAR assays comparing wild-type Col-0 with trxm1 and trxm4 (A) or mlp (D). Leaves of 4-week-old plants were mock inoculated (10 mm MgCl2) or induced for SAR by pressure infiltration with 106 cfu mL−1 Pst DC3000/avrRpt2. Two days later, distant leaves were challenged with 106 cfu mL−1 Pst DC3000, and Pst levels in these leaves were quantified 3 d post inoculation (dpi). Experiments were repeated at least three times with similar results. B and C, Local resistance assays comparing wild-type Col-0 with trxm1 and trxm2 (B) or mlp (C). Local resistance to virulent (VIR; Pst DC3000) and avirulent (AVIR; Pst DC3000/avrRpt2) strains of Pst was assessed by inoculating leaves of 4-week-old plants with 106 cfu mL−1 of either strain. Bacterial densities were determined at 0 and 3 dpi. All values represent means ± sd of three sample replicates. Different letters indicate statistically significant differences (ANOVA, Tukey’s honestly significant difference [HSD], P < 0.05).
Several lipid transfer/binding proteins contribute to the SAR response (Jung et al., 2009; Xia et al., 2012; Champigny et al., 2013; Cecchini et al., 2015). Two lipid-binding proteins were identified in our SAR phloem proteome. Glycosylphosphatidylinositol-anchored LIPID TRANSFER PROTEIN6 (LTPG6) accumulated in phloem exudates collected from leaves induced with virulent and avirulent Pst, and a putative lipid-binding major latex protein (MLP; AT4G23670) accumulated in phloem exudates collected from Pst DC3000/avrRpt2-induced leaves (Table I; Supplemental Table S1). The SAR phenotypes of ltpg6 and mlp mutants were compared with that of wild-type Col-0 to determine if these lipid-binding proteins are involved in SAR. In two independent experiments, the ltpg6-2 mutant displayed a strong SAR response similar to that of Col-0, indicating that LTPG6 is not required for SAR (Supplemental Fig. S3). In contrast, an mlp T-DNA mutant (Supplemental Fig. S4) displayed a 2-fold reduction in Pst levels in distant leaves of SAR-induced compared with mock-inoculated plants, whereas a 22-fold reduction was observed in Col-0 (Fig. 5C), providing evidence that MLP is involved in SAR. Local resistance assays demonstrated that the mlp mutant supports similar levels of virulent and avirulent Pst compared with Col-0 (Fig. 5D), ruling out the possibility that a defect in local resistance is responsible for the SAR-defective phenotype of the mlp mutant. The data support a role for MLP in long-distance SAR signaling.
Expression levels of TRXm1 to TRXm4 and MLP were monitored in wild-type Col-0 plants during local infection with virulent Pst to determine if increases in gene expression explain why these proteins accumulated in phloem exudates during SAR. ACTIN1 (ACT1) and PR1 were monitored as controls for equal loading and defense activation, respectively. No appreciable changes in gene expression were observed for any of the TRXm family members (TRXm1–TRXm4 ), MLP, or ACT1 after Pst inoculation. In contrast, the defense marker PR1 was highly induced at 24 and 48 hpi (Supplemental Fig. S5). These data indicate that the TRXm1 to TRXm4 and MLP genes are not induced during the induction of SAR, suggesting that the increase in protein abundance in phloem exudates may be due to mobilization into the phloem during SAR.
Functional Characterization of SAR-Suppressed Phloem Proteins
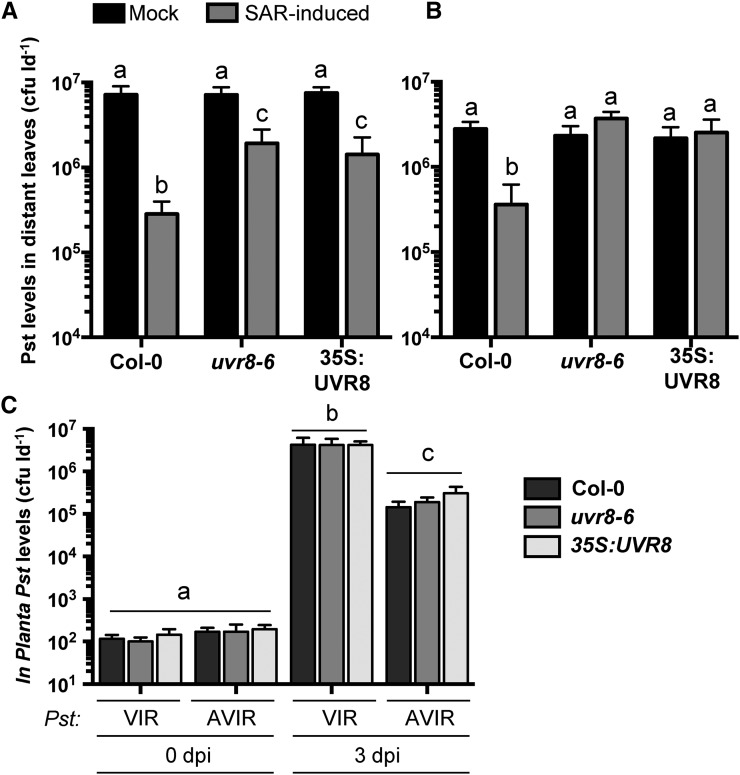
A potential function of proteins whose abundance is reduced in the phloem during SAR may be to act as negative regulators of SAR. To explore this possibility, SAR assays were conducted with mutant and overexpression lines of genes corresponding to two SAR-suppressed phloem exudate proteins. Of the 46 proteins with decreased abundance in SAR phloem exudates, we obtained and tested plant lines with altered expression levels of ANNEXIN1 and UVR8. The ANNEXIN1 overexpression line (35S:AnnAt1) and mutant (annat1-1) were fully SAR competent (Supplemental Fig. S3). In contrast, the 35S:UVR8 overexpression line and the uvr8-6 mutant were defective for SAR compared with wild-type Col-0 (Fig. 6). However, the severity of the defect varied between experiments, such that partial (Fig. 6A) or full (Fig. 6B) defects in the SAR response of uvr8-6 and 35S:UVR8 were observed in three separate experiments. It is possible that environmental conditions, such as variable UV-B radiation, may have impacted the involvement of UVR8 in SAR; however, UV-B radiation was undetectable in our growth chambers. Local resistance responses to virulent and avirulent Pst were unaffected in 35S:UVR8 and uvr8-6 (Fig. 6C), indicating that these lines are specifically impaired in SAR. These data suggest that UVR8 may function as both a positive and negative regulator of SAR.
Figure 6.
The UV-B photoreceptor UVR8 is required for SAR. A and B, Standard SAR assays of 4-week-old Col-0, uvr8-6, and 35S:UVR8 plants. Leaves were mock inoculated (10 mm MgCl2) or induced for SAR by pressure infiltration with 106 cfu mL−1 Pst DC3000/avrRpt2. Two days later, distant leaves were challenged with 106 cfu mL−1 Pst DC3000, and Pst levels in these leaves were quantified 3 dpi. This experiment was performed six times, with similar results observed three times each. C, Local resistance assays of Col-0, uvr8-6, and 35S:UVR8 to virulent (VIR; Pst DC3000) and avirulent (AVIR; Pst DC3000/avrRpt2) strains of Pst. Leaves of 4-week-old plants were inoculated with 106 cfu mL−1 of either strain, and in planta bacterial density was calculated at 0 and 3 dpi. This experiment was performed three times with similar results. All values represent means ± sd of three sample replicates. Different letters indicate statistically significant differences (ANOVA, Tukey’s HSD, P < 0.05).
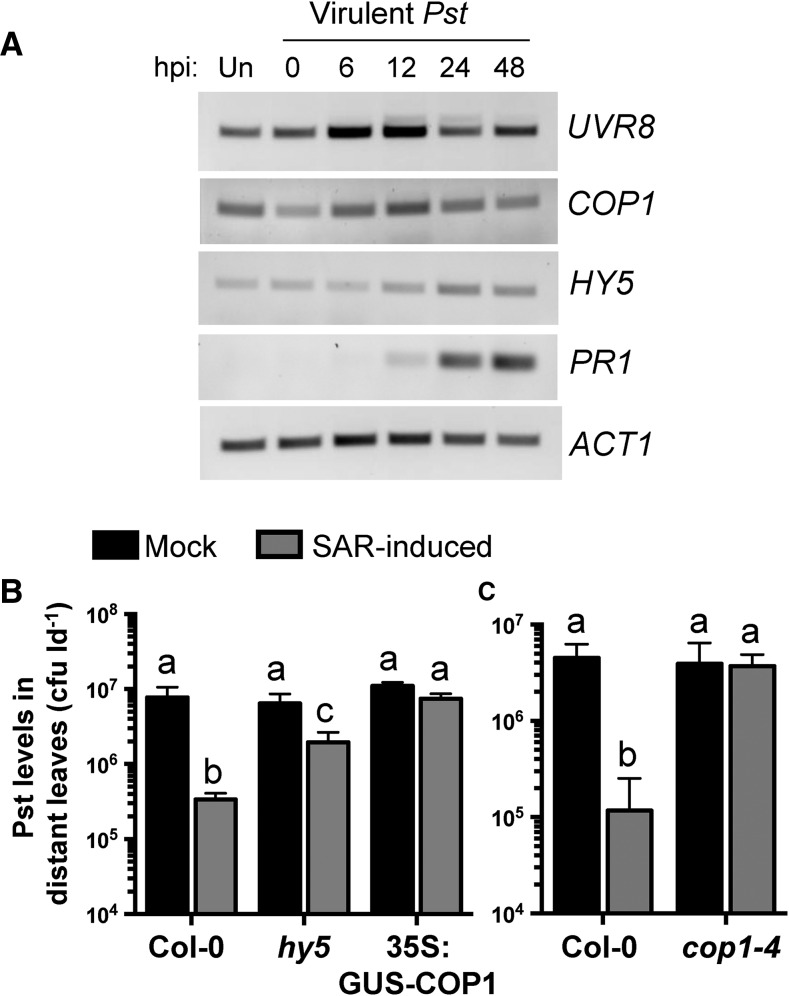
To determine if reduced UVR8 protein in phloem exudates of SAR-induced plants is associated with a decrease in UVR8 mRNA, we monitored UVR8 gene expression during local infection with virulent Pst. The COP1 and HY5 genes also were monitored to determine if the UV-B signaling module is perturbed during infection. In Arabidopsis, COP1 and HY5 are important positive regulators of the UVR8 signaling module (for review, see Tilbrook et al., 2013). ACT1 and PR1 were monitored as loading and defense-activation controls, respectively. As expected, PR1 levels were high at 24 and 48 hpi. Subtle changes in gene expression were observed for UVR8, COP1, and HY5 (Fig. 7A). Since subtle changes in gene expression cannot be quantified using RT-PCR, we queried publicly available gene expression databases (Genevestigator, the Arabidopsis Gene Expression Browser, and the Arabidopsis eFP Browser) for UVR8, COP1, and HY5 expression during local interactions with P. syringae (Winter et al., 2007; Hruz et al., 2008; Zhang et al., 2010). Several studies identified enhanced HY5 expression (4.5-fold maximally) in pathogen-treated compared with mock-treated or untreated controls (Supplemental Figs. S6–S8). Levels of UVR8 and COP1 decreased less than 2-fold during pathogen infection or did not change at all. Altogether, the data suggest that reduced levels of UVR8 in SAR-induced phloem exudates are not associated with reduced UVR8 mRNA levels and that the expression of HY5 is enhanced during local inoculation with virulent Pst.
Figure 7.
The UV-B signaling components COP1 and HY5 are required for the manifestation of SAR. A, Reverse transcription (RT)-PCR of complementary DNA generated from leaves of 4-week-old Col-0 plants that were untreated (Un) or inoculated with 106 cfu mL−1 Pst DC3000 at the indicated time points (hpi). UVR8, HY5, and COP1 expression was compared with that of the ACT1 and PR1 controls. This experiment was performed three times with similar results. B and C, Standard SAR assays comparing wild-type Col-0 with hy5 and 35S:GUS-COP1 (B) or cop1-4 (C). Leaves were mock inoculated (10 mm MgCl2) or induced for SAR by pressure infiltration with 106 cfu mL−1 Pst DC3000/avrRpt2. Two days later, distant leaves were challenged with 106 cfu mL−1 Pst DC3000, and Pst levels in these leaves were quantified 3 dpi. Values represent means ± sd of three sample replicates. Different letters indicate statistically significant differences (ANOVA, Tukey’s HSD, P < 0.05). These experiments were performed three times with similar results.
Given that reduced or elevated levels of UVR8 impair SAR, we hypothesized that altered levels of COP1 and HY5 also may impact systemic immunity. Alternatively, UVR8 function during SAR may be independent of COP1 or HY5. To test these hypotheses, SAR assays were performed with a COP1 mutant and overexpression line as well as a T-DNA insertion mutant of HY5. Wild-type Col-0 displayed a 23-fold decrease in distant leaf Pst levels in SAR-induced compared with mock-inoculated plants, while hy5 displayed a partial defect in SAR (3.3-fold decrease) and 35S:GUS-COP1 was fully defective in SAR (Fig. 7B). The cop1-4 mutant was similarly defective in SAR, as Pst levels were similar in both mock- and SAR-induced plants (Fig. 7C). These data demonstrate that HY5 and COP1 are required for SAR.
DISCUSSION
Phloem Proteomics
Proteomic analysis provides a snapshot of the proteins present in a particular tissue at a given stage of development under a particular set of environmental conditions. The phloem proteome described in this study shared 50% to 63% similarity with two previously published Arabidopsis phloem exudate proteomes. Plants used in this study were short-day grown and young (4 weeks post germination) compared with the older, long-day-grown plants used by Batailler et al. (2012) and Guelette et al. (2012). Despite these differences, 27 common phloem proteins were identified. These included known phloem proteins as well as plastid-targeted proteins that are normally associated with photosynthetic, nonphloem cell types. While this may be indicative of unavoidable contamination caused by cellular leakage from nonphloem cells during exudation, companion cells and sieve elements do contain plastids that could harbor these proteins (Froelich et al., 2011; Cayla et al., 2015). In support of this idea, live imaging of Arabidopsis phloem recently determined that Rubisco-containing plastids occupy a large volume of the companion cell cytoplasm (Cayla et al., 2015). Alternatively, nucleus-encoded proteins with predicted plastid-localization peptides may localize to nonplastid subcellular sites in the phloem. Comparisons with exudate proteomes derived from different plant species yielded fewer similarities, which suggests that protein composition within the phloem is specialized. This also may be due to differences in exudate collection techniques and/or fundamental differences in phloem architecture. This is especially important in comparisons with the cucurbit family, where phloem exudates collected directly from the cut ends of petioles are largely composed of apoplastic fluid mixed with the contents of a specialized extrafascicular phloem system that is not present in other plants (Zhang et al., 2012).
Several groups recently conducted complex comparative proteomics studies of phloem exudates collected during stress. These include comparative phloem proteomes derived from poplar (Populus spp.) and pumpkin upon wounding stress (Dafoe et al., 2009; Gaupels et al., 2012), rice (Oryza sativa) exposed to plant-hopper insects (Du et al., 2015), salt-stressed cucumber (Fan et al., 2015), melon (Cucumis melo) responding to viral infection (Serra-Soriano et al., 2015), and iron-limited Brassica napus (Gutierrez-Carbonell et al., 2015). A common theme among these proteomes, including this study, is the accumulation of redox-related proteins during stress. The presence of a sieve element antioxidant system is well described and is hypothesized to be important for phloem protein regeneration/protection, as enucleate sieve elements cannot easily replace damaged proteins (Walz et al., 2002). Therefore, the accumulation and maintenance of redox-associated proteins is likely essential to maintain phloem function during stress.
The SAR-Induced Phloem Proteome
Inducible, systemic responses such as SAR often rely on the phloem as an avenue for efficient interorgan communication. A number of studies have focused on the identification of SAR-activating small molecules that accumulate in the phloem during SAR (for review, see Dempsey and Klessig, 2012), yet little attention has been given to proteins. This gap in knowledge was addressed by performing comparative proteomics studies to determine the protein profiles of phloem exudates collected from mock-inoculated and SAR-induced plants. To identify SAR-specific phloem proteins, plants were induced for SAR using both virulent and avirulent Pst. These strains induce SAR to the same extent in Arabidopsis Col-0 (Mishina and Zeier; 2007; this study), allowing us to differentiate SAR phloem proteins from those specifically associated with susceptible or resistant interactions. Label-free quantitative LC-MS/MS proteomics of two experimental replicates identified a total of 564 phloem proteins, from which we identified 16 proteins that accumulate and 46 proteins that decrease in abundance in the phloem during SAR induced by both virulent and avirulent Pst. Comparative GO analyses revealed that SAR-enriched proteins were associated with stress-related extracellular terms, while SAR-suppressed proteins were associated with metabolism-related intracellular terms. This result is not surprising, as previous studies demonstrated that pathogen infection modifies host metabolism (Ward et al., 2010) and induces protein secretion to the apoplast (Wang et al., 2005).
Consistent with previous reports, total protein levels were higher in phloem exudates collected from SAR-induced compared with mock-inoculated leaves (Champigny et al., 2013; Carella et al., 2015), which may suggest that the induction of SAR leads to the mass translocation of a number of proteins through the phloem. If this is indeed true, then significant modifications to companion cell plasmodesmatal pore size are likely required to facilitate increased protein loading into the phloem. This idea is consistent with current hypotheses linking plasmodesmata to local and systemic immunity (Lee et al., 2011; Faulkner et al., 2013; Wang et al., 2013; Carella et al., 2015), although the impact of biotic stress on plasmodesmatal permeability in the phloem has yet to be studied. Alternatively, increased protein levels in SAR-induced phloem exudates may result from contamination caused by the deterioration of plant tissues that occurs during infection with pathogens. Indeed, proteins classified as extracellular were enriched in phloem exudates collected during SAR, which may support that cellular contamination is more likely to occur during infection. However, petiole damage was not detected in mock- or SAR-induced leaves in this study. Moreover, extracellular PR proteins are routinely identified in phloem exudate proteomes of healthy plants, including this study (Rodriguez-Celma et al., 2016), suggesting that extracellular proteins access the phloem translocation stream.
Proteins Enriched in SAR-Induced Phloem That Contribute to the SAR Response
We identified 16 proteins that accumulate in phloem exudates during the induction of SAR. Of these, PR1, the putative chitinase AED15, TRXh3, and ACBP6 were associated previously with SAR, demonstrating that SAR-related proteins are present in our SAR phloem proteome. The AED15 and PR1 proteins are known to accumulate in the apoplast during SAR (Moreno et al., 2012; Breitenbach et al., 2014). The localization of these proteins in the phloem suggests that plants produce these antimicrobial and antiherbivory proteins to protect against phloem sap-feeding insects and/or phloem-restricted microbial pathogens.
The SAR-enriched phloem proteins ACBP6 and TRXh3 are required for the manifestation of SAR in Arabidopsis (Tada et al., 2008; Xia et al., 2012). Phloem exudate-swapping experiments with the acbp6 mutant suggest that ACBP6 is required for the production or movement of SAR signals (Xia et al., 2012), similar to the lipid transfer protein DIR1 (Maldonado et al., 2002). In vitro studies indicate that ACBP6 binds acyl-CoA and phosphatidylcholine (Engeseth et al., 1996; Chen et al., 2008) and may be involved in interorganellar lipid transport (Chen et al., 2008), while DIR1 binds monoacylated phospholipids (Lascombe et al., 2008). Accumulation of the ACBP6 (this study) and DIR1 lipid-binding proteins in the phloem during SAR supports the idea that lipid-based long-distance signaling is important for systemic immunity.
TRXh3 contributes to SAR in concert with TRXh5 by regulating the oligomer-to-monomer transition of cytosolic NPR1 via the thiol-disulfide conversion of redox-sensitive Cys residues (Tada et al., 2008). How TRXh3 functions in the phloem during the induction of SAR is unknown, but it may function in the thiol-disulfide conversion of NPR1 or other Cys-containing SAR proteins such as DIR1. Recent evidence demonstrating the effectiveness of phloem-specific AtNPR1 expression in protecting citrus trees against Huanglongbing disease hints that NPR1 function may be important in the phloem (Dutt et al., 2015).
Several redox-related proteins accumulated in the phloem during SAR, including PrxIIE (peroxiredoxin), GRXC2 (glutaredoxin), GSTU5 (glutathione S-transferase), and the m-type thioredoxins TRXm1/2/4. Given that thioredoxins are associated with SAR (Tada et al., 2008), the importance of TRXm1/2/4 function during SAR was investigated. T-DNA mutants in TRXm1 and TRXm4 were partially SAR defective, providing evidence that these thioredoxins are involved in SAR. TRXm1 and TRXm4 belong to the m-type family of plastid-targeted thioredoxins, which also includes TRXm2 and TRXm3 (Collin et al., 2003). Aside from TRXm3, which is involved in mediating intercellular transport during meristem development (Benitez-Alfonso et al., 2009), m-type thioredoxins are thought to play a redundant role in the redox regulation of plastidial enzymes associated with carbon metabolism (Collin et al., 2003). Given their localization in plastids and accumulation in phloem exudates, the function of TRXm1/4 during SAR may involve the redox regulation of target proteins in companion cell and/or sieve element plastids, which is intriguing given that lipidic SAR signals and some Cys-containing SAR proteins (AZI1 and EARLI1) are produced or located in plastids (Chaturvedi et al., 2008; Cecchini et al., 2015).
It is conceivable that TRXm proteins localize to other subcellular compartments in phloem cells during SAR, which would allow for their accumulation in phloem exudates. This idea is supported by observations of dual cytosolic and plastidial localization of TRXm2 (Holscher et al., 2014). Nevertheless, TRXm protein (Guelette et al., 2012; this study) and mRNA (Deeken et al., 2008) accumulate in phloem exudates, and TRXm1 and TRXm4 contribute to SAR (this work). How these proteins contribute to SAR remains to be determined, but recent evidence demonstrating the molecular holdase/foldase activity of NtTRXm in tobacco suggests that TRXm proteins act as molecular chaperones that protect target proteins during stress (Sanz-Barrio et al., 2012). As such, TRXm proteins may protect redox-sensitive proteins important for SAR in the phloem. In addition, TRXm1 was recently shown to bind the defense hormone salicylic acid (SA) using a number of protein-ligand-binding techniques (Manohar et al., 2015). Whether TRXm1 function in the phloem during SAR requires SA remains to be determined.
The putative lipid-binding protein MLP joins a number of lipid-associated proteins important for SAR. Analysis of an mlp T-DNA insertion mutant demonstrated a role for MLP in the SAR response. MLP belongs to a largely uncharacterized family of proteins that contain a BetvI (major birch [Betula spp.] pollen allergen) fold, which produces a forked hydrophobic cavity capable of binding large hydrophobic molecules (Gajhede et al., 1996; Radauer et al., 2008). This protein family includes the defense-associated intracellular PR10 protein, whose molecular function is unknown (Osmark et al., 1998). Since the main feature of MLP appears to be the BetvI fold, we speculate that MLP may bind a hydrophobic SAR signal. The diterpenoid SAR signal dehydroabietinal is a potential MLP ligand, as dehydroabietinal accumulates in the phloem during SAR (Chaturvedi et al., 2012). Future studies to examine if MLP binds dehydroabietinal or other hydrophobic defense activators will shed light on its role during SAR.
Proteins Suppressed in the SAR Phloem Proteome
The accumulation of a number of proteins was suppressed in phloem exudates collected from SAR-induced leaves, some of which were associated previously with plant defense and include TGG1 myrosinase (Barth and Jander, 2006), the jasmonic acid-responsive Man-binding lectin JR1 (León et al., 1998), CALRETICULIN2 (Qiu et al., 2012), the plastidial chaperonin CPN60B (Ishikawa et al., 2003), the fasciclin-like arabinogalactan-protein FLA8 (Gruner et al., 2013), and the Gly-rich RNA-binding protein GRP7 (Fu et al., 2007). Of these proteins, JR1 and FLA8 are down-regulated in distant leaves of SAR-induced plants (Gruner et al., 2013; Bernsdorff et al., 2016), and analysis of cpn60B knockout mutants demonstrated a constitutive SAR-like response to P. syringae pv maculicola (Ishikawa et al., 2003). Interestingly, CPN60, a chloroplastic chaperon protein, also was suppressed in melon phloem during viral infection (Serra-Soriano et al., 2015), hinting that CPN60 may act as a negative regulator of disease resistance responses in the phloem.
The UVR8-Signaling Module Is Important for SAR
Phenotypic analysis of the SAR response in mutant and overexpression lines of a number of SAR-suppressed proteins identified a role for UVR8 in SAR, as both uvr8-6 and 35S:UVR8 plant lines were SAR defective compared with wild-type plants. The UVR8 photoreceptor is a seven-bladed β-propeller protein that perceives UV-B wavelengths using intrinsic Trp residues (Christie et al., 2012). Upon UV-B photoactivation, UVR8 homodimers monomerize and translocate from the cytosol to the nucleus (Kaiserli and Jenkins, 2007). In the nucleus, UVR8 interacts with COP1 to induce the expression of the bZIP transcription factor HY5, which in turn activates UV-B-responsive gene expression (Favory et al., 2009; Rizzini et al., 2011). In this study, we observed reduced levels of UVR8 in phloem exudates of SAR-induced compared with mock-induced plants. It is tempting to speculate that SAR induction causes the accumulation of UVR8 in the nucleus, leading to decreased levels of cytosolic UVR8 available for movement into the phloem translocation stream. Alternatively, UVR8 may be negatively regulated during the induction of SAR. Given that UVR8 gene expression is not affected by inoculation with virulent Pst, we speculate that the suppression of UVR8 involves proteasomal degradation and/or posttranscriptional regulation.
In addition to its well-established role in the UV-B stress response (for review, see Tilbrook et al., 2013), recent evidence demonstrated a positive role for UVR8 in abiotic stress responses (Fasano et al., 2014) as well as UV-B-induced resistance to the necrotrophic fungus Botrytis cinerea (Demkura and Ballaré, 2012). Our analysis of the uvr8-6 mutant and a UVR8 overexpression line suggests that UVR8 plays both a positive and negative role during SAR, which may indicate that UVR8 regulates distinct processes during the SAR response, perhaps in different tissues. Overexpression of wild-type UVR8 protein does not activate UV-B-response gene expression in the absence of UV-B (Heijde et al., 2013). Since UV-B radiation is not detectable in our growth chambers, UVR8 signaling activated by UV-B light probably does not contribute to the SAR defect observed in the UVR8 overexpression line. Rather, increased pools of inactive UVR8 protein in the UVR8 overexpression line may have a dominant-negative effect. In any case, the SAR phenotypes of the UVR8 overexpression and mutant lines indicate that UVR8 is required for SAR, perhaps by regulating core light signaling or UV-response genes.
SAR Utilizes Core Components of Light Signaling Pathways
We further investigated the importance of UVR8 in SAR by assessing the SAR phenotypes of hy5, cop1-4, and 35S:GUS-COP1. Both COP1 and HY5 positively regulate UV-B responses downstream of UVR8 (Tilbrook et al., 2013). SAR was negatively impacted in each of these plant lines, demonstrating that the core members of the UV-B signaling pathway are important for SAR. In addition to their involvement in UV-B signaling, COP1 and HY5 also are central regulators of other light-signaling responses (Jiao et al., 2007), suggesting that core light-signaling machinery is required for SAR. Indeed, several studies indicate an association of light signaling with local and systemic pathogen defense responses (for review, see Roden and Ingle, 2009). The accumulation of SA, PR gene expression, and the manifestation of SAR all require exposure to light (Zeier et al., 2004). Moreover, light signaling components are important for this response, as the red light photoreceptor double mutant phyA/phyB is defective in SAR under typical growth conditions (Griebel and Zeier, 2008) and the blue light photoreceptor CRY1 is required for SAR in continuous light (Wu and Yang, 2010). The duration of light perceived following pathogen infection also impacts SAR, such that plants induced for SAR in the morning are less dependent on methyl salicylate-mediated responses compared with plants induced in the evening (Liu et al., 2011). In addition, exposure to high light intensities induces SA accumulation, the generation of reactive oxygen species, and programmed cell death, resulting in a SAR-like response (Mühlenbock et al., 2008). Recent evidence demonstrated that HY5 is required for light-induced programmed cell death and SA accumulation through the positive regulation of the immune regulator EDS1 (Chai et al., 2015), which itself is required for the generation and perception of mobile SAR signals (Breitenbach et al., 2014). This may suggest that HY5 is a positive regulator of EDS1 and other defense-related genes during the induction of SAR, which is supported by the identification of NPR1, NIMIN2, ADR1, PAD4, and TRXm4 as putative HY5-binding targets (Lee et al., 2007). Furthermore, a recent study identified COP1 as a putative binding target of the SAR transcription factor SARD1 (Sun et al., 2015). Together, these results argue for a central role of light signaling in the establishment of local and systemic immune responses.
CONCLUSION
A comparative proteomics analysis of Arabidopsis phloem exudates collected from mock- and SAR-induced plants identified several proteins with differential abundance. Of these proteins, m-type thioredoxins, a major latex protein-like protein, and UVR8 were discovered to play a role in the SAR response. Further exploration of the UV-B signaling pathway identified COP1 and HY5 as additional regulators of SAR, which is in agreement with several studies that associate light signaling and systemic immunity. Importantly, the proteomics data set obtained in this study bridges fundamental gaps in knowledge by significantly adding to the limited understanding of protein composition in Arabidopsis phloem exudates while providing an in-depth look at phloem proteins associated with SAR long-distance signaling. This study contributes to the emerging field of comparative proteomic analysis of plant vascular sap that will provide insights into interorgan communication during stress.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana ecotype Col-0) and homozygous T-DNA mutant seeds (Supplemental Fig. S4) were surface sterilized and stratified at 4°C in the dark for 2 d. Sterile seeds were plated on Murashige and Skoog plates and germinated for 5 to 7 d under continuous light. Seedlings were transplanted onto soil hydrated with 1 g L−1 20-20-20 fertilizer and grown under short-day photoperiod conditions (9 h of light; 150 μE m−2 s−1) at 22°C with 65% to 85% relative humidity. UV-B levels in growth chambers were undetectable (UV-X radiometer; UVP). Confirmed (homozygous) plant lines were obtained from the Arabidopsis Biological Resource Center or independent research laboratories (Konopka-Postupolska et al., 2009; Tsuchiya et al., 2010; Fasano et al., 2014). Homozygous mlp mutants (Nottingham Arabidopsis Stock Centre; GK-089B08) were confirmed from heterozygous seed stock by germination on Murashige and Skoog medium containing sulfadiazine (5 μg mL−1) followed by molecular characterization of mRNA levels using RT-PCR (Supplemental Fig. S4).
Bacterial Growth, Inoculation, and Quantitation
Standard SAR experiments and local resistance assays were performed as described by Carella et al. (2015) with Pseudomonas syringae pv tomato strains cultured overnight with shaking in King’s B medium (King et al., 1954) supplemented with 50 μg mL−1 kanamycin. For large-scale phloem exudate collection experiments, leaves of 4-week-old Col-0 were pressure infiltrated with 10 mm MgCl2 (mock inoculation) or 106 cfu mL−1 virulent Pst DC3000 (pVSP1) or avirulent Pst DC3000/avrRpt2 (pVSP1 + avrRpt2). In planta Pst levels were quantified by dilution plating as described by Cameron et al. (1999) and Carella et al. (2015). Statistically significant differences in Pst levels were identified by ANOVA (Tukey’s HSD, P < 0.05) using R.
Phloem Exudate Collection
Phloem exudates were collected as described by Carella et al. (2015). At 24 hpi, leaves of mock-inoculated or SAR-induced plants (4-week-old Col-0) were cut at the base of the petiole, surface sterilized quickly (50% ethanol and 0.0006% bleach in 1 mm EDTA), and immediately placed into Eppendorf tubes containing 1 mm EDTA for 1 h. Twelve leaves were placed into each Eppendorf tube. Leaves were then transferred to tubes containing sterile water and allowed to exude in a humidity chamber for 23 h (representing exudation from 25 to 48 hpi). For proteomics analysis, pooled exudates from more than 90 plants per treatment were concentrated using centrifugal concentrators with a 3-kD cutoff (Vivaspin 20; GE Healthcare) according to the manufacturer’s instructions to a final volume of approximately 7 mL. Concentrated exudates were equally subdivided into four tubes, and protein levels were quantified using the Bio-Rad protein reagent with bovine serum albumin as a standard. Samples were then frozen in liquid nitrogen, lyophilized, and stored at −80°C until further use. Phloem exudates used for immunoblotting were collected as described previously (Carella et al., 2015).
LC-MS/MS Measurement, Label-Free Quantitative Analysis, and Database Search
Prior to LC-MS/MS analysis, the samples were centrifuged for 5 min at 4°C. Each approximately 0.5-µg sample was measured on an LTQ OrbitrapXL (Thermo Fisher Scientific) coupled to an Ultimate3000 nano-RSLC device (Dionex) as described previously (Hauck et al., 2010; Molin et al., 2015).
Raw files of each data set were analyzed separately with Progenesis QI software for proteomics as described previously (Hauck et al., 2010; Merl et al., 2012). Briefly, peptide features in the individual runs were aligned to reach a maximum overlay of at least 80%. The samples were assigned to the three individual groups, and all tandem mass spectrometry features with charges +2 to +7 were exported for protein identification using the Mascot search engine (version 2.5.0; Matrix Science) in The Arabidopsis Information Resource database (version 10). Search results were filtered for P < 0.05 and Mascot percolator score ≥ 15 to reach a false discovery rate of 1% (Brosch et al., 2009). Protein identifications were reimported in Progenesis QI software, and normalized abundances of unique peptides were summed for every protein. These values were used for the calculation of abundance ratios between groups and for statistical evaluation by Student’s t test (P < 0.05).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Information Resource under accession numbers AT1G31812 (ACBP6), AT1G35720 (ANNAT1), AT2G32950 (COP1), AT5G48485 (DIR1), AT5G11260 (HY5), AT1G55260 (LTPG6), AT4G23670 (MLP), AT2G14610 (PR1), AT5G42980 (TRXh3), AT1G03680 (TRXm1), AT4G03520 (TRXm2), AT3G15360 (TRXm4), AT5G63860 (UVR8).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SAR assay and phloem exudate collection controls.
Supplemental Figure S2. Complete GO Slim analysis of proteins enriched or suppressed in SAR-induced phloem exudates.
Supplemental Figure S3. Supporting SAR assays.
Supplemental Figure S4. Plant lines used in this study.
Supplemental Figure S5. TRXm and MLP expression analysis.
Supplemental Figure S6. Exploring UVR8/COP1/HY5 expression dynamics in publicly available data obtained from Genevestigator.
Supplemental Figure S7. Exploring UVR8/COP1/HY5 expression dynamics in publicly available data obtained from the Arabidopsis Gene Expression Browser.
Supplemental Figure S8. Exploring UVR8/COP1/HY5 expression dynamics in publicly available data obtained from the Arabidopsis eFP Expression Browser.
Supplemental Table S1. Differentially abundant phloem proteins specific to avirulent Pst treatment.
Supplemental Table S2. Differentially abundant phloem proteins specific to virulent Pst treatment.
Supplemental Table S3. Complete list of proteins suppressed in the phloem during SAR.
Supplemental Table S4. Common Arabidopsis phloem proteins.
Supplemental Table S5. Common phloem proteins in pumpkin, Texas bluebonnet, and Arabidopsis.
Supplemental Table S6. TRXm family similarity matrix.
Supplemental Data S1. Raw proteomics data.
Supplemental Methods S1. Protein isolation and immunoblotting, sample preparation for mass spectrometry, RNA isolation, PCR primers and RT-PCR analysis.
Supplementary Material
Acknowledgments
We thank Dr. Xinnian Dong (Duke University) for the PR1 antibody, Dr. Antonella Leone (University of Salerno) for the uvr8-6 and 35S:UVR8 plant lines, Dr. Jacek Hennig (Polish Academy of Sciences) for the annat1 and 35S:AnnAt1 plant lines, Dr. Peter McCourt (University of Toronto) for the hy5 (SALK_056405), cop1-4, and 35S:GUS-COP1 plants lines, May T.S. Yeo for help with phloem exudate collection, as well as the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre for T-DNA insertion lines.
Glossary
- SAR
systemic acquired resistance
- PAMP
pathogen-associated molecular pattern
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- Pst
Pseudomonas syringae pv tomato
- T-DNA
transfer DNA
- hpi
hours post inoculation
- Col-0
Columbia-0
- GO
Gene Ontology
- dpi
days post inoculation
- RT
reverse transcription
- SA
salicylic acid
- cfu
colony-forming units
- HSD
honestly significant difference
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (Discovery, RTI, and CFI Leadership grants to R.K.C. and scholarship to D.C.W.) and by an Ontario Graduate Scholarship to P.C.
Articles can be viewed without a subscription.
References
- Barth C, Jander G (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46: 549–562 [DOI] [PubMed] [Google Scholar]
- Batailler B, Lemaître T, Vilaine F, Sanchez C, Renard D, Cayla T, Beneteau J, Dinant S (2012) Soluble and filamentous proteins in Arabidopsis sieve elements. Plant Cell Environ 35: 1258–1273 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsdorff F, Doering AC, Gruner K, Brautigam A, Zeier J (2016) Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and independent pathways. Plant Cell 28: 102–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbach HH, Wenig M, Wittek F, Jordá L, Maldonado-Alconada AM, Sarioglu H, Colby T, Knappe C, Bichlmeier M, Pabst E, et al. (2014) Contrasting roles of the apoplastic aspartyl protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY-DEPENDENT1 and LEGUME LECTIN-LIKE PROTEIN1 in Arabidopsis systemic acquired resistance. Plant Physiol 165: 791–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch M, Yu L, Hubbard T, Choudhary J (2009) Accurate and sensitive peptide identification with Mascot Percolator. J Proteome Res 8: 3176–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ (1994) Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J 5: 715–725 [Google Scholar]
- Cameron RK, Paiva NL, Lamb CJ, Dixon RA (1999) Accumulation of salicylic acid and PR-1 gene transcripts in relation to the systemic acquired resistance (SAR) response induced by Pseudomonas syringae pv. tomato in Arabidopsis. Physiol Mol Plant Pathol 55: 121–130 [Google Scholar]
- Carella P, Isaacs M, Cameron RK (2015) Plasmodesmata-located protein overexpression negatively impacts the manifestation of systemic acquired resistance and the long-distance movement of Defective in Induced Resistance1 in Arabidopsis. Plant Biol (Stuttg) 17: 395–401 [DOI] [PubMed] [Google Scholar]
- Cayla T, Batailler B, Le Hir R, Revers F, Anstead JA, Thompson GA, Grandjean O, Dinant S (2015) Live imaging of companion cells and sieve elements in Arabidopsis leaves. PLoS ONE 10: e0118122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini NM, Steffes K, Schläppi MR, Gifford AN, Greenberg JT (2015) Arabidopsis AZI1 family proteins mediate signal mobilization for systemic defence priming. Nat Commun 6: 7658 [DOI] [PubMed] [Google Scholar]
- Chai T, Zhou J, Liu J, Xing D (2015) LSD1 and HY5 antagonistically regulate red light induced-programmed cell death in Arabidopsis. Front Plant Sci 6: 292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champigny MJ, Cameron RK (2009) Action at a distance: long-distance signals in induced resistance. In Van Loon LC, ed, Plant Innate Immunity. Academic Press, London, pp 123–171 [Google Scholar]
- Champigny MJ, Isaacs M, Carella P, Faubert J, Fobert PR, Cameron RK (2013) Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front Plant Sci 4: 230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R, Krothapalli K, Makandar R, Nandi A, Sparks AA, Roth MR, Welti R, Shah J (2008) Plastid ω3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J 54: 106–117 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Venables B, Petros RA, Nalam V, Li M, Wang X, Takemoto LJ, Shah J (2012) An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J 71: 161–172 [DOI] [PubMed] [Google Scholar]
- Chen QF, Xiao S, Chye ML (2008) Overexpression of the Arabidopsis 10-kilodalton acyl-coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiol 148: 304–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278: 23747–23752 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR (2015) Priming for enhanced defense. Annu Rev Phytopathol 53: 97–119 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Dafoe NJ, Zamani A, Ekramoddoullah AKM, Lippert D, Bohlmann J, Constabel CP (2009) Analysis of the poplar phloem proteome and its response to leaf wounding. J Proteome Res 8: 2341–2350 [DOI] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Demkura PV, Ballaré CL (2012) UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol Plant 5: 642–652 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF (2012) SOS: too many signals for systemic acquired resistance? Trends Plant Sci 17: 538–545 [DOI] [PubMed] [Google Scholar]
- Du B, Wei Z, Wang Z, Wang X, Peng X, Du B, Chen R, Zhu L, He G (2015) Phloem-exudate proteome analysis of response to insect brown plant-hopper in rice. J Plant Physiol 183: 13–22 [DOI] [PubMed] [Google Scholar]
- Dutt M, Barthe G, Irey M, Grosser J (2015) Transgenic citrus expressing an Arabidopsis NPR1 gene exhibit enhanced resistance against Huanglongbing (HLB, citrus greening). PLoS ONE 10: e0137134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeseth NJ, Pacovsky RS, Newman T, Ohlrogge JB (1996) Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Arch Biochem Biophys 331: 55–62 [DOI] [PubMed] [Google Scholar]
- Fan H, Xu Y, Du C, Wu X (2015) Phloem sap proteome studied by iTRAQ provides integrated insight into salinity response mechanisms in cucumber plants. J Proteomics 125: 54–67 [DOI] [PubMed] [Google Scholar]
- Fasano R, Gonzalez N, Tosco A, Dal Piaz F, Docimo T, Serrano R, Grillo S, Leone A, Inzé D (2014) Role of Arabidopsis UV RESISTANCE LOCUS 8 in plant growth reduction under osmotic stress and low levels of UV-B. Mol Plant 7: 773–791 [DOI] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ (2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich DR, Mullendore DL, Jensen KH, Ross-Elliott TJ, Anstead JA, Thompson GA, Pélissier HC, Knoblauch M (2011) Phloem ultrastructure and pressure flow: Sieve-Element-Occlusion-Related agglomerations do not affect translocation. Plant Cell 23: 4428–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR (2007) A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447: 284–288 [DOI] [PubMed] [Google Scholar]
- Gajhede M, Osmark P, Poulsen FM, Ipsen H, Larsen JN, Joost van Neerven RJ, Schou C, Løwenstein H, Spangfort MD (1996) X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat Struct Biol 3: 1040–1045 [DOI] [PubMed] [Google Scholar]
- Gaupels F, Sarioglu H, Beckmann M, Hause B, Spannagl M, Draper J, Lindermayr C, Durner J (2012) Deciphering systemic wound responses of the pumpkin extrafascicular phloem by metabolomics and stable isotope-coded protein labeling. Plant Physiol 160: 2285–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel T, Zeier J (2008) Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol 147: 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Griebel T, Návarová H, Attaran E, Zeier J (2013) Reprogramming of plants during systemic acquired resistance. Front Plant Sci 4: 252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes MEM, Richmond S, Kuc J (1980) Induced systemic resistance to anthracnose in cucumber as influenced by the location of the inducer inoculation with Colletotrichum lagenarium and the onset of flowering and fruiting. Physiol Plant Pathol 17: 229–233 [Google Scholar]
- Guelette BS, Benning UF, Hoffmann-Benning S (2012) Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J Exp Bot 63: 3603–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Carbonell E, Lattanzio G, Albacete A, Rios JJ, Kehr J, Abadía A, Grusak MA, Abadía J, López-Millán AF (2015) Effects of Fe deficiency on the protein profile of Brassica napus phloem sap. Proteomics 15: 3835–3853 [DOI] [PubMed] [Google Scholar]
- Hauck SM, Dietter J, Kramer RL, Hofmaier F, Zipplies JK, Amann B, Feuchtinger A, Deeg CA, Ueffing M (2010) Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol Cell Proteomics 9: 2292–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Binkert M, Yin R, Ares-Orpel F, Rizzini L, Van De Slijke E, Persiau G, Nolf J, Gevaert K, De Jaeger G, et al. (2013) Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc Natl Acad Sci USA 110: 20326–20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipper C, Brault V, Ziegler-Graff V, Revers F (2013) Viral and cellular factors involved in phloem transport of plant viruses. Frontiers in Plant Science 4, 54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher C, Meyer T, von Schaewen A (2014) Dual-targeting of Arabidopsis 6-phosphogluconolactonase 3 (PGL3) to chloroplasts and peroxisomes involves interaction with Trx m2 in the cytosol. Mol Plant 7: 252–255 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Tanaka H, Nakai M, Asahi T (2003) Deletion of a chaperonin 60B gene leads to cell death in the Arabidopsis lesion initiation 1 mutant. Plant Cell Physiol 44: 255–261 [DOI] [PubMed] [Google Scholar]
- Jenns AE, Kuc J (1979) Graft transmission of systemic resistance of cucumbers to anthracnose induced by Colletotrichum lagenarium and tobacco necrosis virus. Phytopathology 69: 753–756 [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B-specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloshian I, Walling LL (2005) Hemipterans as plant pathogens. Annu Rev Phytopathol 43: 491–521 [DOI] [PubMed] [Google Scholar]
- Kiefer IW, Slusarenko AJ (2003) The pattern of systemic acquired resistance induction within the Arabidopsis rosette in relation to the pattern of translocation. Plant Physiol 132: 840–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307 [PubMed] [Google Scholar]
- Konopka-Postupolska D, Clark G, Goch G, Debski J, Floras K, Cantero A, Fijolek B, Roux S, Hennig J (2009) The role of annexin 1 in drought stress in Arabidopsis. Plant Physiol 150: 1394–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascombe MB, Bakan B, Buhot N, Marion D, Blein JP, Larue V, Lamb C, Prangé T (2008) The structure of “defective in induced resistance” protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Sci 17: 1522–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattanzio G, Andaluz S, Matros A, Calvete JJ, Kehr J, Abadía A, Abadía J, López-Millán AF (2013) Protein profile of Lupinus texensis phloem sap exudates: searching for Fe- and Zn-containing proteins. Proteomics 13: 2283–2296 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. (2011) A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J, Rojo E, Titarenko E, Sánchez-Serrano JJ (1998) Jasmonic acid-dependent and -independent wound signal transduction pathways are differentially regulated by Ca2+/calmodulin in Arabidopsis thaliana. Mol Gen Genet 258: 412–419 [DOI] [PubMed] [Google Scholar]
- Lin MK, Lee YJ, Lough TJ, Phinney BS, Lucas WJ (2009) Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol Cell Proteomics 8: 343–356 [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Winther JR (2001) Surprisingly high stability of barley lipid transfer protein, LTP1, towards denaturant, heat and proteases. FEBS Lett 488: 145–148 [DOI] [PubMed] [Google Scholar]
- Liu PP, von Dahl CC, Klessig DF (2011) The extent to which methyl salicylate is required for signaling systemic acquired resistance is dependent on exposure to light after infection. Plant Physiol 157: 2216–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Groover A, Lichtenberger R, Furuta K, Yadav SR, Helariutta Y, He XQ, Fukuda H, Kang J, Brady SM, et al. (2013) The plant vascular system: evolution, development and functions. J Integr Plant Biol 55: 294–388 [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Manohar M, Tian M, Moreau M, Park SW, Choi HW, Fei Z, Friso G, Asif M, Manosalva P, von Dahl CC, et al. (2015) Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front Plant Sci 5: 777 10.3389/fpls.2014.00777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merl J, Ueffing M, Hauck SM, von Toerne C (2012) Direct comparison of MS-based label-free and SILAC quantitative proteome profiling strategies in primary retinal Müller cells. Proteomics 12: 1902–1911 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Molin S, Merl J, Dietrich KA, Regauer M, Flaig M, Letulé V, Saucke T, Herzinger T, Ruzicka T, Hauck SM (2015) The hand eczema proteome: imbalance of epidermal barrier proteins. Br J Dermatol 172: 994–1001 [DOI] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A, et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7: e31944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC. (2015) Venny. An interactive tool for comparing lists with Venn's diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
- Osmark P, Boyle B, Brisson N (1998) Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol Biol 38: 1243–1246 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PA (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347–375 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Xi J, Du L, Roje S, Poovaiah BW (2012) A dual regulatory role of Arabidopsis calreticulin-2 in plant innate immunity. Plant J 69: 489–500 [DOI] [PubMed] [Google Scholar]
- Radauer C, Lackner P, Breiteneder H (2008) The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol 8: 286 10.1186/1471-2148-8-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Roden LC, Ingle RA (2009) Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21: 2546–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Celma J, Ceballos-Laita L, Grusak MA, Abadia J, Lopez-Millan AF (2016) Plant fluid proteomics: delving into the xylem sap, phloem sap and apoplastic fluid proteomes. Biochim Biophys Acta http://dx.doi.org/10.1016/j.bbapap.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Sanz-Barrio R, Fernández-San Millán A, Carballeda J, Corral-Martínez P, Seguí-Simarro JM, Farran I (2012) Chaperone-like properties of tobacco plastid thioredoxins f and m. J Exp Bot 63: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurer S, Lauer I, Foetisch K, San Miguel Moncin M, Retzek M, Hartz C, Enrique E, Lidholm J, Cistero-Bahima A, Vieths S (2004) Strong allergenicity of Pru av 3, the lipid transfer protein from cherry, is related to high stability against thermal processing and digestion. J Allergy Clin Immunol 114: 900–907 [DOI] [PubMed] [Google Scholar]
- Serra-Soriano M, Navarro JA, Genoves A, Pallás V (2015) Comparative proteomic analysis of melon phloem exudates in response to viral infection. J Proteomics 124: 11–24 [DOI] [PubMed] [Google Scholar]
- Shah J, Chaturvedi R, Chowdhury Z, Venables B, Petros RA (2014) Signaling by small metabolites in systemic acquired resistance. Plant J 79: 645–658 [DOI] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux JP (1997) Systemic acquired resistance. Annu Rev Phytopathol 35: 235–270 [DOI] [PubMed] [Google Scholar]
- Sun T, Zhang Y, Li Y, Zhang Q, Ding Y, Zhang Y (2015) ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat Commun 6: 10159 10.1038/ncomms10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R (2013) The UVR8 UV-B photoreceptor: perception, signaling and response. The Arabidopsis Book 11: e0164, doi/10.1199/tab.0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P (2010) A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat Chem Biol 6: 741–749 [DOI] [PubMed] [Google Scholar]
- Tuzun S, Kuc J (1985) Movement of a factor in tobacco infected with Peranospora tabacina Adam which systemically protects against blue mold. Physiol Plant Pathol 26: 321–330 [Google Scholar]
- Walz C, Juenger M, Schad M, Kehr J (2002) Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J 31: 189–197 [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wang X, Sager R, Cui W, Zhang C, Lu H, Lee JY (2013) Salicylic acid regulates plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25: 2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JL, Forcat S, Beckmann M, Bennett M, Miller SJ, Baker JM, Hawkins ND, Vermeer CP, Lu C, Lin W, et al. (2010) The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J 63: 443–457 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Yang HQ (2010) CRYPTOCHROME 1 is implicated in promoting R protein-mediated plant resistance to Pseudomonas syringae in Arabidopsis. Mol Plant 3: 539–548 [DOI] [PubMed] [Google Scholar]
- Xia Y, Yu K, Gao QM, Wilson EV, Navarre D, Kachroo P, Kachroo A (2012) Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Front Plant Sci 3: 224 10.3389/fpls.2012.00224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, He SY (2013) Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Yadeta KA, Thomma BPHJ (2013) The xylem as battleground for plant hosts and vascular wilt pathogens. Front Plant Sci 4: 97 10.3389/fpls.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ (2004) A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Soares JM, Mandal MK, Wang C, Chanda B, Gifford AN, Fowler JS, Navarre D, Kachroo A, Kachroo P (2013) A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep 3: 1266–1278 [DOI] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S (2004) Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]
- Zhang C, Yu X, Ayre BG, Turgeon R (2012) The origin and composition of cucurbit “phloem” exudate. Plant Physiol 158: 1873–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhang Y, Liu L, Yu L, Tsang S, Tan J, Yao W, Kang MS, An Y, Fan X (2010) Gene Expression Browser: large-scale and cross-experiment microarray data integration, management, search & visualization. BMC Bioinformatics 11: 433 10.1186/1471-2105-11-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.