The putative catalytic O-GlcNAc domain of SPINDLY is required for stability of the transcription factor TCP14 and cytokinin responses in developing Arabidopsis leaves and flowers.
Abstract
Arabidopsis (Arabidopsis thaliana) SPINDLY (SPY) is a putative serine and threonine O-linked N-acetylglucosamine transferase (OGT). While SPY has been shown to suppress gibberellin signaling and to promote cytokinin (CK) responses, its catalytic OGT activity was never demonstrated and its effect on protein fate is not known. We previously showed that SPY interacts physically and functionally with TCP14 and TCP15 to promote CK responses. Here, we aimed to identify how SPY regulates TCP14/15 activities and how these TCPs promote CK responses. We show that SPY activity is required for TCP14 stability. Mutation in the putative OGT domain of SPY (spy-3) stimulated TCP14 proteolysis by the 26S proteasome, which was reversed by mutation in CULLIN1 (CUL1), suggesting a role for SKP, CUL1, F-box E3 ubiquitin ligase in TCP14 proteolysis. TCP14 proteolysis in spy-3 suppressed all TCP14 misexpression phenotypes, including the enhanced CK responses. The increased CK activity in TCP14/15-overexpressing flowers resulted from increased sensitivity to the hormone and not from higher CK levels. TCP15 overexpression enhanced the response of the CK-induced synthetic promoter pTCS to CK, suggesting that TCP14/15 affect early steps in CK signaling. We propose that posttranslational modification of TCP14/15 by SPY inhibits their proteolysis and that the accumulated proteins promote the activity of the CK phosphorelay cascade in developing Arabidopsis leaves and flowers.
O-linked GlcNAc (O-GlcNAc) modification of Ser and Thr residues by the nucleocytoplasmic O-GlcNAc transferases (OGTs) regulates the posttranslational fate and function of target proteins (Hart et al., 2007; Butkinaree et al., 2010). In mammalian cells, O-GlcNAcylation affects protein localization, phosphorylation, and stability and plays a role in signal transduction, transcription, and proteasomal degradation (Roos and Hanover, 2000; Wells et al., 2001; Hanover, 2001; Hanover et al., 2003, 2010; Zachara and Hart, 2004; Yang et al., 2008; Butkinaree et al., 2010). Although similar modification occurs in plants, only a few O-GlcNAcylated plant proteins have been identified. These include the tobacco (Nicotiana tabacum) histone protein and the nuclear pore protein gp40 (Heese-Peck et al., 1995; Heese-Peck and Raikhel, 1998; Schouppe et al., 2011), the wheat (Triticum aestivum) RNA-binding protein TaGRP2 (Xiao et al., 2014), and the Arabidopsis (Arabidopsis thaliana) GA signaling suppressor DELLAs (Zentella et al., 2016). O-GlcNAcylation of TaGRP2 and DELLA affects their interaction with other proteins (Xiao et al., 2014; Zentella et al., 2016), but other effects typically associated with this modification in mammals, including stability, have yet to be discovered in plants.
The Arabidopsis genome encodes two putative OGTs, SPINDLY (SPY) and SECRET AGENT (SEC). The OGT activity of SEC has been demonstrated (Kim et al., 2011), and a recent study by Zentella et al. (2016) showed that SEC interacts with and O-GlcNAcylates DELLAs, reduces their activity, and promotes GA responses. SPY, similar to SEC, contains multiple tetratricopeptide repeats at the N terminus, which bind substrate proteins, and a putative OGT catalytic domain at the C terminus (Silverstone et al., 2007). Overexpressing SPY’s N terminus without the catalytic OGT domain creates a dominant-negative effect (i.e. a spy-like phenotype; Izhaki et al., 2001; Tseng et al., 2001). It was proposed that the highly expressed tetratricopeptide repeat domain competes with the endogenous intact, but less abundant, SPY for interaction with substrate proteins. The catalytic OGT domain of SPY comprises CD I and CD II, two conserved regions that form a UDP-GlcNAc-binding pocket that catalyzes the transfer of GlcNAc monosaccharide to the substrate proteins in an O-linkage (Roos and Hanover, 2000). Despite the similarity in protein structure, the OGT activity of SPY was never demonstrated; thus, it is still not clear if SPY is a true OGT.
SPY has been implicated in various aspects of plant growth and development, including hormone responses (Olszewski et al., 2010). SPY is a negative regulator of GA signaling (Jacobsen and Olszewski, 1993). Indirect evidence suggested that SPY activates the DELLA proteins, thereby suppressing GA responses (Olszewski et al., 2010). Zentella et al. (2016) showed that SPY interacts with DELLAs, but they did not find evidence for DELLA O-GlcNAcylation by SPY. Thus, the mechanism by which SPY suppresses GA responses is still not clear.
We showed previously that SPY promotes cytokinin (CK) responses in developing leaves and flowers (Greenboim-Wainberg et al., 2005; Maymon et al., 2009). Two spy alleles showing severe (spy-4) and mild (spy-3) GA-associated phenotypes (Filardo and Swain, 2003) exhibited similar resistance to CK, suggesting that SPY enhances CK responses and inhibits GA signaling through distinct pathways (Greenboim-Wainberg et al., 2005). The CK signaling pathway starts with binding of CK to His kinase (HK, or AHK in Arabidopsis) receptors, which are then autophosphorylated. The phosphate group is then transferred by His phosphotransfer proteins (Hpt, or AHP in Arabidopsis) to the nucleus, where it phosphorylates and activates a set of transcriptional regulators known as type B response regulators (RRs, or ARR in Arabidopsis). The phosphorylated type B RRs promote the transcription of various CK-regulated genes, including type A RRs, which, in turn, suppress CK responses (Müller and Sheen, 2007; Hwang et al., 2012). We found that SPY interacts with and activates the class I TCP transcription factors, TCP14 and TCP15, to promote CK responses (Steiner et al., 2012). Since both spy and tcp14 tcp15 loss-of-function mutants suppressed various CK responses in leaves and flowers, including the expression of type A ARR genes, we speculated that SPY and TCPs either promote CK accumulation or increase the activity of components in the CK phosphorelay cascade (Greenboim-Wainberg et al., 2005; Steiner et al., 2012).
TCPs belong to the family of basic helix-loop-helix-type transcription factors. The Arabidopsis genome encodes 24 predicted TCP proteins, 13 of which are grouped as class I and 11 as class II (Martín-Trillo and Cubas, 2010). Class I TCPs have been suggested to promote, and class II to restrict, cell proliferation (Nath et al., 2003; Li et al., 2005, 2012; Efroni et al., 2008). Most class I TCP single mutants have mild or no phenotypes, probably because of genetic redundancy (Martín-Trillo and Cubas, 2010). However, several studies showed a role for these proteins in cell proliferation and organ development. TCP20 binds to the promoters of CYCLIN B1;1 and promotes cell division (Li et al., 2005). TCP15 suppresses endoreduplication by regulating the expression of cell cycle genes (Li et al., 2012). Kieffer et al. (2011) demonstrated the redundant activity of TCP14 and TCP15 in the regulation of cell proliferation during leaf development. Davière et al. (2014) showed that TCP8, TCP14, TCP15, and TCP22 interact with DELLA proteins and suggested that DELLAs inhibit their promoting effect on cell division and stem elongation. Recently, Lucero et al. (2015) showed that TCP15 affects gynoecium development and suggested its role in a feedback loop, regulating the balance between auxin and CK.
Here, we show that SPY activity is required for TCP14 accumulation. Mutation in the putative OGT catalytic domain of SPY promoted TCP14 proteolysis by the 26S proteasome and suppressed CK signaling.
RESULTS
The OGT Catalytic Domain of SPY Is Required for TCP14 Activity
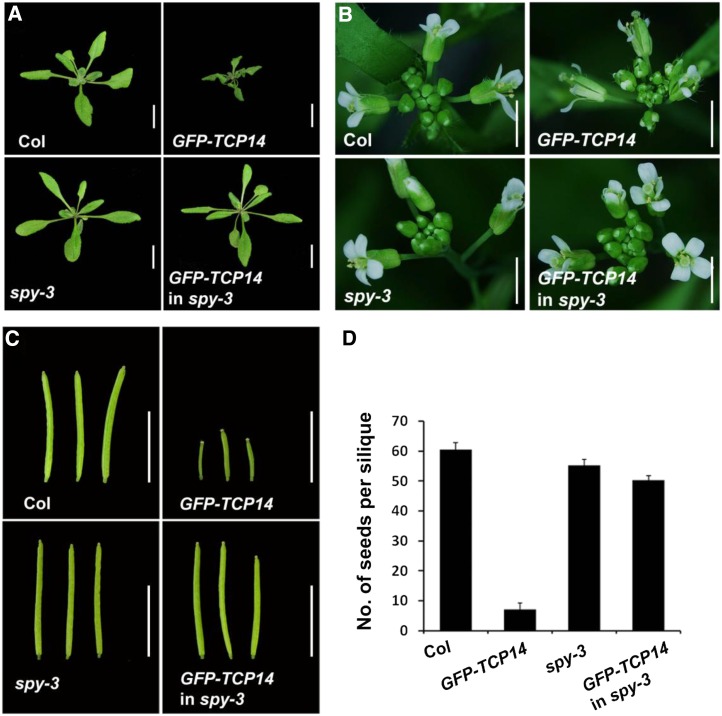
We previously showed that SPY physically and functionally interacts with TCP14 and TCP15 (Steiner et al., 2012). To understand the mechanism by which SPY promotes TCP14/15 activity, we tested the role of SPY’s putative OGT catalytic domain in TCP14 activity. To this end, we overexpressed TCP14 in spy-3, which is a weak allele of SPY, caused by a single amino acid substitution (Gly to Ser, at position 593 at the CD I motif) in the putative OGT catalytic domain (Silverstone et al., 2007). A similar amino acid substitution in the corresponding human OGT completely abolished its OGT catalytic activity (Lazarus et al., 2005). We first expressed GFP-TCP14 under the regulation of the AS1 promoter (Steiner et al., 2012) in wild-type Columbia (Col; pAS1:GFP-TCP14). pAS1:GFP-TCP14 plants had small dark leaves, short narrow petals, an increased number of trichomes on sepals, and small siliques (Fig. 1, A–C). In addition, seed number in pAS1:GFP-TCP14 pods was strongly reduced (Fig. 1D), resulting from the long carpels protruding from the stigma above the stamens (Fig. 1B), which partially prevented self-pollination. Hand pollination of pAS1:GFP-TCP14 wild-type flowers restored normal silique development (Supplemental Fig. S1). We then introgressed the transgene (pAS1:GFP-TCP14) into spy-3 plants. spy-3 strongly suppressed all TCP14 overexpression phenotypes (Fig. 1), suggesting that SPY’s OGT domain is essential for TCP14 activity. GFP-TCP14 mRNA levels in the transgenic wild type and spy-3 were found to be similar (Fig. 2C), suggesting that spy-3 affects TCP14 at the posttranscriptional level.
Figure 1.
spy-3 suppresses TCP14 activity. Phenotypic characterization of wild-type Col and spy-3 as well as transgenic Col (GFP-TCP14) and spy-3 (GFP-TCP14 in spy-3) overexpressing GFP-TCP14 under the regulation of the AS1 promoter. A, Three-week-old plants grown under long-day conditions. B, Inflorescences. C, Siliques. Bars = 1 cm. D, Average seed number per silique, determined from 10 pods, ± se.
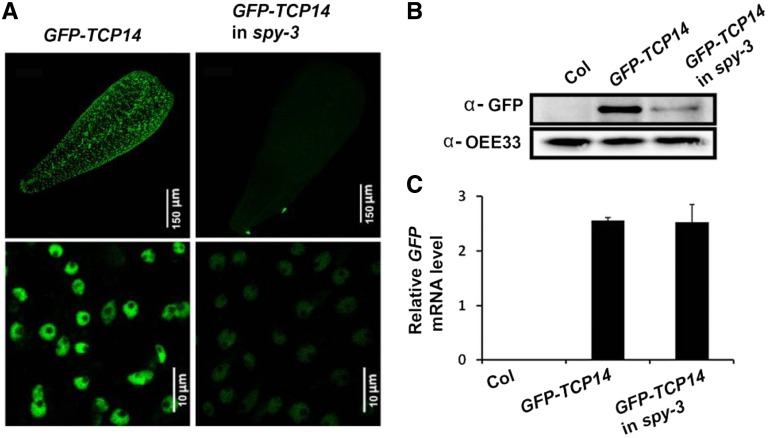
Figure 2.
The SPY OGT domain is required for TCP14 protein accumulation. A, Confocal microscopy images of transgenic pAS1:GFP-TCP14 wild-type Col (GFP-TCP14) and spy-3 (GFP-TCP14 in spy-3) petals. Bottom images show petal nuclei. B, Western-blot analysis of proteins extracted from control nontransgenic Col as well as transgenic pAS1:GFP-TCP14 Col and spy-3 inflorescences. GFP-TCP14 was detected using anti-GFP antibody. OEE33 served as a control to ensure equal loading (Lindahl et al., 1996). C, Quantitative reverse transcription (qRT)-PCR analysis of GFP expression in inflorescences of nontransgenic Col as well as transgenic pAS1:GFP-TCP14 Col and spy-3. Values are averages of three biological replicates ± se.
We then examined whether SPY also affects class II TCPs. To this end, transgenic plants expressing rTCP4-GFP under the control of the BLS promoter (Efroni et al., 2013) were crossed with spy-3 (for introgression of the transgene), and the TCP4 overexpression phenotype in wild-type Col versus spy-3 backgrounds was compared. TCP4 overexpression affecting leaf form was observed in both the wild-type and spy-3 backgrounds (Supplemental Fig. S2), suggesting that SPY has no effect on TCP4 activity.
SPY Affects TCP14 Stability
We next studied how SPY promotes TCP14 activity. O-GlcNAc modifications in mammalian cells regulate the posttranslational fate of target proteins, including cellular localization and stability (Roos and Hanover, 2000; Wells et al., 2001; Hanover et al., 2003; Butkinaree et al., 2010). We examined whether the OGT domain of SPY is required for the regulation of TCP14 localization and/or accumulation. To this end, we imaged the homozygous transgenic pAS1:GFP-TCP14 wild-type and spy-3 plants described above, using confocal microscopy. In both backgrounds, the GFP signal was found in nuclei, but it was much stronger in wild-type than in spy-3 plants (Fig. 2A). These findings suggest that TCP14 accumulation, but not localization, is affected by SPY. To confirm these results, proteins were extracted from young inflorescences of nontransgenic and transgenic wild-type and spy-3 plants expressing GFP-TCP14 to determine the level of the recombinant protein using an anti-GFP antibody. The GFP-TCP14 level was much higher in transgenic wild-type than spy-3 inflorescences (Fig. 2B). Since GFP-TCP14 mRNA levels were similar in transgenic wild-type and spy-3 inflorescences (Fig. 2C), it can be concluded that spy-3 plays a posttranscriptional role in GFP-TCP14 regulation, likely on protein stability.
To rule out the possibility that spy-3 affects GFP and not TCP14 accumulation/stability, we expressed GFP-AHP2 under the control of the 35S promoter in the wild type and spy-3 (35S:GFP-AHP2 was transformed into wild-type Col and then introgressed into spy-3) and analyzed GFP-AHP2 levels. Western-blot analysis using anti-GFP antibody showed similar levels of the detected protein in spy-3 and the wild type (Supplemental Fig. S3), suggesting that SPY specifically affects TCP14 and not GFP accumulation/stability.
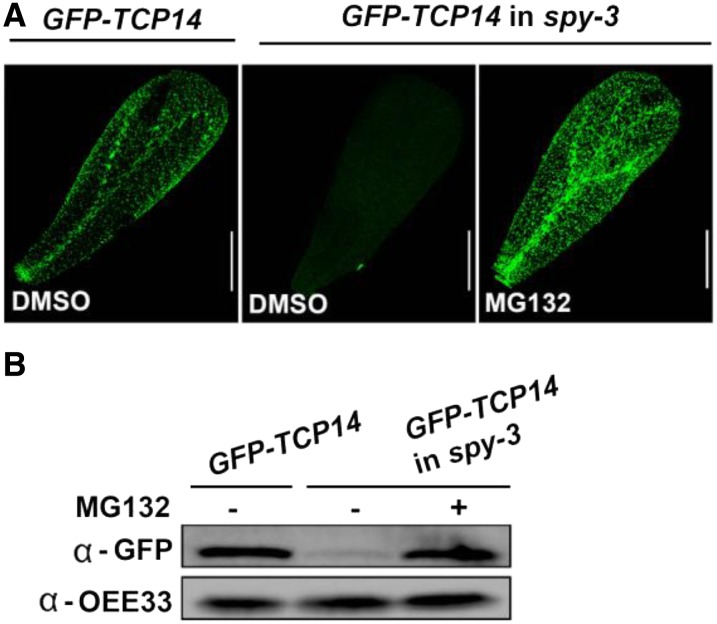
We further investigated whether TCP14 is biodegraded by the 26S proteasome in spy-3. Young transgenic spy-3 flowers were immersed, for 12 h, in a solution containing 50 µm of the 26S proteasome inhibitor MG132 before GFP-TCP14 accumulation was analyzed by confocal microscopy. MG132 treatment promoted the accumulation of GFP-TCP14 in spy-3 petals (Fig. 3A). Immunoblot analysis of proteins extracted from young inflorescences treated as above confirmed the effect of MG132 (Fig. 3B). These results suggest that SPY stabilizes TCP14 and prevents its proteolysis by the 26S proteasome.
Figure 3.
TCP14 is destroyed in spy-3 by the 26S proteasome. A, Confocal analysis of GFP in transgenic pAS1:GFP-TCP14 wild-type Col (GFP-TCP14) and spy-3 (GFP-TCP14 in spy-3) petals treated with 50 µm MG132 or dimethyl sulfoxide (DMSO). Bars = 200 µm. B, Western-blot analysis of proteins extracted from young inflorescences of transgenic pAS1:GFP-TCP14 wild-type Col and spy-3 treated with 50 µm MG132 (+) or dimethyl sulfoxide (−). The recombinant GFP-TCP14 protein was detected using an anti-GFP antibody. OEE33 was used as a loading control.
Since TCP14 activity is promoted by CK (Steiner et al., 2012; Lucero et al., 2015), we examined whether CK also affects TCP14 accumulation. Confocal microscopy analysis of CK-treated pAS1:GFP-TCP14 wild-type petals revealed that the hormone has no effect on GFP-TCP14 level (Supplemental Fig. S4).
To determine whether TCP14 proteolysis is mediated by the SKP, CULLIN (CUL), F-box (SCF)-containing E3 ubiquitin ligase complex, we tested the effect of a mutation in CUL1 on TCP14 stability in spy-3. We introgressed both pAS1:GFP-TCP14 and spy-3 into the weak cul1-6 allele (Moon et al., 2007) background and generated homozygous spy-3 cul1-6-expressing GFP-TCP14 plants. TCP14 overexpression suppressed silique and petal development in the wild type, an effect that was reversed in spy-3, which presented normal siliques and petals (Fig. 4, A and B). pAS1:GFP-TCP14 strongly suppressed silique and petal development in the spy-3 cul1-6 homozygous mutant background, suggesting that TCP14 is active in the cul1-6 spy-3 background. Confocal microscopy analysis showed that GFP-TCP14 accumulation was restored in the cul1-6 spy-3 background (Fig. 4C). These results suggest that TCP14 proteolysis in spy-3 is mediated by the SCF E3 ubiquitin ligase complex.
Figure 4.
cul1-6 restores TCP14 activity and stability in a spy-3 background. A, Mature inflorescences of wild-type Col, spy-3, and cul1-6 and of transgenic pAS1:GFP-TCP14 wild-type Col (GFP-TCP14), spy-3 (GFP-TCP14 in spy-3), and spy-3 cul1-6 (GFP-TCP14 in spy-3 cul1-6). Insets show a representative silique of each plant. Bars = 1 cm. B, Flowers of the different lines. Bars = 1 mm. C, Confocal analysis of GFP in petals of transgenic pAS1:GFP-TCP14 wild-type Col, spy-3, and spy-3 cul1-6.
The Catalytic OGT Domain of SPY Is Required for TCP14-Dependent Enhancement of CK Responses
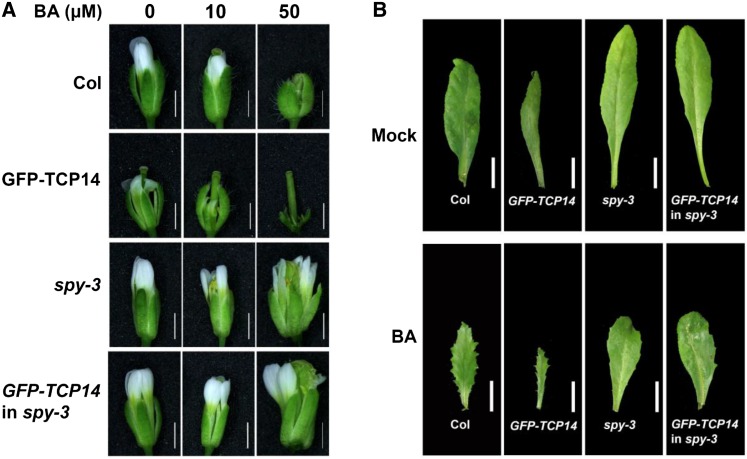
We previously showed that TCP14 and TCP15 promote CK responses (Steiner et al., 2012). Here, we investigated whether the catalytic OGT domain of SPY is required for the promotion of CK responses by TCP14. Seedlings of wild-type Col, pAS1:GFP-TCP14, spy-3, and pAS1:GFP-TCP14 in spy-3 were treated with different concentrations of N6-benzyladenine (BA) twice per week for 1 month to determine the effect of CK on flower and leaf morphology. Treatment with 10 µm BA reduced petal size in wild-type Col and had an even stronger effect in TCP14-overexpressing flowers (Fig. 5A). spy-3 completely blocked this effect in nontransgenic and transgenic pAS1:GFP-TCP14 flowers. Treatment with a higher CK concentration (50 µm BA) strongly suppressed sepal, petal, stamen, and carpel growth in wild-type Col. The effect was stronger in TCP14-overexpressing flowers, except for the carpels, which were hardly affected by the CK treatment. All of these CK effects in the flowers were strongly suppressed by spy-3 (Fig. 5A). BA treatment (50 µm) also affected leaf morphology, as manifested by smaller and serrated wild-type leaves and an even more exaggerated phenotype in pAS1:GFP-TCP14 (Fig. 5B). Again, spy-3 suppressed these CK effects. Taken together, these observations suggest that TCP14 activity in leaves and flowers promotes CK responses (except for the carpels) in a SPY-dependent manner and that the SPY catalytic OGT domain is required for this effect.
Figure 5.
spy-3 suppresses TCP14-stimulated CK responses. A, Flowers of wild-type Col, pAS1:GFP-TCP14 (GFP-TCP14), spy-3, and pAS1:GFP-TCP14 in spy-3 (GFP-TCP14 in spy-3) following treatment with different concentrations of BA (0, 10, and 50 µm). Bars = 1 mm. B, Rosette leaves (leaf 5) of wild-type Col, pAS1:GFP-TCP14, spy-3, and pAS1:GFP-TCP14 in spy-3 following treatment of young seedlings with two true leaves with 50 µm BA or water (Mock) twice per week for 1 month. Bars = 1 cm.
TCP14 and TCP15 Promote Primary CK Responses
To understand how TCP14 and TCP15 promote CK responses, we first analyzed CK levels in flowers of wild-type Col and pAS1:GFP-TCP14 plants. In Arabidopsis, the major active CKs are N6-(Δ2-isopentenyl)adenine and trans-zeatin (Sakakibara, 2006; Frébort et al., 2011). Our analysis showed that the levels of these two active CKs, as well as those of cis-zeatin and dihydrozeatin, were reduced significantly in TCP14-overexpressing inflorescences (Table I). In contrast, the levels of several inactive CKs were increased in the transgenic plants (Supplemental Table S1). These results suggest that TCP14 does not promote CK responses by increasing CK levels but seemingly by promoting hormone signaling. It is likely that the reduced levels of active CK in TCP14-overexpressing flowers result from a negative feedback loop initiated by the increased CK signaling (Kieber and Schaller, 2014).
Table I. Effect of TCP14 overexpression on the accumulation of free-base CKs.
Mass spectrometric measurements were made of free-base CKs in Col and pAS1:GFP-TCP14 (GFP-TCP14) inflorescences. iP, N6-(Δ2-isopentenyl)adenine; tZ, trans-zeatin; cZ, cis-zeatin; DHZ, dihydrozeatin. Values are means of three biological replicates ± se. ND, Not detected
TCP14 and TCP15 may affect CK activity by promoting early CK signaling events or by inducing the transcription of genes downstream to the CK phosphorelay cascade. To distinguish between these possibilities, we analyzed the effect of TCP15 activity on the synthetic CK-responsive promoter-reporter TWO-COMPONENT-OUTPUT SENSOR VERSION2 (TCSv2). This CK reporter is a variant of TCSn (Zürcher et al., 2013) that harbors the concatemerized type B ARR-binding motifs in alternating head-to-head and tail-to-tail orientations and a minimal 35S promoter sequence and, thus, reflects the activity of the CK phosphorelay cascade. We fused TCSv2 to three tandem repeats of the nuclear yellow fluorescent protein (YFP), VENUS (Heisler et al., 2005), to generate TCSv2:3xVENUS-N7, which we then transformed into Landsberg erecta (Ler) plants. Transgenic lines showing moderate YFP signal in the shoot apical meristem were selected for crossing with TCP15-overexpressing plants (pAS1 > >TCP15; Steiner et al., 2012) to generate pAS1 > >TCP15 TCSv2:3xVENUS-N7 plants. One-month-old transgenic TCSv2:3xVENUS-N7 wild-type and pAS1 > >TCP15 plants were sprayed with different CK (BA) concentrations, and 24 h later, TCS activity (YFP signal) was analyzed in young inflorescences by confocal microscopy. TCS activity was promoted in pAS1 > >TCP15 inflorescences following treatment with 1 µm BA and was further augmented in a dose-dependent manner (Fig. 6). In the wild-type background, TCS activity was only visually detectable after treatment with 10 µm BA, and although the signal was enhanced with higher BA concentration, it was much lower than that detected in TCP15-overexpressing inflorescences. These results suggest that TCP15 overexpression increases the sensitivity to CK and that TCP14 and TCP15 promote the early stages of the CK signaling pathway.
Figure 6.
Overexpression of TCP15 increases sensitivity to CK. Confocal microscopy images show TCSv2:3xVENUS-N7 in transgenic wild-type (WT) Ler and TCP15-overexpressing (pAS1 > >TCP15) plants. One-month-old plants were sprayed with different concentrations of BA, and after 24 h, the YFP signal was detected in young inflorescences.
TCSv2 harbors the concatemerized type B ARR-binding motifs (Müller and Sheen, 2008; Zürcher et al., 2013), implying that the promoting effect of TCP15 on CK-induced pTCS activity is mediated by type B ARRs. Since TCPs are transcription factors, at least three scenarios for the interaction between TCP14/15 and type B ARRs are possible: (1) TCPs promote type B ARR gene transcription to promote CK responses; (2) TCP14 and TCP15 interact with type B ARRs and act as transcription coactivators to promote CK responses; and (3) TCPs indirectly promote type B ARR activity. The type B ARR family contains 11 proteins, among which ARR1, ARR10, and ARR12 play central yet redundant roles in CK signal transduction (Mason et al., 2005; Ishida et al., 2008). Expression analysis of ARR1, ARR10, ARR12, and ARR14 in pAS1 > >TCP14 (Steiner et al., 2012) and pAS1 > >TCP15 flowers suggests that both TCPs have no effect on type B ARR transcription (Supplemental Fig. S5). We also performed a yeast two-hybrid assay to test for a possible interaction between TCP14 and ARR1, ARR10, and ARR12. SPY and AHP2 served as positive controls for the interaction with TCP14 and with type B ARRs, respectively (Dortay et al., 2006; Steiner et al., 2012). Indeed, while TCP14 interacted with SPY and all tested type B ARRs interacted with AHP2, no interaction between TCP14 and any tested type B ARRs was observed (Supplemental Fig. S6). These results imply that TCP14 and TCP15 indirectly promote type B ARR activity. To examine if TCP14 and TCP15 affect the transcription of components upstream of type B ARRs in the CK phosphorelay cascade, we analyzed the expression of the CK receptor kinase genes AHK2, AHK3, and AHK4 in wild-type (Col and Ler), TCP14-overexpressing (Col), and TCP15-overexpressing (Ler) inflorescences. qRT-PCR analyses indicated that both TCPs have no effect on the expression of these AHK genes (Supplemental Fig. S7).
DISCUSSION
Previously, we showed that SPY interacts physically and functionally with TCP14 and TCP15 and that the null spy-4 allele suppresses the activity of both TCPs (Steiner et al., 2012). Due to the significant similarity between SPY and animal OGTs (Olszewski et al., 2010), we speculated that SPY O-GlcNAcylates TCP14 and TCP15 to promote their activity (Steiner et al., 2012). This hypothesis was tested here using the weak spy-3 allele, which has a single amino acid substitution (Gly-593 to Ser) in the putative OGT catalytic domain (Silverstone et al., 2007). This specific amino acid (Gly-593) is essential for the catalytic activity of OGTs, as mutation at the corresponding residue of the human OGT completely abolished its OGT activity (Lazarus et al., 2005). In spy-3, TCP14 activity was strongly suppressed, suggesting that the putative OGT catalytic domain of SPY is essential for TCP14 activity. Since the OGT domain is not required for the interaction of SPY with TCP14 or TCP15 (Steiner et al., 2012), it is possible that it modifies them, perhaps with GlcNAc.
O-GlcNAcylation in mammalian cells affects protein stability. For example, O-GlcNAcylation of the tumor suppressor p53 protects the protein from proteolysis by the proteasome (Yang et al., 2006). We found that a mutation in SPY’s OGT catalytic domain dramatically reduces the level of the chimeric GFP-TCP14 protein. Since the accumulation of GFP-TCP14 in spy-3 was recovered by mutation in CUL1 and by the 26S proteasome inhibitor MG132, we suggest that in spy, TCP14 is ubiquitinated by an SCF E3 ubiquitin ligase and consequently destroyed in the proteasome. The results of Peng et al. (2015), showing that TCP14 and TCP15 interact with the ubiquitin receptors DA1, DAR1, and DAR2 for proteolysis, support our suggestion.
We previously showed that SPY promotes CK responses in leaves and flowers (Greenboim-Wainberg et al., 2005) and suggested that this effect is mediated by TCP14 and TCP15 (Steiner et al., 2012). Here, we show that the effect of TCP14 on CK responses is dependent on SPY’s catalytic OGT domain, suggesting that the stabilization of TCP14 by SPY activity is required for the increased CK responses. We also found that the accumulated TCPs act by promoting sensitivity to CK and not by increasing CK content. The level of the central active CK, N6-(Δ2-isopentenyl)adenine, was reduced in TCP14-overexpressing flowers. The reduced levels of active CKs might be the result of a negative feedback regulation, initiated by increased CK signaling (Kieber and Schaller, 2014). High CK activity induces the expression of CYTOKININ OXIDASE/DEHYDROGENASE genes for CK degradation (Brugière et al., 2003; Werner et al., 2003, 2006) and rapidly converts free active CKs to inactive derivatives (Singh et al., 1988; Moffatt et al., 1991; Yonekura-Sakakibara et al., 2004). Indeed, we found higher levels of inactive CKs in TCP14-overexpressing flowers. Altogether, these results suggest that TCP14 overexpression increases CK signaling and not accumulation. This conclusion is supported by our previous observation that leaves and flowers of the double mutant tcp14 tcp15 are less sensitive to exogenous CK (Steiner et al., 2012). We cannot exclude, however, the possibility that TCP14 and TCP15 promote transient increases in CK level at a very early stage of flower development.
TCP14 and TCP15 may promote sensitivity to CK by affecting components of the early phases of CK signaling or by promoting the expression of downstream genes. Since TCP15 overexpression increases the sensitivity of the synthetic CK-induced promoter pTCS to CK, we propose that TCP14/15 affect element(s) in the early stages of CK signaling. pTCS harbors the concatemerized type B ARR-binding motifs (Müller and Sheen, 2008; Zürcher et al., 2013) but not a TCP-binding element, suggesting that TCP15 promotes TCS activity via type B ARRs. As TCP14 and TCP15 did not promote type B ARR gene expression and did not interact with type B ARR proteins in yeast (Saccharomyces cerevisiae), the TCP14/15-driven promotion of type B ARR activity is seemingly indirect.
While it is possible that TCP14 and TCP15 promote the activity of elements upstream of type B ARRs in the CK phosphorelay cascade, we did not find an effect on the expression of the CK receptor genes AHK2, AHK3, and AHK4. It is also possible that TCP14/15 promote type B ARR activity indirectly via the interaction with other signaling pathways. For example, a previous study showed that TCP15 suppresses auxin activity (Lucero et al., 2015). Since auxin interacts negatively with CK (Müller and Sheen, 2008), the inhibition of auxin activity may promote the activity of the CK phosphorelay cascade to promote type B ARR activity. The possible role of type B ARRs in mediating TCP14/15 activity can explain the requirement of CK for TCP14 activity (Steiner et al., 2012); without CK, type B ARRs are not active (Sakai et al., 2001; Müller and Sheen, 2007).
The results of this and other studies suggest that SPY regulates GA responses and CK activity by different mechanisms. While spy-3 has only a mild effect on GA responses (Jacobsen and Olszewski, 1993), it has a strong effect, similar to that of the null spy-4 allele, on CK responses (Greenboim-Wainberg et al., 2005). Thus, while SPY may regulate TCP14/15 and CK responses via glycosylation, the mechanism by which it affects DELLA activity and GA responses is likely to be more complicated and to involve activities other than glycosyltransferase.
A critical question left open is whether SPY indeed O-GlcNAcylates TCP14/15 for stabilization. Since all efforts to demonstrate O-GlcNAcylation by SPY have failed so far (Zentella et al., 2016), it is possible that the catalytic OGT domain of SPY acquired novel glycosyltransferase activity that affects protein fate similar to O-GlcNAcylation. However, we cannot exclude the possibility that steric interference by the catalytic domain of SPY protects TCP14/15 from proteolysis
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) spy-3 mutant plants used in this study, as well as the transgenic pAS1:GFP-TCP14, 35S:GFP-AHP2, and pBLS:rTCP4-GFP plants, were all in the Col background. Transgenic TCSv2:3xVENUS-N7 and pAS1 > >TCP15 TCSv2:3xVENUS-N7 were in the Ler background. Arabidopsis seeds were sterilized, cold treated, and germinated on sterile Murashige and Skoog medium or in pots. The plants were grown in a growth room under controlled temperature (22°C) and long-day (16-h-light/8-h-dark) or short-day (8-h-light/16-h-dark) conditions.
Molecular Cloning/Constructs
The TCP14-encoding region was fused to the 3′ end of the enhanced GFP-encoding sequence at a KpnI site. The GFP-TCP14 fusion product was inserted into a pBJ36 plasmid downstream of the AS1 promoter (Sarojam et al., 2010), between XhoI and XbaI sites, to create pAS1:GFP-TCP14. The construct was subcloned into the pMLBART binary vector, in a NotI site, and was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. The construct was transformed into Col plants by floral dipping (Clough and Bent, 1998), and BASTA-resistant transformants were selected. The DNA sequence of TCSv2 was synthesized with flanking NsiI and BamHI restriction sites. The synthetic promoter was then cloned adjacent to 3xVENUS-N7 in the pBJ36 vector (Heisler et al., 2005). The construct was subcloned into the pGREEN binary vector and introduced into the Arabidopsis Ler background by floral dipping. Kanamycin-resistant transformants were selected.
Immunoblot Analysis
Total protein extracts were obtained by grinding 100 mg of inflorescence tissue in 200 µL of protein extraction buffer (0.2 m Tris-HCl, pH 6.8, 3 m urea, 1% [w/v] glycerol, 8% [w/v] SDS, 0.5 mm dithiothreitol, protease inhibitor cocktail [Sigma-Aldrich], and 5% [w/v] β-mercaptoethanol). Samples were vortexed, boiled for 5 min, and centrifuged at 13,000g for 15 min, and the supernatant was subjected to SDS-PAGE. For detection of the GFP-fused proteins, the samples were separated on a 12% polyacrylamide gel in Tris-Gly buffer and electroblotted onto a polyvinylidene difluoride membrane. Blots were reacted with a commercially available anti-GFP antibody (Covance; catalog no. MMS-118R), diluted 1:5,000, and an anti-mouse horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch), diluted 1:5,000. All blots were developed using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
RNA Purification and qRT-PCR Analyses
Total RNA was extracted from young inflorescences, as described by Shleizer-Burko et al. (2011). qRT-PCR analysis (Table S2) was performed using the Absolute Blue qPCR SYBR Green ROX Mix (AB-4162/B) kit (Thermo Fisher Scientific). Reactions were performed in a Rotor-Gene 6000 cycler (Corbett Research). A standard curve was obtained for each gene, using dilutions of a complementary DNA sample. Each gene was quantified using Corbett Research Rotor-Gene software. At least three independent technical repeats were performed for each complementary DNA sample. The relative expression of each sample was calculated by dividing the expression level of the analyzed gene by that of TUBULIN BETA CHAIN3.
CK Analysis
CKs were isolated and purified as outlined by Novák et al. (2003), with some modifications. Frozen Arabidopsis inflorescences (30 mg) were homogenized using a vibration mill (MM 301) at a frequency of 30 Hz for 3 min (3-mm zirconium oxide beads; Retsch), with 1 mL of ice-cold Bieleski solution (methanol:chloroform:formic acid:water, 12:5:1:2) as the extraction solution. The CKs were then extracted overnight at 4°C using a benchtop laboratory rotator (Stuart SB3; Bibby Scientific), after adding heavy-labeled internal standards (OlChemIm). The samples were further fractionated by two steps of solid-phase extraction. Briefly, passing the extracts, in sequence, through a cation (SCX cartridge) and an anion (DEAE-Sephadex combined with a C18 cartridge) exchanger yielded fraction 1, containing the CK bases, ribosides, and glucosides, and fraction 2, containing the riboside-5′-phosphates. Both fractions were further purified by immunoaffinity extraction based on generic monoclonal anti-CK antibodies, but fraction 2 was first treated with alkaline phosphatase. In fraction 1, the O-glucosides did not bind to the affinity sorbent. The effluent was thus treated with β-glucosidase to yield the O-glucoside fraction (later analyzed as an aglycone). Samples were then redissolved in the mobile phase and analyzed by ultra-HPLC-tandem mass spectrometry (Acquity UPLCTM System, Xevo TQ; Waters MS Technologies) according to Novák et al. (2008). CKs were ionized by electrospray in positive mode and detected using multiple reaction monitoring. Masslynx 4.1 software (Waters) was used to analyze the data.
BA Treatment
Arabidopsis seedlings with two true leaves were sprayed with different concentrations of BA (Sigma-Aldrich) twice per week for approximately 4 weeks. For the TCSv2 experiments, 1-month-old plants were sprayed once with different concentrations of BA.
MG132 Treatment
Young inflorescences were immersed in solution containing 50 µm MG132 (Sigma-Aldrich) and 0.01% Tween 20, for 12 h in the dark, at room temperature. During the first hour, the samples were vacuumed, and the vacuum was released every 15 min.
The Cytokinin Reporter TCSv2
TCSv2 is a variant of TCSn, with alternating head-to-head and tail-to-tail orientations of type B ARR-binding sites compared with the tandem tail-to-tail and head-to-head orientation of sites in TCSn. TCSv2 mediates CK-dependent activation that is comparable to TCSn. Its sequence is 5′-CAAAGATTTTGCAAAATCTTTTAAAGGATTTTGAAAGATCTTTGCAAAGATCTTTATAAATCTTTTCAAAGATTTTTCAAGATCCGATTAAAGATTTTGCAAAATCTTTAGAGAGATCTTTCAAAATCCAACGCTAGTCAAAGATTTTGCAAAATCTTTTAAAGGATTTTGAAAGATCTTTGCAAAGATCTTTATAAATCTTTTCAAAGATTTTTCAAGATCCGATTAAAGATTTTGCAAAATCTTTAGAGAGATCTTTCAAAATCCAAC-3′.
Microscopy and Confocal Imaging
All imaging was done using a confocal laser scanning microscope (Leica TCS SP8; http://www.leica-microsystems.com/) with an HCX PL APO CS 20×/0.70 dry objective (for GFP in petals) or an HC PL FLUOTAR 10×/0.30 dry objective (for VENUS-YFP in flowers). GFP was excited with the 488-nm laser line, and the 505- to 525-nm filter was used for emission. VENUS-YFP was excited using the 514-nm laser line in conjunction with a 520- to 560-nm band-pass filter.
Yeast Two-Hybrid Interaction Assay
The coding regions of ARR1, ARR10, ARR12, and SPY were cloned into pACT2 plasmid. TCP14 and AHP2 were cloned into the pBD-GAL4 CAM plasmid. pACT2-ARR1, pACT2-ARR10, and pACT2-ARR12 were transformed individually into Saccharomyces cerevisiae strain Y190 containing pBD-GAL4-TCP14 or pBD-GAL4-AHP2. Yeast transformants were selected for the presence of plasmids on synthetic dextrose agar plates lacking Leu and Trp. Fusion of the GAL4 activating domain and ARR and translational fusion of the TCP14-GAL4 binding domain were confirmed using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid staining to monitor β-gal reporter gene expression levels. Individual clones were spotted onto synthetic dextrose agar plates lacking Leu and Trp. After a 2-d incubation at 30°C, plates were stained using the chloroform overlay 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid method. Empty pACT2 and pBD-GAL4 were used as negative controls, and pACT2- SPY and pBD-GAL4-AHP2 were used as positive controls.
Accession Numbers
The sequences of genes examined in this study can be found in The Arabidopsis Information Resource data library under the following accession numbers: SPY (AT3G11540), TCP14 (AT3G47620), TCP15 (AT1G69690), CUL1 (AT4G02570), TUB3 (AT5G62700), ARR1 (AT3G16857), ARR10 (AT4G31920), ARR12 (AT2G25180), ARR14 (AT2G01760), AHP2 (AT1G13330), AHK2 (AT5G35750), AHK3 (AT1G27320), and AHK4 (AT2G01830).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Hand pollination of pAS1:GFP-TCP14 flowers restored normal silique development.
Supplemental Figure S2. spy-3 does not suppress the TCP4 overexpression phenotype.
Supplemental Figure S3. spy-3 does not affect GFP-AHP2 accumulation.
Supplemental Figure S4. CK does not affect GFP-TCP14 stability.
Supplemental Figure S5. Overexpression of TCP14 or TCP15 has no effect on type B ARR expression.
Supplemental Figure S6. Type B ARRs do not interact with TCP14 in yeast.
Supplemental Figure S7. Overexpression of TCP14 or TCP15 has no effect on AHK gene expression.
Supplemental Table S1. Effect of TCP14 overexpression on inactive CK levels.
Supplemental Table S2. List of primers.
Supplementary Material
Acknowledgments
We thank Dr. Idan Efroni for valuable suggestions, which improved the article, and Dr. William M. Gray for providing the cul1-6 seeds and for valuable suggestions.
Glossary
- CK
cytokinin
- Col
Columbia
- BA
N6-benzyladenine
- Ler
Landsberg erecta
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the Israel Science Foundation (grant nos. 576–11 and 779/15 to D.W.) and by the National Program of Sustainability I, MEYS, Czech Republic (grant no. LO1204 to P.T. and D.T.).
References
- Brugière N, Jiao S, Hantke S, Zinselmeier C, Roessler JA, Niu X, Jones RJ, Habben JE (2003) Cytokinin oxidase gene expression in maize is localized to the vasculature, and is induced by cytokinins, abscisic acid, and abiotic stress. Plant Physiol 132: 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW (2010) O-Linked beta-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta 1800: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24: 1923–1928 [DOI] [PubMed] [Google Scholar]
- Dortay H, Mehnert N, Bürkle L, Schmülling T, Heyl A (2006) Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 273: 4631–4644 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, Hong JC, Eshed Y, Wagner D (2013) Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell 24: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo FF, Swain SM (2003) SPYing on GA signaling and plant development. J Plant Growth Regul 22: 163–175 [Google Scholar]
- Frébort I, Kowalska M, Hluska T, Frébortová J, Galuszka P (2011) Evolution of cytokinin biosynthesis and degradation. J Exp Bot 62: 2431–2452 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA. (2001) Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J 15: 1865–1876 [DOI] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC (2010) The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta 1800: 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Yu S, Lubas WB, Shin SH, Ragano-Caracciola M, Kochran J, Love DC (2003) Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys 409: 287–297 [DOI] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446: 1017–1022 [DOI] [PubMed] [Google Scholar]
- Heese-Peck A, Cole RN, Borkhsenious ON, Hart GW, Raikhel NV (1995) Plant nuclear pore complex proteins are modified by novel oligosaccharides with terminal N-acetylglucosamine. Plant Cell 7: 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese-Peck A, Raikhel NV (1998) A glycoprotein modified with terminal N-acetylglucosamine and localized at the nuclear rim shows sequence similarity to aldose-1-epimerases. Plant Cell 10: 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Izhaki A, Swain SM, Tseng TS, Borochov A, Olszewski NE, Weiss D (2001) The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J 28: 181–190 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2014) Cytokinins. The Arabidopsis Book 12: e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Udeshi ND, Balsbaugh JL, Shabanowitz J, Hunt DF, Olszewski NE (2011) O-GlcNAcylation of the Plum pox virus capsid protein catalyzed by SECRET AGENT: characterization of O-GlcNAc sites by electron transfer dissociation mass spectrometry. Amino Acids 40: 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus BD, Roos MD, Hanover JA (2005) Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase. J Biol Chem 280: 35537–35544 [DOI] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Li B, Dong AW (2012) The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol Plant 5: 270–280 [DOI] [PubMed] [Google Scholar]
- Lindahl M, Tabak S, Cseke L, Pichersky E, Andersson B, Adam Z (1996) Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J Biol Chem 271: 29329–29334 [DOI] [PubMed] [Google Scholar]
- Lucero LE, Uberti-Manassero NG, Arce AL, Colombatti F, Alemano SG, Gonzalez DH (2015) TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J 84: 267–282 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymon I, Greenboim-Wainberg Y, Sagiv S, Kieber JJ, Moshelion M, Olszewski N, Weiss D (2009) Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J 58: 979–988 [DOI] [PubMed] [Google Scholar]
- Moffatt B, Pethe C, Laloue M (1991) Metabolism of benzyladenine is impaired in a mutant of Arabidopsis thaliana lacking adenine phosphoribosyltransferase activity. Plant Physiol 95: 900–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhao Y, Dai X, Zhang W, Gray WM, Huq E, Estelle M (2007) A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol 143: 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J (2007) Advances in cytokinin signaling. Science 318: 68–69 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Novák O, Hauserová E, Amakorová P, Doležal K, Strnad M (2008) Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 2214–2224 [DOI] [PubMed] [Google Scholar]
- Novák O, Tarkowski P, Tarkowská D, Dolezal K, Lenobel R, Strnad M (2003) Quantitative analysis of cytokinins in plants by liquid chromatography-single-quadrupole mass spectrometry. Anal Chim Acta 480: 207–218 [Google Scholar]
- Olszewski NE, West CM, Sassi SO, Hartweck LM (2010) O-GlcNAc protein modification in plants: evolution and function. Biochim Biophys Acta 1800: 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Chen L, Lu Y, Wu Y, Dumenil J, Zhu Z, Bevan MW, Li Y (2015) The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27: 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos MD, Hanover JA (2000) Structure of O-linked GlcNAc transferase: mediator of glycan-dependent signaling. Biochem Biophys Res Commun 271: 275–280 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman JL (2010) Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22: 2113–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouppe D, Ghesquière B, Menschaert G, De Vos WH, Bourque S, Trooskens G, Proost P, Gevaert K, Van Damme EJM (2011) Interaction of the tobacco lectin with histone proteins. Plant Physiol 155: 1091–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shleizer-Burko S, Burko Y, Ben-Herzel O, Ori N (2011) Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138: 695–704 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Letham DS, Jameson PE, Zhang R, Parker CW, Bandenoch-Jones J, Noodén LD (1988) Cytokinin biochemistry in relation to leaf senescence. IV. Cytokinin metabolism in soybean explants. Plant Physiol 88: 788–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D (2012) The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Swain SM, Olszewski NE (2001) Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol 126: 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW (2001) Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291: 2376–2378 [DOI] [PubMed] [Google Scholar]
- Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T (2006) New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 8: 371–381 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, et al. (2014) O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun 5: 4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol 8: 1074–1083 [DOI] [PubMed] [Google Scholar]
- Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, et al. (2008) Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451: 964–969 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H (2004) Molecular characterization of cytokinin-responsive histidine kinases in maize: differential ligand preferences and response to cis-zeatin. Plant Physiol 134: 1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachara NE, Hart GW (2004) O-GlcNAc a mediator of cellular function: characterizing a family of O-GlcNAc binding proteins. Glycobiology 14: 1063 [Google Scholar]
- Zentella R, Hu J, Hsieh WP, Matsumoto PA, Dawdy A, Barnhill B, Oldenhof H, Hartweck LM, Maitra S, Thomas SG, et al. (2016) O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev 30: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B (2013) A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol 161: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.