Identification by mapping-by-sequencing of the causal mutation underlying a tomato cutin-deficient mutant unravels the central role of GPAT6 in fruit cuticle formation.
Abstract
The thick cuticle covering and embedding the epidermal cells of tomato (Solanum lycopersicum) fruit acts not only as a protective barrier against pathogens and water loss but also influences quality traits such as brightness and postharvest shelf-life. In a recent study, we screened a mutant collection of the miniature tomato cultivar Micro-Tom and isolated several glossy fruit mutants in which the abundance of cutin, the polyester component of the cuticle, was strongly reduced. We employed a newly developed mapping-by-sequencing strategy to identify the causal mutation underlying the cutin deficiency in a mutant thereafter named gpat6-a (for glycerol-3-phosphate acyltransferase6). To this end, a backcross population (BC1F2) segregating for the glossy trait was phenotyped. Individuals displaying either a wild-type or a glossy fruit trait were then pooled into bulked populations and submitted to whole-genome sequencing prior to mutation frequency analysis. This revealed that the causal point mutation in the gpat6-a mutant introduces a charged amino acid adjacent to the active site of a GPAT6 enzyme. We further showed that this mutation completely abolished the GPAT activity of the recombinant protein. The gpat6-a mutant showed perturbed pollen formation but, unlike a gpat6 mutant of Arabidopsis (Arabidopsis thaliana), was not male sterile. The most striking phenotype was observed in the mutant fruit, where cuticle thickness, composition, and properties were altered. RNA sequencing analysis highlighted the main processes and pathways that were affected by the mutation at the transcriptional level, which included those associated with lipid, secondary metabolite, and cell wall biosynthesis.
The cuticle, which serves as a barrier protecting aerial plant organs, is localized on the outer face of primary cell walls of epidermal cells, although in some cases it can extend through the apoplast of additional underlying cell layers. It is composed of cutin, a polyester of glycerol, hydroxy, and epoxy fatty acids, in addition to a range of waxes. In tomato (Solanum lycopersicum), as in many species, the main cutin monomers are C16-based fatty acids (C16 dihydroxy fatty acids, C16 ω-hydroxy fatty acids, and C16 dicarboxylic acids; Mintz-Oron et al., 2008; Isaacson et al., 2009; Girard et al., 2012; Petit et al., 2014). Following their synthesis in the plastids, long-chain fatty acids are transported to the endoplasmic reticulum, where they undergo a series of modifications, including activation to CoA thioesters by long-chain acyl-CoA synthetases (LACS), oxidation by cytochrome P450-dependent fatty acid oxidases (CYP), and esterification to glycerol-based acceptor by glycerol-3-phosphate acyltransferase (GPAT) enzymes to produce acyl glycerols (Li-Beisson et al., 2013). In Arabidopsis (Arabidopsis thaliana), the bifunctional acyltransferase and phosphatase activities of GPAT4 and GPAT6 mainly lead to the production of sn-2 monoacylglycerols (MAGs; Yang et al., 2010). In addition, a recent study from Yang et al. (2012) provided evidence that acyl transfer to glycerol, catalyzed by GPAT, takes place after the oxidation of acyl chains. Following their synthesis in the endoplasmic reticulum, cutin monomers are transported across the plasmalemma and polysaccharide cell wall to the hydrophobic cuticle layer. The monomers are then assembled into a network of linear and branched cutin polymers (Chatterjee et al., 2016; Philippe et al., 2016) by cutin synthases (Girard et al., 2012; Yeats et al., 2012b; Fich et al., 2016).
The cutin matrix is a polyester of polyhydroxy fatty acids and fatty alcohols with varying amounts of phenolic compounds (ferulic, p-coumaric, and cinnamic acids) depending on the species. Cutin may be bound to cell wall polysaccharides; however, the nature of this hypothetical linkage(s) and the coordination of the synthesis of both types of polymers during organ growth are poorly understood (Segado et al., 2016). The cutin matrix is filled with intracuticular waxes and also covered with a thin layer of epicuticular waxes (Nawrath, 2006; Buschhaus and Jetter, 2011). Collectively, the wax fraction is typically a complex mixture composed of derivatives of very-long-chain fatty acids, mainly alkanes and alcohols, but it may also include various secondary metabolites, such as the triterpenol compounds amyrins, flavonoids, and sterols (Bernard and Joubès, 2013). The synthesis of cuticle is tightly coordinated with developmental processes, such as the formation of epidermal cells during plant and fruit growth (Javelle et al., 2011; Lashbrooke et al., 2015). Environmental cues also may affect cuticle formation, as is the case with drought stress triggering changes in the cuticle via abscisic acid signaling (Kosma et al., 2009). The coordination of the various biosynthetic pathways required to build the cuticle and to impart its specific properties occurs at various levels (Hen-Avivi et al., 2014). For example, several transcription factors (TFs) that regulate cuticle formation have been identified. The first characterized example from Arabidopsis was WIN1/SHN1, a member of the AP2/ERF domain family (Aharoni et al., 2004; Broun et al., 2004). Since then, many TFs belonging to several families (AP2/ERF, DREB/CBF, MYB, HD-Zip IV, and WW domain proteins) have been identified in various plant species, including those that regulate cuticle formation in fleshy fruits (Borisjuk et al., 2014; Hen-Avivi et al., 2014). Further studies have revealed the identity of target genes of these TFs, most of which are genes involved in fatty acid, phenylpropanoid, and flavonoid metabolism (Adato et al., 2009; Lashbrooke et al., 2015).
Like many fleshy fruits, tomato fruits are covered by a thick astomatous cuticle that fulfills various functions, including preventing water loss and resisting pathogen infection. It also influences commercially important characteristics of tomato fruit, such as susceptibility to fruit cracking, visual appearance, and postharvest shelf-life (Martin and Rose, 2014). Since the tomato fruit cuticle is so thick and easy to isolate, it has emerged as a model for studying cuticle formation and properties. In recent years, new insights into the regulation, synthesis, assembly, structure, and properties of the cuticle have been obtained using natural (Nadakuduti et al., 2012) or artificially induced genetic variability available in cultivated tomato (Adato et al., 2009; Isaacson et al., 2009; Shi et al., 2013; Petit et al., 2014) and in related wild species (Hovav et al., 2007; Yeats et al., 2012a). However, such studies also have raised many new questions, and many aspects of tomato fruit cuticle formation remain poorly characterized.
To better understand fruit cuticle formation, we recently isolated several cuticle mutants from an ethyl methanesulfonate (EMS)-mutagenized mutant collection in the miniature cv Micro-Tom genetic background and further identified by map-based cloning a mutation in a cutin synthase gene that is responsible for a glossy fruit phenotype (Petit et al., 2014). However, crossing the mutant line with a genetically distant genotype, as required for map-based cloning, introduces a large genetic diversity and, consequently, large phenotypic variability. Since cuticle-related traits are under complex genetic control, the phenotypic analysis of segregating populations used for mapping such traits can be extremely challenging. However, given the recent generation of a high-quality reference tomato genome sequence (Tomato Genome Consortium, 2012), the sequencing of cv Micro-Tom (Kobayashi et al., 2014), and recent advances in deep-sequencing technologies, other strategies based on whole-genome sequencing are now available. Such strategies, called here mapping-by-sequencing, as described in a recent review (Schneeberger, 2014), have been used successfully in the model species Arabidopsis (Schneeberger et al., 2009) and rice (Oryza sativa; Abe et al., 2012), in which they allowed the identification of the causal mutations underlying remarkable mutant traits. The mapping-by-sequencing approach is based on the comparative analysis of the distribution along the genome of single-nucleotide polymorphism (SNP) frequencies in segregating plant bulked populations displaying either the mutant trait or the wild-type trait. The mutant and wild-type parental lines are first crossed in order to generate a segregating population, and since both parents share the same genetic background, with the exception of the EMS-induced polymorphisms, this approach facilitates the study of complex mutant traits.
Here, we demonstrate that the large genetic and phenotypic diversity generated by EMS mutagenesis of tomato can be assessed through the mapping-by-sequencing approach to rapidly identify the causal mutation underlying a specific phenotypic trait. Using this strategy, we established that a point mutation in a GPAT gene (SlGPAT6) is responsible for a previously identified glossy mutation that affects fruit brightness (Petit et al., 2014). Assays using wild-type and mutant recombinant SlGPAT6 proteins produced in yeast (Saccharomyces cerevisiae), together with various chemically synthesized oxidized C16 fatty acyl-CoAs, demonstrated that GPAT activity is abolished in the ΔSlGPAT6 variant. Functional characterization of the mutant further showed that the deficiency in SlGPAT6 activity affects pollen viability, as was reported to be the case in an analogous study of Arabidopsis (Li et al., 2012), but has no effect on fruit set. The most striking phenotype was observed in the mutant fruit, where cuticle thickness, composition, and properties were altered. Remarkably, mutation of this single structural gene, upstream in the cutin biosynthetic pathway, resulted in major changes in exocarp gene expression associated with not only lipid biosynthesis but also other cuticle-related pathways (phenolics and flavonoids) and those related to cell wall synthesis and modeling.
RESULTS
Identification of the Causal Mutation Underlying a Fruit Brightness Mutant via Mapping-by-Sequencing
In a previous study, we isolated from an EMS-mutagenized tomato mutant population (cv Micro-Tom) a set of fruit brightness mutants (named glossy mutants) with abnormal fruit cutin amounts/compositions (Petit et al., 2014). In this study, we focused on one of these glossy mutants (line P23F12), which exhibits enhanced fruit brightness and a reduced cutin load. Genetic mapping of the mutant traits (cuticle permeability and fruit brightness) highlighted a large genomic region (approximately 60 centimorgans) encompassing the centromeric region and comprising 488 annotated genes (Petit et al., 2014). To facilitate the identification of the mutated gene, we adopted a strategy involving whole-genome sequencing of tomato bulked pools of individuals from a segregating BC1F2 population, hereafter referred to as the mapping-by-sequencing approach (Schneeberger, 2014). A general overview of this approach is shown in Supplemental Figure S1. Briefly, once a recessive mutation controlling a phenotypic trait has been identified in a tomato mutant population (e.g. a glossy fruit trait), the mutant line is crossed with the wild-type (nonmutagenized) parental line in order to produce a BC1F2 population, which segregates for both the mutant trait (25% of the plants display the mutant phenotype and 75% of the plants display the wild-type phenotype) and the mutant alleles. Plants from the BC1F2 population showing either phenotype are then pooled to constitute bulks that are submitted to whole-genome sequencing. Sequencing reads are mapped onto the reference tomato genome (Tomato Genome Consortium, 2012), allelic variants are detected, and their frequency in both bulks is analyzed in order to detect the causal mutation. In the mutant bulk, because all BC1F2 individuals exhibiting the mutant phenotype are homozygous for the mutation, the allelic frequency (AF) of the causal mutation should, in principal, be 1. Frequencies of other allelic variants will be either 0.5 < AF < 1 for mutations in linkage disequilibrium with the causal mutation or AF < 0.5 for unrelated mutations. Importantly, the mutant line does not need to undergo successive backcrosses to eliminate SNPs or insertions/deletions (INDELs) unrelated to the trait that is being targeted. Indeed, the mutagenesis-induced SNPs are later used as polymorphic markers to map the causal mutation. It has been reported that a typical cv Micro-Tom EMS mutant plant typically carries 2,000 to 4,000 SNPs (Shirasawa et al., 2016).
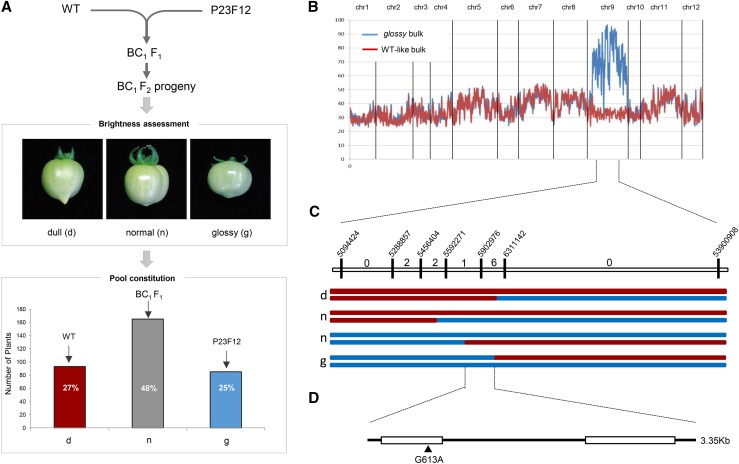
We applied this strategy to identify the causal mutation in the P23F12 glossy mutant line (Petit et al., 2014). As shown in Figure 1A, the homozygous glossy mutant line was back-crossed (BC1) with the wild-type parental line genotype used for generating the EMS mutant collection. The BC1F1 hybrid plant, which displayed a wild-type-like phenotype (dull/normal fruit) because the glossy mutation is recessive, was then selfed. The resulting BC1F2 segregating population (343 plants) was analyzed for the glossy-fruit phenotype. Two bulked pools of plants displaying either glossy fruit or wild-type-like fruit phenotypes were then created, each of which had 80 individuals. Pooled genomic DNA from each bulk was then sequenced to a tomato genome coverage depth of 29× to 37×, the trimmed sequences were mapped onto the tomato reference genome (Supplemental Table S1), and EMS mutation variants were filtered to exclude natural polymorphisms found in cv Micro-Tom (Kobayashi et al., 2014) compared with the cv Heinz 1706 reference genome (Supplemental Table S2). Analysis of the allelic variant frequencies in the two bulks led to the identification of chromosome 9 as the genome region carrying the causal mutation, since it displayed high mutant AFs (AF > 0.95) in the glossy bulk and much lower frequencies (AF < 0.4) in the wild-type-like bulk (Fig. 1B; Supplemental Table S3). Analysis of the putative effects of the mutations on protein functionality highlighted three genes carrying mutations in exons (Table I). One was a synonymous mutation, and the two others were missense mutations. Among these, one affected an annotated AMP deaminase, while the second affected a predicted GPAT (SlGPAT; Solyc09g014350) that was located in the expected chromosomal region according to AF analysis. Given the established function of GPAT proteins in lipid polyester biosynthesis (Li et al., 2007), we considered the latter to be the most likely candidate. To unequivocally associate the gpat mutation with the glossy mutant trait, and to exclude any other mutation, such as the mutation in AMP deaminase, we further analyzed six recombinant plants selected from the BC1F2 progeny. To this end, we used the EMS-induced SNPs surrounding the GPAT gene (Fig. 1C) as genetic markers. This analysis clearly indicated that a single G613A nucleotide transition in the first exon leading to a G163R nonsynonymous mutation was responsible for the glossy fruit phenotype (Fig. 1D).
Figure 1.
Mapping-by-sequencing identification of the gpat6-a mutation responsible for the glossy fruit phenotype. A, The wild-type (WT) parental line was crossed with the glossy mutant. The BC1F2 progeny (343 plants) was screened for glossy fruits (approximately 25% of the plants) and normal/dull fruits (approximately 75% of the plants) to constitute the wild-type-like and the glossy bulks (80 individuals each). B, Variant calling frequency analysis after sequencing the wild-type-like and glossy bulks. Variations in SNP frequencies along the 12 tomato chromosomes are shown using a 20-EMS-mutations sliding window for the wild-type-like bulk (red line) and the glossy bulk (blue line). C, Progeny testing of BC1F2 recombinant plants. Vertical bars indicate the nucleotide positions of SNP markers. The numbers between markers indicate the number of recombinant plants. The red and blue lines represent wild-type-like and glossy homozygous chromosomal segments, respectively. The two haplotypes per individual are represented. d = dull, n = normal, and g = glossy phenotypes. D, A single nucleotide transition, G613A, in the Solyc09g14350 first exon sequence led to the nonsynonymous mutation G163R.
Table I. Annotation of the high-scoring causal mutations identified on chromosome 9 for the glossy fruit mutant.
Filtered EMS mutations were annotated using SnpEff version 4.1 (Cingolani et al., 2012) from build release SL2.50 available on the SGN Web site (http://solgenomics.net). Two EMS missense mutations (mutations at positions 5,902,976 and 6,873,436 on chromosome 9) located in exonic regions may affect protein function. An additional mutation at position 35,265,019 causes a synonymous amino acid change in an exonic region.
| EMS Position | Mutation | Read Depth | Gene Position | Mutation Type | Tomato Gene Identifier | Gene Annotation |
|---|---|---|---|---|---|---|
| 5,902,976 | C to T | 28 | Exonic | Missense (G to R) | Solyc09g014350 | GPAT6 |
| 6,873,436 | C to T | 21 | Exonic | Missense (S to F) | Solyc09g014770 | AMP deaminase |
| 35,265,019 | G to A | 21 | Exonic | Synonymous (L to L) | Solyc09g050010 | Retrotransposon gag protein |
The glossy Mutation Affects a Functional Homolog of GPAT6, Whose Activity Is Abolished in the ΔSlGPAT6 Mutated Variant
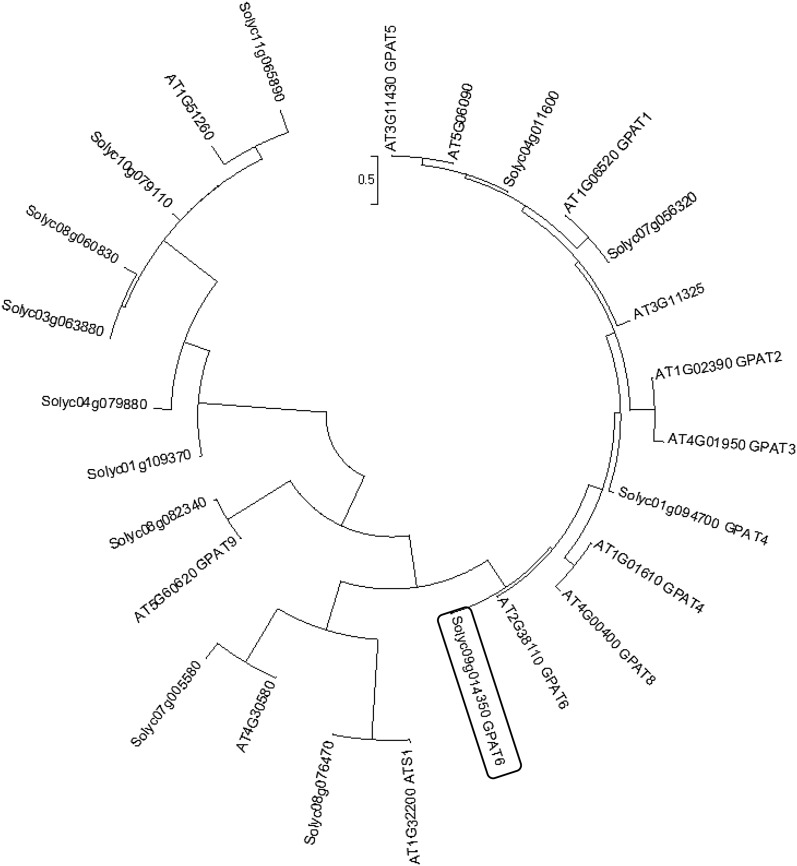
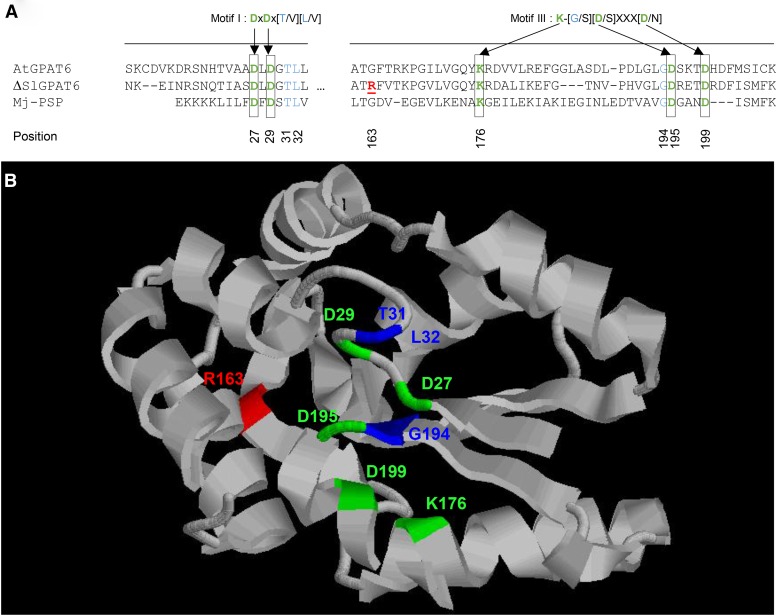
Phylogenetic analysis of tomato and Arabidopsis GPAT protein sequences indicated that the tomato GPAT protein encoded by the Solyc09g014350 gene is homologous to Arabidopsis AtGPAT4, AtGPAT6, and AtGPAT8 (Fig. 2), with the closest homolog being AtGPAT6. Accordingly, the SlGPAT6 glossy mutant (P23F12 line) is hereafter referred to as gpat6-a. We further investigated the effect of mutation position and its possible consequences on SlGPAT6 activity. As shown in Figure 3A, the G163R mutation is predicted to result in an amino acid substitution at position 163 in motif III, one of the two motifs (with motif I) that is known to be required for phosphatase activity. Specifically, the G163R mutation thus introduces an additional charged amino acid with a long side chain (Arg-163) adjacent to the Lys-176 required for phosphatase activity of Arabidopsis AtGPAT4 and AtGPAT6 (Yang et al., 2010). A structural model of SlGPAT6, based on the M. jannaschii phospho-Ser phosphatase template and restricted to the N-terminal 17 to 207 amino acid region, was generated in silico (Fig. 3B). Analysis of the predicted structure revealed that the G-to-R amino acid substitution at position 163 is close to the cluster of residues that are essential for the catalytic activity and Mg2+ binding of GPAT6 (Yang et al., 2010). We inferred from this observation that the G163R mutation in gpat6-a might confer a loss of SlGPAT6-mediated GPAT activity.
Figure 2.
Neighbor-joining phylogenetic tree of the amino acid sequences of GPAT identified from Arabidopsis and tomato (MEGA 6.0; Tamura et al., 2013). The black box surrounds the tomato SlGPAT6 gene (Solyc09g14350).
Figure 3.
Mutation positions in the ΔSlGPAT6 protein. A, Protein sequence alignment of Arabidopsis AtGPAT6, the tomato ΔSlGPAT6 variant, and the Methanococcus jannaschii phospho-Ser phosphatase (Mj-PSP). Black boxes show residues required for PSP activity in motifs I and III, according to Yang et al. (2010). The G163R mutation is indicated as underlined boldface red text. B, In silico ΔGPAT6 modeling based on Protein Data Bank structure 1L7P, restricted to amino acid positions 17 to 207. Critical residues from motifs I and III are indicated in green for D/K amino acids and in blue for other amino acids from canonical motifs. The position of the mutation is indicated in red.
To test this hypothesis, complementary DNAs (cDNAs) encoding the SlGPAT6 wild-type protein and the mutated variant ΔSlGPAT6 carrying the G163R mutation were expressed in a yeast (Saccharomyces cerevisiae) gat1Δ mutant strain in which expression of the major glycerol-3-acyltransferase, GAT1, has been knocked out (Athenstaedt et al., 1999). Microsomes containing recombinant SlGPAT6 and ΔSlGPAT6 proteins were then prepared from the transformed gat1Δ mutant yeast lines as well as from a line transformed with an empty vector as a control. The principal putative substrate of SlGPAT6 is 9(10),16-dihydroxyhexadecanoic acid (C16:0 diOH), the major cutin monomer present in tomato fruit cuticles (Mintz-Oron et al., 2008; Petit et al., 2014). Since this substrate is not commercially available, it was prepared from tomato cuticle extracts using a method adapted from Kawaguchi et al. (1981). Fatty acid methyl esters were first obtained by depolymerization of cutin derived from fresh tomato fruit peel. The C16:0 diOH methyl ester was then purified using thin-layer chromatography (TLC), saponified, and esterified to CoA. The same activation procedure was conducted with commercially available hexadecanoic acid (C16:0), 16-hydroxyhexadecanoic acid (C16:0 ωOH), and hexadecane-1,16-dioic acid (C16:0 DCA). These activated acyl-CoAs were used as substrates in enzymatic assays of recombinant wild-type and variant SlGPAT microsomes, measuring the incorporation of 14C, derived from [14C]G3P, into the acylglycerol groups.
Microsomes from the gat1Δgat2Δ double knockout yeast strain rescued by GAT1 overexpression and supplied with hexadecanoic acid-CoA produced lysophosphatidic acid (LPA), phosphatidic acid (PA), MAG, diacylglycerol (DAG), and lower amounts of triacylglycerol (TAG; Supplemental Fig. S2A). The single knockout mutant gat1Δ transformed with the empty vector control and supplied with C16:0-CoA synthesized PA but also low amounts of LPA, MAG, DAG, and TAG (Supplemental Fig. S2B). These results indicate that the weak activity of the second yeast GAT, GAT2, present in the gat1Δ yeast mutant, is sufficient for the production of low amounts of MAG. Microsomes prepared from the yeast gat1Δ mutant expressing tomato SlGPAT6 were also able to synthesize LPA, PA, MAG, DAG, and TAG (Supplemental Fig. S2C). However, MAG accumulation was much higher than that observed in microsomes from the empty vector control line (Supplemental Fig. S2B), demonstrating that wild-type SlGPAT6 has the dual acyltransferase and phosphatase activities expected from a GPAT enzyme involved in cutin monomer biosynthesis (Yang et al., 2010, 2012).
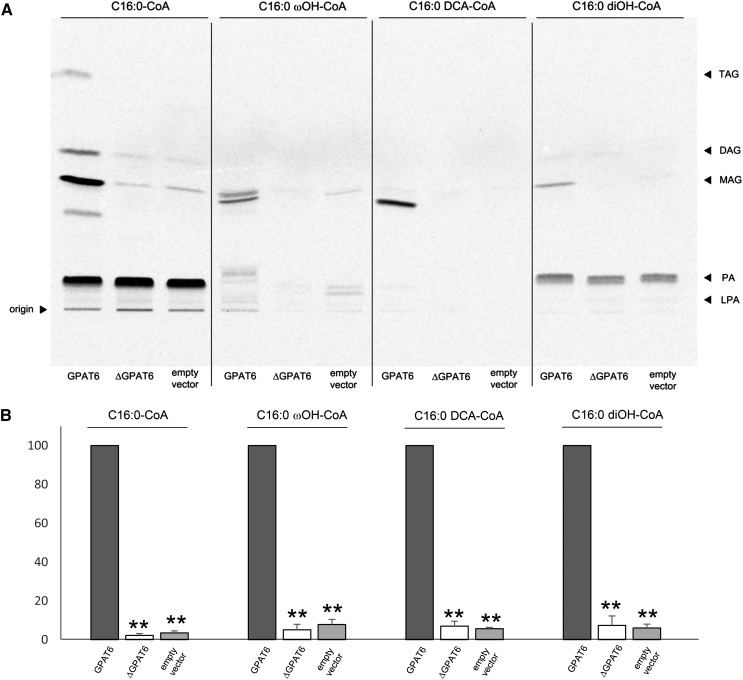
We next assessed SlGPAT6 substrate specificity and the effect of the G163R mutation on its activity. SlGPAT6 was observed to use not only C16:0-CoA as a substrate to produce MAG but also C16:0 ωOH-CoA, C16:0 DCA-CoA, and C16:0 diOH-CoA (Fig. 4A). This is noteworthy because C16:0 diOH is the most abundant cutin monomer in tomato fruit and yet, to date, GPAT activity has not been demonstrated with this natural substrate. This activity was absent in the ΔSlGPAT6 variant, regardless of the substrate used (Fig. 4), indicating that the G163R mutation confers a loss of SlGPAT6 enzyme function.
Figure 4.
SlGPAT6 in vitro assays in yeast, in the presence of [14C]G3P. A, The substrates are CoA-activated hexadecanoic acid (C16:0-CoA), 16-OH hexadecanoic acid (C16:0 ωOH-CoA), hexadecane-1,16 dioic acid (C16:0 DCA-CoA), and 9(10),16 diOH hexadecanoic acid (C16:0 diOH-CoA). The GPAT6- and ΔGPAT6-expressing yeast strains were grown for 24 h at 30°C. After preparation of microsomes and activated substrates, enzyme activities were assayed three times using independent microsome and substrate preparations. Reactions were stopped by adding HClO4, lipids were extracted, and the organic phase was analyzed by TLC. B, MAG signals were quantified using ImageQuant TL (GE Healthcare Life Sciences). The activity of the wild-type SlGPAT6 was arbitrarily set at 100. Values are means ± sd of three independent experiments. Asterisks indicate significant differences from the wild-type value (Student’s t test, P < 0.01).
The Fruit Cuticle of the gpat6-a Mutant Has Reduced Levels of C16 Cutin Monomers
In order to investigate how the loss of SlGPAT6 activity affects cuticle composition, we analyzed the wax and cutin constituents of cuticles from red ripe stage fruits (approximately 45 DPA). No differences were observed in total wax load between wild-type and gpat6-a fruits, but differences in wax composition were apparent (Table II), such as increased or decreased levels of several minor C25 to C30 alkanes in the gpat6-a mutant. Since both even- and odd-numbered alkanes displayed similar trends, these variations were likely independent of the alkane biosynthetic pathways. The most striking wax compositional change in the gpat6-a mutant was an approximately 5-fold increase in δ- and α-amyrin levels and to a lesser extent in β-amyrin levels. In contrast to waxes, the total cutin load in gpat6-a was reduced by almost 3-fold, from 315 to 111 µg cm−2 (Table III). This corresponded to an approximately 3-fold drop in the amount of C16:0 diOH. In addition, differences in the levels of several other major C16 cutin monomers were observed, including C16:0 DCA and C16:0 DCA 9(10) OH (approximately 3- to 4-fold reduction in gpat6-a) as well as C16:0 ωOH and C16:0 ωOH 10 oxo (up to a 10-fold reduction in gpat6-a).
Table II. Wax composition of the fruit cuticles from wild-type and gpat6-a mutant plants.
Mean values (µg cm−2 × 10) of each compound are given with sd (n = 3). The percentage of total wax load is indicated for individual compounds. Unidentified compounds are not included. Letters indicate significant differences from the wild-type composition (Student’s t test: b, P < 0.05; and a, P < 0.01).
| Composition | Classification | Wild Type |
Percentage |
gpat6-a |
Percentage | Significance |
|---|---|---|---|---|---|---|
| Mean ± sd | Mean ± sd | |||||
| Fatty acids | C16 | 0.69 ± 0.24 | 1.2 | 0.64 ± 0.05 | 1.2 | |
| C18 | 0.23 ± 0.14 | 0.4 | 0.40 ± 0.03 | 0.8 | ||
| C24 | 1.84 ± 0.08 | 3.1 | 0.44 ± 0.19 | 0.9 | a | |
| Alkanes | C25 | 0.95 ± 0.09 | 1.6 | 0.15 ± 0.04 | 0.3 | a |
| C27 | 7.11 ± 0.58 | 12.1 | 1.38 ± 0.22 | 2.6 | a | |
| C28 | 0.89 ± 0.22 | 1.5 | 0.11 ± 0.02 | 0.2 | a | |
| C29 | 3.66 ± 0.50 | 6.3 | 7.11 ± 1.73 | 13.6 | b | |
| C30 | 1.94 ± 0.09 | 3.3 | 2.33 ± 0.15 | 4.5 | b | |
| C31 | 13.25 ± 1.94 | 22.7 | 16.22 ± 3.54 | 31.2 | ||
| C32 | 2.35 ± 0.61 | 4.0 | 1.30 ± 0.34 | 2.5 | ||
| C33 | 3.40 ± 1.09 | 5.8 | 1.81 ± 0.43 | 3.5 | ||
| C35 | 0.16 ± 0.06 | 0.3 | 0.30 ± 0.03 | 0.6 | b | |
| iso C29 | 0.16 ± 0.06 | 0.3 | 0.20 ± 0.07 | 0.4 | ||
| iso C30 | 0.25 ± 0.02 | 04 | 0.26 ± 0.02 | 0.5 | ||
| iso C31 | 2.36 ± 0.53 | 4.0 | 1.56 ± 0.24 | 3.0 | ||
| iso C32 | 0.32 ± 0.06 | 0.5 | 0.21 ± 0.11 | 0.4 | ||
| iso C33 | 0.58 ± 0.37 | 1.0 | 0.24 ± 0.08 | 0.5 | ||
| ante iso C29 | 4.03 ± 1.33 | 6.9 | 2.59 ± 0.38 | 5.0 | ||
| ante iso C31 | 0.19 ± 0.06 | 0.3 | 0.11 ± 0.02 | 0.2 | ||
| Amyrins | δ | 1.01 ± 0.13 | 1.7 | 5.87 ± 0.50 | 11.3 | a |
| β | 1.78 ± 0.34 | 3.0 | 2.85 ± 0.29 | 5.5 | b | |
| α | 0.58 ± 0.07 | 1.0 | 2.86 ± 0.25 | 5.5 | a | |
| Lupeol | 0.17 ± 0.05 | 0.3 | 0.28 ± 0.17 | 0.5 | ||
| Multiflorenol | 0.16 ± 0.06 | 0.3 | 0.27 ± 0.03 | 0.5 | ||
| ψ-Taraxasterol | 0.21 ± 0.02 | 0.4 | 0.19 ± 0.03 | 0.4 | ||
| Taraxasterol | 0.30 ± 0.14 | 0.5 | 0.11 ± 0.02 | 0.2 | ||
| Total load | 48.56 ± 5.82 | 49.76 ± 6.83 |
Table III. Cutin composition of the cuticle from fruits of wild-type and gpat6-a mutant plants.
Mean values (µg cm−2) of each compound are given with sd (n = 3). The percentage of total cutin load is indicated for individual compounds. Unidentified compounds are not included. Letters indicate significant differences from the wild-type composition (Student’s t test: b, P < 0.05; and a, P < 0.01).
| Composition | Classification | Wild Type |
Percentage |
gpat6-a |
Percentage | Significance |
|---|---|---|---|---|---|---|
| Mean ± sd | Mean ± sd | |||||
| Fatty acids | C16:0 | 4.5 ± 2.0 | 1.4 | 0.9 ± 0.5 | 0.8 | |
| Dicarboxylic acids | C16:0 DCA | 37.1 ± 3.4 | 11.8 | 8.3 ± 3.0 | 7.5 | a |
| C16:0 DCA (9 or 10) OH | 26.2 ± 2.8 | 8.3 | 7.8 ± 2.3 | 7.0 | a | |
| ω-Hydroxy acids | C16:0 ωOH | 14.1 ± 5.8 | 4.5 | 1.4 ± 0.4 | 1.3 | b |
| C16:0 ωOH 10 oxo | 21.6 ± 3.3 | 6.9 | 2.5 ± 0.9 | 2.3 | a | |
| C18:0 ωOH (9 or 10) OH | 6.6 ± 3.6 | 2.1 | 4.1 ± 1.5 | 3.7 | ||
| C18:0 ωOH (9, 10) epoxy | 17.2 ± 1.6 | 5.5 | 9.0 ± 5.3 | 8.1 | ||
| Coumaric acid | 6.0 ± 3.3 | 1.9 | 10.4 ± 3.6 | 9.4 | ||
| Polyhydroxy acids | C16:0 (9/10, 16) diOH | 181.0 ± 17.7 | 57.5 | 64.5 ± 12.3 | 58.2 | a |
| C18:0(9, 10, 18) triOH | 0.7 ± 0.3 | 0.2 | 1.8 ± 0.6 | 1.6 | ||
| Total load | 314.8 ± 47.0 | 110.7 ± 26.6 | a |
The gpat6-a Mutant Has Abnormal Fruit Cuticle Thickness and Properties
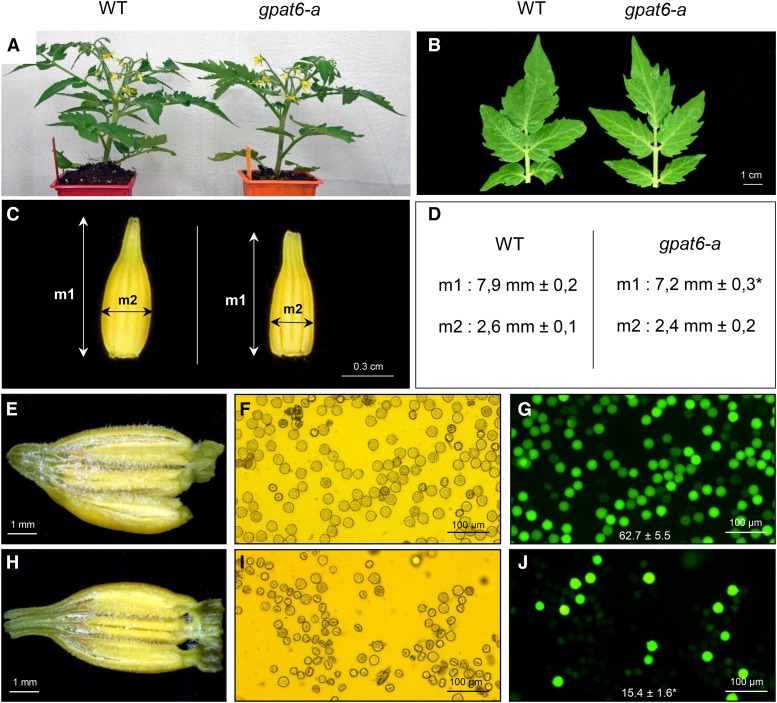
According to digital expression data for SlGPAT6 (Solyc09g014350) available from the BAR database (http://bar.utoronto.ca/efp_tomato/cgi-bin/efpWeb.cgi), the gene is preferentially expressed in reproductive organs and, more specifically, in the unopened flower and the young developing tomato fruit (Supplemental Fig. S3). The Arabidopsis homolog AtGPAT6 also is preferentially expressed in the inflorescences, and its loss of expression causes reduced fertility (Li-Beisson et al., 2009; Li et al., 2012). Consistent with these findings, the whole-plant and leaf phenotypes from the tomato gpat6-a mutant showed no differences compared with the wild type (Fig. 5, A and B). In contrast to Arabidopsis AtGPAT6 knockout plants (Li-Beisson et al., 2009), no abnormal flowers displaying organ fusion were observed in gpat6-a; however, stamen elongation was reduced in the tomato gpat6-a mutant (Fig. 5C). Examination of the anthers revealed that the pollen sacs of gpat6-a had fewer pollen grains than those of the mutant (Fig. 5, E and H). In addition, most mutant pollen grains were small, shrunken, and had an abnormal shape (Fig. 5, F and I), and pollen viability was reduced by approximately 4-fold in the mutant (Fig. 5, G and J). However, the reduced fertility in the gpat6-a mutant had no consequences on fruit size or the number of seeds per fruit (Supplemental Fig. S4).
Figure 5.
Morphological comparison of wild-type (WT) and gpat6-a plants, flowers, and pollen. A, Appearance of the plants 6 weeks after sowing. B, Morphology of the fourth leaf. C, Morphology of the detached anther cone. D, Measurements of the length (measure 1 [m1]) and thickness (m2) of the anther cone. Mean values (in mm) of 20 measures are given with sd. The asterisk indicates a significant difference from wild-type fruit (Student’s t test, P < 0.01). E and H, Macroscopic observations of dissected anthers of wild-type (E) and gpat6-a (H) flowers. F, G, I, and J, Microscopic observations under visible light (F and I) and at λ = 495 nm (G and J) of wild-type (F and G) and gpat6-a (I and J) pollen stained with 2% fluorescein diacetate. The pollen viability is determined by the numeration of fluorescent spots compared with the total number of pollen grains in the acquisition field. Mean values of 10 fields are given in percentage with sd. The asterisk indicates a significant difference from the wild-type fruit (Student’s t test, P < 0.01).
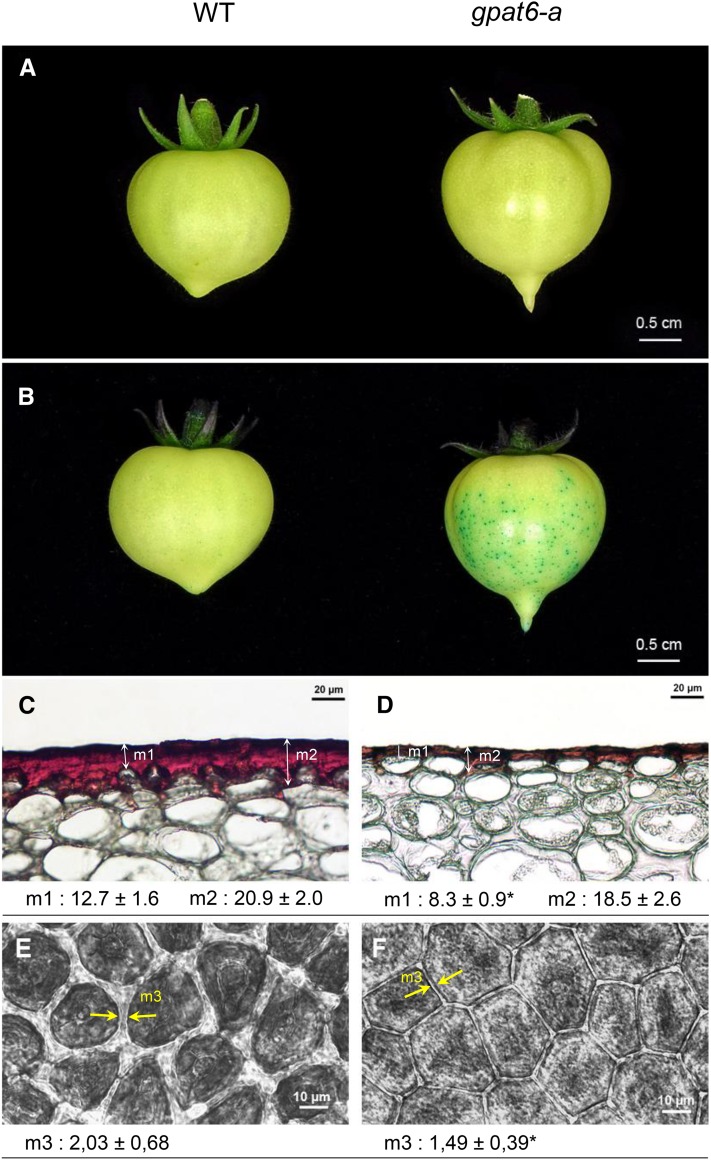
SlGPAT6 expression is highest in young fruit (3-fold more than in flowers), and more specifically in the outer fruit epidermis (Matas et al., 2011). Accordingly, the most striking effect of the gpat6-a mutation was observed on the fruit cuticle. Mutant fruits with enhanced brightness displayed increased permeability to Toluidine Blue stain (Fig. 6, A and B) and altered epidermal cell shape (Fig. 6, C and D). In addition, the thickness of the cuticle covering the fruit epidermal cells was reduced substantially (Fig. 6, C and D), as was the width of the anticlinal cutinized cell wall between adjacent epidermal cells (Fig. 6, E and F). Taken together, these results indicated profound alterations in cuticle load and structure.
Figure 6.
Cuticle phenotypes of wild-type (WT) and gpat6-a fruits. A, Macroscopic observation of the brightness of mature green stage fruits. B, Macroscopic observation of cuticle permeability to 0.5% Toluidine Blue. C and D, Microscopic observations of 8-µm cryosections of cuticles from wild-type (C) and gpat6-a (D) breaker stage fruits after Oil Red O staining. Mean values (µm) of 40 measurements from three different sections are given with sd, in two different positions, on the top of the epidermal cell (measure 1 [m1]) and between epidermal cells (m2). The asterisk indicates a significant difference from the wild-type fruit (Student’s t test, P < 0.01). E and F, Microscopic observations of freshly peeled outer epidermal tissue from wild-type (E) and gpat6-a (F) red ripe stage fruits. The width of the cutinized cell wall was measured between the two yellow arrowheads (m3). Mean values (µm) of 620 measures (20 measurements of 31 sections) are given with sd. The asterisk indicates a significant difference from the wild-type fruit (Student’s t test, P < 0.01).
Genes That Are Expressed at Lower or Higher Levels in gpat6-a Fruit Exocarp Compared with the Wild Type Are Associated with a Wide Range of Regulatory and Biosynthesis Pathways
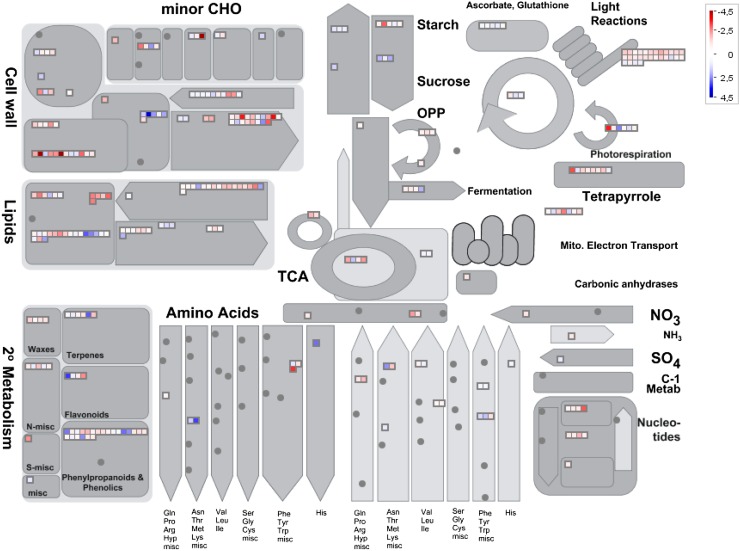
Since the gpat6-a mutation affects not only the levels of cutin monomers in the cuticle but also the formation and accumulation of cuticle compounds synthesized through distantly related (alkanes) or unrelated (amyrins and coumaric acid) pathways, as well as cell wall morphology, we further investigated gene expression changes in the fruit exocarp from wild-type and gpat6-a lines. Fruits at 20 DPA were selected because this stage of fruit development corresponds to the maximum rate of cuticle formation in cv Micro-Tom (Petit et al., 2014) and to the peak of SlGAPT6 expression in the fruit (Supplemental Fig. S3). Transcripts expressed in the fruit skin, which is composed of epidermal cells and underlying cutinized collenchyma cells (Fig. 6C), were subjected to high-throughput RNA sequencing (RNA-seq) analysis using Illumina technology. Among the approximately 23,000 genes detected, 567 were found to be differentially expressed between wild-type and gpat6-a fruit exocarp using a 2-fold cutoff for fold change in transcript abundance (ratios of 1 and −1 on a log2 scale) and P < 0.05 (Supplemental Table S4). Of these, 368 and 199 genes were expressed at higher and lower levels, respectively, in gpat6-a than in the wild type. For the 376 genes belonging to MapMan Gene Ontology classes (Urbanczyk-Wochniak et al., 2006), significant differences were observed not only for genes involved in lipid metabolism, as would be expected, but also for genes belonging to the transport, hormone metabolism, and stress response categories (Supplemental Fig. S5). The largest category was TFs, which contained almost 28% of the differentially expressed genes. An analysis of the metabolism-related functional categories using MapMan revealed differential expression of genes in categories related to cuticle formation, including lipid metabolism and specialized metabolism (terpenes, flavonoids, and phenylpropanoids), but also in categories related to cell wall formation and modification. Relatively few differences were observed in genes associated with central metabolism (Fig. 7).
Figure 7.
MapMan-based general overview of genes differentially expressed between wild-type and gpat6-a mutant fruit. The log2 ratio of the mutant to the wild type for individual genes was plotted onto boxes. Red boxes indicate genes expressed at significantly higher levels in the wild type compared with the gpat6-a mutant; blue boxes indicate genes expressed at significantly higher levels in the gpat6-a mutant compared with the wild type. CHO, Carbohydrate metabolism; OPP, oxidative pentose phosphate; TCA, tricarboxylic acid cycle.
Notably, all the cuticle-associated genes identified, which collectively are involved in various steps in cutin biosynthesis and assembly and in wax biosynthesis, were expressed at significantly lower levels (P ≤ 0.0003) in the gpat6-a mutant than in the wild type. Examples included genes encoding enzymes known to be required for the production of the MAG cutin precursors in Arabidopsis or tomato (Li-Beisson et al., 2013; Shi et al., 2013): LACS2 (Solyc01g109180), putatively required for the activation to CoA thioesters (Schnurr et al., 2004); three cytochrome P450-dependent fatty acid oxidases belonging to protein families required either for in-chain hydroxylation of fatty acids (CYP77A1 and CYP77A2 [Solyc11g007540 and Solyc05g055400]; Li-Beisson et al., 2009) or for terminal chain hydroxylation (CYP86A69 [Solyc08g081220]; Shi et al., 2013); and two GPATs (GPAT4 and GPAT6 [Solyc01g094700 and Solyc09g014350]; Yang et al., 2010; Fig. 2; Table IV). One gene encoded a nonspecific lipid transfer protein, examples of which have been suggested to be involved in the transport of cuticle components across the apoplast to the cuticle, although this has yet to be demonstrated conclusively (Yeats and Rose, 2008). An additional differentially expressed cutin-related gene was predicted to encode an ω-hydroxypalmitate O-feruloyl transferase (Solyc11g008630) that belongs to the BAHD family of acyl-CoA-dependent acyl-transferases, some of which are implicated in cutin biosynthesis (Rautengarten et al., 2012). Finally, 11 GDSL lipase genes expressed at various stages of fruit development (Tomato Genome Consortium, 2012; Tomato eFP browser [http://bar.utoronto.ca/efp_tomato/cgi-bin/efpWeb.cgi]) were expressed at lower levels in gpat6-a. These genes are homologous to the cutin synthase gene CUS1 (Solyc11g006250) encoding the enzyme that catalyzes the extracellular polymerization of cutin (Girard et al., 2012; Yeats et al., 2012b).
Table IV. Genes differentially expressed between the exocarp of gpat6-a mutant and wild-type fruit.
Genes were assigned manually to functional categories.
| Gene Identification | Gene Name | Putative Function | Log2 Fold Change | P |
|---|---|---|---|---|
| Cuticle | ||||
| Solyc02g085870 | CER6 | Fatty acid elongase 3-ketoacyl-CoA synthase | −1.74 | 5.00E-05 |
| Solyc11g072990 | KCS3 | Fatty acid elongase 3-ketoacyl-CoA synthase | −1.85 | 5.00E-05 |
| Solyc08g067260 | KCS10/FDH | Fatty acid elongase 3-ketoacyl-CoA synthase | −1.82 | 5.00E-05 |
| Solyc09g092270 | CER26like | VLCFA elongation | −1.95 | 5.00E-05 |
| Solyc07g006300 | CER3 | Alkane biosynthesis | −1.22 | 3.00E-04 |
| Solyc12g087980 | CER1 | Alkane biosynthesis | −1.32 | 5.00E-05 |
| Solyc01g088400 | CER1like | Alkane biosynthesis | −1.52 | 5.00E-05 |
| Solyc10g080840 | MAH1 | Midchain alkane hydroxylase | −2.1 | 5.00E-05 |
| Solyc01g109180 | LACS2 | Acyl-CoA synthetase | −1.49 | 5.00E-05 |
| Solyc11g007540 | CYP77A1 | Fatty acid hydroxylase | −1.54 | 5.00E-05 |
| Solyc08g081220 | CYP86A69 | Fatty acid hydroxylase | −1.84 | 5.00E-05 |
| Solyc05g055400 | CYP77A2 | Fatty acid hydroxylase | −1.92 | 5.00E-05 |
| Solyc01g094700 | GPAT4 | Glycerol-3-phosphate acyltransferase | −1.41 | 5.00E-05 |
| Solyc09g014350 | GPAT6 | Glycerol-3-phosphate acyltransferase | −1.63 | 5.00E-05 |
| Solyc11g008630 | HHT | ω-Hydroxypalmitate O-feruloyl transferase | −1.35 | 5.00E-05 |
| Solyc05g015490 | LTPG1 | Glycosylphosphatidylinositol (GPI)-anchored lipid protein transfer | −1.8 | 5.00E-05 |
| Solyc02g077330 | GDSL-motif esterase/acyltransferase/lipase | −1.06 | 5.00E-05 | |
| Solyc11g006250 | CUS1 | GDSL-motif esterase/acyltransferase/lipase | −1.33 | 5.00E-05 |
| Solyc03g111550 | GDSL-motif esterase/acyltransferase/lipase | −1.52 | 5.00E-05 | |
| Solyc07g049440 | GDSL-motif esterase/acyltransferase/lipase | −1.56 | 5.00E-05 | |
| Solyc04g081770 | GDSL-motif esterase/acyltransferase/lipase | −1.65 | 5.00E-05 | |
| Solyc03g006240 | GDSL-motif esterase/acyltransferase/lipase | −1.67 | 5.00E-05 | |
| Solyc04g081760 | GDSL-motif esterase/acyltransferase/lipase | −1.72 | 5.00E-05 | |
| Solyc03g121180 | GDSL-motif esterase/acyltransferase/lipase | −1.75 | 5.00E-05 | |
| Solyc11g043110 | GDSL-motif esterase/acyltransferase/lipase | −2.02 | 5.00E-05 | |
| Solyc02g071610 | GDSL-motif esterase/acyltransferase/lipase | −2.19 | 5.00E-05 | |
| Solyc05g043330 | GDSL-motif esterase/acyltransferase/lipase | −2.69 | 5.00E-05 | |
| Secondary metabolism | ||||
| Solyc03g042560 | PAL | Phe ammonia-lyase | 2.76 | 5.00E-05 |
| Solyc09g007910 | PAL | Phe ammonia-lyase | 2.42 | 5.00E-05 |
| Solyc03g097030 | 4CL | 4-Coumarate-CoA ligase | −1.31 | 5.00E-05 |
| Solyc09g091510 | CHS | Chalcone synthase | 3.2 | 5.00E-05 |
| Solyc08g079280 | F3′H | Flavonoid 3′-hydroxylase | −1.36 | 5.00E-05 |
| Solyc01sg107590 | CAD | Cinnamyl alcohol dehydrogenase | −1.07 | 5.00E-05 |
| Solyc02g090350 | CYP76B | Cytochrome P450 | −1.58 | 5.00E-05 |
| Solyc07g062500 | CYP72A | Cytochrome P450 | −1.46 | 5.00E-05 |
| Cell wall | ||||
| Solyc07g043390 | CesA | Cellulose synthase | 2.31 | 5.00E-05 |
| Solyc02g089120 | COBL | COBRA-like | −1.31 | 5.00E-05 |
| Solyc03g083770 | PMEI | Pectin methylesterase inhibitor | 2.33 | 5.00E-05 |
| Solyc05g047590 | PME | Pectin methylesterase | −1.63 | 5.00E-05 |
| Solyc04g077470 | CSLC | Cellulose-synthase-like | 1.59 | 5.00E-05 |
| Solyc02g091920 | XTH | Xyloglucan endotransglucosylase/hydrolase | −2.39 | 5.00E-05 |
| Solyc01g081060 | XTH | Xyloglucan endotransglucosylase/hydrolase | −2.84 | 5.00E-05 |
| Solyc07g009380 | XTH | Xyloglucan endotransglucosylase/hydrolase | −5.03 | 5.00E-05 |
| Solyc09g092520 | XTH | Xyloglucan endotransglucosylase/hydrolase | −6.48 | 5.00E-05 |
| Solyc01g110000 | GLB | β-Galactosidase | −1.61 | 2.80E-03 |
| Solyc03g121540 | GLB | β-Galactosidase | −1.8 | 5.00E-05 |
| Solyc06g076220 | EXP | Expansin | 1.53 | 5.00E-05 |
| Solyc04g081870 | EXP | Expansin | −1.16 | 5.00E-05 |
| Solyc07g053530 | FLA | Fasciclin-like arabinogalactan protein | 1.34 | 2.50E-04 |
| Solyc07g053540 | FLA | Fasciclin-like arabinogalactan protein | −1.31 | 5.00E-05 |
We also observed that the expression of several wax-associated genes involved in very-long-chain fatty acid (VLCFA) synthesis, and modification was reduced in the gpat6-a mutant relative to the wild type (Table IV). These included three genes (KCS3 [Solyc11g072990], KCS10/FDH [Solyc08g067260], and KCS6/CER6 [Solyc02g085870]) encoding β-ketoacyl-CoA synthase enzymes that catalyze the first step in the multienzymatic fatty acid elongase complex generating very-long-chain (VLC) C20-C36 acyl-CoAs. Similarly, the expression of CER26L (Solyc09g092270), which likely controls the chain-length specificities of wax cuticle components (Bernard and Joubès, 2013), also was reduced in gpat6-a, as was the expression of genes encoding enzymes catalyzing successive steps in the alkane biosynthesis pathway (Table IV). CER3 likely catalyzes the conversion of VLC acyl-CoAs to intermediate VLC aldehydes, CER1 (Solyc12g087980) and CER3 (Solyc07g006300) catalyze their conversion to VLC alkanes, while MAH1 (CYP96A15 [Solyc10g080840]) is a midchain alkane hydroxylase responsible for the generation of secondary alcohols and ketone derivatives from alkanes (Bourdenx et al., 2011; Bernard et al., 2012; Bernard and Joubès, 2013).
The RNA-seq data also suggested that the phenylpropanoid and flavonoid biosynthetic pathways were altered in gpat6-a (Table IV). In the phenylpropanoid core pathway, two phenylalanine ammonia-lyase (PAL; Solyc03g042560 and Solyc09g007910) genes were expressed at higher levels in gpat6-a, while a 4-coumarate CoA ligase (4CL; Solyc03g097030), catalyzing the formation of p-coumaroyl-CoA used as a precursor for flavonoids and phenolic compounds, was expressed at lower levels. In the flavonoid biosynthetic pathway, transcript levels of a chalcone synthase (CHS; Solyc09g091510) were higher in gpat6-a, whereas those of a flavonoid 3′-hydroxylase (F3′H; Solyc08g079280) were lower. Two additional genes that were expressed at lower levels in gpat6-a encode an enzyme involved in the formation of cinnamyl alcohols (CAD; Solyc01g107590) and a CYP72A (Solyc07g062500) protein. A member of the CYP72A family was recently shown to catalyze the C-30 oxidation of β-amyrin (Seki et al., 2011).
In addition to the cuticle-associated lipid pathways, the most striking gene expression changes were related to cell wall pathways. For example, a gene encoding cellulose synthase (Solyc07g043390), the catalytic moiety required for the synthesis of cellulose microfibrils, was expressed at higher levels in the gpat6-a mutant. Conversely, the expression of a COBRA-like gene (SlCOBL1; Solyc02g089120), which encodes a GPI-anchored protein regulating epidermal cell wall thickness and cellulose formation in tomato fruit (Cao et al., 2012; Niu et al., 2015), was lower in gpat6-a. We also detected the altered expression of genes related to pectins and hemicelluloses, the matrix polysaccharides of the primary cell wall. A gene encoding a cellulose synthase-like (CSL; Solyc04g077470) gene, a member of a glycosyltransferase family involved in the synthesis of matrix polysaccharides such as xyloglucan (Cosgrove, 2005), was expressed at a higher level in gpat6-a. In contrast, the expression levels of four xyloglucan endotransglucosylase/hydrolase (XTH) genes (Solyc02g091920, Solyc01g081060, Solyc07g009380, and Solyc09g092520) were substantially lower. XTH enzymes are involved in cell wall assembly and restructuring as well as in cell expansion and the integration of newly synthesized xyloglucans in the cell wall (Rose et al., 2002; Cosgrove, 2005; Park and Cosgrove, 2015). Genes encoding a pectin methylesterase (PME; Solyc05g047590) and a pectin methylesterase inhibitor (PMEI; Solyc03g083770) were expressed at lower and higher levels, respectively, in gpat6-a. These proteins have opposite roles, and their interplay likely controls the esterification status of pectins, which in turn is thought to influence the mechanical properties of the cell wall (Reca et al., 2012; Müller et al., 2013). Two genes encoding β-galactosidases (Solyc01g110000 and Solyc03g121540) were expressed at lower levels in the mutant, and these too may play roles in the modification of cell wall polysaccharides (Smith and Gross, 2000). Other differentially expressed genes included two expansin genes (Solyc06g076220 and Solyc04g081870), important modulators of cell wall extensibility (Park and Cosgrove, 2015), which were expressed at higher or lower levels in gpat6-a, and two genes encoding fasciclin-like arabinogalactan proteins (FLAs; Solyc07g053530 and Solyc07g053540), which also showed opposite patterns of relative transcript abundance in the mutant and the wild type. The functions of FLAs are not well understood, although many are GPI anchored to the plasma membrane and play key roles in plant development, including cell wall architecture and composition (MacMillan et al., 2010; Johnson et al., 2011; Seifert et al., 2014).
Not surprisingly, given the large transcriptional changes induced by the GPAT6 mutation deficiency, 53 TFs, belonging to AP2/EREBP (eight), bHLH (three), C2/C2(Zn) (11), C2H2 zinc finger (four), MADS box (four), MYB (six), and other TF categories (17), were expressed at higher (18) or lower (35) levels in gpat6-a (Supplemental Table S5). To our knowledge, most of these have not been associated previously with the fruit epidermis/cuticle; however, several are known to regulate early fruit development, including the MADS box gene TAGL11 (Mejía et al., 2011), or fruit ripening, such as the MADS box gene RIPENING INHIBITOR (RIN [Solyc05g012020]; Vrebalov et al., 2002; Kosma et al., 2010), as well as several ethylene-responsive TFs (Liu et al., 2016). The differentially expressed TFs included those with a possible link to the circadian clock, with two genes (Solyc01g095030 and Solyc10g084370) encoding MYB-related TFs of the REVEILLE family (Farinas and Mas, 2011), one of which was expressed at lower levels in gpat6-a, and six genes encoding CONSTANS-like TFs (Valverde, 2011), one of which (Solyc12g005660) was expressed at much lower levels. Despite the large number of differentially expressed TFs identified, the group did not include TFs that have been reported to regulate cuticle formation in tomato or other species (Shi et al., 2013; Borisjuk et al., 2014; Hen-Avivi et al., 2014), other than a MIXTA-like MYB TF (Oshima and Mitsuda, 2013), whose role in tomato was described recently (Lashbrooke et al., 2015). We observed that the expression of SlMIXTA-like (Solyc02g088190) was lower in the gpat6-a mutant than in the wild type (Supplemental Table S5). Moreover, 10 cutin- and wax-related genes that were differentially expressed between the wild type and the gpat6-a mutant (Table IV) were identified previously as SlMIXTA-like targets (Lashbrooke et al., 2015).
DISCUSSION
Tomato represents an excellent model for studying cuticle formation in fleshy fruits as well as more broadly in plant taxa (Hen-Avivi et al., 2014; Martin and Rose, 2014; Fich et al., 2016). Studies of tomato cuticles span many different fields, ranging from the analysis of cuticle composition and architecture (Mintz-Oron et al., 2008; Buda et al., 2009; Yeats et al., 2012a; Philippe et al., 2016; Segado et al., 2016), biosynthetic pathways, assembly and regulation (Shi et al., 2013; Lashbrooke et al., 2015), interaction with other metabolic pathways and developmental processes (Kosma et al., 2010; Giménez et al., 2015), mechanical properties (Schreiber, 2010; España et al., 2014), as well as the significance of the cuticle for agronomically important traits such as fruit glossiness, postharvest shelf-life, fruit cracking, and resistance to pathogens (Isaacson et al., 2009; Shi et al., 2013; Buxdorf et al., 2014; Petit et al., 2014). Considerable progress has been made through reverse genetics approaches, where the function of a candidate gene previously identified is studied in planta in tomato (Girard et al., 2012). As a complement to reverse genetic approaches, forward genetic strategies can be a highly effective means of gene discovery. In such cases, phenotypic changes in the fruit cuticle can lead to identification of the underlying gene, as was the case with several of the eceriferum Arabidopsis mutants (Negruk et al., 1996). In tomato, these strategies enabled the discovery of new cuticle-associated genes and functions, including genes involved in cutin synthesis and polymerization (Yeats et al., 2012b; Shi et al., 2013; Petit et al., 2014) and in cuticle regulation (Adato et al., 2009; Isaacson et al., 2009; Ballester et al., 2010; Nadakuduti et al., 2012). The diversity in cuticle traits is high in cultivated tomato accessions and in wild related species (Hovav et al., 2007; Yeats et al., 2012a) as well as in artificially produced mutant collections (Isaacson et al., 2009; Kimbara et al., 2013; Shi et al., 2013; Petit et al., 2014; Philippe et al., 2016). Harnessing such genetic and phenotypic diversity will doubtless lead to many new insights into cuticle formation and regulation in tomato and other plant species.
Mapping-by-Sequencing Reveals a gpat6-a Causal Mutation Underlying a glossy Fruit Mutant
All the strategies used to date to identify natural polymorphism or causal mutations responsible for a cuticle phenotype in tomato have been based on genetic mapping of the polymorphism and subsequent map-based cloning of the underlying gene (Isaacson et al., 2009; Yeats et al., 2012b; Kimbara et al., 2013; Shi et al., 2013; Petit et al., 2014). Despite a high-quality tomato reference genome sequence and the development of associated genomic tools (Shirasawa et al., 2010; Sim et al., 2012; Tomato Genome Consortium, 2012; Kobayashi et al., 2014), this remains a time-consuming and laborious task. An attractive alternative strategy is mapping-by-sequencing (Schneeberger, 2014), which is based on a combination of bulk segregant analysis of a BC1F2 segregating population and of whole-genome sequencing followed by SNP frequency analysis to identify the causal mutation. Mapping-by-sequencing has been applied successfully to the model species Arabidopsis (Schneeberger et al., 2009) and rice (Abe et al., 2012). Since the approach involves a simple cross between the mutant and the parental (nonmutagenized) lines, it is well adapted to the study of traits that are highly sensitive to genetic context and/or to environmental conditions, such as the structure and composition of the tomato fruit cuticle. The analysis of a cuticle mutant, therefore, is not restricted to the most severe cuticle alterations, opening the way for the discovery of new cuticle-associated genes and functions. The use of the miniature cv Micro-Tom has an obvious advantage, since it requires far less space for growing populations than for normal-sized genotypes (Meissner et al., 1997). As an example, the culture of the 343-plant BC1F2 population used in this study required as few as 5 m2. Furthermore, the mapping-by-sequencing method described uses simple and widely distributed bioinformatics tools and can be applied to any publicly available tomato mutant collection (Menda et al., 2004; Saito et al., 2011; Just et al., 2013).
SlGPAT6 Plays a Key Role in Cutin Biosynthesis during Early Tomato Fruit Development
MAGs, which form cutin building blocks, are synthesized through the action of long-chain acyl-CoA synthetases (LACS), cytochrome P450 enzymes (CYPs), and acyltransferases (GPATs). The monomers are then exported to the apoplast and polymerized on the outer face of the walls of epidermal cells to form cutin polyesters. In tomato, cutin polymerization was shown to be catalyzed by a member of the GDSL lipase family called SlCUS1 (Girard et al., 2012; Yeats et al., 2012b; Philippe et al., 2016). The following order of the reactions was recently suggested for MAG synthesis: the ω-hydroxylation of fatty acids catalyzed by members of the CYP86A family is followed by in-chain hydroxylation carried out by members of the CYP77A family, after which the GPATs catalyze the transfer to the glycerol backbone (Li-Beisson et al., 2009, 2013; Yang et al., 2012). LACS must precede GPAT to provide the acyl-CoA substrates (Yang et al., 2012; Li-Beisson et al., 2013).
A mutation in a tomato CYP gene, SlCYP86A69, which is regulated by the SlSHN3 TF, has been reported to cause major alterations in the structure of the fruit skin, including a substantial reduction in cuticle thickness (Shi et al., 2013). In a recent study, Lashbrooke et al. (2015) demonstrated that another CYP gene, SlCYP77A1, an SlMIXTA-regulated gene whose expression is reduced in the gpat6-a mutant (Table IV), is responsible for the synthesis of the midchain hydroxylated C16:0 diOH. In this study, we purified C16:0 diOH from tomato peel extracts and used this natural substrate in a yeast system, used previously to assay GPAT activity (Yang et al., 2010, 2012; Dittrich-Domergue et al., 2014), to demonstrate that recombinant SlGPAT6 synthesizes MAG (Fig. 4). It was shown previously that the Arabidopsis GPAT enzymes involved in cutin synthesis are AtGPAT4, AtGPAT6, and AtGPAT8 (Yang et al., 2010, 2012). They possess dual sn2-acyltransferase and phosphatase activities, thus producing predominantly sn2-MAGs (Yang et al., 2010, 2012). AtGPAT6 is essential for the accumulation of cutin in flowers (Li-Beisson et al., 2009). The enzyme has a higher affinity for C16 and C18 acyl-CoA substrates, C16:0 diOH being the dominant monomer in flower cutin (Li-Beisson et al., 2009). Mutation of a key amino acid residue in the phosphatase domain (K178L) of AtGPAT6 abolishes its MAG synthesis activity, due to the loss of phosphatase activity, but keeps the LPA synthesis activity (Yang et al., 2010). We observed that both LPA and MAG did not accumulate when tomato ΔGPAT6 activity was assayed (Fig. 4), suggesting that the G163R mutation, which occurs adjacent to the SlGPAT6 phosphatase domain, impairs both phosphatase and/or acyltransferase activities. As a consequence, the fruit cutin abundance and composition were altered substantially in the gpat6-a mutant, particularly with regard to levels of C16:0 diOH, in agreement with studies of the floral organs of the Arabidopsis gpat6-1 and gpat6-2 mutants (Li-Beisson et al., 2009; Fabre et al., 2016).
AtGPAT6 is expressed preferentially in inflorescences, where it plays diverse roles in the formation of the pollen exine and coat, in tapetum development, and in stamen elongation (Li-Beisson et al., 2009; Li et al., 2012). Accordingly, suppression of AtGPAT6 expression results in reduced fertility (Li-Beisson et al., 2009; Li et al., 2012). While SlGPAT6 also is highly expressed in inflorescence, mostly in unopened flowers, its expression is much higher (3-fold) in young growing fruit. The development of tomato flowers, anthers, and pollen was clearly altered in the gpat6-a mutant, although this had no effect on fruit set and size (Supplemental Fig. S4). The most striking phenotype was in the composition and amount of fruit cutin, as well as cuticle properties (glossiness and permeability to Toluidine Blue). Another GPAT, SlGPAT4 (Fig. 2), the Arabidopsis ortholog of which was shown to be involved in cutin biosynthesis (Yang et al., 2010), has a similar pattern of expression in the fruit, albeit weaker than SlGPAT6 in the fruit but stronger in the flower (http://bar.utoronto.ca/efp_tomato/cgi-bin/efpWeb.cgi). We propose that SlGPAT6 developed a specialized function in the fruit to accommodate the rapid production of polymeric cutin during the early stages of fruit development (Petit et al., 2014), while SlGPAT4 likely has an equivalent function in flowers.
Loss of SlGPAT6 Function Leads to Altered Expression of Genes Involved in Cuticle and Cell Wall Formation and Remodeling
Comparative RNA-seq profiling of the exocarp of gpat6-a and wild-type expanding fruit revealed expression differences in several gene categories, including not only genes involved in cutin or wax biosynthesis but also secondary metabolism and cell wall synthesis and modification. Since SlGPAT6 is necessary for the formation of MAG cutin monomers, it is conceivable that a feedback regulation due to MAG shortage and to the accumulation of MAG precursors in the epidermal cells may affect the expression of genes involved in cutin synthesis as well as in VLCFA synthesis, which shares common precursors with cutin (Li-Beisson et al., 2013). This effect would explain the large reduction in the accumulation of several cutin components as well as the lesser changes in the composition of VLCFAs. The fruit-expressed SlGPAT4 might be expected to compensate for the loss of SlGPAT6 activity, but we observed that SlGPAT4 transcript levels were more than 2-fold lower in the gpat6-a mutant than in the wild type (Table IV). These observations raise the question of why a mutation in a single structural gene triggers such profound alterations in the exocarp transcriptome and the mechanism by which this is mediated.
In recent years, considerable progress has been made in elucidating the factors that influence cuticle formation (Borisjuk et al., 2014), including advances resulting from the study of fleshy fruits (Hen-Avivi et al., 2014; Martin and Rose, 2014). Several families of TFs have been proposed as regulators of cuticle biosynthesis as well as epidermal cell differentiation and patterning. The first such example was the SHN gene family (Aharoni et al., 2004; Broun et al., 2004), whose members control wax and cutin accumulation in various plant species, including Arabidopsis and tomato, and that target genes involved in their synthesis (Aharoni et al., 2004; Shi et al., 2011, 2013). Interestingly, SHN genes have been shown to control epidermal cell elongation and patterning in Arabidopsis flowers (Shi et al., 2011). Moreover, when expressed in rice, the AtSHN2 gene was shown to regulate cell wall and lignin biosynthesis, possibly by targeting NAC and MYB TFs (Ambavaram et al., 2011). Taken together, these results indicate that SHN TFs may coordinate cuticle and cell wall biosynthesis in growing organs. In tomato fruit, members of various TF families have been shown to play prominent roles in cuticle formation and its coordination with (1) flavonoid biosynthesis, including the CD2 HD-Zip IV TF (Isaacson et al., 2009; Nadakuduti et al., 2012) and the MYB12 TF (Adato et al., 2009; Ballester et al., 2010), and (2) fruit ripening, including the FRUITFULL1 (FUL1), FUL2, and TAGL1 MADS box TFs (Bemer et al., 2012; Shima et al., 2013; Giménez et al., 2015). Accordingly, changes in the expression of genes encoding SHN and other known TFs might be expected in the gpat6-a mutant, but we did not observe such changes. However, genes in other TF families that were expressed at higher or lower levels in the gpat6-a mutant than in the wild type may fulfill related roles. These include the MADS box protein RIN, a regulator of fruit ripening that forms DNA-binding complexes with FUL1, FUL2, and TAGL1 (Fujisawa et al., 2014) and links cuticle formation to fruit development (Kosma et al., 2010). Additionally, the MYB SlMIXTA-like gene, which is expressed at lower levels in gpat6-a, was recently shown to play a major role in the control of cuticle formation and of epidermal patterning in tomato fruit (Lashbrooke et al., 2015). Strikingly, among the 17 cuticle-related genes whose expression was reduced in SlMIXTA RNA interference lines, 10 were also expressed at lower levels in the gpat6-a mutant. This suggests that SlMIXTA-like plays a role in the formation of fruit epidermis in response to the cutin deficiency induced by the SlGPAT6 mutation.
During tomato fruit growth, cells in the exocarp maintain a high mitotic activity (Joubès et al., 1999) to accommodate the rapid enlargement of the fruit (Lemaire-Chamley et al., 2005). This is accompanied by a massive increase in the deposition of cutin in the epidermal cell wall (Mintz-Oron et al., 2008; Isaacson et al., 2009) in order to seal the fruit surface. Cuticle properties also undergo profound changes during the cell expansion phase and are affected by cuticle composition, such as the accumulation of flavonoids (España et al., 2014). In addition to the cuticle, the cell wall plays a critical role in the differentiation and maturation of the fruit epidermis, and the cutin-polysaccharide ratio has been suggested to modulate the cuticle viscoelastic deformation necessary for fruit enlargement (Bargel and Neinhuis, 2005; Segado et al., 2016). The changes that we observed in the exocarp transcriptome of gpat6-a were associated with cuticle, phenylpropanoid, flavonoid, and cell wall biosynthesis and modification (Table IV), which is consistent with the remodeling of fruit epidermal cell walls and the overlying cuticle. Such changes may compensate for the cutin deficiency and allow fruit growth to proceed. The question remains how the inactivation of a single structural protein with no known transcriptional regulatory role triggers such a cascade of responses and suggests a mechanism for sensing cutin deficiency or cuticle biophysical properties. This hypothesis may be addressed through future studies of other genotypes, such as single or double mutants affected in various steps of cutin biosynthesis and assembly.
MATERIALS AND METHODS
Plant Materials
The tomato (Solanum lycopersicum) glossy mutant line P23F12 was isolated from an EMS mutant tomato collection generated in the miniature cv Micro-Tom at the Institut National de la Recherche Agronomique in Bordeaux, France, as described previously (Just et al., 2013; Petit et al., 2014). Plant culture conditions were as described by Rothan et al. (2016). To generate the mapping-by-sequencing population, the P23F12 line was first crossed with the wild-type parental line. A single BC1F1 hybrid was then selfed, and the resulting BC1F2 seeds were collected and sown. The 343 plants from the BC1F2 population segregating for the glossy mutant trait were grown in a greenhouse in an area of 5 m2. Fruit brightness was measured at the mature green stage as described by Petit et al. (2014). Fruits were photographed under standardized conditions using a Walimex photographic light box (Walser).
Mapping-by-Sequencing
A mapping BC1F2 population of 343 plants was created by crossing the P23F12 cv Micro-Tom glossy mutant line with a wild-type cv Micro-Tom parental line. Two bulks were then constituted by pooling 80 plants displaying either a glossy fruit phenotype (glossy bulk) or 80 plants with a dull/moderately glossy phenotype (wild-type-like bulk). To this end, five leaf discs (5 mm diameter each) were collected from each BC1F2 plant (approximately 600 mg fresh weight) and pooled into the glossy bulk and in the wild-type-like bulk. The same amount of plant material also was collected from the wild-type parental line. Genomic DNA was extracted from each bulk and the parental line using a cetyl-trimethyl-ammonium bromide method as described by Petit et al. (2014). DNA was suspended in 200 µL of distilled water and quantified by fluorometric measurement with a Quant-it dsDNA assay kit (Invitrogen). Illumina paired-end shotgun-indexed libraries were prepared using the TruSeq DNA PCRF-Free LT Sample Preparation Kit according to the manufacturer’s instructions (Illumina). The libraries were validated using an Agilent High Sensitivity DNA chip (Agilent Technologies) and sequenced using an Illumina HiSeq 2000 at the Institut National de la Recherche Agronomique EPGV facility, operating in a 100-bp paired-end run mode. Raw fastq files were mapped to the tomato reference genome sequence S. lycopersicum build release SL2.50 (ftp://ftp.solgenomics.net/tomato_genome/wgs/assembly/build_2.50/S_lycopersicum_chromosomes.2.50.fa.gz) using BWA version 0.7.12 (Li and Durbin, 2009; http://bio-bwa.sourceforge.net/). Variant calling (SNPs and INDELs) was performed using SAMtools version 1.2 (Li et al., 2009; http://htslib.org). As the tomato reference genome (cv Heinz 1706) used to map the reads is distinct from that of cv Micro-Tom, the variants identified would include both cv Heinz 1706/cv Micro-Tom natural polymorphisms in addition to EMS mutations. In this context, additional sequencing to a minimum depth of 20× of the cv Micro-Tom line was performed to take into account and further remove the cv Heinz 1706/cv Micro-Tom natural polymorphism.
The output file included various quality parameters relevant to sequencing and mapping that were subsequently used to filter the variants. The cv Micro-Tom line output file (.vcf) included all variants (SNPs plus INDELs) corresponding to natural polymorphisms between cv Micro-Tom and cv Heinz 1706. The two .vcf output files obtained from the glossy and wild-type-like bulks included variants (SNPs plus INDELs) corresponding to natural polymorphisms between cv Micro-Tom and cv Heinz 1706 and also to EMS mutations. The .vcf files were annotated using SnpEff version 4.1 (http://snpeff.sourceforge.net/SnpEff.html; Cingolani et al., 2012) using ITAG2.40 gene models (ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/annotation/ITAG2.4_release/ITAG2.4_gene_models.gff3). SNP allelic frequencies between glossy and wild-type-like bulks and the cv Micro-Tom parental line were compared using a custom Python script version 2.6.5 (https://www.python.org).
Once the putative causal mutation was detected using the mapping-by-sequencing procedure, the EMS-induced SNPs flanking the putative mutation were used as markers for genotyping the BC1F2 individuals using a KASP assay (Smith and Maughan, 2015). Specific primer design was performed using batchprimer3 software (Smith and Maughan, 2015; http://probes.pw.usda.gov/batchprimer), and genotyping was done using KASP procedures (LGC Genomics).
SlGPAT6 Phylogenetic Analysis and in Silico Modeling
Arabidopsis (Arabidopsis thaliana) and tomato databases (The Arabidopsis Information Resource [https://www.arabidopsis.org] and SGN [http://solgenomics.net], respectively) were searched for GPAT amino acid sequences using MEGA 6.0 (Tamura et al., 2013). A neighbor-joining tree was then constructed using MEGA 6.0 with default parameter settings.
SlGPAT6 modeling was performed using Rasmol version 2.7.1 (Raswin Molecular Graphics; Sayle and Milner-White, 1995) with the crystal structure of the Methanococcus jannaschii phospho-Ser phosphatase complex (Protein Data Bank [http://www.rcsb.org] entry 1L7P) as a template.
Expression of Recombinant Wild-Type and Mutant SlGPAT6 Proteins
To construct the GPAT6 expression vectors, the cDNAs corresponding to the SlGPAT6 wild-type protein (GPAT6) and the mutated form (ΔGPAT6), with codon optimization for yeast expression as described previously (Dittrich-Domergue et al., 2014), were synthesized and then cloned into the plasmid pDONR221 using the Gene Synthesis & Express Cloning Service (Life Technologies). The forward primer was 1-22 attB1-Express (5′-ACTTTGTACAAAAAAGCAGGCT-3′) and the reverse primer was 1,546-1,566 attB2-Express (5′-ACCCAGCTTTCTTGTACAAAG-3′). The fragments were then introduced into the Gateway destination vector pVT102-U-GW containing a uracil selection cassette (Domergue et al., 2010), through an LR recombination reaction, following the manufacturer’s instructions (Thermo Fisher Scientific).
The recombinant plasmids and the control vector were then introduced into the GPAT GAT1-deficient Saccharomyces cerevisiae strain gat1Δ (Dittrich-Domergue et al., 2014). Transformation with the empty vector (pVT102-U-GW) and the recombinant plasmids was performed using a polyethylene glycol/lithium acetate protocol (Dittrich-Domergue et al., 2014). Selection of positive clones was made on minimal medium agar plates lacking uracil, as described by Ausubel et al. (1995).
Synthesis of Acyl-CoA Substrates and in Vitro Assays of GPAT6 Activity
Samples of 16-hydroxyhexadecanoic acid (98%) and hexadecane-1,16-dioic acid (greater than 98%) were purchased from Sigma-Aldrich France, and C16:0 diOH was purified from about 5 g of red ripe cv M82 tomato peel, using a protocol adapted from Kawaguchi et al. (1981). After 3 h of transmethylation in 5% H2SO4/methanol at 85°C, fatty acid methyl esters were separated by preparative TLC (HPTLC Silica Gel 60 plate [Merck]; diethyl ether:hexane:methanol 40:10:1 [v/v/v]) and eluted from the silica with methanol:water:MTBE 1:1:2 (v/v/v). The purified fatty acid methyl ester was saponified using 11 n potassium hydroxide during 1 h at 80°C. Acyl-CoA substrates were then synthesized from the corresponding fatty acids and purified as described previously (Yang et al., 2010).
Yeast microsomes were prepared as described by Yang et al. (2010), and enzyme activities were assayed as described by Dittrich-Domergue et al. (2014), except that the incubation step was carried out for 10 min at 30°C in 100 µL containing 40 µg of proteins. After lipid extraction, the organic phase was analyzed by TLC using HPTLC Silica Gel 60 plates and chloroform:methanol:water:acetic acid (65:25:3.8:0.2 [v/v/v/v]) as a solvent. Radiolabeled products were identified by comigration with standards, and samples were quantified by autoradiography using a Typhoon FLA9500 Molecular Imager (GE Healthcare) operating in 50-µm resolution mode.
Pollen Morphology and Viability
Pollen was collected from 20 flowers at anthesis stage, and fresh samples were stained on a glass slide with fluorescein diacetate, as described by Mandaokar and Browse (2009). Replicates were collected under similar climatic conditions (time of pollen collection, humidity, and temperature). Microscopic observations were performed under visible light and at λ = 495 nm with a Zeiss Axioplan microscope (Carl Zeiss).
Permeability Measurement of Fruit Cuticle
For measurements of cuticle permeability to stain, we used a protocol adapted from Tanaka et al. (2004). Mature green stage fruits collected from wild-type and gpat6-a mutant plants were placed in 0.1% Toluidine Blue solution for 16 h. Staining of the fruit surface was then visually scored and photographs were taken.
Light Microscopy Analysis of Fruit Cuticle
For cuticle thickness measurements, fruit exocarp (including the cuticle) was obtained from two independent red ripe stage fruits from wild-type and gpat6-a plants. Samples were fixed and embedded in OCT medium as described by Buda et al. (2009). For each fruit, 12-µm cryosections were produced using a Microm HM560 cryostat (Thermo Fisher Scientific) with C-type blade. Oil Red O (Alfa Aesar) stock solution (saturated solution in isopropyl alcohol) was diluted 3:2 with distilled water, mixed well, equilibrated at room temperature for 30 min, and filtered with a 0.45-µm pore size filter. The stain was added to the slides for 30 min in a petri dish containing water to maintain a humid atmosphere. The slides were then exposed to an alcohol series of 50%, 30%, 22%, 15%, and 8% isopropyl alcohol, and the slides were finally mounted in 8% isopropyl alcohol with a coverslip. The width of the cuticle layer was determined by measuring the thickness of the cutinized anticlinal cell wall between two adjacent epidermal cells, as described previously (Petit et al., 2014). Mean cuticle thickness was assessed from 20 measurements each of 31 sections.
Cutin and Wax Analysis
Cuticular waxes were extracted by fruit immersion for 30 s in 6 mL of chloroform containing 6 µg of docosane as an internal standard and subsequently analyzed as described previously (Petit et al., 2014). For the cutin monomer analysis, two 1-cm-diameter discs were isolated from a red ripe fruit epidermal peel, carefully scratched with a scalpel blade to remove exocarp cells, and incubated for 30 min in isopropanol at 85°C. The cutin was then delipidated, depolymerized, and analyzed as described previously (Petit et al., 2014).
RNA-seq Analysis
Twelve 1-cm-diameter discs of epidermal peels were isolated from six 20-DPA fruits collected from three independent wild-type and gpat6-a mutant plants and carefully scratched with a scalpel blade, as above. Two discs per fruit were collected in distinct pools, and three pools were made in order to obtain three biological replicates. RNA was extracted using Trizol reagent (Invitrogen). Samples were ground in liquid nitrogen and stored at −80°C until RNA extraction, as described previously (Mounet et al., 2009). Total RNA integrity and concentration were assessed using an Agilent 2100 Bioanalyzer with RNA Nano Chip (Agilent Technologies). Total RNA samples were converted to cDNA libraries using a TruSeq RNA Sample Preparation Kit (Illumina) according to the manufacturer’s instructions. The final library size distribution was determined using a Fragment Analyzer (Advanced Analytical Technologies). The insert size comprised between 200 and 420 bp, and the average insert size of the library was 274 bp. Three libraries were multiplexed, pooled, and sequenced on a quarter lane of a HiSeq 2500 (Illumina), operating in 125-bp paired-end read mode.
Bioinformatic analysis was performed using the TUXEDO analysis pipeline (Trapnell et al., 2012). Filtered RNA-seq reads were aligned to the tomato reference genome sequence S. lycopersicum build release SL2.50 using TopHat2 (version 2.0.13; Kim et al., 2013). The resulting TopHat alignments were then fed into Cufflinks (version 2.2.1) to assemble the transcripts. Annotated transcripts were obtained using ITAG2.40 gene models. Transcript abundance estimates were measured in fragments per kilobase per million reads. Cuffdiff and CummerRbund were used to determine differential gene expression profiles (Trapnell et al., 2012).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Mapping-by-sequencing strategy.
Supplemental Figure S2. Acylglycerol metabolism in a S. cerevisiae double knockout gat1Dgat2D strain rescued by GAT1.
Supplemental Figure S3. SlGPAT6 gene expression in tomato flower, fruit, leaves, and root.
Supplemental Figure S4. Comparison of fruits (RR stage) and seeds from the wild type and gpat6-a mutant.
Supplemental Figure S5. Functional categories of differentially expressed genes in gpat6-a mutant.
Supplemental Table S1. Sequencing of the BC1F2 individuals from glossy and wild-type-like fruit bulks.
Supplemental Table S2. Number of SNPs in the glossy and wild-type-like bulks.
Supplemental Table S3. Allelic frequency analysis in the glossy and wild-type-like bulks.
Supplemental Table S4. Genes detected before and after the differential analysis.
Supplemental Table S5. Transcription factors showing differential expression in the gpat6-a mutant.
Supplementary Material
Acknowledgments
We thank Marie Christine Le Paslier, Dominique Brunel, and their staff at CEA-IG/CNG, who performed quality control of DNA and Illumina sequencing; Sarah Maman of the Genotoul bioinformatics platform Toulouse Midi-Pyrenees and the Sigenae group for providing help and/or computing and/or storage resources; Frédéric Delmas for helpful comments regarding pollen analysis; Isabelle Atienza for taking care of the plants in the greenhouse; Christel Baudet from the Magendie Neurocentre for the cryostat training; Jonathan Couillet and Ouerdia Hatem for the cytological analysis; and Weili Yang and John Ohlrogge for advice concerning acyl-CoA synthesis.
Glossary
- MAG
monoacylglycerol
- EMS
ethyl methanesulfonate
- SNP
single-nucleotide polymorphism
- AF
allelic frequency
- INDELs
insertions/deletions
- cDNA
complementary DNA
- TLC
thin-layer chromatography
- LPA
lysophosphatidic acid
- PA
phosphatidic acid
- DAG
diacylglycerol
- TAG
triacylglycerol
- VLCFA
very-long-chain fatty acid
- VLC
very-long-chain
- RNA-seq
RNA sequencing
- GPI
glycosylphosphatidylinositol
- TF
transcription factor
Footnotes
This work was supported by the Institut National de la Recherche Agronomique AIP Bioressources, ERA-NET Plant Genomics TomQML, and ANR Bioadapt (grant no. ANR–13–BSV7–0012) Adaptom project, by the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant no. 2015–06803 to J.K.C.R.), and by the U.S. National Science Foundation Plant Genome Research Program (grant no. IOS–1339287 to J.K.C.R.).
Articles can be viewed without a subscription.
References
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, et al. (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30: 174–178 [DOI] [PubMed] [Google Scholar]
- Adato A, Mandel T, Mintz-Oron S, Venger I, Levy D, Yativ M, Domínguez E, Wang Z, De Vos RC, Jetter R, et al. (2009) Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet 5: e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A (2011) Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 155: 916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K, Weys S, Paltauf F, Daum G (1999) Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae. J Bacteriol 181: 1458–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Katagiri F, Mindrinos M, Glazebrook J (1995) Use of Arabidopsis thaliana defense-related mutants to dissect the plant response to pathogens. Proc Natl Acad Sci USA 92: 4189–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester AR, Molthoff J, de Vos R, Hekkert B, Orzaez D, Fernández-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, et al. (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargel H, Neinhuis C (2005) Tomato (Lycopersicon esculentum Mill.) fruit growth and ripening as related to the biomechanical properties of fruit skin and isolated cuticle. J Exp Bot 56: 1049–1060 [DOI] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto PdeB, Angenent GC, de Maagd RA (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure JD, Haslam RP, Napier JA, Lessire R, Joubès J (2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Joubès J (2013) Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog Lipid Res 52: 110–129 [DOI] [PubMed] [Google Scholar]
- Borisjuk N, Hrmova M, Lopato S (2014) Transcriptional regulation of cuticle biosynthesis. Biotechnol Adv 32: 526–540 [DOI] [PubMed] [Google Scholar]
- Bourdenx B, Bernard A, Domergue F, Pascal S, Léger A, Roby D, Pervent M, Vile D, Haslam RP, Napier JA, et al. (2011) Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol 156: 29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buda GJ, Isaacson T, Matas AJ, Paolillo DJ, Rose JKC (2009) Three-dimensional imaging of plant cuticle architecture using confocal scanning laser microscopy. Plant J 60: 378–385 [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R (2011) Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J Exp Bot 62: 841–853 [DOI] [PubMed] [Google Scholar]
- Buxdorf K, Rubinsky G, Barda O, Burdman S, Aharoni A, Levy M (2014) The transcription factor SlSHINE3 modulates defense responses in tomato plants. Plant Mol Biol 84: 37–47 [DOI] [PubMed] [Google Scholar]
- Cao Y, Tang X, Giovannoni J, Xiao F, Liu Y (2012) Functional characterization of a tomato COBRA-like gene functioning in fruit development and ripening. BMC Plant Biol 12: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Matas AJ, Isaacson T, Kehlet C, Rose JK, Stark RE (2016) Solid-state 13C NMR delineates the architectural design of biopolymers in native and genetically altered tomato fruit cuticles. Biomacromolecules 17: 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Dittrich-Domergue F, Joubès J, Moreau P, Lessire R, Stymne S, Domergue F (2014) The bifunctional protein TtFARAT from Tetrahymena thermophila catalyzes the formation of both precursors required to initiate ether lipid biosynthesis. J Biol Chem 289: 21984–21994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, Ono J, Lee JA, Bourdon M, Alhattab R, Lowe C, Pascal S, Lessire R, et al. (2010) Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol 153: 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- España L, Heredia-Guerrero JA, Segado P, Benítez JJ, Heredia A, Domínguez E (2014) Biomechanical properties of the tomato (Solanum lycopersicum) fruit cuticle during development are modulated by changes in the relative amounts of its components. New Phytol 202: 790–802 [DOI] [PubMed] [Google Scholar]
- Fabre G, Garroum I, Mazurek S, Daraspe J, Mucciolo A, Sankar M, Humbel BM, Nawrath C (2016) The ABCG transporter PEC1/ABCG32 is required for the formation of the developing leaf cuticle in Arabidopsis. New Phytol 209: 192–201 [DOI] [PubMed] [Google Scholar]
- Farinas B, Mas P (2011) Histone acetylation and the circadian clock: a role for the MYB transcription factor RVE8/LCL5. Plant Signal Behav 6: 541–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fich EA, Segerson NA, Rose JKC (2016) The plant polyester cutin: biosynthesis, structure, and biological roles. Annu Rev Plant Biol 67: 207–233 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Nakagawa H, Kitagawa M, Kimbara J, Nakano T, Kasumi T, Ito Y (2014) Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez E, Dominguez E, Pineda B, Heredia A, Moreno V, Lozano R, Angosto T (2015) Transcriptional activity of the MADS box ARLEQUIN/TOMATO AGAMOUS-LIKE1 gene is required for cuticle development of tomato fruit. Plant Physiol 168: 1036–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard AL, Mounet F, Lemaire-Chamley M, Gaillard C, Elmorjani K, Vivancos J, Runavot JL, Quemener B, Petit J, Germain V, et al. (2012) Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24: 3119–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hen-Avivi S, Lashbrooke J, Costa F, Aharoni A (2014) Scratching the surface: genetic regulation of cuticle assembly in fleshy fruit. J Exp Bot 65: 4653–4664 [DOI] [PubMed] [Google Scholar]
- Hovav R, Chehanovsky N, Moy M, Jetter R, Schaffer AA (2007) The identification of a gene (Cwp1), silenced during Solanum evolution, which causes cuticle microfissuring and dehydration when expressed in tomato fruit. Plant J 52: 627–639 [DOI] [PubMed] [Google Scholar]
- Isaacson T, Kosma DK, Matas AJ, Buda GJ, He Y, Yu B, Pravitasari A, Batteas JD, Stark RE, Jenks MA, et al. (2009) Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J 60: 363–377 [DOI] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Rogowsky PM, Ingram GC (2011) Epidermis: the formation and functions of a fundamental plant tissue. New Phytol 189: 17–39 [DOI] [PubMed] [Google Scholar]
- Johnson KL, Kibble NA, Bacic A, Schultz CJ (2011) A fasciclin-like arabinogalactan-protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS ONE 6: e25154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Phan TH, Just D, Rothan C, Bergounioux C, Raymond P, Chevalier C (1999) Molecular and biochemical characterization of the involvement of cyclin-dependent kinase A during the early development of tomato fruit. Plant Physiol 121: 857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just D, Garcia V, Fernandez L, Bres C, Mauxion JP, Petit J, Jorly J, Assali J, Bournonville C, Ferrand C, et al. (2013) Micro-Tom mutants for functional analysis of target genes and discovery of new alleles in tomato. Plant Biotechnol 30: 225–231 [Google Scholar]
- Kawaguchi A, Yoshimura T, Okuda S (1981) A new method for the preparation of acyl-CoA thioesters. J Biochem 89: 337–339 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbara J, Yoshida M, Ito H, Kitagawa M, Takada W, Hayashi K, Shibutani Y, Kusano M, Okazaki Y, Nakabayashi R, et al. (2013) Inhibition of CUTIN DEFICIENT 2 causes defects in cuticle function and structure and metabolite changes in tomato fruit. Plant Cell Physiol 54: 1535–1548 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nagasaki H, Garcia V, Just D, Bres C, Mauxion JP, Le Paslier MC, Brunel D, Suda K, Minakuchi Y, et al. (2014) Genome-wide analysis of intraspecific DNA polymorphism in ‘Micro-Tom’, a model cultivar of tomato (Solanum lycopersicum). Plant Cell Physiol 55: 445–454 [DOI] [PubMed] [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA (2009) The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Parsons EP, Isaacson T, Lü S, Rose JK, Jenks MA (2010) Fruit cuticle lipid composition during development in tomato ripening mutants. Physiol Plant 139: 107–117 [DOI] [PubMed] [Google Scholar]
- Lashbrooke J, Adato A, Lotan O, Alkan N, Tsimbalist T, Rechav K, Fernandez-Moreno JP, Widemann E, Grausem B, Pinot F, et al. (2015) The tomato MIXTA-like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiol 169: 2553–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Garcia V, Just D, Baldet P, Germain V, Fagard M, Mouassite M, Cheniclet C, Rothan C (2005) Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol 139: 750–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Zhu J, Yang J, Zhang GR, Xing WF, Zhang S, Yang ZN (2012) Glycerol-3-phosphate acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Mol Plant 5: 131–142 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, doi/10.1199/tab.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Lima Gomes B, Mila I, Purgatto E, Peres LE, Frasse P, Maza E, Zouine M, Roustan JP, Bouzayen M, et al. (2016) Comprehensive profiling of Ethylene Response Factors expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato (Solanum lycopersicum). Plant Physiol 170: 1732–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG (2010) Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J 62: 689–703 [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Browse J (2009) MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LB, Rose JKC (2014) There’s more than one way to skin a fruit: formation and functions of fruit cuticles. J Exp Bot 65: 4639–4651 [DOI] [PubMed] [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, et al. (2011) Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R, Jacobson Y, Melamed S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A (1997) A new model system for tomato genetics. Plant J 12: 1465–1472 [Google Scholar]
- Mejía N, Soto B, Guerrero M, Casanueva X, Houel C, Miccono MdeL, Ramos R, Le Cunff L, Boursiquot JM, Hinrichsen P, et al. (2011) Molecular, genetic and transcriptional evidence for a role of VvAGL11 in stenospermocarpic seedlessness in grapevine. BMC Plant Biol 11: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, et al. (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M, et al. (2009) Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol 149: 1505–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Levesque-Tremblay G, Fernandes A, Wormit A, Bartels S, Usadel B, Kermode A (2013) Overexpression of a pectin methylesterase inhibitor in Arabidopsis thaliana leads to altered growth morphology of the stem and defective organ separation. Plant Signal Behav 8: e26464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti SS, Pollard M, Kosma DK, Allen C Jr, Ohlrogge JB, Barry CS (2012) Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiol 159: 945–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C. (2006) Unraveling the complex network of cuticular structure and function. Curr Opin Plant Biol 9: 281–287 [DOI] [PubMed] [Google Scholar]
- Negruk V, Yang P, Subramanian M, McNevin JP, Lemieux B (1996) Molecular cloning and characterization of the CER2 gene of Arabidopsis thaliana. Plant J 9: 137–145 [DOI] [PubMed] [Google Scholar]
- Niu E, Shang X, Cheng C, Bao J, Zeng Y, Cai C, Du X, Guo W (2015) Comprehensive analysis of the COBRA-like (COBL) gene family in Gossypium identifies two COBLs potentially associated with fiber quality. PLoS ONE 10: e0145725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Mitsuda N (2013) The MIXTA-like transcription factor MYB16 is a major regulator of cuticle formation in vegetative organs. Plant Signal Behav 8: e26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol 56: 180–194 [DOI] [PubMed] [Google Scholar]
- Petit J, Bres C, Just D, Garcia V, Mauxion JP, Marion D, Bakan B, Joubès J, Domergue F, Rothan C (2014) Analyses of tomato fruit brightness mutants uncover both cutin-deficient and cutin-abundant mutants and a new hypomorphic allele of GDSL lipase. Plant Physiol 164: 888–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe G, Gaillard C, Petit J, Geneix N, Dalgalarrondo M, Bres C, Mauxion JP, Franke R, Rothan C, Schreiber L, et al. (2016) Ester cross-link profiling of the cutin polymer of wild-type and cutin synthase tomato mutants highlights different mechanisms of polymerization. Plant Physiol 170: 807–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Ebert B, Ouellet M, Nafisi M, Baidoo EE, Benke P, Stranne M, Mukhopadhyay A, Keasling JD, Sakuragi Y, et al. (2012) Arabidopsis Deficient in Cutin Ferulate encodes a transferase required for feruloylation of ω-hydroxy fatty acids in cutin polyester. Plant Physiol 158: 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reca IB, Lionetti V, Camardella L, D’Avino R, Giardina T, Cervone F, Bellincampi D (2012) A functional pectin methylesterase inhibitor protein (SolyPMEI) is expressed during tomato fruit ripening and interacts with PME-1. Plant Mol Biol 79: 429–442 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Rothan C, Just D, Fernandez L, Atienza I, Ballias P, Lemaire-Chamley M (2016) Culture of the tomato Micro-Tom cultivar in greenhouse. Methods Mol Biol 1363: 57–64 [DOI] [PubMed] [Google Scholar]
- Saito T, Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N, Mizoguchi T, Yamazaki Y, Aoki K, Ezura H (2011) TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol 52: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ (1995) RASMOL: biomolecular graphics for all. Trends Biochem Sci 20: 374. [DOI] [PubMed] [Google Scholar]
- Schneeberger K. (2014) Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat Rev Genet 15: 662–676 [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber L. (2010) Transport barriers made of cutin, suberin and associated waxes. Trends Plant Sci 15: 546–553 [DOI] [PubMed] [Google Scholar]
- Segado P, Domínguez E, Heredia A (2016) Ultrastructure of the epidermal cell wall and cuticle of tomato fruit (Solanum lycopersicum L.) during development. Plant Physiol 170: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Xue H, Acet T (2014) The Arabidopsis thaliana FASCICLIN LIKE ARABINOGALACTAN PROTEIN 4 gene acts synergistically with abscisic acid signalling to control root growth. Ann Bot (Lond) 114: 1125–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Sawai S, Ohyama K, Mizutani M, Ohnishi T, Sudo H, Fukushima EO, Akashi T, Aoki T, Saito K, et al. (2011) Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 23: 4112–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JX, Adato A, Alkan N, He Y, Lashbrooke J, Matas AJ, Meir S, Malitsky S, Isaacson T, Prusky D, et al. (2013) The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol 197: 468–480 [DOI] [PubMed] [Google Scholar]
- Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A (2011) SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet 7: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Kitagawa M, Fujisawa M, Nakano T, Kato H, Kimbara J, Kasumi T, Ito Y (2013) Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol Biol 82: 427–438 [DOI] [PubMed] [Google Scholar]
- Shirasawa K, Hirakawa H, Nunome T, Tabata S, Isobe S (2016) Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol J 14: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa K, Isobe S, Hirakawa H, Asamizu E, Fukuoka H, Just D, Rothan C, Sasamoto S, Fujishiro T, Kishida Y, et al. (2010) SNP discovery and linkage map construction in cultivated tomato. DNA Res 17: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40: W452–W457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Gross KC (2000) A family of at least seven β-galactosidase genes is expressed during tomato fruit development. Plant Physiol 123: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Maughan PJ (2015) SNP genotyping using KASPar assays. Methods Mol Biol 1245: 243–256 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y (2004) A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Usadel B, Thimm O, Nunes-Nesi A, Carrari F, Davy M, Bläsing O, Kowalczyk M, Weicht D, Polinceusz A, et al. (2006) Conversion of MapMan to allow the analysis of transcript data from solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol 60: 773–792 [DOI] [PubMed] [Google Scholar]
- Valverde F. (2011) CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J Exp Bot 62: 2453–2463 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Yang W, Pollard M, Li-Beisson Y, Beisson F, Feig M, Ohlrogge J (2010) A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc Natl Acad Sci USA 107: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB (2012) A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference, and evolution. Plant Physiol 160: 638–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Buda GJ, Wang Z, Chehanovsky N, Moyle LC, Jetter R, Schaffer AA, Rose JKC (2012a) The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J 69: 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Martin LB, Viart HM, Isaacson T, He Y, Zhao L, Matas AJ, Buda GJ, Domozych DS, Clausen MH, et al. (2012b) The identification of cutin synthase: formation of the plant polyester cutin. Nat Chem Biol 8: 609–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC (2008) The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci 17: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.