Putrescine acetylation in Arabidopsis thaliana reduces production of hydrogen peroxide by polyamine oxidases, thereby promoting plant susceptibility to Pseudomonas syringae.
Abstract
Biosynthesis of the polyamines putrescine, spermidine, and spermine is induced in response to pathogen infection of plants. Putrescine, which is produced from Arg, serves as a metabolic precursor for longer polyamines, including spermidine and spermine. Polyamine acetylation, which has important regulatory functions in mammalian cells, has been observed in several plant species. Here we show that Arabidopsis (Arabidopsis thaliana) N-ACETYLTRANSFERASE ACTIVITY1 (NATA1) catalyzes acetylation of putrescine to N-acetylputrescine and thereby competes with spermidine synthase for a common substrate. NATA1 expression is strongly induced by the plant defense signaling molecule jasmonic acid and coronatine, an effector molecule produced by DC3000, a Pseudomonas syringae strain that initiates a virulent infection in Arabidopsis ecotype Columbia-0. DC3000 growth is reduced in nata1 mutant Arabidopsis, suggesting a role for NATA1-mediated putrescine acetylation in suppressing antimicrobial defenses. During infection by P. syringae and other plant pathogens, polyamine oxidases use spermidine and spermine as substrates for the production of defense-related H2O2. Compared to wild-type Columbia-0 Arabidopsis, the response of nata1mutants to P. syringae infection includes reduced accumulation of acetylputrescine, greater abundance of nonacetylated polyamines, elevated H2O2 production by polyamine oxidases, and higher expression of genes related to pathogen defense. Together, these results are consistent with a model whereby P. syringae growth is improved in a targeted manner through coronatine-induced putrescine acetylation by NATA1.
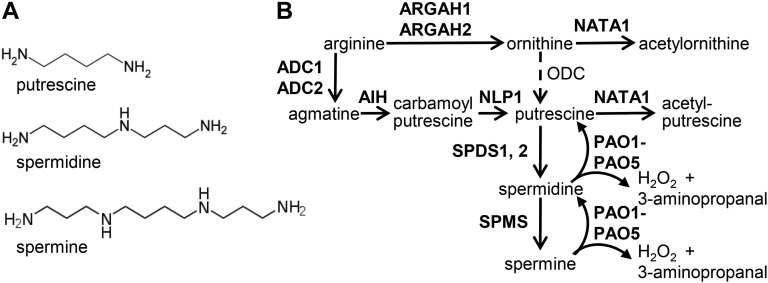
Plant polyamines, including putrescine, spermidine, and spermine (Fig. 1A), accumulate in response to a variety of biotic and abiotic stresses (Walters, 2003a, 2003b; Minocha et al., 2014). Synthesis of putrescine from Arg occurs via two pathways, with either Orn or agmatine as an intermediate (Fig. 1B). Although there are two arginases (ARGAH1 and ARGAH2) in the Arabidopsis (Arabidopsis thaliana) genome (Brownfield et al., 2008), Orn decarboxylase is apparently absent, indicating that, unlike in most other plant species, Orn is not a direct metabolic precursor for putrescine synthesis in Arabidopsis (Hanfrey et al., 2001). Putrescine, in turn, is converted into spermidine through the addition of an aminopropyl group from decarboxylated S-adenosyl-Met by spermidine synthases (SPDS1 and SPDS2). An additional aminopropyl group is added to spermidine to form spermine by spermine synthase (SPMS).
Figure 1.
Polyamine metabolism in Arabidopsis. A, Structures of three common plant polyamines. B, Pathways of polyamine synthesis and catabolism in Arabidopsis. Known Arabidopsis enzymes are indicated in bold. Orn decarboxylase is found in other plants, but is not present in Arabidopsis.
Polyamine acetylation, which has important regulatory functions in mammalian stress responses (Pegg, 2008; Tavladoraki et al., 2012), also has been observed in plants. N-acetylputrescine was detected in Daucus carota, Nicotiana plumbaginifolia, and Datura stramonium (Mesnard et al., 2000; Fliniaux et al., 2004). N1-acetylspermine, N1-acetylspermidine, and N8-acetylspermidine have been found in several plant species (Dufeu et al., 2003; Hennion et al., 2012). In Arabidopsis, N1-acetylspermidine is as abundant as spermidine in both roots and above-ground tissue (Kamada-Nobusada et al., 2008). However, despite the apparently widespread distribution of acetylated polyamines in the plant kingdom, there has been as yet no in-depth investigation of functions of these plant metabolites and their biosynthetic enzymes.
Polyamine catabolism is mediated by two classes of polyamine oxidases: the copper amine oxidases (EC 1.4.3.6) and the flavin-containing polyamine oxidases (PAOs; EC 1.5.3.11; Fig. 1B; Bagni and Tassoni, 2001; Moschou et al., 2008; Alcázar et al., 2010; Moschou et al., 2012). Copper amine oxidases form homodimers and can catalyze the oxidation of putrescine at its primary amino group, producing H2O2, NH4+, and 4-aminobutanal (Cona et al., 2006; Angelini et al., 2010). Arabidopsis PAOs, on the other hand, function in a polyamine back-conversion pathway; oxidizing the carbon at the exo-side of the N4-nitrogen of the triamine spermidine, and the tetraamines spermine and thermospermine, thereby producing putrescine and spermidine, respectively, in addition to 3-aminopropanal and H2O2 (Fig. 1B; Moschou et al., 2012; Ono et al., 2012; Tavladoraki et al., 2012; Ahou et al., 2014; Kim et al., 2014).
Five PAO homologs have been identified in Arabidopsis; whereas PAO1 and PAO5 are cytosolic enzymes, PAO2, PAO3, and PAO4 are targeted to the peroxisomes (Kamada-Nobusada et al., 2008; Moschou et al., 2008; Ahou et al., 2014; Kim et al., 2014). Polyamine oxidation by PAOs is a major source of stress-induced H2O2 accumulation in plant tissue (Yoda et al., 2009). Both necrotrophic and biotrophic pathogens can induce H2O2 production via polyamine oxidation (Marina et al., 2008; Moschou et al., 2009; Angelini et al., 2010), and up-regulation of polyamine biosynthesis has been reported in plants infected with a variety of viral, bacterial, and fungal pathogens (Walters, 2003a, 2003b). Arabidopsis PAOs oxidize spermine and spermidine more efficiently than N1-acetylspermine and N1-acetylspermidine (Moschou et al., 2008; Kim et al., 2014), suggesting that nonacetylated polyamines are more likely to be physiologically relevant substrates of these enzymes.
Coronatine, a toxin produced by some Pseudomonas syringae pathovars, including the commonly studied DC3000 strain (Xin and He, 2013), is a molecular mimic of the jasmonate-Ile conjugate that binds to the COI1 (CORONATINE INSENSITIVE 1) receptor and activates jasmonate-specific defense responses (Brooks et al., 2004; Fonseca et al., 2009). In the case of bacteria on the leaf surface, coronatine can prevent flagellin-induced stomatal closure, thereby enabling bacterial entry into the leaves (Melotto et al., 2006). Arabidopsis NATA1 (N-ACETYLTRANSFERASE ACTIVITY1; At2g39030), which is one of the most strongly induced genes after jasmonate or coronatine treatment, is an Orn acetyltransferase that produces Nδ-acetyl-Orn from Orn (Adio et al., 2011). The NATA2 gene (At2g39020) is directly adjacent to NATA1 in the Arabidopsis genome and encodes a protein that is 80% identical to NATA1. However, unlike NATA1, NATA2 expression is not induced by jasmonic acid or coronatine (Adio et al., 2011). Instead, NATA2 is expressed at a uniformly high level throughout Arabidopsis development (Supplemental Fig. S1).
Recent in vitro assays with NATA1 (Jammes et al., 2014) showed that this enzyme also acetylates 1,3-diaminopropane, which has been reported as a product of polyamine oxidation by maize PAO (Terano and Suzuki, 1978). N-Acetyl-1,3-diaminopropane, in turn, is an antagonist of abscisic acid-induced stomatal closure (Jammes et al., 2014). Thus, it is hypothesized that, by inducing diaminopropane acetylation with coronatine, P. syringae may inhibit stomatal closure and facilitate entry into the leaf. Once P. syringae is growing inside of plant tissue, coronatine promotes virulence by activating jasmonate-regulated defenses and thereby suppressing pathogen-targeted defenses (Cui et al., 2005; Uppalapati et al., 2007).
Salicylic acid and H2O2 act synergistically to initiate both local and systemic plant responses to pathogen infection (Neuenschwander et al., 1995; Sharma et al., 1996; Alvarez et al., 1998; Mukherjee et al., 2010). A rapid increase in H2O2 production, initiated through the activity of NADPH oxidase, occurs as a hypersensitive response to pathogen infection in many plant species (Lamb and Dixon, 1997; Torres et al., 2005). In contrast, more sustained H2O2 accumulation during virulent pathogen infection occurs through degradation of spermidine and spermine by PAO activity and also contributes to plant defense (Marina et al., 2008; Moschou et al., 2009; Angelini et al., 2010). In addition to NADPH oxidase and PAO, Class III peroxidase activity is a source of H2O2 that can limit bacterial growth in Arabidopsis (Mitchell et al., 2015).
After infiltration into Arabidopsis leaves, virulent P. syringae DC3000 propagation was lower in nata1 mutant than in wild-type Columbia-0 (Col-0) plants (Adio et al., 2011), suggesting that NATA1 might function in suppressing antibacterial defenses. We now provide an explanation for this observation by showing that, in addition to using Orn and diaminopropane as substrates (Adio et al., 2011; Jammes et al., 2014), NATA1 functions as a putrescine acetyltransferase (Fig. 1B). Up-regulation of NATA1 expression by coronatine-producing P. syringae DC3000 causes elevated N-acetylputrescine production and reduced H2O2 production by polyamine oxidase. Higher H2O2 accumulation in nata1 mutants leads to increased expression of defense-related genes and reduced bacterial growth. Together, these observations indicate that, by inducing jasmonate-regulated polyamine acetylation, coronatine-producing P. syringae suppress pathogen-specific defenses.
RESULTS
Arabidopsis NATA1 Functions as a Putrescine Acetyltransferase
The Arabidopsis NATA1 and NATA2 genes are the closest homologs of known mammalian polyamine acetyltransferases (Supplemental Fig. S2). This similarity, together with the observation of acetylated polyamines in Arabidopsis and other plants (Mesnard et al., 2000; Fliniaux et al., 2004; Kamada-Nobusada et al., 2008; Hennion et al., 2012), suggested that NATA1 and NATA2 might be polyamine acetyltransferases. Although the predicted catalytic residues of the mammalian enzymes are conserved in the Arabidopsis NATA1 and NATA2 proteins, not all of the known spermine binding residues are present (Supplemental Fig. S3). Thus, Arabidopsis NATA1 and NATA2 likely acetylate amines, but the in vivo substrates might be different from those of the corresponding mammalian enzymes.
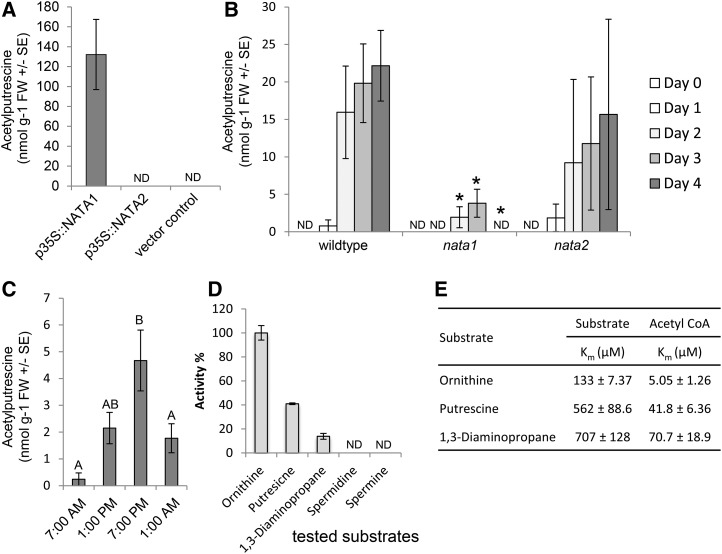
To determine whether NATA1 and/or NATA2 acetylate polyamines, the corresponding genes were expressed transiently in Nicotiana benthamiana. NATA1, but not NATA2, expression caused accumulation of N-acetylputrescine (Fig. 2A). Similar to what was observed in the case of Nδ-acetyl-Orn (Adio et al., 2011), N-acetylputrescine accumulation was induced at 2, 3, and 4 d after methyl jasmonate treatment of wild-type Col-0 plants (Fig. 2B; P < 0.05, t-test relative to day 0 control). Consistent with a role for NATA1 in Arabidopsis polyamine metabolism, this induction was significantly reduced in nata1 mutant plants (Fig. 2B). In contrast, a nata2 T-DNA insertion knockout did not have a significant effect on methyl jasmonate-induced N-acetylputrescine accumulation. Acetylated spermine and spermidine were either not present or accumulated at concentrations below the detection limit of our assay (∼20 nmol g−1 for acetylated spermine and spermidine added to Arabidopsis leaf extracts). Arabidopsis ecotypes with natural nata1 knockout mutations, which were identified through the 1001 Genomes project (Burren-0 [Bur-0], Ameland-1 [Amel-1], and Calver-0 [Cal-0]; http://1001genomes.org/; Supplemental Fig. S4), did not exhibit any detectable amounts of N-acetylputrescine and Nδ-acetyl-Orn accumulation in response to methyl jasmonate treatment. Consistent with the circadian cycle of jasmonate accumulation in plants (Goodspeed et al., 2012; Shin et al., 2012), we observed circadian regulation in the abundance of N-acetylputrescine (Fig. 2C). Maximum levels of N-acetylputrescine accumulation in this experiment were lower than those observed in response to methyl jasmonate or other elicitor treatments. However, given the observed circadian regulation, plant material for all subsequent induction experiments to measure gene expression or metabolite abundance was collected when the background N-acetylputrescine abundance was low, that is, within 3 hours of when the lights were turned on in the growth chambers at 7 am.
Figure 2.
Arabidopsis NATA1 is a putrescine acetyltransferase. A, p35S::NATA1 transient expression in N. benthamiana results in N-acetylputrescine production. Mean ± se of n = 12-14. B, Time course of N-acetylputrescine accumulation in rosette leaves after methyl jasmonate treatment. Mean ± se of n = 3, *P < 0.05, two-tailed t-test relative to wild type at the same time point. C, Accumulation of N-acetylputrescine in Arabidopsis is affected by the circadian clock. Mean ± se of n = 12-23. Plants were grown in a 16-h-light/8-h-dark cycle, with dawn at 7 am. Different letters indicate significant differences, P < 0.05, ANOVA followed by Tukey’s honest significant difference test. D, Relative activity of recombinant NATA1 enzyme, measured by release of CoA, in 100 mm Tris-HCl at pH 7.5 and 30°C. Substrate concentration was fixed at 5 mm and the acetyl-CoA concentration was fixed at 600 μM. Values are mean ± se of n = 3. E, Kinetic constants of substrate acetylation by recombinant NATA1. For Orn, putrescine, and 1,3 diamionopropane, Km was determined by Michaelis-Menten curve with a 1-mm fixed concentration of acetyl-CoA and varying substrate concentration from 25 to 1000 μM. For acetyl-CoA, Km was determined by Michaelis-Menten curve with a 5-mm fixed concentration of substrate and varying acetyl-CoA concentration from 1.25 to 600 μM. All data are mean ± se of three independent experiments. ND, not detected.
To confirm NATA1 catalytic activity and examine substrate preferences, in vitro assays were performed with Arabidopsis NATA1 protein that was purified after expression in Escherichia coli (Supplemental Fig. S5). Orn, putrescine, and 1,3-diaminopropane served as acetyltransferase substrates in vitro (Fig. 2D). Consistent with the absence of known spermine and spermidine binding residues in NATA1 (Supplemental Fig. S3), these two compounds did not serve as acetylation substrates in vitro. The apparent Km values for Orn, putrescine, 1,3-diaminopropane, and acetyl-coA were measured with purified NATA1 by varying the substrate concentrations and graphing Michaelis-Menten curves (Supplemental Fig. S6). Consistent with the relative NATA1 acetylation activity of Orn > putrescine > 1,3-diaminopropane (Fig. 2D), the relative Km values for these substrates were 1,3-diaminopropane > putrescine > Orn (Fig. 2E).
Virulent DC3000 Infection Induces Acetylputrescine Accumulation in a NATA1-Dependent Manner
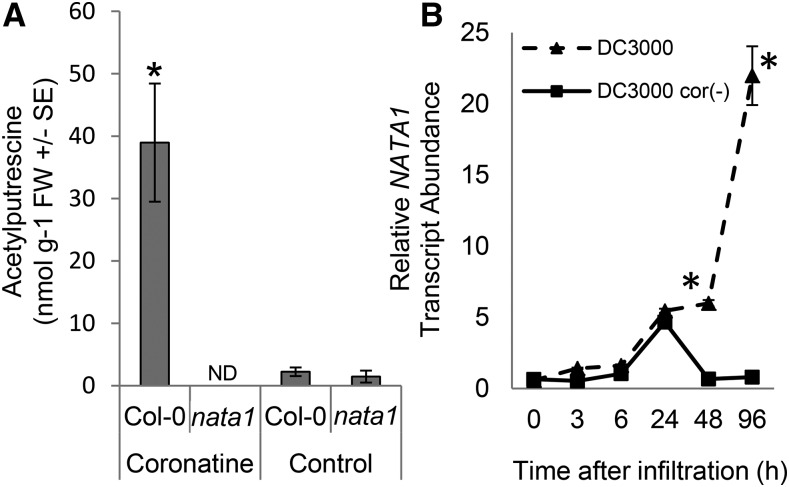
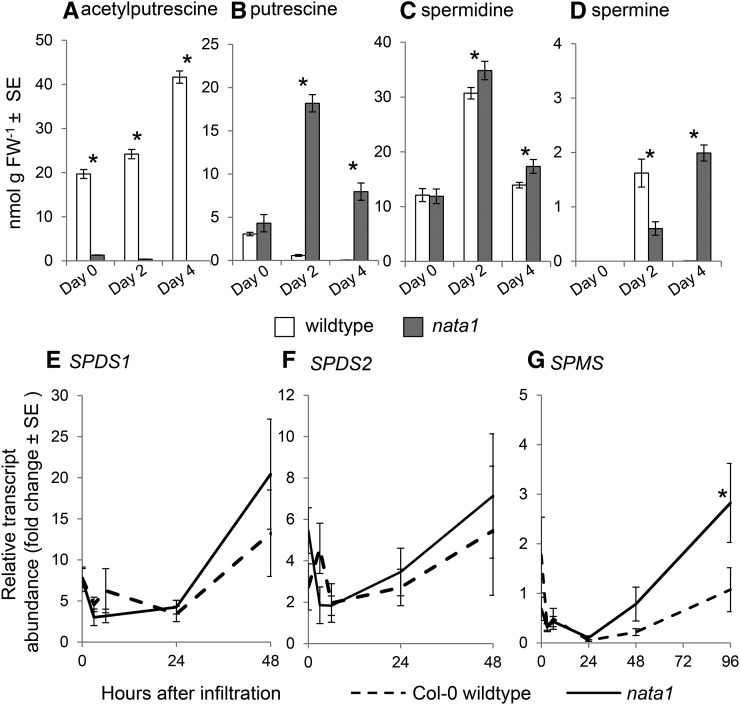
Similar to methyl jasmonate treatment (Fig. 2B), exogenous coronatine addition induced greater N-acetylputrescine accumulation in wild type than in nata1 mutant plants (Fig. 3A). Consistent with a role for coronatine in triggering putrescine acetylation, wild-type P. syringae strain DC3000 infiltration induced Arabidopsis NATA1 gene expression to a higher level than DC3000 cor- (Fig. 3B). DC3000 infection induced N-acetylputrescine accumulation in NATA1-dependent manner (Fig. 4A). Prior research demonstrated that, after methyl jasmonate treatment, putrescine and spermidine are significantly more abundant in nata1 than in wild-type Arabidopsis (Adio et al., 2011). Consistent with the observation that NATA1 converts putrescine to N-acetylputrescine, putrescine accumulated to a higher level during DC3000 infection of nata1 mutant than wild-type plants (Fig. 4B). Spermidine was slightly increased in nata1 relative to wild-type Arabidopsis (Fig. 4C), and spermine showed variable effects, with significantly lower levels in nata1 2 d after P. syringae infiltration and significantly higher levels after 4 d (Fig. 4D). These variable effects may reflect complex regulation of the polyamine pathway in response to stress, as well as the fact that polyamines are pathway intermediates and precursors of other plant metabolites. Thus, the steady-state levels may not be fully indicative of flux through the pathway. The transcript levels of spermidine and spermine synthases, SPDS1, SPDS2, and SPMS, were induced to comparable levels in nata1 mutant and wild-type Arabidopsis 2 d after P. syringae infiltration (Fig. 4, E and F). However, transcript abundance of SPMS was induced to a higher level in nata1 mutant plants 4 d after pathogen challenge (Fig. 4G), which may indicate increased metabolic flux through this pathway.
Figure 3.
P. syringae induces acetylputrescine accumulation in a NATA1-dependent manner. A, N-Acetylputrescine accumulation in rosette leaves 4 d after coronatine treatment. Mean ± se of n = 12 (control) or 9 (coronatine), *P < 0.05, two-tailed t-test comparing Col-0 and nata1. ND, not detected. B, NATA1 gene expression in wild-type Col-0 plants infiltrated with P. syringae DC3000 and DC3000 cor(-). NATA1 gene expression was measured by quantitative RT-PCR. Mean ± se of n = 6-9, *P < 0.05, two-tailed t test comparing mutant and wild type at the same time point.
Figure 4.
Polyamine concentrations and transcript abundance of spermidine and spermine synthases in wild-type Col-0 and nata1 mutant Arabidopsis infiltrated with P. syringae DC3000. N-acetylputrescine (A), putrescine (B), spermidine (C), and spermine (D). FW, fresh weight. Mean ± se of n = 6. *P < 0.05, two-tailed t-test. SPDS1 (E), SPDS2 (F), and SPM (G) expression was measured by quantitative RT-PCR. Mean ± se of n = 8-9, *P < 0.05, two-tailed t-test comparing expression in wild type and nata1 at the same time point.
H2O2 Induction by P. syringae Is Increased in nata1 Mutant Plants
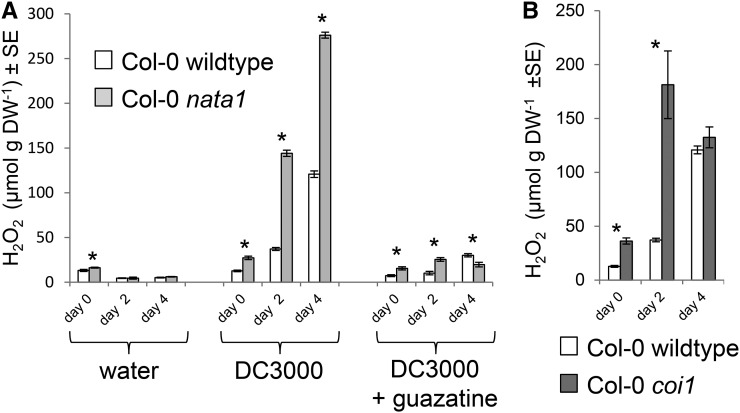
As H2O2 synthesis by polyamine oxidase commonly occurs during pathogen infection (Marina et al., 2008; Moschou et al., 2009; Angelini et al., 2010), we determined whether this response is altered in nata1 mutant Arabidopsis. In contrast to water-only controls, infiltration with virulent P. syringae strain DC3000 induced H2O2 to a higher level in nata1 than in wild-type Arabidopsis (Fig. 5A). H2O2 accumulation was greatly reduced if P. syringae bacteria were coinfiltrated with guazatine, an inhibitor of polyamine oxidase activity (Yoda et al., 2003). The final concentration of guazatine in the leaves was <2 µM, which does not cause significant negative effects on bacterial growth in vitro (Supplemental Fig. S7). Further experiments also showed that in planta P. syringae growth was not inhibited by this level of guazatine infiltration. DC3000 infection induced H2O2 accumulation to a higher level in the coi1 mutant than in wild-type Col-0 (Fig. 5B), indicating that recognition of coronatine by COI1 is required for the suppression of the H2O2 response. Suppression of the H2O2 increase by guazatine suggests that most of the H2O2 that was detected in these experiments is produced from polyamines by PAO.
Figure 5.
H2O2 induction by P. syringae infection. A, H2O2 content of wild-type Col-0 and nata1 mutant Arabidopsis infiltrated with water as a control, wild-type DC3000, and wild-type DC3000 together with guazatine, a polyamine oxidase inhibitor. Mean ± se of n = 6-10. B, H2O2 content in Col-0 wild-type and coi1 mutant Arabidopsis after infiltration with DC3000. Mean ± se of n = 6 for wild type, n = 9 for coi1. *P < 0.05, two-tailed t test. DW, dry weight.
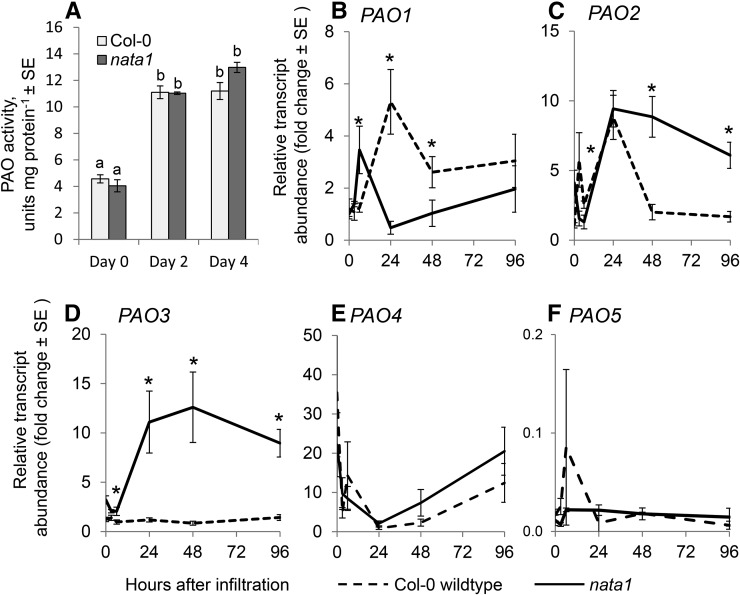
PAO activity was measured in Arabidopsis extracts to determine whether the observed differences in H2O2 accumulation could result from different levels of enzyme activity in wild-type and nata1 plants infiltrated with P. syringae. Two and 4 d after infection, there was a similar induction of polyamine oxidase enzymatic activity by DC3000 infiltration in the two Arabidopsis genotypes (Fig. 6A). Consistent with previously observed differential regulation of the five known Arabidopsis PAO genes (Fincato et al., 2012), the effects of DC3000 infection on PAO expression varied (Fig. 6). Whereas expression of PAO1, PAO2, PAO3, and PAO4 was up-regulated, that of PAO5 was not. However, no clear pattern was observed when comparing PAO transcript levels in the nata1 mutant and wild-type plants. Whereas PAO1 was expressed at a higher level in wild-type Col-0, PAO2 and PAO3 were expressed at a higher level in the nata1 mutant. There were no significant differences in the case of PAO4 and PAO5. Taken together, these data indicate that differential H2O2 accumulation in wild-type and nata1 plants may be caused by substrate availability rather than enzyme abundance.
Figure 6.
Polyamine oxidase responses to P. syringae. A, Col-0 wild-type and nata1 mutant leaves were infiltrated with P. syringae DC3000. Mean ± se of n = 3. Different letters indicate significant differences, P < 0.05, ANOVA followed by Tukey’s honest significant difference test. Transcript abundance of polyamine oxidases PAO1 (B), PAO2 (C), PAO3 (D), PAO4 (E), and PAO5 (F) was measured by quantitative RT-PCR at the indicated times after P. syringae infiltration into leaves. Mean ± se of n = 9, *P < 0.05, two-tailed t-test comparing wild type and nata1 expression at the same time point.
Coronatine-Mediated Induction of NATA1 Improves P. syringae Growth
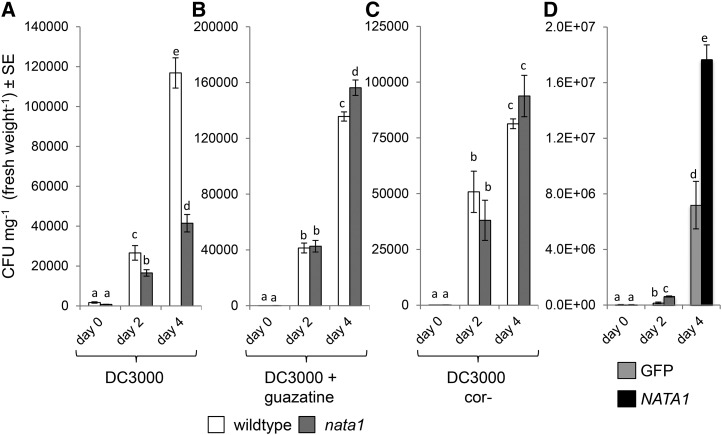
Bacterial growth experiments were conducted to determine whether NATA1 up-regulation by coronatine benefits P. syringae. Four days after infiltration, bacterial counts were 3-fold higher in wild-type Col-0 than in nata1 plants (Fig. 7A), similar to the effect observed by Adio et al. (2011). In contrast, in plants treated with guazatine at the time of P. syringae infiltration, bacteria in nata1 mutants did not grow to a lower final concentration than in wild type (Fig. 7B), showing that H2O2 production by PAO, which is inhibited by guazatine, contributes to plant defense. Consistent with this observation, there was also a significant increase in the bacterial titer in guazatine-treated wild-type Arabidopsis relative to controls 4 d after infiltration (comparing Fig. 7A and 7B; P < 0.05, two-tailed t test). Growth of DC3000 cor- was similar in wild-type and nata1 Arabidopsis, indicating that at least some suppression of plant defenses by coronatine occurs in a NATA1-dependent manner (Fig. 7C).
Figure 7.
Effects of NATA1 on P. syringae growth. Bacterial titers in wild-type Col-0 and nata1 rosette leaves after infiltrating 105 CFU mL−1 culture of DC3000, n = 30 (A); DC3000 and guazatine, n = 20 (B); DC3000 (cor-), n = 22-30 (C); different letters indicate significant differences, P < 0.05, ANOVA followed by Tukey’s honest significant difference test. D, Growth of P. syringae in N. benthamiana was measured over 4 d after infiltrating 108 CFU mL−1 into leaves transiently expressing NATA1 or GFP (control) genes from the constitutive cauliflower mosaic virus 35S promoter. n = 20-30. CFU, colony-forming units. Different letters indicate significant differences, P < 0.05, ANOVA followed by Tukey’s honest significant difference test.
To determine whether NATA1 activity can lead to a suppression of antibacterial defenses in other plants, P. syringae growth was assessed in N. benthamiana leaves that were transiently expressing NATA1. Unlike in the case of Arabidopsis, wild-type DC3000 effectors are recognized by N. benthamiana to mount a defense response. Therefore, to trigger a virulent infection that is comparable to the Arabidopsis experiments, a DC3000 ΔhrcQ-U mutant (Badel et al., 2006) was used to infect N. benthamiana. Consistent with the Arabidopsis results, DC3000 ΔhrcQ-U grew to a higher titer 2 and 4 d after infiltration into N. benthamiana that was transiently expressing NATA1 compared to control plants expressing a GFP gene (Fig. 7D).
Differential Responses to P. syringae in Wild-Type and nata1 Arabidopsis
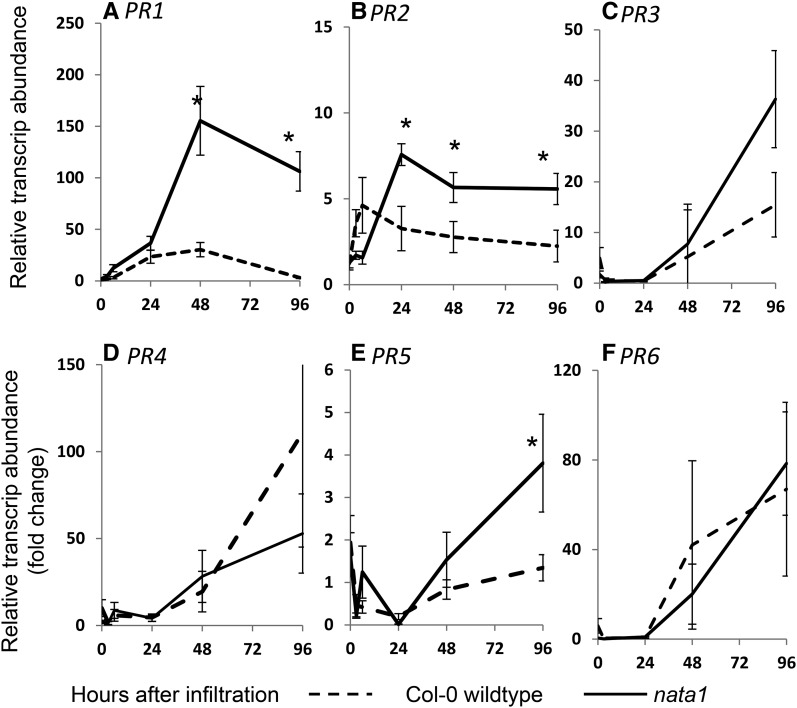
Reduced P. syringae growth in nata1 mutant Arabidopsis could be due to the induction of other defense responses. Therefore, quantitative RT-PCR assays were used to determine whether known defense-related genes are differentially regulated by P. syringae infection of wild-type and nata1 Arabidopsis leaves. The expression levels of PR1, PR2, and PR5 are commonly used as markers for the induction of antimicrobial defense responses in Arabidopsis. Consistent with the elevated P. syringae resistance of nata1 plants (Fig. 7A), P. syringae infiltration induced PR1 expression to a significantly higher level than in wild-type Arabidopsis (Fig. 8A). Induced PR2 and PR5 expression showed a delayed response, but ultimately also reached a higher level in nata1 (Fig. 8, B and E). On the other hand, expression of PR3, PR4, and PR6, common marker genes for jasmonate-induced plant defenses, was up-regulated to comparable levels in nata1 and wild-type Arabidopsis leaves (Fig. 8, C, D, and F).
Figure 8.
Defense-related gene expression in response to P. syringae. Wild-type Col-0 and nata1 were infiltrated with P. syringae DC3000. Expression of PR1 (A), PR2 (B), PR3 (C), PR4 (D), PR5 (E), and PR6 (F) measured by quantitative PCR. Mean ± se of n = 9. *P < 0.05, two-tailed t test comparing Col-0 wild type and nata1 at the same time point.
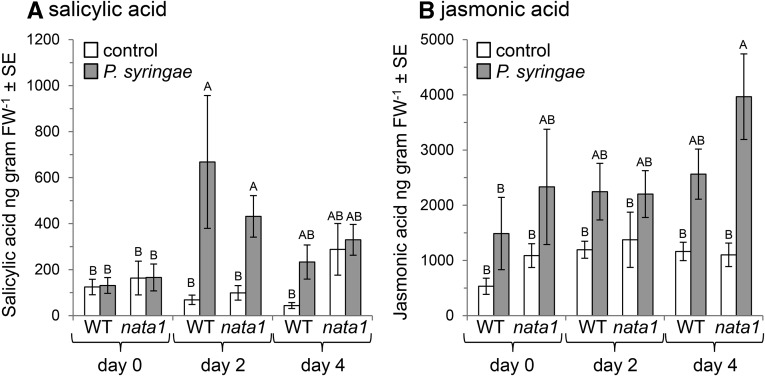
As H2O2 acts synergistically with salicylic acid to induce plant defense responses (Neuenschwander et al., 1995; Sharma et al., 1996; Alvarez et al., 1998; Mukherjee et al., 2010), we measured salicylic acid concentrations in response to DC3000 infection. Although P. syringae infiltration into Arabidopsis leaves significantly increased the concentration of salicylic acid 2 d later, there was no significant difference in the free or total salicylic acid content of wild-type and nata1 leaves (Fig. 9A; Supplemental Fig. S8). Similarly, although there was a jasmonic acid increase in P. syringae-infected nata1 leaves relative to controls on day 4 after infection, this was not significantly higher than in P. syringae-infected wild-type Arabidopsis (Fig. 9B).
Figure 9.
Plant hormone induction by P. syringae infection. Salicylic acid (A) and jasmonic acid (B) levels at 0, 2, and 4 d after P. syringae infiltration into wild-type Col-0 wildtype and nata1 Arabidopsis leaves. Mean ± se of n = 12. Within each figure, different letters indicate significant differences, P < 0.05, ANOVA followed by Tukey’s honest significant difference.
DISCUSSION
Our observation of increased H2O2 accumulation and P. syringae resistance in the Arabidopsis nata1mutant is consistent with prior observations of greater pathogen resistance in plants with elevated polyamines. Overexpression of S-adenosyl-Met decarboxylase, which is required to convert putrescine to spermidine, increased spermidine abundance in Arabidopsis (Marco et al., 2014). These transgenic plants also had elevated defense gene expression (including PR1and PR2) and are more resistant to P. syringae. Similarly, both spermine synthase overexpression and exogenous spermine addition made Arabidopsis more resistant to Pseudomonas viridiflava (Gonzalez et al., 2011). Concomitant addition of a polyamine oxidase inhibitor partly suppressed the plant-protective effect of exogenous spermine, indicating that this oxidation contributes to increased plant resistance.
Induction of NATA1 activity by the P. syringae DC3000 coronatine effector resulted in greater putrescine acetylation and reduced levels of nonacetylated polyamines (Figs. 3A and 4). In nata1 mutant Arabidopsis, there was increased H2O2 accumulation from spermine and/or spermidine oxidation (Fig. 5). PR1, PR2, and PR5 gene expression, which is induced as part of the antimicrobial defense response in Arabidopsis, was increased in nata1 mutant plants relative to wild type (Fig. 8). As it was not possible to show a significant difference in the abundance of salicylic acid (Fig. 9), defense gene expression may be regulated directly or indirectly by changes in H2O2 abundance. Further experiments using Arabidopsis mutants that are deficient in both NATA1 and salicylic acid signaling, for example, nata1 sid2 or nata1 npr1, will make it possible to test this hypothesis of H2O2-mediated defense gene induction in the absence of changes in salicylic acid abundance.
A previous report showed a 30% increase in salicylic acid after infection with coronatine-deficient P. syringae compared to wild-type P. syringae strain Psm ES4326 (Zheng et al., 2012). However, we observed no salicylic acid differences between wild-type and nata1 mutant plants (Fig. 9A), suggesting that the decrease of salicylate abundance in response to P. syringae-derived coronatine is regulated independently of polyamine acetylation. Another recent publication showed that during the second stage of the biotrophic Plasmodiophora brassicae (clubroot) infection, which results in the formation of root galls through cellular hyperplasia and hypertrophia (Mithen and Magrath, 1992), there is enhanced accumulation of salicylate, as well as lower jasmonate pathway induction, in Bur-0, an Arabidopsis ecotype that has a deletion in NATA1 promoter region and start codon (http://1001genomes.org/; Supplemental Fig. S4), but not in Col-0 (Lemarié et al., 2015). The role of NATA1 in the observed plant defense differences is further indicated by the fact that both nata1 mutant Arabidopsis and Bur-0 show enhanced resistance to clubroot compared to Col-0 (Lemarié et al., 2015). Together with our findings, this suggests that NATA1-mediated polyamine acetylation plays a function in the complex cross-talk between salicylate and jasmonate defense signaling. However, more research will be required to further elucidate the molecular mechanisms of this predicted cross-talk.
Our results are consistent with the hypothesis that NATA1-mediated regulation of H2O2 production by PAOs influences plant defense. However, we cannot rule out the possibility that diamine oxidases (EC 1.4.3.6) may also contribute to the H2O2 production by oxidizing putrescine. In an in vitro experiment, guazatine inhibited PAO activity by 75% and diamine oxidase activity by only 30% (Marina et al., 2008). Hence, the greatly diminished differences between nata1 mutant and wild-type Arabidopsis observed after guazatine treatment (Figs. 5 and 7) may indicate that polyamine catabolism via polyamine oxidases is the main source of defense-induced H2O2 production in these experiments.
In addition to regulation of H2O2 production, other NATA1 functions may also contribute to the phenotypes observed in our experiments. For instance, diaminopropane acetylation by NATA1 could counteract abscisic acid-dependent stomatal closure (Jammes et al., 2014). However, the timing of stomatal closure in response P. syringae, which occurs within hours (Melotto et al., 2006), appears to be different from the induction of NATA1 expression and metabolite accumulation, which occurs over a period of days (Fig. 3B). Compared to wild-type plants, nata1 mutants show a smaller attenuation of primary metabolism, particularly sugars and amino acids, after jasmonate treatment (Adio et al., 2011). This alteration in primary metabolism could have as-yet-unknown effects during Arabidopsis defense against P. syringae and other pathogens.
The involvement of diverse metabolic and signaling pathways likely accounts for the fact that coronatine is such an effective suppressor of plant defenses during P. syringae infection. In addition to promoting stomatal opening and reducing salicylic acid levels (Zheng et al., 2012), coronatine also regulates glucosinolate accumulation and thereby inhibits callose induction by indole glucosinolate breakdown (Geng et al., 2012). Coronatine-mediated suppression of innate immune defenses in Arabidopsis roots occurs in a COI1-dependent manner, but does not involve direct interference with salicylic acid signaling (Millet et al., 2010). Future research will determine whether NATA1 acts in parallel or sequentially with these other coronatine-regulated plant defense pathways.
NATA1-mediated polyamine acetylation induced by P. syringae-derived coronatine likely represents the misdirection of a plant defense response. A possible endogenous plant function for jasmonate-induced NATA1 expression may be to attenuate H2O2 production during insect feeding, which typically triggers a strong jasmonate response in Arabidopsis. Nδ-Acetyl-Orn, a metabolic product of NATA1, also has been shown to have direct effects on aphid resistance (Adio et al., 2011). The observation that there are at least three Arabidopsis ecotypes that lack NATA1 (Bur-0, Amel-1, and Cal-0) suggests divergent selection for this enzymatic activity. In certain environments, the avoidance of coronatine-mediated suppression of Arabidopsis defenses may outweigh any benefits that are provided by NATA1 enzymatic activity.
Compared to the circadian regulation of jasmonate accumulation (Goodspeed et al., 2012; Shin et al., 2012), the observed diurnal N-acetylputrescine oscillation is slightly delayed, consistent with the observation that N-acetylputrescine accumulation is jasmonate regulated. Several studies have demonstrated circadian regulation of Arabidopsis susceptibility to pathogen infection (Shin et al., 2012; Zhang et al., 2013; Ingle and Roden, 2014; Zhou et al., 2015). Thus, the circadian cycle of N-acetylputrescine biosynthesis (Fig. 2C) may be a component of a network of plant responses that contribute to diurnal variation in susceptibility to P. syringae infection. In particular, based on the results that we have presented, low constitutive NATA1 activity early in the morning would be expected to promote resistance to virulent P. syringae by increasing the availability of polyamines for the production of H2O2.
It is likely that polyamine-mediated defense regulation is not limited to Arabidopsis, but rather is more widespread in the plant kingdom. Negative cross-talk between the jasmonate- and salicylate-dependent signaling pathways predates the development of vascular plants (Thaler et al., 2002). Similarly, polyamines and their acetylated versions have an ancient evolutionary origin, being present in plants, animals, and microorganisms. Other plant species for which significant amounts of genome data are available contain genes that are predicted to be polyamine acetyltransferases (Supplemental Fig. S2), and expression of Arabidopsis NATA1 in N. benthamiana causes not only acetylputrescine accumulation (Fig. 2A) but also improved P. syringae growth (Fig. 7B). To date, acetylated polyamines have been reported in only a relatively limited number of plant species (Deagazio et al., 1995; Mesnard et al., 2000; Dufeu et al., 2003; Fliniaux et al., 2004; Kamada-Nobusada et al., 2008; Hennion et al., 2012; Kim et al., 2014). However, this is more likely due to the fact that that these metabolites have not been actively searched for, rather than that they are not present.
Whereas most research on cross-talk between plant defense signaling pathways has focused on the role of transcription factors, the pathway described here is mediated by metabolic enzymes and small molecules. Thus, jasmonate-regulated putrescine acetylation may represent a mechanism whereby H2O2 production by polyamine oxidases and the initiation of other antimicrobial plant defense responses can be attenuated. The observation of polyamine-mediated cross-talk between plant defense signaling pathways in Arabidopsis, as well as the evolutionarily ancient origins of the contributing metabolites, suggests that further research is warranted to determine whether similar defense regulation occurs more broadly in other plant species.
MATERIALS AND METHODS
Plants and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) ecotypes Amel-1, Bur-0, Cal-0, Col-0, Pedriza, TDr-1, and Zal-1, as well as the nata1 mutation in the Col-0 background (GK-256F07; Rosso et al., 2003) and nata2 mutation in the Col-0 background (SALK_092319; Alonso et al. 2003), were obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). Plants were grown in Cornell mix (by weight: 56% peat moss, 35% vermiculite, 4% lime, 4% Osmocote slow-release fertilizer [Scotts, Marysville, OH], and 1% Unimix [Scotts]) in 20- x 40-cm nursery flats in Conviron growth chambers with a photosynthetic photon flux density of 200 mmol m−2 s−1 and a 16-h-day/8-h-night photoperiod, with lights being turned on at 7 am at 23°C with a 50% relative humidity.
Methyl Jasmonate and Coronatine Induction
For methyl jasmonate induction, the leaves of 4-week-old Arabidopsis plants were sprayed with an aqueous solution containing 0.01% (v/v) Tween 20 and 0.45 mm methyl jasmonate (diluted from a 1.5-m stock in acetone). Control plants were treated with water containing 0.01% Tween 20 and 0.03% acetone. Plants were covered with plastic domes, and leaves were harvested 4 d after elicitation and immediately frozen in liquid nitrogen. Pulverized tissue was extracted and polyamines were analyzed as described below. For coronatine induction, similar assays were performed, with the sprayed aqueous solution containing 0.01% (v/v) Tween 20 and 10 µM coronatine.
Transient Gene Expression
For transient expression in Nicotiana benthamiana, NATA1 and NATA2 were cloned downstream of the cauliflower mosaic virus 35S promoter in the T-DNA binary vector pMDC32, as described by Adio et al. (2011), and transformed into Agrobacterium tumefaciens strain GV3101 for Arabidopsis transformation (Clough and Bent, 1998). A pMDC32::GFP vector, which was generated by cloning GFP amplified from the plasmid pMDC83 (Curtis and Grossniklaus, 2003) as the gene of interest in the same construct, was used as a control, together with an empty vector control. Primers used for cloning are listed in Supplemental Table S1. GV3101 carrying the transgenic constructs was cultured overnight at 30°C in Luria-Bertani (LB) broth supplemented with 100 mg mL−1 kanamycin. Bacteria were pelleted at 3,600g for 20 min, washed three times with sterile water, and resuspended in sterile 10 mm MgCl2 solution to approximately 0.2 x 108 colony-forming units/mL. To reduce gene expression silencing, cultures were mixed with A. tumefaciens (0.1 x 108 CFU mL−1) transformed with the turnip crinkle virus capsid protein (P38; Thomas et al., 2003). The bacterial solution was infiltrated into N. benthamiana leaves using a 1-mL needleless syringe. Leaf plugs (8-mm diameter) were collected 3 d after infiltration to confirm expression of the transgenes by qRT-PCR and 4 d after infiltration to extract polyamines for high-performance liquid chromatography (HPLC) analysis.
Protein Extraction and in Vitro Assays
For protein purification, Escherichia coli strain M15 was transformed with full-length NATA1 cDNA cloned with a His tag in pQE30 (Qiagen, Hilden, Germany). Expression of recombinant NATA1 was induced by 0.5 mm isopropyl-β-d-1-thiogalactopyranoside at 22°C for 8 h. Cells were centrifuged at 3,000g and 4°C for 10 min and resuspended in 0.5 m NaCl, 50 mm NaH2PO4 (pH 8.0) prior to being incubated with lysozyme on ice for 30 min. Cells were further disrupted by sonication and centrifuged at 15,000g for 15 min at 4°C to obtain a clear supernatant containing the soluble proteins. Recombinant NATA1 was purified with the Ni-NTA Purification System from Invitrogen (Carlsbad, CA). The supernatants were applied to Ni2+-charged resin equilibrated in extraction buffer. After 60 min of binding with gentle agitation, the resin was washed three times with 0.5 m NaCl, 50 mm NaH2PO4 (pH 8.0) with 20 mm imidazole, followed by protein elution with the same buffer containing 250 mm imidazole. After purification, a single protein band of about 27 kD could be observed by SDS-PAGE with silver staining. Enzymatic activity toward potential substrates and the catalytic parameter Km were determined using 2 μg purified protein under the following conditions: 100 mm Tris-HCl (pH 7.5), 2 mm EDTA (pH 7.5), and 10% glycerol at 30°C, with substrate concentration varying from 1.25 to 1000 μM. Orn, putrescine, spermidine, spermine, and 1,3-diaminopropane were used as substrates, while acetyl-CoA was used as the acetyl-group donor, as it is the case for known spermidine/spermine-N1-acetyltransferases (Ignatenko et al., 1996). Two mM 5-5′-dithiobis[2-nitrobenzoic acid] was added at the end of each reaction to allow spectrophotometric measurement of the free CoA that was generated. Samples were further derivitazed and analyzed by HPLC, as described below, for confirmation of the expected products. Km values were determined from Michaelis-Menten plots, and nonlinear least-squares fitting of data were performed using Microsoft Excel with Solver add-in software (www.solver.com).
Quantification of Free Polyamines
Polyamines were extracted and derivatized either with either benzoyl chloride or 6-aminoquinolyl-N-hydroxysuccinimidylcarbamate using an AccQ-Fluor reagent kit (Waters, Millford, MA) prior to HPLC detection. One hundred milligrams of fresh rosette leaves was ground in liquid nitrogen to fine powder with 3-mm steel beads using a Harbil model 5G-HD paint shaker (Fluid Management, Wheeling, IL). Ground tissue was taken up in 20 mm HCl (10 mL mg−1 of leaf tissue) containing 40 µM L-nor-Leu as an internal standard. Extracts were centrifuged at 15,000g at 23°C for 20 min, and the supernatants were saved for analysis. For derivatization, 5-µL extracts were mixed with 35 µL borate buffer prior to initiating the reaction by adding 10 µL 6-aminoquinolyl-N-hydroxysuccinimidylcarbamate solution. The mixture was then immediately incubated at 55°C for 10 min, and 10-µL aliquots of each sample were injected onto a Nova-Pak C18 column (3.9- x 150-mm particle size 4 µm) using a Waters 2695 pump system. A Waters model 2475 fluorescence detector with an excitation wavelength of 250 nm and an emission wavelength of 395 nm was used to detect eluted amino acid derivatives, with the data being recorded by Waters Empower Software. Solvent A (containing sodium acetate and tris-acetate-EDTA at pH 5.05) was purchased premixed from Waters; Solvent B was acetonitrile:water (60:40). The gradient used was 0 to 0.01 min, 100% A; 0.01 to 0.5 min, linear gradient to 3% B; 0.5 to 12 min, linear gradient to 5% B; 12 to 15 min, linear gradient to 8% B; 15 to 40 min, linear gradient to 30% B; 40 to 43 min, linear gradient to 37% B; 43 to 46 min, linear gradient to 42% B; 46 to 54 min, linear gradient to 45% B; 54 to 58 min, linear gradient to 50% B; 58 to 60 min, linear gradient to 60% B; 60 to 67 min, linear gradient to 80% B; 68 to 71 min, linear gradient to 100% B. The flow rate was 1.0 mL min−1.
For improved sensitivity and accuracy in quantifying spermidine and spermine, a benzoylation method was used for derivatization prior to HPLC-based detection. Polyamines were extracted from the ground leaf powder in water with 1% HCl containing 25 µM 1,6-hexanediamine (internal standard), derivatized, and detected by HPLC (Yoda et al., 2003). The extracts were centrifuged at 15,000g at 10°C for 20 min. Then 500 µL of the supernatants was transferred to fresh tubes, 500 µL 2 n NaOH and 10 µL benzoyl chloride were added, and samples were incubated at 48°C for 20 min. Following incubation, 1 mL saturated NaCl was added, samples were mixed, and benzoylated polyamines were extracted by the addition of 1 mL diethyl ether after centrifuging the extract at 5,000g, 25°C for 10 min. The upper phase was collected and evaporated under N2 gas. After complete evaporation, the residue was dissolved in 200 µL diethyl ether and evaporated under N2 gas. Benzoylated polyamines were dissolved in 50 µL of 50% acetonitrile for HPLC analysis with the system described above. The gradient was water (solvent A) and 100% acetonitrile (solvent B), with a flow rate of 1.0 mL min−1 at 28°C with the following gradient: 0 to 1 min 100% A; 1 to 3 min 85% A, 3 to 10 min 70% A,10 to 20 min 47.5% A, 20 to 21 min 100% B; 21 to 27 min 100% B; 27 to 28 min 100% A; and 28 and 35 min 100% A. Separated benzoylated polyamines were detected at 229 nm with Waters 2996 photodiode array detector. Known concentrations of polyamine standards were treated according to both procedures described above, and standard curves were used for polyamine quantification.
Bacterial Growth Assays
Pseudomonas syringae strains DC3000 and DC3000 Cor- [mutant line DB29 cfa- cma- (Millet et al., 2010)] were supplied by Nicole Clay (Yale University, New Haven, CT). DC3000 ΔhrcQ-U was supplied by Alan Collmer (Cornell University, Ithaca, NY). Bacteria were cultured at 30°C in LB broth supplemented with 50 µg mL−1 rifampicin and 50 µg mL−1 kanamycin. For leaf infiltration, bacterial cultures were centrifuged at 3,000g for 10 min, resuspended in water, and diluted to approximately 105 colony forming units mL−1. Three-week-old Col-0 wild-type and nata1 leaves were pressure-infiltrated using a 1-mL syringe, and excess bacterial solution was wiped off with a paper towel. Plants were infiltrated in a staggered manner, so that leaf discs (8-mm diameter) corresponding to 0, 2, and 4 d after infiltration were collected on the same day. Leaf discs were submerged in 1 mL sterile water with 0.01% Tween 20 for 4 h to allow equilibration of the bacteria between the apoplastic space of the leaf disks and the surrounding water, as described previously (Anderson et al., 2006). Ten-µL samples of a dilution series (10−1 to 10−6) prepared in 96-well microtiter plates were spotted on an LB agar plate supplemented with 50 µg mL−1 rifampicin and incubated at 30°C. After 2 d, bacterial colonies were counted at a point in the dilution series where there were fewer than about 50 colonies in the spotted cluster, and the concentration of bacteria relative to the weight of the leaf disks was calculated.
For bacterial growth in N. benthamiana, DC3000 ΔhrcQ-U was cultured at 30°C in LB broth supplemented with 50 µg mL−1 rifampicin and 50 µg mL−1 kanamycin. Overnight bacterial cultures were centrifuged at 3,000g for 10 min, resuspended in water, and diluted to approximately108 CFU mL−1. N. benthamiana leaves were infiltrated with P. syringae solution 2 d after they had been infiltrated with A. tumefaciens carrying the desired overexpression construct. P. syringae growth in N. benthamiana leaves was measured as described above for Arabidopsis.
H2O2 Detection and Quantification
H2O2 was detected and quantified by 3.3′-diaminobenzidine (DAB) staining (Ramel et al., 2009; Daudi et al., 2012). Leaves from mock- or P. syringae-infiltrated plants were placed in 12-well microtiter plates. Two mL of 1 mg mL−1 DAB solution with 0.05% v/v Tween 20 (pH 3.0) was pipetted into the wells and plates were covered with aluminum foil and placed on a platform shaker at 100 rpm for 4 h at 25°C. After incubation, the DAB solution was replaced with 2 mL of bleaching solution (3:1:1 ethanol: acetic acid: glycerol), and plates were placed in water bath at 85 to 90°C for 15 to 20 min. Following bleaching, the solution in the wells was replaced with 1 mL of fresh bleaching solution, and leaves were used for photography or quantification. For quantification assays, DAB-stained leaves were ground in liquid nitrogen and homogenized in 0.2 m HClO4. Following homogenization, samples were centrifuged at 12,000g for 10 min at 4°C. Supernatants were transferred to new tubes and their absorbance was measured at 450 nm. Quantification of H2O2 was calculated by comparing the absorbance values with a standard curve prepared with known H2O2 concentrations (5-200 μmol mL−1).
Gene Expression Analysis
Relative transcript abundances of PR1 (At2g14610), PR2 (At3g57260), PR3 (At3G54420), PR4 (At3G04720), PR5 (At1G75040), PR6 (At2G38900), PAO1 (At5g13700), PAO2 (At2g43020), PAO3 (At3g59050), PAO4 (At1g65840), PAO5 (At4g29720), SPDS1 (AT1G23820), SPDS2 (AT1G70310), SPMS (AT5G53120), and NATA1 (At2g39030) were analyzed and compared by quantitative reverse transcription-PCR (qRT-PCR) using elongation factor 1-α (EF1α, At5g60390) as the internal standard. Leaves (approximately 30 mg) from sample groups were ground in liquid nitrogen, and total RNA was extracted using the SV total RNA isolation system (Promega, www.promega.com) with on-column DNAse treatment. After the verification of the integrity of RNA and quantification with a Nanodrop system (www.nanodrop.com), 1 µg of total RNA was reverse transcribed using AMV Reverse Transcriptase (Promega) and RNasin RNase inhibitor (Promega) using oligo(dT)15 as primer. Following cDNA synthesis, the samples were diluted 10-fold in nuclease free water. Gene-specific primers (Supplemental Table S2) were designed using Primer-Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). qRT-PCR reactions were performed using an Applied Biosystems 7900HT Instrument with the SYBR Green PCR master mix (Applied Biosystems, www.appliedbiosystems.com). The reaction mixture (10 µL total volume) contained 3.3 µL of cDNA solution and 6.7 µL PCR master mix (5 µL of SYBR Green master mix, 0.4 µL of forward and reverse gene specific primers, both at 10 µM, and 0.9 µL nuclease free H2O). Reactions were initiated by the activation of the enzyme at 95°C for 10 min, followed by 40 cycles of: 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s and final extension at 72°C for 2 min. The resulting CT values were used to calculate the relative transcript abundance according to the standard curve method.
Polyamine Oxidase Enzyme Activity Assay
Polyamine oxidase activity was assayed spectrophotometrically (Lim et al., 2006; Tavladoraki et al., 2006). Leaves (200 mg) were ground in liquid nitrogen, homogenized in 0.2 m sodium phosphate buffer, pH 6.5 (1:5 w/v), and centrifuged at 12,000g for 20 min. Supernatants were transferred to fresh tubes, and half of the total volume was used for protein quantification by Bradford assay using bovine serum albumin as the standard (Bradford, 1976). The reaction mixture contained 1 mg mL−1 horseradish peroxidase, 0.1 mm 4-aminoantipyrine, 1 mm 3,5-dichloro-2-hydroxybenzenesulphonic acid, and 2 mm spermine in 0.2 m Na-phosphate buffer (pH 6.5). The reaction was initiated by the addition of 100 µL of the leaf homogenate into the reaction mixture, and A515 was recorded after incubating for 20 min at 25°C. Specific enzyme activity was calculated per milligram protein.
Guazatine Inhibition Assays
For PAO inhibition treatments, 1 mm guazatine acetate solution containing 2.5% Tween 20 was sprayed onto plants 2 d before P. syringae infiltration (Fu et al., 2011). The guazatine concentration in the leaves was determined by HPLC and quantified using a standard curve based on samples with known guazatine concentrations. As guazatine is a mixture of compounds, the two most abundant peaks in the HPLC runs of guazatine solution were used for quantification, preparation of a standard curve, and estimation of leaf guazatine concentrations. Leaf samples (100 mg) were extracted with 20 mm HCl containing 100 pM L-nor-Leu as the internal standard and centrifuged at 12,000g at 25°C for 15 min. Supernatants were transferred to new tubes and used for derivatization with 6-aminoquinolyl-N-hydroxysuccinimidylcarbamate using an AccQ-Fluor reagent kit (Waters). For derivatization, 5 μL extract was mixed with 35 μL borate buffer, and the reaction was initiated by the addition of 10 μL AccQ-Fluor reagent (Waters), followed by mixing and incubation for 10 min at 55°C. Ten microliters of each sample were injected onto a Nova-Pak C18 column (3.9- x 150-mm particle size 4 µm) using a Waters 2695 pump system at 38°C . Eluted guazatine derivatives were detected using a Waters model 2475 fluorescence detector with an excitation wavelength of 250 nm and an emission wavelength of 395 nm, and the data were recorded using Waters Empower software. Solvent A, containing sodium acetate and TAE at pH 5.05, was purchased premixed from Waters; Solvent B was acetonitrile:water (60:40). Linear gradient of the run was as follows with a 1.0-mL min−1 flow rate: 1 to 12 min 97% A; 12 to 15 min 95% A; 15 to 40 min 92% A; 40 to 43 min 70% A, B; 43 to 46 min 63% A; 46 to 54 min 58% A; 54 to 58 min 55% A; 58 to 60 min 50% A; 60 to 67 min 40% A; 67 to 67.1 min 20% A; 67.1 to 71 min 0%A; 71 to 75 min 100% A.
For in planta growth experiments, P. syringae DC3000 was cultured at 30°C in LB broth supplemented with 50 mg mL−1 rifampicin, centrifuged, resuspended in sterile water, and diluted to a concentration of 10−5 CFU mL−1. Bacterial cultures were pressure-infiltrated into leaves with a syringe and samples were collected to measure bacterial growth, as described above. To test the effect of guazatine on P. syringae growth in culture, 0, 1, 10, and 100 µM guazatine acetate was added to LB medium, cultures were inoculated with 1:10 diluted (500 µL into 5 mL) fresh cultures, and optical density at 600 nm was measured at 0, 1, 3, 6, 9, 12, 24, and 48 h after inoculation.
Phytohormone Analysis
For free phytohormone analysis, 100 mg of fresh rosette leaves was ground in liquid nitrogen as described above and extracted in 1 mL of isopropanol:water:HCl (2:1:0.005) buffer. Then 80 ng each of D4-salicylic acid and D5-jasmonic acid (C/D/N Isotopes Inc., www.cdnisotopes.com/) were added as internal standards. After homogenization in a FastPrep homogenizer (MP Biomedicals, www.mpbio.com) at 6 ms−1 for 45 s, samples were extracted with dichloromethane, dried in a rotary evaporator, and dissolved in 200 μL methanol. For free and conjugated salicylic acid measurement, 200 mg of fresh rosette leaves was extracted in 1 mL 90% methanol prior to being reextracted in 0.5 mL 100% methanol. Then 60 ng of D4-salicylic acid was added as an internal standard. Extracts were combined and dried completely prior to being resuspended in 5% trichloroacetate and extracted with an ethylacetate-cyclopentane-isopropanol (50:49:1) buffer. The organic phase, containing free salicylic acid, was collected, dried under nitrogen flow, and dissolved in 200 μL methanol. The aqueous phase was adjusted to pH 1 with concentrated HCl, boiled for 30 min, and extracted again as described above. The resulting organic phase was dried under nitrogen flow and dissolved in 200 μL methanol for total salicylic acid analysis.
Extracted samples were analyzed using a triple quadrupole liquid chromatography-tandem mass spectrometry system (Thermo Scientific, www.thermoscientific.com) on a C18 reverse-phase HPLC column (3 μm, 150 × 2.00 mm; Gemini-NX; Phenomenex, www.phenomenex.com), as described previously (Rasmann et al., 2012). Analytes were separated using a gradient of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) at a flow rate of 300 μL min−1. Phytohormones were analyzed by negative electrospray ionization (spray voltage, 3.5 kV; sheath gas, 15; auxiliary gas, 15; capillary temperature, 350°C), collision-induced dissociation (argon gas pressure, 1.3 mTorr; energy, 16 V), and selected reaction monitoring of compound-specific parent/product ion transitions (salicylic acid, 137→93; D4-SA, 141→97; jasmonic acid, 209→59; D5-JA, 214→62).
Sequence Alignment and Phylogenetic Tree Generation
Arabidopsis NATA1 homologs in the Brassicaceae and other plants were obtained by comparison to genomic and EST data in GenBank and Sol Genomics Network. The closest NATA1 homolog of human (Homo sapiens) was included as an outgroup. Amino acid sequences were aligned using Clustal Omega (Sievers et al., 2011), and rooted phylogenetic trees were constructed with MEGA5 using a maximum-likelihood algorithm (Jones-Taylor-Thornton probability model, gamma distributed rates among sites). To evaluate the tree topology, a bootstrap resampling analysis with 1,000 replicates was performed.
Statistical Analysis
Statistical analysis was conducted using JMP software (www.jmp.com).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. List of primers used for cloning and PCR amplification in this study.
Supplemental Table S2. List of primers used for qRT-PCR.
Supplemental Figure S1. Developmental expression pattern of NATA1 and NATA2.
Supplemental Figure S2. Phylogeny of polyamine acetyltransferase-like proteins in plants.
Supplemental Figure S3. Acetyltransferase sequence alignment.
Supplemental Figure S4. NATA1 natural variants.
Supplemental Figure S5. Protein gel showing purified NATA1.
Supplemental Figure S6. Michaelis-Menten curves for NATA1 enzymatic activity.
Supplemental Figure S7. Effect of guazatine on P. syringae growth in vitro.
Supplemental Figure S8. Total salicylic acid accumulation in wild-type and nata1 plants after P. syringae infection.
Supplementary Material
Glossary
- Amel-1
Ameland-1
- Bur-0
Burren-0
- Col-0
Columbia-0
- DAB
3.3′-diaminobenzidine
- LB
Luria-Bertani
Footnotes
Articles can be viewed without a subscription.
References
- Adio AM, Casteel CL, De Vos M, Kim JH, Joshi V, Li B, Juéry C, Daron J, Kliebenstein DJ, Jander G (2011) Biosynthesis and defensive function of Nδ-acetylornithine, a jasmonate-induced Arabidopsis metabolite. Plant Cell 23: 3303–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahou A, Martignago D, Alabdallah O, Tavazza R, Stano P, Macone A, Pivato M, Masi A, Rambla JL, Vera-Sirera F, et al. (2014) A plant spermine oxidase/dehydrogenase regulated by the proteasome and polyamines. J Exp Bot 65: 1585–1603 [DOI] [PubMed] [Google Scholar]
- Alcázar R, Altabella T, Marco F, Bortolotti C, Reymond M, Koncz C, Carrasco P, Tiburcio AF (2010) Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231: 1237–1249 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 [DOI] [PubMed] [Google Scholar]
- Anderson JC, Pascuzzi PE, Xiao F, Sessa G, Martin GB (2006) Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell 18: 502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini R, Cona A, Federico R, Fincato P, Tavladoraki P, Tisi A (2010) Plant amine oxidases “on the move”: an update. Plant Physiol Biochem 48: 560–564 [DOI] [PubMed] [Google Scholar]
- Badel JL, Shimizu R, Oh HS, Collmer A (2006) A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant Microbe Interact 19: 99–111 [DOI] [PubMed] [Google Scholar]
- Bagni N, Tassoni A (2001) Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids 20: 301–317 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza-Vázquez A, Bender CL, Kunkel BN (2004) Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 17: 162–174 [DOI] [PubMed] [Google Scholar]
- Brownfield DL, Todd CD, Deyholos MK (2008) Analysis of Arabidopsis arginase gene transcription patterns indicates specific biological functions for recently diverged paralogs. Plant Mol Biol 67: 429–440 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cona A, Rea G, Angelini R, Federico R, Tavladoraki P (2006) Functions of amine oxidases in plant development and defence. Trends Plant Sci 11: 80–88 [DOI] [PubMed] [Google Scholar]
- Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM (2005) Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci USA 102: 1791–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, Bolwell GP (2012) The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deagazio M, Zacchini M, Federico R, Grego S (1995) Putrescine accumulation in maize roots treated with spermidine: evidence for spermidine to putrescine conversion. Plant Sci 111: 181–185 [Google Scholar]
- Dufeu M, Martin-Tanguy J, Hennion F (2003) Temperature-dependent changes of amine levels during early seedling development of the cold-adapted subantarctic crucifer Pringlea antiscorbutica. Physiol Plant 118: 164–172 [Google Scholar]
- Fincato P, Moschou PN, Ahou A, Angelini R, Roubelakis-Angelakis KA, Federico R, Tavladoraki P (2012) The members of Arabidopsis thaliana PAO gene family exhibit distinct tissue- and organ-specific expression pattern during seedling growth and flower development. Amino Acids 42: 831–841 [DOI] [PubMed] [Google Scholar]
- Fliniaux O, Mesnard F, Raynaud-Le Grandic S, Baltora-Rosset S, Bienaimé C, Robins RJ, Fliniaux MA (2004) Altered nitrogen metabolism associated with de-differentiated suspension cultures derived from root cultures of Datura stramonium studied by heteronuclear multiple bond coherence (HMBC) NMR spectroscopy. J Exp Bot 55: 1053–1060 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Fu XZ, Chen CW, Wang Y, Liu JH, Moriguchi T (2011) Ectopic expression of MdSPDS1 in sweet orange (Citrus sinensis Osbeck) reduces canker susceptibility: involvement of H2O2 production and transcriptional alteration. BMC Plant Biol 11: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Cheng J, Gangadharan A, Mackey D (2012) The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 24: 4763–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, Marco F, Minguet EG, Carrasco-Sorli P, Blázquez MA, Carbonell J, Ruiz OA, Pieckenstain FL (2011) Perturbation of spermine synthase gene expression and transcript profiling provide new insights on the role of the tetraamine spermine in Arabidopsis defense against Pseudomonas viridiflava. Plant Physiol 156: 2266–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF (2012) Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci USA 109: 4674–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfrey C, Sommer S, Mayer MJ, Burtin D, Michael AJ (2001) Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J 27: 551–560 [DOI] [PubMed] [Google Scholar]
- Hennion F, Bouchereau A, Gauthier C, Hermant M, Vernon P, Prinzing A (2012) Variation in amine composition in plant species: how it integrates macroevolutionary and environmental signals. Am J Bot 99: 36–45 [DOI] [PubMed] [Google Scholar]
- Ignatenko NA, Fish JL, Shassetz LR, Woolridge DP, Gerner EW (1996) Expression of the human spermidine/spermine N1-acetyltransferase in spermidine acetylation-deficient Escherichia coli. Biochem J 319: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle RA, Roden LC (2014) Circadian regulation of plant immunity to pathogens. Methods Mol Biol 1158: 273–283 [DOI] [PubMed] [Google Scholar]
- Jammes F, Leonhardt N, Tran D, Bousserouel H, Véry AA, Renou JP, Vavasseur A, Kwak JM, Sentenac H, Bouteau F, et al. (2014) Acetylated 1,3-diaminopropane antagonizes abscisic acid-mediated stomatal closing in Arabidopsis. Plant J 79: 322–333 [DOI] [PubMed] [Google Scholar]
- Kamada-Nobusada T, Hayashi M, Fukazawa M, Sakakibara H, Nishimura M (2008) A putative peroxisomal polyamine oxidase, AtPAO4, is involved in polyamine catabolism in Arabidopsis thaliana. Plant Cell Physiol 49: 1272–1282 [DOI] [PubMed] [Google Scholar]
- Kim DW, Watanabe K, Murayama C, Izawa S, Niitsu M, Michael AJ, Berberich T, Kusano T (2014) Polyamine oxidase 5 regulates Arabidopsis growth through thermospermine oxidase activity. Plant Physiol 165: 1575–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lemarié S, Robert-Seilaniantz A, Lariagon C, Lemoine J, Marnet N, Jubault M, Manzanares-Dauleux MJ, Gravot A (2015) Both the jasmonic acid and the salicylic acid pathways contribute to resistance to the biotrophic clubroot agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol 56: 2158–2168 [DOI] [PubMed] [Google Scholar]
- Lim TS, Chitra TR, Han P, Pua EC, Yu H (2006) Cloning and characterization of Arabidopsis and Brassica juncea flavin-containing amine oxidases. J Exp Bot 57: 4155–4169 [DOI] [PubMed] [Google Scholar]
- Marco F, Busó E, Carrasco P (2014) Overexpression of SAMDC1 gene in Arabidopsis thaliana increases expression of defense-related genes as well as resistance to Pseudomonas syringae and Hyaloperonospora arabidopsidis. Front Plant Sci 5: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marina M, Maiale SJ, Rossi FR, Romero MF, Rivas EI, Gárriz A, Ruiz OA, Pieckenstain FL (2008) Apoplastic polyamine oxidation plays different roles in local responses of tobacco to infection by the necrotrophic fungus Sclerotinia sclerotiorum and the biotrophic bacterium Pseudomonas viridiflava. Plant Physiol 147: 2164–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mesnard F, Azaroual N, Marty D, Fliniaux MA, Robins RJ, Vermeersch G, Monti JP (2000) Use of 15N reverse gradient two-dimensional nuclear magnetic resonance spectroscopy to follow metabolic activity in Nicotiana plumbaginifolia cell-suspension cultures. Planta 210: 446–453 [DOI] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocha R, Majumdar R, Minocha SC (2014) Polyamines and abiotic stress in plants: a complex relationship. Front Plant Sci 5: 175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K, Brown I, Knox P, Mansfield J (2015) The role of cell wall-based defences in the early restriction of non-pathogenic hrp mutant bacteria in Arabidopsis. Phytochemistry 112: 139–150 [DOI] [PubMed] [Google Scholar]
- Mithen R, Magrath R (1992) A contribution to the life history of Plasmodiophora brassicae: secondary plasmodia development in root galls of Arabidopsis thaliana. Mycol Res 96: 877–885 [Google Scholar]
- Moschou PN, Sanmartin M, Andriopoulou AH, Rojo E, Sanchez-Serrano JJ, Roubelakis-Angelakis KA (2008) Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis. Plant Physiol 147: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou PN, Sarris PF, Skandalis N, Andriopoulou AH, Paschalidis KA, Panopoulos NJ, Roubelakis-Angelakis KA (2009) Engineered polyamine catabolism preinduces tolerance of tobacco to bacteria and oomycetes. Plant Physiol 149: 1970–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou PN, Wu J, Cona A, Tavladoraki P, Angelini R, Roubelakis-Angelakis KA (2012) The polyamines and their catabolic products are significant players in the turnover of nitrogenous molecules in plants. J Exp Bot 63: 5003–5015 [DOI] [PubMed] [Google Scholar]
- Mukherjee M, Larrimore KE, Ahmed NJ, Bedick TS, Barghouthi NT, Traw MB, Barth C (2010) Ascorbic acid deficiency in arabidopsis induces constitutive priming that is dependent on hydrogen peroxide, salicylic acid, and the NPR1 gene. Mol Plant Microbe Interact 23: 340–351 [DOI] [PubMed] [Google Scholar]
- Neuenschwander U, Vernooij B, Friedrich L, Uknes S, Kessmann H, Ryals J (1995) Is hydrogen peroxide a second messenger of salicylic acid in systemic acquired resistance. Plant J 8: 227–233 [Google Scholar]
- Ono Y, Kim DW, Watanabe K, Sasaki A, Niitsu M, Berberich T, Kusano T, Takahashi Y (2012) Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids 42: 867–876 [DOI] [PubMed] [Google Scholar]
- Pegg AE. (2008) Spermidine/spermine-N1-acetyltransferase: a key metabolic regulator. Am J Physiol Endocrinol Metab 294: E995–E1010 [DOI] [PubMed] [Google Scholar]
- Ramel F, Sulmon C, Bogard M, Couée I, Gouesbet G (2009) Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and sucrose-induced tolerance in Arabidopsis thaliana plantlets. BMC Plant Biol 9: 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G (2012) Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol 158: 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sharma YK, Léon J, Raskin I, Davis KR (1996) Ozone-induced responses in Arabidopsis thaliana: the role of salicylic acid in the accumulation of defense-related transcripts and induced resistance. Proc Natl Acad Sci USA 93: 5099–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Heidrich K, Sanchez-Villarreal A, Parker JE, Davis SJ (2012) TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24: 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavladoraki P, Cona A, Federico R, Tempera G, Viceconte N, Saccoccio S, Battaglia V, Toninello A, Agostinelli E (2012) Polyamine catabolism: target for antiproliferative therapies in animals and stress tolerance strategies in plants. Amino Acids 42: 411–426 [DOI] [PubMed] [Google Scholar]
- Tavladoraki P, Rossi MN, Saccuti G, Perez-Amador MA, Polticelli F, Angelini R, Federico R (2006) Heterologous expression and biochemical characterization of a polyamine oxidase from Arabidopsis involved in polyamine back conversion. Plant Physiol 141: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terano S, Suzuki Y (1978) Formation of beta-alanine from spermine and spermidine in maize shoots. Phytochemistry 17: 148–149 [Google Scholar]
- Thaler JS, Karban R, Ullman DE, Boege K, Bostock RM (2002) Cross-talk between jasmonate and salicylate plant defense pathways: effects on several plant parasites. Oecologia 131: 227–235 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Leh V, Lederer C, Maule AJ (2003) Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306: 33–41 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, Mysore KS, Bender CL (2007) The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 20: 955–965 [DOI] [PubMed] [Google Scholar]
- Walters D. (2003a) Resistance to plant pathogens: possible roles for free polyamines and polyamine catabolism. New Phytol 159: 109–115 [DOI] [PubMed] [Google Scholar]
- Walters DR. (2003b) Polyamines and plant disease. Phytochemistry 64: 97–107 [DOI] [PubMed] [Google Scholar]
- Xin XF, He SY (2013) Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Yoda H, Fujimura K, Takahashi H, Munemura I, Uchimiya H, Sano H (2009) Polyamines as a common source of hydrogen peroxide in host- and nonhost hypersensitive response during pathogen infection. Plant Mol Biol 70: 103–112 [DOI] [PubMed] [Google Scholar]
- Yoda H, Yamaguchi Y, Sano H (2003) Induction of hypersensitive cell death by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiol 132: 1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xie Q, Anderson RG, Ng G, Seitz NC, Peterson T, McClung CR, McDowell JM, Kong D, Kwak JM, et al. (2013) Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog 9: e1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wang W, Karapetyan S, Mwimba M, Marqués J, Buchler NE, Dong X (2015) Redox rhythm reinforces the circadian clock to gate immune response. Nature 523: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.