RNA exosome-mediated control of cuticular wax biosynthesis requires participation of SUPERKILLER complex proteins SKI2, SKI3, and SKI8.
Abstract
ECERIFERUM7 (CER7)/AtRRP45B core subunit of the exosome, the main cellular 3′-to-5′ exoribonuclease, is a positive regulator of cuticular wax biosynthesis in Arabidopsis (Arabidopsis thaliana) inflorescence stems. CER7-dependent exosome activity determines stem wax load by controlling transcript levels of the wax-related gene CER3. Characterization of the second-site suppressors of the cer7 mutant revealed that small interfering RNAs (siRNAs) are direct effectors of CER3 expression. To explore the relationship between the exosome and posttranscriptional gene silencing (PTGS) in regulating CER3 transcript levels, we investigated two additional suppressor mutants, wax restorer1 (war1) and war7. We show that WAR1 and WAR7 encode Arabidopsis SUPERKILLER3 (AtSKI3) and AtSKI2, respectively, components of the SKI complex that associates with the exosome during cytoplasmic 3′-to-5′ RNA degradation, and that CER7-dependent regulation of wax biosynthesis also requires participation of AtSKI8. Our study further reveals that it is the impairment of the exosome-mediated 3′-5′ decay of CER3 transcript in the cer7 mutant that triggers extensive production of siRNAs and efficient PTGS of CER3. This identifies PTGS as a general mechanism for eliminating highly abundant endogenous transcripts that is activated when 3′-to-5′ mRNA turnover by the exosome is disrupted. Diminished efficiency of PTGS in ski mutants compared with cer7, as evidenced by lower accumulation of CER3-related siRNAs, suggests that reduced amounts of CER3 transcript are available for siRNA synthesis, possibly because CER3 mRNA that does not interact with SKI is degraded by 5′-to-3′ XRN4 exoribonuclease.
The cuticle is an extracellular hydrophobic structure that coats primary aerial surfaces of land plants. It serves as a key barrier that restricts transpirational water loss, and provides protection from abiotic and biotic stresses (Kunst and Samuels, 2003; Kosma et al., 2009; Reina-Pinto and Yephremov, 2009). It also plays an important role in drought stress signaling (Wang et al., 2011) and prevents organ fusions during plant development (Sieber et al., 2000). The two major cuticle constituents are cutin and cuticular wax. Cutin is an aliphatic polyester composed of C16 and C18 oxygenated fatty acids and glycerol that functions as a structural scaffold. It is overlaid and embedded with cuticular wax, a mixture of very long chain fatty acids and their derivatives, including primary alcohols, wax esters, aldehydes, alkanes, ketones, and secondary alcohols (Samuels et al., 2008). Wax also usually contains small amounts of cyclic components, such as triterpenoids and sterols (Jetter et al., 2006).
Formation of wax components has been well-studied, and enzymes catalyzing wax biosynthetic reactions have been characterized (Bernard and Joubès, 2013). Transport of cuticular wax to the cuticle has been shown to involve ATP-binding cassette transporters and lipid transfer proteins (Pighin et al., 2004; Bird et al., 2007; Debono et al., 2009; Kim et al., 2012), but in contrast to wax biosynthesis, our knowledge of this process is incomplete. Similarly, regulation of wax production is not fully understood. Wax biosynthesis can be induced by environmental stress and hormone treatment (Kosma et al., 2009), and several transcriptional regulators have been shown to affect wax accumulation. These include an APETALA2 (AP2) domain-containing transcription factor WAX INDUCER1/SHINE1, whose overexpression results in glossy leaves with an increased wax load (Aharoni et al., 2004; Kannangara et al., 2007); MYB30, expressed in response to bacterial pathogen attack, which promotes the transcription of the fatty acid elongase complex genes (Raffaele et al., 2008); MYB96, which directly activates the promoters of several wax biosynthetic genes upon abscisic acid treatment or drought stress (Seo et al., 2011); and an AP2/ERF-type transcriptional repressor, DECREASE WAX BIOSYNTHESIS, which regulates wax accumulation during diurnal light/dark cycles and is involved in the organ-specific regulation of total wax loads on plant surfaces (Go et al., 2014; Suh and Go, 2014).
Besides transcription factors, cuticular wax loads are also influenced by two additional proteins: an E3 ubiquitin ligase encoded by the ECERIFERUM9 (CER9) gene that acts as a negative regulator of both wax and cutin monomers (Lü et al., 2012), and CER7/RRP45B core subunit of the exosome that regulates wax production during inflorescence stem development (Hooker et al., 2007). The exosome is an evolutionarily conserved multiprotein complex that mediates cellular RNA processing and degradation in the 3′-to-5′ direction in both the nucleus and the cytoplasm (Houseley et al., 2006; Chekanova et al., 2007; Lange et al., 2014). Extensive studies of the yeast and human exosome complexes indicated that all nine core exosomal subunits were required for its integrity and function (Allmang et al., 1999a, 1999b; Liu et al., 2006). In light of this holistic view of the eukaryotic exosome complex, the discovery that a core subunit of the exosome is involved in a specialized plant process such as cuticular wax deposition (Hooker et al., 2007) was surprising. Further investigation of the plant exosome demonstrated that two additional core exosomal subunits of Arabidopsis (Arabidopsis thaliana) exhibited functional specialization. The RRP41 subunit was shown to be involved in the development of the female gametophyte, whereas the RRP4 subunit was required for embryogenesis (Chekanova et al., 2007). Furthermore, unlike in yeast and metazoa where the intactness of the exosome complex is essential for function, the loss of the CSL4 subunit is not detrimental to plant growth and development (Chekanova et al., 2007).
To gain additional insight into regulation of wax deposition by the CER7/RRP45B subunit, we carried out a screen for second-site suppressors of cer7-associated wax deficiency and isolated a series of wax restorer (war) mutants (Lam et al., 2012). Systematic identification of genes disrupted in these war lines revealed that the majority encode enzymes of the small interfering RNA (siRNA) biosynthetic pathway. We demonstrated that siRNAs negatively regulate wax deposition in Arabidopsis stems by posttranscriptional gene silencing (PTGS) of the key wax biosynthetic gene CER3 and that siRNA levels in turn are determined by CER7/RRP45B-dependent exosome activity (Lam et al., 2012, 2015).
To further explore the relationship between the CER7-dependent exosome and PTGS in regulating CER3 transcript levels, we have characterized two additional cer7 suppressor mutants, war1 and war7. Positional cloning and analysis of WAR1 and WAR7 genes indicated that they carry mutations in Arabidopsis SUPERKILLER3 (AtSKI3; At1g76630) and AtSKI2 (At3g46960), respectively, that encode components of the SKI complex. In Arabidopsis, as in other eukaryotes studied thus far, SKI proteins have been shown to be required for cytoplasmic 3′-to-5′ RNA degradation by the exosome (Houseley et al., 2006; Dorcey et al., 2012; Halbach et al., 2013; Lange et al., 2014; Zhang et al., 2015), but evidence for their functional association into a complex is missing. Detailed analyses of the phenotypes of the ski single and ski cer7 double mutants reported here demonstrate that all SKI proteins participate in the CER7/RRP45B-mediated control of wax deposition in developing Arabidopsis inflorescence stems, and support the notion that they act in the same pathway or complex.
RESULTS
Mutation in AtSKI3 Restores the cer7-1 Wax Deficiency
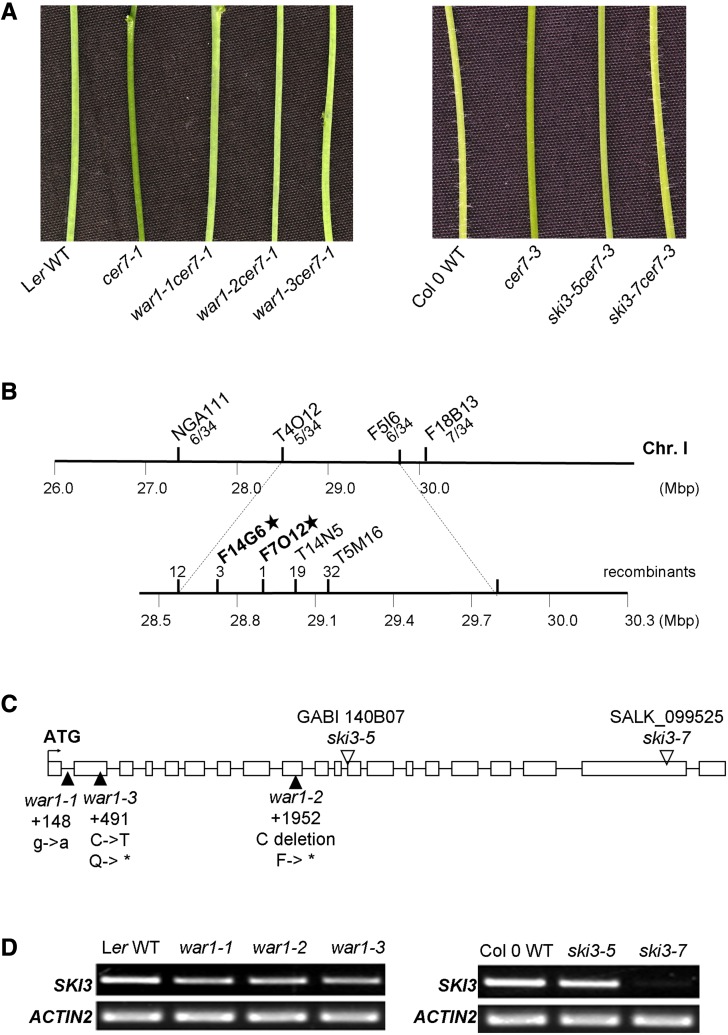
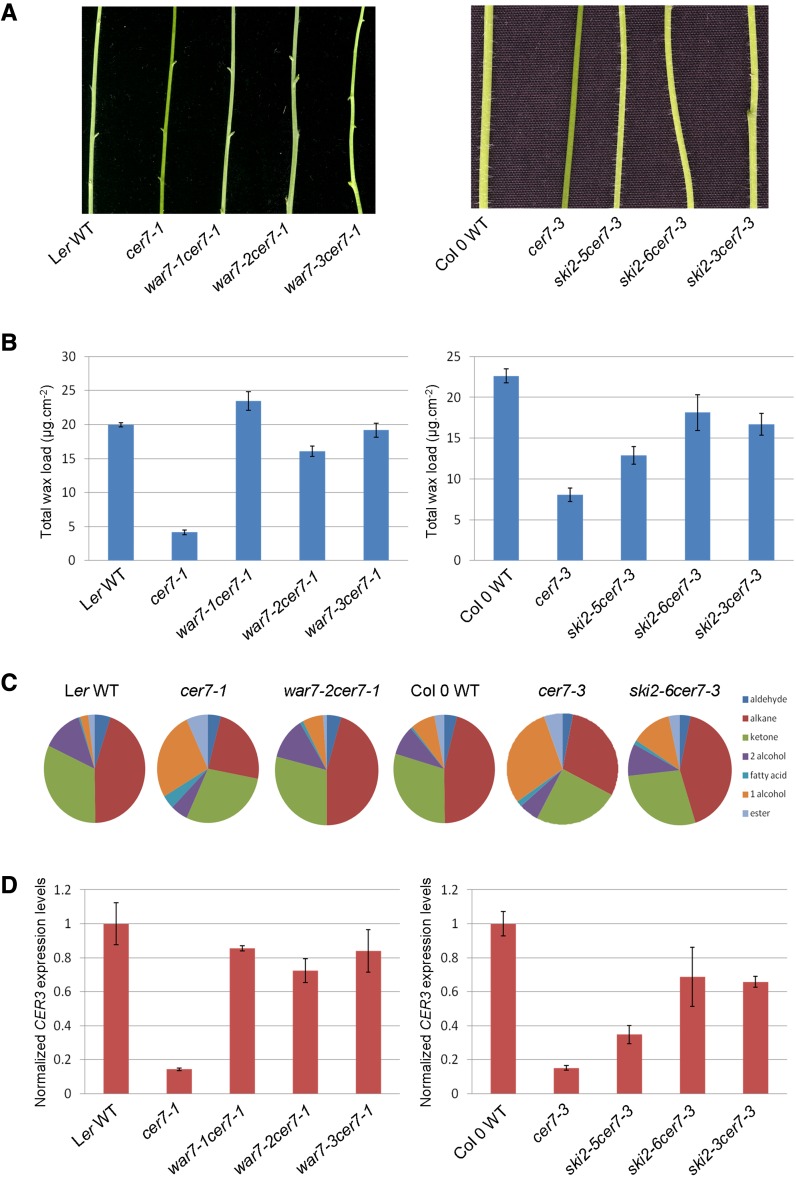
The war1 mutant was identified from a cer7-1 forward genetic screen, as described previously (Lam et al., 2012). This second-site mutation results in recovery of near wild-type cuticular wax load on the cer7-1 inflorescence stems (Figs. 1A and 2A).
Figure 1.
Identification of WAR1 as SKI3 component of the SKI complex required for CER7-mediated control of cuticular wax biosynthesis. A, Stems of 6-week-old Ler wild-type (WT), cer7-1, war1 cer7-1, Col-0 wild-type, cer7-3, and ski3 cer7-3 mutant plants showing the suppression of the cer7 wax-deficient phenotype in the war1 cer7-1 and ski3 cer7-3 mutants as indicated by glaucous stems. B, Schematic representation of the chromosomal location of war1 as determined by fine-mapping. The markers used for mapping and the number of recombinants are indicated. C, Schematic representation of the AtSKI3 gene structure. The exons are indicated as white boxes and introns as black lines. The translational start site is represented by the bent arrow. The positions and types of the mutations in ski3 alleles are also shown. D, RT-PCR analysis of AtSKI3 transcript levels in stems of war1 and ski3 mutants compared with the corresponding wild type.
Figure 2.
Analyses of wax profiles and CER3 transcript levels of war1 cer7-1 and ski3 cer7-3 mutants. A, Stem wax loads of war1 cer7-1 and ski3 cer7-3 double mutants compared with their corresponding wild type (WT) and cer7 single mutant. Values represent means ± sd (n = 3). B, Stem wax composition of war1-3 cer7-1 and ski3-7 cer7-3 double mutants compared with their corresponding wild type and cer7 mutants. Wax compositions for all double mutants are restored to wild-type-like ratios of major wax components. C, CER3 transcript levels of war1 cer7-1 and ski3 cer7-3 double mutants compared with their corresponding wild type and cer7 single mutants. ACTIN2 was used as an internal control, and control samples were normalized to 1. Values represent means ± sd (n = 3).
To pinpoint the mutation in war1, the war1-3 cer7-1 suppressor line was first backcrossed to cer7-1. The approximately 3:1 segregation ratio of glossy mutant to waxy wild-type phenotypes (631:205; χ2 = 0.26, P > 0.5) in the F2 population indicated that wax restoration was caused by a recessive mutation in a single nuclear gene. An outcross of war1-3 cer7-1 in the Landsberg erecta (Ler) background to the Arabidopsis Columbia-0 (Col-0) cer7-3 mutant was performed to create a mapping population for identification of the WAR1 gene. Rough mapping using 17 F2 plants that displayed a waxy stem phenotype indicated that the mutation in war1-3 lies on chromosome 1 between the markers T4O12 and F18B13 (Fig. 1B). An enlarged F2 population containing 714 waxy plants was then used for fine mapping that narrowed down the war1-3 mutation to a 120-kb region, flanked by the markers F14G6 and F7O12, which contains 29 annotated genes. DNA sequencing of this region revealed a C-to-T single nucleotide mutation at position 491 in the second exon of At1g76630, predicted to result in a premature stop codon (Fig. 1C). Mutations in the At1g76630 gene were also detected in two additional war1 alleles, uncovered from the same suppressor screen; war1-2 contains a single base-pair (C) deletion at position 1952 in the ninth exon, leading to a frame shift and an early stop codon at position 1957, whereas war1-1 has a G-to-A mutation at position 148 of the first intron, which may affect the nuclear mRNA processing or preclude the translation of a functional protein product. These data indicate that WAR1 is At1g76630 (Fig. 1C).
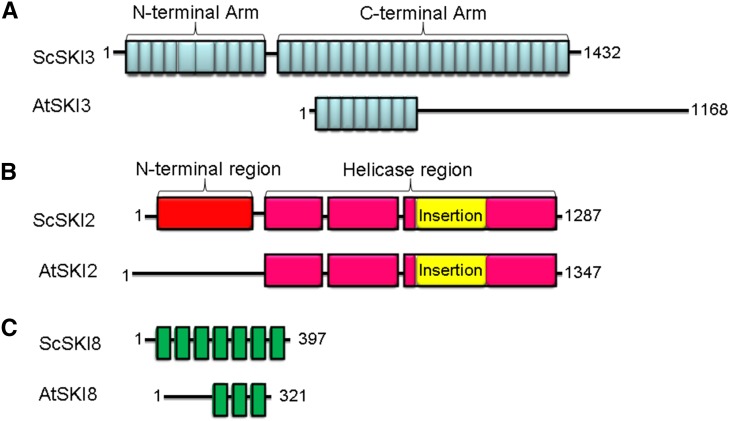
The At1g76630 gene encodes a predicted polypeptide of 1168 amino acids. Sequence analysis with the protein domain prediction tool SMART (http://smart.embl-heidelberg.de/) revealed that the WAR1 protein has six tetratricopeptide repeat domains clustered at the N terminus and exhibits 23% identity and 40% similarity with the Saccharomyces cerevisiae SUPERKILLER3 (ScSKI3) C-terminal arm (Fig. 5A). To confirm that mutations in AtSKI3 can restore wax deficiency of cer7, we obtained two T-DNA insertion alleles, GABI_140B07 and SALK_099525, previously designated ski3-5 and ski3-7 (Yu et al., 2015). T-DNA insertion sites in the 11th intron of AtSKI3 in ski3-5 and in the 19th exon in ski3-7, respectively, were established by DNA sequencing (Fig. 1C). Semiquantitative reverse transcription (RT)-PCR using total RNA extracts of the whole stems demonstrated that the overall abundance of the AtSKI3 mRNA in ski3-5 was similar to the wild type, whereas ski3-7 retained only trace amounts of mRNA, implying that the gene disruption in ski3-7 is more severe (Fig. 1D). In contrast, all three original war1 suppressor mutants, war1-1/ski3-8, war1-2/ski3-9, and war1-3/ski3-10, had only slightly lower AtSKI3 mRNA levels than the wild type. Homozygous ski3-5 and ski3-7 single mutants do not exhibit a stem wax phenotype or any other visible phenotype. However, when crossed into the cer7-3 background, both ski3-5 and ski3-7 can rescue the stem wax phenotype of the cer7 mutant like other war1/ski3 alleles (Fig. 1A).
Figure 5.
Comparison of structures of SKI proteins in S. cerevisiae (Sc) and Arabidopsis (At). A, Domain structure of the subunit SKI3 in yeast and Arabidopsis. Individual TPR motifs are indicated as blue boxes. B, Domain structure of the subunit SKI2 in yeast and Arabidopsis. The N-terminal region is shown in red, the helicase region in pink, and the insertion domain in yellow. C, Domain structure of the subunit SKI8 in yeast and Arabidopsis. WD40 motifs are shown as green rectangles. Structures of ScSKI proteins are derived from the structural analysis reported in Halbach et al. (2013). Structures of AtSKI proteins are based on the alignments to yeast sequences.
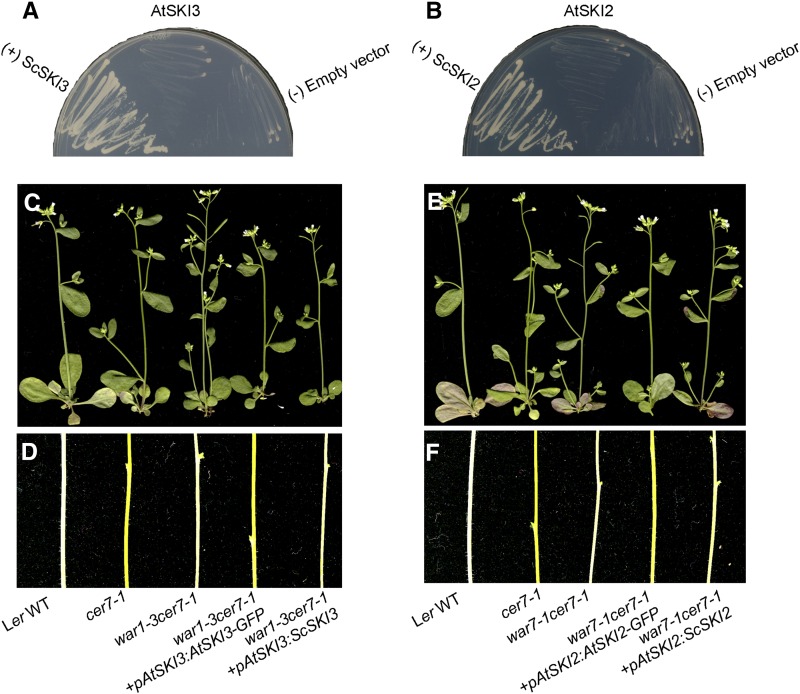
Stem wax analyses by gas chromatography with flame ionization detection (GC-FID) showed that all war1 cer7-1 suppressors and ski3 cer7-3 double mutants contain considerably higher wax loads than the cer7 single mutants (Fig. 2A). Additionally, in these lines, the dramatic reduction in alkane pathway components characteristic of cer7 was also restored to the wild type (Fig. 2B; Supplemental Table S4). CER3 transcript accumulation, examined by real-time PCR, was also partially or completely restored to wild-type levels (Fig. 2C). To further corroborate the conclusion that the mutation in AtSKI3 is responsible for the recovered waxy phenotype of the suppressors, transgene complementation of the war1-3 cer7-1 suppressor line with genomic AtSKI3, fused to green fluorescent protein (GFP) and driven by its native promoter, was carried out. The introduction of AtSKI3-GFP converted wild-type waxy stems of war1-3 cer7-1 to glossy bright green cer7-like stems in transgene recipients (Fig. 9D), indicative of successful complementation. Collectively, these results demonstrate that WAR1 is AtSKI3. The war1 alleles were therefore renamed ski3 (Supplemental Table S1).
Figure 9.
Yeast SKI proteins cannot functionally complement Arabidopsis ski mutants, and Arabidopsis SKI proteins cannot functionally complement yeast ski mutants. A, AtSKI3 cannot restore the growth of the yeast ski3 xrn1 mutant. B, AtSKI2 cannot restore the growth of the yeast ski2 xrn1 mutant. C and D, ScSKI3 cannot restore the wax deficiency of war1-3 cer7-1. E and F, ScSKI2 cannot restore the wax deficiency of war7-1cer7-1. WT, Wild type.
WAR7 Is AtSKI2, the Putative Helicase of the SKI Complex
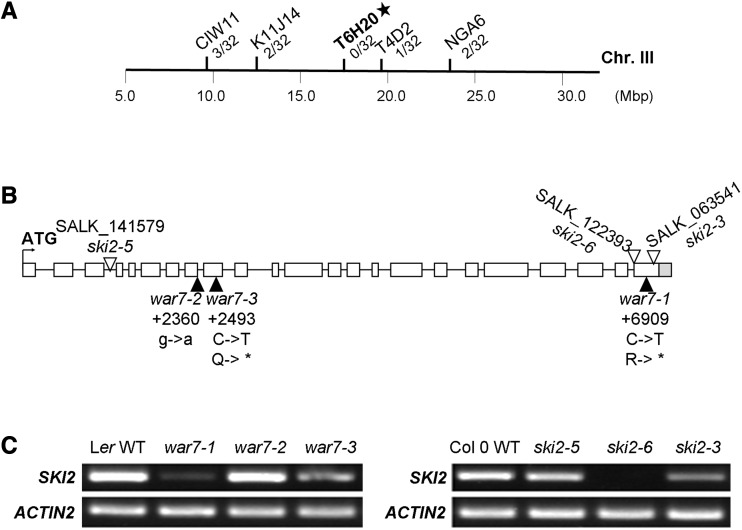
An additional mutation that rescues the cer7-related stem wax deficiency, war7, was mapped to chromosome 3 in the vicinity of marker T6H20 (Fig. 3A) and AtSKI2 (At3g46960), which encodes the RNA helicase subunit of the SKI complex (Jolivet et al., 2006; Dorcey et al., 2012). Because the SKI complex is conserved across many uni- and multicellular organisms, including yeast, Drosophila, Arabidopsis, and humans, we suspected that mutations in AtSKI2, just like those in AtSKI3, might also be able to restore the wax phenotype of the cer7 mutant. We therefore sequenced AtSKI2 in the war7-1 cer7-1 and war7-3 cer7-1 suppressors. Comparisons with the wild-type sequence revealed nonsense C-to-T mutations at nucleotide 6909 of AtSKI2 in war7-1 and at nucleotide 2493 in war7-3. Both these mutations produce a premature stop codon that would truncate the protein and likely inactivate it (Fig. 3B). In contrast, war7-2 contains a G-to-A substitution at position 1 of the eighth intron of the AtSKI2 gene, which might lead to mRNA splicing and protein translation defects.
Figure 3.
Identification of WAR7 as SKI2 component of the SKI complex required for CER7-mediated control of cuticular wax biosynthesis. A, Schematic representation of the chromosomal location of war7 as determined by rough-mapping. The markers used for mapping and the number of recombinants are indicated. B, Schematic representation of the AtSKI2 gene structure. The exons are indicated as white boxes and introns as black lines. The translational start site is represented by the bent arrow. The positions and types of the mutations in ski2 alleles are also shown. C, RT-PCR analysis of AtSKI2 transcript levels in stems of war7 and ski2 mutants compared with the corresponding wild type (WT).
We identified three more alleles of AtSKI2 in T-DNA insertional mutant collections: ski2-5 (SALK_141579), ski2-6 (SALK_122393), and ski2-3 (SALK_063541). The T-DNA insertion was within the third intron in the ski2-5 allele, and in the last exon in both ski2-6 and ski2-3. RT-PCR analysis using stem RNA demonstrated that AtSKI2 mRNA levels in war7-2 and ski2-5 were similar to the wild type, reduced in war7-1, war7-3, and ski2-3, and undetectable in ski2-6 (Fig. 3C). All of the homozygous ski2 T-DNA mutants were indistinguishable from the wild type with respect to their stem wax loads. When these ski2 alleles were crossed into the cer7-3 background, they could restore the cer7-3 stem wax load to near wild-type levels (Fig. 4, A and B). Wax accumulation and composition was verified by GC-FID analysis and confirmed that all war7 cer7-1 suppressors and ski2 cer7-3 double mutants displayed wild-type-like wax profiles (Fig. 4, B and C; Supplemental Table S4). Real-time PCR also demonstrated wild-type or near wild-type CER3 transcript levels in these suppressor and double mutant lines (Fig. 4D). These results indicate that WAR7 is AtSKI2. Hence, the war7 alleles will hereafter be referred to as ski2 (Supplemental Table S2).
Figure 4.
Analyses of stem wax phenotypes, wax profiles, and CER3 transcript levels of war7 cer7-1 and ski2 cer7-3 mutants. A, Stems of 6-week-old Ler wild-type (WT), cer7-1, war7 cer7-1, Col-0 wild-type, cer7-3, and ski2 cer7-3 mutant plants showing the suppression of the cer7 wax-deficient phenotype in the war7 cer7-1 and ski2 cer7-3 mutants as indicated by glaucous stems. B, Stem wax loads of war7 cer7-1 and ski2 cer7-3 double mutants compared with their corresponding wild type and cer7 single mutant. Values represent means ± sd (n = 3). C, Stem wax composition of war7-2 cer7-1 and ski2-6 cer7-3 double mutants compared with their corresponding wild type and cer7 mutants. Wax compositions for all double mutants are restored to wild-type-like ratios of major wax components. D, CER3 transcript levels of war7 cer7-1 and ski2 cer7-3 double mutants compared with their corresponding wild type and cer7 single mutants. ACTIN2 was used as an internal control, and control samples were normalized to 1. Values represent means ± sd (n = 3).
Sequence alignment showed that AtSKI2 and ScSKI2 share 43% identity and 59% similarity over 1000 amino acids at the C terminus (Fig. 5B), mainly covering the entire helicase region that is essential for RNA binding and ATP hydrolysis (Halbach et al., 2013). In contrast, the N terminus of ScSKI2 that is necessary and sufficient for interaction with ScSKI3 and ScSKI8 in yeast does not exhibit significant homology with the Arabidopsis AtSKI2. This does not, however, preclude successful interactions of AtSKI2 with AtSKI3 and AtSKI8 subunits as demonstrated by pull-down experiments in Arabidopsis (Dorcey et al., 2012; Zhang et al., 2015).
AtSKI8, the Third Component of the SKI Complex, Is Also Necessary for Stem Wax Deposition
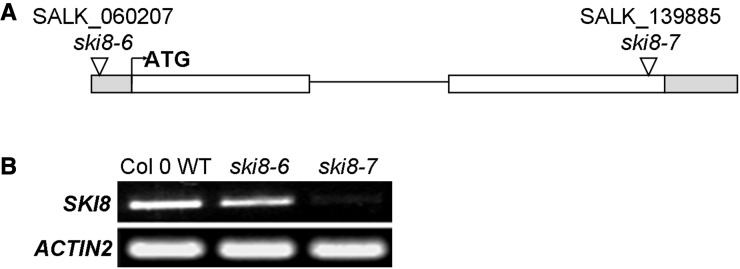
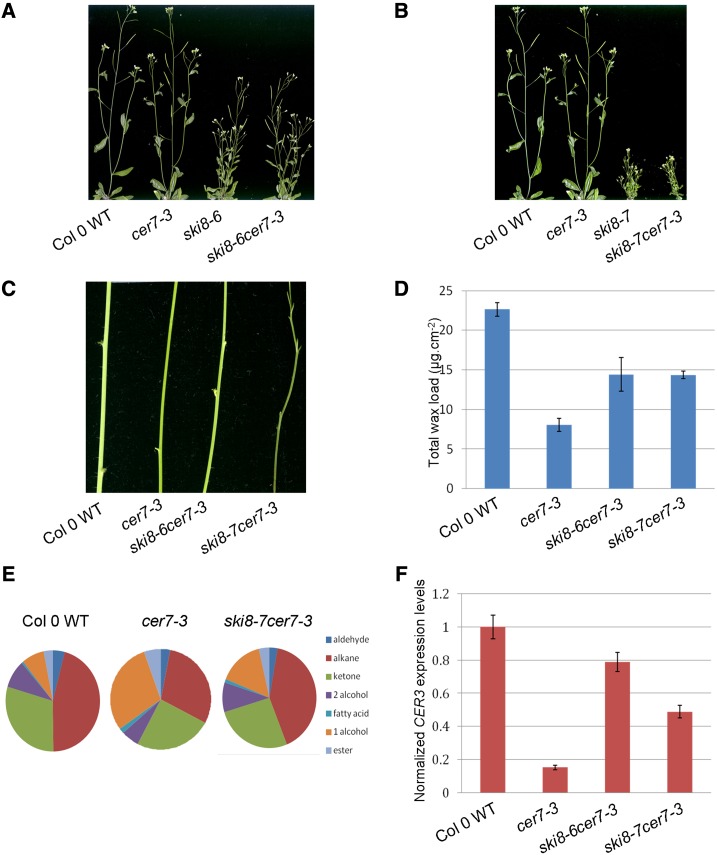
It is well-documented that a third component of the SKI complex, named SKI8, is indispensable for the cytoplasmic functions of the exosome (Brown et al., 2000; Orban and Izaurralde, 2005; Dorcey et al., 2012). To determine whether AtSKI8 works together with AtSKI2 and AtSKI3 to regulate stem wax biosynthesis in Arabidopsis, we obtained two T-DNA insertional mutants in the AtSKI8 gene from the Arabidopsis Biological Resource Center and employed them for reverse genetic analyses. The T-DNA insertion in ski8-6 (SALK_060207) is in the 5′ untranslated region, while ski8-7 (SALK_139885) harbors an insertion in the second exon that strongly affects AtSKI8 mRNA level (Fig. 6; Supplemental Table S3). Both ski8-7 and to a lesser extent ski8-6 display dwarf and bushy phenotypes (Fig. 7, A and B), and the ski8-7 mutation also results in flower abnormalities (Dorcey et al., 2012) absent from ski8-6. When ski8-6 and ski8-7 were crossed into the cer7-3 background, the resulting double mutants showed dramatically improved wax accumulation on inflorescence stems compared with cer7-3 and substantially restored CER3 transcript levels (Fig. 7, C–F; Supplemental Table S4). Based on these data, we conclude that in addition to AtSKI3 and AtSKI2, AtSKI8 also takes part in CER7-mediated regulation of stem wax biosynthesis, perhaps in a complex with AtSKI2 and AtSKI3.
Figure 6.
AtSKI8 gene structure and transcript levels in ski8 mutants. A, Schematic representation of the AtSKI8 gene structure. The 5′ and 3′ untranslated regions are indicated as gray boxes, exons as white boxes, and intron as black line. The translational start site is represented by the bent arrow. The positions of the T-DNA insertion in ski8 alleles are also shown. B, RT-PCR analysis of AtSKI8 transcript levels in stems of ski8 mutants compared with the wild type (WT).
Figure 7.
Analyses of mutant phenotypes, wax profiles, and CER3 transcript levels of ski8 cer7-3 mutants. A, Six-week-old Col-0 wild type (WT), cer7-3, ski8-6, and ski8-7 cer7-3 mutants. B, Six-week-old Col-0 wild type, cer7-3, ski8-7, and ski8-7 cer7-3 mutants. C, Stems of 6-week-old Col-0 wild type, cer7-3, and ski8 cer7-3 mutant plants showing the suppression of the cer7 wax-deficient phenotype ski8 cer7-3 mutants as indicated by glaucous stems. D, Stem wax loads of ski8 cer7-3 double mutants compared with Col-0 wild type and cer7 single mutant. Values represent means ± sd (n = 3). E, Stem wax composition of ski8-7 cer7-3 double mutant compared with Col-0 wild type and cer7-3 mutant. Wax compositions of ski8 cer7-3 double mutants are restored to wild-type-like ratios of major wax components. F, CER3 transcript levels of ski8 cer7-3 double mutants compared with Col-0 wild type and cer7-3 single mutant. ACTIN2 was used as an internal control, and control samples were normalized to 1. Values represent means ± sd (n = 3).
AtSKI3 and AtSKI2 Are Localized to the Cytoplasm and Cytoplasmic Granules
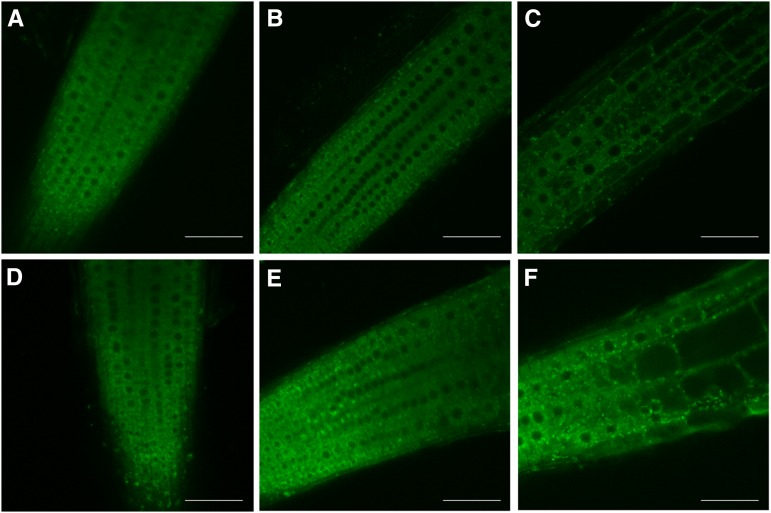
To investigate if AtSKI3 and AtSKI2 function in the cytoplasm like the yeast and metazoan SKI complexes, AtSKI3-GFP and AtSKI2-GFP fusions under the control of the AtSKI3 and AtSKI2 native promoters, respectively, were transformed into war1-3 cer7-1 and war7-1 cer7-1 mutants. Introduction of both transgenes resulted in the conversion of waxy stems into wax-deficient stems (Fig. 9, C–F), indicative of restored AtSKI3 and AtSKI2 functions and correct cellular localization of the AtSKI3-GFP and AtSKI2-GFP proteins. For both proteins, green fluorescence signals were detected in the cytoplasm of root cells (Fig. 8, A, B, D, and E) as well as in cytoplasmic granules of unknown function (Fig. 8, C and F), but were excluded from the nuclei. Cytoplasmic localization of AtSKI2 and AtSKI3, consistent with their roles as auxiliary factors required for exosomal activities in the cytoplasm, has been demonstrated previously (Zhang et al., 2015; Yu et al., 2015), but not their presence in the cytoplasmic granules shown here. This suggests the possibility that one or more activities of the AtSKI proteins might require their association into processing bodies (P bodies), cytoplasmic protein complexes known to be involved in degradation and translational arrest of mRNA (Maldonado-Bonilla, 2014). Further localization studies, including colocalization of AtSKI proteins with confirmed P body components, are needed to address this hypothesis.
Figure 8.
AtSKI3 and AtSKI2 are localized to the cytoplasm, but excluded from the nuclei. A and B, Confocal images roots of transgenic Arabidopsis plants expressing the AtSKI3p:AtSKI3-GFP construct. C, AtSKI3-GFP signal is also found in brightly fluorescing granules throughout the cytoplasm. D and E, Confocal images roots of transgenic Arabidopsis plants expressing the AtSKI2p:AtSK23-GFP construct. F, AtSKI2:GFP fluorescence is also localized to the cytoplasmic granules. Bars = 50 µm.
AtSKI3 and AtSKI2 Cannot Rescue Yeast ski3 xrn1 and ski2 xrn1; ScSKI3 and ScSKI2 Cannot Functionally Complement war1-3 cer7-1 and war7-1 cer7-1
Biochemical analyses carried out by several groups indicate that AtSKI2 and AtSKI3 are part of the cytoplasmic SKI complex in Arabidopsis, which also includes the AtSKI8 subunit (Dorcey et al., 2012; Zhang et al., 2015). Moreover, the yeast ScSKI8 ortholog was shown to rescue the dwarf phenotype of the Atski8 mutant (Dorcey et al., 2012), consistent with the notion that yeast and Arabidopsis SKI8 subunits are functionally equivalent. To determine if this is also the case for the SKI2 and SKI3 components, we performed reciprocal complementation experiments in S. cerevisiae and Arabidopsis. When a construct containing the AtSKI3 coding region driven by the ScSKI3 promoter was introduced into the yeast ski3 xrn1 mutant (Johnson and Kolodner, 1995), it failed to rescue the ski3 xrn1 growth phenotype (Fig. 9A). Similarly, the transgene containing the ScSKI3 coding region under the control of the native AtSKI3 promoter failed to substitute for AtSKI3 in controlling wax biosynthesis on Arabidopsis inflorescence stems (Fig. 9, C and D). These results were not surprising because protein sequence alignments show that AtSKI3 and ScSKI3 are highly divergent, and that AtSKI3 lacks tetratricopeptide repeats 28 to 33, whose deletion causes lethality in a yeast strain lacking XRN1 due to the loss of its ability to interact with ScSKI2 and ScSKI8 (Wang et al., 2005; Fig. 5A).
Even though yeast and Arabidopsis SKI2 subunits show a greater degree of sequence similarity, with the helicase region mostly conserved in both AtSKI2 and ScSKI2 proteins (Fig. 5B), heterologous complementation of the yeast ski2 xrn1 mutant was also unsuccessful (Fig. 9B). In a reciprocal experiment, AtSKI2p::ScSKI2 transgene was also unable to complement the wax phenotype of the war7-1 cer7-1 mutant (Fig. 9, E and F). These results suggest that the observed differences between yeast and Arabidopsis SKI3 and SKI2 proteins lead to distinct organizations of their respective SKI complexes. At present, it is not possible to deduce why this is the case, since the essential interacting domains that are required for tethering SKI2 and SKI8 to SKI3 in yeast have not been identified for the Arabidopsis SKI proteins.
All AtSKI Complex Subunits Are Required for the Exosome-Mediated Regulation of CER3-Related siRNA Levels and Thereby the Determination of CER3 Transcript Accumulation
Previously, we have shown that the CER7/RRP45B core subunit of the exosome determines cuticular wax loads on Arabidopsis stems by affecting levels of siRNAs involved in PTGS of CER3, a key wax biosynthetic gene. To directly assess if all three SKI proteins, AtSKI2, AtSKI3, and AtSKI8, work alongside CER7/RRP45B in the exosome-mediated control of wax deposition by controlling CER3-related siRNA accumulation, we examined the abundance of two such siRNA species, siRNA-1 (5′-UGACAUGUAACAGAUCAGGCU-3′) and siRNA-2 (5′-AACAGAUUGAUCACGAAUGGC-3′; Lam et al., 2015), as well as steady-state CER3 transcript levels, in ski single and ski cer7 double mutants in comparison to cer7.
As shown in Figure 10, ski3, ski2, and ski8 mutants, as well as ski3 cer7-3, ski2 cer7-3, and ski8 cer7-3 double mutants, accumulated considerably lower levels of the two siRNAs of interest than the cer7 mutant. Consequently, the CER3 transcript abundance was greater in all the ski and ski cer7 mutant lines compared with the wax-deficient cer7. However, due to the persistence of the small silencing RNAs in the ski single and double mutants, CER3 transcript concentration in these genotypes generally did not reach wild-type levels (Fig. 10C). Because mutations in all three SKI genes have a similar overall effect on siRNA accumulation, CER3 expression, and cuticular wax accumulation, which is distinct from cer7, it is likely that SKI proteins function as a complex in assisting exosome-mediated CER3 mRNA turnover.
Figure 10.
Accumulation of CER3-related siRNAs and CER3 transcripts in ski single and ski cer7 double mutants in comparison to cer7 and the wild type (WT). A, Detection of siRNA-1 in Col-0 wild type, cer7-3, ski, and ski cer7-3 mutants by real-time PCR. B, Detection of siRNA-2 in Col-0 wild type, cer7-3, ski, and ski cer7-3 mutants by real-time PCR. C, Detection of CER3 transcript levels in Col-0 wild type, cer7-3, ski, and ski cer7-3 mutants by real-time PCR. ACTIN2 was used as an internal control, and control samples were normalized to 1. Values represent means ± sd (n = 3).
DISCUSSION
Characterization of the Arabidopsis wax-deficient cer7 mutant revealed that the CER7/AtRRP45B core subunit of the RNA-degrading exosome complex takes part in a uniquely plant process, the regulation of cuticular wax biosynthesis, by affecting the transcript concentration of wax biosynthetic gene CER3 (Hooker et al., 2007). Whether CER7 carries out this function as part of the intact exosome, or in a different context, is not known. To elucidate the mechanism of CER7-mediated control of wax production, we performed a cer7 suppressor screen that resulted in the identification of siRNAs as direct effectors of CER3 expression (Lam et al., 2012, 2015). Based on these results, we hypothesized that CER7-dependent exosomal degradation of siRNAs, or their precursors, involved in PTGS of the CER3 gene is required to promote wax production in the wild-type stem. To further investigate this intriguing regulatory process, and determine if it involves other players in addition to CER7 and siRNAs, we characterized two novel cer7 suppressors, war1 and war7, and cloned the genes disrupted in these mutant lines. Identification of WAR1 and WAR7 revealed that they encode proteins with sequence similarity to SKI3 and SKI2, respectively, components of the SKI complex known to be required for many cytoplasmic functions of the RNA exosome, the major eukaryotic 3′-to-5′ exoribonuclease (Brown et al., 2000; Orban and Izaurralde, 2005). Results of our reverse genetic analysis indicate that, in addition to AtSKI2 and AtSKI3, efficient regulation of CER3 expression also requires participation of the AtSKI8 protein.
The well-studied SKI complex of S. cerevisiae functions as a tetramer of SKI3, SKI2, and SKI8 subunits that assemble with 1:1:2 stoichiometry (Halbach et al., 2013). The only protein with enzymatic activity in yeast is the SKI2 RNA helicase, whereas SKI3 and SKI8 subunits have structural roles and modulate the activity of SKI2 (Halbach et al., 2013). An additional protein, SKI7, links the SKI complex with the exosome in yeast (Araki et al., 2001). Evidence for structural association of the Arabidopsis SKI proteins was first provided by Dorcey et al., (2012), who successfully coimmunoprecipitated AtSKI3 and AtSKI2 with the GFP-ATSKI8 subunit. Zhang et al. (2015) confirmed direct interaction between AtSKI2 and AtSKI3 proteins by pull-down experiments, and identified the AtSKI7 component. Our analyses of the ski mutants described here further demonstrate that all three SKI proteins of Arabidopsis are required for the exosomal degradation of the CER3 transcript during stem wax deposition. As in CER7, mutations in AtSKI3, AtSKI2, or AtSKI8 result in higher levels of siRNAs that control CER3 expression, and consequently reduced CER3 transcript levels in comparison to wild type. Collectively, these results suggest that SKI proteins likely function in a complex, and that the SKI complex and the CER7-dependent exosome act together in controlling CER3 transcript levels.
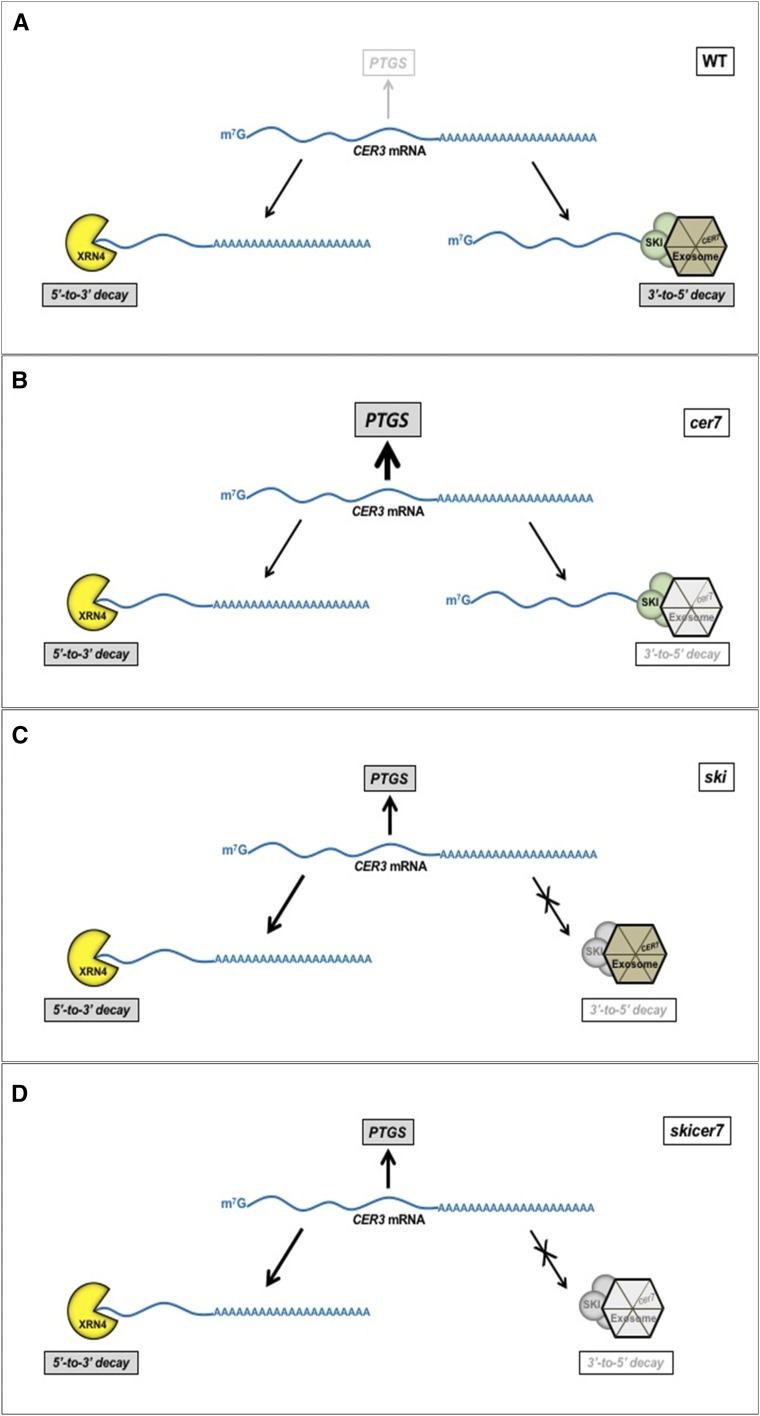
Given that exosomal 3′-to-5′ RNA degradation of CER3 mRNA requires participation of SKI proteins, it is surprising that ski mutants suppress the cer7 phenotype. All ski lines accumulate considerably lower amounts of CER3-related siRNAs than the cer7 mutant, which results in greater CER3 transcript levels, and much higher wax loads in comparison to cer7. This ski phenotype is apparent even in ski cer7 double mutants. Suppression of the cer7-related wax deficiency by ski mutations can be explained if SKI proteins are necessary to trigger the production of high levels of siRNA resulting in strong PTGS of CER3. How could SKI proteins be necessary for siRNA production? Until recently, PTGS and bidirectional cytoplasmic RNA decay pathways, which include the 3′-to-5′ exosome and 5′-to-3′ XRN4 exoribonuclease, have been considered independent mechanisms that eliminate endogenous aberrant mRNAs or exogenous mRNAs. However, new evidence has revealed that bidirectional RNA degradation pathways serve as primary cellular responses that prevent accumulation of undesirable mRNAs, and repress PTGS (Zhang et al., 2015; Yu et al., 2015), perhaps by reducing the amount of available aberrant or transgene mRNA used for siRNA production. Impairment or overload of either one of the bidirectional RNA degradation pathways has been shown to activate siRNA production and PTGS (Yu et al., 2015). In light of this new information, it is possible that in wild-type plants excess CER3 mRNA that does not enter the 3′-to-5′ exosomal decay pathway through the SKI complex is degraded instead by the 5′-to-3′ XRN4 exoribonuclease, and siRNA production is not triggered (Fig. 11A). This results in optimal expression of CER3 and stem wax biosynthesis. In the cer7 mutant, the SKI complex presumably delivers CER3 mRNA to the impaired exosome that cannot degrade it. The association with the SKI complex makes CER3 mRNA unavailable for 5′-to-3′ XRN-mediated decay, and the buildup of CER3 transcript triggers robust PTGS of CER3 that causes wax deficiency (Fig. 11B). In ski mutants, in the absence of the functional SKI complex to thread the CER3 transcript to the exosome, much of the CER3 mRNA that accumulates in the cytoplasm is diverted to the 5′-to-3′ XRN4 pathway for degradation. This leads to a reduced amount of CER3 transcript available for siRNA biosynthesis, diminished PTGS of CER3, and near-wild-type wax loads in ski mutants (Fig. 11C). The same thing happens in the ski cer7 double mutants because the dysfunctional ski complex components still prevent access of the CER3 mRNA to the exosome (Fig. 11D).
Figure 11.
Model illustrating the roles of SKI complex and CER7 in triggering the PTGS to regulate cuticular wax biosynthesis in Arabidopsis. A, In the wild type (WT), both 3′-5′ and 5′-3′ decay pathways are used to degrade excess CER3 mRNA; therefore, PTGS of CER3 is not triggered and CER3 is expressed and cuticular wax production proceeds. B, In the cer7 mutant, 3′-5′ decay is blocked. However, the CER3 mRNA threaded by SKI complex is no longer accessible for 5′-3′ decay; therefore, accumulated CER3 mRNA triggers robust PTGS, resulting in wax deficiency. C, In ski mutants, most of the CER3 mRNA is diverted to the 5′-3′ pathway for degradation. This leads to diminished PTGS of CER3 and near-wild-type wax loads. D, In the ski cer7 double mutants, most of the CER3 mRNA is also channeled to the 5-3′ degradation, leaving a small amount of CER3 transcript for siRNA production.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) mutant lines cer7-1 (Ler background; Koornneef et al., 1989), cer7-3 (Col-0 background; SAIL-747-B08), Atski3-7 (SALK_099525), Atski2-5 (SALK_141579), Atski2-6 (SALK_122393), Atski2-3 (SALK_063541), Atski8-6 (SALK_060207), and Atski8-7 (SALK_139885) were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/), and Atski3-5 (GABI_140B07) was ordered from GABI_Kat (Max Planck Institute for Plant Breeding Research). Wild-type and mutant seeds were stratified for 2 to 3 d at 4°C, and germinated on AT-agar plates (Somerville and Ogren, 1982) for 7 to 10 d and transplanted to soil (Sunshine Mix 4; SunGro). All plants were grown at 20°C under continuous light (90–110 μmol m−2 s−1 photosynthetically active radiation) in an environmental growth chamber.
Mapping of Suppressor Mutations
To identify the mutated genes in suppressor lines, each suppressor line in Ler background was crossed to cer7-3 in Col-0 ecotype and grown to produce the F2 generation. Leaf genomic DNA from 16 or 17 individual plants with the wild-type waxy stem phenotype was extracted on FTA cards (Whatman). These DNA samples were subjected to PCR amplification using simple sequence length polymorphism markers to determine linkage of the mutated genes. To further narrow the mapping interval of the war loci, more than 2000 plants were screened with simple sequence length polymorphism markers until a small region was delineated.
Genotyping
Genomic DNA was extracted as described by Berendzen et al. (2005) and used for PCR amplification. All the T-DNA insertion lines were genotyped using gene-specific primers (http://signal.salk.edu/tdnaprimers.2.html) and LBb1.3 (except GABI 140B07, for which o8409 was used) as listed in Supplemental Table S5.
Cuticular Wax Extraction and Analysis
Cuticular waxes were extracted from 4- to 6-week-old Arabidopsis stems. Stems were immersed in chloroform containing 10 μg of n-tetracosane (internal standard) for 30 s. Samples were blown to dryness under a stream of nitrogen and redissolved in 10 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (Sigma) and 10 μL of pyridine (Fluka). Derivatization of the samples was performed at 80°C for 1 h. After that, excess N,O-bis(trimethylsilyl)trifluoroacetamide and pyridine were dried under a stream of nitrogen, and samples were dissolved in 30 μL of chloroform. Gas-liquid chromatography was performed using an HP 7890A series gas chromatograph equipped with a flame ionization detector and a 30-m HP-1 column using hydrogen as the carrier gas. Gas chromatography was carried out with temperature-programmed on-column injection and oven temperature set at 50°C for 2 min, raised by 40°C min−1 to 200°C, held for 2 min at 200°C, raised by 3°C min−1 to 320°C, and held for 30 min at 320°C.
Quantification of wax components was carried out by comparing the flame ionization detector peak areas with the internal standard. Stem surface area was calculated by photographing stems prior to wax extraction, measuring the number of pixels, converting them to cm2, and multiplying by π.
RT-PCR and Quantitative RT-PCR
RT-PCR was used to analyze the expression levels of AtSKI genes in different ski alleles and their corresponding wild-type ecotypes. Total RNA was extracted from 4- to 5-week-old stems using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA quantification was carried out using Nano-Drop 8000 (Thermo Scientific). One microgram of total RNA was treated with DNaseI (Fermentas) and then used for first-strand cDNA synthesis using iScript RT supermix (Bio-Rad). For RT-PCR, the cycle number and amount of template were optimized for all fragments to generate products in the linear range of the reaction. The actin gene ACTIN2 (At3g18780) was used as a constitutive control with primers ACTIN2-F and ACTIN2-R. Gene-specific primers are listed in Supplemental Table S5.
Quantitative RT-PCR was performed using gene-specific primer sets from Supplemental Table S5 in a 20-μL volume with iQ SYBR Green supermix (Bio-Rad). The reactions were performed in triplicates and run on the iQ5 real-time PCR detection system (Bio-Rad). Data were analyzed using the method of Pfaffl (2001), and control samples were normalized to 1. Small RNA detection was designed and performed according to the protocol provided by Wesley et al. (2001).
Plasmid Construction and Yeast and Plant Transformation
A 6,037-bp DNA genomic fragment containing 696 bp of the upstream region of AtSKI3 minus the stop codon was amplified from wild-type Col-0 plants with primers AtSKI3p-attB1 and AtSKI3-attB2-noSTOP shown in Supplemental Table S5, using Phusion DNA polymerase (New England Biolabs). Gateway adapters were added using the adapter protocol (Invitrogen). This 6037-bp fragment was cloned into pDONR207 using BP Clonase II (Invitrogen) and sequenced to confirm that no mutations were introduced during PCR amplification. The fragment was then recombined into the destination vector pGWB4 (Nakagawa et al., 2007) using LR Clonase II (Invitrogen) to generate pGWB4:ProAtSKI3:AtSKI3-GFP.
An 8077-bp DNA genomic fragment including 1016 bp of the upstream region of AtSKI2 minus the stop codon was amplified from wild-type Col-0 plants with primers AtSKI2p-attB1 and AtSKI2-attB2-noSTOP shown in Supplemental Table S5, using Phusion DNA polymerase (New England Biolabs). Gateway adapters were added using the adapter protocol (Invitrogen). This 8077-bp fragment was cloned into pDONR207 using BP Clonase II (Invitrogen) and sequenced to confirm that no mutations were introduced by PCR. The fragment was recombined into the destination vector pGWB4 (Nakagawa et al., 2007) using LR Clonase II (Invitrogen) to generate pGWB4:ProAtSKI2:AtSKI2-GFP.
To generate a construct of ScSKI3 that can be expressed in Arabidopsis, yeast SKI3 was amplified from pAJ264 (SKI3 subclone in YEp351; Johnson and Kolodner, 1995) using primers ScSKI3-attB1 and ScSKI3-attB2, and cloned into pDONR207 using BP Clonase II (Invitrogen). The 696-bp promoter of AtSKI3 was amplified from pGWB4:ProAtSKI3:AtSKI3-GFP with primers AtSKI3p-attB4 and AtSKI3p-attB1R, and inserted into pDONRGP4-P1R (Nakagawa et al., 2008) using BP Clonase II (Invitrogen). Both fragments were then recombined into the destination vector R4PGWB4_SRDX_HSP (Oshima et al., 2011) using LR Clonase II (Invitrogen) to generate R4PGWB4:ProAtSKI3:ScSKI3:SRDX.
To generate a construct of ScSKI2 that can be expressed in Arabidopsis, yeast SKI2 was amplified from pAJ66 (SKI3 subclone in YEp351; Johnson and Kolodner, 1995) using primers ScSKI2-attB1 and ScSKI2-attB2, and cloned into pDONR207 using BP Clonase II (Invitrogen). The 1016-bp promoter of AtSKI2 was amplified from pGWB4:ProAtSKI2:AtSKI2-GFP with primers AtSKI2p-attB4 and AtSKI2p-attB1R, and inserted into pDONRGP4-P1R (Nakagawa et al., 2008) using BP Clonase II (Invitrogen). Both fragments were then recombined into the destination vector R4PGWB4_SRDX_HSP (Oshima et al., 2011) using LR Clonase II (Invitrogen) to generate R4PGWB4:ProAtSKI2:ScSKI2:SRDX.
These constructs were sequenced to confirm that no errors were present, and then transformed into Agrobacterium tumefaciens strain GV3101, pMP90 (Koncz and Schell, 1986). war1-3 cer7-3 and war7-1 cer7-1 mutants were transformed using the floral dip method (Clough and Bent, 1998).
GFP fluorescence was examined with a Zeiss Pascal Excite laser scanning confocal microscope (Carl Zeiss MicroImaging GmbH). A 488-nm excitation wavelength with the emission filter set at 500 to 530 nm was used for GFP. All confocal images obtained were processed with Adobe Photoshop 5.0 software.
For expression in yeast, AtSKI3 coding regions, amplified from Arabidopsis cDNA with primers AtSKI3-F and AtSKI3-R, were fused to ScSKI3 promoter, amplified by PCR from pAJ264 (SKI3 subclone in YEp351; Johnson and Kolodner, 1995), and inserted into sites of SmaI and XbaI in YEp351. Then, the ScSKI3 terminator was amplified with primers ScSKI3t-F and ScSKI3t-R from pAJ264, and inserted into the sites of XbaI and HindIII in YEp351 to produce YEp351:ScSKI3p:AtSKI3:ScSKI3t. AtSKI2 coding regions, amplified from Arabidopsis cDNA with primers AtSKI2-F and AtSKI2-R, were fused to ScSKI2 promoter and ScSKI2 terminator, amplified by PCR from PAJ66 (SKI2 subclone in YEp351). The generated fragment was inserted into SmaI and PstI sites in YEp351 to generate YEp351:ScSKI2p:AtSKI2:ScSKI2t. These constructs were sequenced and transformed into Saccharomyces cerevisiae strains AJY107 (RDKY2060 with pRDK297) and AJY100 (RKY2053 with pRDK297), respectively (Johnson and Kolodner, 1995), using the method described by Gietz and Woods (2002). Transformed cells were grown on Leu− plates for 4 d. Individual transformed colonies were streaked on Leu− plates containing 5-fluoroorotic acid.
Accession Numbers
Sequence data from this article can be obtained from the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CER7, At3g60500; CER3, At5g57800; SKI3, At1g76630; SKI2, At3g46960; SKI8, At4g29830; and ACTIN2, At1g49240.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Nomenclature and description of the ski3 alleles.
Supplemental Table S2. Nomenclature and description of the ski2 alleles.
Supplemental Table S3. Nomenclature and description of the ski8 alleles.
Supplemental Table S4. Cuticular wax composition of inflorescence stems of Arabidopsis wild type (WT), cer7, suppressors, and double mutants.
Supplemental Table S5. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the Salk Institute for Genomic Analysis Laboratory for providing sequence-indexed Arabidopsis T-DNA insertion mutants, Arlen Johnson for the yeast ski xrn1 mutants and yeast SKI gene subclones, the Bioimaging Facility at the University of British Columbia for assistance with microscopy, and Tegan Haslam and George Haughn for helpful discussions and critical evaluation of the manuscript.
Glossary
- siRNA
small interfering RNA
- PTGS
posttranscriptional gene silencing
Footnotes
This work was supported through funding provided by a Natural Sciences and Engineering Research Council of Canada Discovery Grant to L.K. and by the University of British Columbia Four Year Ph.D. Fellowship to L.Z.
Articles can be viewed without a subscription.
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D (1999a) Functions of the exosome in rRNA, snoRNA and snRNAsynthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P (1999b) The yeast exosome and human PM-Scl are related complexes of 3'–> 5' exonucleases. Genes Dev 13: 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T (2001) Ski7p G protein interacts with the exosome and the Ski complex for 3'-to-5' mRNA decay in yeast. EMBO J 20: 4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen K, Searle I, Ravenscroft D, Koncz C, Batschauer A, Coupland G, Somssich IE, Ulker B (2005) A rapid and versatile combined DNA/RNA extraction protocol and its application to the analysis of a novel DNA marker set polymorphic between Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta. Plant Methods 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Joubès J (2013) Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog Lipid Res 52: 110–129 [DOI] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Brown JT, Bai X, Johnson AW (2000) The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. (2007) Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debono A, Yeats TH, Rose JK, Bird D, Jetter R, Kunst L, Samuels L (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorcey E, Rodriguez-Villalon A, Salinas P, Santuari L, Pradervand S, Harshman K, Hardtke CS (2012) Context-dependent dual role of SKI8 homologs in mRNA synthesis and turnover. PLoS Genet 8: e1002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Go YS, Kim H, Kim HJ, Suh MC (2014) Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26: 1666–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Reichelt P, Rode M, Conti E (2013) The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell 154: 814–826 [DOI] [PubMed] [Google Scholar]
- Hooker TS, Lam P, Zheng H, Kunst L (2007) A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19: 904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D (2006) RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Jetter R, Kunst L, Samuels L (2006) Composition of plant cuticular waxes. In Biology of the plant cuticle. Blackwell Publishing Ltd; Hoboken, NJ [Google Scholar]
- Johnson AW, Kolodner RD (1995) Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol 15: 2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet S, Vezon D, Froger N, Mercier R (2006) Non conservation of the meiotic function of the Ski8/Rec103 homolog in Arabidopsis. Genes Cells 11: 615–622 [DOI] [PubMed] [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Höfte H, Pauly M, Riechmann JL, Broun P (2007) The transcription factor WIN1/SHN1 regulates Cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee SB, Kim HJ, Min MK, Hwang I, Suh MC (2012) Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol 53: 1391–1403 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Koornneef M, Hanhart CJ, Thiel F (1989) A genetic and phenotypic description of eceriferum (cer) mutants in Arabidopsis-thaliana. J Hered 80: 118–122 [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA (2009) The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Samuels AL (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Lam P, Zhao L, Eveleigh N, Yu Y, Chen X, Kunst L (2015) The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol 167: 323–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Zhao L, McFarlane HE, Aiga M, Lam V, Hooker TS, Kunst L (2012) RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol 159: 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Zuber H, Sement FM, Chicher J, Kuhn L, Hammann P, Brunaud V, Bérard C, Bouteiller N, Balzergue S, et al. (2014) The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet 10: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD (2006) Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Lü S, Zhao H, Des Marais DL, Parsons EP, Wen X, Xu X, Bangarusamy DK, Wang G, Rowland O, Juenger T, et al. (2012) Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol 159: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Bonilla LD. (2014) Composition and function of P bodies in Arabidopsis thaliana. Front Plant Sci 5: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nakamura S, Tanaka K, Kawamukai M, Suzuki T, Nakamura K, Kimura T, Ishiguro S (2008) Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci Biotechnol Biochem 72: 624–629 [DOI] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E (2005) Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11: 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Mitsuda N, Nakata M, Nakagawa T, Nagaya S, Kato K, Ohme-Takagi M (2011) Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol 28: 201–210 [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real time RT–PCR. Nucleic Acids Res 29: 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-Pinto JJ, Yephremov A (2009) Surface lipids and plant defenses. Plant Physiol Biochem 47: 540–549 [DOI] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Metraux J-P, Nawrath C (2000) Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12: 721–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1982) Isolation of photorespiratory mutants of Arabidopsis. In M Edelman, RB Hallick, N-H Chua, eds, Methods in Chloroplast Molecular Biology. Elsevier, New York, pp 129–139 [Google Scholar]
- Suh MC, Go YS (2014) DEWAX-mediated transcriptional repression of cuticular wax biosynthesis in Arabidopsis thaliana. Plant Signal Behav 9: e29463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R (2002) Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [DOI] [PubMed] [Google Scholar]
- Wang L, Lewis MS, Johnson AW (2005) Domain interactions within the Ski2/3/8 complex and between the Ski complex and Ski7p. RNA 11: 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Xiong L, Li W, Zhu J-K, Zhu J (2011) The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 23: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Rose JK (2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Saudemont B, Bouteiller N, Elvira-Matelot E, Lepère G, Parent JS, Morel JB, Cao J, Elmayan T, Vaucheret H (2015) Second-site mutagenesis of a hypomorphic argonaute1 allele identifies SUPERKILLER3 as an endogenous suppressor of transgene posttranscriptional gene silencing. Plant Physiol 169: 1266–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu Y, Liu X, Hong X, Xu Y, Zhu P, Shen Y, Wu H, Ji Y, Wen X, et al. (2015) Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 348: 120–123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.