Abstract
Despite a growing number of studies investigating the impact of natural killer (NK) cells on HIV-1 pathogenesis, the exact mechanism by which NK cells recognize HIV-1-infected cells and exert immunological pressure on HIV-1 remains unknown. Previously several groups including ours have introduced autologous HIV-1-infected CD4+ T cells as suitable target cells to study NK-cell function in response to HIV-1 infection in vitro. Here, we re-evaluated and optimized a standardized in vitro assay that allows assessing the antiviral capacity of NK cells. This includes the implementation of HIV RNA copy numbers as readout for NK-cell-mediated inhibition of HIV-1 replication and the investigation of inter-assay variation in comparison to previous methods, such as HIV-1 p24 Gag production and frequency of p24+ CD4+ T cells. Furthermore, we investigated the possibility to hasten the duration of the assay and provide concepts for downstream applications. Autologous CD4+ T cells and NK cells were obtained from peripheral blood of HIV-negative healthy individuals and were separately enriched through negative selection. CD4+ T cells were infected with the HIV-1 strain JR-CSF at an MOI of 0.01. Infected CD4+ T cells were then co-cultured with primary NK cells at various effector:target ratios for up to 14 days. Supernatants obtained from media exchanged at days 4, 7, 11 and 14 were used for quantification of HIV-1 p24 Gag and HIV-1 RNA copy numbers. In addition, frequency of infected CD4+ T cells was determined by flow cytometric detection of intracellular p24 Gag. The assay displayed minimal inter-assay variation when utilizing viral RNA quantification or p24 Gag concentration for the assessment of viral replication. Viral RNA quantification was more rigorous to display magnitude and kinetics of NK-cell-mediated inhibition of HIV-1 replication, longitudinally and between tested individuals. The results of this study demonstrate that NK-cell-mediated inhibition of HIV-1 replication can be reliably quantified in vitro, and that viral RNA quantification is comparable to p24 Gag quantification via ELISA, providing a robust measurement for NK-cell-mediated inhibition of viral replication. Overall, the described assay provides an optimized tool to study the antiviral capacity of NK cells against HIV-1 and an additional experimental tool to investigate the molecular determinants of NK-cell recognition of virus-infected cells.
Keywords: HIV-1, natural killer cell, CD4+ T cell, HIV-1 p24 ELISA, HIV-1 RNA
1. Introduction
Natural killer (NK) cells represent a subset of lymphocytes that is critically involved in the control of viral infections (Jost and Altfeld, 2013; Orange, 2002; Vidal et al., 2011). The function of NK cells is governed by multiple inhibitory and activating receptors balancing self-tolerance and effective responses recognizing cells with “non-self” or “altered-self” phenotypes. NK-cell activation is accompanied by the release of cytotoxic granules (degranulation) to eliminate virus-infected cells and the production of various pro-inflammatory and antiviral cytokines. Accumulating evidence strongly indicate a role of NK cells in HIV-1 pathogenesis (Carrington and Alter, 2012; Jost and Altfeld, 2012). Epidemiological studies have identified several members of the killer-cell immunoglobulin-like receptor (KIR) family expressed by NK cells to be involved in the control of HIV-1 infection (Martin et al., 2007, 2002). Furthermore, it has been shown that NK cells are able to produce cytokines upon stimulation that compete with HIV-1 co-receptor CCR5 (Fauriat et al., 2010; Oliva et al., 1998). However, it is still not fully understood how NK cells recognize HIV-1 infected cells and which cellular host factors would enable efficient NK-cell-mediated control of HIV-1 infection.
The function of NK cells in response to cellular targets has been studied extensively mostly using MHC class I devoid cell lines such as 721.221 (Burlingham et al., 1989) and K-562 (Lisovsky et al., 2015a). However, given the divergent origin of these cells, they display certain limitations with respect to HIV-1 research. This includes the lack of MHC class I molecules that serve as important ligands for activating and inhibitory NK-cell receptors and therefore are potent regulators of NK-cell activation (Nash et al., 2014). Thus, results and conclusions on NK-cell function drawn from experiments with these models should be used with caution in the setting of HIV-1 infection. In contrast, autologous CD4+ T cells are the natural target for HIV-1. The abundant expression of ligands for inhibitory NK-cell receptors, including MHC class I molecules, and low expression of ligands for activating receptors render autologous CD4+ T cells initially not susceptible to NK-cell-mediated killing. However, in vitro HIV-1 infection and subsequent HIV-1-mediated alterations of the cellular phenotype make HIV-1-infected autologous CD4+ T cells a suitable model for HIV-1-specific target-cell recognition by NK cells. Several groups as well as ours have developed assays for the assessment of direct and indirect antiviral functions of NK cells (Bonaparte and Barker, 2003; Ward et al., 2007; Fogli et al., 2008; Davis et al., 2011; Lisovsky et al., 2015b; Norman et al., 2011; Alter et al., 2007; Oliva et al., 1998; Bernstein et al., 2004). This includes the assessment of the ability of NK cells to produce antiviral cyto- and chemokines, to lyse infected target cells or to inhibit HIV-1 replication.
Based on a previous approach of Alter et al. (Alter et al., 2007), we re-evaluated and optimized a standardized in vitro assay that allows the assessment of the antiviral capacity of primary NK cells. The presented approach uses HIV-1-infected autologous CD4+ T cells as target cells and quantification of NK-cell-mediated inhibition of HIV-1 replication as a measure for the ability of NK cells to control HIV-1 infection in vitro.
2. Materials and methods
2.1 Study subjects
A total of 22 HIV-1-negative healthy subjects were recruited for this study. Subjects were enrolled at the Massachusetts General Hospital in Boston, USA and at the Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany. The study was approved by the respective local Institutional Review Boards. All individuals gave written informed consent for participation in this study. Study participants have been randomly selected for each individual experiment.
2.2 Sample processing and isolation of peripheral blood mononuclear cells (PBMC)
50 ml ACD-treated venous peripheral blood was obtained through phlebotomy from all participants. PBMC were isolated by density-gradient centrifugation within 2 hours of sample collection and subsequently resuspended in complete medium (RPMI-1640 medium (Sigma) supplemented with 10% (v/v) fetal bovine serum (Sigma), 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin (All from Corning Cellgro). PBMC yield and viability was assessed using a NucleoCounter (NC-200, ChemoMetec). Recovery of PBMC ranged between 50–100 million cells with an average viability >99%.
2.3 Enrichment of autologous CD4+ T cells and primary NK cells
PBMC were divided at a 1:10 ratio for enrichment of CD4+ T cells and primary NK cells respectively. Negative-selection strategy was applied using CD4+ T cell or NK-cell enrichment kits (Stemcell Technologies) accoring to the manufacturer’s protocol. Purity of the enriched cell populations was verified by multi-parameter flow cytometry using the following antibodies: CD4-APC (clone RPA-T4), CD3-PacificBlue (clone UCHT1), CD56-APC-Cy7 (clone HCD56), CD8-FITC (clone HIT8a, all BD) and CD16 BV510 (clone EG8, Biolegend). Average cell purity for CD4+ T cells was 97.2% and 94.5% for NK cells respectively with less than 0.3% contamination of CD8+ T cells. CD4+ T cells were resuspended in complete media at 5*106 cells/ml and stimulated with 100 IU/ml human recombinant IL-2 (hrIL-2; NIH) and 1 µg/ml phytohaemagglutinin (PHA, Fisher) overnight. NK cells were resuspended in complete media supplemented with 1 ng/ml human recombinant IL-15 (hrIL-15; R&D systems) overnight.
2.4 In vitro infection and long-term NK/CD4+ T cell co-culture
Overnight cultured CD4+ T cells were infected with the laboratory HIV-1 strain JR-CSF at a multiplicity of infection (MOI) of 0.01 in 100 ul of complete media supplemented with 100 IU/ml hrIL-2 for 4 hours at 37°C. Following infection CD4+ T cells were washed twice with 14 ml of complete media to remove free viral particles. CD4+ T cells were then plated in a 96 round-bottom well plate (5*104 cells/well) in the absence or presence of NK cells. NK cells were added at effector:target ratios of 0.1:1, 1:1 and 10:1 in a total volume of 300 ul of complete media supplemented with 50 IU/ml hrIL-2 and 1 ng/ml hrIL-15 for 14 days. Each condition was plated at least in duplicate.
2.5 Sample collection and assessment of HIV-1 replication
Culture supernatants were collected every 3 or 4 days for subsequent batch analysis of HIV-1 p24 Gag concentration and copy numbers of HIV-1 RNA. A total of 150 ul of supernatant was collected from each well, briefly centrifuged (5 min, 350 RCF) to remove residual cells and then stored at −20°C. Replicates for each condition were pooled prior to freezing. In addition, residual cells were used to assess the frequency of HIV-infected CD4+ T cells after 14 days of co-culture.
2.6 Quantification of HIV-1-infected CD4+ T cells by flow cytometry (p24 ICS)
The frequency of HIV-1-infected CD4+ T cells was measured by intracellular detection (Intracellular staining, ICS) of the HIV-1 p24 capsid protein using multi-parameter flow cytometry. After removal of culture supernatants, cells of identical replicates were pooled and washed once with PBS. Surface staining for viability and lineage markers was performed by resuspension of cells in 100 ul PBS supplemented with 2% (v/v) FBS followed by 30 min incubation at room temperature with the respective antibodies and reagents: CD3-Pacific Blue (BD), CD4-APC (BD), LIVE/DEAD Fixable blue dye (Invitrogen). Subsequent intracellular staining was conducted using anti-p24-FITC (clone KC57, Beckman Coulter) and a commercially available cell permeabilization and fixation kit (Invitrogen) according to the manufacturer’s protocol. Labelled cells were washed with PBS and fixed in 1% (w/v) paraformaldehyde (PFA) solution (Affymetrix) until flow cytometric acquisition.
2.7 Quantification of p24 Gag protein in viral supernatants by ELISA (p24 ELISA)
For the assessment of HIV-1 p24 Gag concentration in culture supernatants a commercially available HIV-1 p24 ELISA kit (Perkin Elmer) was used in accordance with the manufacturer’s instructions. Samples were read in duplicate on a TECAN Sunrise ELISA Plate Reader and analysed by Magellan Software Version 6.5. Analytical sensitivity of the ELISA was 4.3 pg/ml; reproducibility within the assay was C.V.: 5.5%.
2.8 Quantification of viral RNA by RT-qPCR (HIV RNA)
Isolation of viral RNA from culture supernatants was performed using QIAamp Viral RNA Kits (Qiagen). Purified viral RNA was quantified through amplification of HIV-1 gag by real-time reverse transcriptase polymerase chain reaction (RT-qPCR) with a detection limit of 10 viral RNA copies per microliter of supernatant. RT-qPCR was performed with the QuantiFast SYBR Green RT-PCR Kit (Qiagen) according to manufacturer's instructions in a 384 well plate on a Roche Lightcycler 480. The protocol utilizes a combination of gag “SK” primers that components of the Amplicor HIV-1 Monitor viral load test: SK145 primer (forward): AGTGGGGGGACATCAAGCAGCCATGCAAAT (30 bp; 72.5 Tm); SK431 primer (reverse): TGCTATGTCACTTCCCCTTGGTTCTCT (27 bp; 61.3 Tm). 2 ul of sample was used and sampling was performed in duplicate with a standard deviation of 0.5 between crossing threshold (Ct) numbers as quality control cut-off. Concentrations were calculated from a 10,000 copy number standard of HIV-1 HxB2.
2.9 Quantification of CD4+ T cell and NK cell numbers
Assessment of absolute cell numbers in the NK/CD4+ T cell co-culture was conducted using fluorescent CountBright absolute counting beads (Invitrogen) and subsequent direct acquisition of cells and beads by flow cytometry. CD4+ T cells and NK cells were stained with fluorescent dyes (cell tracker, Life technologies) prior to the co-culture to allow identification of the respective cell type.
2.10 Assessment of NK cell activation
Levels of NK cell activation have been determined through expression of CD107a on the surface of NK cells (Alter et al., 2004). Enriched primary NK cells were cultured for 3 days in complete media supplemented with 50 IU/ml hrIL-2 and 1 ng/ml hrIL-15 and subsequently co-cultured with differentially stimulated autologous CD4+ T cells. Autologous CD4+ T cells were either cultured with CD3/28 beads (Gibco) or PHA with or without subsequent HIV-1 infection for a total of 3 days. Monensin was added one hour after setup of the co-culture followed by additional 3 hours of incubation. Cells were stained for viability (Live/Dead Blue), expression of CD3, CD4, CD16, CD56 and then fixed with paraformaldehyde (Cell fix, BD).
2.11 Data acquisition and statistical analyses
Acquisition of flow cytometric data was performed on a BD LSR Fortessa (BD Biosciences) and further analysed using FlowJo software v7.6.5 (Tree Star, Inc.). GraphPad Prism 6.0 (GraphPad Software, Inc.) was used for statistical analyses. Comparison between two groups was performed using non-parametric Wilcoxon matched-pairs signed rank test for paired values. Spearman rank was used to test for correlation between parameters.
3. Results
The goal of this study was to optimize and evaluate a standardized in vitro assay which allows the quantitative assessment of the antiviral capacity of primary NK cells against HIV-1. Therefore, we co-incubated purified primary NK cells with autologous HIV-1-infected CD4+ T cells as target cells and monitored levels of HIV-1 replication for up to 14 days using the following three methods: i) frequency of HIV-1-infected CD4+ T cells by detection of intracellular HIV-1 p24 Gag (p24 ICS), ii) concentration of HIV-1 p24 Gag in culture supernatants by ELISA (p24 ELISA) and iii) quantification of copy numbers of HIV-1 RNA in culture supernatants by RT-qPCR (HIV RNA).
3.1 HIV-1 RNA copy numbers are a suitable marker for quantification of HIV-1 replication in vitro
First, we sought to compare the reproducibility of the three methods of HIV-1 quantification by monitoring HIV-1 replication in CD4+ T cells from 3 different healthy donors. CD4+ T cells were plated in at seven replicates and cultured for 14 days. Assessment of HIV-1 replication revealed that the reproducibility as determined by the coefficient of variation (C.V.) varied between each method. In comparison to the intracellular detection of p24 Gag in CD4+ T cells, RT-qPCR and p24 ELISA displayed consistently lower dispersion (Table 1). These results indicate that the frequency of p24+ CD4+ T cells as a measurement of HIV-1 replication is less consistent in the setting of this in vitro assay than HIV-1 RNA copy numbers or p24 Gag concentrations. In contrast, the results showed that HIV-1 RNA copy numbers determined by RT-qPCR are a suitable marker to quantify HIV-1 replication in vitro.
Table 1.
Intra-assay variation for different measurements of HIV-1 replication.
| Donor 1 | Donor 2 | Donor 3 | ||||
| Mean* | C.V. (%)# | Mean* | C.V. (%)# | Mean* | C.V. (%)# | |
| % p24+ CD4+ T cells | 0.15 | 96.1 | 1.99 | 69.8 | 0.81 | 78.7 |
| HIV-1 p24 conc. (pg/ml) | 24,426 | 24.0 | 47,121 | 17.2 | 25,377 | 13.2 |
| HIV-1 RNA (copies/µl) | 47,439 | 14.6 | 122,020 | 29.9 | 24,306 | 23.0 |
values were obtained from at least 7 replicates per donor;
coefficient of variation (C.V.).
3.2 Primary NK cells are able to inhibit HIV-1 replication in vitro in a dose-dependent manner
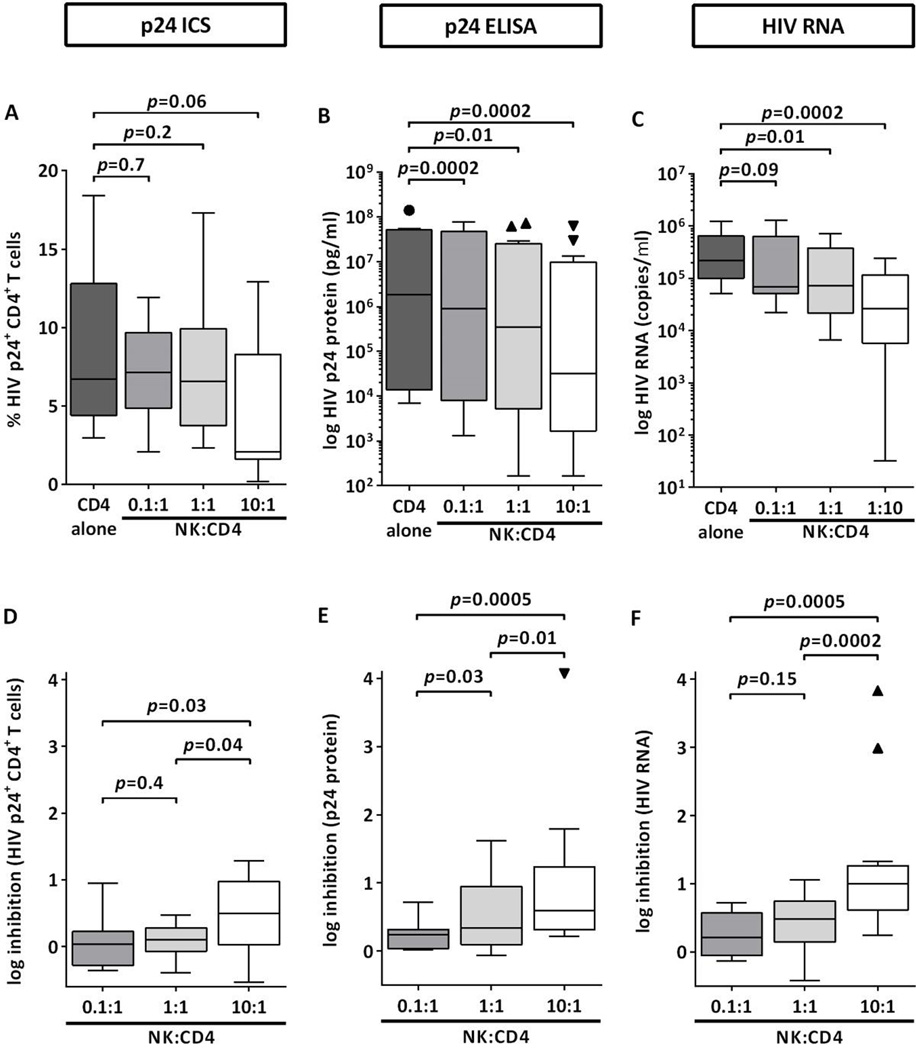
Next, we investigated the ability of the in vitro assay to detect NK-cell-mediated inhibition of HIV-1 replication. Autologous CD4+ T cells from 13 healthy volunteers were infected with HIV-1 at an MOI of 0.01 and co-incubated with primary NK cells at effector:target cell ratios ranging from 0.1:1 to 10:1 for 14 days. Generally, increasing numbers of NK cells in the co-culture were associated with reduced frequencies of p24+ CD4+ T cells as well as decreased p24 Gag concentration and HIV-1 RNA copy numbers in culture supernatants (Figure 1A–C). However, while p24 ELISA and RT-qPCR were able to detect the inhibitory effect of low NK cell numbers on HIV-1 replication, no significant changes in the percentages of p24+ CD4+ T cells were observed at low or equal numbers of NK cells. Similar results were obtained when NK-cell-mediated inhibition of HIV-1 replication was calculated based on the ratio between levels of HIV-1 replication in NK:CD4+ T cell co-cultures and CD4+ T cells alone (Figure 1D–F). Again, p24 ELISA and to a slightly lesser extent RT-qPCR were able to display the effects of increasing E:T ratios on HIV-1 replication. However, differences in NK-cell-mediated inhibition were detected between E:T ratios of 1:1 and 10:1 by all methods, indicating that a minimal number of NK cells is required to exert a measurable antiviral effect. Taken together, the assay was able to efficiently detect quantitative effects of different NK-cell effector:CD4+ T cell target ratios and the differential antiviral capacities of NK cells of donors, in particular when p24 ELISA and RT-qPCR were used as readouts of HIV-1 replication.
Figure 1. NK-cell-mediated inhibition of HIV-1 replication is dependent on the effector:target cell ratio between NK cells and CD4+ T cells.
The figure display levels of HIV-1 replication (A–C) and NK-cell-mediated inhibition (D–F) measured as frequency of p24+ CD4+ T cells (p24 ICS), concentration of p24 Gag (p24 ELISA) and copy numbers of viral RNA (HIV RNA) in the culture supernatant. HIV-1-infected autologous CD4+ T cells from 13 healthy donors were either cultured alone or in the presence of increasing numbers of primary NK cells. Panels A–C show levels of HIV-1 replication after 14 days of co-culture. Panels D–F display NK-cell-mediated inhibition of HIV-1 replication as compared to CD4+ T cells alone. Wilcoxon matched-pairs signed rank test was used to identify differences between two groups.
3.3 HIV-1 RNA copy numbers strongly correlate with production of p24 Gag
Overall detection of NK-cell-mediated inhibition of HIV-1 replication was achieved with all three virological readouts. However, all three methods displayed differential inter-assay variability and varying sensitivity. Therefore, it was further investigated whether the frequency of p24+ CD4+ T cells, p24 Gag concentrations and HIV-1 RNA copy numbers, generally correlated with each other. For this, the values of the three virological parameters were plotted against each other (Figure 2).
Figure 2. Correlations between different methods for quantification of HIV-1 replication.
Frequency of HIV-1 p24+ CD4+ T cells, p24 Gag concentration and HIV-1 RNA copy numbers in the supernatant of all individuals (n=13) and conditions were plotted against each other. Percentage of p24+ CD4+ T cells was assessed by flow cytometry, p24 Gag production was determined by ELISA and viral RNA was quantified by RT-qPCR respectively. Panel A displays absolute values of HIV-1 replication (52 data points); panel B shows log inhibition of HIV-1 replication as compared to CD4+ T cells alone (39 data points). Non-parametric spearman’s rank correlation was used to determine association between two methods.
Statistical analysis showed that HIV-1 RNA copy numbers strongly correlated with p24 Gag concentrations (Rs=0.85, p<0.0001) and to a lesser extent with the percentage of p24+ CD4+ T cells (Rs=0.41, p=0.003) (Figure 2A). However, transformation of the values to NK-cell-mediated inhibition using CD4+ T cells alone as a reference revealed significant correlations between all three readouts (Figure 2B). Again, HIV-1 RNA copy numbers and p24 Gag production showed the strongest co-dependence (Rs=0.896, p<0.0001), followed by HIV-1 RNA copy numbers and frequency of HIV-1-infected CD4+ T cells (Rs=0.71, p<0.0001). These results indicate that HIV-1 viral load as determined by HIV-1 RNA copy numbers and p24 Gag concentrations are interchangeable measures for quantification of NK-cell-mediated inhibition of HIV-1 replication with high co-dependence.
3.4 Levels of NK-cell-mediated inhibition can be detected by day 7 of NK/CD4+ T cell co-culture
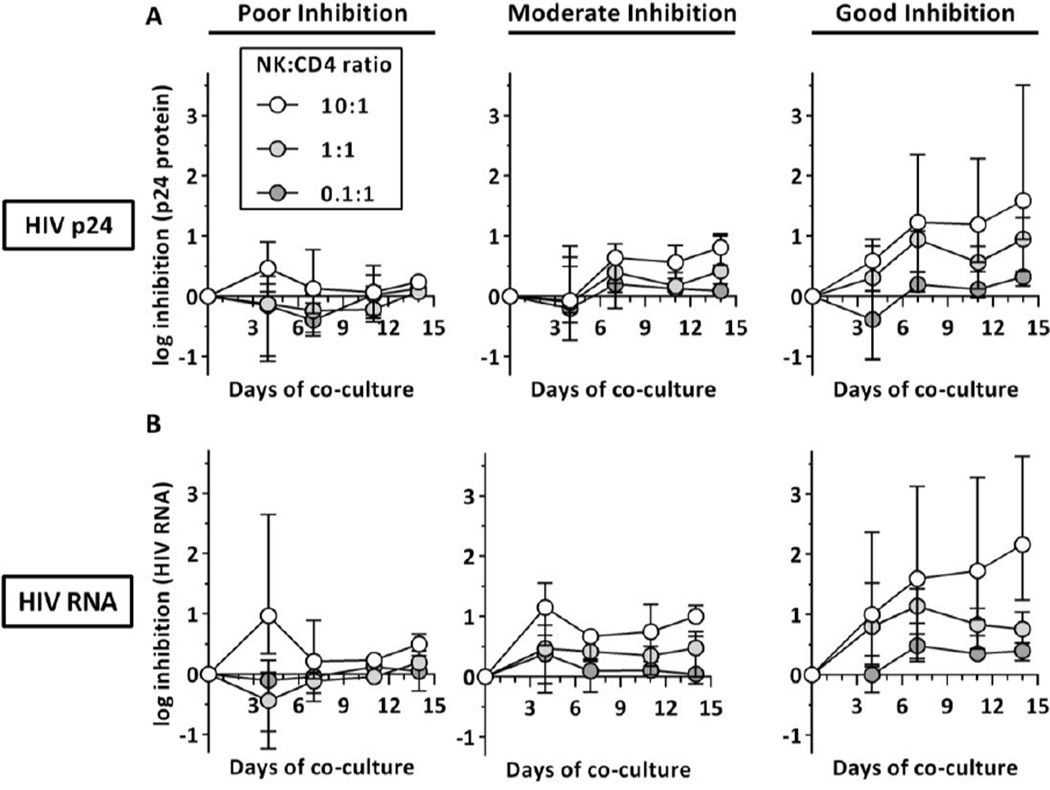
Finally, the kinetics of NK-cell-mediated inhibition of HIV-1 replication were determined over time. Therefore, p24 Gag production (Figure 3A) and HIV-1 viral load (Figure 3B) was monitored for 14 days and stratified in 3 groups depending on the antiviral capacity of the donor NK cells (poor, moderate or good).
Figure 3. Kinetics of NK-cell-mediated inhibition of HIV-1 replication.
The figure displays the kinetics of NK cell-mediated viral inhibition stratified into three donor groups with different antiviral capacities (poor: bottom third (n=4), moderate: middle third (n=5), good: top third (n=4) at day 14 with NK:CD4+ T cell ratio 10:1). p24 Gag production (upper panel, A) and HIV-1 RNA copy numbers (lower panel, B) at different NK:CD4+ T cell ratios (0.1:1, 1:1 and 10:1) were measured longitudinally over the course of 14 days.
For both virological readouts, NK cells with poor antiviral capacity (Figure 3, left panel) displayed low levels of inhibition at all times with one exception. Initial higher levels of inhibition were only observed at high NK-cell numbers at day 4 but were lost over time. In contrast, for donors with moderate and good antiviral capacity (Figure 3, middle and right panel) high E:T ratios were associated with increasing levels of inhibition over time indicating sustained NK-cell-mediated inhibition. For culture conditions with low and medium effector:target ratios (0.1:1, 1:1) inhibition was less consistent, however the association between effector:target ratios and increasing antiviral capacity was observed as early as by day 7 of the co-culture. Correlation analysis (not displayed) between levels of inhibition at day 14 of the co-culture and earlier time points revealed significant correlation for all days using HIV RNA copy numbers with increasing R values for later time points day 7 (n=39; day 4: Rs=0.44, p=0.005; day 7: Rs=0.57, p=0.0001; day 11: Rs=0.62, p<0.0001). For p24 Gag production significant associations between culture days were observed for day 7 and 11; however no association was observed between day 4 and day 14 (n=39; day 4: Rs=0.13, p=0.42; day 7: Rs=0.62 p<0.0001; day 11: Rs=0.55, p=0.0003). Overall, these results show that shorter periods of the co-culture (day 7) are sufficient to detect and discriminate NK-cell-mediated inhibition of HIV-1 replication and confirmed that co-cultures with higher NK-cell numbers show the most consistent results.
3.5 NK cell co-culture results in minimal unspecific NK cell activation and does not affect CD4+ T cell numbers
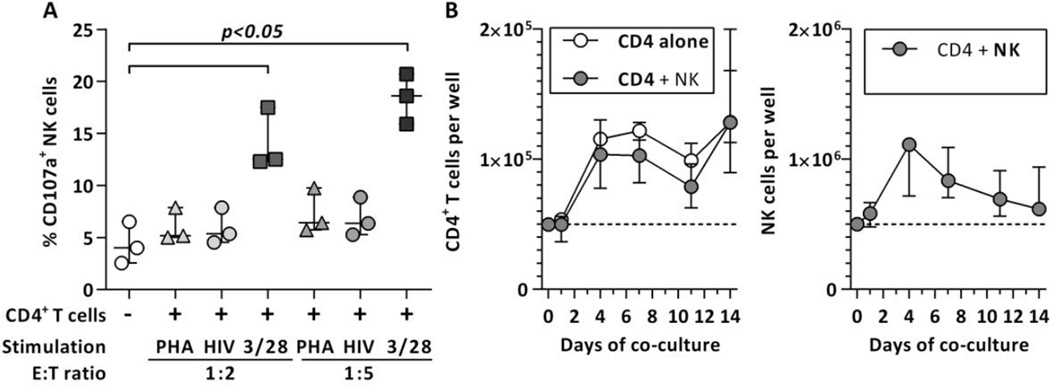
The observed results strongly indicate a donor and dose-dependent inhibition of HIV-1 replication in NK/CD4+ T cell co-cultures. However, factors other than the antiviral function of NK cells may potentially skew the observed results. In that regard, unspecific activation of NK cells und subsequent killing of uninfected bystander cells as well as limited survival of target and effector cell populations in the long-term culture need to be considered. Therefore, levels of NK cell activation as well as absolute cell numbers of CD4+ T cells and NK cells in the co-culture were determined. Our results showed no increase in NK cell degranulation when cultured with HIV-1-infected and uninfected PHA-stimulated CD4+ T cells (Figure 4A). Finally, assessment of absolute cell numbers of both CD4 T cells and NK cells showed an expansion of both effector and target cell populations (Figure 4B). Altogether, our results demonstrate that the inhibition of HIV-1 replication in the co-culture is not significantly impacted by the investigated factors and therefore, most likely attributed to the direct and indirect antiviral functions of NK cells.
Figure 4. NK cell activation and effector/target cells numbers during NK/CD4+ T cell co-culture.
Figure 4A displays the levels of NK cell degranulation (CD107a expression) following co-incubation with differentially stimulated CD4+ T cells at two effector:target cell ratios. CD4+ T cells were either stimulated with CD3/28 beads (squares) or PHA with (circles) or without (triangles) subsequent in vitro HIV-1 infection for 3 days and then co-incubated with enriched NK cells for 4 hours. Figure 4B shows the kinetics of NK cells and CD4+ T cells numbers during the co-culture. CD4+ T cell numbers following infection with HIV-1 (MOI: 0.01) in the presence (grey circles) or absence of NK cells (clear circles) are shown in the left panel of 4B, NK cell numbers on the right panel. The initial effector:target cell ratio was 10:1. Cell numbers were normalized to 5*104 and 5*105 per well respectively for better comparison. Median and IQR of 3 independent experiments (subjects) are displayed for both experiments. Statistical analysis was conducted using Friedman test and Dunn’s multiple comparison test.
4. Discussion
Increasing evidence indicated that NK cells have an important role in the control of HIV-1 infection (Jost and Altfeld, 2013). However, no precise receptor profile associated with efficient elimination of infected cells or control of HIV-1 has been identified so far. Given recent advances in the adoptive transfer of NK cells in tumor therapy (Davies et al., 2014), NK cells may serve as a potential target for immunotherapeutic interventions in HIV-1 infection. Thus, identification and characterization of those NK cells with potent anti-HIV-1 activity would be a promising approach for development of NK-cell-based vaccines or strategies to achieve a functional cure. In this study we describe a protocol to determine the antiviral capacity of primary NK cells. In the described in vitro assay, HIV-1 replication was monitored for up to 14 days in the presence or absence of primary NK cells using HIV-1-infected autologous CD4+ T cells. Furthermore, evaluation of various methods to measure HIV-1 replication showed that quantification of p24 Gag by ELISA and HIV-1 RNA copy numbers by RT-qPCR in culture supernatants gave the most robust and consistent results when compared to the percentage of CD4+ T cells expressing p24 Gag. Overall, using this assay we demonstrate that NK cells are able to inhibit HIV-1 replication in vitro in a dose and donor-dependent manner; thus providing an optimized tool to identify NK cells with high antiviral potential.
Several studies have used autologous HIV-1-infected CD4+ T cells to investigate NK-cell responses upon exposure to HIV-1-infected target cells (Bonaparte and Barker, 2003; Ward et al., 2007; Fogli et al., 2008; Davis et al., 2011; Lisovsky et al., 2015b; Norman et al., 2011). While Lisovsky and co-workers assessed the levels of cytokine production and degranulation in NK-cell subpopulations, the majority of the studies determined the direct cytotoxic potential of NK cells by assessing lysis of HIV-1-infected cells using various killing assays (Bonaparte and Barker, 2003; Fogli et al., 2008; Norman et al., 2011; Ward et al., 2007). Both approaches provided valuable information on the molecular mechanisms of target cell recognition by NK cells and the quality and quantity of NK-cell-mediated effector functions in responses to HIV-1-infected cells. However, cytokine production as well as CD107a expression, which is a well-established marker for NK-cell activation (Alter et al., 2004), cannot be used as suitable surrogate markers for elimination of infected cells or inhibition of HIV-1 replication. In contrast, killing assays such as the 51chromium release cytotoxicity assay (Kiessling et al., 1975) or the Calcein acetoxymethyl ester (CAM) cytotoxicity assay (Neri et al., 2001) are suitable assays to directly measure NK-cell cytotoxicity. However, NK cells are also able to indirectly affect HIV-1 replication by the production and release of CCR5 ligand chemokines that compete with HIV-1 for the co-receptor CCR5 (Fauriat et al., 2010; Oliva et al., 1998; Song et al., 2014). Therefore, longitudinal assessment of HIV-1 replication in the presence of NK cells integratea the direct and indirect antiviral functions of NK cells and would provide additional information. However, factors independent of the direct and indirect antiviral functions of NK cells may potentially skew the results of our and other in vitro inhibition models and therefore require thorough evaluation. Such factors include the survival of effector and target cell populations as well as their modulation by the type and amount of cytokines added in long-term cultures. In the present study, our results show that both cell types undergo early expansion and remain above the initially deployed cell numbers. In that regard the overall numbers of CD4+ T cells were not affected by the presence of NK cells indicating a negligible effect of NK cells on the overall survival of CD4+ T cells. This observation was further confirmed by demonstrating that neither the presence of IL-2 and IL-15 nor the stimulation of autologous CD4+ T cells with PHA significantly induced unspecific activation of NK cells. Lastly, it is possible that the composition of the donor T cell pool may affect infection of the T cell population and subsequently seeding of the infection. In fact, virus production varied by the factor >10 across the study population indicating a differential susceptibility for HIV-1 infection for each donor CD4+ T cell pool. However, correlation analysis (not shown) revealed that levels of NK-cell-mediated inhibition were not associated with the overall virus production (in the absence of NK cells) indicating that the antiviral capacity of NK cell of each donor represents an independent factor and may be only partially affected by the susceptibility of the autologous CD4+ T cells to infection. Altogether, our results show that the viral inhibition assay is able to comprehensively detect NK-cell-mediated inhibition of HIV-1 replication and thus can serve as an additional tool that takes direct and indirect NK-cell-mediated effects on HIV-1 replication into account.
An additional goal of this study was to evaluate various methods that have been applied to quantify HIV-1 replication in vitro. A few studies assessed the antiviral potential of NK cells by assessing NK-cell-mediated inhibition of HIV-1 replication in vitro (Alter et al., 2011, 2007; Oliva et al., 1998; Song et al., 2014). Alter et al. and more recently Song et al. determined the frequency of infected cells by intracellular detection of p24 Gag as well as quantified p24 Gag in culture supernatants. While some of these studies observed a significant correlation between p24 Gag and the frequency of infected cells, systematic correlation analysis of our larger dataset failed to reach statistical significance. However, we observed a significant correlation between levels of NK-cell-mediated inhibition using these two readouts, indicating that the antiviral effects of NK cells are associated with a lower frequency of infected cells. Nevertheless, given the high coefficient of variance for intracellular p24 Gag detection this method should be used in combination with other measures of HIV-1 replication. Apart from the intracellular detection or quantification of p24 Gag other methods have been used to measure viral replication. Activity of the HIV-1 reverse transcriptase was used as a readout by Bernstein et al. investigating the antiviral activity of neonatal NK cells against HIV-1-infected CD4+ T cells (Bernstein et al., 2004) and Oliva et al. exploring the effect of CC chemokine production by NK cells (Oliva et al., 1998). Initially the assay required a facility to handle radioactive isotopes as the assay was originally based on the radiometric quantification of incorporated [32P] nucleotides (Willey et al., 1988). However, modified versions of this method do not require radioactive nucleotides anymore and seem to at least outperform p24 ELISA (Iqbal et al., 2007; Vermeire et al., 2012). As HIV-1 RNA copy numbers are widely established in HIV-1 diagnostics as a surrogate marker for HIV-1 replication and clinical progression of HIV-1 infection (Murray et al., 1999; HIV Surrogate Marker Collaborative Group, 2000), we included this virological parameter in our assay. Our results show that dose and donor-dependent inhibition of HIV-1 replication by NK cells can be detected by HIV-1 RNA copy numbers. In addition, HIV-1 viral load significantly correlated with both frequency of infected cells and levels of p24 Gag in culture supernatant thus representing a suitable method to measure NK-cell-mediated inhibition of HIV-1 replication in vitro. Of note, HIV-1 RNA can be used for downstream applications such as RNA-sequencing to detect in vitro viral escape by NK-cell-mediated immune pressure (Yang et al., 2003; Alter et al., 2011). Furthermore, the assay can be useful to investigate the molecular determinants of NK-cell recognition of HIV-1-infected cells. That would include blocking experiments with NK-cell receptor-specific antibodies, sorting of NK cell sub-populations with distinct receptor profiles (Alter et al., 2007) or generation of NK-cell clones with defined phenotypes. In particular, the contribution of inhibitory NK cell receptors such as NKG2A (Davis et al., 2016), KIR2DL1/L3 (Körner et al., 2014) and KIR3DL1 (Boudreau et al., 2016) to the antiviral capacity of NK cells could be further elucidated. Finally, given the increasing relevance of broadly neutralizing antibodies and their potential to induce subsequent immune functions such as antibody-dependent (NK) cell-mediated cytotoxicity (ADCC) in HIV-1 cure approaches (Caskey et al., 2015; Kim et al., 2015; Stephenson and Barouch, 2016), the assay can be modified through addition of autologous serum or purified antibodies to study ADCC as previously described (Dugast et al., 2014).
Taken together, the in vitro viral replication assay described here provides a versatile tool to study the antiviral capacity of NK cells against HIV-1, thus complementing existing methods to identify NK cells with potent anti-HIV-1 activity for immunotherapeutic interventions in HIV-1 infection.
Highlights.
We developed a standardized assay to assess the antiviral capacity of NK cells.
NK cells from different donors differentially inhibit HIV-1 replication in vitro.
HIV-1 RNA is a sensitive marker for HIV-1 replication in this in vitro assay.
Acknowledgments
The authors gratefully acknowledge the support and assistance of the Ragon Institute Flow Cytometry core and the Ragon Institute Virology platform. This study was supported by the National Institutes of Health (NIH) (R01-AI067031-08), the Ragon Institute of MGH, MIT and Harvard and the National Natural Science Foundation of China (81261120379). Sebastian Lunemann was supported by the German Research Foundation (SFB841).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Xuan He, Email: hexuan09@yahoo.com.
Camille R. Simoneau, Email: csimoneau@mgh.harvard.edu.
Mitchell E. Granoff, Email: mitchell.granoff@gmail.com.
Sebastian Lunemann, Email: sebastian.lunemann@hpi.uni-hamburg.de.
Anne-Sophie Dugast, Email: adugast@merrimack.com.
Yiming Shao, Email: yshao@bjmu.edu.cn.
Marcus Altfeld, Email: marcus.altfeld@hpi.uni-hamburg.de.
Christian Körner, Email: christian.koerner@hpi.uni-hamburg.de.
References
- Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI, Carrington M, Allen TM, Altfeld M. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HB, Kinter AL, Jackson R, Fauci AS. Neonatal natural killer cells produce chemokines and suppress HIV replication in vitro. AIDS Res. Hum. Retroviruses. 2004;20:1189–1195. doi: 10.1089/aid.2004.20.1189. [DOI] [PubMed] [Google Scholar]
- Bonaparte MI, Barker E. Inability of natural killer cells to destroy autologous HIV-infected T lymphocytes. Aids. 2003;17:487–494. doi: 10.1097/00002030-200303070-00003. [DOI] [PubMed] [Google Scholar]
- Boudreau JE, Mulrooney TJ, Le Luduec J-B, Barker E, Hsu KC. KIR3DL1 and HLA-B Density and Binding Calibrate NK Education and Response to HIV. J. Immunol. Baltim. Md. 2016:1950. doi: 10.4049/jimmunol.1502469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlingham WJ, Ceman SS, DeMars R. Secretion and cell surface expression of IgG1 are impaired in human B lymphoblasts that lack HLA-A, -B, and -C antigens. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8005–8009. doi: 10.1073/pnas.86.20.8005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Carrington M, Alter G. Innate immune control of HIV. Cold Spring Harb. Perspect. Med. 2012;2:a007070. doi: 10.1101/cshperspect.a007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JOJ, Stringaris K, Barrett AJ, Rezvani K. Opportunities and limitations of natural killer cells as adoptive therapy for malignant disease. Cytotherapy. 2014;16:1453–1466. doi: 10.1016/j.jcyt.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ZB, Cogswell A, Scott H, Mertsching A, Boucau J, Wambua D, Le Gall S, Planelles V, Campbell KS, Barker E. A Conserved HIV-1-Derived Peptide Presented by HLA-E Renders Infected T-cells Highly Susceptible to Attack by NKG2A/CD94-Bearing Natural Killer Cells. PLoS Pathog. 2016;12:e1005421. doi: 10.1371/journal.ppat.1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ZB, Ward JP, Barker E. Preparation and Use of HIV-1 Infected Primary CD4+T-Cells as Target Cells in Natural Killer Cell Cytotoxic Assays. J. Vis. Exp. 2011 doi: 10.3791/2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugast A-S, Stamatatos L, Tonelli A, Suscovich TJ, Licht AF, Mikell I, Ackerman ME, Streeck H, Klasse PJ, Moore JP, Alter G. Independent evolution of Fc- and Fab-mediated HIV-1-specific antiviral antibody activity following acute infection. Eur. J. Immunol. 2014;44:2925–2937. doi: 10.1002/eji.201344305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauriat C, Long EO, Ljunggren H-G, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115:2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogli M, Mavilio D, Brunetta E, Varchetta S, Ata K, Roby G, Kovacs C, Follmann D, Pende D, Ward J, Barker E, Marcenaro E, Moretta A, Fauci AS. Lysis of Endogenously Infected CD4+ T Cell Blasts by rIL-2 Activated Autologous Natural Killer Cells from HIV-Infected Viremic Individuals. PLoS Pathog. 2008;4:e1000101. doi: 10.1371/journal.ppat.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIV Surrogate Marker Collaborative Group. Human immunodeficiency virus type 1 RNA level and CD4 count as prognostic markers and surrogate end points: a meta-analysis. HIV Surrogate Marker Collaborative Group. AIDS Res. Hum. Retroviruses. 2000;16:123–1133. doi: 10.1089/088922200414965. [DOI] [PubMed] [Google Scholar]
- Iqbal HS, Balakrishnan P, Cecelia AJ, Solomon S, Kumarasamy N, Madhavan V, Murugavel KG, Ganesh AK, Solomon SS, Mayer KH, Crowe SM. Use of an HIV-1 reverse-transcriptase enzyme-activity assay to measure HIV-1 viral load as a potential alternative to nucleic acid-based assay for monitoring antiretroviral therapy in resource-limited settings. J. Med. Microbiol. 2007;56:1611–1614. doi: 10.1099/jmm.0.47456-0. [DOI] [PubMed] [Google Scholar]
- Jost S, Altfeld M. Control of Human Viral Infections by Natural Killer Cells. Annu. Rev. Immunol. 2013;31:163–194. doi: 10.1146/annurev-immunol-032712-100001. [DOI] [PubMed] [Google Scholar]
- Jost S, Altfeld M. Evasion from NK cell-mediated immune responses by HIV-1. Microbes Infect. Inst. Pasteur. 2012;14:904–915. doi: 10.1016/j.micinf.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur. J. Immunol. 1975;5:112–117. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- Kim JH, Excler J-L, Michael NL. Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection. Annu. Rev. Med. 2015;66:423–437. doi: 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- Körner C, Granoff ME, Amero MA, Sirignano MN, Vaidya SA, Jost S, Allen TM, Rosenberg ES, Altfeld M. Increased frequency and function of KIR2DL1-3(+) NK cells in primary HIV-1 infection are determined by HLA-C group haplotypes. Eur. J. Immunol. 2014 doi: 10.1002/eji.201444751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisovsky I, Isitman G, Bruneau J, Bernard NF. Functional analysis of NK cell subsets activated by 721.221 and K562 HLA-null cells. J. Leukoc. Biol. 2015a;97:761–767. doi: 10.1189/jlb.4AB1014-499R. [DOI] [PubMed] [Google Scholar]
- Lisovsky I, Isitman G, Song R, DaFonseca S, Tremblay-McLean A, Lebouché B, Routy J-P, Bruneau J, Bernard NF. A higher frequency of NKG2A+ than NKG2A- NK cells respond to autologous HIV-infected CD4 cells irrespective of whether they co-express KIR3DL1. J. Virol. 2015b doi: 10.1128/JVI.01546-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Gao X, Lee J-H, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002 doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JS, Elashoff MR, Iacono-Connors LC, Cvetkovich TA, Struble KA. The use of plasma HIV RNA as a study endpoint in efficacy trials of antiretroviral drugs. AIDS Lond. Engl. 1999;13:797–804. doi: 10.1097/00002030-199905070-00008. [DOI] [PubMed] [Google Scholar]
- Nash WT, Teoh J, Wei H, Gamache A, Brown MG. Know Thyself: NK-Cell Inhibitory Receptors Prompt Self-Tolerance, Education, and Viral Control. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri S, Mariani E, Meneghetti A, Cattini L, Facchini A. Calcein-Acetyoxymethyl Cytotoxicity Assay: Standardization of a Method Allowing Additional Analyses on Recovered Effector Cells and Supernatants. Clin. Diagn. Lab. Immunol. 2001;8:1131–1135. doi: 10.1128/CDLI.8.6.1131-1135.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JM, Mashiba M, McNamara LA, Onafuwa-Nuga A, Chiari-Fort E, Shen W, Collins KL. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat. Immunol. 2011;12:975–983. doi: 10.1038/ni.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva A, Kinter AL, Vaccarezza M, Rubbert A, Catanzaro A, Moir S, Monaco J, Ehler L, Mizell S, Jackson R, Li Y, Romano JW, Fauci AS. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J. Clin. Invest. 1998;102:223–231. doi: 10.1172/JCI2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange JS. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. 2002;4:1545–1558. doi: 10.1016/s1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- Song R, Lisovsky I, Lebouché B, Routy J-P, Bruneau J, Bernard NF. HIV Protective KIR3DL1/S1-HLA-B Genotypes Influence NK Cell-Mediated Inhibition of HIV Replication in Autologous CD4 Targets. PLoS Pathog. 2014;10:e1003867. doi: 10.1371/journal.ppat.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson KE, Barouch DH. Broadly Neutralizing Antibodies for HIV Eradication. Curr. HIV/AIDS Rep. 2016;13:31–37. doi: 10.1007/s11904-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, Taghon T, Pizzato M, Verhasselt B. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PloS One. 2012;7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SM, Khakoo SI, Biron CA. Natural killer cell responses during viral infections: flexibility and conditioning of innate immunity by experience. Curr. Opin. Virol. 2011;1:497–512. doi: 10.1016/j.coviro.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J, Bonaparte M, Sacks J, Guterman J, Fogli M, Mavilio D, Barker E. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood. 2007;110:1207–1214. doi: 10.1182/blood-2006-06-028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang OO, Sarkis PTN, Ali A, Harlow JD, Brander C, Kalams SA, Walker BD. Determinants of HIV-1 Mutational Escape From Cytotoxic T Lymphocytes. J. Exp. Med. 2003;197:1365–1375. doi: 10.1084/jem.20022138. [DOI] [PMC free article] [PubMed] [Google Scholar]