Abstract
SMP-domains are found in proteins that localize to membrane contact sites. Elucidation of the properties of these proteins gives clues as to the molecular bases underlying processes that occur at such sites. Described here are recent discoveries concerning the structure, function, and regulation of the Extended-Synaptotagmin proteins and ERMES complex subunits, SMP-domain proteins at endoplasmic reticulum (ER)- plasma membrane and ER-mitochondrial contacts, respectively. They act as tethers contributing to the architecture of these sites and as lipid transporters that convey glycerolipids between apposed membranes.
Membrane contact sites (MCSs), regions where the membranes of two different organelles are in close proximity (typically 10–30 nm), are implicated in a variety of physiological functions, such as regulation of Ca2+ dynamics, signal transduction and control of lipid homeostasis (1–3). Processes that take place at these sites have been the focus on intense investigations in recent years. However, the molecular bases for many such processes remain poorly understood. One approach to a better mechanistic understanding is to characterize the properties and functions of the proteins that localize there.
Many proteins which are selectively concentrated at MCSs also participate in their formation and/or stabilization by tethering the two participating membranes. Some of these proteins, in addition, contain modules that harbor lipids in a hydrophobic cavity and mediate lipid transfer between the two adjacent bilayers independently of membrane fusion and fission reactions. One such domain is the so-called SMP (synaptotagmin-like mitochondrial-lipid binding protein) domain (4–7). Here we describe advances regarding the function of SMP family proteins, focusing on the better characterized members of the group. These are the Extended-Synaptotagmins (E-Syt1, 2 3 in mammals and tricalbin1, 2, 3 in yeast), conserved in all eukaryotes, which localize to contact sites between the plasma membrane (PM) and the endoplasmic reticulum (ER) (8, 9), and the ER-mitochondrial encounter structure (ERMES), a complex so far identified across many of the eukaryotic supergroups except metazoans (10, 11 ).
Extended -Synaptotagmins
The E-Syts act as tethers between the ER and plasma membrane (8, 9). Each E-Syt has an N-terminal β-hairpin embedded in the ER membrane (9), which is followed by the SMP domain and three to five C2 modules in the C-terminal portion of the molecule (Fig. 1A). Nomenclature for these proteins derives from their distant resemblance to synaptotagmin, the Ca2+ sensor for exocytosis, which has an N-terminal membrane anchor followed by two C2 domains (12). C2 domains, including those of synaptotagmin, frequently function in membrane binding (13). Accordingly, the C2 domains in the E-Syts mediate their binding to the PM, and thus the recruitment to the PM of the ER to which they are anchored, thereby promoting the formation or expansion of ER-PM contact sites (9, 14, 15). PM binding involves the interaction of their most C-terminal C2 domain (C2C for E-Syts 2/3 and C2E for E-Syt1) to PI(4,5)P2, a phosphoinositide enriched in this membrane (9, 14, 15) (Fig. 1B). This tethering function, however, is further regulated by Ca2+.
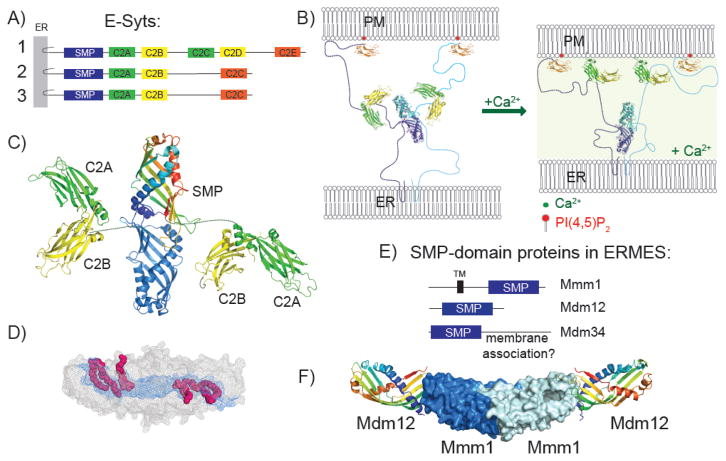
Fig. 1. Architecture and structure of SMP-domain proteins.
(A) E-Syt domain structure. The E-syts are anchored in the ER by an N-terminal beta-hairpin. Their cytosolic portion consists of an SMP domain and 3-5 C2 domains. The most C-terminal C2 domain, which has a type I C2-fold like synaptotagmin, recognizes PI(4,5)P2 at the plasma membrane (9, 14, 15). The other C2 domains, which have a type II fold, are arranged in pairs, with the more N-terminal domain interacting with membrane in the presence of calcium (7, 12, 16). (B) Schematic for membrane tethering by E-Syt2. The distance between contact site membranes may change in response to elevated calcium levels, when C2A is expected to bind membrane. PDB entries 4P42, 2DMG, and 4NPK were used in constructing this cartoon, which is roughly to scale. (C) Structure of an E-Syt2 fragment containing the SMP domain, C2A, and C2B (PDB entry 4P42) (7). One SMP module in the SMP dimer is blue; the other is colored from blue at the N-terminus to red at the C-terminus. (D) Mesh representation of the SMP dimer. Hydrophobic residues lining the channel along the length of the dimer are blue. In the structure, the dimer binds two glycerophospholipids and two detergent molecules, such that hydrophobic and hydrophilic portions are in the channel and extruded into the solvent, respectively. Hydrophilic portions are disordered and were not modeled. (E) Domain architecture of SMP-proteins in the ERMES complex. Mmm1 has a trans-membrane helix anchoring it to the ER; the C-terminal portion of Mdm34 may be required for its association with mitochondria. (F) Organization of the Mmm1-Mdm12 heterotetramer subcomplex of ERMES, based on (30). Mmm1 SMP domains dimerize in the same head-to-head fashion as E-Syt2; Mdm12 associates with Mmm1 head-to-tail.
The C2A and C2B domains of E-Syt2 form an arch-like structure which interacts with membranes when Ca2+ occupies several calcium binding sites in C2A (7, 12, 16) (Fig. 1B–C). Sequence conservation suggests that C2A–C2B in E-Syt3 and C2A–C2B and C2C–C2D in E-Syt1 assemble similarly, with the more N-terminal C2 domain in each pair mediating membrane interaction in a Ca2+ dependent manner. Thus, while E-Syt1 localizes throughout the ER, elevated levels of cytosolic Ca2+ significantly enhance its accumulation at ER-PM contact sites and E-Syt1 over expression leads to a ~7 nm decrease in contact site inter-membrane distance (14). Although E-Syts 2 and 3, when over expressed alone, are constitutively targeted to ER-PM contacts (9), Ca2+ likely also leads to shortened inter-membrane distances at contacts formed by these proteins as C2A engages the membrane (Fig. 1B). Additionally, E-Syt2 and E-Syt3 form heterodimers with E-Syt1, with the resulting heterodimer having properties intermediate between their properties and those of E-Syt1 (9).
The structural analysis of a fragment of E-Syt2 (7), including its SMP-domain and the two adjacent tandem C2 domains (C2A–C2B), demonstrated that the SMP-domain adopts a fold common to proteins in the tubular lipid-binding (TULIP) superfamily (Fig. 1C), validating bioinformatics predictions (5). Like other TULIP modules, the SMP domain of E-Syt2 consists of 6-beta strands and 3 helices arranged to form a barrel, whose interior is lined almost exclusively by hydrophobic residues. The SMP-domain mediates E-Syt dimerization, likely significant physiologically as residues at the interface are highly conserved, with two SMP-domains associating head-to-head into a ~90 Å long, slightly curved tube. The SMP dimer bears a striking resemblance to cholesterol ester-transfer protein (CETP) and bactericidal/permeability-increasing protein (BPI) (17, 18), two of the most studied TULIP proteins. Although both these proteins are monomers, they contains two TULIP domains tandemly arranged in the primary sequence. Such domains assemble into a head-to-head manner to form a tube similar to the SMP dimer of E-Syt2, though in CETP and BPI the two TULIP domains are separated by additional structural elements.
In the E-Syt2 SMP dimer, as well as in CETP and BPI, the hydrophobic cavity is continuous along almost the entire length of tube and connects to solvent via a long “seam”, thus defining an elongated channel. An exciting feature of the crystallographically determined SMP dimer was the presence of glycerophospholipids in the hydrophobic channel, strongly supporting a role of the E-Syts in lipid transfer (Fig. 1D). Accordingly, subsequent mass spectrometry analysis of E-Syt2 expressed in mammalian cells indicated that the protein co-purifies with glycerophospholipids, though without a preference for any particular headgroup, with four lipids bound per SMP dimer. This is in contrast to other intracellular lipid transport proteins that typically prefer one or two lipid species. The lipids presumably bind as observed in the crystal structure: their hydrophobic fatty acid moieties are nested in the hydrophobic channel, and the hydrophilic headgroups protrude through the seam and into solvent, similar to how CETP and BPI interact with glycerophospholipids (17, 18). A role of the SMP domain in lipid transfer between bilayers independently of membrane fusion is supported by cell free studies involving liposomes (unpublished).
Importantly, the property to harbor, and at least in some cases to transport, lipids, appears to be a shared function of members of the TULIP domain family, although many of them function in extracellular media, rather than within the cytosolic space. For example, CETP is an extensively characterized serum protein that exchanges lipids between high density and low density lipoproteins. Its TULIP domain can harbor several different lipids, including cholesterol esters (18). Another serum protein, BPI, plays a role in the innate immune response by extracting lipopolysacharides present in the membrane of GRAM-negative bacteria (19), and a BPI-family protein NRF-5 has been proposed to extract phosphatidylserine from the surface of dead cells for presentation to macrophages during apoptosis (20). In these proteins, the arrangement of two TULIP domains within the same polypeptide sequence may enhance the stability of the TULIP domain dimer in the extracellular environment.
How the E-Syt SMP dimer associates with membranes to extract and subsequently to deliver lipids remains unclear. One possibility is that the SMP tube associates with membranes length-wise, and that lipids enter via the seam near the middle of the dimer. Entry could also be via the tube tips, or still another possibility is that entry and exit routes are different. An interesting idea, given proposed roles of the neuronal protein synaptotagmin in perturbing the plasma membrane bilayer during exocytotic fusion (21), is that membrane binding by the E-Syt C2 domains may similarly affect the lipid bilayers to facilitate lipid extraction and/or delivery by the SMP module. However, we note that a number of SMP-containing proteins, including the ERMES components discussed below, lack C2 domains in favor of other tethering mechanisms (22). Thus, although we cannot exclude that the E-Syt C2 domains may facilitate transport by the SMP dimer, they likely are not required for transport except as tethering modules.
In a model proposed for the lipid transfer between lipoprotein particles by CETP (but see below for a different model), the TULIP domains act as bridges between apposed lipid templates (or membranes in the case of the E-Syts) (18, 22–24). Lipids could then “snorkel” through the aqueous cytosol with their fatty acid moieties protected in the hydrophobic tunnel while their hydrophilic lipid headgroups protrude into the solvent. Implicit in this model is that the SMP dimer associates with the two membranes via its tips and that lipids enter and exit there. One problem with this model is that at ~90 Å in length, the E-Syt dimer is too short to span typical ER-PM contact site distances, and additional multimerization to form longer tetrameric tubes is unlikely given geometrical constraints imposed by linker sequence lengths. Further, there is no evidence that the SMP-dimers associate stably with membrane at all, never mind in an end-on fashion. Electron micrographs of E-Syt induced contact sites do not support the formation of tunnels, as E-Syt density is concentrated midway between the two membranes and not uniformly as would be expected for membrane-bridging tubes (14); further, the residues at the tube tips are not well suited for membrane interaction as there are no hydrophobic loops or extended basic surfaces. Additionally, the SMP dimer does not interact with membranes in vitro (unpublished).
We therefore strongly favor a second model, in which the SMP lipid binding module is tethered between the ER and PM via flanking membrane binding regions and free to move between them, functioning as a shuttle to transfer lipid cargo between membranes (Fig. 1B). Indeed, most characterized lipid transporters act in this way, including members of the ORP and START families (25).
Like other lipid transfer proteins, the E-Syts likely transfer lipid passively along their concentration gradient. This process may result in net lipid transfer if metabolic or thermodynamic traps in the two participating membranes impact the free concentration of the lipid (1). Thus, passive lipid transport does not necessarily leads to an equilibration of the lipid composition of the two membranes and is compatible with the different bulk glycerolipid composition of the ER and the PM. It is also possible that the E-Syts mediate formation of lipid microdomains near contact sites, enriching zones in the PM with ER lipids and vice versa. Or the E-Syts might function in membrane expansion or contraction, carrying glycerophospholipids unidirectionally either to the PM from the ER or back, responding to yet to be identified signals much more quickly than would be possible via vesicular transport. An interesting possibility is that the E-Syts play a role in the transport of metabolic products of glycerophospholipid metabolism from the PM to the ER, in exchange for newly synthesized glycerolipids, as recently shown for the lipid transport protein Nir2 (26). This notion is attractive as the E-Syts respond to changes in calcium levels that accompany signal transduction and satisfying because none of these metabolites have elaborate headgroups, explaining why the E-Syts do not need to recognize glycerophospholipids specifically.
ERMES complex
ER-mitochondria contact sites are very abundant. Among their multiple functions they provide conduits for Ca2+ exchange between the two organelles and for the transfer of specific lipids. As mitochondria are not part of the secretory pathway, it has long been appreciated that they import many of their membrane lipids independently of vesicle trafficking. Additionally, the partnership between ER and mitochondria play a role in the metabolism of specific lipids, most notably PE (27). The ERMES complex was initially identified as a tethering complex that maintains these contact sites (10). ERMES comprises Mdm10, an integral membrane protein in the outer mitochondrial membrane (OMM), and three proteins, Mdm12 and Mdm34 and Mmm1, with an SMP domain (Fig. 1E). Mdm12 is cytosolic, whereas Mdm34 associates with the OMM and Mmm1 is an integral protein of the ER membrane. Additionally, the dynamin-related OMM-anchored GTPase Gem1, which is present substoichiometrically, has been suggested to regulate ERMES activity (28, 29).
Presence of SMP domains in ERMES proteins makes this complex the first identified lipid transporter specifically targeted to ER-mitochondrial contacts. Electron microscopy reconstruction and biochemistry studies have begun to elucidate the stoichiometry and architecture of the ERMES complex at low resolution (30). They indicate that the SMP domain of Mmm1 dimerizes in the same head-to-head fashion observed for E-Syt2, and that a monomer of Mdm12 interacts head-to-tail with each end of the Mmm1 dimer to form a slightly curved cylinder ~210 Å long and ~35 Å wide (Fig. 1F). Most likely, by analogy with the E-Syts, CETP and BPI, there is a long hydrophobic cavity that runs the length of the Mmm1 homodimer and potentially continues into Mdm12 as well. The SMP module of Mdm34 interacts with the Mmm1-Mdm12 heterotetramer via Mdm12, but the interaction is weak (30). Perhaps additional sequences outside the SMP domains are required for a stable interaction, or perhaps the interaction is a transient one. For example, for lipid transport, there may be a hand-off of lipid between Mmm1-Mdm12, anchored to the ER, and Mdm34 at the OMM.
Biochemical analysis of lipids bound by ERMES subcomplexes (Mmm1, Mdm12, Mmm1-Mdm12) indicates an interaction with glycerophospholipids and, among these, a preference for phosphatidylcholine (PC) binding (30). In the absence of high resolution structural data, the molecular basis for PC recognition remains unknown. Importantly, its specificity for PC indicates that one of the functions of ERMES is in PC transport from its site of synthesis in the ER to the mitochondria. Whether ERMES mediates the transport of additional lipids, perhaps in the reverse direction, is not known.
Future directions
Future developments will hopefully address how the E-Syts, ERMES, and other SMP-domain proteins extract, recognize and deliver lipids, whether they transport lipids vectorially and, if so, which mechanisms underlie vectorial transport, and how their activities at contact sites can be regulated. As yeast studies have shown that neither the E-Syts nor the ERMES complex are essential for cellular life (8, 10), their interplay or redundancy with other lipid transport proteins will also have to be elucidated (24, 31–34). This field of research, which is rapidly growing, will likely reveal important new concepts in membrane biology.
Highlights.
SMP-domain proteins tether apposing membranes at contact sites.
SMP-domain proteins bind and likely mediate glycerolipid transfer at contact sites.
Acknowledgments
We thank Dr. Pascal Egea for providing model coordinates for the Mmm1-Mdm12 subcomplex of ERMES (panel F in Fig. 1), and we are grateful to members of our labs for their comments regarding this manuscript. Work in the authors’ labs is supported by grants from the NIH (GM080616, GM114068 to KMR and R37NS036251 and DK082700 to PDC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lahiri S, Toulmay A, Prinz WA. Membrane contact sites, gateways for lipid homeostasis. Curr Opin Cell Biol. 2015 Apr;33:82. doi: 10.1016/j.ceb.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol. 2006 Aug;18:371. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Stefan CJ, Manford AG, Emr SD. ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol. 2013 Aug;25:434. doi: 10.1016/j.ceb.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci. 2012 Jan 1;125:49. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010 Aug 15;26:1927. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee I, Hong W. Diverse membrane-associated proteins contain a novel SMP domain. FASEB J. 2006 Feb;20:202. doi: 10.1096/fj.05-4581hyp. [DOI] [PubMed] [Google Scholar]
- 7.Schauder CM, et al. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014 Jun 26;510:552. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012 Dec 11;23:1129. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Giordano F, et al. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013 Jun 20;153:1494. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009 Jul 24;325:477. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wideman JG, Gawryluk RM, Gray MW, Dacks JB. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol Biol Evol. 2013 Sep;30:2044. doi: 10.1093/molbev/mst120. [DOI] [PubMed] [Google Scholar]
- 12.Min SW, Chang WP, Sudhof TC. E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc Natl Acad Sci U S A. 2007 Mar 6;104:3823. doi: 10.1073/pnas.0611725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jimenez JL, Davletov B. Beta-strand recombination in tricalbin evolution and the origin of synaptotagmin-like C2 domains. Proteins. 2007 Aug 15;68:770. doi: 10.1002/prot.21449. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Busnadiego R, Saheki Y, De Camilli P. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc Natl Acad Sci U S A. 2015 Apr 21;112:E2004. doi: 10.1073/pnas.1503191112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Idevall-Hagren O, Lu A, Xie B, De Camilli P. Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J. 2015 Sep 2;34:2291. doi: 10.15252/embj.201591565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, et al. Structure and Ca(2)(+)-binding properties of the tandem C(2) domains of E-Syt2. Structure. 2014 Feb 4;22:269. doi: 10.1016/j.str.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beamer LJ, Carroll SF, Eisenberg D. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science. 1997 Jun 20;276:1861. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 18.Qiu X, et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat Struct Mol Biol. 2007 Feb;14:106. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 19.Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against Gram-negative bacteria. Biochem Soc Trans. 2003 Aug;31:785. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Wang H, Kage-Nakadai E, Mitani S, Wang X. C. elegans secreted lipid-binding protein NRF-5 mediates PS appearance on phagocytes for cell corpse engulfment. Curr Biol. 2012 Jul 24;22:1276. doi: 10.1016/j.cub.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008 Jul;9:543. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 22.Kopec KO, Alva V, Lupas AN. Bioinformatics of the TULIP domain superfamily. Biochem Soc Trans. 2011 Aug;39:1033. doi: 10.1042/BST0391033. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, et al. Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat Chem Biol. 2012 Apr;8:342. doi: 10.1038/nchembio.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang A, John Peter AT, Kornmann B. ER-mitochondria contact sites in yeast: beyond the myths of ERMES. Curr Opin Cell Biol. 2015 Aug;35:7. doi: 10.1016/j.ceb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Mesmin B, Antonny B, Drin G. Insights into the mechanisms of sterol transport between organelles. Cell Mol Life Sci. 2013 Sep;70:3405. doi: 10.1007/s00018-012-1247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev Cell. 2015 Jun 8;33:549. doi: 10.1016/j.devcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011 Jan 10;192:7. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci U S A. 2011 Aug 23;108:14151. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stroud DA, et al. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol. 2011 Nov 4;413:743. doi: 10.1016/j.jmb.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 30.AhYoung AP, et al. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci U S A. 2015 Jun 23;112:E3179. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbaz-Alon Y, et al. Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep. 2015 Jul 7;12:7. doi: 10.1016/j.celrep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murley A, et al. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J Cell Biol. 2015 May 25;209:539. doi: 10.1083/jcb.201502033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gatta AT, et al. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife. 2015;4 doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez Montoro A, Ungermann C. StARTing to understand membrane contact sites. Trends Cell Biol. 2015 Sep;25:497. doi: 10.1016/j.tcb.2015.07.001. [DOI] [PubMed] [Google Scholar]