Abstract
Small-angle X-ray scattering measurements can provide valuable information about the solvent environment around biomolecules, but it can be difficult to extract solvent-specific information from observed intensity profiles. Intensities are proportional to the square of scattering amplitudes, which are complex quantities. Amplitudes in the forward direction are real, and the contribution from a solute of known structure (and from the waters it excludes) can be estimated from theory; hence, the amplitude arising from the solvent environment can be computed by difference. We have found that this “square root subtraction scheme” can be extended to non-zero q values, out to 0.1 Å−1 for the systems considered here, since the phases arising from the solute and from the water environment are nearly identical in this angle range. This allows us to extract aspects of the water and ion distributions (beyond their total numbers), by combining experimental data for the complete system with calculations for the solutes. We use this approach to test molecular dynamics and integral-equation (3D-RISM (three-dimensional reference interaction site model)) models for solvent structure around myoglobin, lysozyme, and a 25 base-pair duplex DNA. Comparisons can be made both in Fourier space and in terms of the distribution of interatomic distances in real space. Generally, computed solvent distributions arising from the MD simulations fit experimental data better than those from 3D-RISM, even though the total small-angle X-ray scattering patterns are very similar; this illustrates the potential power of this sort of analysis to guide the development of computational models.
I. INTRODUCTION
Ions and water molecules have been long known to play crucial roles in governing biomolecule stability and function.1–7 Elucidating how ions and water molecules distribute themselves around the solutes can provide valuable insights into their function and can also provide experimental tests for theoretical predictions.4–6,8–10 However, there are few experimental methods that directly probe the positions of ions and water molecules in the solution. Ion counting via dialysis can provide a quantitative measure of the ionic atmosphere around the solute, but it provides only a total (excess) number, and not any information the shape of the ion cloud.11,12 The q = 0 limit of small-angle X-ray scattering (SAXS) can provide similar excess counts for both water and ions.13 Anomalous small angle X-ray scattering (ASAXS) data in principle yield additional information about the extent of perturbations of the ion/water environment,14–17 but the ASAXS signal is known to be intertwined with all components in the system, complicating the analysis.13,18 Efforts to extract the contribution from ions to ASAXS profiles date back to 2003 with the work of Ballauff and coworkers.15,19 Using multiple energy ASAXS, they were able to decompose the total scattering intensity into solute-ion and ion-ion contributions, though limited to spherical solutes. Recently, Meisburger et al. proposed a similar approach to separate information about the ion contribution around a DNA duplex, using the heavy ion replacement SAXS rather than ASAXS profiles.20 Both approaches, nonetheless, only show the solute-ion and ion-ion correlations in reciprocal space and are not readily extended to the water distribution.
In this paper, we propose a new method of analysis to extract both water and ion distributions from SAXS profiles provided the scattering intensities are calibrated on the absolute scale. Both the excess number (as in ASAXS and ion-counting experiments12,16) and (low-resolution) information about the actual distribution can be obtained for ions and water molecules. The correlation between solute-ion and solute-water can be displayed in both reciprocal space (as partial intensities) and real space (as interatomic distribution functions). Since the focus is on ions and water, the proposed analysis requires knowledge about the biomolecule structure in advance. Although the proposed method is approximate, we use theoretical models to demonstrate that the errors are rather small for q between 0 and 0.1 Å−1. The resulting ion and water distributions are then used to test predictions from integral equation theory21–25 and explicit MD simulation for relatively rigid proteins and a DNA duplex.
II. THEORY AND BACKGROUND
A. Calculation of SAXS profiles
Here we briefly discuss the procedure of calculating SAXS profiles from MD simulation and 3D-RISM (three-dimensional reference interaction site model), which has been described in more detail elsewhere.13,26–31 (A brief summary of 3D-RISM theory is given in Section V B.) X-ray scattering experiments on biomolecules compare the scattering intensity from the sample of interest to a “blank” with just solvent present, and report the difference, or “excess” intensity,
| (1) |
where the bracket indicates that the intensities are averaged over the measurement time and volume. and are Fourier transforms of the scattering amplitudes for the sample and blank, respectively,
| (2) |
where is the electron density in the system. It has been shown that the total intensity can be approximately (though usefully) rewritten as13,26,29
| (3) |
where and are Fourier transforms for the sample and blank, respectively, but here only considering regions where there is excess/deficit electron density relative to the bulk value. In 3D-RISM, the second and third terms vanish,13 leading to
| (4) |
The approximation made in going from Eqs. (3) to (4) has been shown to be valid up to q = 1.5 Å−1.13 Finally, the angular averaging is performed to obtain the total intensity,
| (5) |
The total excess amplitude can be expressed as the sum of terms arising from the solute (biomolecule) and the solvent,
| (6) |
The separation between Fsolu and Fsolv can be made in different ways and is primarily for convenience in interpreting results. Here we have chosen to include in the Fsolu term the scattering from the excess electron density in the region of space occupied by the solute,
| (7) |
The first term on the right-hand-side represents the scattering from the solute atoms, where is the atomic scattering factor and Bj is the B-factor of atom j. The second term gives the contribution from the (negative) excess solvent density in the volume occupied by the solute; as in earlier work,13,32 we use a “cube method” to compute the scattering from a three-dimensional voxel, so that
| (8) |
where is the excess electron density arising from the solvent; a, b, and c are the grid length, width, and height, respectively, and the integral only goes over all points within the excluded volume of the solute.
With this definition, Fsolu(q) is then the scattering amplitude of a hypothetical system where the solute displaces waters inside its volume, but does not affect the water molecules and ions around it. The details of how to determine the solute excluded volume, and the choice to include the “excluded” waters in Fsolu, are somewhat arbitrary. Here, the excluded volume is computed based on the algorithm of Voss and Gerstein for the 3D grid with the a probe radius of 1.4 Å.33 Points lying inside this volume are assigned to the excluded volume. A key advantage of Eq. (7) is that it is readily calculated for a solute of known structure: the atomic positions and scattering factors are known, and the excess solvent density inside the molecule is just the negative of the bulk solvent density. The “interesting” parts of solvation, i.e., how the solvent in the vicinity of the solute is perturbed, are included in Fsolv(q). The key point of this paper, given in Sec. II B, is to show how Fsolv(q) can be extracted from experimental data.
B. Extracting water and ion distributions from SAXS and anomalous SAXS
We can further divide the solvent scattering into excess terms arising from water and from ions,
| (9) |
As above, the solvent terms come from scattering outside the biomolecule,
| (10) |
Since is proportional to the excess electron density , we can further decompose the solvation shells into contributions of hydration water and ions by considering excess electron density coming from only water or ions . Note that this particular decomposition reflects our interest in studying the waters of hydration and ions around the solute. In principle, any decomposition scheme should work.
Now, as the total intensity is
| (11) |
we define, similarly, the partial intensity for each component i (where i = solu, hyd, counterion, or co-ion),
| (12) |
The square root of the partial intensity will be called , by definition a real quantity. At q = 0, it is nothing but the number of excess electrons from component k, i.e., , where Ni is the excess number of component i coming from the hydration shell, and Z is the number of electrons of component i. Thus we always have , or
| (13) |
and we can extract the number excess of particle Ni from , as shown earlier.13 At non-zero q, is generally complex, but we expect that there should be a small range at low angles where one can still decompose the total into partial amplitudes,
| (14) |
For the above equality to be true, the phases of should be identical (or at least very close to each other) at all small angles. The phase difference α between and is, by definition,
| (15) |
Since depends on all possible orientations of the q vector (with the same magnitude q), the approximation in Eq. (14) holds if and only if cosα is very close to 1 (α ≈ 0) for every q. As we show below, this condition is valid for q less than about 0.1 Å−1. It is not a priori obvious that we can capture enough information from such a narrow region to reconstruct useful real-space distance distributions. We show below that the approximate pair distance distribution functions (PDDFs) extracted in this way from simulated data recapitulate the original PDDFs, and that comparisons to experimental data can be used to discriminate among various theoretical models for the solvent distribution. Comparisons between theory and experiment made using Eq. (14) are easier to interpret than are comparisons of just the total scattered intensity.
1. Solvent is pure water
For systems with only the solute in pure water, the third term in Eq. (9) vanishes; therefore, we can extract directly from SAXS,
| (16) |
where , which can be computed from the (known) structure of the solute. To describe how those water molecules distribute around the solute, we seek an approximation of the cross-term solute-hyd which is the correlation between the hydration water density and the solute. From Eq. (11),
| (17) |
The first two terms are and (as defined in Eq. (12)), respectively; therefore, we can approximate the cross-term (the third term) as
| (18) |
As before, we can compute from the structure of the biomolecule, from Eq. (16), and get the cross-term from Eq. (18). Examples of how this analysis is used are given in Section III.
2. Solvent contains ions and water
In this case, one has more than one unknown (from hyd, counterion (“ion”), and co-ion) in Eq. (9), and additional measurements or assumptions are required to carry out the decomposition. Changing the energy of the incident beam in an anomalous SAXS (ASAXS) experiment is one approach, varying the atomic scattering factor of a given ion.14–16,18,34,35 Another approach uses heavy ion replacement, assuming the ion and water distributions are similar for the same type of ions (alkalies, for example).14,20 By doing this, only is allowed to vary while the co-ion and hydration terms are fixed. Subtracting the square roots of two measured intensities, therefore, gives the contribution from the counterion only,
| (19) |
with Z and Z′ are the atomic scattering factors (q = 0) at two different energy beams, is the normalized (). This can be scaled up to compute back the “full” term
| (20) |
[Note that this includes contributions from those inside the intrinsic (or geometric) volume of the solute (since “inside” the solute there is a deficit density of the ion) and those coming from the ionic cloud (“outside” term). We consider that the “inside” contribution is small enough so that the “outside” term (which we are interested in) can be approximated by the total . For example, if the intrinsic volume of the DNA in this work is around 20 000 Å3, then the deficit number of excluded-volume Na+ is ∼1 (with the bulk concentration of 0.1M). Considering the total excess number of Na+ is ∼37, then the contribution of excluded volume term is less than 3%.] Rewriting Eq. (14), we have
| (21) |
The co-ion contribution in principle could be accurately subtracted from but only at q = 0, since we only know its excess number from electroneutrality: if the total charge of the solute is Z, and the number of excess counterions Nion can be computed as , the number of excess co-ions must be Nco−ion = Nion − Z. However, there is currently no way to obtain the co-ion spatial distribution from experiment. ASAXS experiments of the solute in NaBr or NaI with beam energies close to the absorbance edge of Br or I could potentially provide the answer for this. One primitive way to account for the co-ion distribution is to construct a box around the solute and assume the co-ions are completely depleted inside this box (g = 0), whereas outside this box, its concentration returns back to the bulk value (g = 1). The shape of the box ideally closely resembles the solute, although it is acceptable to use a rectangular box for the DNA here. The size of the box is chosen so that the number of excess co-ions Nco−ion in this box exactly matches the calculation above. With approximately determined, we can now extract the hydration term .
We need to account for the co-ion term because its magnitude is of the same order as the counterion and hydration water terms; the importance of co-ion exclusion was emphasized in a recent “ion counting” study by Herschlag and colleagues.36 A next level of approximation might be to consider a co-ion distribution obeying Poisson–Boltzmann equation, but this is beyond the scope of the current manuscript. In addition, the lack of experimental data of co-ion distribution also prevents us from considering more sophisticated models in this work (since we do not know yet which models are better).
As for the pure water case above, it is generally most useful to compute the cross-terms or . Examples are shown below.
III. RESULTS
A. Validation of the new decomposition scheme
As discussed above, the condition for decomposition of the total scattering intensity into partial intensities is that all the phases should be very close to each other for every orientation of the q vector. To study the behavior of the phases, we must rely on the calculated profiles. We choose to use the calculated SAXS profiles computed by 3D-RISM as they have been shown to match the experimental curves up to wide angle region.13 The phase for each component is computed from the complex amplitude of the corresponding term. The phases are different at each orientation of q (with the same magnitude q), and the average phase is shown in Fig. 1 for lysozyme and a 25-bp DNA in 100 mM NaCl solution. It is expected that those phases start to deviate from 0 as q increases, but at small angles most of the phases are identical, except for the co-ion Cl− in the DNA case, which is not surprising because Cl− is mostly expelled from the DNA and its contribution is expected to be negative. The average phase of the solute closely tracks that of the solvent in the range of q < 0.1 Å−1, but diverges sharply at larger angles. The validation of our principle approximation is shown in Figure 2, which shows that the relative phases of the solute and solvent are actually aligned in all directions, and not just on average. Consider the difference in phase between two amplitudes, α. From Eq. (15) one has
| (22) |
where the solvent amplitude is . Fig. 2 plots the average value of cosα over all orientations of the q vector. The condition of cosα ≈ 1 is valid for q < 0.1 Å−1, with a maximum deviation of 0.005, as shown in the inset of Fig. 2. Above this limit the values of cosα depart from unity rather quickly, as the phases of the solute and solvent diverge. The angle at which cosα begins to deviate significantly from unity determines the limits of this analysis, and the behavior illustrated in Fig. 2 is empirical: in particular, we do not yet understand how the limit q < 0.1 Å depends on the size of the solute. Since the primary goal here is to extract information about the solvent, it would not be sensible to apply our model (at least in its current form) to much larger (and usually more flexible) systems, since uncertainties in those structures are much greater than those for the single-domain proteins and oligonucelotides considered here, and since it would be quite difficult to create solvent models that could be tested by the means described in this paper.
FIG. 1.
Phases (plotted between −180° and +180°) of component amplitudes in lysozyme (left) and DNA (right) from the 3D-RISM scattering profiles. Each value is the average phase for a given magnitude of q, and the error bars report the standard deviation of the distribution of phases for different directions. The phase of the total amplitude is also shown in red. For both systems, at small angle most of the amplitudes are aligned (i.e., in phase), except for the co-ion Cl− in the DNA case, which is excluded from the solute and thus is out of phase.
FIG. 2.
Average values of cos α versus q for lysozyme (left) and DNA (right), where α is the phase difference between and (the standard deviations are also depicted as error bars). At small angles up to 0.1 Å−1, the two amplitudes have nearly the same phase at every orientation of q with very small error bars (see the insets which focus on the region below 0.1 Å−1), thus validating the decomposition of total intensity at small q region.
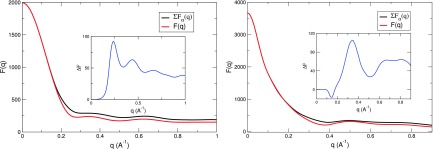
Fig. 3 illustrates the overall accuracy of Eq. (14), comparing the total amplitude and the sum of partial amplitudes for lysozyme and the DNA. The partial amplitudes are defined and calculated as in Section II B. For Lys, k is solu and hyd; while for DNA there are also counterion (Na+) and co-ion (Cl−). One can see that the sum of partial amplitudes is a good approximation of the square root of the measured intensity in the small angle region (q < 0.2 Å−1).
FIG. 3.
Comparison between the “real” excess amplitude (red) with the sum of component amplitudes (black) for lysozyme (left) and DNA (right) using calculated profiles from 3D-RISM. For Lys, k is solu and hyd; while for DNA there are also counter-ion (Na+) and co-ion (Cl−). The difference between these two is plotted in the inset (blue).
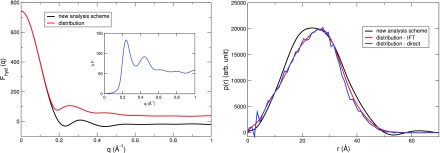
To check whether the extracted above reflects the distribution of water in the reciprocal space, we compare it with computed directly from the (calculated) water distribution around the solute. The Ihyd is calculated by performing the SAXS calculation as usual, but ignoring the solute term (the solute excess form factor is set to 0, i.e., it does not interact with the X-ray beam). The result is plotted in Fig. 4 (left) for lysozyme as the test case. It can be seen that from the new analysis scheme is essentially identical with the one computed from the 3-dimensional distribution of water in the small angle region. We can test the utility of this approximation to by calculating the PDDF in real space (details are in Section V C) and comparing this directly with the 3-dimensional distribution. As can be seen in Fig. 4 (right), the PDDFs from all three approaches have similar shapes, although the peak in the black curve is about 1 Å lower than that of the blue and red curves. This illustrates the ability of the proposed analysis to extract approximate (but useful) solvent distribution around small and rigid biomolecules, but distinctions between models whose differences are smaller than that of the curves in Fig. 4 (right) could not be relied upon.
FIG. 4.
(Left) Scattering amplitude of water around lysozyme extracted from the sqrt subtraction (black) in comparison with those computed directly from the water distribution (red). The difference between those two, as shown in the inset, is negligible at small angle. (Right) Pair distance distribution function (PDDF) of water in the hydration shells computed for two curves in the left by indirect Fourier transformation of . Also shown in blue is the PDDF calculated by using the direct method in real space (see Section V C for more details).
In Secs. III B and III C, we apply this new analysis to both calculated and experimental data for proteins and DNA. The main goal is to study the quality of the predicted ion and water distributions from different theoretical models, to understand their weaknesses, and to provide a framework to develop further approaches to improve the models and force fields.
B. Protein test cases
Fig. 5 plots the “square-root” subtraction from SAXS data and those from 3D-RISM and MD for lysozyme and myoglobin (calculated from Eq. (16)). The total amplitudes for lysozyme are also shown in the inset, which shows little difference between calculated and experimental total intensity profiles. However, applying the decomposition scheme to extract from the data emphasizes the difference between 3D-RISM and MD hydration profiles. This illustrates the potential power of this analysis to guide the development of computational models.
FIG. 5.
for lysozyme (left) and myoglobin (right) from SAXS data (black circles, experimental data from Ref. 26), compared to RISM and MD calculations. The calculation was done as in Eq. (16). The inset in the left shows the total for lysozyme.
There are two main features that can be extracted from those curves. The first is the total excess number of water molecules in the hydration shell, visible at q = 0. Since each water molecule has 10 electrons, should be equal to 10 times the number of excess hydration waters Nhyd. Second, the shape of the curve contains information about the water distribution in the real space. If decays rapidly towards zero, that means the hydration shell is thick. On the other hand, if the curve slowly approaches zero, the hydration water shells are more compact. Of course, it should be easier to explore the latter feature in the real space rather than the reciprocal space using a restricted Fourier transform (IFT or Indirect Fourier Transform) technique.
All RISM closures tend to overestimate , especially in the small angle region. The higher the order of PSE-n closures, the more serious the overestimation is. This indicates that the water attraction to the solute in RISM is too strong. For example, in Lys, RISM-KH overestimates Fhyd(0) by about 200, corresponding to ∼20 water molecules (Table I). The MD results are in much better agreement with the experiment. There are also some discrepancies around q = 0.2 Å−1, where the new analysis scheme may break down (see Fig. 2). It is worth emphasizing that although the computed total SAXS profiles of those two proteins from RISM and MD are nearly identical13 (see the inset in Fig. 5), the extracted here is clearly able to separate MD from RISM results. This demonstrates the power of the proposed analysis and illustrates its potential use in testing new theoretical models for solvent distributions.
TABLE I.
Number of excess hydration water for lysozyme and myoglobin computed as . Those numbers are very close to the values computed by integration all over the hydration shells. For lysozyme, RISM-KH values are 71.7 vs. 71.9.
| Protein | RISM-KH | RISM-PSE2 | RISM-PSE3 | MD | SAXS |
|---|---|---|---|---|---|
| Lysozyme | 71.7 | 83.5 | 92.9 | 48.1 | 50 ± 1 |
| Myoglobin | 86.5 | 101.4 | 113.0 | 53.1 | 60 ± 2 |
To get information about the placement of those excess water molecules in real space, we can compute the cross-term (via Eq. (18)) and transform it to real space to obtain a pair distribution function by using the IFT technique discussed in Section V C. The results for two proteins, lysozyme and myoglobin, are given in Fig. 6. Computed SAXS profiles for MD and 3D-RISM were taken from Ref. 13 and analyzed in the same fashion as described above for the experimental SAXS profiles. The PDDF plot is essentially a distance histogram of hydration water and the solute, weighted by the excess electron density (relative to the bulk solution). 3D-RISM, regardless of closure, overestimates the hydration water interaction with proteins, whereas results from MD simulation are generally much better. The PDDFs for the two proteins show rich structural features, especially at small distances; this is reminiscent of similar structure seen by Kofinger and Hummer29 (although their PDDF arises from a Fourier transform of the total intensity, not of a component as we use here). Since only excess waters that are close to the proteins contribute to the PDDF, for nearly spherical proteins, the peak location correlates well with the protein radius.
FIG. 6.
Pair distance distribution function (PDDF) of water-solute for lysozyme (left) and myoglobin (right). The PDDFs are computed by Fourier-transforming (using IFT) the cross-term for experimental data (black, taken from Ref. 26), MD (orange), and 3D-RISM.
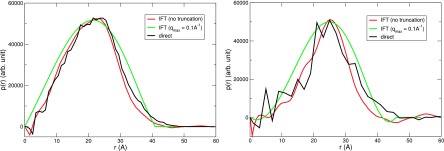
To make sure the features observed in the PDDF are real, and not artifacts of the IFT technique, we compute directly the PDDF in the real space and find that it agrees well with the IFT PDDF. (See Section V C for more details about how we construct the excess density map.) For lysozyme, we observe that using q ≤ 0.1 Å−1 is enough to construct a “coarse” PDDF for the solute-hydration term (see Fig. 7). Using less data leads to the difficulty of converging the IFT procedure, whereas using more data introduces finer features into the calculated PDDF. As shown in Figs. 2–4, however, the validity of the analysis scheme is increasingly suspect at larger angles, so these finer features are probably of little interest.
FIG. 7.
Influence of truncation in Fourier space on the PDDF in real space of lysome-water term . The transform is carried out by IFT using experimentally extracted data.
C. Duplex DNA in salt solutions
If a solute is highly charged, the situation gets more complicated because of the presence of the ionic atmosphere. As we will show below, the contributions of counterions and co-ions can be the same order of magnitude as that of the more numerous hydration water molecules, so that one must account for them in the decomposition. Anomalous SAXS (ASAXS) is one approach to probe the spatial distribution of ions around DNA.14,35 The ASAXS experiment probes the same sample at two different energies, which causes the “effective” number of electron in the interested ion to vary. However, the ASAXS profile does not entirely come from the ion of interest but also has contributions from hydration water-ion cross terms (and solute-ion terms), making it difficult to interpret and draw fruitful conclusions. Another technique is to use heavy ion replacement where instead one varies the ion identity to change the contrast. Using a novel analysis technique (instead of the simple subtraction between the two scattering curves as is done in conventional ASAXS), Meisburger et al.20 were able to separate the ion-DNA term from the water-DNA term and thus could gain insight into the nature of ion cloud around duplex DNA. The method assumes that both the ion and water distributions around DNA are not sensitive to ion type and it has applied successfully to alkali chlorides. (See the Appendix for the relationship between our analysis and the method from Meisburger et al.) It is not clear how accurate this assumption is or whether the same assumption would be valid for highly charged ions (such as Mg2+ and Sr2+) since the interaction between those ions with nucleic acids is expected to be ion-dependent.37–42
Here, we apply our analysis method to a 25 base-pair duplex DNA. Fig. 8 shows the ion-solute cross-terms for experimental and calculated SAXS data of the duplex DNA in 100 mM RbCl or 10 mM SrCl2. As for hydration waters, we extract the total number of excess ions and a qualitative description of their distribution in real space. (Probing the ion cloud around charged biomolecules in a very dilute solution, as the 10 mM SrCl2 solution, using MD simulation is prohibitively expensive,43–46 so we only report 3D-RISM results for Sr2+.) As reported earlier,13 and shown here in Fig. 8 (at q = 0) and Table II, 3D-RISM (with high order closures) and MD simulation (at least for monovalent ions) are able to reproduce accurately the excess number of ions around DNA, including both monovalent and divalent ions.
FIG. 8.
Rb+–DNA (left) and Sr2+–DNA (right) cross-terms from the calculated (color solid lines) and experimental (black dots) ASAXS data of DNA in 100 mM RbCl or 10 mM SrCl2 solutions; experimental data is from Ref. 13. The curves were offset to facilitate visual comparison. From those curves, one could extract the excess number of ions (at q = 0) and qualitatively infer about the ion cloud around the DNA.
TABLE II.
Number of excess ions around a −48 charged duplex DNA in 100 mM RbCl or 10 mM SrCl2 solutions, computed as from the new analysis scheme. Those numbers are very close to the values computed by integration over all space. For example, values computed at RISM-PSE3 for Rb+ are 36.77 (from the q = 0 limit) vs. 36.85 (from directly integrating the distribution).
| DNA/100 mM RbCl | DNA/10 mM SrCl2 | |||||||
|---|---|---|---|---|---|---|---|---|
| System | ASAXS | MD | RISM-PSE3 | RISM-PSE2 | RISM-KH | ASAXS | RISM-PSE2 | RISM-KH |
| Ncation | 37 ± 2 | 37.95 | 36.77 | 35.08 | 30.16 | 20 ± 2 | 22.24 | 19.14 |
We also performed IFT to obtain the PDDFs of the ion-DNA crossterm, which are shown in Fig. 9. The curves for Rb+ look encouraging, especially for the MD simulation and for high order closures in RISM. High order closures tend to place more ions closer to the DNA, which is more consistent with MD and experiment. The errors at large r of the PDDF curves could arise from the way theoretical models approximate ion-solute interaction. The PDDF curves for divalent ions show large deviations from experiment despite having somewhat reasonable agreement in the Fourier space from PSE2 closure. This highlights the fact that the number of excess ions should not be used solely to characterize the ion cloud. Instead, information about the shape of the ion cloud should be also taken into account. The 3D-RISM model, in its current form, is known to have difficulties with divalent ions,47,48 perhaps resulting from the lack of polarization effects.49,50 More work currently is underway to test new ion models in RISM calculations.
FIG. 9.
PDDF of Rb+–DNA (left) and Sr2+–DNA (right) obtained by inverse Fourier transformation of the ion-DNA cross-terms . Results are shown for experimental data (black, taken from Ref. 13), MD (orange) and 3D-RISM. The curves are offset to facilitate visual comparison. No MD data is reported for SrCl2 due to the high cost of simulation for dilute (10 mM) solution.
To determine the water hydration term, the co-ion term (in this case Cl−) needs to be taken into account. We note that the contribution of the solvent to the total intensity is based on the scattering contrast (relative to the bulk concentration), and since the co-ions are entirely excluded from such a highly charged DNA, the contribution of this deficit to the X-ray scattering difference is therefore significant. A simple calculation shows that with the DNA considered here (which has a −48e charge), NRb = 37 leads to NCl = − 11 and therefore vs. . The co-ion term will contribute even more strongly if lighter counter-ions (Na+, K+, …) and/or heavier co-ions (Br−, I−, …) are used. (A recent ion counting experiment emphasizes the importance of co-ion identity to the ion atmosphere around nucleic acids.36) Here, we estimate the co-ion contribution using the simple model described in Section II B 2.
The water term is then determined and plotted in Fig. 10 for DNA in two different salt solutions. The PDDFs of water-DNA in two different salt solutions are computed by performing an IFT of and are shown in Fig. 11. All RISM and MD results overestimate the number of excess waters; this is in contrast to the protein results reported above, where MD simulations led to good estimates for the number of excess waters. (The reason why the number of excess waters is overestimated for nucleic acids by 3D-RISM and MD is currently unclear, and future investigations are certainly needed.) Furthermore, the predicted number of excess waters is nearly the same for RbCl and SrCl2, whereas estimates based on experiment vary a lot. The experimental values for Nwat are ∼70 and 110 for RbCl and SrCl2, respectively, i.e., the difference is around 1.6 water molecules per base pair. By contrast, the 3D-RISM-KH values are 144.0 and 130.0, respectively (the difference is around −0.6 water molecules per base pair, in the opposite direction). This difference could potentially come from the fact that fewer Sr2+ are required to neutralize the DNA than Rb+ (see Table II), leading to fewer ions accumulating near the DNA surface and therefore providing more space for water. Also, Sr2+ is expected to have denser and stronger hydration shells than Rb+, which will be dragged along the ions towards the DNA. The much smaller concentration of SrCl2 compared with RbCl (10 mM vs. 100 mM) is probably another factor leading to fewer ions accumulating near the DNA surface.
FIG. 10.
Water hydration term extracted from SAXS data of the 25-bp DNA in 100 mM RbCl (left) or 10 mM SrCl2 (right) (computed as described in Eq. (21)) for experimental SAXS (black error bars taken from Ref. 13), MD (orange), and 3D-RISM.
FIG. 11.
PDDF of water-DNA in RbCl 100 mM (left) or SrCl2 10 mM (right), calculated by performing IFT for . Black circles are from the decomposition of experimental data reported in Ref. 13.
IV. CONCLUSIONS
Water molecules and ions around biomolecules often play a crucial role in function. Here we propose a new analysis scheme for X-ray scattering data to extract information about how water molecules and ions distribute around the solute. Although the analysis requires some approximation, it is accurate enough to obtain reliable partial scattering intensities in Fourier space as well as distribution functions in real space. The resulting distributions could then be used to study the dynamic nature of the solvation shells, for instance, via time-resolved scattering techniques.51–53 It could also be used to test the accuracy of theoretical predictions, facilitating improvements in how those theories treat water molecules, ions, and cosolvents in general. Comparing theory vs. experiment for individual interaction terms (as in Figs. 6, 9, and 11) is likely to be more helpful in assessing the strengths and weaknesses of theoretical models than just making comparisons to the complete SAXS profile.
The proposed analysis complements recent experimental techniques (such as ion counting12 and anomalous SAXS16) by providing not only the number excess of particles but also their distribution in real space. It is, however, worth restating the fact that our decomposition requires an independent knowledge of the structure of solute, which is assumed to be rigid; it cannot be used (in its current form) for systems with significant conformational heterogeneity or disorder, primarily because uncertainties in computing the scattering from the solute would probably lead to large uncertainties in extracting solvent information. To illustrate the new analysis, we extract the water hydration distribution around two proteins—lysozyme and myoglobin—from regular X-ray scattering profiles. Comparison between those experimental distributions (extracted from SAXS data) and the calculated distributions from 3D-RISM and MD simulation reveals that MD simulation accurately accounts for water in terms of both number of excess water and its real space distribution, whereas 3D-RISM overestimates the number of excess waters leading to the accumulation of water hydration near the proteins. This overestimation comes from the approximate treatments employed in the SSOZ and closure equations of RISM.25
For highly charged systems such as the DNA duplex, both MD simulation and 3D-RISM (with high order closures) are capable of capturing the ion cloud of counterions around the DNA, again both in terms of number excess and the real space distribution. Water molecules, on the other hand, are predicted to be too strongly attracted towards the DNA, presumably the phosphate groups, by both MD simulation and 3D-RISM. This is an unexpected result and highlights the need to recalibrate nucleic acid force fields. There are also some studies in the literature reporting a misbalance between solute-water and solute-solute interactions, which is probably relevant to the situation here.54–58 Our current ongoing work suggests that the distribution of partial atomic charges within the phosphate group needs to be adjusted to calibrate the DNA-water interaction. A full analysis of those errors is beyond the scope of this paper and will be presented elsewhere.
We have chosen in Eq. (7) to include the “excluded volume” effect (the fact that solvent is excluded from the interior of the biomolecule) into our definition of Fsolu(q). This was an arbitrary choice, but driven by the fact that the “excluded volume” effect can be easily computed from a known structure, and by the perspective that the “interesting” parts of hydration are those that take place outside the solute interior. But other choices, such as including only the first term of Eq. (7) in Fsolu(q), are possible and do not change the analysis method here in any fundamental way. Future studies should help to determine the relative strengths and weaknesses of difference decomposition methods.
V. COMPUTATIONAL DETAILS
We took two proteins—lysozyme and myoglobin—and a 25-bp duplex DNA as test cases for validating the new analysis scheme. The coordinates for the proteins were taken from Protein Data Bank with PDB ID 1WLA and 6LYZ for Mb and Lys, respectively. The DNA structure with the sequence of GCATCTGGGCTATAAAAGGGCGTCG was built by Nucleic Acid Builder (NAB) using ideal fiber diffraction models. The all-atom Amber force field ff99-bsc059 was used for the DNA and ff14SB60 for proteins. (Since there is no histidine-bound heme group parameter for Amber force field, we use the cysteine-bound heme parameter for Cytochrome P450 taken from Shahrokh et al.61) Monovalent ion (alkali, halide) parameters were taken from Joung–Cheatham ion model.62 Sr2+ ion was taken from Li et al. (we here report the IOD set results as we found that there was no difference between SAXS calculations using these three sets).63
A. MD simulations
For each system, the solute was immersed in a preequilirated cubic water box with a buffer distance of 20 Å (proteins) or 60 Å (DNA). Counter-ions were added to neutralize the protein systems. For DNA, Na+ (or Rb+) and Cl− ions were added to neutralize the negative charges and reach the concentration of 100 mM. Nonbonded interaction cutoff was set at 9.0 Å. The long-range electrostatic interaction is calculated by using the smooth particle mesh Ewald method.64,65 Equations of motion were integrated by employing the leap-frog Verlet algorithm with a 2 fs time step. Covalent bond lengths involving hydrogen atoms were constrained using SHAKE.66 The system was first minimized with 2000 steps of steepest descent, followed by 3000 steps of conjugate gradient method to remove bad contacts. The system was then equilibrated at 298.15 K and 1 atm with the solute kept fixed (with the restraint of 10.0 kcal/(mol. Å2)) for 25 ns (proteins) or 120 ns (DNA). Temperature was regulated by using Langevin thermostat with a collision frequency of 2.0 ps−1 while pressure was maintained using Berendsen barostat. All simulations were performed using the GPU accelerated pmemd code (pmemd.cuda).67–69 Only the last 20 ns (for proteins) or 80 ns (for DNA) of trajectories were kept and subjected to SAXS calculation.
In order to calculate SAXS intensity for the solute from MD simulation, another “blank” system in which there is only pure solvent (water molecules and ions, and no solute present) is needed.26 We set up the pure solvent boxes that are nearly identical to the “sample” system (box size, running condition) except there is no solute in it. The equilibration step was run for 20 ns (only water) or 50 ns (with ions). Both trajectories of “sample” and “blank” systems were used to calculate SAXS using the saxs_md code in AmberTools, which followed exactly the protocol described in Refs. 26 and 13.
B. RISM and SAXS calculations
3D-RISM is a microscopic approach to calculate the equilibrium distribution of a solvent on a three-dimensional grid around a fixed solute. The theory25 and implementation in Amber70 have been widely reviewed, and only the most important considerations are given here.
3D-RISM provides the solvent structure in the form of a 3D site distribution function, , for each solvent site, γ. With , the solvent density distribution approaches the solvent bulk density ργ. The 3D-RISM integral equation has the form
| (23) |
where superscripts U and V denote the solute and solvent species, respectively; is the site-site total correlation function; is the 3D direct correlation function for solvent site α; and is the site-site susceptibility of the solvent, given by
| (24) |
Here, is the intramolecular correlation function, representing the internal geometry of the solvent molecules while is the site-site radial total correlation function of the pure solvent calculated from the dielectrically consistent version of the 1D-RISM theory (DRISM). Eq. (23) is often complemented with the 3DRISM-KH closure
| (25) |
where
and is the 3D interaction potential of the solute acting on solvent site γ, given by the sum of the pairwise site-site potentials from all the solute interaction sites i located at Ri,
| (26) |
The so-called hypernetted chain (HNC) closure would use just the top line of Eq. (25) and is generally the most accurate common closure for polar systems with strong long-range interactions, but it is often difficult to find converged solutions. The KH closure is easier to converge. The partial series expansion of order-n (3DRISM-PSE-n) offers a way to interpolate between KH and HNC,71
| (27) |
Hence, KH is the special case of PSE closure when n = 1; and when n → ∞ HNC is obtained. For the purposes of this paper, results from different closures offer a way to generate a series of plausible solvent models; we can then use our analysis to test the ability of small-angle scattering data to discriminate among these.
All RISM calculations were performed using the rism1d and rism3d.snglpnt codes from AmberTools.70 The water model used in this study was cSPC/E.70 First, the 1D-RISM was carried out with only the solvent (water + ion if any) to obtain the solvent susceptibility which contained all the information about the bulk solvent. This was subsequently used for 3D-RISM to compute the solvent structure around a solute of choice. Thus one needed to perform only one 1D-RISM step and used the resulting χVV for all subsequent 3D-RISM calculations which were at the same condition (salt concentration, temperature, pressure, etc.). The modified direct inversion of the iterative subspace solver (MDIIS)72 was used to iteratively solve the RISM equations to a residual tolerance of 10−12 and 10−5 for 1D and 3D-RISM, respectively, at 298.15 K. For 1D-RISM, the 0.025 Å grid spacing was used with 16 384 and 32 768 grid points for pure water and 100 mM RbCl solution, respectively. With more diluted solutions (10 mM SrCl2, for example), the grid points were doubled until we got the results converged. For 3D-RISM, a 3D grid with 0.5 Å grid spacing was used with the buffer region of 20 Å for proteins and 40 Å for DNA in 100 mM RbCl and up to 80 Å for 10 mM SrCl2.
The output from 3D-RISM program was 3-dimensional g(r) for each atomic site in solvents (for instance, Hw and Ow in water), reflecting the excess or deficit of each solvent site relative to bulk concentration around the solute in real space. Those were then served as inputs to compute SAXS profiles using the saxs_rism code in AmberTools.13
C. Pair distance distribution functions (PDDF)
1. Indirect Fourier transform (IFT)
The PDDF is the ultimately sought quantity and carries the most information content from SAXS experiment. In principle, a direct Fourier transform of the pseudo-intensity should provide a real space representation of the distribution of hydration water under the form of the PDDF . However, such an approach is of little use since it gives large systematic errors because of the noise, smearing, and truncation of the experimental data. Instead, indirect Fourier transform (IFT) technique was developed long time ago, pioneering by Glatter,73 Moore,74 and Svergun et al.75 to deal with this problem. The main idea of IFT was to express the PDDF as a linear combination of a set of smooth basis functions and fit the coefficients in order to reproduce the intensity in reciprocal space. In this work, we used the IFT method based on Bayesian analysis from Hansen, which was shown to give similar results with Glatter method; all tranformations were performed using the webserver BayesApp.76–78 Since IFT is an underdetermined problem, i.e., several solutions can fit the experimental data equally well, additional constraints must be introduced to obtain unique solution.73–76 The PDDF is first written as a sum of smooth basis functions (cubic B-splines, for instance) . The coefficients ai are then tuned by minimizing the regulation functional S, subjected to the constraint that χ2 takes sensible value, with and σj is the standard deviation at data point j. The regulation functional S usually controls the smoothness of the solution and several forms of it exist. We followed Hansen and used .76–78
2. Direct calculation
The PDDF is related to the density-density correlation by79,80
where is the scattering contrast (difference in electron density relative to the bulk density). To generate an electron density map of the solute, we assume that the solute is composed of isolated atoms (not chemical bonded), and the deformation electron density is negligibly small. (It should be noted that there are lots of work done to incorporate the anisotropicity and asphericity of electron density in order to minimize the deformation density, for example, see Refs. 81–83.) The density is thus computed by summing up the contribution of all the atoms. In practice, since electron is mostly found near the nucleus, a simple cutoff to decide whether an atom contributes electron density is sufficient. One way that is widely used in X-ray crystallography to compute the electron density around an atom is via the analytic Fourier transform of the atomic scattering factor.84,85 Conventionally, the atomic scattering factor was fitted with the sum of some Gaussian terms (with a and b are tabulated constants for each atom, we use here the sum of six Gaussians by Su and Coppens86),
Fourier transform of the above formula gives the electron density at a distance r (Å) from the nucleus,
The PDDF can also be calculated for the cross-term, or . This time the density must come from both solute and water (or counterions),
A comparison between the “direct” and IFT PDDF for lysozyme is shown in Fig. 12. Although there is some slight discrepancy at 30–40 Å distance (the IFT moderately underestimates the PDDF in this range), the two methods agree quantitatively, suggesting that the new analysis proposed here is capable of obtaining a usefully accurate pair distribution. Fig. 13 shows a similar comparison between the direct and IFT PDDF profiles for ion-solute pair distances in DNA.
FIG. 12.
Comparison between water-solute PDDF computed by IFT and “direct” method for lysozyme using RISM-KH (left) and MD (right) data. For the “direct” method, two grids of excess electron density are created separately for the solute and hydration shells. The PDDF is then computed by making a distance histogram between the solute and hydration water, weighted with the excess electron density. See the text for a detailed description of the “direct” method. With IFT, two separate calculations are performed: (i) using all q-data in the reciprocal space (red) or (ii) using only q < 0.1 Å−1 (green). Although the decomposition is strictly “correct” only at q < 0.1 Å−1 (see Figs. 3 and 4), introducing higher q-data helps the extracted PDDF to closely approach the “real” PDDF (computed directly from the distribution).
FIG. 13.
Comparison between ion-DNA PDDF computed by IFT and “direct” method for Rb+ (left, using RISM-PSE3 distribution) and Sr2+ (right, using RISM-PSE2 distribution). See Section V C for a detailed description of the “direct” method.
Acknowledgments
We thank Darrin York, Steve Meisberger, and George Giambasu for many helpful discussions and Jay Bardhan and Lee Makowski for providing the experimental SAXS profiles for two proteins. This work was supported by NIH Grant Nos. P50 GM103297 (D.A.C.) and GM085062 (L.P.).
APPENDIX: COMPARISON TO AN ALTERNATE DECOMPOSITION APPROACH
Instead of using different beam line energies to change the efficient contrast of the interested ion, one may instead vary the ion itself and assume both the ion and water distributions are unchanged. Meisburger et al. express the total SAXS intensity as20
| (A1) |
where δ is the scattering contrast and P is the partial scattering form factor (which has a range from 0 to 1). For the solute, δsolu = Zsolu + NwatZwat; therefore, it includes all the contributions from water coming from both from the excluded volume and hydration shells. The first term in Eq. (A1) is equivalent to the sum of solute and water in our approach,
| (A2) |
with is the normalized () and the number of excess waters Nwat is partitioned into excluded volume contribution Nexcl and hydration contribution Nhyd. If one assumes then the right hand side of Eq. (A2) could be rewritten as , leading to
| (A3) |
The third term in Eq. (A1) is equivalent to our ion term,
| (A4) |
Comparing Eq. (A4) with the last term of Eq. (A1) leads to
The cross-term in Eq. (A1) is then
| (A5) |
Eqs. (A3)-(A5) relate the two different approaches to extract ion distribution from X-ray scattering experiment. It is obvious that and , which are both defined to be within 0 and 1, are basically the same entity. couples with the intensity I, while couples with the partial amplitude . The advantage of using instead of is that every component is separated completely from each other and there is no cross-term; therefore, one only needs instead of to specify a system with n components. In addition, the partial amplitudes are additive while the intensities are not, which is potentially easier to work with.
Fig. 14 compares and from Meisburger et al. and our analysis. from our method is directly related to which is nothing but the normalized . It is apparent that the two methods agree quantitatively despite the fact that Meisburger et al. implicitly assume , which seems to be reasonable (the hydration shell shape should be somewhat similar to the solute shape).
FIG. 14.
(left) and (right) comparison between two analysis schemes for the DNA SAXS data: Black circles are from Meisburger et al.;20 red circles from the method proposed here. Error bars are computed by propagating the errors from experimental intensity data. P(q) is actually a normalized curve (in Fig. 8), so the error bars here are the same error bars in Fig. 8 after normalization. They were computed by propagating the error bars when (square root) subtracting the two experimental intensities Ioff and Ion for , times with .
REFERENCES
- 1.Ball P., Chem. Rev. 108, 74 (2008). 10.1021/cr068037a [DOI] [PubMed] [Google Scholar]
- 2.Levy Y. and Onuchic J. N., Annu. Rev. Biophys. Biomol. Struct. 35, 389 (2006). 10.1146/annurev.biophys.35.040405.102134 [DOI] [PubMed] [Google Scholar]
- 3.Zhong D., Pal S. K., and Zewail A. H., Chem. Phys. Lett. 503, 1 (2011). 10.1016/j.cplett.2010.12.077 [DOI] [Google Scholar]
- 4.Draper D. E., Grilley D., and Soto A. M., Annu. Rev. Biophys. Biomol. Struct. 34, 221 (2005). 10.1146/annurev.biophys.34.040204.144511 [DOI] [PubMed] [Google Scholar]
- 5.Anderson C. F. and Record M. T., Annu. Rev. Phys. Chem. 46, 657 (1995). 10.1146/annurev.pc.46.100195.003301 [DOI] [PubMed] [Google Scholar]
- 6.Lipfert J., Doniach S., Das R., and Herschlag D., Annu. Rev. Biochem. 83, 813 (2014). 10.1146/annurev-biochem-060409-092720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarov V., Pettitt B. M., and Feig M., Acc. Chem. Res. 35, 376 (2002). 10.1021/ar0100273 [DOI] [PubMed] [Google Scholar]
- 8.Record M. T., Anderson C. F., and Lohman T. M., Q. Rev. Biophys. 11, 103 (1978). 10.1017/S003358350000202X [DOI] [PubMed] [Google Scholar]
- 9.Shui X., McFail-Isom L., Hu G. G., and Williams L. D., Biochemistry 37, 8341 (1998). 10.1021/bi973073c [DOI] [PubMed] [Google Scholar]
- 10.Nakano S.-i., Fujimoto M., Hara H., and Sugimoto N., Nucl. Acids Res. 27, 2957 (1999). 10.1093/nar/27.14.2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss U. P., Helfgott C., and Pink H., J. Phys. Chem. 71, 2550 (1967). 10.1021/j100867a024 [DOI] [PubMed] [Google Scholar]
- 12.Bai Y., Greenfeld M., Travers K. J., Chu V. B., Lipfert J., Doniach S., and Herschlag D., J. Am. Chem. Soc. 129, 14981 (2007). 10.1021/ja075020g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen H. T., Pabit S. A., Meisburger S. P., Pollack L., and Case D. A., J. Chem. Phys. 141, 22D508 (2014). 10.1063/1.4896220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das R., Mills T. T., Kwok L. W., Maskel G. S., Millett I. S., Doniach S., Finkelstein K. D., Herschlag D., and Pollack L., Phys. Rev. Lett. 90, 188103 (2003). 10.1103/PhysRevLett.90.188103 [DOI] [PubMed] [Google Scholar]
- 15.Dingenouts N., Patel M., Rosenfeldt S., Pontoni D., Narayanan T., and Ballauff M., Macromolecules 37, 8152 (2004). 10.1021/ma048828j [DOI] [Google Scholar]
- 16.Pabit S. A., Meisburger S. P., Li L., Blose J. M., Jones C. D., and Pollack L., J. Am. Chem. Soc. 132, 16334 (2010). 10.1021/ja107259y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollack L., Annu. Rev. Biophys. 40, 225 (2011). 10.1146/annurev-biophys-042910-155349 [DOI] [PubMed] [Google Scholar]
- 18.Guilleaume B., Ballauff M., Goerigk G., Wittemann M., and Rehahn M., Colloid Polym. Sci. 279, 829 (2001). 10.1007/s003960100521 [DOI] [Google Scholar]
- 19.Dingenouts N., Merkle R., Guo X., Narayanan T., Goerigk G., and Ballauff M., J. Appl. Crystallogr. 36, 578 (2003). 10.1107/S0021889803000256 [DOI] [Google Scholar]
- 20.Meisburger S. P., Pabit S. A., and Pollack L., Biophys. J. 108, 2886 (2015). 10.1016/j.bpj.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovalenko A. and Hirata F., J. Chem. Phys. 110, 10095 (1999). 10.1063/1.478883 [DOI] [Google Scholar]
- 22.Chandler D. and Andersen H. C., J. Chem. Phys. 57, 1930 (1972). 10.1063/1.1678513 [DOI] [Google Scholar]
- 23.Hirata F., in Molecular Theory of Solvation, edited by Hirata F. (Kluwer Acadamic Publishers, Dordrecht, 2003), pp. 1–60. [Google Scholar]
- 24.Luchko T., Joung I., and Case D., in Innovations in Biomolecular Modeling and Simulation, edited by Schlick T. (Royal Society of Chemistry, London, 2012), Vol. 1, pp. 51–86. [Google Scholar]
- 25.Ratkova E. L., Palmer D. S., and Fedorov M. V., Chem. Rev. 115, 6312 (2015). 10.1021/cr5000283 [DOI] [PubMed] [Google Scholar]
- 26.Park S., Bardhan J. P., Roux B., and Makowski L., J. Chem. Phys. 130, 134114 (2009). 10.1063/1.3099611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seki Y., Tomizawa T., Khechinashvili N. N., and Soda K., Biophys. Chem. 95, 235 (2002). 10.1016/S0301-4622(01)00260-5 [DOI] [PubMed] [Google Scholar]
- 28.Oroguchi T., Hashimoto H., Shimizu T., Sato M., and Ikeguchi M., Biophys. J. 96, 2808 (2009). 10.1016/j.bpj.2008.12.3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kofinger J. and Hummer G., Phys. Rev. E 87, 052712 (2013). 10.1103/PhysRevE.87.052712 [DOI] [PubMed] [Google Scholar]
- 30.Chen P.-c. and Hub J. S., Biophys. J. 107, 435 (2014). 10.1016/j.bpj.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merzel F. and Smith J. C., Acta Crystallogr., Sect. D: Biol. Crystallogr. 58, 242 (2002). 10.1107/S0907444901019576 [DOI] [PubMed] [Google Scholar]
- 32.Virtanen J. J., Makowski L., Sosnick T. R., and Freed K. F., Biophys. J. 101, 2061 (2011). 10.1016/j.bpj.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voss N. R. and Gerstein M., Nucl. Acids Res. 38, W555 (2010). 10.1093/nar/gkq395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel M., Rosenfeldt S., Ballauff M., Dingenouts N., Pontoni D., and Narayanan T., Phys. Chem. Chem. Phys. 6, 2962 (2004). 10.1039/b402155j [DOI] [Google Scholar]
- 35.Pabit S., Finkelstein K., and Pollack L., Methods Enzymol. 469, 391 (2009). 10.1016/S0076-6879(09)69019-4 [DOI] [PubMed] [Google Scholar]
- 36.Gebala M., Giambasu G. M., Lipfert J., Bisaria N., Bonilla S., Li G., York D. M., and Herschlag D., J. Am. Chem. Soc. 137, 14705 (2015). 10.1021/jacs.5b08395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu T. K. and Dickerson R. E., J. Mol. Biol. 301, 915 (2000). 10.1006/jmbi.2000.4012 [DOI] [PubMed] [Google Scholar]
- 38.Davey C. A. and Richmond T. J., Proc. Natl. Acad. Sci. U. S. A. 99, 11169 (2002). 10.1073/pnas.172271399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anastassopoulou J., J. Mol. Struct. 651–653, 19 (2003). 10.1016/S0022-2860(02)00625-7 [DOI] [Google Scholar]
- 40.Ahmad R., Arakawa H., and Tajmir-Riahi H. A., Biophys. J. 84, 2460 (2003). 10.1016/S0006-3495(03)75050-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egli M., Chem. Biol. 9, 277 (2002). 10.1016/S1074-5521(02)00116-3 [DOI] [PubMed] [Google Scholar]
- 42.Strick R., Strissel P. L., Gavrilov K., and Levi-Setti R., J. Cell Biol. 155, 899 (2001). 10.1083/jcb.200105026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirmizialtin S., Pabit S. A., Meisburger S. P., Pollack L., and Elber R., Biophys. J. 102, 819 (2012). 10.1016/j.bpj.2012.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giambasu G. M., Luchko T., Herschlag D., York D. M., and Case D. A., Biophys. J. 106, 883 (2014). 10.1016/j.bpj.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoo J. and Aksimentiev A., J. Phys. Chem. B 116, 12946 (2012). 10.1021/jp306598y [DOI] [PubMed] [Google Scholar]
- 46.Savelyev A. and MacKerell A. D., J. Phys. Chem. B 119, 4428 (2015). 10.1021/acs.jpcb.5b00683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joung I. S., Luchko T., and Case D. A., J. Chem. Phys. 138, 044103 (2013). 10.1063/1.4775743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giambasu G. M., Gebala M. K., Panteva M. T., Luchko T., Case D. A., and York D. M., Nucl. Acids Res. 43, 8405 (2015). 10.1093/nar/gkv830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li P. and Merz K. M., J. Chem. Theory Comput. 10, 289 (2014). 10.1021/ct400751u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li P., Song L. F., and Merz K. M., J. Phys. Chem. B 119, 883 (2015). 10.1021/jp505875v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollack L., Tate M. W., Finnefrock A. C., Kalidas C., Trotter S., Darnton N. C., Lurio L., Austin R. H., Batt C. A., Gruner S. M., and Mochrie S. G. J., Phys. Rev. Lett. 86, 4962 (2001). 10.1103/PhysRevLett.86.4962 [DOI] [PubMed] [Google Scholar]
- 52.Cammarata M., Levantino M., Schotte F., Anfinrud P. A., Ewald F., Choi J., Cupane A., Wulff M., and Ihee H., Nat. Methods 5, 881 (2008). 10.1038/nmeth.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J. G., Kim T. W., Kim J., and Ihee H., Acc. Chem. Res. 48, 2200 (2015). 10.1021/acs.accounts.5b00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Best R. B., Zheng W., and Mittal J., J. Chem. Theory Comput. 10, 5113 (2014). 10.1021/ct500569b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piana S., Donchev A. G., Robustelli P., and Shaw D. E., J. Phys. Chem. B 119, 5113 (2015). 10.1021/jp508971m [DOI] [PubMed] [Google Scholar]
- 56.Gotz A. W., Bucher D., Lindert S., and McCammon J. A., J. Chem. Theory Comput. 10, 1631 (2014). 10.1021/ct401049q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henriques J., Cragnell C., and Skepo M., J. Chem. Theory Comput. 11, 3420 (2015). 10.1021/ct501178z [DOI] [PubMed] [Google Scholar]
- 58.Savelyev A. and MacKerell A. D., J. Phys. Chem. B 118, 6742 (2014). 10.1021/jp503469s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez A., Marchan I., Svozil D., Sponer J., Cheatham T. E. III, Laughton C. A., and Orozco M., Biophys. J. 92, 3817 (2007). 10.1529/biophysj.106.097782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., and Simmerling C., J. Chem. Theory Comput. 11, 3696 (2015). 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shahrokh K., Orendt A., Yost G. S., and Cheatham T. E., J. Comput. Chem. 33, 119 (2012). 10.1002/jcc.21922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joung I. S. and Cheatham T. E., J. Phys. Chem. B 112, 9020 (2008). 10.1021/jp8001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P., Roberts B. P., Chakravorty D. K., and Merz K. M., J. Chem. Theory Comput. 9, 2733 (2013). 10.1021/ct400146w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Darden T., York D., and Pedersen L., J. Chem. Phys. 98, 10089 (1993). 10.1063/1.464397 [DOI] [Google Scholar]
- 65.Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., and Pedersen L. G., J. Chem. Phys. 103, 8577 (1995). 10.1063/1.470117 [DOI] [Google Scholar]
- 66.Ryckaert J.-P., Ciccotti G., and Berendsen H. J. C., J. Comput. Phys. 23, 327 (1977). 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- 67.Gotz A. W., Williamson M. J., Xu D., Poole D., Le Grand S., and Walker R. C., J. Chem. Theory Comput. 8, 1542 (2012). 10.1021/ct200909j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salomon-Ferrer R., Gotz A. W., Poole D., Le Grand S., and Walker R. C., J. Chem. Theory Comput. 9, 3878 (2013). 10.1021/ct400314y [DOI] [PubMed] [Google Scholar]
- 69.Le Grand S., Gotz A. W., and Walker R. C., Comput. Phys. Commun. 184, 374 (2013). 10.1016/j.cpc.2012.09.022 [DOI] [Google Scholar]
- 70.Luchko T., Gusarov S., Roe D. R., Simmerling C., Case D. A., Tuszynski J., and Kovalenko A., J. Chem. Theory Comput. 6, 607 (2010). 10.1021/ct900460m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kast S. M. and Kloss T., J. Chem. Phys. 129, 236101 (2008). 10.1063/1.3041709 [DOI] [PubMed] [Google Scholar]
- 72.Kovalenko A., Ten-no S., and Hirata F., J. Comput. Chem. 20, 928 (1999). [DOI] [Google Scholar]
- 73.Glatter O., J. Appl. Crystallogr. 10, 415 (1977). 10.1107/S0021889877013879 [DOI] [Google Scholar]
- 74.Moore P. B., J. Appl. Crystallogr. 13, 168 (1980). 10.1107/S002188988001179X [DOI] [Google Scholar]
- 75.Svergun D. I., Semenyuk A. V., and Feigin L. A., Acta Crystallogr., Sect. A: Found. Adv. 44, 244 (1988). 10.1107/S0108767387011255 [DOI] [Google Scholar]
- 76.Hansen S., J. Appl. Crystallogr. 33, 1415 (2000). 10.1107/S0021889800012930 [DOI] [Google Scholar]
- 77.Hansen S., in Bayesian Methods in Structural Bioinformatics, edited byHamelryck T., Mardia K., and Ferkinghoff-Borg J.,. Statistics for Biology and Health (Springer, Berlin, 2012), pp. 313–342. [Google Scholar]
- 78.Hansen S., J. Appl. Crystallogr. 45, 566 (2012). 10.1107/S0021889812014318 [DOI] [Google Scholar]
- 79.Glatter O., J. Appl. Crystallogr. 12, 166 (1979). 10.1107/S0021889879012139 [DOI] [Google Scholar]
- 80.Glatter O. and Hainisch B., J. Appl. Crystallogr. 17, 435 (1984). 10.1107/S0021889884011894 [DOI] [Google Scholar]
- 81.Volkov A., Li X., Koritsanszky T., and Coppens P., J. Phys. Chem. A 108, 4283 (2004). 10.1021/jp0379796 [DOI] [Google Scholar]
- 82.Dominiak P. M., Volkov A., Li X., Messerschmidt M., and Coppens P., J. Chem. Theory Comput. 3, 232 (2007). 10.1021/ct6001994 [DOI] [PubMed] [Google Scholar]
- 83.Schnieders M. J., Fenn T. D., Pande V. S., and Brunger A. T., Acta Crystallogr., Sect. D: Struct. Biol. 65, 952 (2009). 10.1107/S0907444909022707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Agarwal R. C., Acta Crystallogr., Sect. A: Found. Adv. 34, 791 (1978). 10.1107/S0567739478001618 [DOI] [Google Scholar]
- 85.Afonine P. V. and Urzhumtsev A., Acta Crystallogr., Sect. A: Found. Adv. 60, 19 (2004). 10.1107/S0108767303022062 [DOI] [PubMed] [Google Scholar]
- 86.Su Z. and Coppens P., Acta Crystallogr., Sect. A: Found. Adv. 53, 749 (1997). 10.1107/S0108767397004558 [DOI] [Google Scholar]