Abstract
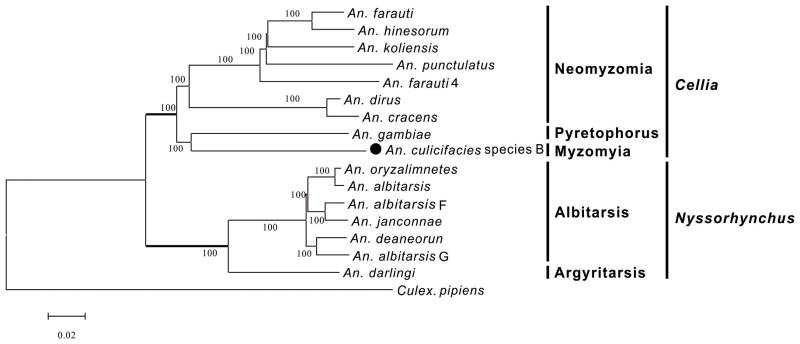
The complete mitogenome sequence of Anopheles culicifacial species B was sequenced in this study. The length of the mitogenome is 15,330 bp, which contains 13 PCGs, 22 tRNA genes, 2 rRNA genes, and a non-coding control region. The gene order and composition are consistent with those previously reported for other mosquito species. The initiation codon of the PCGs complies with the ATN rule except for COI using TCG and ND5 using GTG as a start codon, and the stop codon is TAA or an only T. The total base composition is 40.4% A, 38.1% T, 12.4% C, and 9.1% G. The phylogenetic tree based on the sequences of 13 PCGs showed that these species were classified into two clades, corresponding to the subgenus Cellia and subgenus Nyssorhynchus. An. culicifacies species B of Myzomyia Series was clusterd with An. gambiae of Pyretophorus Series with a 100% bootstrap value.
Keywords: Mitogenome, sequencing analysis, Anopheles culicifacies species B, Culicidae, phylogenetic tree
Anopheles culicifacies Giles is widely distributed in India and neighboring countries, and has been recognized as a complex of five sibling species, A, B, C, D and E. These species are morphologically indistinguishable, whereas exhibit distinct differences in biological characters (Subbarao et al., 1997). All of them except for species B are important vectors of malaria in Southeast Asia (Barik et al., 2009). None of the complete mitogenome of the complex has been reported, and their identification is mainly based on PCR assay that involves in mitochondrial COII. In addition to the use as molecular markers for identification, mitogenomes have been widely used in phylogenetics and molecular evolution studies (Logue et al. 2013). In this paper, we report the complete mitogenome of this species, which can provide an important basis for molecular identification and phylogenetic studies of the complex.
The complete mitogenome of An. culicifacies species B was successfully sequenced in this study (GenBank number KR732656). This sequencing was based on one female adult, collected from Ja Htu Kong village in the Kachin Region of Myanmar (the border of Yunnan Province in China). The PCR method described in Goswami et al. (2006) was used for the species identification. Genomic DNA was extracted from the whole mosquito using the Qiagen DNeasy blood and tissue kit. The complete mitogenome was amplified using 22 primer pairs designed in our Institute, which is in preparation for publication. The control region was cloned with the vector pMD-19T.
The length of the mitogenome of An. culicifacies species B is 15,330 bp, which consists of 13 protein-coding, 22 tRNA, 2 rRNA genes and a non-coding control region. Except for the 9 tRNA genes (Gln, Cys, Tyr, Ser (AGN), Phe, His, Pro, Leu (CUN), Val), 4 PCGs (ND5, ND4, ND4L, ND1) and 2 rRNA genes, all other genes are encoded on the heavy strand. The gene arrangement is consistent with other mosquito species (Hardy et, al. 2014). The overall nucleotide composition is 40.4% A, 38.1% T, 12.4% C, and 9.1% G. With the exception of COI with TCG and ND5 with GTG as a start codon, all other PCGs are initiated with the standard ATN (ATG, ATT, ATA, and ATC) (Beard et al., 1993). For stop codon, 9 PCGs (ND2, ATP8, ATP6, ND3, ND5, ND4L, ND6, Cytb, and ND1) have the complete TAA, and the other 4 genes (COI, COII, COIII, ND4) only have the incomplete T. There are 13 overlap between genes (ranging from 1 to 7 bp) and 9 intergenic spacer regions (ranging from 1 to 17 bp) in the genome. The ribosomal RNAs are relatively conversed as in other insects (Krzywinski et al., 2011). The 16S rRNA is 1,322 bp, located between tRNALeu and tRNAVal and have an AT content of 83.1%, and the 12S rRNA is 791 bp, flanked by tRNAVal and the control region with an AT content of 80.2%. The 22 tRNAs are totally 1,474 bp in length with gene length ranging from 64 to 72 bp. The total A+T content of 22 tRNAs is 78.6% with tRNAGlu having the highest AT content 92.4% and tRNAArg the lowest AT content 68.8%. As reported, the anticodens of 22 tRNAs are identical to other anophelines (Krzywinski et al., 2011; Logue et al., 2013). The control region is 498 bp long and has an A+T content of 94%. Compared with other anophelines, this species also contains a conservative T-stretch structure, which is located in position 14,975 bp, totally 18 bp in size (Krzywinski et al., 2011).
We reconstructed the ML phylogenetic tree of the species and 15 other Anopheles species using MeGa 5.1, based on the sequences of 13 PCGs with the Culex pipiens as outgroup (Figure 1). The 16 Anopheles species were classified into two group, corresponding to subgenera Cellia and Nyssorhynchus. An. culicifacies species B of Myzomyia Series was clusterd with An. gambiae of Pyretophorus Series with a 100% bootstrap values.
Figure 1.
The maximum likelihood (ML) tree based on PCG’s sequences of An. culicifacies species B and 15 other Anopheles species published. The GTR+G+I model suggested by ModelTest was used, and the bootstrap for 1000 replicates was indicated on each node. Species with GenBank accession number in bracket: An. farauti (JX219741), An. hinesorum (JX219734), An. koliensis (JX219743), An. punctulatus (JX219738), An. farauti 4 (JX219735), An. dirus (JX219731), An. cracens (JX219733), An. gambiae (NC002084), An. culicifacies species B (KR732656), An. oryzalimnetes (HQ335345), An. albitarsis (HQ335344), An. albitarsis F (HQ335349), An. janconnae (HQ335348), An. deaneorun (NC020663), An. albitarsis G (HQ335346), An. darlingi (GQ918272), Culex. pipiens (NC015079)
Footnotes
Declaration of interest
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Barik TK, Sahu B, Swain V. A review on Anopheles culicifacies: From bionomics to control with special reference to Indian subcontinent. Acta Tropica. 2009;109:87–97. doi: 10.1016/j.actatropica.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Beard CB, Hamm D, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 1993;2:103–124. doi: 10.1111/j.1365-2583.1993.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Goswami G, Singh OP, Nanda N, Raghavendra K, Gakhar SK, Subbarao SK. Identification of all members of the Anopheles culicifacies complex using allele-specific polymerase chain reaction assays. Am J Trop Med Hyg. 2006;75:454–460. [PubMed] [Google Scholar]
- Hardy CM, Court LN, Morgan MJ, Webb CE. The complete mitochondrial DNA genomes for two lineages of Aedes notoscriptus (Diptera: Culicidae) Mitochondrial DNA. 2014;0:1–2. doi: 10.3109/19401736.2014.974171. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Li C, Morris M, Conn JE, Lima JB, Povoa MM, Wilkerson RC. Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol Phylogenet Evol. 2011;58(3):469–477. doi: 10.1016/j.ympev.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue K, Chan ER, Phipps T, Small ST, Reimer L, Henry-Halldin C, Sattabongkot J, Siba PM, Zimmerman PA, Serre D. Mitochondrial genome sequences reveal deep divergences among Anopheles punctulatus sibling species in Papua New Guinea. Malar J. 2013;12:64. doi: 10.1186/1475-2875-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao SK, Sharma VP. Anopheline species complexes and malaria control. Indian Journal of Medical Research. 1997;106:164–173. [PubMed] [Google Scholar]