Abstract
BACKGROUND
Post hip fracture generalized pain can lead to a progressive decline in function and greater disability.
OBJECTIVES
The purpose of this study was to explore the factors that influence pain among older adults post hip fracture, including genetic variability, and evaluate whether or not pain directly or indirectly influenced upper and lower extremity function.
METHODS
This was a secondary data analysis using data from the first 200 participants in a Baltimore Hip Study (BHS), BHS-7. Assessments were done at 2 months post hip fracture and included age, gender, marital status, education, cognitive status, comorbidities, Body Mass Index (BMI), upper and lower extremity function, single nucleotide polymorphisms (SNPs) from 10 candidate genes, and total areas of pain and pain intensity. Model testing was done using the AMOS statistical program.
RESULTS
The full sample included 172 participants with an average age of 81. Fifty percent were female and the majority was Caucasian (93%). Model testing was done on 144 individuals whom completed 2 month surveys. Across all models age, cognition and BMI were significantly associated with total areas of pain. Thirty SNPs from five genes (BDNF, FKBP5, NTRK2, NTRK3, and OXTR) were associated with areas of pain and/or pain intensity. Together age, cognition, BMI and the SNP from one of the five genes explained 25% of total areas of pain and 15% of pain intensity. Only age and cognition were significantly associated with lower extremity function and only cognition was significantly associated with upper extremity function.
DISCUSSION
The full model was partially supported in this study. Our genetic findings related to pain expand prior reports related to BDNF and NTRK2.
Keywords: Pain, genetics, physical activity, function, resilience, hip fracture
Hip fractures commonly occur among older adults with over 1.5 million occurring each year worldwide (Cheng, Levy, Lefalvre, Guy, Kuramoto, Sobolev, 2011). Less than half of individuals who experience a hip fracture return to prefracture physical function (Magaziner et al., 2003). The return to prefracture physical function is influenced by many factors, including cognitive status, type of surgical interventions, age, other comorbid conditions and course following surgery(Ariza-Vega, Jiménez-Moleón, & Tange Kristensen, 2014; Beaupre, Jones, Johnston, Wilson, & Majumdar, 2012). In addition, pain has been associated with physical disability (Eggermont et al., 2014) and sedentary behavior (Stubbs et al., 2013). In the post hip fracture period, the presence of generalized pain can lead to a progressive decline in function and greater disability (Jensen, Moore, Bockow, Ehde, & Engel, 2011). The relationship between pain and function is also influenced by the anatomical location of the pain, the intensity of the pain and whether there are multiple sources or sites of pain (Eggermont et al., 2014; Yagci, Duymaz, & Cavlak, 2014). Repeatedly, it has been noted that chronic pain in multiple musculoskeletal locations increases the risk of future decline in mobility and function (Eggermont et al., 2014; Fowler-Brown, Wee, Marcantonio, Ngo, & Leveille, 2013).
Age-related changes in the structure, function, and chemistry of the nervous system may alter the perception of pain. These changes include a reduction of myelinated and unmyelinated fibers and a slowing of nerve conduction (Verdú, Ceballos, Vilches, & Navarro, 2000), a reduction in the functional integrity of sensory neurons that impact pain (Khalil, Ralevic, Bassirat, Dusting, & Helme, 1994), changes in brain volume in the prefrontal cortex and hippocampus which further influence pain perception (Farrell, 2012), and reduced functioning of endogenous pain modulatory mechanisms with regard to dopaminergic neurons in the basal ganglia (Cole, Farrell, Gibson, & Egan, 2010). Generally the cumulative effect of these changes and clinical findings suggest that with age there is an increased threshold and decreased tolerance for pain (Gibson & Farrell, 2004).
There is evidence to suggest that single nucleotide polymorphisms (SNPs) in multiple genes influence pain perception and interpretation (Table 1). SNPs are variations in a single DNA building block (A, T, C, G), called a nucleotide. Genetic influences of pain contribute to the modulation of pain in both the central nervous system (CNS) and in the periphery; SNPs in genes that participate in synaptic plasticity or the activation of spinal microglia have been associated with pain. Genetic variation can also influence nerve conduction and synaptic transmission, which could lead to altered pain sensation. To date, candidate gene analyses in pain research have focused mainly on 10 genes that were identified either in animal models or humans to be associated with pain (Belfer et al., 2013; Di Lorenzo et al., 2014; Renn, Leitch, & Dorsey, 2009). These genes include: BDNF, FKBP5, NTRK2, NTRK3, OXTR, NTRK1, DRD4, SLC6A4, COMT and MAOA. Table 1 provides a detailed description of these genes and their mechanisms of action. The candidate genes are believed to influence pain on the basis of their encoded proteins being involved in pathways that are logically expected to affect pain. Details of how specific SNPs from these genes influence pain are not yet established. Replication of the associations tested between the genes, associated SNPs and pain has not been consistent across studies and patient populations. Moreover, there have been no prior studies examining genetic factors that are associated with pain and subsequent function among older adults post hip fracture.
Table 1.
Candidate Genes, Mechanisms of Action and Rational for Inclusion in Model Testing
| Gene: SNPs that were tested in Model testing* | Gene location | Description of Mechanism | Prior Associations of Gene with Pain |
|---|---|---|---|
| FKBP5: rs16878806; rs16879378; rs145774; rs2817032 | Chromosome 6, 35573585- 35728583 bp | The FKBP5 gene is involved in the regulation of plasma cortisol following stress such as that associated with pain. | In prior research (Bortsov, Diatchenko, & McLean, 2014) this gene was associated with musculoskeletal pain following traumatic motor vehicle accidents. |
| BDNF: rs11602246 | Chromosome 11, 27, 654, 892- 27, 722, 057 bp | The BDNF gene is a neurotrophin that promotes the survival and proliferation of nerve cells and is involved in inflammation. BDNF is an important central pain mediator. | In prior research (Cheng et al., 2011; Ding et al., 2014)3 (Forsgren, Grimsholm, Dalén, & Rantapää-Dahlqvist, 2011) this gene was associated with neuropathic pain as well as pain from rheumatoid arthritis. |
| NTRK1 (No SNPs from this gene were significantly associated with pain so were not included in model testing) | Chromosome 1, 156, 815, 749- 156, 881, 849 bp | The NTRK1 gene provides instructions for making a protein that is essential for the development and survival of nerve cells (neurons), especially those that transmit information about sensations such as pain, temperature, and touch (sensory neurons). | In prior research (Mardy et al., 1999) there was evidence that mutations in the NTRK1 gene cause congenital insensitivity to pain with anhidrosis, a condition characterized by the inability to feel pain and decreased or absent sweating. |
| NTRK2: rs4142910; rs1140755; rs12340748; rs10780688; rs7019555; rs4631550; rs1565447; rs1565446; rs1565445; rs10125469; rs7045296; rs7030960; rs7857957; rs1490406; rs1490405; rs1490404; rs10123600; rs41277883; rs2013566 | Chromosome 9, 84, 668, 501- 85, 027, 069 bp | The NTRK2 gene encodes a member of the neurotrophic tyrosine receptor kinase (NTRK) family and is involved with neuron survival. | In prior research (Lin, Ro, Wang, & Chen, 2011; Renn, Leitch, & Dorsey, 2009) this gene was associated with nociception and the ability to feel inflammatory pain. |
| NTRK3: rs177652811; rs11634388; rs16941001; rs16941392; rs8033396; rs1863482; rs11073768; rs7172053 | Chromosone 15, 87, 859, 751–88, 256, 768 bp | The NTRK3 gene is involved with mediating the effects of neurotrophic factor including neuronal differentiation and survival. | In prior research (Ghilardi et al., 2011; Tender, Kaye, Li, & Cui, 2011) this gene was associated with neuropathic pain in animal models and skeletal pain in human models. |
| COMT (No SNPs from this gene were significantly associated with pain or function so were not included in model testing) | Chromosome 22, 19, 941, 739–19, 969, 974 bp | The COMT gene mediates the inactivation of catecholamine neurotransmitters, including dopamine, adrenaline, and noradrenaline. Reduced COMT enzymatic activity results in increased pain sensitivity and sensations of pain. | In prior research (Belfer et al., 2013; Bortsov et al., 2014; Cohen, Neumann, Glazer, Ebstein, & Buskila, 2009; James, 2013; Smith et al., 2014) this gene was consistently associated with chronic pain, musculoskeletal pain and other conditions with pain as a symptom. COMT has been noted to confer protection or decrease pain and help with the modulation of analgesic efficacy. |
| SLC6A4 (No SNPs from this gene were significantly associated with pain or function so were not included in model testing) | Chromosome 17 30194319- 30236002 bp | The SLC6A4 gene is associated with transporting the neurotransmitter serotonin from synaptic spaces into presynaptic neurons. | In prior research (Hooten, Hartman, Black, Laures, & Walker, 2013; Kumar, Ranjan, Mittal, & Ghoshal, 2012; Lindstedt et al., 2011) this gene was associated with the pain that occurred from irritable bowel disease. In addition SLC6A4 was associated with chronic pain disorders and with heat pain. |
| OXTR: rs2270465; rs401015 | Chromosome 3,. 8, 750, 408–8, 769, 628 bp | The OXTR gene belongs to the G- protein coupled receptor family and acts as a receptor for oxytocin. Receptors in the central nervous system modulate behaviors such as stress and anxiety; social memory; sexual aggression and bonding. The OXTR gene has also been associated with optimism, mastery, and self-esteem. | In prior research (Osherovich, 2011; Russo et al., 2012) this gene was associated with hyperalgesia in animal models. OXTR was also associated with the ability to cope with pain and thus is being considered as influential with regard to treatment modalities. |
| DRD4 (No SNPs from this gene were significantly associated with pain or function so were not included in model testing) | Chromosome 11, 637, 305 to 640, 706 bp | The DRD4 gene is associated with the neurotransmitter dopamine. It is linked to many neurological and psychiatric conditions including Parkinson’s Disease, addictive behaviors, eating disorders and schizophrenia as well as being associated with chronic pain. | In prior research (Buskila, Cohen, Neumann, & Ebstein, 2004; Limer, Nicholl, Thomson, & McBeth, 2008; Mercado, Barjola, Fernández-Sánchez, Guerra, & Gómez-Esquer, 2013) this gene was associated with widespread pain, such as that noted in individuals with fibromyalgia. |
| MAOA(No SNPs from this gene were significantly associated with pain or function so were not included in model testing) | X chromosome, 43, 654, 906–43, 746, 823 bp | The MAOA gene encodes mitochondrial enzymes which catalyze the oxidative deamination of amines, such as dopamine, norepinephrine, and serotonin. | In prior research (Di Lorenzo et al., 2014; Kim, Lee, Rowan, Brahim, & Dionne, 2006) this gene was associated with trigeminal pain disorders as well as post-surgical pain. |
Key:
COMT = catecholamine-O-methyltransferase
BDNF = Brain Derived Neurotrophic Factor
NTRK1 = neurotrophic tyrosine kinase, receptor, type 1.
NTRK2 = neurotrophic tyrosine kinase, receptor, type 2.
NTRK3 = neurotrophic tyrosine kinase, receptor, type 3.
SLC6A4= Solute Carrier Family 6 (Neurotransmitter Transporter), Member 41
OXTR= oxytocin receptor
DRD4= Dopamine receptor gene
MAOA= monoamine oxidase A
FKBP5 = K506 binding protein 5
Bolded SNPs are those that remained significant in model testing.
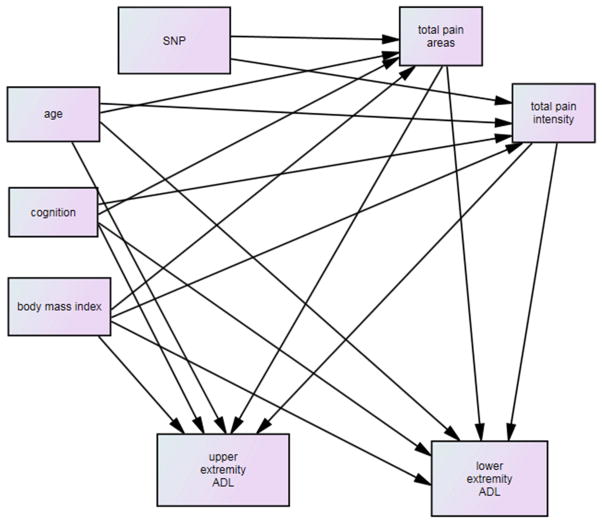
The purpose of this study was to explore the factors that influence pain among older adults post hip fracture and whether or not pain directly or indirectly influenced upper and lower extremity function. As shown in Figure 1, it was hypothesized that relevant demographic and descriptive factors (those that were significantly correlated based on bivariate correlations with total areas of pain, total pain intensity or upper or lower extremity function) and genetic variability would be associated with total areas of pain and total pain intensity and all of these variables would directly and indirectly be associated with upper and lower extremity function at 2 months post hip fracture.
Figure 1.
Full hypothesized Model
Methods
Design and Sample
This was a secondary data analysis using data from the first 200 participants in a Baltimore Hip Study (BHS), BHS-7. The primary focus of the parent study, BHS-7, was to compare men and women, frequency matched (1:1) on calendar time of fracture and hospital, regarding consequences, recovery trajectories and their predictors post hip fracture. Individuals were eligible if they were living in the community prior to fracture, aged 65 years or older and admitted for surgical repair of a hip fracture to one of the eight study hospitals. Individuals were excluded if they had a pathologic fracture, were not ambulating unaided by another person prior to the fracture, did not speak English, resided more than 70 miles from the hospital, weighed more than 300 pounds, or had hardware in the contralateral hip. The first 200 study participants were included in this analysis. To obtain the sample, 911 hip fracture patients were screened, 517 (57%) were eligible (222 males; 295 females), and 105 men and 107 women consented to participate. Twenty-three participants were withdrawn (eight participants failed to provide baseline or two-month data; six were ineligible; and nine were removed as a result of an IRB-requested post procedure audit) leaving 189 participants. Among these participants, 172 (91%) had a DNA sample and were included in this analysis. The parent study was approved by the Institutional Review Boards of the University of Maryland Baltimore and review boards within the participating hospitals.
Measures
Following consent, baseline assessments and a blood draw were performed by study evaluators and repeat assessments were done at two months. A proxy was asked to respond to survey questions focused on function for participants who scored < 36 on the Modified Mini-Mental State Examination (3MS)(MacKnight, Graham, & Rockwood, 1999). All measures used in this study had previously established evidence of reliability and validity.
Demographic information included age, gender, marital status, and education. Cognitive status was evaluated using the 3MS(MacKnight et al., 1999). Scores can range from 0 to 100 with higher numbers indicating less impairment. To describe comorbidities, the Charlson Comorbidity Index (Charlson, Pompei, Ales, & MacKenzie, 1987) was completed using data from medical records. Body Mass Index (BMI) was based on weight in kilograms divided by height in meters.
Function and Pain
Measures of function were based on subjective input from the participant/proxy. Lower extremity function was evaluated using the Physical Activities of Daily Living (PADL) survey, which was modified from the Functional Status Index (Jette, 1980). Scores ranged from 0–12 with higher scores representing greater impairment. Upper extremity Activities of Daily Living were evaluated based on four items (putting on and buttoning a shirt, feeding oneself and grooming). Scores ranged from 0 to 4 with higher scores indicating more impairment. Pain was calculated based on total areas of pain (upper extremity, hip, knee, back or ankle pain with a range of 0 reflecting that the participant had no pain in any of the areas evaluated to 5 indicating that there was pain in all 5 areas evaluated) and an overall sum of pain intensity across each those areas (0–10 rating scale per area and a total range of 0 to 50).
DNA Extraction and Genotyping
DNA was extracted from 3.5–7ml of whole blood drawn into a BD vacutainer® ACD blood collection tube. The sample was transported on ice to the Translational Core Lab (University of Maryland School of Medicine, Baltimore MD) and blood was frozen at −20C within one hour of blood draw. The 64 SNP OpenArray chip was created using the online design software from Life Technologies. Eighteen of the 64 assays were custom assays, designed by the software, the others were off-the-shelf Taqman assays. Assays were run on the QuantStudio 12K Flex with the OpenArray block, following published protocols(Applied Biodsystems QuantSutdio, 2012). Data were analyzed using the provided Genotype software.
Participants were genotyped using the Affymetrix 1M SNP chip (v6.0, Santa Clara, CA USA) by the Genomics Core Laboratory at the University of Maryland according to the manufacturer’s recommendations. Genotype calling was done using Birdseed algorithm (v2.0) which is part of the Birdsuite tools (Korn, Kuruvilla, McCarroll, & et al, 2008). We identified 429 SNP variants in 10 candidate genes that have been previously associated with pain for genotyping in our sample. Twenty-eight of these SNPs were from BDNF, 22 from FKBP5, 18 from NTRK1, 128 from NTRK2, 129 from NTRK3, 41 from OXTR, 5 from DRD4, 12 from SLC6A4, 24 from COMT and 22 from MAOA. Of these, 34 were significantly associated with total pain and/or total pain intensity.
Statistical Analysis
Descriptive analyses were used to describe the sample and structural equation modeling using SPSS and AMOS. For all models tested, the sample covariance matrix was used as input and a full information maximum likelihood (FIML) solution sought. Model fit was evaluated based on the chi-square statistic divided by degrees of freedom and the Stiegers Root Mean Square Error of Approximation (RMSEA). A ratio of ≤ 3 was considered to be a good fit (Bollen, 1989). A RMSEA of < 0.10 was considered good, and <0.05 very good. Path significance was based on the Critical Ratio (CR) (Bollen, 1989). Significance for path estimates was set at p ≤ 0.05. Differences in model fit were based on significant changes between the χ2 and degrees of freedom of each model. Missing data were handled using the FIML procedure(Joreskog & Sorbom, 1993). A squared multiple correlation (R2) was calculated for each of the dependent variables or outcomes (total areas of pain, total pain intensity, and upper and lower extremity function). The R2 indicates the proportion of variance in the dependent variable accounted for by the set of independent variables in the model. There was complete data on 144 participants across all model variables and no differences between those with or without missing data in terms of demographic factors.
Each SNP that was significantly correlated (p≤ 0.05) with either total pain areas or total pain intensity based on bivariate correlations was tested in the model independently. An additive genetic model was tested for these models. Specifically, coding for the SNP variables represented the number of minor alleles carried by each participant (i.e., 0, 1 or 2). Linkage disequilibrium (LD) in the observed regions was calculated for both D′ and r2 using HAPLOVIEW. LD plots are provided in Appendix A for candidate genes with multiple significant SNP findings. Linkage disequilibrium occurs when there is a strong association between having a particular allele (alleles are defined as two alternate forms of a gene that are composed of at least one SNP) at one location of a gene and having another allele at a different location on the gene. The association suggests that the two alleles are not providing separate information. If there is linkage disequilibrium the two loci or positions of the alleles or SNPs being considered are both contributing to the phenotype which in this study is an individual with pain.
Secondary Analysis – NTRK 2 3′UTR
We identified an association signal in NTRK2 which was particularly interesting as it was in the 3′ untranslated region (UTR) of the truncated isoform of the BDNF receptor, termed trkB.T1. We used phylogenetic module complexity analysis(Claussnitzer, Dankel, & Klocke, 2014) to examine the potential functional relevance of the SNPs in this 3′ untranslated region and identified rs41277883 for further study. This SNP was found to overlap with evolutionary conserved elements and was predicted to be a weak enhancer. With this in mind we conducted follow-up genotyping on the full dataset using TaqMan assays (Life Technologies, Grand Island, NY) according to the manufactures specifications.
Full model testing is shown in Figure 1. At two months post hip fracture, age, cognition, BMI, gender and comorbidities were hypothesized to be directly associated with total areas of pain, pain intensity and upper and lower extremity function. Age, cognition, BMI, gender and comorbidities were hypothesized to be indirectly associated with upper and lower extremity function through total pain areas and pain intensity. Each relevant SNP was hypothesized to be directly associated with total number of areas of pain and total pain intensity. Pain intensity and total pain areas were hypothesized to be directly associated with upper and lower extremity function.
Results
As per study design 50% of the participants were female. The majority of the participants were Caucasian (93%), and the average age was 81.09 (SD=7.42). Descriptive statistics are provided in Table 2. As shown in Table 1, 30 SNPs of the 34 SNPs included in model testing were significantly associated with either total pain areas or total pain intensity in the tested models. The 30 SNPs significantly associated with pain were from five genes, BDNF, FKBP5, NTRK2, NTRK3, and OXTR. The path estimates are provided in Table 3 and show whether or not there is a significant association between the SNP and pain. When significant it means that there is a genetic influence of that gene on pain. In addition, the path estimates or relationships between all the other variables in the model are also shown in Table 3. Each of the SNPs conformed to the expectations of Hardy Weinberg equilibrium (p>0.05) and had sufficiently high call rate (>90%) to be included in model testing.
Table 2.
Descriptive Statistics for Model Variables
| Model Variables | Range | Mean | SD |
|---|---|---|---|
| Age | 65 to 96 years of age | 81.09 | 7.42 |
| Comorbidities | 0 to 8 comorbidities | 2.22 | 1.89 |
| Body Mass Index | 15.40 to 48.40 | 25.38 | 5.06 |
| Cognitive Status [The Modified Mini-Mental State (3MS) Test] | 0 to 100 with higher numbers indicating less impairment. 3MS total score of < 79 is indicative of cognitive impairment and scores of < 48 are indicative of severe impairment. |
82.55 | 19.28 |
| Instrumental Activities of Daily Living | 0 to 4 with higher scores indicating more impairment | 3.37 | 1.10 |
| Lower Extremity Activities of Daily Living | 0 to 12 with higher scores indicating more impairment | 7.57 | 3.21 |
| Total pain intensity | 0 to 50 (sum of intensity across all five areas evaluated) | 12.63 | 9.56 |
| Hip pain | 0 to 10 | 2.88 | 3.41 |
| Knee pain | 0 to 10 | 5.34 | 2.71 |
| Back pain | 0 to 10 | 5.47 | 2.64 |
| Foot/ankle pain | 0 to 10 | 5.63 | 2.39 |
| Upper extremity pain | 0 to 10 | 5.30 | 2.12 |
| Total areas of Pain | 0 to 5 | 2.36 | 1.37 |
| Pain Frequency Across 5 Areas (whether or not pain was present in each area evaluated) | Present N (%) | Absent N (%) | Missing N (%) |
|---|---|---|---|
| Hip | 86 (50%) | 40 (23%) | 46(27%) |
| Knee | 65(38%) | 62(36%) | 450(26%) |
| Back | 59(34%) | 68(40%) | 45(26%) |
| Foot/Ankle | 27(22%) | 90(52%) | 45(26%) |
| Upper Extremity | 51(30%) | 76(44%) | 45(26%) |
Table 3.
Model Path Estimates for Genes and significant pain associated SNPs (NS = nonsignificant path)
| Genes | BDNF | FKBP5 | NTRK2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | rs11602246 | rs16878806 | rs16879378 | rs1475774 | rs4142910 | rs11140755 | rs12340748 | rs10780688 | ||||||||||
| Risk allele a | C | G | C | T | A | C | C | G | ||||||||||
| Model Paths | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | ||
| total pain areas | <--- | SNP | −.169 | .016 | .146 | .035 | .170 | .013 | .145 | .038 | NS | NS | −.146 | .036 | −.146 | .036 | .164 | .019 |
| total pain intensity | <--- | SNP | NS | NS | NS | NS | NS | NS | NS | NS | −.143 | .044 | NS | NS | NS | NS | NS | NS |
| total pain areas | <--- | age | −.222 | .002 | −.188 | .007 | −.171 | .012 | −.207 | .003 | −.227 | .001 | −.198 | .005 | −.198 | .005 | −.235 | .001 |
| total pain areas | <--- | cognition | .393 | .001 | .440 | .001 | .464 | .001 | .424 | .001 | .405 | .001 | .423 | .001 | .423 | .001 | .392 | .001 |
| total pain areas | <--- | body mass index | .144 | .001 | .142 | .001 | .136 | .002 | .136 | .002 | .144 | .001 | .143 | .001 | .143 | .001 | .141 | .002 |
| total pain intensity | <--- | age | −.195 | .006 | −.166 | .020 | −.168 | .018 | −.187 | .009 | −.199 | .005 | −.174 | .014 | −.174 | .014 | −.204 | .004 |
| total pain intensity | <--- | cognition | .312 | .001 | .327 | .001 | .329 | .001 | .316 | .001 | .303 | .001 | .321 | .001 | .321 | .001 | .299 | .001 |
| upper extremity ADL | <--- | cognition | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.678 | .001 |
| lower extremity ADL | <--- | cognition | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 |
| lower extremity ADL | <--- | age | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 |
| Gene | NTRK2 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | rs7019555 | rs4631550 | rs1565447 | rs1565446 | rs1565445 | rs10125469 | rs7045296 | rs7030960 | ||||||||||
| Risk Allele | A | A | G | G | T | G | A | C | ||||||||||
| Model Paths | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | ||
| total pain areas | <--- | SNP | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| total pain intensity | <--- | SNP | −.139 | .050 | −.139 | .050 | .139 | .050 | .139 | .050 | .139 | .050 | −.139 | .050 | −.139 | .050 | −.139 | .050 |
| total pain areas | <--- | age | −.228 | .001 | −.228 | .001 | −.228 | .001 | −.228 | .001 | −.228 | .001 | −.228 | .001 | −.228 | .001 | −.228 | .001 |
| total pain areas | <--- | cognition | .404 | .001 | .404 | .001 | .404 | .001 | .404 | .001 | .404 | .001 | .404 | .001 | .404 | .001 | .404 | .001 |
| total pain areas | <--- | body mass index | .144 | .001 | .144 | .001 | .144 | .001 | .144 | .001 | .144 | .001 | .144 | .001 | .144 | .001 | .144 | .001 |
| total pain intensity | <--- | age | −.200 | .005 | −.200 | .005 | −.200 | .005 | −.200 | .005 | −.200 | .005 | −.200 | .005 | −.200 | .005 | −.200 | .005 |
| total pain intensity | <--- | cognition | .302 | .001 | .302 | .001 | .302 | .001 | .302 | .001 | .302 | .001 | .302 | .001 | .302 | .001 | .302 | .001 |
| upper extremity | <--- | cognition | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 |
| lower extremity ADL | <--- | cognition | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 |
| lower extremity ADL | <--- | age | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 |
| Gene | NTRK2 | NTRK3 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | rs7857957 | rs1490406 | rs1490405 | rs1490404 | rs10123600 | rs41277883b | rs17765281 | rs11634388 | ||||||||||
| Risk Allele | G | C | G | A | C | C | G | |||||||||||
| Model Paths | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | ||
| total pain areas | <--- | SNP | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | −.148 | .036 | .180 | .009 |
| total pain intensity | <--- | SNP | .143 | .044 | −.143 | .044 | .143 | .044 | −.143 | .044 | −.143 | .044 | −.139 | .047 | NS | NS | .148 | .036 |
| total pain areas | <--- | age | −.227 | .001 | −.227 | .001 | −.227 | .001 | −.227 | .001 | −.227 | .001 | −.232 | .001 | −.210 | .003 | −.211 | .002 |
| total pain areas | <--- | cognition | .405 | .001 | .405 | .001 | .405 | .001 | .405 | .001 | .405 | .001 | .420 | .001 | .399 | .001 | .430 | .001 |
| total pain areas | <--- | body mass index | .144 | .001 | .144 | .001 | .144 | .001 | .144 | .001 | .144 | .001 | .139 | .002 | .149 | .001 | .139 | .002 |
| total pain intensity | <--- | age | −.199 | .005 | −.199 | .005 | −.199 | .005 | −.199 | .005 | −.199 | .005 | −.204 | .003 | −.190 | .008 | −.188 | .008 |
| total pain intensity | <--- | cognition | .303 | .001 | .303 | .001 | .303 | .001 | .303 | .001 | .303 | .001 | .345 | .001 | .318 | .001 | .319 | .001 |
| upper extremity | <--- | cognition | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 |
| lower extremity ADL | <--- | cognition | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.470 | .001 | −.469 | .001 | −.469 | .001 |
| lower extremity ADL | <--- | age | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 |
| Gene | NTRK3 | OXTR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | rs16941001 | rs16941392 | rs8033396 | rs1863482 | rs11073768 | rs2270465 | ||||||||
| Risk Allele | A | G | G | C | G | G | ||||||||
| Model Paths | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | Path | P* | ||
| total pain areas | <--- | SNP | −.152 | .030 | .141 | .043 | .141 | .046 | NS | NS | NS | NS | NS | NS |
| total pain intensity | <--- | SNP | NS | NS | .141 | .047 | NS | NS | −.138 | .053 | .140 | .048 | .165 | .019 |
| total pain areas | <--- | age | −.239 | .001 | −.193 | .006 | −.217 | .002 | −.224 | .001 | −.239 | .001 | −.225 | .001 |
| total pain areas | <--- | cognition | .395 | .001 | .433 | .001 | .405 | .001 | .407 | .001 | .396 | .001 | .407 | .001 |
| total pain areas | <--- | body mass index | .145 | .001 | .146 | .001 | .145 | .001 | .140 | .002 | .142 | .002 | .143 | .001 |
| total pain intensity | <--- | age | −.208 | .003 | −.168 | .018 | −.190 | .008 | −.196 | .006 | −.213 | .003 | −.196 | .005 |
| total pain intensity | <--- | cognition | .314 | .001 | .330 | .001 | .315 | .001 | .307 | .001 | .296 | .001 | .315 | .001 |
| upper extremity | <--- | cognition | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 | −.679 | .001 |
| lower extremity ADL | <--- | cognition | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 | −.469 | .001 |
| lower extremity ADL | <--- | age | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 | .178 | .016 |
p significant at ≤ .05
Note:
Risk allele: (the more copies of this allele, the more pain intensity or areas reported)
rs41277883, a potentially functional variant, was tested as part of a secondary analysis and not part of the GWAS platform
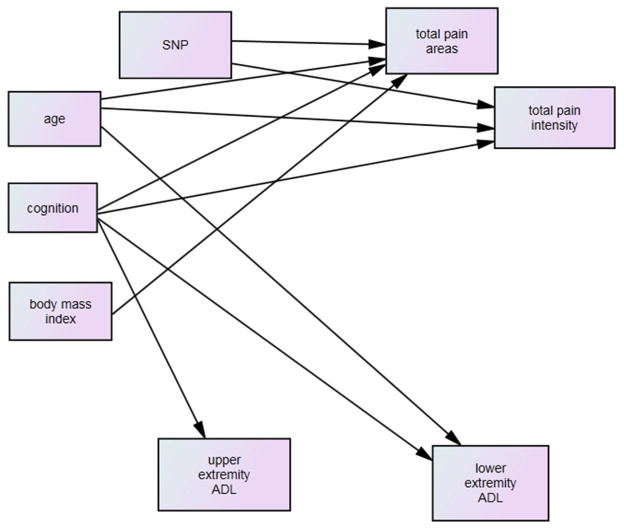
Full hypothesized model testing indicated that 10 of the 18 hypothesized paths were significant and there was a fair fit of the model to the data (χ2 / df = 3.82, NFI=.84, and RMSEA = .13). The model was revised with all non-significant paths removed (Figure 2) and testing of the revised model showed a non-significant improvement in fit with χ2 / df = 2.52, NFI=.88, and RMSEA = .09 (χ2 / df difference between the two models = 1.30, df difference of 1, p > .05).
Figure 2.
Model: Significant Paths Only
As shown in Table 3, the SNPs tested from BDNF (rs11602246) and FKBP5 (rs16878806; rs16879378; rs145774) were significantly associated with total number of areas of pain and three SNPs from NTRK2 (rs11140755; rs12340748; rs10780688) and five SNPs from NTRK3 (rs17765281; rs11634388; rs16941001; rs16941392; rs8033396) were significantly associated with total areas of pain. The single significant SNP from OXTR (rs2270465) was associated with total pain intensity and 15 SNPs from NTRK2 (rs4142910; rs7019555; rs4631550; rs1565447; rs1565446; rs1565445; rs10125469; rs7045296; rs7030960; rs7857957; rs1490406; rs1490405; rs1490404; rs10123600; including rs41277883 from the secondary data analysis) and three SNPs from NTRK3 (rs11634388; rs16941392; rs11073768) were also associated with total pain intensity. There were two SNPs from NTRK3 (rs11634388 and rs16941392) that were significantly associated with both total areas of pain and total pain intensity. The risk allele associate with each significant SNP is shown in Table 3.
In each of the models tested, the R2 for total areas of pain was 25% indicating that age, cognition, BMI and the associated SNPs explained 25% of the variance in total areas of pain. Those who were younger, more cognitively intact, had higher BMIs and had a genetic predisposition in the significant SNPs reported more areas of pain. The R2 for total pain intensity was 15% indicating that age, cognition and significantly associated SNPs explained 15% of the variance in pain intensity. Those who were younger, more cognitively intact and had a genetic predisposition in the significant SNPs reported more intense pain.
As shown in Table 3, in the revised model, only age and cognition were directly associated with lower extremity function and based on the R2 explained 25% of the variance in lower extremity function. Those who were older and less cognitively intact had greater impairment in lower extremity function. Cognition was the only variable associated with upper body function such that more cognitively impaired individuals had greater impairment in function. Based on the R2, cognition explained 46% of the variance in upper body function.
Genes with multiple significant SNPs were tested for linkage disequilibrium (Appendix A). All the SNPs from FKBP5 were in high LD (r2>0.92), and all of the SNPs in the NTRK2 gene had high pair-wise LD (r2 > 0.86) except for rs10780688The LD pattern for NTRK3 was less apparent. Two SNPs (rs1863482 and rs11073768) were in perfect LD and two other SNPs (rs8033396 and rs17765281) were in high LD (r2=0.81). Modest, but not high LD was observed between the other significant SNPs. For the SNPs with high LD it is likely that they are similar SNPs from the same location within the gene and do not contribute differently to the interpretation of pain or other outcomes.
Discussion
Across all of the models tested there was at most 10 significant paths out of the 18 hypothesized paths tested. Therefore, the hypothesized model was only partially supported in this study. The lack of association noted in our findings between pain and function has also been reported among patients with non-cancer associated pain (Kauppila, Pesonen, Tarkkila, & Rosenberg, 2007) as well as prior samples of patients post hip fracture (Ekström et al., 2013). When pain was associated with function among hip fracture patients, it was between 4 and 12 months post fracture (Morrison, Sean, Fischberg, & Cintron, 2009) rather than at the 2 month time point. Although it is possible that among older adults pain does not influence function and that resilience or other factors not tested in the model may be more important. Conversely, it is also possible that the lack of association between pain and function in this study may have been due to the fact that participants did not note pain as they were sedentary and receiving help with activities of daily living at the 2 month follow up period.
From five of the candidate genes a total of 30 SNPs were associated with pain based on model testing: BDNF (1 SNP), FKBP5 (3 SNPs), NTRK2 (18 SNPs), NTRK3 (7 SNPs), and OXTR (1 SNPs). Our findings expand prior reports related to BDNF and NTRK2. The neurotrophin BDNF has been shown to be a potent modulator of pain processing in the CNS (Merighi, Salio, Ghirri, & al., 2008). Noxious stimulation increases BDNF production in the spinal dorsal horn (SDH)(Coull, et al., 2005) and brainstem (Renn, Lin, Thomas, & Dorsey, 2006), leading to hyperalgesia and the formation of mechanical allodynia. This is presumably because BDNF signaling participates in the development of windup and central sensitization in the SDH, using mechanisms similar to those in the development of long-term potentiation in the hippocampus (Latremoliere & Woolf, 2009).
The NTRK2 SNP (rs41277883) tested in the secondary analysis is particularly interesting because it is in the 3′ untranslated region (UTR) of the truncated isoform of the BDNF receptor, termed trkB.T1. We have shown that trkB.T1, but not the full-length kinase active receptor, is significantly upregulated in a variety of rodent models of pain including persistent inflammation, acute heat pain, and spinal cord injury hyperpathia (Renn, Leitch, & Dorsey, 2009; Wu, Renn, Faden, & Dorsey, 2013). Mice in which we genetically deleted the trkB.T1 receptor (Dorsey et al., 2006) developed significantly less hyperalgesia and allodynia in these models. It is possible, therefore, that reducing trkB.T1 expression reduces pain (Renn et al., 2009; Wu et al., 2013). Thus, this study is among the first to provide evidence for a genotype/pain phenotype association with SNPs in the 3′UTR of the trkB.T1 receptor gene NTRk2. Future research needs to focus on replicating this work in humans as it may be possible to delete the trkB.T1 receptor and decrease pain sensation among humans.
In addition to specific SNPs from the 5 candidate genes, age, cognition, and BMI were significantly associated with total number of areas of pain and age and cognition were associated with total pain intensity. Participants who were older reported more areas of pain but overall less intensity of pain. This is consistent with prior reports suggesting that pain sensitivity may decrease with increasing age (Alves, et al., 2012). Physiologically pre-clinical studies (Gagliese, 2009) have noted that with age there tends to be an increase in the pain threshold and it takes more of a stimulus to get a response. Despite a decrease in pain sensitivity with increased age, older adults tend to have decreased tolerance to pain (Molton & Terrill, 2014). Our BMI findings support prior work noting the negative impact of body weight in terms of causing pain in the weight bearing joints of the musculoskeletal system (Karlsson, Magnusson, Cöster, Karlsson, & Rosengren, 2015).
Cognition was the only variable to be associated with upper extremity function and this may reflect the frontal lobe function needed to plan and complete bathing and dressing (Liu-Seifert et al., 2015). Age was associated with lower extremity but not upper extremity function. Taken together, only a small percentage of the variance in function was explained. It is likely that many other factors are involved with performance of these basic functional tasks such as availability of a caregiver to complete the tasks thus eliminating the older adult’s opportunity to perform the given activity.
Implications for Nursing Practice
The findings from this study can help guide nurses in the evaluation of factors that may be associated with pain among older adults post hip fracture. As shown in Table 3 there is some indication that there is a genetic influence associated with experiencing pain. Although currently there is insufficient research to be able to identify specific genotypes that will be more likely to have musculoskeletal pain post hip fracture, nurses should recognize that there is a relationship between genetic makeup and pain sensation. Further, nurses should expect that those who are older and have higher BMIs may be more likely to have multiple areas of pain following a hip fracture but actually less pain intensity than a younger individual. When providing care to older adults post hip fracture it should not be assumed that pain reported is only from the fracture site. It is possible that many other commonly experienced areas of musculoskeletal pain due to chronic illnesses are contributing to the pain experienced by the older individual. The treatment for these areas of pain may be different than the acute management of fracture or post-surgical pain. Likewise, recognizing that older patients may not have as intense pain for the same clinical problem as a younger individual is important to avoid over treatment of the pain with excessive opiod use.
Our findings suggest that nurses providing care to patients in the post-acute recovery period (i.e., at 2 months post hip fracture) should recognize that pain does not significantly influence function among these individuals. Rather, evaluation of cognition is critical to anticipating the patient’s ability to engage in functional tasks. Interventions to optimize function among those who are cognitively impaired include such things as modeling the behavior versus providing verbal cues, giving simple one step commands, and practicing functional tasks repeatedly (Galik, Resnick, Hammersla, Brightwater, 2014).
Study Limitations
One of the strengths of this study was the inclusion of equal numbers of males and females. The study was limited, however, by inclusion of community dwelling older adults from a single state. In addition, data were obtained based on verbal report at a single time point. We had a small percentage of African American participants which may have biased the genetic findings. Given the exploratory nature of this study we did not correct for the testing of multiple models. Repeat testing of the proposed model with a larger sample is needed to establish the reliability of the noted associations between SNPs and outcome variables.
Conclusion
The purpose of this study was to explore the factors that influence pain among older adults post hip fracture, including genetic variability, and evaluate whether or not pain directly or indirectly influenced upper and lower extremity function. Despite stated limitations this study provides some support for previously identified candidate genes for pain among older adults and supports prior work suggesting that pain may not strongly influence function in the early post hip fracture period. Further exploration of the association between pain and function, gene expression and protein synthesis is needed to elucidate the mechanistic relationships with musculoskeletal pain in older adults.
Acknowledgments
This work has been supported through the National Institute on Aging via K08 AG043548, R01 AG046217-01, NR013736-01A1, P30 AG028747, and R37 AG009901, R01 AG029315.
Appendix A
Linkage Disequilibrium (LD) Plot for FKBP5
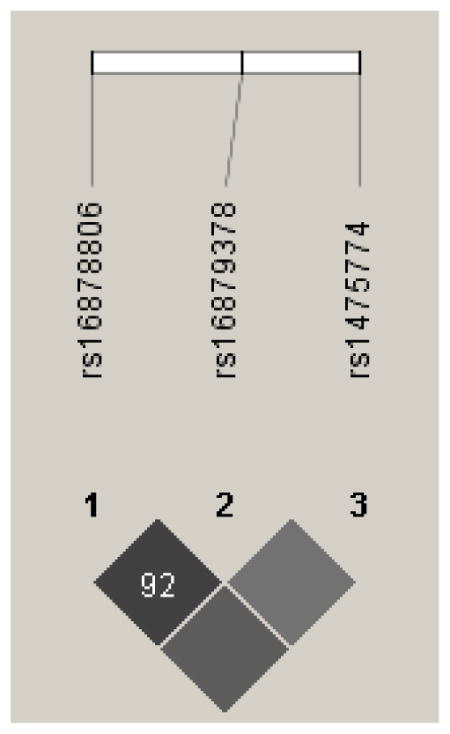
Darker shading indicates higher r2 (black indicates r2=1) and numerical values are calculated for D′.
Linkage Disequilibrium (LD) Plot for NTRK2
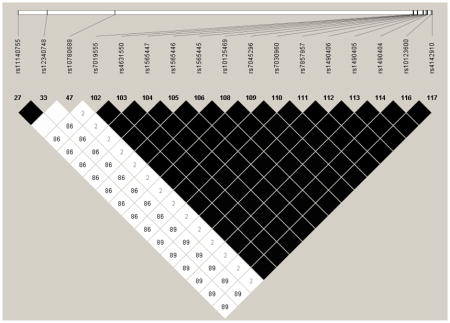
Darker shading indicates higher r2 (black indicates r2=1) and numerical values are calculated for D′.
Linkage Disequilibrium (LD) Plot for NTRK3
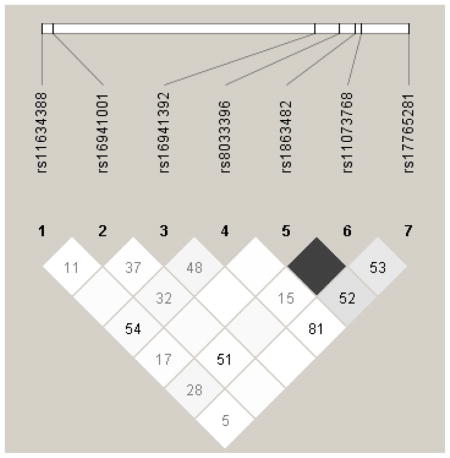
Darker shading indicates higher r2 (black indicates r2=1) and numerical values are calculated for D′
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alves NFE, Bergmann A, do Amaral e Silva B, Padula-Rabeiro AC, de Souza-Abrahao K, da Costa Leite Ferreira MG, … Santos Thuler LC. Post-mastectomy pain syndrome: incidence and risks. Breast. 2012;21(3):321–325. doi: 10.1016/j.breast.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Applied Biodsystems QuantSutdio. [Last accessed October, 2015];Applied Biosystems™ QuantStudio™ 12K Flex Real-Time PCR System OpenArray® Experiments. 2012 Available at: http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_102213.pdf.
- Ariza-Vega P, Jiménez-Moleón J, Tange Kristensen M. Non-Weight-Bearing Status Compromises the Functional Level Up to 1 yr After Hip Fracture Surgery. American Journal of Physical Medicine & Rehabilitation. 2014;93(8):641–648. doi: 10.1097/PHM.0000000000000075. [DOI] [PubMed] [Google Scholar]
- Beaupre L, Jones C, Johnston D, Wilson D, Majumdar S. Recovery of function following a hip fracture in geriatric ambulatory persons living in nursing homes: Prospective cohort study. Journal of the American Geriatrics Society. 2012;60(7):1268–1273. doi: 10.1111/j.1532-5415.2012.04033.x. [DOI] [PubMed] [Google Scholar]
- Belfer I, Segall S, Lariviere W, Smith S, Dai F, Slade G, … Diatchenko L. Pain modality- and sex-specific effects of COMT genetic functional variants. Pain. 2013;154(8):1368–1376. doi: 10.1016/j.pain.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen KA. Structural equations with latent variables. John Wiley & Sons; New York, NY: 1989. [Google Scholar]
- Bortsov A, Diatchenko L, McLean S. Complex multilocus effects of catechol-O-methyltransferase haplotypes predict pain and pain interference 6 weeks after motor vehicle collision. Neuromolecular Medicine. 2014;16(1):83–93. doi: 10.1007/s12017-013-8255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskila D, Cohen H, Neumann L, Ebstein R. An association between fibromyalgia and the dopamine D4 receptor exon III repeat polymorphism and relationship to novelty seeking personality traits. Molecular Psychiatry. 2004;9(8):730–731. doi: 10.1038/sj.mp.4001506. [DOI] [PubMed] [Google Scholar]
- Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: Prognostic development and validation. Journal of Chronic Disease. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- Cheng S, Levy a, Lefalvre K, Guy P, Kuramoto L, Sobolev B. Geographic trends in incidence of hip fracture: A comprehensive literature review. Osteoporosis International. 2011;22:2575–2586. doi: 10.1007/s00198-011-1596-z. [DOI] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel S, Klocke B. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156(1–2):343–358. doi: 10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H, Neumann L, Glazer Y, Ebstein R, Buskila D. The relationship between a common catechol-O-methlytransferase (COMT) polymorphism val (158)met and firbromyalgia. Clinical Experiments in Rheumatology. 2009;27(supplement 56):S51–S56. [PubMed] [Google Scholar]
- Cole L, Farrell M, Gibson S, Egan G. Age-related differences in pain sensitivity and regional brain activity evoked by noxious pressure. Neurobiology of Aging. 2010;31:494–503. doi: 10.1016/j.neurobiolaging.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Coull JA, Beggs S, Boudreau D, Boivin D, Tsuda M, Inoue K, … De Koninck Y. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 2005;438:1017–1021. doi: 10.1038/nature04223. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo C, Daverio A, Pasqualetti P, Coppola G, Giannoudas I, Barone Y, … Di Lorenzo G. The upstream variable number tandem repeat polymorphism of the monoamine oxidase type A gene influences trigeminal pain-related evoked responses. European Journal of Neuroscience. 2014;39(3):501–507. doi: 10.1111/ejn.12458. [DOI] [PubMed] [Google Scholar]
- Ding X, Cai J, Song L, Liu X, Wan L, Xing G. BDNF contributes to the development of neuropathic pain by induction of spinal long-term potentiation via SHP2 associated GluN2B-containing NMDA receptors activation in rats with spinal nerve ligation. Neurobiology of Disease. 2014;11(73C):428–451. doi: 10.1016/j.nbd.2014.10.025. [DOI] [PubMed] [Google Scholar]
- Dorsey S, Renn C, Carim-Todd L, Barrick C, Bambrick L, Krueger B, … Tessarollo T. In vivo restoration of physiological levels of truncated trkBT1 receptor rescues neuronal cell death in a trisomic mouse model. Neuron. 2006;51:21–28. doi: 10.1016/j.neuron.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Eggermont L, Leveille S, Shi L, Kiely D, Shmerling R, Jones R, … Bean J. Pain characteristics associated with the onset of disability in older adults: The maintenance of balance, independent living, intellect, and zest in the elderly Boston Study. Journal of the American Geriatrics Society. 2014;62(6):1007–1016. doi: 10.1111/jgs.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekström W, Al-Ani A, Sääf M, Cederholm T, Ponzer S, Hedström M. Health related quality of life, reoperation rate and function in patients with diabetes mellitus and hip fracture—A 2 year follow-up study. Injury. 2013;44(6):769–775. doi: 10.1016/j.injury.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Farrell M. Age-related changes in the structure and function of brain regions involved in pain processing. Pain Medicine. 2012;13:S37–S43. doi: 10.1111/j.1526-4637.2011.01287.x. [DOI] [PubMed] [Google Scholar]
- Forsgren S, Grimsholm O, Dalén T, Rantapää-Dahlqvist S. Measurements in the blood of BDNF for RA patients and in response to anti-TNF treatment help us to clarify the magnitude of centrally related pain and to explain the relief of this pain upon treatment. [Last accessed January, 2016];International Journal of Inflammation. 2011 doi: 10.4061/2011/650685. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3132632/pdf/IJI2011-650685.pdf. [DOI] [PMC free article] [PubMed]
- Fowler-Brown A, Wee C, Marcantonio E, Ngo L, Leveille S. The mediating effect of chronic pain on the relationship between obesity and physical function and disability in older adults. Journal of the American Geriatrics Society. 2013;61(12):2079–2086. doi: 10.1111/jgs.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliese L. Pain and aging: The emergence of a new subfield of pain research. Journal of Pain. 2009;10:343–353. doi: 10.1016/j.jpain.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Galik E, Resnick B, Hammersla M, Brightwater J. Optimizing function and physical activity among nursing home residents with dementia: Testing the impact of Function-Focused Care. Gerontologist. 2014;54(6):930–43. doi: 10.1093/geront/gnt108. [DOI] [PubMed] [Google Scholar]
- Ghilardi J, Freeman K, Jimenez-Andrade J, Mantyh W, Bloom A, Bouhana K, … Mantyh P. Sustained blockade of neurotrophin receptors TrkA, TrkB and TrkC reduces non-malignant skeletal pain but not the maintenance of sensory and sympathetic nerve fibers. Bone. 2011;48(2):389–398. doi: 10.1016/j.bone.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SJ, Farrell M. A review of age differences in the neurophysiology of nociception and the perceptual experience of pain. Clinical Journal of Pain. 2004;20:227–239. doi: 10.1097/00002508-200407000-00004. [DOI] [PubMed] [Google Scholar]
- Hooten W, Hartman W, Black JR, Laures H, Walker D. Associations between serotonin transporter gene polymorphisms and heat pain perception in adults with chronic pain. BMC Medical Genetics. 2013;14(78):1114–1178. doi: 10.1186/1471-2350-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. Human pain and genetics: Some basics. [Last accessed January, 2016];British Journal of Pain. 2013 doi: 10.1177/2049463713506408. Available at: http://bjp.sagepub.com/content/early/2013/09/24/2049463713506408.full.pdf+html. [DOI] [PMC free article] [PubMed]
- Jensen M, Moore M, Bockow T, Ehde D, Engel J. Psychosocial factors and adjustment to persistent pain in persons with physical disabilities: A systematic review. Archives of Physical Medicine & Rehabilitation. 2011;92:146–160. doi: 10.1016/j.apmr.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette AM. Functional Status Index: Reliability of a chronic disease evaluation instrument. Archives of Physical Medicine and Rehabilitation. 1980;61(September):395–401. [PubMed] [Google Scholar]
- Joreskog K, Sorbom D. PRELIS 2 user’s reference guide. Chicago: Scientific Software International; 1993. [Google Scholar]
- Karlsson M, Magnusson H, Cöster M, Karlsson C, Rosengren B. Patients with knee osteoarthritis have a phenotype with higher bone mass, higher fat mass, and lower lean body mass. Clinical Orthopaedics & Related Research. 2015;473(1):258–264. doi: 10.1007/s11999-014-3973-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila T, Pesonen A, Tarkkila P, Rosenberg P. Cognitive dysfunction and depression may decrease activities in daily life more strongly than pain in community-dwelling elderly adults living with persistent pain. Pain Practice. 2007;7(3):241–247. doi: 10.1111/j.1533-2500.2007.00133.x. [DOI] [PubMed] [Google Scholar]
- Khalil Z, Ralevic V, Bassirat M, Dusting G, Helme RD. Effects of ageing on sensory nerve function in rat skin. Brain Research. 1994;641:265–272. doi: 10.1016/0006-8993(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Kim H, Lee H, Rowan J, Brahim J, Dionne R. Genetic polymorphisms in monoamine neurotransmitter systems show only weak association with acute post-surgical pain in humans. Molecular Pain. 2006;2:2–24. doi: 10.1186/1744-8069-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn J, Kuruvilla F, McCarroll S, Wysoker A, Nemesh J, Cawley S, … Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nature Genetics. 2008;40(10):1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Ranjan P, Mittal B, Ghoshal U. Serotonin transporter gene (SLC6A4) polymorphism in patients with irritable bowel syndrome and healthy controls. Journal of Gastrointestinal and Liver Disease. 2012;21(1):31–38. [PubMed] [Google Scholar]
- Latremoliere A, Woolf C. Central sensitization: A generator of pain hypersensitivity by central neural plasticity. Pain. 2009;10:895–926. doi: 10.1016/j.jpain.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limer K, Nicholl B, Thomson W, McBeth J. Exploring the genetic susceptibility of chronic widespread pain: The tender points in genetic association studies. Rheumatology. 2008;47(5):572–577. doi: 10.1093/rheumatology/ken027. [DOI] [PubMed] [Google Scholar]
- Lin Y, Ro L, Wang H, Chen J. Up-regulation of dorsal root ganglia BDNF and trkB receptor in inflammatory pain: An in vivo and in vitro study. Journal of Neuroinflammation. 2011;30(8):126. doi: 10.1186/1742-2094-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt F, Berrebi J, Greayer E, Lonsdorf T, Schalling M, Ingvar M, Kosek E. Conditioned pain modulation is associated with common polymorphisms in the serotonin transporter gene. [Last accessed January, 2016];PLoS One. 2011 6(3):e18252. doi: 10.1371/journal.pone.0018252. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3065474/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Seifert H, Siemers E, Sundell K, Price K, Han B, Selzler K, … Mohs R. Cognitive and functional decline and their relationship in patients with mild Alzheimer’s dementia. Journal of Alzheimer’s Disease and Associated Dementias. 2015;43(3):949–955. doi: 10.3233/JAD-140792. [DOI] [PubMed] [Google Scholar]
- MacKnight C, Graham J, Rockwood K. Factors associated with inconsistent diagnosis of dementia between physicians and neuropsychologists. Journal of the American Geriatrics Society. 1999;47(11):1294–1299. doi: 10.1111/j.1532-5415.1999.tb07428.x. [DOI] [PubMed] [Google Scholar]
- Magaziner J, Fredman L, Hawkes W, Hebel J, Zimmerman S, Orwig D, … Wehren L. Changes in functional status attributable to hip fracture: A comparison of hip fracture patients to community-dwelling aged. American Journal of Epidemiology. 2003;157:1023–1031. doi: 10.1093/aje/kwg081. [DOI] [PubMed] [Google Scholar]
- Mardy S, Miura Y, Fumio Endo F, Matsuda I, Sztriha L, Frossard P, … Indo Y. Congenital insensitivity to pain with anhidrosis: Novel mutations in the TRKA (NTRK1) gene encoding a high-affinity receptor for nerve growth factor. The American Journal of Human Genetics. 1999;64(6):1570–1579. doi: 10.1086/302422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado F, Barjola P, Fernández-Sánchez M, Guerra V, Gómez-Esquer F. [Last accessed January, 2016];Brain function in fibromyalgia: Altered pain processing and cognitive dysfunction. 2013 Available at: http://cdn.intechopen.com/pdfs-wm/45276.pdf.
- Merighi A, Salio C, Ghirri G, Lossi L, Ferrini F, Betelli C, … Bardoni R. BDNF as a pain modulator. Prognostic Neurobiology. 2008;85:297–317. doi: 10.1016/j.pneurobio.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Molton I, Terrill A. Overivew of persistent pain in older adults. American Psychologist. 2014;69(2):197–207. doi: 10.1037/a0035794. [DOI] [PubMed] [Google Scholar]
- Morrison R, Sean F, Fischberg D, Cintron A, Siu AL. A novel interdisciplinary analgesic program reduces pain and improves function in older adults after orthopedic surgery. Journal of the American Geriatrics Society. 2009;57(1):1–10. doi: 10.1111/j.1532-5415.2008.02063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich L. Optimism about oxytocin. [Last accessed January, 2016];Analysis: Targets and Mechanisms - Neurology. 2011 4(37) Available at: http://www.nature.com/scibx/journal/v4/n37/full/scibx.2011.1031.html. [Google Scholar]
- Renn C, Leitch C, Dorsey S. In vivo evidence that truncated trkB.T1 participates in nociception. [Last accessed October, 2015];Molecular Pain. 2009 5(61) doi: 10.1186/1744-8069-5-61. Available at: http://www.molecularpain.com/content/pdf/1744-8069-5-61.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn C, Lin L, Thomas S, Dorsey S. Full-length tropomyosin-related kinase B expression in the brainstem in response to persistent inflammatory pain. Neuroreport. 2006;17:1175–1179. doi: 10.1097/01.wnr.0000215771.61355.e1. [DOI] [PubMed] [Google Scholar]
- Russo R, D’Agostino G, Mattace Raso G, Avagliano C, Cristiano C, Meli R, Calignano A. Central administration of oxytocin reduces hyperalgesia in mice: implication for cannabinoid and opioid systems. Peptides. 2012;38(1):81–88. doi: 10.1016/j.peptides.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Smith S, Reenilä I, Männistö P, Slade G, Maixner W, Diatchenko L, Nackley A. Epistasis between polymorphisms in COMT, ESR1, and GCH1 influences COMT enzyme activity and pain. Pain. 2014;155(11):2390–2399. doi: 10.1016/j.pain.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs B, Binnekade T, Soundy A, Schofield P, Huijen I, Eggermonth L. Are older adults with chronic musculoskeletal pain less active than older adults without pain? A systematic review and meta-analysis. Pain Medicine. 2013;14:1316–1331. doi: 10.1111/pme.12154. [DOI] [PubMed] [Google Scholar]
- Tender G, Kaye A, Li Y, Cui J. Neurotrophin-3 and tyrosine kinase C have modulatory effects on neuropathic pain in the rat dorsal root ganglia. Neurosurgery. 2011;68(4):1048–1055. doi: 10.1227/NEU.0b013e318208f9c4. [DOI] [PubMed] [Google Scholar]
- Verdú E, Ceballos D, Vilches J, Navarro X. Influence of aging on peripheral nerve function and regeneration. Journal of the Peripheral Nervous System. 2000;5:191–208. doi: 10.1046/j.1529-8027.2000.00026.x. [DOI] [PubMed] [Google Scholar]
- Wu J, Renn C, Faden A, Dorsey S. TrkB.T1 contributes to neuropathic pain following spinal cord injury through regulation of cell cycle pathways. Journal of Neuroscience. 2013;33:12447–12463. doi: 10.1523/JNEUROSCI.0846-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagci N, Duymaz T, Cavlak U. How does pain localization affect physical functioning, emotional status and independency in older adults with chronic musculoskeletal pain? Journal of Physical Therapy Science. 2014;26(8):1189–1192. doi: 10.1589/jpts.26.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]