Abstract
Background
We investigated the molecular mechanism of ß-lactam resistance in extended-spectrum ß-lactamase (ESBL)-producing Enterobacterial strains isolated in neonatal units of different hospitals in Anatnanarivo, Madagascar.
Methods
Bacteria were identified by standard biochemical methods, disc diffusion antibiograms and Etest. Resistance genes were sought by PCR. Strains were characterized by Rep-PCR (Diversilab), plasmid analysis and rep-typing.
Results
From April 2012 to March 2013, 29 ESBL-producing E. cloacae and 15 K. pneumoniae were isolated from blood culture (n = 32) or gastric samples (n = 12) performed at day 0 or 2 from 39/303 newborns suspected of early neonatal infection. These infants were treated with expanded spectrum cephalosporins, due to lack of carbapenems, leading to a high mortality rate (45 %). Isolates recovered were all, but 4, multidrug resistant, particularly to fluoroquinolones (FQ) except for 21 E. cloacae isolates. Isolates produced TEM-1 and CTX-M-15 ß-lactamases and their genes were located on several self-transferable plasmids of variable sizes sizes that could not be linked to a major plasmid incompatibility group. E. cloacae isolates belonged to 6 Rep-types among which two counted for 11 isolates each. The FQ resistant E. cloacae isolates belonged to one clone, whereas the FQ susceptible E. cloacae isolates belonged to four clones. The K. pneumoniae isolates belonged to 9 Rep-types among which one included five isolates.
Conclusion
This study is the first molecular characterization of ESBL-producing isolates from neonatology units in Madagascar, a country with limited epidemiological data. It revealed an important multi-clonal dissemination of CTX-M-15-producing isolates reflecting both the high community carriage and the very early nosocomial contamination of the neonates.
Keywords: ESBL, Outbreak, Madagascar, Neonatology
Background
Extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae represent a major worldwide threat among drug-resistant bacteria in both hospital and community settings [1]. They are mostly associated with urinary tract infections, but may also cause significant bloodstream-associated infections [1]. ESBLs are often located on large plasmids that also harbor resistant genes to other antimicrobial classes resulting in multidrug-resistant isolates [1, 2]. Plasmid-encoded ESBLs of the CTX-M-type are reported increasingly worldwide in Gram-negative rods and account now for most of the ESBLs found in Enterobacteriaceae [1, 3–5]. CTX-Ms form a rapidly growing family that comprises currently up to 154 variants that are divided into five groups according to amino-acid sequence identity (CTX-M-1, −2, −8, −9 and −25 groups) with CTX-M-15 being the most prevalent in most countries [3–5]. BlaCTX-M-15 genes are often encoded on plasmids belonging to the incompatibility group IncF [2–4]. In the upstream region of CTX-M genes an insertion sequence element, ISEcp1, is commonly present and is likely responsible for the transposition process of the genes [6].
Over recent years, the importance of community-acquired infections due to ESBL-producing isolates has been increasingly demonstrated [3–5]. As a consequence, fecal carriage of ESBL-producing isolates is now widely studied in hospitals but also in healthy populations in the community [5]. Surveys since 2000 have shown an alarming trend of associated resistance to other classes of antimicrobial agents among ESBL-producing organisms isolated from community sites [3, 4, 7].
ESBL-producing Enterobacteriae were first isolated in Madagascar between 2005 and 2006 from community-acquired urinary tract infections in 9.7 % of isolated Enterobacteriaceae [8]. More recently, 21.3 % of clinical isolates from patients in surgery and intensive care units and 21.2 % of intestinal carriage isolates from children hospitalized in a pediatric department of a large teaching hospital were ESBL-producers [9, 10]. The prevalence of carriage of ESBL in the community of Antananarivo was estimated at 10 % in healthy people in 2011 [11]. The most frequenly involved bacteria being Escherichia coli, Klebsiella pneumoniae and Enterobacter cloacae. More recently, Rasamiravaka et al. reported in 2015, a rate 12 % of ESBL- producing Enterobacteriacae in urine samples [12]. The same year, Chereau et al., studying the fecal carriage in pregnant women, reported a rate of 18.5 % of ESBL-producing Enterobacteriacae [13].
Infections are a major contributor to newborn deaths in developing countries and are responsible for an estimated 35 % of all neonatal deaths [14]. In resource-poor countries, sepsis due to resistant Gram-negative bacilli is an emerging problem and the currently recommended first-line (penicillin/ampicillin plus gentamicin) or second-line antibiotics (a third-generation cephalosporin) do not provide adequate cover. Laboratory tests are often not available and the diagnosis of neonatal infections is based only on clinical signs leading to an antimicrobial treatment often not adapted to the local epidemiology. For this reason, we decided to conduct a study in Madagascar to identify the epidemiology of bacterial early neonatal infections. This study allowed us to show a predominance of Enterobacteriaceae and especially of ESBL-producing E.cloacae and K. pneumoniae. None of the Escherichia coli strains isolated here were ESBL-producers. Therefore, the present work focused on the molecular mechanism of ß-lactam resistance in E. cloacae and K. pneumoniae isolates identified in these neonatal units of two different hospital at Antananarivo, Madagascar.
Methods
Study design
During April 2012 and March 2013, newborns from the two hospitals of Antananarivo with a suspected neonatal infection determined based on a clinical score according to the International and French consensus guidelines (fever (>37°8C) or hypothermia (<35 °C), tachycardia or bradycardia, arterial hypotension, poor perfusion, respiratory distress, apnea, seizure, floppy infant, bulging fontanel, irritability, lethargy, purpura were included [15, 16]. The French guidelines for materno-fetal infections were used to categorize the clinical situations, as proved infection when the blood culture was positive, probable infection when the gastric fluid sample was positive with a pathogen bacteria and a positive C-reactive protein (CRP) [15].
Bacterial strains, antimicrobial agents and susceptibility testing
Bacterial identification was performed using the API 20E system (bioMérieux, Marcy-l’Etoile, France). Antibiograms for 32 antibiotics (amoxicillin, amoxicillin/clavulanic acid, aztreonam, ceftazidime, cefalotine, cefmandole, cefotaxime, cefepime, cefoxitine, imipenem, meropenem, Moxalactam, pipéracilline, ticarcilline, ticarcilline/acide clavulanique, piperacilline/tazobactam, fosfomycine, colistine, rifampicine, cotrimxazole, ciprofloxacine, pefloxacine; norfloxacine, nalidixic acid, tetracycline, tigecycline, chloramphenicol, kanamycine, amikacine, netilmicine, tobramycine, gentamicine) were determined by the disc diffusion method on Mueller-Hinton agar (BioRad, Marnes-La-Coquette, France) and the susceptibility breakpoints were determined and interpreted as recommended by the Clinical and Laboratory Standards Institute [17]. All plates were incubated at 37 °C for 18 h. Minimum inhibitory concentrations (MICs) of given ß-lactams were determined using the Etest technique (bioMérieux, Marcy l’Etoile, France) for the following β-lactam antibiotics: amoxicillin ± clavulanic acid, cefoxitin, cefotaxime, ceftazidime, aztreonam, cefepime and imipenem. Confirmation of ESBL producers was performed by double disc synergy testing between ticarcillin/clavulanate and aztreonam and/or ceftazidime and/or cefepime [18].
Nucleic acid extractions, PCR and DNA sequencing
Whole-cell DNAs were extracted using QIAamp DNA Mini Kit (Qiagen, Les Ulis, France). The blaCTX-M, blaSHV, blaTEM, minor ESBL and quinolone resistant qnrA, B and S genes were searched for by PCR and subsequently characterized by Sanger sequencing. PCR experiments were performed on an ABI 2700 thermocycler (Applied Biosystems, Les Ulis, France) using laboratory-designed primers [19]. Both strands of the PCR products, were sequenced using laboratory-designed primers with an automated sequencer (ABI PRISM 3100; Applied Biosystems). The nucleotide and the deduced protein sequences were analyzed using software available at the National Center of Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Plasmid content, mating out and electroporation experiments
Direct transfer of resistance into azide-resistant E. coli J53 was attempted by liquid and solid mating-out assays at 30 and 37 °C [19]. Transconjugant selection was performed on trypticase soy agar plates (bioMérieux) containing ciprofloxacin (3 mg/ml) and either ceftazidime (10 mg/ml) or ticarcillin (150 mg/ml).
Plasmids were introduced by electroporation into E. coli TOP10 using a Gene Pulser II (BioRad). Natural plasmids were extracted using Kieser extraction method and subsequently analyzed by electrophoresis on a 0.7 % agarose gel [20].
Replicon typing
PCR-based replicon typing (PBRT) of the main plasmid incompatibility groups reported in Enterobacteriaceae was performed as described [21] using the respective PBRT controls. The obtained amplicons were sequenced to confirm their identity. Genetic structures surrounding the blaCTX-M gene were determined by PCR using primers specific of the known genetic environment of group 1 CTX-M variants [19].
Fingerprinting analysis
Genomic relatedness of the K. pneumoniae and E. cloacae isolates was investigated by semi-automated rep-PCR typing (Diversilab®, bioMérieux) as recommended by the manufacturer. Isolates were considered belonging to a same clone if they shared at least 95 % similarity.
Results
Patients
Between April 1st, 2012 and March 31st 2013, 8500 newborn (NB) were born in the two hospitals (Befeletanana and Soavinandriana). Among them, 303 NB (3.6 %) had clinical signs of maternal infections and were included in the study, 164 were from Befelatanana (B) and 139 from Soavinandriana (S). The mean gestational age of the NB was 38 ± 3.5 weeks gestationnal age (WA) comprised between 24 and 43 (Table 1). Out of the 303 NB, 60 (19.5 %) were born before 38 weeks, among which 20 were born before 32 weeks and 40 between 32 and 36 weeks. The sex ratio of the NB was 172 (56.8 %) males and 131 (43.2 %) females. Gastric fluid was sampled from 282 NB, blood culture from 254 NB, and CRP determination from 272 NB. Pregnancy monitoring results are indicated in Table 1.
Table 1.
ᅟ
| Total n (%) | Befelatanana n (%) | Soavinandriana n (%) | p-value | |
|---|---|---|---|---|
| Neonates | ||||
| Number | 303 | 164 | 139 | |
| Sex Male Female |
||||
| 172 (56.8) | 90 (54.9) | 82 (59.0) | ||
| 131 (43.2) | 74 (45.1) | 57 (41.0) | 0.54 | |
| Sex-ratio | 1.3 | 1.2 | 1.4 | 0.54 |
| Gestational age (weeks) [mean] | 38 [24–43] | 38 [24–43] | 38 [26–42] | 0.25 |
| Birth weight (g) [mean, 95%CI] |
2663 [2576–2750] |
2559 [2441–2677] |
2785 [2658–2912] |
0.01 |
| Antimicrobial treatment | 154 (50.8) | 120 (73.2) | 34 (24.5) | <0.01 |
| Death | 47 (15.5) | 41 (25) | 6 (4.3) | <0.01 |
| Gastric samples Positive Gastric samples True Positives (contamination excluded) Bacteria Escherichia coli ESBLa-producer Enterobacter cloacae ESBL-producer Klebsiella pneumoniae ESBL-producer Group B Streptococcus Other organismsb |
||||
| 282 (93.1) | 143 (87.2) | 139 (100) | ||
| 60 (21.3) | 34 (23.8) | 26 (18.7) | <0.01 | |
| 72 | 39 | 33 | ||
| 32 (44.4) 0 |
15 (38.5) 0 |
17 (51.5) 0 |
0.68 | |
| 11 (15.3) 7 |
7 (17.9) 5 |
4 (12.1) 2 |
||
| 6 (8.3) 5 |
4 (10.2) 3 |
2 (6.1) 2 |
||
| 4 (5.5) | 2 (5.1) | 2 (6) | ||
| 19 (26.3) | 11 (28.2) | 8 (24.2) | ||
| Blood cultures Positive blood cultures |
||||
| 254 (83.8) | 145 (88.4) | 109 (78.4) | ||
| True Positives (contamination excluded) Bacteria |
76 (29.9) | 62 (42.8) | 14 (12.8) | <0.01 |
| 99 | 79 | 20 | ||
|
Escherichia coli
ESBL-producer Enterobacter cloacae ESBL-producer Klebsiella pneumonia ESBL-producer Acinetobacter baumannii Group B Streptococcus Enterococcus sp Other organismsc |
7 (7.1) 0 |
4 (5.14) 0 |
3 (15) 0 |
|
| 28 (28.3) 28 |
27 (34.2) 27 |
1 (5) 1 |
<0.01 | |
| 14 (14.1) 11 |
13 (16.4) 10 |
1 (5) 1 |
||
| 8 (8.1) | 6 (7.6) | 2 (10) | ||
| 9 (9) | 5 (6.3) | 4 (20) | ||
| 14 (14.1) | 12 (15.1) | 2 (10) | ||
| 19 (19.1) | 12 (19.2) | 7 (35) | ||
| Mothers | ||||
| Antibiotic before delivery | 36 (11.9) | 5 (3) | 31 (22.3) | <0.01 |
| Pregnancy monitoring Hospital Health center General practitioner Mid-wife No follow up |
||||
| 101 (33.3) | 10 (6.1) | 91 (65.5) | <0.01 | |
| 97 (32) | 69 (42.1) | 28 (20.1) | ||
| 35 (11.6) | 25 (15.2) | 10 (7.2) | 0.33 | |
| 38 (12.5) | 33 (20.1) | 5 (3.6) | ||
| 22 (7.3) | 20 (12.2) | 2 (1.4) | <0.01 | |
| Intrapartum antibiotic | 77 (25.4) | 11 (6.7) | 66 (47.5) | <0.01 |
Bacterial epidemiology
During the study period, 168/254 (66.1 %) blood cultures, 60/282 (21.3 %) gastric fluids, and 102/272 (37.5 %) CRP (cutoff > 6 mg/L) were positive. For 58 NB the CRP was >20 mg/L. Results obtained in each of the two hospitals are summarized in Table 1.
Among the 168 positive blood cultures, 92 were positive with Staphylococcus coagulase negative or Corynebacteriae and were therefore considered as contaminated. Finally, 76 were positives with 99 bacterial isolates and therefore corresponded to a proven infection. Enterobacteriaceae (51/99, 51.5 %) among which Enterobacter cloacae 28/51 (54.9 %), Klebsiella pneumoniae 14/51 (27.5 %), Escherichia coli 7/51 (13.7 %) and Proteus mirabilis 2/51 (3.9 %) were the most predominant isolates (Table 1). Group B Streptococcus, Acinetobacter baumannii and Enterococcus sp. represented respectively 9, 8 and 14 % of the isolates.
Among these 76 positive blood culture, 13 were positive with two bacteria: Enterobacter cloacae with Klebsiella pneumoniae (five cases), E. cloacae with Enterococcus faecalis (one case), Acinetobacter baumanii was associated with E. faecalis or Streptococcus sp or Proteus mirabilis (three cases), E. faecalis was associated with E. coli (three cases), and Staphylococcus aureus (one case).
Twenty four NB were considered as probably infected since they had a positive gastric fluid culture, a negative blood culture and an elevated CRP according to the French guidelines. Bacteria involved were mostly Gram-negative bacteria (12 E. coli, 6 E. cloacae, one K. pneumoniae, and four group B Streptococcus and one S. aureus). All other NB were considered as only colonized.
In all, 47/303 (15.5 %) NB died. The mortality rate was 41/164 (25 %) at B and 6/139 (4.3 % at S). Among these, 25 died of neonatal infection (20 with positive blood culture and five with positive gastric fluid and elevated CRP). Among these 25, 24 were born at B of which eight had a positive blood culture with an ESBL-producing E. cloacae, four with an ESBL-producing K. pneumoniae and two with the both. Others had a positive blood culture with A. baumannii (n = 1), Haemophilus influenzae (n = 2), E. coli (n = 2) and Streptococcus anginosus (n = 1).
Bacterial isolates and antibiotic susceptibility
Bacterial isolates and results are summarized in Table 1. Gram negative were the most prevalent bacteria (115/171 isolated bacteria). Among these, 108 were identified as E. coli (n = 39), K. pneumoniae (n = 20), E. cloacae (n = 39), or Acinetobacter baumannii (n = 10). Looking at the antimicrobial susceptibility, 35/39 E. cloacae and 16/20 K. pneumoniae were ESBL-producers as revealed by a typical synergy image between clavulanic acid and expanded-spectrum cephalosporins and were selected for further analysis. Their distribution by hospitals is described Table 1. None of the E. coli isolates were ESBL-producers.
Antibiograms revealed that all ESBL-producing isolates were multidrug-resistant and most of them were resistant to first line antibiotics (ampicillin, cefotaxime or gentamicin) used to treat neonatal infections. In addition, high rates of resistance to gentamicin (88.9 %) tobramycin (81.5 %), ciprofloxacin (35 %) and to trimethoprim-sulfamethoxazole (87 %), were observed They were susceptible to imipenem while K. pneumoniae isolates were also susceptible to cefoxitin. Resistance to cefoxitin in all E. cloacae isolates was due to the inducible production of AmpC β-lactamase from a chromosomal gene. Among ESBL-producers, 16/17 K. pneumoniae and 10/39 E. cloacae isolated from blood or gastric samples were resistant to fluoroquinolones (FQR). The finding of multidrug resistance among ESBL-producing isolates is of great clinical relevance due to the limited therapeutic options and the high risk of treatment failure in patients infected with these strains.
Molecular epidemiology of ESBL-producers
Among the 35 ESBL- producing E. cloacae 29 were further studied, 21 were isolated from blood culture (20 from B and one from S) and eight were isolated from gastric fluid samples. Eight out of the 29 were resistant to fluoroquinolones. Rep-PCR analyses (Fig. 1) revealed that E. cloacae isolates belonged to 6 Rep-PCR-types among which two counted for 11 isolates each. Clone 1 (P1) included 11 strains (one from gastric fluid and ten from blood culture) either resistant to fluoroquinolones (n = 7) or susceptible (n = 4) and was present all along the study period (Fig. 2). Conversely, clone 6 (P6) included also 11 strains (six from blood culture and five from gastric fluis sample), all resitant to fluoroquinolones and was present for a shorter period of 6 months. One NB exhibited the same strain in blood and gastric fluid and died.
Fig. 1.
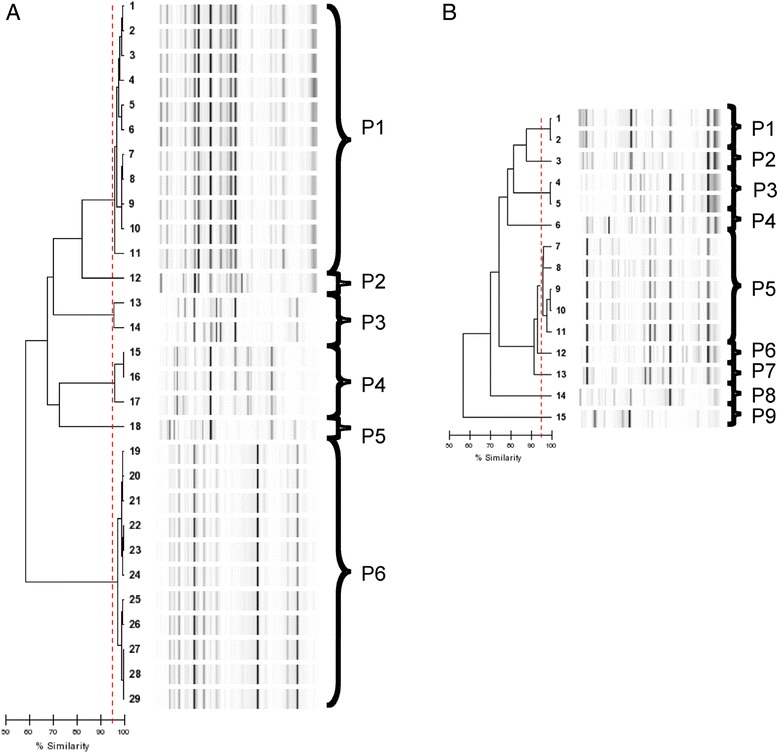
Rep-PCR results showing genetic relatedness between the 29 ESBL-producing E. cloacae (a) and the 15 ESBL-producing K. pneumoniae (b) isolates
Fig. 2.
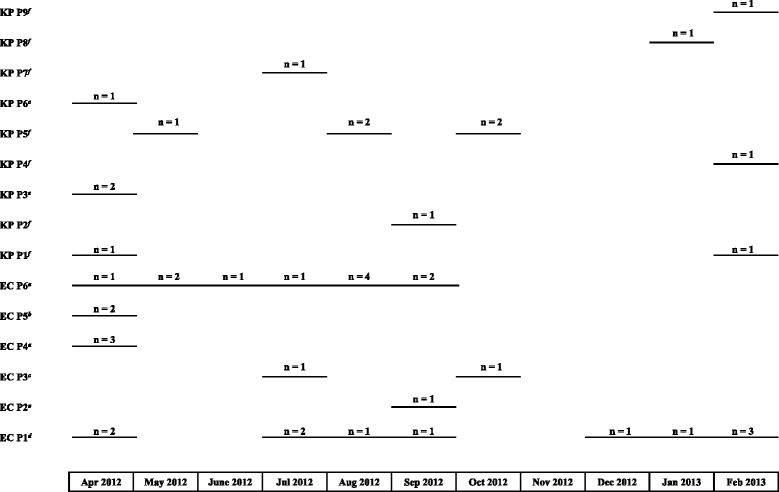
Isolation of the different clones of ESBL-producing E. cloacae (n = 6) and K. pneumoniae (n = 9) over the study period. a all strains were isolated at B and were susceptible to fluroquinolones; b all strains were isolated at S, one susceptible and one resistant to fluroquinolones respectively; c all strains were isolated at B, one susceptible and one resistant to fluroquinolones respectively; d all strains were isolated at B, five susceptible and six resistant to fluroquinolones respectively; e all strains were isolated at S and resistant to fluroquinolones; f all strains were isolated at B and resistant to fluroquinolones
The seven remaining strains were divided into four different clones. Clones 2 (P2), isolated at B was represented each by only one strain from a gastric fluid. Clone 5 (P5) was isolated only at S in gastric fluid and blood from two different NB. In April 2012, three clones were present at the same time at B (Fig. 2).
Among the 16 ESBL-producing K. pneumonia, 15 were studied, 11 were isolated from blood culture (ten from B and one from S) and four were isolated from gastric fluid samples. K. pneumoniae isolates were of more diverse origin (Fig. 1): they belonged to 9 Rep-PCR-types among 1 (P5) included five isolates sharing more than 95 % similarity. Two other clones (P1 and P3) were represented each by two isolates and the six remaining clones included only one strain (Fig. 1). Clone 1 (P1) included two strains and was present at B hospital at the beginning and at the end of the study. Clone 3 (P3) (including two strains, one from blood culture and one from fluid gastric sample from the same NB) was present only in S hospital. The clone 5 (P5) was present at Befelatanana hospital in May to October. Other isolates were isolated from gastric samples, belonged to three or four different profiles and were all isolated at B.
Interestingly, four blood cultures were positive with both K. pneumoniae and E. cloacae isolates. In two cases, the Rep-PCR profiles were different for both bacteria. In two other cases, the blood cultures isolated from two different NB born two days apart exhibited the same profile for E. cloacae (P1) but different profiles for K. pneumoniae (P4 and P9).
Molecular analysis
ESBL producing isolates harboured blaCTX-M and blaTEM genes as revealed by PCR. Sequencing of the PCR products revealed that blaCTX-M-15 and blaTEM-1 genes were present in all isolates. PCR-sequencing of the genetic environment of blaCTX-M gene revealed the presence of ISEcp1 insertion sequence located upstream of all blaCTX-M-15 gene. The ESBL-producing isolates contained several plasmids of different sizes, ranging from less than 5 kb to more than 150 kb (Fig. 3). Both resistance genes were carried on several self-transferable plasmids of variable sizes. For E. cloacae isolates, clone P1 exhibited only one plasmid whereas clone P6 exhibited at least five plasmids of different size. Transformants into E. coli DH10B could be obtained with most of the isolates. The E. coli transformants had a ß-lactam resistance pattern that correspond to the expression of an ESBL-like gene. Transformants obtained with E. cloacae P1 and K. pneumoniae P5 were also resistant to gentamicin, tobramycin, tetracycline, cotrimoxazole and chloramphenicol. Transformants obtained with E. cloacae P6 harbored the same profile but were susceptible to tetracycline and cotrimoxazole. The blaCTX-M-15 gene carrying plasmid could also be transferred by conjugation to E. coli J53 for all the isolates. PCR-based replicon typing of the main plasmid incompatibility groups reported in Enterobacteriaceae was not able to identify the inc group of the plasmids carrying blaCTX-M-15 gene. The quinolones resistant isolates were tested negative for qnrA, B and S.
Fig. 3.
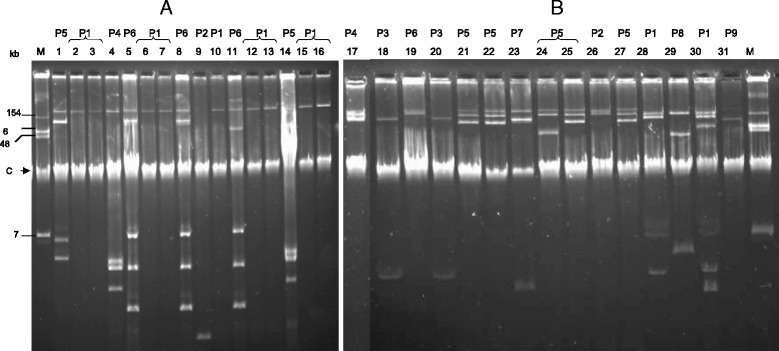
Plasmid analysis by Kieser method and corresponding clones obtained by Rep-PCR (P): (a) 15 ESBL-producing E. cloacae, 5/6 clones are represented with the two main ones P1 and P6; (b) 15 ESBL-producing K. pneumoniae isolates, 9/9 clones are represented. M: E. coli 50192, reference strain harboring four plasmids (molecular size plasmid marker). “C” indicated the chromosomal DNA band
Discussion
Between April 2012 to March 2013, 35 E. cloacae and 16 K. pneumoniae isolates producing an ESBL were isolated from blood cultures (n = 39) or gastric fluid samples (n = 12) performed at day 0 or 1 from 39/165 newborns suspected of early-onset neonatal infection. These NB were treated with ceftriaxone, due to lack of carbapenems, and resulted in a high mortality rate. Molecular studies could be performed on 29 ESBL-producing E. cloacae and 15 ESBL-producing K. pneumoniae. All strains exhibited blaCTX-M-15 ESBL gene. The CTXM-15 ESBL is considered to be the most prevalent ESBL worldwide [1, 3, 4]. Our findings confirm the predominance of blaCTX-M-15 among ESBL producing isolates. Previously in 2009, the blaCTX-M-15 gene has already been reported to be the most prevalent ESBL in Madagascar, as it was detected in 75.5 % of Enterobacteriaceae isolated from feces [11]. These results were confirmed in 2015 showing an increase in the prevalence of colonization by ESBL-producing Enterobacteriaceae (18 %, among which 68 % carried the blaCTX-M-15 gene), consistent with the worldwide increase of ESBL-producing Enterobacteriaceae carriage in the community [13]. This predominance is is highlighted by a study involving nine Asian countries reported that blaCTX-M-15 gene was highly prevalent among ESBL-producing K. pneumoniae isolates (60 %, 55/92) [22].
CTX-M genes may disseminate through clonal expansion or horizontal gene transfer [3, 4]. In our study, ISEcp1 was found upstream from blaCTX-M-15 at variable distances, as was previously described [7, 23]. ISEcp1 was found to be in the vicinity of many blaCTX-M genes (including blaCTX-M-15) and was reported to be responsible of the expression of downstream located genes [6]. The plasmids carrying blaCTX-M-15 were assigned to the IncFII, IncFIA or IncHI2 incompatibility group replicons [2]. Association of the blaCTX-M-15 gene with IncF plasmids carrying the FII replicon in association with the FIA or FIB replicon has been reported previously for isolates in Canada, France, Spain, Tunisia, and the United Kingdom [2–4, 24]. The IncHI2 plasmid, frequently associated with blaCTX-M-2 or blaCTX-M-9, was first identified in Serratia marcescens [2], but rarely reported in association with blaCTX-M-15. In our study blaCTX-M-15 gene could not be linked to a major plasmid incompatibility group by PCR.
Our study presents some limitations, especially the lack of vaginal samples of the mother prior birth, which would have allowed to distinguish nosocomial and a maternal transmission. The hypothesis of nosocomial infection is more likely with regard E. cloacae as two Rep-PCR-types represented each 11 identical isolates from the same hospital, one isolated throughout the study (EC-P1) and another only the first 6 months (EC-P6). The clone EC-P4, including three isolates, was present only in April. Conversely, the K. pneumoniae are of much more diverse origin since they belonged to 9 Rep-PCR-types among two included only two isolates and the six other clones only one strain. In two cases, the E. cloacae strains isolated both from the gastric sample and blood sample belonged to the same clone (EC-P5 at S hospital and EC-P6 at B hospital).
A community origin and therefore a mother-to-child transmission can then be discussed in this case. This hypothesis was evocated by Cherau et al. who investigated the ESBL-PE rectal colonization among pregnant women at delivery in the community in Madagascar and estimated a prevalence ranging from 14.5 to 22.6 % according to the place [13]. Conversely, the nosocomial hypothesis is supported by the data of a previous study, reporting a clonal outbreak of K. pneumoniae harboring blaCTX-M-15 and blaSHV-2 genes described in the neonatal units of two hospitals in Antananarivo and highlighting the role of contaminated aspiration tubes [25]. The precocity of neonatal infections due to E. cloacae and K. pneumoniae occurring between D0 and D1 in this study study does not allow to discriminate between the two hypothesis. It is likely that the two causes of infection have coexisted: the nosocomial hypotheses is more plausible for E. clocae (two major clones) and community hypothesis for K. pneumoniae. Interestingly, no relationship between the two hospitals has been demonstrated. This is confirmed by the fact that the strains of E. cloacae were isolated mainly to the hospital B, except two belonging to a different clone. Interestingly, B hospital supports more deliveries per year (about 7000 vs 1500) with a population of a lower socioeconomic level.
Our study further underscores that in developing countries, neonatal infections are mainly due to Gram-negative bacteria and especially K. pneumoniae (respectively 57.4 and 26.4 %) as reported by Zaidi et al. [26]. We found also the same percentage of A. baumannii (8 %), Group B Streptococcus (8 %) and a lower percentage of E. coli (7 %) similarly to previous studies [26, 27].
Neonates are exposed to external risks factors, particularly deficient hygiene that put them at high risk of neonatal infection and if neonatal culture confirmed sepsis rates is of 1–3 per 1000 live births reported from industrialized countries [28], this rate can reached 37 per 1000 live births in developing countries [29, 30]. Poor quality of care in developing countries are a major source of neonatal infections for hospital-born infants. Lack of infection-control procedures, inadequate sterilization of multiuse instruments, understaffing and overcrowded nurseries are responsible for nosocomial infections in most hospitals in developing countries and promotes neonatal infections due to environmental pathogens as reflected in this study by the positivity of the gastric samples cultures with E. cloacae and K. pneumoniae [31]. A supplementary burden being the high antimicrobial drug resistance rates due to a combination of several factors, including irrational antimicrobial drug use [5]. In addition, imipenem was not marketed in Madagascar at the time of the study explaining, in part, this elevated mortality.
Conclusion
This study confirmed the global emergence of blaCTX-M-15 genes from a country with limited epidemiological data. In addition, this is the first molecular characterization of ESBL-producing isolates from Neonatology ward in Madagascar. We confirmed a high prevalence and the multi-clonal dissemination of blaCTX-M-15 genes among these isolates thus reflecting both high community carriage and a very early nosocomial contamination of the neonates. These findings underline the need for a rational use of antibiotic and for preventive hygiene strategies, which can reduce the burden of neonatal infections in these countries.
Abbreviations
CRP, c reactive protein; CTX, Cefotaxime; DNA, desoxyribonucleic acid; DO and D1, day zero and day one; ESBL, extended spectrum beta-lactamase; FQ, fluoroquinolones; MIC, minimum inhibitory concentration; NB, newborn; PCR, polymerase chain reaction; Rep-PCR, repetitive extragenic palindromic sequence polymerase chain reaction; WA, weeks gestationam age
Acknowledgments
With acknowledge “Jeremi Rhônes- Alpes” for its financial support
Funding
This work was partially funded by a grant from the INSERM (U914) and the Université Paris Sud, France. TN and GC are members of the Laboratory of Excellence LERMIT supported by a grant from ANR (ANR-10-LABX-33).
Availability of data and materials
Raw data of the Rep-PCR results that served for the phylogenetic tree contraction are available upon request to the corresponding author.
Authors’ contributions
JR and PI conceived of the study and participated to its design and coordination. JR drafted the manuscript. ALR, ZA, ER, ZNR were responsible for coordination and acquisition of data. ER carried out the bacteriological cultures. TN and GC carried out the molecular genetic studies and drafted the manuscript. PN was involved in the manuscript revision. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not Applicable.
Ethics approval and consent to participate
The study protocols were approved by the National Ethics Committee of Madagascar. Written informed consents were obtained from at least one parent of each child before enrolment.
Contributor Information
T. Naas, Email: thierry.naas@bct.aphp.fr
J. Raymond, Phone: 33-1-58-41-15-25, Email: josette.raymond@cch.aphp.fr
References
- 1.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peirano G, Pitout JD. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents. 2011;35:316–21. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 5.Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev. 2013;26:744–58. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Naas T, Nordmann P. Genetic support of extended-spectrum beta-lactamases. Clin Microbiol Infect. 2008;14(Suppl 1):75–81. doi: 10.1111/j.1469-0691.2007.01865.x. [DOI] [PubMed] [Google Scholar]
- 7.Vardakas KZ, Tansarli GS, Rafailidis PI, Falagas ME. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum β-lactamases: a systematic review and meta-analysis. J Antimicrob Chemother. 2012;67(12):2793–803. doi: 10.1093/jac/dks301. [DOI] [PubMed] [Google Scholar]
- 8.Randrianirina F, Soares JL, Carod JF, Ratsima E, Thonnier V, Combe P, et al. Antimicrobial resistance among uropathogens that cause community-acquired urinary tract infections in Antananarivo, Madagascar. J Antimicrob Chemother. 2007;59:309–12. doi: 10.1093/jac/dkl466. [DOI] [PubMed] [Google Scholar]
- 9.Randrianirina F, Vaillant L, Ramarokoto CE, Rakotoarijaona A, Andriamanarivo ML, Razafimahandry HC, et al. Antimicrobial resistance in pathogens causing nosocomial infections in surgery and intensive care units of two hospitals in Antananarivo, Madagascar. J Infect Dev Ctries. 2010;4:74–82. doi: 10.3855/jidc.454. [DOI] [PubMed] [Google Scholar]
- 10.Andriatahina T, Randrianirina F, Hariniana ER, Talarmin A, Raobijaona H, Buisson Y, et al. High prevalence of fecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a pediatric unit in Madagascar. BMC Infect Dis. 2010;10:204. doi: 10.1186/1471-2334-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herindrainy P, Randrianirina F, Ratovoson R, Ratsima Hariniana E, Buisson Y, Genel L, et al. Rectal carriage of extended-spectrum Beta-lactamase-producing gram-negative bacilli in community settings in madagascar. PLoS ONE. 2011;6:e22738. doi: 10.1371/journal.pone.0022738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasamiravaka T, Shaista Sheila HS, Rakotomavojaona T, Rakoto-Alson AO, Rasamindrakotroka A. Changing profile and increasing antimicrobial resistance of uropathogenic bacteria inMadagascar. Med Mal Infect. 2015;45(5):173–6. doi: 10.1016/j.medmal.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Chereau F, Herindrainy P, Garin B, Huynh BT, Randrianirina F, Padget M, et al. Colonization of extended-spectrum-β-lactamase- and NDM-1-producing Enterobacteriaceae among pregnant women in the community in a low-income country: a potential reservoir for transmission of multiresistant Enterobacteriaceae to neonates. Antimicrob Agents Chemother. 2015;59(6):3652–5. doi: 10.1128/AAC.00029-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawn JE, Kerber K, Enweronu-Laryea C, Cousens S. 3.6 million neonatal deaths-what is progressing and what is not? Semin Perinatol. 2010;34:371–86. doi: 10.1053/j.semperi.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Diagnostic et traitement curatif de l’infection bactérienne précoce du nouveau-né. ANAES 2002; Haute Autorité de santé, France. http://www.has-sante.fr/portail/upload/docs/application/pdf/recos-_inn-_mel_2006.pdf. Accessed 4 Jun 2016.
- 16.Haque KN. Definitions of bloodstream infection in the newborn. Pediatr Crit Care Med. 2005;6(3 Suppl):S45–9. doi: 10.1097/01.PCC.0000161946.73305.0A. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: CLSI document M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA; 2015.
- 18.Naas T, Coignard B, Carbonne A, Blanckaert K, Bajolet O, Bernet C, et al. VEB-1 Extended-spectrum beta-lactamase-producing Acinetobacter baumannii, France. Emerg Infect Dis. 2006;12:1214–22. doi: 10.3201/eid1708.051547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djamdjian L, Naas T, Tandé D, Cuzon G, Hanrotel-Saliou C, Nordmann P. CTX-M-93, a CTX-M variant lacking penicillin hydrolytic activity. Antimicrob Agents Chemother. 2011;55:1861–6. doi: 10.1128/AAC.01656-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieser T. Factors affecting the isolation of cccDNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. doi: 10.1016/0147-619X(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 21.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 22.Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. High prevalence of CTX-M-15- producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents. 2011;38:160–3. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 23.Rakotonirina HC, Garin B, Randrianirina F, Richard V, Talarmin A, Arlet G. Molecular characterization of multidrug-resistant extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiol. 2013;13(1):85. doi: 10.1186/1471-2180-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novais A, Canton R, Moreira R, Peixe L, Baquero F, Coque TM. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, −3, and −32) plasmids. Antimicrob Agents Chemother. 2007;51:796–9. doi: 10.1128/AAC.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randrianirina F, Vedy S, Rakotovao D, Ramarokoto CE, Ratsitohaina H, Carod JF, et al. Role of contaminated aspiration tubes in nosocomial outbreak of Klebsiella pneumoniae producing SHV-2 and CTX-M-15 extended-spectrum beta-lactamases. J Hosp Infect. 2009;72:23–9. doi: 10.1016/j.jhin.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Zaidi AK, Huskins WC, Thaver D, Bhutta ZA, Abbas Z, Goldmann DA. Hospital-acquired neonatal infections in developing countries. Lancet. 2005;365:1175–88. doi: 10.1016/S0140-6736(05)71881-X. [DOI] [PubMed] [Google Scholar]
- 27.Waters D, Jawad I, Ahmad A, Lukšić I, Nair H, Zgaga L, et al. Aetiology of community- acquired neonatal sepsis in low and middle income countries. J Glob Health. 2011;1:154–70. [PMC free article] [PubMed] [Google Scholar]
- 28.Morken NH, Kallen K, Jacobsson B. Outcomes of preterm children according to type of delivery onset: a nationwide population-based study. Paediatr Perinat Epidemiol. 2007;21:458–64. doi: 10.1111/j.1365-3016.2007.00823.x. [DOI] [PubMed] [Google Scholar]
- 29.Thaver D, Zaidi AK. Burden of neonatal infections in developing countries: a review of evidence from community-based studies. Pediatr Infect Dis J. 2009;28:S3–9. doi: 10.1097/INF.0b013e3181958755. [DOI] [PubMed] [Google Scholar]
- 30.Seale AC, Blencowe H, Zaidi A, Ganatra H, Syed S, Engmann C, et al. Neonatal Infections Estimation Team. Neonatal severe bacterial infection impairment estimates in South Asia, sub-Saharan Africa, and Latin America for 2010. Pediatr Res. 2013;74:73–85. doi: 10.1038/pr.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stapleton PJ, Murphy M, McCallion N, Brennan M, Cunney R. Drew RJ Outbreaks of extended spectrum beta-lactamase-producing Enterobacteriaceae in neonatal intensive care units: a systematic review. Arch Dis Child Fetal Neonatal. 2015;101:72–8. doi: 10.1136/archdischild-2015-308707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data of the Rep-PCR results that served for the phylogenetic tree contraction are available upon request to the corresponding author.


