Abstract
More than 70 years ago, the filamentous ascomycete Trichoderma reesei was isolated on the Solomon Islands due to its ability to degrade and thrive on cellulose containing fabrics. This trait that relies on its secreted cellulases is nowadays exploited by several industries. Most prominently in biorefineries which use T. reesei enzymes to saccharify lignocellulose from renewable plant biomass in order to produce biobased fuels and chemicals. In this review we summarize important milestones of the development of T. reesei as the leading production host for biorefinery enzymes, and discuss emerging trends in strain engineering. Trichoderma reesei has very recently also been proposed as a consolidated bioprocessing organism capable of direct conversion of biopolymeric substrates to desired products. We therefore cover this topic by reviewing novel approaches in metabolic engineering of T. reesei.
Keywords: Trichoderma reesei, Cellulase, Recombinant protein production, Biorefinery, Lignocellulose, Gene expression, Consolidated bioprocessing
Background
A brief history of T. reesei
The most ancient biotechnological practices involving fungi for the production of beer, wine and cheese might date back several millennia, i.e. to the very beginning of literate civilization itself [1–3]. In contrast, the discovery of the filamentous mesophilic ascomycete Trichoderma reesei (then Trichoderma viride) for its astonishing potential to produce extracellular cellulases took place just over 70 years ago. Initially the destructive potential of the original Trichoderma sp. isolated from rotting US Army equipment on the Solomon Islands during World War II was seen rather problematic. Nevertheless, it was not long before researchers at the Natick Army Research Laboratories led by Mary Mandels and Elwyn T. Reese, the name giving researcher, sought to turn this problematic potential into purposive products [4]. In a screening of 14,000 moulds of the Quartermaster Collection Trichoderma sp. QM6a showed an outstanding ability to degrade native crystalline cellulose. The designation “QM6a” for the last remaining original Trichoderma isolate, which identified it as the sixth of six cultures of the fungus stored at the Quartermaster Collection at Natick, remained. This particular strain is not only regarded as the T. reesei reference strain, but also is the one strain from which all the mutants used in industry today have been derived.
Then and now, research on T. reesei was propelled by the idea that its secreted cellulases could have a game changing impact on the 200 year-old struggle to economically produce fuels from renewable, lignocellulosic biomass [5]. T. reesei research has since pioneered the concept of enzymatic saccharification of cellulose by a synergistic combination of different cellulase activities [6] and laid the groundwork for our current understanding of the regulation of the involved enzymes [7–9]. Its major cellobiohydrolase CBH1 (CEL7a) was also the first eukaryotic cellulase to be cloned and the first cellulase whose structure was solved [10, 11]. An important step towards applying T. reesei cellulases industrially was the development of efficient strain mutagenesis and screening procedures in the 1970s. In the following two decades, the titer of extracellular protein produced by the original strain QM6a could be increased by up to 20 fold through mutagenesis programs at Natick and Rutgers University. The latter culminated in the isolation of strain RUT-C30 [12, 13] (where “RUT” stands for Rutgers). Although the gold standard for cellulase production in industry was reported to be higher than 100 g/L, this strain still is the prototype cellulase hyperproducer available to the public domain [14] with titers of extracellular protein reaching 30 g/L on the cellulase inducing substrate lactose [15].
At first, however, other commercial applications for cellulases were developed [16–18] as it became clear that efficient and complete saccharification of lignocellulosic biomass to fermentable sugars requires many more enzymatic activities than initially anticipated. Again, research involving T. reesei continued to break new ground in the enzymology of both cellulose and hemicellulose degradation [19–25] and still does so today [26–28]. High-speed atomic force microscopy has now also visualized cellulose degradation demonstrating how the major cellobiohydrolase CBH1/CEL7A slides unidirectionally along the cellulose surface [29]. By the early 1990s, transformation techniques facilitating genetic engineering of T. reesei had become available [30, 31]. During the coming decade these technologies were instrumental in gaining new insights into the regulation of its enzymes and in altering the enzyme profile secreted by the fungus [32, 33]. At the time, T. reesei was also among the first hosts for the expression of mammalian proteins as exemplified by the expression of calf chymosin under cbh1 (cel7a) expression signals [34].
By the end of the 1990s, Kuhls et al. [35] discovered that Hypocrea jecorina is in fact the sexual form of T. reesei which is the reason why a number of subsequent publications used H. jecorina as species name instead of T. reesei. A more detailed study on sexual development led to the hypothesis that strain QM6a is in fact female sterile [36] which could subsequently be linked to a mutation in the MAP-kinase scaffold encoding gene ham5 [37].
The turn of the millennium, which for genetics can be viewed as a turn away from the study of isolated genes and pathways towards the study of entire genomes, saw the arrival of T. reesei in the so called Genomic Era. The first global approach to study T. reesei gene expression in 2003 was a transcriptomic study by Foreman et al. [38], who constructed DNA microarrays based on cDNAs that corresponded to over 5000 different transcripts of the T. reesei genome. Five years later, the genome sequencing and analysis of the original T. reesei isolate QM6a [39] laid the basis for the broad scale application of genome wide studies. In the following years, comparative genomic analysis of a number of cellulase hyper- and nullproducer strains led to the discovery of potential novel factors involved in cellulase hyperproduction such as nucleocytoplasmic transport, vacuolar protein trafficking and mRNA turnover [40–42]. These comparative analyses benefited from the fact that all T. reesei strains used in academia and industry are derived from strain QM6a. Sixty-five years after Elwyn Reese’s initial studies on cellulose degradation by T. reesei [43], the world-wide installed cellulosic biofuel production capacity is now 480.5 million liters per year (MMLY) of ethanol of which 380.5 MMLY (or roughly 80 %) are produced using T. reesei enzyme formulations such as Accellerase and Cellic (Fig. 1a). Without doubt, this effort required maturation of the underlying technology at several levels, including process engineering and substrate pretreatment. Still, the production and optimization of enzyme formulations for the biomass saccharification step was and remains to be one of the key factors determining the cost performance of cellulosic ethanol processes [44]. Moreover, enzyme production using T. reesei is by no means limited to the production of biorefinery enzymes. In fact, roughly 11 % of all technical enzyme formulations registered by the Association of Manufacturers and Formulators of Enzyme Products are produced using T. reesei as the expression host (Fig. 1b, c). Last but not least, T. reesei still maintains its importance in research, exemplified by more than 100 research articles dealing with the fungus or its enzymes published each year (Fig. 1d).
Fig. 1.
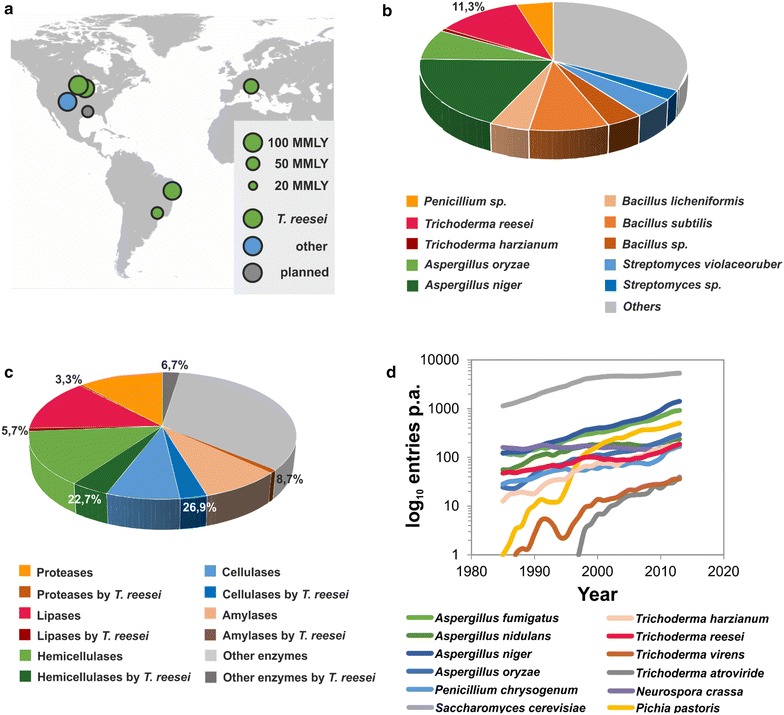
a Installed and planned cellulosic ethanol production as of April 2015 in million liter per year (MMLY). Capacity data were compiled from different specialist publications on cellulosic biofuels and press releases of involved consortia and companies. b Number of different technical enzyme preparations produced by individual species. c Number of a given type of enzyme produced by T. reesei (darker color) or other fungi (lighter color). In both cases (B + C) data were retrieved from the list of technical enzymes (2014 version) with kind permission from the Association of Manufacturers and Formulators of enzyme products (http://www.amfep.org). d Number of research papers per year for different fungi retrieved by a Scopus search with the species name as the entry. Results were averaged over 3 year intervals to reduce the effect of random fluctuation. When a second name for the species exists, control searches with both names were performed and the numbers compiled
The T. reesei biomass enzyme mix: new insights and limitations
In nature lignocellulose deconstruction is rarely accomplished by a single organism. It is rather achieved through the sequential order and collective effort of several organism that produce multiple carbohydrate-active enzymes (CAZymes) [45] to degrade the different polymers. Hence, it is not surprising that the secreted cellulase mix of T. reesei had to be significantly adapted to deliver cost competitive enzyme formulation for complete lignocellulose saccharification. Early on researchers realized that T. reesei formulations lacked sufficient β-glucosidase activity because most of the activity is bound to the fungal cell wall [46–48]. Consequently, increased β-glucosidase activity improved cellulose degradation because it counteracts product inhibition through cellobiose, which, in turn, is released by the cooperative action of endoglucanases and cellobiohydrolases [49]. This feedback mechanism otherwise seriously slows down cellulose saccharification. Likewise, hemicellulose-derived xylo- and mannooligosaccharides inhibit cellobiohydrolases of T. reesei [50–52] which strongly indicates that sufficient β-xylosidase and β-mannosidase activities are required to efficiently degrade lignocellulose. In 2003, Foreman et al. [38] described two proteins that are co-induced with the major cellulases and named them cellulose induced protein 1 and 2 (CIP1 and CIP2). They were since shown to be important to efficiently degrade lignocellulose [53]. Recent results show that CIP1 has structural similarities with lyases, although lyase activity could not be demonstrated [54], and that CIP2 is a glucuronoylesterase of the CE15 family [55]. Another important secreted protein is the swollenin SWO1 which contains a carbohydrate binding module (CBM) linked to an expansin like domain [56]. Despite cellulose disrupting activity [57, 58], SWO1 synergistically enhances endoxylanase rather than endoglucanase or cellobiohydrolase activity during enzymatic hydrolysis of pretreated corn stover [59]. One proposed mode-of-action is that it renders the xylan portion of lignocellulose more accessible for degradation by xylanases and thereby indirectly promotes the action of cellulases. Arguably the biggest revolution in recent years in cellulose degradation was the discovery of the lytic polysaccharide monooxygenases (LPMO). These enzymes introduced a new, oxidative mechanism to polysaccharide degradation. In cellulose degradation it is assumed that LPMOs act on the surface of crystalline cellulose fibrils, thereby rendering them more accessible to cellulases [60–62]. Intriguingly, these enzymes can derive the electrons needed for this process from plant cell wall lignin via long-range electron transfer, thereby turning the plant’s defense mechanisms against it [63]. Alternatively, GMC oxidoreductases or cellobiose dehydrogenases can work as electron donors [64]. This finding could well explain how T. reesei fuels its LPMOs as it was previously shown that several such GMC oxidoreductases are indeed induced by wheat straw [65]. However, this oxidative mechanism is by no means limited to cellulose depolymerization. Originally demonstrated for chitin [66, 67], LPMOs also play a role in xyloglucan [68] and amylose degradation [69]. In the CAZy database, enzymes belonging to this group have been reclassified to “auxiliary activities” (AA) as opposed to their previous classification as glycoside hydrolases (e.g. GH61) and are found in AA families 9–11 and 13 [45]. As outlined before, hemicellulolytic activities are important for complete lignocellulose saccharification (reviewed by Harris et al. [70]). Different amounts of individual activities are required depending on the hemicellulose types present in the substrate [71]. It is therefore noteworthy that the enzymatic repertoire of T. reesei has some clear limitations for certain types of hemicellulose specific linkages. One such missing activity is α-xylosidase. Adding α-xylosidase to a commercial T. reesei enzyme formulation enhanced the xylose and glucose release from pretreated corn stover [72]. Similarly, adding a GH family 5 cellulase with activity against glucomannan and xylan significantly improved a synthetic T. reesei enzyme preparation [73]. Other activities that are absent or very limiting in the T. reesei cellulase mix include endo-arabinase [74] and several pectinase activities [39, 75]. Supplementing commercial cellulase mixes with these enzyme activities consequently improved the saccharification of different substrates [76–78]. Another yet to be answered question is the in vivo function of secreted laccase-like multicopper oxidases encoded in the T. reesei genome [79].
Improving T. reesei as protein production host
Given the fact that the cellulases and the majority of the other lignocellulose degrading enzymes are coordinately and conditionally expressed [33, 80], their transcriptional regulation represents a logical engineering target to improve cellulase production by the fungus. One of the master regulators is the carbon catabolite repression mediating C2H2 type transcription factor CRE1. CRE1 shuts down the transcription of its target genes when more favorable carbon sources such as glucose are present. Its truncation is one of the main causes for the improved cellulase production achieved by random mutagenesis programs of T. reesei QM6a leading to strain RUT-C30, which shows both a higher basal level and induced level of cellulase production [81–83]. Similarly, replacing CRE1 binding motifs in the promoter region of the major cellobiohydrolase cel7a with those of a known cellulase activator reduces carbon catabolite repression and elevates cel7a transcription under activating and repressing condition [84]. Additionally, the transcription of cellulase, xylanase and a number of other genes encoding enzymes involved in lignocellulose degradation strictly depends on the Zn(II)2Cys6 type transcriptional activator XYR1 [74, 85]. This is in contrast to other fungi including the Sordariomycetes Neurospora crassa and Fusarium fujikuroi, where the XYR1 orthologue exclusively modulates xylanase gene expression [80]. A mutation leading to a truncated form of XYR1 was found to cause the cellulase-negative phenotype of strain QM9136 which originates from the mutagenesis program at Natick [86]. Accordingly, cellulase over- and hyperproducing mutants have elevated levels of mRNA for the transcriptional activators xyr1 [87]. It has also been proven that overexpression of XYR1 leads to a higher expression of cellulases and abolishes their catabolite repression in the presence of glucose [88, 89]. Furthermore, a point mutation within a putative regulatory region of XYR1 leads to a similarly deregulated expression pattern [90]. Besides XYR1 and CRE1, three transcription factors ACE1, ACE2 and ACE3 regulate cellulase and xylanase expression in T. reesei [91–93]. Similarly to CRE1, ACE1 is a C2H2 zinc finger repressor and its deletion therefore improves the production of both cellulases and xylanases [92]. ACE2 and ACE3, just like XYR1, are Zn(II)2Cys6 type transcription activators [87, 91, 93]. When ace2 is absent, transcription of cellulases and xylanases is reduced accordingly, although cellulase induction by sophorose apparently remains unaffected [91]. Deletion of ace3 completely abolishes cellulase transcription but merely reduces that of xylanases. While overexpression of ACE2 has not yet been attempted, overexpression of ACE3 leads to increased activities of both types of enzymes, as does the overexpression of six other so far uncharacterized regulators [93]. These include two further Zn(II)2Cys6 type transcription factors, as well as two WD40 proteins, a bromodomain protein and a gcn5 related acetyltransferase. All three of the Zn(II)2Cys6 transcriptional activators (XYR1, ACE2 and ACE3) resemble the well characterized Gal4 protein of S. cerevisiae. It is well established that Gal4 recruits the Gcn5 containing SAGA complex [94, 95] and thereby promotes the transcription of its target genes through histone acetylation and euchromatin formation. As a matter of fact the Gcn5 orthologue of T. reesei is indispensable for cellulase expression and involved in the acetylation of histones in the cbh1 promoter [96]. In recent years, a picture emerged which shows that the transcription of cellulases and related CAZymes in fungi is governed by a combination of many transcription factors representing a complex transcriptional-regulatory network influenced by counteracting activators and repressors. In Penicillium oxalicum, for instance, twenty transcription factors modulate the activation or repression of cellulase genes. Amongst these ClrB was identified as key integrator of all other regulators with their target genes [97]. Homologues of these regulators are found in T. reesei, but given the diversity of adaptations in plant cell wall regulation [80], it is expected that other and different regulators will also play significant roles. Another key player in cellulase regulation is the T. reesei orthologue of the enigmatic Aspergillus LaeA, which is involved in the regulation of secondary metabolite gene clusters in different fungi [98, 99]. While the deletion of this putative protein methyltransferase leads to a strong downregulation of different cellulase and other CAZyme genes, its overexpression can strongly promote their expression [88]. Similar effects were found for the LAE1 interacting VEL1 protein of the VELVET complex [100].
Most of the aforementioned studies provide fundamental insights into the regulation of cellulase formation. As most of these studies were carried out either in the original T. reesei isolate QM6a or the moderately overproducing strain QM9414, it remains unclear whether and to what extent these effects can be implemented in hyperproducing strains. In these strains processes such as translation, secretion and turnover of secreted enzymes, rather than transcription, might limit a further increase of cellulase production. It will be interesting to see whether or not several of the reported ways to improve cellulase gene expression can be stacked and how such strains would perform when compared to the hyperproducers derived by random mutagenesis.
Simply boosting the transcription of the gene of interest does not always lead to improved product formation especially in the case of non-fungal proteins. One successful strategy to circumvent low product formation is the fusion gene approach which uses besides promoter and terminator region of a highly expressed gene also the encoded protein as expression enhancer. For T. reesei this is the cellobiohydrolase encoding cel7A which is the most strongly expressed protein under cellulase inducing conditions [34]. It is believed that these gene fusions generally increase mRNA stability, import into the ER and passage through the secretory pathway. To this end the modular structure of CEL7A consisting of a catalytic module, a linker and a CBM is often exploited thereby replacing the C-terminal CBM by the gene of interest. Variations of this gene fusion approach are now available which target the protein to the ER for correct folding, disulfide bridge formation and glycosylation but subsequently aim for intracellular protein accumulation to avoid degradation of the desired product by extracellular proteases. One such strategy uses hydrophobins with an attached ER retention signal as carrier. These fusion proteins self-assemble to micelle-like structures and can be purified using a surfactant-based aqueous two-phase system [101]. A further strategy to target proteins to the ER uses the γ-zein peptide (ZERA) derived from the maize storage protein. Analogously, these self-assembling fusion proteins form protein bodies surrounded by ER membrane which protect them from proteolysis [102]. Developing fungi as efficient production hosts for mammalian proteins also requires the inactivation of the frequently encountered proteases in the fermentation broth. In a systematic study different secreted proteases related to degradation of biopharmaceuticals including antibodies, interferon α 2b, and insulin like growth factor were identified and several inactivated [103]. This led not only to a drastic reduction of protease activity but also to a strong increase of stability of all three recombinant proteins, with the antibody showing the most pronounced effect. Although engineering of the N-glycosylation pattern for the production of therapeutic proteins of high value was attempted before, creating an authentic human glycosylation pattern in a fungal expression host does not seem feasible at the moment. It is therefore questionable whether such biopharmaceuticals will be produced by fungal cell factories in the future, especially given the rapid development of CHO cells [104].
The above mentioned hydrophobins are another group of proteins which received considerable attention due to their surface-active properties [105]. These small, extracellular proteins self-assemble into protein layers at hydrophobic/hydrophilic interfaces due to their amphiphilic properties and make hydrophobic surfaces wettable or hydrophilic surfaces hydrophobic. They have a broad potential in food- and medical applications to disperse hydrophobic materials, stabilize foams or target different molecules to surfaces [106]. Cerato-platanins are another group of small, secreted proteins with four conserved cysteines. They bind to chitin and N-acetylglucosamine oligosaccharides and possess self-assembling properties at hydrophobic/hydrophilic interfaces. In contrast to hydrophobins, cerato-platanins rather enhance the polarity/apolarity properties of surfaces [107]. A targeting function is also attributed to CBMs present in different CAZymes. They can improve the hydrolytic activity of the catalytic domain they are attached to and lead to a more favorable pH and temperature optimum [108, 109]. Their carbohydrate binding properties can furthermore be exploited for affinity purification of fusion proteins using e.g. cellulose columns [110]. The broad range of other applications of recombinant CBMs has recently been reviewed elsewhere [111].
Trichoderma reesei for consolidated bioprocessing and whole cell catalysis
Consolidated bioprocessing (CBP) is classically understood as the integration of the cellulolytic enzyme production, enzymatic hydrolysis and fermentation steps of a cellulosic ethanol production process into a single unit operation. Therefore, a single organism with good cellulolytic properties and an efficient ethanol fermentation pathway would be desirable. Nevertheless, using microbial consortia has also received some attention [112–114]. Unfortunately, no single organism that fulfils both requirements is currently available (Fig. 2). Consequently, efforts to engineer ethanologens to become cellulolytic [115, 116] or cellulolytic organisms to become ethanologenic [117, 118] have been undertaken. In the context of the first scenario, T. reesei has often served as a CAZyme gene donor, especially for cel5a and cel7b encoding two of its endoglucanases and for its two cellobiohydrolases cel6a and cel7a (Table 1). In all published studies the relatively low secretory capacity of S. cerevisiae limited substrate conversion by the secreted or membrane anchored heterologous CAZymes. Hence, high ethanol yields with realistic cellulosic substrates required supplementation with commercial T. reesei enzyme cocktails. Nevertheless, while efforts for improving the secretion and surface display capacity of engineered S. cerevisiae strains are now ongoing [119–121], the currently available cellulase displaying strains already have the potential to lead to substantial reductions of required enzyme loadings for biomass saccharification [122]. In the context of the second scenario, T. reesei itself represents a promising target organism. But although this fungus naturally possesses the ability to metabolize all the biomass related sugars and convert them to ethanol, yields are low and acetic acid is formed as an unwanted byproduct [123]. On the other hand, large scale fermentation regimes for T. reesei are well established due to its wide application in commercial enzyme production, and the molecular tools for its genetic engineering are very well developed too [124]. One of the remaining big challenges to employ T. reesei as a CBP organism is that several of its cellulases and glycolytic genes are repressed by hypoxia [125, 126], which is necessary for ethanol production. Additionally, the transcriptional repression of cellulases in the presence of ethanol [127] represents a further challenge to be overcome. However, the cellulase hyperproducing strain RUT-C30 has a higher ethanol tolerance as compared to strain QM9414 from the Natick lineage [127], and therefore represents a perfect platform strain to develop T. reesei as a CBP organism, especially since it is also carbon catabolite derepressed [81]. Furthermore, in the presence of a lignocellulosic pulp, RUT-C30 exhibits a pellet like morphology during the early stages of fermentation [128], which can be achieved in a growth independent manner by addition of the surfactant Triton X-100 and then also leads to a higher enzyme production [129]. This is important because poor mixability and low maximum cell densities as a consequence of filamentous growth so far hampered the use of T. reesei as a CBP organism. More generally speaking, consolidated bioprocessing could also be used to describe other integrated processes in which the substrate is not necessarily lignocellulosic biomass but another bio-polymer such as chitin or starch and the product is not ethanol, but any other metabolite [130]. To this end, a number of recent studies aimed at overproducing different metabolites via engineering of T. reesei (Table 2). However, the yields that could be achieved were in most cases far from commercialization, hence requiring further optimization.
Fig. 2.
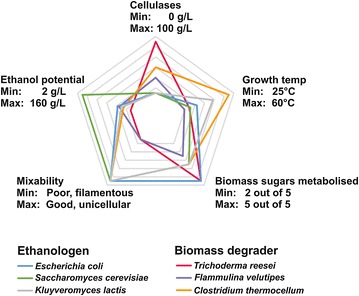
Radar chart showing the potential of different fungal and bacterial organisms as CBP organisms. Data were compiled from different reviews and original publications [123, 151–159]. The five biomass sugars are the hexoses glucose, mannose and galactose as well as the pentoses xylose and arabinose
Table 1.
T. reesei genes used to engineer the ethanologenic yeast S. cerevisiae to a cellulose or hemicellulose degrader
| Genes expresseda | Enzyme localization | Substrate | Ethanol (g/l) | YEtOH (% theory)b | Reference |
|---|---|---|---|---|---|
| cel5a + bglA A. aculeatus | Cell surface display (CSD) | 45 g/l β-glucan | 16.5 | 93 | [160] |
| xyn2 + xylA A. oryzae | CSD | 100 g/l birchwood xylan | 7.1 | 59 | [161] |
| cel6a, cel5a + bglA A. aculeatus | CSD | 10 g/l phosphoric acid swollen cellulose (PASC) | 2.9 | 89 | [162] |
| cel7a | Secretion | PASC, bacterial microcrystalline cellulose | – | – | [163] |
| cel6a, cel5a + bglA A. aculeatus | CSD | 20 g/l PASC | 7.1 | 75 | [164] |
| cel6a, cel5a + bglA A. aculeatus | CSD | 100 g/l pretreated rice straw | 7.5 | 33 | [164] |
| cel6a, cel7a + 20 other cellobiohydrolases | Secretion | 20 g/l avicel cellulose | 3 | 33 | [165] |
| cel6a, cel5a + bglA A. aculeatus + swo1 | CSD | 20 g/l PASC | 3.4 | 36 | [166] |
| cel6a, cel5a + bglA A. aculeatus + cdt1 N. crassa | CSD | 20 g/l PASC | 4.3 | 45 | [115] |
| cel6a, cel5a + bglA A. aculeatus + commercial cellulase | CSD | 100 g/l avicel cellulose | 48.9 | 87 | [167] |
| cel6a, cel5a + bglA A. aculeatus + commercial cellulase | CSD | 200 g/l rice straw | 42.2 | 86 | [167] |
| xyn2, xylA A. oryzae , bglA A. aculeatus | CSD | Slurry from rice straw pretreatment containing ~20 g/l poly-, oligo- and monomeric sugars | 4.0 | 39 | [151] |
| cel5a + cbh1 T. emersonii fused to CBM of cel7a + bglA A. aculeatus | CSD and secretion | 10 g/l PASC | 2.6 and 2.9 | 87 and 82 | [168] |
a Genes are derived from T. reesei or from the species given in subscript
b Based on a stochiometry of 0.51 g ethanol per g consumed sugar. Residual sugars in non-hydrolyzed substrates were considered when the data were available
Table 2.
Examples for the genetically engineering of T. reesei towards overproduction of a metabolite or interesting molecule
| Substance produced | Substrate employed | T. reesei strain background | Genetic alteration | Maximum titer | Reference |
|---|---|---|---|---|---|
| Ethylene | Wheat straw, cellulose | T. reesei (viride) TL124 | Ethylene forming enzyme of Pseudomonas syringae expressed under the cel7a promoter | 1.06 µl h−1 g−1 dry weight | [169] |
| Ethylene | Wheat straw | QM9414 | Ethylene forming enzyme of P. syringae expressed under the cel7a, pgk1 and A. nidulans gpdA promoters | 4.012 nl h−1 l−1 | [170] |
| N-acetyl-neuraminic acid | Chitin | QM9414 | Expression of codon optimized N-acetyl glucosamine-2-epimerase from Anabaena sp. and N-acetyl neuramininc acid synthase from Campylobacter jejuni under the pki1 and xyn1 promoters | 13 µg g−1 mycelium | [171] |
| Xylitol | Barley straw | QM9414 | Deletion of xylitol dehydrogenase, double deletion of xylitol dehydrogenase and l-arabinitol-4-dehydrogenase | 13.2 g l−1 | [172] |
| Xylitol | Xylose and glucose | QM9414 | Knockdown (antisense RNA) of xylulokinase; deletion of xylitol dehydrogenase, overexpression of d-xylose reductase | 3.7 g l−1 | [173] |
| Erythritol | Wheat straw | RUT-C30, QM6a | Erythrose reductase expressed under the pki1 and bxl1 promoters | 5 mg l−1 | [174] |
Tools for cell design and engineering
While two comprehensive reviews on the molecular tool box of T. reesei and other Trichoderma species have only been given recently [124, 131], we want to update these with the most recent advances in the field. Targeted strain engineering towards improved cellulase production or for metabolic engineering requires efficient methods to introduce directed genetic alterations into the organism. The generally low efficiency of gene targeting has for a long time been a major challenge to obtain a reasonable number of transformants by homologous integration of a deletion or expression cassette. This problem has been mainly solved by inactivating components of the non-homologous end joining (NHEJ) pathway of DNA repair such as tku70 [132] or tmus53 [133]. Tku70 deleted strains do show improved gene targeting although the efficiency of homologous integration in these strains can still vary depending on the targeted locus and thus might drop to 30 % [134]. Based on this improvement, a number of novel approaches were developed to insert expression cassettes at a defined genomic region, thereby avoiding pleiotropic effects caused by their random integration. Using a tku70 background Jorgensen et al. [135] developed an expression platform that employs the easy to screen ade2 locus as the preferred site of integration. Upon integration of the expression cassette into this locus, ade2 is destroyed and the resulting transformants develop a distinct red pigmentation. In another study, the pyr4 and asl1 loci were chosen to develop a strain with uridine and l-arginine auxotrophy that allows site directed integration at these sites [136]. Ouaedraogo et al. [137] followed a different strategy and expressed the S. cerevisiae I-SceI meganuclease in T. reesei. I-SceI generates artificial double-strand breaks at an I-SceI recognition site which was introduced beforehand at a predefined locus and improved both transformation and homologous integration efficiencies. In a follow up study I-SceI mediated double strand breaks were combined with a tku70 deletion [138]. Here, the inability to repair double strand breaks via NHEJ favors integration of the cassette leading to homologous recombination efficiencies of up to 100 %. A revolution for genetic engineering or genome editing was introduced with the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system [139]. Underscoring both the need of such a technology and the rising importance as an enzyme producer, this system was firstly tested for filamentous fungi in T. reesei [140]. It introduces specific DNA double strand breaks to stimulate gene targeting and depends only on a Cas9 (CRISPR associated) nuclease which uses a single chimeric guide RNA for targeting. Accurate targeting of this RNA-guided Cas9 to a specific DNA sequence is achieved by the protospacer sequence of the guide RNA via simple base pairing. Using 200 bp up- and downstream flanking regions for the gene deletion construct, HR frequencies of higher than 90 % could be reached. Double deletions and triple deletions occurred at a frequency of 45 and 4 %, respectively, following a single round of transformation. While in this study in vitro transcribed guide RNAs were cotransformed with the deletion cassette into a Cas9 expressing T. reesei, Nødvig et al. [141] used two flanking ribozyme sequences to liberate guide RNAs from a larger transcript that also encodes the Cas9 enzyme in different Aspergilli. Another essential tool needed in both recombinant protein expression and strain engineering are promoters that allow gene expression in a controllable fashion. Although a number of inducible and repressible promoters are available with the different cellulase promoter regions, they usually have drawbacks as their expression is linked to the host metabolism and activators might be limiting due to promoter titration effects. Useful alternatives include a set of l-methionine repressible T. reesei genes with different basal expression strength [65]. It was shown that one of these promoters can drive repressible expression of different reporter genes on several carbon sources including wheat straw. In a similar study, the promoter of a copper permease gene of T. reesei was used to control expression of the major cellulase and hemicellulase regulator xyr1 in the absence of copper [89]. However, given the fact that copper containing enzymes from AA family 9 are important components of the T. reesei cellulase mix [142], it is, however, doubtful whether this system can be applied in a cellulase production scenario.
Beside these exciting molecular tools, some older tricks from classical genetics are now available too. Strain development of T. reesei was for a long time hampered by the fact that the fungus was thought to be asexual preventing strain crossing. A milestone in this respect was the finding that QM6a has a MAT1-2 mating type locus and can be readily crossed with certain MAT1-1T. reesei wild-type isolates [36]. But when the MAT1-2 locus of QM6a was replaced by its MAT1-1 counterpart, no stromata were formed upon confrontation with the original MAT1-2 QM6a strain. Consequently, it was impossible to exploit mating for strain engineering in the different academic and industrial T. reesei strains derived from QM6a. Using a systems biological approach the lacking gene responsible for female sterility was identified as ham5 [37]. In N. crassa, ham-5 encodes for a protein which serves as a MAP kinase scaffold during cell fusion [143, 144]. Reintroduction of a functional ham5 restores stromata formation in strain QM6a and enables the restoration of female fertility in other strains originating from QM6a backgrounds [37]. This finding is especially important because crossing with the aforementioned H. jecorina isolates can lead to segmentally aneuploid progenies [145]. With this tool in hand, the basis for identifying relevant mutations leading to e.g. cellulase hyperproduction is laid. This is important as traditional complementation approaches to identify gene(s) causing mutant phenotypes were largely unsuccessful in species such as T. reesei. And although high-throughput sequencing and comparative genome analysis can easily identify mutations in the QM6a strain line of cellulase hyper- and nullproducers, only in a few cases this has already led to the linking of a mutation to a particular phenotype [86, 90, 146]. But in cases where a high number of mutations was found or the mutations affected genes with unknown function, comparative sequence analysis did not reveal the nature of the desired target genes [42]. Several researchers [147–150] have therefore successfully applied bulk segregant analysis in combination with next-generation sequencing to identify relevant mutations. In this approach, the mutant is crossed with a reference strain and the genomic DNAs of the segregants displaying the wanted phenotype are pooled and sequenced. Genome comparison of this pool of sequenced DNA to the genomes of the parental strains can then reveal conserved mutations relevant for the phenotype. Mutations which are unrelated to the phenotype will be underrepresented. Although it cannot be expected that this approach will lead to the identification of a single mutation because mutations close to the relevant mutation usually co-segregate, the number of targets for further investigation is considerably reduced.
Conclusion
Research on T. reesei is highly driven by the industrial application of this fungus for the production of cellulases, other related CAZymes and recombinant proteins. Although a number of relevant key factors to improve protein production were discovered in the past decades, the underlying reason for its high potential to overproduce cellulases remained largely unidentified. Today, strain crossing in combination with different high throughput methods including comparative genomics in combination with tools such as the CRISPR/Cas9 system, provide efficient ways to pinpoint relevant mutations and thereby accelerate not only our understanding of cellulase regulation and expression but also to rational engineer and design this hyperproducing cell factory for a broader range of enzymes. Metabolic engineering of T. reesei for the direct conversion of lignocellulose to useful chemicals is still very much in its infancy. Future studies aimed in this direction will therefore need to push beyond a mere proof of principle and need to address and overcome inherent limitations of filamentous fungi as cell factories.
Authors’ contributions
BS conceived of the manuscript. All authors were involved in the design of the manuscript. RB and JR drafted the abstract and main part of the review to equal parts and prepared the tables and figures. BS drafted the conclusions and edited the whole manuscript. All authors read and approved the final manuscript.
Acknowledgements
Illaria Valentini is acknowledged for assisting in generating the enzyme dataset, Elisabeth Fitz, Alexander Lichius and Theresa Bischof for critical reading the manuscript.
Availability of data and supporting materials
The datasets supporting the conclusions of Fig. 1 are publicly available and the sources are stated in the figure legend. Data were collected from http://www.ethanolproducer.com, http://www.biofuelsdigest.com, http://www.iea.gov, www.abengoa.com, http://www.abengoabioenergy.com, http://www.novozymes.com, http://www.iogen.ca.
Competing interests
The authors declare that they have no competing interests.
Funding
This work has been supported by the Austrian Science Fund (P24219-B21) and by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol, the Government of Lower Austria and Business Agency Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG to BS. JR is member of the Ph.D. school program MEIBio, financed by TU Wien.
Abbreviations
- AA
auxiliary activities
- Cas9
CRISPR associated protein 9
- CAZymes
carbohydrate-active enzymes
- CBM
carbohydrate binding module
- CBP
consolidated bioprocessing
- CHO
Chinese hamster ovary
- CIP
cellulose induced protein
- CRISPR
clustered regularly interspaced short palindromic repeats
- CSD
cell surface display
- GH
glycoside hydrolase
- LPMO
lytic polysaccharide monooxygenase
- MAT
mating type
- MMLY
million liters per year
- NHEJ
non-homologous end joining
- PASC
phosphoric acid swollen cellulose
- QM
quartermaster
- MAP
mitogen-activated protein
Footnotes
Robert H. Bischof and Jonas Ramoni contributed equally to the manuscript
Contributor Information
Robert H. Bischof, Email: robert.bischof@acib.at
Jonas Ramoni, Email: jonas.ramoni@tuwien.ac.at.
Bernhard Seiboth, Email: bernhard.seiboth@tuwien.ac.at.
References
- 1.Michel RH, McGovern PE, Badler V. Chemical evidence for ancient beer. Nature. 1992;360:24. doi: 10.1038/360024b0. [DOI] [Google Scholar]
- 2.McGovern PE, Glusker DL, Exner LJ, Voigt MM. Neolithic resinated wines. Nature. 1996;281:480–481. doi: 10.1038/381480a0. [DOI] [Google Scholar]
- 3.Salque M, Bogucki PI, Pyzel J, Sobkowiak-Tabaka I, Grygiel R, Szmyt M, Evershed RP. Earliest evidence for cheese making in the sixth millennium BC in northern Europe. Nature. 2013;493:522–525. doi: 10.1038/nature11698. [DOI] [PubMed] [Google Scholar]
- 4.Allen F, Andreotti R, Eveleigh DE, Nystrom J. Mary Elizabeth Hickox Mandels, 90, bioenergy leader. Biotechnol Biofuels. 2009;2:22. doi: 10.1186/1754-6834-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeffries T, Lindblad P. We march backwards into the future. Curr Opin Biotechnol. 2009;20:255–256. doi: 10.1016/j.copbio.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Reese ET. A microbiological process report; enzymatic hydrolysis of cellulose. App Microbiol. 1956;4:39–45. doi: 10.1128/am.4.1.39-45.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandels M, Reese ET. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J Bacteriol. 1957;73:269–278. doi: 10.1128/jb.73.2.269-278.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandels M, Reese ET. Induction of cellulase in fungi by cellobiose. J Bacteriol. 1960;79:816–826. doi: 10.1128/jb.79.6.816-826.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandels M, Parrish FW, Reese ET. Sophorose as an inducer of cellulase in Trichoderma viride. J Bacteriol. 1962;83:400–408. doi: 10.1128/jb.83.2.400-408.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoemaker S, Schweickart V, Ladner M, Gelfand D, Kwok S, Myambo K, Innis M. Molecular cloning of exo-cellobiohydrolase I derived from Trichoderma reesei strain L27. Nat Biotech. 1983;1:691–696. doi: 10.1038/nbt1083-691. [DOI] [Google Scholar]
- 11.Divne C, Stahlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JK, Teeri TT, Jones TA. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994;265:524–528. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- 12.Mandels M, Weber J, Parizek R. Enhanced cellulase production by a mutant of Trichoderma viride. App Microbiol. 1971;21:152–154. doi: 10.1128/am.21.1.152-154.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eveleigh DE, Montenecourt BS. Increasing yields of extracellular enzymes. Adv Appl Microbiol. 1979;25:57–74. doi: 10.1016/S0065-2164(08)70146-1. [DOI] [PubMed] [Google Scholar]
- 14.Peterson R, Nevalainen H. Trichoderma reesei RUT-C30–thirty years of strain improvement. Microbiology. 2012;158:58–68. doi: 10.1099/mic.0.054031-0. [DOI] [PubMed] [Google Scholar]
- 15.Durand H, Clanet M, Tiraby G. Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme Microb Technol. 1988;10:341–346. doi: 10.1016/0141-0229(88)90012-9. [DOI] [Google Scholar]
- 16.Galante Y, De Conti A, Monteverdi R. Applications of Trichoderma reesei enzymes in the food and feed industry. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. London: Taylor & Francis Ltd; 1998. pp. 338–342. [Google Scholar]
- 17.Galante Y, De Conti A, Monteverdi R. Applications of Trichoderma reesei enzymes in the textile industry. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. London: Taylor & Francis Ltd; 1998. pp. 311–326. [Google Scholar]
- 18.Buchert J, Oksanen J, Pere J, Siika-aho M, Suurnäkki A, Viikari L. Application of Trichoderma reesei enzymes in the pulp and paper industry. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. London: Taylor & Francis Ltd; 1998. pp. 343–364. [Google Scholar]
- 19.Dekker RFH. Bioconversion of hemicellulose: aspects of hemicellulase production by Trichoderma reesei QM 9414 and enzymic saccharification of hemicellulose. Biotechnol Bioeng. 1983;25:1127–1146. doi: 10.1002/bit.260250419. [DOI] [PubMed] [Google Scholar]
- 20.Biely P, Puls J, Schneider H. Acetyl xylan esterases in fungal cellulolytic systems. FEBS Lett. 1985;186:80–84. doi: 10.1016/0014-5793(85)81343-0. [DOI] [Google Scholar]
- 21.Saloheimo M, Lehtovaara P, Penttilä M, Teeri TT, Ståhlberg J, Johansson G, Pettersson G, Claeyssens M, Tomme P, Knowles JKC. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene. 1988;63:11–21. doi: 10.1016/0378-1119(88)90541-0. [DOI] [PubMed] [Google Scholar]
- 22.Törrönen A, Mach RL, Messner R, Gonzalez R, Kalkkinen N, Harkki A, Kubicek CP. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Nat Biotechnol. 1992;10:1461–1465. doi: 10.1038/nbt1192-1461. [DOI] [PubMed] [Google Scholar]
- 23.Tenkanen M, Buchert J, Viikari L. Binding of hemicellulases on isolated polysaccharide substrates. Enzyme Microb Technol. 1995;17:499–505. doi: 10.1016/0141-0229(94)00050-2. [DOI] [Google Scholar]
- 24.Tenkanen M, Luonteri E, Teleman A. Effect of side groups on the action of β-xylosidase from Trichoderma reesei against substituted xylo-oligosaccharides. FEBS Lett. 1996;399:303–306. doi: 10.1016/S0014-5793(96)01313-0. [DOI] [PubMed] [Google Scholar]
- 25.Biely P, Côté GL, Kremnický L, Greene RV, Tenkanen M. Action of acetylyxlan esterase from Trichoderma reesei on acetylated methyl glycosides. FEBS Lett. 1997;420:121–124. doi: 10.1016/S0014-5793(97)01500-7. [DOI] [PubMed] [Google Scholar]
- 26.Tenkanen M, Vršanská M, Siika-Aho M, Wong DW, Puchart V, Penttilä M, Saloheimo M, Biely P. Xylanase XYN IV from Trichoderma reesei showing exo- and endo-xylanase activity. FEBS J. 2013;280:285–301. doi: 10.1111/febs.12069. [DOI] [PubMed] [Google Scholar]
- 27.Biely P, Cziszárová M, Agger JW, Li XL, Puchart V, Vršanská M, Eijsink VGH, Westereng B. Trichoderma reesei CE16 acetyl esterase and its role in enzymatic degradation of acetylated hemicellulose. Biochim Biophys Acta. 2014;1840:516–525. doi: 10.1016/j.bbagen.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Biely P, Puchart V, Stringer MA, Krogh KBRM. Trichoderma reesei XYN VI—a novel appendage-dependent eukaryotic glucuronoxylan hydrolase. FEBS J. 2014;281:3894–3903. doi: 10.1111/febs.12925. [DOI] [PubMed] [Google Scholar]
- 29.Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, Okamoto T, Penttilä M, Ando T, Samejima M. Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science. 2011;333:1279–1282. doi: 10.1126/science.1208386. [DOI] [PubMed] [Google Scholar]
- 30.Gruber F, Visser J, Kubicek CP, De Graaff LH. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr Genet. 1990;18:71–76. doi: 10.1007/BF00321118. [DOI] [PubMed] [Google Scholar]
- 31.Penttilä M, Nevalainen H, Ratto M, Salminen E, Knowles J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene. 1987;61:155–164. doi: 10.1016/0378-1119(87)90110-7. [DOI] [PubMed] [Google Scholar]
- 32.Kubicek CP, Messner R, Gruber F, Mach RL, Kubicek-Pranz EM. The Trichoderma cellulase regulatory puzzle: from the interior life of a secretory fungus. Enzyme Microb Technol. 1993;15:90–99. doi: 10.1016/0141-0229(93)90030-6. [DOI] [PubMed] [Google Scholar]
- 33.Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels. 2009;2:1–14. doi: 10.1186/1754-6834-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harkki A, Uusitalo J, Bailey M, Penttilä M, Knowles JKC. A novel fungal expression system: secretion of active calf chymosin from the filamentous fungus Trichoderma reesei. Bio Technol. 1989;7:596–603. doi: 10.1038/nbt0689-596. [DOI] [Google Scholar]
- 35.Kuhls K, Lieckfeldt E, Samuels GJ, Kovacs W, Meyer W, Petrini O, Gams W, Börner T, Kubicek CP. Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina. Proc Natl Acad Sci USA. 1996;93:7755–7760. doi: 10.1073/pnas.93.15.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seidl V, Seibel C, Kubicek CP, Schmoll M. Sexual development in the industrial workhorse Trichoderma reesei. Proc Natl Acad Sci USA. 2009;106:13909–13914. doi: 10.1073/pnas.0904936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linke R, Thallinger GG, Haarmann T, Eidner J, Schreiter M, Lorenz P, Seiboth B, Kubicek CP. Restoration of female fertility in Trichoderma reesei QM6a provides the basis for inbreeding in this industrial cellulase producing fungus. Biotechnol Biofuels. 2015;8:155. doi: 10.1186/s13068-015-0311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, Goedegebuur F, Houfek TD, England GJ, Kelley AS, et al. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem. 2003;278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- 39.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 40.Kubicek CP. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J Biotechnol. 2013;163:133–142. doi: 10.1016/j.jbiotec.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bischof R, Seiboth B. Systems biology of carbohydrate active enzyme production in Trichoderma reesei. In: Hallenbeck PC, editor. Advances in enzymatic conversion of biomass to biofuels. Montreal: Future Medicine Ltd; 2015. pp. 6–19. [Google Scholar]
- 42.Le Crom S, Schackwitz W, Pennacchio L, Magnuson JK, Culley DE, Collett JR, Martin J, Druzhinina IS, Mathis H, Monot F, et al. Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:16151–16156. doi: 10.1073/pnas.0905848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levinson HS, Mandels GR, Reese ET. Products of enzymatic hydrolysis of cellulose and its derivatives. Arch Biochem Biophys. 1951;31:351–365. doi: 10.1016/0003-9861(51)90150-6. [DOI] [PubMed] [Google Scholar]
- 44.Wyman CE. What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol. 2007;25:153–157. doi: 10.1016/j.tibtech.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Messner R, Hagspiel K, Kubicek CP. Isolation of a β-glucosidase binding and activating polysaccharide from cell walls of Trichoderma reesei. Arch Microbiol. 1990;154:150–155. doi: 10.1007/BF00423325. [DOI] [Google Scholar]
- 47.Ryu DDY, Mandels M. Cellulases: biosynthesis and applications. Enzyme Microb Technol. 1980;2:91–102. doi: 10.1016/0141-0229(80)90063-0. [DOI] [Google Scholar]
- 48.Mach RL, Seiboth B, Myasnikov A, Gonzalez R, Strauss J, Harkki AM, Kubicek CP. The bgl1 gene of Trichoderma reesei QM 9414 encodes an extracellular, cellulose-inducible β-glucosidase involved in cellulase induction by sophorose. Mol Microbiol. 1995;16:687–697. doi: 10.1111/j.1365-2958.1995.tb02430.x. [DOI] [PubMed] [Google Scholar]
- 49.Sørensen A, Lübeck M, Lübeck PS, Ahring BK. Fungal beta-glucosidases: a bottleneck in industrial use of lignocellulosic materials. Biomolecules. 2013;3:612–631. doi: 10.3390/biom3030612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qing Q, Yang B, Wyman CE. Xylooligomers are strong inhibitors of cellulose hydrolysis by enzymes. Bioresour Technol. 2010;101:9624–9630. doi: 10.1016/j.biortech.2010.06.137. [DOI] [PubMed] [Google Scholar]
- 51.Xin D, Ge X, Sun Z, Viikari L, Zhang J. Competitive inhibition of cellobiohydrolase I by manno-oligosaccharides. Enzyme Microb Technol. 2015;68:62–68. doi: 10.1016/j.enzmictec.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Viikari L. Xylo-oligosaccharides are competitive inhibitors of cellobiohydrolase I from Thermoascus aurantiacus. Bioresour Technol. 2012;117:286–291. doi: 10.1016/j.biortech.2012.04.072. [DOI] [PubMed] [Google Scholar]
- 53.Banerjee G, Car S, Scott-Craig JS, Borrusch MS, Aslam N, Walton JD. Synthetic enzyme mixtures for biomass deconstruction: production and optimization of a core set. Biotechnol Bioeng. 2010;106:707–720. doi: 10.1002/bit.22741. [DOI] [PubMed] [Google Scholar]
- 54.Jacobson F, Karkehabadi S, Hansson H, Goedegebuur F, Wallace L, Mitchinson C, Piens K, Stals I, Sandgren M. The crystal structure of the core domain of a cellulose induced protein (Cip1) from Hypocrea jecorina, at 1.5 Å resolution. PLoS One. 2013;8:e70562. doi: 10.1371/journal.pone.0070562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pokkuluri PR, Duke NEC, Wood SJ, Cotta MA, Li XL, Biely P, Schiffer M. Structure of the catalytic domain of glucuronoyl esterase Cip2 from Hypocrea jecorina. Prot Struct Funct Bioinform. 2011;79:2588–2592. doi: 10.1002/prot.23088. [DOI] [PubMed] [Google Scholar]
- 56.Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttilä M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur J Biochem. 2002;269:4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 57.Andberg M, Penttilä M, Saloheimo M. Swollenin from Trichoderma reesei exhibits hydrolytic activity against cellulosic substrates with features of both endoglucanases and cellobiohydrolases. Bioresour Technol. 2015;181:105–113. doi: 10.1016/j.biortech.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 58.Jäger G, Girfoglio M, Dollo F, Rinaldi R, Bongard H, Commandeur U, Fischer R, Spiess AC, Büchs J. How recombinant swollenin from Kluyveromyces lactis affects cellulosic substrates and accelerates their hydrolysis. Biotechnol Biofuels. 2011;4:33. doi: 10.1186/1754-6834-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gourlay K, Hu J, Arantes V, Andberg M, Saloheimo M, Penttilä M, Saddler J. Swollenin aids in the amorphogenesis step during the enzymatic hydrolysis of pretreated biomass. Bioresour Technol. 2013;142:498–503. doi: 10.1016/j.biortech.2013.05.053. [DOI] [PubMed] [Google Scholar]
- 60.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 61.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eibinger M, Ganner T, Bubner P, Rošker S, Kracher D, Haltrich D, Ludwig R, Plank H, Nidetzky B. Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J Biol Chem. 2014;289:35929–35938. doi: 10.1074/jbc.M114.602227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westereng B, Cannella D, Agger JW, Jorgensen H, Andersen ML, Eijsink VG, Felby C. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep. 2015;5:18561. doi: 10.1038/srep18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kracher D, Scheiblbrandner S, Felice AK, Breslmayr E, Preims M, Ludwicka K, Haltrich D, Eijsink VG, Ludwig R. Extracellular electron transfer systems fuel cellulose oxidative degradation. Science. 2016;352(6289):1098–1101. doi: 10.1126/science.aaf3165. [DOI] [PubMed] [Google Scholar]
- 65.Bischof R, Fourtis L, Limbeck A, Gamauf C, Seiboth B, Kubicek CP. Comparative analysis of the Trichoderma reesei transcriptome during growth on the cellulase inducing substrates wheat straw and lactose. Biotechnol Biofuels. 2013;6:127. doi: 10.1186/1754-6834-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10:122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, Eijsink VGH. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 68.Agger JW, Isaksen T, Várnai A, Vidal-Melgosa S, Willats WGT, Ludwig R, Horn SJ, Eijsink VGH, Westereng B. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci USA. 2014;111:6287–6292. doi: 10.1073/pnas.1323629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leggio LL, Simmons TJ, Poulsen JC, Frandsen KE, Hemsworth GR, Stringer MA, von Freiesleben P, Tovborg M, Johansen KS, De Maria L, et al. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun. 2015;6:5961. doi: 10.1038/ncomms6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harris PV, Xu F, Kreel NE, Kang C, Fukuyama S. New enzyme insights drive advances in commercial ethanol production. Curr Opin Chem Biol. 2014;19:162–170. doi: 10.1016/j.cbpa.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 71.Hu J, Arantes V, Pribowo A, Saddler JN. The synergistic action of accessory enzymes enhances the hydrolytic potential of a “cellulase mixture” but is highly substrate specific. Biotechnol Biofuels. 2013;6:112. doi: 10.1186/1754-6834-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jabbour D, Borrusch MS, Banerjee G, Walton JD. Enhancement of fermentable sugar yields by α-xylosidase supplementation of commercial cellulases. Biotechnol Biofuels. 2013;6:58. doi: 10.1186/1754-6834-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye Z, Zheng Y, Li B, Borrusch MS, Storms R, Walton JD. Enhancement of synthetic Trichoderma-based enzyme mixtures for biomass conversion with an alternative family 5 glycosyl hydrolase from Sporotrichum thermophile. PLoS One. 2014;9:e109885. doi: 10.1371/journal.pone.0109885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akel E, Metz B, Seiboth B, Kubicek CP. Molecular regulation of arabinan and l-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei) Eukaryot Cell. 2009;8:1837–1844. doi: 10.1128/EC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ries L, Pullan ST, Delmas S, Malla S, Blythe MJ, Archer DB. Genome-wide transcriptional response of Trichoderma reesei to lignocellulose using RNA sequencing and comparison with Aspergillus niger. BMC Genom. 2013;14:541. doi: 10.1186/1471-2164-14-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ravalason H, Grisel S, Chevret D, Favel A, Berrin JG, Sigoillot JC, Herpoel-Gimbert I. Fusarium verticillioides secretome as a source of auxiliary enzymes to enhance saccharification of wheat straw. Bioresour Technol. 2012;114:589–596. doi: 10.1016/j.biortech.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 77.Kracher D, Oros D, Yao W, Preims M, Rezic I, Haltrich D, Rezic T, Ludwig R. Fungal secretomes enhance sugar beet pulp hydrolysis. Biotechnol J. 2014;9:483–492. doi: 10.1002/biot.201300214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laothanachareon T, Bunterngsook B, Suwannarangsee S, Eurwilaichitr L, Champreda V. Synergistic action of recombinant accessory hemicellulolytic and pectinolytic enzymes to Trichoderma reesei cellulase on rice straw degradation. Bioresour Technol. 2015;198:682–690. doi: 10.1016/j.biortech.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 79.Levasseur A, Saloheimo M, Navarro D, Andberg M, Pontarotti P, Kruus K, Record E. Exploring laccase-like multicopper oxidase genes from the ascomycete Trichoderma reesei: a functional, phylogenetic and evolutionary study. BMC Biochem. 2010;11:32. doi: 10.1186/1471-2091-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glass LN, Schmoll M, Cate JHD, Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 81.Ilmén M, Thrane C, Penttilä M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full length and a truncated mutant form. Mol Gen Genet. 1996;251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 82.Mello-de-Sousa TM, Gorsche R, Rassinger A, Pocas-Fonseca MJ, Mach RL, Mach-Aigner AR. A truncated form of the carbon catabolite repressor 1 increases cellulase production in Trichoderma reesei. Biotechnol Biofuels. 2014;7:129. doi: 10.1186/s13068-014-0129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seidl V, Gamauf C, Druzhinina IS, Seiboth B, Hartl L, Kubicek CP. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genom. 2008;9:327. doi: 10.1186/1471-2164-9-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zou G, Shi S, Jiang Y, van den Brink J, de Vries RP, Chen L, Zhang J, Ma L, Wang C, Zhou Z. Construction of a cellulase hyper-expression system in Trichoderma reesei by promoter and enzyme engineering. Microb Cell Fact. 2012;11:1. doi: 10.1186/1475-2859-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stricker AR, Grosstessner-Hain K, Würleitner E, Mach RL. Xyr1 (Xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot Cell. 2006;5:2128–2137. doi: 10.1128/EC.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lichius A, Bidard F, Buchholz F, Le Crom S, Martin J, Schackwitz W, Austerlitz T, Grigoriev IV, Baker SE, Margeot A, et al. Genome sequencing of the Trichoderma reesei QM9136 mutant identifies a truncation of the transcriptional regulator XYR1 as the cause for its cellulase-negative phenotype. BMC Genom. 2015;16:326. doi: 10.1186/s12864-015-1526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Portnoy T, Margeot A, Seidl-Seiboth V, Le Crom S, Chaabane FB, Linke R, Seiboth B, Kubicek CP. Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot Cell. 2011;10:262–271. doi: 10.1128/EC.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seiboth B, Karimi RA, Phatale PA, Linke R, Hartl L, Sauer DG, Smith KM, Baker SE, Freitag M, Kubicek CP. The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol Microbiol. 2012;84:1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lv X, Zheng F, Li C, Zhang W, Chen G, Liu W. Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction-independent production of cellulases in Trichoderma reesei. Biotechnol Biofuels. 2015;8:67. doi: 10.1186/s13068-015-0249-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Derntl C, Gudynaite-Savitch L, Calixte S, White T, Mach RL, Mach-Aigner AR. Mutation of the Xylanase regulator 1 causes a glucose blind hydrolase expressing phenotype in industrially used Trichoderma strains. Biotechnol Biofuels. 2013;6:62. doi: 10.1186/1754-6834-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aro N, Saloheimo A, Ilmén M, Penttilä M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xvlanase genes of Trichoderma reesei. J Biol Chem. 2001;276:24309–24314. doi: 10.1074/jbc.M003624200. [DOI] [PubMed] [Google Scholar]
- 92.Aro N, Ilmén M, Saloheimo A, Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003;69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Häkkinen M, Valkonen M, Westerholm-Parvinen A, Aro N, Arvas M, Vitikainen M, Penttilä M, Saloheimo M, Pakula T. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels. 2014;7:14. doi: 10.1186/1754-6834-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xin Q, Gong Y, Lv X, Chen G, Liu W. Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression. Curr Microbiol. 2013;67:580–589. doi: 10.1007/s00284-013-0396-4. [DOI] [PubMed] [Google Scholar]
- 97.Li Z, Yao G, Wu R, Gao L, Kan Q, Liu M, Yang P, Liu G, Qin Y, Song X, et al. Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum. PLoS Genet. 2015;11:e1005509. doi: 10.1371/journal.pgen.1005509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sarikaya-Bayram Ö, Palmer JM, Keller N, Braus GH, Bayram Ö. One Juliet and four Romeos: VeA and its methyltransferases. Front Microbiol. 2015;6:1. doi: 10.3389/fmicb.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aghcheh RK, Németh Z, Atanasova L, Fekete E, Paholcsek M, Sándor E, Aquino B, Druzhinina IS, Karaffa L, Kubicek CP. The VELVET a orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS One. 2014;9:e112799. doi: 10.1371/journal.pone.0112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Linder MB, Qiao M, Laumen F, Selber K, Hyytia T, Nakari-Setälä T, Penttilä ME. Efficient purification of recombinant proteins using hydrophobins as tags in surfactant-based two-phase systems. Biochemistry. 2004;43:11873–11882. doi: 10.1021/bi0488202. [DOI] [PubMed] [Google Scholar]
- 102.Smith W, Jantti J, Oja M, Saloheimo M. Comparison of intracellular and secretion-based strategies for production of human alpha-galactosidase A in the filamentous fungus Trichoderma reesei. BMC Biotechnol. 2014;14:91. doi: 10.1186/s12896-014-0091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Landowski CP, Huuskonen A, Wahl R, Westerholm-Parvinen A, Kanerva A, Hanninen AL, Salovuori N, Penttilä M, Natunen J, Ostermeier C, et al. Enabling low cost biopharmaceuticals: a systematic approach to delete proteases from a well-known protein production host Trichoderma reesei. PLoS One. 2015;10:e0134723. doi: 10.1371/journal.pone.0134723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nevalainen H, Peterson R. Making recombinant proteins in filamentous fungi—are we expecting too much? Front Microbiol. 2014;5:75. doi: 10.3389/fmicb.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Linder MB, Szilvay GR, Nakari-Setälä T, Penttilä ME. Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev. 2005;29:877–896. doi: 10.1016/j.femsre.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 106.Wösten HA, Scholtmeijer K. Applications of hydrophobins: current state and perspectives. Appl Microbiol Biotechnol. 2015;99:1587–1597. doi: 10.1007/s00253-014-6319-x. [DOI] [PubMed] [Google Scholar]
- 107.Gaderer R, Bonazza K, Seidl-Seiboth V. Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl Microbiol Biotechnol. 2014;98:4795–4803. doi: 10.1007/s00253-014-5690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thongekkaew J, Ikeda H, Masaki K, Iefuji H. Fusion of cellulose binding domain from Trichoderma reesei CBHI to Cryptococcus sp. S-2 cellulase enhances its binding affinity and its cellulolytic activity to insoluble cellulosic substrates. Enzyme Microb Technol. 2013;52:241–246. doi: 10.1016/j.enzmictec.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 109.Tang CD, Li JF, Wei XH, Min R, Gao SJ, Wang JQ, Yin X, Wu MC. Fusing a carbohydrate-binding module into the Aspergillus usamii β-mannanase to improve its thermostability and cellulose-binding capacity by in silico design. PLoS One. 2013;8:e64766. doi: 10.1371/journal.pone.0064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sugimoto N, Igarashi K, Samejima M. Cellulose affinity purification of fusion proteins tagged with fungal family 1 cellulose-binding domain. Protein Expr Purif. 2012;82:290–296. doi: 10.1016/j.pep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 111.Oliveira C, Carvalho V, Domingues L, Gama FM. Recombinant CBM-fusion technology—applications overview. Biotechnol Adv. 2015;33:358–369. doi: 10.1016/j.biotechadv.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 112.Brethauer S, Studer MH. Consolidated bioprocessing of lignocellulose by a microbial consortium. Energy Environ Sci. 2014;7:1446–1453. doi: 10.1039/c3ee41753k. [DOI] [Google Scholar]
- 113.Minty JJ, Singer ME, Scholz SA, Bae CH, Ahn JH, Foster CE, Liao JC, Lin XN. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci USA. 2013;110:14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zuroff TR, Curtis WR. Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol. 2012;93:1423–1435. doi: 10.1007/s00253-011-3762-9. [DOI] [PubMed] [Google Scholar]
- 115.Yamada R, Hasunuma T, Kondo A. Endowing non-cellulolytic microorganisms with cellulolytic activity aiming for consolidated bioprocessing. Biotechnol Adv. 2013;31:754–763. doi: 10.1016/j.biotechadv.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 116.den Haan R, van Rensburg E, Rose SH, Görgens JF, van Zyl WH. Progress and challenges in the engineering of non-cellulolytic microorganisms for consolidated bioprocessing. Curr Opin Biotechnol. 2015;33:15–22. doi: 10.1016/j.copbio.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 117.van Zyl W, den Haan R, la Grange D. Developing cellulolytic organisms for consolidated bioprocessing of lignocellulosics. In: Gupta VK, Tuohy MG, editors. Biofuel Technologies. Berlin: Springer; 2013. pp. 189–220. [Google Scholar]
- 118.Amore A, Faraco V. Potential of fungi as category I consolidated bioprocessing organisms for cellulosic ethanol production. Renew Sust Energ Rev. 2012;16:3286–3301. doi: 10.1016/j.rser.2012.02.050. [DOI] [Google Scholar]
- 119.Kroukamp H, den Haan R, van Wyk N, van Zyl WH. Overexpression of native PSE1 and SOD1 in Saccharomyces cerevisiae improved heterologous cellulase secretion. Appl Energ. 2013;102:150–156. doi: 10.1016/j.apenergy.2012.05.062. [DOI] [Google Scholar]
- 120.Inokuma K, Hasunuma T, Kondo A. Efficient yeast cell-surface display of exo- and endo-cellulase using the SED1 anchoring region and its original promoter. Biotechnol Biofuels. 2014;7:8. doi: 10.1186/1754-6834-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lambertz C, Garvey M, Klinger J, Heesel D, Klose H, Fischer R, Commandeur U. Challenges and advances in the heterologous expression of cellulolytic enzymes: a review. Biotechnol Biofuels. 2014;7:135. doi: 10.1186/s13068-014-0135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McBride J, Brevnova E, Ghandi C, Mellon M, Froehlich A, Deleault K, Rajgharia V, Flatt J, Van ZE, Den HR. Yeast expressing cellulases for simultaneous saccharification and fermentation using cellulose. 2010. International patent WO2010060056.
- 123.Xu Q, Singh A, Himmel ME. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr Opin Biotechnol. 2009;20:364–371. doi: 10.1016/j.copbio.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 124.Bischof R, Seiboth B. Molecular tools for strain improvement of Trichoderma spp. In: Gupta VG, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohy M, editors. Biotechnology and biology of Trichoderma. Oxford: Elsevier; 2014. pp. 179–191. [Google Scholar]
- 125.Rautio JJ, Smit BA, Wiebe M, Pentillä M, Saloheimo M. Transcriptional monitoring of steady state and effects of anaerobic phases in chemostat cultures of the filamentous fungus Trichoderma reesei. BMC Genom. 2006;7:247. doi: 10.1186/1471-2164-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bonaccorsi ED, Ferreira AJS, Chambergo FS, Ramos ASP, Mantovani MC, Farah JPS, Sorio CS, Gombert AK, Tonso A, El-Dorry H. Transcriptional response of the obligatory aerobe Trichoderma reesei to hypoxia and transient anoxia: implications for energy production and survival in the absence of oxygen. Biochemistry. 2006;45:3912–3924. doi: 10.1021/bi052045o. [DOI] [PubMed] [Google Scholar]
- 127.Haab D, Gassner B, Kubicek CP. Protein hypersecretory Trichoderma reesei mutant RUT C-30 displays increased ethanol and polyene resistance. J Biotechnol. 1993;29:97–108. doi: 10.1016/0168-1656(93)90043-M. [DOI] [Google Scholar]
- 128.Peciulyte A, Anasontzis GE, Karlström K, Larsson PT, Olsson L. Morphology and enzyme production of Trichoderma reesei Rut C-30 are affected by the physical and structural characteristics of cellulosic substrates. Fungal Genet Biol. 2014;72:64–72. doi: 10.1016/j.fgb.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 129.Callow NV, Ju LK. Promoting pellet growth of Trichoderma reesei Rut C30 by surfactants for easy separation and enhanced cellulase production. Enzyme Microb Technol. 2012;50:311–317. doi: 10.1016/j.enzmictec.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 130.Lynd LR, Van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 131.Steiger MG. Molecular tools in Trichoderma genetic studies. In: Mukherjee PK, Horwitz BA, Singh US, Mukherjee M, Schmoll M, editors. Trichoderma: biology and applications. Oxfordshire: CABI; 2013. pp. 128–143. [Google Scholar]
- 132.Guangtao Z, Hartl L, Schuster A, Polak S, Schmoll M, Wang T, Seidl V, Seiboth B. Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J Biotechnol. 2009;139:146–151. doi: 10.1016/j.jbiotec.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 133.Steiger MG, Vitikainen M, Uskonen P, Brunner K, Adam G, Pakula T, Penttilä M, Saloheimo M, Mach RL, Mach-Aigner AR. Transformation system for Hypocrea jecorina (Trichoderma reesei) that favors homologous integration and employs reusable bidirectionally selectable markers. Appl Environ Microbiol. 2011;77:114–121. doi: 10.1128/AEM.02100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schuster A, Bruno KS, Collett JR, Baker SE, Seiboth B, Kubicek CP, Schmoll M. A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol Biofuels. 2012;5:1. doi: 10.1186/1754-6834-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jørgensen M, Skovlund D, Johannesen P, Mortensen U. A novel platform for heterologous gene expression in Trichoderma reesei (Teleomorph Hypocrea jecorina) Microb Cell Fact. 2014;13:33. doi: 10.1186/1475-2859-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Derntl C, Kiesenhofer DP, Mach RL, Mach-Aigner AR. Novel strategies for genomic manipulation of Trichoderma reesei with the purpose of strain engineering. Appl Environ Microbiol. 2015;81:6314–6323. doi: 10.1128/AEM.01545-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ouedraogo JP, Arentshorst M, Nikolaev I, Barends S, Ram AF. I-SceI-mediated double-strand DNA breaks stimulate efficient gene targeting in the industrial fungus Trichoderma reesei. Appl Microbiol Biotechnol. 2015;99:10083–10095. doi: 10.1007/s00253-015-6829-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ouedraogo JP, Arentshorst M, Nikolaev I, Barends S, Ram AF. I-SceI enzyme mediated integration (SEMI) for fast and efficient gene targeting in Trichoderma reesei. J Biotechnol. 2016;222:25–28. doi: 10.1016/j.jbiotec.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 139.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu R, Chen L, Jiang Y, Zhou Z, Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015;1:15007. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nødvig CS, Nielsen JB, Kogle ME, Mortensen UH. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One. 2015;10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Banerjee G, Car S, Scott-Craig JS, Borrusch MS, Walton JD. Rapid optimization of enzyme mixtures for deconstruction of diverse pretreatment/biomass feedstock combinations. Biotechnol Biofuels. 2010;3:22. doi: 10.1186/1754-6834-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dettmann A, Heilig Y, Valerius O, Ludwig S, Seiler S. Fungal communication requires the MAK-2 pathway elements STE-20 and RAS-2, the NRC-1 adapter STE-50 and the MAP kinase scaffold HAM-5. PLoS Genet. 2014;10:e1004762. doi: 10.1371/journal.pgen.1004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jonkers W, Leeder AC, Ansong C, Wang Y, Yang F, Starr TL, Camp DG, 2nd, Smith RD, Glass NL. HAM-5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa. PLoS Genet. 2014;10:e1004783. doi: 10.1371/journal.pgen.1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chuang YC, Li WC, Chen CL, Hsu PW, Tung SY, Kuo HC, Schmoll M, Wang TF. Trichoderma reesei meiosis generates segmentally aneuploid progeny with higher xylanase-producing capability. Biotechnol Biofuels. 2015;8:30. doi: 10.1186/s13068-015-0202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nitta M, Furukawa T, Shida Y, Mori K, Kuhara S, Morikawa Y, Ogasawara W. A new Zn(II)2Cys6-type transcription factor BglR regulates β-glucosidase expression in Trichoderma reesei. Fungal Genet Biol. 2012;49:388–397. doi: 10.1016/j.fgb.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 147.Pomraning KR, Smith KM, Freitag M. Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1. Eukaryot Cell. 2011;10:724–733. doi: 10.1128/EC.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Niu J, Arentshorst M, Nair PDS, Dai Z, Baker SE, Frisvad JC, Nielsen KF, Punt PJ, Ram AFJ. Identification of a classical mutant in the industrial host Aspergillus niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 (Bethesda) 2016;6:193–204. doi: 10.1534/g3.115.024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nowrousian M, Teichert I, Masloff S, Kück U. Whole-genome sequencing of Sordaria macrospora mutants identifies developmental genes. G3 (Bethesda) 2012;2:261–270. doi: 10.1534/g3.111.001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bok JW, Wiemann P, Garvey GS, Lim FY, Haas B, Wortman J, Keller NP. Illumina identification of RsrA, a conserved C2H2 transcription factor coordinating the NapA mediated oxidative stress signaling pathway in Aspergillus. BMC Genom. 2014;15:1011. doi: 10.1186/1471-2164-15-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hasunuma T, Hori Y, Sakamoto T, Ochiai M, Hatanaka H, Kondo A. Development of a GIN11/FRT-based multiple-gene integration technique affording inhibitor-tolerant, hemicellulolytic, xylose-utilizing abilities to industrial Saccharomyces cerevisiae strains for ethanol production from undetoxified lignocellulosic hemicelluloses. Microb Cell Fact. 2014;13:145. doi: 10.1186/s12934-014-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cherry JR, Fidantsef AL. Directed evolution of industrial enzymes: an update. Curr Opin Biotechnol. 2003;14:438–443. doi: 10.1016/S0958-1669(03)00099-5. [DOI] [PubMed] [Google Scholar]
- 153.Freier D, Mothershed CP, Wiegel J. Characterization of Clostridium thermocellum JW20. Appl Environ Microbiol. 1988;54:204–211. doi: 10.1128/aem.54.1.204-211.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Goshima T, Negi K, Tsuji M, Inoue H, Yano S, Hoshino T, Matsushika A. Ethanol fermentation from xylose by metabolically engineered strains of Kluyveromyces marxianus. J Biosci Bioeng. 2013;116:551–554. doi: 10.1016/j.jbiosc.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 155.Rodrussamee N, Lertwattanasakul N, Hirata K, Suprayogi, Limtong S, Kosaka T, Yamada M. Growth and ethanol fermentation ability on hexose and pentose sugars and glucose effect under various conditions in thermotolerant yeast Kluyveromyces marxianus. Appl Microbiol Biotechnol. 2011;90:1573–1586. doi: 10.1007/s00253-011-3218-2. [DOI] [PubMed] [Google Scholar]
- 156.Kubicek CP. Lignocellulose biorefinery. Hoboken: Wiley-Blackwell; 2013. [Google Scholar]
- 157.Maehara T, Ichinose H, Furukawa T, Ogasawara W, Takabatake K, Kaneko S. Ethanol production from high cellulose concentration by the basidiomycete fungus Flammulina velutipes. Fungal Biol. 2013;117:220–226. doi: 10.1016/j.funbio.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 158.El-Batal AI, Farahat LM, El-Rehim HA. Ethanol production by Kluyveromyces lactis immobilized cells in copolymer carriers produced by radiation polymerization. Acta Microbiol Pol. 2000;49:157–166. [PubMed] [Google Scholar]
- 159.Ohta K, Beall DS, Mejia JP, Shanmugam KT, Ingram LO. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991;57:893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fujita Y, Takahashi S, Ueda M, Tanaka A, Okada H, Morikawa Y, Kawaguchi T, Arai M, Fukuda H, Kondo A. Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Appl Environ Microbiol. 2002;68:5136–5141. doi: 10.1128/AEM.68.10.5136-5141.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Katahira S, Fujita Y, Mizuike A, Fukuda H, Kondo A. Construction of a xylan-fermenting yeast strain through codisplay of xylanolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. Appl Environ Microbiol. 2004;70:5407–5414. doi: 10.1128/AEM.70.9.5407-5414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fujita Y, Ito J, Ueda M, Fukuda H, Kondo A. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl Environ Microbiol. 2004;70:1207–1212. doi: 10.1128/AEM.70.2.1207-1212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Den Haan R, McBride JE, Grange DCL, Lynd LR, Van Zyl WH. Functional expression of cellobiohydrolases in Saccharomyces cerevisiae towards one-step conversion of cellulose to ethanol. Enzyme Microb Technol. 2007;40:1291–1299. doi: 10.1016/j.enzmictec.2006.09.022. [DOI] [Google Scholar]
- 164.Yamada R, Taniguchi N, Tanaka T, Ogino C, Fukuda H, Kondo A. Direct ethanol production from cellulosic materials using a diploid strain of Saccharomyces cerevisiae with optimized cellulase expression. Biotechnol Biofuels. 2011;4:1–8. doi: 10.1186/1754-6834-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ilmén M, den Haan R, Brevnova E, McBride J, Wiswall E, Froehlich A, Koivula A, Voutilainen SP, Siika-aho M, la Grange DC, et al. High level secretion of cellobiohydrolases by Saccharomyces cerevisiae. Biotechnol Biofuels. 2011;4:1–15. doi: 10.1186/1754-6834-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nakatani Y, Yamada R, Ogino C, Kondo A. Synergetic effect of yeast cell-surface expression of cellulase and expansin-like protein on direct ethanol production from cellulose. Microb Cell Fact. 2013;12:66. doi: 10.1186/1475-2859-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Matano Y, Hasunuma T, Kondo A. Simultaneous improvement of saccharification and ethanol production from crystalline cellulose by alleviation of irreversible adsorption of cellulase with a cell surface-engineered yeast strain. Appl Microbiol Biotechnol. 2013;97:2231–2237. doi: 10.1007/s00253-012-4587-x. [DOI] [PubMed] [Google Scholar]
- 168.Liu Z, Inokuma K, Ho S-H, Haan Rd, Hasunuma T, Zyl WH, Kondo A. Combined cell-surface display- and secretion-based strategies for production of cellulosic ethanol with Saccharomyces cerevisiae. Biotechnol Biofuels. 2015;8:1–12. doi: 10.1186/s13068-014-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tao L, Dong HJ, Chen X, Chen SF, Wang TH. Expression of ethylene-forming enzyme (EFE) of Pseudomonas syringae pv. glycinea in Trichoderma viride. Appl Microbiol Biotechnol. 2008;80:573–578. doi: 10.1007/s00253-008-1562-7. [DOI] [PubMed] [Google Scholar]
- 170.Chen X, Liang Y, Hua J, Tao L, Qin W, Chen S. Overexpression of bacterial ethylene-forming enzyme gene in Trichoderma reesei enhanced the production of ethylene. Int J Biol Sci. 2010;6:96–106. doi: 10.7150/ijbs.6.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Steiger MG, Mach-Aigner AR, Gorsche R, Rosenberg EE, Mihovilovic MD, Mach RL. Synthesis of an antiviral drug precursor from chitin using a saprophyte as a whole-cell catalyst. Microb Cell Fact. 2011;10:1. doi: 10.1186/1475-2859-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dashtban M, Kepka G, Seiboth B, Qin W. Xylitol production by genetically engineered Trichoderma reesei strains using barley straw as feedstock. Appl Biochem Biotechnol. 2013;169:554–569. doi: 10.1007/s12010-012-0008-y. [DOI] [PubMed] [Google Scholar]
- 173.Hong Y, Dashtban M, Kepka G, Chen S, Qin W. Overexpression of d-xylose reductase (xyl1) gene and antisense inhibition of d-xylulokinase (xyiH) gene increase xylitol production in Trichoderma reesei. BioMed Res Int. 2014;2014:8. doi: 10.1155/2014/169705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jovanović B, Mach RL, Mach-Aigner AR. Erythritol production on wheat straw using Trichoderma reesei. AMB Expr. 2014;4:1–12. doi: 10.1186/s13568-014-0034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


