Abstract
Background
Wnt proteins are secreted signalling molecules found in all animal phyla. In bilaterian animals, including humans, Wnt proteins play key roles in development, maintenance of homeostasis and regeneration. While Wnt gene repertoires and roles are strongly conserved between cnidarians and bilaterians, Wnt genes from basal metazoans (sponges, ctenophores, placozoans) are difficult or impossible to assign to the bilaterian + cnidarian orthologous groups. Moreover, dramatic differences in Wnt numbers among basal metazoan exist, with only three present in the genome of Amphimedon queenslandica, a demosponge, and 21 in the genome of Sycon ciliatum, a calcisponge. To gain insight into the ancestral Wnt repertoire and function, we have chosen to investigate Wnt genes in Halisarca dujardini, a demosponge with relatively well described development and regeneration, and a very distant phylogenetic relationship to Amphimedon.
Results
Here we describe generation of a eukaryotic contamination-free transcriptome of Halisarca dujardini, and analysis of Wnt genes repertoire and expression in this species. We have identified ten Wnt genes, with only one orthologous to Amphimedon Wnt, and six appearing to be a result of a lineage specific expansion. Expression analysis carried out by in situ hybridization of adults and larvae revealed that two Halisarca Wnts are expressed in nested domains in the posterior half of the larvae, and six along the adult body axis, with two specific to the osculum. Strikingly, expression of one of the Wnt genes was elevated in the region undergoing regeneration.
Conclusions
Our results demonstrated that the three Poriferan lineages (Demospongiae, Calcarea and Homoloscleromorpha) are characterized by highly diverse Wnt gene repertoires which do not display higher similarity to each other than they do to the non-sponge (i.e. ctenophore, cnidarian and bilaterian) repertoires. This is in striking contrast to the uniform Wnt repertoires in Cnidarians and Bilaterians, suggesting that the Wnt family composition became “fixed” only in the last common ancestor of Cnidarians and Bilaterians. In contrast, expression of Wnt genes in the apical region of sponge adults and the posterior region of sponge larvae suggests conservation of the Wnt role in axial patterning across the animal kingdom.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-016-0700-6) contains supplementary material, which is available to authorized users.
Background
Wnt genes encode secreted glycoproteins acting as signalling molecules to direct cell proliferation, migration, differentiation and survival during animal development, maintenance of homeostasis and regeneration [1–6]. While some Wnt pathway components have been identified outside of the animal kingdom, Wnt genes themselves are a conserved metazoan innovation [7, 8].
Representatives of the Wnt family have been identified in all animals studied so far, including so-called “basally branching” or non-bilaterian clades: cnidarians [9, 10], placozoans [11], ctenophores [12, 13] and sponges [14–18]. Wnt repertoires are surprisingly conserved between cnidarians and bilaterians, with 12 of 13 bilaterian orthologs present in the sea anemone, Nematostella vectensis [9]. This conservation appears to extend to function, as demonstrated by involvement of Wnts in segregation of germ layers during gastrulation, in embryonic and adult axial patterning and in restoration of lost body parts in both cnidarians and bilaterians [19–22]. Conservation of the blastoporal Wnt expression in cnidarians and chordates is particularly striking [5]. In cnidarian polyps such as Hydra, this expression persists in the oral region located in the apical part of the adult body [10]. In line with this, over-activation of Wnt signalling in Hydra results in formation of additional structures with head identity [23]. In chordates, where the blastopore gives rise to the anus, Wnt expression and activity confers posterior identity to developing structures [24, 25]. In line with this, over-activation of the Wnt signalling in vertebrates results in loss of anterior structures, while loss of Wnt function results in posterior truncation [26, 27].
Wnt genes identified in placozoans, ctenophores and sponges are difficult or impossible to assign to the bilaterian + cnidarian orthologous groups [12, 16–18]. Yet, Wnt expression in ctenophores and sponges is consistent with conserved involvement in axial patterning [13, 15, 18]. In particular, Wnt genes have been found to be expressed in the larval posterior pole of two major sponge model species: the demosponge Amphimedon queenslandica [15] and the calcisponge Sycon ciliatum [18]. In addition, Wnt expression is associated with osculum (the major exhalant opening of adult sponges, located at the apical pole) of Sycon ciliatum [18]. Such expression is consistent with homology of the larval and adult body axes between sponges and cnidarians, supporting homologous relationship between the cnidarian mouth and the sponge osculum [18, 28, 29].
While Wnt expression in adult demosponges has not been reported, pharmacological over-activation of the Wnt pathway in a freshwater species, Ephydatia mulleri, resulted in multiplication of the body axis, as evidenced by formation of multiple oscula [30]. This outcome is strikingly similar to Wnt over-activation experiments in cnidarians, resulting in formation of ectopic head structures [23]. Moreover, experiments involving transplantation of oscula demonstrated their organizer properties, in line with organizer properties of the cnidarian head, and animal blastopores in general [10, 31].
Halisarca dujardini (Chondrillida) is a marine demosponge which is very distantly related to Amphimedon queenslandica (Haplosclerida) [32, 33]. Halisarca embryonic development, metamorphosis and regeneration are well described at morphological level [34–36], but sequence resources have been lacking. Here we report generation of a transcriptome dataset and identification of a surprisingly rich Wnt repertoire (ten genes, in contrast to only three present in the genome of Amphimedon). Two of these genes are expressed in nested domains in the posterior half of the larvae, and six along the adult body axis, with two specific to the osculum. Moreover, Wnt expression is elevated in the region undergoing regeneration, suggesting conservation of the Wnt role in axial patterning and restoration of lost body parts across the animal kingdom.
Methods
Samples
No permits were required to collect sponge specimens in Norwegian waters. Total RNA and gDNA were isolated from wild-collected adult sponges and several hundred larvae freshly released in laboratory conditions. To avoid eukaryotic contaminations, the larvae were washed in sterile-filtered sea water and visually inspected under dissecting microscope. Nucleic acids were isolated using Allprep Mini kit (Qiagen) following manufacturer’s instructions, and the RNA yield and quality were determined using the NanoDrop spectrophotometer (Thermo Scientific) and the Agilent 2100 BioAnalyzer RNA 6000 Nano chip (Agilent Technologies).
Sequencing
Two RNA-Seq libraries were prepared using Illumina TruSeq RNA Library Prep Kit: one from the wild-collected adult specimen and another one from eukaryotic contaminations-free larvae. An additional gDNA library was prepared from the same larvae using Illumina TruSeq DNA Library Prep Kit. The libraries were paired-end sequenced on Illumina HiSeq 2000 with read length of 100.
Transcriptome assembly
The transcriptome was assembled de-novo from the two RNA-Seq libraries. The assembly was performed with Trinity 2.1.1 [37] including reads’ pre-processing with Trimmomatic [38]. We have modified Trinity’s final step called Butterfly to use read pairing information: The fasta sequence files prepared for Butterfly runs were supplemented to include both ends for all the fragments (missing-pair reads were added) and option ‘run_as_paired’ was added to all Butterfly commands. Assembled transcriptome was screened to exclude eukaryotic contaminations by aligning reads from the clean juvenile gDNA library. The alignments were done using bowtie with default parameters. Transcriptome contigs not aligned to any of the clean read were removed from the assembly. Assembly was screened for sequencing vectors using blastn against UniVec database. Transcripts of Wnt ligands were identified by sequence homology using tblastn and Wnt proteins from other organisms and are available in TSA under ids HADA01000001 – HADA01000010.
Phylogenetic analysis
Wnt protein sequences were aligned with Mafft v7.123 using option L-INS-i. Alignment was then manually trimmed to remove poorly aligned and divergent regions. Phylogenetic tree was built using Mr Bayes 3.1 [39] which we modified to incorporate the LG model (as LG was selected as best fit substitution model by ProTest 3) [40]. Mr Bayes was run with two sets of 4 Markov chains each, till standard deviation of split frequencies dropped below 0.01.
In situ hybridization
In situ hybridization has been carried as described for Sycon ciliatum [41], except that proteinase treatment was 10 min at 37 °C.
Results and discussion
Ten Wnt genes are present in Halisarca dujardini
We have generated transcriptome dataset for Halisarca dujardini representing genes expressed in adult specimens and free-swimming larvae (see Methods for details). Using a variety of sponge, cnidarian and bilaterian sequences we have BLAST-searched this dataset for Wnt genes and recovered ten complete coding protein sequences (Additional file 1). This stands in contrast with only three Wnt genes present in another demosponge, Amphimedon queenslandica, and also differs from the number of 21 genes identified in Sycon ciliatum (Calcarea, Calcaronea) [16, 18]. For comparison, at least eight Wnt genes are present in Homoscleromorph sponges, e.g. Oscarella sp. [18].
We next wanted to know whether the ten newly identified Halisarca Wnts are orthologous to other sponge (or other metazoan) Wnt genes. We have thus carried out Bayesian analysis adding these new sequences to the previously constructed comprehensive Wnt sequence dataset [18]. Surprisingly, only one Halisarca sequence appeared to be in orthologous relationship with previously described Wnt genes, namely the Amphimedon WntC sequence, while no AmqWntA or AmqWntB orthologues could be identified in Halisarca (Fig. 1 and Additional file 2). We have named this gene HduWntC, and the remaining nine sequences HduWntD-WntL according to the order in which they were identified. Of these, six clustered together in our analysis with high support, suggesting they are likely a result of independent subfamily expansion in the Halisarca lineage (Fig. 1). Another pair of Halisarca Wnts was associated with Sycon ciliatum WntS, although with very weak support, and the last one showed no particular affinity to any of previously identified Wnt genes (Fig. 1).
Fig. 1.
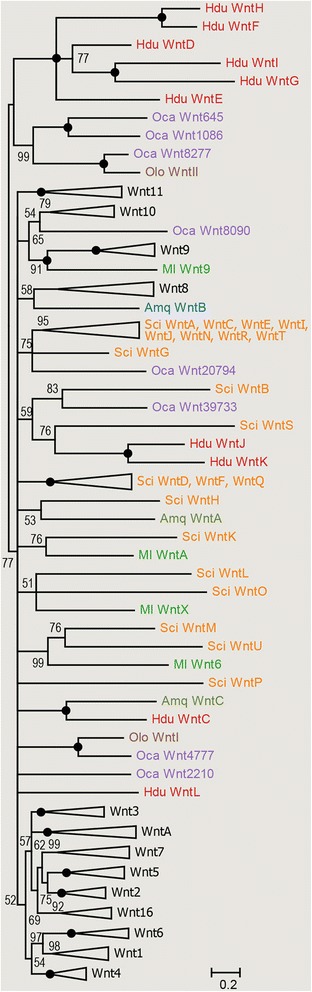
Bayesian inference gene tree of Wnt ligands. The values at the tree nodes are posterior probabilities for each split defined over the range [0, 100]. Black circles denote 100 % support (posterior probability of 1.00). Species acronyms: Amq, Amphimedon queenslandica, Hdu, Halisarca dujardini, Ml, Mnemiopsis leidyi, Nv, Nematostella vectensis, Oca, Oscarella carmela, Olo, Oscarella lobularis, Sci, Sycon ciliatum. Sub-trees not containing sponge sequences were collapsed; the complete tree is available as Additional file 2
Thus, all so-far studied sponges, representing three Poriferan lineages (Demospongiae, Calcarea and Homoloscleromorpha) are characterized by highly diverse Wnt gene repertoires which do not display higher similarity to each other than they do to the non-sponge (i.e. ctenophore, cnidarian and bilaterian) repertoires. This is in striking contrast to the uniform Wnt repertoires in Cnidarians and Bilaterians, suggesting that the Wnt family composition became “fixed” only in the last common ancestor of Cnidarians and Bilaterians.
Halisarca Wnts are expressed along the adult and larval axes and during regeneration
In ctenophores, cnidarians and calcareous sponges Wnt genes are expressed along the major (oral-aboral or apical-basal) body axis in sets of nestled domains, suggesting existence of a “Wnt code” possibly conveying positional information [4, 9, 10, 13, 18]. In Oscarella lobularis, a homoscleromorph sponge, two Wnt genes are expressed in a complementary fashion with domains in the ostia (multiple openings in the inhalant canals on the surface of the body) and in exopinacoderm surrounding the ostia of adult specimens [17]. It is important to note here that at least six other Wnt genes are present in Oscarella sp. [18], expression of which has not been reported so far.
We have attempted cloning and expression analysis of all ten Halisarca Wnts. Of these, six genes were expressed in the adult specimens in four unique patterns:
WntD (Fig. 2b) and WntE (Fig. 2c) at the tip of the osculum; WntG (Fig. 2d) throughout the entire exopinacoderm, with particularly high concentration of positive cells in the osculum; WntF (Fig. 2e) and WntH (Fig. 2f, f’) in the peripheral exopinacoderm and basopinacoderm; and finally WntK (Fig. 2g, see also Fig. 2l) was prominently expressed within the oscular chimney, and weakly throughout the exopinacoderm.
Fig. 2.
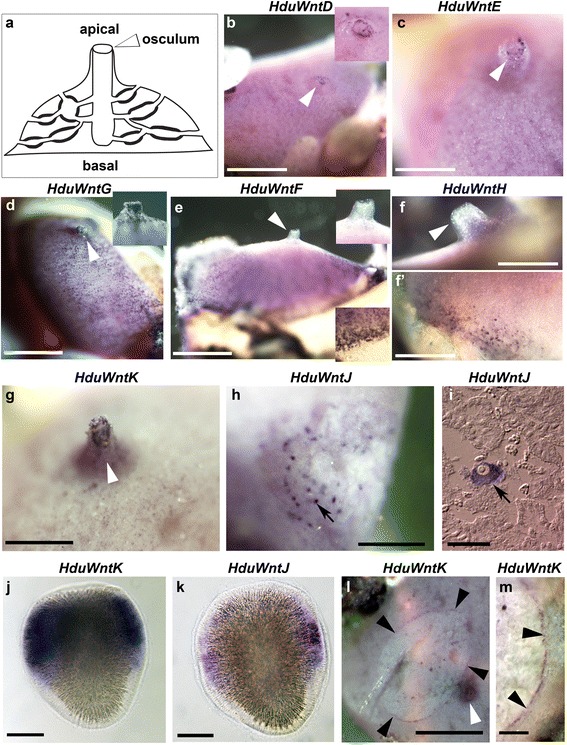
Expression of Wnt genes in Halisarca dujardini. a schematic representation of the adult body plan of Halisarca, apical and basal regions and the osculum are labelled; b, c, HduWntD and HduWntE transcripts are localized around the osculum; d, HduWntG transcripts are present throughout the exopinacoderm and particularly in the oscular chimney; e, f, f’, HduWntF and HduWntH transcripts are absent from the osculum and the apical region, but strong along the base; g, HduWntK transcripts are present along the oscular chimney; h and i, HduWntJ transcripts are present in the oocytes; j, HduWntK transcripts are present in the posterior half of the larva except the polar region; l, HduWntJ transcripts are present in cells distributed along the larval equator; l and m, HduWntK transcripts are conspicuously present along the wound margin. White arrowheads indicate the osculum; insets in the upper and lower corners are enlargements of the apical and basal regions, respectively, black arrowheads indicate wound margin; black arrows indicate oocytes; posterior pole of the larvae is towards the top. Note that the specific staining is dark purple, while the uniform pink coloration of some samples is background staining. Scale bars: b, d, e – 5 mm; c – 2.5 mm; f, f’ – 2 mm; g, h, l, m – 3 mm; i – 30 μm; j, k – 50 μm
Detection of WntJ expression revealed positive large cells within the mesohyl of some specimens, which were identified as young oocytes upon sectioning (Fig. 2h, i). While during our collections we have not found any specimens with embryos, we have been able to carry out in situ hybridization on larvae released from adults briefly maintained in laboratory conditions. Two Halisarca Wnt genes revealed robust expression in the larvae: WntK (Fig. 2j) throughout most of the posterior hemisphere, except of the polar cells themselves, and WntJ in a band of equatorial cells (Fig. 2k).
Thus, the identified Wnt expression domains encompass the entire apical-basal axis of the adult Halisarca body, with majority of the genes expressed uniquely or predominantly in the osculum. At the same time, Wnt expression is associated with the posterior region of the larvae. These nested patterns, and the prevalence of apical and posterior expression are consistent with the postulated conservation of Wnt role in axial patterning throughout the metazoans.
In addition to the conserved role in axial patterning, Wnt genes are also known to be involved in wound healing and regeneration in many animal lineages [3, 6, 10, 19–22]. We have recently described cellular processes leading to regeneration of the ectosome in H. dujardini [36], and we wanted to know whether Wnt genes might be involved in these processes. While majority of the Wnt genes did not display detectable expression changes in the regeneration zone, HduWntK expression was prominent in the exopinacocytes surrounding the wound at 12 h after wounding (Fig. 2l, m). These cells are actively involved in the regeneration, as they temporarily dedifferentiate, phagocyte the debris and contribute to restoration of the ectosome by migration and re-differentiation [36]. Thus, as in other animal lineages, the demosponge Wnt pathway is implicated in the regeneration processes.
Conclusions
Transcriptome sequencing of Halisarca dujardini allowed us the first insight into gene repertoire of a demosponge from a previously unexplored order. We have identified ten Wnt genes, nine of which are without orthologs in any previously reported species. While the diversity of Wnt subfamilies is striking, expression of the identified genes suggests conservation of roles in axial patterning and regeneration. We wonder what mechanisms are responsible – or permissive – for the apparent lack of constraints on Wnt protein sequences in sponges (as well as ctenophores and possibly placozoans) as opposed to cnidarians and bilaterians.
Abbreviations
Amq, Amphimedon queenslandica; BLAST, Basic Local Alignment Search Tool; ENA, European Nucleotide Archive; gDNA, genomic deoxyribonucleic acid; Hdu, Halisarca dujardini; LG model, Le-Gascuel substitution model; Ml, Mnemiopsis leidyi; Nv, Nematostella vectensis; Oca, Oscarella carmela; Olo, Oscarella lobularis; RNA, ribonucleic acid; Sci, Sycon ciliatum; SRA, Sequence Read Archive; TSA, Transcriptome Shotgun Assembly.
Acknowledgments
We thank Sven Leininger for providing adult Halisarca RNA. We thank the “Molecular and cell technologies” research resource center of Saint-Petersburg State University for technical support. The authors are grateful to Dr. A. A. Sukhotin, the director of the White Sea Biological Station of the Zoological Institute of the Russian Academy of Sciences, and The Sven Lovén Centre for Marine Sciences for providing facilities for field research.
Funding
We acknowledge funding from the Sars International Centre for Marine Molecular Biology to M. Adamska and grants from the Russian Foundation for Basic Research (n° 13-04-01084 and 16-04-00084), research grant № 1.38.209.2014 awarded by Saint-Petersburg State University and European Community grant ASSEMBLE, agreement no. 227799. I. Borisenko was supported by funds from the Master student research training program at the Sars Centre in 2010 and 2011, and a fellowship of French Government for master students in 2013.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the ENA-SRA and ENA-TSA repositories. Reads were deposited in under study ERP013253, biosamples: SAMEA3672117 for RNA-Seq from larvae, SAMEA3672118 for the RNA-Seq from the adult specimen and SAMEA3672119 for gDNA from larvae. Final transcriptome was deposited in ENA-TSA database under accession numbers HADA01000001 – HADA01138992 http://www.ebi.ac.uk/ena/data/view/HADA01000001-HADA01138992.
Authors’ contributions
I.B. carried out database searches and laboratory work and participated in data analysis. Mar. A. participated in sample collection and carried out sequence assembly and phylogenetic analyses. A.E. participated in study design and data analysis. Maj. A. participated in study design, sample collection and data analysis and drafted the manuscript. All authors contributed to the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent to publish
Not applicable.
Ethical approval and consent to participate
Not applicable.
Additional files
wnt.alignment.nex.txt: trimmed alignment of Wnt protein sequences used to generate Bayesian inference trees shown in Additional file 1 and Fig. 1. Species: Amq, Amphimedon queenslandica, Ate, Achaearanea (Parasteatoda) tepidariorum, Bf, Branchiostoma floridae, Cte, Capitella teleta, Hdu, Halisarca dujardini, Hs, Homo sapiens, Lgi, Lottia gigantea, Ml, Mnemiopsis leidyi, Nv, Nematostella vectensis, Oca, Oscarella carmela, Olo, Oscarella lobularis, Sci, Sycon ciliatum, Sko, Saccoglossus kowalewski, Spu, Strongylocentrotus purpuratus, Tc, Tribolium castaneum. (TXT 39 kb)
wnt.tree.nwk.txt: Newick format Bayesian inference gene tree of Wnt ligands. Species: Amq, Amphimedon queenslandica, Ate, Achaearanea (Parasteatoda) tepidariorum, Bf, Branchiostoma floridae, Cte, Capitella teleta, Hdu, Halisarca dujardini, Hs, Homo sapiens, Lgi, Lottia gigantea, Ml, Mnemiopsis leidyi, Nv, Nematostella vectensis, Oca, Oscarella carmela, Olo, Oscarella lobularis, Sci, Sycon ciliatum, Sko, Saccoglossus kowalewski, Spu, Strongylocentrotus purpuratus, Tc, Tribolium castaneum. (TXT 3 kb)
References
- 1.Willert K, Nusse R. Wnt proteins. Cold Spring Harb Perspect Biol. 2012;4:a007864. doi: 10.1101/cshperspect.a007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 3.De Robertis EM. Wnt signaling in axial patterning and regeneration: lessons from planaria. Sci Signal. 2010;3(127):e21. doi: 10.1126/scisignal.3127pe21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guder C, Philipp I, Lengfeld T, Watanabe H, Hobmayer B, Holstein TW. The Wnt code: cnidarians signal the way. Oncogene. 2006;25(57):7450–60. doi: 10.1038/sj.onc.1210052. [DOI] [PubMed] [Google Scholar]
- 5.Petersen CP, Reddien PW. Wnt Signaling and the Polarity of the Primary Body Axis. Cell. 2009;139:1056–68. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 7.Holstein TW. The evolution of the Wnt pathway. Cold Spring Harb Perspect Biol. 2012;4(7):a007922. doi: 10.1101/cshperspect.a007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adamska M. Developmental signalling and emergence of animal multicellularity. In: Ruiz-Trillo I, Nedelcu AM, editorss. Evolutionary Transitions to Multicellular Life, Advances in Marine Genomics 2, Springer Science + Business Media. ISBN: 978-94-017-9641-5.
- 9.Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–60. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 10.Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol. 2009;330:186–99. doi: 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, Signorovitch AY, Moreno MA, Kamm K, Grimwood J, Schmutz J, Shapiro H, Grigoriev IV, Buss LW, Schierwater B, Dellaporta SL, Rokhsar DS. The Trichoplax genome and the nature of placozoans. Nature. 2008;454(7207):955–60. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 12.Pang K, Ryan JF, Comparative Sequencing Program NISC, Mullikin JC, Baxevanis AD, Martindale MQ. Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi. Evodevo. 2010;1:10. doi: 10.1186/2041-9139-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jager M, Dayraud C, Mialot A, Quéinnec E, le Guyader H, Manuel M. Evidence for involvement of Wnt signalling in body polarities, cell proliferation, and the neuro-sensory system in an adult ctenophore. PLoS One. 2013;8:e84363. doi: 10.1371/journal.pone.0084363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci U S A. 2006;103(33):12451–6. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamska M, Degnan SM, Green KM, Adamski M, Craigie A, Larroux C, Degnan BM. Wnt and TGF-beta expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. P. L. o. S. One. 2007;2:e1031. doi: 10.1371/journal.pone.0001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamska M, Larroux C, Adamski M, Green K, Lovas E, Koop D, Richards GS, Zwafink C, Degnan BM. Structure and expression of conserved Wnt pathway components in the demosponge Amphimedon queenslandica. Evol Dev. 2010;12(5):494–518. doi: 10.1111/j.1525-142X.2010.00435.x. [DOI] [PubMed] [Google Scholar]
- 17.Lapébie P, Gazave E, Ereskovsky A, Derelle R, Bézac C, Renard E, Houliston E, Borchiellini C. WNT/beta-catenin signalling and epithelial patterning in the homoscleromorph sponge Oscarella. PLoS One. 2009;4:e5823. doi: 10.1371/journal.pone.0005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leininger S, Adamski M, Bergum B, Guder C, Liu J, Laplante M, Bråte J, Hoffmann F, Fortunato S, Jordal S, Rapp HT, Adamska M. Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nature Comm. 2014;5:3905. doi: 10.1038/ncomms4905. [DOI] [PubMed] [Google Scholar]
- 19.Whyte JL, Smith AA, Helms JA. Wnt signaling and injury repair. Cold Spring Harb Perspect Biol. 2012;4(8):a008078. doi: 10.1101/cshperspect.a008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chera S, Ghila L, Dobretz K, Wenger Y, Bauer C, Buzgariu W, Martinou J-C, Galliot B. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive Hydra head regeneration. Dev Cell. 2009;17:279–89. doi: 10.1016/j.devcel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Trevino M, Stefanik DJ, Rodriguez R, Harmon S, Burton PM. Induction of canonical Wnt signaling by alsterpaullone is sufficient for oral tissue fate during regeneration and embryogenesis in Nematostella vectensis. Dev Dyn. 2011;240:2673–9. doi: 10.1002/dvdy.22774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DuBuc TQ, Nikki Traylor-Knowles N, Martindale MQ. Initiating a regenerative response; cellular and molecular features of wound healing in the cnidarian Nematostella vectensis. BMC Biol. 2014;12:24. doi: 10.1186/1741-7007-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broun M, Gee L, Reinhardt B, Bode HR. Formation of the head organizer in hydra involves the canonical Wnt pathway. Development. 2005;132(12):2907–16. doi: 10.1242/dev.01848. [DOI] [PubMed] [Google Scholar]
- 24.Holland LZ, Holland NN, Schubert M. Developmental expression of AmphiWnt1, an amphioxus gene in the Wnt1/wingless subfamily. Dev Genes Evol. 2000;210:522–4. doi: 10.1007/s004270000089. [DOI] [PubMed] [Google Scholar]
- 25.Onai T, Lin HC, Schubert M, Koop D, Osborne PW, Alvarez S, Alvarez R, Holland ND, Holland LZ. Retinoic acid and Wnt/beta-catenin have complementary roles in anterior/posterior patterning embryos of the basal chordate amphioxus. Dev Biol. 2009;332:223–33. doi: 10.1016/j.ydbio.2009.05.571. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C, Chen L, Tsukui T, Gomer L, Dorward DW, Glinka A, Grinberg A, Huang SP, Niehrs C, Izpisúa Belmonte JC, Westphal H. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell. 2001;1:423–34. doi: 10.1016/S1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- 27.Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407(6803):535–8. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 28.Haeckel E. On the organization of sponges and their relationship to the corals. Ann Mag Nat Hist. 1870;5(1–13):107–20. doi: 10.1080/00222937008696118. [DOI] [Google Scholar]
- 29.Adamska M. Sponges as the Rosetta Stone of Colonial-to-Multicellular Transition. In: Niklas KJ, Newman SA, editors. Multicellularity. Origins and Evolution. Cambridge, Massachusetts; London, England: The MIT Press; 2016. [Google Scholar]
- 30.Windsor PJ, Leys SP. Wnt signaling and induction in the sponge aquiferous system: evidence for an ancient origin of the organizer. Evol Dev. 2010;12:481–590. doi: 10.1111/j.1525-142X.2010.00434.x. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert SF. Developmental Biology. 6th edition. Sunderland: Sinauer Associates; 2000. Axis Formation in Amphibians: The Phenomenon of the Organizer. Available from: http://www.ncbi.nlm.nih.gov/books/NBK10101/.
- 32.Ereskovsky AV, Lavrov DV, Boury-Esnault N, Vacelet J. Molecular and morphological description of a new species of Halisarca (Demospongiae: Halisarcida) from Mediterranean Sea with redescription of the type species Halisarca dujardini. Zootaxa. 2011;2768:5–31. [Google Scholar]
- 33.Morrow C, Cárdenas P. Proposal for a revised classification of the Demospongiae (Porifera) Front Zool. 2015;12:7. doi: 10.1186/s12983-015-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ereskovsky AV, Gonobobleva EL. New data on embryonic development of Halisarca dujardini Johnston, 1842 (Demospongiae: Halisarcida) Zoosystema. 2000;22:355–68. [Google Scholar]
- 35.Gonobobleva EL, Ereskovsky AV. Metamorphosis of the larva of Halisarca dujardini (Demospongiae, Halisarcida). Bull.inst. Roy. Sci. nat. Belgique. Biol. 2004;74:101–15. [Google Scholar]
- 36.Borisenko IE, Adamska M, Tokina DB, Ereskovsky AV. Transdifferentiation is a driving force of regeneration in Halisarca dujardini (Demospongiae, Porifera) PeerJ. 2015;3:e1211. doi: 10.7717/peerj.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 40.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fortunato S, Adamski M, Bergum B, Guder C, Jordal S, Leininger S, Zwafink C, Rapp HT, Adamska M. Genome-wide analysis of the Sox family in the calcareous sponge Sycon ciliatum: multiple genes with unique expression patterns. EvoDevo. 2012;3:14. doi: 10.1186/2041-9139-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the ENA-SRA and ENA-TSA repositories. Reads were deposited in under study ERP013253, biosamples: SAMEA3672117 for RNA-Seq from larvae, SAMEA3672118 for the RNA-Seq from the adult specimen and SAMEA3672119 for gDNA from larvae. Final transcriptome was deposited in ENA-TSA database under accession numbers HADA01000001 – HADA01138992 http://www.ebi.ac.uk/ena/data/view/HADA01000001-HADA01138992.


