Abstract
Background
Zimbabwe set up 12 sentinel sites to monitor HIV drug resistance (HIVDR) following the international standards for prevention of HIVDR from 2008 to 2010.
Methods
Participants were consecutively enrolled. Blood was collected and used for CD4 count, viral load (VL) and pre-treatment DR (PDR) tests besides routine baseline tests. We analyzed the characteristics of participants enrolled into the survey and estimated the point prevalence of PDR and its associated factors among ART initiators in a cross-sectional analysis using the baseline data collected from a prospective cohort in 12 purposefully selected sentinel sites.
Results
A total of 1728 participants (96 % response rate) were enrolled and 1610 had complete data. Of the 1610 there were more females (68.7 %) than males (31.3 %). The median CD4 count was 168 cells/mm3 with males having lower values (P = 0.003). Ninety-six percent of participants had a VL ≥ 1000 copies/ml and the median VL was 128,000. Previous exposure to antiretroviral drugs (ARVs) was mainly through PMTCT (5 % of the participants). Overall, PDR mutations were detected in 6.3 % (95 % CI 5.2–7.7) of the 1480 successfully genotyped participants. However, the prevalence of PDR mutations was double for those with previous exposure (12.1 %) to ARVs compared with those without previous exposure (5.7 %, P = 0.002).
Conclusions
The results show a moderate level of PDR prevalence among ART initiators. To maintain the efficacy of the current first-line regimens, there is need to strengthen all HIVDR prevention efforts and to conduct further studies to investigate optimal strategies that can prolong the efficacy of first-line ARV regimens in the country.
Keywords: Prevalence, HIVDR, PDR, Surveillance, Genotyping
Background
As the global scale-up of antiretroviral therapy (ART) increases there are growing concerns over the increase in the prevalence of human immunodeficiency virus (HIV) drug resistance (HIVDR). This is inevitable because of HIV’s rapid and error-prone replication, a high mutation rate in the presence of drug selective pressure, sub-optimal therapy and/or poor adherence to treatment schedules, and viral recombination [1]. HIVDR mutations may also be transmitted to ART naïve patients from HIV-infected patients who have HIV drug resistant viruses. Thus, some degree of HIVDR should be anticipated in patients starting first-line ART. Factors contributing to the development of HIVDR are regimen- or drug-specific, virus-related, patient-specific and programmatic factors [2, 3]. The provision of high-quality care and treatment, early diagnosis, and consistent follow-up are necessary to minimize the emergence of drug-resistant HIV.
In response to the concerns over patients in resource-limited countries developing HIVDR, the World Health Organization (WHO) developed a global HIVDR prevention strategy. This strategy recommends countries to conduct surveys that focus on identifying program and site-specific factors associated with HIVDR development, thus enabling site and program interventions to alter these factors. This HIVDR prevention strategy helps to maximize the long-term efficacy and durability of available antiretroviral regimens [4].
Routine testing of transmitted HIVDR is an important strategy in monitoring HIVDR in resource-rich countries to ensure the HIV-positive patients are prescribed the efficacious ART regimens [5]. Unfortunately, this is not available for most patients in resource-limited countries. Thus addressing this issue using public health approach has become a mammoth task to ensure the efficacy of the current first-line regimens in these countries.
In support of the ART scaling up and prevention of HIVDR emergence and transmission, Zimbabwe established 12 sentinel ART sites to monitor the prevalence of HIVDR in the period of 2008–2010. The aim of our surveys was to establish the prevalence and distribution of HIVDR, and to identify risk factors associated with HIVDR emergence. These surveys are critical to the Zimbabwe ART program in providing information on the appropriateness of the non-nucleoside reverse transcriptase inhibitors (NNRTI) and nucleoside reverse transcriptase inhibitors (NRTI) based first-line regimens recommended by WHO and interventions for preserving these affordable regimens for the years to come. In the current report, we analyzed the characteristics of participants enrolled into the surveys, and estimated the point prevalence of Pre-treatment DR (PDR) and its associated factors among ART initiators in a cross-sectional analysis using the baseline data collected from a prospective cohort in 12 purposefully selected sentinel sites.
Methods
ART site selection and participant enrolment
The ART sites were purposefully selected in a phased approach (three sites in 2008, five sites in 2009 and four sites in 2010) following the WHO-recommended methods [6], for a total of 12 sites. After signing the volunteer participation consent, participants were recruited sequentially as they initiated ART until the pre-determined sample size of 150 was reached at each site.
Demographic and clinical data collection
Baseline patient socio-demographic and clinical data were obtained using a standard interviewer-administered questionnaire. Clinical data were assessed for correctness and accuracy by the HIVDR technical working group while at the same time support visits were provided to the sites to verify data submissions.
Specimen collection
Ten millilitres of blood were collected from each participant using two ethylene-diamine-tetra-acetic acid (EDTA) tubes. One tube was used for routine baseline liver function, full blood and CD4 cell count tests and preparation of dried blood spot (DBS) specimens for genotyping on-site. Each participant had two DBS cards prepared (One was prepared for backup purpose) and each card had five spots containing 50 µl of blood on each spot. These DBS specimens were dried overnight, packaged following standard operating procedures (SOPs) and transported to the National Microbiology Reference Laboratory (NMRL) at ambient temperature where they were stored at −80 °C. The other tube was centrifuged on site to obtain plasma which was then aliquoted for viral load (VL) testing and HIVDR genotyping.
Viral load measurement and HIVDR genotyping
Viral load testing was performed at NMRL using the Roche COBAS Taqman HIV-1 version 2 test with High Pure extraction system following the manufacturer`s instructions. An aliquot of plasma/DBS from each participant was shipped on dry ice to the WHO-designated DR laboratories, including the national DR laboratory at Medical Research Council of Uganda, Entebbe, Uganda; the National Institute of Communicable Diseases (NICD), South Africa and the specialized DR laboratory at CDC, Atlanta, United States for testing using validated and quality-assured in-house genotyping methods [7–9]. Genotyping analyses included both the protease (Pro) and reverse transcriptase (RT) gene regions and data quality was ensured by following SOPs established in the laboratories which include phylogenetic analysis to eliminate cross-contaminations. The quality-assured consensus sequences generated were then used to identify DR mutations interpreted using the Stanford HIV Drug Resistance Interpretations Algorithm (http://hivdb.stanford.edu/). The newly obtained HIV-1 pol sequences are submitted in the GeneBank and with accession numbers pending.
Statistical analyses
Prevalence, frequencies, proportions, odds ratios, stratified analysis and Chi square tests were calculated using Epi info 3.5.4 (CDC, Atlanta, 2013) and Microsoft Excel ®2007 was used to draw graphs. All calculations were made at 95 % confidence intervals (CI). Univariate analysis was initially done fitting information into previously prepared shell tables and graphs. This was followed by bivariate analysis to examine two variables in association with risk factors and outcomes. Stratified analysis was done for factors found to be statistically significant in the bivariate analysis to control for confounding and identifying effect modification. The analysis was completed in Stata v13, with svyset commands to apply inverse probability weights that account for oversampling of provincial PSUs, and to adjust for clustering of observations within PSUs and stratification by site.
Results
Site characteristics, and participant demographic and clinical characteristics
Participants were enrolled from a Central Hospital, Provincial Hospital, Private Clinic, District Hospitals, Mission Hospital, and a Local Authority Clinic. All 12 sites were offering comprehensive Opportunistic Infection (OI)/ART services. Pre-study site assessment had showed that none of the 12 sites had adequate staff based on the Zimbabwe OI/ART Standard Operating Procedures (December 2006), although they all had the core staff for the purpose of initiating and managing participants on ART. There were no reported drug stock-outs within 6 months prior to the survey period. Ninety-six percent (1728) of the expected 1800 participants were enrolled at baseline with a median site enrolment of 97 %.
The 93.2 % (1610) of enrolled participants had complete data which were used for the analysis. Females constituted 68.7 % (1106) of all participants. Of the 87 reporting previous exposure to ARVs, 72.4 % (63) were women who had used single-dose nevirapine (NVP) for prevention of mother-to-child transmission of HIV (PMTCT). Most (70.8 %) participants had no previous exposure to ARVs while 5 % had known exposure status, 23.9 % had missing information on exposure and 0.4 % were classified as other.
Median CD4 count was 168 cells/mm3 with males having lower CD4 count than females (P = 0.003). Median VL was 128,000 copies/ml with no significant difference between males and females (P = 0.724) (Table 1). Table 1 highlights the WHO clinical staging for the 1610 participants (63.9 % were eligible for ART). Fifteen percent of participants had a CD4 count less than 100 cells/mm3. Of all participants enrolled and commenced on ART, 53.6 % were eligible for ART based on a CD4 count <350 cells/mm3 alone according to the National ART guidelines in place during the period of the study. Males were more likely to have advanced HIV infection (CD4 count <200) at ART initiation than females (OR 1.59; 95 % CI 1.22–2.07).
Table 1.
Characteristics of the 1610 participants in the study among the 12 sentinel ART sites in Zimbabwe between 2008 and 2010
| Characteristic | N (%) |
|---|---|
| Gender | |
| Female | 1106 (68.7) |
| Male | 487 (30.3) |
| Missing data | 17 (1.1) |
| Age | |
| Median age (IQR) | 36 (30;44) |
| WHO clinical staging | |
| Stage 1 | 106 (6.6) |
| Stage 2 | 289 (18.0) |
| Stage 3 | 951 (59.1) |
| Stage 4 | 78 (4.8) |
| Missing data | 186 (11.5) |
| Previous Antiretroviral drugs (ARVs) exposure | |
| PMTCT | 63 (3.9) |
| ARVs prior to initiation | 18 (1.1) |
| Others | 6 (0.4) |
| No exposure | 1139 (70.7) |
| Missing exposure status | 384 (23.9) |
| CD4 count | |
| Median baseline CD4 count (IQR) | 168 (94;253) |
| Females | 180 (106;265) |
| Males | 136 (71;222) |
| Viral loads (copies/ml) | |
| ≥1000 | 1417 (95.67) |
| Females | 982 (66.31) |
| Males | 435 (29.36) |
| <1000 | 46 (4.33) |
| Drug resistance mutations | |
| No. with genotyping results | 1483 |
| No resistance | 1389 |
| NNRTI resistance only | 73 (4.9) |
| NRTI resistance only | 18 (1.2) |
| NRTI + NNRTI resistance | 5 (0.3) |
| PI resistance | 10 (0.7) |
| Any resistance mutation | 94 (6.3) |
We also analyzed ART initiation criteria based on the WHO clinical stages in comparison to CD4 count. There were 515 patients in the WHO stage 3 or 4 with a CD4 count <350 cell/mm3, 514 with CD4 count <350 cells/mm3 in the WHO stage 1 or 2, 100 in the WHO stage 3 or 4 with CD4 count >350 cells/mm3, and 294 patients in the WHO stage 1 or 2 with CD4 count >350. According to the 2010 Zimbabwe National ART guidelines 294 patients were not eligible for initiation of ART based on CD4 count or WHO clinical staging.
HIV-1 viral load determination and drug resistance genotyping
Among the 1610 participants who had completed dataset, VL data were available for 1463 (90.9 %) participants while 147 (9.1 %) were missing. Among the 1463 participants with VL measurements, 1417 (95.7 %) had VL levels greater than or equal to 1000 copies/ml while 46 (4.3 %) were less than 1000 copies/ml. There was no difference on VL levels between men and women (P = 0.72) (Table 1).
HIV-1 drug resistance genotyping was performed for all 1610 specimens and 92 % of the samples (1483) were successfully genotyped. Among the genotyped participants, 93 [6.3 % (95 % CI 5.2–7.7)] had one or more DR mutations (Figs. 1, 2). The proportion of participants with mutations ranged from 3.0 to 10.3 % at the 12 participating sites. The prevalence of PDR mutations was comparable for men (5.5 %) and women (6.7 %) (P = 0.402). The PDR mutations were mainly those against NNRTIs (4.9 %). Among the 73 NNRTI mutations identified, the most common mutations are K103N (2.7 %) and Y181C (0.9 %). These mutations can cause a high level of resistance to efavirenz (EFV) and NVP, the two ARVs that are components of the WHO-recommended first-line regimens. Twenty NRTI mutations were also identified and the most common ones were K219E/N (0.3 %) and M184 V (0.3 %). These mutations also can cause high level of resistance to NRTI drugs: lamivudine (3TC) and emtricitabine (FTC); and a low level of resistance to abacavir (ABC) and didanosine (DDI). The former two ARVs are part of the current first-line regimens recommended by WHO. In Addition, 10 PI mutations were identified and the most common ones were N88D (0.2 %) and M46I (0.2 %) which can lead to resistance to indinavir (IDV), nelfinavir (NFV), lopinavir (LPV) and atazanavir (ATV), in which ritonavir-boosted LPV (LPV/r) and ATV (ATV/r) are the components of the 2nd-line regimens recommended by WHO (Fig. 1).
Fig. 1.
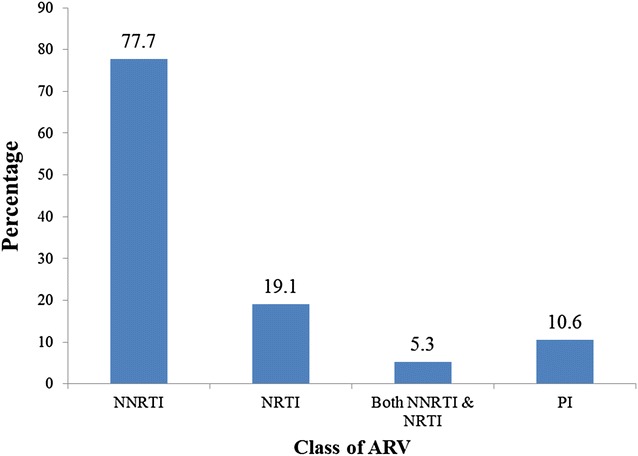
Distribution of baseline antiretroviral drug class mutations among the 93 patients enrolled into the pre-treatment HIV Drug Resistance survey in Zimbabwe: 2008–2010
Fig. 2.
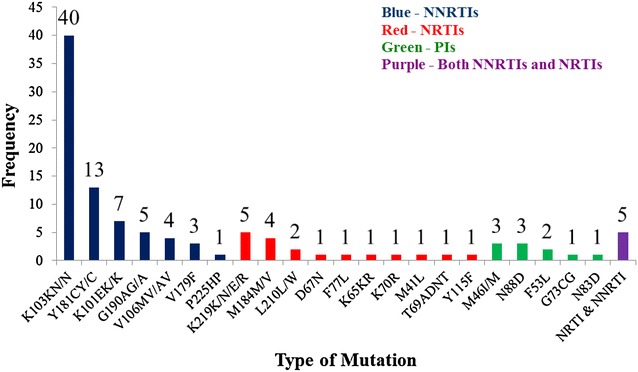
Prevalence of pre-treatment drug resistance mutations among the participants initiating ART and enrolled in the 2008–2010 HIV drug resistance survey
Factors associated with PDR
Among those 93 participants with PDR mutations, 10 participants had previous exposure and 65 had no previous exposure to ARVs while 18 had missing data. Prevalence of PDR was significantly higher among those with previous exposure (12.6 %, 95 % CI 5.9–19.8) than those without exposure (5.7 %, 95 % CI 4.0–6.2) to ARVs (P = 0.002) and nine of the 10 who had previous exposure were women (Table 4). All three participants who had been exposed to ART had resistance to NNRTI (K103 N) while one had an additional PI mutation (M46I). Similarly, among the 7 women exposed in PMTCT, six had NNRTI and one had PI mutations. Among those exposed to ARVs there was no statistically significant difference in the prevalence of PDR mutations between those exposed to ARVs [16.7 %; CI 5.4–57.2] and those exposed to PMTCT [11 %; CI 4.9–22.9] (P = 0.467). Analysis of PDR by CD4 count values (Table 2) showed that 53.8 % had CD4 count below 350 cells/mm3. The prevalence of PDR among participants with different CD4 counts was not different (P = 0.997).
Table 4.
Prevalence of Pre-treatment drug resistance mutations among enrolled participants, 2008–2010
| Variable | HIV DR | 95 % CIs |
|---|---|---|
| Sex | ||
| Female | 6.7 | 5.2–8.4 |
| Male | 5.5 | 3.6–8.1 |
| Previous exposure to any form of antiretroviral drugs | ||
| Yes | 11.6 | 5.9–19.8 |
| No | 5.0 | 4.0–6.2 |
| WHO clinical stage | ||
| 3 or 4 | 6.8 | 5.3–8.6 |
| 1 or 2 | 6.0 | 3.8–9.0 |
| CD4 count (cells/mm 3 ) | ||
| <350 | 6.3 | 4.7–8.2 |
| ≥350 | 6.3 | 4.6–8.2 |
| Viral load (copies/ml) | ||
| ≥1000 | 12.5 | 2.7–32.4 |
| <1000 | 6.3 | 5.2–79.1 |
Table 2.
Distribution of pre-treatment drug resistance mutations by CD4 count among enrolled patients, 2008–2010
| CD4 count (cells/mm3) | Drug resistance mutations (N = 1483) | |
|---|---|---|
| With mutation (%) | Without mutation (%) | |
| <200 | 33 (2.2) | 491 (33) |
| 200–349 | 17 (1.1) | 261 (17.6) |
| ≥350 | 43 (2.9) | 635 (42.8) |
| Missing | 0 (0) | 3 (0.2) |
| Total | 93 (6.3) | 1390 (93.7) |
Bivariate analysis showed that only previous exposure to ARVs was significantly associated with PDR (OR 2.58, 95 % CI 1.3–5.12). Other factors which were not significantly associated with PDR were female sex (OR 1.22), WHO clinical stage 3 or 4 (OR 1.14) and CD4 count <350 (OR 1.02) (Tables 3, 4).
Table 3.
Factors associated with pre-treatment drug resistance mutations among enrolled participants, 2008–2010
| Variable | Mutation | No mutation | OR | 95 % CI |
|---|---|---|---|---|
| Sex | ||||
| Female | 68 | 947 | 1.22 | 0.76–1.96 |
| Male | 25 | 426 | ||
| Previous exposure to ARVs | ||||
| Yes | 11 | 65 | 2.58 | 1.30–5.12* |
| No | 65 | 991 | ||
| WHO clinical stage | ||||
| 3 or 4 | 65 | 887 | 1.14 | 0.69–1.88 |
| 1 or 2 | 22 | 343 | ||
| CD4 count (cells/mm 3 ) | ||||
| <350 | 43 | 635 | 1.02 | 0.6–1.55 |
| ≥350 | 50 | 752 | ||
| Viral load (copies/ml) | ||||
| ≥1000 | 86 | 1288 | 0.47 | 0.14–1.60 |
| <1000 | 3 | 21 | ||
* Statistically significant
Discussion
A moderate level (6.3 %) of PDR was identified among the participants newly initiating ART in the 12 sentinel ART sites in Zimbabwe. The prevalence of PDR was significantly higher among those who had previously exposed to ARVs than those without. Exposure of ARVs and the most previous exposure to ARVs were women enrolled into the PMTCT program. Most participants initiated ART were in the WHO clinical stage III or IV while more than half of all participants had CD4 count below 350 cells/mm3. Majority of the participants (95.7 %) had a VL ≥ 1000 copies/ml. The CD4 count, WHO clinical stage, VL, age and sex were not significantly associated with the detection of PDR among those with these variables recorded. In this study, men were more likely to have advanced HIV infection at the initiation of ART than women, despite the likelihood of having PDR being comparable between men and women.
The results of this study are consistent with the findings of a multi-centred observational study in six countries, including Zimbabwe, which showed drug class-specific resistance prevalence of 2.5 % for NRTIs, 3.3 % for NNRTIs, 1.3 % for protease inhibitors, and 1.2 % for dual-class resistance to NRTIs and NNRTIs [5]. In South Africa provincial level studies reported prevalence rates of 4.2 % in Gauteng Province, 2.5 % in Cape Town and 3.6 % in Free State Province between 2002 and 2004 [10]. These values almost mirror what we found in this study. The South African study also suggested rising levels of transmitted drug resistance in the region from 3 % (2005/6) to 7 % (2007/8) [10]. Other studies in different parts of the world have also shown prevalence of HIVDR ranging from 3 to 15 % [3, 11, 12].
In our study, there was no association between age, sex, clinical stage and CD4 count with PDR. Similarly in a study by Hunt et al. [13], there was no association between resistance and CD4 percentage, sex, and WHO clinical stage. However, there was an association with younger age. Therefore, these factors may not be relevant in considering factors associated with having PDR in our population despite the non-random distribution of the study sites.
The resistance mutations described herein were observed after more than 4 years of the availability of treatment in the public health sector in Zimbabwe. It should be noted that, although access to treatment in the public sector began in 2004, ARVs were already available in the private sector, notwithstanding the higher cost, limited access and non-standardized regimens.
The most common PDR mutations detected in the current study were against NNRTIs, and this finding is consistent with the widespread use of this drug class as part of Zimbabwe`s National ART Guidelines as the standard first-line ART regimens, as well as single-dose NVP for PMTCT. As Zimbabwe has adopted the Option B+ strategy where all HIV-positive pregnant women and lactating mothers are commenced on NNRTI-based ART and the finding of an association on detecting PDR with previous ARV exposure mainly through PMTCT in the current study (Tables 3, 4), there is reason to be cautious/alert to the possibility that women/patients having resistance to ARVs prior to initiating the standard first-line ART may be negatively impacted on their treatment outcomes. A study by Hammers and others also showed that 13 % of patients with previous exposure to ARVs had PDR. The authors then proposed still starting patients on the standard first-line regimens with closely monitoring of VL and/or genotyping at 6 months into the therapy [14]. In Zimbabwe this is not possible considering the limited resources, such as laboratory capacity where CD4 count and VL are not even routinely available to patient services.
Irrespective of whether the observed frequency of PDR mutations is predominantly caused by transmission or by selection of undisclosed medication with ARVs, PDR mutations in patients who are starting ART may translate into reduced efficacy of the first-line and second-line ART regimens. As higher ART coverages are achieved as projected, the risk of transmitted HIVDR is likely to be increased.
There are limitations of the current study. Firstly, the sites were not randomly selected to represent all people living with HIV and AIDS in their respective regions. Thus caution is warranted when extrapolating the results to different subpopulations or regions. Secondly, participants were chronically infected and eligible for ART initiation, thus observed PDR may have been transmitted or acquired during earlier undisclosed ART or PMTCT. Thirdly, due to the design of study to enrol chronically HIV infected participants, the PDR prevalence reported here may be underestimated, thus the true prevalence of PDR might be higher than reported here. However, given the challenges of identifying individuals with recent HIV infections in resource-limited countries, there are values in surveying PDR in populations initiating ART in which at least a proxy of PDR in pre-treatment populations can be estimated using national representative sampling method which will provide important information about the probable effectiveness of currently available first-line regimens for each country/region to ensure the effectiveness of national/regional ART programs.
Conclusions
In conclusion, this cross-sectional PDR survey in ART initiators at 12 purposefully selected sentinel ART sites reveals a moderate level of PDR among the survey participants. To maintain the efficacy of the current first-line regimens there is need to strengthen all HIVDR prevention efforts and to conduct further studies to investigate optimal strategies that can prolong the efficacy of the current first-line ARV regimens in Zimbabwe and beyond.
Authors’ contributions
MM, MM2, EG, JD, JD2, PM, SN, CY, MW: conception, design, acquisition, data collection, analysis and interpretation of data and drafting the manuscript. JS and CY: protocol design, laboratory testing of samples, interpretation of the laboratory results, data analysis; TA, OM, PHK, CC, GS, MT had oversight of all stages of the research and critically reviewed the final draft for important intellectual content. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by the following people and organisations both intellectually and financially: Provincial Medical Directors in Zimbabwe, Healthcare workers in the various centres participating in the survey and the participants who spend their time participating.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Raw data can be made available on request to the corresponding author.
Disclaimer
Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease control and Prevention. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of [the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry].
Ethics approval and consent to participate
The protocol was approved by the Medical Research Council of Zimbabwe. The anonymous testing of plasma/DBS samples for the current study at the Centers for Disease Control and Prevention (CDC) was determined as non-human subjects research by the Associate Director for Science at the Center for Global Health, CDC, Atlanta, GA, USA.
Funding
This project was supported by the President’s Emergency Plan for AIDS Relief (PEPFAR) through US Centers for Disease Control and Prevention Cooperative Agreement Number 1U2GGH000315-03.
Abbreviations
- 3TC
lamivudine
- ABC
abacavir
- ART
antiretroviral therapy
- ARVs
antiretroviral drugs
- ATV
atazanavir
- CD4
cluster of differentiation
- CDC
Centers for Disease Control and Prevention
- CI
confidence interval
- DBS
dried blood spot
- DDI
didanosine
- EDTA
ethylene-diamine-tetra-acetic acid
- EFV
efavirenz
- FTC
emtricitabine
- HIV
human immunodeficiency virus
- HIVDR
human immunodeficiency virus drug resistance
- LPV
lopinavir
- LPV/r
ritonavir-boosted lopinavir
- NFV
nelfinavir
- NICD
National Institute of Communicable Diseases
- NMRL
National Microbiology Reference Laboratory
- NNRTI
non-nucleoside reverse transcriptase inhibitors
- NRTI
nucleoside reverse transcriptase inhibitors
- NVP
nevirapine
- OI/ART
opportunistic infection/antiretroviral therapy
- OR
odds ratio
- PDR
pre-treatment drug resistance
- PI
protease inhibitor
- PMTCT
prevention of mother-to-child transmission of HIV
- RT
reverse transcriptase
- SOP
standard operating procedures
- VL
viral load
- WHO
World Health Organization
Contributor Information
More Mungati, Phone: +263 4 792157, Phone: +263 739 572350, Email: mungatim@yahoo.com.
Mutsa Mhangara, Email: mutsa.mhangara@gmail.com.
Elizabeth Gonese, Email: hol9@cdc.gov.
Owen Mugurungi, Email: atp.director@ymail.com.
Janet Dzangare, Email: janet.dzangare@gmail.com.
Stella Ngwende, Email: stngwende@gmail.com.
Patience Musasa, Email: musasap@nmrl.org.zw.
Maureen Wellington, Email: dr.maureenw@gmail.com.
Gerald Shambira, Email: gshambira@yahoo.com.
Tsitsilina Apollo, Email: tsitsiapollo@gmail.com.
Chunfu Yang, Email: cxy0@cdc.gov.
Joshua DeVos, Email: ext8@cdc.gov.
Jennifer Sabatier, Email: fvv9@cdc.gov.
Peter Kilmarx, Email: peter.kilmarx@nih.gov.
Christine Chakanyuka-Musanhu, Email: musanhuc@who.int.
Mufuta Tshimanga, Email: tshimangamufuta@gmail.com.
References
- 1.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267(5197):483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 2.Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 3.WHO . WHO HIV drug resistance report 2012. Geneva: World Health Organisation; 2012. [Google Scholar]
- 4.Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. The World Health Organisation’s global strategy for prevention and assessment on HIV drug resistance. Antivir Ther. 2008;13(2):1–13. [PubMed] [Google Scholar]
- 5.Hamers RL, Wallis CL, Kityo C, Siwale M, Mandaliya K, Conradie F, Botes ME, Wellington M, Osiboqun A, Siqaloff KC, Nankya I, Schuurman R, Wit FW, Stevens WS, van Vuqt M, de Wit TF. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: a multicentre observational study. Lancet Infect Dis. 2011;11:750–759. doi: 10.1016/S1473-3099(11)70149-9. [DOI] [PubMed] [Google Scholar]
- 6.Jordan MR, Bennet DE, Winberg MA, Havlir D, Hammer S, Yang C, Morris L, Peeters M, Wensing AM, Parkin N, Nachega JB, Phillips A, De Luca A, Geng E, Calmy A, Raizes E, Sandstrom P, Archibald CP, Perriens J, McClure CM, Hong SY, McMahon JH, Dedes N, Sutherland D, Bertagnolio S. Update on World Health Organization HIV drug resistance prevention and assessment strategy: 2004–2011. Clin Infect Dis. 2012;54(Suppl 4):S245–S249. doi: 10.1093/cid/cis206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z, Wagar N, DeVos JR, Rottinghaus E, Karidia D, Nguyen DB, Bassey O, Ugbena R, Wadonda-Kabondo N, McConnell MS, Zulu I, Chilima B, Nkengasong J, Yang C. Optimization of a low cost and broadly sensitive genotyping assay for HIV-1 drug resistance surveillance and monitoring in resource-limited settings. PLoS One. 2011;6(11):e28184. doi: 10.1371/journal.pone.0028184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillay V, Ledwaba J, Hunt G, Rakgotho M, Signh B, Makubalo L, Bennett DE, Morris L. Antiretroviral drug resistance surveillance among drug-naive HIV-1 infected individuals in Gauteng Province, South Africa in 2002 and 2004. Antivir Ther. 2008;13:101–107. [PubMed] [Google Scholar]
- 9.Wallis CL, Papathanasopoulos MA, Lakhi S, Karita E, Kamali A, Kaleebu P, Sanders E, Anzala O, Bekker LG, Stevens G, de Wit TF, Stevens W. Affordable in-house antiretroviral drug resistance assay with good performance in non-subtype B HIV-1. J Virol Methods. 2010;163:505–508. doi: 10.1016/j.jviromet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nwobegahay J, Bessong P, Masebe T, Mavhandu L, Manhaeve C, Ndjeka N, Selabe G. Prevalence of drug-resistant mutations in newly diagnosed drug-naïve HIV-1 infected individuals in a treatment site in the Waterberg district, Limpopo province. S Afr Med J. 2011;101:335–337. doi: 10.7196/SAMJ.4391. [DOI] [PubMed] [Google Scholar]
- 11.Daar ES, Richman DD. Confronting the emergence of drug resistant HIV type 1: impact of antiretroviral therapy on individual and population resistance. AIDS Res Hum Retrovir. 2005;21:343–357. doi: 10.1089/aid.2005.21.343. [DOI] [PubMed] [Google Scholar]
- 12.Pillay D. Current patterns in the epidemiology of primary HIV drug resistance in North America and Europe. Antivir Ther. 2004;9:695–702. [PubMed] [Google Scholar]
- 13.Hunt GH, Coovadia A, Abrams AJ, Sherman G, Meyers T, Morris L, et al. HIV-1 drug resistance at antiretroviral treatment initiation in children previously exposed to single-dose nevirapine. AIDS. 2011;25(12):1461–1469. doi: 10.1097/QAD.0b013e3283492180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kambugu A. Pre-ART HIV resistance testing in Africa: are we there yet? Lancet Infect Dis. 2012;12:261. doi: 10.1016/S1473-3099(11)70280-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data can be made available on request to the corresponding author.


