Abstract
Recurrent, reciprocal genomic disorders resulting from non-allelic homologous recombination (NAHR) between near-identical segmental duplications (SDs) are a major cause of human disease, often producing phenotypically distinct syndromes. The genomic architecture of flanking SDs presents a significant challenge for modeling these syndromes; however, the capability to efficiently generate reciprocal copy number variants (CNVs) that mimic NAHR would represent an invaluable modeling tool. We describe here a CRISPR/Cas9 genome engineering method, Single-guide-CRISPR/Cas-targeting-Of-Repetitive-Elements (SCORE), to model reciprocal genomic disorders and demonstrate its capabilities by generating reciprocal CNVs of 16p11.2 and 15q13.3, including alteration of one copy-equivalent of the SDs that mediate NAHR in vivo. The method is reproducible and RNAseq reliably clusters transcriptional signatures from human subjects with in vivo CNV and their corresponding in vitro models. This new approach will provide broad applicability for the study of genomic disorders and, with further development, may also permit efficient correction of these defects.
Keywords: CRISPR, Cas9, Genomic Disorder, 16p11.2, NAHR, Genome Editing, Copy Number Variation (CNV), Structural Variation, Homology
Recurrent microdeletion and microduplication syndromes (rMDS) are among the most common causes of human neurodevelopmental and psychiatric disorders. These recurrent rearrangements are mediated by NAHR, which occurs between two highly homologous SDs that flank a genomic segment and can result in either copy loss (microdeletion) or the reciprocal copy gain (microduplication) of this segment1. In one study, the prevalence of five common NAHR-mediated CNVs was estimated to be 0.47% of consecutive newborns2, and the rate of recurrent rMDS in prenatal samples referred for diagnostic screening has been estimated to be 1.5%, emphasizing the relevance of the rearrangements to developmental abnormalities3.
Reciprocal CNV of a small segment of chromosome 16p11.2 (OMIM #611913) is a common rMDS that has been associated with intellectual disability, autism spectrum disorder (ASD), schizophrenia and other neuropsychiatric disorders, as well as anthropometric traits, including obesity4. Like all such NAHR-mediated genomic disorders, 16p11.2 rMDS involves gain or loss of a unique genic segment and one copy-equivalent of the SD. The unique genic segment of the 16p11.2 CNV spans 593 kb5, containing 47 genes, of which 28 are annotated as protein coding (based on Ensembl GRCh37 V.716). It is flanked by parallel and highly homologous (>99% identity) SDs, each spanning 147 kb5 and containing six duplicated genes (five annotated as protein coding6). The mechanism of NAHR-mediated CNV formation in vivo involves the mispairing of the flanking SDs, which can result in either the loss or gain of a 740 kb segment encompassing one copy of the unique genic segment and one copy-equivalent of the SD7. Disentangling the effects of the entire CNV and of individual genes within it are confounded by several factors in studies of humans and their cells, including variable genetic background, the uncertain relevance of available peripheral tissues, and the inability to collect large cohorts. Meanwhile, animal models may be limited in their generalizability to humans8, 9. Novel methods to model rMDS alterations in induced pluripotent stem cells (iPSCs) that can then be differentiated into relevant tissues, including neural precursors and derivative neural cells, would open access to innovative approaches to understanding the impact of rMDS in human neurodevelopment.
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated proteins (Cas) was discovered as a ribonucleoprotein interfering complex involved in bacterial adaptive immunity10, and recently it evolved into a new programmable and broadly applicable genome engineering tool11. Like zinc finger nucleases (ZFNs)12 and transcription activator-like effector nucleases (TALENs)13, CRISPR/Cas has inherent endonuclease activity that can generate DNA a double strand break when it encounters its target along with a proto-spacer-associated motif (PAM) sequence, 5′-NGG, where N is any nucleotide14. Most current applications of CRISPR/Cas rely on a single Cas protein in the type II CRISPR/Cas system15, Cas9, which in its host bacteria forms a complex with CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA)14. In the laboratory, a single engineered RNA (a guide RNA, or gRNA) as a chimera of a crRNA and a fixed tracrRNA can direct Cas9 to a specified target sequence that is complementary to crRNA14. One of the most common applications for the CRISPR/Cas system is targeted genome editing in cells16 and model organisms8, 17 to generate point mutations or deletion of a genic segment. Repair of gRNA-directed Cas9 nuclease-induced double-strand breaks through non-homologous end-joining (NHEJ) can induce indel mutations that abolish normal expression of the target gene16. To introduce precise point mutations or insertions at a target gene locus cleaved by Cas9, a donor template carrying a mutation can be delivered into the cell to permit repair by homologous recombination (HR)11. In addition, pairs of gRNAs can be used in a dual-guide RNA (dgRNA) strategy to produce deletions or other genomic rearrangements16, 18–21
In this study, we present a new method, SCORE, to efficiently produce microdeletions and microduplications comparable to those associated with reciprocal genomic disorders in humans as a consequence of NAHR between flanking SDs. To accomplish this, SCORE uses a single-guide RNA (sgRNA) strategy to directly target perfectly homologous sequences within the SDs. We demonstrate the capability of the method to rapidly and efficiently generate reciprocal CNVs with the SCORE method using two proof-of-principle rMDS regions of differing size and composition, 16p11.2 and 15q13.3. In each region, the method generates reciprocal CNVs that mimic the products of the NAHR mechanism, including deletion or duplication of one copy-equivalent of the SDs, with reproducible rates of efficiency. These data suggest that SCORE can open widespread access to modeling these common causes of human developmental anomalies.
RESULTS
Generation of reciprocal 16p11.2 CNVs using CRISPR/Cas9
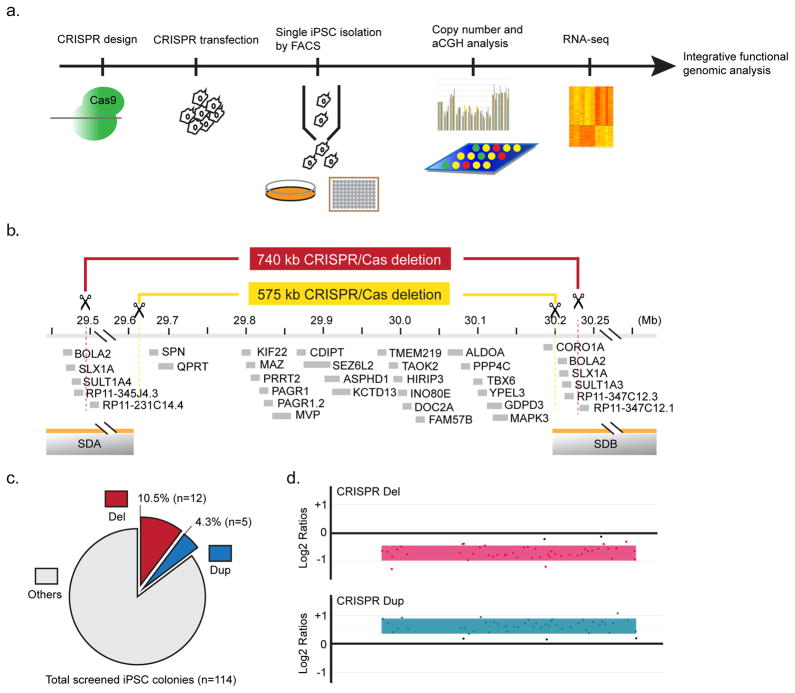
We postulated that optimizations in emerging genome engineering technologies could be applied to human iPSCs to generate reciprocal CNV directly comparable to those in recurrent NAHR-mediated rMDS. We initially tested this hypothesis on the common 16p11.2 rMDS. The overview of our experimental design is shown in Fig. 1a. All studies used a previously described control iPSC line (8330)22 as the test sample. As positive controls, we iPSC lines generated from subjects harboring 16p11.2 rMDS that were previously detected from clinical screening (four subjects with microdeletion, three subjects with microduplication) as well as family-based controls (See Online Methods; Supplementary Fig. 1). We refer herein to the distal SD (closest to the telomere) as ‘SDA’ and the proximal SD (closest to the centromere) as ‘SDB’. It has been previously shown that the CRISPR/Cas system can efficiently generate predictable deletions and inversions using a dgRNA strategy18. We first used this proven approach to create heterozygous deletions of a contiguous 575 kb sequence containing all genes within the unique genic segment as an initial comparison group (Fig. 1b). This initial experiment used two independent and uniquely mapping dgRNAs targeted within the contiguous genic segment between the SDs (See Online Methods). This experiment generated the predicted 575,414 bp microdeletion while leaving the flanking SDs fully intact (GRCh37 chromosome 16: 29,624,445–30,199,859) (Fig. 1b). Sanger sequencing confirmed the precise breakpoint and revealed NHEJ as the likely repair mechanism (Supplementary Fig. 2). However, this dgRNA strategy to delete the unique genic segment does not accurately model reciprocal rMDS in humans, where NAHR alters this unique segment and one copy-equivalent of the SDs.
Figure 1. Generation of 16p11.2 rMDS in human iPSC by CRISPR/Cas9.
(a) Overview and design of the experimental procedures. (b) Illustration of targeted 16p11.2 rMDS segment and/or SDs. For simplicity, only protein coding genes are shown from the Ensembl GRCh37 V.71 annotation6. The targeted unique genomic segment for the dual-guide 575 kb deletion is indicated in yellow, and the single-guide RNA targeting the SDs to promote a model of NAHR-mediated CNV is indicated in red (Target sequence: 5′-ACATGCCTATATCGCATAG -3′, chromosome 16: 29,487,572–29,487,590 and 30,226,917–30,226,935). (c) Efficiency of CRISPR/Cas9 generation of the 740 kb rMDS using the single-guide SCORE method that targets the SDs is shown, as determined by copy number screening assay for six genes. Further characterization by microarray and RNAseq was performed for a subset of microdeletion clones (6), and all microduplication clones (5). (d) Microarray analyses showing deletion (CRISPR Del) and duplication (CRISPR Dup) of the 16p11.2 region in CRISPR-treated lines is shown. Gains or losses of 16p11.2 region were determined by normalized log2 ratios. No off-target CNVs were observed in CRISPR-generated clones.
To generate a method that would mimic the consequences of the in vivo NAHR mechanism, we hypothesized that transfecting a sgRNA that targets perfectly homologous sites in each flanking SD could generate a 740 kb microdeletion of the 16p11.2 rMDS that is mediated by NAHR, i.e., encompassing the unique genic segment and one copy-equivalent of each gene in the SDs, mirroring the size of the CNV in humans. We evaluated the potential of this new single-guide method SCORE (Single-guide CRISPR/Cas targeting Of Repetitive Elements) to model rMDS lesions (Fig. 1b). We targeted a homologous site in each of SDA and SDB to promote simultaneous DNA breaks in both SDs, but not elsewhere in the genome (Fig. 1b; see Online Methods for detailed methods on sgRNA design). We predicted this strategy would promote formation of a 739,346 bp deletion (chromosome 16: 29,487,574–30,226,919) comprising all of the 593 kb unique genic segment and one copy-equivalent of the SDs, comparable to the in vivo mechanism. We used the sgRNA CRISPR transfection methods described in Online Methods, followed by FACS sorting to isolate single iPSCs into individual wells of Matrigel-coated 96-well plates. Once the clonal iPSC colonies were formed, relevant gene dosage was measured by copy number screening. This screening readily identified CRISPR/Cas modified clones that showed decreased dosage consistent with the expected microdeletion. The assay also identified clones with increased 16p11.2 dosage, consistent with apparent microduplication, suggesting that our SCORE method can promote predictable reciprocal dosage imbalances that mimic the in vivo consequences of NAHR.
Replication and characterization of 16p11.2 CNV iPSCs
To estimate the efficiency of the approach, the entire experiment was repeated and 114 clones were systematically screened by copy number analysis (Fig. 1c). In this replicate experiment, 12 clones harbored the expected 740 kb deletion, suggesting an overall efficiency of 10.5% for microdeletion (12 of 114 clones screened), and 4.3% of clones harbored a putative microduplication by copy number analysis (5 of 114 clones screened). Further analyses of select dosage-altered clones was then performed by strand-specific transcriptome sequencing (RNAseq; 23 total lines, see Supplementary Fig. 3 for RNAseq overview) and genome-wide DNA microarray analysis to screen for on-target and off-target CNVs (see Supplementary Fig. 3). Dosage microarray and RNAseq confirmed the observed microdeletions and microduplications (Fig. 1 and 2). Notably, no off-target CNVs were detected from genome-wide analyses (Supplementary Fig. 4). We did not obtain homozygous deletions of the 16p11.2 rMDS region, suggesting that total absence of one or more genes in this segment may be incompatible with iPSC viability in our system.
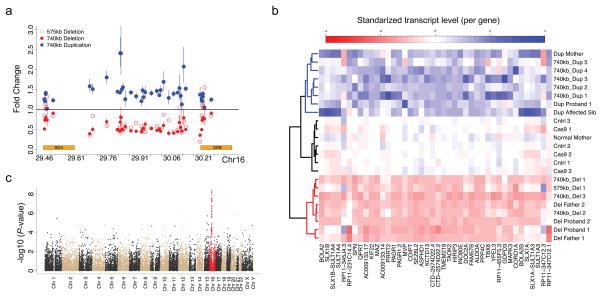
Figure 2. Gene Expression Characterization of Putative CNV Lines.
(a) Relative fold change of gene expression based on RNAseq within the 16p11.2 rMDS and SD regions is shown for the selected CRISPR/Cas9 575kb deletion line (open red circles), CRISPR/Cas9 740Kb microdeletion lines (solid red circle), and CRISPR/Cas9 740 kb microduplication lines (solid blue circle). Fold changes were calculated as the mean difference (MD) of expression for each gene between the CNV lines and all controls based on contrasts from the GLM and error bars represent the back transformed MD ±1 SEMD. (b) Heatmap of expression, estimated as counts per million (CPM), for all genes within the 16p11.2 rMDS and SD regions and hierarchical clustering, using average linkage, for all CRISPR/Cas9 treated lines and patient iPSC lines. (c) Genome wide p-value distribution of genes from linear model of expression as a linear function of the CNV. Chromosome 16 is highlighted in red and we see the strongest signal from the genes within the 16p11.2 rMDS region, as reported in previous studies9.
To provide insight into the rearrangement mechanism associated with SCORE-generated iPSCs, we attempted to clone and sequence the breakpoints generated by CRISPR/Cas within the SDs. To our knowledge, identification of the precise exchange points in SDs of 16p11.2 rMDS subjects has not been accomplished due to the inability to distinguish the SDA and SDB sequence at most sites. However, in our experiments, we were able to explore the predicted sgRNA targeting sites for potential alterations by performing cloning and Sanger sequencing of the region. We found clear junctions among 12 CRISPR-treated 740 kb microdeletion lines and 5 microduplication lines, which are shown in Supplementary Fig. 5a,b. Three different types of indel mutation were found in three microdeletion lines (Del 1~3), two mutations were found in one deletion line (Del 4), a single-nucleotide T insertion was found in four deletion lines (Del 5~8), and four deletion junctions contained no evidence of new mutations at the breakpoints, suggesting potential homology-mediated repair (Del 9~12) (Supplementary Fig. 5a). Intriguingly, only a single-nucleotide T insertion was found in four of five CRISPR-treated 740 kb microduplication lines (Supplementary Fig. 5b). These data suggest disparate mechanisms of repair for these CRISPR-treated microdeletion and microduplication lines, which clearly involve non-homologous or microhomology mediated mechanisms, but likely also involve homology mediated mechanisms, potentially including NAHR.
RNAseq was performed for 23 lines, including CRISPR-treated 575 kb and 740 kb microdeletion lines, CRISPR-treated 740 kb microduplication lines, 16p11.2 patient lines with either microdeletion23 or microduplication, and two types of control lines (those from CRISPR-treatment that did not produce a CNV and those from family member controls of 16p11.2 CNV carriers) (Supplementary Fig. 3). RNAseq analyses revealed reduced expression and overexpression of all genes within the unique genic segment for the 740 kb SCORE-generated microdeletion and microduplication lines, respectively (Fig. 2a). Moreover, we observed expression patterns reflecting the loss of one copy of the genes localized to the SDs from the 740 kb deletion lines (reduction from 4 to 3 total copies of the SDs across both homologous chromosomes) and the gain of one additional copy of the SDs in the microduplication lines (increase from 4 to 5 copies) (Fig. 2a). These in vitro results were consistent with the transcriptional patterns described previously in lymphoblastoid cell lines from families harboring 16p11.2 rMDS CNV9. As expected, there was no reduction of genes in the SDs from RNAseq of the 575 kb microdeletion line. Hierarchical clustering from the transcriptome data of the 16p11.2 rMDS region revealed that all CRISPR-treated deletion and duplication lines clustered together with the corresponding patient deletion and duplication lines, respectively, while control lines clustered together with the control family member (Fig. 2b). In addition, western blot analysis of MAPK3, encoded within the unique genic segment, also confirmed altered levels of protein expression in our CRISPR-treated iPSC lines with 16p11.2 CNVs (Supplementary Fig. 6).
Statistical analyses using general linear models evaluated gene-level contrasts of each genotype (deletion or duplication) compared to controls. We also assessed results from a linear model requiring gene-dosage to be positively or negatively correlated with copy state (e.g., a linear trend in gene expression is observed with copy state = 1, 2, or 3), analogous to a previous study in families harboring 16p11.2 CNV and in the cortex of mouse models9. We observed largely identical results in the CRISPR-treated iPSCs to those previous detected; all 32 genes detected by RNAseq (25 Protein coding) within the region showed a linear increase in expression with copy number. In the 740kb microdeletion and microduplication lines, 28 genes (22 protein coding) and 25 genes (20 protein coding) were significantly down-regulated and up-regulated, respectively, relative to controls (nominal p-value < 0.05) (Fig. 2a). Genome-wide differential expression was most significant for genes within the 16p11.2 rMDS region (Fig. 2c), again replicating previous findings in humans and mouse models5, 9, 24. These results imply that in vitro models yield comparable expression profiles to human iPSCs harboring the in vivo 16p11.2 CNV.
Replication of the SCORE approach in 15q13.3 rMDS
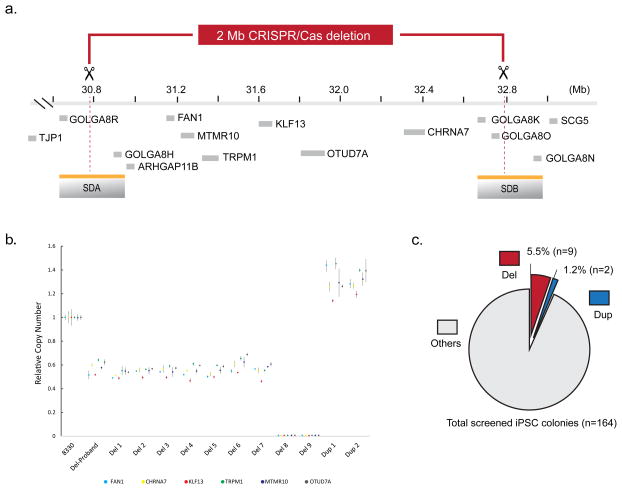
To demonstrate generalizability of the SCORE method, we performed a second replication study, targeting another common, yet much larger, rMDS region encompassing a 1.771 Mb unique genic segment and 218 kb flanking SDs within 15q13.3. Recurrent rMDS of this region have been associated with mental retardation, epilepsy, schizophrenia, autism, obesity and variable facial and digital dysmorphisms (OMIM #612001)25–27. The unique genic segment contains seven protein encoding genes (Ensembl GRCh37 chromosome 15: 30,910,306–32,445,406), and when combined with one copy-equivalent of the flanking SVs, the total genomic imbalance due to NAHR in affected subjects is 1.989 Mb28. To model this rMDS, we designed a sgRNA to uniquely target these flanking SDs (see Fig. 3a and online methods for sgRNA sequence and localization). We used the same transfection approach as in the 16p11.2 CNV, followed by single cell isolation and clonal expansion. Copy number screening of 164 CRISPR-treated iPSC colonies expanded from single cells in the initial experiment revealed 9 microdeletions and 2 microduplications (Fig. 3b). In contrast to the 16p11.2 CNV, we identified 2 clones that were consistent with homozygous microdeletion of the 15p13.3 segment. We observed no dosage increase beyond a single duplication, and no dosage changes were detected for genes flanking the CRISPR targeted region (Supplementary Fig. 7). As shown in Fig. 3c, we found the overall efficiency of the SCORE approach for this larger ~2 Mb CNV to be 5.5% for microdeletion (9 of 164 clones screened) and 1.2 % for microduplication (2 of 164 clones screened). Notably, these relatively high efficiencies for such large CNVs were detected based on a single initial experiment without prior optimization. Further development in this or other regions could result in still higher efficiencies.
Figure 3. Generation of 15q13.3 rMDS in human iPSC by CRISPR/Cas9.
(a) Illustration of targeted 15q13.3 rMDS segment and flanking SDs, with all protein coding genes shown (Ensembl GRCh37. The single-guide RNA targeting the SDs to promote a model of NAHR-mediated CNV is indicated in red (Target sequence: 5′-CCTTAGGGGATTGCGGGAC -3′, chromosome 15: 30,792,593–30,792,611 and 32,799,503–32,799,521). (b) Copy number screening of all genes within the segment identified eleven iPSC lines harboring 15q13.3 rMDS (deletion and duplication). (c) Efficiency of CRISPR/Cas9 generation of 2 Mb microdeletion and microduplication using the SCORE strategy that targets the SDs is shown.
DISCUSSION
This Technical Report reveals that carefully designed CRISPR/Cas9 genome engineering can emulate the genetic architecture of NAHR-mediated recurrent genomic disorders by generating lesions that mimic those produced in vivo. Previous studies have shown that dual-guide CRISPR/Cas9 genome editing can generate multiple classes of structural variation mediated by non-homologous repair within unique genic segments across a range of sizes18–20, 29. This SCORE approach demonstrates that it is possible to model the outcome of NAHR-mediated rMDS with relatively high efficiency, including reciprocal dosage imbalance of one copy-equivalent of the flanking SDs. SCORE thereby opens access to genome-wide modeling of these common causes of human congenital anomalies, including interrogation the unique genic segment and the SDs in any rMDS region.
The junction sequences observed in the CRISPR/Cas-generated CNVs suggest that different molecular mechanisms may predominate in the generation of microdeletions and microduplications using SCORE (Supplementary Fig. 5). A mechanism based purely on NAHR that produced coincident deletion and duplication events in a single cell would not be detected in our screen for altered dosage. However, if NAHR-mediated alterations affected sister chromatids, segregation of the events in mitosis prior to FACS sorting would then lead to individual cells with either deletion or duplication, which might be expected to be recovered in roughly equal numbers. However the junction sequences recovered suggest that non-NAHR mechanisms also operate in this system. We observed evidence of both NHEJ and microhomology-mediated repair for the microdeletions, as well as wild-type sequence for some microdeletions or a single insertion of a T nucleotide at the breakpoint of all SCORE generated microduplication lines (Supplementary Fig. 5b). These data suggest that homology-mediated repair, possibly including NAHR, may be important in the production of microduplications and of a fraction of microdeletions generated using this method. Notably, this efficiency and the presence of multiple breakpoint compositions could be influenced by mosaicism and other factors. We performed FACS sorting to obtain single cells, followed by clonal expansion to minimize the potential for mosaicism. Nonetheless, sister chromatid exchange could occur during clonal expansion leading to low levels of mosaicism in the final cell population, which would be challenging to detect. Scrutiny of the RNAseq and microarray results did not suggest mosaicism in a meaningful fraction of cells, though we cannot exclude this possibility. It is also possible that deletion occurs on one chromosome and duplication occurs on the other, leading to a balanced dosage state that would not be detected by our dosage screening, though such events would be expected to be rare. Further experiments will be needed to tease apart these varied mechanisms of repair and their relative efficiencies.
The generation of 15q13.3 rMDS CNV also confirms that relatively large reciprocal CNV models can be derived with the SCORE approach with an efficiency that was comparable to the much smaller 740 kb 16p11.2 CNVs initially generated. Previous studies suggest that dual-guide CRISPR that is reliant upon NHEJ has less than 1% efficiency for deletions greater than 100 kb19. By contrast, we find that the 10.5% SCORE efficiency to generate the 740 kb 16p11.2 microdeletion with a single cut site is only modestly reduced to 5.5% when the deletion size is increased to the 2 Mb of 15q13. For microduplication, the corresponding efficiency decrease was from 4.3% to 1.2%. This suggests that relatively efficient modeling rMDS will be tractable across a broad size distribution. It may also indicate that the participation of homology-mediated repair contributes to a greater efficiency of CNV formation than methods relying upon non-homologous repair alone. The precise size limitations of the SCORE approach remains to be determined, but we note that the 15q13.3 CNV is one of the larger recurrent, reciprocal genomic disorders commonly observed in humans (1/40000 for population incidence25, 0.48% of cases with neurodevelopmental abnormalities30).
In conclusion, we describe here a novel method to use a single-guide RNA that targets highly homologous SDs that flank unique genomic segments and derive recurrent, reciprocal dosage imbalances, thus modeling the consequences of NAHR, which drives many of the most common causes of human congenital anomalies. Our experiments reveal that SCORE can be rapidly implemented to produce rMDS CNVs of different size and copy state, as we observed 0, 1, 2, and 3 copies of the rMDS regions. Genome-wide analyses suggest that the method generates models with high fidelity, as we did not observe off-target CNVs following genome-wide screening, and transcriptome studies of the in vitro edited lines recapitulated previous findings in rMDS families and mouse models. We provide complete details on the design and application of this method, which will enable modeling of rMDS in multiple tissue types and, with further development and optimization, could provide a tractable route to in vitro correction of these common genomic imbalances.
METHODS
Guide RNA design and preparation
We used the CRISPR Design Tool (http://tools.genome-engineering.org)31 to obtain the sequences of guide RNAs targeting the 16p11.2 rMDS segment. The design of guide RNAs and reference sequence are based on genome assembly GRCh37. To generate 575kb microdeletion that includes all unique genes within the rMDS segment, the sequence of dual-guide RNAs were as follows:
gRNA427: 5′-GCAGTGGCAGGCCATGAGCT -3′, chromosome 16: 29,624,428–29,624,447;
gRNA841: 5′-GCCTGGACACCGGGCGCAGG -3′, chromosome 16: 30,199,842–30,199,861.
To design the sgRNAs targeting 16p11.2 SDs and 15q13.3 SDs with the SCORE approach, we first identified all possible 18–25mer guides with Jellyfish32 and performed a degenerate Blast search to identify sequences that would uniquely target the 16p11.2 and 15q13.3 SDs respectively, with no predicted off-target effects. Additionally, for the 15q13.3 guides, we refined our original guide design methodology by utilizing a recently released tool, Off-Spotter33, to screen out candidate guides with less than four mismatches across non-target regions, further mitigating any possible off-target effects. Finally, we cross-verified the target sites and regions with the new assembly (GRCh38) to ensure there were no artifacts arising from updates to the reference genome.
For generation of the 740 kb microdeletion and microduplication in 16p11.2, the following single-guide RNA was used:
sg569: 5′-GACATGCCTATATCGCATAG -3′, chromosome 16: 29,487,572–29,487,590 and 30,226,917–30,226,935.
For generation of the 2 Mb microdeletion and microduplication in 15q13.3, the following single-guide RNA was used:
sg387: 5′-GCCTTAGGGGATTGCGGGAC -3′, chromosome 15: 30,792,593–30,792,611 and 32,799,503–32,799,521.
All gRNAs were cloned into pSpCas9(BB)-2A-Puro plasmid with a puromycin resistance marker (pX459, Addgene plasmid 48139) using a BbsI restriction site31. gRNA841 was cloned into gRNA_Cloning Vector (pGuide, Addgene plasmid 41824) using a BbsI restriction site34. Validation of the guide sequence in the gRNA vector was confirmed by Sanger Sequencing. Before transfection, all plamids were purified from EndoFree Plasmid Maxi Kit according to the manufacturer’s instruction (Qiagen).
Cell culture and DNA transfection
Human induced pluripotent stem cells (iPSCs), derived from adult fibroblasts, are maintained on Matrigel-coated dish (Corning) with Essential 8 medium (Invitrogen) and incubated at 37 °C in a humidified atmosphere with 5% CO2. For the dual-guide approach, we transfected human iPSCs with pSpCas9(BB)-2A-Puro plasmid and gRNA_Cloning Vectors which carry both guide RNAs and collected individual iPSC colonies after puromycin selection (see Online Methods for details). At 72 hours post-transfection, iPSCs were dissociated and single cells were isolated by fluorescence-activated cell sorting (FACS) gated for a high level of GFP expression. Genomic DNAs from iPSC colonies were screened by PCR for the presence of an expected ~1 kb junction product (Supplementary Fig. 2). For the SCORE method, the Human iPSCs (5 × 105 cells) were transfected with 2 μg total DNA plasmid, Cas9-sgRNA expression vector (pX459) with the chosen guide RNA (sg569: 5′-GACATGCCTATATCGCATAG-3′) and EGFP vector or pGuide-sgRNA vectors. Transfection utilized Human Stem Cell Nucleofector Kit 1 (Lonza) and Amaxa Nucleofection II device (Lonza) with programs B-016, according to the manufacturer’s instructions. After Nucleofection, the iPSCs were cultured in two different settings: i) on Matrigel-coated wells using Essential 8 medium (Invitrogen) supplemented with 10 μM ROCK inhibitor (Santa Cruz Biotech), ii) co-cultured on a monolayer of feeder cells (mouse embryonic fibroblasts, MEF) using MEF conditioned media (CM) supplemented with 10 ng/ml bFGF (R&D) and 10 μM ROCK inhibitor (Santa Cruz Biotech), which was made by incubating human iPS KnockOut Serum Replacement (KOSR) media with feeder cells overnight. For Puromycin selection, 24 hours post-Nucleofection iPSCs were harvested in fresh Essential 8 medium with Puromycin (0.1μg/mL). After 24 hours, surviving colonies are then collected into Matrigel-coated 96-well plates individually (one colony per well), for subsequent screenings. For cell sorting, the iPSC were recovered in fresh Essential 8 medium for three days after Nucleofection.
Single cell isolation by fluorescence-activated cell sorting (FACS)
To obtain isogenic iPSC colonies following CRISPR/Cas9 treatment, single cell isolation was obtained by FACS. At 72 hours post-nucleofection, the iPSCs were dissociated into a single cell suspension with Accutase and resuspended in PBS with 10 μM ROCK inhibitor (Santa Cruz Biotech). All samples were filtered through 5mL polystyrene tubes with 35um mesh cell strainer caps (BD Falcon 352235) immediately before being sorted. After adding the viability dye TO-PRO-3 (Invitrogen), the GFP+ TO-PRO-3- iPSCs were sorted and plated, with one cell placed into each well of Matrigel-coated 96-well plates by BD FACSAriaII with 100-mm nozzle under sterile conditions. Once multi-cellular colonies were clearly visible (2~3 days after sorting), they were collected into individual wells of Matrigel-coated 96-well plates by manually picking. Once individual iPSC colonies were available (~14 days after sorting), the genomic DNA from those colonies were characterized by copy number assay.
Genomic DNA extraction
For high throughput extraction in 96-well format, the genomic DNA was isolated from cells transfected by the Cas9-gRNA expression vector (pX459) using a Rapid DNA extraction method35. Cells were lysed by adding DNA extraction buffer containing Proteinase K (0.2 mg/ml). Samples were digested at 55 °C for 30 min followed by Proteinase K inactivation at 95 °C for 10 min, followed by RNase A incubation at room temperature for 10 min. For genomic DNA extraction from 16p11.2 patient iPSC lines, we performed extraction by DNeasy Blood & Tissue Kit (Qiagen) and followed the manufactural instruction. For 15q13.3 patient genomic DNA, we obtained it from Simons Foundation Autism Research Initiative.
Screening of individual iPSC colonies
To isolate genomic DNA from the iPSC colonies, iPSCs were detached with ReLeSR (Stem Cell Technologies) and then prepared by using Rapid DNA extraction method35. For detection of deletion, the genomic region flanking the CRISPR target site was amplified by PCR (Supplementary Fig. 2a). The primers were synthesized by Integrated DNA Technologies (IDT) with the following sequences:
Forward, 5′-CAACTGCAATTTCCTTTTCAA-3′;
Reverse, 5′-CCCTCAGGTCATCCTCTCAT-3′.
PCR reactions were performed using 2 μl of genomic DNA and Phusion High Fidelity Master Mix (NEB), with the following cycling conditions: 95 °C for 2 min; 95 °C for 15 s, 63 °C for 30 s, 72 °C for 1 min (45 cycles); 72 °C for 2 min. PCR products were visualized, followed by Sanger sequencing to determine the exact modification that occurred (for the 575 kb deletions) (Supplementary Fig. 2b). To determine the heterozygosity of 16p11.2 region in CRISPR targeting iPSC lines, we followed the same PCR protocol performed in the detection of deletion to amplify part of 16p11.2 region which was deleted in 16p11.2 rMDS. Also, the PCR was performed using the same forward primer in detection of deletion and a new-design reverse primer targeting on the unique rMDS segment.
Identification of the junction sequences in 16p11.2 rMDS iPSC lines
For isolation the sequences, the genomic region around the CRISPR target site was amplified by nested PCR. The primers were synthesized by Integrated DNA Technologies (IDT) with the following sequences:
Forward, 5′-TGCAAGTTTCAGGAAACTTGG -3′;
Reverse-1, 5′-CGTGGTGGGAGACATGCACCA -3′; Reverse-2, 5′-GTAGAGGCGGGCTGGCCAG -3′. First PCR reaction was performed using 1 μl of genomic DNA, forward and reverse-1 primers and Phusion High Fidelity Master Mix (NEB), with the following cycling conditions: 95 °C for 2 min; 95 °C for 15 s, 61 °C for 30 s, 72 °C for 1 min (45 cycles); 72 °C for 1 min. Second PCR reaction was performed using 1 μl of First PCR product, forward and reverse-2 primers and Phusion High Fidelity Master Mix (NEB), with the following cycling conditions: 95 °C for 2 min; 95 °C for 15 s, 61 °C for 30 s, 72 °C for 1 min (45 cycles); 72 °C for 1 min. PCR products were visualized, followed by Zero Blunt PCR cloning (ThermoFisher Scientific) and Sanger sequencing to determine the exact mutation that occurred (Supplementary Fig. 5).
Copy Number Analysis
Quantitative real-time PCR (qRT-PCR)
For initial copy number screening of CRISPR treated iPSC lines, qRT-PCR was performed using six sets of TaqMan probes targeting six genes at 16p11.2 microdeletion region and a reference primer/probe set targeting the RNaseP gene at chromosome 14 (TaqMan Human RNaseP gene, Applied Biosystems). Reactions were carried out in triplicate in 20 μl total volume containing 1 μl of genomic DNA sample, LightCycler 480 Probes Master (Roche) and 20× primer/probe mix. The 16p11.2-region primers and FAM-labeled probes were synthesized by IDT with the following sequences: QPRT-F, 5′-CTAAACCGGAAGAGGATGACAC -3′ QPRT-R, 5′-CATTGGCCACTGACCCTAAA -3′ QPRT-probe, 5′-CATGGGTTACGTGGCTCCTCAGG -3′ KCTD13-F, 5′-CTCAGACTGTGGTGATGTCAG -3′ KCTD13-R, 5′-CCAGCTGGTTAAGAGGGATTTA -3′ KCTD13-probe, 5′-CACACTTTGGCATGGACGATGCAC -3′ CDIPT-F, 5′-GGCCCTGAGCCAAGAATATC -3′ CDIPT-R, 5′-AAGCCATCCACAGCCTTC-3′ CDIPT-probe, 5′-CCAAGTCAAATAGTGAAGTCCCGCCA-3′ BOLA2-F, 5′-ATCAGAGCCTGAGGAAGGT-3′ BOLA2-R, 5′-TGGAGGCGGGTGTAGAA-3′ BOLA2-probe, 5′-TACCATTCAGCATCGGCTCCGC-3′ SLX1A-F, 5′-AGGCTGCCTCCGGAT AG -3′ SLX1A-R, 5′-GGTTGATTGCCGTGACCTA-3′ SLX1A-probe, 5′-CCCGAGAGCTTGTTCCGAAGCA-3′ SULT1A4-F, 5′-AGCCTGTATTGGAAAGGAAGAG-3′ SULT1A4-R, 5′-TTACTTCTCCAAACCCTTCTCC-3′ SULT1A4-probe, 5′-TCTGAGCTGAAAGAGTGAATGCCCG-3′.
For the copy number screening in 15q13.3 CRISPR work, qRT-PCR was performed using six sets of TaqMan probes targeting six genes (FAN1, MTMR10, TRPM1, KLF13, OTUD7A and CHRNA7) at 15q13.3 microdeletion region, two sets of TaqMan probes targeting two genes (SCG5 and TJP1) outside of 15q13.3 CRISPR targeting SDs and a reference primer/probe set targeting the RNaseP gene. For all the primer/probe sets were purchased from ThermoFisher Scientific with the following assay ID: FAN1 (Hs07226355_cn), MTMR10 (Hs03916555_cn), TRPM1 (Hs00942380_cn), KLF13 (Hs02255264_cn), OTUD7A (Hs00989444_cn), CHRNA7 (Hs03904338_cn), SCG5 (Hs03900067_cn) and TJP1 (Hs03893363_cn).
Amplification was performed by using the LightCycler 480 Real Time PCR system (Roche) under the following cycling conditions: pre-incubation step, 95 °C for 10 min; amplification step, 95 °C for 10 s, 55 °C for 30 s and 72 °C for 1 s (50 cycles); cooling step, 40 °C for 10 s. Relative copy number of genes was determined by using the 2−ΔΔCt method36, where ΔCt is the difference of cycle threshold values for the test gene and reference gene RNaseP. For each gene, experiments are in triplicate. Statistical Analysis for Copy Number was performed using one-way analysis of variance (ANOVA) followed by Dunnett’s adjustment for multiple comparisons.
Chromosomal Microarray Analysis
Array-based comparative genomic hybridization (aCGH) was performed on the Agilent 4×180K SurePrint G3 Human CGH Microarray (Design #022060) according to the protocol provided by the manufacturer. The assay assessed for imbalances (i.e., gains/losses) in the genomic DNA sample being tested by comparing the test DNA to a Promega male control DNA sample obtained from a pool of karyotypically normal individual(s). This array platform contains 180,880 probes taken from throughout the human genome. A genomic imbalance is noted when six or more oligos show a minimum average log ratio of 0.25 for one-copy gains and −0.50 for one- copy losses; Oligonucleotide information is based on the March 2006, NCBI 36.3 (hg18) build of the Human Genome (UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway). However, the data reports were generated using the genome build hg19. This assay does not exclude chromosome anomalies smaller than the assay’s effective resolution. The assay is also not specifically designed to detect mosaicism, uniparental disomy, methylation abnormalities, or other chromosomal rearrangements (including chromosomal translocations, insertions, and inversions).
Library preparation and RNA-sequencing (RNA-seq)
RNA samples were prepared from 23 iPSC lines (Supplementary Fig. 3). There are 5 iPSC lines derived from patients harboring 16p11.2 microdeletion (two families) as well as 3 lines derived from patients with 16p11.2 microduplication (one family) (Supplementary Fig. 3). Other lines were created using CRISPR/Cas9 system with or without guide RNAs. All RNA samples were extracted with Trizol reagent according to the manufacturer’s instruction (Invitrogen). RNA-seq libraries were prepared from 200 ng of total RNA using a TruSeq Stranded mRNA Sample Prep Kit (Illumina cat# RS-122-2102) Each library also included 1 μl of a 1:100 dilution of ERCC RNA Control Spike-Ins (Ambion) containing 92 synthetic RNA standards of known concentration and sequence. These synthetic RNAs cover a 106 range of concentration, as well as varying in length and GC content to allow for validation of dose response and the fidelity of the procedure in downstream analyses37. Libraries were multiplexed, pooled, and sequenced on multiple lanes of an Illumina HiSeq2500, generating an average of 40M paired-end 50-cycle reads for each sample.
RNA-seq data analysis
RNAseq data was processed using a standard workflow, which includes quality control of fastq reads using FastQc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were aligned to the Ensembl GRCh37 Human reference using GSNAP ver 2014/12/1938 and quality control of the alignments was performed with custom script wrappers for multiple utilities viz., PicardTools, the RNAseqQC39 module from GenePattern and RSeqQC40. Read counts were quantified using the BedTools Suite41, taking into account strand-specificity (described in9) and ERCC spike-ins42 were used to threshold genes within the lower limits of detection (described in9) and genes with > 3 reads across all samples were chosen for subsequent analysis and normalized by the library size. We also filtered out rRNA and tRNA genes as well as genes < 250nt in length, which is the minimum length for the ERCC spike-in transcripts. Read counts were analyzed using Generalized Linear models (GLMs) assuming a negative binomial distribution within the R environment and significance was assessed based on contrasts for hypothesis testing using, both, nominal and FDR p-value cut-offs. For details of the model fitting procedures and the statistical analysis procedures the readers are referred to our previous study9.
iPSC whole-cell lysate preparation and western blot
iPSC lysate was prepared in RIPA buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 1% IGEPAL CA-630, 1 mM phenymethylsulfonyl fluoride (PMSF), 20 mg/ml pepstatin A, 20 mg/ml leupeptin, 20 mg/ml aprotinin, 50 mM NaF and 1 mM Na3VO4). The lysate was resolved by 10% Bis–Tris NuPAGE (ThermoFisher Scientific). The proteins resolved by SDS–PAGE were transferred to the Nitrocellulose (NC) membrane (Satorious) and western blot analysis was conducted by using the following antibodies: mouse anti-MAPK3 (12D11, cat# MA1-13041, ThermoFisher Scientific), rabbit anti-β actin (cat# ab8227, Abcam)43. The secondary antibodies used were HRP-conjugated goat-anti mouse IgG antibody (Abcam) and HRP-conjugated goat-anti rabbit IgG antibody (Abcam). Membrane was developed by reacting with chemiluminescence HRP substrate (Millipore) and exposed to the Amersham Hyperfilm ECL (GE Healthcare) for visualization of protein bands. The protein bands were quantified by using the NIH Image J Software. Statistical Analysis for western blot was performed using one-way analysis of variance (ANOVA) followed by Dunnett’s adjustment for multiple comparisons.
A supplementary methods checklist is available.
Supplementary Material
Acknowledgments
These studies were supported by funding from the Simons Foundation for Autism Research (SFARI #328656 and 346042 (M.E.T.) and SFARI # 308955 (J.F.G.)), the Nancy Lurie Marks Family Foundation (J.F.G. and M.E.T.), the National Institutes of Health (R01NS093200 (J.F.G. and M.E.T.), R00MH095867 (M.E.T.), P01GM061354 (J.F.G. and M.E.T.)), the March of Dimes (M.E.T.), NARSAD (M.E.T.), and Autism Speaks (J.F.G.). Research reported in this publication was also partially supported by NCI award P30CA034196 (C.L.) and NHGRI U41HG007497 (C.L.). We thank S. Haggarty and S. Sheridan (Center for Human Genetic Research, Massachusetts General Hospital) for generously providing the control iPSC line. We gratefully acknowledge the resources provided by the AGRE consortium and the participating AGRE families. AGRE is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). C.L. is an Ewha Womans University Distinguished Professor, and D.J.C.T. is a recipient of the Postdoctoral Research Abroad Program sponsored by Ministry of Science and Technology of Taiwan R.O.C. (102-2917-I-564-012).
Footnotes
AUTHOR CONTRIBUTIONS
M.E.T., J.F.G., D.J.C.T., A.S. and I.B. conceived of and designed the studies, D.J.C.T., P.M., C.M.S., A.S. performed molecular studies, A.R., R.L.C., S.E. performed computational and statistical analyses, X.C., Y.S., and M.S. obtained 16p11.2 family tissue samples, C.Z. and C.L. designed and performed microarray studies. D.J.C.T., J.F.G., and M.E.T. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends in genetics : TIG. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 2.Tucker T, et al. Prevalence of selected genomic deletions and duplications in a French-Canadian population-based sample of newborns. Molecular genetics & genomic medicine. 2013;1:87–97. doi: 10.1002/mgg3.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konialis C, et al. Uncovering recurrent microdeletion syndromes and subtelomeric deletions/duplications through non-selective application of a MLPA-based extended prenatal panel in routine prenatal diagnosis. Prenatal diagnosis. 2011;31:571–577. doi: 10.1002/pd.2750. [DOI] [PubMed] [Google Scholar]
- 4.Maillard AM, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Molecular psychiatry. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. The New England journal of medicine. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 6.Flicek P, et al. Ensembl 2013. Nucleic acids research. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters RG, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golzio C, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumenthal I, et al. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. American journal of human genetics. 2014;94:870–883. doi: 10.1016/j.ajhg.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature biotechnology. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nature biotechnology. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 13.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 14.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nature reviews Microbiology. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal PK, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell stem cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canver MC, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. The Journal of biological chemistry. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao A, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic acids research. 2013;41:e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veres A, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell stem cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheridan SD, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PloS one. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, et al. Intra-family phenotypic heterogeneity of 16p11.2 deletion carriers in a three-generation Chinese family. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156:225–232. doi: 10.1002/ajmg.b.31147. [DOI] [PubMed] [Google Scholar]
- 24.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Human molecular genetics. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 25.Sharp AJ, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nature genetics. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Bon BW, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. Journal of medical genetics. 2009;46:511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antonacci F, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nature genetics. 2014;46:1293–1302. doi: 10.1038/ng.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, et al. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. Journal of molecular cell biology. 2015 doi: 10.1093/jmcb/mjv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassos E, et al. Penetrance for copy number variants associated with schizophrenia. Human molecular genetics. 2010;19:3477–3481. doi: 10.1093/hmg/ddq259. [DOI] [PubMed] [Google Scholar]
- 31.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pliatsika V, Rigoutsos I. “Off-Spotter”: very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs. Biology direct. 2015;10:4. doi: 10.1186/s13062-015-0035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McClive PJ, Sinclair AH. Rapid DNA extraction and PCR-sexing of mouse embryos. Molecular reproduction and development. 2001;60:225–226. doi: 10.1002/mrd.1081. [DOI] [PubMed] [Google Scholar]
- 36.Yaffe MB, et al. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nature biotechnology. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 37.Jiang L, et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 41.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang L, et al. Synthetic spike-in standards for RNA-seq experiments. Genome research. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verbovsek U, et al. Expression analysis of all protease genes reveals cathepsinK to be overexpressed in glioblastoma. PloS one. 2014;9:e111819. doi: 10.1371/journal.pone.0111819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.