Abstract
Pre-steady-state kinetic analysis is a powerful and widely used method to obtain multiple kinetic parameters. This protocol provides a step-by-step procedure for pre-steady-state kinetic analysis of single-nucleotide incorporation by a DNA polymerase. It describes the experimental details of DNA substrate annealing, reaction mixture preparation, handling of the RQF-3 rapid quench-flow instrument, denaturing polyacrylamide DNA gel preparation, electrophoresis, quantitation, and data analysis. The core and unique part of this protocol is the rationale for preparation of the reaction mixture (the ratio of the polymerase to the DNA substrate) and methods for conducting pre-steady-state assays on an RQF-3 rapid quench-flow instrument, as well as data interpretation after analysis. In addition, the methods for the DNA substrate annealing and DNA polyacrylamide gel preparation, electrophoresis, quantitation and analysis are suitable for use in other studies.
Keywords: DNA polymerase, Pre-steady-state kinetics
We present a general protocol for pre-steady-state kinetic analysis of single-nucleotide incorporation by DNA polymerases with a model system in which human DNA polymerase η (hpol η) R61M mutant (N-terminal 1–432 amino acids) inserts dCTP opposite a DNA lesion, 7,8-dihydro-8-oxo-2´-deoxyguanosine (8-oxodG)(Su et al., 2015). This protocol lists a step-by-step procedure of pre-steady-state kinetic measurements conducted using a RQF-3 rapid quench-flow instrument and the corresponding data analysis using GraphPad Prism software. In addition, we include commonly applied procedures for DNA duplex substrate annealing and preparation, electrophoresis, imaging, and quantitation of polyacrylamide DNA gels.
Materials
40% Acrylamide/bis, 19:1, w/v, 5% crosslinker (Bio-Rad Laboratories)
Ammonium persulfate (APS) (Bio-Rad Laboratories)
Bovine serum albumin (BSA), 2 mg/mL (Pierce Protein Biology Products)
Bromophenol blue (Sigma-Aldrich)
dNTP, 100 mM (New England Biolabs)
DL-Dithiothreitol (DTT) (Research Products International)
DNA primer: 5´-/FAM/CGG GCT CGT AAG CGT CAT-3´ (Integrated DNA Technologies)
DNA template: 5´-TCA T(8-oxodG)A TGA CGC TTA CGA GCC CG-3´ (Integrated DNA Technologies)
EDTA
Eppendorf tubes, 1.5 mL
Formamide (Roche)
Gel loading buffer (see recipe)
Glycerol
Human DNA polymerase η (hpol η) R61M (N-terminal 1–432 amino acids, expressed and purified as described (Biertumpfel et al., 2010; Su et al., 2015)
Nuclease-free H2O
Magnesium chloride solution, 25 mM
N,N,N,N-Tetramethylethylenediamine (TEMED) (Bio-Rad Laboratories)
Potassium chloride
Tris-HCl
-
5X TBE buffer (0.445M Tris-HCl, 0.445M boric acid, and 0.01 M EDTA)
Urea (electrophoresis grade, Sigma-Aldrich)
Xylene cyanol FF (Sigma-Aldrich)
Plastic cling wrap
Dry block heater and thermometer (VWR International)
GraphPad Prism software (GraphPad)
ImageJ software (National Institutes of Health)
Luer Lock disposable syringes, 1 mL and 5 mL (Bio-Rad Laboratories)
Microcentrifuge
RQF-3 Rapid Quench-Flow Instrument (KinTek Corporation)
Sequi-Gen GT Nucleic Acid Sequencing Cell, 38 × 50 cm (Bio-Rad Laboratories)
Typhoon System (GE Healthcare Life Sciences)
DNA Substrate Preparation
-
1.Dissolve the single-stranded DNA primer or template respectively to a final concentration of 1 mM in nuclease-free H2O.The primer contains a fluorescent dye, FAM (6-carboxyfluorescein), at its 5´ end. The template contains a DNA lesion, 7,8-dihydro-8-oxo-2´-deoxyguanosine (8-oxodG). The H2O used in this protocol is nuclease-free. Alternatively, the primer can be labeled by another fluorescent dye, e.g. cy3 or cy5, or 32P. The template may be an unmodified oligonucleotide or it may contain a DNA lesion.
-
2.
Mix 6 µL DNA primer stock solution, 6 µL DNA template stock solution, and 18 µL H2O in a nuclease-free Eppendorf tube. The final concentration of DNA duplex will be 200 µM.
-
3.
Heat the mixture at 95 °C for 5 min in a dry block heater, and slowly cool to room temperature (~25 °C).
-
4.
After the temperature drops to room temperature, spin the Eppendorf tube briefly in a mini-centrifuge to make sure that all the sample is at the bottom of the tube. The annealed DNA duplex substrate is ready for use; it can be stored at 4 °C for as long as 2 weeks.
Preparation for the Reaction
-
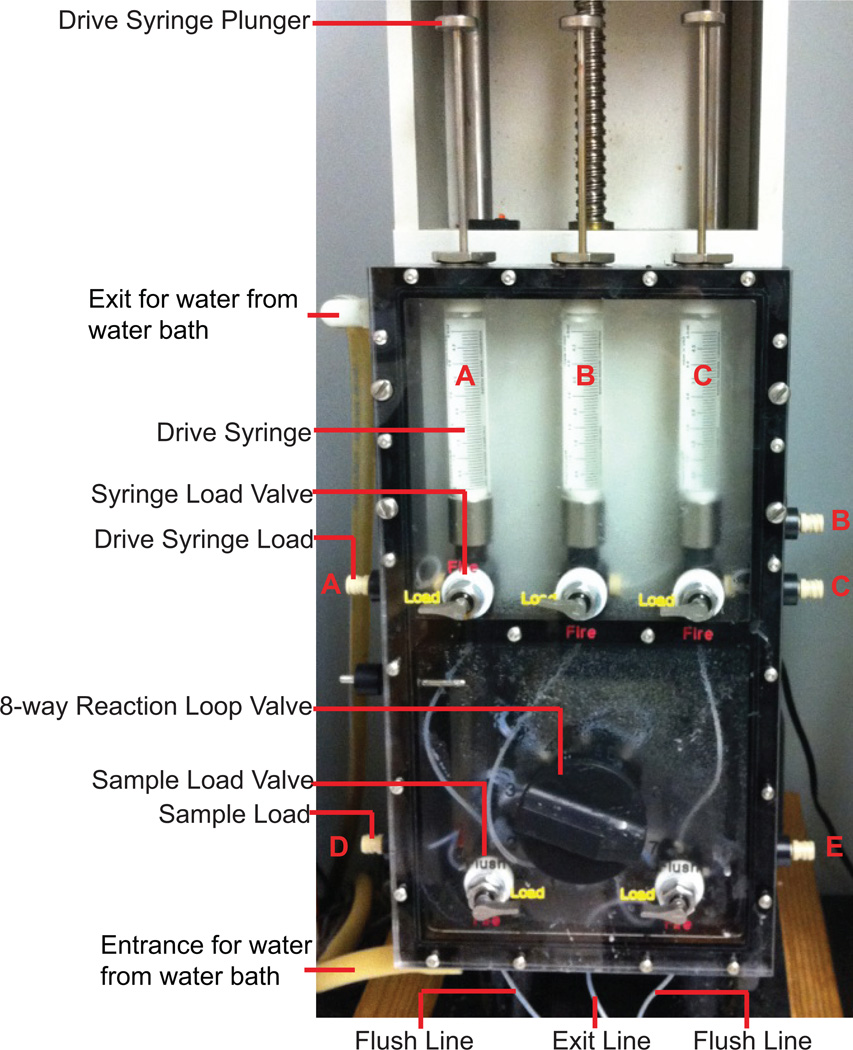
5.Turn on the water bath (connected with the RQF-3 Rapid Quench-Flow Instrument, but not shown in Fig 1, setting at 25 °C) and pump to equilibrate the whole system 30 min before use.25 °C is used here as the reaction temperature. Also, 37 °C is another commonly used temperature, particularly for mammalian enzymes.
-
6.
Use the Keypad to move the Step Motor up (the Step Motor is above the Drive Syringe Plungers, not shown in Fig. 1), to allow enough space for the Drive Syringe Plungers to move freely.
-
7.
Set the Drive Syringe Load Valves in the Load position. Fill the 5-mL Luer Lock disposable syringes with H2O and attach to Drive Syringe Loads. Wash the Drive Syringes with H2O (Fig. 1).
-
8.After removing H2O by pushing the Drive Syringe Plungers, fill Drive Syringes A and C with 25 mM Tris-HCl buffer (pH 7.5) and Drive Syringe B with 500 mM EDTA (Fig. 1).If any bubbles appear in the Drive Syringes, remove them by pumping the solution back and forth.
-
9.
Use the Keypad to move the Step Motor to attach the tops of all three Drive Syringe Plungers.
-
10.
Set the Sample Load Valve on the left at the Load position and the right one at the Flush position, connect the Exit Line to the vacuum, inject H2O through the Sample Load D to wash the Sample Loop (the Sample Loop is the line between the Sample Load Valve and the 8-way Reaction Loop Valve, not shown in Fig. 1.) rinse with methanol, and dry.
-
11.
Wash and dry the other side of Sample Loop and Sample Load as in step 10.
-
12.
Set both Sample Load Valves to the Flush position, set the 8-way Reaction Loop Valve to the 1 position, connect the Exit Line to the vacuum, and put the ends of both Flush Lines in H2O to wash the Reaction Loop 1.
-
13.
Put the ends of Flush Lines into methanol to remove H2O. Dry the Reaction Loop, Flushing Lines, and the Exit Line by continuing vacuum after moving the ends of Flush Lines into the air.
-
14.
Change the 8-way Reaction Loop Valve to the 2–7 positions, individually, wash and dry as Steps 12 and 13.
-
15.
Label empty Eppendorf tubes for later use.
-
16.
Prepare Pre-mixture I on ice as Table 1. Gently mix the mixture by inverting the tube 5 times.
-
17.
Prepare Pre-mixture II on ice as indicated in Table 2 and mix gently by inverting the tube 5 times.
-
18.
Place the two pre-mixtures on ice; the samples are ready for use.
Fig. 1.
RQF-3 Rapid Quench-Flow Instrument.
Table 1.
Pre-mixture I. The stock concentrations, volumes to add, and final concentrations of reagents are listed.
| Reagent | Stock conc. | Volume to add | Final conc. |
|---|---|---|---|
| Tris-HCl, pH7.5 | 500 mM | 72 µL | 40 mM |
| BSA | 2 mg/mL | 45 µL | 0.1 mg/mL |
| DTT | 100 mM | 90 µL | 10 mM |
| Glycerol | 50 % (v/v) | 90 µL | 5% (v/v) |
| KCl | 2.5 M | 36 µL | 100 mM |
| hpol η (R61M) | 22 µM | 20 µL | 500 nM |
| Annealed DNA duplex | 200 µM | 4.5 µL | 1 µM |
| H2O | 542 µL | ||
| Total | 900 µL |
Table 2.
Pre-mixture II. Stock concentrations, volumes to add, and final concentrations of reagents are shown.
| Reagent | Stock conc. | Volume to add | Final conc. |
|---|---|---|---|
| dNTP | 100 mM | 9.0 µL | 1 mM |
| MgCl2 | 25 mM | 360 µL | 10 mM |
| H2O | 531 µL | ||
| Total | 900 µL |
Pre-Steady-State Kinetic Reaction Conducted on RQF-3 Rapid Quench-Flow Instrument
-
19.Enter the reaction time on the Keypad and press Enter. The Keypad will show the corresponding Reaction Loop number.The time can be as short as 0.005 s.
-
20.Set the 8-way Reaction Loop Valve to the number of the Reaction Loop accordingly.Note: if the Reaction Loop number is in the range of 4–7, add more 25 mM Tris-HCl buffer (pH 7.5) to Drive Syringe A and C to the tops of the two plungers attached to the Step Motor.
-
21.
Fill 1-mL Luer Lock disposable syringes with Pre-mixtures I and II; attach the syringes to Sample Load D and E, respectively.
-
22.
Set the Sample Load Valve on the left to the Load position and the one on the right to the Flush position.
-
23.Load Pre-mixture I through Sample Load D to the edge of the 8-way Reaction Loop Valve.Be sure that no bubbles are in the sample loop and that Pre-mixture I does NOT cross the edge of the 8-way Reaction Loop Valve. This is critical for experimental accuracy.
-
24.
Load Pre-mixture II though Sample Load E up to the edge of the 8-way Reaction Loop Valve, as in Steps 22 and 23.
-
25.
Set all Syringe Load Valves and Sample Load Valves at the Fire positions and insert the end of the Exit Line into a labeled empty Eppendorf tube (Step 15).
-
26.
Press G or Start on the Keypad.
-
27.
Wait until the reaction mixture flows out from the Exit Line into the Eppendorf tube.
-
28.
Wash the Sample Loops, Reaction loop, Flushing Lines, and Exit Line with H2O, rinse with methanol and dry them, as in Steps 12 and 13.
-
29.
Conduct experiments with different reaction times (0.005, 0.0075, 0.01, 0.015, 0.02, 0.03, 0.04, 0.06, 0.08, 0.1, 0.2, 0.5, 1, and 5 s) as in Steps 22–28.
Note: the instrument is used solely for accurate timing, and the product from each time point is collected in a separate tube for further analysis.
-
30.For a control experiment, replace Pre-mixture II with H2O and set the reaction time at 0.005 s.Be sure to remove all of the Pre-mixture II from the parts of the instrument (Sample Load E to the Sample Loop) and dry thoroughly, before loading H2O.
-
31.
Conduct each reaction at least twice for good data quantitation.
-
32.Clean and dry the Sample Loads, Sample Loops, Reaction Loops, Flush Lines, and the Exit Line as in Steps 10–14.If necessary, wash with the following reagents: 2 M NaOH, H2O, 2 M HCl, H2O, and methanol. Then dry the loops and lines.
-
33.
Use the Keypad to move the Step Motor up. Wash the Drive Syringe with H2O as in Step 7.
-
34.
Turn off the water bath and pump.
-
35.
Mix 10 µL of each reaction product with 10 µL of gel loading buffer, respectively.
Prepare, Run and Image 18% Polyacrylamide Gel
-
36.
Dissolve 45 mL of 40% acrylamide/bis (19:1, 5% crosslinker, w/v), 10 mL 5X TBE buffer, and 45 g urea in H2O. The final volume is 100 mL.
-
37.
Add 500 µL 15% ammonium persulfate (APS, w/v) and 40 µL TEMED, stir the solution briefly, and cast the gel.
-
38.
Allow the gel to polymerize for about 2 h.
-
39.
Pre-warm the gel in 0.5X TBE buffer by applying power at 100 W for about 30 min.
-
40.
Heat the samples from Step 35 at 95 °C for 5 min in a dry block heater and put them on ice immediately for 2 min. The samples are ready to load.
-
41.Load 7.5 µL of each sample in the pre-warmed gel. Run at 100 W for about 2 h.The acrylamide concentration of the gel and running time can be changed according to the size of the oligonucleotide.
-
42.
Separate the two glass plates casting the gel; the gel should stick to one of them. Cover the gel with plastic cling wrap.
-
43.Put the gel on the glass platen of the Typhoon instrument. [*Copy Editor: Per the author “It should be “platen”, according to the instrument of Typhoon machine: http://bascompalmer.org/documents/ge_healthcare_typhoon_operating_instructions.pdf”]Make sure that the plastic cling wrap side is attached to the glass platen of the Typhoon instrument.
-
44.Select the scan area, select “fluorescence” as the acquisition mode, and choose “526 SP Fluorescein, AlexaFluor488” as the emission filter.Alternatively, if the primer (in Step 1) is labeled by another fluorescent dye or 32P, the acquisition mode and the emission filter should be changed accordingly.
-
45.
Scan the gel. The result is ready for data quantitation.
Data Quantitation and Analysis
-
46.
Open the “.gel” file in the ImageJ software.
-
47.
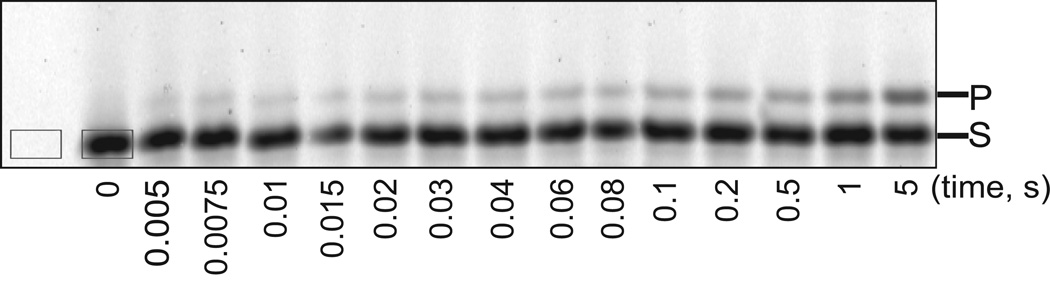
Open the ROI manager function. Quantify bands and the corresponding background as shown in Fig. 2.
-
48.
Calculate the “IntDen” (integrated density, total values of the pixels in the selected image) for substrate (S) and product (P): band IntDen minus background IntDen. The results from the two time points are shown in Table 3 as SIntDen and PIntDen.
-
49.
The results and calculations of the two time points are shown as examples in Table 3. The calculations are applied to all of the data set.
-
50.
Open GraphPad Prism, select XY under the NEW TABLE & GRAPH heading, and enter replicate values in adjoining columns.
-
51.
Enter the reaction time in the X column and the calculated product concentration in the Y column.
-
52.Analyze the data by fitting them to the following burst equation:
-
53.
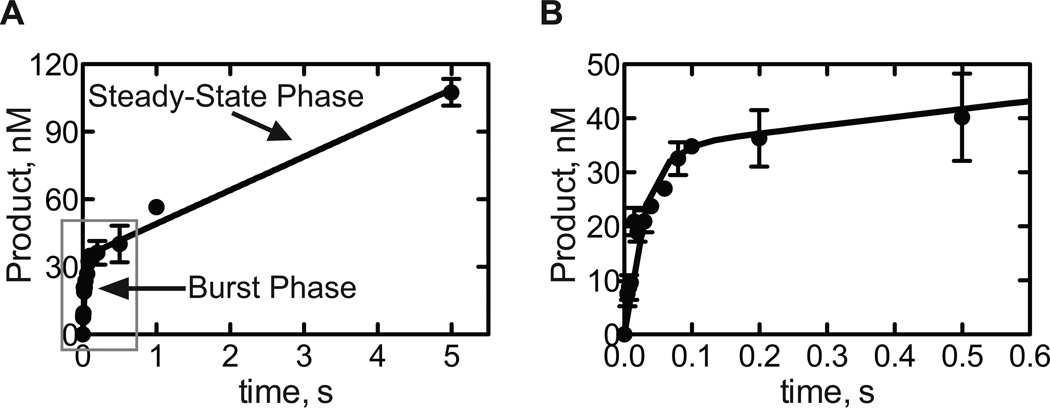
A graph is shown in Fig. 3 and the resulting parameters are in Table 4. “A” is the burst amplitude (the apparent concentration of the active form of the enzyme), “b” is the burst rate, and v is the steady-state rate times the total enzyme concentration. In this case, the burst amplitude is 34 ± 2 nM, the burst rate is 35 ± 5 s−1, and the steady-state rate is 0.060 ± 0.003 s−1.
The total enzyme concentration=Final concentration of DNA polymerase/2=250 nM.
Fig. 2.
Denaturing polyacrylamide gel image of the pre-steady-state reaction products of hpol η R61M incorporation of dCTP opposite 8-oxodG. The gray rectangles indicate how to select a band and a control for quantitation.
Table 3.
An example of gel quantification (data derived from the ImageJ software).
| Time, s | SIntDen | PIntDen | Ra | R-R0b | Product Conc, nMc |
|---|---|---|---|---|---|
| 0 | 1422 | 52.9 | 0.0359 | 0 | 0 |
| 0.005 | 1148 | 65.8 | 0.0542 | 0.0183 | 9.2 |
R=PIntDen/(PIntDen+SIntDen)
R0 is the calculated result from the control reaction conducted in Step 30 (the reaction time lists as 0 s in the table).
Product Concentration=(R-R0) × 500 nM (500 nM is the reaction concentration of the annealed DNA duplex). The reaction concentration is one-half of the final concentration in Table 1 because Pre-mixes I and II are mixed in equal volumes in the reaction.
Fig. 3.
A. Pre-steady-state analysis of dCTP insertion opposite 8-oxodG by hpol η R61M. The data are fit to the burst equation. B. Close view of the early time points.
Table 4.
The analysis results, as parameters from GraphPad Prism (to two significant digits)
| Best-fit values | |
|---|---|
| A | 34 |
| b | 35 |
| v | 15 |
| Std. Error | |
| A | 2 |
| b | 5 |
| v | 1 |
Reagents and Solutions
Gel Loading buffer
90% (v/v) formamide
10 mM EDTA, pH 8.0
0.01% (w/v) bromophenol blue
0.01% (w/v) xylene cyanol FF
COMMENTARY
Background Information
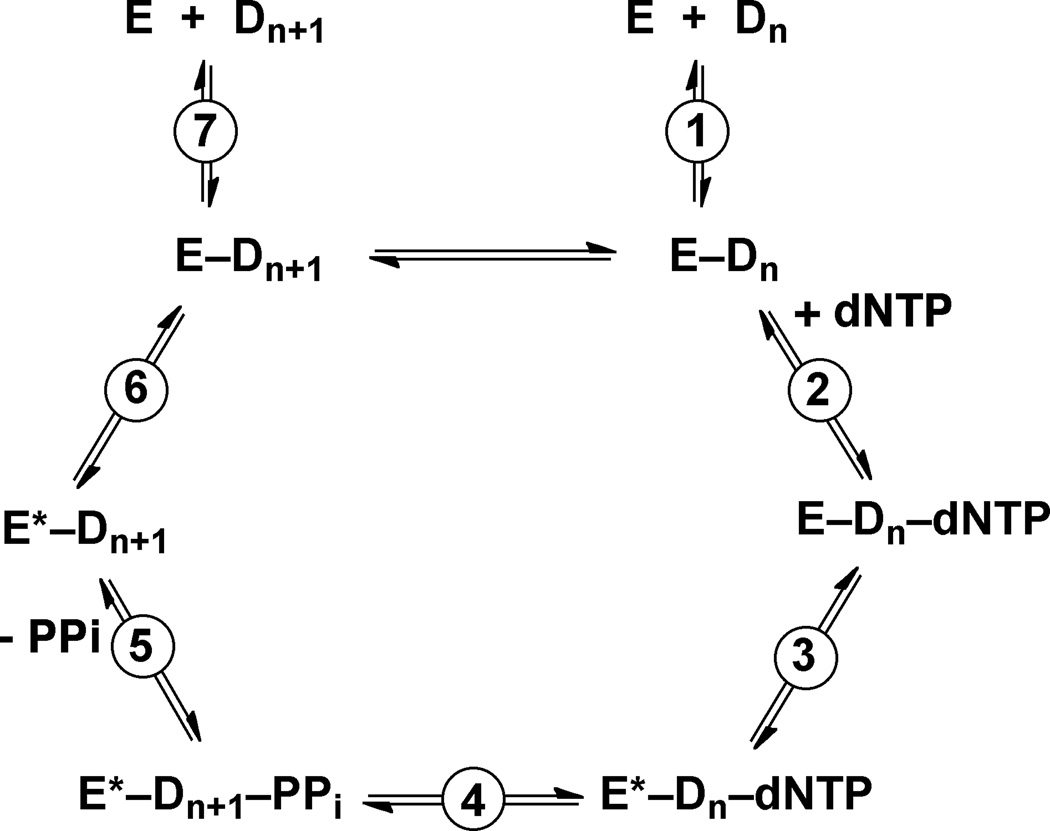
In general, the major role of DNA polymerases is to extend the primer strand by incorporating dNTP opposite the template base (or the lesion). The catalytic cycle is composed of several steps: the DNA polymerase (E) binds with DNA (D) to form a binary complex; the binary complex further interacts with dNTP to generate a ternary complex; the chemical bond formation for the polymerization occurs (Reaction 4 in Fig 4); the generated pyrophosphate is released from the complex; and the binary complex dissociates or goes into the next catalytic cycle (Fig 4). For Y-family DNA polymerases Dpo4 and hpol κ, it has been shown that the rate-limiting step is the release of pyrophosphate (Beckman et al., 2008; Zhao et al., 2014). The slowest step for other DNA polymerases is also generally believed to occur after the chemical bond formation, as revealed by burst kinetic results, and could be either the conformational relaxation or product release (Fig 4) (Guengerich, 2006; Johnson, 1993).
Fig. 4.
A model for the catalytic cycle of a DNA polymerase reaction. The figure is adapted from Beckman et al. (Beckman et al., 2008).
Pre-steady-state kinetics is a powerful method used to obtain kinetic parameters in both the burst phase (transient phase or the first catalytic cycle without the steady-state rate-limiting step) and the steady-state phase (multiple catalytic cycles with the rate-limiting step). In the reaction, the polymerase concentration is usually less than the DNA concentration. Ideally, the polymerase is saturated with DNA substrate, and the extra DNA is present for the reaction in subsequent catalytic cycles (the steady state). The polymerase concentration should not be too low; otherwise the product signal will be hard to detect. Pre-steady-state assays are conducted by varying the reaction time, and data can be fit to the burst equation:
“A” represents the apparent concentration of the active form of the enzyme, also called burst amplitude, “b” is the burst rate, and v is the steady-state rate times the total enzyme concentration, as shown in Steps 52 and 53 (Guengerich, 2006; Sassa et al., 2013;Su et al., 2015;Patra et al., 2014; Patra et al., 2015). The fitted curve includes both the burst phase (exponential) and the steady-state phase (linear) (Fig. 3). This log-linear analysis is only suitable when the slowest step is after the formation E*-Dn+1-PPi for single nucleotide incorporation by a DNA polymerase; otherwise the reaction will follow a linear course (Johnson, 1986;Johnson, 1995;Johnson, 1998).
Critical Parameters and Troubleshooting
Some possible problems and their solutions follow:
The data do not fit well to the burst equation, or the fitted curve is almost linear and the steady-state phase curve goes through the origin. Such a result indicates that the rate-limiting step occurs prior to the chemical bond formation of E*-Dn+1-PPi in the catalytic cycle of the DNA polymerase reaction (Fig. 4). In this case, the data obtained from the pre-steady-state kinetic assays are nearly identical to those in the steady-state measurements (see UNIT 7.21). This behavior is seen in some cases with bulky DNA adducts in the template.
In the fitted curve, almost all the data points are located in the steady-state phase, and the y-intercept of the linear course of the steady-state phase is positive. The reaction time should be reduced, if possible. The shortest time for an RQF-3 rapid quench-flow system is 0.005 s, and reducing the reaction time may not be enough for some DNA polymerases with high catalytic efficiency. Because the enzymatic reaction depends on temperature, reducing the reaction temperature from 37 °C to 25 °C (as shown in the model system in this protocol), 16 °C, or even 4 °C may be useful in making the analysis possible.
No product mixture flows out from the Exit Line into the Eppendorf tube in Step 27. If the Reaction Loop number is in the range of 4–7, be sure to add additional 25 mM Tris-HCl buffer (pH 7.5) to the Drive Syringes A and C to the tops of the two plungers attached to the Step Motor (note in the Step 20).
No DNA product band is observed after imaging the gel in Typhoon. There are two possible reasons: the amount of product is too small to be detected, or the product and the substrate are not separated in the polyacrylamide gel. If the reason is the former, increasing the reaction time or the polymerase concentration (but still keeping it less than the DNA concentration) may solve this problem. However, to separate the substrate and the product in the gel, one can decrease the percentage of acrylamide in the gel, increase the running time, or heat the samples at 95 °C before loading in the gel.
Anticipated Results
Three parameters can be obtained after the successful measurement and analysis of the pre-steady-state data of single nucleotide insertion by a DNA polymerase. The burst amplitude (the apparent concentration of the active form of the enzyme, 34 ± 2 nM in this model) is usually smaller than the ideal total enzyme concentration (250 nM in this model), because of the impurity of the purified DNA polymerase or the formation of the inactive form(s) of E-Dn, E-Dn-dNTP, or E*-Dn-dNTP complex(es) during the catalytic cycle in Fig 4 (Furge and Guengerich, 1999). In addition, the burst rate and the steady-state rate can be obtained. The burst rate is for the exponential burst phase, and the steady-state rate is the slope of the steady-state phase divided by the total enzyme concentration (Fig 3).
Time Considerations
The timeline for pre-steady-state kinetic assays of single-nucleotide incorporation by a DNA polymerase is about 1 to 2 days for one complete set of experiments, and this depends on the efficiency of the user of the RQF-3 rapid quench-flow instrument. About 2 h is required for the DNA duplex substrate annealing, 5 – 6 h (dissolving 0.5 h, polymerizing 2 h, pre-warming 0.5 h, and loading and running 2 – 3 h) for preparation and electrophoresis of the polyacrylamide DNA gel, and 1 h for gel imaging, quantitation, and data analysis. During the 2 h for DNA duplex annealing, the RQF-3 rapid quench-flow system can be warmed and washed, the reagents for mixtures can be prepared on ice, and the tubes labeled. The reactions can be conducted during the 2 h for gel polymerization. Thus, the total time is about 8 – 9 h, approximately one workday. However, since it is critical and challenging to control the loading of pre-mixtures to the edges of the 8-way Reaction Loop Valve (as shown in Step 23), a first-time user may need some time to become familiar with the RQF-3 rapid quench-flow system. We suggest that a first-time user become familiar with the instrument and conduct the reactions the day before running the polyacrylamide DNA gel.
Acknowledgments
The example of this work is taken in part from Su et al. (2015). This work was supported by National Institutes of Health Grants R01 ES010375 and R01 ES010546.
Literature Cited
- Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman JW, Wang Q, Guengerich FP. Kinetic analysis of correct nucleotide insertion by a Y-family DNA polymerase reveals conformational changes both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence. J. Biol. Chem. 2008;283:36711–36723. doi: 10.1074/jbc.M806785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge LL, Guengerich FP. Explanation of pre-steady-state kinetics and decreased burst amplitude of HIV-1 reverse transcriptase at sites of modified DNA bases with an additional, nonproductive enzyme-DNA-nucleotide complex. Biochemistry. 1999;38:4818–4825. doi: 10.1021/bi982163u. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Advances in transient-state kinetics. Curr. Opin. Biotechnol. 1998;9:87–89. doi: 10.1016/s0958-1669(98)80089-x. [DOI] [PubMed] [Google Scholar]
- Patra A, Zhang Q, Su Y, Egli M, Guengerich FP. Structural and kinetic analysis of nucleoside triphosphate incorporation opposite an abasic site by human translesion DNA polymerase. J. Biol. Chem. 2015;290:8028–8038. doi: 10.1074/jbc.M115.637561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra A, Nagy LD, Zhang Q, Su Y, Muller L, Guengerich FP, Egli M. Kinetics, structure, and mechanism of 8-oxo-7,8-dihydro-2´-deoxyguanosine bypass by human DNA polymerase. J. Biol. Chem. 2014;289:16867–16882. doi: 10.1074/jbc.M114.551820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa A, Beard WA, Shock DD, Wilson SH. Steady-state, pre-steady-state, and single-turnover kinetic measurement for DNA glycosylase activity. J. Visualized Expts. 2013:e50695. doi: 10.3791/50695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Patra A, Harp JM, Egli M, Guengerich FP. Roles of residues Arg-61 and Gln-38 of human DNA polymerase η in bypass of deoxyguanosine and 7,8-dihydro-8-oxo-2´-deoxyguanosine. J. Biol. Chem. 2015;290:15921–15933. doi: 10.1074/jbc.M115.653691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Pence MG, Eoff RL, Yuan S, Fercu CA, Guengerich FP. Elucidation of kinetic mechanisms of human translesion DNA polymerase κ using tryptophan mutants. FEBS J. 2014;281:4394–4410. doi: 10.1111/febs.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]