Abstract
Background:
There are now over 100 established genetic risk variants for schizophrenia; however, their influence on brain structure and circuitry across the human lifespan are not known.
Methods:
We examined healthy individuals 8–86 years of age, from the Centre for Addiction and Mental Health, the Zucker Hillside Hospital, and the Philadelphia Neurodevelopmental Cohort. Following thorough quality control procedures, we investigated associations of established genetic risk variants with heritable neuroimaging phenotypes relevant to schizophrenia, namely thickness of frontal and temporal cortical regions (n = 565) and frontotemporal and interhemispheric white matter tract fractional anisotropy (FA) (n = 530).
Results:
There was little evidence for association of risk variants with imaging phenotypes. No association with cortical thickness of any region was present. Only rs12148337, near a long noncoding RNA region, was associated with white matter FA (splenium of corpus callosum) following multiple comparison correction (corrected p = .012); this single nucleotide polymorphism was also associated with genu FA and superior longitudinal fasciculus FA at p <.005 (uncorrected). There was no association of polygenic risk score with white matter FA or cortical thickness.
Conclusions:
In sum, our findings provide limited evidence for association of schizophrenia risk variants with cortical thickness or diffusion imaging white matter phenotypes. When taken with recent lack of association of these variants with subcortical brain volumes, our results either suggest that structural neuroimaging approaches at current resolution are not sufficiently sensitive to detect effects of these risk variants or that multiple comparison correction in correlated phenotypes is too stringent, potentially “eliminating” biologically important signals.
Key words: imaging genetics, cortical thickness, DTI, polygenic risk, asymmetry
Introduction
More than 100 genetic risk loci were identified in the largest schizophrenia genome-wide association study (GWAS) to date.1 An important next step is to identify the neural circuitry associated with these variants to help detect genetically mediated risk mechanisms of this devastating brain disorder. A recent GWAS in a large number of healthy individuals identified a small number of variants associated with subcortical brain volumes, including hippocampus and striatum; however, none of those loci were any of the 100+ schizophrenia risk loci.2 While there is evidence for involvement of the hippocampus, striatum, and other subcortical structures in schizophrenia,3,4 there is perhaps even better evidence for alterations in cortical brain regions and related circuitry.5,6 Frontal and temporal cortical thickness reductions are a well-replicated finding in schizophrenia.6,7 In addition, diffusion anisotropy reductions in frontotemporal and interhemispheric white matter tracts are often found in people with schizophrenia compared to healthy controls,8,9 including in high-risk individuals.10 Therefore, a focus on cortical brain phenotypes may provide some insight into the neural mechanisms via which these genetic variants confer risk for schizophrenia.
Consistent with these proposed neural mechanisms, the first GWAS-single nucleotide polymorphism (SNP) imaging study relevant to schizophrenia demonstrated effects of a ZNF804A allele on functional frontotemporal and interhemispheric dysconnectivity phenotypes in healthy adult controls.11 Others have also used the approach of studying healthy controls to help “validate” the potential effects of risk variants on structural and/or functional neural circuitry, along with neurocognitive performance.12,13 However, not all studies demonstrate an association,14 and a recent systematic review has highlighted a number of inconsistencies in imaging genetics studies using white matter phenotypes,15 in part because phenotypes tested are often not the same.
An underutilized aspect of intermediate phenotype neuroimaging studies is the dimension of time. The use of neurodevelopmental samples, along with adult samples, can help to identify “when” in the lifespan genetic risk variants confer effects on brain structure or function, in addition to the more commonly assessed question of “where” in the brain genetic risk occurs.16 An effect observed early in life might be interpreted as more relevant for psychosis or schizophrenia disease risk while an effect seen during adulthood may be more important for disease course or progression.17
The primary objective of the present study was to assess the effects of genome-wide significant variants from the largest GWAS in schizophrenia to date1 on intermediate phenotypes of cortical thickness and white matter fractional anisotropy (FA). The secondary objective was to assess the effects of polygenic risk score (PRS) on these same phenotypes. Three datasets (including the Philadelphia Neurodevelopmental Cohort [PNC]), that each assessed individuals at different stages of the lifespan, were used for examination of SNP-imaging phenotype associations, providing the largest known assessment of established genetic risk variants for schizophrenia on neuroimaging phenotypes of cortical thickness and white matter FA to date. Based on published individual GWAS SNP-neuroimaging phenotype findings at much smaller sample sizes, we hypothesized that a sizeable number of schizophrenia risk variants would be associated with cortical thickness and white matter FA phenotypes.
Methods
Participant Recruitment, Assessment, Imaging and Genetics
Centre for Addiction and Mental Health Sample.
A total of 112 healthy volunteers were recruited at the Centre for Addiction and Mental Health (CAMH; Toronto, Ontario, Canada). All individuals (age 18–86) completed extensive clinical assessment protocols including the Structured Clinical Interview for DSM-IV Disorders (SCID-I) and the Mini-Mental State Examination,18 to rule out the presence of any psychiatric disorder or a dementia, as previously described.19,20 Genotyping for all subjects was performed using the Illumina HumanOmniExpress-12v1.0 BeadChips assay. T1-weighted and diffusion-weighted magnetic resonance imaging (MRI) acquisitions were as previously described21; see supplementary methods for details. The study was approved by the Research Ethics Board of CAMH, and all participants provided informed, written consent. After excluding all non-Caucasian subjects (those not matched to the HapMap 3 reference populations CEU or TSI; http://www.broadinstitute.org/~debakker/p3.html), ethnic outliers, and highly related individuals, n = 96 subjects remained for cortical thickness analyses and n = 93 for diffusion tensor imaging (DTI) analyses.
Zucker Hillside Hospital Sample.
A total of 107 healthy Caucasian subjects (age 8–68) were examined from an ongoing study at the Zucker Hillside Hospital (ZHH), Glen Oaks, NY, recruited by advertisement and word of mouth. Exclusion criteria included serious medical illness and any history of psychosis or major mood disorders, as determined by structured and semistructured assessments as previously published.22–24 Genotyping for all subjects was performed using the Illumina HumanOmniExpress-12v1.0 BeadChips assay. T1-weighted and diffusion-weighted MRI acquisitions were as previously described25,26—see supplementary methods for details. This study was approved by the Institutional Review Board of the North Shore—Long Island Jewish Health System. After excluding all non-Caucasian subjects, ethnic outliers, and highly related individuals, n = 99 subjects remained for cortical thickness analyses and n = 99 for DTI analyses.
PNC Sample.
A total of 802 subjects with quality-controlled T1-weighted MRI and 705 subjects with diffusion-weighted MRI scans were analyzed from the PNC cohort.27 Genome-wide genotyping was performed on Affymetrix (6.0 Genechip and Axiom) and Illumina (Human610, HumanHap550 v1.0, and HumanHap550 v3.0) platforms. T1-weighted and diffusion-weighted MRI acquisitions were as previously described28—see supplementary methods for details. The institutional review boards of both the University of Pennsylvania and the Children’s Hospital of Philadelphia approved all study procedures. After excluding all non-Caucasian subjects, ethnic outliers, and highly related individuals, n = 370 subjects remained for cortical thickness analyses and n = 338 for DTI analyses (table 1).
Table 1.
Sample Demographics Summary
| Demographic | CAMH (n = 96) | ZHH (n = 99) | PNC (n = 370) | Diff. (p)* | Combined (n = 565) |
|---|---|---|---|---|---|
| Cortical thickness analysis | |||||
| Age, mean (y) (SD) | 46 (18.8) | 33 (16) | 15 (3.6) | <.0001 | 23 (16.4) |
| Sex | 50M, 46 F | 53M, 47 F | 178M, 193 F | .58 | 281M, 286 F |
| Handedness | 89 R, 7 L | 89R, 10 L | 328 R, 42 L | .57 | 506 R, 59 L |
| IQ, mean score (SD)a | 118 (8.5)b | 106 (9.6)c | 106 (13.2)d | — | — |
| CAMH (n = 93) | ZHH (n = 99) | PNC (n = 338) | Diff. (p)* | Combined (n = 530) | |
| Fractional anisotropy analysis | |||||
| Age, mean (y) (SD) | 46 (18.7) | 32 (16) | 15 (3.5) | <.0001 | 23 (16.4) |
| Sex | 48M, 45 F | 52M, 47 F | 164M, 178 F | .66 | 264M, 270 F |
| Handedness | 87 R, 6 L | 88R, 10L, 1 NA | 303 R, 38L, 1 NA | .44 | 478 R, 54L, 2 NA |
| IQ, mean score (SD)a | 118 (8.5)e | 106 (9.6)f | 107 (13.2)g | — | — |
Note: CAMH, Centre for Addiction and Mental Health; IQ, Intelligence Quotient; NA, data not available; PNC, Philadelphia Neurodevelopmental Cohort; ZHH, Zucker Hillside Hospital. R, right handed; L, left handed; M, male; F, female.
aIQ was evaluated using different tests in each sample subset; for CAMH sample, the Wechsler Test of Adult Reading (WTAR); for ZHH, the Wide-Range Achievement Test v3 (WRAT-3); and for PNC, the Wide-Range Achievement Test v4 (WRAT-4). Therefore, no cross-sample or combined-sample statistical comparison of IQ score was performed.
IQ data were only available for a subset of each sample: b n = 85, c n = 88, d n = 370, e n = 84, f n = 87, g n = 337.
*p values for sample differences are 2 sided and were calculated using ANOVA for continuous data (age) and Fisher’s Exact Test for factor data (sex, handedness).
Image Analysis
Cortical Thickness Processing.
To calculate vertex-wise cortical thickness in the 3 samples, the T1-weighted images were submitted to the CIVET pipeline (version 1.1.10, Montreal Neurological Institute at McGill University, Montreal, Québec, Canada) to generate cortical models, as described previously.26 Quality control criteria for each subject were generated by examining quantitative metrics associated with the quality of image registration and surface extraction. For each subject, vertex-wise cortical thickness maps were parcellated using cortical regions defined in the LONI Probabilistic Brain Atlas29 (LPBA40; available online at http://www.loni.usc.edu/atlases/), with additional manual segmentation of the middle frontal gyri.26 Average cortical thickness was computed in a total of 52 distinct cortical regions; 24 frontal and temporal regions were then chosen from these for analysis (listed in supplementary table 1).
DTI Processing and White Matter Parcellation.
Correction for head motion and eddy currents by affine registration between each diffusion weighted image and b=0 reference volume was performed using FSL (v4.1.9)30 was performed. Diffusion gradient orientations were also corrected for rotational motion.31,32 In the CAMH dataset, the three 1.5T diffusion repetitions were averaged, to increase signal to noise, prior to FA calculation.
Tract-based spatial statistics (TBSS) is a commonly used technique to investigate white matter differences between populations.33,34 The TBSS pipeline provides a framework to align FA images from multiple subjects to a template FA image in MNI152-space for subsequent voxelwise analysis. Nonlinear transformations are first calculated between each FA image to the standard FA template. Each subject’s registered image was manually inspected for gross anatomical inconsistencies and/or any apparent warping. These transformations are then applied to each subject FA image and an average FA is generated from these registered FA maps. The mean FA image is utilized to generate a white matter skeleton onto which the maximum FA from each of the subjects is projected.
White matter parcellation analysis was then performed using regions of interest (ROI) defined in the ENIGMA protocol.35 A set of FA maps were first registered to a common space defined by the MNI152 FA template. The mean FA values were calculated over each white matter region as defined by intersection of the Mori John Hopkins white matter atlas in MNI152-space and the rigid white matter skeleton resulting from the TBSS pipeline. A total of 20 ROIs were selected for analysis (listed in supplementary table 2).
Genetics Analysis
Quality control of genetic data was performed according a standard protocol as outlined by Anderson et al (2010).36 Imputation was performed for all samples and platforms separately with PLINK (v1.90b)37 and IMPUTE2 (v2.3.1)38,39 software packages, using the 1000 Genomes Phase I integrated haplotypes (b37) reference panel (see supplementary methods). Ethnicity was determined from the genetic data using multidimensional scaling, and degree of cryptic relatedness between subjects was calculated using estimates of identity-by-descent, both using PLINK (see supplementary methods).
Statistical Analysis
Power Analysis.
Due to the uncommonly large sample size analyzed in the Ripke et al (2014)1 paper, several SNPs found to be genome-wide significant for schizophrenia have low minor allele frequencies (MAFs; 9 SNPs have MAF <0.1); this results in inadequate statistical power when analyzed in relatively smaller samples, especially when considering continuous traits and additive genetic models (for a SNP with MAF = 0.1, in sample of n = 1000, only 10 subjects would be expected to possess the rare homozygous genotype). Therefore, Quanto software (v1.2.4, http://biostats.usc.edu/Quanto.html) was used to determine the MAF threshold for inclusion in our study for cortical thickness and white matter microstructure analyses separately. For cortical thickness, in order to detect small effect sizes (Cohen’s D = 0.2) in the combined sample (CAMH/ZHH/PNC) of n = 565, SNPs with MAF <0.22 were excluded from analysis. For white matter microstructure, in the combined sample of N = 530, the corresponding MAF threshold was 0.27.
Primary Analyses.
Cortical Thickness
Statistical analysis was conducted using R software (v3.1.1)40 with Frank Harrel’s “rms” statistical package.41 After excluding SNPs with MAF <0.22, 60 of the 91 SNPs remained. Of these 60 SNPs, 2 (rs77502336 and rs6434928) showed slight deviations from Hardy-Weinberg equilibrium (HWE), with chi-squared p = .009 and p = .045, respectively, and were excluded from further analyses (final number of SNPs = 58). We conducted combined-sample analyses in 2 steps for each SNP/cortical ROI combination: first, between-sample variance in thickness was accounted for using ordinary least squares regression, by modeling average thickness as a product of study site. Second, to identify the effects of each SNP, residuals from this first model were entered into a second linear regression model that included additive genotype, age, sex, and handedness as predictors (supplementary methods). In total, 1392 SNP × ROI combinations were tested, resulting in a Bonferroni multiple-correction significance threshold of p = 3.59×10−5 (2 sided). Corresponding models including age-by-SNP interaction terms were also tested in the combined sample. For models where the combined-sample genetic effect had a p value below this threshold, post hoc tests of dominant and recessive models were tested within each subsample separately. To test robustness of our findings, linear mixed-effects regression42 was also performed, with study site modeled as a random effect and genotype, sex, age, and handedness modeled as fixed effects, for all primary analyses. Results according to this method were not different (supplementary methods).
White Matter Microstructure
The same 2-step modeling approach was used for analyzing the effect of each SNP on average FA for each white matter ROI. After excluding SNPs with MAF <0.27 and HWE p <.05, 43 remained, resulting in a total of 860 tests and a Bonferroni multiple-correction significance threshold of p = 5.81×10−5. Due to significant observed nonlinear effects of age on all average FA measures, as well as the known trajectory of FA change across the lifespan,43,44 age was modeled using a second degree polynomial. Again, corresponding models including age-by-SNP interaction terms were also tested in the combined sample.
Secondary Analyses.
Polygene Risk Score
In order to assess the additive effect of genome-wide significant SNPs from the Ripke et al (2014) paper,1 we calculated a single PRS for each subject in all 3 study samples and evaluated the effect of this score on cortical thickness and white matter microstructure. The polygene score for each subject was calculated by multiplying the number of risk alleles possessed at each SNP by the natural logarithm of its respective combined-sample OR (for schizophrenia risk, from Ripke et al [2014]1) and summing the products. Association of the PRS with each imaging phenotype was tested in the combined sample using the same 2-step regression technique as for the individual SNP analyses.
Hemispheric Laterality
To explore potential genetic effects on hemispheric asymmetry (a potential neuroimaging phenotype for schizophrenia45–49) in our measures of cortical thickness and FA, we calculated a laterality index50 for each bilateral ROI using the following formula:
These indices were then used as outcomes in regression models as specified above.
Results
Primary Analyses
Associations of Genetic Risk Variants for Schizophrenia With Cortical Thickness.
No SNP was significantly associated with cortical thickness of any region tested, after correction for 1392 tests (58 SNPs × 24 cortical ROIs). Each tested cortical thickness ROI and top-associated SNP is presented in supplementary table 1. No age-by-SNP interaction was significant after correction.
Associations of Genetic Risk Variants for Schizophrenia With White Matter FA.
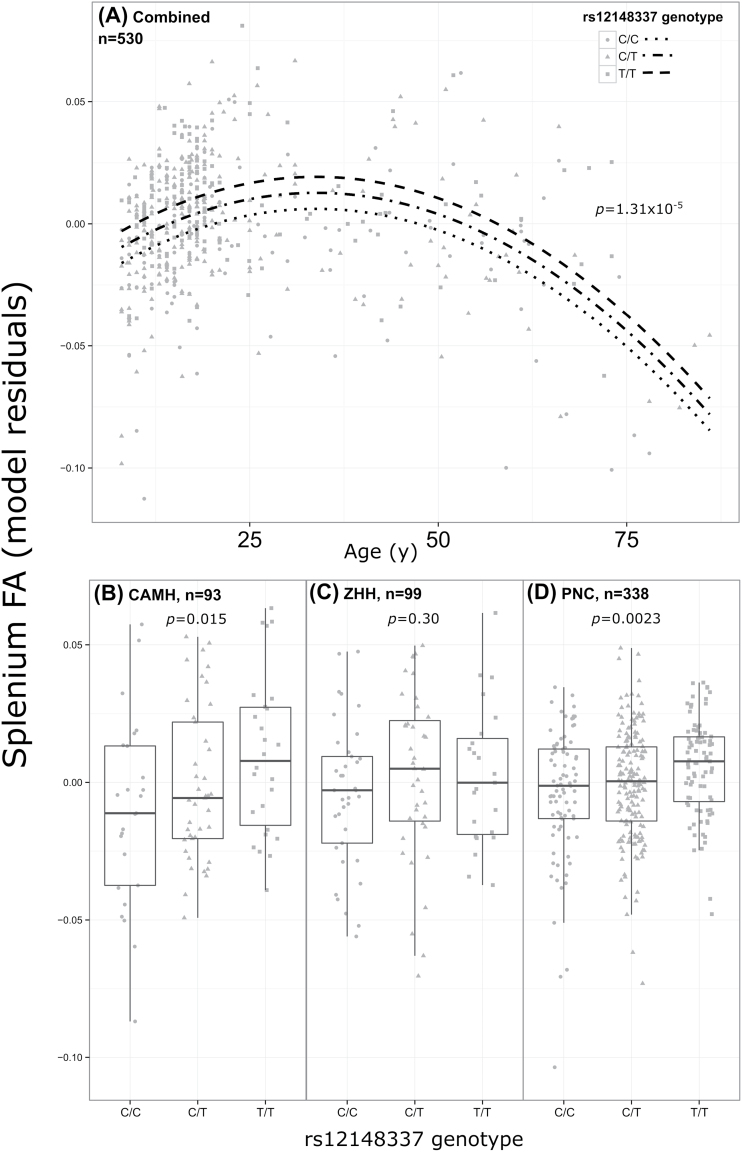
In the combined sample of 530 subjects, the T allele of rs12148337 was significantly associated with an increase in FA of the splenium, after correction for 860 tests (additive t = 4.40, p unadjusted = 1.31×10−5, p Bonferroni = .011). While the allelic direction of effect was consistent across subsamples, post hoc testing of the genotype effect within each subsample separately yielded significant effects at p <.05 in the CAMH (n = 93, additive t = 2.47, p = .015) and PNC (n = 338, additive t = 3.07, p = .0023) datasets only (figure 1). The direction of effect between homozygote groups in the ZHH sample was consistent with the other samples (ie, T/T subjects had greater thickness than C/C subjects), however, the heterozygote group showed the greatest cortical thickness, which is inconsistent with an additive model; the additive effect was not statistically significant (n = 99, t = 1.04, p = .30). The rs12148337 SNP also showed an association with a number of other white matter tracts (at uncorrected p < .005) including genu of the corpus callosum, and the left and right superior longitudinal fasciculus among others. Only one other SNP (rs10791097) was associated with more than one white matter tract at uncorrected p <.005. Each tested FA ROI and top-associated SNP is presented in supplementary table 2. No age-by-SNP interaction was significant after correction.
Fig. 1.
Significant effect of rs12148337 on splenium fractional anisotropy in (A) combined sample of N = 530 (p = 1.31×10−5) and subsamples: (B) CAMH (p = .015), (C) ZHH (p = .30), and (D) PNC (p = .0023). Model residuals in panel (A) are derived from a different model than in (B), (C), and (D), as they are from regression predicting site-normalized residuals (residuals from final model were manually verified as being approximately normally distributed). The curves in (A) were drawn based on predicted values from the model for each genotypic group (Levene’s test for equal variance among genotype groups p = .14). Residuals plotted in (B), (C), and (D) are from regression models predicting raw splenium FA, covarying for nonlinear effects of age, as well as sex. Center lines of boxplots (panels B, C, and D) are medians, and error bars extend ±1.5 × interquartile range; p values are 2 sided.
Secondary Analyses
PRS Analysis.
The additive PRS, derived from 91 SNPs imputed with high quality, was normally distributed in both the cortical thickness (n = 565, Shapiro-Wilk p = .43) and FA (n = 530, Shapiro-Wilk p = .63) analysis. After correction for multiple testing, PRS was not associated with any neuroimaging phenotype in the combined sample (all p > .05), and no within-subsample associations survived multiple correction independently (all p Bonferroni > .05) (see supplementary figure 1).
Hemispheric Laterality.
For cortical thickness ROIs, there were no significant associations of laterality index found with any SNP in the combined sample. For tract-wise FA, one SNP showed significant association with laterality index after correction for multiple testing: the A allele of rs11139497 was associated with increased laterality index (greater left vs right asymmetry) of uncinate fasciculus FA in the combined sample, controlling for handedness (n = 528, additive t = 4.0, p unadjusted = 6.9×10−5, p Bonferroni = .047). The directionality of this effect was consistent across study subsamples (CAMH: t = 3.01, p = .003; ZHH: t = 2.0, p = .049; PNC: t = 1.6, p = .11).
Discussion
We examined the association of recently established genome-wide significant SNPs for schizophrenia with frontotemporal and interhemispheric cortical thickness and white matter FA neuroimaging phenotypes in healthy individuals across the lifespan. No SNP was associated with cortical thickness following multiple comparison correction. The rs12148337 SNP was associated with FA of the splenium of the corpus callosum following multiple comparison correction, while the rs11139497 SNP was associated with the laterality index of FA of the uncinate fasciculus. PRS was also not associated with cortical thickness or white matter FA, following correction for multiple comparisons. When taken together, our findings provide limited evidence for association of GWAS-significant SNPs for schizophrenia with neuroimaging phenotypes of cortical thickness and white matter FA.51,52
We began with all 114 SNPs from the PGC2 study, not including insertion/deletion variants. After imputation across the 3 samples, 91 remained. Based on our power calculations, we proceeded to assess 60 SNPs in relation to neuroimaging phenotypes. With primarily negative results, our findings stand in contrast to the predominantly positive findings in imaging genetics studies of GWAS-significant SNPs for schizophrenia and cortical neuroimaging phenotypes,12,53–56 including recent studies assessing PRS on cortical phenotypes, typically in much smaller samples.57–59 Our results are surprising, because a number of aspects of our study design might have biased us toward positive association. First, it might be expected that our decision to focus on frontal and temporal cortical thickness, and frontotemporal and interhemispheric white matter FA would have biased the likelihood toward positive association based on the extant literature. Our selected neuroimaging phenotypes (eg, cortical thickness) are separable from other cortical phenotypes (surface area) in their genetic underpinnings.52 There is also a wealth of evidence for alterations in frontal and temporal brain gene expression60–62 and brain structure3,63,64 and function65,66 in schizophrenia. Second, the collective sample size of our study was larger than most published imaging genetics studies, particularly those examining cortical thickness and white matter FA and those that have employed polygenic risk approaches.58,67,68 Third, the sampling of participants across the lifespan provided an opportunity to assess SNP-neuroimaging associations with the dimension of time, which might have revealed effects of SNPs on neuroimaging phenotypes at a given phase of the lifespan.
Our results were not entirely negative, however. The association of the rs12148337 SNP with FA of the splenium of the corpus callosum that survived Bonferroni correction (for 900 comparisons) was consistent in direction across all 3 samples. The splenium of the corpus callosum provides interhemispheric temporal, parietal, and occipital connections. A meta-analysis showed consistent alterations in FA in the splenium of the corpus callosum in people with schizophrenia compared to healthy controls, supporting a role for this structure in schizophrenia.69 The trajectory of splenium “maturation” as indexed by FA across our 3 samples is consistent with the literature, where a peak is reached at approximately 25 years of age.44 The effect of this SNP on splenium FA appears to be consistent across the lifespan, supporting an “early hit” model. As shown in supplementary table 2, the allelic direction of effect indicates that the rs12148337 T risk allele from Ripke et al (2014)1 is associated with higher FA in our study samples. Association at an uncorrected p <.005 of this same SNP with FA of the genu of the corpus callosum, along with left and right superior longitudinal fasciculus, which are all tracts strongly implicated in schizophrenia,5 provides added confidence for our primary finding, given that association with the splenium alone might be due to registration error, or some other spurious effect.
The rs12148337 SNP is located on chromosome 15q23 approximately 25kb 5′ of the long noncoding RNA gene RP11-543G18.1 (HaploReg v270). The SNP is also near genes MIR629, a microRNA gene and TLE3, transducin-like enhancer of split 3 which functions in the Notch signaling pathway that regulates determination of cell fate during development. Rs12148337 is in high linkage disequilibrium (r 2 = .93, D′ = 0.98; HaploReg v270) with SNP rs1971791 which has been associated with the neurodegenerative disorder, amyotrophic lateral sclerosis.71 Long noncoding RNAs play regulatory roles during transcription and translation, as well as in epigenetic regulation, and are known to play an important role in brain development, neuron function and maintenance, and neuropsychiatric disorders.72 Less is known about the rs11139497 SNP, which was associated with laterality of uncinate fasciculus FA, following multiple comparison correction. Although less typically a focus of neuroimaging investigations, good evidence for altered laterality of brain structure in schizophrenia exists.45,73 The rs11139497 SNP is located on chromosome 9, approximately 63kb 5′ of the FAM75B (also SPATA31B1P or C9orf36B) gene within an uncharacterized locus (LOC105376107, ncRNA). The NCBI Gene database lists 2 Gene Ontology biological process terms for this pseudogene: cell differentiation and spermatogenesis, although there are no supporting publications (evidence inferred from electronic annotation).
Some potential limitations deserve consideration. It is possible that some of the SNPs examined might be associated with parietal or occipital brain structure, given reports of alterations in these brain regions74–76 or of more anatomically widespread alterations in brain structure77 in schizophrenia. We did not study patients with schizophrenia: effects of genetic variants in patient populations may be different than those of controls. A previous study17 by our group found no effect of the MIR137 GWAS-significant SNP on these same neuroimaging phenotypes in healthy individuals but found a prominent effect in patients with schizophrenia or schizoaffective disorder, in addition to effects on age at onset. Different scanners were used at each site. Despite rigorous statistical modeling that took potential study site effects into account, it remains possible that there was an effect of site or an effect of the scanner at that site which may have influenced our results. Ours is not the first study, however, to consider imaging genetics data from multiple sites.78 The main advantage of a multisite analysis, which is also true of the present study, is the increase in sample size and accompanying power. Some have argued that with the appropriate methodological approaches, sample sizes much smaller than those used in the present study should be able to detect effects of genetic variation on brain structure or function.79,80 In contrast, others have argued that much larger sample sizes are needed. However, 2 studies that included >10000 participants2,81 revealed no significant association of any schizophrenia GWAS-significant variant with subcortical brain volumes, which suggests that an especially large sample size may not be the answer. The SNPs that we were underpowered to examine might have effect on our neuroimaging phenotypes of interest, especially since more rare variants may have disproportionately large effect sizes.
When taken together, our findings provide limited evidence for effects of genome-wide significant risk variants for schizophrenia or for PRS on well-established frontotemporal and interhemispheric neuroimaging phenotypes of cortical thickness and white matter FA. Even larger sample sizes may not be the answer for discovery of neural mechanisms of established schizophrenia risk variants, although such studies would need to be conducted to be certain. Since schizophrenia is a brain disorder, it may be that structural neuroimaging phenotypes at current resolution82 are not sufficiently sensitive to reliably detect associations with these risk variants in healthy individuals. Notably, most PGC-derived susceptibility SNPs are thought to be regulatory in nature. It may be that variants which play a direct role in protein conformation (eg, BDNF Val66Met21,83,84 and APOE ε420,85,86 important in neuropsychiatric disorders) may have greater impact on brain, which is detectable using current structural MRI approaches. Alternatively, the statistical threshold for detection in studies with many comparisons may be too stringent, “drowning out” important biological clues and mechanisms, particularly given that thickness of brain regions tend to covary, as does FA of different white matter tracts,8 and as such may not be truly “independent.” Relaxing Bonferroni thresholds for imaging genetics studies, possibly in combination with plausible in vitro or complementary analyses, may provide a clearer window into novel genetically mediated risk mechanisms for schizophrenia and therapeutic opportunities for this debilitating disorder.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
Canadian Institutes of Health Research Vanier Canada Graduate Scholarship and the Peterborough K.M. Hunter Graduate Scholarship to D.F.; the Canadian Institutes of Health Research, the CAMH Foundation, Ontario Ministry of Research and Innovation, Brain and Behavior Research Foundation, and Canada Foundation for Innovation to A.N.V.; and the National Institute of Mental Health (grant numbers R01MH099167 and R01MH102324 to A.N.V., R01MH079800 and P50MH080173 to A.K.M.). No sponsor or funder played any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Supplementary Material
Acknowledgments
A.K.M. is a consultant to Forum Pharmaceuticals and Genomind Inc. J.L.K. has received honoraria from Roche, Novartis, and Eli Lilly Co. He also serves in an unpaid role on the advisory board to Assure Rx. A.N.V., D.F., A.L.W., D.J.R., M.L., S.P., P.R.S., and T.L. reported no biomedical financial interests or potential conflicts of interest.
References
- 1. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hibar DP, Stein JL, Renteria ME, et al. ; Alzheimer’s Disease Neuroimaging Initiative; CHARGE Consortium; EPIGEN; IMAGEN; SYS Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shenton ME, Dickey CC, Frumin M, McCarley RW. A review of MRI findings in schizophrenia. Schizophr Res. 2001;49:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adriano F, Caltagirone C, Spalletta G. Hippocampal volume reduction in first-episode and chronic schizophrenia: a review and meta-analysis. Neuroscientist. 2012;18:180–200. [DOI] [PubMed] [Google Scholar]
- 5. Wheeler AL, Voineskos AN. A review of structural neuroimaging in schizophrenia: from connectivity to connectomics. Front Hum Neurosci. 2014;8:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Voineskos AN, Foussias G, Lerch J, et al. Neuroimaging evidence for the deficit subtype of schizophrenia. JAMA Psychiatry. 2013;70:472–480. [DOI] [PubMed] [Google Scholar]
- 7. Rimol LM, Nesvåg R, Hagler DJ, Jr, et al. Cortical volume, surface area, and thickness in schizophrenia and bipolar disorder. Biol Psychiatry. 2012;71:552–560. [DOI] [PubMed] [Google Scholar]
- 8. Voineskos AN, Lobaugh NJ, Bouix S, et al. Diffusion tensor tractography findings in schizophrenia across the adult lifespan. Brain. 2010;133:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kubicki M, Styner M, Bouix S, et al. Reduced interhemispheric connectivity in schizophrenia-tractography based segmentation of the corpus callosum. Schizophr Res. 2008;106:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. von Hohenberg CC, Pasternak O, Kubicki M, et al. White matter microstructure in individuals at clinical high risk of psychosis: a whole-brain diffusion tensor imaging study. Schizophr Bull. 2014;40:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esslinger C, Walter H, Kirsch P, et al. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. [DOI] [PubMed] [Google Scholar]
- 12. Voineskos AN, Lerch JP, Felsky D, et al. The ZNF804A gene: characterization of a novel neural risk mechanism for the major psychoses. Neuropsychopharmacology. 2011;36:1871–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bigos KL, Mattay VS, Callicott JH, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry. 2010;67:939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cousijn H, Eissing M, Fernández G, et al. No effect of schizophrenia risk genes MIR137, TCF4, and ZNF804A on macroscopic brain structure. Schizophr Res. 2014;159:329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gurung R, Prata DP. What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol Med. 2015;45:2461–2480. [DOI] [PubMed] [Google Scholar]
- 16. Felsky D, Szeszko P, Yu L, et al. The SORL1 gene and convergent neural risk for Alzheimer’s disease across the human lifespan. Mol Psychiatry. 2014;19:1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lett TA, Chakravarty MM, Chakavarty MM, et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol Psychiatry. 2013;18:443–450. [DOI] [PubMed] [Google Scholar]
- 18. Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. [DOI] [PubMed] [Google Scholar]
- 19. Voineskos AN, O’Donnell LJ, Lobaugh NJ, et al. Quantitative examination of a novel clustering method using magnetic resonance diffusion tensor tractography. Neuroimage. 2009;45:370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Felsky D, Voineskos AN. APOE ϵ 4, aging, and effects on white matter across the adult life span. JAMA Psychiatry. 2013;70:646–647. [DOI] [PubMed] [Google Scholar]
- 21. Voineskos AN, Lerch JP, Felsky D, et al. The brain-derived neurotrophic factor Val66Met polymorphism and prediction of neural risk for Alzheimer disease. Arch Gen Psychiatry. 2011;68:198–206. [DOI] [PubMed] [Google Scholar]
- 22. Peters BD, Szeszko PR, Radua J, et al. White matter development in adolescence: diffusion tensor imaging and meta-analytic results. Schizophr Bull. 2012;38:1308–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaufman J, Birmaher B, Brent D, et al. Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry. 1997;36:980–988. [DOI] [PubMed] [Google Scholar]
- 24. First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Non-Patient Edition. (SCID-I/NP). 2002.
- 25. Toteja N, Guvenek-Cokol P, Ikuta T, et al. Age-associated alterations in corpus callosum white matter integrity in bipolar disorder assessed using probabilistic tractography. Bipolar Disord. 2015;17:381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wheeler AL, Wessa M, Szeszko PR, et al. Further neuroimaging evidence for the deficit subtype of schizophrenia: a cortical connectomics analysis. JAMA Psychiatry. 2015;72:446–455. [DOI] [PubMed] [Google Scholar]
- 27. Satterthwaite TD, Connolly JJ, Ruparel K, et al. The Philadelphia Neurodevelopmental Cohort: a publicly available resource for the study of normal and abnormal brain development in youth. Neuroimage. 2016;124:1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Satterthwaite TD, Elliott MA, Ruparel K, et al. Neuroimaging of the Philadelphia Neurodevelopmental Cohort. Neuroimage. 2014;86:544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shattuck DW, Mirza M, Adisetiyo V, et al. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. [DOI] [PubMed] [Google Scholar]
- 31. Jeurissen B, Leemans A, Sijbers J. Automated correction of improperly rotated diffusion gradient orientations in diffusion weighted MRI. Med Image Anal. 2014;18:953–962. [DOI] [PubMed] [Google Scholar]
- 32. Leemans A, Jones DK. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn Reson Med. 2009;61:1336–1349. [DOI] [PubMed] [Google Scholar]
- 33. Smith SM, Jenkinson M, Johansen-Berg H, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. [DOI] [PubMed] [Google Scholar]
- 34. Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(suppl 1):S208–S219. [DOI] [PubMed] [Google Scholar]
- 35. Jahanshad N, Kochunov PV, Sprooten E, et al. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the ENIGMA-DTI working group. Neuroimage. 2013;81:455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. [DOI] [PubMed] [Google Scholar]
- 40. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation For Statistical Computing; 2014. [Google Scholar]
- 41. Harrel F, Jr. Regression modeling strategies, “rms” package for R Statistical Program (v4.2-1). 2014. [Google Scholar]
- 42. Bates D, Machler M, Bolker B, et al. Linear mixed-effects models using eigen and S4, “lme4” package for R Statistical Program (v1.1-7). 2014. [Google Scholar]
- 43. Westlye LT, Walhovd KB, Dale AM, et al. Life-span changes of the human brain white matter: diffusion tensor imaging (DTI) and volumetry. Cereb Cortex. 2010;20:2055–2068. [DOI] [PubMed] [Google Scholar]
- 44. Lebel C, Gee M, Camicioli R, Wieler M, Martin W, Beaulieu C. Diffusion tensor imaging of white matter tract evolution over the lifespan. Neuroimage. 2012;60:340–352. [DOI] [PubMed] [Google Scholar]
- 45. Ribolsi M, Daskalakis ZJ, Siracusano A, Koch G. Abnormal asymmetry of brain connectivity in schizophrenia. Front Hum Neurosci. 2014;8:1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Niznikiewicz M, Donnino R, McCarley RW, et al. Abnormal angular gyrus asymmetry in schizophrenia. Am J Psychiatry. 2000;157:428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oertel-Knöchel V, Linden DE. Cerebral asymmetry in schizophrenia. Neuroscientist. 2011;17:456–467. [DOI] [PubMed] [Google Scholar]
- 48. Sommer IE, Ramsey NF, Kahn RS. Language lateralization in schizophrenia, an fMRI study. Schizophr Res. 2001;52:57–67. [DOI] [PubMed] [Google Scholar]
- 49. Gur RE, Chin S. Laterality in functional brain imaging studies of schizophrenia. Schizophr Bull. 1999;25:141–156. [DOI] [PubMed] [Google Scholar]
- 50. Catani M, Allin MP, Husain M, et al. Symmetries in human brain language pathways correlate with verbal recall. Proc Natl Acad Sci USA. 2007;104:17163–17168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kochunov P, Jahanshad N, Marcus D, et al. Heritability of fractional anisotropy in human white matter: a comparison of Human Connectome Project and ENIGMA-DTI data. Neuroimage. 2015;111:300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eyler LT, Chen CH, Panizzon MS, et al. A comparison of heritability maps of cortical surface area and thickness and the influence of adjustment for whole brain measures: a magnetic resonance imaging twin study. Twin Res Hum Genet. 2012;15:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wassink TH, Epping EA, Rudd D, et al. Influence of ZNF804a on brain structure volumes and symptom severity in individuals with schizophrenia. Arch Gen Psychiatry. 2012;69:885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schultz CC, Nenadic I, Riley B, et al. ZNF804A and cortical structure in schizophrenia: in vivo and postmortem studies. Schizophr Bull. 2014;40:532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rose EJ, Hargreaves A, Morris D, et al. Effects of a novel schizophrenia risk variant rs7914558 at CNNM2 on brain structure and attributional style. Br J Psychiatry. 2014;204:115–121. [DOI] [PubMed] [Google Scholar]
- 56. Thong JY, Qiu A, Sum MY, et al. Effects of the neurogranin variant rs12807809 on thalamocortical morphology in schizophrenia. PLoS One. 2013;8:e85603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Papiol S, Mitjans M, Assogna F, et al. Polygenic determinants of white matter volume derived from GWAS lack reproducibility in a replicate sample. Transl Psychiatry. 2014;4:e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Walton E, Turner J, Gollub RL, et al. Cumulative genetic risk and prefrontal activity in patients with schizophrenia. Schizophr Bull. 2013;39:703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Terwisscha van Scheltinga AF, Bakker SC, van Haren NE, et al. ; Psychiatric Genome-wide Association Study Consortium Genetic schizophrenia risk variants jointly modulate total brain and white matter volume. Biol Psychiatry. 2013;73:525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68; image 5. [DOI] [PubMed] [Google Scholar]
- 61. Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. [DOI] [PubMed] [Google Scholar]
- 62. Guillozet-Bongaarts AL, Hyde TM, Dalley RA, et al. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2014;19:478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hulshoff Pol HE, Kahn RS. What happens after the first episode? A review of progressive brain changes in chronically ill patients with schizophrenia. Schizophr Bull. 2008;34:354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kasai K, Shenton ME, Salisbury DF, et al. Progressive decrease of left superior temporal gyrus gray matter volume in patients with first-episode schizophrenia. Am J Psychiatry. 2003;160:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Spoletini I, Cherubini A, Di Paola M, et al. Reduced fronto-temporal connectivity is associated with frontal gray matter density reduction and neuropsychological deficit in schizophrenia. Schizophr Res. 2009;108:57–68. [DOI] [PubMed] [Google Scholar]
- 66. Gur RE, Gur RC. Functional magnetic resonance imaging in schizophrenia. Dialogues Clin Neurosci. 2010;12:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Whalley HC, Sprooten E, Hackett S, et al. Polygenic risk and white matter integrity in individuals at high risk of mood disorder. Biol Psychiatry. 2013;74:280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Oertel-Knöchel V, Lancaster TM, Knöchel C, et al. Schizophrenia risk variants modulate white matter volume across the psychosis spectrum: evidence from two independent cohorts. Neuroimage Clin. 2015;7:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Patel S, Mahon K, Wellington R, Zhang J, Chaplin W, Szeszko PR. A meta-analysis of diffusion tensor imaging studies of the corpus callosum in schizophrenia. Schizophr Res. 2011;129:149–155. [DOI] [PubMed] [Google Scholar]
- 70. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2011:40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ahmeti KB, Ajroud-Driss S, Al-Chalabi A, et al. Age of onset of amyotrophic lateral sclerosis is modulated by a locus on 1p34.1. Neurobiol Aging. 2013;34:357, e7–357.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69–80. [DOI] [PubMed] [Google Scholar]
- 73. Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162:2233–2245. [DOI] [PubMed] [Google Scholar]
- 74. Kuperberg GR, Broome MR, McGuire PK, et al. Regionally localized thinning of the cerebral cortex in schizophrenia. Arch Gen Psychiatry. 2003;60:878–888. [DOI] [PubMed] [Google Scholar]
- 75. Narr KL, Toga AW, Szeszko P, et al. Cortical thinning in cingulate and occipital cortices in first episode schizophrenia. Biol Psychiatry. 2005;58:32–40. [DOI] [PubMed] [Google Scholar]
- 76. Cheung V, Chiu CP, Law CW, et al. Positive symptoms and white matter microstructure in never-medicated first episode schizophrenia. Psychol Med. 2011;41:1709–1719. [DOI] [PubMed] [Google Scholar]
- 77. Anderson VM, Goldstein ME, Kydd RR, Russell BR. Extensive gray matter volume reduction in treatment-resistant schizophrenia. Int J Neuropsychopharmacol. 2015;18:pyv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thompson PM, Stein JL, Medland SE, et al. ; Alzheimer’s Disease Neuroimaging Initiative, EPIGEN Consortium, IMAGEN Consortium, Saguenay Youth Study (SYS) Group The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Meyer-Lindenberg A, Nicodemus KK, Egan MF, Callicott JH, Mattay V, Weinberger DR. False positives in imaging genetics. Neuroimage. 2008;40:655–661. [DOI] [PubMed] [Google Scholar]
- 80. Silver M, Montana G, Nichols TE; Alzheimer’s Disease Neuroimaging Initiative False positives in neuroimaging genetics using voxel-based morphometry data. Neuroimage. 2011;54:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Stein JL, Medland SE, Vasquez AA, et al. ; Alzheimer’s Disease Neuroimaging Initiative; EPIGEN Consortium; IMAGEN Consortium; Saguenay Youth Study Group; Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium; Enhancing Neuro Imaging Genetics through Meta-Analysis Consortium Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Weinberger DR, Radulescu E. Finding the elusive psychiatric “lesion” with 21st-century neuroanatomy: a note of caution [published online ahead of print August 28, 2015]. Am J Psychiatry. doi: appiajp201515060753. [DOI] [PubMed] [Google Scholar]
- 83. Toro R, Chupin M, Garnero L, et al. Brain volumes and Val66Met polymorphism of the BDNF gene: local or global effects? Brain Struct Funct. 2009;213:501–509. [DOI] [PubMed] [Google Scholar]
- 84. Yang X, Liu P, Sun J, et al. Impact of brain-derived neurotrophic factor Val66Met polymorphism on cortical thickness and voxel-based morphometry in healthy Chinese young adults. PLoS One. 2012;7:e37777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Madhyastha T, Borghesani P, Aylward E, et al. Effect of the APOE-ɛ4 allele on longitudinal changes in cortical thickness in normal aging. Alzheimers Dement J Alzheimers Assoc. 2012;8:P4. [Google Scholar]
- 86. Nyberg L, Salami A. The APOE ε4 allele in relation to brain white-matter microstructure in adulthood and aging. Scand J Psychol. 2014;55:263–267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.