Abstract
Mutated CpG sites (CpG-SNPs) are potential hotspots for human diseases because in addition to the sequence variation they may show individual differences in DNA methylation. We performed methylome-wide association studies (MWAS) to test whether methylation differences at those sites were associated with schizophrenia. We assayed all common CpG-SNPs with methyl-CpG binding domain protein-enriched genome sequencing (MBD-seq) using DNA extracted from 1408 blood samples and 66 postmortem brain samples (BA10) of schizophrenia cases and controls. Seven CpG-SNPs passed our FDR threshold of 0.1 in the blood MWAS. Of the CpG-SNPs methylated in brain, 94% were also methylated in blood. This significantly exceeded the 46.2% overlap expected by chance (P-value < 1.0×10−8) and justified replicating findings from blood in brain tissue. CpG-SNP rs3796293 in IL1RAP replicated (P-value = .003) with the same direction of effects. This site was further validated through targeted bisulfite pyrosequencing in 736 independent case-control blood samples (P-value < 9.5×10−4). Our top result in the brain MWAS (P-value = 8.8×10−7) was CpG-SNP rs16872141 located in the potential promoter of ENC1. Overall, our results suggested that CpG-SNP methylation may reflect effects of environmental insults and can provide biomarkers in blood that could potentially improve disease management.
Key words: DNA methylation, psychosis, SNPs, MBD-seq, methylome-wide association study, postmortem brain samples
Introduction
CpG sequences are highly mutable.1 This is partly caused by the cytosine methylation that can occur at the carbon 5 position. Hydrolytic deamination of unmethylated cytosines produces uracil, which is a foreign base in the DNA sequence that is fixed by the DNA repair system. Methylated cytosines, however, are hydrolytically deaminated to thymines that are not identified as foreign. These mutations are therefore less likely to be repaired, resulting in a C→T transition.2
Point mutated CpG sites, called CpG-SNPs,3 are potential hotspots for human diseases because in addition to the sequence variation they may show individual differences in DNA methylation. Indeed, although few CpG-SNP studies have been conducted, associations have already been identified with outcomes such as type 2 diabetes4 and alcohol dependence.5 CpG-SNP methylation may impact gene function through a variety of mechanisms. DNA methylation is a critical gene-silencing mechanism that protects the integrity of the genome by inactivating DNA elements.6,7 For example, repetitive DNA sequences are often methylated to avoid an impact on gene expression.8 In addition, methylation itself may impact gene expression. For example, promoter methylation can suppress transcription9 (eg, by inhibiting the binding of transcription factors to their recognition elements10), exon methylation may aid the spliceosome in distinguishing exons from introns11 and intragenic methylation can affect transcription elongation efficiency via alternative promoters located within gene bodies12 or regulate noncoding transcripts that may then alter the transcription of the associated genes.13,14
In this article, we study methylation differences at CpG-SNP sites between schizophrenia cases and controls. Whereas previous studies focused on a few sites in candidate genes,4,5 we will test all common germline mutations that create CpG-SNPs. Using genome-wide SNP genotyping in combination with imputation we first determined the CpG-SNPs. Next, the methylation at these sites was assayed using methyl-CpG binding domain protein-enriched genome sequencing (MBD-seq).15,16 With this approach, DNA is fragmented after which the methylated fragments are captured and sequenced. MBD-seq has demonstrated to be sensitive and capable of identifying differentially methylated regions,16–21 detect previously reported robust associations,22 and produce findings that replicate using “gold standard” technologies.23
Methylation can be tissue specific and studies of psychiatric disorders are therefore ideally performed in brain tissue.24 However, because procurement of brain tissue is not possible in living human beings, it is equally critical to capitalize on observations that parts of the brain CpG methylome may be mirrored in blood.25–27 To obtain sufficient statistical power, a CpG-SNP methylome-wide association studies (MWAS) was first performed using DNA extracted from blood in a large “discovery” sample of 1408 case-control samples. Next, methylome-wide significant findings were replicated in 66 postmortem brain samples. Finally, sites that replicated in brain with the same direction of effect after accounting for multiple testing, were further validated using blood from 736 independent case-control samples through targeted bisulfite pyrosequencing.
Methods
Table 1 shows descriptive statistics for the samples used in this study.
Table 1.
Descriptive Statisticsa for the CpG-SNP MWAS Sample, Replication Postmortem Brain Sample, and Validation Blood and Brain Samples
| Controls | Cases | All | Difference Test | ||||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Statistic | P-value | ||
| CpG-SNP MWAS | N = 696 | N = 712 | N = 1408 | ||||
| Age | 55.3 | 11.8 | 53.2 | 11.5 | 11.54 | .001 | |
| Sex | 0.46 | 0.45 | 0.19 | .659 | |||
| Non-nordic | 0.07 | 0.10 | 3.01 | .083 | |||
| Replication brain | N = 27 | N = 39 | N = 66 | ||||
| Age | 43.1 | 7.4 | 43.3 | 9.9 | 0.01 | .909 | |
| Sex | 0.70 | 0.59 | 0.47 | .493 | |||
| PMI | 29.0 | 14.1 | 33.1 | 15.4 | 1.21 | .276 | |
| pH | 6.6 | 0.3 | 6.4 | 0.3 | 7.34 | .009 | |
| Validation blood | N = 377 | N = 370 | N = 736 | ||||
| Age | 58.4 | 10.4 | 54.7 | 11.0 | 21.87 | 3.5E-06 | |
| Sex | 0.38 | 0.38 | 0.01 | .908 | |||
| Non-nordica | 0.0 | 0.0 | NA | NA | |||
Note: MWAS, methylome-wide association studies. N indicates number of individuals. Age is given in years. Sex is coded 0 = female and 1 = male. Postmortem interval (PMI) is the time that elapsed between mortem and the collection of the biological sample. Because of screening, 0% of the validation samples have Finnish or non-Nordic Parents. Different tests are done on contingency table (chi-square test) for categorical variables and means for continuous variables (T test).
aThis table is limited to a set of overlapping variables and supplementary table S1 provides more details in each of the samples.
CpG-SNP MWAS
Samples. Subjects are part of a larger study28,29 and initially ascertained using the Hospital Discharge Register in Sweden, which has high agreement with medical30,31 and psychiatric diagnoses,32 and further validated by a phone interview (see Ripke et al28 for details). DNA was extracted from the buffy coat of whole blood.
Genotyping and Imputation. Genotyping was performed at Broad Institute (92.6% with Genome-wide SNP Array 6.0 from Affymetrix and 7.4% with the earlier 5.0 chip). QC exclusionary measures for subjects included: genotype call rates <95%; ancestry outliers identified after principal component analysis (PCA) of the SNP data33; a randomly selected member of any pair of subjects with high relatedness; and suspected sample contamination. SNPs were excluded for departure from Hardy–Weinberg equilibrium (P-value < 1×10−6) and nonrandom genotyping failure inferred from the flanking haplotype background.
We imputed SNPs from 1000 genomes data (phase I version 3) using minimac.34 After selecting SNPs with minor allele frequency > 0.05 and imputation quality R 2 > 0.5, 5 567 610 imputed and genotyped SNPs remained. This set included 1 437 103 (30.2%) SNPs that created/destroyed a CpG site in the reference genome.
Methylation Assay. We used MethylMiner (Invitrogen) that employs MBD protein-based enrichment of the methylated DNA fraction, followed by single end sequencing (50bp reads) on the SOLiD platform (Life Technologies). We eluted the captured methylated fraction with 0.5M NaCl to increase the relative number of fragments from CpG poor regions,35 which otherwise would not be as well covered.36 Compared to other MBD kits, this protocol has a favorable noise to signal ratio and coverage of the methylome.37
Details on the QC can be found elsewhere.35 In summary, we obtained an average of 68 million (SD = 26.8) reads per sample. Reads were aligned (build hg19/GRCh37) using BioScope 1.2. Subjects were excluded either because <40% alignment or <15 million reads remained after all QC, withdrawn consent, or missing genotype information. For the remaining 1408 samples, the average percentage of mapped reads was 69.2% (SD = 6.2). We eliminated 32.1% of the mapped reads because they were low quality multi-reads (reads aligning to multiple locations) or duplicate-reads (reads with identical start positions). To quantify methylation we estimated the number of fragments covering each CpG-SNP.38 CpG-SNPs in loci showing alignment problems (eg, in repeats) were discarded. This reduced the number of CpG-SNPs by 33.5% to 954 383. Similar to selecting only common SNPs, we also eliminated sites that were unmethylated in the majority of subjects. This further reduced the number of sites to 598 974.
Model and Interpretation. Significance tests were performed by fitting the following regression equation:
Y are the methylation measurements and b 0 is the intercept of the regression line, and E are the residual effects. The CpG-SNP is coded as 0, 1, and 2 corresponding to having 0, 1, or 2 CpGs. A nonzero value of parameter b 1 indicates that the CpG is methylated. Disease is coded 0 and 1 for controls and cases, respectively. By taking expectations E(Y) using this coding for CpG-SNP and disease, we obtain the expected means reported on table 2. The null-hypothesis, b 2 = 0, states that there are no case control differences in methylation. The final row shows that b 2 is zero in subjects without CpGs, and that with 1 copy the expected difference will be half of the difference of the group with 2 CpGs. Parameters b 3…b k represent the effect of covariates and possible confounders.
Table 2.
Expect Means of Disease Groups by CpG-SNP Status
| # CpGs | 0 | 1 | 2 |
|---|---|---|---|
| Disease status | |||
| Control | b 0 | b 0 + b 1 | b 0 + 2b 1 |
| Case | b 0 | b 0 + b 1 + b 2 | b 0 + 2b 1+ 2b 2 |
| Difference | 0 | b 2 | 2b 2 |
Note: b 0 is the intercept, b 1 is the effect of the SNP and b 2 the effect of the disease on methylation levels.
If the null-hypothesis, b 2 = 0, is rejected, there are several possible interpretations. First, the methylation could have a causal effect. If in this scenario there is also a main effect of the CpG-SNP (b 1 ≠ 0), one would predict the SNP to be associated in, eg, GWAS studies. Although the SNP is an imperfect “tag” because it may not be methylated in all subjects, this is because methylation levels will generally be higher in subjects that have the cytosine.
Second, DNA methylation serves to protect the integrity of the genome by inactivating DNA elements.6,7 Failure to silence a deleterious SNP (or in linkage disequilibrium [LD] with a deleterious SNP) would result in a pattern where methylation increases with the number of C alleles (ie, b 1 > 0) in all subjects. However, cases would show reduced methylation levels if the cytosine (C allele) is present (ie, b 2 < 0) because the locus not silenced in subsets of cells or patients.
Third, the methylation change may itself not be causal but the result of a causal disease processes or confounder. To minimize effects of confounding variables, we included assay-related variables such as the quantity of genomic starting material, the quantity of methylation-enriched DNA captured, and sample batch. We also controlled for age and sex and performed PCA was performed on coverage estimates of all QC’ed CpG-SNPs to capture the major remaining unmeasured confounders (see Chen et al39). Based on a screen test, the first 7 principal components (PCs) were selected. We correlated PC scores with a variety of variables (see table 2 in Aberg et al35) to check that no additional covariates were required. These analyses showed, eg, that ancestry (estimated from the genome-wide SNP data) did not contribute substantially to variation in the methylome and was therefore not included as a covariate.
Blood and brain consists of different cell types. This can produce false positives if 2 conditions hold simultaneously (1) the relative abundance of (common) cell types differs between cases and controls, and (2) methylation patterns of these cell types differ. Ideally, we would have methylation data obtained from separated cells40 to identify sites that are at risk for being false positives. However, the use PC scores as covariates provides some protection against such false positive findings.40–42 This is because (similar to how false positives due to ancestry subgroups are handled in GWAS) cell type differences will create correlated multi-locus methylation patterns that will be captured by PCA. In addition, because blood and brain will have different cell types, cell type related findings in blood are unlikely to replicate in our brain samples.
Replication in Postmortem Brain Samples
Postmortem prefrontal cortex brain tissue (Brodmann area 10) was obtained from the Stanley Medical Research Institute (SMRI).43 Subjects were diagnosed with schizophrenia (N = 26), bipolar disorder (N = 22) or were controls (N = 27). Diagnosing disease in subjects providing postmortem brain samples is a challenge.44 The SMRI uses DSM-IV diagnoses made by 2 senior psychiatrists using medical records and, when necessary, telephone interviews with family members. Diagnoses of unaffected controls are based on structured interviews by a senior psychiatrist with family member(s) to rule out Axis I diagnoses. To increase power as much as possible, in the main analysis we focused on the 39 patients with psychosis (26 schizophrenia and 13 bipolar) and the 27 controls. Studies suggest considerable etiological overlap between BP and SZ45 thereby suggesting that psychosis may reflect similar disease processes in both groups of patients.
Genotype data generated by Liu et al46 were used. The genotyping was conducted using Affymetrix GeneChip Mapping 5.0K Array (Affymetrix) at Translational Genomics Research Institute. BRLMM-p was used as the genotype calling algorithm. SNPs with call rates <99% and showing departure from Hardy–Weinberg equilibrium (P-value < .001) were filtered. SNPs were again imputed with 1000 genomes data (see section CpG-SNP MWAS).
Using MBD-seq we obtained an average of 51.3 million (SD = 15.6) reads per sample. The average percentage of mapped reads was 78.0% (SD = 5.9). After eliminating low quality multi-reads (reads aligning to multiple locations) or duplicate-reads (reads with identical start positions), an average of 34.1 million reads/sample were left.
The number of brain samples available for the main analysis was 66. Compared to the analysis of regular CpGs, statistical power will be lower for CpG-SNPs. This is because there cannot be differences in subjects with no CpGs, and the expected case-control difference in subjects with 1 CpG is only half of the difference in subjects with 2 CpGs. We therefore confined the use of brain to replicating significant findings from the MWAS in the blood. This improves statistical power because rather than testing many sites, we now only test a few sites that are more likely to be relevant for schizophrenia and where we can require the same direction of effect (ie, perform 1-sided test). In addition to age, sex and PCs, pH and postmortem interval were regressed out in the MWAS.
Validation
CpG-SNPs that replicated in brain were further validated in independent samples using a different technology. The 736 blood samples came from different subjects from the same overarching study (see Ripke et al28 and Bergen et al29). Table 1 shows that compared to the CpG-SNP MWAS samples, these samples did not include individuals with Finnish or non-Nordic parents. SNP genotyping was performed through pyrosequencing. Methylation measurements were obtained through targeted pyrosequencing of bisulfite converted DNA.47,48
Results
CpG-SNP MWAS
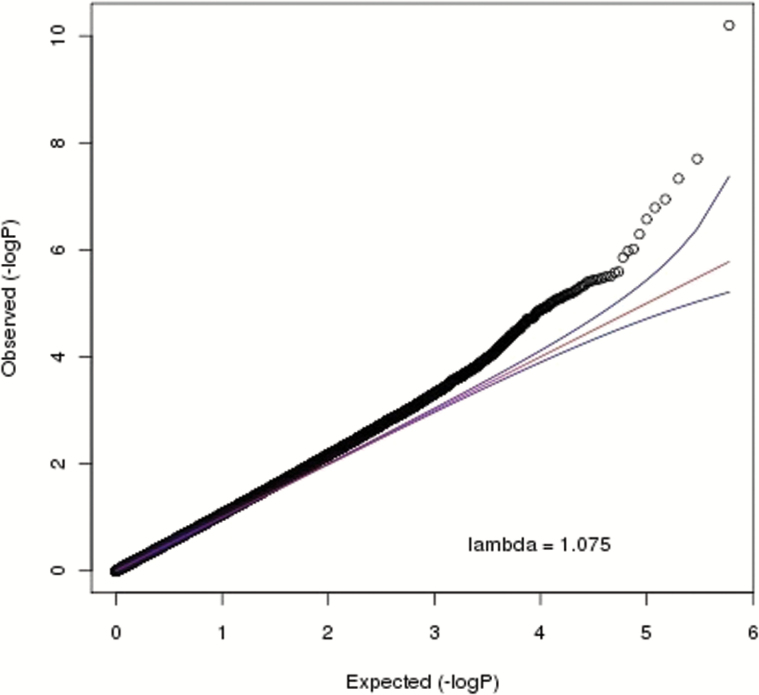
The quantile–quantile plot in figure 1 shows an enrichment of small P-values with a test statistic inflation parameter λ being only slightly higher (λ = 1.075) compared to what is commonly observed in GWAS. This inflation is unlikely an artifact and reflects that methylation studies are more akin to gene expression studies that typically show many correlated and relatively large effects.
Fig. 1.
Quantile–quantile plot for methylome-wide association studies (MWAS) in blood.
Table 3 shows that 7 CpG-SNPs reached methylome-wide significance49 by passing our false discovery rate threshold of 0.1 (ie, q-values < 0.1). This threshold ensures that only 10% of the significant findings are expected to be false discoveries. Using a more liberal threshold of q-value < 0.25 resulted in 97 additional “suggestive” findings. We use GenomeRunner50 to test for enrichment of genomic features among these 97 sites (supplementary table 2). We found a significant enrichment of sites that bind to CEBPB (q-value = 0.022). In addition, considering all ENCODE cell types, we found significant enrichment for H3K4me1 (an activating mark associated with possible upregulation of gene expression) in blood (q-value = 0.032).
Table 3.
Replication of MWAS Association Findings (H0: b 2 = 0) in Blood With q-Value < 0.1 in Postmortem Brain Samples
| Chr | SNP | Position (bp) | MWAS | Replication | Gene | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| T | P-value | q-value | Effect | T | P-value | Effect | ||||
| 2 | rs605832 | 31454545 | −5.65 | 2.0E-08 | 0.010 | −0.301 | −0.91 | 1.8E-01 | −0.232 | |
| 3 | rs9827299 | 71353372 | −6.59 | 6.3E-11 | 0.000 | −0.351 | 0.79 | 7.8E-01 | 0.202 | FOXP1 |
| 3 | rs3796293 | 190332093 | −5.49 | 4.7E-08 | 0.015 | −0.292 | −2.88 | 3.0E-03 | −0.732 | IL1RAP |
| 15 | rs56153788 | 86170992 | −5.33 | 1.1E-07 | 0.028 | −0.284 | 0.70 | 7.5E-01 | 0.177 | AKAP13 |
| 16 | rs2542671 | 54209836 | −5.26 | 1.6E-07 | 0.032 | −0.280 | −0.06 | 4.8E-01 | −0.015 | |
| 17 | rs9912900 | 71057060 | −5.17 | 2.7E-07 | 0.044 | −0.276 | 0.69 | 7.5E-01 | 0.175 | SLC39A11 |
| 17 | rs2589133 | 78793476 | −5.05 | 5.1E-07 | 0.071 | −0.296 | −1.52 | 6.8E-02 | −0.386 | RPTOR |
Note: “Chr.” is chromosome. Position is for reference genome build hg19/GRCh37. Signs of the T statistics (T) give the direction of effect in the MWAS and replication, respectively, where a negative value indicates a decreased methylation level in the SZ cases. Effect is effects size calculated by Cohen’s d (mean difference between the cases and controls divided by a pooled estimate of the standard deviation) is calculated for the replication sample only. Gene is name of the gene allowing for a ±20 Kb flanking region. CpG-SNPs in bold reach significance in the replication after a Bonferroni correction for multiple testing.
Replication in Postmortem Brain Samples
We found that 67.7% of CpG-SNPs were generally methylated in brain. This was very comparable to the 68.3% found in blood. Of the sites generally methylated in brain, 94% were also generally methylated in blood. This significantly exceeded the 46.2% (=67.7% × 68.3%) expected by chance (P-value < 1.0×10−8). This overlap suggests that the methylation statuses of many CpG-SNP sites in brain are partly mirrored in blood, and justifies making comparisons across the 2 tissues.
Table 3 shows the replication results for the 7 MWAS findings with q-value < 0.1. For this replication, we required the same direction of effects. Allowing for a type I error of 0.05, the threshold for declaring significance after Bonferroni correction was 0.007. CpG-SNP rs3796293 replicated with a P-value of .003. The significant main effect of rs3796293 (H 0: b 1 = 0, P-value = 4.07E-08) suggested this indeed a CpG-SNP and located in the interleukin 1 receptor accessory protein (IL1RAP) gene. The effects size of rs3796293 was calculated using Cohen’s d (the mean difference between the cases and controls divided by a pooled estimate of the standard deviation) and was 0.292 in the MWAS and 0.732 in the replication. Smoking was not included as a covariate in the MWAS because of the many missing values (31.1%) but because it did not have a significant effect on the methylation of rs3796293 (T = 0.672, P-value = .51), it unlikely caused this association.
We performed exploratory analyses to examine associations between CpG-SNP rs3796293 and specific diagnoses. Association results reached significance for SZ (P-value = .014), BP (P-value = .029), as well as the combined BP/SZ sample (P-value = .002) implying that our result was not diagnosis specific and reflected a shared disease cause.
Validation
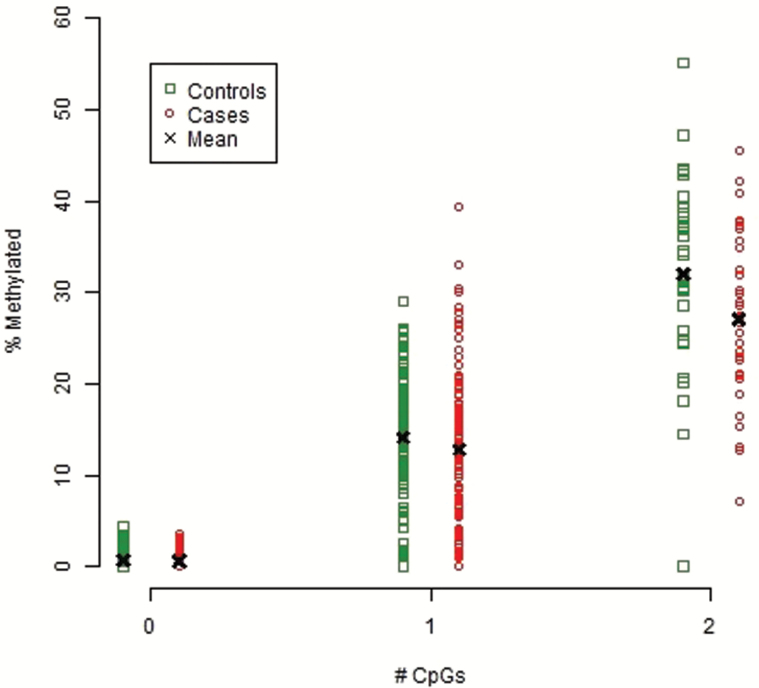
CpG-SNP rs3796293 was further validated in 736 independent blood samples using targeted bisulfate pyrosequencing. Figure 2 shows that the amount of methylation as well as the case control difference was essentially zero in groups with no CpGs. In groups with 1 CpG, the site was 14.1% methylated in controls and 12.7% in cases. In groups with 2 CpGs, the site was 32.0% methylated in controls and 27.1% in cases. Thus, consistent with the direction of effects in the CpG-SNP MWAS and brain samples, the site was less methylated in cases vs controls and this difference was again significant (P-value < 9.5×10−4). These effect sizes were very comparable for bisulfite pyrosequencing/MBD-seq data and equaled 0.17/0.21 in groups with 1 CpG and 0.48/0.39 in groups with 2 CpGs.
Fig. 2.
Methylation levels (x-axis) for rs3796293 grouped by number of CpGs (x-axis) and disease status.
Discussion
We performed a CpG-SNP MWAS on DNA extracted from blood of 1408 schizophrenia case-control samples. Of the 7 CpG-SNPs that reached methylome-wide significance, CpG-SNP rs3796293 in IL1RAP replicated (P-value = .003) in 66 postmortem brain samples after accounting for multiple testing and with the same direction of effects. This finding was further validated using a different technology in blood samples from 736 independent cases and controls (P-value < 9.5×10−4).
IL1RAP encodes the interleukin 1 receptor accessory protein. Interleukin 1 induces synthesis of acute phase and proinflammatory proteins during infection, tissue damage, or stress, by forming a complex at the cell membrane with an interleukin 1 receptor and an accessory protein.51 There is an extensive body of evidence linking inflammation in general52,53 and interleukin 1 in specific54,55 to schizophrenia. Several mechanisms could explain this link. First, increased levels of proinflammatory cytokines could be a consequence of mental stress associated with the disease. Second, a growing body of evidence links prenatal infection and maternal immune alterations during pregnancy to risk of schizophrenia. Because most viruses do not cross the placenta, the damaging effects to the fetus may be the result of maternal responses to infection that includes the proinflammatory cytokines that are known to influence neural development. Indeed, magnetic resonance imaging (MRI) studies, eg, have shown structural neuroanatomic alterations among schizophrenia cases.56 Finally, inflammatory cytokines affect synaptic transmission.57,58 Animal experiments, eg, illustrate that an inflammatory cytokine challenge can induce psycho-behavioral and/or cognitive impairments59 that mimic aspects of schizophrenia.
IL1RAP has 19 known splice variants (Ensembl release 77). Smith et al60 found that mice with a specific IL1RAP splice variant were more vulnerable to local inflammatory challenge in the central nervous system and suffer enhanced neuronal degeneration. A meta-analysis in postmortem brain samples from 153 schizophrenia cases and 153 controls,61 did not find significant differences in the expression of IL1RAP at the time of death. This does not necessarily exclude a functional role. The gene could exert its effects at a more developmental stage of the disease, and other factors such as the somewhat different brain regions or that relevant variants splice variants may not have been assayed could play a role. Using ENCODE data, we found that in both blood (B cells) and brain (astrocytes), CpG-SNP rs3796293 overlapped with an EZH2 locus. EZH2 (Enhancer of Zeste homolog 2) is a subunit of a protein complex that represses gene expression by binding to chromatin and locally altering the chromatin structure.
CpG-SNP rs3796293 showed a pattern where methylation increased with the number of CpGs (ie, b 1 > 0) in all subjects but where cases had reduced methylation levels (ie, b 2 < 0). Because methylation levels are generally higher in subjects that have the CpG, such a pattern predicts that the SNP itself is associated with the disease assuming the methylation is causal. However, Rs3796293 was not significant in GWAS studies of the Swedish samples used in the present study (P-value .397; 5001 cases and 6243 controls),28 nor the recent PGC2 meta-analysis (P-value .373; 36 989 cases and 113 075 controls).62 Thus, the methylation change at rs3796293 was unlikely causal. A model assuming that rs3796293 may be a deleterious mutation that is not properly silenced in schizophrenia cases, is in principle consistent with the observed methylation pattern. However, although this explanation cannot be excluded entirely, the above evidence linking IL1RAP to inflammation or stress suggests that in the methylation change may be more likely the result of such disease processes. The successful replication in brain tissue is relevant in this context as it means that the process that caused the methylation change in blood affected brain as well. Assuming that the majority of schizophrenia disease processes occur in brain, this would be a necessary requirement for the methylation to potentially tag an actual disease process. The fact that we see the methylation difference in blood is still important as this may provide a biomarker for that disease process.
Our finding could be spurious and the result of a confounder that is not a direct cause of the disease. To minimize effects of possible confounder we regressed out a variety of covariates (possible assay-related technical artifacts, demographic variables, as well as unmeasured confounders through PC scores). In addition, we were able to detect effects of processes that have previously been linked to schizophrenia and were able to replicate marks of these processes in brain tissue where the majority of the disease processes are likely to occur. However, ultimately functional studies, eg, assessing the impact of inflammation on the methylation of IL1RAP or using epigenome editing,63–65 would be needed to validate our findings and completely rule out artifacts caused by confounding factors. We note that this is somewhat akin to (SNP) studies of sequence variation. Due to LD it is generally not possible to pinpoint the actual causal mutation and functional studies are needed to address this question.
Thematically consistent with a possible role of IL1RAP, our 97 “suggestive” findings in blood were enriched for sites that bind transcription factor CEBPB (CCAAT/enhancer-binding protein beta) that is known to regulate the expression of genes involved in immune and inflammatory responses.66 In addition, CEBPB is capable of increasing the expression of central nervous system genes such as Choline O-Acetyltransferase (ChAT) that encodes an enzyme which catalyzes the biosynthesis of the neurotransmitter acetylcholine.67
Given the small samples sizes our findings should be followed up in larger cohorts of postmortem brain samples. Furthermore, to obtain sufficient statistical power, we tested for association in the blood and then replicated those findings in brain. A limitation of this order is that sites that are not mirrored in blood will be missed. As an exploratory analysis we reversed the order. The top finding in the brain MWAS was CpG-SNP rs16872141 (P-value = 8.8×10−7). Although it did not reach methylome-wide significance (q-value = 0.36), it did replicate in blood with the same direction of effects in the MBD-seq data from 1408 subjects (P-value = .049) and validated in the 736 independent case-control samples (P-value = 4.1×10−6) using targeted bisulfite pyrosequencing. Rs16872141 is in an intron of HEX1 and overlaps with a CpG shore located within 3kb upstream of ENC1.The protein encoded by ENC1 (ectodermal-neural cortex 1) plays a role in the oxidative stress response as a regulator of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor). This finding is consistent with studies suggesting that the etiology of schizophrenia can be partly explained by oxidative stress.68 Furthermore, Nrf2 is essential for neuronal differentiation of neural stem/progenitor cells, regulates injury-induced neurogenesis, and provides protection against amyloid-beta-induced toxicity.69 Being in the potential promoter of ENC1, the methylation of rs16872141 could potentially suppress transcription of this gene. However, rs16872141 was not significant in GWAS studies of the Swedish samples used in the present study (P-value = .303; 5001 cases and 6243 controls),28 nor the recent PGC2 meta-analysis (P-value = .739; 36 989 cases and 113 075 controls).62 This suggested that rather than playing a causal role, the methylation of rs16872141 may not play a causal role but reflect a process, eg, oxidative stress, that affected both blood and brain.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
This study was supported by grants from the National Institute of Mental Health (RC2MH089996, R03MH102723, and R01MH097283) and the Stanley Medical Research Institute (#08R-1959).
Supplementary Material
Acknowledgments
The present study is part of a larger project entitled “A Large-Scale Schizophrenia Association Study in Sweden.” Institutions involved in this project are: Karolinska Institutet, Icahn School of Medicine at Mount Sinai, University of North Carolina at Chapel Hill, and the Broad Institute. Brain tissue for this study was provided by the Stanley Medical Research Institute. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–1357. [DOI] [PubMed] [Google Scholar]
- 2. Bird A. The dinucleotide CG as a genomic signalling module. J Mol Biol. 2011;409:47–53. [DOI] [PubMed] [Google Scholar]
- 3. Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dayeh TA, Olsson AH, Volkov P, Almgren P, Rönn T, Ling C. Identification of CpG-SNPs associated with type 2 diabetes and differential DNA methylation in human pancreatic islets. Diabetologia. 2013;56:1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taqi MM, Bazov I, Watanabe H, et al. Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict Biol. 2011;16:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. [DOI] [PubMed] [Google Scholar]
- 7. Kapoor A, Agius F, Zhu JK. Preventing transcriptional gene silencing by active DNA demethylation. FEBS Lett. 2005;579:5889–5898. [DOI] [PubMed] [Google Scholar]
- 8. Hsieh J, Fire A. Recognition and silencing of repeated DNA. Annu Rev Genet. 2000;34:187–204. [DOI] [PubMed] [Google Scholar]
- 9. Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. [DOI] [PubMed] [Google Scholar]
- 11. Shukla S, Kavak E, Gregory M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deaton AM, Webb S, Kerr AR, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mohn F, Weber M, Schübeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP). Methods Mol Biol. 2009;507:55–64. [DOI] [PubMed] [Google Scholar]
- 16. Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li N, Ye M, Li Y, et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52:203–212. [DOI] [PubMed] [Google Scholar]
- 18. Harris RA, Wang T, Coarfa C, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hogart A, Lichtenberg J, Ajay SS, Anderson SM, Margulies EH, Bodine DM. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal over-representation of ETS transcription factor binding sites. Genome Res. 2012;22:1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lan X, Adams C, Landers M, et al. High resolution detection and analysis of CpG dinucleotides methylation using MBD-Seq technology. PLoS One. 2011;6:e22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nair SS, Coolen MW, Stirzaker C, et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011;6:34–44. [DOI] [PubMed] [Google Scholar]
- 22. McClay JL, Aberg KA, Clark SL, et al. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum Mol Genet. 2014;23:1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aberg KA, McClay JL, Nerella S, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. [DOI] [PubMed] [Google Scholar]
- 25. Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ripke S, O’Dushlaine C, Chambert K, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergen SE, O'Dushlaine CT, Ripke S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 2012;17:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kristjansson E, Allebeck P, Wistedt B. Validity of the diagnosis of schizophrenia in a psychiatric inpatient register. Nordisk Psykiatrik Tidsskrift. 1987;41:229–234. [Google Scholar]
- 31. Dalman C, Broms J, Cullberg J, Allebeck P. Young cases of schizophrenia identified in a national inpatient register--are the diagnoses valid? Soc Psychiatry Psychiatr Epidemiol. 2002;37:527–531. [DOI] [PubMed] [Google Scholar]
- 32. Ekholm B, Ekholm A, Adolfsson R, et al. Evaluation of diagnostic procedures in Swedish patients with schizophrenia and related psychoses. Nord J Psychiatry. 2005;59:457–464. [DOI] [PubMed] [Google Scholar]
- 33. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 34. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aberg KA, McClay JL, Nerella S, et al. MBD-seq as a cost-effective approach for methylome-wide association studies: demonstration in 1500 case-control samples. Epigenomics. 2012;4:605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aberg KA, Xie L, Chan RF, et al. Evaluation of methyl-binding domain based enrichment approaches revisited. PLoS One. 2015;10:e0132205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van den Oord EJ, Bukszar J, Rudolf G, et al. Estimation of CpG coverage in whole methylome next-generation sequencing studies. BMC Bioinformatics. 2013;14:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen W, Gao G, Nerella S, et al. methylPCA: a toolkit for principal component analysis in methylome-wide association studies [published online ahead of print March 2, 2013]. BMC Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y, Aryee MJ, Padyukov L, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun YV, Turner ST, Smith JA, et al. Comparison of the DNA methylation profiles of human peripheral blood cells and transformed B-lymphocytes. Hum Genet. 2010;127:651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–155. [DOI] [PubMed] [Google Scholar]
- 44. Deep-Soboslay A, Iglesias B, Hyde TM, et al. Evaluation of tissue collection for postmortem studies of bipolar disorder. Bipolar Disord. 2008;10:822–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smoller JW, Craddock N, Kendler K, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu C, Cheng L, Badner JA, et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry. 2010;15:779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35:152–156. [DOI] [PubMed] [Google Scholar]
- 48. Aparicio A, North B, Barske L, et al. LINE-1 methylation in plasma DNA as a biomarker of activity of DNA methylation inhibitors in patients with solid tumors. Epigenetics. 2009;4:176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19:537–542. [DOI] [PubMed] [Google Scholar]
- 50. Dozmorov MG, Cara LR, Giles CB, Wren JD. GenomeRunner: automating genome exploration. Bioinformatics. 2012;28:419–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang D, Zhang S, Li L, Liu X, Mei K, Wang X. Structural insights into the assembly and activation of IL-1beta with its receptors. Nat Immunol. 2010;11:905–911. [DOI] [PubMed] [Google Scholar]
- 52. Fineberg AM, Ellman LM. Inflammatory cytokines and neurological and neurocognitive alterations in the course of schizophrenia. Biol Psychiatry. 2013;73:951–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Potvin S, Stip E, Sepehry AA, Gendron A, Bah R, Kouassi E. Inflammatory cytokine alterations in schizophrenia: a systematic quantitative review. Biol Psychiatry. 2008;63:801–808. [DOI] [PubMed] [Google Scholar]
- 54. Katila H, Appelberg B, Hurme M, Rimón R. Plasma levels of interleukin-1 beta and interleukin-6 in schizophrenia, other psychoses, and affective disorders. Schizophr Res. 1994;12:29–34. [DOI] [PubMed] [Google Scholar]
- 55. Akiyama K. Serum levels of soluble IL-2 receptor alpha, IL-6 and IL-1 receptor antagonist in schizophrenia before and during neuroleptic administration. Schizophr Res. 1999;37:97–106. [DOI] [PubMed] [Google Scholar]
- 56. Ellman LM, Deicken RF, Vinogradov S, et al. Structural brain alterations in schizophrenia following fetal exposure to the inflammatory cytokine interleukin-8. Schizophr Res. 2010;121:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nawa H, Takahashi M, Patterson PH. Cytokine and growth factor involvement in schizophrenia-support for the developmental model. Mol Psychiatry. 2000;5:594–603. [DOI] [PubMed] [Google Scholar]
- 58. Müller N, Ackenheil M. Psychoneuroimmunology and the cytokine action in the CNS: implications for psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry. 1998;22:1–33. [DOI] [PubMed] [Google Scholar]
- 59. Tohmi M, Tsuda N, Watanabe Y, Kakita A, Nawa H. Perinatal inflammatory cytokine challenge results in distinct neurobehavioral alterations in rats: implication in psychiatric disorders of developmental origin. Neurosci Res. 2004;50:67–75. [DOI] [PubMed] [Google Scholar]
- 60. Smith DE, Lipsky BP, Russell C, et al. A central nervous system-restricted isoform of the interleukin-1 receptor accessory protein modulates neuronal responses to interleukin-1. Immunity. 2009;30:817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mistry M, Gillis J, Pavlidis P. Genome-wide expression profiling of schizophrenia using a large combined cohort. Mol Psychiatry. 2013;18:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Maeder ML, Angstman JF, Richardson ME, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013;31:1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Voigt P, Reinberg D. Epigenome editing. Nat Biotechnol. 2013;31:1097–1099. [DOI] [PubMed] [Google Scholar]
- 65. Chen H, Kazemier HG, de Groote ML, Ruiters MH, Xu GL, Rots MG. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res. 2014;42:1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sahin E, Haubenwallner S, Kuttke M, et al. Macrophage PTEN regulates expression and secretion of arginase I modulating innate and adaptive immune responses. J Immunol. 2014;193:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Robert I, Sutter A, Quirin-Stricker C. Synergistic activation of the human choline acetyltransferase gene by c-Myb and C/EBPbeta. Brain Res Mol Brain Res. 2002;106:124–135. [DOI] [PubMed] [Google Scholar]
- 68. Emiliani FE, Sedlak TW, Sawa A. Oxidative stress and schizophrenia: recent breakthroughs from an old story. Curr Opin Psychiatry. 2014;27:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Karkkainen V, Pomeshchik Y, Savchenko E, et al. Nrf2 regulates neurogenesis and protects neural progenitor cells against Abeta toxicity. Stem Cells. 2014;32:1904–1916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.