Abstract
The transcriptional coactivator peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC-1α) has been linked to multiple neurological and psychiatric disorders including schizophrenia, but its involvement in the pathophysiology of these disorders is unclear. Experiments in mice have revealed a set of developmentally-regulated cortical PGC-1α-dependent transcripts involved in calcium buffering (parvalbumin, PV), synchronous neurotransmitter release (synaptotagmin 2, Syt2; complexin 1, Cplx1) and axonal integrity (neurofilamaent heavy chain, Nefh). We measured the mRNA expression of PGC-1α and these transcripts in postmortem cortical tissue from control and schizophrenia patients and found a reduction in PGC-1α-dependent transcripts without a change in PGC-1α. While control subjects with high PGC-1α expression exhibited high PV and Nefh expression, schizophrenia subjects with high PGC-1α expression did not, suggesting dissociation between PGC-1α expression and these targets in schizophrenia. Unbiased analyses of the promoter regions for PGC-1α-dependent transcripts revealed enrichment of binding sites for the PGC-1α-interacting transcription factor nuclear respiratory factor 1 (NRF-1). NRF-1 mRNA expression was reduced in schizophrenia, and its transcript levels predicted that of PGC-1α-dependent targets in schizophrenia. Interestingly, the positive correlation between PGC-1α and PV, Syt2, or Cplx1 expression was lost in schizophrenia patients with low NRF-1 expression, suggesting that NRF-1 is a critical predictor of these genes in disease. These data suggest that schizophrenia involves a disruption in PGC-1α and/or NRF-1-associated transcriptional programs in the cortex and that approaches to enhance the activity of PGC-1α or transcriptional regulators like NRF-1 should be considered with the goal of restoring normal gene programs and improving cortical function.
Key words: transcriptional regulation, interneuron, maturation
Introduction
Schizophrenia is a debilitating psychiatric disorder that affects approximately 1% of the population. Currently, antipsychotic medications alleviate the positive symptoms of the disease, but cognitive symptoms are particularly unresponsive to treatment.1,2 While the molecular basis underlying cognitive deficits in schizophrenia have not been well-defined, a large body of evidence points to dysfunction of parvalbumin-positive interneurons (PV-INs).3 PV-INs are responsible for the entrainment of pyramidal neurons (PNs) at the γ frequency for higher cognitive functioning,4 and abnormalities in cortical γ oscillations are observed in patients with schizophrenia.5,6 A prevalent finding in postmortem schizophrenia cortex is a reduction in PV expression,7–10 suggesting incomplete interneuron maturation.7,11 Electrophysiological investigations of the cortical circuitry underlying the generation of γ oscillations have revealed involvement of PNs as well12 and there is extensive evidence for alterations in PN structure and gene expression in schizophrenia,13–16 potentially reflecting a contribution of altered PN maturation to cognitive deficits.11
Studies from our laboratory indicate that peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), a transcriptional regulator linked to schizophrenia and bipolar disorder,17–21 regulates PV expression. PGC-1α expression is upregulated during postnatal cortical maturation, peaking at postnatal day 14 in rodents; while PGC-1α is predominantly expressed in GABAergic populations, it is also expressed to a lesser extent in cortical pyramidal neurons.22 Loss of PGC-1α greatly impacts the excitatory:inhibitory balance within the cortex; PV-INs receive reduced glutamatergic input from cortical pyramidal neurons and specific deletion of PGC-1α in PV-INs reduces PV expression and PV-IN firing rate in the cortex23–25 without a loss of neurons.24 Further, PGC-1α may be involved in synaptic plasticity in PNs, as siRNA-mediated knockdown of PGC-1α in hippocampal neurons reduces dendritic length, complexity and spine density independent of cell loss.26
Synaptotagmin 2 (Syt2) and complexin 1 (Cplx1), 2 calcium-sensitive regulators of synchronous neurotransmitter release, and neurofilament heavy chain (Nefh), an axonal structural protein, are localized to both PV-positive and negative soma and terminals, upregulated during the postnatal period of cortical maturation and significantly reduced in the PGC-1α null cortex.23 These data indicate that a generalized disruption in cortical circuitry by a loss of PGC-1α impacts both inhibitory and excitatory transmission and potentially transcriptional regulation within both populations to influence cognitive function. In fact, deletion of PGC-1α specifically from PV-INs causes asynchronous GABA release and long-term memory deficits.23
Considering that PV expression is frequently reduced in the cortex of schizophrenia patients,7–9 we hypothesized that the newly identified PGC-1α-dependent genes Syt2, Cplx1, and Nefh could be reduced in cortical neuronal populations in parallel with the interneuron marker PV in schizophrenia patients. We found that transcript levels for PV, Syt2, Cplx1 and Nefh are significantly reduced in the anterior cingulate cortex (ACC) of schizophrenia patients, in the absence of changes in PGC-1α expression. The expression of the PGC-1α-interacting transcription factor nuclear respiratory factor 1 (NRF-1) is also reduced, and the association between PGC-1α and its dependent genes in schizophrenia is disrupted when NRF-1 expression is low. These data suggest that disruption in PGC-1α and/or NRF-1 signaling may contribute to the pathophysiology of schizophrenia.
Materials and Methods
q-RT-PCR
RNA from frontal pole and ACC of patients with schizophrenia and controls was procured from the Stanley Consortium27 (n = 32–33/group; see demographics and RNA integrity values in table 1). Reverse transcription and q-RT-PCR were conducted as previously described.24,28 After RNA samples arrived in the laboratory, concentrations were measured using a NanoDrop 2000 (Thermo Fisher Scientific) and equivalent amounts of RNA (1 μg) were treated with DNase I (Promega) at 37°C for 30 minutes followed by DNAse Stop solution (Promega) 65°C for 15 minutes, and reverse transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems).
Table 1.
Patient Demographics
| Control | Schizophrenia | |
|---|---|---|
| N | 32 | 33 |
| Sex | 23m/9f | 24m/9f |
| Tissue pH | 6.6±0.2 (6.1–6.9) | 6.5±0.2 (5.9–6.9) |
| PMI (h) | 29.7±13.4 (9–58) | 31.4±15.7 (9–80) |
| Age (y) | 44±7 (31–60) | 42±9 (19–59) |
| RNA integrity number | 8.3±0.7 (6.6–9.7) | 8.4±0.6 (7.2–9.6) |
Note: f, female; m, male; PMI, post-mortem interval. Values presented as mean ± SD, with range in parentheses.
Transcript measurement was performed in duplicate using JumpStart Taq Readymix (Sigma) and Applied Biosystems primers with an initial ramp time of 2 minutes at 50°C and 10 minutes at 95°C and 40 subsequent cycles of 15 seconds at 95°C and 1 minute at 60°C. Relative concentrations of the genes of interest were calculated in comparison to a standard curve made from pooling cDNA from all samples together and diluting 1:5, 1:10 and 1:20 (ie, the calibrator method). Taqman primer/probe sets spanned areas lacking known single nucleotide polymorphisms, and all primer/probe sets produced a single band of expected size visualized by agarose gel. Primer/probe sets were specific for 18s rRNA (Hs_99999901_s1), actin-beta (Hs99999903_m1), beta2-microglobulin (Hs99999907_m1), glyceraldehyde-3-phosphate dehydrogenase (Hs99999905_m1), cyclophilin A (Hs99999904_m1), PGC-1α (Hs00173304_m1, Hs01208831_m1, Hs01016722_m1), PV (Hs01075686_m1), Syt2 (Hs_00980604_m1), Cplx1 (Hs00362510_m1), Nefh (Hs00606024_m1), NRF-1 (Hs00192316_m1), or p65/Rela (NF-kB; Hs00153294_m1). Values were normalized to the geometric mean of 3–4 control genes that did not differ between experimental groups29 (FP: 18S, GAPDH, and beta 2 microglobulin; ACC: 18S, beta actin, beta 2 microglobulin, and GAPDH). Any samples with less than 0.18 arbitrary unit values for the geometric mean of control genes were removed from analysis (2 control samples, 1 schizophrenia sample) as individual control genes were at the lower limit of the standard curve.
Haldol Treatment
Haloperidol treatments were conducted as described in Drummond et al30. Briefly, rats were injected with either haldol decanoate (28.5mg/kg) or vehicle (sesame oil) intramuscularly in house-paired male Sprague–Dawley rats (Charles River) once every 3 weeks for 9 months for a total of 12 injections. Animals were sacrificed by decapitation, and tissue was immediately dissected by region and stored at −80°C. One microgram of tissue from the frontal cortex was stabilized with RNAlater-ICE (Life Technologies), and RNA was isolated using an RNeasy Mini RNA isolation kit (Qiagen) and reverse transcribed using a High-Capacity cDNA RT Kit (Applied Biosystems). Transcripts were measured as described above using the following primers: PGC-1α (Rn00580241_m1), Syt2 (Rn00561994_m1), Cplx1 (Rn02396766_m1), Nefh (Rn00709325_m1), and Nrf-1 (Rn01455958_m1). Transcripts were normalized to actin-beta (Rn00667869_m1) and expressed as fold of vehicle-treated control values.
Promoter Analysis
Analysis was performed with the Genomatix Software Suite (http://genomatix.de, ElDorado version 12–2010, Matrix Family Library Version 8.3, October 2010).31 Gene2Promoter was used to identify the predicted transcription start sites for Pvalb (GXP_1492890), Syt2 (GXP_180812), Cplx1 (GXP_149838), and Nefh (GXP_90358). RegionMiner was used to predict transcription factor consensus binding sites within the promoter region (2kb 5′ to 1kb 3′ of transcription start site) and within the entire gene region and to compare the observed number of sites with genome-wide frequencies in each of those regions. Enrichment of consensus binding sites was ranked by descending z-score.
Statistics
Postmortem data were analyzed with hierarchical regression or ANCOVA using SPSS 22.0. Variables displaying a non-normal distribution or unequal variances were transformed (log: Syt2, Cplx1, Nefh; square root: PV). Potential covariates (from pH, age, sex, postmortem interval, and brain weight) were identified using backwards step-wise variable selection (with P < .05 as the criteria for model inclusion); pH was the only covariate selected for inclusion for the dependent variables PV, Syt2, Cplx1, and Nefh. No covariates were identified for PGC-1α, NF-kB or NRF-1. For hierarchical regression, data were stratified according to diagnosis, and covariates (ie, pH) were entered in the first step; change in R 2 was used determine whether predictor variables contributed to any additional variance above and beyond the covariate(s). Significant interactions between diagnosis and high vs low levels of PGC-1α (or NRF-1) on the expression of PGC-1α target genes was determined with 2-way ANOVA using a median split for high and low expression groups (for PGC-1α or NRF-1); pH was included as a covariate when appropriate. P values equal or less than .05 were considered statistically significant.
Results
Expression of Cortical PGC-1α-Dependent Transcripts Is Reduced in Patients With Schizophrenia
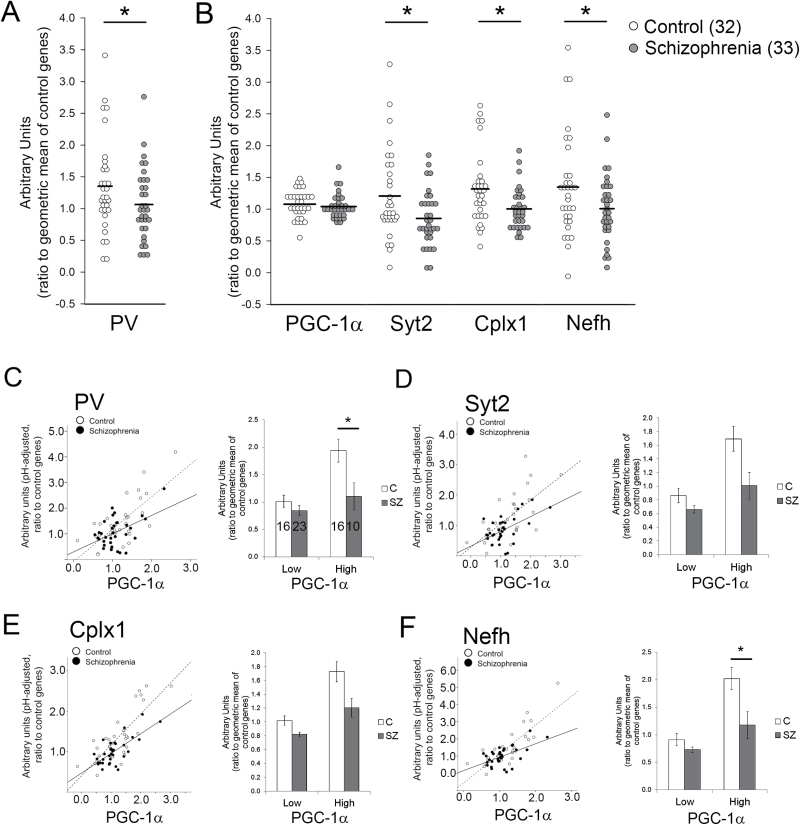
Considering the linkage of PGC-1α to schizophrenia17,18,20 and our previous findings of PGC-1α’s roles in PV-IN function,23,25 we sought to determine whether PGC-1a and its downstream genes Syt2, Cplx1 and Nefh are reduced in the cortex of schizophrenia patients. Multiple regions of the frontal cortex show transcriptional and functional abnormalities in schizophrenia patients32,33; thus, available RNA samples from the ACC and frontal pole (FP) of schizophrenia patients and healthy controls were acquired from the Stanley Consortium.27 As our hypothesis was that PGC-1α-dependent genes would be downregulated in parallel with PV, we first assessed the transcript levels of PV in the ACC and FP using q-RT-PCR, as reduced PV expression has been consistently reported in schizophrenia tissue.8,9,34,35 Schizophrenia patients exhibited a significant down-regulation of PV transcript in the ACC compared to controls while transcript was unaffected in the FP in this sample set (n = 32–33/group; ANCOVA with pH as a covariate, P < .05; figure 1A). Therefore, transcript levels for remaining targets were measured in the ACC in subsequent experiments.
Fig. 1.
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) dependent gene expression is reduced in anterior cingulate cortex of schizophrenia patients. Expression of parvalbumin (PV) transcript was significantly reduced in individuals with schizophrenia in the anterior cingulate cortex (ACC, A). Synaptotagmin 2 (Syt2), complexin 1 (Cplx1), and Nefh transcripts were also reduced in the ACC, but PGC-1α was not (B). PGC-1α expression was positively correlated with all target genes in schizophrenia patients and controls (C–F; left panels). However, when samples were divided into groups with high and low PGC-1α expression, patients with schizophrenia had significantly reduced PV and Nefh expression compared to controls with high PGC-1α expression (C–F, right panels). *P ≤ .05.
Syt2, Cplx1, and Nefh expression levels were significantly reduced in the ACC of the schizophrenia group compared to healthy controls (n = 32–33/group; ANCOVA with pH as a covariate, P < .05), but transcript levels for PGC-1α remained unaffected (figure 1B). Results with 2 other primer/probe sets recognizing PGC-1α gene exon-exon borders also revealed no difference between groups (data not shown). Based on our observation that target gene expression in the cortex depends on PGC-1α in mice,23,24 we used hierarchical regression analysis to investigate whether PGC-1α was significantly correlated with its targets in ACC. We found significant positive correlations between the expression of PGC-1α and its target genes PV (figure 1C; R 2 = .306, P < .005 for control; R 2 = .212, P < .005 for schizophrenia), Syt2 (figure 1D; R 2 = .378, P < .005 for control; R 2 = .176, P < .05 for schizophrenia), Cplx1 (figure 1E; R 2 = .659, P < .005 for control; and R 2 = .532, P < .005 for schizophrenia), and Nefh (figure 1F; R 2 = .464, P < .005 for control; R 2 = .273, P < .005 for schizophrenia) in both controls and patients with schizophrenia (n = 32–33/group). Given the reduced correlation slopes between PGC-1α and its targets in patients with schizophrenia compared to controls (figures 1C–F), we divided subjects into groups displaying high and low PGC-1α expression using a median split. We found that while control subjects with high PGC-1α expression also exhibited high PV (figure 1C) and Nefh expression (figure 1F), schizophrenia subjects with high PGC-1α did not show a high level of PV or Nefh gene expression (2-way ANCOVA; main effect of diagnosis, P < .05; main effect of PGC-1α expression group, P < .05; diagnosis x PGC-1α expression group interaction, P = .05). The interaction between PGC-1α and diagnosis on the expression of Syt2 or Cplx1 failed to reach significance (P = .15 and .27), although overall expression levels of both genes were significantly lower (P < .05). These data suggest that there is dissociation between PGC-1α and some downstream targets in the cortex of schizophrenia patients.
The Expression of the PGC-1α-Interacting Factor NRF-1 Is Reduced in Patients With Schizophrenia
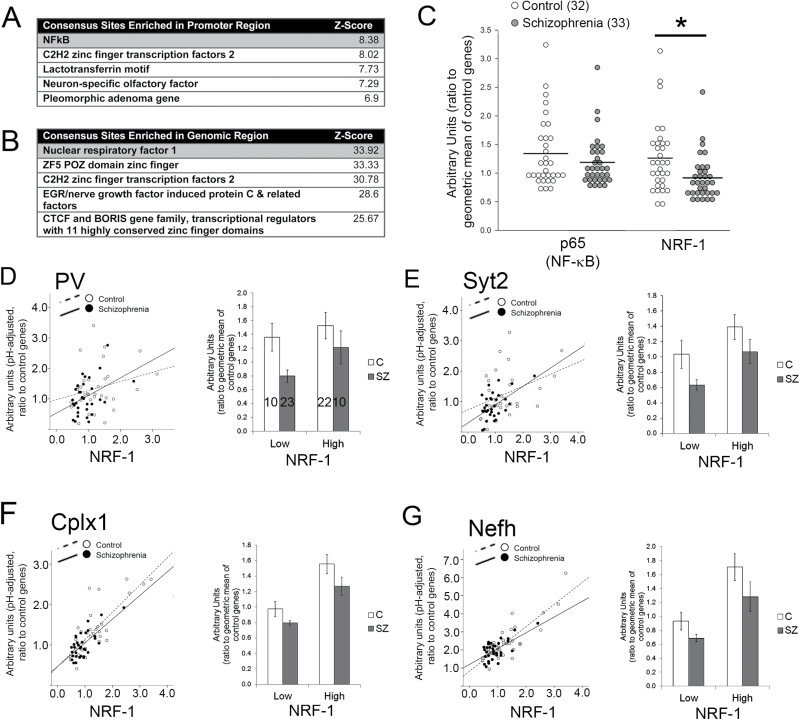
Considering that PGC-1α cannot directly bind DNA, a possible contributor to dissociation between PGC-1α and target gene expression could be a reduction in the expression and/or function of PGC-1α-associated transcription factors. To identify the putative transcription factors that could mediate PGC-1α’s control of PV, Syt2, Cplx, and Nefh transcription, we used the Genomatix Software Suite,31 to determine enrichment for consensus transcription factor binding sites ± 2kb around their default transcription start sites (figure 2A) or in the genomic region (figure 2B). This analysis revealed an enrichment for nuclear factor kappa B (NFκB) binding sites within 2kb of the transcription start sites and enrichment for binding sites for Nrf-1 within the respective genes. We then measured the expression of the NFκB subunit responsible for gene activation (p65;36) and NRF-1 in the ACC from controls and patients with schizophrenia. While there was not a statistically significant reduction in p65, NRF-1 was significantly reduced in the ACC of schizophrenia patients compared to controls (ANOVA, P < .05, figure 2C).
Fig. 2.
Nuclear respiratory factor 1 (NRF-1) expression is reduced in the cortex of schizophrenia patients and is associated with the expression of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) dependent genes in both control and schizophrenia samples. Sequence analysis of parvalbumin (PV), synaptotagmin 2 (Syt2), complexin 1 (Cplx1), and Nefh genes revealed transcription factor binding site enrichment ±2kb around transcription start sites (A) or in the entire gene region (B). Expression of NRF-1 was reduced in the anterior cingulate cortex (ACC) of patients with schizophrenia while p65 expression was unchanged (C). In general, NRF-1 gene expression was positively correlated with PGC-1α-target gene expression in both schizophrenia patients and controls (see stats in text; D–G; left panels), and no statistically significant differences were observed between controls and schizophrenics, when NRF-1 data were binned into low and high categories with a median split (D–G, right panels) *P < .05.
With our prediction that alterations in the expression of PGC-1α-interacting factors could be contributing to the dissociation between PGC-1α and its targets, we then investigated whether NRF-1 expression was associated with the expression of PGC-1α targets in control and schizophrenia patients. In general, NRF-1 expression was associated with PGC-1α target gene expression (figures 2D–G); however, in contrast to what was seen with PGC-1α and PV/Nefh in schizophrenia, there was no statistically significant difference between controls and schizophrenics in the relationship between NRF-1 and PV (figure 2D; R 2 = .044, P = .192 for control; R 2 = .205, P < .01 for schizophrenia), Syt2 (figure 2E; R 2 = .086, P = .083 for control; R 2 = .294, P = .001 for schizophrenia), Cplx1 (figure 2F; R 2 = .525, P < .001 for control; and R 2 = .545, P < .001 for schizophrenia), or Nefh (figure 2G; R 2 = .311, P < .005 for control; R 2 = .493, P < .005 for schizophrenia). Interestingly, PV and Syt2 were not associated with NRF-1 in controls, but were in schizophrenia patients (above).
The Associations Between PGC-1α and Target Gene Expression Are Differentially Influenced by NRF-1 Levels
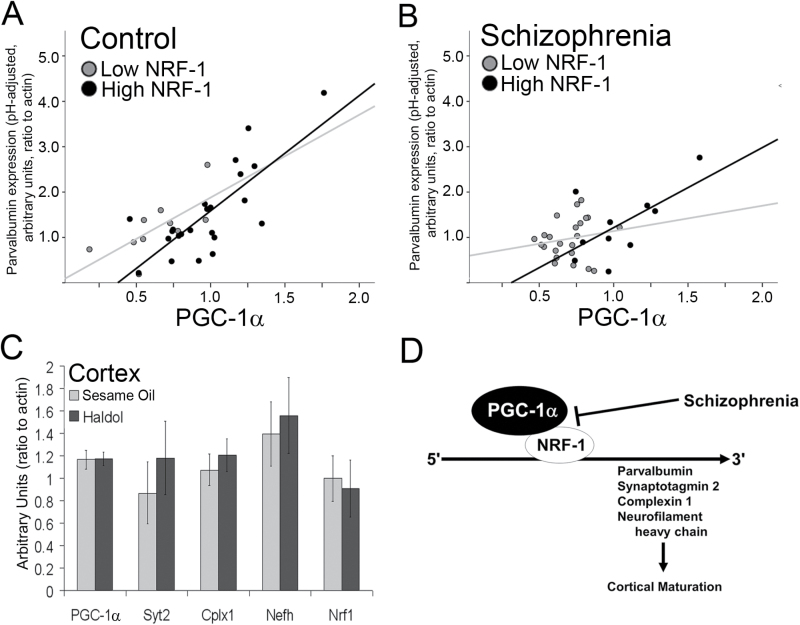
These data suggest that NRF-1 could be a better predictor of gene expression in schizophrenia than PGC-1α. To determine whether this is the case, we looked at the association between PGC-1α and its targets in high and low NRF-1 conditions (PV shown as an example, figures 3A and 3B). While there was a positive correlation between PGC-1α and PV expression in controls, irrespective of NRF-1 level (PV: R 2 = .793 and .336, respectively, P < .01 for both; figure 3A), and in schizophrenia patients with high NRF-1 (PV: R 2 = .436, P < .05; figure 3B), the positive association between PGC-1α and PV was abolished in schizophrenia patients with low NRF-1 expression levels (R 2 = .02, P = .463; figure 3B). A similar result was found with PGC-1α targets Cplx1 and Syt2, where significant associations were found for control patients with high or low NRF-1 expression (Cplx1: R 2 = .739 and .492, respectively, P < .01 for both; Syt2: R 2 = .733 and .244, respectively) and schizophrenia patients with high NRF-1 expression (Cplx1: R 2 = .579, P < .05; Syt2: R 2 = .387, P < .05) but not schizophrenia patients with low NRF-1 expression (Cplx1: R 2 = .062, P = .243; Syt2: R 2 = .079, P = .782). Interestingly, while Nefh expression was associated with NRF-1 expression in all controls (P ≤ .01 for both; NEFH: R 2 = .711 and .337, respectively, P < .01), no significant associations were found between PGC-1α and Nefh in schizophrenia (R 2 = .005 and .281, respectively, P ≥ .11 for both).
Fig. 3.
The association between peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and Parvalbumin (PV) expression is influenced by nuclear respiratory factor 1 (NRF-1) levels. PV expression data are displayed for controls (A) and patients with schizophrenia (B), with respect to PGC-1α and NRF-1 expression. The positive relationship between the expression of PGC-1α and its targets is no longer evident when NRF-1 expression is low. Rats treated for 36 weeks with haldol showed no difference in transcript for PGC-1α, its putative targets, or NRF-1 compared to rats treated with sesame oil (n = 10/group; C; mean ± SEM). We propose that a reduction in NRF-1 could contribute to a decrease in the expression of PGC-1α-dependent genes in schizophrenia (D).
No Effect of Haldol Treatment on Gene Expression in Rats
We then evaluated whether changes in gene expression could be caused by long-term exposure to antipsychotic medications. To model chronic treatment with antipsychotics, we treated rats for 36 weeks with Haldol,30 and mRNA expression was measured for PGC-1α, its novel targets, and NRF-1. All were unchanged in the cortex of rats treated with haldol compared to sesame oil treated controls (figure 3C). This suggests that long-term exposure to antipsychotic medications alone cannot account for the observed reductions in NRF-1 and PGC-1α-dependent transcripts in patients with schizophrenia. Based on previously published evidence for functional interactions between PGC-1α and NRF-137,38 and our results suggesting a role for PGC-1α and its targets in cortical maturation,23 we propose a model by which alterations in PGC-1α/NRF-1-mediated transcriptional programs impair neuronal maturation, leading to neurotransmitter release and structural abnormalities in patients with schizophrenia (figure 3D).
Discussion
In this study, we show that the PGC-1α-dependent genes PV, Syt2, Cplx1, and Nefh are significantly reduced in the ACC of schizophrenia patients, while PGC-1α expression is unchanged. As a coactivator, PGC-1α regulates downstream targets via interaction with numerous transcription factors and thus, PGC-1α-dependent transcription may be compromised through reductions in these interacting factors. In fact, we demonstrate that the aforementioned transcripts share binding sites within their genomic region for NRF-1 and that NRF-1 expression is associated with the expression of PGC-1α-dependent transcripts in subjects with schizophrenia. Based on the known function of these proteins, we predict that the buffering of calcium (PV), the synchronous release of neurotransmitter (Syt2 and Cplx1), and axonal integrity (Nefh) are compromised in cortical neurons in schizophrenia. These transcripts are upregulated during a time of synaptic maturation in animal models4,7,11,39 and the publicly available BrainCloud database40 demonstrates that these transcripts are developmentally regulated in a similar way in the human cortex. Thus, reductions in these transcripts could represent a disruption in this process, which is consistent with identified candidate genes41 and reductions in maturational gene expression in schizophrenia.42
Based on our previously published work, we propose that any interference with PGC-1α activity could cause the deficiency in PV, Syt2, Cplx1, and Nefh. We found that the associations between PGC-1α and the targets PV and Nefh are significantly reduced in schizophrenia and investigated whether this could be attributable to levels of NRF-1, a transcription factor that can mediate PGC-1α’s actions on gene expression.43 Interestingly, while there was a positive association between PGC-1α and PV/Syt2/Cplx1 in all control patients and schizophrenia patients with high NRF-1 expression, there was no association between PGC-1α and PV/Syt2/Cplx1 in schizophrenia patients with low NRF-1. However, the association between PGC-1α and Nefh, while statistically significant in all controls, was absent in schizophrenia patients, irrespective of NRF-1 levels. While this supports the general idea that the transcriptional activity of PGC-1α complexes is disrupted in schizophrenia, it highlights the complexity of gene regulation and suggests that Nefh is regulated by different mechanisms (potentially other proteins within the transcriptional complex) than PV, Syt2, and Cplx1. While PGC-1α shows robust localization in the rodent brain to GABAergic interneurons,44 it is possible that PGC-1α also has a role in transcriptional regulation within pyramidal neurons (PNs). We have found that Syt2, Cplx1, and Nefh are highly expressed by PV-INs,23 but we have found expression of Syt2 and Nefh also in pyramidal neurons by immuno-electron microscopy (unpublished data). Thus, a reduction in transcript for PGC-1α and its target genes may be due to changes within a single neuronal population or multiple cell types. Heterogeneity in cell-specific responses could contribute, at least in part, to the differential dependencies of Nefh and PV/Syt2/Cplx1 on NRF-1 expression. While mice lacking PGC-1α in neurons expressing calcium calmodulin kinase II (CaMKII; deletion from cortical, hippocampal, and striatal neurons) exhibit evidence for cortical degeneration,45 no studies have evaluated the effects of PGC-1α deletion specifically from cortical PNs. Such studies have the potential to elucidate the relative contribution of PV-INs and PNs to cortical dysfunction in the context of PGC-1α deficiency.
In mice, disrupted PGC-1α function and/or expression can impair transcription and neurotransmitter release in a cell- and region-specific manner.23,24,28,46 PGC-1α null mice exhibit profound motor deficits, cerebral vacuolizations,28 reductions in metabolic, structural, and neurotransmitter release transcripts23,47,48 and reduced GABA release in the cortex.25 Mice lacking PGC-1α from PV-INs exhibit long-term memory deficits and asynchronous cortical GABA release,23 potentially due to decreases in Syt2 and Cplx1.49 Electrophysiological recordings from Syt2 -/- mice and Cplx1 -/- mice show asynchronous calcium-mediated vesicle release.50,51 These data indicate that both Syt2 and Cplx1 have a prominent role in neurotransmitter release, a loss of which may contribute to cognitive dysfunction. In fact, Cplx1 has previously been reported to be reduced in postmortem cortical and hippocampal samples from schizophrenic and bipolar patients.10,52–54
In addition to regulating the non-metabolic transcripts tested here, PGC-1α can interact directly with NRF-1 to drive transcription of mitochondrial genes.37,38 Interestingly, NRF-1 regulates neuronal Na+/K+ ATPases in an activity-dependent manner55 suggesting that PGC-1α/NRF-1 may provide a link between neuronal activity and the metabolic machinery necessary for neuronal activity. Transcriptional programs for oxidative phosphorylation, metabolism, mitochondrial translocation and glycolysis are among the top gene programs downregulated in the cortex of schizophrenia patients.56 Additionally, redox dysregulation and oxidative stress, both of which are controlled by PGC-1α-driven antioxidant programs57 have been proposed as a contributor to schizophrenia pathology.58 Thus, it is possible metabolic PGC-1α /NRF-1-regulated metabolic gene programs are also compromised in schizophrenia.
It is attractive to speculate that that PGC-1α interacts with NRF-1 to coordinate the developmental expression of genes for neurotransmission and metabolism and that disruption of these paralleled transcriptional programs in schizophrenia would impair PV-IN and/or PN maturation and function. Identification of transcription factors that associate with PGC-1α in specific cell types may provide information that can be utilized to enhance activity of transcriptional programs to promote proper synaptic function in schizophrenia in a cell-selective manner.
Funding
This work was supported by National Institute of Mental Health (MH077955-05) and National Institute of Neurological Disorders and Stroke (NS070009), and Civitan Emerging Scholar (E.K.L.) and McNulty Investigator (R.M.C.) awards. Bioinformatics analyses were supported by UAB Center for Clinical and Translational Science Grant Number UL1TR001417 from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH).
Acknowledgments
We thank Juan Molina and Rosalinda Roberts for helpful discussions related to the manuscript, Farah Lubin for discussions related to NF-kB, and the Stanley Foundation. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. Citrome L. Unmet needs in the treatment of schizophrenia: new targets to help different symptom domains. J Clin Psychiatry. 2014;75:21–26. [DOI] [PubMed] [Google Scholar]
- 2. Gray JA, Roth BL. The pipeline and future of drug development in schizophrenia. Mol Psychiatry. 2007;12:904–922. [DOI] [PubMed] [Google Scholar]
- 3. Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gonzalez-Burgos G, Hashimoto T, Lewis DA. Alterations of cortical GABA neurons and network oscillations in schizophrenia. Curr Psychiatry Rep. 2010;12:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2010;11:100–113. [DOI] [PubMed] [Google Scholar]
- 7. Lewis DA, Cruz D, Eggan S, Erickson S. Postnatal development of prefrontal inhibitory circuits and the pathophysiology of cognitive dysfunction in schizophrenia. Ann N Y Acad Sci. 2004;1021:64–76. [DOI] [PubMed] [Google Scholar]
- 8. Volk DW, Matsubara T, Li S, et al. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry. 2012;169:1082–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beasley CL, Reynolds GP. Parvalbumin-immunoreactive neurons are reduced in the prefrontal cortex of schizophrenics. Schizophr Res. 1997;24:349–355. [DOI] [PubMed] [Google Scholar]
- 10. Torrey EF, Barci BM, Webster MJ, Bartko JJ, Meador-Woodruff JH, Knable MB. Neurochemical markers for schizophrenia, bipolar disorder, and major depression in postmortem brains. Biol Psychiatry. 2005;57:252–260. [DOI] [PubMed] [Google Scholar]
- 11. Hoftman GD, Lewis DA. Postnatal developmental trajectories of neural circuits in the primate prefrontal cortex: identifying sensitive periods for vulnerability to schizophrenia. Schizophr Bulletin. 2011;37:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pafundo DE, Miyamae T, Lewis DA, Gonzalez-Burgos G. Cholinergic modulation of neuronal excitability and recurrent excitation-inhibition in prefrontal cortex circuits: implications for gamma oscillations. J Physiol. 2013;591:4725–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. [DOI] [PubMed] [Google Scholar]
- 14. Kolluri N, Sun Z, Sampson AR, Lewis DA. Lamina-specific reductions in dendritic spine density in the prefrontal cortex of subjects with schizophrenia. Am J Psychiatry. 2005;162:1200–1202. [DOI] [PubMed] [Google Scholar]
- 15. Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: a postmortem morphometric study of schizophrenia and Huntington disease. Arch Gen Psychiatry. 1998;55:215–224. [DOI] [PubMed] [Google Scholar]
- 16. Pierri JN, Volk CL, Auh S, Sampson A, Lewis DA. Decreased somal size of deep layer 3 pyramidal neurons in the prefrontal cortex of subjects with schizophrenia. Arch Gen Psychiatry. 2001;58:466–473. [DOI] [PubMed] [Google Scholar]
- 17. Christoforou A, Le Hellard S, Thomson PA, et al. Association analysis of the chromosome 4p15-p16 candidate region for bipolar disorder and schizophrenia. Mol Psychiatry. 2007;12:1011–1025. [DOI] [PubMed] [Google Scholar]
- 18. Christoforou A, McGhee KA, Morris SW, et al. Convergence of linkage, association and GWAS findings for a candidate region for bipolar disorder and schizophrenia on chromosome 4p. Mol Psychiatry. 2011;16:240–242. [DOI] [PubMed] [Google Scholar]
- 19. Blackwood DH, He L, Morris SW, et al. A locus for bipolar affective disorder on chromosome 4p. Nat Genet. 1996;12:427–430. [DOI] [PubMed] [Google Scholar]
- 20. Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le Hellard S, Lee AJ, Underwood S, et al. Haplotype analysis and a novel allele-sharing method refines a chromosome 4p locus linked to bipolar affective disorder. Biol Psychiatry. 2007;61:797–805. [DOI] [PubMed] [Google Scholar]
- 22. Jiang Z, Rompala GR, Zhang S, Cowell RM, Nakazawa K. Social isolation exacerbates schizophrenia-like phenotypes via oxidative stress in cortical interneurons. Biol Psychiatry. 2013;73:1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lucas EK, Dougherty SE, McMeekin LJ, et al. PGC-1alpha provides a transcriptional framework for synchronous neurotransmitter release from parvalbumin-positive interneurons. J Neurosci. 2014;34:14375–14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lucas EK, Markwardt SJ, Gupta S, et al. Parvalbumin deficiency and GABAergic dysfunction in mice lacking PGC-1alpha. J Neurosci. 2010;30:7227–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dougherty SE, Bartley AF, Lucas EK, Hablitz JJ, Dobrunz LE, Cowell RM. Mice lacking the transcriptional coactivator PGC-1alpha exhibit alterations in inhibitory synaptic transmission in the motor cortex. Neuroscience. 2014;271:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng A, Wan R, Yang JL, et al. Involvement of PGC-1α in the formation and maintenance of neuronal dendritic spines. Nat Commun. 2012;3:1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–155. [DOI] [PubMed] [Google Scholar]
- 28. Lucas EK, Dougherty SE, McMeekin LJ, Trinh AT, Reid CS, Cowell RM. Developmental alterations in motor coordination and medium spiny neuron markers in mice lacking pgc-1α. PLoS One. 2012;7:e42878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drummond JB, Simmons M, Haroutunian V, Meador-Woodruff JH. Upregulation of cornichon transcripts in the dorsolateral prefrontal cortex in schizophrenia. Neuroreport. 2012;23:1031–1034. [DOI] [PubMed] [Google Scholar]
- 31. Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. [DOI] [PubMed] [Google Scholar]
- 32. Tamminga CA, Vogel M, Gao X, Lahti AC, Holcomb HH. The limbic cortex in schizophrenia: focus on the anterior cingulate. Brain Res Brain Res Rev. 2000;31:364–370. [DOI] [PubMed] [Google Scholar]
- 33. Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165:479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang ZJ, Reynolds GP. A selective decrease in the relative density of parvalbumin-immunoreactive neurons in the hippocampus in schizophrenia. Schizophr Res. 2002;55:1–10. [DOI] [PubMed] [Google Scholar]
- 35. Beasley CL, Zhang ZJ, Patten I, Reynolds GP. Selective deficits in prefrontal cortical GABAergic neurons in schizophrenia defined by the presence of calcium-binding proteins. Biol Psychiatry. 2002;52:708–715. [DOI] [PubMed] [Google Scholar]
- 36. Schmitz ML, Baeuerle PA. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO J. 1991;10:3805–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knutti D, Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol Metab. 2001;12:360–365. [DOI] [PubMed] [Google Scholar]
- 38. Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. [DOI] [PubMed] [Google Scholar]
- 39. Uhlhaas PJ, Roux F, Rodriguez E, Rotarska-Jagiela A, Singer W. Neural synchrony and the development of cortical networks. Trends Cogn Sci. 2010;14:72–80. [DOI] [PubMed] [Google Scholar]
- 40. Colantuoni C, Lipska BK, Ye T, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Farrell MS, Werge T, Sklar P, et al. Evaluating historical candidate genes for schizophrenia [published online ahead of print March 10, 2015]. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gandal MJ, Nesbitt AM, McCurdy RM, Alter MD. Measuring the maturity of the fast-spiking interneuron transcriptional program in autism, schizophrenia, and bipolar disorder. PLoS One. 2012;7:e41215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cowell RM, Blake KR, Russell JW. Localization of the transcriptional coactivator PGC-1alpha to GABAergic neurons during maturation of the rat brain. J Comp Neurol. 2007;502:1–18. [DOI] [PubMed] [Google Scholar]
- 45. Ma D, Li S, Lucas EK, Cowell RM, Lin JD. Neuronal inactivation of peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1alpha) protects mice from diet-induced obesity and leads to degenerative lesions. J Biol Chem. 2010;285:39087–39095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lucas EK, Reid CS, McMeekin LJ, Dougherty SE, Floyd CL, Cowell RM. Cerebellar transcriptional alterations with Purkinje cell dysfunction and loss in mice lacking PGC-1alpha. Front Cell Neurosci. 2014;8:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin J, Wu PH, Tarr PT, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. [DOI] [PubMed] [Google Scholar]
- 48. Leone TC, Lehman JJ, Finck BN, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin MY, Rohan JG, Cai H, Reim K, Ko CP, Chow RH. Complexin facilitates exocytosis and synchronizes vesicle release in two secretory model systems. J Physiol. 2013;591:2463–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun J, Pang ZP, Qin D, Fahim AT, Adachi R, Sudhof TC. A dual-Ca2+-sensor model for neurotransmitter release in a central synapse. Nature. 2007;450:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pang ZP, Melicoff E, Padgett D, et al. Synaptotagmin-2 is essential for survival and contributes to Ca2+ triggering of neurotransmitter release in central and neuromuscular synapses. J Neurosci. 2006;26:13493–13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eastwood SL, Harrison PJ. Decreased expression of vesicular glutamate transporter 1 and complexin II mRNAs in schizophrenia: further evidence for a synaptic pathology affecting glutamate neurons. Schizophr Res. 2005;73:159–172. [DOI] [PubMed] [Google Scholar]
- 53. Knable MB, Barci BM, Webster MJ, Meador-Woodruff J, Torrey EF; Stanley Neuropathology Consortium Molecular abnormalities of the hippocampus in severe psychiatric illness: postmortem findings from the Stanley Neuropathology Consortium. Mol Psychiatry. 2004;9:609–620, 544. [DOI] [PubMed] [Google Scholar]
- 54. Sawada K, Barr AM, Nakamura M, et al. Hippocampal complexin proteins and cognitive dysfunction in schizophrenia. Arch Gen Psychiatry. 2005;62:263–272. [DOI] [PubMed] [Google Scholar]
- 55. Johar K, Priya A, Wong-Riley MT. Regulation of Na(+)/K(+)-ATPase by nuclear respiratory factor 1: implication in the tight coupling of neuronal activity, energy generation, and energy consumption. J Biol Chem. 2012;287:40381–40390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Prabakaran S, Swatton JE, Ryan MM, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697, 643. [DOI] [PubMed] [Google Scholar]
- 57. St-Pierre J, Drori S, Uldry M, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. [DOI] [PubMed] [Google Scholar]
- 58. Do KQ, Cabungcal JH, Frank A, Steullet P, Cuenod M. Redox dysregulation, neurodevelopment, and schizophrenia. Curr Opin Neurobiol. 2009;19:220–230. [DOI] [PubMed] [Google Scholar]