SUMMARY
Autophagic turnover of intracellular constituents is critical for cellular housekeeping, nutrient recycling, and various aspects of growth and development in eukaryotes. Here we show that autophagy impacts the other major degradative route involving the ubiquitin-proteasome system by eliminating 26S proteasomes, a process we termed proteaphagy. Using Arabidopsis proteasomes tagged with GFP, we observed their deposition into vacuoles via a route requiring components of the autophagy machinery. This transport can be initiated separately by nitrogen starvation and chemical or genetic inhibition of the proteasome, implying distinct induction mechanisms. Proteasome inhibition stimulates comprehensive ubiquitylation of the complex, with the ensuing proteaphagy requiring the proteasome subunit RPN10, which can simultaneously bind both ATG8 and ubiquitin. Collectively, we propose that Arabidopsis RPN10 acts as a selective autophagy receptor that targets inactive 26S proteasomes by concurrent interactions with ubiquitylated proteasome subunits/targets and lipidated ATG8 lining the enveloping autophagic membranes.
INTRODUCTION
All cellular organisms use a variety of degradative routes for cellular housekeeping and to selectively regulate the abundance of their internal constituents. The main routes in eukaryotes are the ubiquitin-26S proteasome system (UPS) and autophagy, which, together, enable nutrient recycling, turnover of organelles and aberrant or aggregated proteins, and the precise control of regulators necessary for proper growth and development. The UPS involves the covalent attachment of multiple ubiquitins to selected target proteins, which facilitates their recognition and degradation by the 26S proteasome (Finley, 2009; Vierstra, 2009; Bhattacharyya et al., 2014).
Because the UPS cannot degrade organelles and is limited in its ability to remove large protein complexes and insoluble protein aggregates, eukaryotes also engage autophagy, in which cytoplasmic material is encapsulated and delivered in bulk to the vacuole or lysosome for breakdown (Li and Vierstra, 2012; Klionsky and Schulman, 2014). Autophagy occurs at a basal level but is upregulated when extensive nutrient remobilization is required. The initial step is the formation of a double membrane-bound autophagosome that traps appropriate cargo. This then fuses with the limiting membrane of the vacuole or lysosome and releases the internal vesicle, termed an autophagic body, which is degraded by resident hydrolases. Central to this process is the attachment of the ATG8 protein to the lipid phos-phatidylethanolamine (PE) via a conjugation cascade mechanistically similar to, but distinct from, ubiquitylation. The ATG8-PE adduct decorates the expanding autophagic membranes and provides a docking site for proteins that encourage vesicle closure and receptors that recruit specific cargo. Through such receptors, autophagy can selectively remove large protein complexes, insoluble protein aggregates, organelles, and even invading pathogens (Rogov et al., 2014). One class of receptors, which includes NBR1, has affinity for ubiquitin as well as ATG8, therefore allowing ubiquitylation and autophagy to work in concert (Kirkin et al., 2009; Svenning et al., 2011).
The 26S proteasome is a 2.5-MDa, self-compartmentalized proteolytic machine located in the cytosol and nucleus. It is composed of two functionally distinct sub-particles, the 20S core protease (CP) and the 19S regulatory particle (RP) (Finley, 2009; Bhattacharyya et al., 2014). The CP is created by assembly of four stacked heptameric rings of α and β subunits (Arabidopsis PAA-PAG and PBA-PBG, respectively) that generate a central proteolytic chamber housing catalytic sites provided by the β1 (PBA), β2 (PBB) and β5 (PBE) subunits. The axial channels that access this chamber are gated so that only proteins that are deliberately recognized, unfolded, and imported are degraded. The CP is capped at one or both ends by the RP. The RP consists of two sub-complexes; the base, which includes a hexameric ring of ATPases (RPT1–6) plus two non-ATPase subunits, RPN1 and RPN2; and the lid, which incorporates at least 11 additional non-ATPase subunits (RPN3, RPN5–13, and SEM1/DSS1; Finley, 2009; Bhattacharyya et al., 2014). Substrates are recognized by several ubiquitin receptors intrinsic to the RP lid, including RPN10, RPN13, and SEM1/DSS1 (van Nocker et al., 1996a; Finley, 2009; Fatimababy et al., 2010; Lin et al., 2011; Paraskevopoulos et al., 2014), along with several extrinsic ubiquitin-binding factors that shuttle ubiquitylated cargo to the RP (Finley, 2009).
Given the central importance of the 26S proteasome to the UPS, its activity and abundance are controlled at numerous levels. To meet proteolytic demand, many proteasome subunit genes are transcriptionally upregulated when 26S proteasomes become limiting. This regulon is sensitive to chemical and genetic inhibition of the complex and is controlled by dedicated transcription factors (Nguyen et al., 2013; Sha and Goldberg, 2014). Subsequent construction of the 26S particle is driven by a suite of chaperones that mediate the sequential assembly of the α and β subunit rings of the CP and the RPT ring of the RP (Tomko and Hochstrasser, 2013). In addition, the 26S proteasome is subject to numerous post-translational modifications (Besche et al., 2014; Cui et al., 2014). Particularly pervasive is ubiquitylation, with many subunits conjugated at multiple sites (Book et al., 2010; Kim et al., 2011; Besche et al., 2014; Piterman et al., 2014). In Arabidopsis, this ubiquitylation is upregulated upon proteasome inhibition, with at least 35 of the 52 subunit isoforms becoming modified (Book et al., 2010; Kim et al., 2013). Although the function(s) of proteasome ubiquitylation are mostly unknown, roles in regulating the receptor functions of RPN10, RPN13, and shuttle factors bearing ubiquitin-like (UBL) domains have been proposed (Besche et al., 2014; Piterman et al., 2014).
Here we show that the UPS and autophagy converge in Arabidopsis, with the turnover of 26S proteasomes being driven by autophagy. This degradation is induced separately by nutrient starvation and proteasome inhibition, implying that both bulk and selective routes exist. Proteasome inhibitors in particular stimulate extensive ubiquitylation of the complex, with subsequent autophagic clearance engaging RPN10 to tether the particle to enveloping autophagic vesicles through its ability to bind both ubiquitin and ATG8. This “proteaphagy” likely plays a key role in modulating proteasome abundance by removing inactive or excess particles.
RESULTS
26S Proteasome Subunits Are More Abundant in Autophagy Mutants
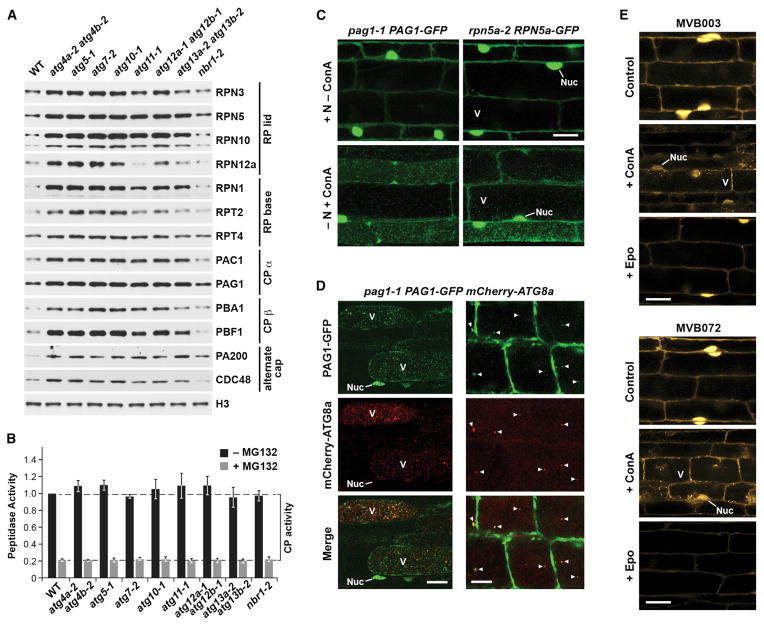
During our studies of the interplay between the UPS and autophagy in Arabidopsis, we noticed that autophagy mutants have elevated levels of many 26S proteasome subunits, even under well fed conditions (Figure 1A). This was observed in mutants compromising either ATG8 lipidation (atg4a-2 atg4b-2, atg5-1, atg7-2, atg10-1, and atg12a-1 atg12b-1) or autophagic induction by the ATG1 kinase (atg11-1 and atg13a-2 atg13b-2), with the latter set generally having a weaker effect (Figure 1A; Table S1). Only the nbr1-2 mutant eliminating the autophagic ubiquitin receptor NBR1 had no effect. Although some of these increases could reflect accumulation of unassembled polypeptides, the fact that the atg mutants showed normal 26S proteasome assembly and that the pool of PBA1 in the mutants was almost exclusively the processed form that appears only upon CP assembly (Figure 1A; Figures S1B and S1C; Book et al., 2010) argued that the abundance of the entire particle rose in the absence of autophagy.
Figure 1. Arabidopsis 26S Proteasome Abundance Is Controlled by Autophagy.
(A) Proteasome subunit levels rise in autophagy mutants. Shown is immunoblot detection of various proteasome subunits and accessory factors in total protein extracts from the wild-type (WT) or a collection of autophagy mutants.
(B) 26S proteasome activity is unchanged in autophagy mutants. Total protein extracts were assayed for CP peptidase activity using succinyl-leucyl-leucyl-valyl-tyroyl-7-amido-4-methylcoumarin (succinyl-LLVY-AMC). Bars represent the mean (± SD) of three biological replicates, each generated with three technical replicates.
(C) Proteasome subunits are delivered to the vacuole upon nitrogen starvation. Seedlings expressing PAG1-GFP or RPN5a-GFP were either kept on nitrogen (N)-rich medium or switched to N-free medium plus 1 μM ConA for 16 hr. Root cells were imaged by confocal microscopy. Scale bar, 10 μm. Nuc, nucleus; V, vacuole.
(D) PAG1-GFP co-localizes with mCherry-ATG8a in autophagic bodies. Seedlings were exposed to N starvation and 1 μM ConA for either 16 or 4 hr (left and right, respectively). Scale bars, 5 μm (left) and 10 μm (right).
(E) Assembled proteasomes accumulate in autophagic vesicles. WT seedlings were incubated for 16 hr with 20 μM of the fluorescent epoxomycin (Epo) probes MVB003 or MVB072, which label active proteasomes. Where indicated, seedlings were treated simultaneously with 1 μM ConA or pre-treated with 20 μM Epo for 4 hr. Scale bar, 5 μm.
See also Figures S1 and S2.
Despite the increased abundance of 26S proteasomes, the panel of autophagy mutants did not elevate the levels of the corresponding subunit mRNAs (Figure S1A), strongly suggesting that this effect was not driven transcriptionally. Surprisingly, elevated subunit levels did not increase the pool of enzymatically active proteasomes (Figure 1B), nor did we observe a significant change in the levels of ubiquitin conjugates (Figure S1D), as might occur when active proteasome levels are altered. Therefore, the increased abundance of 26S proteasomes in atg mutants appeared to reflect attenuated turnover of inactive particles.
The 26S Proteasome Is Degraded by ATG8-Mediated Autophagy
To directly test whether 26S proteasomes are cleared by autophagy, we generated Arabidopsis lines in which either the CP subunit PAG1 (α7) or the RP lid subunit RPN5a were replaced with GFP-tagged counterparts. Both reporters rescued the corresponding mutant phenotypes (embryo lethality for pag1-1 and the severe dwarf phenotype for rpn5a-2), were effectively integrated into the 26S particle, and remained mostly intact in planta (Figure S2), demonstrating that they could reliably report proteasome location. Confocal fluorescence microscopy of pag1-1 PAG1-GFP and rpn5a-2 RPN5a-GFP seedlings grown under nutrient-rich conditions revealed that Arabidopsis 26S proteasomes are located in the nucleus and cytoplasm (Figure 1C; Figure S2B), in agreement with prior cytological studies (Kolodziejek et al., 2011). However, following induction of autophagy by nitrogen starvation and pre-treatment with concanamycin A (ConA), an inhibitor that stabilizes autophagic bodies by raising the vacuolar pH, numerous ~1-μm fluorescent puncta resembling autophagic bodies accumulated within vacuoles (Figure 1C). To support their identity, we co-localized PAG1-GFP with the mCherry-ATG8a marker, which decorates autophagic vesicles upon lipidation (Suttangkakul et al., 2011). After nitrogen starvation and extended ConA exposure, numerous vacuolar puncta were observed that displayed both GFP and mCherry fluorescence (Figure 1D). To better visualize this, we examined seedlings starved for nitrogen and treated only briefly with ConA. Under these conditions, near complete coincidence of the PAG1-GFP and mCherry-ATG8a signals was observed in the reduced number of vacuolar vesicles that accumulated (Figure 1D).
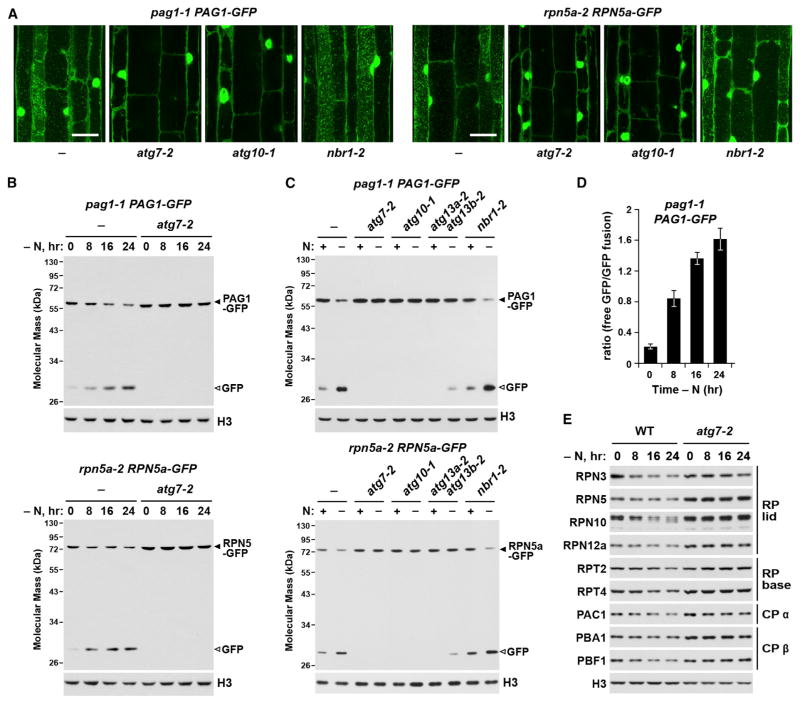
For additional confirmation that the vacuolar puncta labeled with PAG1-GFP or RPN5a-GFP were indeed of autophagic origin, we examined their accumulation in the strong autophagy mutants atg7-2 and atg10-1 (Phillips et al., 2008; Chung et al., 2010). Although wild-type roots readily accumulated these puncta after nitrogen starvation and ConA treatment, none were evident in the mutant lines (Figure 2A). However, these vacuolar puncta were still evident in nbr1-2 seedlings, ruling out a role for NBR1 in this process (Figure 2A).
Figure 2. 26S Proteasomes Are Delivered to the Vacuole upon Nitrogen Starvation.
(A) Delivery of vesicles containing PAG1-GFP and RPN5a-GFP to the vacuole is blocked in strong autophagy mutants. WT, atg7-2, atg10-1, and nbr1-2 seedlings were switched to N-free medium containing 1 μM ConA for 16 hr. Root cells were examined for autophagic bodies by confocal microscopy. Scale bars, 10 μm.
(B) Time course of free GFP release from PAG1-GFP and RPN5a-GFP during N starvation. WT and atg7-2 seedlings expressing each reporter were starved for the indicated times, and total protein extracts were immunoblotted with anti-GFP antibodies. The GFP fusions and free GFP are indicated by closed and open arrowheads, respectively.
(C) Effect of autophagy mutants on the cleavage of PAG1-GFP and RPN5a-GFP during N starvation. WT and mutant seedlings were either kept on N-rich medium or switched to N-free medium for 16 hr. Immunoblotting was performed as in (B).
(D) Quantification of the free GFP/PAG1-GFP ratio during N starvation by densitometric scans of the immunoblots shown in (B). Bars represent the mean (± SD) of three biological replicates.
(E) N starvation induces the loss of many 26S proteasome components. WT and atg7-2 seedlings were N-starved for the indicated times. Total protein extracts were probed with antibodies against various proteasome subunits.
It was possible that these puncta contained unincorporated PAG1-GFP or RPN5a-GFP rather than complete proteasomes. To demonstrate that assembled proteasomes were indeed engulfed, we exploited two fluorescent derivatives of the proteasome inhibitor epoxomycin (MVB003 and MVB072) that form covalent adducts with assembled CPs, therefore allowing us to track active proteasomes in planta (Kolodziejek et al., 2011). Fluorescence from both probes was observed in the cytosol and nucleus of root cells (Figure 1E), and its intensity was reduced, but not abolished, by pre-incubation with epoxomycin, implying that the signal was genuinely from labeled proteasomes. When MVB003 and MVB072 labeling was accompanied by ConA treatment, fluorescence also became visible in cytosolic and vacuolar puncta (Figure 1E), reminiscent of the autophagic vesicles labeled with PAG1-GFP or RPN5a-GFP. Taken together, we concluded that whole proteasomes are cleared by an autophagic process we termed proteaphagy.
To assess proteaphagy more quantitatively, we exploited observations that the GFP moiety often becomes detached from GFP-tagged autophagic substrates inside the vacuole and accumulates because of its resistance to further digestion (Chung et al., 2010; Suttangkakul et al., 2011). Consequently, the ratio of free GFP to the GFP-tagged parent can be used to measure autophagic flux. We found here that this cleavage assay works well for the PAG1-GFP and RPN5a-GFP reporters. In wild-type Arabidopsis, small amounts of released free GFP were detected in extracts from pag1-1 PAG1-GFP and rpn5a-2 RPN5a-GFP seedlings well fed with nitrogen, with its levels relative to the fusion parents rising substantially when the seedlings were nitrogen-starved (Figure 2B). In contrast, this released free GFP was absent in the strong atg7-2 and atg10-1 mutants, even upon nitrogen starvation, and was substantially reduced in the weaker atg13a-2 atg13b-2 mutant (Figures 2B and 2C). To demonstrate how this assay could detect changes in autophagic flux, we quantified the free GFP/tagged-GFP ratio in pag1-1 PAG1-GFP plants during nitrogen deprivation. This ratio rose steadily as proteasomes were cleared (Figure 2D). Importantly, this increasing ratio coincided with a steady decline of many other 26S proteasome subunits in wild-type but not atg7-2 plants, further con-firming that the entire particle was being recycled (Figure 2E).
Proteaphagy Is Induced by Proteasome Inhibition
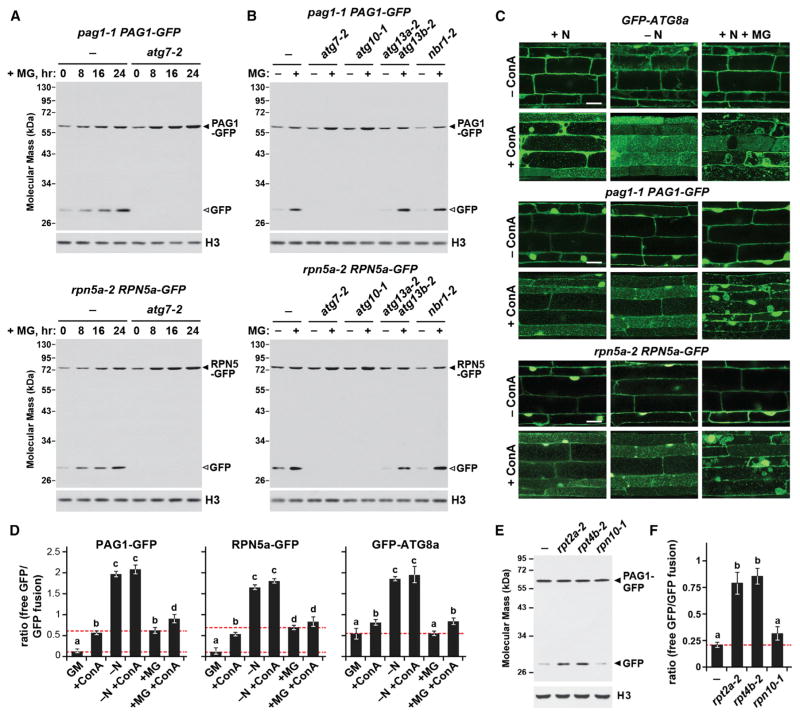
We next examined other conditions that might affect proteaphagy, using the GFP cleavage assay to monitor autophagic flux. Neither heat, tunicamycin, or arsenite affected free GFP accumulation in pag1-1 PAG1-GFP and rpn5a-2 RPN5a-GFP plants (Figures S3A and S3B) despite evidence that the treatments were effective in generating proteotoxic stress (Figures S3C and S3D). By contrast, inhibiting proteasomes with MG132 stimulated proteaphagy substantially, even in seedlings well supplied with nitrogen. The 50 μM dose of MG132 used here was judged to effectively inhibit proteasomes by its ability to increase the steady-state levels of ubiquitin conjugates, accumulate the unprocessed form of PBA1, decrease the pool of active 26S proteasomes, and upregulate the expression of proteasome genes through the proteasome stress regulon (Figures S4A–S4C). When a 24-hr course of MG132 treatment was examined, a continuous release of free GFP from either PAG1-GFP or RPN5a-GFP was seen, which was not evident in the atg7-2 and atg10-1 mutants (Figures 3A and 3B). However, in contrast to its noticeable impact on nitrogen stress-induced proteaphagy, the atg13a-2 atg13b-2 mutant failed to slow MG132-induced proteaphagy, implying that a second route exists (Figure 3B). The levels of PAG1-GFP or RPN5a-GFP, along with most other proteasome subunits, actually rose during MG132 exposure because of activation of the proteasome stress regulon (Figure 3A; Figures S4B and S4D). This rise was accentuated in the atg7-2 mutant, reflecting the fact that inhibited particles could not be degraded (Figures 3A and 3B; Figure S4D).
Figure 3. Proteaphagy Is Stimulated by Chemical or Genetic Inhibition of the Proteasome.
(A) Time course of free GFP release from PAG1-GFP and RPN5a-GFP following MG132 exposure. WT and atg7-2 seedlings were exposed to 50 μM MG132 (MG) for the indicated times, and total protein extracts were immunoblotted with anti-GFP antibodies. The GFP fusions and free GFP are indicated by closed and open arrowheads, respectively.
(B) Effects of various autophagy mutants on the cleavage of PAG1-GFP and RPN5a-GFP during MG132 exposure. WT, atg7-2, atg10-1, atg13a-2 atg13b-2, and nbr1-2 seedlings were incubated with or without 50 μM MG132 for 16 hr. Immunoblotting was performed as in (A).
(C) MG132 induces the delivery of autophagic vesicles to the vacuole. WT seedlings were either N-fed or starved and/or exposed to 50 μM MG132 and/or 1 μM ConA for 16 hr. Root cells were examined for autophagic bodies by confocal microscopy. Scale bar, 10 μm.
(D) Quantification of the free GFP/tagged-GFP ratio during N starvation or MG132 treatment of the seedlings analyzed in (C). Corresponding immunoblots are shown in Figure S6C. Bars represent the mean (± SD) of three biological replicates. Letters indicate values that are statistically different from one another (p < 0.05).
(E) The proteasome mutants rpt2a-2 and rpt4b-2, but not rpn10-1, stimulate proteaphagy. Well fed seedlings were assayed for the release of free GFP by immunoblotting as in (A).
(F) Quantification of the free GFP/PAG1-GFP ratio in the proteasome mutants by densitometric scans of the immunoblot shown in (E). Bars represent the mean (± SD) of three biological replicates. Letters indicate values that are statistically different from one another (p < 0.05).
See also Figures S3–S6.
MG132-induced proteaphagy could also been seen by fluorescence microscopy of pag1-1 PAG1-GFP and rpn5a-2 RPN5a-GFP seedlings. Although a few autophagic bodies decorated with the reporters were detected in MG132-treated root cells, they became abundant when exposed to both MG132 and ConA (Figure 3C). Surprisingly, we also detected large aggregates decorated with PAG1-GFP or RPN5a-GFP inside vacuoles when both inhibitors were applied (Figure 3C; Figure S5A). Comparable structures were seen in similarly treated wild-type plants expressing the GFP-ATG8a reporter but not in wild-type plants expressing free GFP alone, nor when these reporters were expressed in the atg7-2 background, suggesting that they were of autophagic origin (Figure 3C; Figure S5B).
Using the free GFP/tagged-GFP ratio, we then directly monitored proteaphagy during nitrogen starvation and proteasome inhibition and compared it to bulk autophagy measured using the GFP-ATG8a reporter (Figure 3D; Figure S6). The average free GFP/tagged-GFP ratio for GFP-ATG8a in well fed, untreated Arabidopsis seedlings was ~0.5, with this value slightly elevated by ConA-induced stabilization of free GFP inside vacuoles, therefore providing a baseline readout of overall autophagic flux. Consistent with nitrogen limitation activating bulk autophagy (Li and Vierstra, 2012), this ratio increased significantly to ~2 (up ~4-fold) following nitrogen starvation, revealing how strongly nutrient stress upregulates such recycling (Figure 3D). By contrast, MG132 treatment did not alter the free GFP/GFP-ATG8 ratio, implying that proteasome inactivation does not enhance bulk autophagy (Figure 3D). When we analyzed pro-teaphagy specifically based on the PAG1-GFP and RPN5a-GFP reporters, the free GFP/tagged-GFP ratios were only ~0.15 for well fed plants, increasing to ~0.5 with ConA, indicating that 26S proteasomes undergo autophagic turnover even under basal conditions (Figure 3D). As with GFP-ATG8a, the free GFP/tagged-GFP ratio for these reporters increased dramatically following nitrogen starvation to a ratio of ~2 (up 13-fold), indicating that proteaphagy is strongly controlled by nutrient supply (Figure 3D). As expected, proteaphagy was also highly sensitive to proteasome inhibition, with MG132 increasing the free GFP/tagged-GFP ratio for both proteasome reporters ~5 fold (from ~0.15 to ~0.7; Figure 3D).
As further proof that inactivation of 26S proteasomes stimulates proteaphagy, we examined the effect of three Arabidopsis proteasome mutants. The rpt2a-2 and rpt4b-2 alleles compromise assembly (Lee et al., 2011; R.S.M., K.H. Lee, and R.D.V., unpublished data), whereas the rpn10-1 allele expresses a truncated RPN10 unable to bind ubiquitylated substrates (Smalle et al., 2003). Upon introgression into the pag1-1 PAG1-GFP background, it was clear that both the rpt2a-2 and rpt4b-2 mutations stimulated proteaphagy based on a significant increase in the free GFP/tagged-GFP ratio, even for well fed plants in the absence of MG132 (Figures 3E and 3F). By contrast, no such increase was observed in rpn10-1 plants, implying either that 26S proteasomes are not sufficiently impaired by this mutation or, more interestingly, that RPN10 directs proteaphagy.
Inhibitor-Induced Proteaphagy Coincides with Proteasome Ubiquitylation
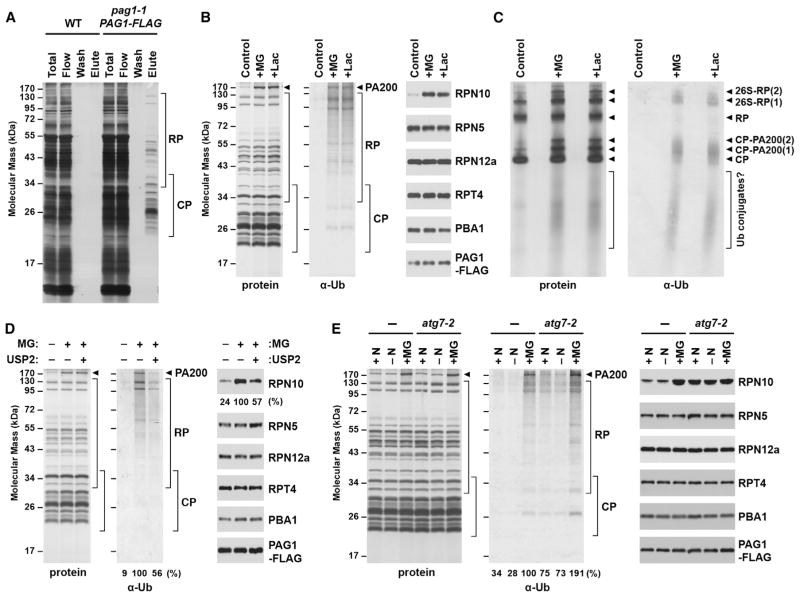
Several types of selective autophagy are driven by specific ubiquitylation of the cargo (Kirkin et al., 2009; Rogov et al., 2014). Given that many subunits of the 26S proteasome from Arabidopsis and other eukaryotes are ubiquitylated (Book et al., 2010; Besche, et al., 2014; Cui et al., 2014) and that this modification is stimulated by proteasome inhibitors (Kim et al., 2011; Kim et al., 2013), we examined whether this inhibitor-triggered ubiquitylation is relevant to proteaphagy. For this we took advantage of our ability to rapidly purify proteasomes from Arabidopsis seedlings using a transgenic line in which the PAG1 subunit was replaced with a FLAG-tagged variant (Figure 4A; Book et al., 2010).
Figure 4. Proteasome Inhibition Stimulates Ubiquitylation of the Particle and Increased Association of RPN10.
(A) Affinity purification of 26S proteasomes from Arabidopsis seedlings expressing PAG1-FLAG. Total protein extracts from WT and pag1-1 PAG1-FLAG seedlings were incubated with anti-FLAG beads, washed, and eluted with FLAG peptide. Fractions were subjected to SDS-PAGE and stained for protein with silver.
(B) Composition of 26S proteasomes upon inhibition. Proteasomes were affinity-purified from seedlings treated with 50 μM MG132 (MG) or clasto-lactacystin β-lactone (Lac). Preparations were separated by SDS-PAGE and stained for protein with silver (left) or immunoblotted with anti-ubiquitin (Ub) antibodies (center) or antibodies against proteasome subunits and the FLAG epitope (right). The arrowhead identifies PA200.
(C) Analysis of proteasome preparations by native-PAGE. Proteasomes were affinity-purified from seedlings treated as in (B) and separated by native PAGE. Gels were either stained for total protein with silver (left) or immunoblotted with anti-ubiquitin antibodies (right). Migration positions of the CP, the CP-PA200 complex, the RP, singly and doubly capped 26S proteasomes (26S-RP(1) and 26S-RP(2), respectively), and dissociated Ub conjugates are indicated by the arrowheads and bracket.
(D) Treatment with the deubiquitylating enzyme USP2 releases RPN10. Proteasomes were affinity-purified from control and MG132-treated seedlings as in (A), but, prior to elution, samples were incubated in the presence or absence of 10 nM USP2. Following elution, samples were separated by SDS-PAGE and analyzed as in (B). The amounts of associated ubiquitin and RPN10 were quantified and expressed as a percentage of the MG132-treated samples before digestion.
(E) Treatment with MG132, but not N starvation, induces proteasome ubiquitylation and increased RPN10 association. Proteasomes were affinity-purified from pag1-1 PAG1-FLAG seedlings with or without the atg7-2 mutation after treatment with or without N starvation or 50 μM MG132. Samples were analyzed as in (B). Ubiquitylation levels were quantified and expressed as a percentage of the MG132-treated WT sample.
Analysis of proteasome preparations isolated from seedlings pre-treated with MG132 or a second proteasome inhibitor, clasto-lactacystin β-lactone, confirmed that Arabidopsis proteasomes become heavily ubiquitylated upon inhibition. At effective concentrations (as judged by suppressed PBA1 processing, increased levels of ubiquitin conjugates, and upregulation of the transcriptional regulon; Figure S4), these inhibitors did not impact 26S proteasome composition overall except for a greater association with PA200 (Figure 4B). However, in agreement with prior mass spectrometric studies (Kim et al., 2013), proteasomes became heavily decorated with ubiquitin (Figures 4B and 4D). Conspicuous bands likely reflecting ubiquitylated forms of RPN1, RPN2, and PA200 were evident, in addition to diffuse staining that could represent modification of other subunits, as well as co-purification of ubiquitylated substrates awaiting degradation. Immunoblot analysis of these preparations following native gel electrophoresis confirmed these possibilities, with the detection of ubiquitin associated with 26S proteasomes, either singly or doubly capped with RP, and with free CP particles (Figure 4C). A smear of lower mass species was also prominent, which might represent ubiquitylated substrates, disintegrating ubiquitylated proteasomes, and/or ubiquitylated but stable CP sub-complexes. The inhibitors also induced the accumulation of two particles with native masses greater than the CP that likely represent CP capped with PA200 (Book et al., 2010). Surprisingly, little ubiquitin signal co-migrated with the free RP (Figure 4C), which suggested either that inhibitor-induced RP ubiquitylation is less prevalent, that the RP is rapidly deubiquitylated, and/or that ubiquitylated RP releases from the CP and is therefore poorly enriched via the PAG1-FLAG tag.
When CP and RP subunits were examined individually in these purified proteasome samples, the levels of most were not impacted by proteasome inhibition (Figure 4B). Strikingly, the lone exception was RPN10, which became dramatically more prevalent. Prior studies showed that RPN10 is unique among proteasome subunits because it also exists in a free form (van Nocker et al., 1996b) and is often bound to the 26S particle in substoichiometric amounts (Finley, 2009). Consequently, the inhibitor-induced increase in proteasome-associated RPN10 could reflect a greater stable incorporation of RPN10 into the particle or increased binding of free RPN10 to ubiquitin moieties now associated with the particle. To distinguish between these possibilities, we purified proteasomes from MG132-treated seedlings, trimmed the attached ubiquitins with the deubiquitylating enzyme USP2 (Besche et al., 2014), and assessed the amount of RPN10 remaining bound. Immunoblot analyses showed that USP2 treatment reduced the amount of associated ubiquitin by ~44% (Figure 4D). This reduction did not affect the amount of most CP and RP subunits but did commensurately reduce the amount of RPN10 (down by ~43%; Figure 4D). Taken together, the elevated RPN10 levels observed upon proteasome inhibition were likely driven by its binding to ubiquitylated proteasome subunits/targets rather than its increased integration into the RP.
Based on the scenario that ubiquitylated proteasomes are intermediates for autophagic elimination, we also examined the ubiquitylation status of proteasomes affinity-purified from autophagy-defective seedlings pre-treated with MG132. As predicted, proteasomes isolated from atg7-2 pag1-1 PAG1-FLAG seedlings had a similar subunit profile but were more heavily ubiquitylated after MG132 exposure than similarly treated seedlings wild-type for ATG7 (up 1.9-fold; Figure 4E). A similar increase in ubiquitylated proteasomes and RPN10 occupancy was also seen in atg7-2 seedlings in the absence of MG132, which could reflect basal autophagy of naturally disabled proteasomes.
RPN10 Interacts with ATG8 and Undergoes Autophagic Transport
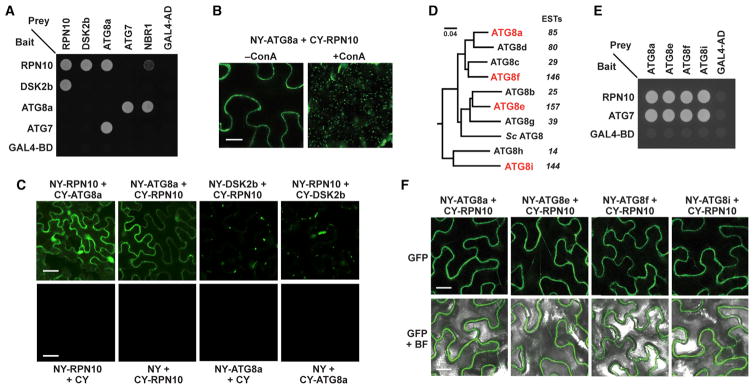
Our observations that RPN10 levels are increased dramatically in atg mutants; the rpn10-1 mutation, unlike other proteasome mutations, does not upregulate proteaphagy; and that proteaphagy is accompanied by increased particle ubiquitylation and RPN10 association led us to hypothesize that RPN10 is an autophagy receptor for ubiquitylated proteasomes. Because autophagy receptors typically bind both their target(s) and ATG8 (Klionsky and Schulman, 2014; Rogov et al., 2014), we examined whether Arabidopsis RPN10 and ATG8 interact even though RPN10 did not contain an obvious ATG8-interacting motif (Rogov et al., 2014). In our first test, interaction between ATG8a and RPN10 was observed by yeast two-hybrid assays (Y2H), with known interactions between ATG8a and ATG7 or NBR1 and between RPN10 and DSK2b used as controls (Lin et al., 2011). Binding of RPN10 to ATG8a was next observed in planta by bimolecular fluorescence complementation (BiFC). BiFC signals were detected throughout the cytoplasm and nucleus in untreated cells (Figure 5C) and became strongly visible in vacuolar puncta following pre-treatment with ConA, as expected for a cargo receptor binding to ATG8 in autophagic bodies (Figure 5B).
Figure 5. The 26S Proteasome Ubiquitin Receptor RPN10 Interacts with ATG8.
(A) Y2H interactions between Arabidopsis RPN10 and ATG8a. The indicated full-length proteins fused to either the GAL4 activating (AD) or binding (BD) domains were co-expressed and selected for growth on medium lacking tryptophan, leucine, and histidine and containing 3-AT. The known interactions between RPN10 and DSK2b, and ATG8a and ATG7 or NBR1, were included as positive controls, whereas combinations involving the GAL4 AD and BD domains alone were included as negative controls.
(B and C) Arabidopsis RPN10 and ATG8a interactions detected in planta by BiFC. Tobacco leaf epidermal cells were co-infiltrated with the indicated plasmid combinations, and the fluorescent signals were detected by confocal microscopy 36 hr after infiltration. The known interaction between RPN10 and DSK2b was included as a control. For B, the infiltrated leaf sections were excised, vacuum-infiltrated with 1 μM ConA, and incubated in the dark for 16 hr prior to imaging. Scale bars, 10 μm (B) and 5 μm (C).
(D) Phylogenetic analysis of the nine Arabidopsis ATG8 isoforms and their ortholog from Saccharomyces cerevisiae. The sequences were aligned in Clustal Omega and subjected to tree analysis using MrBayes. The expressed sequence tag (EST) value for each Arabidopsis isoform is indicated.
(E) Y2H interactions between RPN10 and members from each ATG8 subclade. The assays were performed as in (A).
(F) RPN10 interactions with members of the ATG8 family detected in planta by BiFC. Tobacco leaf epidermal cells were co-infiltrated with the indicated plasmid combinations and imaged as in (C). Scale bar, 10 μm.
Arabidopsis expresses nine isoforms of ATG8, which can be subdivided into four distinct clades (Figure 5D). Given that members of the human ATG8 family (MAP1LC3 and GABARAP) have subfunctionalized their cargo receptor binding preferences (Behrends et al., 2010), we tested whether the Arabidopsis ATG8 isoforms might also have different affinities for RPN10. However, examination of representatives from each of the four clades (ATG8a, ATG8e, ATG8f, and ATG8i) by Y2H and BiFC assays revealed that each has a similarly robust affinity for RPN10 (Figures 5E and 5F).
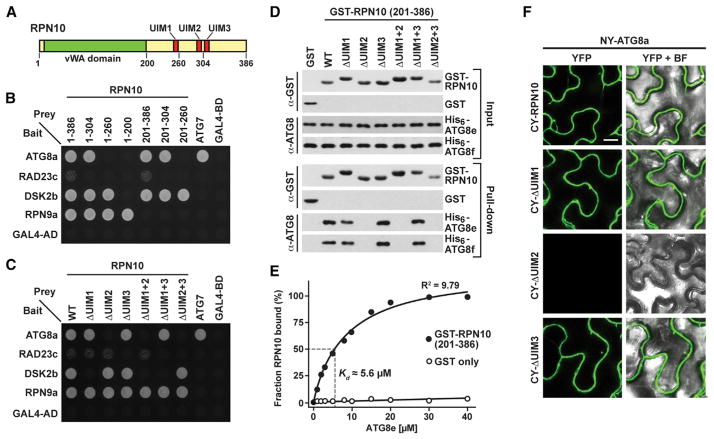
RPN10 Binds ATG8 via a Specific Ubiquitin-Interacting Motif
Arabidopsis RPN10 possesses a von Willebrand factor A (vWA) domain at its N terminus that tethers RPN10 to the RP, followed by a C-terminal domain harboring three ubiquitin-interacting motifs (UIMs) that bind ubiquitin and/or UBL domains present in shuttle factors such as RAD23 and DSK2 (Figure 6A; Fu et al., 1998; Fatimababy et al., 2010; Lin et al., 2011). Extensive mapping studies showed that UIM1 binds ubiquitin and the UBL domain of DSK2 and that UIM3 prefers the UBL domain of RAD23, but the binding preferences of UIM2 were unclear (Lin et al., 2011).
Figure 6. ATG8 Interacts with the UIM2 Region of RPN10.
(A) Diagram of the Arabidopsis RPN10 protein. The positions of the vWA domain and the three UIMs are indicated. The residue numbers define the various C-terminal truncations used in (B).
(B and C) Y2H interactions of Arabidopsis ATG8a with various truncations and site-directed substitutions of RPN10. The known interactions between the vWA domain and RPN9, UIM1 and DSK2b, UIM3 and RAD23c, and ATG8a and ATG7 were included as positive controls.
(D) In vitro binding assays demonstrate an interaction between UIM2 of RPN10 and ATG8. Equal amounts (5 μg) of purified GST or GST-RPN10 variants (residues 201–386 bearing single and combination mutants of UIM1-3) were incubated together with His6-ATG8e or His6-ATG8f and pulled down with GST-Bind resin. Bound proteins were visualized by immunoblotting with anti-ATG8 or anti-GST antibodies.
(E) Quantification of the binding affinity between RPN10 and ATG8. Varying concentrations of His6-ATG8e were incubated with 1 μM RPN10(201–386) and pulled down with Ni-NTA beads. RPN10 remaining in the supernatant was quantified by SDS-PAGE and immunoblotting with anti-GST antibodies and expressed as a percentage of the RPN10 input.
(F) ATG8a interacts with UIM2 of RPN10 in planta by BiFC. Tobacco leaf epidermal cells were co-infiltrated with the indicated plasmid combinations, and fluorescent signals were detected by confocal microscopy 36 hr after infiltration. Scale bar, 10 μm.
See also Figure S7.
To locate the site(s) in RPN10 that bind(s) ATG8, we tested a series of C-terminal truncations by Y2H, using the known interactions between DSK2b and UIM1, RAD23c and UIM3, and RPN9a and the vWA domain as controls. Strong interactions were seen between ATG8a and full-length RPN10 (amino acids 1–386) or a C-terminal truncation of RPN10 missing the last 82 amino acids, including UIM3 (1–304). In contrast, binding was absent with a version missing 44 more residues bracketing UIM2 (1–260), implying that the ATG8 interaction site is within this region (Figure 6B). As a direct test, we modified the hydrophobic pocket of each UIM with a cluster of amino acid substitutions shown previously to disrupt UIM/partner interactions (Lin et al., 2011). RPN10s bearing these mutations, either alone or in combination, were then examined for their ability to bind DSK2b (UIM1), ATG8a (unknown), and RAD23c (UIM3) by Y2H, BiFC, and in vitro pull-down assays. Although mutation of UIM1 and UIM3 predictably abolished binding of RPN10 to DSK2b and RAD23c, respectively (Figure 6C), only mutation of UIM2 abolished the interaction between RPN10 and ATG8 (Figures 6C–6F). Quantitative equilibrium binding assays revealed that RPN10 (201–386) and ATG8e interacted with a Kd of ~5.6 μM, which is comparable with the known affinities of RPN10 for ubiquitin chains and autophagy receptors for ATG8 (Rozenknop et al. 2011).
The RPN10 family displays a strong sequence similarity within the plant kingdom, especially in the vWA domain and the core hydrophobic patches that define the three UIMs (Figure S7A). To test whether RPN10/ATG8 binding is conserved across plants, we performed Y2H assays with RPN10 and ATG8 orthologs from Physcomitrella patens, Brachypodium distachyon, and Zea mays. In all three cases, full-length RPN10 strongly bound ATG8 (Figure S7B). Taken together, our data clearly show that RPN10 proteins from plants have affinity for both ubiquitin and ATG8 and, therefore, likely function as autophagic cargo receptors in addition to their roles as ubiquitin and UBL domain receptors within the 26S proteasome.
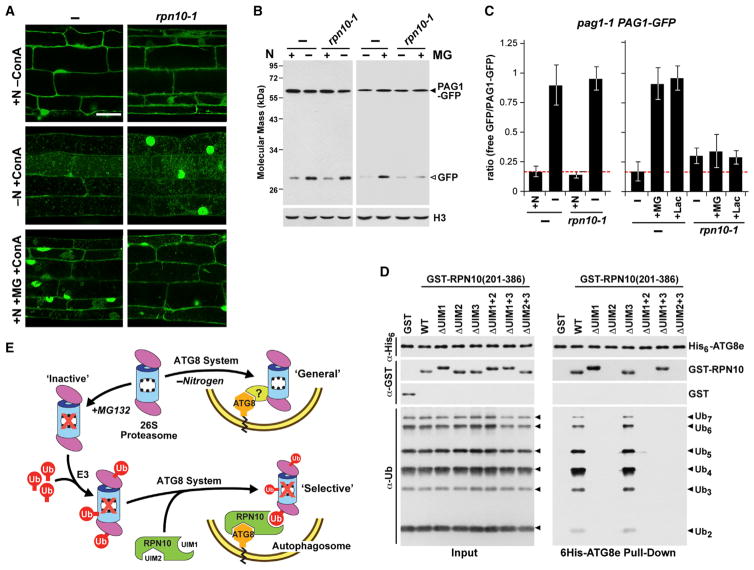
Inhibitor-Induced Proteaphagy Is Blocked in an rpn10 Mutant
To further support RPN10 acting as a proteaphagy receptor, we examined the autophagic turnover of proteasomes in the rpn10-1 mutant that expresses the vWA domain of RPN10 (amino acids 1–186) without the UIMs (Smalle et al., 2003). Consequently, the truncated protein can still integrate into the 26S particle but is unable to bind ubiquitin, UBL domains, and ATG8.
When rpn10-1 pag1-1 PAG1-GFP seedlings were nitrogen-starved, an increased accumulation of vacuolar puncta labeled with PAG1-GFP and elevated free GFP relative to the intact PAG1-GFP reporter were observed, indicating that RPN10 is not required for proteaphagy induced by nitrogen deficiency (Figures 7A–7C). By contrast, when proteaphagy was induced by proteasome inhibition, the rpn10-1 mutation had a dramatic effect. Using the free GFP/PAG1-GFP ratio as a measure of proteaphagic flux, the increased ratio typically seen in wild-type plants upon MG132 or clasto-lactacystin β-lactone exposure was attenuated significantly (Figures 7B and 7C). Fluorescence microscopic images of rpn10-1 pag1-1 PAG1-GFP root cells treated with MG132 and ConA also failed to show a strong increase in vacuolar puncta labeled with PAG1-GFP and did not accumulate the larger vacuolar aggregates seen in similarly treated wild-type seedlings (Figure 7A). Taken together, RPN10 appears to be non-essential for bulk proteaphagy induced by nutrient stress but is required for robust inhibitor-induced selective proteaphagy.
Figure 7. RPN10 Is Essential for Inhibitor-Induced, but Not Nitrogen Starvation-Induced, Proteaphagy.
(A) The rpn10-1 mutation blocks autophagic transport of PAG1-GFP upon proteasome inhibition but not N starvation. WT and rpn10-1 seedlings expressing PAG1-GFP were treated as in Figure 3C, and root cells were examined for autophagic bodies by confocal fluorescence microscopy. Scale bar, 10 μm.
(B and C) Release of free GFP from PAG1-GFP in N-starved or MG132-treated pag1-1 PAG1-GFP plants with or without the rpn10-1 mutation. Seedlings were treated as in Figures 2C or 3B.
(B) Immunoblot detection of PAG1-GFP and free GFP in total protein extracts with anti-GFP antibodies.
(C) Quantification of the free GFP/PAG1-GFP ratio following N starvation or MG132 or Lac treatment. Bars represent the mean (± SD) of three biological replicates. Letters indicate values that are statistically different from one another (p < 0.05).
(D) A model for proteaphagy in Arabidopsis. The MG132-induced selective route involves initial ubiquitylation of inhibited proteasomes by one or more E3s, association of RPN10 with the ubiquitin moieties, and binding of RPN10 to ATG8-PE lining the enveloping autophagic vesicles. The N starvation-induced bulk route involves non-selective transport into the vacuole with or without the help of a hypothetical proteaphagy receptor responsive to N stress.
(E) ATG8, RPN10, and ubiquitin form a tripartite complex. Equal amounts (1 μg) of purified His6-ATG8e, either GST or GST-RPN10 harboring UIM1-3 mutations, and poly-ubiquitin chains were incubated together and pulled down with Ni-NTA beads. Input and bound proteins were visualized by immunoblotting with anti-ATG8, anti-GST, or anti-ubiquitin antibodies.
Our model of inhibitor-induced proteaphagy envisions free RPN10 acting as a bridge to tether ubiquitylated proteasomes to ATG8-PE lining expanding autophagic membranes (Figure 7D). To confirm this tripartite assembly, we performed pull-down assays for free poly-ubiquitin chains using His6-ATG8e attached to nickel-nitriloacetic acid (Ni-NTA) beads together with the C-terminal fragment of RPN10 harboring mutant combinations of UIM1–3. ATG8e in combination with wild-type RPN10(201–386) readily pulled down poly-ubiquitin, but this activity was abolished when the UIMs necessary for ubiquitin (UIM1) or ATG8 (UIM2) interaction were mutated (Figure 7E).
DISCUSSION
Given the central role of the UPS in controlling the abundance of key regulators and maintaining cellular homeostasis by removing damaged or misfolded proteins, it is not surprising that 26S proteasome activity is tightly regulated at multiple levels. Here we demonstrated that proteasome abundance in Arabidopsis is controlled by ATG8-mediated autophagy. Autophagy inactivation under well fed conditions increases the levels of most proteasome subunits but not the peptidase activity of the assembled CP, suggesting that a main role for proteaphagy under basal conditions is to remove inactive particles. Like other selective autophagy events (Rogov et al., 2014), this proteaphagy is upregulated by nutrient demand (bulk) and by conditions that impair target function (selective). Although our work involved Arabidopsis, prior studies have implied that such proteaphagy is universal among eukaryotes. For example, Cuervo et al. (1995) detected the accumulation of CP in rat liver lysosomes upon nutrient starvation or treatment with the lysosomal peptidase inhibitor leupeptin, leading to the proposal that proteasomes are degraded by this compartment. More recent proteomic studies have detected 26S proteasome subunits in enriched human autophagosome preparations (Dengjel et al., 2012), whereas preliminary studies by us have also detected proteaphagy in yeast, which appears to be similarly induced by nitrogen stress and proteasome inhibition (R.S.M. and R.D.V., unpublished data).
Our studies with Arabidopsis detected two mechanistically distinct proteaphagic routes (Figure 7D). One is stimulated by nitrogen starvation and, presumably, by general nutrient stress. Remarkably, this route strongly reduced proteasome levels, with over 50% cleared after 24 hr. This “bulk” proteaphagy is dependent on the core autophagic machinery, proceeds via ATG8-decorated autophagic bodies, and is at least partially controlled by the regulatory ATG1 kinase complex, which is consistent with the role of this kinase in integrating starvation signals from TOR and other nutrient sensors (Li and Vierstra, 2012; Xiong and Sheen, 2014). However, this bulk proteaphagy is independent of autophagy receptors like NBR1 or RPN10 and is not coincident with proteasome ubiquitylation. Whether proteasomes are engulfed non-discriminately or captured deliberately remains unclear.
The second proteaphagic route responds to particle inactivation. This “selective” route is also dependent on the core autophagy machinery but appears to be independent of ATG1. Stimulation of this selective route is concomitant with substantial ubiquitylation of the particle, which can be seen by proteomic studies of the entire Arabidopsis ubiquitylome (Kim et al., 2013), detection of ubiquitin attachment sites (Book et al., 2010), and by immunoblotting purified proteasome preparations (Figure 4B). Although some of the ubiquitylated species detected immunologically reflect direct modification of individual CP and RP subunits, others might represent ubiquitylated targets bound to chemically stalled proteasomes.
The discovery that Arabidopsis RPN10 has the capacity to bind both ubiquitin and ATG8 has led us to propose that RPN10 serves as a dual receptor that not only helps 26S pro-teasomes recognize ubiquitylated targets when integrated into the RP lid but also directs proteaphagy of ubiquitylated proteasomes in its free form. This second role was directly confirmed by the ability of RPN10 to form a stable tripartite complex with ubiquitin and ATG8. (Figure 7E) and might explain why RPN10 is one of the few 26S proteasome subunits for which a substantial free pool exists (van Nocker et al., 1996b; Finley, 2009). Although a direct connection between ubiquitylation and proteaphagy awaits, it is supported by the facts that inhibitors stimulate both proteasome ubiquitylation and proteaphagy, the UIM region of RPN10 is needed for inhibitor-induced proteaphagy, ubiquitylation of inhibited proteasomes is increased in autophagy deficient backgrounds, and RPN10 increases its association with proteasomes under conditions that stimulate particle ubiquitylation. Clearly an unequivocal link between inhibitor-induced, RPN10-dependent proteaphagy and ubiquitylation will require the identification of the responsible E3(s) and the subunits that become modified. Several studies have identified E3s associated with proteasomes (Xie and Varshavsky, 2000; Besche et al., 2014; A.J.B. and R.D.V., unpublished data), but their roles in proteaphagy, if any, remain to be determined.
Another question is whether these Arabidopsis proteaphagic routes are relevant in other eukaryotes. The affinity for ATG8 through UIM2 might be restricted to the plant version of RPN10 because its yeast counterpart does not include this motif (Figure S7A), whereas human RPN10 (PSMD4) binds only weakly, if at all, to the human versions of ATG8 (Behrends et al., 2010). In confirmation, we failed to detect binding of yeast and human RPN10 to their ATG8 counterparts (R.S.M. and R.D.V., unpublished data). However, given that proteaphagy is likely conserved, inhibitor-induced proteaphagy outside of the plant kingdom could require different autophagy receptors that may or may not bind ubiquitin. Whatever the mechanisms, proteaphagy adds yet another facet to the ever-expanding repertoire of quality control and recycling processes influenced by autophagy. These also include the selective turnover of mitochondria, peroxisomes, chloroplasts, ER, ribosomes, protein aggregates, and invading pathogens (Li and Vierstra, 2012; Klionsky and Schulman, 2014; Rogov et al., 2014).
One logistical complication to proteaphagy is that most proteasomes are localized to the nucleus, whereas the autophagic capture system is cytoplasmic. Assuming that proteasomes are lost equally from the cytoplasm and nucleus, mechanism(s) should exist that shuttle nuclear proteasomes back to the cytosol, especially to clear disabled particles. Intriguingly, examples of direct nuclear autophagy have been reported, and mechanisms that export 26S proteasomes or other large protein complexes out of the nucleus have been proposed (Shibata and Morimoto, 2014).
In conclusion, we report here an autophagic route for the control of 26S proteasome levels that likely plays an important role in regulating the UPS by removing excess or damaged proteasomes. This turnover occurs via two pathways in Arabidopsis that are separately induced by nutrient starvation and proteasome inhibition, with the second pathway involving the dual ubiquitin/ATG8 receptor RPN10. Our finding adds yet another layer to the complex interplay between the UPS and autophagy and their collective attempts to control protein abundance, recycle nutrients, and mitigate proteotoxic stress.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Col-0 was used as the wild-type control. Transfer DNA (T-DNA) insertion mutants were obtained from the Arabidopsis Biological Resource Center or the GABI-Kat collection. Details of growth conditions, generation of transgenic lines, antibodies, and other methods are provided in the Supplemental Experimental Procedures. Information about mutant and transgenic lines and all PCR primers is provided in Tables S1, S2, and S3.
RNA Extraction and Quantitative Real-Time PCR Analysis
Total RNA was extracted from seedlings using the RNeasy plant mini kit (QIAGEN), treated with DNase I (Invitrogen), and then converted to cDNA using the SuperScript III first strand synthesis system (Invitrogen) and oligo(dT)20 primers. Quantitative real-time PCR was performed using a LightCycler 480 and SYBR Green I master mix(Roche Diagnostics) with threetechnical replicates for each reaction. Relative transcript abundance was determined by the comparative threshold cycle method using the ACT2 and PP2A genes as controls.
Proteasome Fractionation, Purification, and Activity Assays
Affinity purification, activity assays, and glycerol gradient fractionation of Arabidopsis 26S proteasomes were performed as described previously (Book et al., 2010; Lee et al., 2011).
Confocal Fluorescence Microscopy and Image Analysis
Root cells from the lower elongation zone of Arabidopsis seedlings were visualized with a Zeiss 510 Meta confocal laser-scanning microscope. Images were processed using the Zeiss LSM 510 image browser and/or Adobe Photoshop CC.
Yeast Two-Hybrid and In Vitro Binding Assays
Y2H assays were performed using the ProQuest two-hybrid system (Life Technologies). Pairwise gene combinations in pDEST22 and pDEST32 (or empty vector controls) were co-transformed into the Saccharomyces cerevisiae strain MaV203. Protein-protein interactions were identified by growth at 30ºC on medium lacking leucine, tryptophan, and histidine and containing 25 mM 3-amino-1,2,4-triazole (3-AT).
Proteins for in vitro binding assays were expressed from the pDEST15 or pDEST17 plasmids (Life Technologies) in Escherichia coli strain BL21(DE3) pLysS. Recombinant proteins were affinity-purified from lysed cells using GST-Bind resin (EMD4 Biosciences) or Ni-NTA beads (QIAGEN). Interactions were tested by a 2-hr incubation at 4ºC in 500 μl of binding buffer (150 mM Tris-HCl [pH 7.4], 150 or 300 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 5% [v/v] glycerol, and 0.01% [v/v] Triton X-100), followed by pull-down with appropriate pre-washed affinity beads.
Supplementary Material
Acknowledgments
We thank Sebastian Y. Bednarek, Hongyong Fu, Hermen S. Overkleeft, Elizabeth Vierling, Alessandro Vitale, and Daniel P. Woods for providing various reagents. We also thank Sarah J. Swanson for assistance with confocal microscopy and the Vierstra lab for advice and technical support. This work was funded by grants from the Department of Energy Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division (DE-FG02-88ER13968) and the National Science Foundation Plant Genome Program (IOS-1339325).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.04.023.
AUTHOR CONTRIBUTIONS
R.S.M. and R.D.V. conceived the project, R.S.M. performed most experiments, F.L. provided materials and performed confocal microscopy, D.C.G. designed experiments and assisted with proteasome purifications, A.J.B. generated transgenic lines, and R.S.M. and R.D.V. analyzed data and wrote the manuscript.
References
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Sha Z, Kukushkin NV, Peth A, Hock EM, Kim W, Gygi S, Gutierrez JA, Liao H, Dick L, Goldberg AL. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33:1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Book AJ, Gladman NP, Lee SS, Scalf M, Smith LM, Vierstra RD. Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem. 2010;285:25554–25569. doi: 10.1074/jbc.M110.136622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010;62:483–493. doi: 10.1111/j.1365-313X.2010.04166.x. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- Cui Z, Scruggs SB, Gilda JE, Ping P, Gomes AV. Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond. J Mol Cell Cardiol. 2014;71:32–42. doi: 10.1016/j.yjmcc.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J, Høyer-Hansen M, Nielsen MO, Eisenberg T, Harder LM, Sckehandorff S, Farkas T, Kirkegaard T, Becker AC, Schröder S, et al. The identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.014035. 014053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatimababy AS, Lin YL, Usharani R, Radjacommare R, Wang HT, Tsai HL, Lee Y, Fu H. Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J. 2010;277:796–816. doi: 10.1111/j.1742-4658.2009.07531.x. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Sadis S, Rubin DM, Glickman M, van Nocker S, Finley D, Vierstra RD. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell. 2013;25:1523–1540. doi: 10.1105/tpc.112.108613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol. 2014;21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejek I, Misas-Villamil JC, Kaschani F, Clerc J, Gu C, Krahn D, Niessen S, Verdöes M, Willems LI, Overkleeft HS, et al. Proteasome activity imaging and profiling characterizes bacterial effector syringolin A. Plant Physiol. 2011;155:477–489. doi: 10.1104/pp.110.163733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Minami A, Marshall RS, Book AJ, Farmer LM, Walker JM, Vierstra RD. The RPT2 subunit of the 26S proteasome directs complex assembly, histone dynamics, and gametophyte and sporophyte development in Arabidopsis. Plant Cell. 2011;23:4298–4317. doi: 10.1105/tpc.111.089482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17:526–537. doi: 10.1016/j.tplants.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Lin YL, Sung SC, Tsai HL, Yu TT, Radjacommare R, Usharani R, Fatimababy AS, Lin HY, Wang YY, Fu H. The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell. 2011;23:2754–2773. doi: 10.1105/tpc.111.086702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HM, Schippers JH, Gõni-Ramos O, Christoph MP, Dortay H, van der Hoorn RA, Mueller-Roeber B. An upstream regulator of the 26S proteasome modulates organ size in Arabidopsis thaliana. Plant J. 2013;74:25–36. doi: 10.1111/tpj.12097. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulos K, Kriegenburg F, Tatham MH, Rösner HI, Medina B, Larsen IB, Brandstrup R, Hardwick KG, Hay RT, Kragelund BB, et al. Dss1 is a 26S proteasome ubiquitin receptor. Mol Cell. 2014;56:453–461. doi: 10.1016/j.molcel.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178:1339–1353. doi: 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piterman R, Braunstein I, Isakov E, Ziv T, Navon A, Cohen S, Stanhill A. VWA domain of S5a restricts the ability to bind ubiquitin and Ubl to the 26S proteasome. Mol Biol Cell. 2014;25:3988–3998. doi: 10.1091/mbc.E13-11-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Rozenknop A, Rogov VV, Rogova NY, Löhr F, Güntert P, Dikic I, Dötsch V. Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. J Mol Biol. 2011;410:477–487. doi: 10.1016/j.jmb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Morimoto RI. How the nucleus copes with proteotoxic stress. Curr Biol. 2014;24:R463–R474. doi: 10.1016/j.cub.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD. The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell. 2003;15:965–980. doi: 10.1105/tpc.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23:3761–3779. doi: 10.1105/tpc.111.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7:993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S, Deveraux Q, Rechsteiner M, Vierstra RD. Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc Natl Acad Sci USA. 1996a;93:856–860. doi: 10.1073/pnas.93.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996b;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci USA. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014;164:499–512. doi: 10.1104/pp.113.229948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.