The physical properties of micro- and nanoparticles – size, shape and stiffness – play an important role in determining their function in applications such as medical diagnostics and drug delivery.[1–3] Changing a synthetic particle's shape and elasticity can influence the way the particle moves through the body and alter its interactions with individual cells.
Recent advances in fabrication techniques for non-spherical, soft particles, including microfluidic techniques,[4,5] non-wetting template molding (PRINT®),[6] electrohydrodynamic jetting,[7] and film stretching,[8] have enabled systematic studies of how physical properties of microparticles affect circulation in the body,[3,9] cellular internalization,[10] and passage through epithelial layers.[11] These studies have outlined important new design criteria for effective drug delivery vehicles. For example, flexible particles are more effective at squeezing through small capillaries in the bloodstream, increasing their overall circulation time and enhancing biodistribution compared to their rigid counterparts.[9,12] Other studies have shown that a particle's shape affects its interactions with individual cells[10,13] – for example, antibody displaying rod-shaped particles show increased specific uptake by cancer cells compared to spherical particles.[14] In a similar vein, recent work has demonstrated the importance of rationally designing the shape of pillars used in deterministic lateral displacement (DLD) microfluidic devices to improve the label-free separation of non-spherical biological entities (e.g. red blood cells and bacteria)[15,16]. Unconventional I-shaped pillars were shown to improve the separation efficiency of disk and rod shaped cells by inducing rotational movements[15,16]. These studies highlight the importance of designing material shape and elasticity for interfacing with biological cells.
Despite significant advances in this field with regards to designing particle shape for drug delivery and pillar shape for cell separation, the role of both shape and elasticity on particle function in many other biological environments remains to be studied. Recent reviews have highlighted the importance of designing the physical and chemical properties of material interfaces for cell capture, the first step in many diagnostic applications[17,18]. However, to date, there have been no controlled studies that examine the advantages of custom-shape, flexible particles for affinity-based capture of specific cell populations.
A strong motivation for designing new cell capture strategies is the detection and characterization of circulating tumor cells (CTCs), cells that are shed from tumors and move through the bloodstream, contributing to cancer metastasis.[19,20] Ideally, it would be possible to isolate viable CTCs from a patient's blood sample, characterize the cells with molecular diagnostics and subsequently culture the cells for analysis of drug sensitivity. This would enable less invasive alternatives to biopsies for cancer diagnosis and optimization of treatment regimens. Unfortunately, achieving this goal remains a challenge because CTCs are extremely rare (0.3-100 CTCs/mL of whole blood in cancer patients, amongst millions of white blood cells and billions of red blood cells), heterogeneous in nature, and difficult to keep viable for analysis after isolation.[17,21–23] Techniques that currently exist for CTC detection and separation include cell-affinity chromatography (i.e. capture using antibodies immobilized in microfluidic channels),[24–26] immunomagnetic sorting (i.e. capture by antibody-coated magnetic particles suspended in solution),[27] size-based sorting (i.e. separation based on the larger size of CTCs compared to blood cells),[28] and dielectrophoretic techniques (i.e. separation based on differing cell responses to electric fields).[27,29,30] Microfluidic negative depletion of blood cells to isolate CTCs has also been reported.[31,32]
Recently, microfluidic approaches for CTC capture have been gaining popularity because of large surface area-to-volume ratios and multiplexing capabilities.[30,33] However, particle-based approaches can offer more flexibility for transport and manipulation of cells, eliminate the need for functionalization of individual channels, and be used synergistically with microfluidic systems.[34–36] Customizable hydrogel microparticles have additional advantages: they are biocompatible, easy to functionalize – with antibodies, aptamers,[37] DNA,[38] magnetic nanoparticles,[39] or drug-loaded nanoemulsions[40] – and can be fabricated with tailored shape and size to maximize available capture area.
In this work, we explore how to leverage particle shape to improve capture efficiency of cells expressing epithelial cell adhesion molecule (EpCAM), a protein that is frequently expressed by CTCs.[41] We fabricate hydrogel microparticles with mask-defined shapes via stop flow lithography,[4,42] and demonstrate specific capture of EpCAM-expressing cancer cells in solution using functionalized particles. By systematically varying particle shape, we demonstrate how surface area, hydrodynamic effects, and steric constraints affect cell capture efficiency. Drawing upon previous work describing the influence of particle shape on mechanical flexibility,[43] we proceed to investigate how cell-laden particles of different shapes traverse through microfluidic constrictions, and show the effect of shape-induced flexibility on the retention of captured cells. The microfluidic constrictions act as in vitro models to allow study of particle passage in simulated biological environments.[44]
This study is a demonstration of how microparticle shape can be exploited to increase cell capture efficiency and improve cell retention in flow through microfluidic constrictions. Our results indicate that the physical parameters of shape and flexibility should be considered when designing microparticles for cell-capture-based diagnostic applications.
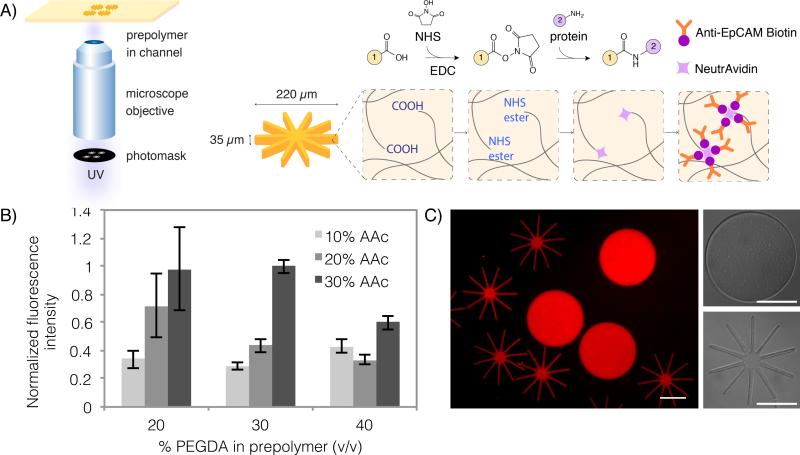
The particle synthesis and functionalization procedure is illustrated in Figure 1A. We fabricate poly(ethylene)glycol (PEG)-based hydrogel particles via stop flow lithography (SFL). In SFL, a prepolymer flowing through a microfluidic channel is polymerized by UV light in a mask-defined shape.[4,42] Using this technique, it is possible to make custom-shape microparticles with uniform shape and size, in a high-throughput manner. We fabricate particles using poly(ethylene glycol) diacrylate (PEGDA, Mn = 700 g/mol) co-polymerized with acrylic acid (AAc). Our prepolymer solution also consists of a photoinitiator, and PEG (MW = 200 g/mol), which acts as a porogen to increase hydrogel porosity and improve mechanical flexibility.[45]
Figure 1. Fabrication of antibody-functionalized hydrogel microparticles.
A) Poly(ethylene glycol) diacrylate (PEGDA) is copolymerized with acrylic acid (AAc) to fabricate particles with photomask-defined shapes via stop flow lithography (SFL). Carboxylic acid groups of PEGDA-AAc particles are functionalized with NeutrAvidin using carbodiimide (EDC) chemistry, which can then be conjugated with biotinylated anti-EpCAM. In the schematic, side group 1 corresponds to the rest of the PEGDA particle, while side group 2 is NeutrAvidin. B) The optimal prepolymer composition (30% PEGDA, 30% AAc) was chosen to achieve maximum antibody functionalization with good particle uniformity. Functionalization capacity was determined by measuring the intensity of a biotinylated fluorophore used to label the functionalized particles. C) Fluorescent and bright field images of disk and octopus-shaped particles fabricated using this method. The fluorescence of these particles is due to the addition of rhodamine acrylate in the prepolymer. Scale bars are 100 μm.
The carboxylic acid groups from the co-polymerized AAc allow the hydrogel to be easily functionalized with proteins using carbodiimide chemistry.[46] The carboxylic acid groups are activated to NHS ester groups, which can then couple with primary amines present in the lysine residues of any protein. We covalently conjugated NeutrAvidin, a neutral form of avidin with less non-specific binding, to our hydrogel particles. The particles were then functionalized with a biotinylated antibody against EpCAM (Anti-EpCAM biotin). Surface-bound anti-EpCAM biotin enables interactions between cells and particles. This two-step functionalization strategy enables particle functionalization by any biotinylated probe, improves protein uniformity throughout the hydrogel, and ensures no changes to antibody function due to direct covalent conjugation. Antibodies conjugated to hydrogel particles in this manner retain their activity after more than two months of storage (Figure S1).
To ensure optimal particle functionalization, we tested the effect of different particle prepolymer compositions on the resulting degree of NeutrAvidin conjugation. In Figure 1B, we varied the volume percent of PEGDA and AAc present in the prepolymer solution and tested the level of functionalization by labeling with excess biotinylated fluorophore. The fluorescence intensity observed should correlate to the amount of Anti-EpCAM biotin bound to the particles. As expected, increasing the amount of AAc in the prepolymer increased the level of functionalization by increasing the number of carboxylic acid groups available for conjugation. For particles fabricated using more than 20% (v/v) AAc in the prepolymer, increasing the amount of PEGDA decreased the level of functionalization. This may be due to the tighter mesh of the hydrogel, which sterically hinders the conjugation of NeutrAvidin, or the diffusion of the fluorescent probe into the gel. Based on our results, we chose an optimized prepolymer composition of 30% PEGDA and 30% AAc to achieve highly functionalized particles with good reproducibility.
In Figure 1C, we show particles synthesized using our chosen prepolymer composition, demonstrating excellent uniformity in particle size and shape. We were able to fabricate multi-arm particles with finely patterned in-plane features (arm length to width ratio of 8:1), with the same nominal outer diameter as more conventional disk-shaped particles (diameter = 220 μm). Our multi-arm particle geometry was inspired by shapes found in nature – both in animals and plants, as well as microscale organisms and tissues. For example, octopus and squid use suction cup-covered arms for capturing prey and some plants (e.g. sundews) use sticky tentacles to catch insects. In our own bodies, finger-like protrusions on cell membranes of some epithelial cells called microvilli, are useful for absorbing nutrients and improving cellular adhesion.[47] In all of these examples, the flexible, mobile nature of the arms or protrusions is critical for their function. Inspired by these biological motifs, we designed our “octopus particles” with thin arms arranged radially around a central core. It is worth noting that our synthetic multi-arm particle geometry resembles much smaller star polymers, a common polymer architecture consisting of many linear polymer chains (“arms”) connected to a central core. Star polymers demonstrate improved response to stimuli due to high functionalization density.[48] We predicted that the high aspect ratio arms on our particles would be well-suited for cell capture due to their high surface area and mechanical flexibility.
In order to test whether our functionalized octopus particles could capture cells of interest, we performed experiments using two cell lines: SKBR3s – breast cancer cells with high expression of EpCAM (EpCAM+, dyed blue for all experiments) and SKMEL28s – melanoma cells that do not express the protein (EpCAM−, dyed green for all experiments).[49,50] Figure 2A shows that functionalized particles effectively capture EpCAM+ cells, while demonstrating no non-specific adherence to EpCAM- cells. As another negative control, we incubated plain PEGDA-AAc particles (before functionalization) with the cells under identical conditions and found no capture of either cell type. This confirms the bio-inert nature of PEGDA particles,[51] and shows that the copolymerized AAc does not lead to any non-specific interactions. Other groups have also shown PEG-based and PEG-functionalized hydrogels to be suitable materials for antibody-based cell capture, and have demonstrated triggered cell release by breaking photolabile or physical crosslinks within the gel.[52–54] Previously, photopatterned PEG structures have also been used to capture cells from flowing solutions in microfluidic devices.[55]
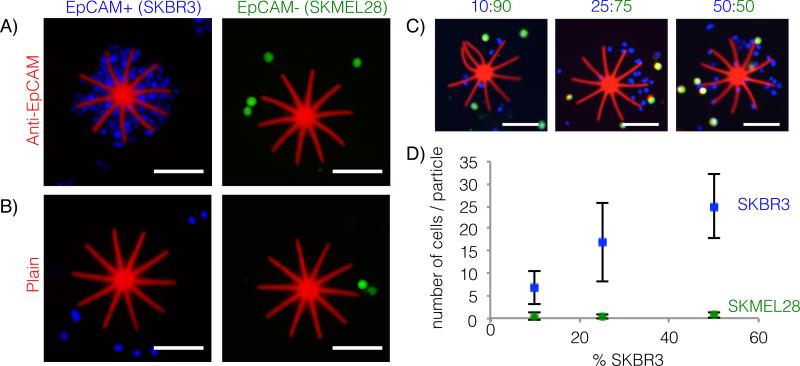
Figure 2. Cell capture by octopus particles.
A) EpCAM-expressing cells (SKBR3s, blue) are captured by anti-EpCAM functionalized particles, while non-EpCAM expressing cells (SKMEL28s, green) do not adhere. B) Plain, non-functionalized particles (PEGDA-AAc) do not capture either cell type. C) Particles successfully capture SKBR3s in mixed cell solutions (ratio of SKBR3 to SKMEL28 are indicated above images, total cell concentration = 100 000 cells/mL). D) Quantification of the number of SKBR3s and SKMEL28s captured per particle in the mixed cell solutions corresponding to the representative images in part C (averages are plotted ± standard deviation, N = 20). All scale bars are 100μm. Figure S2 shows multiple particles in the same image for the 50:50 mixed cell incubation condition.
To determine quantitative trends between solution cell concentration and number of cells captured, we incubated the octopus particles in mixed cell solutions containing different ratios of SKBR3s to SKMEL28s. We counted the cells attached to the top and side surfaces of the particles, ignoring the hidden bottom surfaces, and distinguished between the two cells types by fluorescence color. It should be noted that images in Figure 2C show a higher number of SKBR3s compared to SKMEL28s, regardless of cell incubation concentrations, due to the imaging setup. By imaging particles in a small drop of solution immediately after they settle on a microscope slide, cells attached to the particles are in focus, while cells suspended in solution are not. In the cell concentration range between 10 000 – 50 000 SKBR3s/mL (10-50% SKBR3s, total cell concentration: 100 000 cells/mL), we saw an almost linear correlation between the number of cells attached to our particles and the concentration of cells in solution. The data plotted in Figure 2D corresponds to a SKBR3 capture efficiency of approximately 40%, 40%, and 30% for the 10:90, 25:75 and 50:50 incubation conditions, respectively. While the concentrations used in this experiment are not representative of clinical CTC concentrations expected in patient blood samples, this proof-of-concept demonstration indicates that our particles are able to selectively capture cells of interest in mixed cell populations and are quantitatively sensitive to changes in cell concentration. Optimization of incubation conditions is expected to increase the recovery rate of target cells, bringing us closer to clinically relevant cell concentrations. Increasing the particle-to-cell ratio will also help to capture rare cells; in future work, particle production can be scaled-up using the recently developed contact flow lithography technique.[56]
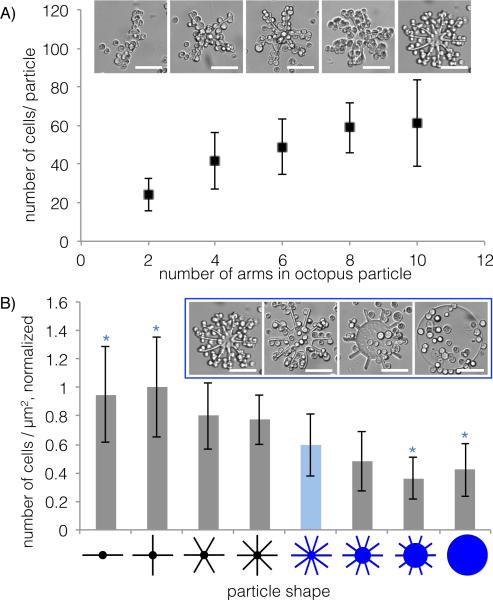
Motivated by our observations that octopus particles were effective at capturing SKBR3s, we proceeded to investigate the effect of particle shape on cell capture. We designed shapes such that all of them could be circumscribed by a 220 μm diameter circle, and polymerized all particles using the same pre-polymer, microfluidic channel design, and UV exposure conditions. All particles were approximately 35 μm tall and expected to have similar surface chemistry and degree of functionalization. First, we made particles with different number of arms: 2, 4, 6, 8 and 10. As expected, the number of cells captured on the particles increased with number of arms (Figure 3A), which also corresponds to increasing total surface area (Table S1). However, this curve plateaus when the number of arms exceeds 8. At this point, steric hindrance prevents more cells from attaching to the particles as the gap between neighboring arms becomes comparable in dimension to the cell diameter.
Figure 3. Cell capture by different shapes.
A) Number of SKBR3s captured by octopus particles with 2, 4, 6, 8, and 10 arms. Representative brightfield micrographs are shown for each data point. B) Number of captured SKBR3s normalized by individual particle total surface area (and normalized across all shapes) are plotted for 8 different shapes with the same nominal outer diameter (including the 5 shapes from part A). Representative brightfield micrographs are shown for the particle shapes highlighted in blue. Averages are plotted ± standard deviation, N = 13. (* = P < 0.05 by Student's t-test with Bonferroni correction, compared to the 10-arm particle indicated by the light blue bar). Scale bars are 100 μm.
We proceeded to normalize the number of cells attached to each particle by the particle total surface area (Figure 3B; surface areas shown in Table S1, data before normalization shown in Figure S3), and confirmed that cell capture depends on more than just surface area – other shape effects must also play a role. We tested three additional shapes, where we progressively increased the core diameter at the expense of arm length, maintaining the same outer particle diameter. In the extreme case, the arms were eliminated altogether, and we tested the cell capture ability of a solid disk. In Figure 3B, the particle shapes are arranged by increasing in-plane area (or top surface area, shown in Table S1). We see a general trend where cells captured per μm2 decreases as top surface area increases. We predict that hydrodynamic effects are responsible for this trend. From a simplified point of view, we can say that the in-plane particle area alters the streamlines of the passing cell-containing fluid when particles and cells are mixed gently in solution. The disk particle displaces the most fluid as it moves in solution, making it less likely for cells to come into contact with the particle surface. This is the same underlying concept as strategies that vary microfluidic device geometry to maximize contact between flowing CTCs and antibody-coated surfaces (e.g. microposts,[25,57] herringbone structures[23]). Based on our results, it appears that octopus particles with a small core and high aspect ratio arms that are far enough apart to allow cell-containing fluid to flow between them are better suited to capturing cells in solution than disk particles.
After capturing cells on particles of different shape, we compared the behavior of 10-arm octopus particles and disk particles during flow through a microfluidic constriction. Microfluidic channels are often used as models for blood capillaries to test the deformability of red blood cells and synthetic particles in order to predict their behavior in the bloodstream.[44] We designed microfluidic constrictions that are just less than half the width of our particle diameter – these relative dimensions are similar to what red blood cells may experience when passing through narrow capillaries.
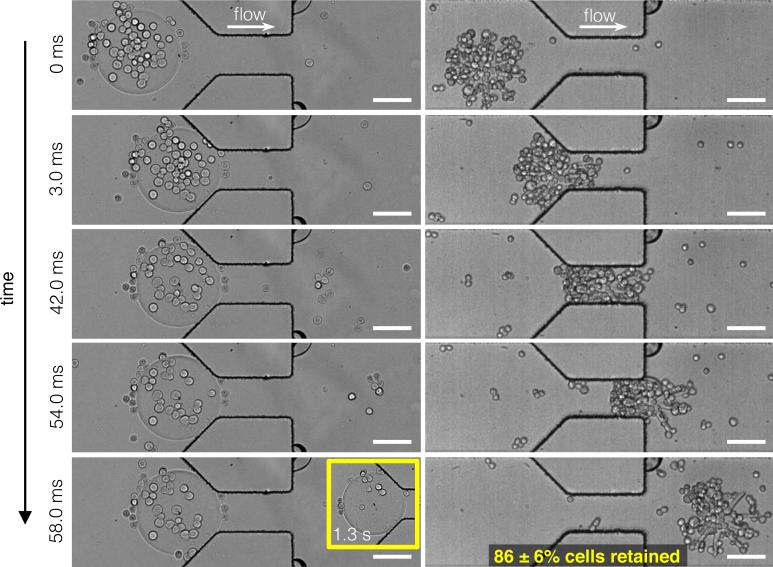
Figure 4 shows a series of frames from two representative high-speed videos (Video S1 and S2) of a disk and an octopus particle (both 220 μm in diameter, 35 μm tall) passing through a channel containing a narrow constriction (100 μm wide × 200 μm long × 60 μm tall). At a syringe pump-driven flow rate of 20 μl/min, the disk was unable to pass through the constriction, while the octopus passed through easily. The thin arms of the octopus are able to bend, allowing the particle to move through the narrow constriction in less than 1/10th of a second, while holding onto the majority of cells. The disk, on the other hand, was stuck at the entrance of the constriction and the continuous shear force of the fluid flowing past the particle stripped away most of the attached cells after one second. The disk remained stuck at the entrance of the constriction after several minutes of flow. We found that octopus particles retained an average of 86% of captured cells after passing through the constriction and that the direction of passage did not influence this behavior (Figure S4).
Figure 4. Particle flow through microfluidic constrictions.
Still images from representative high-speed videos (Video S1 and S2) of cell-laden disk and octopus particles flowing through a microfluidic constriction (narrow part of channel is 100 μm wide). Flow was driven by a syringe pump at a rate of 20 μl/min. Disks are unable to pass through the constriction while octopus particles pass through easily. The inset shows that after 1.3 s, the disk remains stuck at the constriction entrance with few cells remaining. Octopus particles retain 86% of captured cells after passing through the constriction (N = 8). Scale bars are 100 μm.
We also studied the passage of taller disks (220 μm diameter, 55 μm tall) through the constrictions at increased flow rates of 1 ml/min (Figure S5). At this flow rate, the disks compressed to pass through the constriction in approximately 5 seconds, but the majority of cells were stripped off in the process (6% cells were retained on average).
We expect that the cells attached to the octopus particles remain viable after passage through the constriction at low to moderate flow rates. First, PEG hydrogels are biocompatible and do not affect the viability of captured cells.[53,55] Second, cell capture and release on EpCAM-functionalized hydrogels in PDMS microfluidic channels has been previously shown to have no significant effect on cell viability or proliferative potential.[24] Lastly, at a flow rate of 20 μl/min, we estimate the wall shear stress in the constriction to be on the order of 50 dyne/cm2, within the range of physiological shear stress in the arterial vascular network (10-70 dyne/cm2).[58] The constriction only causes deformation of the flexible particles, not of the cells.
It is evident that geometry affects a particle's overall deformability when passing through a microfluidic constriction. This is supported by a previous study showing that particle shape affects mechanical flexibility as well as mode of passage for particles travelling through a microfluidic constriction.[43] Our observations on the effect of particle shape on retention of captured cells may be useful for recovery of specific cell populations in microfluidic systems, or future in-vivo applications involving particles travelling through the bloodstream.
In conclusion, we demonstrate the use of custom-shape, flexible hydrogel microparticles for specific antibody-based cell capture. We believe this approach has many advantages, including high-throughput particle synthesis,[56] easy functionalization with various probes, potential for synergistic use with microfluidic devices for downstream analysis of captured cells, and subsequent culture of isolated cells directly on particle substrates.[59] As one example, our system may be advantageous for addressing the ongoing challenge of CTC heterogeneity, which makes efficient capture difficult and results in isolated cells that play different functional roles.[17] Using customizable microparticles that can be identified by a geometric or color-defined barcode,[38,60,61] we could functionalize several groups of particles using different CTC biomarkers (e.g. EpCAM, HER2, EGFR, MUC1), enabling downstream characterization of CTC populations with different biochemical properties.
In this work, we show that particle shape is one important design criterion for maximizing cell capture efficiency. Specifically, we show that octopus particles can capture more cells than similarly sized disks, by increasing cell-particle interactions through increased total surface area as well as improved hydrodynamic geometry. In addition, the effect of shape on overall particle deformability and retention of captured cells is demonstrated using a model microfluidic constriction. This work highlights the importance of shape and flexibility for particle-based cell-capture applications.
Experimental Section
Materials
Poly(ethylene glycol) diacrylate (Mn = 700 g/mol), acrylic acid (anhydrous, ≥ 99.0%), poly(ethylene glycol) (Mn = 200 g/mol), 2-hydroxy-2-methyl-1-phenyl-propan-1-one (Darocur® 1173), N-(3-Dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC, ≥ 99.0%) and N-hydroxysuccinimide (NHS, 98%) were purchased from Sigma-Aldrich and used as received. Methacryloxyethyl thiocarbomoyl rhodamine B (λex/λem = 548/570nm, Polysciences) was dissolved in poly(ethylene glycol) at a concentration of 1 mg/mL. NeutrAvidin™ (Thermo Fisher) was diluted with PBS to a concentration of 5 mg/mL and the stock solution was stored at 4°C. Anti-Human CD326 (EpCAM) Biotin (0.5 mg/mL, Affymetrix) was used as received. PBST was made with 1X phosphate buffered saline (without calcium & magnesium, Corning) and 0.05% (v/v) Tween® 20 (Sigma-Aldrich).
Particle Synthesis
All particles were fabricated via stop flow lithography as previously described,[4,42] using a pre-polymer composition of 30% poly(ethylene glycol) diacrylate (PEGDA), 30% acrylic acid (AAc), 30% poly(ethylene glycol) (PEG), 5% rhodamine acrylate solution and 5% photoinitiator (Darocur® 1173), by volume. All particles were synthesized in 30 μm tall rectangular PDMS (Sylgard 184, Dow Corning) microfluidic channels bonded on PDMS-coated glass slides. Particles were polymerized by ultraviolet (UV) light (Lumen 200 metal arc lamp, Prior Scientific) through a UV filter set (11000v3-UV, Chroma Technology, 365nm, 150ms exposure time, 2200mW/cm2) in a mask-defined shape (designed using AutoCAD, printed by Fineline Imaging), and the particles were collected in a microcentrifuge tube filled with PBST. Particles were rinsed 8 times with PBST by centrifugation and stored in PBST at 4°C.
Particle Functionalization
All particle functionalization steps were carried out at a particle concentration between 5000-10000 particles/mL in PBS. EDC and NHS were dissolved separately in PBS and stock solutions were added to the washed particles to achieve final concentrations of 3.3 mg/mL each. The solution was vortexed for 30 seconds and placed on a horizontal shaker (650 rpm) at room temperature for 30 minutes. After activation, particles were rinsed 4 times in PBST by centrifugation. After washing, NeutrAvidin stock solution was added to the particles to achieve a final protein concentration of 0.83 mg/mL and the solution was incubated on a horizontal shaker for 2.5 hours (650 rpm, room temperature). The particles were rinsed 4 times in PBST and anti-EpCAM biotin was added to achieve a concentration of 0.083 mg/mL. The solution was incubated on a horizontal shaker for 30 minutes (650 rpm, room temperature). Particles were rinsed 4 times and stored in PBST at 4°C.
Cell Culture
High-EpCAM-expressing breast cancer (SKBR3) and non-EpCAM-expressing melanoma (SKMEL28) cell lines were obtained from the American Type Culture Collection (ATCC) and cultured according to standard culture protocols. Standard culture media comprised McCoy's 5A (ATCC) or Eagle's Minimum Essential Medium (EMEM; ATCC) supplemented with 10% fetal bovine serum (Life Technologies) and 1% penicillin-streptomycin (Life Technologies), for SKBR3 and SKMEL28 lines, respectively. Cells were grown in 25 cm2 rectangular canted-neck cell culture flasks with vent-caps (Corning Life Sciences) at 37° C and 5% carbon dioxide, and were either passaged or used at 85% confluency. For cell-particle experiments, cells were stained using CellTracker™ Green CMFDA (λex/λem = 492/517nm, Life Technologies), CellTracker™ Blue CMAC (λex/λem = 353/466nm, Life Technologies), or Hoechst 33342 (λex/λem = 350/461nm, Life Technologies) following manufacturer's instructions. Cells were subsequently trypsinized using 0.05% trypsin-EDTA (Life Technologies), and then resuspended in media at a concentration of 1 million cells/mL.
Cell-particle Experiments
Particles (600 particles/mL) and cells (400 000 cells/mL in 50% media, 50% PBST, by volume for all experiments, except Figures 2C and D which use 100 000 total cells/mL) were incubated together for two hours at room temperature, under gentle agitation. Total solution volumes were ~1 mL. Before imaging, solutions were vortexed gently to remove non-adherent cells from particles.
Imaging and Flow Experiments
Epifluorescence and brightfield images were taken using an inverted microscope (Axio Observer.A1, Zeiss; 10x and 20x objectives) connected to a cooled interline CCD camera (Clara, Andor). Videos of microfluidic flow experiments were taken using high-speed cameras (Phantom v4.2 and Phantom Miro M310, Vision Research) at frame rates ranging from 2000-8500fps and exposure times ranging from 2-10 μs.
Supplementary Material
Acknowledgments
LC and HZA contributed equally to this work. This work is supported in part by the MRSEC Program of the National Science Foundation under award number DMR – 1419807, the National Science Foundation CMMI – 1120724, and Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the U.S. Army Research Office. The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred. Additional support was provided by the US National Institutes of Health (NIH) P41 Resource Center and a NIH National Institute of Biomedical Imaging and Bioengineering Quantum Grant. We thank Octavio Hurtado for help with device microfabrication, and Aimal H. Khankhel for help with cell culture. LC is supported in part by a postgraduate scholarship from Natural Sciences and Engineering Research Council (NSERC) of Canada.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Lynna Chen, Department of Biological Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA.
Dr. Harry Z. An, Department of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA
Dr. Ramin Haghgooie, General Fluidics, 34 Anderson St., Ste 5, Boston, MA 02114, USA
Aaron T. Shank, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School, Building 114, 16th Street, Charlestown, MA 02129, USA
Dr. Joseph M. Martel, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School, Building 114, 16th Street, Charlestown, MA 02129, USA
Dr. Mehmet Toner, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School, Building 114, 16th Street, Charlestown, MA 02129, USA
Prof. Patrick S. Doyle, Department of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA
References
- 1.Mitragotri S, Lahann J. Adv. Mater. 2012;24:3717. doi: 10.1002/adma.201202080. [DOI] [PubMed] [Google Scholar]
- 2.Best JP, Yan Y, Caruso F. Adv. Healthc. Mater. 2012;1:35. doi: 10.1002/adhm.201100012. [DOI] [PubMed] [Google Scholar]
- 3.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Nat. Nanotechnol. 2007;2:249. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dendukuri D, Pregibon DC, Collins J, Hatton TA, Doyle PS. Nat. Mater. 2006;5:365. doi: 10.1038/nmat1617. [DOI] [PubMed] [Google Scholar]
- 5.Shum HC, Abate AR, Lee D, Studart AR, Wang B, Chen C-H, Thiele J, Shah RK, Krummel A, Weitz DA. Macromol. Rapid Commun. 2010;31:108. doi: 10.1002/marc.200900590. [DOI] [PubMed] [Google Scholar]
- 6.Rolland J, Maynor B, Euliss L, Exner A, Denison G, DeSimone J. J. Am. Chem. Soc. 2005;127:10096. doi: 10.1021/ja051977c. [DOI] [PubMed] [Google Scholar]
- 7.Bhaskar S, Pollock KM, Yoshida M, Lahann J. Small. 2010;6:404. doi: 10.1002/smll.200901306. [DOI] [PubMed] [Google Scholar]
- 8.Champion JA, Katare YK, Mitragotri S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11901. doi: 10.1073/pnas.0705326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merkel TJ, Jones SW, Herlihy KP, Kersey FR, Shields AR, Napier M, Luft JC, Wu H, Zamboni WC, Wang AZ, Bear JE, DeSimone JM. Proc. Natl. Acad. Sci. U. S. A. 2011;108:586. doi: 10.1073/pnas.1010013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11613. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uskoković V, Lee K, Lee PP, Fischer KE, Desai TA. ACS Nano. 2012;6:7832. doi: 10.1021/nn3019865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anselmo AC, Zhang M, Kumar S, Vogus DR, Menegatti S, Helgeson ME, Mitragotri S. ACS Nano. 2015;9:3169. doi: 10.1021/acsnano.5b00147. [DOI] [PubMed] [Google Scholar]
- 13.Champion JA, Mitragotri S. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4930. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barua S, Yoo J, Kolhar P, Wakankar A, Gokarn YR, Mitragotri S. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3270. doi: 10.1073/pnas.1216893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeming KK, Ranjan S, Zhang Y. Nat. Commun. 2013;4:1625. doi: 10.1038/ncomms2653. [DOI] [PubMed] [Google Scholar]
- 16.Ranjan S, Zeming KK, Jureen R, Fisher D, Zhang Y. Lab Chip. 2014;14:4250. doi: 10.1039/c4lc00578c. [DOI] [PubMed] [Google Scholar]
- 17.Li Y-Q, Chandran BK, Lim CT, Chen X. Adv. Sci. 2015 doi: 10.1002/advs.201500118. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bole AL, Manesiotis P. Adv. Mater. 2015 doi: 10.1002/adma.201503962. n/a. [DOI] [PubMed] [Google Scholar]
- 19.Steeg PS. Nat. Med. 2006;12:895. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 20.Pantel K, Brakenhoff RH. Nat. Rev. Cancer. 2004;4:448. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 21.den Toonder J. Lab Chip. 2011;11:375. doi: 10.1039/c0lc90100h. [DOI] [PubMed] [Google Scholar]
- 22.Yu ZTF, Aw Yong KM, Fu J. Small. 2014;10:1687. doi: 10.1002/smll.201302907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stott SL, Hsu C-HC-H, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18392. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M. Anal. Chem. 2012;84:3682. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagrath S, V Sequist L, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei KI, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Angew. Chemie - Int. Ed. 2009;48:8970. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plouffe BD, Murthy SK, Lewis LH. Reports Prog. Phys. 2015;78:016601. doi: 10.1088/0034-4885/78/1/016601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS-W, Lim W-T, Han J, Bhagat AAS, Lim CT. Sci. Rep. 2013;3:1. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Li J, Sun Y. Lab Chip. 2012;12:1753. doi: 10.1039/c2lc21273k. [DOI] [PubMed] [Google Scholar]
- 30.Qian W, Zhang Y, Chen W. Small. 2015:3850. doi: 10.1002/smll.201403658. [DOI] [PubMed] [Google Scholar]
- 31.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen P, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, V Sequist L, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Sci. Transl. Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen P, Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA, Toner M. Nat. Protoc. 2014;9:694. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. J. Cell Biol. 2011;192:373. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horak D, Svobodova Z, Autebert J, Coudert B, Plichta Z, Kralovec K, Bílková Z, Viovy JL. J. Biomed. Mater. Res. - Part A. 2013;101 A:23. doi: 10.1002/jbm.a.34297. [DOI] [PubMed] [Google Scholar]
- 35.Arya C, Kralj JG, Jiang K, Munson MS, Forbes TP, DeVoe DL, Raghavan SR, Forry SP. J. Mater. Chem. B. 2013;1:4313. doi: 10.1039/c3tb20818d. [DOI] [PubMed] [Google Scholar]
- 36.Hoshino K, Huang Y-Y, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang X. Lab Chip. 2011;11:3449. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivas RL, Chapin SC, Doyle PS. Anal. Chem. 2011;83:9138. doi: 10.1021/ac202335u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pregibon DC, Toner M, Doyle PS. Science. 2007;315:1393. doi: 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]
- 39.Suh SK, Yuet K, Hwang DK, Bong KW, Doyle PS, Hatton TA. J. Am. Chem. Soc. 2012;134:7337. doi: 10.1021/ja209245v. [DOI] [PubMed] [Google Scholar]
- 40.An HZ, Helgeson ME, Doyle PS. Adv. Mater. 2012;24:3838. doi: 10.1002/adma.201200214. [DOI] [PubMed] [Google Scholar]
- 41.Went PTH, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Hum. Pathol. 2004;35:122. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 42.Dendukuri D, Gu SS, Pregibon DC, Hatton TA, Doyle PS. Lab Chip. 2007;7:818. doi: 10.1039/b703457a. [DOI] [PubMed] [Google Scholar]
- 43.Haghgooie R, Toner M, Doyle PS. Macromol. Rapid Commun. 2010;31:128. doi: 10.1002/marc.200900302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Björnmalm M, Yan Y, Caruso F. J. Control. Release. 2014;190:139. doi: 10.1016/j.jconrel.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 45.Choi NW, Kim J, Chapin SC, Duong T, Donohue E, Pandey P, Broom W, Hill WA, Doyle PS. Anal. Chem. 2012;84:9370. doi: 10.1021/ac302128u. [DOI] [PubMed] [Google Scholar]
- 46.Nakajima N, Ikada Y. Bioconjug. Chem. 1995;6:123. doi: 10.1021/bc00031a015. [DOI] [PubMed] [Google Scholar]
- 47.Friederich E, Louvard D. In: Encycl. Ref. Genomics Proteomics Mol. Med. Ganten D, Ruckpaul K, editors. Springer; Berlin Heidelberg: 2006. pp. 1116–1121. [Google Scholar]
- 48.Wu W, Wang W, Li J. Prog. Polym. Sci. 2015;46:55. [Google Scholar]
- 49.Joshi P, Jacobs B, Derakhshan A, Moore LR, Elson P, Triozzi PL, Borden E, Zborowski M. Oncotarget. 2014;5:2450. doi: 10.18632/oncotarget.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martowicz A, Spizzo G, Gastl G, Untergasser G. BMC Cancer. 2012;12:501. doi: 10.1186/1471-2407-12-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.An HZ, Eral HB, Chen L, Chen MB, Doyle PS. Soft Matter. 2014;10:7595. doi: 10.1039/c4sm01400f. [DOI] [PubMed] [Google Scholar]
- 52.Fischer P, Tibbitt M, Kloxin A, Anseth KS, Oakey J. Biomed. Sci. Instrum. 2014;50:62. [PubMed] [Google Scholar]
- 53.Shin DS, You J, Rahimian A, Vu T, Siltanen C, Ehsanipour A, Stybayeva G, Sutcliffe J, Revzin A. Angew. Chemie - Int. Ed. 2014;53:8221. doi: 10.1002/anie.201404323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hatch A, Hansmann G, Murthy SK. Langmuir. 2011;27:4257. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khademhosseini A, Yeh J, Jon S, Eng G, Suh KY, Burdick JA, Langer R. Lab Chip. 2004;4:425. doi: 10.1039/b404842c. [DOI] [PubMed] [Google Scholar]
- 56.Le Goff GC, Lee J, Gupta A, Hill WA, Doyle PS. Adv. Sci. 2015;2 doi: 10.1002/advs.201500149. DOI 10.1002/advs.201500149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Lab Chip. 2010;10:27. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malek AM, Alper SL, Izumo S. J. Am. Med. Assoc. 1999;282:2035. doi: 10.1001/jama.282.21.2035. [DOI] [PubMed] [Google Scholar]
- 59.Bong KW, Kim JJ, Cho H, Lim E, Doyle PS, Irimia D. Langmuir. 2015;31:13165. doi: 10.1021/acs.langmuir.5b03501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee J, Bisso PW, Srinivas RL, Kim JJ, Swiston AJ, Doyle PS. Nat. Mater. 2014;13:524. doi: 10.1038/nmat3938. [DOI] [PubMed] [Google Scholar]
- 61.Zheng F, Cheng Y, Wang J, Lu J, Zhang B, Zhao Y, Gu Z. Adv. Mater. 2014;26:7333. doi: 10.1002/adma.201403530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.