Abstract
Animal defense strategies against microbes are most often thought of as a function of the immune system, the primary function of which is to sense and kill microbes through the execution of resistance mechanisms. However, this antagonistic view creates complications for our understanding of beneficial host-microbe interactions. Pathogenic microbes are described as employing a few common behaviors that promote their fitness at the expense of host health and fitness. Here, a complementary framework is proposed to suggest that in addition to pathogens, beneficial microbes have evolved behaviors to manipulate host processes in order to promote their own fitness and do so through the promotion of host health and fitness. In this Perspective, I explore the idea that patterns or behaviors traditionally ascribed to pathogenic microbes are also employed by beneficial microbes to promote host tolerance defense strategies. Such strategies would promote host health without having a negative impact on microbial fitness and would thereby yield cooperative evolutionary dynamics that are likely required to drive mutualistic co-evolution of hosts and microbes.
Introduction
Our interactions with microbes are primarily thought of as antagonistic. This perspective is not limited to professional pathogenic microbes. The emerging interest in the intestinal microbiota – the trillions of microbes inhabiting the intestine that are essential for our health – has been accompanied by a rise in the perspective that these microbes are the cause of diseases of interest in the developed world (Longman et al., 2013). Thus, understanding how these microbes may cause disease has taken priority over understanding how our health may benefit from these microbial interactions. A consequence of this perspective is that it has biased our view of host defense strategies against microbes. Traditionally, the defense response in animals against microbes has been most often thought of as a consequence of the immune response; the primary function of which is to sense and eradicate microbes through the engagement of microbial killing pathways, collectively referred to as “resistance mechanisms.” In this perspective, I consider evidence that the response to microbes is not exclusively antagonistic. Beneficial microbes can induce host defense responses that promote both host and microbe fitness and will lead to mutualistic host-microbe interactions.
An examination of the evolutionary implications that resistance mechanisms have on host-microbe interactions reveals an important complication of this antagonistic perspective. Resistance protects the host by having a negative impact on microbial fitness, and leads to the coevolution of antagonistic traits in both the host and microbial populations (Svensson and Raberg, 2010). Negative selective pressures placed on the host population by microbes drives the selection for adaptive defense strategies, which places selective pressures on the microbe population to drive the selection for counter-attack defense strategies, which in turn leads to “new” selective pressures on the host population. These interactions have the potential to lead to open-ended evolutionary dynamics causing the oscilliation of resistance alleles in both populations, called the Red Queen effect (Figure 1) as well as the selection of new resistance traits. Many host-viral interactions provide excellent examples of these principles (Daugherty and Malik, 2012). For example, tetherin blocks the release of HIV-1 virions from infected cells and is a target of several viral factors. This antagonistic trait has driven the evolution of a mutant tetherin, resistant to these viral antagonists, which has then driven the evolution of an alternative method to antagonize tetherin (Daugherty and Malik, 2012).
Figure 1. Evolutionary dynamics of host-microbe interactions.
(A) Resistance traits in the host population place negative selective pressures on a microbial population leading to selection of a counter-attack strategy in the microbial population. This response places a new selective pressure on the host population, driving the selection for a “new” resistance trait and a decline in the presence of the previous resistance trait. An oscillation of antagonistic traits in both host and microbe population, or the Red Queen Effect, results. Graph adapted from (Svensson and Raberg, 2010). (B) A tolerance trait in the host population will have a neutral to positive selective pressure on a microbial population. This balance will maintain the presence of the microbe population and associated selective pressures on the host population that will drive the selection and spread of the tolerance trait in the host population, eventually leading to fixation of that trait. In addition to host-encoded tolerance mechanisms, beneficial microbes likely have evolved traits that promote tolerance of their host and are predicted to yield similar evolutionary dynamics.
Considering only pathogenic host-microbe interactions, these evolutionary dynamics may make sense. However, our microbial interactions that yield pathogenic outcomes make up only a small fraction of our total interactions with microbes, which are largely benign and often beneficial. The best example illustrating this principle is the microbiota, which performs essential functions for host physiology, including shaping the immune response, harvesting energy, and sustaining brain health. While it is established that resistance mechanisms help to shape the microbiota ecology (Strowig et al., 2012), there is little evidence to suggest that a loss of resistance mechanisms alone can trigger pathogenicity by the microbiota. It would be maladaptive to mount unnecessary resistance responses to our microbiota, as this would result in pathological consequences for the host. Thus, there must be other mechanisms in addition to killing mechanisms that enable a host to co-evolve beneficial microbial relationships.
Several models have been proposed to explain how we can have mutualistic microbial relationships with the microbiota while still responding to pathogenic threats. Originally, it was thought that the microbiota remains sequestered within the intestinal lumen with minimal interaction with the host. However, it is now well established that that microbiota participates in a dynamic dialog with the host. Pathogens can be considered highly adapted organisms that have the capacity to cause disease in their host. In 1989, Finlay and Falkow described common “themes” that pathogens have evolved (Finlay and Falkow, 1989). These themes can be broadly described as behaviors or “patterns of pathogenesis” that pathogens utilize to establish infection (Vance et al., 2009). As both pathogenic and beneficial microbes encode microbial-associated molecular patterns (MAMPs), innate immune recognition of patterns of pathogenesis combined with the recognition of MAMPs, have been useful to describe in part how a host can distinguish a pathogen from a beneficial microbe and mount an appropriate response to a microbial threat (Vance et al., 2009).
An issue that further complicates host-microbial relationships is that they are seldom binary. It is rare that a single microorganism, particularly in the case of those composing the microbiota, can be classified strictly as pathogenic or beneficial to the host. Rather pathogenicity or beneficial effects of a single microbe on host health is dependent on individual microbial behaviors and the context in which these behaviors occur. The capacity of microbes to cause disease is dependent on immune status, genetics and diet of a particular host as well as microbial location within the body. For example, Bacteroides thetaiotaomicron is considered a mutualist in most hosts. However, in genetically predisposed immunocompromised animals, B. thetaiotaomicron colonization leads to colonic inflammation (Bloom et al., 2011; Hickey et al., 2015). As such, it is likely that the microbial strategies that lead to beneficial outcomes for the host will likely be highly analogous to the ones that result in pathogenic outcomes.
In this perspective, I explore the idea that beneficial microbes have evolved strategies to manipulate host processes in order to promote the fitness of both host and microbe, framing the discussion around the intestinal microbiota of mammals. Such strategies would yield cooperative evolutionary dynamics that are required to drive mutualistic co-evolution of hosts and microbes.
Tolerance defenses in host-microbiota interactions
In a mutualistic relationship, if a microbe can promote its own fitness by promoting host health, and thus host fitness then there must be host-encoded mechanisms that are induced during microbial interactions and that enhance host health without killing the microbe. Such mechanisms were initially described in the plant literature in the context of infections and herbivore interactions. Ecologists have long recognized that in addition to resistance, plants rely on a distinct defense strategy called ‘tolerance’ to protect against pathogens and pests. Tolerance promotes plant fitness in the presence of given levels of pathogen or herbivore (Bent et al., 1993; Caldwell et al., 1958). In recent years, the concept of tolerance defenses has been introduced into the field of animal host-microbe interactions (Ayres and Schneider, 2011; Medzhitov et al., 2012; Raberg et al., 2007; Schneider and Ayres, 2008). In this context, tolerance is a defense strategy that minimizes the physiological damage that occurs during interactions with microbes without having a negative impact on microbial numbers. Thus, tolerance protects the host during microbial interactions by promoting health (and fitness) while having a neutral to positive impact on microbial fitness, suggesting implications for the co-evolution of host-microbe interactions that are distinct from resistance. The example of B. thetaiotaomicron described above provides an excellent example of tolerance defenses in mutualistic interactions. In immunocompromised mice, the loss of IL-10/TGFb signaling results in colonic inflammation in B. thetaiotaomicron-colonized mice (Bloom et al., 2011; Hickey et al., 2015). Thus the induction of IL-10/TGFb in immunocompetent mice promotes tolerance of this mutualist. As long as a microbe remains present in the host population, it will drive the selection for tolerance traits to spread in the host population and will go to fixation. Tolerance defenses would therefore yield the predicted evolutionary dynamics required to drive host-microbe mutualistic relationships and can help to explain how we co-evolved with our microbiota (Ayres, 2013) (Figure 1).
The field of disease tolerance in animals is in its infancy and as a result we do not know the full spectrum of the underlying mechanisms, however tolerance defenses in host-microbiota interactions theoretically fall into four main classes: repair, regulation of inflammation, neutralization of toxins and metabolic homeostasis (Ayres, 2013). Although the microbiota is beneficial for host health, the close proximity of this abundant microbial community to host tissues poses potential health risks to the host if homeostasis is disrupted. For example, a compromise in the barrier integrity of the gut epithelium that separates the community from extraintestinal sites can cause excessive inflammation, defects in nutrient and electrolyte absorption and disruptions in fluid homeostasis – all of which are detrimental to the host-microbiota mutualism. The induction of repair mechanisms to promote barrier integrity would therefore promote tolerance. Components of the innate immune system, such as the inflammasome, have been shown to be important for inducing repair of the intestinal epithelium in chemical models of intestinal injury. Further, when the barrier is compromised, the induction of anti-inflammatory mechanisms promotes tolerance by minimizing the costs incurred by an immune response, regulating the degree and duration of the response such that the collateral damage incurred is minimized (Ayres, 2013).
Host-microbe interplay can cause the release of host-derived and microbial-derived effectors or toxins, resulting in tissue damage. Mechanisms that prevent such damage by neutralizing the potentially dangerous effects of these toxins would promote tolerance as they protect host health without having a negative impact on microbial fitness. For example, the generation of heme from host-derived substrates in a variety of infection states, including sepsis, is toxic to host tissues. Detoxification of heme by heme-oxygenase-1 promotes tolerance in a model of mouse model of sepsis caused by cecal ligation and puncture (Larsen et al., 2010).
Maintenance of metabolic homeostasis also contributes to host tolerance of the microbiota. Immune and inflammatory responses impose significant energetic costs to the host (Ayres and Schneider, 2011). The unnecessary activation of inflammatory responses triggered by the microbiota can cause dysregulated energy allocation to fuel the host response resulting in physiological damage. The role the microbiota plays in energy harvesting and regulation of appetite as well as systemic metabolism will also likely influence tolerance (Ayres, 2013).
Tolerance inducing behaviors of the microbiota
Patterns of pathogenesis can be viewed as behaviors that pathogens utilize to promote their fitness by overcoming host barriers, responding to environmental stimuli, and growing within their host, which ultimately results in disease of the host. Broadly speaking, the pathogenic state and the associated behaviors that facilitate this state are the product of dynamic selective pressures on microbial populations that drive their adaptation to the host niche. This same logic can be applied to beneficial microbes that have evolved behaviors to promote their fitness and adaptation to the host niche through the induction of host physiologies that promote tolerance. These behaviors are distinct from virulence strategies employed by pathogens in that they induce a host response that promotes health rather than disease, but the outcome on microbial fitness is the same – positive impact on fitness by stabilizing a niche and mode of transmission. Tolerance can be determined experimentally as illustrated in Figure 2.
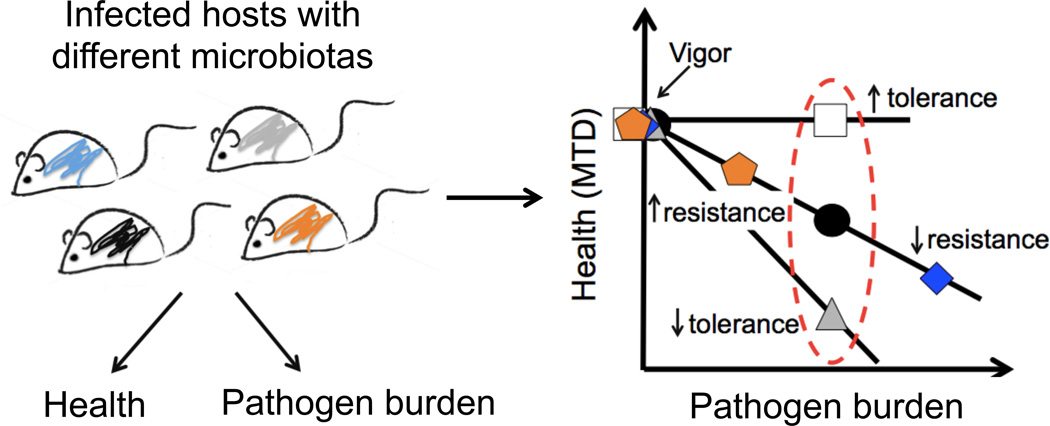
Figure 2. Measuring resistance and tolerance.
Resistance and tolerance can be measured in a host-microbe system by examining the relationship between a selected parameter of host health and microbial levels in target tissues. Using these parameters, and assuming the health of the host when uninfected is equivalent between different host populations (vigor), a dose response curve can be generated to determine how host health changes as microbial levels change. Changes along the diagonal indicate health is changing as microbial levels change and would identify hosts that vary in resistance defenses. Changes along the y-axis would indicate health is changing without a change in microbe levels and would identify hosts that vary in tolerance. The more tolerant a host, the shallower the slope of the dose response curve would be. This method, which assumes a linear response, can be used to determine how different factors including environmental and genetic factors can influence host defenses. These relationships however are likely more complex and further experimentation and data points, for example measuring pathogen burden over the course of the infection, would reveal how differences in host populations influence resistance and tolerance at different stages of the infection. Adapted from (Ayres and Schneider, 2008).
Tolerance mechanisms are either constitutively active or inducible (Medzhitov et al., 2012) and while the microbiota likely has evolved strategies to positively regulate both, these behaviors are most evident experimentally when homeostasis is disrupted. The microbiota may be confronted by rapid environmental changes that have the potential to be harmful to both the host and the microbiota. Most microbiota studies have involved disease models and selective pressures that are relatively new on the evolutionary scale, such as antibiotic treatment, metabolic diseases, high fat diets, and autoimmunity. While these are useful for revealing how microbes can behave in a non-adapted disease state, typically revealing pathogenic effects, they do not elucidate cooperative microbial tolerance behaviors, as the microbiota has not evolved under these conditions. The best way to reveal these cooperative behaviors would be in the context of diseases that mimic the evolutionary pressures in which hosts and their microbiotas evolved, such as under-nutrition, infectious diseases, hibernation, and microbial competition.
Pathogens use only a few common behaviors that trigger disease, including occupation of specialized niches, meeting metabolic demands to support growth, and manipulation of host processes including the inflammatory response. Some of these behaviors will be conceptually similar in the application of tolerance mechanisms to mutualism and are here termed "cooperative microbial tolerance behaviors." Below is a broad consideration of some major microbial behaviors that promote either pathogenesis or host tolerance along with examples in the context of disease states in which homeostasis of host-microbiota systems is compromised (Figure 3).
Figure 3. Microbial adaptation behaviors leading to pathogenicity or tolerance.
(A) Microbes occupy specialized niches that can cause pathogenicity or promote tolerance defenses. Left, upon entry into the intestine S. Typhimurium invades Peyer’s Patches and infects lamina propria phagocytes, which then enter the lymphatics and bloodstream and infected distal organs including the liver. Right, during intestinal and extraintestinal infections, the microbe E. coli O21:H+ translocates to white adipose tissue deposits to induce tolerance via induction of innate immune – endocrine interactions. (B) Meeting metabolic demands of microbes to promote growth. Left, the opportunistic pathogen C. difficle forages on glycans liberated by members of the microbiota, supporting pathogen growth upon induction of infection with antibiotics. Right, under conditions in which nutrients are limited, beneficial microbes alter their foraging to utilize fucosylated host species. In the context of LPS systemic injection and oral infection with Citrobacter rodentium, fucosylation and flexible foraging of microbes promotes tolerance by preventing the microbes from becoming pathogenic (Pickard et al. 2014). In other contexts, for example during chronic malnutrition, this change in foraging behavior may become pathogenic to the host. (C) Regulation of host inflammation. Many pathogens induce an inflammatory response that can facilitate infection and pathogenicity. Left, H. hepaticus induces inflammation in the colon. Right, PSA generated by B. fragilis restores inflammatory balance in the host through the induction of an IL-10 dependent anti-inflammatory response.
Pattern I. Occupation of specialized niches
Many intestinal pathogens are not capable of establishing residency in an established microbiota upon infection of a host. To overcome this barrier, these microbes have evolved mechanisms to facilitate their invasion and occupancy of specialized niches to avoid competition with members of the microbiota ultimately yielding a pathogenic outcome for the host. For example, Salmonella Typhimurium penetrates the gut epithelial layer by invasion of Peyer’s Patches. After penetration of the epithelial barrier, S. Typhimurium preferentially infects phagocytes within the lamina propria, effectively creating a new niche. In host-adapted salmonellosis, these cells can gain access to the bloodstream and lymphatics, facilitating the spread of the pathogen to distal organs including the liver and spleen and cause disease (Monack et al., 2004) (Figure 3).
The majority of the microbiota is compartmentalized within the lumen of the intestine, with smaller proportions occupying the mucus layer that overlays the gut epithelium and niches close to the epithelium. We are now gaining insight into the heterogeneity and spatial organization that appears to exist within the luminally bound gut microbiota (Earle et al., 2015; Nava et al., 2011). A common feature of a variety of disease states is localization changes of the microbiota within the intestine. For example, in inflammatory bowel disease (IBD) the normally luminal microbiota moves closer to host tissues (Sanchez de Medina et al., 2014). While these new niches are thought to be a mislocalization of the microbiota that potentiates disease, this may instead represent a cooperative microbial tolerance behavior that constituents of the microbiota have acquired to induce tolerance defenses to return to homeostasis. In a mouse model of IBD, oral administration of the sulphated polysaccharide dextran sulphate sodium (DSS) causes mucus erosion and localization of the microbiota closer to the gut epithelium. Recognition of the microbiota by Toll like receptors (TLRs) that occurs as a result induces cytoprotective and tissue repair factors to restore barrier integrity (Rakoff-Nahoum et al., 2004). Thus, the new niche occupied by the microbiota may be essential for the induction of repair mechanisms encoded by the host to promote tolerance.
Spatial re-organization and new niches also arise when nutrient supplies are limited. Microbiota accessible carohydrates (MACs) are abundant in dietary fiber and a primary metabolic input for the intestinal microbiota. On MAC-deficient diets, gnotobiotic models with simplified microbiomes and mice with complex humanized microbiomes, show changes in the spatial organization of the microbiota (Earle et al., 2015). It remains to be determined how this change in niche influences host health and tolerance defenses.
Although it was long thought that extraintestinal spatial relocation of the intestinal microbiota could only result in pathogenic interactions with the host, recent data suggests that mutualists can in fact occupy extraintestinal niches and in doing so promote tolerance defenses of the host (Schieber et al., 2015). When mice were infected intranasally with the pneumonic pathogen Burkholderia thailandensis or orally with the pathogen S. Typhimurium, they exhibited skeletal muscle wasting. A member of the intestinal microbiota, E. coli O21:H+ antagonized muscle wasting during these infections. This protection was associated with the translocation of E. coli O21:H+ from the intestine to white adipose tissue (WAT) deposits during infection and occurred without a negative impact on B. thailandensis or S. Typhimurium fitness. Thus, E. coli O21:H+ evolved a behavioral strategy involving physical relocation to a distinct site to induce tolerance defenses in the host, promoting health of the host and fitness of E. coli O21:H+ (Figure 3). As invasive oral microbes were the only microbes believed to translocate from the intestine to occupy extraintestinal niches, the finding that a constituent of the microbiota occupies an extraintestinal niche to promote health of the host blurs our traditional distinction of what is a pathogen and what is a mutualist.
Pattern II. Meeting metabolic demands for growth
Upon infection or colonization of a host, microbes must meet their metabolic demands to support their replication. For pathogens, this involves adapting their foraging behavior to the changing environment and nutrient supplies within the host during the infection process. For example, in hosts colonized with Clostridium difficile spores, antibiotic treatment triggers an active C. difficile infection as this pathogen has evolved to utilize microbiota-liberated carbohydrates as an energy source during its expansion in the antibiotic-treated gut (Ng et al., 2013) (Figure 3).
Conditions that affect the energy homeostasis of the host, including infection-induced anorexia and undernutrition, will also compromise energy homeostasis of the microbiota as it reduces substrate availability. A host has finite energy levels and the energetic demands of the microbiota in large part have to be satisfied by this supply. Emerging evidence suggests that some constituents of the microbiota have evolved mechanisms to adapt to the changes in nutrient status in the host. B. thetaiotaomicron preferentially feeds on liberated hexose sugars when the host is fed a polysaccharide rich diet. However, when gnotobiotic animals monocolonized with B. thetaiotaomicron were given a simple sugar diet, this symbiont changed its foraging behavior to consume host mucus glycans, revealing the capacity for certain mutualists to have flexible foraging (Sonnenburg et al., 2005). This specific change in foraging behavior is associated with B. thetaiotaomicorn occupying a new niche within the mucus layer. There are now early indications that the outcome flexible foraging confers on the host is contextual and depends on the particular microbiota constituent involved.
During certain life stages, this kind of foraging behavior may become pathogenic to the host. In addition to infections, the microbiota influences body size and postnatal animal and human growth in chronically undernourished conditions (Blanton et al., 2016; Schwarzer et al., 2016). Gut microbes that are present in the neonate gut and that consume human milk oligosaccharides (HMOs) induce the same genes during HMO consumption that are used to harvest host mucus glycans, which are structurally similar to HMOs (Marcobal et al., 2011). Two recent studies have demonstrated that the intestinal microbiota can exacerbate malnutrition in children (Smith et al., 2013; Trehan et al., 2013). One study showed that breast milk consumed by malnourished infants had reduced levels of fucosylated and sialylated forms of HMOs (Charbonneau et al., 2016), suggesting that under conditions in which HMOs are scarce, gut microbes present in the neonate gut alter their foraging to utilize host mucus glycans, which may be detrimental to the growing neonate. Another possible explanation is that similar to E. coli O21:H+, there are protective microbes in the neonate gut that translocate to WAT deposits to induce protective innate-endocrine host responses to promote growth (Schieber et al., 2015). Indeed, the microbiota constituent Lactobacillus plantarum was found to maintain body size of neonate mice in a mouse model of chronic undernutrition (Schwarzer et al., 2016); however the exact mechanisms by which this and other intestinal microbiota members can protect from growth impairment under conditions of chronic undernutrition remain to be determined.
In the context of infection, flexible foraging by the microbiota has been shown to be beneficial for the host through the induction of tolerance defenses. Systemic administration of lipopolysaccharide (LPS) to mice induces an acute anorexic response that resembles what is seen with infections and causes rapid α(1,2)-fucosylation of small intestinal epithelial cells (IECs). This fucosylation is dependent on TLR sensing of microbial agonists and ultimately the production of IL-22 that induces upregulation of a fucosyltransferase in IECs, resulting in increased fucosylation of proteins that are expressed on cell surface or secreted into the lumen. Fucosidase-expressing members of the microbiota direct their foraging to these fucosylated species liberating fucose residues that can be metabolized by them or other constituents of the flora (Pickard et al., 2014) (Figure 3). In a mouse model of infectious colitis, the absence of fucosylation results in increased pathology without a change in pathogen levels, suggesting that fucosylation promotes tolerance during conditions in which food consumption and energy balance of the microbiota are compromised (Pickard et al., 2014).
This sort of microbial cooperative behavior to promote mutualism is reminiscent of the ant-aphid relationship. Aphids are phloem-sucking insects that can be prey for ants. To limit their risk of predation by the ants, they excrete honeydew (sugary aphid feces), which the ants consume rather than the aphids (Barton and Ives, 2014). The aphid, in essence, bribes the ant. During sickness-induced anorexia, the host may utilize increased fucosylation as a bribery tactic to prevent the microbiota from feeding on host nutrients and by promoting this behavior of the microbiota, tolerance of the system is benefits. Despite the diverse metabolic capacity of the microbiota members, the key point here is that changes in foraging and metabolic behavior are common behaviors microbes employ and are likely evolved adaptation strategies to promote both host and microbe fitness to maintain mutualism.
Pattern III. Regulation of inflammation
In the context of infection, many microbes are thought to intentionally induce host responses, such as the inflammatory response, to aid in their niche establishment resulting in increased pathogenicity. In the context of mutualism, microbes can regulate or optimize host inflammatory responses to prevent tissue damage as sustained inflammatory responses triggered by the microbiota or during infection can result in physiological damage from dysregulated energy allocation to fuel the host response. Excessive inflammation can also disrupt energy balance by causing tissue damage that can cause dehydration, electrolyte imbalances and disruptions in proper nutrient absorption.
Mutualism between the host and the intestinal microbiota requires that the immune system maintain a balance between immunity to pathogenic threats and hyporesponsiveness to beneficial microbes. Part of this optimization is to allow the host to mount a response when there is an infection, but to balance it so that it remains hypo-responsive to the resident microbiota. One mechanism by which this can occur is through chemical signaling through small-molecule metabolites produced by bacteria that typically serve for communication between microbes. Accumulating evidence suggests that the immune system can sense microbial metabolites (Brestoff and Artis, 2013). Short chain fatty acids, such as n-butyrate, are produced by anaerobic fermentation of dietary fiber by gut microbes. Treatment of LPS-stimulated colonic lamina propria macrophages with n-butyrate reduced production of pro-inflammatory mediators, and similarly, when mice were treated orally with n-butyrate, isolated colonic lamina propria macrophages exhibited reduced transcription of pro-inflammatory mediators (Chang et al., 2014). These observations suggest that n-butyrate modulates the inflammatory response of colonic lamina propria macrophages under homeostatic conditions.
Mutualists can also optimize host responses under infectious conditions to promote tolerance. Helicobacter hepaticus is a pathobiont. In immunocompetent hosts it behaves as a commensal – a microbe that has no known beneficial or pathogenic effect to the host - but in immunodeficient animals it causes disease. In a T cell model of chronic colitis, oral infection of mice with H. hepaticus causes elevated intestinal levels of TNFα and severe colitis. However, when mice were colonized with both H. hepaticus and the mutualist Bacteroides fragilis, animals were protected from colitis. This protection on host health had no effect on H. hepaticus levels in the intestine, demonstrating that B. fragilis promotes tolerance defenses for H. hepaticus infection (Mazmanian et al., 2008). Polysaccharide A (PSA) produced by B. fragilis is necessary and sufficient for the tolerance response in H. hepaticus-infected mice by promoting an IL-10 dependent anti-inflammatory response. Thus, the production of PSA by B. fragilis optimizes the host response by inducing an anti-inflammatory mechanism.
Virulence factors as cooperative factors
Pathogens utilize a number of factors called virulence factors in order to occupy specialized niches, meet metabolic demands, and regulate inflammatory responses. Access to the cytosol of host cells has traditionally been described as a pattern of pathogenesis that is utilized by many pathogens to maintain fitness within the host (Vance et al., 2009). However, many of the described studies suggest that this may also be a behavior of mutualists for adaptation to the host niche. For example, the protection described above mediated by E. coli O21:H+ requires activation of an intracellular sensor (Schieber et al., 2015). Similarly, the antiinflammatory response of n-butyrate involves an intracellular target (Chang et al., 2014). One possibility to explain how these intracellular responses are induced is that the activating ligands are passively transported into the cytosol, or the host has evolved transporters to facilitate their delivery. Another possibility is that mutualists actively deliver these products to the cytosol through specialized secretion systems that facilitate host-microbe interactions; although these have traditionally been classified as virulence factors. Indeed, genome sequencing of E. coli O21:H+ revealed a Type Three Secretion System (TTSS) – a multi-protein syringe like complex that that delivers bacterial proteins to host cell cytosol to manipulate host cell processes (Ayres et al., 2012; Schieber et al., 2015). How or whether this delivery mechanism is employed, remains to be determined.
Delivery of ligands to the cytosol can also be achieved by intestinal microbes through the occupation an intracellular niche. Emerging evidence suggests that a unique subset of commensal bacteria can live in intestinal lymphoid cells of healthy animals (Fung et al., 2016). The translocation of E. coli O21:H+ to WAT during infections may also occur via a cellular trafficking mechanism that would involve this microbe inhabiting an intracellular niche (Schieber et al., 2015). Thus, the current viewpoint that the access to the cytosol is associated with virulence is complicated by the discovery of microbes that can also access the cytosol, which is associated with beneficial effects on host health.
There likely will be additional patterns utilized by beneficial microbes to promote tolerance that have traditionally been associated with virulence revealed as the field of host-mutualistic interactions progresses. For example, interacting with the host cell cytoskeleton is an additional behavior believed to be specific to pathogens (Vance et al., 2009). Segmented filamentous bacteria (SFB) is a commensal that shapes the host immune system and makes physical contact with gut epithelial cells, creating an invagination which likely alters the cytoskeleton (Ivanov et al., 2009). The role of cytoskeletal rearrangements in the context of SFB and other beneficial microbes and how these microbes induce these cytoskeletal changes remains to be explored.
The immune system mediates tolerance behaviors induced by the microbiota
Discrimination of pathogens from nonpathogenic microbes by the innate immune system has been proposed to occur in part by recognition of both MAMPs and pathogen specific behaviors to guide the appropriate resistance responses (Vance et al., 2009). An exciting theme that is emerging in the context of beneficial microbial interactions is that cooperative microbial tolerance behaviors require innate immune mechanisms, extending the function of these pathways beyond resistance mechanisms. For example, recognition of commensal-derived ligands, including LPS, is required for the induction of tissue repair pathways in a murine model of intestinal injury (Rakoff-Nahoum et al., 2004). Many inflammasome cytosolic sensors, including NLRP3, NLRP6 and NLRP12, have also emerged as critical regulators of homeostasis and tissue repair in response to intestinal insult, although a role for recognition of microbiota constituents in mediating protection has not been established (Allen et al., 2010; Bauer et al., 2010; Chen et al., 2011; Dupaul-Chicoine et al., 2010; Elinav et al., 2011; Normand et al., 2011; Siegmund et al., 2001; Takagi et al., 2003; Zaki et al., 2010; Zaki et al., 2011). The role of innate immune sensors in tissue homeostasis and repair likely extends beyond gut tissue. Older studies demonstrated that while germ free mice were defective in skin wound repair, both conventional and the germ-free rodents had accelerated skin repair when exposed to the bacterium Staphylococcus aureus (Levenson et al., 1983; Okada, 1994).
Immune cells of the lymphoid lineage and adaptive immune system appear to be important for tolerance defenses in host-microbiota interactions. The induction of the fucosylation response triggered by sickness-induced anorexia requires crosstalk between CD11c+ dendritic cells (DCs) and RORgt-dependent innate lymphoid cells (ILCs) to secrete IL-22, which ultimately results in increased fucosylation (Pickard et al., 2014). The anti-inflammatory response triggered by B. fragilis requires recognition of PSA by TLR2 and the production of IL-10 by CD4+ T cells (Mazmanian et al., 2008; Wang et al., 2006). The fact that host pathways that we have traditionally thought of as microbial killing mechanisms, such as inflammasomes and TLRs, appear to also be important for the induction of tolerance defenses by beneficial microbes complicates our traditional perspective of how the immune system distinguishes between pathogenic and nonpathogenic microbes.
The context in which these signals are sensed combined with variations in cellular/tissue responses of the host will dictate the outcome of these interactions. For example, the protection against muscle wasting mediated by E. coli O21:H+ requires the NLRC4 inflammasome and the downstream effector IL-18. Yet, in models of sepsis, an E. coli isolate of the same serotype can colonize the lung, liver, spleen and kidney (Ayres et al., 2012). Despite protection against wasting in this model, colonization of these tissues is associated with inflammasome activation leading to IL-1β-mediated pathology. Thus the beneficial effects triggered by the E. coli O21:H+ - inflammasome interaction are overridden in contexts in which this same microbe leads to IL-1β-dominated pathogenic responses (Ayres et al., 2012). Thus, it will be important to move from host-microbe interactions at the cellular level and study these interactions at the organismal level to reveal how these interactions affect health and disease.
Open questions and future perspectives
The relationship between hosts, microbes and health are profound and intricate. While professional pathogenic microbes differ from beneficial microbes in their influence on host health, both pathogen and mutualist fitness is dependent on the microbe’s ability to replicate, meet metabolic demands and transmit to a new host. The long-standing interest in understanding how pathogenic microbes cause disease has resulted in a great understanding of behaviors and features that are important for virulence. The traditional distinction between pathogenic and benign/beneficial microbes holds that beneficial microbes lack such features and behaviors. However, “virulence” factors may be more broadly construed as microbial adaptation factors that pathogenic microbes utilize to promote their fitness at the expense of host health and fitness. Here I have considered the hypothesis that, in addition to pathogens, beneficial microbes have also evolved strategies to manipulate host processes that, in the true sense of mutualism, promotes their fitness through the promotion of their host’s fitness.
The evidence presented indicates that there is considerable overlap between the evolved strategies that pathogens and beneficial microbes use, including the occupation of specialized niches, strategies to meet metabolic demands, and manipulation of host processes, including the inflammatory response. The involvement of the innate and adaptive immune systems in mediating cooperative microbial tolerance behaviors raises the interesting question of which function these immune pathways evolved first – tolerance functions to promote host-microbe mutualism or resistance functions leading to antagonistic host-microbe interactions.
The discussion presented here is clearly not exhaustive, and the full spectrum of microbiota behaviors that mutualists employ has only begun to emerge. For example, many pathogens such as attaching and effacing (A/E) pathogens occupy a niche that involves their adherence to host cells, which ultimately causes disease. Several intestinal commensals including SFB and Prevotella are found to adhere to the epithelial layer of the intestine and it is possible that these microbes provide a beneficial function to host health. Although Prevotella species have been associated with inflammatory conditions, these disease states have only been revealed under conditions in which the microbiota have not evolved, including high fat diet (Elinav et al., 2011; Henao-Mejia et al., 2012). Studying these adherent microbes in the context of diseases in which the microbiota have evolved may be important for revealing beneficial functions of these microbes. Thus, it will be important to study these processes in the host at the organismal level to reveal the contexts in which these behaviors are positive or negative for host health.
Currently, we know very few host encoded tolerance mechanisms. Incorporating the concept of tolerance into our host-microbiota studies will be important for appreciating the full spectrum of mechanisms in this defense strategy, and will also be informative for understanding how to induce these mechanisms to treat disease. This knowledge will lead to a better understanding of how we interact with microbes – pathogenic, commensal, and mutualist.
Acknowledgments
JSA is supported by NIH grant R01AI114929 and CA014195, the NOMIS Foundation, the Searle Scholar Foundation, The Ray Thomas Edward Foundation and a DARPA Young Faculty Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS. Inflammasome-microbiota interplay in host physiologies. Cell host & microbe. 2013;14:491–497. doi: 10.1016/j.chom.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Ayres JS, Schneider DS. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol. 2008;6:2764–2773. doi: 10.1371/journal.pbio.0060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Schneider DS. Tolerance of infections. Annu Rev Immunol. 2011;30:271–294. doi: 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton BT, Ives AR. Direct and indirect effects of warming on aphids, their predators, and ant mutualists. Ecology. 2014;95:1479–1484. doi: 10.1890/13-1977.1. [DOI] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, Tschopp J, Endres S, Latz E, Schnurr M. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192–1199. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Innes RW, Staskawicz BJ. Use of Arabidopsis thaliana and Pseudomonas syringae in the Study of Plant Disease Resistance and Tolerance. J Nematol. 1993;25:519–525. [PMC free article] [PubMed] [Google Scholar]
- Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351 doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell RM, Schafer JF, Compton LE, Patterson FL. Tolerance to Cereal Leaf Rusts. Science. 1958;128:714–715. doi: 10.1126/science.128.3326.714. [DOI] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, et al. Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell. 2016;164:859–871. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. Journal of immunology. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell host & microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Falkow S. Common themes in microbial pathogenicity. Microbiol Rev. 1989;53:210–230. doi: 10.1128/mr.53.2.210-230.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, Wang K, Ziegler CG, Goc J, Shima T, et al. Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity. 2016;44:634–646. doi: 10.1016/j.immuni.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, et al. Colitogenic Bacteroides thetaiotaomicron Antigens Access Host Immune Cells in a Sulfatase-Dependent Manner via Outer Membrane Vesicles. Cell Host Microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen R, Gozzelino R, Jeney V, Tokaji L, Bozza FA, Japiassu AM, Bonaparte D, Cavalcante MM, Chora A, Ferreira A, et al. A central role for free heme in the pathogenesis of severe sepsis. Sci Transl Med. 2010;2:51ra71. doi: 10.1126/scitranslmed.3001118. [DOI] [PubMed] [Google Scholar]
- Levenson SM, Kan-Gruber D, Gruber C, Molnar J, Seifter E. Wound healing accelerated by Staphylococcus aureus. Arch Surg. 1983;118:310–320. doi: 10.1001/archsurg.1983.01390030042007. [DOI] [PubMed] [Google Scholar]
- Longman RS, Yang Y, Diehl GE, Kim SV, Littman DR. Microbiota: host interactions in mucosal homeostasis and systemic autoimmunity. Cold Spring Harb Symp Quant Biol. 2013;78:193–201. doi: 10.1101/sqb.2013.78.020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell host & microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nature reviews Microbiology. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, Lemoine Y, Hot D, Chamaillard M. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A. 2011;108:9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M. The influence of intestinal flora on wound healing in mice. Surg Today. 1994;24:347–355. doi: 10.1007/BF02348566. [DOI] [PubMed] [Google Scholar]
- Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Sanchez de Medina F, Romero-Calvo I, Mascaraque C, Martinez-Augustin O. Intestinal inflammation and mucosal barrier function. Inflamm Bowel Dis. 2014;20:2394–2404. doi: 10.1097/MIB.0000000000000204. [DOI] [PubMed] [Google Scholar]
- Schieber AM, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, Evans RM, Ayres JS. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science. 2015;350:558–563. doi: 10.1126/science.aac6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, Martino ME, Balmand S, Hudcovic T, Heddi A, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016;351:854–857. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- Siegmund B, Lehr HA, Fantuzzi G, Dinarello CA. IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. Proc Natl Acad Sci U S A. 2001;98:13249–13254. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- Svensson EI, Raberg L. Resistance and tolerance in animal enemy-victim coevolution. Trends Ecol Evol. 2010;25:267–274. doi: 10.1016/j.tree.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Takagi H, Kanai T, Okazawa A, Kishi Y, Sato T, Takaishi H, Inoue N, Ogata H, Iwao Y, Hoshino K, et al. Contrasting action of IL-12 and IL-18 in the development of dextran sodium sulphate colitis in mice. Scand J Gastroenterol. 2003;38:837–844. doi: 10.1080/00365520310004047. [DOI] [PubMed] [Google Scholar]
- Trehan I, Goldbach HS, LaGrone LN, Meuli GJ, Wang RJ, Maleta KM, Manary MJ. Antibiotics as part of the management of severe acute malnutrition. N Engl J Med. 2013;368:425–435. doi: 10.1056/NEJMoa1202851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell host & microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, McLoughlin RM, Cobb BA, Charrel-Dennis M, Zaleski KJ, Golenbock D, Tzianabos AO, Kasper DL. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J Exp Med. 2006;203:2853–2863. doi: 10.1084/jem.20062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. Journal of immunology. 2010;185:4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Vogel P, Malireddi RK, Body-Malapel M, Anand PK, Bertin J, Green DR, Lamkanfi M, Kanneganti TD. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20:649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]