Abstract
Septins are a family of cytoskeletal GTP-binding proteins that assemble into membrane-associated hetero-oligomers and organize scaffolds for recruitment of cytosolic proteins or stabilization of membrane proteins. Septins have been implicated in a diverse range of cancers, including gastric cancer, but the underlying mechanisms remain unclear. The hypothesis tested here is that septins contribute to cancer by stabilizing the receptor tyrosine kinase ErbB2, an important target for cancer treatment. Septins and ErbB2 were highly over-expressed in gastric cancer cells. Immunoprecipitation followed by mass-spectrometry analysis identified ErbB2 as a septin-interacting protein. Knockdown of septin-2 or cell exposure to forchlorfenuron (FCF), a well-established inhibitor of septin oligomerization, decreased surface and total levels of ErbB2. These treatments had no effect on EGFR, emphasizing the specificity and functionality of the septin-ErbB2 interaction. The level of ubiquitylated ErbB2 at the plasma membrane was elevated in cells treated with FCF, which was accompanied by a decrease in co-localization of ErbB2 with septins at the membrane. Cathepsin B inhibitor, but not bafilomycin or lactacystin, prevented FCF-induced decrease in total ErbB2 by increasing accumulation of ubiquitylated ErbB2 in lysosomes. Therefore, septins protect ErbB2 from ubiquitylation, endocytosis, and lysosomal degradation. The FCF-induced degradation pathway is distinct from and additive with the degradation induced by inhibiting ErbB2 chaperone HSP90. These results identify septins as novel regulators of ErbB2 expression that contribute to the remarkable stabilization of the receptor at the plasma membrane of cancer cells and may provide a basis for the development of new ErbB2-targeting anti-cancer therapies.
Keywords: ubiquitylation, gastric cancer, septin, ErbB2, endocytosis, lysosomal degradation
Introduction
Human septins are a family of 13 cytoskeletal proteins that have important roles in a variety of cellular processes in both health and disease [1, 2]. Septins have been implicated in a diverse range of human cancers, including gastric cancer [3–7]. Nine of the13 septins are overexpressed or downregulated in various cancers, and experimentally altered levels of septins correlate with increased cell growth, motility, and invasiveness (reviewed in [3]). Both septin-2 and septin-9 are overexpressed in gastric cancer [3–7], which is the fourth most common cancer and the second most common cause of cancer death worldwide [8]. Septins function as small GTPases that assemble into hetero-oligomeric complexes and higher-order structures, such as filaments and rings. They directly interact with membranes and link them to both actin and tubulin cytoskeletal networks [1, 9]. By interacting also with transmembrane and cytosolic proteins, septins organize segregated membrane microdomains and form scaffolds for protein recruitment [1, 9]. At the cellular level, septins contribute to regulation of numerous processes, including cell division, cell migration, ubiquitylation, endocytosis, and degradation [10–12]. The involvement of septins in tumorigenesis is thought to be associated with their role in cytokinesis [13], microtubule-dependent cell migration and polarization [14, 15], Rho signaling [16] and HIF-1 (hypoxia-inducible factor-1) stabilization [17]. In addition, septins are known to cluster and stabilize integral membrane proteins [18, 19], including cancer-associated receptor tyrosine kinases. Particularly, septin-2 and septin-11 regulate surface expression of Met [20], and septin-9 stabilizes epidermal growth factor receptor (EGFR) [21], suggesting a possible role for septins in the abnormal persistence of other receptor tyrosine kinases at the membrane of cancer cells.
The Erb receptor tyrosine kinase 2 (ErbB2 or HER-2/neu) belongs to the EGFR family of proteins (ErbB1 or EGFR, ErbB2, ErbB3, ErbB4), which control growth and differentiation in normal cells and promote cancer progression when overexpressed. Mutation or overexpression of ErbB2 is seen in multiple cancers [22], including 22% of gastric cancers [23], making the receptor an important target for cancer treatment. Specific treatment models, such as humanized ErbB2 antibody [24, 25], have proven to be effective, but many cancer patients develop resistance to these agents due to compensatory overexpression of other membrane proteins that limit binding to ErbB2 or activation of competing signaling pathways [26, 27]. The evolving need to develop new synergistic treatment strategies [25, 28–31] highlights the importance of gaining additional knowledge of ErbB2 stability, trafficking and degradation. In cancer cells, ErbB2 is localized almost exclusively at the plasma membrane likely due to inefficient internalization and rapid recycling [32, 33]. Ubiquitylation of ErbB2 may be critical for its endocytic downregulation as the recombinant addition of 4 ubiquitin residues linearly connected to the C-terminus of the receptor induced constitutive endocytosis and lysosomal degradation of ErbB2 [34]. The ubiquitin ligases, including c-Cbl, cullin5, Lnx1, and CHIP (carboxy terminus Hsp70 interacting protein), and de-ubiquitylating enzymes, including Usp8 and Usp9x, interact with ErbB2, and the balance between ubiquitylation and deubiquitylation appears to play a critical role in surface expression of the protein [35–40]. Leucine rich repeats and immunoglobulin-like domains protein-1 (LRIG1) is a membrane protein identified as a negative regulator of ErbB receptors, including ErbB2. Mice deficient in LRIG1 develop highly penetrant duodenal adenomas with gastric metaplasia and increased ErbB signaling [41]. Initial reports suggested that LRIG1 promotes c-Cbl-dependent ubiquitylation and increases the degradation rate of ErbB receptor [42, 43]. However, more recent studies demonstrated that LRIG1 promotes degradation of the receptors independent of c-Cbl (reviewed in [44]), leaving the mechanism of LRIG1 action unclear. The chaperone Hsp90 has been identified as a positive regulator of ErbB2 required for stable surface expression of the receptor [45, 46]. The inhibitors of Hsp90, such as geldanamycin, disrupt the association between ErbB2 and Hsp90, which allows for recruitment of Hsp70 and the ubiquitin ligases CHIP and/or cullin5, leading to ErbB2 ubiquitylation [35, 39, 47], proteasome-dependent internalization, and lysosomal degradation [48, 49]. Despite these advances in knowledge on ErbB2 degradation, the mechanism of persistence of ErbB2 at the plasma membrane in cancer cells remains incompletely understood, and identification of additional regulators of ErbB2 stability could lead to development of novel treatment targets.
In the present study, ErbB2 was identified as a septin-binding partner in gastric cancer cells by mass spectrometry analysis of both septin and ErbB2 interactomes. The interaction and co-localization between septins and ErbB2 was confirmed by immunoprecipitation/immunoblotting and immunofluorescence. Depletion of septins by specific siRNA or disruption of septin oligomerization by forchlorfenuron (FCF) decreased co-localization and interaction of septins with ErbB2, resulting in specific ubiquitylation of ErbB2 followed by its endocytosis and lysosomal degradation. The data demonstrate that septins contribute to the abnormal persistence of ErbB2 in the plasma membrane of cancer cells, presenting a potential link between these cancer-associated proteins.
Materials and methods
Cell culture
HGE-20 cells [50] were grown in 50:50 DMEM:F12 (DMEM-Cellgro Mediatech, Manassas, VA, USA, F12-Life Technologies, Carlsbad, CA, USA) with 10% FBS (Gemini Bio-Products, West Sacremento, CA, USA) and 100 units/ml penicillin, 0.1 mg/ml streptomycin (Sigma, St. Louis, MO, USA). HGE-20 cells were provided by Dr. Daniel Mènard, who kindly granted permission to use the cells for this work. AGS, HEK-293, A549 and MDCK cells (ATCC, Manassas, VA, USA) and HGT-1 cells [51] were grown in DMEM medium (Cellgro Mediatech) containing 4.5 g/L glucose, 2 mM L-glutamine, 8 mg/L phenol red, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 10% FBS. The growth media for A549 cells was supplemented with 20 mM HEPES. PC12 cells (ATCC) were grown in RPMI 1640 with 2 mM GlutaMAX™, 10 mM HEPES, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma) supplemented with 10% FetalPlex animal serum complex (Gemini Bio-Products). Human mammary epithelial (HB2) cells [52] were maintained in DMEM supplemented with 10% FBS, 5 μg/ml hydrocortisone and 10 μg/ml insulin (Santa Cruz Biotechnology, Dallas, TX, USA). Human gastric antrum tissue was obtained through the UCLA TPCL program with a non-human subjects IRB exemption.
Antibodies
The following primary antibodies were used: ErbB2 (mouse, Ab-17, clone E2-4001+3B5, 1:1000 western blot, 1:100 immunofluorescence, ThermoFisher, Waltham, MA, USA), β-actin (mouse, clone C4, 1:1000, Milipore, Billarica, MA, USA, rabbit, 1:1000, Cell Signaling Technologies, Beverly, MA, USA), septin-2 (Sigma prestige, 1:2000 western blot, 1:100 immunofluorescence), septin-9 (Sigma prestige, 1:1000 western blot, 1:100 immunofluorescnece), septin-7 (rabbit, H-120, Santa Cruz, 1:1000), EGFR (1:1000, NeoMarkers, Fremont, CA), ubiquitin (mouse, clone P4D1, 1:500, Santa Cruz), NSF (mouse, clone NSF-1, 1:1000, Millipore), Na,K-ATPase β subunit (mouse, 1:1000, Affinity Bioreagents, Golden, CO, USA), LAMP2 (rabbit, Sigma prestige, 1:50), PSMC-1 (rabbit, Sigma prestige, 1:50), caveolin-1 (mouse, 1:100 immunofluorescence, BD Transduction Laboratories, Franklin Lakes, NJ, USA), EEA-1 (rabbit, 1:100 immunofluorescence, Abcam, San Francisco, CA, USA), cofilin (rabbit, Abcam, Cambridge, MA, USA, 1:1000), CIN85 (clone 179.1.E1, mouse, Upstate Cell Signaling Solutions, Lake Placid, NY, 1:1000), green fluorescent protein (rabbit, Clontech, Mountain View, CA, USA, 1:1000), human influenza hemagglutinin (HA) (mouse, clone F-7, Santa Cruz, 1:1000). The following secondary antibodies were used: goat anti-rabbit horseradish peroxidase (American Qualex, San Clemente, CA, USA, 1:10,000), goat anti-mouse horseradish peroxidase (American Qualex, 1:10,000), clean blot horseradish peroxidase against rabbit (used for western blots on immunoprecipitated samples to avoid interference from antibodies, ThermoFisher, 1:3000), goat anti rabbit or goat anti mouse AlexaFluor 488 or 633nm (for immunofluorescence, ThermoFisher, 1:200).
Inhibitors, biotinylation, and cell lysis
HGE-20 cells were plated on transwells and allowed to reach confluency with a measureable transepithelial electrical resistance (TEER) [53]. Inhibitors were added to the apical and basolateral sides. The following inhibitors and concentrations were used: FCF 200 μM for HGE-20 cells (based on preliminary immunofluorescence data with septin antibodies, data not shown) and 100 μM for AGS cells (Sigma), cycloheximide 20 μg/mL (sigma), lactacystin 2.5 nM (sigma), batimastat (BB-94) 5 μM (Sigma), bafilomycin 50 nM (Sigma), geldanamycin 1 μM (Invivogen, San Diego, CA, USA), cathepsin B inhibitor CA074-me (Sigma) 100 μM. For experiments with cycloheximide, this was added 2 hours before other inhibitors. As a wide range of concentrations for lactacystin are reported in the literature, effectiveness of the 2.5 nM concentration, as demonstrated previously in other cell lines [11], was confirmed in HGE-20 cells by incubation with lactacystin, lysis, and western blot with ubiquitin antibodies (data not shown). After incubation with inhibitors, cells were washed twice with ice cold phosphate buffered saline containing 1mM MgCl2 and 1 mM CaCl2 (PBS++) on ice, then lysed with 50 mM Tris pH 7.5 containing 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate and Complete Protease Inhibitor Cocktail, 1 tablet/50 ml, (Roche Diagnostics, Indianapolis, IN, USA) at 4°C for 30 min. For samples intended for use in experiments where ubiquitylation would be studied, 10 mM iodoacetamide (Sigma) was added to lysis buffer. Cells were scraped from the plates and cell extracts were clarified by centrifugation (20,000 g, 15 min) at 4°C. For biotinylated samples, inhibitor incubations were as above. Samples were washed twice with ice cold PBS on ice, then biotin (EZ-Link Sulfo-NHS-SS-Biotin, ThermoFisher) at 1 mg/mL in PBS was added to the basolateral chamber for 30 minutes. Biotin was quenched by 2 × 15 minute incubations with 20 mM ammonium chloride. Cells were washed and lysed with 1% triton X-100 in PBS. Protein concentration was determined using BCA (Thermo). Biotinylated proteins were extracted with streptavidin-agarose (Sigma). Identical amounts of protein were incubated overnight with beads, washed x 3 with 0.5% triton X-100 in PBS, then extracted by 5 minutes boiling in 4% SDS, 20% glycerol, 1% β-mercaptoethanol in 0.1 M Tris pH 6.8.
Immunoprecipitation
HGE-20 cells were incubated with FCF or vehicle and lysed as described above (in the presence of 10 mM iodoacetamide when indicated). ErbB2 and septin-2 were immunoprecipitated from total cell lysates (1–2 mg protein) using 2 μl of appropriate antibody as described previously [54, 55]. Protein G or A-agarose suspension (Roche Diagnostics, Indianapolis, IN, USA) was used as appropriate. For successive immunoprecipitation reactions, Dynabeads Protein G or A (Life Technologies) were used. Adherent proteins were eluted from the beads by incubation in 35 μl SDS-PAGE sample buffer (4% SDS, 0.05% bromophenol blue, 20% glycerol, 1% β-mercaptoethanol in 0.1 M Tris pH 6.8) for 5 min at 80°C. Negative controls for immunoprecipitation included controls with no lysate to allow delineation of IgG bands from bands of interest and controls with beads, cell lysate and unrelated antibody or no antibody to assure there was no nonspecific binding to the beads.
Western blotting
Cell lysates, biotinylated protein, or immunoprecipitated samples were size fractionated by SDS-PAGE and transferred to nitrocellulose membranes for western blotting. Primary antibodies were used as described above. Secondary antibodies were horseradish peroxidase linked goat anti-mouse or goat anti-rabbit (1:10,000, American Qualex, San Clemente, CA, USA). Bands were detected using SuperSignal West Pico Chemiluminescence Kit (Thermo). Immunoblots were quantified by densitometry using Image Studio Software (LI-COR Inc., Lincoln, NE, USA). β-actin was used as a loading control for total lysates and Na,K-ATPase β subunit for biotinylated samples.
Determination of ErbB2 ubiquitylation levels
To determine the level of ubiquitylation, cells were lysed as above, in the presence of 10 mM iodoacetamide to preserve ubiquitinated forms by inhibiting de-ubiquitylating enzymes. Clarified lysate was used for immunoprecipitation with ErbB2 antibody as described above; 10 mM iodoacetamide was present in all washing buffers. Immunoprecipitated proteins were analyzed by Western blot using ubiquitin antibodies. Blots were stripped and re-probed using ErbB2 antibodies to ensure the increase in ubiquitylated forms was not due to unequal loading. To determine the level of ErbB2 ubiquitylation in the plasma membrane, cells were biotinylated from the basolateral side as described above. After lysis, ErbB2 was immunoprecipitated, proteins were eluted from the beads by boiling in 0.5% SDS in PBS for 5 min to denature proteins and disrupt protein-protein interactions. A portion of boiled eluates was diluted 34 times with 1% Triton X-100 in PBS, followed by a second immunoprecipitation with ErbB2 antibody and elution by boiling in 0.5% SDS in PBS for 5 min. Aliquots of eluted proteins after first and second immunoprecipitation were analyzed by western blot using antibodies against ubiquitin, ErbB2 and ErbB2 binding partners septin-9 and cofilin to confirm that the boiling procedure in SDS was sufficient to disrupt protein-protein interactions. Proteins eluted after the second immunoprecipitation were diluted 34 times with 1% Triton X-100 in PBS, followed by precipitation of the biotinylated ErbB2 on SA beads, elution and western blot analysis using antibodies against ubiquitin and ErbB2.
Nano-liquid chromatography with tandem mass spectrometry (nLC-MS/MS)
To analyze proteins co-immunoprecipitated with ErbB2 or septin-2 by nLC-MS/MS, immunoprecipitation was performed as described above. Proteins were eluted from the beads by incubation in 35 μL 0.5% sodium deoxycholate, 12 mM sodium lauryl sarcosine, and 50 mM triethylammonium bicarbonate for 5 min at 95°C. Bicinchoninic acid protein assay was used for quantitation. Proteins were sequentially reduced with 5 mM tris(2-caboxyethyl)-phosphine (final concentration) for 30 minutes at room temperature and alkylated with 10 mM iodoacetamide (final concentration) for 30 minutes at room temperature in the dark. Samples were diluted 1:5 (v/v) with 50 mM triethylammonium bicarbonate and enzymatically digested with 1:50 (protein:enzyme) trypsin (Promega, Madison, WI, USA) overnight at 37°C. Samples were acidified with 0.5% trifluoroacetic acid (final concentration). Sodium deoxycholate was phase-transferred using 1:1 (v/v) ethyl acetate, centrifuged at 16,000xg for 5 minutes at room temperature, and organic phase was decanted. Peptides were desalted and fractionated using C18-SCX StageTips [56] and analyzed by nLC-MS/MS with Collision Induced Dissociation (CID). This was performed on the Orbitrap XL mass spectrometer (Thermo) integrated with an Eksigent 2D nano-LC. A prepacked reverse-phase C18 75 μm x 20 cm column containing C18 5-μm particle-size, 300-Å pore-size resin (Acutech Scientific, San Diego, CA, USA) was used for peptide chromatography and subsequent CID analyses. ESI conditions using the nano-spray source (Thermo) for the Orbitrap were set as follows: capillary temperature of 210°C, tube lens 125 V and a spray voltage of 2.3kV. The flow rate for reverse-phase chromatography was 300 nL/min for loading and analytical separation (buffer A: 0.1% formic acid, 3% acetonitrile; buffer B: 0.1% formic acid, 100% acetonitrile). Peptides were resolved by a linear gradient of 3 40% buffer B over 180 min. The LTQ Orbitrap was operated in data-dependent mode with a full precursor scan at high-resolution (60000 at m/z 400) and twelve MS/MS experiments at low resolution on the linear trap while the full scan was completed. For CID, the intensity threshold was set to 500, where mass range was 350 2000. Dynamic exclusion was set to a repeat count of 1, repeat duration of 30s, and exclusion duration of 30s. Spectra were searched using Mascot software (v2.4, Matrix Science, UK) in which results with p<0.05 (95% confidence interval) were considered significant and indicating identity.
Immunofluorescent staining
HGE-20 cells were grown to confluency. Cells were incubated with FCF, inhibitors, or vehicle when applicable and fixed for 30 minutes with 3.7% formaldehyde in PBS at room temperature. Immunostaining was done as described previously [54, 55]. Staining was detected by confocal microscopy. Confocal images were acquired using the Zeiss LSM 510 laser scanning confocal microscope (Carl Zeiss MicroImaging GmbH, Germany) and analyzed using the ZEN 2009 software platform.
RNA extraction, cDNA construction, and qPCR
RNA was isolated from HGE-20 cells following 12 hour incubation with FCF or vehicle using a combination of the Trizol® method (ThermoFisher) and the RNeasy mini kit (Qiagen, Valencia, CA, USA) as described previously [57]. RNA concentration was measured on a nanodrop machine (Thermo Scientific) and RNA quality was confirmed on an Agilent Bioanalyzer using the RNA nano 600 kit (Agilent Technologies, Santa Clara, CA, USA). All RNA used in this study had an RNA integrity number of 10. The qScript one-step SYBR® green qRT-PCR kit was used for cDNA construction and qPCR (Quanta Biosciences, Gaithersburg, MD, USA). qPCR was completed on an Applied Biosystems 7500 thermo cycler (ThermoFisher) with annealing temperature of 60°. All samples (vehicle and FCF) were run in triplicate, and PCRs were run in triplicate with each sample for a total of 3 biologic replicates and 3 technical replicates for each condition. GAPDH was used as a housekeeping gene to normalize results. Results were analyzed using the comparative CT method [58]. Primers for qPCR were as follows: erbB2-s 5′-AATTCCAGTGGCCATCAAAG-3′; erbB2-as 5′-TTTCCCGGACATGGTCTAAG-3′; GAPDH-s 5′-ACCCAGAAGACTGTGGATGG-3′; GAPDH-as 5′-TTCTAGACGGCAGGTCAGGT-3′.
siRNA silencing of septin-2
Expression of septin-2 was knocked down in AGS cells using a mixture of two Ambion pre-designed siRNA duplexes: Sense: 5′-GAAAAUCGACUCUCAUAAATT-3′, Antisense: 5′-UUUAUGAGAGUCGAUUUUCCT-3′ (duplex 1) and Sense: 5′-CAAUCAAGUUCACCGAAAATT-3′, Antisense: 5′-UUUUCGGUGAACUUGAUUGGG-3′ (duplex 2) (Life Technologies). The Ambion Silencer® Negative Control No. 1 siRNA (Life Technologies) was used as a negative control. AGS cells in a 6-well plate (50% confluent) were transfected with siRNA using Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturers′ protocols. 48 hours after the first transfection, cells were transfected again with the mixture of the siRNA that was used in the first transfection. Cells were lysed 24 hours after the second transfection.
Results
ErbB2 interacts and co-localizes with septins
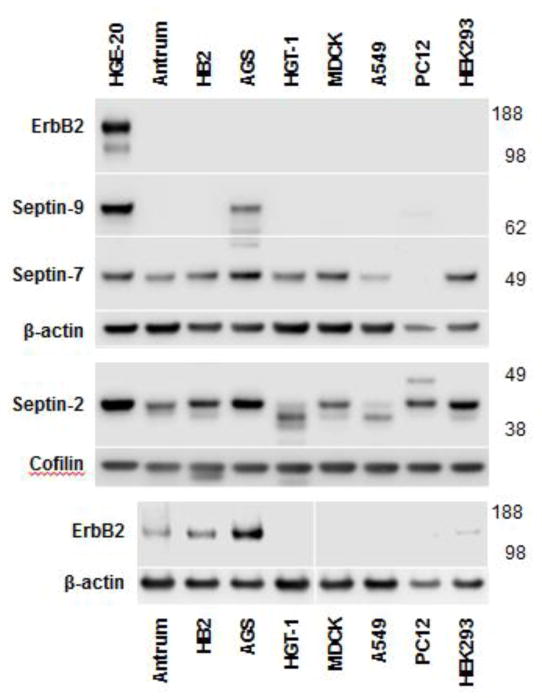
Expression levels of septins and ErbB2 were analyzed in various cell lines and human gastric antrum. ErbB2 was highly expressed in two gastric cancer cell lines, HGE-20 and AGS cells (Fig. 1), and also, as expected, in HB2 breast cancer cells [52]. These cells all demonstrated higher ErbB2 expression than human gastric antrum. ErbB2 was so highly expressed in HGE-20 cells compared to other cells tested that an additional gel needed to be run without the HGE-20 cell lysate to allow visualization of the ErbB2 band in other lanes. Septin-9 was found almost exclusively in HGE-20 and AGS cells and was not seen in normal human gastric antrum. Both septin-2 and septin-7 had increased expression in the gastric cancer cells but were also detected in gastric antrum and other cell lines (Fig. 1).
Figure 1. ErbB2 and septin-9 are highly expressed in gastric cancer cells.
ErbB2, septin-9, septin-2, and septin-7 expression were determined by western blot analysis of total lysates of various cell lines and human gastric antrum, β-actin was used as a control. Cofilin was used as a control for septin-2 given the size similarity between septin-2 and β-actin. ErbB2 is very highly expressed in HGE-20 gastric cancer cells, to the extent that the band is not visible in other cell types with reasonable exposure of the HGE-20 cell band. Once HGE-20 cells were removed from the ErbB2 blot, expression was also seen in human gastric antrum, HB2 breast cancer cells, and AGS gastric cancer cells. Septin-9 similarly showed increased expression in gastric cancer cells compared to other cell types.
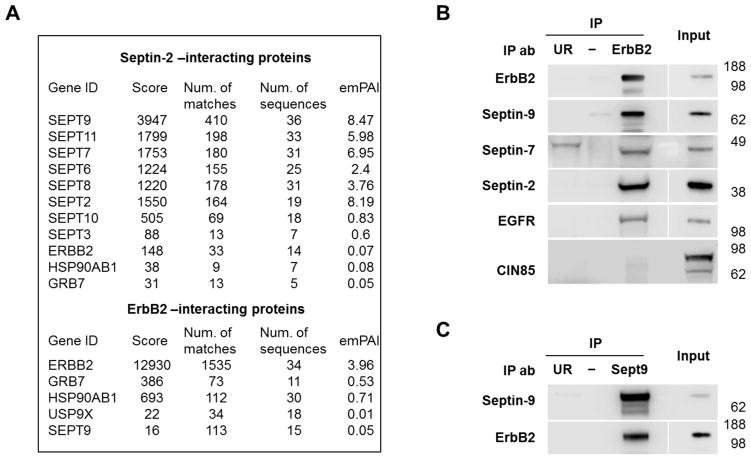
To determine whether ErbB2 and septins interact with each other, proteins that co-immunoprecipitate with septin-2 or with ErbB2 were analyzed in HGE-20 cells, which have the highest expression level of ErbB2 and septins (Fig. 1). Analysis of septin-2 immunoprecipitate by mass spectrometry revealed ErbB2 and two known ErbB2-interacting partners, GRB7 and Hsp90 [59–61] (Fig. 2A). As expected, immunoprecipitation of septin-2 also resulted in co-precipitation of several other septin proteins (Fig. 2A), which raises the possibility of ErbB2 interaction with any of these septin proteins rather than specific interaction with septin-2 alone. Mass spectrometry analysis of the proteins co-immunoprecipitated with ErbB2 identified septin-9, ErbB2 itself, and other known ErbB2-binding partners, GRB7, Hsp90, and USP9x [37] (Fig. 2A). Western blot analysis using specific antibodies confirmed the presence of septin-9 and also demonstrated the presence of septin-2 and septin-7 in the ErbB2-immunoprecipitates (Fig. 2B). As expected, EGFR, a known dimerization partner of ErbB2, was also seen in the ErbB2 immunoprecipitates. CIN85, an adapter protein for the ubiquitylating enzyme c-cbl that is known to mediate the interaction between EGFR and septin-9 [21], was not seen in ErbB2 immunoprecipitates (Fig. 2B). This suggests a different mechanism of interaction between ErbB2 and septins as compared to EGFR. Immunoprecipitation of HGE-20 cell lysate using septin-9 antibodies confirmed interaction with ErbB2 (Fig. 2C). ErbB2 was localized almost exclusively to the basolateral plasma membrane domain in HGE-20 cell monolayers (Fig. 3A), co-localizing with septin-2 and septin-9 (Fig. 3B and Fig. 5B, left panels). Coordinated over-expression of ErbB2 and septins in gastric cancer cell lines, as well as their interaction and co-localization, suggest a possible functional importance of septin-ErbB2 complexes in gastric cancer.
Figure 2. ErbB2 interacts with septins in HGE-20 gastric cancer cells.
Proteins immunoprecipitated using antibodies against septin-2 or ErbB2 from HGE-20 cell lysates were analyzed by mass spectrometry. ErbB2, known ErbB2-associated proteins, were detected in septin-2 immunoprecipitate and septin-9 was detected in ErbB2 immunoprecipitate (A). Interaction of ErbB2 with septins and a known dimerization partner, EGFR, was confirmed by western blot, following immunoprecipitation of ErbB2 in HGE-20 cell lysate. Ubiquitin ligase c-cbl binding protein, CIN85, a protein that mediates the interaction between septin-9 and EGFR, was not seen in ErbB2 IP (B). Interaction between septin-9 and ErbB2 was again confirmed by western blot following IP of HGE-20 cell lysates with septin-9 antibody (C). emPAI-exponentially modified protein abundance index, UR-unrelated antibody negative control (anti-human influenza hemagglutinin for ErbB2 IP and anti-green fluorescent protein for septin-9 IP).
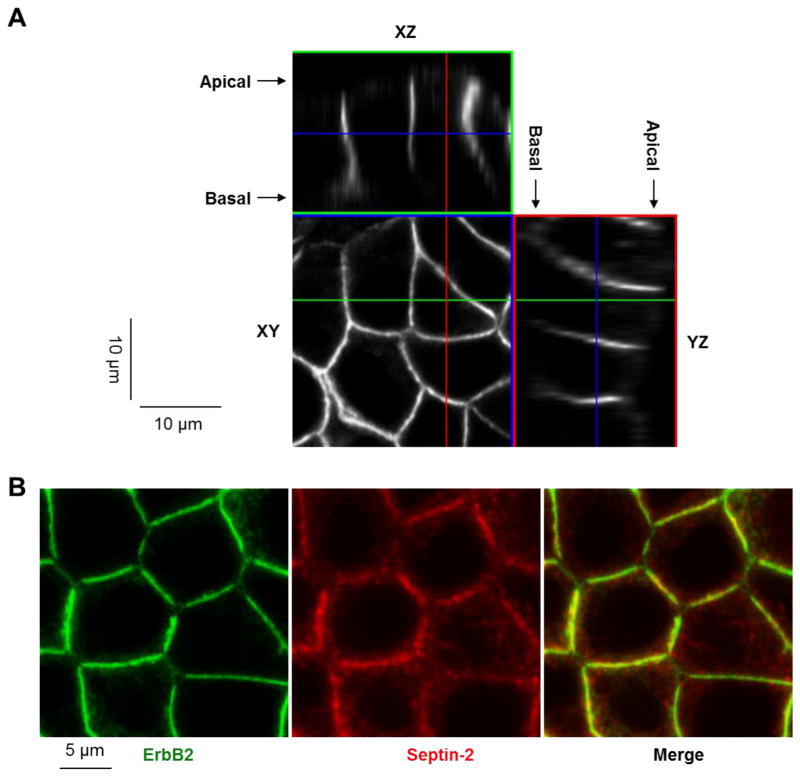
Figure 3. ErbB2 is localized to the plasma membrane and co-localizes with septins.
ErbB2 protein is localized mainly to the basolateral membrane of HGE-20 cells as shown by confocal microscopy (A). ErbB2 and septin-2 co-localize at the lateral membrane in HGE-20 cells, as shown by immunofluorescence using antibodies against ErbB2 and septin-2 (B).
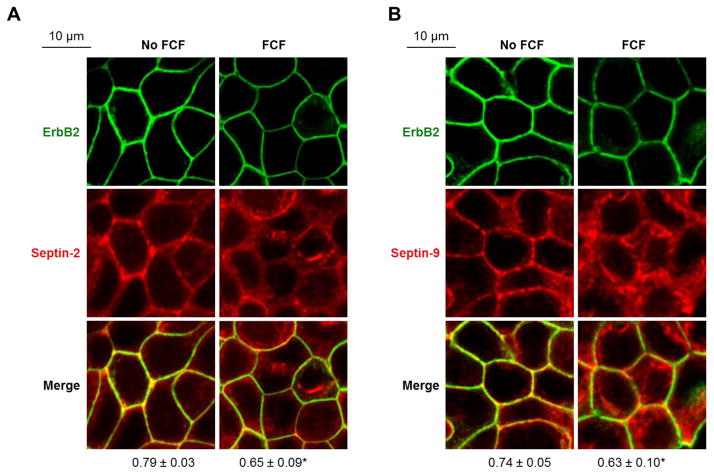
Figure 5. FCF decreases co-localization and co-immunoprecipitation of ErbB2 with septins.
HGE-20 cells were incubated with FCF or vehicle for 12 hours, followed by fixation and immunofluorescence. The images show an altered distribution of septin-2 (A) and septin-9 (B) and a decrease in co-localization between septins and ErbB2 (A-B) in the presence of FCF. Coefficients of co-localization as calculated using Zen 2009 software are shown below the images. Errors bars represent s.d., n=3 independent experiments, at least 12 fields were analyzed for each condition; *-significant difference from no FCF control, P <0.05, Student’s t-test.
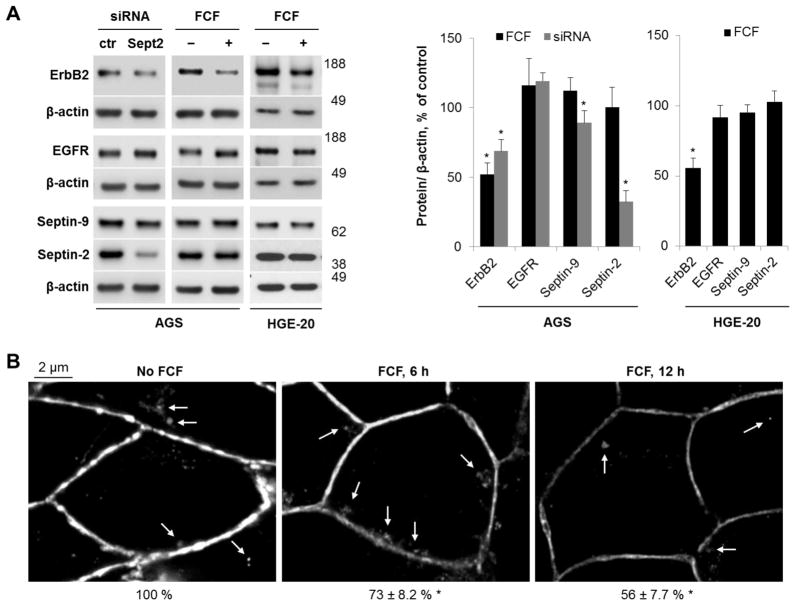
Disruption of septin organization by knockdown of septin-2 or cell exposure to FCF decreases ErbB2 levels in gastric cancer cells
Septin expression was silenced by transient transfection with septin-2-specific siRNA to investigate the significance of the interaction between septins and ErbB2. Septin-2 was chosen for silencing because it is the major ubiquitously expressed septin, and down-regulation of septin-2 affects both levels and intracellular distribution of other septins, including septin-7 and septin-9 [62]. AGS cells were used for these experiments due to better efficiency of transfection as compared to HGE-20 cells. Transient transfection with septin-2-specific siRNA decreased septin-2 levels by ~70%, demonstrating that silencing was effective. Septin-9 levels were also moderately decreased (Fig. 4A), presumably due to posttranslational down-regulation, consistent with known coordinated regulation of levels of individual septins [63]. Septin knockdown resulted in an almost 2-fold decrease in ErbB2 protein without affecting EGFR levels (Fig. 4A).
Figure 4. siRNA silencing and interruption of septin dynamics with FCF decrease ErbB2 levels in gastric cancer cells.
AGS cells were transiently transfected with septin-2-specific or control (ctr) siRNA or AGS and HGE-20 cells were incubated with FCF for 12 hours. Western blot was performed on clarified lysates. ErbB2 protein levels were decreased with siRNA silencing of septin-2. Use of FCF led to a similar decrease in ErbB2 protein in both cell lines. The structurally similar EGFR was not affected, demonstrating specificity of response. β-actin was used as a loading control (A). Immunofluroescence with ErbB2 antibodies in HGE-20 cells showed accumulation of ErbB2 in intracellular compartments following 6 hour incubation with FCF, with overall decreased expression and minimal intracellular appearance of ErbB2 at 12 hours, suggestive of intracellular degradation (B). Arrows in the images point to intracellular accumulation of ErbB2. Numbers below images indicate the intensity of total fluorescence as % of control as calculated by analyzing at least 10 microscopic fields for each condition using Zen 2009 software. n=3 independent experiments, errors or error bars represent standard deviation (s.d.), *-significant difference from condition without siRNA or FCF, p<0.05, Student’s t-test.
As cytoskeletal proteins, septins constantly undergo dynamic re-organization of hetero-oligomeric complexes, which can be interrupted by FCF, a compound known to specifically inhibit assembly-disassembly of septin oligomers without affecting actin or tubulin dynamics [64]. Previous data showing similar effects of FCF and septin silencing on various cellular processes, including mitosis and cell migration [64], protein stability [11], glucose transport [65], store-operated Ca2+ influx [66], ciliogenesis [63], epithelial junctions [67], gastrulation during embryogenesis [68], and bacterial invasion [69], demonstrate that FCF is a valuable tool to interfere with septin dynamics. Exposure of AGS and HGE-20 cells to FCF for 12 hours resulted in about a 2-fold decrease in ErbB2 protein levels in both cell lines (Fig. 4A). However, FCF did not alter levels of EGFR (Fig. 4A), the β1 subunit of the highly abundant basolateral plasma membrane protein, Na,K-ATPase (data not shown), or a cytosolic septin-interacting protein, NSF [62] (data not shown), showing the effect of FCF is specific to ErbB2 in both cell lines. There was no difference in the levels of septin-2 or septin-9 with and without the inhibitor (Fig. 4A), demonstrating that the effect of FCF was not due to changes in the expression levels of septins. The results demonstrate that interference with septin biosynthesis or oligomerization leads to a targeted decrease in ErbB2 levels in two distinct gastric cancer cell lines.
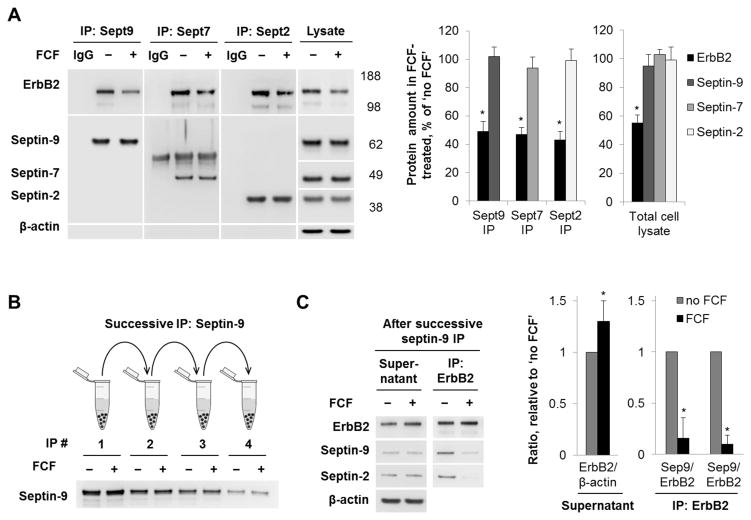
Immunofluorescence of ErbB2 in HGE-20 cells exposed to FCF for 6 or 12 hours showed a stepwise decrease in the ErbB2 expression at the plasma membrane (Fig. 4B). A minor intracellular accumulation of ErbB2 was seen after 6 hours but was scarcely detected after 12 hours (Fig. 4B, arrows). As expected, incubation of HGE-20 cells with FCF altered cellular distribution of septins by decreasing association of both septin-2 and septin-9 with the plasma membrane and resulting in a more diffuse distribution of the proteins in the cytoplasm (Fig. 5A and 5B). As a result, the degree of co-localization between ErbB2 and septin proteins was decreased in the presence of FCF (Fig. 5A and 5B). Cell exposure to FCF for 12 hours decreased the amount of co-precipitated ErbB2 with septin-9, septin-7 and septin-2 by 51%, 53% and 57 %, respectively (Fig. 6A). The diminished association of septins with ErbB2 is likely to reflect the FCF-induced decrease in the levels of ErbB2 and septins at the plasma membrane (Fig. 5), with no effect on total levels of septins (Fig 6A and Fig. 4). The data might suggest that FCF impairs the association of septins with the membrane and membrane-resident ErbB2, resulting in decreased stability of ErbB2 in the plasma membrane, followed by its internalization and degradation. To explore such a possibility, we depleted septin-9 and hence septin-9-bound ErbB2 to determine whether FCF-induced mislocalization of septins increases the amount of cytosolic septin-free ErbB2. This was achieved by successive septin-9 immunoprecipitation performed 4 times. After each step, the supernatant was used for the next immunoprecipitation. Septin-9 was partially depleted by the fourth immunoprecipitation, as shown by western blot (Fig. 6B). The level of ErbB2 was greater in the final supernatant from FCF-treated cells as compared to control, indicating that FCF increases the amount of septin-unbound ErbB2 (Fig. 6C). When ErbB2 was immunoprecipitated from this final supernatant, septin-9 and septin-2 were co-precipitated in a control sample but hardly were seen in a sample from FCF-treated cells (Fig. 6C), further supporting the conclusion that FCF increases the amount of septin-unbound ErbB2. Collectively, the results suggest that association with septins is important for stable plasma membrane expression of ErbB2.
Figure 6. FCF decreases the interaction between septins and ErbB2.
HGE-20 cells were incubated with FCF for 12 hours, followed by immunoprecipitation with antibodies against septin proteins. ErbB2 was decreased in septin immunoprecipitates and cell lysates in the presence of FCF, while septin protein levels were unchanged. IgG refers to immunoprecipitation procedure without cell lysate (A). HGE-20 cells were incubated with or without FCF for 12 hours and lysed. The level of septin-9 was depleted in HGE-20 cell lysates by 4 consecutive immunoprecipitations with septin-9 antibodies, each using the supernatanat of the prior preparation. The level of septin-9 was clearly depleted by the 4th immunoprecipitation (B). Western blot analysis of the final supernatant from (B) detects more ErbB2 in a sample from FCF-treated cells as compared to control. Western blot analysis of ErbB2 immunoprecipitates from the final supernatant from (B) shows less septin-2 and septin-9 bound to ErbB2 in the sample from FCF-treated cells (C). n=3 independent experiments, error bars represent s.d., *-significant difference from no FCF condition, P<0.05, Student’s t-test.
FCF induces ubiquitylation of ErbB2 at the plasma membrane
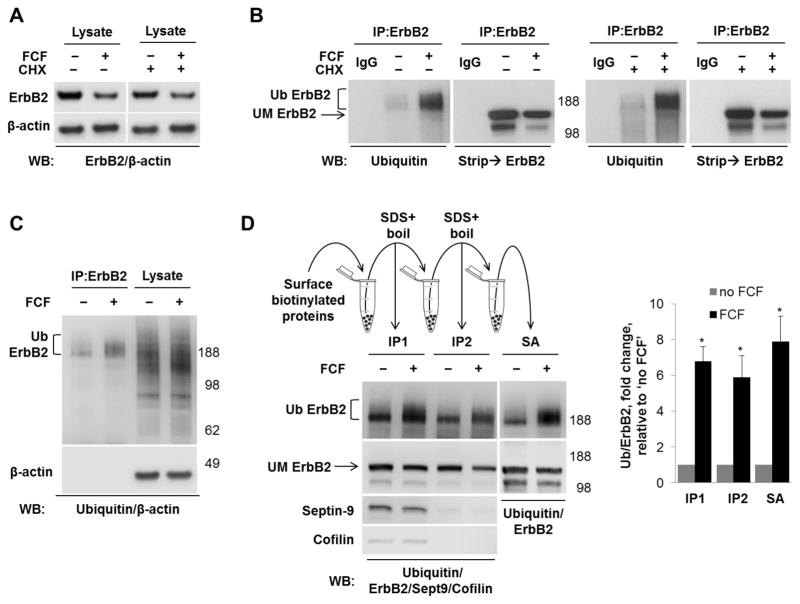
The decrease in ErbB2 in the presence of FCF could be due to decreased transcription, decreased biosynthesis of ErbB2 or increased degradation of the mature ErbB2. qPCR showed the fold change of ErbB2 gene expression in cells treated with FCF compared to untreated controls was minor, 0.84 ± 0.059 (mean ± standard deviation, n=3). A similar FCF-induced decrease in ErbB2 levels observed in the absence and in the presence of the inhibitor of protein synthesis, cycloheximide (Fig. 7A), indicates that FCF increases the rate of degradation of the mature ErbB2. To determine whether FCF-induced degradation of ErbB2 is related to ubiquitylation, ErbB2 was immunoprecipitated from HGE-20 cells after incubation with or without FCF for 12 hours. Western blot analysis using antibodies against ubiquitin detected a diffuse band above 188 kDa that significantly increased in intensity in the FCF-exposed sample, while the amount of immunoprecipitated ErbB2 was decreased in this sample, as shown by stripping and re-probing the same blot with ErbB2 antibodies (Fig. 7B). These results suggest that FCF induces ubiquitylation of ErbB2. The effect of FCF was unaffected by inhibition of protein synthesis by cycloheximide (Fig. 7B), showing that FCF-induced ubiquitylation of ErbB2 is not related to ER-associated degradation of misfolded, newly synthesized ErbB2 and suggesting that it is the mature ErbB2 that is ubiquitylated. The lack of increased ubiquitin signal in total cell lysates (Fig. 7C) demonstrates that the effect of FCF is specific to ErbB2. However, this does not exclude the possibility that ErbB2-interacting partner(s) present in the immunoprecipitated fraction rather than ErbB2 itself is (are) ubiquitylated.
Figure 7. FCF induces ubiquitylation of ErbB2 at the plasma membrane.
There was no difference in FCF-induced decrease in ErbB2 in the presence or absence of cycloheximide, as shown by western blot of total HGE-20 cell lysate, suggesting the effect of FCF is on mature ErbB2 protein (A). Incubation with FCF or vehicle was completed for 12 hours with or without cycloheximide, followed by immunoprecipitation with ErbB2 antibodies. Western blot using ubiquitin antibodies, followed ErbB2 antibodies, showed increased ubiquitylation of ErbB2 in the presence of FCF, independent of new protein synthesis (B). Ubiquitylation induced by FCF is specific to ErbB2, as shown by western blot comparison of ErbB2 immunoprecipitate and total cell lysate using ubiquitin antibodies; β-actin was used as a loading control (C). Cells were incubated with FCF or vehicle, surface proteins were biotinylated from basolateral side, followed by immunoprecipitation with ErbB2 antibodies (IP1), then boiling of eluted proteins in SDS to remove interacting proteins, followed by dilution in non-ionic detergent and repeat immunoprecipitation with ErbB2 antibodies (IP2). Loss of septin-9 and cofilin signals on western blot in IP2 confirms dissociation of ErbB2 from interacting proteins. Increased ubiquitylation of ErbB2 is still seen in the absence of interacting proteins, demonstrating that ErbB2 itself is ubiquitylated. Proteins eluted from the second IP by boiling in SDS were diluted in non-ionic detergent and biotinylated ErbB2 was isolated by streptavidin extraction (SA). As shown by western blot with ubiquitin antibodies, the increased ubiquitylation of ErbB2 induced by FCF is seen in this fraction, demonstrating ubiquitylation occurs at the plasma membrane. The graph shows the FCF-increased ubiquitylation seen in each experiment as compared to control (D). Ub-ubiquitylated ErbB2, UM-unmodified ErbB2, IgG-immunoprecipitation control without cell lysate, SA-streptavidin.
To determine if ErbB2 itself or interacting partners were ubiquitylated, proteins were immunoprecipitated using ErbB2 antibodies, eluted from the beads with SDS, boiled to denature proteins and disrupt protein-protein interactions, and re-precipitated with anti-ErbB2 antibodies. As seen from the loss of co-precipitated septin-9 after the second immunoprecipitation (Fig. 7D, compare IP1 and IP2, left lower panels), boiling of eluted proteins in SDS was sufficient to separate ErbB2 from its interacting partners. This conclusion was also supported by the detection of cofilin, another ErbB2-interacting protein identified by MS-MS (data not shown) in the ErbB2-immunoprecipitated fraction before (Fig. 7D, IP1), but not after boiling of samples in SDS (Fig. 7D, IP2). Importantly, disruption of protein-protein interactions did not abolish the FCF-induced increase in ubiquitin signal (Fig. 7D, left top panel), indicating that ErbB2 itself was ubiquitylated. An increase in ubiquitin signal in the presence of FCF was also observed when surface biotinylated ErbB2 was precipitated with streptavidin beads (SA) from the interactor-free fraction of ErbB2 (IP2) (Fig. 7D, right panels), demonstrating that the plasmalemma-resident ErbB2 was ubiquitylated. Quantification of the results showed a similar FCF-induced increase in ubiquitin signal in plasmalemma-located ErbB2 (SA) and total ErbB2 isolated before (IP1) or after disruption of protein-protein interactions (IP2) (Fig. 7D, bar graph), indicating that FCF induces ubiquitylation of ErbB2 at the basolateral plasma membrane.
Ubiquitylated forms of ErbB2 are protected by degradation inhibitors CA074-me and lactacystin
Previous data suggested the involvement of both lysosomes and proteasomes in ErbB2 degradation [34, 37, 48, 70, 71]. In addition, cleavage of the extracellular domain of ErbB2 by matrix metalloproteinases (MMPs) is critical for its subsequent targeting for degradation (reviewed in [47]). To determine whether inhibition of any of these degradation pathways prevents FCF-induced decrease in ErbB2 or affects ErbB2 ubiquitylation, lysosomal cathepsin B inhibitor CA074-me, proteasome inhibitor lactacysin, lysosomal H+-ATPase inhibitor bafilomycin, or MMP inhibitor BB-94 were added to cells incubated with or without FCF for 6 hours. Basolateral biotinylation was performed prior to cell lysis as indicated.
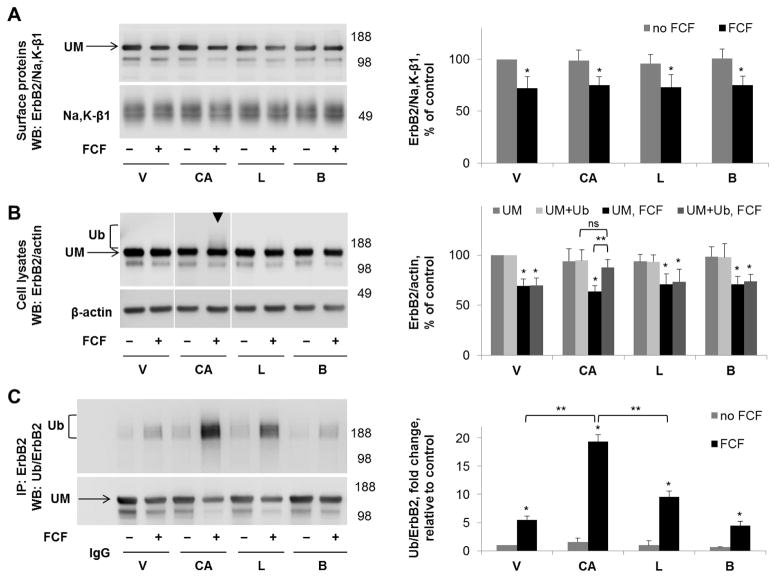
None of the inhibitors prevented or diminished the FCF-induced reduction in the intensity of the main ErbB2 band in the plasma membrane or cell lysates (Fig. 8A,B). In cell lysates, but not biotinylated samples, the presence of CA074-me led to an increased density above the ErbB2 band on western blot (Fig. 8B, arrowheads), suggesting that the inhibitor prevents degradation of FCF-induced ubiquitylated forms of ErbB2. In the presence of CA074-me, in total cell lysates, there was no significant difference in total ErbB2 protein levels (combined density of ErbB2 bands corresponding to unmodified and ubiquitylated fractions) between control and FCF-exposed samples, indicating that the cathepsin B inhibitor rescued ErbB2 from FCF-induced lysosomal degradation. Immunoprecipitation with ErbB2 antibody followed by western blot with anti-ubiquitin antibody confirmed that FCF-induced ubiquitylated forms of ErbB2 were protected by CA074-me and showed a similar, but lesser, protective effect of lactacystin (Fig. 8C). These results demonstrate that even though FCF-induced ubiquitylation of ErbB2 occurs at the plasma membrane, degradation of ubiquitylated ErbB2 occurs in intracellular compartments and is dependent on cathepsin B activity and possibly also on proteasomal activity. The inhibitor of H+-ATPase, bafilomycin, did not have the same effect, indicating that limiting acidification of lysosomes does not inhibit the action of cathepsin B. The MMP inhibitor BB-94 did not protect ErbB2 from the effects of FCF (data not shown). There was no change in the results of any of the above experiments in the presence of cycloheximide (data not shown). Taken together, these results suggest that FCF-induced ubiquitylation of ErbB2 at the plasma membrane triggers endocytosis of ubiquitylated forms, followed by intracellular degradation.
Figure 8. Cathepsin B inhibitor increases the amount of FCF-induced ubiquitylated forms of ErbB2 in cell lysates but not in the plasma membrane fraction.
HGE-20 cells were incubated with the indicated inhibitors for 6 hours, followed by basolateral biotinylation of surface proteins, streptavidin extraction, and western blot using anti-ErbB2 antibodies. Na+,K+-ATPase β1 subunit was used as a loading control. ErbB2 protein levels in the membrane fraction were not protected by any of the indicated inhibitors in the presence of FCF (A). In total cell lysates, ubiquitylated ErbB2, seen as an increased density running above the main ErbB2 band (arrowhead), was increased in the presence of cathepsin B inhibitor CA074-me (B). ErbB2 was immunoprecipitated, followed by western blot using anti-ubiquitin, then anti-ErbB2 antibodies, confirming a significant increase in ubiquitylated forms of ErbB2 in the presence of CA074-me and also showing a moderate increase in the presence of lactacystin (C). Quantification for each blot is shown to the right. Error bars, s.d., n=4 independent experiments, statistics done by Student’s t-test, ns-not significant, * - significant difference from the no-FCF control, P<0.05, ** - significant difference between indicated conditions, p<0.05, CA-CA074-me, L-lactacystin, B-bafilomycin, V-vehicle, Ub-ubiquitylated form of ErbB2, UM-unmodified form of ErbB2.
FCF induces ErbB2 internalization followed by trafficking to lysosomes
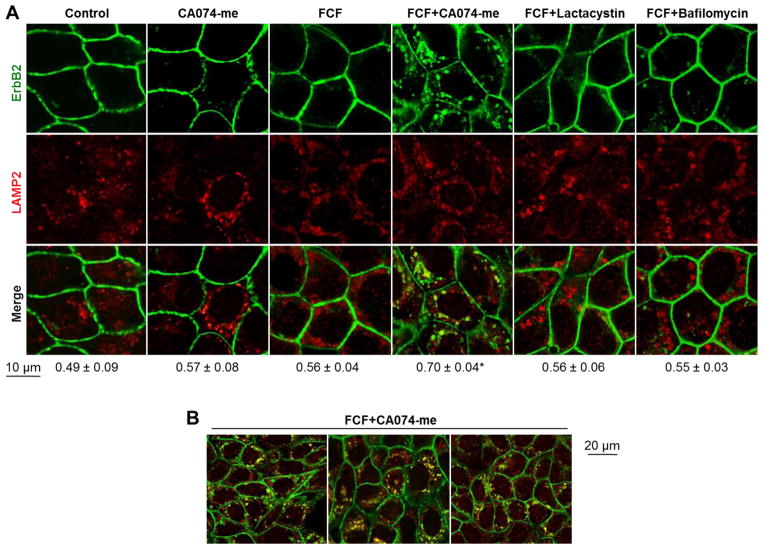
To determine whether FCF induces lysosomal degradation of ErbB2, immunofluorescence was performed using antibodies against ErbB2 and lysosomal protein LAMP2 after cell exposure to FCF and the inhibitors of degradation. The cathepsin B inhibitor CA074-me significantly increased accumulation of ErbB2 in lysosomes upon incubation with FCF (Fig. 9A,B). In conjunction with the data on the protective effect of CA074-me on FCF-induced ubiquitylated forms of ErbB2 in cell lysates (Fig. 8B), these results suggest that FCF induces ErbB2 internalization, followed by lysosomal degradation. Lysosomal accumulation of ErbB2 was not seen when cells were incubated with CA074-me and without FCF (Fig. 9A). The lysosomal acidification inhibitor bafilomycin did not increase accumulation of ErbB2 in lysosomes in the presence of FCF, in agreement with the lack of protective effect of bafilomycin on FCF-induced ubiquitylated forms of ErbB2 (Fig. 8A,B). As expected, the proteasome inhibitor lactacystin did not increase accumulation of ErbB2 in lysosomes. Also, co-localization of ErbB2 and proteasomal protein PSMC1 (26s subunit) was not seen following incubation with lactacystin in the presence or absence of FCF (data not shown). These results indicate ubiquitylated forms of ErbB2 are degraded in lysosomes, but not proteasomes. The moderate protective effect of lactacystin on ubiquitylated forms of ErbB2 (Fig. 8C) suggests that ErbB2 endocytosis and/or trafficking to lysosomes requires proteasomal activity.
Figure 9. Incubation of HGE-20 cells with FCF and cathepsin B inhibitor leads to accumulation of ErbB2 in lysosomes.
HGE-20 cells were incubated with the indicated inhibitors for 6 hours, then fixed. Immunofluorescence was performed using antibodies against ErbB2 and the lysosomal protein LAMP2. There was an increase in co-localization of ErbB2 with lysosomes in the presence of FCF and cathepsin B inhibitor CA074-me. Coefficient of overlap between ErbB2 and LAMP2 as calculated using Zen 2009 software with the confocal microscope is shown below the images (A). Low magnification images of HGE-20 cells incubated with FCF and CA074-me show that lysosomal accumulation of ErbB2 occurs in the majority of cells in a cell monolayer (B). Error bars, s.d., n=3 independent experiments, at least 12 fields were analyzed for each condition, * - significant difference from no-FCF condition, P = 0.0007, Student’s t-test.
To determine whether FCF induces accumulation of ErbB2 in endocytic compartments en route to lysosomes, immunofluorescence was performed using antibodies against the marker of early endosomes, EEA-1, and caveolae marker, caveolin-1. Minor co-localization of ErbB2 with EEA-1 or caveolin-1 was found not only in the presence, but also in the absence of FCF (Fig. Supp. 1, arrows), suggesting no or minor involvement of classical clathrin-dependent or caveolae-mediated pathways in FCF-induced internalization of ErbB2.
FCF augments the effect of geldanamycin on ErbB2 and works via a distinct mechanism
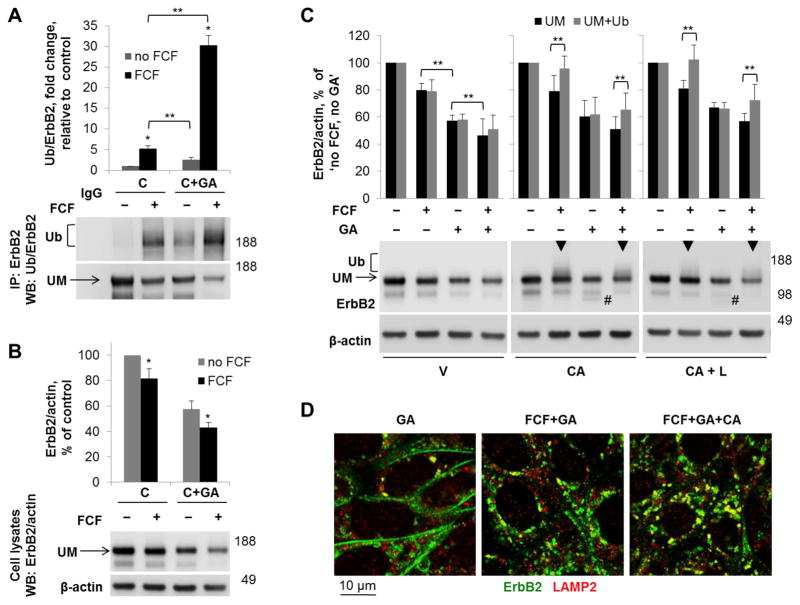
Geldanamycin induces ubiquitylation and degradation of ErbB2 via inhibition of Hsp90, which stabilizes ErbB2 at the plasma membrane [71]. Geldanamycin was used in combination with FCF to determine if the effects were additive. Cells were incubated with or without FCF or geldanamycin as indicated for 6 hours, and ubiquitylation and total levels of ErbB2 were determined. To study the effect of the inhibitors on the mature rather than newly synthesized ErbB2, all experiments were performed in the presence of cycloheximide. In agreement with previous data [49, 71], geldanamycin increased ubiquitylation and decreased the total amount of ErbB2 (Fig. 10A, B). However, the extent of ubiquitylation in the presence of geldanamycin was significantly lower as compared to that in the presence of FCF. Moreover, ubiquitylation was greatly augmented in the presence of both geldanamycin and FCF (Fig. 10A). Similarly, the effect of FCF in combination with geldanamycin on the total amount of ErbB2 was greater than that of either agent alone both in the presence and absence of cycloheximide (Fig. 10B and Fig. 10C, left panels).
Figure 10. FCF augments the effect of Hsp90 inhibitor geldanamycin on ubiquitylation and degradation of ErbB2 and works via a distinct pathway.
HGE-20 cells were incubated for 6 hours with cycloheximide and with or without FCF and geldanamycin. ErbB2 was immunoprecipitated, followed by western blot with antibodies against ubiquitin, and then ErbB2. Ubiquitylation of ErbB2 in the presence of FCF and geldanamycin was greater than with either agent alone (A). Western blot of total cell lysates obtained following the same incubation conditions demonstrates an additive effect of both agents on the level of mature ErbB2. β-actin was used as a loading control (B). HGE-20 cells were incubated with FCF and/or geldanamycin and the indicated inhibitors of protein degradation for 6 hours, followed by western blot using antibodies against ErbB2, with β-actin as a loading control. FCF-induced ubiquitylated forms of ErbB2, shown as an increased density above the main band (arrowheads), are increased by CA074-me in the presence and absence of geldanamycin. CA074-me protected geldanamycin-induced ErbB2 degradation products (indicated by # on the blot) independent of FCF. Lactacystin did not augment the effects of CA074-me on the FCF or geldanamycin-induced changes, demonstrating that proteasomes are not directly involved in ErbB2 degradation (C). HGE-20 cells were incubated with the indicated inhibitors for 6 hours, fixed, and immunofluorescence was completed using antibodies against ErbB2 and LAMP2. Co-localization of ErbB2 with lysosomes was increased in the presence of both FCF and geldanamycin (D). Error bars, s.d., n=3 independent experiments, * - significant difference from no-FCF condition, Student’s t-test, P<0.05, ** - significant difference between indicated conditions, Student’s t-test, P<0.05, C-cycloheximide, GA-geldanamycin, UM-unmodified ErbB2, V-vehicle, CA-CA074-me, L-lactacystin.
Cell incubations with FCF and/or geldanamycin were repeated in the presence of CA074-me or lactacystin. CA074-me protected FCF-induced ubiquitylated forms of ErbB2 in the presence or absence of geldanamycin, as indicated by the increased density above the main ErbB2 band in cell lysates (Fig. 10C, arrowheads). In contrast, CA074-me did not cause an increase in the density above the main band in geldanamycin-treated samples. Instead, a degradation product was detected when cells were incubated with geldanamycin and CA074-me (indicated by # in Fig. 10C), suggesting that this form is protected by CA074-me. Lactacystin did not augment the effects of CA074-me on cells incubated with FCF or geldanamycin, demonstrating that neither FCF nor geldanamycin induce proteasomal degradation of ErbB2, but not excluding the possibility that proteasomal activity is needed for ErbB2 endocytosis and/or trafficking to lysosomes. Internalization of ErbB2 and its minor co-localization with lysosomes following incubation with geldanamycin was confirmed by immunofluorescence (Fig. 10D). The presence of FCF in combination with geldanamycin increased both internalization and co-localization with lysosomes, and co-localization with lysosomes was further increased in the presence of CA074-me (Fig. 10D). Therefore, both FCF and geldanamycin induce ubiquitylation, internalization and lysosomal degradation of ErbB2. However, the pathways are distinct and effects are additive.
Discussion
The data presented here identify septins as interacting partners and regulators of the persistent expression of an important oncoprotein, ErbB2. The results demonstrate that septins stabilize ErbB2 at the plasma membrane in gastric cancer cells, and interference with septin dynamics leads to ubiquitylation of ErbB2 at the plasma membrane, followed by internalization and intracellular degradation mainly in lysosomes. Both siRNA knockdown of septin-2/septin-9 and inhibition of septin filament assembly-disassembly with FCF induce ErbB2 degradation (Fig. 4A), confirming that degradation of ErbB2 is a result of interference with septin dynamics rather than an unknown, alternate target of FCF [72]. By re-organizing septin oligomers, FCF disrupts ErbB2-septin complexes at the plasma membrane and induces internalization and degradation of ErbB2. The effect is specific to ErbB2 since other cytosolic and membrane proteins, and, more importantly, the closely related EGFR, are not affected. Impairment of septin dynamics induces ubiquitylation and degradation of the mature plasmalemma-resident ErbB2 rather than the intracellular, newly synthesized receptor as evident from identical effects in total cell lysates and plasma membrane fractions, both in the absence and in the presence of cycloheximide. The FCF-induced increase in the ubiquitylation of ErbB2 is not prevented by removal of ErbB2-interacting proteins by boiling samples prior to immunoprecipitation, confirming that it is ErbB2 itself that is modified with ubiquitin chains in the presence of FCF (Fig. 7).
Ubiquitylation can trigger degradation of plasma membrane proteins either directly at the membrane, or by inducing endocytosis followed by intracellular degradation [47, 73, 74]. The data presented here indicates that FCF induces degradation of ErbB2 by the latter mechanism. In the presence of FCF, the specific inhibitor of the lysosomal protease cathepsin B, CA074-me, prevented degradation of ubiquitylated, but not unmodified, ErbB2 and resulted in accumulation of ErbB2 in lysosomes. A CA074-me-induced increase in ubiquitylated ErbB2 was observed in total cell lysates and ErbB2-immunoprecipitates but not in the plasma membrane fraction (Fig. 8). These results demonstrate that FCF-induced ubiquitylated forms of ErbB2 are selectively internalized and delivered to lysosomes, where they are degraded by intra-lysosomal proteases, including cathepsin B. FCF-induced ubiquitylated forms of ErbB2 were also protected, though to a lesser extent, by lactacystin. However, no accumulation of ErbB2 in proteasomes was observed, and no additional accumulation was found when lactacystin was added together with CA074-me as compared to CA074-me alone. Taken together, these findings suggest proteasomes are not directly involved in FCF-induced degradation of ErbB2, but proteasomal activity appears to be necessary for endocytosis and/or sorting of ErbB2 to lysosomes. Proteasomal activity is required for efficient trafficking of EGFR to lysosomes via multivesicular bodies [75], so a similar process could be involved with ErbB2 degradation pathways.
The H+-ATPase inhibitor bafilomycin had no effect on FCF-induced ubiquitylation, degradation or lysosomal trafficking of ErbB2. Bafilomycin and CA074-me are both considered inhibitors of lysosomal degradation, but their mechanism of action is different. The proteolytic activity of lysosomes is generally dependent on acidic pH, and bafilomycin raises the pH via inhibition of the H+-ATPase. In addition, bafilomycin-induced increase in pH inside EEA-1-positive early endosomes prevents lysosomal trafficking of proteins internalized by clathrin-mediated endocytosis [76]. The protective effect of CA074-me but not of bafilomycin on ubiquitylated ErbB2 suggests that the bafilomycin-induced increase in pH is insufficient to prevent the action of lysosomal protease cathepsin B in breaking down ubiquitylated forms of ErbB2 following incubation with FCF. Cathepsin B activity is partially preserved at neutral pH [77]. In addition, the lack of effect of bafilomycin might suggest that FCF-induced endocytosis of ErbB2 is clathrin-independent. Supporting this interpretation, no increase in ErbB2 co-localization with EEA-1 was observed in the presence of FCF. The lack of significant FCF-induced co-localization of ErbB2 and caveolin-1 suggests that FCF-induced endocytosis is also caveolae-independent. This interpretation is in agreement with previously published results on clathrin- and caveolae-independent endocytosis of ErbB2 in the presence of geldanamycin [78]. However, it is also possible that rapid trafficking of internalized ErbB2 to lysosomes does not allow its detection in early endocytic compartments.
The effect of FCF was compared to geldanamycin, which is known to down-regulate ErbB2 via inhibition of Hsp90 [45, 48, 71, 79]. Both geldanamycin and FCF induce ubiquitylation and degradation of ErbB2, and the effects are additive. The pathway taken after ubiquitylation differs based on the results shown here. Capthesin B inhibitor CA074-me protects polyubiquitylated forms of ErbB2 in the presence of FCF, but not in the presence of geldanamycin. On the other hand, CA074-me protects the cleavage product induced by geldanamycin, which is absent in cells exposed to FCF. As the effects of FCF and geldanamycin both promote lysosomal degradation of ErbB2, yet are distinct, additive and complementary, this presents a mechanism for potential augmented targeting of ErbB2.
The impairment of septin dynamics decreases association of septins with ErbB2 (Fig. 5) and increases the level of ErbB2 ubiquitylation (Fig. 7). These results imply that normally organized septin filaments protect ErbB2 from ubiquitylation. Septins may limit access of ubiquitin ligases to ErbB2, perhaps via formation of organized microdomains around the protein or via formation of protein complexes with ubiquitin ligases. Analysis of septin interactomes by yeast-two-hybrid analysis and mass spectrometry identified several ubiquitin ligases as septin-interacting proteins [62, 80]. Mass spectrometry analysis also identified several ubiquitin ligases in septin-2 and ErbB2 interactomes in HGE-20 cells, and one of the E3 ubiquitin-ligases, UBR4, was found in both septin-2 and ErbB2 interactomes (data not shown). Alternatively, septins may recruit de-ubiquitylating enzymes to ErbB2, resulting in a lower steady state level of ErbB2 ubiquitylation and consequently, increased ubiquitylation in FCF-treated cells. A recent study implicates de-ubiqiutylating enzyme USP9x in protecting ErbB2 from PS341-induced lysosomal degradation [37]. Notably, this enzyme was identified by mass spectrometry in ErbB2 immunoprecipitates in HGE-20 cells (Fig. 2A). The involvement of USP9x and other identified ubiquitylation-related proteins in the protective effect of septins from ErbB2 ubiquitylation remains to be determined. In addition, septins could prevent ubiquitylation of ErbB2 by regulating the activity of ubiquitin ligases. Such regulation has recently been demonstrated for EGFR, which was stabilized at the membrane by septin-9-mediated inhibition of the interaction between c-Cbl and its adaptor protein CIN85, with consequent decrease in EGFR ubiquitylation by c-Cbl [21]. However, the FCF-induced ubiquitylation of ErbB2 is unlikely to be due to septin-mediated regulation of c-Cbl because, as shown here, ErbB2 does not interact with CIN85 and FCF does not affect levels of EGFR. In addition, as demonstrated previously, recruitment of c-Cbl by ErbB2 is less efficient compared with EGFR [36, 81]. Of note, c-Cbl or CIN85 was not detected in the septin interactome [80], or among septin-2 or ErbB2-co-immunoprecipitated proteins in the present study.
While ubiquitylation is a prominent contributor based on the data presented here, the effect of septin organization on ErbB2 may be multifactorial. Contributing mechanisms unrelated to ubiquitylation are not ruled out. For example, septin oligomers could attenuate endocytosis of ErbB2 by organizing the relatively rigid membrane microdomains, thus decreasing the mobility of ErbB2 within a bilayer, which is critical for internalization of ErbB2 [48, 82]. Since septins are known to be involved in activation of various kinases [83–85], it is possible that septins regulate ErbB2 endocytosis indirectly by triggering unidentified signaling cascades. Septin dynamics play a critical role in exocytosis [62], so use of FCF may delay or prevent ErbB2 recycling pathways, leading in part to the observed decrease in protein levels at the plasma membrane.
Due to existence of primary or secondary resistance mechanisms to ErbB2-targeted therapy, combined and complimentary medication regimens need to be used in synergy [24, 25, 28–31, 86], and better understanding of mechanisms responsible for the persistence of ErbB2 at the plasma membrane in cancer cells is critical for discovery of new ways to downregulate ErbB2. The data presented here demonstrate that septins create a dynamic barrier to ErbB2 ubiquitylation at the plasma membrane, preventing endocytosis and targeting the protein for degradation, most significantly via lysosomal pathways. This mechanism provides another potential treatment target for aggressive malignancies.
Supplementary Material
Summary Statement.
Cytoskeletal septin proteins interact with ErbB2, a cell surface receptor overexpressed in aggressive malignancies. Interference with septin dynamics in gastric cancer cells induces ubiquitylation, endocytosis and lysosomal degradation of ErbB2, indicating the importance of septins in persistent expression of ErbB2.
Acknowledgments
The authors thank Dr. Daniel Mènard for allowing use of the HGE-20 cell line for this work and Dr. Amir Orian, Dr. Donald Kohn, Dr. Katherine Wesseling-Perry, and Sergio Solorzano, MS for helpful discussion.
Funding information: Supported by K08DK100661 (EAM), UCLA CDI (EAM), USVA 2I01BX001006 (GS), 1R01DK105156-01 (GS), R01HL113350 (OV).
Abbreviations
- FCF
forchlorfenuron
- EGFR
epidermal growth factor receptor
- LRIG1
Leucine rich repeats and immunoglobulin-like domains protein-1
- Hsp
heat shock protein
- EEA1
early endosome antigen 1
- FBS
fetal bovine serum
Footnotes
Declaration of interest: The authors declare that they have no conflicts of interest with the contents of this article.
Author Contributions: EAM and OV designed the study, performed experiments, analyzed data, and wrote the manuscript. FB aided in conceptual experimental design and edited the manuscript. ET designed and performed experiments, analyzed data, and edited the manuscript. ST performed experiments. JC and JPW performed the mass spectrometry work. DRS performed experiments and edited the manuscript. GS edited the manuscript. All authors reviewed the results and approved the final version of the manuscript.
References
- 1.Hall PA, Russell SE. Mammalian septins: dynamic heteromers with roles in cellular morphogenesis and compartmentalization. The Journal of pathology. 2012;226:287–299. doi: 10.1002/path.3024. [DOI] [PubMed] [Google Scholar]
- 2.Russell SE, Hall PA. Do septins have a role in cancer? British journal of cancer. 2005;93:499–503. doi: 10.1038/sj.bjc.6602753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connolly D, Abdesselam I, Verdier-Pinard P, Montagna C. Septin roles in tumorigenesis. Biol Chem. 2011;392:725–738. doi: 10.1515/BC.2011.073. [DOI] [PubMed] [Google Scholar]
- 4.Kocevar N, Odreman F, Vindigni A, Grazio SF, Komel R. Proteomic analysis of gastric cancer and immunoblot validation of potential biomarkers. World J Gastroenterol. 2012;18:1216–1228. doi: 10.3748/wjg.v18.i11.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott M, Hyland PL, McGregor G, Hillan KJ, Russell SE, Hall PA. Multimodality expression profiling shows SEPT9 to be overexpressed in a wide range of human tumours. Oncogene. 2005;24:4688–4700. doi: 10.1038/sj.onc.1208574. [DOI] [PubMed] [Google Scholar]
- 6.Stanbery L, D’Silva NJ, Lee JS, Bradford CR, Carey TE, Prince ME, Wolf GT, Worden FP, Cordell KG, Petty EM. High SEPT9_v1 Expression Is Associated with Poor Clinical Outcomes in Head and Neck Squamous Cell Carcinoma. Transl Oncol. 2010;3:239–245. doi: 10.1593/tlo.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Kang B, Tan X, Bai Z, Liang Y, Xing R, Shao J, Xu N, Wang R, Liu S, Lu Y. Comparative analysis of the protein profiles from primary gastric tumors and their adjacent regions: MAWBP could be a new protein candidate involved in gastric cancer. J Proteome Res. 2007;6:4423–4432. doi: 10.1021/pr0703425. [DOI] [PubMed] [Google Scholar]
- 8.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 9.Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: architecture and dynamics. Nat Rev Mol Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- 10.Traikov S, Stange C, Wassmer T, Paul-Gilloteaux P, Salamero J, Raposo G, Hoflack B. Septin6 and Septin7 GTP binding proteins regulate AP-3- and ESCRT-dependent multivesicular body biogenesis. PloS one. 2014;9:e109372. doi: 10.1371/journal.pone.0109372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vagin O, Tokhtaeva E, Garay PE, Souda P, Bassilian S, Whitelegge JP, Lewis R, Sachs G, Wheeler L, Aoki R, Fernandez-Salas E. Recruitment of septin cytoskeletal proteins by botulinum toxin A protease determines its remarkable stability. Journal of cell science. 2014;127:3294–3308. doi: 10.1242/jcs.146324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vardi-Oknin D, Golan M, Mabjeesh NJ. Forchlorfenuron disrupts SEPT9_i1 filaments and inhibits HIF-1. PloS one. 2013;8:e73179. doi: 10.1371/journal.pone.0073179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiliotis ET, Kinoshita M, Nelson WJ. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science (New York, NY. 2005;307:1781–1785. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chacko AD, Hyland PL, McDade SS, Hamilton PW, Russell SH, Hall PA. SEPT9_v4 expression induces morphological change, increased motility and disturbed polarity. The Journal of pathology. 2005;206:458–465. doi: 10.1002/path.1794. [DOI] [PubMed] [Google Scholar]
- 15.Spiliotis ET, Hunt SJ, Hu Q, Kinoshita M, Nelson WJ. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. The Journal of cell biology. 2008;180:295–303. doi: 10.1083/jcb.200710039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagata K, Inagaki M. Cytoskeletal modification of Rho guanine nucleotide exchange factor activity: identification of a Rho guanine nucleotide exchange factor as a binding partner for Sept9b, a mammalian septin. Oncogene. 2005;24:65–76. doi: 10.1038/sj.onc.1208101. [DOI] [PubMed] [Google Scholar]
- 17.Amir S, Wang R, Simons JW, Mabjeesh NJ. SEPT9_v1 up-regulates hypoxia-inducible factor 1 by preventing its RACK1-mediated degradation. The Journal of biological chemistry. 2009;284:11142–11151. doi: 10.1074/jbc.M808348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagiwara A, Tanaka Y, Hikawa R, Morone N, Kusumi A, Kimura H, Kinoshita M. Submembranous septins as relatively stable components of actin-based membrane skeleton. Cytoskeleton (Hoboken) 2011;68:512–525. doi: 10.1002/cm.20528. [DOI] [PubMed] [Google Scholar]
- 19.Kinoshita N, Kimura K, Matsumoto N, Watanabe M, Fukaya M, Ide C. Mammalian septin Sept2 modulates the activity of GLAST, a glutamate transporter in astrocytes. Genes to cells : devoted to molecular & cellular mechanisms. 2004;9:1–14. doi: 10.1111/j.1356-9597.2004.00696.x. [DOI] [PubMed] [Google Scholar]
- 20.Mostowy S, Janel S, Forestier C, Roduit C, Kasas S, Pizarro-Cerda J, Cossart P, Lafont F. A role for septins in the interaction between the Listeria monocytogenes INVASION PROTEIN InlB and the Met receptor. Biophys J. 2011;100:1949–1959. doi: 10.1016/j.bpj.2011.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diesenberg K, Beerbaum M, Fink U, Schmieder P, Krauss M. SEPT9 negatively regulates ubiquitin-dependent downregulation of EGFR. Journal of cell science. 2015;128:397–407. doi: 10.1242/jcs.162206. [DOI] [PubMed] [Google Scholar]
- 22.Roskoski R., Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Ou SH. Towards the goal of personalized medicine in gastric cancer--time to move beyond HER2 inhibition. Part I: Targeting receptor tyrosine kinase gene amplification. Discov Med. 2013;15:333–341. [PubMed] [Google Scholar]
- 24.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Ruschoff J, Kang YK, To GATI. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Martin C, Lopez-Rios F, Aparicio J, Barriuso J, Garcia-Carbonero R, Pazo R, Rivera F, Salgado M, Salud A, Vazquez-Sequeiros E, Lordick F. A critical review of HER2-positive gastric cancer evaluation and treatment: from trastuzumab, and beyond. Cancer Lett. 2014;351:30–40. doi: 10.1016/j.canlet.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 27.Bailey TA, Luan H, Clubb RJ, Naramura M, Band V, Raja SM, Band H. Mechanisms of Trastuzumab resistance in ErbB2-driven breast cancer and newer opportunities to overcome therapy resistance. J Carcinog. 2011;10:28. doi: 10.4103/1477-3163.90442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–328. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 29.Nam HJ, Ching KA, Kan J, Kim HP, Han SW, Im SA, Kim TY, Christensen JG, Oh DY, Bang YJ. Evaluation of the antitumor effects and mechanisms of PF00299804, a pan-HER inhibitor, alone or in combination with chemotherapy or targeted agents in gastric cancer. Mol Cancer Ther. 2012;11:439–451. doi: 10.1158/1535-7163.MCT-11-0494. [DOI] [PubMed] [Google Scholar]
- 30.Nam HJ, Kim HP, Yoon YK, Hur HS, Song SH, Kim MS, Lee GS, Han SW, Im SA, Kim TY, Oh DY, Bang YJ. Antitumor activity of HM781-36B, an irreversible Pan-HER inhibitor, alone or in combination with cytotoxic chemotherapeutic agents in gastric cancer. Cancer Lett. 2011;302:155–165. doi: 10.1016/j.canlet.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Wainberg ZA, Anghel A, Desai AJ, Ayala R, Luo T, Safran B, Fejzo MS, Hecht JR, Slamon DJ, Finn RS. Lapatinib, a dual EGFR and HER2 kinase inhibitor, selectively inhibits HER2-amplified human gastric cancer cells and is synergistic with trastuzumab in vitro and in vivo. Clin Cancer Res. 2010;16:1509–1519. doi: 10.1158/1078-0432.CCR-09-1112. [DOI] [PubMed] [Google Scholar]
- 32.Roepstorff K, Grovdal L, Grandal M, Lerdrup M, van Deurs B. Endocytic downregulation of ErbB receptors: mechanisms and relevance in cancer. Histochem Cell Biol. 2008;129:563–578. doi: 10.1007/s00418-008-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Experimental cell research. 2008;314:3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vuong TT, Berger C, Bertelsen V, Rodland MS, Stang E, Madshus IH. Preubiquitinated chimeric ErbB2 is constitutively endocytosed and subsequently degraded in lysosomes. Experimental cell research. 2013;319:32–45. doi: 10.1016/j.yexcr.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Ehrlich ES, Wang T, Luo K, Xiao Z, Niewiadomska AM, Martinez T, Xu W, Neckers L, Yu XF. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20330–20335. doi: 10.1073/pnas.0810571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levkowitz G, Klapper LN, Tzahar E, Freywald A, Sela M, Yarden Y. Coupling of the c-Cbl protooncogene product to ErbB-1/EGF-receptor but not to other ErbB proteins. Oncogene. 1996;12:1117–1125. [PubMed] [Google Scholar]
- 37.Marx C, Held JM, Gibson BW, Benz CC. ErbB2 trafficking and degradation associated with K48 and K63 polyubiquitination. Cancer research. 2010;70:3709–3717. doi: 10.1158/0008-5472.CAN-09-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meijer IM, van Leeuwen JE. ERBB2 is a target for USP8-mediated deubiquitination. Cell Signal. 2011;23:458–467. doi: 10.1016/j.cellsig.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 39.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young P, Nie J, Wang X, McGlade CJ, Rich MM, Feng G. LNX1 is a perisynaptic Schwann cell specific E3 ubiquitin ligase that interacts with ErbB2. Mol Cell Neurosci. 2005;30:238–248. doi: 10.1016/j.mcn.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Shi C, Lu Y, Poulin EJ, Franklin JL, Coffey RJ. Loss of Lrig1 leads to expansion of brunner glands followed by duodenal adenomas with gastric metaplasia. The American journal of pathology. 2015;185:1123–1134. doi: 10.1016/j.ajpath.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, Wides R, Yarden Y. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, Sweeney C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. The Journal of biological chemistry. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 44.Simion C, Cedano-Prieto ME, Sweeney C. The LRIG family: enigmatic regulators of growth factor receptor signaling. Endocr Relat Cancer. 2014;21:R431–443. doi: 10.1530/ERC-14-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedersen NM, Madshus IH, Haslekas C, Stang E. Geldanamycin-induced down-regulation of ErbB2 from the plasma membrane is clathrin dependent but proteasomal activity independent. Mol Cancer Res. 2008;6:491–500. doi: 10.1158/1541-7786.MCR-07-0191. [DOI] [PubMed] [Google Scholar]
- 46.Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 47.Bertelsen V, Stang E. The Mysterious Ways of ErbB2/HER2 Trafficking. Membranes (Basel) 2014;4:424–446. doi: 10.3390/membranes4030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lerdrup M, Hommelgaard AM, Grandal M, van Deurs B. Geldanamycin stimulates internalization of ErbB2 in a proteasome-dependent way. Journal of cell science. 2006;119:85–95. doi: 10.1242/jcs.02707. [DOI] [PubMed] [Google Scholar]
- 49.Tikhomirov O, Carpenter G. Geldanamycin induces ErbB-2 degradation by proteolytic fragmentation. The Journal of biological chemistry. 2000;275:26625–26631. doi: 10.1074/jbc.M003114200. [DOI] [PubMed] [Google Scholar]
- 50.Chailler P, Menard D. Establishment of human gastric epithelial (HGE) cell lines exhibiting barrier function, progenitor, and prezymogenic characteristics. Journal of cellular physiology. 2005;202:263–274. doi: 10.1002/jcp.20124. [DOI] [PubMed] [Google Scholar]
- 51.Carmosino M, Procino G, Casavola V, Svelto M, Valenti G. The cultured human gastric cells HGT-1 express the principal transporters involved in acid secretion. Pflugers Arch. 2000;440:871–880. doi: 10.1007/s004240000363. [DOI] [PubMed] [Google Scholar]
- 52.Berdichevsky F, Alford D, D’Souza B, Taylor-Papadimitriou J. Branching morphogenesis of human mammary epithelial cells in collagen gels. Journal of cell science. 1994;107(Pt 12):3557–3568. doi: 10.1242/jcs.107.12.3557. [DOI] [PubMed] [Google Scholar]
- 53.Marcus EA, Vagin O, Tokhtaeva E, Sachs G, Scott DR. Helicobacter pylori impedes acid-induced tightening of gastric epithelial junctions. American journal of physiology. 2013;305:G731–739. doi: 10.1152/ajpgi.00209.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tokhtaeva E, Munson K, Sachs G, Vagin O. N-glycan-dependent quality control of the Na,K-ATPase beta(2) subunit. Biochemistry. 2010;49:3116–3128. doi: 10.1021/bi100115a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tokhtaeva E, Sachs G, Vagin O. Diverse pathways for maturation of the Na,K-ATPase beta1 and beta2 subunits in the endoplasmic reticulum of Madin-Darby canine kidney cells. The Journal of biological chemistry. 2010;285:39289–39302. doi: 10.1074/jbc.M110.172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 57.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G. Acid-adaptive genes of Helicobacter pylori. Infection and immunity. 2003;71:5921–5939. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 59.Janes PW, Lackmann M, Church WB, Sanderson GM, Sutherland RL, Daly RJ. Structural determinants of the interaction between the erbB2 receptor and the Src homology 2 domain of Grb7. The Journal of biological chemistry. 1997;272:8490–8497. doi: 10.1074/jbc.272.13.8490. [DOI] [PubMed] [Google Scholar]
- 60.Stein D, Wu J, Fuqua SA, Roonprapunt C, Yajnik V, D’Eustachio P, Moskow JJ, Buchberg AM, Osborne CK, Margolis B. The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBO J. 1994;13:1331–1340. doi: 10.1002/j.1460-2075.1994.tb06386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, Yarden Y, Neckers L. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. The Journal of biological chemistry. 2001;276:3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 62.Tokhtaeva E, Capri J, Marcus EA, Whitelegge JP, Khuzakhmetova V, Bukharaeva E, Deiss-Yehiely N, Dada LA, Sachs G, Fernandez-Salas E, Vagin O. Septin dynamics are essential for exocytosis. The Journal of biological chemistry. 2015;290:5280–5297. doi: 10.1074/jbc.M114.616201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghossoub R, Hu Q, Failler M, Rouyez MC, Spitzbarth B, Mostowy S, Wolfrum U, Saunier S, Cossart P, Jamesnelson W, Benmerah A. Septins 2, 7 and 9 and MAP4 colocalize along the axoneme in the primary cilium and control ciliary length. Journal of cell science. 2013;126:2583–2594. doi: 10.1242/jcs.111377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu Q, Nelson WJ, Spiliotis ET. Forchlorfenuron alters mammalian septin assembly, organization, and dynamics. The Journal of biological chemistry. 2008;283:29563–29571. doi: 10.1074/jbc.M804962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wasik AA, Polianskyte-Prause Z, Dong MQ, Shaw AS, Yates JR, 3rd, Farquhar MG, Lehtonen S. Septin 7 forms a complex with CD2AP and nephrin and regulates glucose transporter trafficking. Molecular biology of the cell. 2012;23:3370–3379. doi: 10.1091/mbc.E11-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma S, Quintana A, Findlay GM, Mettlen M, Baust B, Jain M, Nilsson R, Rao A, Hogan PG. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature. 2013;499:238–242. doi: 10.1038/nature12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sidhaye VK, Chau E, Breysse PN, King LS. Septin-2 mediates airway epithelial barrier function in physiologic and pathologic conditions. American journal of respiratory cell and molecular biology. 2011;45:120–126. doi: 10.1165/rcmb.2010-0235OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SK, Shindo A, Park TJ, Oh EC, Ghosh S, Gray RS, Lewis RA, Johnson CA, Attie-Bittach T, Katsanis N, Wallingford JB. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science (New York, NY. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mostowy S, Danckaert A, Tham TN, Machu C, Guadagnini S, Pizarro-Cerda J, Cossart P. Septin 11 restricts InlB-mediated invasion by Listeria. The Journal of biological chemistry. 2009;284:11613–11621. doi: 10.1074/jbc.M900231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cortese K, Howes MT, Lundmark R, Tagliatti E, Bagnato P, Petrelli A, Bono M, McMahon HT, Parton RG, Tacchetti C. The HSP90 inhibitor geldanamycin perturbs endosomal structure and drives recycling ErbB2 and transferrin to modified MVBs/lysosomal compartments. Molecular biology of the cell. 2013;24:129–144. doi: 10.1091/mbc.E12-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. The Journal of biological chemistry. 1996;271:22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 72.Heasley LR, Garcia G, 3rd, McMurray MA. Off-target effects of the septin drug forchlorfenuron on nonplant eukaryotes. Eukaryot Cell. 2014;13:1411–1420. doi: 10.1128/EC.00191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciechanover A. The unravelling of the ubiquitin system. Nat Rev Mol Cell Biol. 2015;16:322–324. doi: 10.1038/nrm3982. [DOI] [PubMed] [Google Scholar]
- 74.Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. Journal of cell science. 2012;125:265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- 75.Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. The Journal of cell biology. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leo MD, Bulley S, Bannister JP, Kuruvilla KP, Narayanan D, Jaggar JH. Angiotensin II stimulates internalization and degradation of arterial myocyte plasma membrane BK channels to induce vasoconstriction. American journal of physiology Cell physiology. 2015 doi: 10.1152/ajpcell.00127.2015. ajpcell 00127 02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Repnik U, Starr AE, Overall CM, Turk B. Cysteine Cathepsins Activate ELR Chemokines and Inactivate Non-ELR Chemokines. The Journal of biological chemistry. 2015;290:13800–13811. doi: 10.1074/jbc.M115.638395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barr DJ, Ostermeyer-Fay AG, Matundan RA, Brown DA. Clathrin-independent endocytosis of ErbB2 in geldanamycin-treated human breast cancer cells. Journal of cell science. 2008;121:3155–3166. doi: 10.1242/jcs.020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lerdrup M, Bruun S, Grandal MV, Roepstorff K, Kristensen MM, Hommelgaard AM, van Deurs B. Endocytic down-regulation of ErbB2 is stimulated by cleavage of its C-terminus. Molecular biology of the cell. 2007;18:3656–3666. doi: 10.1091/mbc.E07-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakahira M, Macedo JN, Seraphim TV, Cavalcante N, Souza TA, Damalio JC, Reyes LF, Assmann EM, Alborghetti MR, Garratt RC, Araujo AP, Zanchin NI, Barbosa JA, Kobarg J. A draft of the human septin interactome. PloS one. 2010;5:e13799. doi: 10.1371/journal.pone.0013799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer research. 2000;60:3384–3388. [PubMed] [Google Scholar]
- 82.Hommelgaard AM, Lerdrup M, van Deurs B. Association with membrane protrusions makes ErbB2 an internalization-resistant receptor. Molecular biology of the cell. 2004;15:1557–1567. doi: 10.1091/mbc.E03-08-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Joo E, Surka MC, Trimble WS. Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev Cell. 2007;13:677–690. doi: 10.1016/j.devcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 84.Li CR, Au Yong JY, Wang YM, Wang Y. CDK regulates septin organization through cell-cycle-dependent phosphorylation of the Nim1-related kinase Gin4. Journal of cell science. 2012;125:2533–2543. doi: 10.1242/jcs.104497. [DOI] [PubMed] [Google Scholar]
- 85.Merlini L, Fraschini R, Boettcher B, Barral Y, Lucchini G, Piatti S. Budding yeast dma proteins control septin dynamics and the spindle position checkpoint by promoting the recruitment of the Elm1 kinase to the bud neck. PLoS Genet. 2012;8:e1002670. doi: 10.1371/journal.pgen.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25:282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.