Abstract
Using a compendium of cell-state-specific gene expression data, we identified genes that uniquely define cell states, including those thought to represent various developmental stages. Our analysis sheds light on human cell fate through the identification of core genes that are altered over several developmental milestones, and across regional specification. Here we present cell-type specific gene expression data for 17 distinct cell states and demonstrate that these modules of genes can in fact define cell fate. Lastly, we introduce a web-based database to disseminate the results.
1. Introduction
If each cell type is defined by the genes it expresses, then one would expect every cell type to show a distinct pattern of expression, characterizing that cell type. Such cell type-specific knowledge is important for advancing our basic understanding of biology and as a useful starting point for drug discovery. Such knowledge also sheds light on how one might reprogram one cell type in to another—a major hurdle in the process of direct reprogramming (Vierbuchen et al., 2010). However, elucidating a unique expression pattern for each cell type requires comparisons across a broad set of cell types. If one were to compare only fibroblasts to neurons, for example, one would find unique signatures distinguishing these cell types from each other, but not from other cells. Therefore, data-derived comparative signatures are context-dependent—subject to the diversity of cell types included in the comparison. Ignoring the context-dependency has lead previous analyses astray—many genes that were identified as being expressed specifically in a particular cell type (i.e., markers for that cell type), were later found to be expressed in several different cell types (Juuri et al., 2012).
One would expect that with an increasing number and variety of cell types, recovered cell-specific signatures would become more refined, and eventually plateau as the amount of data within each cell type added becomes less informative. Herein, we show that as we add more and more data from various cell states to our analysis, the set of identified core expression factors initially changed rapidly, and then became more stable. Our goal here was to define cell states, including those representing various developmental stages, in a context-independent manner, by using a newly-generated, cell-state specific compendium of gene expression.
A secondary goal was to find the unique regulatory core for each cell state—the elements which drive and maintain cell fate (Kim et al., 2008; Wang et al., 2006). Direct reprogramming of cells (e.g., from fibroblast to PSC), has shown that overexpression of a small number of transcription factors can drive a cell to become a completely different type of cell (Takahashi and Yamanaka, 2006). This direct reprogramming approach is quickly serving to identify robust transcriptional networks that drive particular cell fates, even when introduced into cells of a different germ layer. However, identifying these core expression factors has typically taken years of painstaking effort. Normally, the first step in identifying these small groups of cell fate drivers is to compare the gene expression of just two types of cells (or against several others in aggregate), and then to select the most upregulated transcription factors in the desired cell type. Next, through trial and error, cocktails of successful reprogramming factors (not necessarily unique) can sometimes be identified (Takahashi and Yamanaka, 2006; Vierbuchen et al., 2010). This overall approach has been hampered by the selection of factors based on expression differences between just two cell types (or based on comparing one cell type to several others in aggregate). Thus, our second goal was to streamline this type of reprogramming pre-selection process by obtaining a more refined comparison as a result of using a broader data set. We have named our overall approach, CEMA, for Core Expression Module Analysis.
Using a cell-state-by-cell-state logistic regression-based approach (similar in spirit to a one way analysis-of-variance with post hoc analysis), we identified putative core elements of cell-specific transcription for 17 cell states representing nine unique purified human cell types from different germ layers, degree of specification, and developmental age (including neural progenitor cells, fibroblasts, keratinocytes, hepatocytes, mesothelial cells, myopepithelial cells, kidney epithelial cells, pluripotent stem cells, definitive endoderm, smooth muscle cells, and endothelial cells) (Chin et al., 2009; Chin et al., 2010; Patterson et al., 2012). This collection of data represents an improvement over previously described databases (e.g., BioGPS (Wu et al., 2009)) in that we used strictly purified cells from tissue as opposed to whole tissues, and all the analyses were carried out in the same lab to minimize batch effect. In addition, data from cells differentiated from human pluripotent stem cells were included along with tissue-derived counterparts, opening the possibility of identification of gene expression patterns that change across developmental stages. Finally, our collection also included the same cell type (endothelial) derived from different locations within the body to provide information on regional specialization. The detailed list of cell states is provided in Table 1, along with corresponding shorthand notation used throughout the paper. We also make use of an independent and publically available dataset consisting of 84 different cell types and tissues to validate our results.
Table 1.
Each of the 17 cell states in our compendium, along with the shorthand used in figures and on the website (in parentheses). Also denoted in parentheses is the number of independent biological replicates analyzed.
| 1 | tissue endothelial cell | EC (blood vessel)(×10) |
| 2 | pluripotent derived definitive endoderm cell | PSC-Endo (endodermal progenitor)(×2) |
| 3 | pluripotent derived fibroblast cell | PSC-FB (paraxial mesoderm)(×8) |
| 4 | pluripotent derived hepatocyte cell | PSC-HEP (hepatocyte)(×4) |
| 5 | pluripotent derived neuron cell | PSC-NEU (neuron)(×3) |
| 6 | pluripotent derived neural progenitor cell | PSC-NPC (neural tube progenitor)(×13) |
| 7 | tissue smooth muscle cell | SMC (smooth muscle)(×4) |
| 8 | squamous cell carcinoma cell lines | SSC (squamous cell carcinoma)(×4) |
| 9 | tissue derived fibroblast cell | tis-FB (paraxial)(×9) |
| 10 | tissue derived hepatocyte cell | tis-HEP (hepatocyte)(×3) |
| 11 | tissue derived keratinocyte cell | tis-KER (keratinocyte)(×6) |
| 12 | tissue derived kidney epithelial cell | tis-KID (kidney proximal tubule)(×2) |
| 13 | tissue derived mesothelial cells | tis-MESO (mesothelial cell)(×2) |
| 14 | tissue derived myoepithelial cell | tis-MYOEP (myoepithelial)(×4) |
| 15 | tissue derived neural progenitor cell 6–8 PCW | tis-NPC-early (early gestational)(×6) |
| 16 | tissue derived neural progenitor cell 15–19 PCW | tis-NPC-late (mid-gestational)(×5) |
| 17 | undifferentiated pluripotent stem cells | Undiff (model of epiblast)(12) |
We found that many common “marker” genes typically used to define various cell types of the nervous system were in fact expressed in many cells not associated with brain or spinal cord (Pankratz et al., 2007; Zhao et al., 2004). For example, NESTIN was highly expressed in 7 out of 17 cell states. We also show how identified core expression modules changed during development or as a result of spatial specification in different tissues. Using results generated from this approach, we built an interactive web-based application for dissemination and exploration of our results, yielding a valuable resource with a novel perspective on human cell fate, as well as potential leads for inducing one cell state from another. As validation that our approaches can yield factors important for particular cell fates, we provide evidence that CEMA-predicted factors can indeed drive cell fate.
2. Results
2.1. Applying CEMA
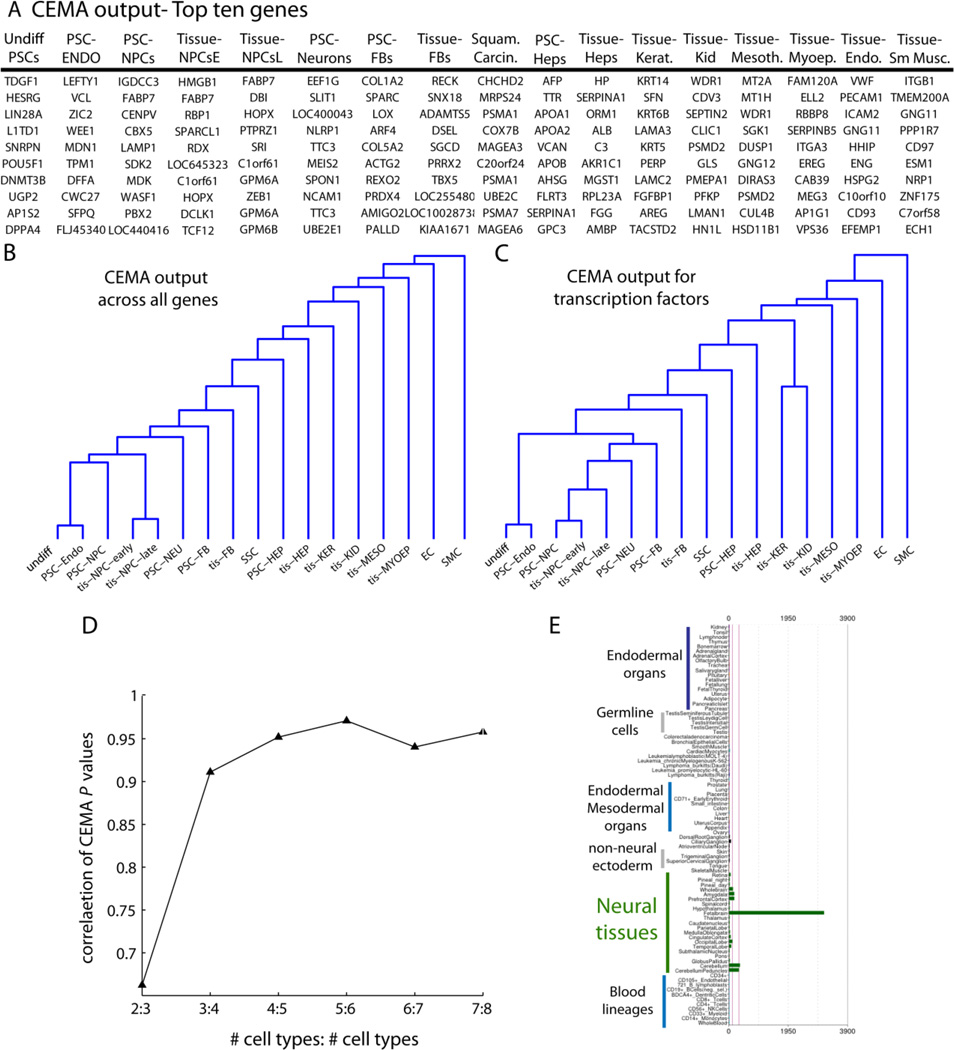
Application of our approach, CEMA (see Statistical Methods, Methods Section, allowed for the identification of a relatively small number of genes that serve to uniquely distinguish each type of cell from every other (the top 10 displayed in Fig. 1A; also shown in Supplemental Table 1 with relative expression values (RMA normalized, log2)). We present the output of results as a dendrogram (see Methods) comparing the CEMA output from all the different cell states (Fig. 1B). The CEMA profiles appeared to cluster almost as expected considering their developmental background and state of differentiation. This was also true when we restricted the analysis to transcription factors (Fig. 1C), raising the possibility that cells can be distinguished by their core transcription factor (TF) expression, an idea consistent with the fact that most proteins in “reprogramming” cocktails identified to date are TFs (e.g. the Yamanaka factors for re-programming fibroblasts to pluripotent cells(Takahashi and Yamanaka, 2006).
Figure 1. Analysis of 17 cell states produces unique expression modules.
(A) a table of the top 10 genes from each cell state shown. (B) and (C) Dendrograms showing the relationship between all tissues in the global analysis, using the correlation between their p-values for all genes (B) or just transcription factors (C). Note that, in general, the CEMA patterns for similar cell states appeared to be more similar for expected cell states. In other words, the list of genes ranked by their specificity for a particular cell state reflected the similarity that would be uncovered by a Pearson-type correlation. (D) Example of how the p-values stabilized as we added more and more cell types In particular, each entry on the x-axis shows the correlation between tis-HEP p-values, when X tissues, or X+1 tissues were used. For example, the 2:3 entry shows that when only two tissues were used, as compared to three tissues, that the p-values have a correlation of around 0.75. However, when we compared the p-values when 8 tissues were used, as compared to 7 (7:8) we see the correlation approaches 1. Here, the order of tissues added for comparison with tis-HEP were: tis-FB then tis-HEP, tis-KER, tis-MYOEP, tis-KID, tis-MESO, tis-NPCe, tis-NPC. (E) To assess the specificity of CEMA-identified genes, an example gene for NPCs was probed across the BioGPS database of profiled cell types from across dozens of cell types. FABP7, identified by CEMA as specific to tis-NPCs, only showed up in fetal brain samples in the Novartis dataset. Note that just a subset of cell types are labelled in the image (blue text), while the analysis was performed on all cell types. A broader analysis is available in Figure S1.
It should also be noted that the vertical lines of the dendrograms are quite long, indicating low similarity—to be expected when a broad compendium is used including quite distinct cell states. Furthermore, although CEMA implicitly is designed to focus on finding genes that are expressed in a single cell state (one vs all), the approach is still flexible enough to allow for the same gene showing up in multiple cell states. As an example of this, FABP7 was represented on all three lists of neural progenitor cells (NPC), but only found with HMGB1 in one NPC state (Tissue-NPC early).
As increasingly diverse cell states were added to the analysis, we expected that the specificity of each list would be refined. An illustrative example of this refinement can be seen in Fig. 1C, which shows a summary plot of how the analysis changed as we increased the number of cell types in the comparison from two through to eight. When just two cell types were compared, they were distinguished by 1000’s of genes, but once five cell types were included in the comparison, the number of unique genes plateaued, presumably owing to a fairly broad compendium having already been used at that point. In our final analysis, we used 17 distinct cell states and produced sets of 20–700 genes expressed in unique combinations in each cell state. These results stand in stark contrast to the roughly 3000–7000 genes we found differentially expressed between any two of these cell states.
To determine the relative specificity of the CEMA output, we first looked at the pattern of the most highly expressed genes in a particular cell type (NPC), in an expression database containing data for 84 cell types and tissues (BioGPS (Wu et al., 2009)). Such an examination revealed that simply taking the magnitude of gene expression into account is ineffective at uncovering cell state specific genes (Fig. 1D, and Supplemental Fig. 1). Instead, sorting for high gene expression within the CEMA refined gene list, produced genes with very high specificity when analyzed on an independent data set (BioGPS) (Supplemental Fig. 1). For instance, FABP7 was a top CEMA gene in PSC- and tissue- derived NPCs (Fig. 1D), and the only positive signal from the BioGPS database that was found in brain (Supplemental Fig. 1). Using a similar analysis for hepatocytes further demonstrated the specificity of the CEMA output. In this case, the top genes identified by CEMA in pluripotent derived hepatocytes showed up as more specific to fetal liver than any other cell type in the database (Supplemental Fig. 2A). Furthermore, CEMA analysis on hepatocytes taken from adult liver yielded genes that appeared as a group to be more specific to adult liver than fetal liver (Supplemental Fig. 2B).
2.2. Temporal and Spatial Analyses
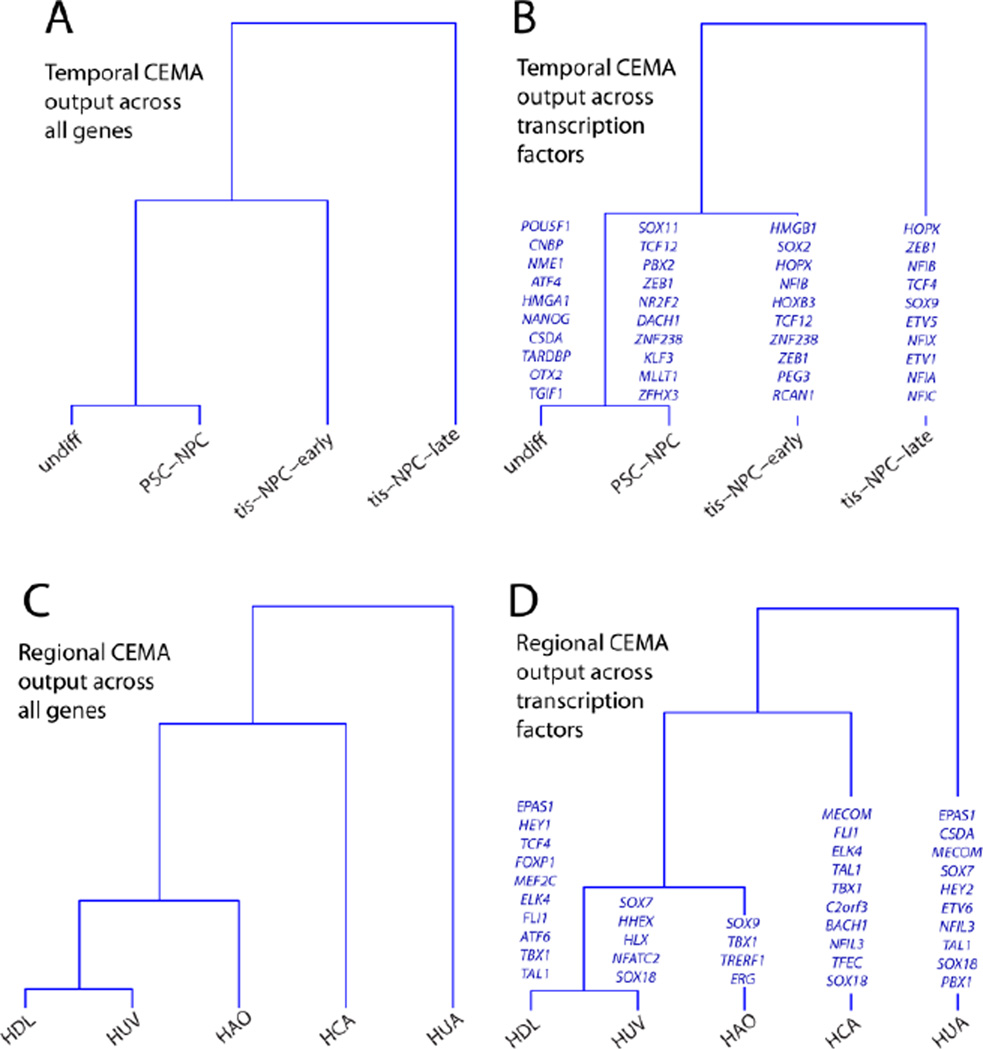
As cells develop from a pluripotent stage through various levels of specification and differentiation, it is likely that their core expression factors change. To uncover such factors for a particular cell lineage, we analyzed different cell types in the neural lineage, each representing a different developmental time point. These cell states were described previously to represent different stages of gestational development based on gene expression and functional criteria (neurogenic versus gliogenic)(Patterson et al., 2012). This gestational timing model was further validated by work showing that PSC-NPCs develop in vitro at a similar rate as they would in vivo (Marin, 2013; Maroof et al., 2013; Nicholas et al., 2013).
We first identified all genes using CEMA and found that the resulting CEMA profiles follow the expected segregation (Fig. 2A). To enrich for genes likely to play a roles in development, we also present results only for transcription factors (Fig. 2B). Note that each time point is characterized by a different set of TFs, rather than by the same sets changing in magnitude (Fig. 2B), indicating a change in mechanism rather than amount, consistent with what is known to happen over time as Yamanaka factors are induced in fibroblasts. Although the statistical analysis seeks out such differences, it is nevertheless interesting to note that it in this case it finds them. While some core expression factors are found across multiple time points, each time stage is, in its entirety, distinct. In addition, while all the NPCs analyzed expressed significant levels of typical NPC markers (SOX2, NESTIN, MUSASHI, CD133, SOX1 and PAX6) (Patterson et al., 2012), they are distinguished by key sets of transcription factors which presumably endow them with different functional capacities. This is typified by the fact that the PSC-NPCs profiled are highly neurogenic, while the tis-NPCs analyzed are mostly gliogenic (Patterson et al., 2012).
Figure 2. Temporal and Spatial CEMA analyses.
(A) using correlation based on all genes, (B) using only those genes labeled as transcription factors (see Methods). In green, the top 10 transcription factors that distinguish each cell type are highlighted. Note that TFs shown in italics were uncovered in at least two of different types of NPCs. Spatial CEMA analysis (C) Dendrograms showing the relationship between all tissues in the spatial analysis, using the correlation between their p-values using correlation based on all genes or, (D) using only those genes labeled as transcription factors (see Methods). In green, the top transcription factors that distinguish each cell type are highlighted. Note that TFs shown in italics were uncovered in at least two of the endothelial cells taken from different tissues.
To uncover differences between core expression modules in the same cell type purified from different portions of the human body, we isolated endothelial cells from the vasculature of various regions (Coronary (HCA), Umbilical (HUV and HUA), Aortic (HAO), and Dermal Lymphatic (HDL) vessels). Applying CEMA to endothelial cells from various tissues highlighted a spatial dependence of core expression factors (Fig. 2C), pointing to variability in their physiology. While all these endothelial cells express very high levels of markers such as VWF and PECAM, there are significant differences in both their total gene and TF specific core expression factors that distinguish them not only from non-endothelial cells, but also from each other (Fig. 2D). Here, CEMA identified key factors that distinguished endothelial cells taken from different tissues with high resolution.
2.3. Finding more consistent constitutive genes
CEMA analysis yielded identification of genes with unique expression patterns. However, we postulated that this compendium of cell types should allow for the identification of genes that are the least variant across many cell types. These genes could then potentially be used to serve as housekeeping control for RT-PCR, western blotting, and so on. First, a list of typically-used housekeeping genes (Gur-Dedeoglu et al., 2009) was analyzed for variability of expression (standard deviation) across the 17 cell types. Ranking just these typical housekeeping genes to find those with the least variance of expression across the 17 cell types, we found that RPL41, coding for a ribosomal protein was the least variant across the data set, more so than either GAPDH or βACTIN, the most frequently used housekeeping genes in the literature (Supplemental Fig. 3A). Furthermore, when instead all genes on the array were assayed for the least variability across these cell types, RPL41 again came up as the least variant gene (Supplemental Figure. 3B). For some applications it could be more accurate to use a housekeeping gene for normalization that is closer in expression level to the gene one is studying. Thus, we separated out the aforementioned analyses to provide candidate housekeeping genes within different levels of absolute expression (low, medium and high expression) (Supplemental Fig. 3B–D).
2.4. Dissemination to the scientific community
To provide general access to our results, a web-based application was developed, intended both for dissemination of our results, as well as to provide a data-driven roadmap for human cell states (www.cemagenes.com, username=preview, password=preview). As of now, the website contains template nodes for most known cell types, with populated nodes for those cell types that have been analyzed thus far. Results restricted to only transcription factors were also generated (Supplemental Fig. 4A and B). The application also allows one to display histograms of the relative expression of individual genes of interest across all cell types analyzed (Supplemental Fig. 4C). At regular intervals we expect to add data for additional cell types, which will generate refined CEMA lists for all cell types analyzed to that point, while maintaining previous versions of the analysis to monitor change over various iterations.
2.5. Evidence that CEMA-identified modules can drive cell fate
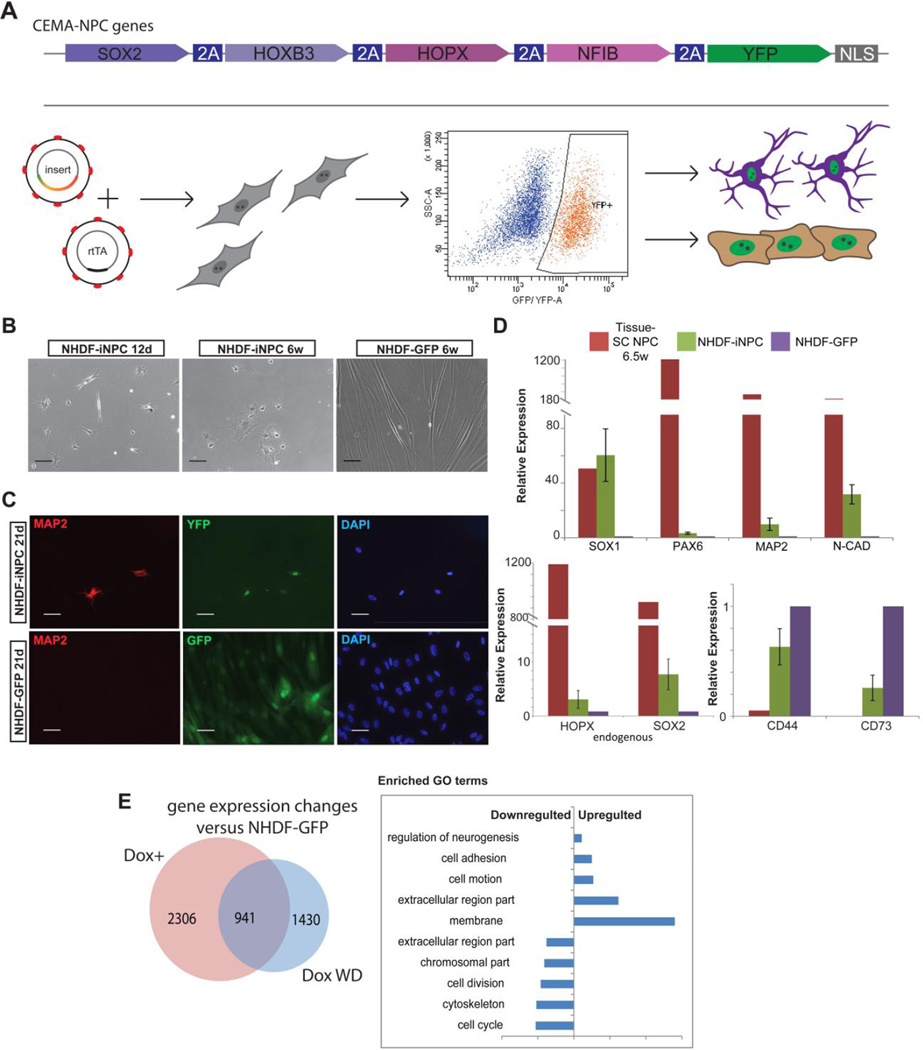
To demonstrate that CEMA identified factors can define cell fate, we designed a system that would allow for a determination of whether these factors can either reprogram cell fate between somatic cell types, or program the cell fate of human pluripotent stem cells. The factors that were used for cell fate experiments were first chosen based on their specificity to the target cell type as identified by the CEMA algorithm. We then sorted them by relative expression level in the target cell. Finally, we identified those that are bona fide TFs, as TFs are well established to play a profound role in reprogramming(Takahashi and Yamanaka, 2006). Polycistronic inducible lentiviral vectors bearing 4 genes identified by the CEMA algorithm for two cell types: early (6 to 8 weeks gestation) neural progenitor cells (eNPC) and endothelial cells were created. The vectors also included a puromycin selection gene and a reporter YFP allele. Early passage primary neonatal dermal fibroblasts (NHDF) were driven to express the reverse tetracycline transactivator (rtTA) upon doxycycline (dox) by lentiviral infection and were stably selected with G418. Similarly, hESC and iPSC rtTA expressing clones were also derived (see schematic in Fig. 3A).
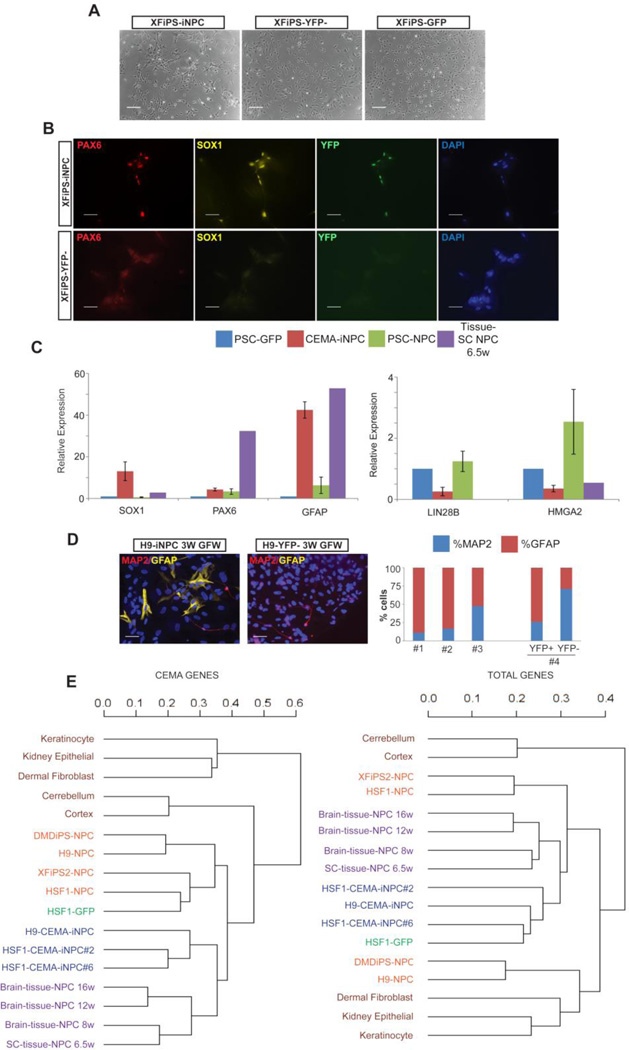
Figure 3. CEMA selected gene sets can induce at least partial reprogramming.
(A) Schematic illustration of experimental design and CEMA selected genes cassettes (B–E), Reprogrammed NHDF–iNPC (B) NHDF-iNPC cells, sorted for YFP+ and treated with dox exhibit distinct morphology changes. The images shown are from a representative experiment, selected from over four separate experiments. (C) Some NHDF-iNPCs acquired neuronal morphology and stained positive for MAP2. Bars represent 50 microns. (D) NHDF-iNPCs expressed various neuronal and NPC markers as measured by quantitative real time PCR relative to NHDF-GFP; GAPDH expression was used for normalization. Error bars represent standard error of the mean of four separate reprogramming experiments, one tissue-NPC sample was used for comparison (E) Venn diagram of genes of which expression level changed 2 fold in NHDF-iNPC, NHDF-iNPC in which Dox was withdrawn (wd) compared to NHDF-GFP.
Following infection and activation of ectopic expression of the polycistron containing the eNPC factors predicted by CEMA, YFP positive fibroblasts were sorted and subsequently cultured conditions supporting reprogramming (Fig. 3B). Induced early-NPC (iNPC) fibroblasts exhibited distinct morphology changes apparent as early as 10 days post induction of ectopic expression (Fig. 3C); upregulated expression of neural markers such as SOX1, PAX6 and MAP2; downregulated fibroblast genes (CD44, CD73); and activated endogenous versions of the CEMA-predicted genes (SOX2, HOPX), which would be a key step of reprogramming (Fig. 3D and E). However, the iNPCs were unable to induce expression of NPC markers to a similar level found in tissue-derived NPCs. Moreover, the iNPCs appeared to proceed spontaneously to a neuronal fate as judged by staining for MAP2, suggesting that reprogramming to a stable NPC state was not achieved (Fig. 3D). These results were consistent across at least three independent reprogramming attempts. To further analyze the degree of cell fate conversion achieved, we compared the gene expression of reprogrammed cells to control fibroblasts using microarray, and found significant changes in gene expression, some of which were retained upon dox withdrawal, a hallmark for reprogramming (Fig. 3E). Differentially expressed genes were related to several biological Gene Ontology (GO) terms categories: cell structure, cell adhesion, regulation of neurogenesis and cell division.
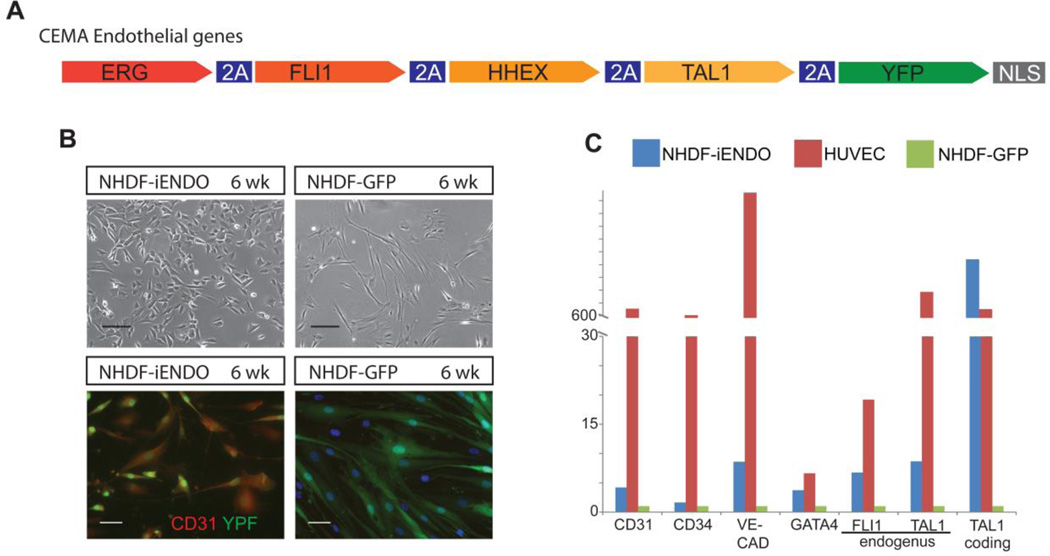
Three to four weeks following CEMA endothelial gene set induction (Fig. 4A), YFP+ fibroblasts began to decrease their cytoplasmic volume, exhibited a “cobblestone” like morphology (Fig. 4B), and upregulated endothelial markers such as CD31 and VE-cad (Fig. 4C) as well as the endogenous versions of CEMA-predicted genes (TAL1, FLI1). Again, the degree of induction of endogenous endothelial markers, across three independent experiments, was significantly lower than that found in bona fide endothelial cells (HUVEC), consistent with results from reprogramming to NPCs from FBs with CEMA-predicted factors. Taken together, these results suggest that at least partial reprogramming can be achieved using CEMA selected genes, but that complete reprogramming is not possible without potentially significant modifications to the protocol or addition of more factors.
Figure 4. CEMA selected genes for NHDF–endothelial reprogramming.
(A) CEMA endothelial selected genes. (B) NHDF- endo 8 Weeks of Doxycycline treatment, Some NHDF-endo stained positive for the endothelial marker CD31. Images of one experiment that is representative of three separate experiments are presented. Bars represent 50 microns. (C) NHDF-endo upregulate various endothelial related genes, though in lower levels compared to primary HUVECS as measured by quantitative real time PCR relative to NHDF-GFP. These results are representative of three independent experiments.
2.6. Using CEMA predicted factors to program cell fate
Reprogramming cells from one somatic fate to another requires massive molecular changes. It is quite possible that the CEMA-selected factors are important for cell fate, but cannot reprogram fate from an alternate somatic state. Therefore, we hypothesized that these factors could drive fate more robustly when starting with cells at the pluripotent state. We generated human PSC clones with stable expression of rtTA and infected them with cocktails of CEMA selected gene sets through polycistronic lentiviral induction. After selection for infected cells, and subsequent doxycycline induction, YFP+/YFP− populations were sorted out and cultured in spontaneous differentiation conditions (without bFGF). After 2 weeks, medium was replaced to target cell culturing conditions.
In concordance with our hypothesis, hPSC engineered to express CEMA-selected NPC factors, differentiated homogenously to NPCs exhibiting typical NPC morphology 2–3 weeks post ectopic induction (Fig. 5A and B), and expressed NPC markers in a level similar to that of early tissue- derived NPC as shown by RT-PCR (Fig. 5C). In stark contrast with typical hPSC derived NPCs, which tend to differentiate mainly towards the neuronal lineage (Patterson et al., 2012), NPCs generated by forced expression of CEMA predicted factors instead exhibited a high tendency to differentiate to astrocytes as measured by GFAP upon growth factor withdrawal (Fig. 5D). This is a very important distinction as the CEMA factors were generated from NPCs from tissue with a glial bias. RT-PCR for gene expression in the induced clones demonstrated that CEMA-predicted factors promoted robust induction of typical NPC markers, and to a degree more similar to that found in tissue-derived NPCs (Fig. 5C). Remarkably, these iNPCs even appeared to suppress expression of let-7 target genes, as would be expected for NPCs derived from tissue, as opposed to those differentiated from pluripotent cells (Fig. 5C)(Patterson et al., 2014). In addition, this pattern of let-7 target expression is consistent with their terminal differentiation pattern (Fig. 5D) (Patterson et al., 2014). This indicates that CEMA-predicted factors not only induced cell fate, but also drove a specific cell fate, relevant to the cell types from which the CEMA factors were identified. Programming cell fate to the neural lineage with these CEMA defined factors was highly reproducible, as the data presented are representative of three independent experiments.
Figure 5. CEMA selected genes expression derive specific cell fates.
(A) 3 weeks post ectopic induction PSC-iNPC clones exhibited homogenous NPC morphology, (B) and expressed NPC markers. Bars represent 50 microns. (C) CEMA-PSC-iNPC expressed various NPC markers as measured by RT-PCR relative to H9-GFP and compared to 6.5w tissue derived NPCs and PSC-NPC derived by standard protocol. Error bars represent standard error of the mean of three samples. GAPDH was used for normalization. (D) Followed growth factor withdrawal, PSC-iNPC exhibited high tendency to differentiate towards the glial lineage (GFAP positive cells) quantification of three separate experiments with different PSC clones is shown. For experiment #4, CEMA-XFiPS cells were chemically induced to differentiate towards NPC using small molecules(Chambers et al., 2009). YFP+/− NPCs were sorted out followed by growth factor withdrawal. (E), Hierarchical clustering of gene expression profiles as measured by strand-specific RNA-seq, shown as dendrograms. Left panel: CEMA selected early tissue NPC genes (see supplementary Figure S1); right panel: total gene expression.
Genome-wide profiling of iNPCs in comparison to NPCs isolated from tissue or differentiated under standard conditions from PSCs by RNA-seq further demonstrated the robustness of the CEMA driven approach. For comprehensive comparison, RNA-seq reads from 5 additional primary cells from the ENCODE project were added to the analyses: hair follicular keratinocyte (ENCFF236EYN), dermis fibroblast (ENCFF000HWI), kidney epithelial cell (ENCFF109IUU), frontal cortex (ENCFF001RNU), cerebellum (ENCFF001ROK). We performed unsupervised hierarchical clustering using cummeRbund and found that CEMA derived iNPCs clustered closer to tissue-NPCs compared to PSC-NPCs made by traditional differentiation (Fig. 5E), particularly when analyzing CEMA identified genes expression (Fig. 5E, left panel).
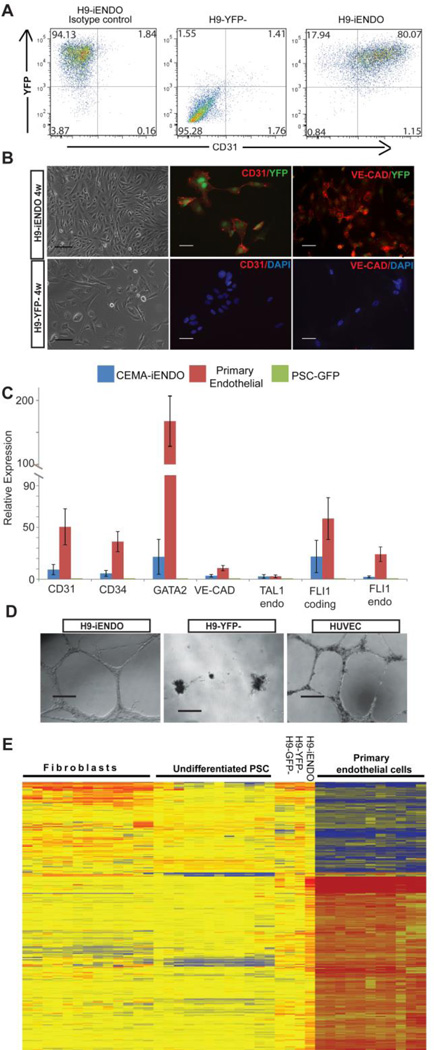
Differentiation of hESCs to the endothelial state is notoriously difficult, typically with a low yield. Here, we used H9-hESCs engineered to inducibly express the CEMA-predicted endothelial factors (same factors as used for reprogramming) to drive fate. Two days post dox induction YFP+/− cells were sorted and grown for 5 days in ESC differentiation medium which was then replaced to EGM2 endothelial growth medium. As shown in Figure 6A, CEMA-factor induced cultures exhibited high differentiation capacity toward the endothelial lineage. At early time points, nearly 50% of YFP positive cells expressed endothelial markers such as CD31, while at later time points greater that 80% of YFP positive cells expressed CD31 (Fig. 6A) compared to 5–10% reported for standard spontaneous differentiation protocols (Levenberg et al., 2010), as well as other endothelial related genes (Fig.6B and 6C). In contrast, YFP− or GFP control cells, induced to spontaneously differentiate, had very rare CD31 positive cells (Fig.6A and 6B) in similar conditions. The programming data shown were typical of at least three independent experiments. To further functionally test the CEMA derived endothelial-like cells, we tested their ability to form tube-like structures. A standard tube formation assay in matrigel demonstrated that CD31 sorted iEndothelial cells were able to form robust vessels, to a degree similar to that observed for HUVEC cells. On the other hand, YFP− or GFP infected cells showed no ability to form tubular structures. (Fig. 6D). Finally, gene expression profiling of the differentiated H9-endo cells was analyzed by microarray and compared against CEMA gene expression database, showing high similarity to primary endothelial cells (Fig. 6E).
Figure 6. CEMA selected genes expression enhance differentiation of human PSCs.
(A) Flow cytometry analysis of CD31 positive cells in sorted YFP+/YFP− populations cultured in endothelial conditions for 8 weeks or 10 weeks (B) CEMA-H9-iendo cells exhibit homogenous cobble-stone morphology and endothelial marker expression. Bars represent 50 microns. (C) CEMA-PSC-iENDO expressed various endothelial related genes in similar levels compared to primary endothelial cells as measured by RT-PCR relative to H9-GFP (PSC-GFP). Error bars represent standard error of the mean of three experiments. (D) Matrigel tube formation assay: H9-endo-YFP+ formed tube like structure similarly to HUVECs. (E) Heat map of H9-endo gene expression compared to H9 controls and other cells in the CEMA gene expression database.
3. Discussion
We have generated a compendium of 17 cell-state-specific gene expression data, and analyzed it to identify unique gene expression patterns. This approach focused on (1) cell-state-specific data, and (2) data from a single laboratory and platform. The latter is an important distinction because of well-known issues with inter-laboratory effects that plague meta-analyses (Guenther et al., 2010). The focus on data from a single laboratory can be viewed either as a restriction, or as a benefit. In general, integration of independent microarray studies is challenging and there is increasing acceptance that only data from the same platform can be integrated (Lukk et al., 2010). It has, however, been shown that when data are combined from different laboratories, and where biological experiments are replicated across laboratories, that the biological effects are stronger than the laboratory effects. Nevertheless, for such a merger to be informative, one must have the same biological condition across several labs, otherwise, lab-specific effects cannot be distinguished from biological effects because both are being changed at the same time. Because our focus was to investigate cell-specificity through developmental stages and across regional specification, it was difficult to amass such redundant public data across laboratories. Additionally, when we tried including new cell types generated in other laboratories, apparent artifacts were introduced in to the analysis. As such, we focused our analysis on the rich cell-state-specific compendium of data generated in our laboratory, for which no such artifacts were apparent.
Using murine ESCs, Correa-Cerro and colleagues (Correa-Cerro et al., 2011) screened the effect of single overexpressed TFs from a panel of 137 transcription factors and selected for those which had the ability to induce a transcriptome shift towards specific lineages at 48 hours post induction. They followed that report by testing some of their selected TF by directly differentiating four distinct cell types and verifying some successful differentiation (Yamamizu et al., 2013). The authors suggested that upon identification and expression of a downstream cell-specific gene combination of TF, rapid specific differentiation to mature cell subtypes can be achieved.
As this manuscript was in preparation, two groups published important studies showing that a similar type of algorithm applied to identify transcription factors specific to particular cell types profiled and deposited into public databases. Both studies also showed that cell state expression patterns can be used to predict transcription factors able reprogram cell fate from fibroblasts to retinal pigment epithelial cells (RPE) or keratinocytes (D'Alessio et al., 2015; Rackham et al., 2016). Both of these studies took advantage of large public databases containing data from at least 100 cell states to compile lists transcription factors with cell-type specific expression patterns. Rackham et al also made a tool to allow simple prediction of TFs that could effectively reprogram cell fate, and used it to demonstrate the accuracy of prediction (Mogrify)(Rackham et al., 2016). Together, these studies, demonstrated that reprogramming factors can be identified solely on the basis of their gene expression pattern.
In the current study, we narrowed the pool of cell-type specific transcription factors to those that are particular to individual cell states, such as different developmental stages of neural progenitors. In doing so, we identified cocktails of transcription factors that can be introduced into the pluripotent state and drive nearly uniform cell fate towards particular states. This allowed for the generation of neural progenitors that were more advanced developmentally at the transcriptional level, and in addition, we more prone to generating astrocytes.
Some examples for human ESC directed differentiation have been reported as well. Ectopic expression of a cocktail of neurogenin-2 (Ngn2) or NeuroD1 (Zhang et al., 2013) and ASCL1 (Chanda et al., 2014) result in efficient rapid induction of mature specific subtype of neuronal cells. Similarly, the 4 endothelial factors selected by CEMA (ERG- FLI1- HHEX- TAL1) were previously tested individually for their ability to induce hemato-endothelial programs in hPSC (Elcheva et al., 2014), however only induction of ERG was successful in promoting endothelial-like cells. The authors further reported that a single factor induction was not sufficient for a faithful induction of mature endothelial fate as witnessed by the failure of the differentiated cells to turn off their pluripotent gene expression program. In contrast, expression of combination of four endothelial CEMA-selected genes drove homogeneous differentiation toward the endothelial fate.
The results presented here point to factors that not only define particular cell lineages such as neural progenitor cells and endothelial cells but also characterize both a temporal (i.e., developmental) and regional patterns of core genes. Lastly, we have provided an interactive web-based tool to allow users to query unique factors of expression, or, simply to obtain the pattern of expression of a particular gene of interest across many human cell types. Our expectation is that the results of the CEMA output will be refined as more cell types are added. We hope that the scientific community will benefit as a result from these analyses that provide information on the types of gene expression patterns that define individual cell states.
Beyond reprogramming and direct differentiation, we expect users will benefit from having the ability to phenotype cells derived from tissue or created from PSCs in their own lab using the new groups of genes defined here. For instance, if one simply uses SOX2 and NESTIN as neural progenitor markers, it is clear from the CEMA output, that a wide array of different types of NPC express these markers. Instead, one could look to CEMA to provide a more complete picture of a cell of interest, from the tissue from which it was derived to the stage of development it might represent.
Our findings suggest that expression of CEMA selected factors can improve standard PSC differentiation protocols and facilitate generation of mature functional differentiated cells with high homogeneity. These data validate CEMA as a tool to identify gene modules important for particular cell fates, and point towards methods that could be used to generate any cell type desired from human pluripotent stem cells as long as a gene expression profile has been obtained.
4. Materials and Methods
4.1. Statistical Analysis
To determine the core set of genes uniquely expressed within a given cell type, one statistical approach would be to apply a t-test for the one cell type of interest against all others, or use analysis of variance with post-hoc tests. In fact, there are many statistical approaches which would serve a similar purpose (Cavalli et al., 2011; Liu et al., 2008; Lukk et al., 2010; McCall et al., 2011; Ogasawara et al., 2006; Su et al., 2002). We used a statistical test that compares the transcriptome of one cell type to that of every other cell type, on a cell-type-by-cell-type basis, rather than in a one-against-rest approach. That is, we queried which genes in one cell type were being expressed differently from every other cell type, and only then aggregated these pair-wise comparisons into a single test statistic.
More formally, our null hypothesis for one cell type, i, and one gene, g, was that the mean gene expression in cell type i was similar to that in at least one other cell type, j. The test statistic was computed by making use of the maximum likelihood of a set of logistic regression models for predicting the cell type from Robust Multiarray Averaged (RMA) expression values. Within each logistic regression we re-weighted each cell type so as to mitigate the effect of the number of replicates available for each class (which differed). This re-weighting ensured that cell types with more available replicates did not dominate the computation (a similar re-weighting approach was taken in (McCall et al., 2011)).
In particular, our test statistic, , for gene g and cell type of interest i, was given by , where was the change in log likelihood between two logistic regression models for predicting cell type i: (a) one which uses the expression levels for gene g and an offset term as features (the alternative model), and (b) another which used just an offset term (the null model). Furthermore, the data used to compute was restricted to only those arrays from cell types i and j, and then the test statistic for cell type i was aggregated over all cell types j. As mentioned, we used an array-weighted logistic regression in order to appropriate equal weight to each cell type regardless of the number of replicates available. We additionally set if the mean gene expression was not higher in the cell type of interest (i) as compared to cell type j, thereby creating a one-tailed test. This enabled us to find genes that were not only uniquely expressed in cell type i, but also expressed more highly as compared to all of the other cell types, which prioritized genes for follow-up.
To compute p-values for our test statistic, we estimated the null distribution by way of a permutation test, assuming that the null distribution of the test statistic was the same for each gene when examining a particular cell type, i. In particular, we estimated the null distribution for by using 100 permutations of the mapping from array to cell type. Note that these 100 permutations yielded 5,467,500 empirical null-distributed test statistics because there were 54,675 probes. Because our null hypothesis is that the mean gene expression is similar to at least one other cell type, we permuted the mapping for only one cell type j, that is, for only one term in the test statistic (where j is chosen at random from among the other cell types for each permutation). (Using a null hypothesis in which one instead permuted the mapping for all other cell types would have yielded more significant hypotheses, likely with a similar rank order to the analysis actually used, but would not have adhered as well to our analysis goals of finding uniquely-expressed genes.) We estimated the p-values from this empirical null distribution of the test statistic in the usual manner, that is, by counting the proportion of times a test statistic of equal or greater value appears in the permuted data. From these p-values, we computed the False Discovery Rate (FDR) for any p-value threshold by way of q-values (Storey and Tibshirani, 2003), setting π0 = 1 which yields a conservative FDR estimate. When calling a gene significant, we used q<0.2. Dendrograms were constructed in Matlab using a euclidean distance with complete linkage for the hierarchical clustering algorithm.
Our goal here was not to improve upon existing statistical approaches in the literature, nor to compare and contrast them, only to use one that was reasonable for the problem at hand. It is likely that many other approaches would have yielded similar results, and ultimately, it would in any case be difficult to assess, in silico, which, if any, is superior. We chose the current approach because: (a) it allowed us to re-weight each class to mitigate the difference in number of replicates available within each class (a similar re-weighting was done in (McCall et al., 2011) using ANOVA with a re-sampling-based scheme, (b) it enabled us to avoid complications in estimating P values from multiple ANOVA posthoc tests, and (c), it allowed us to contrast each cell type against all other cell types on a cell-type by cell-type basis, rather than against all the rest in aggregate.
4.2. General cell culture
For tissue derived cell types, primary cells were derived and cultured in appropriate culture medium for up to 3 weeks. For pluripotent derived cell types, hESCs and hiPSCs were cultured under standard conditions with feeders and driven to differentiate under conditions as described previously. For both tissue and PSC derived cells, the indicated cell types were purified from mixed cultures either by expression of reporter construct (such as AFP-GFP for hepatocytes) and FACS, or by manual dissection based on morphology (such as rosettes for NPCs) (Patterson et al., 2012). All purified cell types were judged to be pure if > 90% positive by immunostaining for at least 3 appropriate marker genes. Many of these cell types were also assayed for appropriate function (Patterson et al., 2012). Acronyms for all cell types profiled are listed in Table 1.
4.3. Pluripotent stem cells
hESCs and hiPSCs were generated as described previously (Chin et al., 2009; Lowry et al., 2008) and cultured in standard growth conditions on immortalized feeders in DMEM containing Knockout Serum Replacer. The following lines were profiled, and some were used to make differentiated progeny described below: H1, HSF1, H9, XFiPSC2(Karumbayaram et al., 2011), hiPSC1, hiPSC2, hiPSC18(Lowry et al., 2008), hiPSC19, and hiPSC21(Chin et al., 2009; Chin et al., 2010). Each line analyzed was extensively characterized by: immunostaining for pluripotency factors (OCT4 and NANOG) and cell surface markers such as Tra1-81; Embryoid body (EB) formation and Teratoma analysis was conducted to demonstrate pluripotency; and Karyotyping analysis was conducted by Cell Line Genetics to ensure a stable karyotype. Immunostaining and gene expression analysis demonstrated that these cells were at least 95% pure(Chin et al., 2009; Lowry et al., 2008).
4.4. Pluripotent stem cell derived definitive endoderm
hESCs and hiPSCs were starved for serum replacer and administered Activin A. Four days later, these cultures showed a dramatic morphological change, and the cells induced expression of definitive endoderm markers such as SOX17 and FOXA2. Immunostaining and gene expression analysis demonstrated that these cells were at least 95% pure(Chan et al., 2012). These endodermal cultures were assayed for their ability to be further differentiated into endodermal cell types, such as hepatocytes ((Patterson et al., 2012) and below).
4.5. Pluripotent stem cell derived neural progenitor cells
hESCs and hiPSCs were driven towards the neural lineage by addition of Neural induction medium (DMEM + B27, N2, EGF, FGF, Retinoic Acid, and Shh). Two weeks later, neural rosettes formed, and were manually dissected from the culture and plated onto Laminin/Ornithine coated plates. This purification scheme produced cultures that were at least 92% pure as judged by immunostaining for a variety of NPC markers (SOX2, PAX6, SOX1, MUSASHI etc)(Patterson et al., 2012). The differentiation capacity of these NPCs was demonstrated upon growth factor withdrawal, where two weeks later, neurons and glia were generated(Patterson et al., 2012).
4.6. Pluripotent stem cell derived neurons
PSC-NPCs were subjected to growth factor withdrawal and allowed to differentiate towards neurons and glia. The cultures were transfected with DCX-GFP, which shows high specificity to neurons in culture. Neurons were then isolated by FACS for GFP positive cells, and collected into RNA lysis buffer.
4.7. Pluripotent stem cell derived fibroblasts
hESCs and hiPSCs were treated with collagenase to produce floating embryoid bodies in non-adherent plates. Two days later, the EBs were allowed to reattach to culture dishes in Fibroblast medium (DMEM + 10% FBS). Colonies with fibroblast morphology were manually isolated and plated into new dishes with FB medium. Cells were passaged four more times to generate pure cultures morphologically. Cell purity was assessed by immunostaining and judged to be 99% pure(Patterson et al., 2012).
4.8. Pluripotent stem cell derived hepatocytes
hESC and hiPSC derived definitive endoderm was further differentiated towards hepatocytes by the addition of various growth factors (HGF, KGF etc). Additionally, the cultures were transfected with AFP-GFP construct to highlight the hepatocyte lineage. Hepatocytes were then isolated by FACS and lysed for gene expression analysis(Patterson et al., 2012). These hepatocytes were judged to be functional by both PAS assay and their ability to secrete albumin(Patterson et al., 2012).
4.9. Pluripotent stem cell derived endothelial cells
PSC differentiating cells were FACS sorted for CD31 (PECAM) expression by CD31-PE conjugated antibody(Levenberg et al., 2010). Cells were grown in EGM2 media (Lonza) for further analysis. Matrigel tube formation assay was performed as described previously (Levenberg et al., 2010).
4.10.Tissue-derived cells
Tissue-NPC- Fetal brain and spinal cord specimens were obtained as discarded, anonymized tissues (IRB exempt). These specimens were physically and chemically dissociated to single cells and placed into NPC induction medium (see above) on Laminin/Ornithine coated plates. These cells were judged to be 100% pure NPCs by immunostaining(Patterson et al., 2012).
Tissue-fibroblasts, keratinocytes and hepatocytes were isolated from discarded anonymized human dermis or liver obtained from Lonza (FBs and Heps) or Invitrogen (Keratinocytes). Fibroblasts were grown in fibroblast media (DMEM + FBS) for two weeks prior to lysis for RNA extraction. Keratinocytes were grown in KSFM (Invitrogen). Hepatocytes were grown in Bullet Kit (Lonza). Hepatocytes were shown to be functional by albumin secretion and PAS assays, and purity was assayed by staining for Albumin(Patterson et al., 2012). Fibroblasts were assayed for their ability to secrete appropriate collagens (Patterson et al., 2012), and keratinocytes were assayed for their function in a calcium switch assay as described(Blanpain et al., 2006).
Tissue-endothelial cells, and smooth muscle cells were isolated and prepared from appropriate human tissue by Promocell. Each were judged by the manufacturer to be at least 95% pure by immunostaining for various markers.
Tissue-mesothelial, kidney epithelial, and myoepithelial cells were isolated, grown and prepared as described previously(Rheinwald et al., 1987).
4.1. Microarray profiling
Each of these cell types were lysed in the same buffer and RNA was isolated using the same method (Stratagene). All RNA samples were submitted to the same facility and hybridized to the same type of microarray chip (HUG133 2.0plus, Affymetrix) as described previously (Patterson et al., 2012). Initial standard expression analysis demonstrated that all cell types were isolated appropriately (Patterson et al., 2012) and data not shown). The expression data presented reflect the average of at least two biologically independent replicates. Functional annotation was performed using DAVID (Huang da et al., 2009)
4.12. Lentiviral constructs and infection
Polycistronic segments of four CEMA selected genes were custom synthesized by BioMatik. Reading frames were separated by the 2A self-cleaving peptide sequence. CEMA selected genes were followed by a YFP transcript engineered to be expressed in the nucleus. CEMA polycistrons were cloned into the pLVX-Tight-Puro (Lenti-X™ Tet-On, Clontech) under the P-Tight composite promoter. Lentiviral particles were generated in 293T cells using stranded protocols followed by concentration with Amicon Ultra-15 centrifugal units (100K; Millipore). For constitutive expression of the tetracycline-controlled transactivator (rtTA), cells were first infected with pLVX-Tet-On. Advanced lentivirus and stable clones were selected by G418.
4.13. qRT-PCR analysis
Total RNA was extracted using an RNeasy Mini Kit (QIAGEN). cDNA synthesis was performed using the Superscript III first-strand cDNA synthesis kit (Invitrogen). Real-time PCR was performed in triplicate using the SYBR green real-time PCR MIX (Roche) in the Roche lightcycler 480 machine. Primers are listed in supplementary table S2.
4.14. RNA-sequencing
Libraries were constructed according to manufacturer instructions (TruSeq Stranded Total RNA with Ribo-Zero; Illumina). Followed second strand PCR amplification, ~200bp sized libraries were excised from agarose gel and pooled together in 10nM concentration each. Samples were sequenced using Illumina HiSeq2000 on single-end 50-bp reads and aligned to human reference genome (Hg19) using Tophat (Trapnell et al., 2009). Processing using Cufflinks and Cuffdiff (Trapnell et al., 2012) was performed to obtain differential fragments per kilobase of transcript per million mapped reads (FPKM). Further analysis was performed using the cummeRbund suite (Trapnell et al., 2012). Hierarchical distance clustering dendrograms were based on Jensen-Shannon clustering metric. Only genes in which FPKM was over 0.5 in at least one sample were included.
Supplementary Material
(A) First, the top ten genes by expression level were queried on the Novartis data set (BioGPS) representing 84 distinct cell types and tissues. As shown, most of these genes were expressed at very high levels across nearly all cell types indicating that simply looking at genes by expression level does not yield cell-type specificity. (B) Taking the top ten genes to come out of CEMA analysis for tis-NPCs showed that in general these genes are highly specific to the brain. Note that one of the top ten CEMA genes was not represented on the platform used by Novartis and was instead replaced by the #11 gene on the list.
As in Figure 2, the top ten CEMA genes were queried on the Novartis data set to determine specificity. (A) CEMA output for PSC-Heps, which have been described as more immature, appeared to be more similar to fetal liver than adult. (B) The opposite was true for the CEMA output for tis-HEPs which were isolated from adult liver. Note that one of the top ten CEMA genes was not represented on the platform used by Novartis and was instead replaced by the #11 gene on each list.
(A) analysis of previously identified housekeeping genes demonstrates their relative variance across the data sets. (B–D) taking the relative expression levels of all genes across all cell types analyzed, the standard deviation of expression for potential housekeeping genes was calculated. By this analysis, RPL41 would be predicted to be the best housekeeping gene across multiple cell types because it was the least variable. These results were further divided to identify potential housekeeping genes at high (B), mid (C) and low (D) levels of expression.
(A) a screen shot taken from the CEMA website depicting the developmental tree of cell types that emerge in various tissues. (B), a screen shot from the CEMA website depicting a particular cell and the information available. (C), on all screens, genes of interest can be typed in and a histogram depicting the expression of individual genes across all cell types will be displayed.
Highlights.
Development of an algorithm, CEMA, for identification of cell specific expression modules.
Ectopic expression of CEMA selected transcription factors can derive partial reprogramming of fibroblasts.
hPSC engineered to express CEMA factors display robust differentiation capacity towards a mature progeny.
Acknowledgments
We would like to thank the Gene Expression Core Facility in the Department of Pathology at UCLA for array analysis; BSCRC Flow Core staff Jessica Scholes and Felicia Codrea for their help with FACS sorting; Jonathan Carlson for useful discussion; and Jason Ernst for thoughtful comments on the manuscript. This work was supported by an Innovation Award from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, NIH (NIGMS-P01GM99134), and a gift from Microsoft Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blanpain C, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli FM, Bourgon R, Vaquerizas JM, Luscombe NM. SpeCond: a method to detect condition-specific gene expression. Genome biology. 2011;12:R101. doi: 10.1186/gb-2011-12-10-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DN, Azghadi SF, Feng J, Lowry WE. PTK7 marks the first human developmental EMT in vitro. PloS one. 2012;7:e50432. doi: 10.1371/journal.pone.0050432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S, Ang CE, Davila J, Pak C, Mall M, Lee QY, Ahlenius H, Jung SW, Sudhof TC, Wernig M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem cell reports. 2014;3:282–296. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010 doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Cerro LS, Piao Y, Sharov AA, Nishiyama A, Cadet JS, Yu H, Sharova LV, Xin L, Hoang HG, Thomas M, et al. Generation of mouse ES cell lines engineered for the forced induction of transcription factors. Scientific reports. 2011;1:167. doi: 10.1038/srep00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio AC, Fan ZP, Wert KJ, Baranov P, Cohen MA, Saini JS, Cohick E, Charniga C, Dadon D, Hannett NM, et al. A Systematic Approach to Identify Candidate Transcription Factors that Control Cell Identity. Stem Cell Reports. 2015 doi: 10.1016/j.stemcr.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcheva I, Brok-Volchanskaya V, Kumar A, Liu P, Lee JH, Tong L, Vodyanik M, Swanson S, Stewart R, Kyba M, et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat Commun. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur-Dedeoglu B, Konu O, Bozkurt B, Ergul G, Seckin S, Yulug IG. Identification of endogenous reference genes for qRT-PCR analysis in normal matched breast tumor tissues. Oncology research. 2009;17:353–365. doi: 10.3727/096504009788428460. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, Klein OD, Thesleff I, Michon F. Sox2+ Stem Cells Contribute to All Epithelial Lineages of the Tooth via Sfrp5+ Progenitors. Developmental cell. 2012 doi: 10.1016/j.devcel.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karumbayaram S, Lee P, Azghadi S, Cooper AR, Patterson M, Kohn DB, Pyle A, Clark AT, Bryrne J, Zack JA, et al. From Skin Biopsy to Neurons Through a Pluripotent Intermediate Under Good Manufacturing Practice Protocols. Stem Cells Translational Medicine. 2011;1 doi: 10.5966/sctm.2011-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenberg S, Ferreira LS, Chen-Konak L, Kraehenbuehl TP, Langer R. Isolation, differentiation and characterization of vascular cells derived from human embryonic stem cells. Nature protocols. 2010;5:1115–1126. doi: 10.1038/nprot.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu X, Zack DJ, Zhu H, Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC bioinformatics. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nature biotechnology. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O. Human cortical interneurons take their time. Cell stem cell. 2013;12:497–499. doi: 10.1016/j.stem.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell stem cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MN, Uppal K, Jaffee HA, Zilliox MJ, Irizarry RA. The Gene Expression Barcode: leveraging public data repositories to begin cataloging the human and murine transcriptomes. Nucleic acids research. 2011;39:D1011–1015. doi: 10.1093/nar/gkq1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, Arnold CM, Chen YJ, Stanley EG, Elefanty AG, et al. Functional Maturation of hPSC-Derived Forebrain Interneurons Requires an Extended Timeline and Mimics Human Neural Development. Cell stem cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara O, Otsuji M, Watanabe K, Iizuka T, Tamura T, Hishiki T, Kawamoto S, Okubo K. BodyMap-Xs: anatomical breakdown of 17 million animal ESTs for cross-species comparison of gene expression. Nucleic acids research. 2006;34:D628–D631. doi: 10.1093/nar/gkj137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, Chan DN, Ha I, Case D, Cui Y, Van Handel B, Mikkola HK, Lowry WE. Defining the nature of human pluripotent stem cell progeny. Cell Res. 2012;22:178–193. doi: 10.1038/cr.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, Gaeta X, Loo K, Edwards M, Smale S, Cinkornpumin J, Xie Y, Listgarten J, Azghadi S, Douglass SM, et al. let-7 miRNAs Can Act through Notch to Regulate Human Gliogenesis. Stem cell reports. 2014 doi: 10.1016/j.stemcr.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham OJ, Firas J, Fang H, Oates ME, Holmes ML, Knaupp AS, Consortium F, Suzuki H, Nefzger CM, Daub CO, et al. A predictive computational framework for direct reprogramming between human cell types. Nat Genet. 2016 doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- Rheinwald JG, Jorgensen JL, Hahn WC, Terpstra AJ, O'Connell TM, Plummer KK. Mesosecrin: a secreted glycoprotein produced in abundance by human mesothelial, endothelial, and kidney epithelial cells in culture. The Journal of cell biology. 1987;104:263–275. doi: 10.1083/jcb.104.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizu K, Piao Y, Sharov AA, Zsiros V, Yu H, Nakazawa K, Schlessinger D, Ko MS. Identification of transcription factors for lineage-specific ESC differentiation. Stem cell reports. 2013;1:545–559. doi: 10.1016/j.stemcr.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Nichols J, Smith AG, Li M. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol Cell Neurosci. 2004;27:332–342. doi: 10.1016/j.mcn.2004.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) First, the top ten genes by expression level were queried on the Novartis data set (BioGPS) representing 84 distinct cell types and tissues. As shown, most of these genes were expressed at very high levels across nearly all cell types indicating that simply looking at genes by expression level does not yield cell-type specificity. (B) Taking the top ten genes to come out of CEMA analysis for tis-NPCs showed that in general these genes are highly specific to the brain. Note that one of the top ten CEMA genes was not represented on the platform used by Novartis and was instead replaced by the #11 gene on the list.
As in Figure 2, the top ten CEMA genes were queried on the Novartis data set to determine specificity. (A) CEMA output for PSC-Heps, which have been described as more immature, appeared to be more similar to fetal liver than adult. (B) The opposite was true for the CEMA output for tis-HEPs which were isolated from adult liver. Note that one of the top ten CEMA genes was not represented on the platform used by Novartis and was instead replaced by the #11 gene on each list.
(A) analysis of previously identified housekeeping genes demonstrates their relative variance across the data sets. (B–D) taking the relative expression levels of all genes across all cell types analyzed, the standard deviation of expression for potential housekeeping genes was calculated. By this analysis, RPL41 would be predicted to be the best housekeeping gene across multiple cell types because it was the least variable. These results were further divided to identify potential housekeeping genes at high (B), mid (C) and low (D) levels of expression.
(A) a screen shot taken from the CEMA website depicting the developmental tree of cell types that emerge in various tissues. (B), a screen shot from the CEMA website depicting a particular cell and the information available. (C), on all screens, genes of interest can be typed in and a histogram depicting the expression of individual genes across all cell types will be displayed.