Abstract
Mild stress from ischemia, seizure, hypothermia, or infection can produce a transient neuroprotected state in the brain. In the neuroprotected state, the brain responds differently to a severe stress and sustains less injury. At the genomic level, the response of the neuroprotected brain to a severe stress is characterized by widespread differential regulation of genes with diverse functions. This reprogramming of gene expression observed in the neuroprotected brain in response to a stress is consistent with an epigenetic model of regulation mediated by changes in DNA methylation and histone modification. Here, we summarize our evolving understanding of the molecular basis for endogenous neuroprotection and review recent findings that implicate DNA methylation and protein mediators of histone modification as epigenetic regulators of the brain’s response to injury.
Keywords: ischemia, seizure, neuroprotection, epigenetic regulation, PcG proteins, DNA methylation
Modulation of gene expression regulates injury
The brain has endogenous protective mechanisms that modulate the degree of injury sustained in response to a given stress. In ischemic tolerance, exposure to a sublethal ischemic event (preconditioning) protects the brain against a subsequent, severe ischemic challenge, producing tolerance [1, 2]. The preconditioned brain shows reduced activation of cell death pathways in response to a severe, normally injurious ischemic event [3]. Classic ischemic tolerance is a gene-based mechanism that takes 1–3 days to develop in vivo [4, 5] or 24 hours to develop in vitro [6–9]. Classic tolerance also requires new protein synthesis [7, 10], as demonstrated by the ability of cycloheximide to block ischemic tolerance. Analogously, several studies have suggested that transient ischemic attacks (TIAs) in humans are associated with improved clinical outcome after stroke, perhaps because they induce ischemic tolerance [11–14].
The preconditioned brain, when challenged by severe ischemic stress, reprograms its genomic response from one of injury induction to one of neuroprotection. Multiple studies have focused on identifying the critical survival genes upregulated by ischemic tolerance. For example, upregulation of the pro-survival protein Bcl-2 is necessary for delayed tolerance to focal ischemia [5, 9]. However, when we used DNA microarrays to measure RNA expression in mice exposed to severe ischemia, with or without preconditioning, we found that numerous genes with diverse functions are differentially regulated in the tolerant brain and that most are downregulated rather than upregulated [15]. This suggests that gene silencing allows endogenous pro-survival mechanisms to predominate and as such represents a potent neuroprotective strategy. Thus, the transcriptional response to a stress dictates outcome (injury vs. protection), and the genomic response to an injurious stress can be reprogrammed to produce endogenous neuroprotection (tolerance).
The tolerance phenomenon is not limited to ischemia. Prolonged epileptic seizures cause brain injury. In epileptic tolerance, brief seizures (or kindling) protect the brain against injury from subsequent prolonged seizures [16]; [17]. Epileptic tolerance is also associated with large-scale transcriptional suppression, specifically involving genes associated with calcium regulation and excitability pathways [18]. Epileptic seizures also protect against ischemia, and preconditioning ischemia protects against epileptic brain injury. Adenosine A1 receptors and KATP channels have been implicated in such “cross tolerance” [19]. Other examples of cross-tolerance include hypothermia protecting against subsequent ischemia [20] and ischemia protecting against subsequent traumatic brain injury [21].
Small signaling molecules can also induce tolerance; many of these molecules target pathways involved in inflammation, such as the NF-κB and TNF pathways. These molecules engage Toll-like receptors (TLR), specifically TLR4 and TLR9 [22]. Lipopolysaccharide (LPS), a TLR4 receptor agonist, has been used as an inflammatory stimulus to produce ischemic tolerance in numerous systems. LPS pretreatment also protects against reperfusion injury in the heart [23]. In the brain [24], the characteristics of preconditioning LPS administration are intriguingly similar to those of preconditioning induced by ischemia: delayed development [25] [4], protein synthesis dependence, and a reprogramed cellular response to a potentially injurious stimulus. Doses of LPS that produce tolerance do not result in brain injury, although they do induce inflammation. Higher doses of LPS preclude tolerance [25, 26]. LPS does not cause acute changes in cerebral blood flow [10, 23]. Preconditioning with LPS also stimulates NO synthesis [23].
Thus, the brain can protect itself from a severe insult though endogenous mechanisms that reprogram the transcriptional response. In the next sections, we summarize our current understanding of the role of epigenetic mechanisms in regulating the brain’s response to injury.
DNA methylation as an epigenetic mediator of neuroprotection
Methylation of DNA plays an important role in the epigenetic modulation of gene expression [27]. It may play a role in regulating the differential responses observed in ischemic injury and ischemic tolerance. Preconditioning ischemia upregulates methyl CpG binding protein 2 (MECP2) [28], a transcriptional repressor that binds methylated DNA and recruits histone deacetylases to repress transcription [29]. In addition, one study has shown that global DNA methylation increases in the brain after ischemic injury. Blocking DNA methylation with 5′-azacytidine, which inhibits DNA methyltransferase activity, attenuated ischemic brain injury [30], and heterozygous knockout of DNA methyltransferase (DNMT) reduced infarct size [30]. Furthermore, another study has shown that upregulation of methylation, induced with the DNA methylation agent methylazoxymethanol, blocks the neuroprotective effects of ischemic tolerance [31]. These findings suggest that reduced DNA methylation confers neuroprotection in the ischemic brain.
While there is evidence that a global reduction in DNA methylation mediates the neuroprotective effects of ischemic tolerance, additional studies suggest that the changes in methylation that mediate ischemic injury and protection are nuanced, the effect dependent on the pattern of methylation changes. In an in vitro ischemia model, an overall attenuation of methylation was observed in ischemic preconditioning, ischemic injury, and ischemic tolerance (mild preconditioning ischemia followed by severe ischemia) [32]. The decrease in global methylation was more pronounced in ischemic tolerance, consistent with the idea that a reduction in methylation contributes to tolerance. At the same time, foci of increased methylation on chromosomes 1, 7, and 17 were observed in ischemic tolerance. In ischemic injury, foci of increased methylation occurred on chromosomes 2, 12, and 13. Thus, in injury and tolerance, methylation was selectively and differentially enhanced at specific chromosomes. The incidence of methylation followed a similar pattern of distribution in different genomic regions in ischemic injury and tolerance. However, in preconditioned cells, the incidence of exon- and intron-associated methylation increased and the incidence of intergenic methylation decreased, relatively. In addition, near transcription start sites, methylation clustered at similar locations in preconditioning and injury, but at a different location in tolerance [32]. Thus, changes in DNA methylation are regional and multifactorial.
In several model systems, changes in DNA methylation appear to regulate tolerance. Both preconditioning ischemia and preconditioning seizure induce neuroprotection through genomic reprograming. As in ischemic tolerance, a reduction in gene expression occurs in seizure tolerance (mild preconditioning seizure followed by severe seizure). Thus, gene repression may mediate tolerance in both the epileptic and ischemic brain [18]. In turn, altered DNA methylation might mediate gene repression. As in ischemic tolerance, the DNA methylation pattern in seizure tolerance shows a global pattern of alteration, with increases and decreases in methylation[33]. In ischemia and seizure, different stresses produce distinct patterns of chromatin methylation on specific chromosomes [33], but it is uncertain how individual chromosomes are targeted for differential methylation. Consistently, however, chromosome-specific gene expression in excitotoxicity has been reported [34]. Chromosomal transcriptional bias is an evolving molecular mechanism that may play a role in the differential transcriptional responses to stress. Attenuation of transcription is associated with the movement of chromosomes into active nuclear territories [35]. A pertinent example is that of ChrX movement in modeled epilepsy [36].
Accordingly, studies of different models of tolerance have shown that dynamic changes in DNA methylation occur in the settings of preconditioning, injury, and tolerance. In cells subject to ischemic preconditioning, ischemic injury, and ischemic tolerance, DNA hypomethylation predominates as the response. However, selective and regionally specific increases in methylation, resulting in hypermethylation, are also observed. The effect of these events upon transcription awaits definition.
Proteins as epigenetic mediators of neuroprotection
Although the transcriptional signature of ischemic tolerance is transcriptional suppression, tolerance is nonetheless protein synthesis dependent. These facts appear to be somewhat contradictory, until the protein-dependence of gene silencing is taken into account. The genes uniquely regulated in tolerance have diverse functions, including regulation of metabolism, transport, participation in immune/defense responses, and control of cell cycle [15]. An epigenetic mechanism is compatible with the regulation of a broad range of genes. Consistently, when we used an unbiased, quantitative proteomic approach coupled with biochemical and physiological studies to characterize the proteomes of sham control, ischemic-preconditioned, ischemic-injured, and ischemic-tolerant mouse brains [37], we found that: 1) Ischemic-tolerant brains are enriched with repressive epigenetic regulators, including polycomb group (PcG) proteins and the histones that PcG proteins modify; 2) PcG proteins are essential for ischemic tolerance (as shown by over- and under-expressing several PcG proteins in vitro and in vivo); and 3) PcG proteins regulate physiological activities that are repressed in tolerance, particularly potassium channel function [15]. These results support an epigenetic mechanism in endogenous neuroprotection, one involving PcG proteins as central mediators of brain ischemic tolerance. Other studies have shown that PcG proteins target genes throughout the genome, including genes regulated in ischemic-tolerant brains [38–41]. Thus, a PcG protein-mediated, endogenous neuroprotective mechanism against ischemic injury explains the genomics and the physiology of ischemic tolerance in the brain.
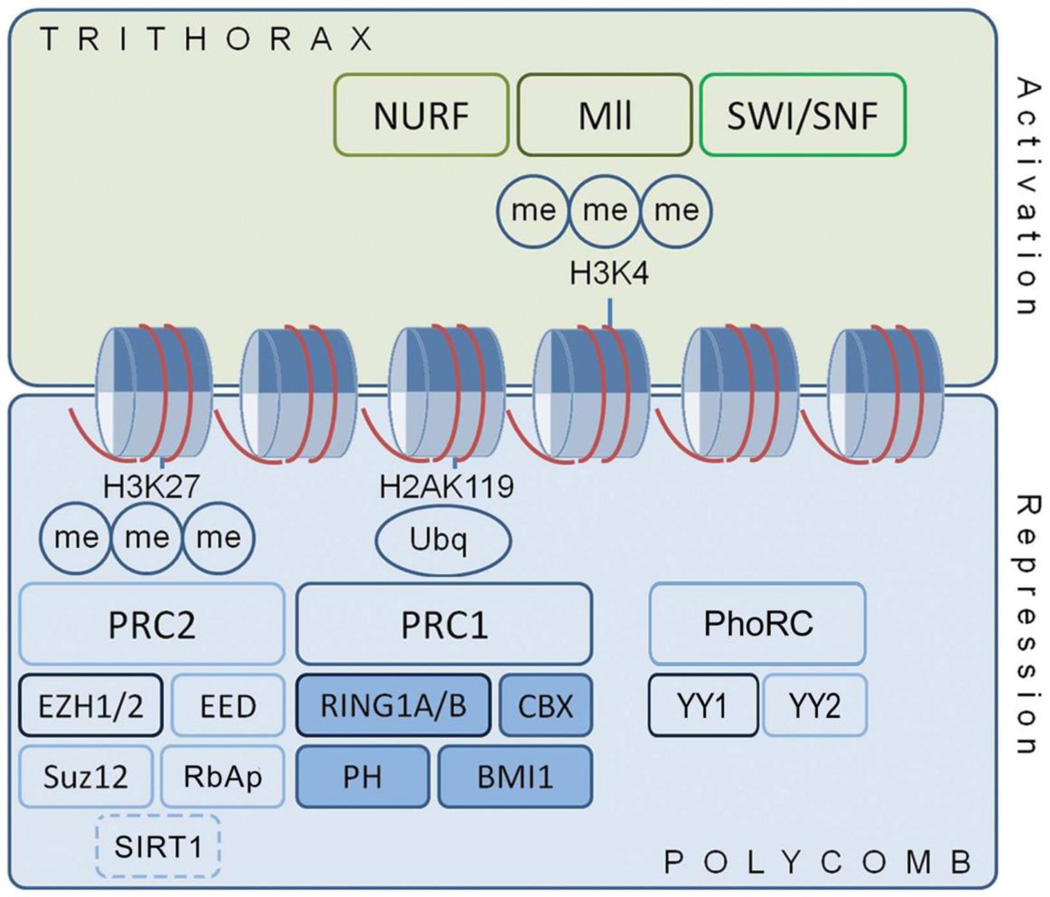
When PcG proteins were examined in the epileptic brain, bidirectional changes in the expression of genes associated with the polycomb repressive complexes (PRC1 and PRC2) and Pho repressive complex (PhoRC) (Figure 1) were observed after injurious seizure. The expression of several genes, including Bmi1, Ring1b, Ezh2, Suz12, Sirt1, and Yy1, increased rapidly (1 h) and then decreased (4–24 h post injurious seizure). Tolerance, induced by a brief seizure the day before injurious status epileptics (resulting in ~50% damage to the vulnerable CA3 subfield), resulted in immediate, regional, and temporal changes in the transcription of the polycomb genes Bmi1, Ezh1, Ezh2, Suz12, Yy1, and Yy2. PcG transcripts were downregulated in injury and tolerance, but to different extents and in a differential manner for genes associated with PRC1 and PRC2. Expression of PRC2-associated genes was higher in tolerance than in injury, while expression of PRC1-associated genes was lower in tolerance [42].
Figure 1. Regulation of chromatin structure and gene expression by PcG and TrxG proteins.
PcG proteins regulate chromatin structure through post-translational modifications, particularly histone tail methylation. PcG proteins and their functional antagonists, TrxG proteins, produce a bivalent state that is responsive to signaling events. Ancillary DNA-binding proteins, such as YY1 in the Pho repressive complex, aid recruitment of PRC1 and PRC2 to target genes. They interact with DNA and chromatin modifiers, including DNA methyl transferases, histone deacetylases, histone acetyltransferases, and the Jarid pathway proteins. H2A, histone 2A; H3, histone 3; Me, methyl group; PRC, polycomb repressive complex; Ubq, ubiquitin; PhoRC, Pho repressive complex. (From Reynolds et al., Transcriptional response of polycomb group genes to status epilepticus in mice is modified by prior exposure to epileptic preconditioning, Frontiers in Neurology, 6:46, 2015)
PcG proteins assemble into at least three major complexes that work in concert. Each has a distinct role in epigenetic regulation and distinct histone modification activities (Figure 1). The catalytic component of PRC2, Ezh1/2, is a histone methyltransferase that trimethylates histone H3 at lysine 27, a histone modification associated with gene repression. Histone H3 trimethylation in turn recruits PRC1. In PRC1, the E3 ubiquitin ligases RING1A and RING1B monoubiquitinate histone H2A at lysine 119. PcG proteins also interact directly with genomic DNA at polycomb response elements (PRE). PcG protein target genes include, but are not limited to, those that encode electron transporters, glucose transporters, endopeptidases, oxidoreductases, and G-protein coupled receptors [41, 43, 44]. An anti-oxidative role of the PcG protein Bmi1 has also been reported [45].
PcG proteins are not the only epigenetic regulators. For many genes, the repressive effect of PcG proteins is countered by the activity of trithorax group proteins (TrxG), which modify different histone residues, namely lysine 4 in histone H3, and thereby activate, rather than repress, target genes (Figure 1). Thus, PcG and TrxG proteins together represent a “bivalent” model of epigenetic regulation in which PcG proteins act as transcriptional silencers and TrxG proteins antagonize PcG silencing. Many target genes carry hallmarks of both active and silenced chromatin, suggesting they have the potential to be activated or silenced at a later stage (“bivalent”). In response to transcriptional cues that trigger cell fate commitment, bivalent loci are thought to switch to a fully silenced state or a fully active state [46]. This model implies that increased activity of PcG proteins in ischemic tolerance is accompanied by decreased activity of TrxG proteins. Consistently, treatment of mice with a peptide that disrupts interactions in the TrxG MLL complex and inhibits histone H3 methylation at lysine 4, reduces ischemic injury[47].
The components of the PcG and TrxG regulatory machineries that are uniquely regulated in brain ischemic tolerance include Scmh1, Bmi1, and Ring1B, three PRC1 proteins that function as interacting proteins, linking PRC1 to other proteins, including a facilitator of histone H2A monoubiquitination and a ligase for H2A ubiquitination [48, 49]. Compositional changes in core PcG complexes have been implicated in cell fate determination [50, 51]. In ischemic-tolerant brains and neuronal cultures, Scmh1, Bmi1, and Ring1B are robustly upregulated. On the other hand, TrxG proteins, which also form multi-protein complexes, may change in the opposite direction. The expression and roles of other PcG and TrxG proteins in ischemic tolerance remain to be elucidated.
Data indicate that PcG and TrxG proteins are regulated during the induction of brain ischemic tolerance, that differential changes in individual PcG and TrxG proteins result in dynamic changes in the composition of PcG and TrxG complexes, and that ratios between different PcG and TrxG proteins ultimately determine whether a target gene is activated or silenced. We do not yet know which components of the regulatory complexes are most responsive in the settings of injury and tolerance, nor do we understand the regulatory mechanisms that differentially regulate PcG and TrxG proteins during the induction of brain ischemic tolerance. Given that the expression of PcG genes changes during development [52], that PcG protein levels increase under tolerant conditions, and that the induction of ischemic tolerance depends on new protein synthesis [10], it is reasonable to assume that increased PcG protein levels are a result of increased PcG gene expression. However, other mechanisms might also regulate PcG protein levels and play a prominent role in the development of ischemic tolerance, such as regulation at the translational level. Indeed, the timing of changes in PcG protein expression and biosynthesis under ischemic-tolerant conditions suggests robust, early upregulation, consistent with transcription-independent translational regulation. In addition, emerging evidence supports a role for microRNAs in the regulation of PcG protein expression [53, 54].
In summary, the development of tolerance involves a rapid increase in the biosynthesis of new PcG proteins (likely accompanied by a decrease in TrxG biosynthesis), altering the stoichiometry of PcG and TrxG complexes and the balance between gene silencing and gene activation. Proteome-wide characterization of newly synthesized proteins offers the ability to identify novel proteins with therapeutic potential.
Conclusion
The brain’s intrinsic mechanisms for protecting itself from insults involve widespread changes in gene expression and DNA methylation, suggesting that successful interventions to reduce brain injury will arise not from targeting an isolated molecule or pathway but from inducing a coordinated transcriptional response. To this end, understanding the role of broad-based epigenetic regulation in establishing endogenous neuroprotection offers a new, promising avenue for the development of treatments for stroke, epilepsy, and other neurological disorders.
Abbreviations
- LPS
lipopolysaccharide
- PcG
polycomb group
- PRC
polycomb repressive complex
- TLR
Toll-like receptor
- TrxG
trithorax group
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dirnagl U, Simon RP, Hallenbeck JM. Ischemic tolerance and endogenous neuroprotection. Trends Neurosci. 2003;26(5):248–254. doi: 10.1016/S0166-2236(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 2.Gidday JM. Cerebral preconditioning and ischaemic tolerance. Nature reviews. Neuroscience. 2006;7(6):437–448. doi: 10.1038/nrn1927. [DOI] [PubMed] [Google Scholar]
- 3.Dave KR, et al. Ischemic preconditioning preserves mitochondrial function after global cerebral ischemia in rat hippocampus. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2001;21(12):1401–1410. doi: 10.1097/00004647-200112000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, et al. Stress proteins and tolerance to focal cerebral ischemia. J Cereb Blood Flow Metab. 1996;16(4):566–577. doi: 10.1097/00004647-199607000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Shimizu S, et al. bcl-2 Antisense treatment prevents induction of tolerance to focal ischemia in the rat brain. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2001;21(3):233–243. doi: 10.1097/00004647-200103000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Bossenmeyer-Pourie C, Daval JL. Prevention from hypoxia-induced apoptosis by pre-conditioning: a mechanistic approach in cultured neurons from fetal rat forebrain. Brain research. Molecular brain research. 1998;58(1–2):237–239. doi: 10.1016/s0169-328x(98)00123-5. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Zulueta M, et al. Requirement for nitric oxide activation of p21(ras)/extracellular regulated kinase in neuronal ischemic preconditioning. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(1):436–441. doi: 10.1073/pnas.97.1.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tauskela JS, et al. Preconditioning of cortical neurons by oxygen-glucose deprivation: tolerance induction through abbreviated neurotoxic signaling. American journal of physiology. Cell physiology. 2003;285(4):C899–C911. doi: 10.1152/ajpcell.00110.2003. [DOI] [PubMed] [Google Scholar]
- 9.Meller R, et al. CREB-mediated Bcl-2 protein expression after ischemic preconditioning. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2005;25(2):234–246. doi: 10.1038/sj.jcbfm.9600024. [DOI] [PubMed] [Google Scholar]
- 10.Barone FC, et al. Ischemic preconditioning and brain tolerance: temporal histological and functional outcomes, protein synthesis requirement, and interleukin-1 receptor antagonist and early gene expression. Stroke; a journal of cerebral circulation. 1998;29(9):1937–1950. doi: 10.1161/01.str.29.9.1937. discussion 1950-1. [DOI] [PubMed] [Google Scholar]
- 11.Weih M, et al. Attenuated stroke severity after prodromal TIA: a role for ischemic tolerance in the brain? Stroke; a journal of cerebral circulation. 1999;30(9):1851–1854. doi: 10.1161/01.str.30.9.1851. [DOI] [PubMed] [Google Scholar]
- 12.Moncayo J, et al. Do transient ischemic attacks have a neuroprotective effect? Neurology. 2000;54(11):2089–2094. doi: 10.1212/wnl.54.11.2089. [DOI] [PubMed] [Google Scholar]
- 13.Sitzer M, et al. Transient ischaemic attack preceding anterior circulation infarction is independently associated with favourable outcome. Journal of neurology, neurosurgery, and psychiatry. 2004;75(4):659–660. doi: 10.1136/jnnp.2003.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaller B. Ischemic preconditioning as induction of ischemic tolerance after transient ischemic attacks in human brain: its clinical relevance. Neuroscience letters. 2005;377(3):206–211. doi: 10.1016/j.neulet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Stenzel-Poore MP, et al. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: similarity to neuroprotective strategies in hibernation and hypoxiatolerant states. Lancet. 2003;362(9389):1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- 16.Sasahira M, et al. Epileptic tolerance: prior seizures protect against seizure-induced neuronal injury. Neuroscience letters. 1995;185(2):95–98. doi: 10.1016/0304-3940(94)11233-9. [DOI] [PubMed] [Google Scholar]
- 17.Kelly ME, McIntyre DC. Hippocampal kindling protects several structures from the neuronal damage resulting from kainic acid-induced status epilepticus. Brain research. 1994;634(2):245–256. doi: 10.1016/0006-8993(94)91927-5. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez-Mateos EM, et al. Hippocampal transcriptome after status epilepticus in mice rendered seizure damage-tolerant by epileptic preconditioning features suppressed calcium and neuronal excitability pathways. Neurobiology of disease. 2008;32(3):442–453. doi: 10.1016/j.nbd.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Plamondon H, et al. Mutually protective actions of kainic acid epileptic preconditioning and sublethal global ischemia on hippocampal neuronal death: involvement of adenosine A1 receptors and K(ATP) channels. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1999;19(12):1296–1308. doi: 10.1097/00004647-199912000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Nishio S, et al. Hypothermia-induced ischemic tolerance. Annals of the New York Academy of Sciences. 1999;890:26–41. doi: 10.1111/j.1749-6632.1999.tb07978.x. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Pinzon MA, et al. Induction of tolerance against traumatic brain injury by ischemic preconditioning. Neuroreport. 1999;10(14):2951–2954. doi: 10.1097/00001756-199909290-00014. [DOI] [PubMed] [Google Scholar]
- 22.Marsh BJ, Williams-Karnesky RL, Stenzel-Poore MP. Toll-like receptor signaling in endogenous neuroprotection and stroke. Neuroscience. 2009;158(3):1007–1020. doi: 10.1016/j.neuroscience.2008.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson DA, et al. Cerebrovascular hemodynamics and ischemic tolerance: lipopolysaccharide-induced resistance to focal cerebral ischemia is not due to changes in severity of the initial ischemic insult, but is associated with preservation of microvascular perfusion. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1999;19(6):616–623. doi: 10.1097/00004647-199906000-00004. [DOI] [PubMed] [Google Scholar]
- 24.Tasaki K, et al. Lipopolysaccharide pre-treatment induces resistance against subsequent focal cerebral ischemic damage in spontaneously hypertensive rats. Brain research. 1997;748(1–2):267–270. doi: 10.1016/s0006-8993(96)01383-2. [DOI] [PubMed] [Google Scholar]
- 25.Bordet R, et al. Increase in endogenous brain superoxide dismutase as a potential mechanism of lipopolysaccharide-induced brain ischemic tolerance. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2000;20(8):1190–1196. doi: 10.1097/00004647-200008000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed SH, et al. Effects of lipopolysaccharide priming on acute ischemic brain injury. Stroke; a journal of cerebral circulation. 2000;31(1):193–199. doi: 10.1161/01.str.31.1.193. [DOI] [PubMed] [Google Scholar]
- 27.Hamidi T, Singh AK, Chen T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015;7(2):247–265. doi: 10.2217/epi.14.80. [DOI] [PubMed] [Google Scholar]
- 28.Lusardi TA, et al. Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J Cereb Blood Flow Metab. 2010;30(4):744–756. doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature genetics. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 30.Endres M, et al. DNA methyltransferase contributes to delayed ischemic brain injury. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20(9):3175–3181. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maysami S, et al. Proliferating progenitor cells: a required cellular element for induction of ischemic tolerance in the brain. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2008;28(6):1104–1113. doi: 10.1038/jcbfm.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meller R, Pearson A, Simon RP. Dynamic changes in DNA methylation in ischemic tolerance. Frontiers in neurology. 2015;6:102. doi: 10.3389/fneur.2015.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller-Delaney SF, et al. Differential DNA methylation patterns define status epilepticus and epileptic tolerance. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(5):1577–1588. doi: 10.1523/JNEUROSCI.5180-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamova B, et al. The X-chromosome has a different pattern of gene expression in women compared with men with ischemic stroke. Stroke; a journal of cerebral circulation. 2012;43(2):326–334. doi: 10.1161/STROKEAHA.111.629337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447(7143):413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 36.Borden J, Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242(4886):1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- 37.Stapels M, et al. Polycomb group proteins as epigenetic mediators of neuroprotection in ischemic tolerance. Science signaling. 2010;3(111):ra15. doi: 10.1126/scisignal.2000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bracken AP, et al. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20(9):1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohler C, Villar CB. Programming of gene expression by Polycomb group proteins. Trends in cell biology. 2008;18(5):236–243. doi: 10.1016/j.tcb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Otte AP, Kwaks TH. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Current opinion in genetics & development. 2003;13(5):448–454. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 41.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature reviews. Cancer. 2006;6(11):846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds JP, et al. Transcriptional Response of Polycomb Group Genes to Status Epilepticus in Mice is Modified by Prior Exposure to Epileptic Preconditioning. Frontiers in neurology. 2015;6:46. doi: 10.3389/fneur.2015.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Current opinion in cell biology. 2006;18(3):275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Tolhuis B, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nature genetics. 2006;38(6):694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 45.Chatoo W, et al. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29(2):529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA biology. 2009;6(2):129–137. doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- 47.Liu K, et al. Neuronal necrosis is regulated by a conserved chromatin-modifying cascade. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(38):13960–13965. doi: 10.1073/pnas.1413644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Molecular cell. 2005;20(6):845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Takada Y, et al. Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development. 2007;134(3):579–590. doi: 10.1242/dev.02747. [DOI] [PubMed] [Google Scholar]
- 50.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136(21):3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 51.Kerppola TK. Polycomb group complexes--many combinations, many functions. Trends in cell biology. 2009;19(12):692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel T, Stoykova A, Gruss P. Differential expression of polycomb repression complex 1 (PRC1) members in the developing mouse brain reveals multiple complexes. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235(9):2574–2585. doi: 10.1002/dvdy.20876. [DOI] [PubMed] [Google Scholar]
- 53.Friedman JM, Jones PA, Liang G. The tumor suppressor microRNA-101 becomes an epigenetic player by targeting the polycomb group protein EZH2 in cancer. Cell cycle. 2009;8(15):2313–2314. doi: 10.4161/cc.8.15.9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim DH, et al. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]