Abstract
Chemical shift anisotropy (CSA) tensors offer a wealth of information for structural and dynamics studies of a variety of chemical and biological systems. In particular, CSA of amide protons can provide piercing insights into hydrogen-bonding interactions that vary with the backbone conformation of a protein and dynamics. However, the narrow span of amide proton resonances makes it very difficult to measure 1H CSAs of proteins even by using the recently proposed 2D 1H/1H anisotropic/isotropic chemical shift (CSA/CS) correlation technique. Such difficulties due to overlapping proton resonances can in general be overcome by utilizing the broad span of isotropic chemical shifts of low-gamma nuclei like 15N. In this context, we demonstrate a proton-detected 3D 15N/1H/1H CS/CSA/CS correlation experiment at fast MAS frequency (70 kHz) to measure 1H CSA values of unresolved amide protons of N-acetyl-15N-L-valyl-15N-L-leucine (NAVL).
Keywords: solid-state NMR, ultrafast MAS, 1H CSA, amide proton, 3D NMR, peptide, 1H detection
Graphical abstract
Introduction
Chemical shift anisotropy (CSA) tensors measured using solution and solid-state NMR methods have always played a pivotal role in getting deeper insights into inter and intra molecular H-bonding, electrostatic, and π-electron interactions.1-12 All these interactions heavily rely on the relative position of protons with respect to each other and/or other atoms. Subsequently, measurement of 1H CSA has been of considerable interest in various research groups.13-20 In particular, amide protons (NH) are mostly involved in H-bonding interactions providing structural strength to enormous chemical and biological systems and, consequently, 1H CSA which is extremely sensitive to the local electronic environment and dynamics can lead to a comprehensive understanding of these interactions. In general, 15N isotropic/anisotropic chemical shifts of amide groups11,21,22 are used to study H-bonding interactions. Since 15N nuclei indirectly participate in H-bonding interactions, therefore measurement of 1H isotropic/anisotropic chemical shifts of amide protons that are directly involved in H-bonding is of a greater significance. Nevertheless, the distribution of isotropic chemical shift of amide protons is rather limited and its relationship with the strength of H-bonding interaction is not so obvious. This brings a strong motivation to develop methods to measure 1H CSA of H-bonded amide protons.
Usually, the determination of 1H CSA has always been challenging due to its small size and the presence of strong homonuclear dipolar interactions between 1Hs. In the past several methods using rotor-synchronous pulse sequences are reported that work well for its extraction either at slow magic angle spinning (MAS) or fast MAS. In a previous report, site-resolved 1H CSA measurement of amide protons in proteins is already demonstrated through 13C-detected 3D 1H/15N/13C CSA/CS/CS experiment at a moderate MAS rate of 14 kHz.15 Herein, the overlapped 1H resonances were resolved in the 15N/13C dimensions, allowing a site specific determination of 1H CSA of amide groups. While 1H CSA can be measured at moderate MAS rates, the development of methods for its measurement at fast MAS (> 60 kHz)18 are essential in order to avoid complexities associated with the experimental setup at slow MAS. Recently we demonstrated a 2D 1H/1H CSA/CS correlation method utilizing fast MAS and composite-180° pulse-based rotor-synchronized γ-encoded CSA recoupling sequences that result in undistorted 1H CSA lineshapes with excellent efficiency.23 In continuation to this, we also presented a 3D 1H/1H/1H CSA/CSA/CS correlation experiment mediated through 1H/1H radio frequency-driven recoupling (RFDR) to extract relative orientation between two interacting 1H CSA tensors at fast MAS.24 However, 2D 1H/1H CSA/CS experiment cannot be utilized for CSA measurements of amide protons due to the lack of resolution in the 1H CS dimension. In this regard, we present a 3D 15N/1H/1H CS/CSA/CS correlation experiment on N-acetyl-15N-L-valyl-15N-L-leucine (NAVL) that results in site-resolved 1H CSAs measured through 15N isotropic chemical shifts. Manifold benefits of the 3D 1H CSA measurement at fast MAS include: 1) enhanced sensitivity by 1H detection,25-37 2) requirement of a tiny amount (0.3 – 1 mg) of sample due to small rotor volume (micro-coil probe design),38-44 3) direct correlation between 1H CSA and 1H CS which otherwise cannot be obtained due to overlap of 1H CS resonances, 4) efficient and robust 1H CSA recoupling,23,24,45 and 5) low-power sequences can be implemented to avoid sample heating.46-50
Pulse sequence
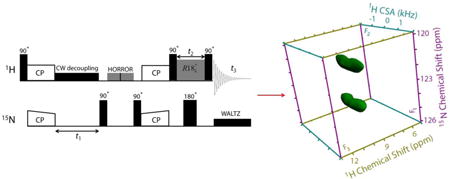
The pulse sequence used for the proton-detected 3D 15N/1H/1H CS/CSA/CS correlation experiment is shown in Figure 1. 1H magnetization is transferred to 15N nuclei using ramped-amplitude cross-polarization (RAMP-CP).51 After which 15N nuclei are allowed to evolve during t1 under their isotropic chemical shifts in the presence of 15N-1H heteronuclear low-power CW decoupling on the 1H channel. 15N magnetization is then back transferred to 1H by the second RAMP-CP. Residual 1H transverse magnetization is removed by the use of the homonuclear rotary resonance (HORROR) sequence52-54 on the 1H channel just before the application of second CP.55 While the 15N magnetization is stored along the z-axis by the first 90° pulse on 15N just before the HORROR irradiation to avoid time-evolution and transversal relaxation, the second 90° pulse on 15N prepares magnetization for the second CP step. 1H magnetization is stored along the z-axis by the use of a 90° pulse just after the second CP step and is allowed to evolve under the recoupled 1H CSA using the symmetry-based sequence with the composite-180° pulse element during the t2 period.23 A 180° pulse in the middle of CSA recoupling period (t2) is applied on the 15N channel to decouple 15N-1H hetronuclear couplings in contrast to the previously reported pulse sequences wherein 180° pulses are applied in the middle of every cycle time of the CSA recoupling pulses.15-17,56 Finally, 1H magnetization is prepared for detection by the application of a 90° read pulse and data is acquired in the presence of 15N-1H WALTZ decoupling on the 15N channel. While the States-TPPI (Time Proportional Phase Incrementation)57 approach is applied to the t1 period to obtain pure absorption line shape, the amplitude modulated signals are recorded in the t2 period.
Figure 1.
Proton-detected 3D 15N/1H/1H CS/CSA/CS pulse sequence. 15N chemical shifts are expressed during t1 under the low-power 1H CW decoupling, while 1H CSAs are recoupled during t2. Simultaneous recoupling of 15N-1H hetronuclear interactions are avoided by the application of 180° pulse on the 15N channel in the middle of CSA recoupling pulses. The phase cycling scheme used in the 3D pulse sequence is as follows: ϕ1 = {4 (0), 4 (180)}, ϕ2 = {90}, ϕ3 = {0}, ϕ4 = {0}, ϕ5 = {90}, ϕ6 = {2 (0), 2 (180)}, ϕ7 = {90}, ϕ8 = {270}, ϕ9 = {{0, 180}, {90, 270}}, ϕ10= {90}, ϕ11 = {270}, ϕ12 = {0}, ϕ13 = {0}, ϕ14 = {0}, ϕacq = {0, 180, 180, 0, 180, 0, 0, 180}.
Experimental
All NMR measurements were carried out at a 600 MHz solid-state NMR spectrometer (JEOL RESONANCE Inc., JNM-ECZ600R) equipped with a 0.75 mm double-resonance ultrafast MAS probe (JEOL RESONANCE Inc.) operating at 1H and 15N Larmor frequencies of 599.67 MHz and 60.76 MHz, respectively. Approximately 0.3 mg of NAVL was packed into a 0.75 mm zirconia rotor and all data were collected at an ambient temperature under 70 kHz MAS. The 1H and 15N 90° pulse lengths were set to 0.6 and 1.4 μs, respectively. A recycle delay of 5 s and an acquisition time of 8.19 ms were used in the 3D experiment. 8 transients were coadded for each 32 t1 and t2 increments. While a longer contact time of 4 ms was used for the first CP (1H to 15N) to maximize the magnetization transfer efficiency from protons to 15N nuclei, a short contact time of 0.4 ms was used for the second CP (15N to 1H) to select only those protons that are directly bonded to nitrogens in NAVL. The double quantum CP condition in both CP steps was achieved using 15 and 55 kHz RF field strengths for 1H and 15N, respectively.47,49,50,58 The use of double quantum CP condition becomes essential especially for heat-sensitive samples like proteins to avoid sample degradation due to rf heating during the experiment. However, such matching condition can be sensitive to chemical shift offsets due to the limited rf power. In the present study as the 1H chemical shift range of amide groups is limited, we used a weak rf field on 1H and, consequently, a relatively stronger rf field on 15N during CP was applied to cover the 15N chemical shift dispersion. A 15N-1H heteronuclear continuous-wave (CW) decoupling of 9.5 kHz was applied on the 1H channel during the t1 evolution period, whereas 15N-1H WALTZ decoupling with 13 μs pulse of RF field strength 18.9 kHz was applied on the 15N channel during t3. The phase-alternated HORROR sequence was applied for 8 ms duration with 35 kHz 1H RF field. The γ-encoded symmetry-based pulse sequence was used for the 1H CSA recoupling. The composite-180° pulse in each R element of pulse sequence is a combination of 270° and 90° RF pulses with corresponding phases alternating between (70°, 250°) and (-70°, -250°). The amplitude modulated t2 signal was obtained at every 8τr period. Delta NMR software (JEOL RESONANCE Inc.) was used to process all the NMR data. While a Fourier transformation after zero filling was applied in the t1 and t3 dimensions, a DC balance to remove DC offset effects (the average of the final 1/8th FID points in t2 is subtracted from total data points) followed by zero filling and real Fourier transformation were applied in the CSA recoupling dimension (t2) to process the 3D 15N/1H/1H CS/CSA/CS correlation data. The CSA parameters were extracted through numerical fittings using SIMPSON program.59 Powder averaging was achieved using 678 (α, β) crystal orientations and 26 γ angles.60
Results and Discussion
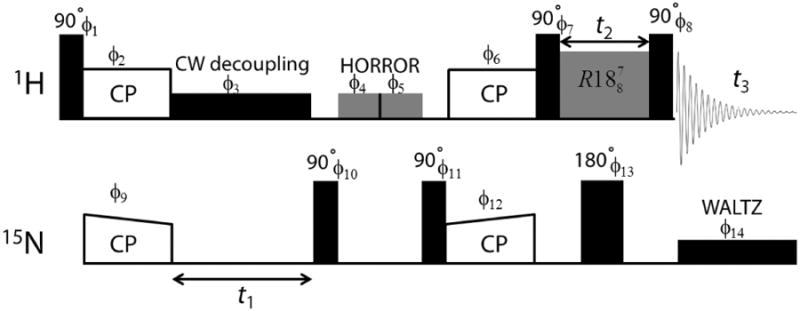
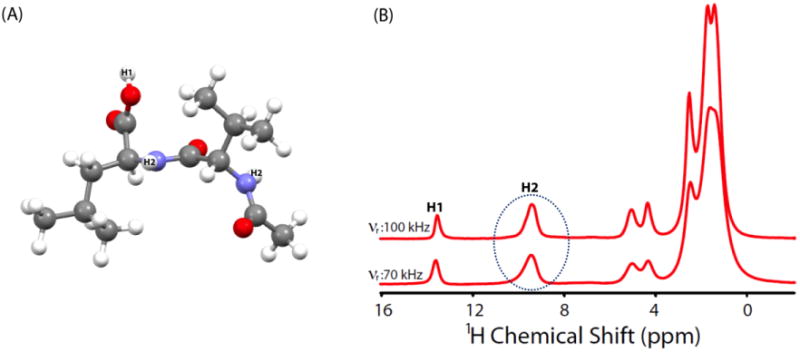
Molecular structure and 1D 1H spectra of NAVL collected at 70 and 100 kHz MAS are shown in Figure 2. While the resolution and sensitivity are seen to be significantly improved for protons H3, H4, H5, H6 and H7 (peaks in the 0 - 6 ppm range) with increasing the MAS rate due to better suppression of 1H/1H homonuclear dipolar interactions, proton resonances from amide groups of Val and Leu remain unresolved even with a MAS rate as fast as 100 kHz. In other words, spectral resolution provided by 100 kHz MAS is not sufficient enough to resolve amide proton resonances with similar isotropic chemical shift values. This problem is crucial especially for the CSA measurement of structurally relevant protons from the 2D 1H/1H CSA/CS correlation experiments that require well-resolved 1H resonances for the precise determination of CSA. To demonstrate this limitation further, we carried out 2D 1H/1H CSA/CS correlation measurement on NAVL at 70 kHz MAS.23 Since CSA and heteronuclear dipolar coupling have the same spatial and spin symmetry, the previously reported pulse sequence simultaneously recouples both interactions. Subsequently, in order to accomplish 15N-1H heteronuclear decoupling such that any recoupling of heteronuclear dipolar interactions is avoided, we applied a 180° pulse on the 15N channel in the middle of the CSA recoupling pulses. It is to be noted that NH proton CSA can also be measured in natural abundant samples wherein 14N-1H heteronuclear interactions are decoupled through on-resonance 14N CW irradiation at ultrafast MAS.45 The fast MAS spectrum of NAVL (Figure 3A) shows well-resolved resonances barring NH peaks, as a result CSA of amide protons cannot be determined. While the 2D 1H/1H CSA/CS correlation spectrum fails to provide any site-specific information about CSA of amide protons of NAVL, it can still be used to extract CSA of OH proton of –COOH group in NAVL. A spectral slice taken parallel to CSA dimension at the 1H isotropic chemical shift of OH resonance is simulated using SIMPSON59 (Figure 3B) and 1H CSA parameters obtained from the line shape fitting are: δaniso = δzz-δiso = 19.1 ppm and η = (δyy-δxx)/δaniso = 0.3. Here, δiso is the isotropic chemical shift calculated using the relation (δxx+δyy+δzz)/3, and the principal components of the chemical shift tensor (δxx, δyy and δzz) are defined as |δzz-δiso| ≥ |δxx-δiso| ≥ |δyy-δiso|.
Figure 2.
Molecular structure and 1H NMR spectra of a powder sample of NAVL recorded at 600 MHz under 70 and 100 kHz MAS frequencies.
Figure 3.
(A) Two-dimensional 1H/1H CSA/CS correlation spectrum of NAVL at 600 MHz under 70 kHz MAS obtained using a previously reported pulse sequence23 with a 180° pulse in the middle of the CSA recoupling period on the 15N channel to decouple 15N-1H heteronuclear interactions. In the 2D 1H/1H CSA/CS correlation experiment 6 scans were collected every 32 t1 increments and a recycle delay of 5 s was used (total experimental time = 16 minutes). Dotted circle indicates the two overlapped amide proton resonances. (B) The experimental CSA lineshape extracted by taking a slice (green line) parallel to the 1H CSA dimension of the 2D spectrum at the 1H isotropic chemical shift frequency of OH resonance of NAVL. Best fitting simulated lineshape (brown) was obtained using SIMPSON simulations.
The straightforward solution to overcome the issue of overlapping 1H resonances of amide groups of NAVL is to carry out CSA measurement in a 3D manner by adding a high-resolution 15N dimension so as to get well-resolved 15N/1H correlations. To this end, we implemented 3D 15N/1H/1H CS/CSA/CS pulse sequence shown in Figure 1 to obtain amide proton CSA of Val and Leu residues of NAVL. As discussed earlier, 15N chemical shifts are allowed to evolve during the incrementable t1 period, and 15N filtered 1H signal after the two CP steps then evolves under recoupled 1H CSA during the t2 period in the absence of 15N-1H heteronuclear couplings. The 2D F1/F3 (15N/1H CS/CS) projection (Figure 4B) extracted from the 3D 15N/1H/1H CS/CSA/CS spectrum of NAVL (Figure 4A) shows well-separated 15N/1H chemical shift correlations for Val and Leu residues of NAVL. Additionally, 1H isotropic chemical shifts of amide protons are more precisely determined and can be used for the structural refinement using the quantum chemical calculations. To determine CSA of amide protons, the 2D F1/F2 (15N/1H CS/CSA) projection (Figure 5A) was extracted as the first step from the 3D 15N/1H/1H CS/CSA/CS spectrum of NAVL (Figure 4A). In the subsequent step, spectral slices were taken parallel to the 1H CSA dimension of the 2D F1/F2 (15N/1H CS/CSA) projection at the 15N isotropic chemical shifts of Val and Leu residues of NAVL (Figure 5A). Finally, numerical simulations were performed to get the best fit of experimental 1H CSA lineshapes (Figure 5B). The 1H chemical shift parameters obtained are as follows: Val (δiso = 9.0 ppm, δaniso = 9.1 ppm, η = 0.7) and Leu (δiso = 9.2 ppm, δaniso = 9.1 ppm, η = 0.7). While we observed similar CSA parameters for the amide protons of NAVL, 3D 15N/1H/1H CS/CSA/CS correlation experiment presented in this study is necessary for the cases where CSA of amide protons with unresolved peaks in the proton CS dimension are different and in principle leads to a direct correlation between 1H CSA and 1H CS. Subsequently, a set of CS and CSA of each amide proton can be extracted simultaneously.
Figure 4.
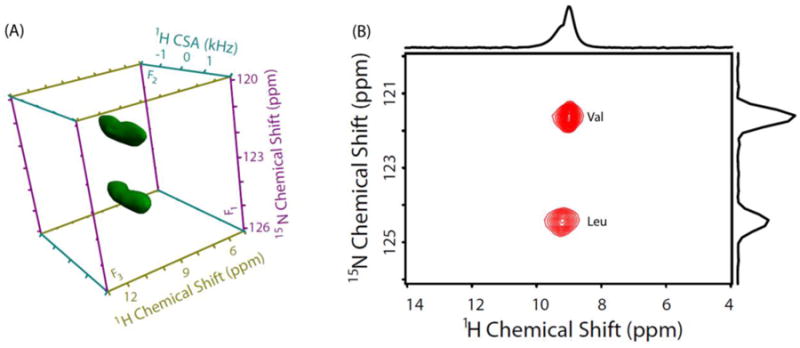
(A) 3D 15N/1H/1H CS/CSA/CS correlation spectrum of NAVL at 70 kHz MAS frequency. (B) 2D F1/F3 (15N/1H CS/CS) projection extracted from the 3D 15N/1H/1H CS/CSA/CS spectrum of NAVL. Total experimental time = 22.5 hours.
Figure 5.
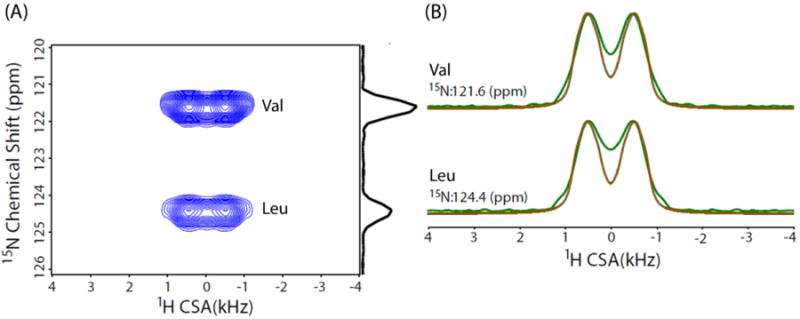
(A) Two-dimensional F1/F2 (15N/1H CS/CSA) projection extracted from the 3D 15N/1H/1H CS/CSA/CS spectrum of NAVL. (B) Spectral slices (green) taken parallel to the 1H CSA dimension at the 15N chemical shifts of Val and Leu residues of NAVL, and corresponding simulated lineshapes (brown).
Conclusion
In summary, we have presented a proton-detected 3D 15N/1H/1H CS/CSA/CS correlation experiment at fast MAS (70 kHz) on NAVL. The main objective of this experiment is to determine site-resolved 1H CSA of overlapped amide proton resonances through 15N isotropic chemical shifts. The measured CSA parameters of amide protons of Val and Leu residues of NAVL are found to be identical. In addition, we have also reported 1H CSA of well-resolved OH resonance of NAVL obtained from 2D 1H/1H CSA/CS correlation experiment. We believe that the proton-detected 3D pulse sequence presented in this study can be implemented for studies relying on 1H CSAs in more complex chemical and biological systems wherein a limited or no resolution is a major problem to overcome. Additionally, a combination of partially-deuterated samples, higher magnetic field, and faster spinning speed (>100 kHz) can further reduce the difficulties due to spectral resolution from macromolecular systems.26,31,34,61,62
Acknowledgments
This work was supported by the funds from National Institutes of Health (GM084018 and GM095640 to A. R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cornilescu G, Bax A. Measurement of Proton, Nitrogen, and Carbonyl Chemical Shielding Anisotropies in a Protein Dissolved in a Dilute Liquid Crystalline Phase. J Am Chem Soc. 2000;122:10143–10154. [Google Scholar]
- 2.Tjandra N, Bax A. Solution NMR Measurement of Amide Proton Chemical Shift Anisotropy in 15N-Enriched Proteins. Correlation with Hydrogen Bond Length. J Am Chem Soc. 1997;119:8076–8082. [Google Scholar]
- 3.Tjandra N, Bax A. Large Variations in 13C(Alpha) Chemical Shift Anisotropy in Proteins Correlate with Secondary Structure. J Am Chem Soc. 1997;119:9576–9577. [Google Scholar]
- 4.Yao LS, Grishaev A, Cornilescu G, Bax A. The Impact of Hydrogen Bonding on Amide 1H Chemical Shift Anisotropy Studied by Cross-Correlated Relaxation and Liquid Crystal NMR Spectroscopy. J Am Chem Soc. 2010;132:10866–10875. doi: 10.1021/ja103629e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao LS, Grishaev A, Cornilescu G, Bax A. Site-Specific Backbone Amide 15N Chemical Shift Anisotropy Tensors in a Small Protein from Liquid Crystal and Cross-Correlated Relaxation Measurements. J Am Chem Soc. 2010;132:4295–4309. doi: 10.1021/ja910186u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G, Freure CJ, Verdurand E. Proton Chemical Shift Tensors and Hydrogen Bond Geometry: A 1H-2H Dipolar NMR Study of the Water Molecule in Crystalline Hydrates. J Am Chem Soc. 1998;120:13187–13193. [Google Scholar]
- 7.Wei YF, de Dios AC, McDermott AE. Solid-State 15N NMR Chemical Shift Anisotropy of Histidines: Experimental and Theoretical Studies of Hydrogen Bonding. J Am Chem Soc. 1999;121:10389–10394. [Google Scholar]
- 8.Pandey MK, Vivekanandan S, Ahuja S, Pichumani K, Im SC, Waskell L, Ramamoorthy A. Determination of 15N Chemical Shift Anisotropy from a Membrane-Bound Protein by NMR Spectroscopy. J Phys Chem B. 2012;116:7181–7189. doi: 10.1021/jp3049229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey MK, Vivekanandan S, Ahuja S, Huang R, Im SC, Waskell L, Ramamoorthy A. Cytochrome-P450-Cytochrome-b5 Interaction in a Membrane Environment Changes 15N Chemical Shift Anisotropy Tensors. J Phys Chem B. 2013;117:13851–13807. doi: 10.1021/jp4086206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey MK, Ramamoorthy A. Quantum Chemical Calculations of Amide-15N Chemical Shift Anisotropy Tensors for a Membrane-Bound Cytochrome-b5. J Phys Chem B. 2013;117:859–867. doi: 10.1021/jp311116p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DK, Wittebort RJ, Ramamoorthy A. Characterization of 15N Chemical Shift and 1H-15N Dipolar Coupling Interactions in a Peptide Bond of Uniaxially Oriented and Polycrystalline Samples by One-Dimensional Dipolar Chemical Shift Solid-State NMR Spectroscopy. J Am Chem Soc. 1998;120:8868–8874. [Google Scholar]
- 12.Loth K, Pelupessy P, Bodenhausen G. Chemical Shift Anisotropy Tensors of Carbonyl, Nitrogen, and Amide Proton Nuclei in Proteins through Cross-Correlated Relaxation in NMR Spectroscopy. J Am Chem Soc. 2005;127:6062–6068. doi: 10.1021/ja042863o. [DOI] [PubMed] [Google Scholar]
- 13.Gerald R, Bernhard T, Haeberlen U, Rendell J, Opella S. Chemical-Shift and Electric-Field Gradient Tensors for the Amide and Carboxyl Hydrogens in the Model Peptide N-Acetyl-D,L-Valine - Single-Crystal Deuterium NMR-Study. J Am Chem Soc. 1993;115:777–782. [Google Scholar]
- 14.Duma L, Abergel D, Tekely P, Bodenhausen G. Proton Chemical Shift Anisotropy Measurements of Hydrogen-Bonded Functional Groups by Fast Magic-Angle Spinning Solid-State NMR Spectroscopy. Chem Commun. 2008:2361–2363. doi: 10.1039/b801154k. [DOI] [PubMed] [Google Scholar]
- 15.Hou GJ, Paramasivam S, Yan S, Polenova T, Vega AJ. Multidimensional Magic Angle Spinning NMR Spectroscopy for Site-Resolved Measurement of Proton Chemical Shift Anisotropy in Biological Solids. J Am Chem Soc. 2013;135:1358–1368. doi: 10.1021/ja3084972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou GJ, Gupta R, Polenova T, Vega AJ. A Magic-Angle-Spinning NMR Spectroscopy Method for the Site-Specific Measurement of Proton Chemical-Shift Anisotropy in Biological and Organic Solids. Isr J Chem. 2014;54:171–183. doi: 10.1002/ijch.201300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou GJ, Byeon IJL, Ahn J, Gronenborn AM, Polenova T. Recoupling of Chemical Shift Anisotropy by R-Symmetry Sequences in Magic Angle Spinning NMR Spectroscopy. J Chem Phys. 2012;137 doi: 10.1063/1.4754149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miah HK, Bennett DA, Iuga D, Titman JJ. Measuring Proton Shift Tensors with Ultrafast MAS NMR. J Magn Reson. 2013;235:1–5. doi: 10.1016/j.jmr.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Brouwer DH, Ripmeester JA. Symmetry-Based Recoupling of Proton Chemical Shift Anisotropies in Ultrahigh-Field Solid-State NMR. J Magn Reson. 2007;185:173–178. doi: 10.1016/j.jmr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Wu CH, Ramamoorthy A, Gierasch LM, Opella SJ. Simultaneous Characterization of the Amide 1H Chemical Shift, 1H-15N Dipolar, and 15N Chemical-Shift Interaction Tensors in a Peptide-Bond by 3-Dimensional Solid-State NMR-Spectroscopy. J Am Chem Soc. 1995;117:6148–6149. [Google Scholar]
- 21.Sack I, Macholl S, Wehrmann F, Albrecht J, Limbach HH, Fillaux F, Baron MH, Buntkowsky G. A 15N-1H Dipolar CSA Solid-State NMR Study of (-CO-CD2-15NH-)n. Appl Magn Reson. 1999;17:413–431. [Google Scholar]
- 22.Hou G, Paramasivam S, Byeon IJ, Gronenborn AM, Polenova T. Determination of Relative Tensor Orientations by Gamma-Encoded Chemical Shift Anisotropy/Heteronuclear Dipolar Coupling 3D NMR Spectroscopy in Biological Solids. Phys Chem chem Phys. 2010;12:14873–14883. doi: 10.1039/c0cp00795a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey MK, Malon M, Ramamoorthy A, Nishiyama Y. Composite-180° Pulse-Based Symmetry Sequences to Recouple Proton Chemical Shift Anisotropy Tensors under Ultrafast MAS Solid-State NMR Spectroscopy. J Magn Reson. 2015;250:45–54. doi: 10.1016/j.jmr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey MK, Nishiyama Y. Determination of Relative Orientation between 1H CSA Tensors from a 3D Solid-State NMR Experiment Mediated through 1H/1H RFDR Mixing under Ultrafast MAS. Solid State Nucl Magn Reson. 2015;70:15–20. doi: 10.1016/j.ssnmr.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Ishii Y, Tycko R. Sensitivity Enhancement in Solid State 15N NMR by Indirect Detection with High-Speed Magic Angle Spinning. J Magn Reson. 2000;142:199–204. doi: 10.1006/jmre.1999.1976. [DOI] [PubMed] [Google Scholar]
- 26.Wickramasinghe A, Wang S, Matsuda I, Nishiyama Y, Nemoto T, Endo Y, Ishii Y. Evolution of CPMAS under Fast Magic-Angle-Spinning at 100 kHz and Beyond. Solid State Nucl Magn Reson. 2015 doi: 10.1016/j.ssnmr.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey MK, Nishiyama Y. Proton-Detected 3D 14N/14N/1H Isotropic Shift Correlation Experiment Mediated through 1H-1H RFDR Mixing on a Natural Abundant Sample under Ultrafast MAS. J Magn Reson. 2015;258:96–101. doi: 10.1016/j.jmr.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Mroue K, Nishiyama Y, Pandey MK, Gong B, McNerny E, Kohn D, Morris M, Ramamoorthy A. Proton-Detected Solid-State NMR Spectroscopy of Bone with Ultrafast Magic Angle Spinning. Scientific Reports. 2015;5:11991. doi: 10.1038/srep11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Pandey MK, Nishiyama Y, Ramamoorthy A. A Novel High-Resolution and Sensitivity-Enhanced Three-Dimensional Solid-State NMR Experiment under Ultrafast Magic Angle Spinning Conditions. Scientific Reports. 2015;5:11810. doi: 10.1038/srep11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asami S, Rakwalska-Bange M, Carlomagno T, Reif B. Protein-Rna Interfaces Probed by 1h-Detected MAS Solid-State NMR Spectroscopy. Angew Chem. 2013;52:2345–2349. doi: 10.1002/anie.201208024. [DOI] [PubMed] [Google Scholar]
- 31.Asami S, Reif B. Proton-Detected Solid-State NMR Spectroscopy at Aliphatic Sites: Application to Crystalline Systems. Acc Chem Res. 2013;46:2089–2097. doi: 10.1021/ar400063y. [DOI] [PubMed] [Google Scholar]
- 32.Asami S, Schmieder P, Reif B. High Resolution 1H-detected Solid-State NMR Spectroscopy of Protein Aliphatic Resonances: Access to Tertiary Structure Information. J Am Chem Soc. 2010;132:15133–15135. doi: 10.1021/ja106170h. [DOI] [PubMed] [Google Scholar]
- 33.Huber M, Hiller S, Schanda P, Ernst M, Bockmann A, Verel R, Meier BH. A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination. ChemPhysChem. 2011;12:915–918. doi: 10.1002/cphc.201100062. [DOI] [PubMed] [Google Scholar]
- 34.Barbet-Massin E, Pell AJ, Retel JS, Andreas LB, Jaudzems K, Franks WT, Nieuwkoop AJ, Hiller M, Higman V, Guerry P, Bertarello A, Knight MJ, Felletti M, Le Marchand T, Kotelovica S, Akopjana I, Tars K, Stoppini M, Bellotti V, Bolognesi M, Ricagno S, Chou JJ, Griffin RG, Oschkinat H, Lesage A, Emsley L, Herrmann T, Pintacuda G. Rapid Proton-Detected NMR Assignment for Proteins with Fast Magic Angle Spinning. J Am Chem Soc. 2014;136:12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi T, Mao K, Paluch P, Nowak-Krol A, Sniechowska J, Nishiyama Y, Gryko DT, Potrzebowski MJ, Pruski M. Study of Intermolecular Interactions in the Corrole Matrix by Solid-State NMR under 100 KHz MAS and Theoretical Calculations. Angew Chem. 2013;52:14108–14111. doi: 10.1002/anie.201305475. [DOI] [PubMed] [Google Scholar]
- 36.Nishiyama Y, Endo Y, Nemoto T, Utsumi H, Yamauchi K, Hioka K, Asakura T. Very Fast Magic Angle Spinning 1H-14N 2d Solid-State NMR: Sub-Micro-Liter Sample Data Collection in a Few Minutes. J Magn Reson. 2011;208:44–48. doi: 10.1016/j.jmr.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Nishiyama Y, Malon M, Ishii Y, Ramamoorthy A. 3D 15N/15N/1H Chemical Shift Correlation Experiment Utilizing an RFDR-Based 1H/1H Mixing Period at 100 KHz MAS. J Magn Reson. 2014;244:1–5. doi: 10.1016/j.jmr.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda K. Microcoils and Microsamples in Solid-State NMR. Solid State Nucl Magn Reson. 2012;47-48:1–9. doi: 10.1016/j.ssnmr.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Hoult DI, Richards RE. Signal-to-Noise Ratio of Nuclear Magnetic-Resonance Experiment. J Magn Reson. 1976;24:71–85. doi: 10.1016/j.jmr.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Minard KR, Wind RA. Solenoidal Microcoil Design. Part I: Optimizing RF Homogeneity and Coil Dimensions. Concept Magnetic Res. 2001;13:128–142. [Google Scholar]
- 41.Minard KR, Wind RA. Solenoidal Microcoil Design - Part II: Optimizing Winding Parameters for Maximum Signal-to-Noise Performance. Concept Magnetic Res. 2001;13:190–210. [Google Scholar]
- 42.Janssen H, Brinkmann A, van Eck ERH, van Bentum PJM, Kentgens APM. Microcoil High-Resolution Magic Angle Spinning NMR Spectroscopy. J Am Chem Soc. 2006;128:8722–8723. doi: 10.1021/ja061350+. [DOI] [PubMed] [Google Scholar]
- 43.Kentgens APM, Bart J, van Bentum PJM, Brinkmann A, Van Eck ERH, Gardeniers JGE, Janssen JWG, Knijn P, Vasa S, Verkuijlen MHW. High-Resolution Liquid- and Solid-State Nuclear Magnetic Resonance of Nanoliter Sample Volumes Using Microcoil Detectors. J Chem Phys. 2008;128 doi: 10.1063/1.2833560. [DOI] [PubMed] [Google Scholar]
- 44.Sakellariou D, Le Goff G, Jacquinot JF. High-Resolution, High-Sensitivity NMR of Nanolitre Anisotropic Samples by Coil Spinning. Nature. 2007;447:694–697. doi: 10.1038/nature05897. [DOI] [PubMed] [Google Scholar]
- 45.Pandey MK, Nishiyama Y. Determination of NH Proton Chemical Shift Anisotropy with 14N-1H Heteronuclear Decoupling Using Ultrafast Magic Angle Spinning Solid-State NMR. J Magn Reson. 2015 doi: 10.1016/j.jmr.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 46.Ernst M, Samoson A, Meier BH. Low-Power Decoupling in Fast Magic-Angle Spinning NMR. Chem Phys Lett. 2001;348:293–302. [Google Scholar]
- 47.Meier BH. Cross Polarization under Fast Magic Angle Spinning - Thermodynamical Considerations. Chem Phys Lett. 1992;188:201–207. [Google Scholar]
- 48.Lange A, Scholz I, Manolikas T, Ernst M, Meier BH. Low-Power Cross Polarization in Fast Magic-Angle Spinning NMR Experiments. Chem Phys Lett. 2009;468:100–105. [Google Scholar]
- 49.Laage S, Marchetti A, Sein J, Pierattelli R, Sass HJ, Grzesiek S, Lesage A, Pintacuda G, Emsley L. Band-Selective 1H-13C Cross-Polarization in Fast Magic Angle Spinning Solid-State NMR Spectroscopy. J Am Chem Soc. 2008;130:17216–17217. doi: 10.1021/ja805926d. [DOI] [PubMed] [Google Scholar]
- 50.Amoureux JP, Pruski M. Theoretical and Experimental Assessment of Single- and Multiple-Quantum Cross-Polarization in Solid State NMR. Mol Phys. 2002;100:1595–1613. [Google Scholar]
- 51.Metz G, Wu XL, Smith SO. Ramped-Amplitude Cross-Polarization in Magic-Angle-Spinning NMR. J Magn Reson Ser A. 1994;110:219–227. [Google Scholar]
- 52.Levitt MH, Oas TG, Griffin RG. Rotary Resonance Recoupling in Heteronuclear Spin Pair Systems. Isr J Chem. 1988;28:271–282. [Google Scholar]
- 53.Oas TG, Griffin RG, Levitt MH. Rotary Resonance Recoupling of Dipolar Interactions in Solid-State Nuclear Magnetic-Resonance Spectroscopy. J Chem Phys. 1988;89:692–695. [Google Scholar]
- 54.Nielsen NC, Bildsoe H, Jakobsen HJ, Levitt MH. Double-Quantum Homonuclear Rotary Resonance - Efficient Dipolar Recovery in Magic-Angle-Spinning Nuclear-Magnetic-Resonance. J Chem Phys. 1994;101:1805–1812. [Google Scholar]
- 55.Ishii Y, Yesinowski JP, Tycko R. Sensitivity Enhancement in Solid-State 13C NMR of Synthetic Polymers and Biopolymers by 1H NMR Detection with High-Speed Magic Angle Spinning. J Am Chem Soc. 2001;123:2921–2922. doi: 10.1021/ja015505j. [DOI] [PubMed] [Google Scholar]
- 56.Hou GJ, Lu XY, Vega AJ, Polenova T. Accurate Measurement of Heteronuclear Dipolar Couplings by Phase-Alternating R-Symmetry (PARS) Sequences in Magic Angle Spinning NMR Spectroscopy. J Chem Phys. 2014;141 doi: 10.1063/1.4894226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marion D, Ikura M, Tschudin R, Bax A. Rapid Recording of 2D NMR-Spectra without Phase Cycling - Application to the Study of Hydrogen-Exchange in Proteins. J Magn Reson. 1989;85:393–399. [Google Scholar]
- 58.Pandey MK, Qadri Z, Ramachandran R. Understanding Cross-Polarization (CP) NMR Experiments through Dipolar Truncation. J Chem Phys. 2013;138 doi: 10.1063/1.4794856. [DOI] [PubMed] [Google Scholar]
- 59.Bak M, Rasmussen JT, Nielsen NC. Simpson: A General Simulation Program for Solid-State NMR Spectroscopy. J Magn Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 60.Bak M, Nielsen NC. Repulsion, a Novel Approach to Efficient Powder Averaging in Solid-State NMR. J Magn Reson. 1997;125:132–139. doi: 10.1006/jmre.1996.1087. [DOI] [PubMed] [Google Scholar]
- 61.Pandey MK, Zhang R, Hashi K, Ohki S, Nishijima G, Matsumoto S, Noguchi T, Deguchi K, Goto A, Shimizu T, Maeda H, Takahashi M, Yanagisawa Y, Yamazaki T, Iguchi S, Tanaka R, Nemoto T, Miyamoto T, Suematsu H, Saito K, Miki T, Ramamoorthy A, Nishiyama Y. 1020 Mhz Single-Channel Proton Fast Magic Angle Spinning Solid-State NMR Spectroscopy. J Magn Reson. 2015 doi: 10.1016/j.jmr.2015.10.003. DOI: http://dx.doi.org/10.1016/j.jmr.2015.10.003. [DOI] [PMC free article] [PubMed]
- 62.Bertini I, Emsley L, Lelli M, Luchinat C, Mao J, Pintacuda G. Ultrafast MAS Solid-State NMR Permits Extensive 13C and 1H Detection in Paramagnetic Metalloproteins. J Am Chem Soc. 2010;132:5558–5559. doi: 10.1021/ja100398q. [DOI] [PubMed] [Google Scholar]


