Abstract
For decades, seeking common, consistent and corresponding anatomical/functional regions across individual brains via cortical parcellation has been a longstanding challenging problem. In our opinion, two major barriers to solve this problem are determining meaningful cortical boundaries that segregate homogeneous regions and establishing correspondences among parcellated regions of multiple brains. To establish a corresponding system across subjects, we recently developed the Dense Individualized and Common Connectivity-based Cortical Landmarks (DICCCOL) system which possesses group-wise consistent white matter fiber connection patterns across individuals and thus provides a dense map of corresponding cortical landmarks. Despite this useful property, however, the DICCCOL landmarks are still far from covering the whole cerebral cortex and do not provide clear structural/functional cortical boundaries. To address the above limitation while leveraging the advantage of DICCCOL, in this paper, we present a novel approach for group-wise consistent parcellation of the cerebral cortex via a hierarchical scheme. In each hierarchical level, DICCCOLs are used as corresponding samples to automatically determine the cluster number so that other cortical surface vertices are iteratively classified into corresponding clusters across subjects within a group-wise classification framework. Experimental results showed that this approach can achieve consistent fine-granularity cortical parcellation with intrinsically-established structural correspondences across individual brains. Besides, comparisons with resting-state and task-based fMRI datasets demonstrated that the group-wise parcellation boundaries segregate functionally homogeneous areas.
Keywords: Cortical parcellation, connectivity, group-wise, dMRI
Graphical Abstract
1. INTRODUCTION
Identification of common and corresponding anatomical/functional cortical regions across different human brains via cortical parcellation has been a longstanding challenge and thus has attracted tremendous efforts from the neuroimaging and brain mapping fields (e.g., Thompson and Toga, 1996; Shen and Davatzikos, 2002; Johansen-Berg et al., 2004; Corouge et al., 2004; Fischl, et al., 1999; Cohen et al., 2008; Blumensath, et al., 2013). Current cortical parcellation methodologies can be broadly divided into three categories based on the features including morphological, connectional and functional features. The first group of methodologies decomposes the cortex into a collection of regions of interest (ROIs) based on an atlas or morphological features. For example, many such works project different brain templates/atlases onto the cortex to achieve parcellation via image registration methods (e.g., Thompson and Toga, 1996; Shen and Davatzikos, 2002; Fischl et al., 2004; Desikan et al., 2006; Gong et al., 2009). Other works are in favor of developing morphological features (e.g., Fischl et al., 1999; Lohmann and von Cramon, 2000; Rettmann, et al., 2002; Yang and Kruggel, 2008; Zhang et al., 2009; Li et al., 2010a) based on the folding pattern of cortical surface, so that vertices on surface with homogeneous features will be grouped into the same parcel (e.g., Zhang et al., 2009; Li et al., 2010a). The second group of methodologies focus on using properties of white matter fibers (e.g., Behrens et al., 2003; Anwander et al., 2007; Hagmann et al., 2007; Klein et al., 2007; Tomassini et al., 2007; Perrin et al., 2008; Beckmann et al., 2009; Honey et al., 2009; Roca et al., 2009; Clarkson et al., 2010; Roca et al., 2010; Zhang et al., 2010; Bastiani et al, 2012; Wang et al., 2013), which are usually derived from diffusion magnetic resonance imaging (dMRI) data. Most of those works were in favor of using fiber bundles estimated from dMRI data as connectivity of brain network and then cortical locations with similar connectivity patterns were grouped into the same region. Importantly, the foundation of those works is the belief that the connectivity pattern of a brain largely determines its function (Passingham et al., 2002). Therefore, functional interpretation based on parcellation results were widely conducted in those works and the results were promising (e.g., Behrens et al., 2003; Johansen-Berg et al., 2004; Beckmann et al., 2009). Instead of seeking help from structural information, the third group of methods devoted effort in using functional features to parcellate the cortex (e.g., Cordes et al., 2000; Hampson et al., 2002; Beckmann et al., 2005; Cohen et al., 2008; van den Heuvel et al., 2008; Nelson et al., 2010; Blumensath, et al., 2013). The important advantage of those works is the easier functional interpretation of the parcellation.
Despite remarkable progresses made in those works, however, consistent fine-granularity parcellation of the whole cortex is still very difficult due to the complexity and variability of the cerebral cortex (Zhu, et al., 2012b). In particular, methods in most previous works were developed on an individual basis and it has been difficult in establishing correspondences between finely-parcellated cortical regions across individuals (Zhu, et al., 2012b). Many efforts have been devoted to establish cross-subject structural/functional correspondence (Corouge et al., 2004; Shen, et al., 2013) by either warping an atlas or developing group-wise parcellation. However, they still have difficulty in providing a stable and consistent parcellation-based cortical reference system, onto which relationship between structure and function (Passingham, et al., 2002; Honey et al., 2009) can be explored. For example, because of high variability across subjects, functional homogeneity based on cortical parcellation result across populations cannot be guaranteed (Shen, et al., 2013) and the choice of cluster number is still an open question that needs to be carefully addressed. Furthermore, because of the unavailability of a stable, common and consistent parcellation across subjects, it also has been difficult in obtaining clear and meaningful functional and structural interpretation of the parcellation boundaries (e.g., Fischl, et al., 1999; Johansen-Berg et al., 2004; Cohen, et al., 2008). In Jbabdi, et al., 2009, the authors proposed a method to cluster brain regions on the basis of their connections, with no prior knowledge on the number of clusters. This method also allowed combining data from different subjects. However, this work was only applied to one brain region, because it needs a couple of targets in other brain regions, to which connections of the region to be segmented, are estimated via probabilistic tracking methods. It seems not easy to apply this method to segment a larger brain region, because more targets are needed to provide connectivity profiles to segment the larger brain region. However, if a set of dense target covering the full brain is chosen, the method might meet a dimensionality reduction problem.
Recently, in order to establish a common and consistent brain reference system, we developed and publicly released a dense map of discrete cortical landmarks named DICCCOL (dense individualized and common connectivity-based cortical landmarks) (Zhu et al., 2012b), and demonstrated that the DICCCOL landmarks possess group-wise consistent white matter fiber connection patterns across individuals and populations. From the functional point of view, the neuroscience foundation of the DICCCOL system is that each brain's cytoarchitectonic area has unique underlying inputs and outputs which largely determine the functions the area performs (Passingham et al., 2002), and thus group-wise consistent structural and connectional profile has predictive power of brain function (Zhu et al., 2012b; Passingham et al., 2002; Honey et al., 2009; Li et al., 2010b). These DICCCOLs have exhibited relatively accurate structurally and functionally connectional correspondences across different brains (Zhu et al., 2012b) and demonstrated the promise of the connectional fingerprint concept (Passingham et al., 2002). Despite that they are still far from far from covering the whole cortex and providing clear structural/functional boundaries, they provide a preferable initialization for the parcellation framework developed in this paper.
The proposed framework in this paper is conducted via a hierarchical scheme to gradually and group-wisely parcellate the cortices into consistent fine-granularity parcels. Conceptually, this framework leverages the proven principles in the DICCCOL system, and advances it to whole-cortex fine-granularity parcellation. The major novelties of the framework are summarized as three-folds. 1) The cortical parcellation is performed via a hierarchical scheme so that cortical surfaces are gradually parcellated into finer-granularity parcels in deeper hierarchical level (detailed in section 2.3). This hierarchical scheme provides a spectrum of multi-scale parcellations, which are more flexible and accurate since it is typically difficult to determine a ‘correct’ parcellation resolution (Blumensath, et al., 2013); 2) DICCCOLs are used as samples to automatically determine the cluster numbers via an adaptive affinity propagation (AP) algorithm (Wang et al., 2007) (detailed in section 2.3), and this step fundamentally guarantees the parcel correspondence across subjects; 3) The group-wise implementation of a classification algorithm (section 2.4), accompanied with group-wise hidden Markov random field (HMRF) smoothing factor, is used to propagate cluster labels obtained in 2) onto whole-brain vertices of different subjects in a group-wise fashion, so that vertices within clustered parcels possess structural homogeneity and the parcels have correspondence across subjects as well (section 2.5).
2. METHODS
2.1 Overview
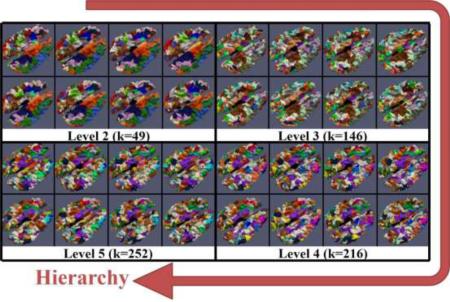
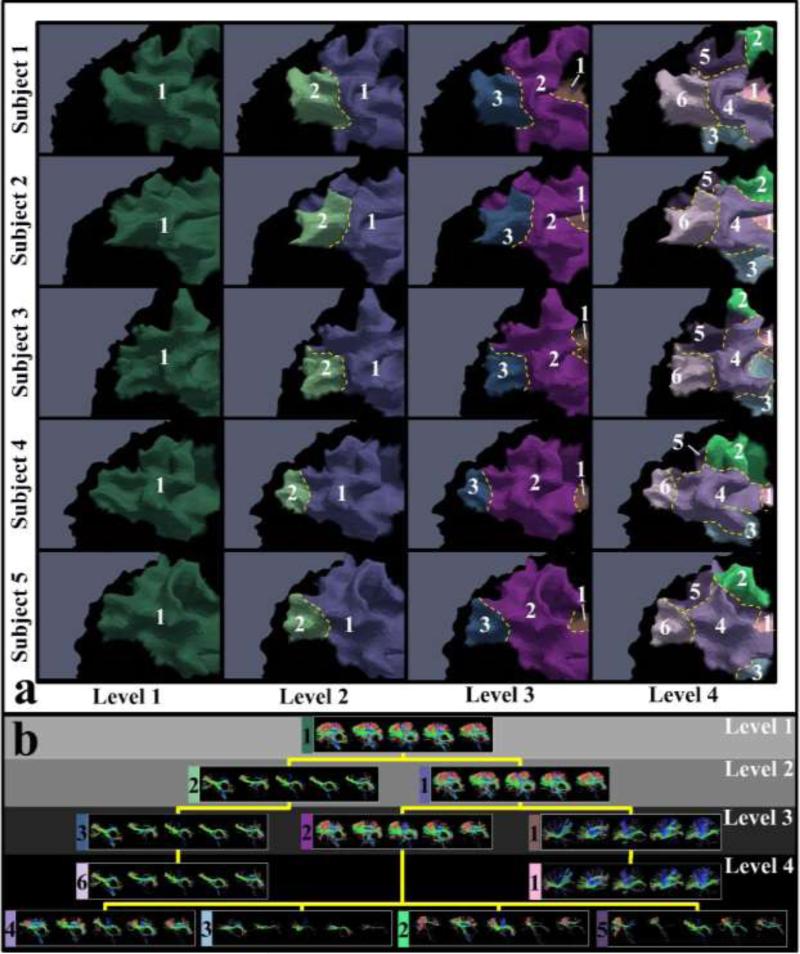
The overview of our framework is illustrated and summarized in Figure 1. Here, we used one corresponding parcel of five subjects as an example to intuitively illustrate how the parcel was gradually divided into finer ones in the hierarchical scheme. Generally, we provided a spectrum of parcellations on cortical surfaces. In each hierarchical level (columns in Figure 1(a)), parcels possess correspondence across subjects. Moreover, the fiber bundles extracted from the corresponding parcels possess group-wise consistent patterns (Figure 1(b)). Therefore, to achieve this goal, we need to solve several key problems: 1) establishing a mapping between cortical surface and white matter, so that cortical surface parcellations can be the equivalent of white matter parcellations (in section 2 in Supplemental Materials); 2) automatically determining the cluster numbers in a group-wise manner (in section 2.3); 3) Group-wisely clustering the entire cortical surface vertices into the corresponding clusters (in section 2.4 & 2.5).
Figure 1.
Overview of the framework and illustration of hierarchically parcellating one parcel into finer ones. Five subjects are used as examples in this illustration. (a) Parcellated cortical parcels in four hierarchical levels. Corresponding parcels in each level are associated with a unique number. Borders are highlighted by the yellow dash curves; (b) Fiber bundles extracted from the parcels in (a). Fiber bundles extracted from the corresponding parcels of the five subjects are displayed in the same box, associated with the number and color of the parcel in (a). The fiber bundle boxes of all levels are organized in a hierarchical tree structure according to the results in (a).
2.2 Data Acquisition and Preprocessing
Dataset 1
Eight healthy young adults were scanned in a GE 3T Signa MRI system using an 8-channel head coil at the Bioimaging Research Center (BIRC) of the University of Georgia (UGA) under IRB approval. The subjects participated in DTI, resting-state fMRI (rs-fMRI) scans and task-fMRI scans and their datasets were used in this paper. DTI data was acquired using the spatial resolution 2mm×2mm×2mm; parameters were TR 15.5s and TE min-full, b-value = 1000 with 30 DWI gradient directions and 3 B0 volumes acquired. All scans were aligned to the AC-PC line. Rs-fMRI acquisition parameters were as follows: 64×64 matrix, 4mm slice thickness, 220mm Field of View (FOV), 30 slices, repetition time (TR)=1.5s, echo time (TE)=25ms, ASSET=2. One in-house verified paradigms fear task (Zhu et al., 2012b) was used in this paper to validate the results. Task-fMRI were acquired using a T2*-weighted single shot echo planar imaging (EPI) sequence and were aligned to the intercommisural line (AC–PC line); TE = 25 ms, TR = 1500 ms, 90° RF pulse, 30 interleaved slices, acquisition matrix = 64 × 64, slice thickness = 4 mm, FOV = 240 × 240 mm, and ASSET factor = 2.
Dataset 2 (Human Connectome Project Data, HCP for short)
Diffusion MRI data of 68 subjects were obtained from the Q1 release of the HCP dataset package. In brief, the imaging parameters are: spin-echo EPI sequence; TR=5520 ms; TE=89.5 ms; flip angle=78 deg; refocusing flip angle=160 deg; FOV=210×180(RO×PE); matrix=168×144(RO×PE); spatial resolution=1.25mm×1.25mm×1.25mm; echo spacing= 0.78 ms. Particularly, a full dMRI session includes 6 runs (each approximately 9 minutes and 50 seconds), representing 3 different gradient tables, with each table acquired once with right-to-left and left-to-right phase encoding polarities, respectively. Each gradient table includes approximately 90 diffusion weighting directions plus 6 b=0 acquisitions interspersed throughout each run. Diffusion weighted data consisted of 3 shells of b=1000, 2000, and 3000 s/mm2 interspersed with an approximately equal number of acquisitions on each shell within each run.
Standard preprocessing for DTI, rs-fMRI and task-fMRI in Dataset 1 was performed in a way similar to the one in Zhu et al., 2012b. Importantly, DTI space was used as intra-subject standard space. For the full-set dMRI in HCP Dataset, multi-shell mode in FDT toolkit from FSL 5.0 (Jbabdi, et al., 2012) was used to estimate orientation distribution functions (ODFs), and DSI Studio (Yeh, et al., 2013) was used to perform deterministic fiber tracking. Details about preprocessing can be found in the Supplemental Materials.
Finally, as we parcellated the cortex via connectional profiles, we extracted fibers for each vertex and adopted trace-map to depict the connectional patterns of the extracted fibers (Zhu et al., 2012a). Also, based on trace-maps, a group of discrete vertices (358) distributed across the whole brain, defined as DICCCOLs (Zhu et al., 2012b) were identified. As the corresponding DICCCOLs across subjects share the similar connectional patterns, they were used as initial corresponding landmarks for our group-wise parcellation methods. More details about mapping connectional profiles to cortical surface and establishment of DICCCOLs can be found in section 2 and 3 in the Supplemental Materials.
2.3 Hierarchical Scheme
After we associated vertices with trace-map feature vectors, the cortical parcellation problem was converted into a feature clustering problem on triangular mesh space. That is to group vertices with similar trace-maps into the same clusters. We also aimed to establish correspondence between the vertices clusters across subjects.
When dealing with the clustering problem, we have to determine the cluster number. In fact, the cluster number could be optimally obtained if one clustering algorithm was applied on trace-maps extracted from all vertices. However, the huge number of vertices causes very heavy computation load, making this optimal solution impractical. Moreover, it is our goal to determine a cluster number which can be group-wisely applied onto multiple subjects, but it is difficult to establish pre-defined correspondence of cortical surface vertices across subjects. Therefore, we use the DICCCOLs as sampled cortical-cortex vertices, in that DICCCOL possess intrinsic correspondence, that is, they exhibit common structural connectivity patterns across subjects and the DICCCOLs cover the whole cortex. As a result, using DICCCOLs as samples will greatly reduce the computation load (358 DICCCOLs vs. 6×104 cortical-cortex vertices).
Based on DICCCOLs, we adopted a hierarchical scheme to automatically generate a spectrum of clusters. Generally, we built a divisive-style hierarchy of parcellations, through which the entire surface were gradually split into finer parcels. In Figure 2(a), we used the brown color patches as an example to represent the entire surface. Once they are used in the upper level to drive clustering, their cluster labels are propagated to all vertices on the surface to segment it into parcels (Figure 2(d)). Then, in the next lower level, taking red parcel in Figure 2(d) for example, those red DICCCOLs (DICCCOL #7~#9) will be used to drive clustering and the cluster labels are propagated throughout the red parcel. This procedure is performed recursively.
Figure 2.
Illustration of clustering steps within one hierarchical level. Four subjects are used as examples. (a) The corresponding parcels in brown color are the parcels in consideration at the current level. The numbered bubbles denote nine DICCCOLs; (b) Adaptive AP clustering results based on DICCCOL samples. DICCCOLs are clustered into four classes and they are of the same color if they are within the same class; (c) Propagating clustering labels obtained from DICCCOLs onto other vertices within the parcels in consideration via a group-wise classification algorithm. Surface vertices within the same class are of the same color; (d) Updating DICCCOLs classification results based on vertices propagation results. DICCCOL #2 highlighted in (c) and (d) are assigned to the green-color class after updating.
Specifically, supposing the orange color parcels in Figure 2(a) are the surfaces in hierarchical level #1 to be classified. In order to group-wisely determine the cluster numbers and cluster centers of these patches, sampled vertices were extracted from those. DICCCOLs are one of the preferable choices to be used as sampled vertices, as they are relatively dense to cover the parcels and, more importantly, corresponding DICCCOLs across subjects have similar white matter connectional profiles. The outer hierarchical loop of the framework consists of the following steps:
We computed the feature (trace-map in this paper) distance matrix of the p sampled vertices (DICCCOLs) for the ith subject of the n subjects. In Figure 2(a), p=9 and n=4. The distance mij is defined as , where is the trace-map (connectional) feature. As the p sampled vertices have correspondences across subjects and corresponding vertices have similar connectional features, we used the average as the group-wise distance matrix. Currently, we considered that each individual equally contributes to determining the group-wise cluster number.
We applied adaptive affinity propagation (AP) algorithm (Wang et al., 2007) on group-wise M̄ to automatically classify the sampled vertices into K clustersStep* (see Figure 2(b)). Details of adaptive AP in Step* can be found in Supplemental Material.
A group-wise classification algorithm was proposed in this work (section 2.4 and 2.5) and applied to propagate classification results of the sampled vertices to the entire orange parcels (see Figure 2(c)). The classification results on sampled vertices were modified according to the parcellation results (see Figure 2(d)).
The four parcels in Figure 2(c)&(d) comprise the hierarchical level #2. The abovementioned three steps were executed on the four parcels in a recursive manner. Each parcel was treated as the one in Figure 2(a). For example, in the red parcels, we can find three DICCCOLs and they are used as sampled vertices on red parcels to construct average distance matrix M̄. Adaptive AP is applied on it to classify the sampled vertices into clusters and the group-wise classification algorithm is applied to propagate classification results of the sampled vertices to the entire red parcel. The hierarchical scheme terminates when the total parcel numbers of the two successive levels are the same, meaning that no finer-granularity parcellation can be obtained.
The AP algorithm (Frey and Dueck, 2007) is an automatic clustering method which takes as input measures of similarity between pairs of data points. The adaptive AP was adopted rather than the original AP because it is difficult to determine the value of parameter ‘preference’ in the original AP algorithm. The parameter ‘preference’ roughly determines a range for cluster number. The adaptive AP ‘adaptively’ scan the preferences to search space of the number of clusters so that it can automatically find the optimal clustering solution.
It is worth noting that, at the end of each hierarchical level, we updated the information that which DICCCOLs locate in which corresponding parcel, because DICCCOLs are just discrete samples of vertices and the clustering results based on them are only used as initial information for the group-wise classification algorithm based propagation. Therefore, the final parcel clustering results on the surface can possibly vary from the initialization provided by DICCCOLs. As an example, DICCCOL #2 in Figure 2(b) are grouped into the blue-color class. But after class labels are propagated onto parcels in Figure 2(c), the DICCCOL #2 locations are invaded by vertices with the green-color labels, and DICCCOL #2 class labels are updated as the green-color accordingly (Figure 2(d)). Therefore, the two steps, cluster number determination based on DICCCOLs (Figure 2(b)) and propagating clustering results onto parcels (Figure 2(c)), are mutually updated and it is not possible that we independently construct a complete hierarchical tree simply based on DICCCOLs at the very beginning and then propagate the hierarchical clustering tree onto the surface accordingly. Therefore, the parcellation results via this framework will remain relatively stable given a small disturbance of initialization of DICCCOLs (see Supplemental Figure 3 and related context in Supplemental Materials for more details).
2.4 Classification Result Propagation via a Group-wise Framework
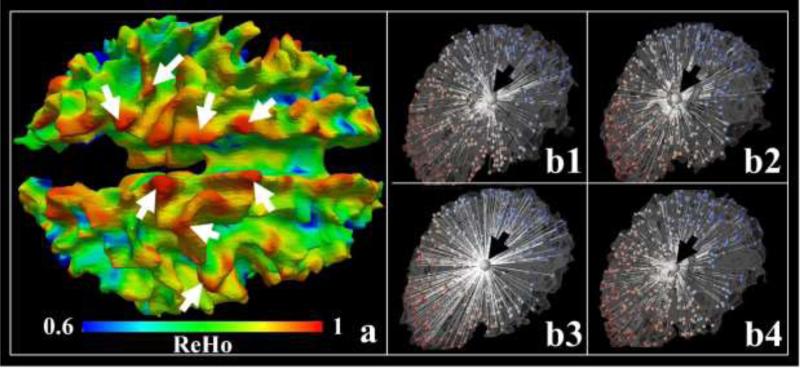
In this section, we detail how to propagate the DICCCOLs classification result in Figure 2(b) onto the entire parcels in Figure 2(c). In general, this propagation problem is equivalent to a parcel parcellation one, which can be formulated as a vertex clustering problem, that is, to cluster cortical vertices with homogeneous connectional profiles (trace-map) into the same parcel. Supposing that we have already determined the cluster number K by classifying DICCCOLs, we then assume each cortical surface parcel, k=1, 2 ... K, approximates a Gaussian distribution of trace-maps and the entire cortex consists of K Gaussian distributions with , where is the mean trace-map of the kth Gaussian distribution and Σk is the variance of trace-maps within the distribution. A straightforward rationale is illustrated in Figure 3(a), in which the Kendall's coefficient concordance (KCC), used to measure the similarity of the trace-map of a vertex to those of its neighbors (similar to brain functional regional homogeneity (ReHo) in Zang et al., 2004), is mapped onto the cortical surface. More specifically, vertices within 3-ring neighborhood on the cortical surface are extracted from one vertex, and then the KCC is applied on trace-maps associated with all those vertices and the value is assigned to the center vertex. As the KCC measures the homogeneity of features within a region, we also use regional homogeneity (ReHo) to denote it in this paper. In Figure 3(a), we illustrated one ReHo map. Higher values (red color) indicate that trace-map patterns at those locations are more consistent. The Gaussian-like patterns visually observed in the ReHo map provide an intuitive support of using Gaussian models in the group-wise classification algorithm.
Figure 3.
(a) ReHo (regional homogeneity) map of trace-maps on the cortical surface. White arrows highlight some areas with high ReHo values. (b1-b4) The four vertices (larger bubbles highlighted by the black arrows) are cross-subject neighbor vertices. Smaller bubbles are DICCCOLs on each subject and the white lines illustrate the Euclidean distances (In fact, geodesic distances is used in the methods and Euclidean distances used in the figure is simply an illustration) between the highlighted vertex and DICCCOLs. The colors of DICCCOLs represent their indices.
Mathematically, for subject i which has M vertices, we used to denote the trace-map feature of vertex m, and used Ti to denote the collection of M vertices’ trace-map features, i.e., . p(ymk = 1) = πmk (Y = {ymk, m = 1, ..., M, k = 1, ..., K}) is the prior probability of assigning class label k for vertex m, where ymk is a binary label and equals to 1 if vertex m is assigned with label k. Y is an M×K indicator matrix consisting of ymk. We used π/w to denote prior/posterior probability of obtaining Y. As mentioned previously, a Gaussian distribution is estimated within each class and the parameters of all Gaussian distributions θ are unknown. Therefore, if we are given the class labels Y = {ymk, m = 1, ..., M, k = 1, ..., K}, then the log-likelihood of the unknown parameters θ can be formulated based on the known trace-maps features Ti as follows:
| (1) |
In other words, the Gaussian distribution θk can be estimated based on the current classification results given by πmk(Y).
However, the class labels Y is also unknown. To find the maximum likelihood estimate of θ, we devised an technique to iteratively apply two steps as follows.
Step I
Calculate the posterior probability distribution of labeling vertex m as parcel k under the current estimate of parameters θt. That is:
| (2) |
Step II
Based on the probability of classification in Step I, we maximize the likelihood function in Equation (1) to estimate the Gaussian distribution parameters for each class:
| (3) |
More specifically, in Step II, we used estimated from Step I as the prior probability () to provide classification probability, based on which parameter θt+1 was estimated. After Step I, we assigned label k to each vertex m based on . We took the ‘winner-takes-all’ approach for each row of to find the smallest value in each row and assign its index k to the mth vertex. Based on the classification information provided by , we can update the parameters θt+1 for all Gaussian distributions:
| (4) |
| (5) |
The above classification algorithm is designed for a single subject i. In order to parcellate N cortical surfaces simultaneously, θ is updated by considering all N subjects in Step II by appending , to form wmk. No vertex correspondence between subjects is needed. After obtaining the posterior probability distributions for all subjects wmk, we split it into {wimk, i = 1, ..., N} accordingly and assign the vertices with label k. We use them as a group-wise prior probabilities by collecting all vertices classified into the kth class from all N subjects. Then, we update the parameters of class k based on the trace-maps from those kth class vertices of all N subjects.
2.5 Group-wise Hidden Markov Random Field (HMRF) Smoothing
In the above procedure, cortical surface vertices were clustered in the associated trace-map feature space. So the surface vertices from distant brain areas can possibly be assigned with the same class label. Therefore, we adopted the hidden Markov random field (HMRF) to prevent this situation by considering that neighboring vertices typically have higher probability to share the same cluster label (Zhang et al., 2001).
Briefly, we estimate the labels in Step I by Equation (6).
| (6) |
In Equation (6), encodes the similarity of other vertices’ labels within the neighborhood system Vm to labeling vertex m as class k and it can be written in the form:
| (7) |
where y is the label of vertices within the neighborhood. The element in Vm is 1 if a vertex is within the neighborhood.
The neighborhood on one subject surface can be established straightforwardly by including all surface vertices linked to the vertex in consideration by surface edges. In order to extend the neighborhood system to a group-wise domain, we used the DICCCOLs as references and computed the geodesic distance of the vertex m from subject i to all DICCCOLs of the same subject, so that we obtained a distance vector . Then, the neighbor vertex of on subject j is defined as the one with the most similar by measuring the Euclidean distance between the distance vectors. As an example, we randomly selected a vertex from one subject highlighted by the black arrow in Figure 3(b1), and its neighboring vertices on other three subjects are shown in Figure 3(b2-b4), as highlighted by the black arrows.
3. RESULTS
3.1 Structural Validation of the Cortical Parcellation Results
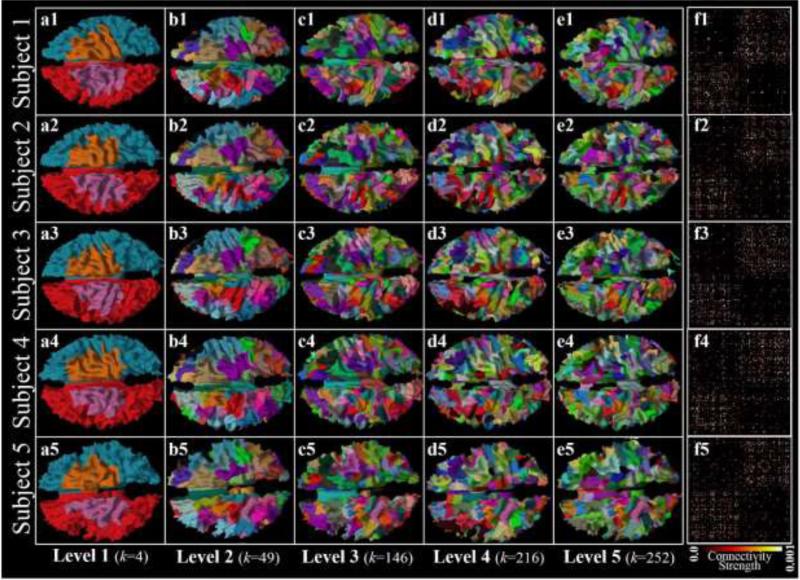
In this section, our methods will be evaluated in the structure aspects by comparing the results with several other methods. We used the eight subjects from Dataset 1 to evaluate the group-wise parcellation framework. The cluster number stops increasing after level 5 in the hierarchical scheme and the group-wise classification algorithm converges after around 50 iterations on most parcels at each level. As an example, the parcellated cortical surfaces of five example subjects were visualized in the first five columns in Figure 4 (the results within the same hierarchical level are in the same column). Structural connectivity matrices based on the level-5 parcellation results were also illustrated in column f. The consistency of parcel location distributions and patterns of structural connectivity matrices across subjects can be qualitatively appreciated in Figure 4.
Figure 4.
(a1-e5) Group-wise surface parcellation results of five example subjects (rows) in five levels (columns). Corresponding parcels in each column share the same color. (f1-f5) Structural connectivity matrices derived from the parcellation results in (e1-e5). The color bar indicates the connectivity strength.
Group-wise Results vs. Individual Based Results
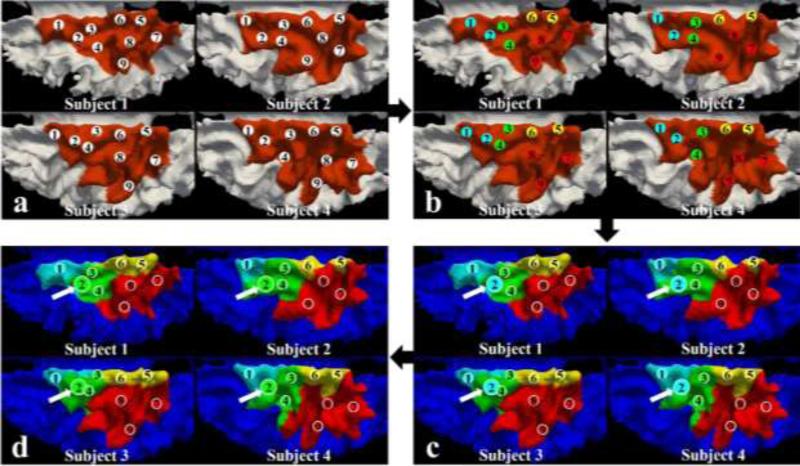
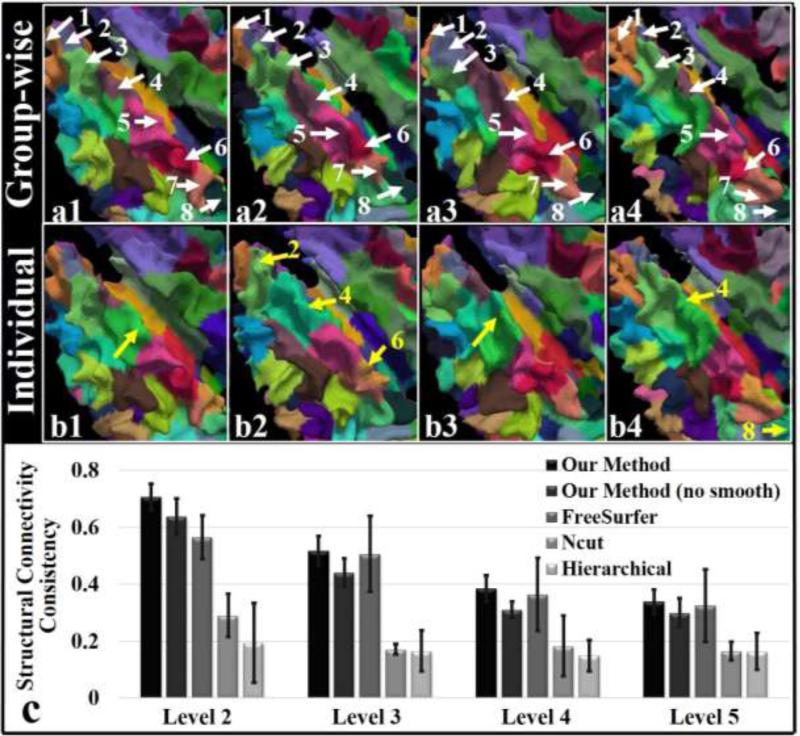
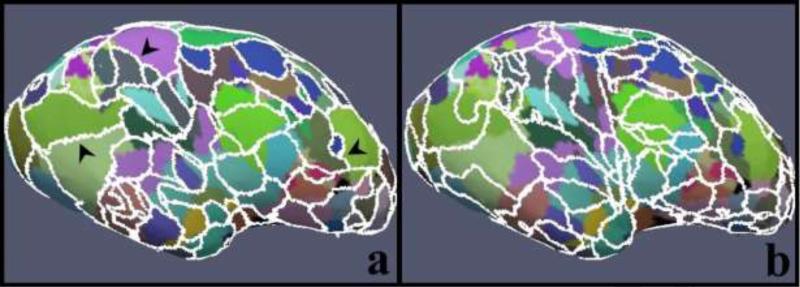
We also visualized the comparison between the group-wise parcellation results (level 5) with those obtained from a single subject (the same subjects as those used in the group-wise framework), and the results are shown in Figure 5. The parcellation for a single subject is accomplished by using the same framework, except that it is applied on one subject each time. In order to make the individual results comparable to the group-wise results, the DICCCOL clustering results from the group-wise results at each level were directly used as the initialization for single subject parcellation.
Figure 5.
(a1-b4) Zoomed-in views of parcellation results on the left frontal lobes of four example subjects via the group-wise classification algorithm (a1-a4) and the one for individuals (b1-b4). The white arrows highlight the corresponding parcels. The yellow arrows highlight missing parcels or invading parcels. (c) Consistency measure of structural connectivity across subjects. The bins and bars show the average consistency and the standard deviation.
We zoomed in the left frontal lobes and identified 8 parcels in Figure 5. We can observe from Figure 5(a1-a4) that the eight parcels (highlighted by the white arrows) parcellated by the group-wise classification algorithm follow exactly the same order from the anterior frontal lobe to the posterior frontal lobe on different subjects. In comparison, the parcellated parcels based on single subject are much less consistent (Figure 5(b1-b4)). The yellow arrows with parcel IDs highlight the missing parcels, while the ones without parcel IDs indicate some other invading parcels. This consequently leads to missing or variant fiber bundles and thus further reduces the structural and functional connectivity consistency across subjects.
Quantitatively, we compared the whole-cortex structural connectivity matrices based on the level-5 group-wise/individual parcellation results. On average, the structural consistency measurement via the group-wise parcellation is 0.34, which is significantly greater than the one (0.26) via individual parcellation (p = 6.57×10−6 by one tailed t-test). Similarly, functional connectivity consistency was also measured based on rs-fMRI data. On average, the consistency measure among functional connectivity matrices via group-wise parcellation is 0.48, which is significantly greater than the one (0.41) via individual parcellation (p = 0.003 by one tailed t-test).
HMRF Smoothing Factor
In order to demonstrate the effectiveness of group-wise HMRF smoothing, we computed the structural connectivities obtained by our methods with/without group-wise HMRF smoothing based on the parcellations on different levels and compared the connectivity consistency in Figure 5(c). Taking level-5 results for example, the consistency decreases (0.30±0.05 without smoothing vs. 0.34±0.04 with smoothing) when group-wise HMRF smoothing was removed from the method. The rationale is that two distant and disconnected parcels are likely to be classified into one cluster if our method is only applied in the structural feature space with no surface-based spatial smoothing constraint, and this will consequently reduce the structural connectivity consistency across subjects.
Comparison with Other Methods
We compared the structural connectivity consistency of the parcellations derived from our group-wise method at the resolution of different levels with those by the Ncut method (Von Luxburg, 2007) and the hierarchical method using the Ward's linkage rule (Ward, 1963), and reported the results in Figure 5(c). Unlike our method that can generate correspondence of parcels (the color parcels in Figure 5(a)), the Ncut and hierarchical methods, however, have difficulty in establishing correspondence across subjects. We used the Dice similarity method detailed in the Supplemental Materials to establish the parcel correspondence across subjects for those two individual based methods. Then the structural connectivity matrices and the structural connectivity matrices consistency were measured based on the corresponding parcellations. Generally, our methods outperform the two methods. For example, the structural connectivity consistency derived from our method is 0.34±0.04 at level 5, which is significantly higher than 0.16±0.03 and 0.16±0.06 for the Ncut and hierarchical methods. The reason that the latter two methods generate low structural consistency is that these two methods are individual based ones, so that there is no intrinsic correspondence of parcels across subjects.
We also compared our method with the registration method. Specifically, one subject of those used in our group-wise parcellation method was defined as the ‘template’ subject. Then, the registration method in FreeSurfer (CVS registration, Postelnicu et al., 2009, Zöllei et al., 2010) was used to warp the surface of the ‘template’ subject to every other subject one by one. The parcellation of the ‘template’ subject was mapped to other subjects accordingly. The parcellation of the ‘template’ subject was originally obtained from our group-wise parcellation method, and by this parcellation method every subject thus has the same number of parcels (n parcels). Therefore, in the registration method, every subject also has n corresponding warped parcels as well. We measured the structural connectivity consistency of the parcellation among subjects for the registration method (Figure 5(c)). On average, the consistency is 0.30±0.05, which is lower than our method (0.34±0.04).
3.2 Comparison Between Cortical Parcellation and fMRI Data
Although the effectiveness of our methods has been validated via several structure-based measurements in section 3.1, the functional interpretation of the structural parcellation has not been studied. In this section, we will use rs-fMRI and task-fMRI derived maps and clusters to study if the structural parcellation also has meaningful functional interpretation.
Parcellation Borders Aligned with Homogeneous Maps and ICA Clusters from Rs-fMRI Data
In this section, the structural parcellation boundaries were analyzed by comparison with the rs-fMRI derived ReHo maps and ICA clusters. The functional correspondence based on structural parcellation was also discussed.
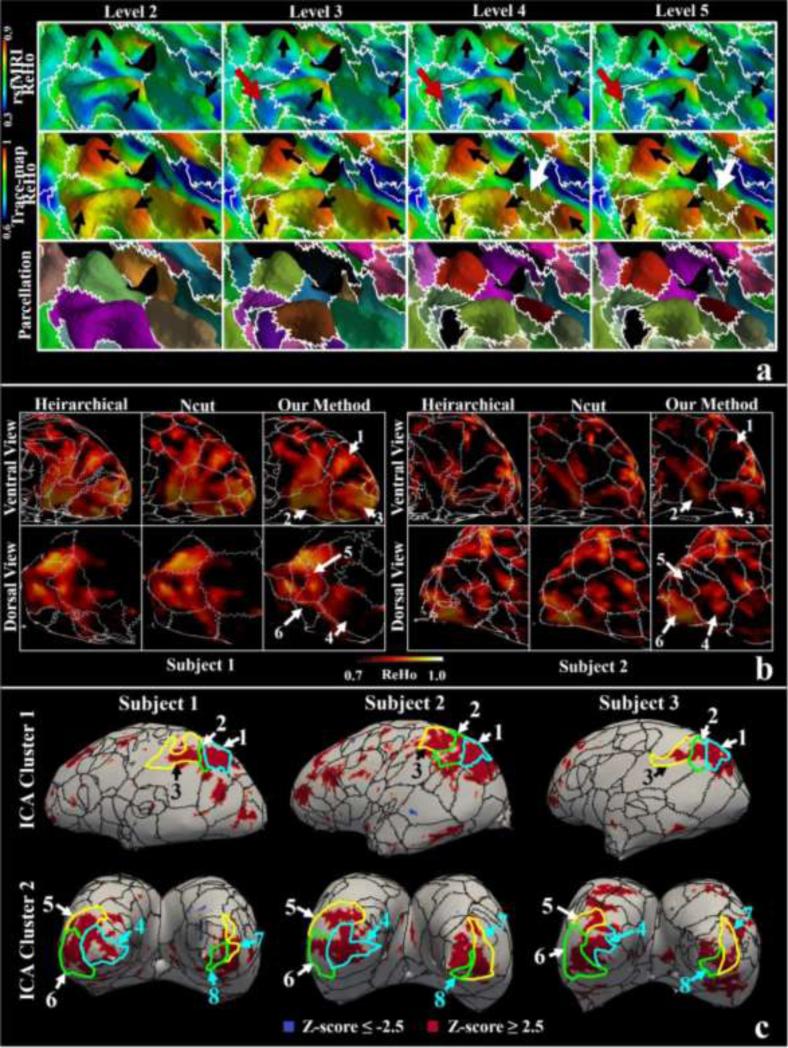
Generally, ReHo assumed that voxels/vertices within a functional brain area were more temporally homogeneous when this area is involved in a specific condition. Therefore, brain regions with higher ReHo values suggest that a homogeneous brain activity would likely occur (Liu et al., 2008), and the regions with lower ReHo values might suggest a possible boundary segregating different brain regions engaged in different brain activities. We overlaid the parcellation boundaries of one subject in different hierarchical levels on the ReHo maps and showed them in a zoomed-in view in the first row of Figure 6(a). Corresponding ReHo maps based on the trace-map (the second row) and the parcellation results (the bottom row) are also shown as reference. We can observe four structural ReHo peaks (black arrows in the second row). When the algorithm runs into finer levels (#3, #4 and #5), the four peaks are completely separated by boundaries and no over-parcellation occurs except one hidden peak (the white arrows), suggesting the effectiveness of the stopping criterion of the hierarchical scheme. As for the functional ReHo derived from rs-fMRI data, the structural parcellation borders also roughly match the areas with low functional ReHo values and ReHo peaks are also found within parcels (the black arrows in the first row of Figure 6(a)). The average functional ReHo value obtained from the vertices that the parcellation borders of the four levels in Figure 6(a) pass by is 0.52, 0.54, 0.54 and 0.55, which are less than the overall average functional ReHo values (0.56). This result suggests that the structural borders may also segregate functionally homogeneous regions. It should be noted that it is possible that no functional ReHo peaks can be found in the parcels highlighted by the red arrows while structural ReHo peaks exist.
Figure 6.
Comparison of structural parcellation borders and rs-fMRI derived ReHo maps. (a) Joint visualization of parcellation borders (the white curves) and ReHo maps of one subject. The parcellation borders are overlaid on rs-fMRI derived ReHo map (the top row), trace-map-derived ReHo map (the middle row) and parcellated surfaces (the bottom row). The interpretation of arrows is referred to the main text; (b) Parcellation borders (at a resolution of 252 parcels) via different parcellation methods are overlaid over ReHo maps (visual cortices) on a scale of 0.7 to 1. Numbered arrows indicate correspondent parcels obtained from our methods. (c) Parcellation borders (at a resolution of 252 parcels) via our methods are overlaid over two ICA clusters derived from rs-fMRI. Corresponding parcels, indicated by numbered arrows, are highlighted by the borders with the same color.
We also compared our method with the Ncut and hierarchical methods by overlaying different parcellation borders onto the rs-fMRI ReHo maps. In Figure 6(b), we visualized the comparison made on visual cortices. Based on visual observation, structural boundaries based on three methods somewhat match the low functionally homogeneous regions. The Ncut borders perform worse to segregate functionally homogeneous regions in the visual cortex, while the hierarchical method borders are more redundant than others at the same parcellation resolution. On the contrary, borders based on our methods provide preferable functional segregation performance and simultaneously possess parcel correspondence (the numbered arrows in Figure 6(b)). The structural correspondence may suggest a similar correspondence of functionally homogeneous regions across subjects (the functionally homogeneous regions in #2, #3 and #4 parcels), though functional variation across subjects can also be spotted. For example, #1 parcels share similar structural profiles but no functionally homogeneous feature can be observed in subject 2. The same situation happens on #5 and #6 parcels as well.
Quantitatively, we overlaid the structural borders via our method over ICA analysis derived clusters in Figure 6(c). The two ICA clusters in rows may have correspondences across subjects based on the component correspondence identification method introduced in the Supplemental Materials (please refer to the numbered corresponding parcels the ICA clusters overlapped with).
Parcellations Possessing Group-wise Functional Connectivity Consistence on Rs-fMRI Data
We computed the functional connectivity consistency based on the parcellations of different level. Ncut method, hierarchical method and FreeSurfer registration method are also used for comparison. Similarly, we used the Dice similarity method to establish the parcel correspondence across subjects for those two individual based methods. We reported in Table 1 the average consistency measure of functional connectivity derived from rs-fMRI across subjects. The intrinsic correspondence of parcels across subjects obtained via our methods results in much better functional connectivity consistency than other ones.
Table 1.
Average consistency measure ± standard deviation of functional connectivity across subjects.
| LEVEL 2 | LEVEL 3 | LEVEL 4 | LEVEL 5 | |
|---|---|---|---|---|
| OUR METHOD | 0.65±0.06 | 0.56±0.05 | 0.52±0.05 | 0.48±0.04 |
| FREESURFER | 0.61±0.07 | 0.50±0.13 | 0.45±0.11 | 0.42±0.10 |
| NCUT | 0.31±0.15 | 0.15±0.22 | 0.21±0.26 | 0.15±0.23 |
| HIERARCHICAL | 0.01±0.07 | 0.17±0.13 | 0.09±0.13 | 0.10±0.15 |
Parcellation Borders Aligned with Task-fMRI Activation Clusters
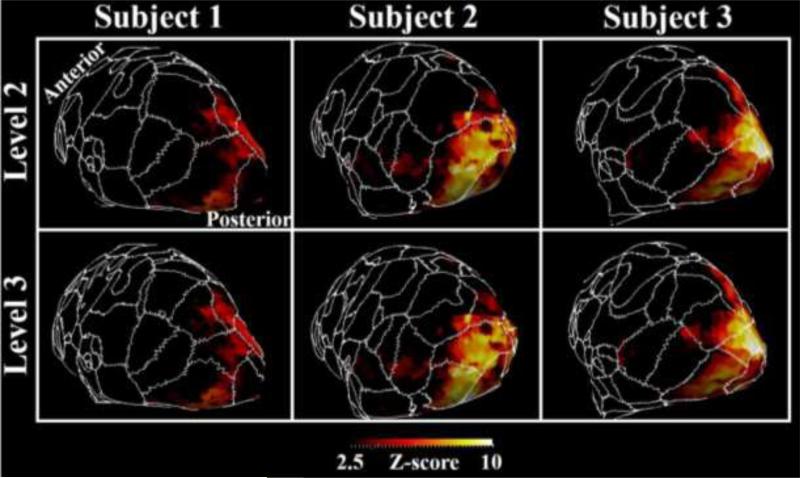
We showed in Figure 7 a zoomed-in view of the parcellation borders at the resolution of 49&146 parcels overlaid over the fear task in Dataset 1. The alignment of parcellation borders with task activation cluster boundaries can be observed in Figure 7.
Figure 7.
Level 2&3 (at the resolution of 49&146 parcels) parcellation borders overlaid over fear task activation maps of three subjects’ primary visual cortices. Some correspondent parcels are highlighted by the dashed curves of the same color.
3.3 Reproducibility on HCP Dataset
In this section, we demonstrated the reproducibility of our methods on an independent dataset. It is noted that we used full-set (multi-shell) dMRI data from 8 HCP subjects to explore if richer connectional information provided by HCP data can produce similar parcellation results.
It is straightforward that we directly applied our methods on the 8 HCP subjects to produce group-wise parcellations. However, it is difficult to compare the parcellation results with Dataset 1 derived ones, as there are not overlapping subjects from the two datasets, and we have to conduct inter-subjects registration to warp parcellation from Dataset 1 to HCP dataset. This is subject to registration errors. Therefore, we adopted strategy #1 as follows:
Strategy #1 is using combined Dataset 1 and HCP Dataset. However, the computation load and memory cost would be very heavy on 16 subjects (8 from Dataset 1 and 8 from HCP Dataset) for the current method. Therefore, we conducted two experiments using our group-wise parcellation method on two data groups: 1) the 8 subjects in Dataset 1 plus one HCP subject (e.g., subject #i); 2) only the 8 subjects in HCP Dataset (including subject #i). Then, the parcellation results of the HCP subject #i from experiments #1 and #2 are compared. As subjects in Dataset 1 dominate the data group in experiments #1, the parcellation on HCP subject #i encodes the group-wise information from Dataset 1. For experiments #2, parcellation on HCP subject #i purely encodes the group-wise information from HCP Dataset. Therefore, by this way, HCP subject #i is the intermedia one for which parcellations from Dataset 1 and HCP Dataset are compared without introducing inter-subject registration errors. The comparison between the two parcellations was measured via Dice similarity. This strategy is performed on every HCP subject.
As a comparison, we also adopted strategy #2, a registration strategy:
Strategy #2 is using the methods in FreeSurfer to register subjects in Dataset 1 to HCP subjects and warp the parcellations accordingly. Then the warped parcellations were compared to those derived from HCP subjects themselves via Dice similarity measurements. This strategy is subject to inter-subjects registration errors.
The Dice similarity measures at different resolutions of parcellation levels were reported in Table 2. On average, Dice similarity is (0.33±0.12) for strategy #1, suggesting that independent datasets can produce similar parcellations, and this similarity is more pronounced than using registration method (Dice similarity is 0.24±0.03 for strategy #2). Figure 8 shows one HCP subject as an example. The parcellation on it was obtained via applying our group-wise method only on HCP Dataset (experiment #2 via strategy #1). To this subject, the parcellation boundaries of one subject from Dataset 1 via registration in strategy #2 were superimposed (Figure 8(b)). The parcellation boundaries of the same HCP subject via experiment #1 in strategy #1 were also superimposed and shown in Figure 8(a). Many matched parcellation boundaries can be found in Figure 8(a) (black arrows highlighted ones), giving an intuitive explanation of the reasons that Dice similarity for strategy #1 is higher than strategy #2.
Table 2.
Average Dice similarities between parcellations via different comparison strategies.
| LEVEL 2 | LEVEL 3 | LEVEL 4 | LEVEL 5 | |
|---|---|---|---|---|
| STRATEGY #1 | 0.28±0.14 | 0.34±0.10 | 0.35±0.11 | 0.35±0.11 |
| STRATEGY #2 | 0.25±0.06 | 0.24±0.04 | 0.24±0.03 | 0.24±0.03 |
Figure 8.
(a) The parcellation boundaries (white curves) of one HCP subject via experiment #1 in strategy #1 were superimposed to the same HCP subject parcellated via experiment #2 in strategy #1; (b) The parcellation boundaries (white curves) of one subject from Dataset 1 were superimposed to the same parcellation in (a) via registration in strategy #2; Black arrows highlight some matching boundaries.
In summary, the full-set HCP dMRI data derived deterministic fiber tracts can produce parcellation results relatively similar to the conventional DTI data. This preliminary cross-modality comparison results show the promise to conduct an upgraded parcellation framework based on the full-set HCP data. In the future work, to fully taking the advantages of the HCP data, we plan to develop new connectional profile descriptors and look for denser landmarks on the full-set HCP data, which might be helpful to promote the parcellation framework to a higher level.
4. DISCUSSION
In general, the advantages of our work over other structural connectivity based cortical parcellation methods (e.g., Anwander et al., 2007; Tomassini et al., 2007; Perrin et al., 2008; Beckmann et al., 2009; Roca et al., 2009; Clarkson et al., 2010; Roca et al., 2010; Wang et al., 2013) are three-folds. (1) Many connectivity-based methods need a large group of pre-defined seeds/ROIs to establish the connectivity matrix for vertices/voxels which need to be classified (e.g., Perrin et al., 2008; Roca et al., 2010). The main challenge to deal with is the huge dimension. Although different approaches have been proposed to reduce the dimension, it still might be a barrier to prevent those methods from being applied to the entire cortex and across populations; (2) It is difficult to establish correspondences of parcellations across subjects. Some works (e.g., Roca et al., 2009) directly provided a solution by group-wisely clustering the connectivity matrices, while some other methods proposed a solution by integrating connectivity information into registration (e.g., Wang et al., 2013). Those methods, however, may have difficulty in dealing with huge dimension problem or be lacking the capacity of considering and preserving individual variance; (3) It is difficult to automatically determine a suitable cluster number. In contrast, our framework used the trace-map model to encode connectivity profiles so that connectivity parcellation is converted into a parcellation problem based on features, which greatly reduce the dimension. DICCCOLs used in this paper provides not only an individual but also consistent and corresponding reference across subjects and populations, which guarantees the parcellations across subjects to have correspondence. Meantime, based on DICCCOLs correspondence, the group-wise classification algorithm in this paper also allows the vertices on different subjects clustered into the same corresponding parcels in their own way. This can preserve a certain level of individual variance. At last, a fully automatic hierarchical scheme is proposed to provide a spectrum of parcellations. Each finer parcellation level is based on the previous coarser level. That is, the parcels in coarser level are further split into sub-parcels. In each hierarchical level, the algorithm automatically determines the cluster numbers. Therefore, it is feasible to use this spectrum of parcellations with hierarchical structure. For example, in Figure 7, we superimposed level 2&3 parcellation levels because they are good enough to match the granularity of the functional clusters. If we overlaid level 5 parcellation, the finer parcellation boundaries will look messy to highlight the boundary consistency. But in Figure 6, ICA and ReHo clusters based on resting state fMRI have higher granularity and the consistency of their borders and parcellation boundaries can be better appreciated in the 5th level. In summary, different structural/functional clusters have different granularity levels. It is more flexible to use a spectrum of parcellation to conduct comparison studies.
In the future, the framework can be improved in the following directions. First, at the current stage, our framework takes 4 hours to obtain a group-wise parcellation on the basis of eight subjects using a typical PC (Intel(R) Xeon(R) CPU model; CPU MHz=1600; Memory 50 GB). However, the computation load is an important limitation to overcome in the future to scale the current framework to a larger dataset. Second, conceptually, our framework is of group-wise fashion so that if we include more subjects, the identified consistent parcels will be more commonly present across subject. Meanwhile, including more subjects will also introduce more variability. Therefore, although we are looking for solutions to identify common and consistent parcellations (like those on the frontal lobe in Figure 5(a)), introducing more variability may decrease the group-wise consistency, which should be dealt with in the future. Third, currently, our framework can generate as dense as three hundreds of parcels. However, higher resolution dMRI data (Sotiropoulos et al., 2013) may potentially and fundamentally bring novel insights to finer-granularity parcellations in the future, though we have already demonstrated that common parcels identified in relatively lower resolution dMRI data (Dataset 1) can be successfully propagated onto the one with high resolution (HCP Dataset). Also, more powerful and descriptive connectional features should be developed consequently based on higher resolution dMRI data to depict more detailed information. Fourth, the current group-wise framework can still be improved in the future. For example, multi-kernel (Blekas, et al., 2012) concepts might be integrated into the group-wise classification algorithm, so that structural patterns from each individual will be dealt with as heterogeneous sources. Also, in a parcel, multiple Gaussian kernels may exist and may be modeled at the same time in the updated classification algorithm. Those improvements may result in more accurate and consistent parcellation when variability within subject and across subjects is considerably taken into consideration. Fifth, application of the method to disease group. Currently, it was not discussed whether the methods in this paper can be applied to diseased brains, though the methods are not ad hoc for normal brains. The parcellation in this paper is based on dMRI-derived white matter structural connectivity, and the fiber bundle shape descriptor, trace-map, was used as a surrogate feature to depict the structural connectivity. Aberrant axonal pathways in disordered brains may lead to dMRI measurements different from controls (e.g., Kubicki et al., 2005; Alexander et al., 2007)). For example, abnormal FA/MD values are found in corpus callosum of autism brains (Alexander et al., 2007). Those abnormal measurements may consequently lead to abnormal structural connectivity profiles, such as fiber bundle shapes. Therefore, it might be risky to obtain parcellation on diseased brains by applying our methods to a combined group of controls and patients. Another possible approach is applying the method on a dataset only consisting of patients with the same brain disease, which will produce more reasonable and specific parcellation results for that disease. Then, comparison between such parcellations with control group may be helpful to identify biomarker (abnormal parcels) on cortices. Those possibilities will be investigated in our future works.
In summary, this paper proposed a novel framework for hierarchical group-wise consistent cortical parcellation based on a group-wise classification algorithm that integrates spatial constraints and uses DICCCOLs as sample landmarks to generate cluster numbers and initial centers via an adaptive AP algorithm. The effectiveness of the group-wise fashion is demonstrated in comparisons with individual-based version of the framework and other methods like the Ncut and hierarchical clustering. Structural and functional connectivity based on group-wise parcellation is consistent across subjects. Importantly, the hierarchical scheme provides a spectrum of parcellation, which is more flexible to be used in scenarios with different scales. The structure derived parcellation is also compared with fMRI-derived maps, like ReHo maps, ICA clusters and activation maps. The comparison result suggests that the structural parcellation may be used as reference to identify functional correspondence, and the parcel borders have the ability to segregate functional homogeneous regions. The framework will be applied and evaluated in cognitive and clinical neuroscience applications in a similar way as other cortical parcellation approaches were applied in the literature. It will be extensively investigated in the future whether the group-wise consistent fine-granularity cortical parcellation framework would be able to reveal novel neuroscientific insights in such applications.
Supplementary Material
Highlights.
DICCCOL provides consistent and corresponding reference across subjects and populations;
Group-wise EM finds consistent corresponding parcels and preserves a certain level of individual variance;
Use of a spectrum of parcellations is flexible;
Structural parcellation boundaries segregate functionally homogeneous areas.
Acknowledgements
T Liu was supported by the NIH R01 DA-033393, NIH R01 AG-042599, NSF CAREER Award IIS-1149260, and NSF CBET-1302089. L Guo was supported by NSFC 61273362 and 61333017. T Zhang was supported by supported by NSFC 31500798, the fundamental research funds for the central universities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander AL, Lee JE, Lazar M, Boudos R, DuBray MB, Oakes TR, Miller JN, Lu J, Jeong EK, McMahon WM, Bigler ED, Lainhart JE. Diffusion tensor imaging of the corpus callosum in Autism. Neuroimage. 2007;34(1):61–73. doi: 10.1016/j.neuroimage.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Anwander A, Tittgemeyer M, von Cramon DY, Friederici AD, Knösche TR. Connectivity-Based Parcellation of Broca's Area. Cereb Cortex. 2007;17(4):816–25. doi: 10.1093/cercor/bhk034. [DOI] [PubMed] [Google Scholar]
- Bastiani M, Shah N, Goebel R, Roebroeck A. Human cortical connectome reconstruction from diffusion weighted MRI: the effect of tractography algorithm. NeuroImage. 2012;62(3):1732–1749. doi: 10.1016/j.neuroimage.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Beckmann C, DeLuca M, Devlin J, Smith S. Investigations into resting-state connectivity using independent component analysis. Phil Trans of Royal Soc. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann M, Johansen-Berg H, Rushworth MF. Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization. J Neurosci. 2009;29(4):1175–90. doi: 10.1523/JNEUROSCI.3328-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Johansen-Berg H, Woolrich MW, Smith SM, Wheeler-Kingshott CA, Boulby PA, Barker GJ, Sillery EL, Sheehan K, Ciccarelli O, Thompson AJ, Brady JM, Matthews PM. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat Neurosci. 2003;6(7):750–7. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- Blekas k, Likas A. The mixture of multi-kernel relevance vector machines model. Data Mining (ICDM) 2012:111–120. [Google Scholar]
- Blumensath T, Jbabdi S, Glasser MF, Van Essen DC, Ugurbil K, Behrens TE, Smith SM. Spatially constrained hierarchical parcellation of the brain with resting-state fMRI. Neuroimage. 2013;76:313–24. doi: 10.1016/j.neuroimage.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson MJ, Malone IB, Modat M, Leung KK, Ryan N, Alexander DC, Fox NC, Ourselin S. A Framework for using diffusion weighted imaging to improve cortical parcellation. MICCAI 2010. 2010;13(Pt1):534–41. doi: 10.1007/978-3-642-15705-9_65. [DOI] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NU, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. Defining functional areas in individual human brains using resting functional connectivity MRI. NeuroImage. 2008;41(1):45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D, Haughton VM, Arfanakis K, Wendt GJ, Turski PA, Moritz CH, Quigley MA, Meyerand ME. Mapping functionally related regions of brain with functional connectivity MR imaging. AJNR Am J Neuroradiol. 2000;21(9):1636–44. [PMC free article] [PubMed] [Google Scholar]
- Corouge I, Gouttard S, Gerig G. Biomedical imaging: nano to macro, 2004: IEEE International Symposium on. IEEE; 2004. Towards a shape model of white matter fiber bundles using diffusion tensor MRI. pp. 344–347. [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- Faraco CC, Unsworth N, Langley J, Terry D, Li K, Zhang D, Liu T, Miller LS. Complex span tasks and hippocampal recruitment during working memory. Neuroimage. 2011;55(2):77–87. doi: 10.1016/j.neuroimage.2010.12.033. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis II: inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Van Der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D. Automatically parcellating the human cerebral cortex. Cereb. Cortex.s. 2004;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Frey BJ, Dueck D. Clustering by Passing Messages between Data Points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- Gerig G, Gouttard S, Corouge I. Analysis of brain white matter via fiber tract modeling. Engineering in Medicine and Biology Society, 2004: IEMBS'04. 26th Annual International Conference of the IEEE. 2004;2:4421–4424. doi: 10.1109/IEMBS.2004.1404229. [DOI] [PubMed] [Google Scholar]
- Gong G, He Y, Concha L, Lebel C, Gross D, Evans A, Beaulieu C. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb. Cortex. 2009;19(3):524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Kurant M, Gigandet X, Thiran P, Wedeen V, Meuli R, Thiran J. Mapping human whole-brain structural networks with diffusion MRI. PLoS One. 2007;2(7):597. doi: 10.1371/journal.pone.0000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Peterson BS, Skudlarski P, Gatenby JC, Gore JC. Detection of functional connectivity using temporal correlations in MR images. Hum Brain Mapp. 2002;15(4):247–62. doi: 10.1002/hbm.10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. PNAS. 2009;106(6):2035–40. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbabdi S, Woolrich MW, Behrens TEJ. Multiple-subjects connectivity-based parcellation using hierarchical Dirichlet process mixture models. NeuroImage. 2009;44(2):373–384. doi: 10.1016/j.neuroimage.2008.08.044. [DOI] [PubMed] [Google Scholar]
- Jbabdi S, Sotiropoulos SN, Savio AM, Graña M, Behrens TE. Model-based analysis of multishell diffusion MR data for tractography: how to get over fitting problems. Magn Reson Med. 2012;68(6):1846–55. doi: 10.1002/mrm.24204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE, Robson MD, Drobnjak I, Rushworth MF, Brady JM, Smith SM, Higham DJ, Matthews PM. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13335–13340. doi: 10.1073/pnas.0403743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JC, Behrens TE, Robson MD, Mackay CE, Higham DJ, Johansen-Berg H. Connectivity-based parcellation of human cortex using diffusion MRI: establishing reproducibility, validity and observer independence in Ba 44/45 and SMA/pre-sma. NeuroImage. 2007;34(1):204–211. doi: 10.1016/j.neuroimage.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Kubicki M, Park H, Westin CF, Nestor PG, Mulkern RV, Maier SE, Niznikiewicz M, Connor EE, Levitt JJ, Frumin M, Kikinis R, Jolesz FA, McCarley RW, Shenton ME. DTI and MTR abnormalities in schizophrenia: Analysis of white matter integrity. Neuroimage. 2005;26(4):1109–1118. doi: 10.1016/j.neuroimage.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Guo L, Li G, Nie J, Faraco C, Cui G, Zhao Q, Miller LS, Liu T. Gyral folding pattern analysis via surface profiling. 2010a;52(4):1202–14. doi: 10.1016/j.neuroimage.2010.04.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Guo L, Zhu D, Hu X, Han J, Liu T. Individualized ROI optimization via maximization of group-wise consistency of structural and functional profiles. NIPS. 2010b;23 doi: 10.1007/s12021-012-9142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Li H, Wong K, Tarokh A, Guo L, Wong STC. Brain tissue segmentation based on DTI data. NeuroImage. 2007;38(1):114–123. doi: 10.1016/j.neuroimage.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Nie J, Tarokh A, Guo L, Wong STC. Reconstruction of central cortical surface from brain MRI images: Method and application. NeuroImage. 2008;40(3):991–1002. doi: 10.1016/j.neuroimage.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang K, Yu C, He Y, Zhou Y, Liang M, Wang L, Jiang T. Regional homogeneity, functional connectivity and imaging markers of Alzheimer's disease: a review of resting-state fMRI studies. Neuropsychologia. 2008;46(6):1648–56. doi: 10.1016/j.neuropsychologia.2008.01.027. [DOI] [PubMed] [Google Scholar]
- Lohmann G, von Cramon DY. Automatic labelling of the human cortical surface using sulcal basins. Med Image Anal. 2000;4(3):179–88. doi: 10.1016/s1361-8415(00)00024-4. [DOI] [PubMed] [Google Scholar]
- Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, Velanova K, Donaldson DI, Phillips JS, Schlaggar BL, Petersen SE. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010;67(1):156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell LJ, Golby AJ, Westin CF. Neuroimage. Fiber clustering versus the parcellation-based connectome. 2013;80:283–9. doi: 10.1016/j.neuroimage.2013.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, Kötter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3(8):606–16. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Perrin M, Cointepas Y, Cachia A, Poupon C, Thirion B, Rivière D, Cathier P, El Kouby V, Constantinesco A, Le Bihan D, Mangin JF. Connectivity-based parcellation of the cortical mantle using q-ball diffusion imaging. Int J Biomed Imaging. 2008;2008:368406. doi: 10.1155/2008/368406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postelnicu GM, Zöllei L, Fischl B. Combined Volumetric and Surface Registration. IEEE Transactions on Medical Imaging. 2009;28(4):508–522. doi: 10.1109/TMI.2008.2004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettmann ME, Han X, Xu C, Prince JL. Automated sulcal segmentation using watersheds on the cortical surface. NeuroImage. 2002;15(2):329–44. doi: 10.1006/nimg.2001.0975. [DOI] [PubMed] [Google Scholar]
- Shen X, Tokoglu F, Papademetris X, Constable RT. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage. 2013;82:403–15. doi: 10.1016/j.neuroimage.2013.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca P, Rivière D, Guevara P, Poupon C, Mangin JF. Tractography-based parcellation of the cortex using a spatially-informed dimension reduction of the connectivitymatrix. Med Image Comput Comput Assist Interv. 2009;12(Pt 1):935–42. doi: 10.1007/978-3-642-04268-3_115. [DOI] [PubMed] [Google Scholar]
- Roca P, Tucholka A, Rivière D, Guevara P, Poupon C, Mangin JF. Inter-subject connectivity-based parcellation of a patch of cerebral cortex. Med Image Comput Comput Assist Interv. 2010;13(Pt 2):347–54. doi: 10.1007/978-3-642-15745-5_43. [DOI] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21(11):1421–39. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Smith SM, Beckmann CF, Andersson J, Auerbach EJ, Bijsterbosch J, Douaud G, Duff E, Feinberg DA, Griffanti L, Harms MP, Kelly M, Laumann T, Miller KL, Moeller S, Petersen S, Power J, Salimi-Khorshidi G, Snyder AZ, Vu AT, Woolrich MW, Xu J, Yacoub E, Uğurbil K, Van Essen DC, Glasser MF WU-Minn HCP Consortium Resting-state fMRI in the Human Connectome Project. Neuroimage. 2013;80:144–68. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos SN, Jbabdi S, Xu J, Andersson JL, Moeller S, Auerbach EJ, Glasser MF, Hernandez M, Sapiro G, Jenkinson M, Feinberg DA, Yacoub E, Lenglet C, Van Essen DC, Ugurbil K, Behrens TE. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage. 2013;80:125–43. doi: 10.1016/j.neuroimage.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P, Toga AW. A surface-based technique for warping three-dimensional images of the brain. IEEE Trans Med Imaging. 1996;15(4):402–17. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- Tomassini V, Jbabdi S, Klein JC, Behrens TE, Pozzilli C, Matthews PM, Rushworth MF, Johansen-Berg H. Diffusion-weighted imaging tractography-based parcellation of the human lateral premotor cortex identifies dorsal and ventral subregions with anatomical and functional specializations. J. Neurosci. 2007;27(38):10259–10269. doi: 10.1523/JNEUROSCI.2144-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56(2):209–25. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Mandl R, Hulshoff Pol H. Normalized cut group clustering of resting-state fMRI data. PLoS One. 2008;3(4):e2001. doi: 10.1371/journal.pone.0002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Luxburg U. A tutorial on spectral clustering. Statistics and Computing. 2007;17(4):395–416. [Google Scholar]
- Wang KJ, Zhang JY, Li D, Zhang XN, Guo T. Adaptive Affinity Propagation Clustering. Acta Automatica Sinica. 2007;33(12):1242–1246. [Google Scholar]
- Wang Q, Yap PT, Wu G, Shen D. Diffusion Tensor Image Registration Using Hybrid Connectivity and Tensor Features. Human Brain Mapping. 2013 doi: 10.1002/hbm.22419. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JHJ. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. [Google Scholar]
- Yang F, Kruggel F. Automatic segmentation of human brain sulci. Med Image Anal. 2008;12(4):442–51. doi: 10.1016/j.media.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Verstynen TD, Wang Y, Fernández-Miranda JC, Tseng WY. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS One. 2013;8(11):380713. doi: 10.1371/journal.pone.0080713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Jiang T, Lu Y, He Y, Tian L. Regional homogeneity approach to fMRI data analysis. Neuroimage. 2004;22(1):394–400. doi: 10.1016/j.neuroimage.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhang T, Guo L, Li G, Nie J, Liu T. Parametric Representation of Cortical Surface Folding based on Polynomials. Medical Image Computing and Computer Assisted Intervention (MICCAI) 2009;12(Pt 2):184–91. doi: 10.1007/978-3-642-04271-3_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Guo L, Li G, Nie J, Deng F, Li K, Hu X, Zhang T, Jiang X, Zhu D, Zhao Q, Liu T. Automatic cortical surface parcellation based on fiber density information. 2010 IEEE International Symposium on Biomedical Imaging. 2010:1133–1136. [Google Scholar]
- Zhu D, Li K, Faraco CC, Deng F, Zhang D, Guo L, Miller LS, Liu T. Optimization of functional brain ROIs via maximization of consistency of structural connectivity profiles. Neuroimage. 2012a;59(2):1382–93. doi: 10.1016/j.neuroimage.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Li K, Guo L, Jiang X, Zhang T, Zhang D, Chen H, Deng F, Faraco C, Jin C, Wee CY, Yuan Y, Lv P, Yin Y, Hu X, Duan L, Hu X, Han J, Wang L, Shen D, Miller LS, Li L, Liu T. DICCCOL: dense individualized and common connectivity-based cortical landmarks. Cereb Cortex. 2012b;23(4):786–800. doi: 10.1093/cercor/bhs072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllei L, Stevens A, Huber K, Kakunoori S, Fischl B. Improved Tractography Alignment Using Combined Volumetric and Surface Registration. NeuroImage. 2010;51(1):206–213. doi: 10.1016/j.neuroimage.2010.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.