Abstract
A fully automated and robust method was developed to quantify β-III-tubulin-stained retinal ganglion cells, combining computational recognition of individual cells by CellProfiler and a machine-learning tool to teach phenotypic classification of the retinal ganglion cells by CellProfiler Analyst. In animal models of glaucoma, quantification of immunolabeled retinal ganglion cells is currently performed manually and remains time-consuming. Using this automated method, quantifications of retinal ganglion cell images were accelerated tenfold: 1800 images were counted in 3 hours using our automated method, while manual counting of the same images took 72 hours. This new method was validated in an established murine model of microbead-induced optic neuropathy. The use of the publicly available software and the method’s user-friendly design allows this technique to be easily implemented in any laboratory.
1. Introduction
Glaucoma is the leading cause of irreversible blindness worldwide, affecting an estimated 60 million people (Tham et al., 2014). While there are many forms of glaucoma, all are associated with an optic neuropathy characterized by the loss of retinal ganglion cells (RGCs) and their axons, resulting in optic nerve degeneration and irreversible vision loss. Animal models of glaucoma that simulate the optic neuropathy observed in human disease facilitate the elucidation of possible mechanisms of RGC loss and enable researchers to develop and evaluate neuroprotective therapies. The ability to specifically identify and accurately count RGCs is essential to assess the death or survival of RGCs in models of the disease.
Various techniques for visualizing and quantifying RGCs have been reported, including retrograde labeling and immunolabeling (Buckingham et al., 2008; Huihui et al., 2011). In a healthy, non-diseased retina, fluorogold retrograde labeling is a specific and accurate method to label and automatically quantify RGCs (Buckingham et al., 2008; Danias et al., 2003). However, this method is technically challenging, requiring intracranial surgery and, in a glaucomatous retina, retrograde labeling can result in the labeling of retinal microglia in addition to RGCs, due to the phagocytosis of degenerating RGCs (Peinado-Ramon et al., 1996; Thanos, 1991a, b). An alternative quantification method of RGC viability may be carried out with immunolabeling of RGCs in retinal flatmounts using an RGC-specific antibody directed against β-III-tubulin, which labels the RGC somata and nerve fiber extensions. The specificity of the β-III-tubulin antibody has been confirmed by colocalization of β-III-tubulin staining with fluorogold-labeled RGCs (Huihui et al., 2011).
While the immunolabeling method is technically simpler than fluorogold labeling, manual quantification of immunolabeled murine RGCs is onerously time-consuming. In a small-scale pre-clinical project with only 2 experimental groups, an investigator would be required to manually quantify RGCs in approximately 1000 images, necessitating at least 48 hours of effort. In addition, manual counting can be inconsistent, with significant inter- and even intra-observer variability, due to the use of differing quantification techniques and/or inhomogeneous staining.
2. Materials and Supplies
Animals
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institute of Health. Wild-type mice bred on a 129S6 background (129S6/SvEvTac, Taconic Farms Inc.) were used in this study. All animals were treated in accordance with the Institutional Animal Care and Use Committees (IACUC) of Massachusetts General Hospital (Subcommittee on Research Animal Care), and the Schepens Eye Research Institute.
Processing and imaging of retinas
Animals were sacrificed under CO2. Eyes were enucleated and retinas dissected from the anterior segments at the conclusion of the study: day 32 post-injection. Resultant retinal cups were incised to create four quadrants of similar size and were fixed in 4% paraformaldehyde at 4°C overnight. Retinas were then treated with 1% Triton-X-100 and 5% fetal BSA in PBS for 1 hour, followed by a 2 hour incubation with DAPI (1:500) and the primary antibody against an RGC marker, β-III-tubulin (anti-TUJ1+, Millipore, Billerica, MA, 1:500), and 1 hour incubation with the Alexa Fluor® 594 Goat Anti-Mouse IgG secondary antibody (Life Technologies, Carlsbad, CA, 1:500) at room temperature. Retinal whole-mounts were then flattened on SuperFrost Plus slides (VWR, Batavia, IL), coverslipped with mounting medium for fluorescence (VectaShield®, Vector Laboratories, Burlingame, CA) and imaged under the Leica TSC SP5 confocal microscope at x63 magnification. Imaging was performed on the mid-peripheral area of the retina (around 0.5 mm distal from the optic nerve head) divided into 4–5 distinct areas across all four quadrants. Each retina was imaged in 20–25 frames of 0.0696 mm2.
Microbead injections
Mice were anesthetized by intraperitoneal injection of a mixture of ketamine (100 mg/kg; Ketaset; Fort Dodge Animal Health, Fort Dodge, IA) and xylazine (9 mg/kg; TranquiVed; Vedco, Inc., St. Joseph, MO) and eyes were dilated by topical application of proparacaine (0.5%; Bausch & Lomb, Tampa, FL). Elevation of IOP was induced unilaterally in adult 129S6 mice by injection of polystyrene microbeads (FluoSpheres; Invitrogen, Carlsbad, CA; 15 μm diameter) into the anterior chamber of the right eye of each animal under a surgical microscope. Microbeads were reformulated at a concentration of 5.0 × 106 beads/ml in phosphate-buffered saline (PBS). The right cornea was gently punctured near the center using a sharp 30-gauge needle (World Precision Instruments Inc., Sarasota, FL). An air bubble was injected via the micropipette connected with a Hamilton syringe, itself coupled to a syringe pump to avoid overflow of the AqH from the anterior chamber prior to injection of microbeads. A precise volume (2 μl) of microbeads was injected through the pre-formed hole into the anterior chamber using the micropipette. Mice were placed on a heating pad for recovery after the injection, and antibiotic Vetropolycin ointment (Dechra Veterinary Products, Overland Park, KS) was applied topically onto the injected eye to prevent infection.
IOP measurement
Mice were anesthetized with isoflurane inhalation (2%), which was delivered in 95% oxygen with a precision vaporizer. IOP measurement was initiated within 2 min after animal lost toe pinch and blink reflex. IOPs were acquired with a TonoLab rebound tonometer (iCare, Franconia, NH, USA). Five TonoLab readings were averaged to obtain a single IOP value per eye. IOP measurements were carried out at days 4, 10, 15, 18, 22 and 28 of the 32-day study.
Statistical analyses
All statistical analyses were performed using GraphPad Prism (version 6.0). Statistical tests included the Student’s t-test, Pearson correlation, or multivariate linear regression. In order to compare correlation coefficients, the Fisher method of r-to-z transformation was employed, using the following equation:
Pairwise correlations with their respective samples sizes (n1 and n2) were compared with the following test statistic:
Correlation dispersion was used to quantify differences between automated and manual counts for each retina. Correlation dispersion was determined using the following equation:
where y represents the cell count in RGC/mm2.
P<0.05 was considered significant throughout the study. Data are presented as mean ± s.d.
3. Detailed methods
Generation of a rapid and robust automated quantification technique
To minimize variability and enable faster and more efficient quantification of immunolabeled RGCs, we sought to develop and validate a reproducible automated quantification method. The approach uses the freely-available softwares, CellProfiler and CellProfiler Analyst (CPA) (Carpenter et al., 2006; Kamentsky et al., 2011; Lamprecht et al., 2007), and can be used for high-throughput image analysis. The CellProfiler image analysis template was designed to recognize all DAPI-positive (nuclear marker) and β-III-tubulin-labeled cylindrical RGC somata (Box 1).
Box 1.
The CellProfiler software is freely available to download at www.cellprofiler.org. The image analysis template used for the automated quantification presented in the manuscript, the latest software updates and the source code are available to download at http://www.cellprofiler.org/published_pipelines.shtml under the “B3-tubulin_RetinalGanglionCell_Assay.cpproj” nomenclature.
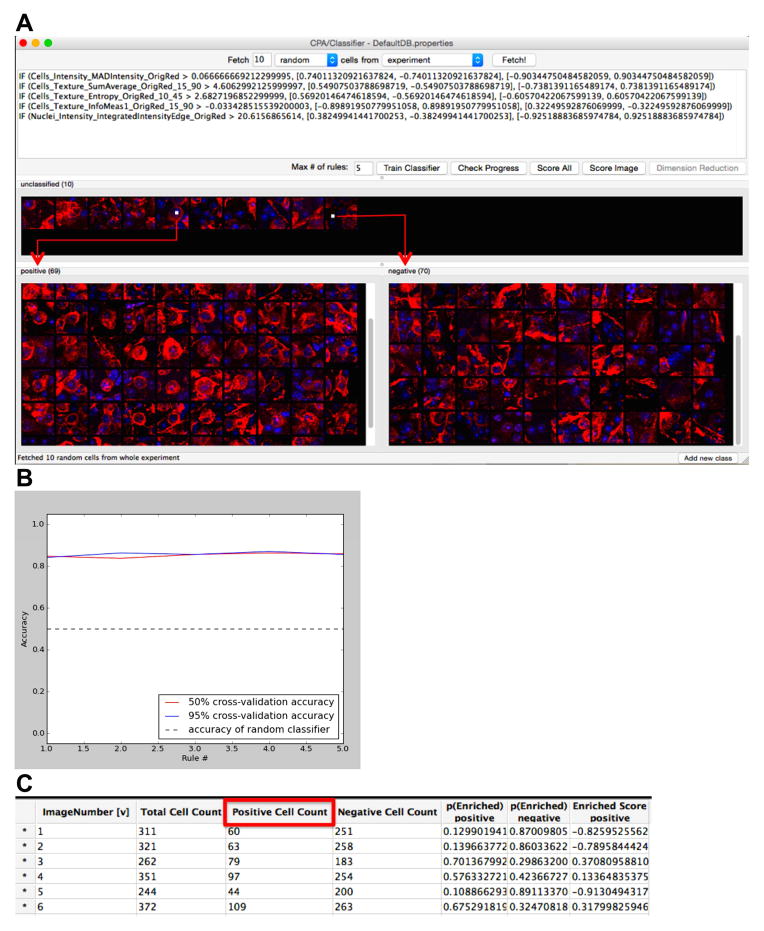
The workflow was as follows: the CellProfiler template was loaded and images from experimental set-ups were fetched into the “File list”. Automated processing of each image was initiated (“Analyze Images”): the morphological features for each cell, including the nuclei and cellular shapes, as well as intensity-based and textural features from both the DAPI and β-III-tubulin channels were recognized and measured by CellProfiler; the complete set of these measurements for each cell is defined as the “cytoprofile” (Carpenter et al., 2006). Once the measurements were collected, the supervised machine-learning tool “Classifier” in CPA (Jones et al., 2008) was used to perform phenotypic classification of the RGCs. In brief, a “training set” was initialized by the researcher, first “fetching” a small selection of sample cells drawn at random from the experiment that exhibit the desired RGC phenotype (i.e. β-III-tubulin-stained cells co-labeled with DAPI: “positives”), as well as cells that did not present that phenotype (“negatives”). The researcher then assigned images of cells that were deemed to be “RGC-positive” to the “positive” window and images of cells that were deemed “RGC-negative” to the “negative” window (Fig. 1A). Following classification of each “fetched” image, the “Train Classifier” tool was activated and a tentative “rule” was formed based on the cytoprofiles of the positive and negative example cells (Fig. 1A). The researcher was then presented with new sample cells to score as positive or negative, the score of which was used by the CPA to improve the training set by correcting errors (Fig. 1A) and refine the quantification “rule”. Following each classification step, the “Train Classifier” tool was initiated to save recognition refinements to the “rule”. This iterative process allowed the researcher to produce a classifier specific to the RGC phenotype of interest within a few (on average, three to five) rounds of corrections and refinements. Overall, the training step generally takes 20 minutes. An example of a “rule” classifier is provided in Figure 1A; each line represents a measurement deemed useful by the CPA classifier in distinguishing the phenotype, with the more valuable measurements located towards the top. The accuracy of the classifier’s ability to correctly identify positive and negative cells was assessed at each training step using cross-validation (“Check Progress”), until training based on 50% of the training set was equally effective as 95% of the training set, i.e. until the training set was large enough that adding more samples would not increase accuracy (Fig. 1B). An accuracy above 80% was considered adequate (Jones et al., 2009). The final step was to apply the classifier to the experimental groups, scoring all cells as positive or negative (“Score All” Fig. 1A; engendering a results table, Fig. 1C).
Figure 1. Automated scoring of cell morphologies using the CellProfiler Analyst machine learning and iterative feedback system.
(A) The software system presents the researcher with individual cells to classify from segments of images, sampled randomly from the set of images. After classification of a few cells, the researcher begins the iterative machine-learning phase (“Train Classifier”), in which the software generates a tentative rule based on the classified cells and presents the researcher with cells classified according to that rule. (B) The accuracy of the CPA classifier rule may be checked after each training set (“Check Progress”) and accuracy above 80% is considered suitable. (C) Following 4 training iterations, all cells from the experiment of interest are classified in order to calculate the number of positive (RGC) and negative (non-RGC) cells. “Positive Cell Count” denotes the cells recognized as RGCs. “Negative Cell Count” refers to image regions, in which the cells’ morphological features were not recognized as RGCs. “Total Cell Count” refers to the total number of cells recognized per image. “Image Number” represents the number of the image associated with the cell counts.
To test the workflow, 72 retinas (1800 images: 25 confocal images per retina) were first quantified using manual and automated techniques. Manual quantifications were carried out using ImageJ (Box 2).
Box 2.
The (Fiji is Just) ImageJ software is freely available to download at http://fiji.sc/Fiji. The Cell Counter plugin information used for manual quantifications of RGCs in this study is available at http://fiji.sc/Cell_Counter.
Images were loaded consecutively through the (Fiji is Just) ImageJ software. Using the “Cell Counter” plugin (Supplementary Fig. 1A), the image loaded into ImageJ was “initialized” and the color counter type was chosen (Supplementary Fig. 1B). “Type 3” was our counter of choice for its green color, enabling clear partition with the β-III-tubulin-positive RGC cells.
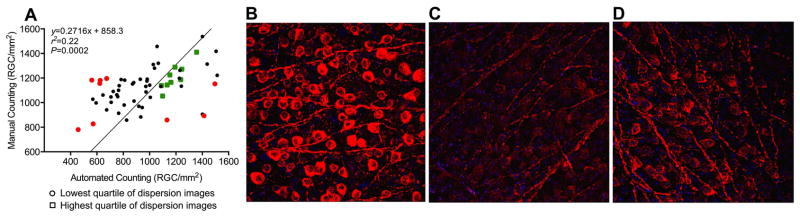
A linear regression model, comparing manual and automated quantifications, initially revealed only a weak correlation between results from the automated quantification method and those obtained by manual quantification (Fig. 2A). We hypothesized that image quality affected the ability of an observer (manual counting) and/or the image processing algorithms (automated counting) to accurately identify and count RGCs. To probe this assumption, we first assessed the quality of the images visually (Figs. 2B, 2C and 2D). While some images displayed consistent staining, homogeneous exposure, and clean RGC delineations (Fig. 2B), other images were of lower quality, due to heterogenous immunostaining (Fig. 2C), increased density of nerve fibers, or uneven imaging (Fig. 2D).
Figure 2.
(A) Correlation between manual and automated quantifications of 72 murine retinas. Each count represents the averages of 20–25 images per retina. Counts are presented as number of RGC/mm2. Green square symbols: 9 retinas with highest quartiles of correlation dispersion (well-correlated manual and automated counts). Red round symbols: 9 retinas with lowest quartiles of correlation dispersion (poorly-correlated manual and automated counts). (B) “High-quality” image with clean RGC body delineations. (C) Image with discordant/low staining or underexposure to highlight the RGCs. (D) Image of a fiber-rich uneven exposure, possibly due to the retinal structure not being fully flattened onto the imaging slide.
Quality control using ImageJ
A guideline in ImageJ was developed to select images of sufficient quality to be reproducibly counted (manually or automatically) based on the averages of circularity and pixel thresholds of RGCs in each image. Specifically, for each image, a binary contrast enhancement was carried out: images were converted from the original color (red-green-blue (RGB), Supplementary Figs. 2A and 2D) to an 8-bit grayscale, set to a threshold intensity of 89 and inverted so that fluorescent cells would appear as black objects on a white background (Supplementary Figs. 2B and 2E). Particles were then analyzed to assess circular areas of pixels (with thresholds set to include Circularity 0.05–1.00 and Size in pixel units 200-Infinity), denoting circular RGCs within same-sized frames in “high-quality/included” (Supplementary Fig. 2C) and “low-quality/excluded” images (Supplementary Fig. 2F). This method highlighted the significant differences in circular pixilation areas between high-quality and low-quality images (Supplementary Figs. 2C and 2F and Supplementary Table 1), irrespective of the RGC counts obtained per image. Images with a circularity score ≥ 0.13 (selected cut-off value; Supplementary Fig. 2 and Supplementary Table 1) were deemed of insufficient quality.
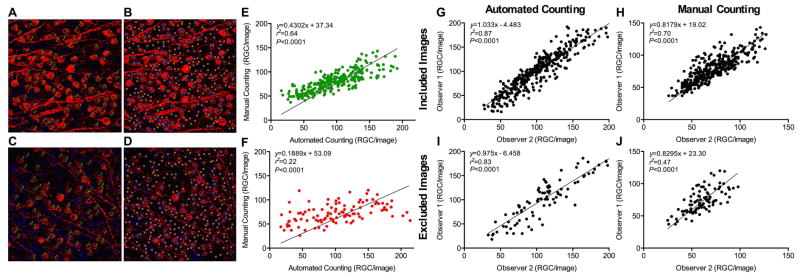
The impact of image quality on the reproducibility of both manual and automated methods was studied in 18 selected retinas (21 images per retina): nine retinas representing the highest quartile of correlation dispersions (Δ398 ± 80.8 RGC/mm2) and nine retinas representing the lowest quartile of correlation dispersions (Δ70.1 ± 68.6 RGC/mm2) (Fig. 2A, see ‘Statistical analyses’ for a description of correlation dispersions calculations). Using the unbiased ImageJ quality assessment platform, 272 images of the total 378 images from 18 retinas were categorized as included images (Figs. 3A and 3B), while 28% (106 images) were labeled as excluded images (Figs. 3C and 3D). The linear regression fit between manual and automated quantifications improved markedly for included images (r2 = 0.64, Fig. 3E) as compared to excluded images (r2 = 0.22, Fig. 3F), highlighting the advantage of a quality control step prior to RGC quantifications, whether performed manually or automatically.
Figure 3.
(A–D) Examples of an included image (Figs. 3A and 3B) and an excluded image (Figs. 3C and 3D, based on ImageJ exclusion criteria described in ‘Materials and Methods’). Cells were counted either manually (Figs. 3A and 3C) or automatically with CellProfiler followed by training with CellProfiler Analyst (Figs. 3B and 3D). Green dots represent RGCs identified by the observer. The number 3 refers to the Cell Counter type chosen, an arbitrary label from the software. Blue dots represent RGCs recognized by the software. Orange dots represent structures identified by a step in the image-processing pipeline but not counted as RGCs by the software. (E) Correlation between the two quantification methods of included images (n = 272). Counts are presented as raw number of cells per image. (F) Correlation between the two quantification methods of excluded images (n = 106). Counts are presented as raw number of cells per image. Markedly greater correlations were observed for included (Fig. 3E) than excluded (Fig. 3F) images (Fisher r-to-z transformation with Z factor = 4.62). (G–J) Correlations of manual (Figs. 3H and 3J) and automated (Figs. 3G and 3I) quantification methods of RGC images carried out by two independent observers. Counts are presented as raw number of cells per image. Image groups are presented as included (Figs. 3G and 3H) and excluded (Figs. 3I and 3J) images, as defined by the pre-selection of image quality carried out using ImageJ.
Additionally, to investigate whether observer bias may have affected the supervised classification quality, two researchers independently counted images manually, and carried out automated quantification using CellProfiler and the machine-learning tool in CPA. The two observers’ counts were strongly correlated for included (r2 = 0.87) and excluded (r2 = 0.83) image groups using automated counting (Z = 1.25, Figs. 3G and 3I, respectively), while correlations were lower for manual quantification, with r2 = 0.70 for included images and r2 = 0.47 for the excluded images (Z = 3.08, Figs. 3H and 3J, respectively). Consistent quantification of RGCs was achieved using both techniques. Importantly, the automated counting method provided greater reproducibility for images of lower quality as compared to the manual quantification method (Figs. 3I and 3J), highlighting the robustness of our automated quantification method. Particularly advantageous is the fact that the automated approach was considerably faster than the manual method; manual counting of 1800 images took one observer 72 hours to complete, while automated quantification was achieved in 3 hours.
In vivo validation of the computational quantification method
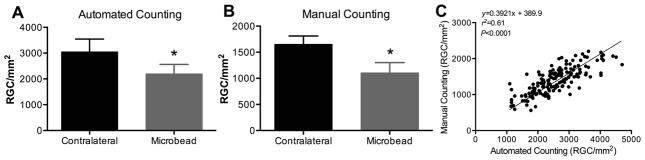
To validate our approach, the automated counting method was applied to study RGC loss in an established mouse model of optic neuropathy, the microbead occlusion model (Huihui et al., 2011; Sappington et al., 2010). Injection of 15 μm-diameter microbeads into the anterior chamber blocks the outflow of aqueous humor, resulting in a significant increase in intraocular pressure (IOP) for up to three weeks post-injection and a 25–40% loss of RGCs (Huihui et al., 2011). The microbead-injected eyes exhibited a significant increase in IOP over a period of 18 days compared to uninjected contra-lateral eyes (average changes in IOP over 18 days: 14 ± 0.8 mmHg to 22 ± 1.1 mmHg, n = 6, P < 0.05). Both our automated approach and manual quantification were able to identify significant neuropathy at the conclusion of the 32-day study, with comparable RGC loss (29.2 ± 9.1% vs. 29.8 ± 10.3% loss of RGC/mm2, Figs. 4A and 4B, respectively). Manual and automated quantifications were also correlated (r2 = 0.61, Fig. 4C), highlighting the efficacy and translatability of our fully automated quantification method.
Figure 4. Automated quantification method successfully captures microbead-induced optic neuropathy.
(A) Number of RGCs/mm2 counted using the CellProfiler automated quantification method in retinas isolated from non-injected (1616 ± 333 RGC/mm2, n = 6) and microbead-injected (1144 ± 452 RGC/mm2, n = 6) eyes. Values are presented as mean ± s.d. *P<0.05 vs. uninjected contralateral eye. (B) Number of RGCs/mm2 counted manually in retinas isolated from non-injected (3019 ± 643 RGC/mm2, n = 6) and microbead-injected (2404 ± 438 RGC/mm2, n = 6) eyes. (C) Results are correlated between manual and automated quantification methods of images (n = 162) from 12 murine retinas.
4. Potential pitfalls and troubleshooting
The CellProfiler software provides a platform to recognize cells according to texture, size and color; the training set in CPA is the tool enabling reproducible quantifications. Since CPA is initialized and overseen by the researcher, setting up a template to accurately recognize and measure β-III-tubulin-positive RGC somata and training for recognition of “positive” and “negative” cells to “include” or “exclude” in quantifications remain relatively subjective. However, subjectivity is also inherent to manual quantifications, in which the researcher arbitrarily decides which cells to count. The CPA training set will consistently quantify cells assigned as “positive” throughout images of experiments, while, during manual quantifications, human error may inadvertently occur and subjectivity may fluctuate between 2 researchers and/or from one set of images to another. The greater reproducibility of the CPA was highlighted with greater correlations between automated quantifications from 2 independent researchers than those between manual quantifications from the same 2 investigators.
Quantifications were also influenced by the quality of the image. In an experimental setting, heterogeneous immunolabeling of β-III-tubulin-positive cells occurs, affecting the ability of the CPA tool to accurately count cells. An ImageJ-based quality control was established to filter high-quality images for quantification. It is important to note that, while performing the ImageJ-based quality control improved the correlation between manual and automated counts, foregoing the use of ImageJ prior to quantifying images using CellProfiler and CPA would not corrupt results obtained, as automated quantifications of cells using poor quality images remained more reliable than those from manual counting. Moreover, the number of training sets within CPA may be increased to refine the “RGC recognition rule” for image sets that present with higher numbers of lower quality images.
In the validation cohort (Fig. 4), absolute RGC numbers were higher when counted using CellProfiler and CPA than when counted manually. This discrepancy may be the result of a combination of subjectivity in manual counts throughout images and definition of inclusion and exclusion criteria in the CPA training phase. Previous studies reported RGC counts similar to the average of 3019 RGC/mm2 we obtained using the automated method, with various staining methods in various strains of mice (Table 1). Our data, together with published data, suggest that our manual count may have underestimated the number of RGCs present. In general, it remains challenging to determine absolute numbers of RGCs: manual quantifications are affected by inter-observer variability due to subjective observer error. Importantly, the reproducibility of the automated method was validated in an established model of glaucoma, in which relative differences in RGC counts (a primary endpoint in several studies (Chen et al., 2011; Huihui et al., 2011; Yang et al., 2012)) between microbead-injected and uninjected eyes were comparable using the presented automated and the manual methods. Taken together, our data suggest that automated counting is at least as reliable as, but much less time consuming and prone to inter-observer variability, than manual counting.
Table 1.
Retinal ganglion cell numbers.
| Reference | Species | Strain | Cell Type | Labeling Method | Count/mm2 | Count/Retina |
|---|---|---|---|---|---|---|
| (Khan et al., 2015) | Mouse | C57Bl/6J | RGC | Neurobiotin | 2867±445 | |
| (Templeton et al., 2014) | Mouse | C57Bl/6J | Axon | PPD | 44,846±3,980 | |
| (Jeon et al., 1998) | Mouse | C57Bl/6J | GCL neurons | EH-stained nuclei | 5510–5918 | |
| (Chen et al., 2011) | Mouse | C57Bl/6J | RGC | Fluorogold | 3828±355 | |
| (Danias et al., 2003) | Mouse | C57Bl/6J | RGC | Fluorogold | 4,212±323 | 69,824±7,943 |
| (Cone et al., 2010) | Mouse | C57Bl/6J | RGC/Axon | Dapi/1% T | 4,196±406 | 51,064±5,045 |
| (Nusbaum et al., 2015) | Mouse | CD-1 | RGC | β-III-Tubulin | 3713±262 | |
| (Cone et al., 2010) | Mouse | CD-1 | RGC/Axon | Dapi/1% T | 3820±232 | 60,995±6,994 |
| (Sanchez- Migallon et al., 2016) | Mouse | Crl:CD-1 | RGC | Brn3a | 49,631±4,633 | |
| (Nadal- Nicolas et al., 2015) | Rat | Sprague Dawley | RGC | Brn3a | 2889±80 | |
| (Danias et al., 2003) | Mouse | DBA/2NNia | RGC | Fluorogold | 3,882±295 | 72,322±5,192 |
| (Cone et al., 2010) | Mouse | DBA/2J | RGC/Axon | Dapi/1% toluidine | 4,121±262 | 60,938±5,553 |
Summary of rodent ganglion cell numbers segmented by species and strain. EH: Ethidium homodimer; T: Toluidine.
Finally, while the presented automated counting platform was designed for β-III-tubulin immunolabeled RGC recognition, the CellProfiler template features may recognize and process additional cell sizes and shapes or be modified to do so as necessary. The use of the machine-learning tool in CPA may then be taught to implement the new cell features. For instance, staining with Brn3a, an RGC-specific protein and alternative to β-III-tubulin staining, has been highlighted as a dependable identification method of RGC somata alone (Leahy et al., 2004; Schlamp et al., 2013). The current pipeline set up in CellProfiler was altered to recognize Brn3a-stained RGCs and cells were successfully recognized and quantified by CellProfiler (Supplementary Fig. 3), with RGC numbers averaging 2546 ± 425 RGC/mm2 (172.1 ± 27.9 RGC/image) and 2586 ± 413 RGC/mm2 (174.8 ± 28.7 RGC/image) for manual and automated quantifications, respectively, which are similar to previous reports of Brn3a-stained RGC counts (2889 RGC/mm2) (Nadal-Nicolas et al., 2015).
In summary, a reliable investigator-supervised counting tool was developed for automatically quantifying immunolabeled RGC somata in a highly time-efficient manner. This freely available protocol enabled rigorous quantification of β-III-tubulin-stained RGCs for unbiased assessments of relative changes in RGC numbers in pathological conditions (e.g. between control animals and animals treated with an experimental therapy). This approach may also serve to and reconcile intra- and inter-observer variability.
Supplementary Material
Supplementary Table 1. Average measurements for included images vs. excluded images.
(A) Individual images are loaded into the software and the “Cell Counter” plug-in chosen for the manual quantification. (B) Each image is then “initialized” into ImageJ and a color counter “type” is selected.
(A) Representative red-green-blue (RGB) “included” image. (B) Binary image of the same “included” image, created following conversion to an 8-bit grayscale and inversion to render fluorescent cells as black objects. (C) Representative outlined cells from the “included” binary image. (D) Representative RGB “excluded” image. (E) Representative binary image of the same “excluded” image. (F) Representative outlined cells from the “excluded” binary image.
(A) Correlation between manual and automated quantification of 25 images of Brn3a-stained murine RGCs using CellProfiler. Counts are presented as number of RGC/image. (B) Manual counting of Brn3a-stained RGCs using the Cell Counter plug-in in ImageJ. (C) Output image following CellProfiler processing and quantification of the same Brn3a-stained RGCs. The Brn3a CellProfiler image analysis template and source code are available to download at http://www.cellprofiler.org/published_pipelines.shtml under the “Brn3a_RetinalGanglionCell_Assay” nomenclature.
Acknowledgments
This work was supported by NIH grants: [F.F. and M.G.K.] EY021543, [A.C.D., K.A. and E.S.B.] EY022746-01 and [M.A.B. and A.C.] GM089652. Jie Ma provided assistance with Brn3a staining.
Footnotes
Author Contributions
A.C.D. and E.S.B. conceived the project. M.A.B., D.L., A.C.D. and A.E.C. developed the cell recognition pipeline. A.C.D. and K.A. carried out manual and automated quantifications. F.F. and M.G.K. carried out microbead injections. R.M. advised on statistical analyses and data interpretation. A.C.D. analyzed all the data. A.C.D. wrote the manuscript. E.S.B., M.G.K and A.E.C. reviewed the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Buckingham BP, Inman DM, Lambert W, Oglesby E, Calkins DJ, Steele MR, Vetter ML, Marsh-Armstrong N, Horner PJ. Progressive ganglion cell degeneration precedes neuronal loss in a mouse model of glaucoma. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:2735–2744. doi: 10.1523/JNEUROSCI.4443-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wei X, Cho KS, Chen G, Sappington R, Calkins DJ, Chen DF. Optic neuropathy due to microbead-induced elevated intraocular pressure in the mouse. Invest Ophthalmol Vis Sci. 2011;52:36–44. doi: 10.1167/iovs.09-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone FE, Gelman SE, Son JL, Pease ME, Quigley HA. Differential susceptibility to experimental glaucoma among 3 mouse strains using bead and viscoelastic injection. Experimental eye research. 2010;91:415–424. doi: 10.1016/j.exer.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danias J, Lee KC, Zamora MF, Chen B, Shen F, Filippopoulos T, Su Y, Goldblum D, Podos SM, Mittag T. Quantitative analysis of retinal ganglion cell (RGC) loss in aging DBA/2NNia glaucomatous mice: comparison with RGC loss in aging C57/BL6 mice. Invest Ophthalmol Vis Sci. 2003;44:5151–5162. doi: 10.1167/iovs.02-1101. [DOI] [PubMed] [Google Scholar]
- Huihui C, Xin W, Kin-Sang C, Guochun C, Rebecca S, David JC, Dong FC. Optic neuropathy due to microbead-induced elevated intraocular pressure in the mouse. Investigative ophthalmology & visual science. 2011;52:36–44. doi: 10.1167/iovs.09-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1826–1831. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC bioinformatics. 2008;9:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MAA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics (Oxford, England) 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AK, Tse DY, van der Heijden ME, Shah P, Nusbaum DM, Yang Z, Wu SM, Frankfort BJ. Prolonged elevation of intraocular pressure results in retinal ganglion cell loss and abnormal retinal function in mice. Experimental eye research. 2015;130:29–37. doi: 10.1016/j.exer.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. BioTechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Leahy KM, Ornberg RL, Wang Y, Zhu Y, Gidday JM, Connor JR, Wax MB. Quantitative ex vivo detection of rodent retinal ganglion cells by immunolabeling Brn-3b. Experimental eye research. 2004;79:131–140. doi: 10.1016/j.exer.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Nadal-Nicolas FM, Sobrado-Calvo P, Jimenez-Lopez M, Vidal-Sanz M, Agudo-Barriuso M. Long-Term Effect of Optic Nerve Axotomy on the Retinal Ganglion Cell Layer. Invest Ophthalmol Vis Sci. 2015;56:6095–6112. doi: 10.1167/iovs.15-17195. [DOI] [PubMed] [Google Scholar]
- Nusbaum DM, Wu SM, Frankfort BJ. Elevated intracranial pressure causes optic nerve and retinal ganglion cell degeneration in mice. Experimental eye research. 2015;136:38–44. doi: 10.1016/j.exer.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado-Ramon P, Salvador M, Villegas-Perez MP, Vidal-Sanz M. Effects of axotomy and intraocular administration of NT-4, NT-3, and brain-derived neurotrophic factor on the survival of adult rat retinal ganglion cells. A quantitative in vivo study. Invest Ophthalmol Vis Sci. 1996;37:489–500. [PubMed] [Google Scholar]
- Sanchez-Migallon MC, Valiente-Soriano FJ, Nadal-Nicolas FM, Vidal-Sanz M, Agudo-Barriuso M. Apoptotic Retinal Ganglion Cell Death After Optic Nerve Transection or Crush in Mice: Delayed RGC Loss With BDNF or a Caspase 3 Inhibitor. Invest Ophthalmol Vis Sci. 2016;57:81–93. doi: 10.1167/iovs.15-17841. [DOI] [PubMed] [Google Scholar]
- Sappington RM, Carlson BJ, Crish SD, Calkins DJ. The microbead occlusion model: a paradigm for induced ocular hypertension in rats and mice. Invest Ophthalmol Vis Sci. 2010;51:207–216. doi: 10.1167/iovs.09-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlamp CL, Montgomery AD, Mac Nair CE, Schuart C, Willmer DJ, Nickells RW. Evaluation of the percentage of ganglion cells in the ganglion cell layer of the rodent retina. Molecular vision. 2013;19:1387–1396. [PMC free article] [PubMed] [Google Scholar]
- Templeton JP, Struebing FL, Lemmon A, Geisert EE. ImagePAD, a novel counting application for the Apple iPad, used to quantify axons in the mouse optic nerve. Experimental eye research. 2014;128:102–108. doi: 10.1016/j.exer.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Thanos S. The Relationship of Microglial Cells to Dying Neurons During Natural Neuronal Cell Death and Axotomy-induced Degeneration of the Rat Retina. The European journal of neuroscience. 1991a;3:1189–1207. doi: 10.1111/j.1460-9568.1991.tb00054.x. [DOI] [PubMed] [Google Scholar]
- Thanos S. Specific transcellular carbocyanine-labelling of rat retinal microglia during injury-induced neuronal degeneration. Neuroscience letters. 1991b;127:108–112. doi: 10.1016/0304-3940(91)90906-a. [DOI] [PubMed] [Google Scholar]
- Yang Q, Cho KS, Chen H, Yu D, Wang WH, Luo G, Pang IH, Guo W, Chen DF. Microbead-induced ocular hypertensive mouse model for screening and testing of aqueous production suppressants for glaucoma. Invest Ophthalmol Vis Sci. 2012;53:3733–3741. doi: 10.1167/iovs.12-9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Average measurements for included images vs. excluded images.
(A) Individual images are loaded into the software and the “Cell Counter” plug-in chosen for the manual quantification. (B) Each image is then “initialized” into ImageJ and a color counter “type” is selected.
(A) Representative red-green-blue (RGB) “included” image. (B) Binary image of the same “included” image, created following conversion to an 8-bit grayscale and inversion to render fluorescent cells as black objects. (C) Representative outlined cells from the “included” binary image. (D) Representative RGB “excluded” image. (E) Representative binary image of the same “excluded” image. (F) Representative outlined cells from the “excluded” binary image.
(A) Correlation between manual and automated quantification of 25 images of Brn3a-stained murine RGCs using CellProfiler. Counts are presented as number of RGC/image. (B) Manual counting of Brn3a-stained RGCs using the Cell Counter plug-in in ImageJ. (C) Output image following CellProfiler processing and quantification of the same Brn3a-stained RGCs. The Brn3a CellProfiler image analysis template and source code are available to download at http://www.cellprofiler.org/published_pipelines.shtml under the “Brn3a_RetinalGanglionCell_Assay” nomenclature.