Abstract
Food-borne campylobacteriosis is caused mainly by the handling or consumption of undercooked chicken meat or by the ingestion of contaminated raw milk. Knowledge about the contributions of different food sources to gastrointestinal disease is fundamental to prioritize food safety interventions and to establish proper control strategies. Assessing the genetic diversity among Campylobacter species is essential to our understanding of their epidemiology and population structure. We molecularly characterized 56 Campylobacter jejuni isolates (31 from patients hospitalized with gastroenteritis, 17 from raw milk samples, and 8 from chicken samples) using multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) in order to trace the source of the disease. We also used a population genetic approach to investigate the source of the human cases from six different reservoirs of infection. MLST identified 25 different sequence types and 11 clonal complexes (CCs) (21, 658, 206, 353, 443, 48, 61, 257, 1332, 354, 574) and these included several alleles not cited previously in the PubMLST international database. The most prevalent CCs were 21, 206, and 354. PFGE showed 34 pulsotypes divided between 28 different clusters. At the fine scale, by means of PFGE and MLST, only two human cases were linked to raw milk, while one case was linked to chicken meat. The investigation revealed the presence of several genotypes among the human isolates, which probably suggests multiple foci for the infections. Finally, the source attribution model we used revealed that most cases were attributed to chicken (69.75%) as the main reservoir in Italy, followed to a lesser extent by the following sources: cattle (8.25%); environment (6.28%); wild bird (7.37%); small ruminant (5.35%), and pork (2.98%). This study confirms the importance of correlating epidemiological investigations with molecular epidemiological data to better understand the dynamics of infection.
Keywords: Campylobacter, PFGE, population genetic approach, epidemiological investigations, MLST genotyping
Introduction
Human campylobacteriosis in the European Union (EFSA, 2014) continues to be the most commonly reported zoonosis, and with 214,268 confirmed cases, this disease has considerable socio-economic impact. The Campylobacter species most commonly associated with human infections are Campylobacter jejuni followed by C. coli and C. lari, but other species, including the non-thermophilic C. fetus, also occasionally cause human diseases (EFSA, 2014; Rodrigues et al., 2015). In Italy, 1252 cases of human campylobacteriosis were reported in 2014 (EFSA and ECDC, 2015). However, the data are likely to grossly underestimate the real number of cases (EFSA and ECDC, 2015) because the Italian reporting system for human infectious illnesses does not differentiate between gastroenteritis caused by Campylobacter and gastroenteritis caused by the other agents listed in the National Legislation in Italy (Calistri and Giovannini, 2008). Therefore, campylobacteriosis is not a statutory notification illness and the only data available on these infections are those reported voluntarily by Enter-Net, the international network for the surveillance of human gastrointestinal infections.
Generally, campylobacteriosis infections are self-limiting and only last for a few days. However, post-infection complications or extra-intestinal infections such as reactive arthritis and neurological manifestations can also arise (Haddad et al., 2010). Outbreaks caused by thermophilic Campylobacter species are most commonly connected with dairy and poultry products, as well as Campylobacter-contaminated food and untreated water (Meldrum et al., 2005; EFSA, 2008, 2014; Taylor et al., 2013). Unpasteurized or inadequately pasteurized cow’s milk has been implicated as the sources of infection in some outbreaks (Schildt et al., 2006; Heuvelink et al., 2009), and recently, the European Union summary report on food-borne disease outbreaks confirmed the importance of milk as a source of human campylobacteriosis (EFSA, 2013).
In a recent survey conducted in Northern Italy, C. jejuni was detected in 12% of the bulk tank milk samples that were examined (Bianchini et al., 2014), and some disease outbreaks have been reported in the same Italian regions (two in Emilia Romagna, one in Veneto, and one in Marche) following the consumption of raw milk from self-service automatic vending machines (Amato et al., 2007; Arrigoni et al., 2009; Petruzzelli et al., 2011). Despite no official data existing for the incidence of campylobacteriosis associated with raw milk consumption, quantitative risk assessment modeling has estimated that the worst possible scenario for human infections with C. jejuni linked to the consumption of raw milk in Italy would exceed 300,000 cases per year (Giacometti et al., 2015).
The high genome diversity and plasticity within the Campylobacter genus hampers the surveillance and the outbreak detection. Nevertheless, multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) are important and well-established tools that can be used to elucidate the epidemiology of Campylobacter cases. MLST is the most common genotyping method successfully applied in population genetic models for clarify the reservoirs of infection, while PFGE is a well standardized technique very useful in localized outbreak investigation (Dingle et al., 2001; Fitzgerald et al., 2001; Sheppard et al., 2009).
The objective of this study was to undertake an epidemio logical investigation of Campylobacter infections in 31 patients hospitalized in the summer of 2012 from the Piemonte region in Northern west Italy. Source investigation using MLST and PFGE comprised C. jejuni isolates from raw bovine milk and chicken meat. A polymorphism in the hip0 gene was also identified in some Campylobacter strains from raw bovine milk, and new PCR primer set for PCR identification were designed and successfully implemented in this study.
Materials and Methods
C. jejuni Isolates
A total of 31 cases of campylobacteriosis were registered in August, 2012 in the local health district of Turin, Northern Italy. A temporal and spatial proximity was suspected because of a cluster of infections; consequently, active surveillance was carried on suspicious retail food items. The monitoring found 17 raw milk samples from vending machines and 8 chicken meat samples from a retail market were positive for Campylobacter. Here, the 56 C. jejuni isolates we investigated were cultured on Columbia blood agar and incubated at 42°C for 48 h in a microaerobic atmosphere.
DNA Isolation
Genomic DNA was extracted using an UltraClean Microbial DNA Isolation Kit (MO BIO Laboratories, CA, USA) according to the manufacturer’s instructions. The DNA concentration was quantified using a Nanodrop spectrophotometer (Nanodrop Technologies, Italy).
Strain Identification
The isolates were typed using a PCR method described previously (Marshall et al., 1999; Di Giannatale et al., 2014).
Six strains isolated from raw milk were non-typable using the PCR method described above. Nevertheless, the strains were determined to be C. jejuni by 16S-rRNA gene sequencing (Zhou et al., 1997). Furthermore, a new PCR protocol targeting the hip0 gene was developed to check for miss-typed isolates. The sequences of the primers, which were designed against the regions upstream and downstream of the original amplification sites, were as follows: P3Fs: 5′-GGAAAAACAGGCGTTGTGGGGG-3′ and P3Rs: 5′-CCGAAGAAGCCATCATCGCACC-3′; P3Fi: 5′-CCTGCTTGAAGAGGGTTTGGGTGG-3′ and P3Ri: 5′-TGCAACCTCACTAGCAAAATCCACA-3′. The PCR was perfor med using Master Mix 2X (Promega, Italy), the final concentration of the primers was 1 μM, the annealing temperature was 55°C, and the procedure involved 35 cycles in a thermal cycler 9700 Applied Biosystems (Applied Biosystems, Foster City, CA, USA). Sequencing of the amplified fragment was performed using an ABI PRISM BigDye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions, and the sequences were analyzed with an ABI PRISM 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
PFGE
Pulsed-field gel electrophoresis was conducted according to the instructions from the 2009 USA PulseNet protocol for Campylobacter (CDC, 2009). The isolates previously identified by PCR were grown on Columbia agar (48 h at 42°C) in a microaerophilic atmosphere and embedded in agarose blocks (Seakem Gold agarose, Lonza, Rockland, MD, USA). Following DNA purification, 1 mm of agar plugs slices was digested 18 h with SmaI restriction enzyme (Promega, Milan, Italy) and the DNA fragments were separated by PFGE (Chef Mapper II, Biorad Laboratories, Hercules, CA, USA) in 1% agarose gel (Seakem Gold agarose, Lonza).
Salmonella Braenderup H9812, digested with XbaI enzyme (Promega, Milan, Italy), was used as the standard molecular weight marker. The gel was stained with SYBR Safe DNA gel stain (Invitrogen, Cergy Pontoise, France) and photographed on a UV transilluminator (Alpha Innotech Corporation, San Leandro, CA, USA). The image analysis was performed using Bionumerics program v. 6.6 (Applied Maths NV, Sint-Martens-Latem, Belgium). The similarity analysis was carried out using the Dice coefficient (position tolerance, 1%). The unweighted pair group mathematical average was used to cluster patterns. Isolates with <90% similarity were clustered as separate pulsotypes.
MLST Analysis
Multilocus sequence typing was performed as reported by Dingle et al. (2001) for all C. jejuni isolates. MLST amplified a segment of seven housekeeping genes: aspA (aspartase, 477 bp), glnA (glutamine synthase, 477 bp), gltA (citrate synthase, 402 bp), glyA (serine hydroxyl methyl transferase, 507 bp), pgm (phosphor glucomutase, 498 bp), tkt (transketolase, 459 bp), and uncA (ATP synthase, alpha subunit, 489 bp) to yield a total composite sequence length (all seven loci) of 3309 bp. PCRs and sequencing reactions were carried out according to the guidelines on the Campylobacter MLST website. Briefly, purified PCR products were sequenced using an ABI PRISM BigDye® Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions and then analyzed by the ABI PRISM 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The alleles, STs, and CCs were identified using the MLST database1 Novel alleles were submitted to the PubMLST C. jejuni/C. coli database curators for number assignment.
Source Attribution Analysis
According to a previous study conducted in the Netherlands (Mughini Gras et al., 2012), Campylobacter isolates from humans were found to share significant similar ST frequency distribution with those from Europe (the Netherlands, the UK, and Switzerland), but were genetically dissimilar from abroad isolates (from the USA and New Zealand). We have assumed this situation is also true for Italy and the details in our reference database included 558 C. jejuni strains from previously published Italian data; Piccirillo et al., 2014) and from our previous survey (Marotta et al., 2015), as well as the 6854 C. jejuni from European countries accessible in PubMLST2
The reservoir data we identified were pooled and arranged in six groups: (i) chicken, (ii) cattle, (iii) environment, (iv) wild bird, (v) small ruminant, and (vi) pork. Environmental strains comprised those from water, sand, and soil. The population genetics approach used for attributing human campylobacteriosis isolates to the six putative reservoirs was the asymmetric island model described by Wilson et al. (2008). This model estimates the recombination rates within the reservoirs, between the reservoirs and from each reservoir in the human population to estimate the posterior distribution used to infer the fraction of human cases attributable to each source.
Diversity Index and Evaluation of the Combined Typing Methods
The discriminatory ability of the two typing systems (PFGE and MLST) was measured according to Simpson’s diversity index. The concordance of the methods was determined by calculating the adjusted Rand and Wallace coefficients. The first coefficient is used to evaluate the extent of agreement between two typing methods, and the second method is an estimate of how much new information is obtained from a typing method in comparison with another one (Behringer et al., 2011). The adjusted Rand and Wallace coefficients were used to calculate the index from the Online Tool for Quantitative Assessment of Classification Agreement3
Results
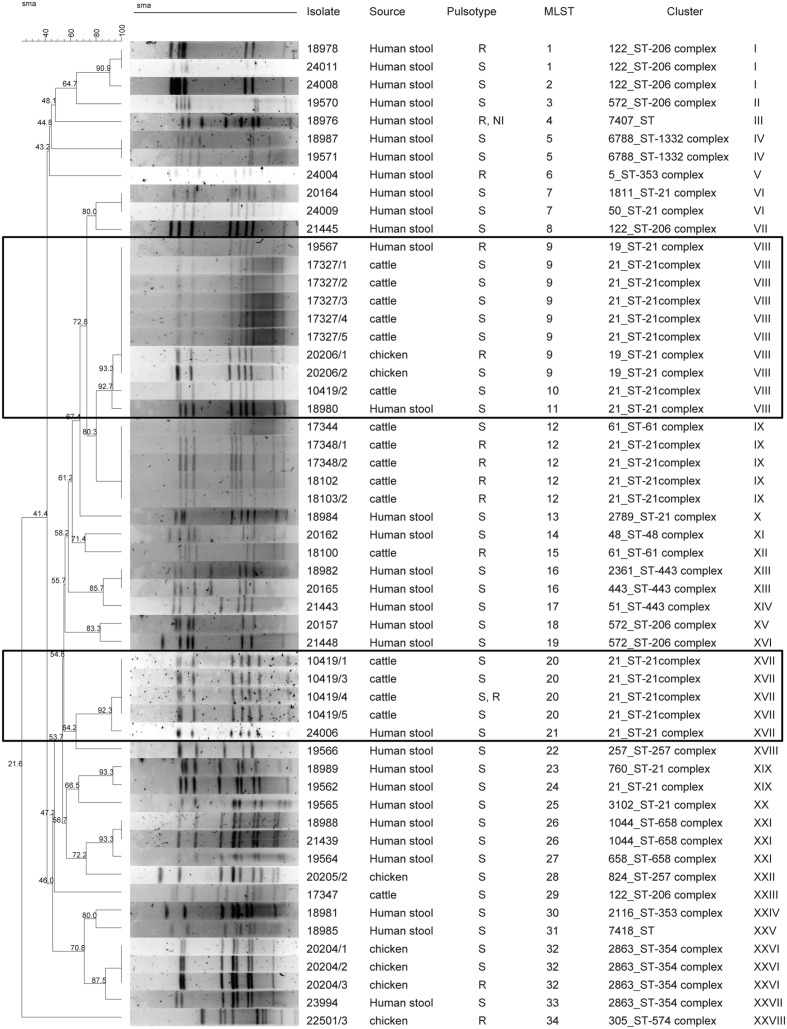
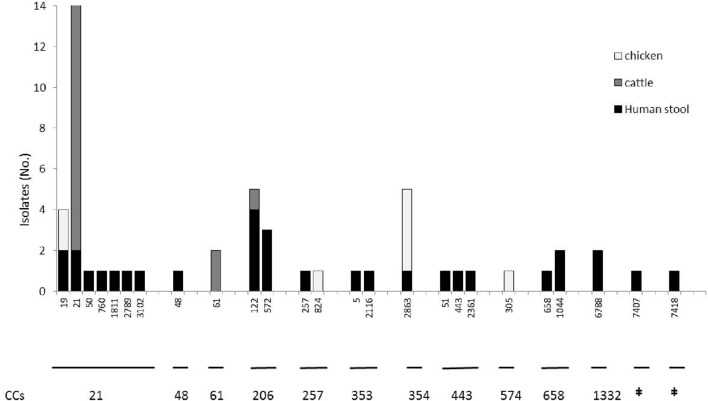
A total of 55 C. jejuni isolates were typed with PFGE using SmaI restriction enzyme. One chicken isolate could not be typed. Thirty-four different pulsotypes were obtained from the isolates typed (Figure 1). The cluster analysis distinguished the isolates according to 28 main clusters, with a similarity of 90% (Figure 1). Among these, only two clusters included non-human sources and three clinical isolates. Cluster VIII contained two human isolates, six raw milk isolates and two chicken meat isolates, and cluster XVII, contained one human and four raw milk isolates (Figure 1). MLST typing for the 56 isolates identified 25 STs featured in 11 clonal CCs (Figure 2). The most prevalent CCs were 21, 206, and 354. CC21 was the largest one shared by humans, cattle, and chickens, while CC206 and CC354 included human and cattle isolates or human and chicken isolates, respectively. At the fine scale, in three cases human isolates were linked to chicken sources sharing the STs 19 and 2863, and in six cases the human isolates were linked to raw milk sources sharing STs 122 and 21. MLST revealed the presence of 18 STs that were not attributable to any of the sources we investigated, including three new STs (STs 7407, 7418, and 6788), probably suggesting multiple foci of infections (Figure 2, Table 1).
FIGURE 1.
Comparison of pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) among Campylobacter jejuni isolates from different sources.
FIGURE 2.
Frequency distribution of the C. jejuni clonal complexes (CCs) isolated from chickens, human stool and cattle samples (n = 56) in the Piemonte region of Italy. STs belonging to the CCs listed below are reported on the x axis, while the y axis represents the number of isolates. ‡No clonal complex applicable.
Table 1.
Genetic diversity and frequency distribution of 56 Campylobacter jejuni strain isolates from human stool, chicken, and cattle sources.
| Clonal complex | ST | Multilocus sequence typing (MLST) allelic profile | Pulsed-field gel electrophoresis (PFGE) profilea | Source and number of isolates | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | SmaI cluster | |||
| 21 | 21 | 2 | 1 | 1 | 3 | 2 | 1 | 5 | VIII–XVII | Human stool (3) |
| Cattle (14) | ||||||||||
| 19 | 2 | 1 | 5 | 3 | 2 | 1 | 5 | VIII | Human stool (1) | |
| Chicken (2) | ||||||||||
| 1811 | 2 | 4 | 12 | 3 | 2 | 1 | 5 | VI | Human stool (1) | |
| 2789 | 2 | 1 | 12 | 3 | 2 | 3 | 5 | X | Human stool (1) | |
| 3102 | 2 | 84 | 12 | 3 | 11 | 1 | 5 | XX | Human stool (1) | |
| 50 | 2 | 1 | 12 | 3 | 2 | 1 | 5 | VI | Human stool (1) | |
| 760 | 2 | 1 | 52 | 3 | 2 | 100 | 5 | XIX | Human stool (1) | |
| 658 | 658 | 2 | 4 | 2 | 4 | 19 | 3 | 6 | XXI | Human stool (1) |
| 1044 | 2 | 10 | 2 | 4 | 19 | 3 | 6 | XXI | Human stool (2) | |
| 206 | 122 | 6 | 4 | 5 | 2 | 2 | 1 | 5 | XXIII | Cattle (1) |
| I–II | Human stool (4) | |||||||||
| 572 | 62 | 4 | 5 | 2 | 2 | 1 | 5 | II–XV–XVI | Human stool (3) | |
| 353 | 2116 | 7 | 17 | 52 | 10 | 89 | 3 | 6 | XXIV | Human stool (1) |
| 5 | 7 | 2 | 5 | 2 | 10 | 3 | 6 | V | Human stool (1) | |
| 443 | 443 | 24 | 17 | 2 | 15 | 23 | 3 | 12 | XIII | Human stool (1) |
| 2361 | 7 | 254 | 2 | 15 | 23 | 3 | 12 | XIII | Human stool (1) | |
| 51 | 7 | 17 | 2 | 15 | 23 | 3 | 12 | XIV | Human stool (1) | |
| 48 | 48 | 2 | 4 | 1 | 2 | 7 | 1 | 5 | XI | Human stool (1) |
| 61 | 61 | 1 | 1 | 4 | 2 | 2 | 6 | 3 | XII–IX | Cattle (2) |
| 257 | 257 | 9 | 2 | 4 | 62 | 4 | 5 | 6 | XVIII | Human stool (1) |
| 824 | 9 | 2 | 2 | 2 | 11 | 5 | 6 | XXII | Chicken (1) | |
| ‡ | 7407§ | 24 | 28 | 2 | 28 | 10 | 1 | 125 | III | Human stool (1) |
| ‡ | 7418§ | 24 | 17 | 52 | 10 | 89 | 418 | 6 | XXV | Human stool (1) |
| 1332§ | 6788§ | 2 | 1 | 4 | 28 | 58 | 29 | 58 | IV | Human stool (2) |
| 354 | 2863 | 198 | 2 | 2 | 2 | 11 | 61 | 6 | XXVI | Chicken (4) |
| XXVII | Human stool (1) | |||||||||
| 574 | 305 | 9 | 53 | 2 | 10 | 11 | 3 | 3 | XXVIII | Chicken (1) |
aPFGE profiles for the restriction enzyme SmaI. Bold font: the two identified clusters shared among humans and cattle or humans and chicken.
§ New allelic combination.
‡No clonal complex applicable.
Overall, the 31 human samples were assigned to 22 STs belonging to 11 CCs; among them, two clonal complexes have not been assigned to the international database yet. By means of the two techniques employed here and as shown in Figure 1, only 3 out of 31 cases were truly traceable back to the two cases linked to raw milk from vending machines, to chicken meat and one human case linked to raw milk.
The Simpson’s index and the confidence intervals for PFGE (0.949, 95% CI = 0.920–0.979) were higher than for MLST (0.897, 95% CI = 0.834–0.962), p = 0.044. PFGE is a more discriminatory typing method than MLST; however, when comparing the 95% confidence intervals, it is noteworthy that they overlap. Therefore, we cannot exclude the hypothesis that both methods have similar discriminatory power (at the 95% confidence level).
The concordance of the methods based on the calculation of the adjusted Rand coefficient and 95% CI shows a concordance between MLST and PFGE of 0.32 (95% CI = 0.209–0.437). The concordance based on the Wallace coefficient was also higher for the PFGE–MLST combination (Wallace = 0.505, 95% CI = 0.290–0.714), with respect to the MLST–PFGE combination (Wallace = 0.236, 95% CI = 0.120–0.343), p = 0.019.
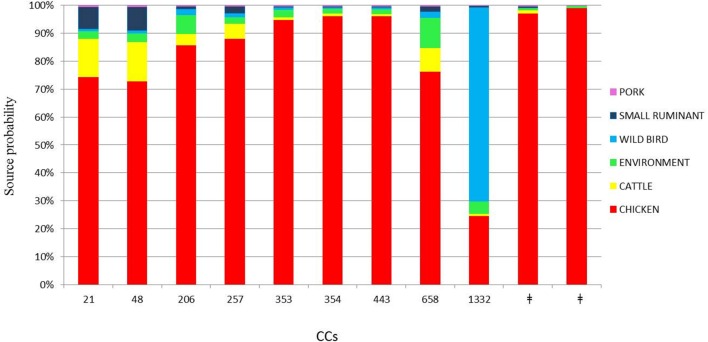
The attribution analysis revealed that 69.75% of the human cases were predominantly related to chicken (Figure 3). Interestingly, only two human cases belonging to CC1332 were predominantly related to wild bird (62.42%, Figure 3). In the asymmetric island model, most human cases (69.7%) were attributed to the chicken reservoir (95% CI 47.6–87.5%). Cattle were the second most probable source reservoir (8.2%, 95% CI 0–25.6%) followed by wild birds (7.3% CI 0–19.9%), the environment (6.2% CI 0–22%), small ruminants (5.3% CI 0–17.8%) and pork (2.9% CI 0–10.7%). Although there was a large credibility interval for the chicken source, the model gave the most probable source at a probability of 99.8%.
FIGURE 3.
Attributed probability (%) for the six most represented clonal complexes originating from chicken, cattle, environment, wild bird, small ruminant, and pork sources. ‡No clonal complex applicable.
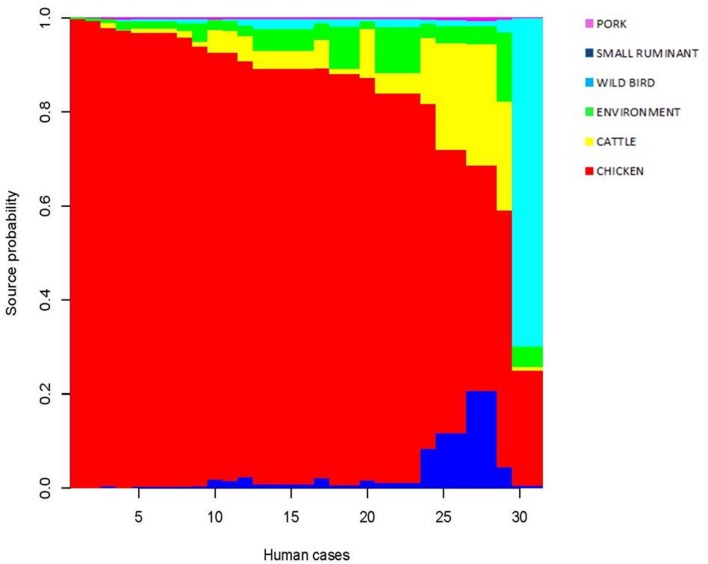
For each patient, the model restituted the posterior probability for the source of infection as shown in Figure 4. One dominant color confirmed that chicken is the likely source for the cases under consideration. It is worth noting that, for the generalist CC21 and CC48 regularly isolated from multiple reservoirs, the model showed a minor probability of chicken in favor of ruminants (cattle and small ruminants). CC1332 featured two clinical isolates and resulted with wild birds as the most probable source of infection. Overall, the 31 human cases investigated were probably infected in 29 instances by chicken and in the last two cases by wild birds. A minor probable source was confirmed for pork.
FIGURE 4.
Posterior probability of the source for each isolate.
Discussion
The percentage of human cases with campylobacteriosis ascribed to eating or handling raw poultry differs between countries and studies. Estimates of cases of a food-borne origin range from 30 to 58% (Mullner et al., 2009), and up to 80% can be attributed to the chicken sources as a whole (EFSA, 2011; Mughini Gras et al., 2012). Furthermore, the sources of the notified cases have not been able to be determined, indicating that the real number of food-borne cases is unknown (Meinersmann et al., 2005; EFSA, 2013). It is well known that the presence of C. jejuni in bovine raw milk samples and fresh dairy products is increasing (Fernandes et al., 2015). Recent data show that the consumption of undercooked chicken meat and raw milk are the most important sources of human campylobacteriosis in Europe (EFSA, 2014), accounting for hundreds of cases. Outbreaks associated with the consumption of raw or pasteurized milk contaminated with Campylobacter were recently recorded in Italy (Giacometti et al., 2012). A quantitative risk assessment focused on one region of Northern Italy evaluated that 1–2% of the population were consumers of raw milk from vending machines and only 57% of users boiled the raw milk before consumption; therefore, the estimated proportion of the people consuming unboiled raw milk is 0.5–0.9% (4548–9096 people; Giacometti et al., 2012).
The present study aimed to investigate an outbreak of campylobacteriosis among humans and to define the infection sources. An active surveillance was undertaken that sampled raw milk from vending machines and chicken meat from a retail market. The PFGE and MLST results suggested that the human cases we studied were not caused by a single-point source, but were likely triggered by multiple foci of infection instead. The evidence supporting this conclusion was the presence of 26 different pulsotypes discovered by PFGE and 22 STs for MLST. The MLST results found nine clinical cases with similar profiles for the raw milk and chicken samples. In particular, ST21 and ST122 were shared among humans and cattle, while ST19 and ST 354 were shared between humans and chickens.
Calculation of the two coefficients (adjusted Rand and Wallace coefficients) and their confidence intervals permits quantification of the congruence between the two different method results (Wallace, 1983; Pinto et al., 2008). In our study, the adjusted Rand coefficient gave low congruence between the different methods. The PFGE analysis showed that isolates with the same MLST profile were partially congruent and sometimes grouped inside different clusters, as was also revealed by the adjusted Rand coefficient between MLST and PFGE of 0.32 (95% CI = 0.209–0.437). However, the Wallace coefficient for PFGE and MLST gave a value of 0.5, which is higher than the 0.2 value from the MLST–PFGE combination, suggesting that PFGE performs better in an outbreak scenario.
Nevertheless, only three cases featured in VIII and XVII groups clustered together with chicken and raw milk samples, a finding in agreement with the MLST results. In contrast, ST 122 and ST 354 did not cluster together in the PFGE profiles, suggesting a possible diverse origin for them. In this study, the clustering concordance of the two molecular techniques is not similar, but we propose that complementation of the results could lead to a higher level of isolate discrimination.
The data suggest that genomic-based techniques like MLST and PFGE can provide useful insight for outbreak investigations. PFGE has been standardized for Campylobacter spp., and has been used in localized outbreak investigations (Fitzgerald et al., 2001), while MLST is useful for investigating the DNA sequence diversity via the index of variations in housekeeping genes (Dingle et al., 2001).
Recently, MLST has commonly been employed to increase the analysis resolution of outbreak-associated isolates, leading to quicker and much more accurate source identification, as well as investigating different epidemiological hypotheses.
Our findings revealed three human cases that had similar genetic profiles to chicken and cattle isolates. However, in this study, there were no clues for tracing the remaining isolates investigated and we did not retrieve similar profiles from the source strains isolated during the active surveillance. Therefore, to try to understand the source of infection, we applied a genetic population approach using MLST data from our database and the data from other studies conducted in Europe.
From our analysis, chicken was estimated to be the most dominant source of infection for the human isolates we studied. Using asymmetric island model, the MLST profiles associated with human disease were most similar to those from a chicken source in 69.75% of the cases of C. jejuni infection. Ruminants (cattle and small ruminants) contributed far fewer cases of C. jejuni infection (13.6%) and the contribution from environment, wild bird, and pig sources was very low (6.28, 7.37, and 2.98%, respectively). This finding is in line with many other studies performed in industrialized countries, even if divergence in the percentage of cases attributable to the chicken source varied among these studies (Wilson et al., 2008; Mullner et al., 2009; Sheppard et al., 2009). Our results should indicate the importance of chicken as the main reservoir of human campylobacteriosis and lend to the suggestion that this disease could be greatly reduced by focusing interventions on chickens. The cases studied are not representative of the Italian epidemiological context but seem to be congruent with other European scenarios.
Notably, inspecting the three cases that were traced back to a potential source revealed that the model estimated chicken as the most probable source of infection for all the cases as well. Contrastingly, our investigation found that one case featured in PFGE cluster VIII and ST 21 was attributable to raw milk instead of chicken. In fact, CC21 is a multi-host lineage shared among different sources, and this can lead to an incorrect assignment in the genetic model. The model calculates the likely source of the infection but cannot rule out completely the minor reservoir as the actual source. In this case, the back-tracing analysis using PFGE and MLST is likely to be the best approach for drawing the correct reconstruction of the food-borne illness. In this scenario, whole-genomeı sequencing (WGS) would probably provide the best fit for the epidemiological analysis, but this approach is not yet ready to be applied routinely because of the lack of standardization of the analysis of the WGS data in the context of the molecular epidemiology of food-borne pathogens.
Conclusion
The number of clinical isolates, albeit limited to only 56 samples from a restricted area and in a short time window, confirms the high genetic diversity present in Campylobacter. Genetic assessment of Campylobacter spp. is fundamental to our understanding of its epidemiology. Additional analyses of isolates from various sources will consent major progresses in the knowledge of the epidemiology and population structure of C. jejuni in Italy. This study confirms the interest of molecular epidemiology as a powerful support for epidemiological investigations.
Author Contributions
EG has contributed to the conception and design of the work and wrote the paper. AA performed experiments. FM performed experiments, analyzed data and wrote the paper. GG performed the source attribution analysis, analyzed data and wrote the paper. LC performed the source attribution analysis. GD helped draft the manuscript. WV and LD contributed strains.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
- Amato S., Maragno M., Mosele P., Sforzi M., Mioni R., Barco L., et al. (2007). “An outbreak of Campylobacter jejuni linked to the consumption of raw milk in Italy,” in Proceedings of the Zoonoses and Public Health Conference (Oxford: Blackwell Publishing; ), 23–23. [Google Scholar]
- Arrigoni N., Scavia G., Tamba M. (2009). “Raw milk: experiences and hygiene-sanitary problems in Emilia Romagna Region,” in Proceedings of the Conference on Large Animal Review: SIVAR-Societa Italiana Veterinari per Animali da Reddito, Cremona, 215–219. [Google Scholar]
- Behringer M., Miller W. G., Oyarzabal O. A. (2011). Typing of Campylobacter jejuni and Campylobacter coli isolated from live broilers and retail broiler meat by flaA-RFLP, MLST, PFGE and REP-PCR. J. Microbiol. Methods 84 194–201. 10.1016/j.mimet.2010.11.016 [DOI] [PubMed] [Google Scholar]
- Bianchini V., Luini M., Borella L., Parisi A., Jonas R., Kittl S., et al. (2014). Genotypes and antibiotic resistances of Campylobacter jejuni isolates from cattle and pigeons in dairy farms. Int. J. Environ. Res. Public Health 11 7154–7162. 10.3390/ijerph110707154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calistri P., Giovannini A. (2008). Quantitative risk assessment of human campylobacteriosis related to the consumption of chicken meat in two Italian regions. Int. J. Food Microbiol. 128 274–287. 10.1016/j.ijfoodmicro.2008.08.021 [DOI] [PubMed] [Google Scholar]
- CDC (2009). Standard Operating Procedure for PulseNet PFGE of Campylobacter jejuni. Available at: http://www.cdc.gov/pulsenet/pdf/campylobacter-pfge-protocol-508c.pdf [Google Scholar]
- Di Giannatale E., Di Serafino G., Zilli K., Alessiani A., Sacchini L., Garofolo G., et al. (2014). Characterization of antimicrobial resistance patterns and detection of virulence genes in Campylobacter isolates in Italy. Sensors 14 3308–3322. 10.3390/s140203308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle K., Colles F., Wareing D., Ure R., Fox A., Bolton F., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39 14–23. 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (2008). Analysis of the baseline survey on the prevalence of Campylobacter in broiler batches and of Campylobacter and Salmonella on broiler carcasses in the EU. EFSA J. 8 1503. [Google Scholar]
- EFSA and ECDC (2015). The european union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 13 3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (2011). The european union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2009. EFSA J. 9 2090. [Google Scholar]
- EFSA (2013). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J. 11 3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (2014). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 12 3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A. M., Balasegaram S., Willis C., Wimalarathna H. M., Maiden M. C., McCarthy N. D. (2015). Partial failure of milk pasteurization as a risk for the transmission of campylobacter from cattle to humans. Clin. Infect. Dis. 61 903–909. 10.1093/cid/civ431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald C., Helsel L. O., Nicholson M. A., Olsen S. J., Swerdlow D. L., Flahart R., et al. (2001). Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39 2386–2390. 10.1128/JCM.39.7.2386-2390.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti F., Bonilauri P., Amatiste S., Arrigoni N., Bianchi M., Losio M. N., et al. (2015). Human campylobacteriosis related to the consumption of raw milk sold by vending machines in Italy: quantitative risk assessment based on official controls over four years. Prev. Vet. Med. 121 151–158. 10.1016/j.prevetmed.2015.06.009 [DOI] [PubMed] [Google Scholar]
- Giacometti F., Serraino A., Bonilauri P., Ostanello F., Daminelli P., Finazzi G., et al. (2012). Quantitative risk assessment of verocytotoxin-producing Escherichia coli O157 and Campylobacter jejuni related to consumption of raw milk in a province in Northern Italy. J. Food Prot. 75 2031–2038. 10.4315/0362-028X.JFP-12-163 [DOI] [PubMed] [Google Scholar]
- Haddad N., Marce C., Magras C., Cappelier J.-M. (2010). An overview of methods used to clarify pathogenesis mechanisms of Campylobacter jejuni. J. Food Prot. 73 786–802. [DOI] [PubMed] [Google Scholar]
- Heuvelink A. E., van Heerwaarden C., Zwartkruis-Nahuis A., Tilburg J. J., Bos M. H., Heilmann F. G., et al. (2009). Two outbreaks of campylobacteriosis associated with the consumption of raw cows’ milk. Int. J. Food Microbiol. 134 70–74. 10.1016/j.ijfoodmicro.2008.12.026 [DOI] [PubMed] [Google Scholar]
- Marotta F., Garofolo G., Di Donato G., Aprea G., Platone I., Cianciavicchia S., et al. (2015). Population diversity of campylobacter jejuni in poultry and its dynamic of contamination in chicken meat. Biomed Res. Int. 2015 859845 10.1155/2015/859845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall S. M., Melito P. L., Woodward D. L., Johnson W. M., Rodgers F. G., Mulvey M. R. (1999). Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 37 4158–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann R. J., Phillips R. W., Hiett K. L., Fedorka-Cray P. (2005). Differentiation of Campylobacter populations as demonstrated by flagellin short variable region sequences. Appl. Environ. Microbiol. 71 6368–6374. 10.1128/AEM.71.10.6368-6374.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldrum R., Griffiths J., Smith R., Evans M. R. (2005). The seasonality of human campylobacter infection and Campylobacter isolates from fresh, retail chicken in Wales. Epidemiol. Infect. 133 49–52. 10.1017/S0950268804003188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughini Gras L., Smid J. H., Wagenaar J. A., de Boer A. G., Havelaar A. H., Friesema I., et al. (2012). Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: a combined case-control and source attribution analysis. PLoS One 7:e42599 10.1371/journal.pone.0042599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner P., Spencer S. E., Wilson D. J., Jones G., Noble A. D., Midwinter A. C., et al. (2009). Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect. Genet. Evol. 9 1311–1319. 10.1016/j.meegid.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Petruzzelli A. A. G., Foglini M., Orazietti N., Mancini P., Omiccioli E., Brandi G., et al. (2011). “Identificazione di patogeni in un allevamento di bovine da latte mediante metodo in multiplex real-time pcr,” in Proceedings of the Convegno: XIII Congresso Nazionale S.I.Di.L.V., Trani, 322–323. [Google Scholar]
- Piccirillo A., Giacomelli M., Salata C., Bettanello S., De Canale E., Palu G. (2014). Multilocus sequence typing of Campylobacter jejuni and Campylobacter coli from humans and chickens in North-Eastern Italy. New Microbiol. 37 557–562. [PubMed] [Google Scholar]
- Pinto F. R., Melo-Cristino J., Ramirez M. r. (2008). A confidence interval for the Wallace coefficient of concordance and its application to microbial typing methods. PLoS ONE 3:e3696 10.1371/journal.pone.0003696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues R. C., Bronnec V., Tresse O., Cappelier J.-M., Haddad N. (2015). “A review of Campylobacter jejuni pathogenesis: main virulence factors and their use as biomarkers,” in Campylobacter Infections: Epidemiology, Clinical Management and Prevention, ed. Bertucci B. A. (New York, NY: Nova Science Publishers Inc.). [Google Scholar]
- Schildt M., Savolainen S., Hänninen M. L. (2006). Long-lasting Campylobacter jejuni contamination of milk associated with gastrointestinal illness in a farming family. Epidemiol. Infect. 134 401–405. 10.1017/S0950268805005029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S. K., Dallas J. F., Strachan N. J., MacRae M., McCarthy N. D., Wilson D. J., et al. (2009). Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48 1072–1078. 10.1086/597402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. V., Herman K. M., Ailes E., Fitzgerald C., Yoder J., Mahon B., et al. (2013). Common source outbreaks of Campylobacter infection in the USA, 1997-2008. Epidemiol. Infect. 141 987–996. 10.1017/S0950268812001744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. L. (1983). Comment. J. Am. Stat. Assoc. 78 569–576. 10.1080/01621459.1983.10478009 [DOI] [Google Scholar]
- Wilson D. J., Gabriel E., Leatherbarrow A. J., Cheesbrough J., Gee S. S, Bolton E., et al. (2008). Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203 10.1371/journal.pgen.1000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Bowler L. D., Spratt B. G. (1997). Interspecies recombination, and phylogenetic distortions, within the glutamine synthetase and shikimate dehydrogenase genes of Neisseria meningitidis and commensal Neisseria species. Mol. Microbiol. 23 799–812. 10.1046/j.1365-2958.1997.2681633.x [DOI] [PubMed] [Google Scholar]