Abstract
The emergence of carbapenem-resistant Gram-negative pathogens poses a serious threat to public health worldwide. In particular, the increasing prevalence of carbapenem-resistant Klebsiella pneumoniae is a major source of concern. K. pneumoniae carbapenemases (KPCs) and carbapenemases of the oxacillinase-48 (OXA-48) type have been reported worldwide. New Delhi metallo-β-lactamase (NDM) carbapenemases were originally identified in Sweden in 2008 and have spread worldwide rapidly. In this review, we summarize the epidemiology of K. pneumoniae producing three carbapenemases (KPCs, NDMs, and OXA-48-like). Although the prevalence of each resistant strain varies geographically, K. pneumoniae producing KPCs, NDMs, and OXA-48-like carbapenemases have become rapidly disseminated. In addition, we used recently published molecular and genetic studies to analyze the mechanisms by which these three carbapenemases, and major K. pneumoniae clones, such as ST258 and ST11, have become globally prevalent. Because carbapenemase-producing K. pneumoniae are often resistant to most β-lactam antibiotics and many other non-β-lactam molecules, the therapeutic options available to treat infection with these strains are limited to colistin, polymyxin B, fosfomycin, tigecycline, and selected aminoglycosides. Although, combination therapy has been recommended for the treatment of severe carbapenemase-producing K. pneumoniae infections, the clinical evidence for this strategy is currently limited, and more accurate randomized controlled trials will be required to establish the most effective treatment regimen. Moreover, because rapid and accurate identification of the carbapenemase type found in K. pneumoniae may be difficult to achieve through phenotypic antibiotic susceptibility tests, novel molecular detection techniques are currently being developed.
Keywords: carbapenemase, Klebsiella pneumoniae, epidemiology, KPC, NDM, OXA-48-like
Introduction
The increasing prevalence of antibiotic resistance and the lack of new antibiotic drug development has gradually reduced the treatment options for bacterial infections (Lee et al., 2013a; Nathan and Cars, 2014). In 2013, the Centers for Disease Control and Prevention (CDC) named three microorganisms that pose an urgent threat to public health: carbapenem-resistant (CR) Enterobacteriaceae (CRE), drug-resistant Neisseria gonorrhoeae, and Clostridium difficile (Zowawi et al., 2015). Carbapenems (imipenem, meropenem, biapenem, ertapenem, and doripenem) are antibiotics used for the treatment of severe infections caused by multi-resistant Enterobacteriaceae, such as Klebsiella pneumoniae and Escherichia coli (Nordmann et al., 2009). However, over the past 10 years, CRE have increasingly been reported worldwide (Nordmann et al., 2011a). In particular, K. pneumoniae have acquired carbapenemases, which are enzymes capable of breaking down most β-lactams including carbapenems, and thus conferring resistance to these drugs (Jeon et al., 2015). High mortality rates have been reported in patients with bloodstream infections caused by CR K. pneumoniae (Munoz-Price et al., 2013). Carbapenemases can be divided according to their dependency on divalent cations for enzyme activation into metallo-carbapenemases (zinc-dependent class B) and non-metallo-carbapenemases (zinc-independent classes A, C, and D; Jeon et al., 2015). The class A carbapenemases, such as the K. pneumoniae carbapenemase (KPC) enzymes, have been identified worldwide in K. pneumoniae (Tangden and Giske, 2015). Various class B and D carbapenemases have also been detected in hospital-acquired multi-resistant K. pneumoniae (Nordmann et al., 2011a), whereas class C carbapenemases have rarely been reported. In this review, we summarize the epidemiology of the major four classes of carbapenemases and discuss their molecular genetics, methods used for their detection, and the therapeutic options available for their treatment.
The Epidemiology, Genetic Context, Treatment Options, and Detection Methods of Carbapenem-Resistant K. pneumoniae
Class A Carbapenemases
Epidemiology
Various class A carbapenemases forming six distantly related branches have been identified (Jeon et al., 2015). While some carbapenemases are chromosome-encoded (IMI-1, NMC-A, SME enzymes, SHV-38, and SFC-1), others are plasmid-encoded (KPC enzymes, GES enzymes, and IMI-2). KPCs have been the most frequently observed class A carbapenemases since their first description in the eastern the USA in 1996 (Yigit et al., 2001). Of the many different KPC family variants (KPC-1 to KPC-22), the most well-characterized variants are KPC-2 and KPC-3. KPCs are mostly plasmid-encoded enzymes and bacteria producing these enzymes are susceptible to only a few antibiotics such as colistin, aminoglycosides, and tigecycline. Therefore, the mortality of the patient’s bloodstream infections caused by these bacteria is very high (Munoz-Price et al., 2013).
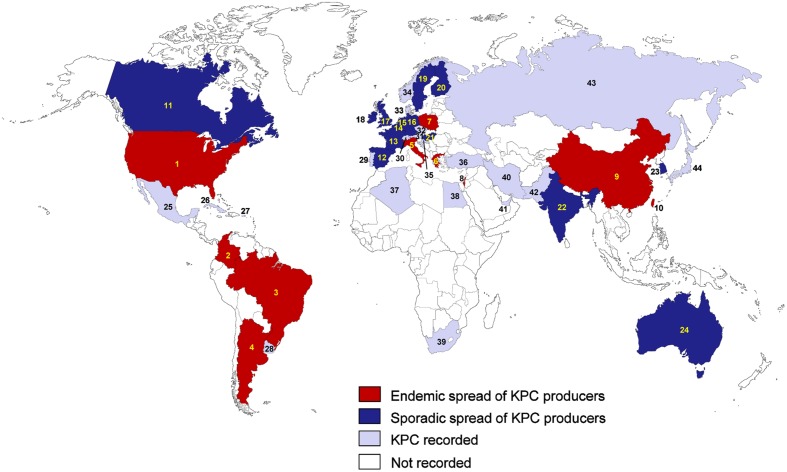
The epidemiology of K. pneumoniae producing KPCs varies geographically. The endemic spread of these bacteria has been reported in the USA, China, Italy, Poland, Greece, Israel, Brazil, Argentina, Colombia, and Taiwan (Munoz-Price et al., 2013; Figure 1). Sporadic spread of KPC-producing K. pneumoniae has also been observed in many European countries including Spain, France, Germany, the Netherlands, the UK, Ireland, Belgium, Sweden, and Finland, and in several countries in the Asia-Pacific region, including India, South Korea, and Australia (Munoz-Price et al., 2013; Nordmann and Poirel, 2014). In the USA, the transmission of CR K. pneumoniae is primarily driven by the spread of organisms carrying KPC enzymes (Kaiser et al., 2013), but other carbapenemase enzymes, such as the New-Delhi metallo-β-lactamase (NDM), have also emerged (Lascols et al., 2013). Within the USA, the prevalence of KPC-positive isolates was relatively stable between 2007 and 2009 (5.9% in 2007, 4.9% in 2008, and 5.7% in 2009; Kaiser et al., 2013), and KPC-2 and KPC-3 were the most frequently identified carbapenemases in K. pneumoniae (Deshpande et al., 2006).
FIGURE 1.
Epidemiological features of KPC-producing Klebsiella pneumoniae. (1) USA; (2) Colombia; (3) Brazil; (4) Argentina; (5) Italy; (6) Greece; (7) Poland; (8) Israel; (9) China; (10) Taiwan; (11) Canada; (12) Spain; (13) France; (14) Belgium; (15) Netherlands; (16) Germany; (17) UK; (18) Ireland; (19) Sweden; (20) Finland; (21) Hungary; (22) India; (23) South Korea; (24) Australia; (25) Mexico; (26) Cuba; (27) Puerto Rico; (28) Uruguay; (29) Portugal; (30) Switzerland; (31) Austria; (32) Czech Republic; (33) Denmark; (34) Norway; (35) Croatia; (36) Turkey; (37) Algeria; (38) Egypt; (39) South Africa; (40) Iran; (41) United Arab Emirates; (42) Pakistan; (43) Russia; (44) Japan.
The outbreaks caused by KPC-producing K. pneumoniae have been reported in the USA (Woodford et al., 2004) and Israel (Leavitt et al., 2007), but recently, similar outbreaks associated with patients traveling to endemic areas have also been reported in many European counties. Since KPC-producing K. pneumoniae was identified in France, Italy, and Greece (Naas et al., 2005; Tsakris et al., 2008; Giani et al., 2009), the sporadic spread of KPC-producing K. pneumoniae has been observed in many European countries including Spain (Robustillo Rodela et al., 2012), France (Carbonne et al., 2010), Germany (Wendt et al., 2010; Steinmann et al., 2011), the Netherlands (Weterings et al., 2015), the UK (Woodford et al., 2008; Virgincar et al., 2011), Ireland (Roche et al., 2009; Morris et al., 2012), Belgium (Bogaerts et al., 2010), Sweden (Samuelsen et al., 2009), and Finland (Osterblad et al., 2012; Kanerva et al., 2015). KPC-producing K. pneumoniae were also recently detected in eastern European countries including the Czech Republic (Hrabak et al., 2013b), Hungary (Toth et al., 2010), and Croatia (Bedenic et al., 2012).
In Greece, KPC-producing K. pneumoniae was first isolated in August 2007 (Tsakris et al., 2008), and the prevalence of KPC-producers among carbapenemase-producing K. pneumoniae isolates collected at a tertiary Greek hospital increased from 0% in 2003 to 38.3% in 2010 (Zagorianou et al., 2012). Most of the genotyped KPC-producing K. pneumoniae in Greece harbored KPC-2 (Zagorianou et al., 2012). While many carbapenemase-producing K. pneumoniae in the USA and Greece had KPC enzymes (Nordmann et al., 2009; Zagorianou et al., 2012), several studies in Spain showed that most carbapenemase-producing K. pneumoniae harbored OXA-48-like or class B carbapenemases, and the rate of KPC- producing K. pneumoniae was very low (2–3%; Oteo et al., 2013b; Palacios-Baena et al., 2016). These results indicate that the prevalent genotype of carbapenemase-producing K. pneumoniae varies geographically. For example, in Italy which is a representative southern European country where KPC is becoming endemic, 89.5% of carbapenemase producers have been reported to have KPC-type enzymes, followed by VIM-1 (9.2%) and OXA-48 (1.3%; Giani et al., 2013).
In America, the endemic spread of KPCs has been reported in Colombia (Villegas et al., 2006; Rojas et al., 2013), Brazil (Peirano et al., 2009; Fehlberg et al., 2012), and Argentina (Pasteran et al., 2008; Gomez et al., 2011). In Canada, KPC-producing K. pneumoniae has sporadically been reported (Goldfarb et al., 2009; Lefebvre et al., 2015), and since plasmid-mediated KPC-producing K. pneumoniae was first detected in Ottawa in Goldfarb et al. (2009), a laboratory surveillance program found a high frequency (89.3%) of KPC-type enzymes among carbapenemase producers between 2010 and 2012 (Lefebvre et al., 2015). The emergence of KPCs in Argentina was characterized by two patterns of dispersion: the first was the irregular occurrence of diverse Enterobacteriaceae harboring blaKPC-2 in the IncL/M transferable plasmid in distant regions and the second was the sudden clonal spread of K. pneumoniae ST258 harboring blaKPC-2 in Tn4401a (Gomez et al., 2011). KPC-producing K. pneumoniae was recently also detected in Cuba (Quinones et al., 2014), Mexico (Garza-Ramos et al., 2014), Uruguay (Marquez et al., 2014), and Puerto Rico (Gregory et al., 2010).
In the Asia-Pacific region, the endemic dissemination of KPC-producing K. pneumoniae has been reported in China (Li et al., 2014) and Taiwan (Tseng et al., 2015), and the sporadic spread has been reported in India (Shanmugam et al., 2013), South Korea (Yoo et al., 2013), and Australia (Partridge et al., 2015). A novel KPC-15 variant which is closely homologous with KPC-4 was discovered in China (Wang et al., 2014b) and its enzymatic activity and phenotype was characterized (Wang et al., 2014a). In China, the frequency of KPC-type enzymes among carbapenemase producers was high (63%; Li et al., 2014). While ST258 is the predominant clone observed in European countries and the USA (Giani et al., 2013; Chen et al., 2014e; Bowers et al., 2015), ST11, which is closely related to ST258, is the prevalent clone associated with the spread of KPC-producing K. pneumoniae in Asia (particularly in China and Taiwan; Qi et al., 2011; Yang et al., 2013; Tseng et al., 2015). KPC-producing ST11 strain has also been reported in Latin America (Munoz-Price et al., 2013). Although it is unknown why ST11 is prevalent, a recent report showed that the KPC-producing K. pneumoniae ST11 clone was resistant to serum killing (Chiang et al., 2016). In a Chinese hospital, another nosocomial outbreak of KPC-2-producing K. pneumoniae was caused by multiple K. pneumoniae strains including ST37, ST392, ST395, and ST11, implying the horizontal transfer of blaKPC-2 gene between different K. pneumoniae clones in China (Yang et al., 2013). In Taiwan, two novel KPC variants were identified; KPC-16 and KPC-17 differed from KPC-2 by two (P202S and F207L) and a single (F207L) amino acid substitutions, respectively (Yu et al., 2015). A nationwide survey in Taiwan between 2011 and 2013 reported the national spread of KPC-2 and KPC-17 (Tseng et al., 2015). KPC-producing K. pneumoniae was recently also detected in Japan (Saito et al., 2014), Pakistan (Pesesky et al., 2015), Iran (Nobari et al., 2014), and United Arab Emirates (Sonnevend et al., 2015a). In the Arabian Peninsula, the prevalence of KPC-producing K. pneumoniae was very low in comparison to NDM-1 and OXA-48-like carbapenemases (Sonnevend et al., 2015b). Sonnevend et al. (2015a), two K. pneumoniae ST14 strains producing KPC-2 were first identified in the United Arab Emirates of the Arabian Peninsula. In Africa, several countries such as South Africa (Brink et al., 2012), Algeria (Bakour et al., 2015b), and Egypt (Metwally et al., 2013), have also isolated KPC-producing K. pneumoniae.
The coexistence of KPCs and other carbapenemases in K. pneumoniae was frequently reported worldwide, including in Italy (KPC-3/VIM-2 and KPC-2/VIM-1; Richter et al., 2012; Perilli et al., 2013), Colombia (KPC-2/VIM-24; Rojas et al., 2013), Brazil (KPC-2/NDM-1; Pereira et al., 2015), China (KPC-2/NDM-1, KPC-2/CMY-2, and KPC-2/IMP-4; Hu et al., 2014; Dong et al., 2015; Liu et al., 2015), Canada (KPC-3/CMY-2; Leung et al., 2012), and Greece (KPC-2/VIM-1; Giakkoupi et al., 2009), indicating the worldwide prevalence of K. pneumoniae co-harboring two carbapenemases.
Aside from KPC-type carbapenemases, other class A carbapenemases, such as GES-2, GES-4, GES-5, GES-6, GES-11, GES-14, GES-18, SFC-1, SHV-38, NMC-A, SME-1, and IMI-type enzymes, were rarely found in K. pneumoniae (Table 1).
Table 1.
The epidemiology of various carbapenemases in Klebsiella pneumoniae.
Molecular and Genetic Context
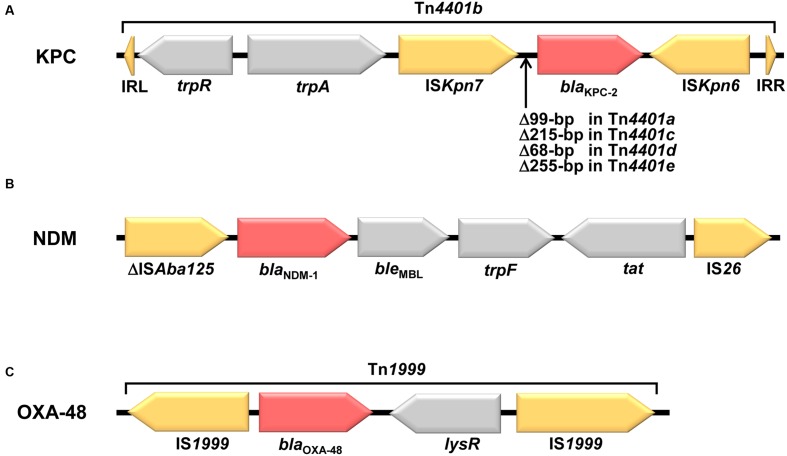
The blaKPC in K. pneumoniae has been reported on numerous plasmid types, such as IncF, IncI2, IncX, IncA/C, IncR, and ColE1 (Garcia-Fernandez et al., 2012; Chen et al., 2014e; Pitout et al., 2015), but the predominant plasmid type is IncF with FIIK replicons (Pitout et al., 2015). IncF often contains several additional genes responsible for resistance to other antibiotics, including aminoglycosides, tetracyclines, quinolones, trimethoprim, and sulfonamides (Pitout et al., 2015). Many blaKPC genes are associated with a promiscuous transposon-related structure Tn4401, which is approximately 10 kb in size and consists of a transposase gene, a resolvase gene, the blaKPC gene, and two insertion sequences, ISKpn6 and ISKpn7 (Figure 2A; Naas et al., 2008). This transposon has jumped to numerous plasmids that are commonly conjugative (Chen et al., 2014e). In China, a novel genetic environment was detected (Shen et al., 2009). It contains an integration structure consisting of a Tn3-based transposon and partial Tn4401 segment, with the gene order Tn3-transposase, Tn3-resolvase, ISKpn8, the blaKPC-2 gene, and the ISKpn6-like element (Shen et al., 2009). This genetic structure is the chimera form of several transposon-associated elements. This transposon was also identified in many other countries (Chen et al., 2014e), and several variants with various fragment insertions between the ISKpn8 and blaKPC gene have been found among Enterobacteriaceae in China (Shen et al., 2009; Li et al., 2011; Qi et al., 2011). Tn4401 has five isoforms which differ by deletions (68–255 bp) just upstream of the blaKPC gene [(a) deletion of 99 bp; (b) no deletion; (c) deletion of 215 bp; (d) deletion of 68 bp; (e) deletion of 255 bp; Chen et al., 2014e]. Notably, in many cases, different Tn4401 isoforms was associated with different blaKPC-harboring plasmids. Tn4401a was frequently found in the blaKPC-3-harboring IncFIIK2 plasmids (Leavitt et al., 2010; Garcia-Fernandez et al., 2012; Chen et al., 2014d), and Tn4401b and Tn4401d were often associated with the IncN and IncFIA plasmids, respectively (Chen et al., 2013a, 2014c,e). Up to now, more than 30 blaKPC-harboring plasmids obtained from K. pneumoniae have been sequenced (Gootz et al., 2009; Shen et al., 2009; Jiang et al., 2010; Leavitt et al., 2010; Almeida et al., 2012; Chen et al., 2013a,b, 2014b,c,e). One of common features shared by these sequenced plasmids is the presence of the tra operon, which encodes the plasmid conjugation machinery proteins that induce the spread of plasmids (Chen et al., 2014e). These genes may be important for the successful dissemination of blaKPC-harboring plasmids. The IncFII plasmids are one of predominant blaKPC-harboring plasmids. pKpQIL, which is an IncFIIK2 plasmid harboring Tn4401a, was initially identified in Israel in 2006 (Leavitt et al., 2010), and then this plasmid and its variants are believed to have spread to Italy, Poland, the UK, Colombia, the Czech Republic, the USA, and other countries (Baraniak et al., 2011; Garcia-Fernandez et al., 2012; Hidalgo-Grass et al., 2012; Warburg et al., 2012; Hrabak et al., 2013b; Chen et al., 2014d,e), suggesting the wide dissemination of this plasmid. The blaKPC gene has also been identified in other non-Tn4401 mobile elements that mostly have partial ISKpn6 genes (Shen et al., 2009; Gomez et al., 2011). Based on the insertion sequence upstream of the blaKPC gene, they can be divided into three groups: group I, no insertion (Shen et al., 2009; Liu et al., 2012; Chen et al., 2014g); group II, insertion of truncated blaTEM (Gomez et al., 2011); group III, insertion of Tn5563/IS6100 (Wolter et al., 2009). These non-Tn4401 genetic elements harboring blaKPC sometimes have an IS26 transposon (Liu et al., 2012; Chen et al., 2014g).
FIGURE 2.
Structural features of representative genetic environments of blaKPC-2, blaNDM-1, and blaOXA-48 genes. (A) The blaKPC-2-containing Tn4401 transposon from the plasmid pNYC (GenBank accession no. EU176011) is shown in horizontal arrows. Two inverted repeat sequences (IRL and IRR) of Tn4401 are depicted in triangles at either end. Tn4401 has five isoforms which differ by deletions (68–255 bp) just upstream of the blaKPC gene [(a) deletion of 99 bp; (b) no deletion; (c) deletion of 215 bp; (d) deletion of 68 bp; (e) deletion of 255 bp]. (B) The blaNDM-1 genetic context of pNDM_MGR194 (GenBank accession no. KF220657) is shown in horizontal arrows. (C) The blaOXA-48-containing Tn1999 transposon from the plasmid pOXA-48 (GenBank accession no. JN626286) is shown in horizontal arrows.
Carbapenemase genes often spread worldwide through clonal expansion in several successful pathogenic strains (Chen et al., 2014e). For example, the dissemination of KPC-producing K. pneumoniae in most countries including the USA and European countries is largely caused by expansion of a single dominant strain, ST258 (Giani et al., 2013; Chen et al., 2014e; Bowers et al., 2015). This strain is a prototype of a high-risk clone of K. pneumoniae. Recent data from Israel showed that the KPC-producing K. pneumoniae ST258 clone remains the predominant clone, representing 80% of the KPC-producing K. pneumoniae population (Adler et al., 2015). ST258 may be a hybrid clone that was created by a large recombination event between ST11 and ST442 (Pitout et al., 2015). It is unknown why the ST258 lineage is the most prevalent clone of KPC-producing Klebsiella species. The ST258 clone is highly susceptible to serum killing in animal models and lacks well-known K. pneumoniae virulence factors, such as aerobactin genes, K1, K2, and K5 capsular antigen genes, and the regulator of the mucoid phenotype gene rmpA (Tzouvelekis et al., 2013; Pitout et al., 2015). Two recent reports revealed that the ST258 strains consist of two distinct genetic clades and genetic differentiation between the two clades (-1 and cps-2) results from an approximately 215-kb region of divergence that includes cps genes involved in capsule polysaccharide synthesis (Chen et al., 2014e; Deleo et al., 2014). Multiplex PCR for the identification of two capsular types in K. pneumoniae ST258 strains revealed a significant association between the cps type and KPC variant: the cps-1 clade is largely associated with KPC-2, while the cps-2 clade is primarily associated with KPC-3 (Chen et al., 2014a). Because the capsule polysaccharide can help K. pneumoniae to evade phagocytosis, the global success of this strain may involve the capsule polysaccharide biosynthesis regions cps-1 and cps-2. A recent report revealed a relationship between the integrative conjugative element ICEKp258.2 and the global success of the ST258 clone (Adler et al., 2014). ICEKp258.2 contains two specific gene clusters, a type IV pilus gene cluster (i.e., pilV) associated with the uptake and exchange of plasmids and adherence to living and non-living surfaces, and a gene cluster of a type III restriction-modification system determining host specificity in the exchange of certain compatible plasmids or mobile elements (Adler et al., 2014). Because these genes associated with the restriction of plasmids and specific mobile elements were present only in ST258 and genetically related sequence types, this difference may explain the divergence of ST258 predominantly harboring KPC and ST11, another high-risk clone that lacks ICEKp258.2, harboring a broad range of plasmids and carbapenemases, including KPC, NDM, OXA-48, VIM, and IMP (Chen et al., 2014f; Pitout et al., 2015). Although the ICEKp258.2 of ST258 strains may contribute to global success, the precise reason for the predominance of the ST258 strain in KPC-producing K. pneumoniae is still not entirely understood. Recently, an outbreak of non-ST258 KPC-producing K. pneumoniae clones has been reported in the USA and Europe (Ruiz-Garbajosa et al., 2013; Bonura et al., 2015; Garbari et al., 2015).
The habitat of K. pneumoniae is not limited to humans but extends to the ecological environment, such as soil, water, and sewage, and K. pneumoniae can survive in extreme environments for long periods of time (Pitout et al., 2015). Therefore, K. pneumoniae producing KPCs were detected in various nosocomial environments, such as gowns and gloves (Rock et al., 2014) and wastewater (Chagas et al., 2011; Galler et al., 2014). The frequency of KPC-producing K. pneumoniae contamination of gowns and gloves of healthcare workers is similar to that of contamination with methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus (Rock et al., 2014), indicating fast transmission of KPC-producing Klebsiella species in a nosocomial environment. A long-term observation in a hospital with low-frequency outbreaks of KPC-producing K. pneumoniae suggested the possible role of a persisting environmental reservoir of resistant strains in the maintenance of this long-term outbreak (Tofteland et al., 2013). After discharge from the hospital, long-term (>3 years) carriage of KPC-producing K. pneumoniae is also possible (Lübbert et al., 2014), and lateral gene transfer of KPC among Enterobacteriaceae colonizing the human intestine appears frequent, for example from K. pneumoniae to E. coli (Richter et al., 2011; Gona et al., 2014). Therefore, reservoirs in healthcare workers, patients, or the hospital environment may be a principle mode of spread in nosocomial outbreaks.
Treatment Options
Carbapenemase-producing K. pneumoniae strains are currently one of the most important nosocomial pathogens. Hospital outbreaks of KPC-producing K. pneumoniae mainly affect severely ill patients and are associated with an increased risk of death (Ducomble et al., 2015; Tumbarello et al., 2015). KPC-producing K. pneumoniae bloodstream infections in intensive care unit (ICU) have also been associated with increased mortality (Chang et al., 2015).
Because carbapenemase-producing K. pneumoniae are mostly resistant to several important antibiotic classes (β-lactam drugs, fluoroquinolones, and aminoglycosides), antibiotics, such as polymyxin B, colistin (polymyxin E), fosfomycin, tigecycline, and sometimes selected aminoglycosides, are the last-resort agents. KPC-producing K. pneumoniae are usually resistant to all β-lactam antibiotics, but temocillin can be active against some KPC-producing K. pneumoniae, particularly in the case of lower urinary tract infections (Adams-Haduch et al., 2009). To maximize bacterial killing and minimize bacterial resistance, combined therapy is sometimes recommended. Combination therapy including a carbapenem, such as a combination of tigecycline, colistin, and meropenem, was strongly effective in the treatment of KPC-producing K. pneumoniae, including colistin-resistant isolates (Tumbarello et al., 2012, 2015; Giamarellou et al., 2013; Hong et al., 2013; Daikos et al., 2014). The synergistic combination of colistin and rifampin was also effective in the treatment of colistin-resistant KPC-producing K. pneumoniae by slowing the selection of heteroresistant subpopulations during colistin therapy (Tascini et al., 2013). However, several reports have shown that combination therapy was not superior to monotherapy (de Oliveira et al., 2015; Toledo et al., 2015). Thus, extensive studies will be required to assess the effectiveness of combination therapy. A triple combination of colistin-doripenem-ertapenem was effective only in isolates with high levels of OmpK35 and OmpK36 porin expression (Hong et al., 2013). The expression level of OmpK36 was also involved in the rapid induction of high-level carbapenem resistance in heteroresistant KPC-producing K. pneumoniae populations (Adams-Sapper et al., 2015). Therefore, molecular characterization of the KPC-producing K. pneumoniae strain, such as the determination of the expression level of OmpK35 and OmpK36, can be used to identify effective combination regimens. However, as a minor effect of OmpK35 and OmpK36 on carbapenem resistance of K. pneumoniae was also reported (Zhang et al., 2014), more extensive studies on the role of these proteins on K. pneumoniae carbapenem resistance are also required.
Colistin (polymyxin E), an agent discovered more than 60 years ago, is a key component of the combination of antimicrobial regimens used for the treatment of severe KPC-producing K. pneumoniae infections (Cannatelli et al., 2014b). Since the global spread of KPC-producing K. pneumoniae, the emergence of colistin resistance in KPC-producing K. pneumoniae have been reported in many countries, including Italy (Cannatelli et al., 2014b; Giani et al., 2015), the USA (Bogdanovich et al., 2011), Greece (Kontopoulou et al., 2010), Hungary (Toth et al., 2010), and Turkey (Labarca et al., 2014). The increasing prevalence of colistin-resistant K. pneumoniae producing KPC poses a threat to public health because colistin resistance increases the mortality due to KPC-producing K. pneumoniae bloodstream infections and reduces therapeutic options. A multicenter case-control-control study in Italy showed that the rate of colistin resistance among KPC-producing K. pneumoniae blood isolates increased more than threefold during the 4.5-years study period, and the 30-days mortality due to colistin-resistant KPC-producing K. pneumoniae bloodstream infections was approximately 51% (Giacobbe et al., 2015). Data collected from 21 hospital laboratories in Italy between 2013 and 2014 also showed that 43% of carbapenemase-producing K. pneumoniae were resistant to colistin, 6% were resistant to tigecycline, 16% were resistant to gentamicin, 82% were resistant to trimethoprim-sulfamethoxazole, and 1% were resistant to all four antibiotics, and colistin-resistant isolates were detected in all participating hospital laboratories (Monaco et al., 2014). The progressive increase of colistin resistance was also reported elsewhere (Pena et al., 2014; Bonura et al., 2015; Parisi et al., 2015). These results indicate that the strict rules for colistin use are required to diminish the spread of colistin resistance in the endemic regions of KPC-producing K. pneumoniae.
Molecular and biochemical studies have shown that insertional inactivation of the mgrB gene, encoding a negative-feedback regulator of the PhoQ–PhoP signaling system, can be responsible for colistin resistance in KPC-producing K. pneumoniae, due to the resulting up-regulation of the Pmr lipopolysaccharide modification system (Cannatelli et al., 2013, 2014b). A recent study analyzing a series of colistin-resistant K. pneumoniae isolates of worldwide origin identified a single amino acid change (T157P) in the PmrB protein as being responsible for the overexpression of pmrCAB and pmrHFIJKLM operons involved in lipopolysaccharide modification, leading to colistin resistance (Jayol et al., 2014). The relationship between colistin resistance and inactivation of the mgrB gene was further supported by analysis of clinical colistin-resistant K. pneumoniae isolates producing KPC (Bonura et al., 2015; Giani et al., 2015). The emergence of colistin resistance was also associated with low-dosage colistin treatment (Cannatelli et al., 2014a). A recent report showed that the plasmid carrying the mcr-1 gene, which encodes a phosphoethanolamine transferase enzyme catalyzing the addition of phosphoethanolamine to lipid A, is a major contributor to colistin resistance in Gram-negative bacteria and is spread through horizontal gene transfer (Liu et al., 2016). This mcr-1-harboring plasmid was also detected in E. coli isolates collected from 78 (15%) of 523 samples of raw meat, 166 (21%) of 804 animals, and 16 (1%) of 1322 samples from inpatients with infection, indicating the emergence of this plasmid-mediated colistin resistance mechanism (Liu et al., 2016).
Fosfomycin is a broad-spectrum antibiotic that inhibits bacterial cell wall biogenesis by inactivating the enzyme UDP-N-acetylglucosamine-3-enolpyruvyltransferase, also known as MurA (Brown et al., 1995). Fosfomycin has been used to treat KPC-producing K. pneumoniae, but recently, a high fosfomycin resistance rate was observed is in countries with higher usage (Giske, 2015). Only 43.4% of KPC-producing K. pneumoniae strains retained susceptibility to fosfomycin in a Chinese university hospital (Li et al., 2012) and a similar fosfomycin susceptibility rate (39.2%) was observed in KPC-producing K. pneumoniae collected from 12 hospitals in China (Jiang et al., 2015). Like colistin, fosfomycin resistance seems to be associated with the plasmid containing the fosA3 gene which encodes glutathione S-transferase to modify fosfomycin and was characterized first in CTX-M-producing E. coli in Japan (Wachino et al., 2010). In China, the fosA3-harboring plasmid was attributed to 55.6% of fosfomycin-resistant KPC-producing K. pneumoniae strains (Jiang et al., 2015). Although the fosA3 gene is mainly associated with the blaCTX-M gene, the fosA3 gene has also been characterized in atypical blaKPC-harboring plasmids (Jiang et al., 2015; Li et al., 2015). In pFOS18 (Jiang et al., 2015) and pKP1034 (Xiang et al., 2015), the fosA3 and blaKPC-2 genes were located on different transposon systems, whereas in pHS102707 belonging to the IncP1 group (Li et al., 2015), two genes were co-located in the same Tn1721-Tn3-like transposon.
Tigecycline, a derivative of minocycline, is the first member of the glycylcycline class that acts as a protein synthesis inhibitor by blocking the interaction of aminoacyl-tRNA with the A site of the ribosome (Rose and Rybak, 2006). Due to the increased clinical use of tigecycline for treatment of KPC-producing K. pneumoniae, increased tigecycline resistance was reported (Zagorianou et al., 2012; Papadimitriou-Olivgeris et al., 2014; Weterings et al., 2015). In the Netherlands, all KPC-producing K. pneumoniae isolates exhibited reduced susceptibility to tigecycline (Weterings et al., 2015). Another report showed that during ICU stay, 17.9% (39/257) of patients became colonized by tigecycline-resistant KPC-producing K. pneumoniae during their stay (Papadimitriou-Olivgeris et al., 2014). In a Greek tertiary hospital during 2004 to 2010, 11.3% (34/301) of KPC-producing isolates were resistant to tigecycline (Zagorianou et al., 2012). Overproduction of efflux pumps such as AcrAB and overexpression of RamA, a positive regulator of the AcrAB efflux system, seem to be major factors for decreased sensitivity of K. pneumoniae strains to tigecycline (Ruzin et al., 2005; Rosenblum et al., 2011; Sun et al., 2013). A recent report in China showed that the OqxAB efflux pump was also contributed to tigecycline resistance in K. pneumoniae isolates (Zhong et al., 2014).
Because KPC-producing K. pneumoniae sometimes remains susceptible to several aminoglycosides such as gentamicin (Tzouvelekis et al., 2014), aminoglycosides can be used alone or in combination therapy to treat KPC-producing K. pneumoniae infections. Actually, gentamicin monotherapy or in combination with tigecycline was recently reported to reduce the mortality from sepsis caused by K. pneumoniae ST512 clone producing KPC-3, SHV-11, or TEM-1 (Gonzalez-Padilla et al., 2015). New weapons are always indispensable for combating KPC-producing K. pneumoniae infections (Lee et al., 2007, 2015a). The effectiveness of some antibiotics in development was also estimated against KPC-producing K. pneumoniae. Potent inhibitors of serine β-lactamases, such as avibactam and MK7655, were effective against KPC-producing K. pneumoniae infections (Temkin et al., 2014). Combination therapy with avibactam and ceftazidime exhibited significant synergetic effects against organisms with combinations of extended-spectrum β-lactamases (ESBLs), AmpCs, and KPC-2 (Wang et al., 2014e). Plazomicin (a novel aminoglycoside) also exhibited significant activity against KPC-producing K. pneumoniae (Temkin et al., 2014). The novel polymyxin derivatives with lower nephrotoxicity are under development (Vaara, 2010). A recent report suggested that synthetic peptides with antimicrobial and antibiofilm activities are a promising strategy in the treatment of infections caused by KPC-producing K. pneumoniae (Ribeiro et al., 2015). The in vitro activity of the next-generation aminoglycoside plazomicin alone and in combination with colistin, meropenem, fosfomycin or tigecycline was tested against carbapenemase-producing Enterobacteriaceae (CPE) strains. When plazomicin was combined with meropenem, colistin or fosfomycin, synergy was observed against CPE isolates (Rodriguez-Avial et al., 2015).
Detection Methods
Because a delay in the appropriate antibiotic therapy for severe infections of KPC-producing K. pneumoniae is strongly associated with unfavorable prognosis and increased mortality rates (Karaiskos and Giamarellou, 2014), rapid detection of CR strains is essential for the effective management of these infections (Lee et al., 2005b, 2006b, 2016). Various methods for the identification of KPCs have been developed, including multiplex PCR assay (Spanu et al., 2012; Adler et al., 2014), real-time PCR assay (Wang et al., 2012; Lee et al., 2015c), DNA microarray (Peter et al., 2012; Braun et al., 2014), Raman spectroscopic analysis (Willemse-Erix et al., 2012), single-colony whole-genome sequencing (Koser et al., 2014), matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS; Carvalhaes et al., 2014), loop-mediated isothermal amplification (LAMP) method (Nakano et al., 2015), chromogenic medium (Vrioni et al., 2012), and a new phenotypic test, called the carbapenem inactivation method (CIM; van der Zwaluw et al., 2015). False positive results can occur when the modified Hodge test is used to detect carbapenemases in carbapenemase-negative K. pneumoniae clinical isolates (Wang et al., 2011). Therefore, to improve the efficiency in the phenotypic detection of KPC-producing K. pneumoniae isolates, the modified Hodge test can be combined with an EDTA disk test (Yan et al., 2014) or a disk test using boronic acid compounds (Pournaras et al., 2010a). These methods enhanced the sensitivity and specificity of KPC detection in K. pneumoniae isolates. In a new developed phenotypic test, called the CIM, a susceptibility-testing disk containing carbapenem was immersed in the suspension made by suspending an inoculation loop of bacterial culture (van der Zwaluw et al., 2015). After incubation, the disk was placed on an agar plate inoculated with a susceptible E. coli indicator strain. If the bacterial isolate produces carbapenemase, the susceptibility-testing disk will allow the growth of the susceptible indicator strain. This method showed high concordance with results obtained by PCR (van der Zwaluw et al., 2015). Nevertheless, these culture-based phenotypic tests are time-consuming and cannot easily detect ESBLs and carbapenemases produced by Enterobacteriaceae, owing to varied levels of enzyme expression and the poor specificity of some antibiotic combinations (Okeke et al., 2011; Swayne et al., 2013). To overcome these limitations of phenotypic methods, various molecular-based diagnostic methods have been developed. Especially, the direct detection of the carbapenemase gene using multiplex PCR, real-time PCR, and DNA microarray method can increase the speed and accuracy of detecting CR strains (Okeke et al., 2011; Solanki et al., 2014). Because of the high genetic diversity of genes coding for carbapenemase, the precise design of primers or probes is necessary for correctly amplifying or detecting only expected carbapenemase genes. Therefore, already developed methods for bla gene detection are restricted to the detection of only limited types of bla genes (Lee et al., 2015b). However, the large-scaleblaFinder was recently developed on the basis of multiplex PCR, and this large-scale detection method can detect almost all bla genes, including KPCs, NDMs, OXA-48-likes, present in bacterial pathogens (Lee et al., 2015b). Recently, mass spectrometry-based methods, such as MALDI-TOF MS and ultra-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS), have been shown to be capable of characterizing carbapenemase-producing bacteria (Carricajo et al., 2014; Carvalhaes et al., 2014). These methods are fast and accurate to routinely identify bacterial isolates with great specificity and sensitivity (Hrabak et al., 2011; Patel, 2013; Carvalhaes et al., 2014; Lasserre et al., 2015), but these systems do not accurately provide the carbapenem minimum inhibitory concentrations (MICs) for carbapenemase-producing K. pneumoniae (Patel, 2013). Several experiments showed that this method is more rapid and accurate for detection of carbapenemase activity in Gram-negative bacteria than some methods including the modified Hodge test (Lee et al., 2013b; Chong et al., 2015). The LAMP method has emerged as a powerful gene amplification assay for the rapid identification of microbial infections (Notomi et al., 2000). This method employs a DNA polymerase and a set of four specially designed primers that recognize a total of six distinct sequences on the target DNA. The assay amplifies the DNA under isothermal conditions (63–65°C) with high degrees of specificity, efficiency, and speed (Reddy et al., 2010). The cycling reaction continues with accumulation of 109 copies of target in less than an hour. The assay can be conducted in a water bath or heating block instead of the thermal cycling using a PCR machine (Notomi et al., 2000). The LAMP assay can be applied for detection of KPC producers in the clinical laboratory (Nakano et al., 2015) and has greater sensitivity, specificity, and rapidity compared to the phenotypic methods and PCR for the detection of KPC-producing K. pneumoniae (Solanki et al., 2013).
Colistin has often been used as a therapeutic option for the treatment of CR K. pneumoniae infections. However, the imprudent use of colistin has caused rapid spread of colistin resistance in K. pneumoniae producing carbapenemases, particularly the KPC-type carbapenemases (Monaco et al., 2014; Giacobbe et al., 2015). This situation demonstrates the need for the development of accurate and reliable methods for detecting colistin resistance. Recently, several methods for the identification of colistin resistance were reported, including various routine colistin MIC testing methods, such as BMD, BMD-P80, AD, Etest, MTS, and Vitek2 (Dafopoulou et al., 2015; Humphries, 2015); capillary electrophoresis method according to characteristic surface properties of bacteria (Sautrey et al., 2015); and the micromax assay based on evaluation of the efficacy of antibiotics that affect cell wall integrity (Tamayo et al., 2013). Because a recent report showed that the mcr-1 gene, involved in the modification of lipid A, is a major contributor to colistin resistance in Gram-negative bacteria (Liu et al., 2016), detection of this gene may be important in the detection of colistin resistance.
Class B Carbapenemases
Epidemiology
Class B β-lactamases are metallo-β-lactamases that require zinc or another heavy metal for catalysis. Class B β-lactamases have a broad substrate spectrum and can catalyze the hydrolysis of virtually all β-lactam antibiotics including carbapenems except for monobactams (Jeon et al., 2015). Class B carbapenemases were mostly identified in Enterobacteriaceae and include VIMs, IMPs, and the emerging NDM group (Jeon et al., 2015). Among them, NDM (New Delhi metallo-β-lactamase) is one of the most clinically significant carbapenemases. NDM-1 was first detected in 2008 in K. pneumoniae and E. coli in a patient returning to Sweden from India and has since spread worldwide (Yong et al., 2009; Jeon et al., 2015). Thus far, 15 NDM variants have been assigned (Jeon et al., 2015), and most of them originated from Asia (Nordmann and Poirel, 2014). NDMs shares very little identity with other metallo-β-lactamases (Nordmann and Poirel, 2014).
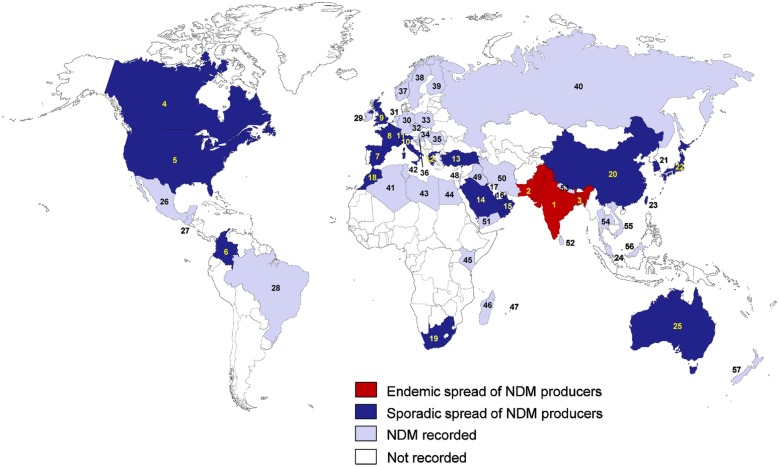
Since 2008, K. pneumoniae producing NDMs rapidly spread in many countries (Berrazeg et al., 2014; Figure 3). NDM-producing K. pneumoniae are considered to be endemic in the Indian subcontinent, including India, Pakistan, and Bangladesh (Nordmann et al., 2011b; Nordmann and Poirel, 2014). The sporadic spread has been reported in the USA (Rasheed et al., 2013; Centers for Disease Control and Prevention, 2014; Doi et al., 2014), Canada (Mulvey et al., 2011; Borgia et al., 2012; Lowe et al., 2013), Colombia (Escobar Perez et al., 2013; Ocampo et al., 2015), Spain (Oteo et al., 2013b; Seara et al., 2015), France (Arpin et al., 2012; Robert et al., 2014), Switzerland (Poirel et al., 2011g; Spyropoulou et al., 2016), Italy (Gaibani et al., 2011), the UK (Kumarasamy et al., 2010; Giske et al., 2012), Greece (Voulgari et al., 2014; Spyropoulou et al., 2016), Turkey (Poirel et al., 2014; Kilic and Baysallar, 2015), Morocco (Poirel et al., 2011b; Barguigua et al., 2013), South Africa (Brink et al., 2012; de Jager et al., 2015), Singapore (Chen et al., 2012; Balm et al., 2013; Ling et al., 2015), Saudi Arabia (Shibl et al., 2013; Zowawi et al., 2014), Oman (Poirel et al., 2011a; Zowawi et al., 2014), United Arab Emirates (Sonnevend et al., 2013; Dash et al., 2014), Kuwait (Jamal et al., 2012, 2013; Sonnevend et al., 2015b), China (Qin et al., 2014; Jin et al., 2015; Liu et al., 2015), Japan (Yamamoto et al., 2013; Nakano et al., 2014), Taiwan (Chiu et al., 2013; Tseng et al., 2015), South Korea (Kim et al., 2012; Cho et al., 2015), and Australia (Shoma et al., 2014; Wailan et al., 2015). In India, NDM-1 was the most common carbapenemase type detected and accounted for 75.22% of the carbapenemase-producing isolates (Kazi et al., 2015). In Singapore and the United Arab Emirates, NDM-1 also was the most common carbapenemase type observed (44.4 and 100%, respectively; Dash et al., 2014; Ling et al., 2015). The endemic spread of NDM-producing K. pneumoniae has also been reported in the UK, which has close relationships with India and Pakistan (Nordmann and Poirel, 2014). In China, NDM-1 has been found mostly in Acinetobacter spp., but data obtained from patients between June 2011 and July 2012 showed that 33.3% of the CRE isolates, including K. pneumoniae, had NDM-1, suggesting the possible transmission of blaNDM-1-containing sequences from Acinetobacter spp. to Enterobacteriaceae (Qin et al., 2014). These findings reveal the emergence and active transmission of NDM-1-producing K. pneumoniae in China. Comparative analyses of the conserved NDM-1-encoding region among different plasmids from K. pneumoniae and E. coli suggested that the transposable elements and two unknown inverted repeat-associated elements flanking the NDM-1-encoding region aided the spreading of this resistance determinant (Chen et al., 2012). Recently, in China, eight K. pneumoniae isolates producing NDM-1 were identified in the neonatal ward of a teaching hospital (Zhang et al., 2015), and four diverse types (NDM-1, KPC-2, VIM-2, and IMP-4) of carbapenemase of K. pneumoniae clones were identified in a single hospital in China (Liu et al., 2015).
FIGURE 3.
Epidemiological features of NDM-producing K. pneumoniae. (1) India; (2) Pakistan; (3) Bangladesh; (4) Canada; (5) USA; (6) Colombia; (7) Spain; (8) France; (9) UK; (10) Italy; (11) Switzerland; (12) Greece; (13) Turkey; (14) Saudi Arabia; (15) Oman; (16) United Arab Emirates; (17) Kuwait; (18) Morocco; (19) South Africa; (20) China; (21) South Korea; (22) Japan; (23) Taiwan; (24) Singapore; (25) Australia; (26) Mexico; (27) Guatemala; (28) Brazil; (29) Ireland; (30) Germany; (31) Netherlands; (32) Czech Republic; (33) Poland; (34) Hungary; (35) Romania; (36) Croatia; (37) Norway; (38) Sweden; (39) Finland; (40) Russia; (41) Algeria; (42) Tunisia; (43) Libya; (44) Egypt; (45) Kenya; (46) Madagascar; (47) Mauritius; (48) Israel; (49) Iraq; (50) Iran; (51) Yemen; (52) Sri Lanka; (53) Nepal; (54) Thailand; (55) Vietnam; (56) Malaysia, (57) New Zealand.
The Balkan states (Livermore et al., 2011; Voulgari et al., 2014), the Arabian Peninsula (Nordmann and Poirel, 2014), and North African countries (Dortet et al., 2014c), have also been recently considered as an additional reservoir of NDM producers. In the Arabian Peninsula, NDM-1 was the most frequently encountered carbapenemase (46.5%) followed by OXA-48-like carbapenemases (32.5%; Sonnevend et al., 2015b). In Greece, among 132 non-repetitive CRE isolates between 2010 and 2013, 78 K. pneumoniae isolates with the blaNDM-1 gene were identified (Voulgari et al., 2014). In the USA, KPC-producing K. pneumoniae have been responsible for much of the increase in carbapenemase-producing bacteria detection, but recent increases in NDM-producing K. pneumoniae have the potential to add to this burden (Rasheed et al., 2013; Centers for Disease Control and Prevention, 2014).
The movement of patients between countries may be a trigger for the international spread of carbapenemase-producing K. pneumoniae (Berrazeg et al., 2014). Carbapenemase-producing Gram-negative bacteria including K. pneumoniae, obtained from patients that had recently traveled outside Canada between 2010 and 2013, were found to be NDM-producing K. pneumoniae, belonging to various sequence types (ST15, ST16, ST147, ST258, ST340, ST512, and ST972) with different plasmids (IncF, IncA/C, and IncL/M), and were imported from India to Canada (Peirano et al., 2014). Therefore, more careful attention is required when treating patients with a recent history of foreign hospitalization in countries where carbapenemase-producing bacteria are prevalent. The international transportations of patients between countries recently resulted in the detection of NDM-producing K. pneumoniae in Mexico (Barrios et al., 2014), Guatemala (Pasteran et al., 2012), Brazil (Quiles et al., 2015), the Netherlands (Bathoorn et al., 2015), Ireland (McDermott et al., 2012), Poland (Baraniak et al., 2016), Czech Republic (Studentova et al., 2015), Croatia (Kocsis et al., 2016), Russia (Ageevets et al., 2014), Tunisia (Ben Nasr et al., 2013), Romania (Lixandru et al., 2015), Egypt (Bathoorn et al., 2013), Kenya (Poirel et al., 2011f), Madagascar (Chereau et al., 2015), Iraq (Poirel et al., 2011e), Yemen (Gharout-Sait et al., 2014), Iran (Shahcheraghi et al., 2013), Mauritius (Poirel et al., 2012b), Sri Lanka (Dortet et al., 2013), Thailand (Rimrang et al., 2012), Nepal (Tada et al., 2013), Vietnam (Hoang et al., 2013), Malaysia (Al-Marzooq et al., 2015), and New Zealand (Williamson et al., 2012).
The global dissemination of NDM-producing K. pneumoniae also has a serious impact on neonatal mortality rates, particularly in low-income countries where the burden of neonatal sepsis is high (Zaidi et al., 2005). Colonization of NDM-producing K. pneumoniae isolates in pregnant women in the community in Madagascar highlighted the potential for mother-to-child transmission (Chereau et al., 2015). In India, analysis of Enterobacteriaceae, including K. pneumoniae, isolated from the blood of septicaemic neonates, indicted that 14% of the isolates possessed blaNDM-1, and there was a significantly higher incidence of sepsis caused by NDM-1-harboring isolates (Datta et al., 2014). In China, an outbreak of blaNDM-1-producing K. pneumoniae ST20 and ST17 isolates was identified in a neonatal unit (Jin et al., 2015). In Turkey, the spread of NDM-1-producing K. pneumoniae in a neonatal ICU was reported (Poirel et al., 2014). In Colombia, NDM-1-producing K. pneumoniae strains were identified from an outbreak that affected six neonatal patients (Escobar Perez et al., 2013).
As with KPC, coexistence of NDMs and other carbapenemases in K. pneumoniae has also been reported worldwide, in Brazil (NDM-1/KPC-2; Pereira et al., 2015), Malaysia (NDM-1/OXA-232; Al-Marzooq et al., 2015), South Korea (NDM-5/OXA-181; Cho et al., 2015), China (NDM-1/IMP-4; Chen et al., 2015), India (NDM-1/OXA-232; Al-Marzooq et al., 2015), Turkey (NDM-1/OXA-48; Kilic and Baysallar, 2015), Pakistan (NDM-1/KPC-2; Sattar et al., 2014), Switzerland (NDM-1/OXA-48; Seiffert et al., 2014), United Arab Emirates (NDM-1/OXA-48-like; Dash et al., 2014), Australia (NDM-1/OXA-48; Sidjabat et al., 2015), Morocco (NDM-1/OXA-48; Barguigua et al., 2013), Singapore (NDM-1/OXA-181 and NDM-5/OXA-181; Balm et al., 2013), and the USA (NDM-1/OXA-232; Doi et al., 2014).
Besides NDM-type carbapenemases, the IMP and VIM groups have also been detected worldwide in K. pneumoniae, but other carbapenemases, such as GIM-1, KHM-1, and SPM-1, have been not found in K. pneumoniae (Table 1). Since IMP and VIP were first detected in Serratia marcescens in 1991 and in Pseudomonas aeruginosa in 1996, respectively (Osano et al., 1994; Lauretti et al., 1999), IMP- and VIM-producing K. pneumoniae have spread in Europe and Asia, but were rarely reported in other regions, such as America and Africa (Table 1). Although in this review we have focused only on carbapenemase-producing K. pneumoniae, the VIM group is one of the most commonly reported carbapenemases worldwide if we consider all bacteria species (the VIM groups have been mainly identified in P. aeruginosa; Poirel et al., 2007).
Molecular and Genetic Context
The blaNDM gene is frequently observed in the transposon Tn125 (with two flanking ISAba125 elements) within NDM-producing species of the genus Acinetobacter (Partridge and Iredell, 2012; Wailan et al., 2015). The blaNDM gene was hypothesized to originate in the genus Acinetobacter (Toleman et al., 2012). In Enterobacteriaceae, the ISAba125 elements of the Tn125 structure carrying blaNDM are frequently truncated (Tn125) at various lengths and the Tn125 structure frequently has different IS elements (Figure 2B; Partridge and Iredell, 2012; Wailan et al., 2015). The blaNDM genes in K. pneumoniae have been reported on numerous broad-host-range plasmid types, including IncA/C (Hudson et al., 2014), IncF (Hishinuma et al., 2013), IncR (Studentova et al., 2015), IncH (Villa et al., 2012), IncN (Chen et al., 2014a), IncL/M (Peirano et al., 2014), and IncX types (Wang et al., 2014d). The predominant plasmid type responsible for spreading blaNDM-1 is the IncA/C type plasmids (Poirel et al., 2011d; Pitout et al., 2015). Many IncA/C plasmids with blaNDM-1 also carry various antibiotic resistance genes including 16S rRNA methylases (RmtA and RmtC), associated with aminoglycoside resistance; CMY-type β-lactamases, associated with broad-spectrum cephalosporin resistance; and QnrA, associated with quinolone resistance (Pitout et al., 2015). Consequently, many NDM-producing K. pneumoniae were susceptible only to colistin, fosfomycin, and tigecycline (Nordmann and Poirel, 2014). A novel NDM-1 variant (NDM-9) located on a novel IncH variant plasmid was recently identified in a clinical K. pneumoniae isolate in China (Wang et al., 2014c). Because the blaNDM genes are located in numerous broad-host-range plasmids, the spread of NDM-1 is facilitated by horizontal gene transfer between bacteria. In China, CRE 21 strains harboring the blaNDM-1 gene were found to consist of multiple Enterobacteriaceae species including nine Enterobacter cloacae, three E. coli, three Citrobacter freundii, two K. pneumoniae, two K. oxytoca, and two Morganella morganii strains (Wang et al., 2015).
Unlike KPC, the blaNDM genes were detected in various K. pneumoniae clones. ST11, a major high-risk sequence type of KPC-producing K. pneumoniae in Asia, has also been associated with blaNDM-1 in K. pneumoniae identified in many countries, such as the USA (Rasheed et al., 2013), Greece (Voulgari et al., 2014), Australia (Shoma et al., 2014), Switzerland (Seiffert et al., 2014), the Czech Republic (Studentova et al., 2015), Spain (Oteo et al., 2013b), and Thailand (Netikul et al., 2014). The recent outbreak of NDM-producing K. pneumoniae ST11 in Poland was caused by a clone similar to an isolate identified in the Czech Republic in 2013 (Baraniak et al., 2016), indicating the local spread of this clone. ST11 has also been associated with OXA-48-like enzymes from isolates found in Argentina, Turkey, and Spain (Lascols et al., 2013; Oteo et al., 2013b). ST14, ST147, and ST340 have sometimes been associated with blaNDM in K. pneumoniae in many countries (Peirano et al., 2011, 2014; Poirel et al., 2011d; Giske et al., 2012; Osterblad et al., 2012; Lascols et al., 2013; Gharout-Sait et al., 2014; Lee et al., 2014; Shoma et al., 2014; Wang et al., 2014d; Izdebski et al., 2015; Sonnevend et al., 2015b). Over 50% of NDM-producing K. pneumoniae isolates from India belonged to either ST11 or ST147 (Lascols et al., 2013). The analysis of clinical isolates of NDM-1-producing K. pneumoniae from India, the UK, and Sweden, showed that the most frequently detected sequence types were ST14, ST11, ST149, ST231, and ST147 (Giske et al., 2012). Although ST258 is a high-risk KPC-producing K. pneumoniae clone, ST258 harboring blaNDM has never been reported, to the best of our knowledge. This phenomenon may result from the integrative conjugative element ICEKp258.2, present only in ST258 and genetically related sequence types (Adler et al., 2014). This genetic locus contains a type IV pilus gene cluster and a type III restriction-modification system. Because these genes are associated with the restriction of plasmids and specific mobile elements, most ST258 may predominantly harbor plasmids with the blaKPC gene. Therefore, PCR for this unique region (ICEKp258.2) can provide a reliable tool for the rapid detection of the ST258 clone.
High-resolution genomic analysis of multidrug-resistant hospital outbreaks of K. pneumoniae through whole-genome sequencing revealed the emergence of a capsule switching NDM-1 bearing K. pneumoniae ST15 strain, suggesting that further studies should concentrate on the diversity and spread of this specific clone (Chung The et al., 2015). ST15 harboring blaNDM-1 has often been reported in many countries, including Spain (Ruiz-Garbajosa et al., 2013), Croatia (Kocsis et al., 2016), Thailand (Netikul et al., 2014), Canada (Peirano et al., 2014), China (Hu et al., 2013), France (Arpin et al., 2012), and Morocco (Poirel et al., 2011b). In Bulgaria, this clone was responsible for the clonal dissemination of KPC-2-producing K. pneumoniae (Markovska et al., 2015). Another whole-genome sequencing analysis of CR K. pneumoniae strains, which were isolated from 26 individuals involved in infections in a Nepali neonatal unit, showed that three temporally separated cases were caused by a single NDM-producing K. pneumoniae strain with four conserved plasmids including a plasmid carrying blaNDM-1 (Stoesser et al., 2014). The plasmids contained a large number of antimicrobial resistance and plasmid maintenance genes, which may explain their persistence (Stoesser et al., 2014). These reports suggest that whole-genome sequencing analysis play an important role in the elucidation of the factors that allow emergence and persistence of resistance.
Treatment Options
New-Delhi metallo-β-lactamase-producing K. pneumoniae are usually resistant to most β-lactam antibiotics but remain susceptible to aztreonam (Nordmann et al., 2012a). As with the case of KPC-producing K. pneumoniae, the effect of combination therapy was tested in the treatment of NDM-producing K. pneumoniae infections. When double- and triple-antibiotic combinations of aztreonam, ciprofloxacin, colistin, daptomycin, fosfomycin, meropenem, rifampin, telavancin, tigecycline, and vancomycin were used in patients infected with two NDM-producing K. pneumoniae strains susceptible to colistin, the combination of rifampin-meropenem-colistin was the most effective regimen against these strains (Tangden et al., 2014). The in vitro synergetic effect of the combination therapy of colistin and fosfomycin against NDM-producing K. pneumoniae has also been reported (Bercot et al., 2011). The combination of polymyxin B and chloramphenicol used against NDM-producing K. pneumoniae substantially enhanced bacterial killing and suppressed the emergence of polymyxin resistance (Abdul Rahim et al., 2015). Combination therapy including aztreonam and avibactam (a novel inhibitor of serine β-lactamases under development) was effective in the treatment of metallo-β-lactamase-producing bacterial infections (Wang et al., 2014e).
In the case of NDM-1-producing Enterobacteriaceae infections, carbapenems have been suggested to still represent a viable treatment option (Wiskirchen et al., 2014b). Despite unfavorable in vitro MICs of NDM-producing K. pneumoniae, recent in vivo studies have demonstrated the efficacy of carbapenems against NDM-1-producing isolates in immunocompetent-mouse and neutropenic-mouse thigh infection models (Wiskirchen et al., 2013, 2014b; MacVane et al., 2014). Although high-dose, prolonged infusions of ertapenem or doripenem induced reduction in bacterial density, bacterial density was also reduced in standard infusions of ertapenem at 1 g every 24 h or of doripenem at 500 mg every 8 h (Wiskirchen et al., 2013, 2014b). Notably, these efficacies were observed only against NDM-1-producing K. pneumoniae (Wiskirchen et al., 2013, 2014b). In addition to carbapenems, this discordance between in vitro and in vivo activities against NDM-1-producing K. pneumoniae was also observed in human simulated regimens of ceftazidime at 2 g every 8 h or ceftazidime/avibactam at 2,000/500 mg every 8 h (MacVane et al., 2014). Despite experiments in immunocompetent-mouse and neutropenic-mouse thigh infection models, these results show that standard infusions of ertapenem and doripenem could reduce bacterial density. Therefore, further experiments in human are required to determine whether carbapenems are sometimes a viable treatment option for NDM-1-producing K. pneumoniae infections.
The copy number of blaNDM-1 was assessed using Southern blotting and quantitative PCR under different conditions. The blaNDM-1 sequence was maintained under antibiotic selection; however, removal of the antibiotic selection led to the emergence of susceptible bacterial populations with a reduced copy number or even the complete loss of the blaNDM-1 gene (Huang et al., 2013). The dynamic nature of the copy number of blaNDM-1 provides a strong argument for the prudent use of clinically important antibiotics to reduce the development and dissemination of antibiotic resistance among pathogenic bacteria (Huang et al., 2013).
Detection Methods
Because NDM-producing K. pneumoniae infections are also associated with significant in-hospital mortality (de Jager et al., 2015), the rapid and accurate detection of NDM-producing K. pneumoniae is becoming a major issue in limiting the spread of CR bacteria. Several methods recently developed to detect NDM-producing K. pneumoniae include Xpert® Carba-R based on real time PCR (Anandan et al., 2015), the Carba NP test based on the color change of a pH indicator (Nordmann et al., 2012b), and its derivatives (Pires et al., 2013; Dortet et al., 2014b), and a method based on MALDI-TOF (Hrabak et al., 2013a). Xpert® Carba-R based on real time PCR effectively identified various carbapenemases of KPC, NDM, IMP, and VIM, with 100% sensitivity and 77% specificity (Anandan et al., 2015). However, this method failed to detect OXA-48-like carbapenemases, in contrast to multiplex PCR (Anandan et al., 2015).
The Carba NP test using chromogenic medium is based on the color change of a pH indicator (Nordmann et al., 2012b). The enzymatic hydrolysis of the β-lactam ring of a carbapenem (usually imipenem) causes the acidification of an indicator solution (phenol red for the Carba NP test) that changes its color due to pH modification (Garcia-Fernandez et al., 2016). This method could rapidly detect KPC, IMP, VIM, NDM, and OXA-48-like producers with sensitivity and specificity of 97.9 and 100%, respectively, directly from spiked blood cultures (Dortet et al., 2014a). Comparative evaluation of the Carba NP test with other detection methods was tested in many reports (Vasoo et al., 2013; Huang et al., 2014; Yusuf et al., 2014; Gallagher et al., 2015; Lifshitz et al., 2016). When the Carba NP test and the modified Hodge test were compared, the Carba NP test was more specific (100% versus 80%) and faster (Vasoo et al., 2013; Huang et al., 2014; Yusuf et al., 2014). When it was compared the performance of the Carba NP test and the commercially available imipenem hydrolysis-based rapid test (the Rosco Rapid CARB screen kit) for detecting CPE and P. aeruginosa, the Carba NP test showed superior specificity and sensitivity (Huang et al., 2014; Yusuf et al., 2014; Gallagher et al., 2015). A novel simplified protocol of the Carba NP test designed for carbapenemase detection direct from bacterial cultures (instead of bacterial extracts) showed enhanced detection of carbapenemase producers (Pasteran et al., 2015).
However, several reports showed that false-negative results in the Carba NP test were associated with mucoid strains or linked to enzymes with low carbapenemase activity, particularly OXA-48-like (Tijet et al., 2013; Osterblad et al., 2014). To overcome these problems, several derivatives of the Carba NP test were developed, such as the Rapidec Carba NP test (bioMérieux; Poirel and Nordmann, 2015), the CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. (Dortet et al., 2014b), the Rapid CARB Screen (Rosco Diagnostica; Dortet et al., 2015), the Blue-Carba test using bromothymol blue as a pH indicator solution (Garcia-Fernandez et al., 2016), a modified Carba NP test (Bakour et al., 2015a), and the BYG Carba test based on an electro-active polymer biosensing technology (Bogaerts et al., 2016). Many studies evaluated the performance of the Rapidec Carba NP test (bioMérieux), which was introduced into the market for the detection of carbapenemase production (Dortet et al., 2015; Hombach et al., 2015; Kabir et al., 2016; Lifshitz et al., 2016). These report showed that this method was user-friendly and had a high overall performance, making it an attractive option for clinical laboratories (Kabir et al., 2016). Recently, performance evaluation of two biochemical rapid tests commercialized (the Rapidec Carba NP test and the Rapid CARB Screen) was reported and compared with the home-made Carba NP test (Dortet et al., 2015). The Rapidec CARBA NP test possesses the best performance for rapid and efficient detection of CPE (Dortet et al., 2015). The BYG Carba test based on a new and original electrochemical method detects the variations of conductivity of a polyaniline (an electro-sensing polymer)-coated electrode which is highly sensitive to the modifications of pH and of redox activity occurring during the imipenem enzymatic hydrolysis reaction (Bogaerts et al., 2016). In comparison with PCR results, the BYG Carba test displayed sensitivity of 95% and specificity of 100% versus 89% and 100%, respectively, for the Carba NP test (Bogaerts et al., 2016). The development of these detection methods based on inexpensive and affordable techniques can limit the spread of CR bacteria.
Class D Carbapenemases
Epidemiology
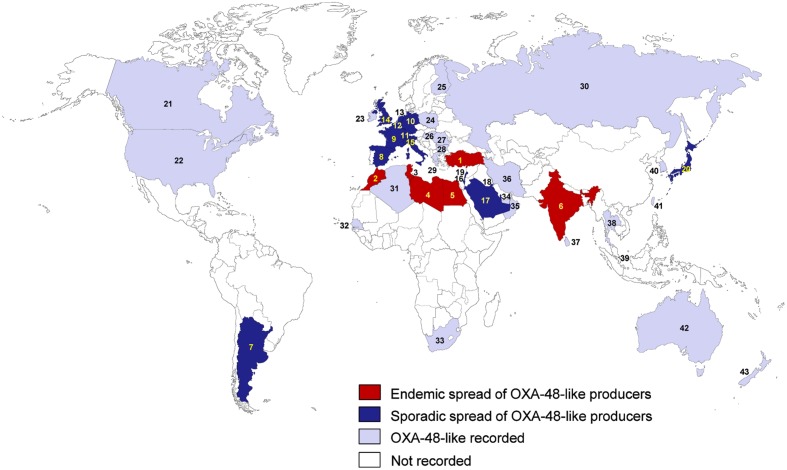
Class D β-lactamases were referred to as oxacillinases (OXAs) because they commonly hydrolyze isoxazolylpenicillins (oxacillin, cloxacillin, and dicloxacillin) much faster than benzylpenicillin (Jeon et al., 2015). Of over 400 Class D β-lactamases, only some variants actually possess carbapenemase activity. Based on their amino acid sequence, class D carbapenemases were recently reclassified into 12 subgroups: OXA-23, OXA-24/40, OXA-48, OXA-51, OXA-58, OXA-134a, OXA-143, OXA-211, OXA-213, OXA-214, OXA-229, and OXA-235 (Jeon et al., 2015). Among them, only several subgroups such as OXA-23, OXA-48, OXA-51, and OXA-58 are reported in K. pneumoniae (Evans and Amyes, 2014). OXA-48 is the most efficient class D carbapenemase for imipenem and is one of the most prevalent class D carbapenemases (Jeon et al., 2015). The OXA-48 was first identified in K. pneumoniae in Turkey in 2003 (Poirel et al., 2004), and thus far, 10 variants of the blaOXA-48 gene have been identified (Jeon et al., 2015). Turkey may be one of the main reservoirs of OXA-48-producing K. pneumoniae (Nordmann and Poirel, 2014). Since 2003, the endemic spread of these bacteria has been reported in countries such as Turkey, Morocco, Libya, Egypt, Tunisia, and India (Nordmann and Poirel, 2014; Figure 4). The sporadic spread has been reported in France (Liapis et al., 2014; Semin-Pelletier et al., 2015), Spain (Oteo et al., 2013b; Pena et al., 2014), Italy (Giani et al., 2013, 2014), Belgium (Cuzon et al., 2008; Huang et al., 2013), the Netherlands (Kalpoe et al., 2011; Dautzenberg et al., 2014), the UK (Dimou et al., 2012; Thomas et al., 2013), Germany (Pfeifer et al., 2012; Kola et al., 2015), Switzerland (Potron et al., 2012; Seiffert et al., 2014), Argentina (Poirel et al., 2011c; Lascols et al., 2013), Lebanon (Beyrouthy et al., 2014), Israel (Adler et al., 2013, 2015), Kuwait (Poirel et al., 2012a; Zowawi et al., 2014), Saudi Arabia (Shibl et al., 2013; Liu et al., 2015), and Japan (Nagano et al., 2013; Hashimoto et al., 2014). The prevalence of OXA-48 carbapenemases among carbapenemase-producing K. pneumoniae in Spain and France was particularly high (74 and 78%, respectively; Robert et al., 2014; Palacios-Baena et al., 2016). In Africa, OXA-48-producing K. pneumoniae have been mainly reported in the northern countries, such as Morocco, Libya, Egypt, Tunisia, and Algeria (Figure 4). In the Arabian Peninsula, the prevalence of OXA-48-like carbapenemases among carbapenemase-producing K. pneumoniae was also high (32.5–56%; Zowawi et al., 2014; Sonnevend et al., 2015b). Among CRE isolates in Lebanon, 88% produced OXA-48 carbapenemase (Beyrouthy et al., 2014). In Saudi Arabia, 78% of carbapenemase-producing K. pneumoniae isolates harbored blaOXA-48, and three strains of 47 blaOXA-48-positive K. pneumoniae isolates were resistant to colistin, suggesting that colistin resistance is emerging in Saudi Arabia (Shibl et al., 2013).
FIGURE 4.
Epidemiological features of OXA-48-like-producing K. pneumoniae. (1) Turkey; (2) Morocco; (3) Tunisia; (4) Libya; (5) Egypt; (6) India; (7) Argentina; (8) Spain; (9) France; (10) Germany; (11) Switzerland; (12) Belgium; (13) Netherlands; (14) UK; (15) Italy; (16) Israel; (17) Saudi Arabia; (18) Kuwait; (19) Lebanon; (20) Japan; (21) Canada; (22) USA; (23) Ireland; (24) Poland; (25) Finland; (26) Hungary; (27) Romania; (28) Bulgaria; (29) Greece; (30) Russia; (31) Algeria; (32) Senegal; (33) South Africa; (34) United Arab Emirates; (35) Oman; (36) Iran; (37) Sri Lanka; (38) Thailand; (39) Singapore; (40) South Korea; (41) Taiwan; (42) Australia; (43) New Zealand.
In France, examination of the epidemiologic features of an outbreak of OXA-48-producing K. pneumoniae in an ICU revealed that the outbreak was caused by environmental persistence of OXA-48-producing K. pneumoniae over 20 months (Pantel et al., 2016). This report emphasizes the importance of early environmental screening to interrupt the transmission of carbapenemase-producing K. pneumoniae (Pantel et al., 2016). Similarly, a large outbreak of OXA-48 carbapenemase-producing K. pneumoniae in a French university hospital was recently attributed to the late implementation of successive cohort units and a high level of staff movement between the infectious diseases and internal medicine ward (Semin-Pelletier et al., 2015). These results suggest that practical guidelines are required to help hospitals confronting uncontrolled outbreaks. Because the gut of colonized patients is the main source of CPE, accurate and stringent hygiene of endoscopic instruments is also important. A recent report partially attributed an outbreak of OXA-48-producing K. pneumoniae in a German University hospital to complex instruments such as the duodenoscope (Kola et al., 2015). Strict hygiene regulations for various nosocomial environments, including endoscopic instruments as well as gowns and gloves, are required.
OXA-48-producing K. pneumoniae was recently also detected in Canada (Ellis et al., 2013), the USA (Mathers et al., 2013), Ireland (Wrenn et al., 2014), Poland (Izdebski et al., 2015), Hungary (Janvari et al., 2014), Greece (Voulgari et al., 2013), Romania (Lixandru et al., 2015), Bulgaria (Markovska et al., 2015), Finland (Osterblad et al., 2012), Russia (Fursova et al., 2015), Algeria (Cuzon et al., 2015), United Arab Emirates (Ahn et al., 2015), Iran (Azimi et al., 2014), South Africa (Brink et al., 2013), Senegal (Moquet et al., 2011), Taiwan (Ma et al., 2015), Singapore (Ling et al., 2015), South Korea (Jeong et al., 2015), and Australia (Espedido et al., 2013; Figure 4). In North America, the frequency of OXA-48-like enzymes among carbapenemase-producing K. pneumoniae isolates was very low (11%; Lascols et al., 2013). A recent report in Romania showed that among 65 carbapenemase-producing K. pneumoniae, the most frequently identified gene was the blaOXA-48 gene (78%), 12% were positive for blaNDM-1 gene, 6% had the blaKPC-2 gene (Lixandru et al., 2015). The recent spread of OXA-48 and OXA-244 carbapenemase genes in Russia was reported among Proteus mirabilis, E. aerogenes, and E. cloacae as well as K. pneumoniae (Fursova et al., 2015). In South Africa, the emergence of a colistin-resistant OXA-181-producing K. pneumoniae isolate was also reported (Brink et al., 2013), and in the Netherlands, an OXA-48-producing K. pneumoniae was reported to infect two patients (Kalpoe et al., 2011), and a hospital-wide outbreak was successfully controlled (Dautzenberg et al., 2014).
OXA-181, a derivative of OXA48 with the substitution of a single amino acid, was first identified in India in Potron et al. (2011), and then has been spread to many different countries, such as the UK (Dimou et al., 2012), Romania (Székely et al., 2013), Canada (Peirano et al., 2014), Oman (Potron et al., 2011), Singapore (Balm et al., 2013), Sri Lanka (Hall et al., 2014), South Korea (Cho et al., 2015), Australia (Sidjabat et al., 2015), Japan (Kayama et al., 2015), and New Zealand (Williamson et al., 2011). However, in many cases, the infections have been associated with India (Potron et al., 2011; Williamson et al., 2011; Dimou et al., 2012; Hall et al., 2014; Peirano et al., 2014). Other OXA-48-derivatives, such as OXA-204 (Potron et al., 2013), OXA-232 (Potron et al., 2013), and OXA-163 (Poirel et al., 2011b), were recently identified in Tunisia, France, and Argentina, respectively, and OXA-244 and OXA-245 were first reported in Spain (Oteo et al., 2013a). All of these countries are regions with a high prevalence of OXA-48-producing K. pneumoniae (Figure 4). In addition, OXA-232-producing K. pneumoniae has been reported in various countries, such as the USA (Doi et al., 2014), Singapore (Teo et al., 2013), India (Al-Marzooq et al., 2015), and South Korea (Jeong et al., 2015). These results indicate that besides OXA-48, its many derivatives also spread worldwide. OXA-163, which differs from OXA-48 by a four amino acid deletion and a single amino acid substitution, has lower carbapenemase activity than OXA-48, but this enzyme exhibits extended activity against expanded-spectrum cephalosporins, and its activity is partially inhibited by clavulanic acid, a β-lactamase inhibitor (Poirel et al., 2011c). OXA-247, a derivative of OXA-163 with a single amino acid substitution, was recently reported in Argentina (Gomez et al., 2013), where OXA-163-producing K. pneumoniae were often reported (Lascols et al., 2013).
The coexistence of OXA-48-like and other carbapenemases in K. pneumoniae was also frequently reported worldwide, such as in Turkey (OXA-48/NDM-1; Kilic and Baysallar, 2015), Switzerland (OXA-48/NDM-1; Seiffert et al., 2014), United Arab Emirates (OXA-48-like/NDM-1; Dash et al., 2014), Australia (OXA-48/ NDM-1; Sidjabat et al., 2015), Morocco (OXA-48/NDM-1; Barguigua et al., 2013), India (OXA-181/VIM-5; Castanheira et al., 2011), Singapore (OXA-181/NDM-1 and OXA-181/NDM-5; Balm et al., 2013), the USA (OXA-232/NDM-1; Doi et al., 2014), and India (OXA-232/NDM-1; Al-Marzooq et al., 2015).
Other class D carbapenemases, such as OXA-23, OXA-24/40, OXA-51, OXA-58, OXA-134a, OXA-143, OXA-211, OXA-213, OXA-214, OXA-229, and OXA-235, were mainly identified in Acinetobacter species such as A. baumannii but not in K. pneumoniae (Table 1; Evans and Amyes, 2014).
Molecular and Genetic Context
Unlike KPCs and NDMs, one highly transferable IncL group plasmid (pOXA-48a) was mainly responsible for the spread of the blaOXA-48 gene in K. pneumoniae (Figure 2C; Pitout et al., 2015). The molecular epidemiology of OXA-48 in European and North African countries showed that in 92.5% of the isolates, the blaOXA-48 gene was located on this self-conjugative IncL/M type plasmid (Potron et al., 2013). The blaOXA-48 gene was found only in the IncL group of IncL/M type plasmids (Carattoli et al., 2015). In contrast to the IncA/C plasmids of NDM-1, the pOXA-48a plasmid contains blaOXA-48, a unique antibiotic resistance gene (Pitout et al., 2015). The conjugation rate of the pOXA-48a plasmid was very high (1 × 10-1); therefore, this self-conjugative plasmid can conjugate at a very high frequency to Enterobacteriaceae (Potron et al., 2014). Inactivation of the tir gene, which is known to encode a transfer inhibition protein, was recently reported to be responsible for a 50- to 100-fold increase in the efficiency of transfer of the pOXA-48a plasmid (Potron et al., 2014), explaining the spread of the pOXA-48a plasmid with blaOXA-48. Recently, the blaOXA-48-like gene has also been reported on other plasmids and genetic elements, such as IncA/C types (Ma et al., 2015), IncH types (Wang et al., 2014c), and Tn1999 (Poirel et al., 2012a). In comparison to the pOXA-48a plasmid, the transmission frequency of the Tn1999 composite transposon was very low (<1.0 × 10-7; Aubert et al., 2006).
OXA-48 has most often been found in K. pneumoniae, but OXA-48 was also found in various Enterobacteriaceae, because of the high conjugation rate of the pOXA-48a (Potron et al., 2014). Molecular and epidemiological analyses in a German hospital showed the horizontal gene transfer of the OXA-48-containing plasmid from K. pneumoniae to E. coli (Gottig et al., 2015). Besides E. coli, OXA-48 has also been identified in K. oxytoca, Enterobacter spp., Providencia rettgeri, C. freundii, and S. marcescens (Poirel et al., 2012c; Berger et al., 2013). The blaOXA-48 gene was identified in all Enterobacteriaceae isolates from the index case in Spain, indicating the capacity of OXA-48 carbapenemase to spread among Enterobacteriaceae by the horizontal gene transfer (Arana et al., 2015).
Similar to NDM, the blaOXA-48-like genes were detected in various K. pneumoniae clones. ST11 has often been associated with blaOXA-48-like in K. pneumoniae isolated in many countries, such as Spain (Oteo et al., 2013a,b, 2015; Ruiz-Garbajosa et al., 2013; Branas et al., 2015), Taiwan (Ma et al., 2015), Libya (Lafeuille et al., 2013), Turkey (Lascols et al., 2013), Argentina (Lascols et al., 2013), and Greece (Voulgari et al., 2013). In Spain, a large outbreak was initiated in 2013 by a OXA-48-producing K. pneumoniae ST11 clone, and this strain was detected in 44 patients (Branas et al., 2015). The ST11 isolates carried various carbapenemases, including NDM-1, VIM-1, OXA-48, KPC-2, and OXA-245 (Lascols et al., 2013; Oteo et al., 2013b, 2015). In addition, ST14, ST15, ST101, ST147, and ST405 harboring blaOXA-48-like have often been reported in many countries, such as the USA (Lascols et al., 2013), Spain (Oteo et al., 2013b, 2015; Ruiz-Garbajosa et al., 2013; Arana et al., 2015; Cubero et al., 2015), the Czech Republic (Hrabak et al., 2015), Libya (Lafeuille et al., 2013), India (Lascols et al., 2013), Germany (Gottig et al., 2015), Finland (Osterblad et al., 2012), France (Liapis et al., 2014), and Japan (Hashimoto et al., 2014). Recent results from 83 hospitals in Spain showed that OXA-48 (71.5%) and VIM-1 (25.3%) were the most frequently detected carbapenemases, and the most prevalent sequence types were ST11 and ST405 for K. pneumoniae (Oteo et al., 2015). However, the molecular epidemiology of OXA-48-producing enterobacterial isolates collected from European and north-African countries between 2001 and 2011 indicated that ST101 was the most commonly observed sequence type in K. pneumoniae isolates, accounting for 17 out of 67 isolates (25.4%), followed by ST395 and ST15 (each seven isolates, 10.5%; Potron et al., 2013). Two outbreaks of OXA-48-producing K. pneumoniae ST101 clones were reported in Spain (Pitart et al., 2011; Cubero et al., 2015). OXA-48-producing K. pneumoniae appear to vary geographically. As with NDM, to the best of our knowledge, ST258 harboring blaOXA-48-like has never been reported.
Treatment Options
OXA-48-like-producing K. pneumoniae are usually resistant to most β-lactam antibiotics, but OXA-48-producing K. pneumoniae without ESBLs remain susceptible to the expanded-spectrum cephalosporins (Nordmann et al., 2012a; Munoz-Price et al., 2013). In addition, OXA-48-like-producing K. pneumoniae sometimes remains susceptible to several aminoglycosides such as gentamicin (Tzouvelekis et al., 2014). Unlike for NDMs (Wiskirchen et al., 2013, 2014b; MacVane et al., 2014), carbapenems may not be a reliable treatment option for OXA-48 producer infection (Wiskirchen et al., 2014a). Combination therapy with sulbactam, meropenem, and colistin was more effective in isolates producing NDM carbapenemase than those producing OXA-48-like carbapenemases, suggesting that the identification of the carbapenemase type helps determine the combination most likely to clear the infection (Laishram et al., 2015). The combination of fosfomycin with imipenem, meropenem, and tigecycline was also synergistic against OXA 48-positive K. pneumoniae strains in vitro with the ratios of 42, 33, and 33%, respectively (Evren et al., 2013). Similarly, the in vitro assays indicate that imipenem-containing combinations were effective against serine-β-lactamase producers (KPC, OXA-48), while no synergy was observed for all NDM-1 producers (Poirel et al., 2016). However, because carbapenems were effective against NDM-1-producing isolates in vivo (Wiskirchen et al., 2013, 2014a; MacVane et al., 2014), the effect of carbapenem combination therapy on carbapenemase-producing isolates should be determined in vivo, particularly in the case of NDM-1-producing bacteria.
Despite the effectiveness of combination therapies, the prognosis for bloodstream infections caused by OXA-48-producing Enterobacteriaceae remains poor, and the 30-days mortality reached 50% (Navarro-San Francisco et al., 2013). A similar result was reported in OXA-48-producing K. pneumoniae infections in a tertiary hospital in Spain (Pano-Pardo et al., 2013). Although OXA-48-producing K. pneumoniae were susceptible to several antibiotics, including amikacin (97.2% susceptible), colistin (90.1%), tigecycline (73%), and fosfomycin (66.2%), in-hospital mortality among patients with OXA-48-producing K. pneumoniae infections was 43.5% (Pano-Pardo et al., 2013). Therefore, to prevent delay in diagnosis and initiation of optimal antimicrobial therapy, rapid identification of OXA-48-producing isolates is required.
Detection Methods
As with KPCs and NDMs, various detection methods were developed to identify OXA-48-like carbapenemases (Tsakris et al., 2010; Giske et al., 2011; Girlich et al., 2013; Naas et al., 2013; Lee et al., 2015c). Accurate differentiation of the various carbapenemase types, such as KPC-type, NDM-type, and OXA-48-type enzymes, is crucial for controlling the spread of carbapenem resistance among Enterobacteriaceae (Nordmann et al., 2012a). Many phenotypic detection methods to allow differentiation between class A and class B carbapenemases were developed using boronic acid derivatives and EDTA or dipicolinic acid (Tsakris et al., 2010; Giske et al., 2011). Recently, a specific phenotypic method to differentiate a single OXA-48 producer from those producing other carbapenemase types (e.g., KPC-types, NDM-types) was also developed (Tsakris et al., 2015). This method was based on an imipenem disk and two blank disks adjacent to the imipenem disk, loaded with the tested strain and impregnated with EDTA and EDTA plus phenyl boronic acid, respectively (Tsakris et al., 2015). This novel method exhibited 96.3% sensitivity and 97.7% specificity (Tsakris et al., 2015). ChromID OXA-48, based on chromogenic media, have been commercialized for the direct isolation of CPE from clinical samples (Girlich et al., 2013), with an estimated 91% sensitivity and 100% specificity (Girlich et al., 2013).
Emerging Class C Carbapenemases
Class C β-lactamases confer resistance to penicillins, cephalosporins, and cephamycins (cefoxitin and cefotetan) and are not significantly inhibited by clinically applied β-lactamase inhibitors such as clavulanic acid (Jeon et al., 2015). Although four class C carbapenemases (ACT-1, CMY-2, CMY-10, and ADC-68) have been reported, ACT-1 and CMY-2 exhibit reduced susceptibility to carbapenems, particularly ertapenem, only when combined with permeability defects, due to their low catalytic efficiencies (Kcat/Km) for imipenem (0.007 and 0.04 M-1⋅S-1, respectively; Mammeri et al., 2010). CMY-10 with the catalytic efficiency of 0.14 M-1⋅S-1 for imipenem was the first reported carbapenemase among plasmidic class C β-lactamases (Kim et al., 2006), and this enzyme was also a class C extended-spectrum β-lactamase with extended substrate specificity for extended-spectrum cephalosporins (Lee et al., 2009, 2012a; Jeon et al., 2015). Among the chromosomal class C β-lactamases, ADC-68 identified in A. baumannii was the first reported enzyme possessing both class C extended-spectrum β-lactamase and carbapenemase activities (Jeon et al., 2014, 2015), and its catalytic efficiency for imipenem is 0.17 M-1 S-1. CMY-10-producing K. pneumoniae was identified only in South Korea (Lee et al., 2005a, 2006a) and ADC-68-producing K. pneumoniae was never reported.
Many reports showed that carbapenem resistance can be triggered by the loss of two major porins, OmpK35 and OmpK36, in combination with ESBLs or Ambler class C AmpC cephalosporinases, and the production of carbapenemase (Wang et al., 2009; Shin et al., 2012; Tsai et al., 2013). A recently genetically engineered mutant of K. pneumoniae showed that several carbapenems (imipenem, meropenem, and doripenem) remain effective against these carbapenemase-independent CR strains (Tsai et al., 2013). Therefore, laboratory testing for susceptibility to imipenem, meropenem, and doripenem can improve the accuracy of identification of these isolates (Tsai et al., 2013).
Conclusion
We analyzed the epidemiology of K. pneumoniae producing true carbapenemases (Ambler molecular class A, B, D, and several carbapenemases of class C) responsible for non-susceptibility to carbapenems without additional permeability defects. Many types of CR K. pneumoniae have been identified worldwide (Figures 1, 3, and 4). During the past 3 years, many countries have reported the arrival of carbapenemases previously unreported in those countries. For example, although K. pneumoniae producing KPC- and NDM-type carbapenemases have been extensively reported in the USA, a K. pneumoniae producing OXA-48-type carbapenemases was recently detected in the USA (Mathers et al., 2013). Since a first report of NDM-1 in 2008, this carbapenemase has rapidly spread worldwide, and NDM-producing K. pneumoniae still continues to be found in new countries, implying that NDM-producing K. pneumoniae is still spreading quickly. Despite the global dissemination of KPC, NDM, and OXA-48, the prevalence of carbapenemases varies geographically. The frequency of KPC- and NDM-producing strains was significantly higher in the USA, Canada, Greece, Taiwan, Colombia, and China, whereas OXA-48-producing strains were rarely found in those countries (Figures 1, 3, and 4). In Argentina, despite the extensive spread of KPC- and OXA-48-producing strains, no NDM-producing K. pneumoniae has been reported. Although NDM- and OXA-48-producing K. pneumoniae significantly spread in Turkey, KPC-producing strains have rarely been reported there. In Brazil, KPC-producing K. pneumoniae has been mainly reported. In the Arabian Peninsula, OXA-48 and NDM producers are common, whereas KPC-type, VIM-type, or IMP-type producers are rare. In India, Spain, France, Italy, and the UK, all three types of carbapenemases have been frequently reported (Figures 1, 3, and 4).
ST258 is an important strain responsible for the extensive global spread of KPC-producing K. pneumoniae. Although the precise reason for the predominance of the ST258 strain in KPC-producing K. pneumoniae is not fully understood, recent molecular studies unveiled the genetic characteristics of this strain. The ST258 strains consists of two distinct genetic clades (cps-1 and cps-2) derived from genetic differentiation in genes involved in capsule polysaccharide biosynthesis. In addition, the integrative conjugative element ICEKp258.2, present only in ST258 and genetically related sequence types, may be linked to the global success of the ST258 clone. ICEKp258.2 contains a type IV pilus gene cluster and a type III restriction-modification system. The type IV pilus gene cluster ICEKp258.2, particularly pilV, may contribute to the global success of ST258 clone. The type III restriction-modification system associated with the restriction of plasmids and specific mobile elements may explain the differences observed between ST258 predominantly harboring KPC and ST11, another high-risk clone that lacks ICEKp258.2, harboring various carbapenemases, such as NDM-1, OXA-48, KPC-2, VIM-1, and OXA-245. IncF with FIIK replicons, a plasmid most commonly identified in ST258 with blaKPC, often contains several genes associated with resistance to other antibiotics, such as aminoglycosides, tetracyclines, quinolones, trimethoprim, and sulfonamides. The features of this plasmid may also play an important role in the current global success of ST258.
The rapid global spread of NDM-type carbapenemases may be partly attributed to the dissemination of various epidemic broad-host-range plasmids bearing the blaNDM genes. NDM-type carbapenemases were found in various plasmids such as IncA/C, IncF, IncR, IncH, IncN, IncL/M, and IncX types. The IncA/C type plasmids, most common plasmids associated with spread of the blaNDM genes, often have various antibiotic resistance genes, such as 16S rRNA methylases associated with aminoglycoside resistance, CMY-type β-lactamases associated with broad-spectrum cephalosporin resistance, and QnrA associated with quinolone resistance. These features may be linked to the current global success of NDM-producing K. pneumoniae.
The current spread of OXA-48-producing bacteria is attributed to the pOX-48a plasmid, which belongs to the IncL group of IncL/M type plasmids. Although the pOXA-48a plasmid contains blaOXA-48, a unique antibiotic resistance gene, the conjugation rate of the pOXA-48a plasmid was very high, which may be responsible for its global spread in K. pneumoniae. The high pOXA-48a conjugation rate was recently attributed to mutations in the tir gene known to encode a transfer inhibition protein, which may lead to a 50- to 100-fold increase in the efficiency of transfer of the pOXA-48a plasmid. Furthermore, the pOXA-48a plasmid is self-conjugative. Therefore, these specific features of the pOXA-48a plasmid may explain the global dissemination of OXA-48-type carbapenemases.
ST11, ST14, ST101, ST147, and ST258 are major carbapenemase-producing K. pneumoniae clones. ST258 was mainly found in KPC-producing K. pneumoniae, whereas other clones were found in various carbapenemase-producing K. pneumoniae regardless of carbapenemase types. Well-designed epidemiological and molecular studies will be required to understand the dynamics of transmission, risk factors, and reservoirs of these K. pneumoniae clones. This will provide information essential for preventing infections and the spread of these risky sequence types.
Most currently available antibiotics may be not sufficiently effective for the treatment of all types of carbapenemase producers in monotherapy. Combination therapy of carbapenems with polymyxin B, colistin, rifampin, fosfomycin, or tigecycline has been reported to effectively treat carbapenemase-producing K. pneumoniae. Despite these data supporting the use of combination therapy for treatment of severe carbapenemase-producing K. pneumoniae infections, current clinical evidence for treatment guidelines are limited and more accurate randomized controlled in vivo studies are required. Moreover, considerable caution is required when applying these therapies. For example, temocillin can actively treat against some KPC-producing K. pneumoniae, particularly lower urinary tract infections, and NDM-producing K. pneumoniae is often susceptible to aztreonam. OXA-48-producing K. pneumoniae remain susceptible to the expanded-spectrum cephalosporins in approximately 20% of cases without ESBLs. In the case of NDM-1-producing K. pneumoniae, carbapenems were recently reported to represent a viable treatment option for infections caused by these bacteria, despite unfavorable in vitro MICs. Because these results imply that carbapenems can sometimes be a viable treatment option for infection with carbapenemase producers, more extensive studies on the effect of carbapenem monotherapy will be required in the case of NDM-producing K. pneumoniae infections.
The accurate and rapid detection of the genotype of carbapenemases can minimize the delay to appropriate prescription of antibiotics. Many detection kits based on various phenotypic or molecular techniques, such as multiplex PCR assay, real-time PCR assay, DNA microarray, Raman spectroscopic analysis, single-colony whole-genome sequencing, MALDI-TOF MS, loop-mediated isothermal amplification method, chromogenic medium, and new phenotypic test methods, have been developed. Through the imprudent use of colistin which is a key component used for the treatment of severe carbapenemase-producing K. pneumoniae infections, the rapid spread of colistin resistance was recently reported in K. pneumoniae producing carbapenemases, particularly KPC-type carbapenemases. This situation strongly demonstrates the need for the development of novel accurate and reliable methods for detecting resistance to clinically important antimicrobial agents, such as colistin. Hospital interventions can effectively reduce the spread of carbapenemase-producing K. pneumoniae. Standard infection control guidelines should be implemented upon the detection of carbapenemase-producing K. pneumoniae, and carbapenemase-producing K. pneumoniae positive patients should be individually isolated and treated according to strict standard guidelines.
Author Contributions
C-RL, JL, and SL contributed to the conception and the design of the review and C-RL, JL, KP, YK, BJ, and SL researched and wrote the review.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This review was supported by the National Research Lab Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (No. 2011-0027928); the Cooperative Research Program for Agriculture Science & Technology Development (No. PJ01103103) of Rural Development Administration in South Korea; and the National Research Foundation of the Ministry of Education, South Korea (NRF-2015R1C1A1A02037470).
References
- Abdul Rahim N., Cheah S. E., Johnson M. D., Yu H., Sidjabat H. E., Boyce J., et al. (2015). Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two ‘old’ antibiotics-polymyxin B and chloramphenicol. J. Antimicrob. Chemother. 70 2589–2597. 10.1093/jac/dkv135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Haduch J. M., Potoski B. A., Sidjabat H. E., Paterson D. L., Doi Y. (2009). Activity of temocillin against KPC-producing Klebsiella pneumoniae and Escherichia coli. Antimicrob. Agents Chemother. 53 2700–2701. 10.1128/AAC.00290-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Sapper S., Nolen S., Donzelli G. F., Lal M., Chen K., Justo da Silva L. H., et al. (2015). Rapid induction of high-level carbapenem resistance in heteroresistant KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 59 3281–3289. 10.1128/AAC.05100-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler A., Hussein O., Ben-David D., Masarwa S., Navon-Venezia S., Schwaber M. J., et al. (2015). Persistence of Klebsiella pneumoniae ST258 as the predominant clone of carbapenemase-producing Enterobacteriaceae in post-acute-care hospitals in Israel, 2008-13. J. Antimicrob. Chemother. 70 89–92. 10.1093/jac/dku333 [DOI] [PubMed] [Google Scholar]
- Adler A., Khabra E., Chmelnitsky I., Giakkoupi P., Vatopoulos A., Mathers A. J., et al. (2014). Development and validation of a multiplex PCR assay for identification of the epidemic ST-258/512 KPC-producing Klebsiella pneumoniae clone. Diagn. Microbiol. Infect. Dis. 78 12–15. 10.1016/j.diagmicrobio.2013.10.003 [DOI] [PubMed] [Google Scholar]
- Adler A., Solter E., Masarwa S., Miller-Roll T., Abu-Libdeh B., Khammash H., et al. (2013). Epidemiological and microbiological characteristics of an outbreak caused by OXA-48-producing Enterobacteriaceae in a neonatal intensive care unit in Jerusalem. Israel J. Clin. Microbiol. 51 2926–2930. 10.1128/JCM.01049-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ageevets V. A., Partina I. V., Lisitsyna E. S., Ilina E. N., Lobzin Y. V., Shlyapnikov S. A., et al. (2014). Emergence of carbapenemase-producing Gram-negative bacteria in Saint Petersburg. Russ. Int. J. Antimicrob. Agents 44 152–155. 10.1016/j.ijantimicag.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Ahn C., Butt A. A., Rivera J. I., Yaqoob M., Hag S., Khalil A., et al. (2015). OXA-48-producing Enterobacteriaceae causing bacteremia, United Arab Emirates. Int. J. Infect. Dis. 30 36–37. 10.1016/j.ijid.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Marzooq F., Ngeow Y. F., Tay S. T. (2015). Emergence of Klebsiella pneumoniae producing dual carbapenemases (NDM-1 and OXA-232) and 16S rRNA methylase (armA) isolated from a Malaysian patient returning from India. Int. J. Antimicrob. Agents 45 445–446. 10.1016/j.ijantimicag.2014.12.013 [DOI] [PubMed] [Google Scholar]
- Almeida A. C., de Sa Cavalcanti F. L., Vilela M. A., Gales A. C., de Morais M. A., Jr. (2012). Escherichia coli ST502 and Klebsiella pneumoniae ST11 sharing an IncW plasmid harbouring the blaKPC-2 gene in an intensive care unit patient. Int. J. Antimicrob. Agents 40 374–376. 10.1016/j.ijantimicag.2012.05.022 [DOI] [PubMed] [Google Scholar]
- Anandan S., Damodaran S., Gopi R., Bakthavatchalam Y. D., Veeraraghavan B. (2015). Rapid screening for carbapenem resistant organisms: current results and future approaches. J. Clin. Diagn. Res. 9 DM01–DM03 10.7860/JCDR/2015/14246.6530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana D. M., Saez D., Garcia-Hierro P., Bautista V., Fernandez-Romero S., Angel de la Cal M., et al. (2015). Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clin. Microbiol. Infect. 21 148e141–148.e144. 10.1016/j.cmi.2014.07.008 [DOI] [PubMed] [Google Scholar]
- Arpin C., Noury P., Boraud D., Coulange L., Manetti A., Andre C., et al. (2012). NDM-1-producing Klebsiella pneumoniae resistant to colistin in a French community patient without history of foreign travel. Antimicrob. Agents Chemother. 56 3432–3434. 10.1128/AAC.00230-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert D., Naas T., Heritier C., Poirel L., Nordmann P. (2006). Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of b-lactam resistance genes. J. Bacteriol. 188 6506–6514. 10.1128/JB.00375-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi L., Nordmann P., Lari A. R., Bonnin R. A. (2014). First report of OXA-48-producing Klebsiella pneumoniae strains in Iran. GMS Hyg. Infect. Control. 9:Doc07 10.3205/dgkh000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae I. K., Lee Y. N., Jeong S. H., Hong S. G., Lee J. H., Lee S. H., et al. (2007). Genetic and biochemical characterization of GES-5, an extended-spectrum class A b-lactamase from Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 58 465–468. 10.1016/j.diagmicrobio.2007.02.013 [DOI] [PubMed] [Google Scholar]
- Bakour S., Garcia V., Loucif L., Brunel J. M., Gharout-Sait A., Touati A., et al. (2015a). Rapid identification of carbapenemase-producing Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii using a modified Carba NP test. New Microbes New Infect. 7 89–93. 10.1016/j.nmni.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakour S., Sahli F., Touati A., Rolain J. M. (2015b). Emergence of KPC-producing Klebsiella pneumoniae ST512 isolated from cerebrospinal fluid of a child in Algeria. New Microbes New Infect. 3 34–36. 10.1016/j.nmni.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balm M. N., La M. V., Krishnan P., Jureen R., Lin R. T., Teo J. W. (2013). Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin. Microbiol. Infect. 19 E421–E423. 10.1111/1469-0691.12247 [DOI] [PubMed] [Google Scholar]
- Baraniak A., Grabowska A., Izdebski R., Fiett J., Herda M., Bojarska K., et al. (2011). Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob. Agents Chemother. 55 5493–5499. 10.1128/AAC.05118-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak A., Izdebski R., Fiett J., Gawryszewska I., Bojarska K., Herda M., et al. (2016). NDM-producing Enterobacteriaceae in Poland, 2012-14: inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J. Antimicrob. Chemother. 7 85–91. 10.1093/jac/dkv282 [DOI] [PubMed] [Google Scholar]
- Barguigua A., El Otmani F., El Yaagoubi F. L., Talmi M., Zerouali K., Timinouni M. (2013). First report of a Klebsiella pneumoniae strain coproducing NDM-1, VIM-1 and OXA-48 carbapenemases isolated in Morocco. APMIS 121 675–677. 10.1111/apm.12034 [DOI] [PubMed] [Google Scholar]
- Barrios H., Silva-Sanchez J., Reyna-Flores F., Sanchez-Perez A., Sanchez-Francia D., Aguirre-Torres J. A., et al. (2014). Detection of a NDM-1-producing Klebsiella pneumoniae (ST22) clinical isolate at a pediatric hospital in Mexico. Pediatr. Infect. Dis. J. 33:335 10.1097/INF.0000000000000173 [DOI] [PubMed] [Google Scholar]
- Bathoorn E., Friedrich A. W., Zhou K., Arends J. P., Borst D. M., Grundmann H., et al. (2013). Latent introduction to the Netherlands of multiple antibiotic resistance including NDM-1 after hospitalisation in Egypt, August 2013. Euro. Surveill. 18:20610 10.2807/1560-7917.ES2013.18.42.20610 [DOI] [PubMed] [Google Scholar]
- Bathoorn E., Rossen J. W., Lokate M., Friedrich A. W., Hammerum A. M. (2015). Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro Surveill. 20:30040 10.2807/1560-7917.ES.2015.20.41.30040 [DOI] [PubMed] [Google Scholar]
- Bedenic B., Mazzariol A., Plecko V., Bosnjak Z., Barl P., Vranes J., et al. (2012). First report of KPC-producing Klebsiella pneumoniae in Croatia. J. Chemother. 24 237–239. 10.1179/1973947812Y.0000000017 [DOI] [PubMed] [Google Scholar]
- Ben Nasr A., Decre D., Compain F., Genel N., Barguellil F., Arlet G. (2013). Emergence of NDM-1 in association with OXA-48 in Klebsiella pneumoniae from Tunisia. Antimicrob. Agents Chemother. 57 4089–4090. 10.1128/AAC.00536-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercot B., Poirel L., Dortet L., Nordmann P. (2011). In vitro evaluation of antibiotic synergy for NDM-1-producing Enterobacteriaceae. J. Antimicrob. Chemother. 66 2295–2297. 10.1093/jac/dkr296 [DOI] [PubMed] [Google Scholar]
- Berger S., Alauzet C., Aissa N., Henard S., Rabaud C., Bonnet R., et al. (2013). Characterization of a new blaOXA-48-carrying plasmid in Enterobacteriaceae. Antimicrob. Agents Chemother. 57 4064–4067. 10.1128/AAC.02550-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrazeg M., Diene S., Medjahed L., Parola P., Drissi M., Raoult D., et al. (2014). New Delhi Metallo-b-lactamase around the world: an eReview using Google Maps. Euro. Surveill. 19:20809 10.2807/1560-7917.ES2014.19.20.20809 [DOI] [PubMed] [Google Scholar]
- Beyrouthy R., Robin F., Dabboussi F., Mallat H., Hamze M., Bonnet R. (2014). Carbapenemase and virulence factors of Enterobacteriaceae in North Lebanon between 2008 and 2012: evolution via endemic spread of OXA-48. J. Antimicrob. Chemother. 69 2699–2705. 10.1093/jac/dku181 [DOI] [PubMed] [Google Scholar]
- Birgy A., Doit C., Mariani-Kurkdjian P., Genel N., Faye A., Arlet G., et al. (2011). Early detection of colonization by VIM-1-producing Klebsiella pneumoniae and NDM-1-producing Escherichia coli in two children returning to France. J. Clin. Microbiol. 49 3085–3087. 10.1128/JCM.00540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaerts P., Montesinos I., Rodriguez-Villalobos H., Blairon L., Deplano A., Glupczynski Y. (2010). Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing KPC-2 carbapenemase in Belgium. J. Antimicrob. Chemother. 65 361–362. 10.1093/jac/dkp453 [DOI] [PubMed] [Google Scholar]
- Bogaerts P., Yunus S., Massart M., Huang T. D., Glupczynski Y. (2016). Evaluation of the BYG Carba test, a new electrochemical assay for rapid laboratory detection of carbapenemase-producing enterobacteriaceae. J. Clin. Microbiol. 54 349–358. 10.1128/JCM.02404-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovich T., Adams-Haduch J. M., Tian G. B., Nguyen M. H., Kwak E. J., Muto C. A., et al. (2011). Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin. Infect. Dis. 53 373–376. 10.1093/cid/cir401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonura C., Giuffre M., Aleo A., Fasciana T., Di Bernardo F., Stampone T., et al. (2015). An Update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple Non-ST258 Clones. PLoS ONE 10:e0132936 10.1371/journal.pone.0132936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgia S., Lastovetska O., Richardson D., Eshaghi A., Xiong J., Chung C., et al. (2012). Outbreak of carbapenem-resistant enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin. Infect. Dis. 55 e109–17 10.1093/cid/cis737 [DOI] [PubMed] [Google Scholar]
- Bowers J. R., Kitchel B., Driebe E. M., MacCannell D. R., Roe C., Lemmer D., et al. (2015). Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae Pandemic. PLoS ONE 10:e0133727 10.1371/journal.pone.0133727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branas P., Villa J., Viedma E., Mingorance J., Orellana M. A., Chaves F. (2015). Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital in madrid: successful establishment of an OXA-48 ST11 clone. Int. J. Antimicrob. Agents 46 111–116. 10.1016/j.ijantimicag.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Braun S. D., Monecke S., Thurmer A., Ruppelt A., Makarewicz O., Pletz M., et al. (2014). Rapid identification of carbapenemase genes in gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS ONE 9:e102232 10.1371/journal.pone.0102232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink A. J., Coetzee J., Clay C. G., Sithole S., Richards G. A., Poirel L., et al. (2012). Emergence of New Delhi metallo-b-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J. Clin. Microbiol. 50 525–527. 10.1128/JCM.05956-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink A. J., Coetzee J., Corcoran C., Clay C. G., Hari-Makkan D., Jacobson R. K., et al. (2013). Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J. Clin. Microbiol. 51 369–372. 10.1128/JCM.02234-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. D., Vivas E. I., Walsh C. T., Kolter R. (1995). MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 177 4194–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A., D’Andrea M. M., Giani T., Di Pilato V., Arena F., Ambretti S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57 5521–5526. 10.1128/AAC.01480-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A., Di Pilato V., Giani T., Arena F., Ambretti S., Gaibani P., et al. (2014a). In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob. Agents Chemother. 58 4399–4403. 10.1128/AAC.02555-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A., Giani T., D’Andrea M. M., Di Pilato V., Arena F., Conte V., et al. (2014b). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58 5696–5703. 10.1128/AAC.03110-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Seiffert S. N., Schwendener S., Perreten V., Endimiani A. (2015). Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS ONE 10:e0123063 10.1371/journal.pone.0123063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonne A., Thiolet J. M., Fournier S., Fortineau N., Kassis-Chikhani N., Boytchev I., et al. (2010). Control of a multi-hospital outbreak of KPC-producing Klebsiella pneumoniae type 2 in France, September to October 2009. Euro. Surveill 15 19734. [DOI] [PubMed] [Google Scholar]
- Carricajo A., Verhoeven P. O., Guezzou S., Fonsale N., Aubert G. (2014). Detection of carbapenemase-producing bacteria by using an ultra-performance liquid chromatography-tandem mass spectrometry method. Antimicrob. Agents Chemother. 58 1231–1234. 10.1128/AAC.01540-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhaes C. G., Cayo R., Visconde M. F., Barone T., Frigatto E. A., Okamoto D., et al. (2014). Detection of carbapenemase activity directly from blood culture vials using MALDI-TOF MS: a quick answer for the right decision. J. Antimicrob. Chemother. 69 2132–2136. 10.1093/jac/dku094 [DOI] [PubMed] [Google Scholar]
- Castanheira M., Deshpande L. M., Mathai D., Bell J. M., Jones R. N., Mendes R. E. (2011). Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob. Agents Chemother. 55 1274–1278. 10.1128/AAC.01497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2014). Notes from the Field: New Delhi metallo-β-lactamase-producing Escherichia coli associated with endoscopic retrograde cholangiopancreatography - Illinois, 2013. MMWR Morb. Mortal. Wkly. Rep. 62:1051. [PMC free article] [PubMed] [Google Scholar]
- Chagas T. P., Seki L. M., da Silva D. M., Asensi M. D. (2011). Occurrence of KPC-2-producing Klebsiella pneumoniae strains in hospital wastewater. J. Hosp. Infect. 77:281 10.1016/j.jhin.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Chang Y. Y., Chuang Y. C., Siu L. K., Wu T. L., Lin J. C., Lu P. L., et al. (2015). Clinical features of patients with carbapenem nonsusceptible Klebsiella pneumoniae and Escherichia coli in intensive care units: a nationwide multicenter study in Taiwan. J. Microbiol. Immunol. Infect. 48 219–225. 10.1016/j.jmii.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Chen C. J., Wu T. L., Lu P. L., Chen Y. T., Fung C. P., Chuang Y. C., et al. (2014a). Closely related NDM-1-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. PLoS ONE 9:e104899 10.1371/journal.pone.0104899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chavda K. D., Al Laham N., Melano R. G., Jacobs M. R., Bonomo R. A., et al. (2013a). Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 57 5019–5025. 10.1128/AAC.01397-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chavda K. D., Findlay J., Peirano G., Hopkins K., Pitout J. D., et al. (2014b). Multiplex PCR for identification of two capsular types in epidemic KPC-producing Klebsiella pneumoniae sequence type 258 strains. Antimicrob. Agents Chemother. 58 4196–4199. 10.1128/AAC.02673-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chavda K. D., Fraimow H. S., Mediavilla J. R., Melano R. G., Jacobs M. R., et al. (2013b). Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57 269–276. 10.1128/AAC.01648-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chavda K. D., Melano R. G., Hong T., Rojtman A. D., Jacobs M. R., et al. (2014c). Molecular survey of the dissemination of two blaKPC-harboring IncFIA plasmids in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 58 2289–2294. 10.1128/AAC.02749-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chavda K. D., Melano R. G., Jacobs M. R., Koll B., Hong T., et al. (2014d). Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob. Agents Chemother. 58 2871–2877. 10.1128/AAC.00120-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Mathema B., Chavda K. D., DeLeo F. R., Bonomo R. A., Kreiswirth B. N. (2014e). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22 686–696. 10.1016/j.tim.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Mathema B., Pitout J. D., DeLeo F. R., Kreiswirth B. N. (2014f). Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio. 5 e1355–14 10.1128/mBio.01355-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. T., Lin A. C., Siu L. K., Koh T. H. (2012). Sequence of closely related plasmids encoding blaNDM-1 in two unrelated Klebsiella pneumoniae isolates in Singapore. PLoS ONE 7:e48737 10.1371/journal.pone.0048737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. T., Lin J. C., Fung C. P., Lu P. L., Chuang Y. C., Wu T. L., et al. (2014g). KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J. Antimicrob. Chemother. 69 628–631. 10.1093/jac/dkt409 [DOI] [PubMed] [Google Scholar]
- Chen Z., Wang Y., Tian L., Zhu X., Li L., Zhang B., et al. (2015). First report in China of Enterobacteriaceae clinical isolates coharboring blaNDM-1 and blaIMP-4 drug resistance genes. Microb. Drug Resist. 21 167–170. 10.1089/mdr.2014.0087 [DOI] [PubMed] [Google Scholar]
- Chereau F., Herindrainy P., Garin B., Huynh B. T., Randrianirina F., Padget M., et al. (2015). Colonization of extended-spectrum-β-lactamase- and NDM-1-producing Enterobacteriaceae among pregnant women in the community in a low-income country: a potential reservoir for transmission of multiresistant Enterobacteriaceae to neonates. Antimicrob. Agents Chemother. 59 3652–3655. 10.1128/AAC.00029-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang T. T., Yang Y. S., Yeh K. M., Chiu S. K., Wang N. C., Lin T. Y., et al. (2016). Quantification and comparison of virulence and characteristics of different variants of carbapenemase-producing Klebsiella pneumoniae clinical isolates from Taiwan and the United States. J. Microbiol. Immunol. Infect. 49 83–90. 10.1016/j.jmii.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Chiu S. K., Wu T. L., Chuang Y. C., Lin J. C., Fung C. P., Lu P. L., et al. (2013). National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS ONE 8:e69428 10.1371/journal.pone.0069428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. Y., Huh H. J., Baek J. Y., Chung N. Y., Ryu J. G., Ki C. S., et al. (2015). Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg. Infect. Dis. 21 1088–1089. 10.3201/eid2106.150048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong P. M., McCorrister S. J., Unger M. S., Boyd D. A., Mulvey M. R., Westmacott G. R. (2015). MALDI-TOF MS detection of carbapenemase activity in clinical isolates of Enterobacteriaceae spp., Pseudomonas aeruginosa, and Acinetobacter baumannii compared against the Carba-NP assay. J. Microbiol. Methods 111 21–23. 10.1016/j.mimet.2015.01.024 [DOI] [PubMed] [Google Scholar]
- Chung The H., Karkey A., Pham Thanh D., Boinett C. J., Cain A. K., Ellington M., et al. (2015). A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol. Med. 7 227–239. 10.15252/emmm.201404767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero M., Cuervo G., Dominguez M. A., Tubau F., Marti S., Sevillano E., et al. (2015). Carbapenem-resistant and carbapenem-susceptible isogenic isolates of Klebsiella pneumoniae ST101 causing infection in a tertiary hospital. BMC Microbiol. 15:177 10.1186/s12866-015-0510-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzon G., Bentchouala C., Vogel A., Hery M., Lezzar A., Smati F., et al. (2015). First outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Constantine, Algeria. Int. J. Antimicrob. Agents 46 725–727. 10.1016/j.ijantimicag.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Cuzon G., Naas T., Bogaerts P., Glupczynski Y., Huang T. D., Nordmann P. (2008). Plasmid-encoded carbapenem-hydrolyzing β-lactamase OXA-48 in an imipenem-susceptible Klebsiella pneumoniae strain from Belgium. Antimicrob. Agents Chemother. 52 3463–3464. 10.1128/AAC.00543-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafopoulou K., Zarkotou O., Dimitroulia E., Hadjichristodoulou C., Gennimata V., Pournaras S., et al. (2015). Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 59 4625–4630. 10.1128/AAC.00868-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikos G. L., Tsaousi S., Tzouvelekis L. S., Anyfantis I., Psichogiou M., Argyropoulou A., et al. (2014). Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob. Agents Chemother. 58 2322–2328. 10.1128/AAC.02166-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash N., Panigrahi D., Zarouni M. A., Darwish D., Ghazawi A., Sonnevend A., et al. (2014). High incidence of New Delhi metallo-β-lactamase-producing Klebsiella pneumoniae isolates in Sharjah, United Arab Emirates. Microb. Drug Resist. 20 52–56. 10.1089/mdr.2013.0040 [DOI] [PubMed] [Google Scholar]
- Datta S., Roy S., Chatterjee S., Saha A., Sen B., Pal T., et al. (2014). A five-year experience of carbapenem resistance in Enterobacteriaceae causing neonatal septicaemia: predominance of NDM-1. PLoS ONE 9:e112101 10.1371/journal.pone.0112101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautzenberg M. J., Ossewaarde J. M., de Kraker M. E., van der Zee A., van Burgh S., de Greeff S. C., et al. (2014). Successful control of a hospital-wide outbreak of OXA-48 producing Enterobacteriaceae in the Netherlands, 2009 to 2011. Euro. Surveill. 19:20723 10.2807/1560-7917.ES2014.19.9.20723 [DOI] [PubMed] [Google Scholar]
- de Jager P., Chirwa T., Naidoo S., Perovic O., Thomas J. (2015). Nosocomial outbreak of New Delhi Metallo-β-Lactamase-1-Producing Gram-Negative BACTERIA in South Africa: a case-control study. PLoS ONE 10:e0123337 10.1371/journal.pone.0123337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleo F. R., Chen L., Porcella S. F., Martens C. A., Kobayashi S. D., Porter A. R., et al. (2014). Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 111 4988–4993. 10.1073/pnas.1321364111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira M. S., de Assis D. B., Freire M. P., Boas do Prado G. V., Machado A. S., Abdala E., et al. (2015). Treatment of KPC-producing Enterobacteriaceae: suboptimal efficacy of polymyxins. Clin. Microbiol. Infect. 21 179.e1–179.e7 10.1016/j.cmi.2014.07.010 [DOI] [PubMed] [Google Scholar]
- Deshpande L. M., Rhomberg P. R., Sader H. S., Jones R. N. (2006). Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States medical centers: report from the MYSTIC Program (1999-2005). Diagn. Microbiol. Infect. Dis. 56 367–372. 10.1016/j.diagmicrobio.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Dimou V., Dhanji H., Pike R., Livermore D. M., Woodford N. (2012). Characterization of Enterobacteriaceae producing OXA-48-like carbapenemases in the UK. J. Antimicrob. Chemother. 67 1660–1665. 10.1093/jac/dks124 [DOI] [PubMed] [Google Scholar]
- Doi Y., O’Hara J. A., Lando J. F., Querry A. M., Townsend B. M., Pasculle A. W., et al. (2014). Co-production of NDM-1 and OXA-232 by Klebsiella pneumoniae. Emerg. Infect. Dis. 20 163–165. 10.3201/eid2001.130904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D., Liu W., Li H., Wang Y., Li X., Zou D., et al. (2015). Survey and rapid detection of Klebsiella pneumoniae in clinical samples targeting the rcsA gene in Beijing, China. Front. Microbiol. 6:519 10.3389/fmicb.2015.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L., Agathine A., Naas T., Cuzon G., Poirel L., Nordmann P. (2015). Evaluation of the RAPIDECR CARBA NP, the Rapid CARB ScreenR and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 70 3014–3022. 10.1093/jac/dkv213 [DOI] [PubMed] [Google Scholar]
- Dortet L., Brechard L., Grenet K., Nguessan M. S., Nordmann P. (2013). Sri Lanka, another country from the Indian subcontinent with NDM-1-producing Enterobacteriaceae. J. Antimicrob. Chemother. 68 2172–2173. 10.1093/jac/dkt145 [DOI] [PubMed] [Google Scholar]
- Dortet L., Brechard L., Poirel L., Nordmann P. (2014a). Rapid detection of carbapenemase-producing Enterobacteriaceae from blood cultures. Clin. Microbiol. Infect. 20 340–344. 10.1111/1469-0691.12318 [DOI] [PubMed] [Google Scholar]
- Dortet L., Poirel L., Errera C., Nordmann P. (2014b). CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J. Clin. Microbiol. 52 2359–2364. 10.1128/JCM.00594-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L., Poirel L., Nordmann P. (2014c). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res. Int. 2014 249856 10.1155/2014/249856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducomble T., Faucheux S., Helbig U., Kaisers U. X., Konig B., Knaust A., et al. (2015). Large hospital outbreak of KPC-2-producing Klebsiella pneumoniae: investigating mortality and the impact of screening for KPC-2 with polymerase chain reaction. J. Hosp. Infect. 89 179–185. 10.1016/j.jhin.2014.11.012 [DOI] [PubMed] [Google Scholar]
- Ellis C., Chung C., Tijet N., Patel S. N., Desjardins M., Melano R. G., et al. (2013). OXA-48-like carbapenemase-producing Enterobacteriaceae in Ottawa. Canada. Diagn. Microbiol. Infect. Dis. 76 399–400. 10.1016/j.diagmicrobio.2013.04.017 [DOI] [PubMed] [Google Scholar]
- Escobar Perez J. A., Olarte Escobar N. M., Castro-Cardozo B., Valderrama Marquez I. A., Garzon Aguilar M. I., Martinez de la Barrera L., et al. (2013). Outbreak of NDM-1-producing Klebsiella pneumoniae in a neonatal unit in Colombia. Antimicrob. Agents Chemother. 57 1957–1960. 10.1128/AAC.01447-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espedido B. A., Steen J. A., Ziochos H., Grimmond S. M., Cooper M. A., Gosbell I. B., et al. (2013). Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution. PLoS ONE 8:e59920 10.1371/journal.pone.0059920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B. A., Amyes S. G. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27 241–263. 10.1128/CMR.00117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evren E., Azap O. K., Colakoǧlu S., Arslan H. (2013). In vitro activity of fosfomycin in combination with imipenem, meropenem, colistin and tigecycline against OXA 48-positive Klebsiella pneumoniae strains. Diagn. Microbiol. Infect. Dis. 76 335–338. 10.1016/j.diagmicrobio.2013.04.004 [DOI] [PubMed] [Google Scholar]
- Fehlberg L. C., Carvalho A. M., Campana E. H., Gontijo-Filho P. P., Gales A. C. (2012). Emergence of Klebsiella pneumoniae-producing KPC-2 carbapenemase in Paraiba, Northeastern Brazil. Braz. J. Infect. Dis. 16 577–580. 10.1016/j.bjid.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Fursova N. K., Astashkin E. I., Knyazeva A. I., Kartsev N. N., Leonova E. S., Ershova O. N., et al. (2015). The spread of blaOXA-48 and blaOXA-244 carbapenemase genes among Klebsiella pneumoniae, Proteus mirabilis and Enterobacter spp. isolated in Moscow, Russia. Ann. Clin. Microbiol. Antimicrob. 14:46 10.1186/s12941-015-0108-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaibani P., Ambretti S., Berlingeri A., Cordovana M., Farruggia P., Panico M., et al. (2011). Outbreak of NDM-1-producing Enterobacteriaceae in northern Italy, July to August 2011. Euro. Surveill. 16:20027. [PubMed] [Google Scholar]
- Gales A. C., Castanheira M., Jones R. N., Sader H. S. (2012). Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008-2010). Diagn. Microbiol. Infect. Dis. 73 354–360. 10.1016/j.diagmicrobio.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Gallagher L. C., Roundtree S. S., Lancaster D. P., Rudin S. D., Bard J. D., Roberts A. L., et al. (2015). Performance of the CLSI Carba NP and the rosco carb screen assays Using North American Carbapenemase-Producing Enterobacteriaceae and Pseudomonas aeruginosa isolates. J. Clin. Microbiol. 53 3370–3373. 10.1128/JCM.01547-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galler H., Feierl G., Petternel C., Reinthaler F. F., Haas D., Grisold A. J., et al. (2014). KPC-2 and OXA-48 carbapenemase-harbouring Enterobacteriaceae detected in an Austrian wastewater treatment plant. Clin. Microbiol. Infect. 20 O132–O134. 10.1111/1469-0691.12336 [DOI] [PubMed] [Google Scholar]
- Garbari L., Busetti M., Dolzani L., Petix V., Knezevich A., Bressan R., et al. (2015). pKBuS13, a KPC-2-encoding plasmid from Klebsiella pneumoniae sequence type 833 carrying Tn4401b inserted into an Xer site-specific recombination locus. Antimicrob. Agents Chemother. 59 5226–5231. 10.1128/AAC.04543-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez A., Villa L., Carta C., Venditti C., Giordano A., Venditti M., et al. (2012). Klebsiella pneumoniae ST258 producing KPC-3 identified in italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56 2143–2145. 10.1128/AAC.05308-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez S., Morosini M. I., Gijon D., Beatobe L., Ruiz-Garbajosa P., Dominguez L., et al. (2016). Detection of carbapenemase production in a collection of Enterobacteriaceae with characterized resistance mechanisms from clinical and environmental origins by use of both Carba NP and Blue-Carba Tests. J. Clin. Microbiol. 54 464–466. 10.1128/JCM.02580-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Ramos U., Barrios H., Reyna-Flores F., Sanchez-Perez A., Tamayo-Legorreta E., Ibarra-Pacheco A., et al. (2014). Characteristics of KPC-2-producing Klebsiella pneumoniae (ST258) clinical isolates from outbreaks in 2 Mexican medical centers. Diagn. Microbiol. Infect. Dis. 79 483–485. 10.1016/j.diagmicrobio.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Gharout-Sait A., Alsharapy S. A., Brasme L., Touati A., Kermas R., Bakour S., et al. (2014). Enterobacteriaceae isolates carrying the New Delhi metallo-β-lactamase gene in Yemen. J. Med. Microbiol. 63 (Pt 8), 1316–1323. 10.1099/jmm.0.073767-0 [DOI] [PubMed] [Google Scholar]
- Giacobbe D. R., Del Bono V., Trecarichi E. M., De Rosa F. G., Giannella M., Bassetti M., et al. (2015). Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case-control-control study. Clin. Microbiol. Infect. 21 1106.e1–1106.e8 10.1016/j.cmi.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Giakkoupi P., Pappa O., Polemis M., Vatopoulos A. C., Miriagou V., Zioga A., et al. (2009). Emerging Klebsiella pneumoniae isolates coproducing KPC-2 and VIM-1 carbapenemases. Antimicrob. Agents Chemother. 53 4048–4050. 10.1128/AAC.00690-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellou H., Galani L., Baziaka F., Karaiskos I. (2013). Effectiveness of a double-carbapenem regimen for infections in humans due to carbapenemase-producing pandrug-resistant Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57 2388–2390. 10.1128/AAC.02399-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani T., Arena F., Vaggelli G., Conte V., Chiarelli A., Henrici De Angelis L., et al. (2015). Large nosocomial outbreak of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae traced to clonal expansion of an mgrB deletion mutant. J. Clin. Microbiol. 53 3341–3344. 10.1128/JCM.01017-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani T., Conte V., Mandala S., D’Andrea M. M., Luzzaro F., Conaldi P. G., et al. (2014). Cross-infection of solid organ transplant recipients by a multidrug-resistant Klebsiella pneumoniae isolate producing the OXA-48 carbapenemase, likely derived from a multiorgan donor. J. Clin. Microbiol. 52 2702–2705. 10.1128/JCM.00511-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani T., D’Andrea M. M., Pecile P., Borgianni L., Nicoletti P., Tonelli F., et al. (2009). Emergence in Italy of Klebsiella pneumoniae sequence type 258 producing KPC-3 carbapenemase. J. Clin. Microbiol. 47 3793–3794. 10.1128/JCM.01773-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani T., Pini B., Arena F., Conte V., Bracco S., Migliavacca R., et al. (2013). Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro. Surveill. 18:20489. [PubMed] [Google Scholar]
- Girlich D., Anglade C., Zambardi G., Nordmann P. (2013). Comparative evaluation of a novel chromogenic medium (chromID OXA-48) for detection of OXA-48 producing Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 77 296–300. 10.1016/j.diagmicrobio.2013.08.015 [DOI] [PubMed] [Google Scholar]
- Giske C. G. (2015). Contemporary resistance trends and mechanisms for the old antibiotics colistin, temocillin, fosfomycin, mecillinam and nitrofurantoin. Clin. Microbiol. Infect. 21 899–905. 10.1016/j.cmi.2015.05.022 [DOI] [PubMed] [Google Scholar]
- Giske C. G., Froding I., Hasan C. M., Turlej-Rogacka A., Toleman M., Livermore D., et al. (2012). Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob. Agents Chemother. 56 2735–2738. 10.1128/AAC.06142-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giske C. G., Gezelius L., Samuelsen O., Warner M., Sundsfjord A., Woodford N. (2011). A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin. Microbiol. Infect. 17 552–556. 10.1111/j.1469-0691.2010.03294.x [DOI] [PubMed] [Google Scholar]
- Goldfarb D., Harvey S. B., Jessamine K., Jessamine P., Toye B., Desjardins M. (2009). Detection of plasmid-mediated KPC-producing Klebsiella pneumoniae in Ottawa, Canada: evidence of intrahospital transmission. J. Clin. Microbiol. 47 1920–1922. 10.1128/JCM.00098-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez S., Pasteran F., Faccone D., Bettiol M., Veliz O., De Belder D., et al. (2013). Intrapatient emergence of OXA-247: a novel carbapenemase found in a patient previously infected with OXA-163-producing Klebsiella pneumoniae. Clin. Microbiol. Infect. 19 E233–E235. 10.1111/1469-0691.12142 [DOI] [PubMed] [Google Scholar]
- Gomez S. A., Pasteran F. G., Faccone D., Tijet N., Rapoport M., Lucero C., et al. (2011). Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin. Microbiol. Infect. 17 1520–1524. 10.1111/j.1469-0691.2011.03600.x [DOI] [PubMed] [Google Scholar]
- Gona F., Barbera F., Pasquariello A. C., Grossi P., Gridelli B., Mezzatesta M. L., et al. (2014). In vivo multiclonal transfer of blaKPC-3 from Klebsiella pneumoniae to Escherichia coli in surgery patients. Clin. Microbiol. Infect. 20 O633–O635. 10.1111/1469-0691.12577 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Padilla M., Torre-Cisneros J., Rivera-Espinar F., Pontes-Moreno A., Lopez-Cerero L., Pascual A., et al. (2015). Gentamicin therapy for sepsis due to carbapenem-resistant and colistin-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 70 905–913. 10.1093/jac/dku432 [DOI] [PubMed] [Google Scholar]
- Gootz T. D., Lescoe M. K., Dib-Hajj F., Dougherty B. A., He W., Della-Latta P., et al. (2009). Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53 1998–2004. 10.1128/AAC.01355-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottig S., Gruber T. M., Stecher B., Wichelhaus T. A., Kempf V. A. (2015). In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin. Infect. Dis. 60 1808–1815. 10.1093/cid/civ191 [DOI] [PubMed] [Google Scholar]
- Gregory C. J., Llata E., Stine N., Gould C., Santiago L. M., Vazquez G. J., et al. (2010). Outbreak of carbapenem-resistant Klebsiella pneumoniae in Puerto Rico associated with a novel carbapenemase variant. Infect. Control Hosp. Epidemiol. 31 476–484. 10.1086/651670 [DOI] [PubMed] [Google Scholar]
- Hall J. M., Corea E., Sanjeewani H. D., Inglis T. J. (2014). Molecular mechanisms of β-lactam resistance in carbapenemase-producing Klebsiella pneumoniae from Sri Lanka. J. Med. Microbiol. 63(Pt 8), 1087–1092. 10.1099/jmm.0.076760-0 [DOI] [PubMed] [Google Scholar]
- Hamzan N. I., Yean C. Y., Rahman R. A., Hasan H., Rahman Z. A. (2015). Detection of blaIMP4 and blaNDM1 harboring Klebsiella pneumoniae isolates in a university hospital in Malaysia. Emerg. Health Threats J. 8 26011 10.3402/ehtj.v8.26011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto A., Nagamatsu M., Ohmagari N., Hayakawa K., Kato Y., Kirikae T. (2014). Isolation of OXA-48 carbapenemase-producing Klebsiella pneumoniae ST101 from an overseas traveler returning to Japan. Jpn. J. Infect. Dis. 67 120–121. 10.7883/yoken.67.120 [DOI] [PubMed] [Google Scholar]
- Hidalgo-Grass C., Warburg G., Temper V., Benenson S., Moses A. E., Block C., et al. (2012). KPC-9, a novel carbapenemase from clinical specimens in Israel. Antimicrob. Agents Chemother. 56 6057–6059. 10.1128/AAC.01156-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishinuma A., Yoshida A., Suzuki H., Okuzumi K., Ishida T. (2013). Complete sequencing of an IncFII NDM-1 plasmid in Klebsiella pneumoniae shows structural features shared with other multidrug resistance plasmids. J. Antimicrob. Chemother. 68 2415–2417. 10.1093/jac/dkt190 [DOI] [PubMed] [Google Scholar]
- Hoang T. H., Wertheim H., Minh N. B., Duong T. N., Anh D. D., Phuong T. T., et al. (2013). Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae strains containing New Delhi metallo-β-lactamase isolated from two patients in Vietnam. J. Clin. Microbiol. 51 373–374. 10.1128/JCM.02322-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach M., von Gunten B., Castelberg C., Bloemberg G. V. (2015). Evaluation of the rapidec carba NP Test for detection of carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 53 3828–3833. 10.1128/JCM.02327-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. H., Clancy C. J., Cheng S., Shields R. K., Chen L., Doi Y., et al. (2013). Characterization of porin expression in Klebsiella pneumoniae Carbapenemase (KPC)-producing K. pneumoniae identifies isolates most susceptible to the combination of colistin and carbapenems. Antimicrob. Agents Chemother. 57 2147–2153. 10.1128/AAC.02411-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak J., Chudackova E., Walkova R. (2013a). Matrix-assisted laser desorption ionization-time of flight (maldi-tof) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin. Microbiol. Rev. 26 103–114. 10.1128/CMR.00058-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak J., Papagiannitsis C. C., Studentova V., Jakubu V., Fridrichova M., Zemlickova H., et al. (2013b). Carbapenemase-producing Klebsiella pneumoniae in the Czech Republic in 2011. Euro. Surveill. 18:20626 10.2807/1560-7917.ES2013.18.45.20626 [DOI] [PubMed] [Google Scholar]
- Hrabak J., Studentova V., Jakubu V., Adamkova V., Dvorakova L., Balejova M., et al. (2015). Prevalence study on carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolates in Czech hospitals–results from Czech Part of European Survey on Carbapenemase–Producing Enterobacteriaceae (EuSCAPE). Epidemiol. Mikrobiol. Imunol. 64 87–91. [PubMed] [Google Scholar]
- Hrabak J., Walkova R., Studentova V., Chudackova E., Bergerova T. (2011). Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49 3222–3227. 10.1128/JCM.00984-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Zhong Q., Shang Y., Wang H., Ning C., Li Y., et al. (2014). The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol. Infect. 142 1972–1977. 10.1017/S0950268813002975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Zhong Q., Tu J., Xu Y., Qin Z., Parsons C., et al. (2013). Emergence of blaNDM-1 among Klebsiella pneumoniae ST15 and novel ST1031 clinical isolates in China. Diagn. Microbiol. Infect. Dis. 75 373–376. 10.1016/j.diagmicrobio.2013.01.006 [DOI] [PubMed] [Google Scholar]
- Huang T. D., Berhin C., Bogaerts P., Glupczynski Y. (2014). Comparative evaluation of two chromogenic tests for rapid detection of carbapenemase in Enterobacteriaceae and in Pseudomonas aeruginosa isolates. J. Clin. Microbiol. 52 3060–3063. 10.1128/JCM.00643-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. W., Chen T. L., Chen Y. T., Lauderdale T. L., Liao T. L., Lee Y. T., et al. (2013). Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS ONE 8:e62774 10.1371/journal.pone.0062774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C. M., Bent Z. W., Meagher R. J., Williams K. P. (2014). Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS ONE 9:e99209 10.1371/journal.pone.0099209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries R. M. (2015). Susceptibility testing of the polymyxins: where are we now? Pharmacotherapy 35 22–27. 10.1002/phar.1505 [DOI] [PubMed] [Google Scholar]
- Izdebski R., Bojarska K., Baraniak A., Literacka E., Herda M., Zabicka D., et al. (2015). NDM-1- or OXA-48-producing Enterobacteriaceae colonising Polish tourists following a terrorist attack in Tunis, March 2015. Euro. Surveill. 20:20489 10.2807/1560-7917.ES2015.20.23.21150 [DOI] [PubMed] [Google Scholar]
- Jamal W., Rotimi V. O., Albert M. J., Khodakhast F., Nordmann P., Poirel L. (2013). High prevalence of VIM-4 and NDM-1 metallo-β-lactamase among carbapenem-resistant Enterobacteriaceae. J. Med. Microbiol. 62 1239–1244. 10.1099/jmm.0.059915-0 [DOI] [PubMed] [Google Scholar]
- Jamal W., Rotimi V. O., Albert M. J., Khodakhast F., Udo E. E., Poirel L. (2012). Emergence of nosocomial New Delhi metallo-β-lactamase-1 (NDM-1)-producing Klebsiella pneumoniae in patients admitted to a tertiary care hospital in Kuwait. Int. J. Antimicrob. Agents 39 183–184. 10.1016/j.ijantimicag.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Janvari L., Damjanova I., Lazar A., Racz K., Kocsis B., Urban E., et al. (2014). Emergence of OXA-162-producing Klebsiella pneumoniae in Hungary. Scand. J. Infect. Dis. 46 320–324. 10.3109/00365548.2013.879993 [DOI] [PubMed] [Google Scholar]
- Jayol A., Poirel L., Brink A., Villegas M. V., Yilmaz M., Nordmann P. (2014). Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 58 4762–4766. 10.1128/AAC.00084-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J. H., Hong M. K., Lee J. H., Lee J. J., Park K. S., Karim A. M., et al. (2014). Structure of ADC-68, a novel carbapenem-hydrolyzing class C extended-spectrum b-lactamase isolated from Acinetobacter baumannii. Acta Crystallogr. D Biol. Crystallogr. 70 2924–2936. 10.1107/S1399004714019543 [DOI] [PubMed] [Google Scholar]
- Jeon J. H., Lee J. H., Lee J. J., Park K. S., Karim A. M., Lee C. R., et al. (2015). Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int. J. Mol. Sci. 16 9654–9692. 10.3390/ijms16059654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S. H., Bae I. K., Kim D., Hong S. G., Song J. S., Lee J. H., et al. (2005). First outbreak of Klebsiella pneumoniae clinical isolates producing GES-5 and SHV-12 extended-spectrum β-lactamases in Korea. Antimicrob. Agents Chemother. 49 4809–4810. 10.1128/AAC.49.11.4809-4810.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S. H., Lee K. M., Lee J., Bae I. K., Kim J. S., Kim H. S., et al. (2015). Clonal and horizontal spread of the blaOXA-232 gene among Enterobacteriaceae in a Korean hospital. Diagn. Microbiol. Infect. Dis. 82 70–72. 10.1016/j.diagmicrobio.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Shen P., Wei Z., Liu L., He F., Shi K., et al. (2015). Dissemination of a clone carrying a fosA3-harbouring plasmid mediates high fosfomycin resistance rate of KPC-producing Klebsiella pneumoniae in China. Int. J. Antimicrob. Agents 45 66–70. 10.1016/j.ijantimicag.2014.08.010 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Yu D., Wei Z., Shen P., Zhou Z., Yu Y. (2010). Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob. Agents Chemother. 54 3967–3969. 10.1128/AAC.00137-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Shao C., Li J., Fan H., Bai Y., Wang Y. (2015). Outbreak of multidrug resistant NDM-1-producing Klebsiella pneumoniae from a neonatal unit in Shandong Province, China. PLoS ONE 10:e0119571 10.1371/journal.pone.0119571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir M. H., Meunier D., Hopkins K. L., Giske C. G., Woodford N. (2016). A two-centre evaluation of RAPIDECR CARBA NP for carbapenemase detection in Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter spp. J. Antimicrob. Chemother. 71 1213–1216. 10.1093/jac/dkv468 [DOI] [PubMed] [Google Scholar]
- Kaiser R. M., Castanheira M., Jones R. N., Tenover F., Lynfield R. (2013). Trends in Klebsiella pneumoniae carbapenemase-positive K. pneumoniae in US hospitals: report from the 2007-2009 SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 76 356–360. 10.1016/j.diagmicrobio.2013.03.032 [DOI] [PubMed] [Google Scholar]
- Kalpoe J. S., Al Naiemi N., Poirel L., Nordmann P. (2011). Detection of an ambler class D OXA-48-type β-lactamase in a Klebsiella pneumoniae strain in the netherlands. J. Med. Microbiol. 60 677–678. 10.1099/jmm.0.028308-0 [DOI] [PubMed] [Google Scholar]
- Kanerva M., Skogberg K., Ryynanen K., Pahkamaki A., Jalava J., Ollgren J., et al. (2015). Coincidental detection of the first outbreak of carbapenemase-producing Klebsiella pneumoniae colonisation in a primary care hospital, Finland, 2013. Euro. Surveill. 20:21172 10.2807/1560-7917.ES2015.20.26.21172 [DOI] [PubMed] [Google Scholar]
- Karaiskos I., Giamarellou H. (2014). Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Exp. Opin. Pharmacother. 15 1351–1370. 10.1517/14656566.2014.914172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayama S., Koba Y., Shigemoto N., Kuwahara R., Kakuhama T., Kimura K., et al. (2015). Imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae producing OXA-181 in Japan. Antimicrob. Agents Chemother. 59 1379–1380. 10.1128/AAC.04330-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi M., Drego L., Nikam C., Ajbani K., Soman R., Shetty A., et al. (2015). Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur. J. Clin. Microbiol. Infect. Dis. 34 467–472. 10.1007/s10096-014-2249-x [DOI] [PubMed] [Google Scholar]
- Kilic A., Baysallar M. (2015). The First Klebsiella pneumoniae isolate Co-Producing OXA-48 and NDM-1 in Turkey. Ann. Lab. Med. 35 382–383. 10.3343/alm.2015.35.3.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Jung H. I., An Y. J., Lee J. H., Kim S. J., Jeong S. H., et al. (2006). Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase. Mol. Microbiol. 60 907–916. 10.1111/j.1365-2958.2006.05146.x [DOI] [PubMed] [Google Scholar]
- Kim M. N., Yong D., An D., Chung H. S., Woo J. H., Lee K., et al. (2012). Nosocomial clustering of NDM-1-producing Klebsiella pneumoniae sequence type 340 strains in four patients at a South Korean tertiary care hospital. J. Clin. Microbiol. 50 1433–1436. 10.1128/JCM.06855-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsis E., Guzvinec M., Butic I., Kresic S., Crnek S. S., Tambic A., et al. (2016). bla Carriage on IncR Plasmid in Enterobacteriaceae Strains. Microb. Drug Resist. 22 123–128. 10.1089/mdr.2015.0083 [DOI] [PubMed] [Google Scholar]
- Kola A., Piening B., Pape U. F., Veltzke-Schlieker W., Kaase M., Geffers C., et al. (2015). An outbreak of carbapenem-resistant OXA-48 - producing Klebsiella pneumoniae associated to duodenoscopy. Antimicrob. Resist. Infect. Control. 4:8 10.1186/s13756-015-0049-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontopoulou K., Protonotariou E., Vasilakos K., Kriti M., Koteli A., Antoniadou E., et al. (2010). Hospital outbreak caused by Klebsiella pneumoniae producing KPC-2 β-lactamase resistant to colistin. J. Hosp. Infect. 76 70–73. 10.1016/j.jhin.2010.03.021 [DOI] [PubMed] [Google Scholar]
- Koser C. U., Fraser L. J., Ioannou A., Becq J., Ellington M. J., Holden M. T., et al. (2014). Rapid single-colony whole-genome sequencing of bacterial pathogens. J. Antimicrob. Chemother. 69 1275–1281. 10.1093/jac/dkt494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca J., Poirel L., Ozdamar M., Turkoglu S., Hakko E., Nordmann P. (2014). KPC-producing Klebsiella pneumoniae, finally targeting Turkey. New Microbes New Infect. 2 50–51. 10.1002/nmi2.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafeuille E., Decre D., Mahjoub-Messai F., Bidet P., Arlet G., Bingen E. (2013). OXA-48 carbapenemase-producing Klebsiella pneumoniae isolated from Libyan patients. Microb. Drug Resist. 19 491–497. 10.1089/mdr.2012.0219 [DOI] [PubMed] [Google Scholar]
- Laishram S., Anandan S., Devi B. Y., Elakkiya M., Priyanka B., Bhuvaneshwari T., et al. (2015). Determination of synergy between sulbactam, meropenem and colistin in carbapenem-resistant Klebsiella pneumoniae and Acinetobacter baumannii isolates and correlation with the molecular mechanism of resistance. J. Chemother 10.1179/1973947815Y.0000000079 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Lascols C., Peirano G., Hackel M., Laupland K. B., Pitout J. D. (2013). Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob. Agents Chemother. 57 130–136. 10.1128/AAC.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre C., De Saint Martin L., Cuzon G., Bogaerts P., Lamar E., Glupczynski Y., et al. (2015). Efficient detection of carbapenemase activity in enterobacteriaceae by matrix-assisted laser desorption ionization-time of flight mass spectrometry in less than 30 minutes. J. Clin. Microbiol. 53 2163–2171. 10.1128/JCM.03467-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauretti L., Riccio M. L., Mazzariol A., Cornaglia G., Amicosante G., Fontana R., et al. (1999). Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43 1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A., Chmelnitsky I., Carmeli Y., Navon-Venezia S. (2010). Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 54 4493–4496. 10.1128/AAC.00175-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A., Navon-Venezia S., Chmelnitsky I., Schwaber M. J., Carmeli Y. (2007). Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51 3026–3029. 10.1128/AAC.00299-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. R., Cho I. H., Jeong B. C., Lee S. H. (2013a). Strategies to minimize antibiotic resistance. Int. J. Environ. Res. Public Health 10 4274–4305. 10.3390/ijerph10094274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Vasoo S., Hu F., Patel R., Doi Y. (2014). Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J. Clin. Microbiol. 52 4109–4110. 10.1128/JCM.01404-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Bae I. K., Lee S. H. (2012a). New definitions of extended-spectrum β-lactamase conferring worldwide emerging antibiotic resistance. Med. Res. Rev. 32 216–232. 10.1002/med.20210 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Jeong S. H., Cha S. S., Lee S. H. (2007). A lack of drugs for antibiotic-resistant Gram-negative bacteria. Nat. Rev. Drug Discov. 6 938–939. 10.1038/nrd2201-c1 [DOI] [Google Scholar]
- Lee J. H., Jeong S. H., Cha S. S., Lee S. H. (2009). New disturbing trend in antimicrobial resistance of gram-negative pathogens. PLoS Pathog. 5:e1000221 10.1371/journal.ppat.1000221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Lee J. J., Park K. S., Lee S. H. (2015a). Urgent need for b-lactam-β-lactamase inhibitors. Lancet Infect. Dis. 15 876–877. 10.1016/S1473-3099(15)00143-7 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Park A. M., Karim C. R., Lee C. R., Lee S. H. (2016). How to minimise antibiotic resistance. Lancet Infect. Dis 16 17–18. 10.1016/S1473-3099(15)00467-3 [DOI] [PubMed] [Google Scholar]
- Lee J. J., Lee J. H., Kwon D. B., Jeon J. H., Park K. S., Lee C. R., et al. (2015b). Fast and accurate large-scale detection of β-lactamase genes conferring antibiotic resistance. Antimicrob. Agents Chemother. 59 5967–5975. 10.1128/AAC.04634-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Hong S. G., Park Y. J., Lee H. S., Song W., Jeong J., et al. (2005a). Evaluation of phenotypic screening methods for detecting plasmid-mediated AmpC β-lactamases-producing isolates of Escherichia coli and Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 53 319–323. 10.1016/j.diagmicrobio.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Lee K., Lee M., Shin J. H., Lee M. H., Kang S. H., Park A. J., et al. (2006a). Prevalence of plasmid-mediated AmpC β-lactamases in Escherichia coli and Klebsiella pneumoniae in Korea. Microb. Drug Resist. 12 44–49. 10.1089/mdr.2006.12.44 [DOI] [PubMed] [Google Scholar]
- Lee N. Y., Wu J. J., Lin S. H., Ko W. C., Tsai L. H., Yan J. J. (2012b). Characterization of carbapenem-nonsusceptible Klebsiella pneumoniae bloodstream isolates at a Taiwanese hospital: clinical impacts of lowered breakpoints for carbapenems. Eur. J. Clin. Microbiol. Infect. Dis. 31 1941–1950. 10.1007/s10096-011-1525-2 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Jeong S. H., Cha S. S. (2005b). Minimising antibiotic resistance. Lancet Infect. Dis. 5 668–670. 10.1016/S1473-3099(05)70247-4 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Jeong S. H., Cha S. S. (2006b). Screening for carbapenem-resistant Gram-negative bacteria. Lancet Infect. Dis. 6 682–684. 10.1016/S1473-3099(06)70607-7 [DOI] [PubMed] [Google Scholar]
- Lee T. D., Adie K., McNabb A., Purych D., Mannan K., Azana R., et al. (2015c). Rapid Detection of KPC, NDM, and OXA-48-Like carbapenemases by Real-Time PCR from rectal swab surveillance samples. J. Clin. Microbiol. 53 2731–2733. 10.1128/JCM.01237-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Chung H. S., Lee Y., Yong D., Jeong S. H., Lee K., et al. (2013b). Comparison of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry assay with conventional methods for detection of IMP-6, VIM-2, NDM-1, SIM-1, KPC-1, OXA-23, and OXA-51 carbapenemase-producing Acinetobacter spp., Pseudomonas aeruginosa, and Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 77 227–230. 10.1016/j.diagmicrobio.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Lefebvre B., Levesque S., Bourgault A. M., Mulvey M. R., Mataseje L., Boyd D., et al. (2015). Carbapenem non-susceptible enterobacteriaceae in Quebec, Canada: results of a laboratory surveillance program (2010-2012). PLoS ONE 10:e0125076 10.1371/journal.pone.0125076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung V., Loo V. G., Frenette C., Domingo M. C., Bourgault A. M., Mulvey M. R., et al. (2012). First Canadian outbreak of Enterobacteriaceae-expressing Klebsiella pneumoniae carbapenemase type 3. Can. J. Infect. Dis. Med. Microbiol. 23 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wei Q., Wang Y., Du X., Zhao Y., Jiang X. (2011). Novel genetic environment of the plasmid-mediated KPC-3 gene detected in Escherichia coli and Citrobacter freundii isolates from China. Eur. J. Clin. Microbiol. Infect. Dis. 30 575–580. 10.1007/s10096-010-1124-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zhang Y., Bi D., Shen P., Ai F., Liu H., et al. (2015). First report of a clinical, multidrug-resistant Enterobacteriaceae isolate coharboring fosfomycin resistance gene fosA3 and carbapenemase gene blaKPC-2 on the same transposon, Tn1721. Antimicrob. Agents Chemother. 59 338–343. 10.1128/AAC.03061-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhang J., Liu Y., Zheng R., Chen H., Wang X., et al. (2014). Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn. Microbiol. Infect. Dis. 78 63–65. 10.1016/j.diagmicrobio.2013.10.002 [DOI] [PubMed] [Google Scholar]
- Li J. J., Sheng Z. K., Deng M., Bi S., Hu F. S., Miao H. F., et al. (2012). Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese University Hospital. BMC Infect. Dis. 12:373 10.1186/1471-2334-12-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liapis E., Pantel A., Robert J., Nicolas-Chanoine M. H., Cavalie L., van der Mee-Marquet N., et al. (2014). Molecular epidemiology of OXA-48-producing Klebsiella pneumoniae in France. Clin. Microbiol. Infect. 20 O1121–O1123. 10.1111/1469-0691.12727 [DOI] [PubMed] [Google Scholar]
- Lifshitz Z., Adler A., Carmeli Y. (2016). Comparative study of a novel biochemical assay, the rapidec carba NP Test, for detecting carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 54 453–456. 10.1128/JCM.02626-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbago B. M., Rasheed J. K., Anderson K. F., Zhu W., Kitchel B., Watz N., et al. (2011). IMP-producing carbapenem-resistant Klebsiella pneumoniae in the United States. J. Clin. Microbiol. 49 4239–4245. 10.1128/JCM.05297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling M. L., Tee Y. M., Tan S. G., Amin I. M., How K. B., Tan K. Y., et al. (2015). Risk factors for acquisition of carbapenem resistant Enterobacteriaceae in an acute tertiary care hospital in Singapore. Antimicrob. Resist. Infect. Control. 4:26 10.1186/s13756-015-0066-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li P., Jiang X., Bi D., Xie Y., Tai C., et al. (2012). Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 194 1841–1842. 10.1128/JB.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wan L. G., Deng Q., Cao X. W., Yu Y., Xu Q. F. (2015). First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol. Infect. 143 376–384. 10.1017/S0950268814000995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Walsh T. R., Toleman M., Woodford N. (2011). Balkan NDM-1: escape or transplant? Lancet Infect. Dis. 11:164 10.1016/S1473-3099(11)70048-2 [DOI] [PubMed] [Google Scholar]
- Lixandru B. E., Cotar A. I., Straut M., Usein C. R., Cristea D., Ciontea S., et al. (2015). Carbapenemase-Producing Klebsiella pneumoniae in Romania: a Six-Month survey. PLoS ONE 10:e0143214 10.1371/journal.pone.0143214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe C. F., Kus J. V., Salt N., Callery S., Louie L., Khan M. A., et al. (2013). Nosocomial transmission of New Delhi metallo-b-lactamase-1-producing Klebsiella pneumoniae in Toronto, Canada. Infect. Control Hosp. Epidemiol. 34 49–55. 10.1086/668778 [DOI] [PubMed] [Google Scholar]
- Lübbert C., Lippmann N., Busch T., Kaisers U. X., Ducomble T., Eckmanns T., et al. (2014). Long-term carriage of Klebsiella pneumoniae carbapenemase-2-producing K. pneumoniae after a large single-center outbreak in Germany. Am. J. Infect. Control 42 376–380. 10.1016/j.ajic.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Ma L., Wang J. T., Wu T. L., Siu L. K., Chuang Y. C., Lin J. C., et al. (2015). Emergence of OXA-48-producing Klebsiella pneumoniae in Taiwan. PLoS ONE 10:e0139152 10.1371/journal.pone.0139152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVane S. H., Crandon J. L., Nichols W. W., Nicolau D. P. (2014). Unexpected in vivo activity of ceftazidime alone and in combination with avibactam against New Delhi metallo-β-lactamase-producing Enterobacteriaceae in a murine thigh infection model. Antimicrob. Agents Chemother. 58 7007–7009. 10.1128/AAC.02662-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammeri H., Guillon H., Eb F., Nordmann P. (2010). Phenotypic and biochemical comparison of the carbapenem-hydrolyzing activities of five plasmid-borne AmpC b-lactamases. Antimicrob. Agents Chemother. 54 4556–4560. 10.1128/AAC.01762-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovska R., Stoeva T., Schneider I., Boyanova L., Popova V., Dacheva D., et al. (2015). Clonal dissemination of multilocus sequence type ST15 KPC-2-producing Klebsiella pneumoniae in Bulgaria. APMIS 123 887–894. 10.1111/apm.12433 [DOI] [PubMed] [Google Scholar]
- Marquez C., Ingold A., Echeverria N., Acevedo A., Vignoli R., Garcia-Fulgueiras V., et al. (2014). Emergence of KPC-producing Klebsiella pneumoniae in Uruguay: infection control and molecular characterization. New Microbes New Infect. 2 58–63. 10.1002/nmi2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers A. J., Hazen K. C., Carroll J., Yeh A. J., Cox H. L., Bonomo R. A., et al. (2013). First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the new world. J. Clin. Microbiol. 51 680–683. 10.1128/JCM.02580-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott H., Morris D., McArdle E., O’Mahony G., Kelly S., Cormican M., et al. (2012). Isolation of NDM-1-producing Klebsiella pnemoniae in Ireland, July 2011. Euro. Surveill 17 20087. [PubMed] [Google Scholar]
- Melegh S., Kovacs K., Gam T., Nyul A., Patko B., Toth A., et al. (2014). Emergence of VIM-4 metallo-b-lactamase-producing Klebsiella pneumoniae ST15 clone in the Clinical Centre University of Pecs, Hungary. Clin. Microbiol. Infect. 20 O27–O29. 10.1111/1469-0691.12293 [DOI] [PubMed] [Google Scholar]
- Metwally L., Gomaa N., Attallah M., Kamel N. (2013). High prevalence of Klebsiella pneumoniae carbapenemase-mediated resistance in K. pneumoniae isolates from Egypt. East Mediterr. Health J. 19 947–952. [PubMed] [Google Scholar]
- Monaco M., Giani T., Raffone M., Arena F., Garcia-Fernandez A., Pollini S., et al. (2014). Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro. Surveill. 19:20939 10.2807/1560-7917.ES2014.19.42.20939 [DOI] [PubMed] [Google Scholar]
- Moquet O., Bouchiat C., Kinana A., Seck A., Arouna O., Bercion R., et al. (2011). Class D OXA-48 carbapenemase in multidrug-resistant enterobacteria, Senegal. Emerg. Infect. Dis. 17 143–144. 10.3201/eid1701.100244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D., Boyle F., Morris C., Condon I., Delannoy-Vieillard A. S., Power L., et al. (2012). Inter-hospital outbreak of Klebsiella pneumoniae producing KPC-2 carbapenemase in Ireland. J. Antimicrob. Chemother. 67 2367–2372. 10.1093/jac/dks239 [DOI] [PubMed] [Google Scholar]
- Morris D., O’Connor M., Izdebski R., Corcoran M., Ludden C. E., Mc G. E., et al. (2016). Dissemination of clonally related multidrug-resistant Klebsiella pneumoniae in Ireland. Epidemiol. Infect. 144 443–448. 10.1017/S0950268815001041 [DOI] [PubMed] [Google Scholar]
- Mulvey M. R., Grant J. M., Plewes K., Roscoe D., Boyd D. A. (2011). New Delhi metallo-β-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada. Emerg. Infect. Dis. 17 103–106. 10.3201/eid1701.101358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price L. S., Poirel L., Bonomo R. A., Schwaber M. J., Daikos G. L., Cormican M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13 785–796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Cotellon G., Ergani A., Nordmann P. (2013). Real-time PCR for detection of blaOXA-48 genes from stools. J. Antimicrob. Chemother. 68 101–104. 10.1093/jac/dks340 [DOI] [PubMed] [Google Scholar]
- Naas T., Cuzon G., Villegas M. V., Lartigue M. F., Quinn J. P., Nordmann P. (2008). Genetic structures at the origin of acquisition of the β-lactamase bla KPC gene. Antimicrob. Agents Chemother. 52 1257–1263. 10.1128/AAC.01451-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Nordmann P., Vedel G., Poyart C. (2005). Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49 4423–4424. 10.1128/AAC.49.10.4423-4424.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano N., Endoh Y., Nagano Y., Toyama M., Matsui M., Shibayama K., et al. (2013). First report of OXA-48 carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in Japan from a patient returned from Southeast Asia. Jpn. J. Infect. Dis. 66 79–81. 10.7883/yoken.66.79 [DOI] [PubMed] [Google Scholar]
- Nakano R., Nakano A., Hikosaka K., Kawakami S., Matsunaga N., Asahara M., et al. (2014). First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob. Agents Chemother. 58 7611–7612. 10.1128/AAC.04265-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano R., Nakano A., Ishii Y., Ubagai T., Kikuchi-Ueda T., Kikuchi H., et al. (2015). Rapid detection of the Klebsiella pneumoniae carbapenemase (KPC) gene by loop-mediated isothermal amplification (LAMP). J. Infect. Chemother. 21 202–206. 10.1016/j.jiac.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Naseer U., Eriksen B. O., Sundsfjord A., Samuelsen O. (2012). Fecal colonization of VIM-1-producing Klebsiella pneumoniae and in vivo transfer of multidrug-resistant IncN plasmid in a renal transplant patient. Diagn. Microbiol. Infect. Dis. 72 363–366. 10.1016/j.diagmicrobio.2011.12.010 [DOI] [PubMed] [Google Scholar]
- Nathan C., Cars O. (2014). Antibiotic resistance–problems, progress, and prospects. N. Engl. J. Med. 371 1761–1763. 10.1056/NEJMp1408040 [DOI] [PubMed] [Google Scholar]
- Navarro-San Francisco C., Mora-Rillo M., Romero-Gómez M. P., Moreno-Ramos F., Rico-Nieto A., Ruiz-Carrascoso G., et al. (2013). Bacteraemia due to OXA-48-carbapenemase-producing Enterobacteriaceae: a major clinical challenge. Clin. Microbiol. Infect. 19 E72–E79. 10.1111/1469-0691.12091 [DOI] [PubMed] [Google Scholar]
- Netikul T., Sidjabat H. E., Paterson D. L., Kamolvit W., Tantisiriwat W., Steen J. A., et al. (2014). Characterization of an IncN2-type blaNDM-1-carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J. Antimicrob. Chemother. 69 3161–3163. 10.1093/jac/dku275 [DOI] [PubMed] [Google Scholar]
- Nobari S., Shahcheraghi F., Rahmati Ghezelgeh F., Valizadeh B. (2014). Molecular characterization of carbapenem-resistant strains of Klebsiella pneumoniae isolated from Iranian patients: first identification of blaKPC gene in Iran. Microb. Drug Resist. 20 285–293. 10.1089/mdr.2013.0074 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Cuzon G., Naas T. (2009). The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9 228–236. 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Dortet L., Poirel L. (2012a). Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18 263–272. 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Naas T., Poirel L. (2011a). Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17 1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Poirel L. (2014). The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 20 821–830. 10.1111/1469-0691.12719 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Poirel L., Dortet L. (2012b). Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18 1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Poirel L., Walsh T. R., Livermore D. M. (2011b). The emerging NDM carbapenemases. Trends Microbiol. 19 588–595. 10.1016/j.tim.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A. M., Chen L., Cienfuegos A. V., Roncancio G., Chavda K. D., Kreiswirth B. N., et al. (2015). High frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics: a two-year surveillance in five Colombian tertiary care hospitals. Antimicrob. Agents Chemother. 60 332–342. 10.1128/AAC.01775-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke I. N., Peeling R. W., Goossens H., Auckenthaler R., Olmsted S. S., de Lavison J. F., et al. (2011). Diagnostics as essential tools for containing antibacterial resistance. Drug Resist. Updat. 14 95–106. 10.1016/j.drup.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Osano E., Arakawa Y., Wacharotayankun R., Ohta M., Horii T., Ito H., et al. (1994). Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38 71–78. 10.1128/AAC.38.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterblad M., Hakanen A. J., Jalava J. (2014). Evaluation of the Carba NP test for carbapenemase detection. Antimicrob. Agents Chemother. 58 7553–7556. 10.1128/AAC.02761-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterblad M., Kirveskari J., Hakanen A. J., Tissari P., Vaara M., Jalava J. (2012). Carbapenemase-producing Enterobacteriaceae in Finland: the first years (2008–2011). J. Antimicrob. Chemother. 67 2860–2864. 10.1093/jac/dks299 [DOI] [PubMed] [Google Scholar]
- Oteo J., Hernandez J. M., Espasa M., Fleites A., Saez D., Bautista V., et al. (2013a). Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J. Antimicrob. Chemother. 68 317–321. 10.1093/jac/dks383 [DOI] [PubMed] [Google Scholar]
- Oteo J., Saez D., Bautista V., Fernandez-Romero S., Hernandez-Molina J. M., Perez-Vazquez M., et al. (2013b). Carbapenemase-producing enterobacteriaceae in Spain in 2012. Antimicrob. Agents Chemother. 57 6344–6347. 10.1128/AAC.01513-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteo J., Ortega A., Bartolome R., Bou G., Conejo C., Fernandez-Martinez M., et al. (2015). Prospective multicenter study of carbapenemase-producing Enterobacteriaceae from 83 hospitals in Spain reveals high in vitro susceptibility to colistin and meropenem. Antimicrob. Agents Chemother. 59 3406–3412. 10.1128/AAC.00086-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Baena Z. R., Oteo J., Conejo C., Larrosa M. N., Bou G., Fernandez-Martinez M., et al. (2016). Comprehensive clinical and epidemiological assessment of colonisation and infection due to carbapenemase-producing Enterobacteriaceae in Spain. J. Infect. 72 152–160. 10.1016/j.jinf.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Pano-Pardo J. R., Ruiz-Carrascoso G., Navarro-San Francisco C., Gomez-Gil R., Mora-Rillo M., Romero-Gomez M. P., et al. (2013). Infections caused by OXA-48-producing Klebsiella pneumoniae in a tertiary hospital in Spain in the setting of a prolonged, hospital-wide outbreak. J. Antimicrob. Chemother. 68 89–96. 10.1093/jac/dks364 [DOI] [PubMed] [Google Scholar]
- Pantel A., Richaud-Morel B., Cazaban M., Bouziges N., Sotto A., Lavigne J. P. (2016). Environmental persistence of OXA-48-producing Klebsiella pneumoniae in a French intensive care unit. Am. J. Infect. Control 44 366–368. 10.1016/j.ajic.2015.09.021 [DOI] [PubMed] [Google Scholar]
- Papadimitriou-Olivgeris M., Christofidou M., Fligou F., Bartzavali C., Vrettos T., Filos K. S., et al. (2014). The role of colonization pressure in the dissemination of colistin or tigecycline resistant KPC-producing Klebsiella pneumoniae in critically ill patients. Infection 42 883–890. 10.1007/s15010-014-0653-x [DOI] [PubMed] [Google Scholar]
- Parisi S. G., Bartolini A., Santacatterina E., Castellani E., Ghirardo R., Berto A., et al. (2015). Prevalence of Klebsiella pneumoniae strains producing carbapenemases and increase of resistance to colistin in an Italian teaching hospital from January 2012 To December 2014. BMC Infect. Dis. 15:244 10.1186/s12879-015-0996-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. J., Kim T. K., Song W., Kim J. S., Kim H. S., Lee J. (2013). An Increase in the clinical isolation of acquired AmpC β-lactamase-producing Klebsiella pneumoniae in Korea from 2007 to 2010. Ann. Lab. Med. 33 353–355. 10.3343/alm.2013.33.5.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Ginn A. N., Wiklendt A. M., Ellem J., Wong J. S., Ingram P., et al. (2015). Emergence of blaKPC carbapenemase genes in Australia. Int. J. Antimicrob. Agents 45 130–136. 10.1016/j.ijantimicag.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Partridge S. R., Iredell J. R. (2012). Genetic contexts of blaNDM-1. Antimicrob. Agents Chemother. 56 6065–6067. 10.1128/AAC.00117-12 author reply 6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteran F., Albornoz E., Faccone D., Gomez S., Valenzuela C., Morales M., et al. (2012). Emergence of NDM-1-producing Klebsiella pneumoniae in Guatemala. J. Antimicrob. Chemother. 67 1795–1797. 10.1093/jac/dks101 [DOI] [PubMed] [Google Scholar]
- Pasteran F., Tijet N., Melano R. G., Corso A. (2015). Simplified protocol for carba NP Test for enhanced detection of carbapenemase producers directly from bacterial cultures. J. Clin. Microbiol. 53 3908–3911. 10.1128/JCM.02032-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteran F. G., Otaegui L., Guerriero L., Radice G., Maggiora R., Rapoport M., et al. (2008). Klebsiella pneumoniae carbapenemase-2, buenos aires, argentina. Emerg. Infect. Dis. 14 1178–1180. 10.3201/eid1407.070826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. (2013). Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin. Infect. Dis. 57 564–572. 10.1093/cid/cit247 [DOI] [PubMed] [Google Scholar]
- Peirano G., Ahmed-Bentley J., Fuller J., Rubin J. E., Pitout J. D. (2014). Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: the first 3 years. J. Clin. Microbiol. 52 1575–1581. 10.1128/JCM.00162-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano G., Pillai D. R., Pitondo-Silva A., Richardson D., Pitout J. D. (2011). The characteristics of NDM-producing Klebsiella pneumoniae from Canada. Diagn. Microbiol. Infect. Dis. 71 106–109. 10.1016/j.diagmicrobio.2011.06.013 [DOI] [PubMed] [Google Scholar]
- Peirano G., Seki L. M., Val Passos V. L., Pinto M. C., Guerra L. R., Asensi M. D. (2009). Carbapenem-hydrolysing β-lactamase KPC-2 in Klebsiella pneumoniae isolated in Rio de Janeiro. Braz. J. Antimicrob. Chemother. 63 265–268. 10.1093/jac/dkn484 [DOI] [PubMed] [Google Scholar]
- Pena I., Picazo J. J., Rodriguez-Avial C., Rodriguez-Avial I. (2014). Carbapenemase-producing Enterobacteriaceae in a tertiary hospital in Madrid, Spain: high percentage of colistin resistance among VIM-1-producing Klebsiella pneumoniae ST11 isolates. Int. J. Antimicrob. Agents 43 460–464. 10.1016/j.ijantimicag.2014.01.021 [DOI] [PubMed] [Google Scholar]
- Pereira P. S., Borghi M., Albano R. M., Lopes J. C., Silveira M. C., Marques E. A., et al. (2015). Coproduction of NDM-1 and KPC-2 in Enterobacter hormaechei from Brazil. Microb. Drug Resist. 21 234–236. 10.1089/mdr.2014.0171 [DOI] [PubMed] [Google Scholar]
- Perilli M., Bottoni C., Grimaldi A., Segatore B., Celenza G., Mariani M., et al. (2013). Carbapenem-resistant Klebsiella pneumoniae harbouring blaKPC-3 and blaVIM-2 from central Italy. Diagn. Microbiol. Infect. Dis. 75 218–221. 10.1016/j.diagmicrobio.2012.10.008 [DOI] [PubMed] [Google Scholar]
- Pesesky M. W., Hussain T., Wallace M., Wang B., Andleeb S., Burnham C. A., et al. (2015). KPC and NDM-1 genes in related Enterobacteriaceae strains and plasmids from Pakistan and the United States. Emerg. Infect. Dis. 21 1034–1037. 10.3201/eid2106.141504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter H., Berggrav K., Thomas P., Pfeifer Y., Witte W., Templeton K., et al. (2012). Direct detection and genotyping of Klebsiella pneumoniae carbapenemases from urine by use of a new DNA microarray test. J. Clin. Microbiol. 50 3990–3998. 10.1128/JCM.00990-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer Y., Schlatterer K., Engelmann E., Schiller R. A., Frangenberg H. R., Stiewe D., et al. (2012). Emergence of OXA-48-type carbapenemase-producing Enterobacteriaceae in German hospitals. Antimicrob. Agents Chemother. 56 2125–2128. 10.1128/AAC.05315-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picao R. C., Santos A. F., Nicoletti A. G., Furtado G. H., Gales A. C. (2010). Detection of GES-5-producing Klebsiella pneumoniae in Brazil. J. Antimicrob. Chemother. 65 796–797. 10.1093/jac/dkq024 [DOI] [PubMed] [Google Scholar]
- Pires J., Novais A., Peixe L. (2013). Blue-carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J. Clin. Microbiol. 51 4281–4283. 10.1128/JCM.01634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitart C., Sole M., Roca I., Fabrega A., Vila J., Marco F. (2011). First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 β-lactamase in Klebsiella pneumoniae in Spain. Antimicrob. Agents Chemother. 55 4398–4401. 10.1128/AAC.00329-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout J. D., Nordmann P., Poirel L. (2015). Carbapenemase-Producing Klebsiella pneumoniae, a Key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59 5873–5884. 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Al Maskari Z., Al Rashdi F., Bernabeu S., Nordmann P. (2011a). NDM-1-producing Klebsiella pneumoniae isolated in the Sultanate of Oman. J. Antimicrob. Chemother. 66 304–306. 10.1093/jac/dkq428 [DOI] [PubMed] [Google Scholar]
- Poirel L., Benouda A., Hays C., Nordmann P. (2011b). Emergence of NDM-1-producing Klebsiella pneumoniae in Morocco. J. Antimicrob. Chemother. 66 2781–2783. 10.1093/jac/dkr384 [DOI] [PubMed] [Google Scholar]
- Poirel L., Castanheira M., Carrer A., Rodriguez C. P., Jones R. N., Smayevsky J., et al. (2011c). OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 55 2546–2551. 10.1128/AAC.00022-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Dortet L., Bernabeu S., Nordmann P. (2011d). Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55 5403–5407. 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Fortineau N., Nordmann P. (2011e). International transfer of NDM-1-producing Klebsiella pneumoniae from Iraq to France. Antimicrob. Agents Chemother. 55 1821–1822. 10.1128/AAC.01761-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Revathi G., Bernabeu S., Nordmann P. (2011f). Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 55 934–936. 10.1128/AAC.01247-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Schrenzel J., Cherkaoui A., Bernabeu S., Renzi G., Nordmann P. (2011g). Molecular analysis of NDM-1-producing enterobacterial isolates from Geneva, Switzerland. J. Antimicrob. Chemother. 66 1730–1733. 10.1093/jac/dkr174 [DOI] [PubMed] [Google Scholar]
- Poirel L., Carbonnelle E., Bernabeu S., Gutmann L., Rotimi V., Nordmann P. (2012a). Importation of OXA-48-producing Klebsiella pneumoniae from Kuwait. J. Antimicrob. Chemother. 67 2051–2052. 10.1093/jac/dks167 [DOI] [PubMed] [Google Scholar]
- Poirel L., Lascols C., Bernabeu S., Nordmann P. (2012b). NDM-1-producing Klebsiella pneumoniae in mauritius. Antimicrob. Agents Chemother. 56 598–599. 10.1128/AAC.05639-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Potron A., Nordmann P. (2012c). OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67 1597–1606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- Poirel L., Heritier C., Podglajen I., Sougakoff W., Gutmann L., Nordmann P. (2003). Emergence in Klebsiella pneumoniae of a chromosome-encoded SHV β-lactamase that compromises the efficacy of imipenem. Antimicrob. Agents Chemother. 47 755–758. 10.1128/AAC.47.2.755-758.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Heritier C., Tolun V., Nordmann P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48 15–22. 10.1128/AAC.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Kieffer N., Nordmann P. (2016). In vitro evaluation of dual carbapenem combinations against carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 71 156–161. 10.1093/jac/dkv294 [DOI] [PubMed] [Google Scholar]
- Poirel L., Nordmann P. (2015). Rapidec carba NP Test for rapid detection of carbapenemase producers. J. Clin. Microbiol. 53 3003–3008. 10.1128/JCM.00977-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Pitout J. D., Nordmann P. (2007). Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2 501–512. 10.2217/17460913.2.5.501 [DOI] [PubMed] [Google Scholar]
- Poirel L., Yilmaz M., Istanbullu A., Arslan F., Mert A., Bernabeu S., et al. (2014). Spread of NDM-1-producing Enterobacteriaceae in a neonatal intensive care unit in Istanbul, Turkey. Antimicrob. Agents Chemother. 58 2929–2933. 10.1128/AAC.02047-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron A., Nordmann P., Lafeuille E., Al Maskari Z., Al Rashdi F., Poirel L. (2011). Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55 4896–4899. 10.1128/AAC.00481-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron A., Poirel L., Nordmann P. (2014). Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob. Agents Chemother. 58 467–471. 10.1128/AAC.01344-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron A., Poirel L., Rondinaud E., Nordmann P. (2013). Intercontinental spread of OXA-48 β-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro. Surveill. 18:20549 10.2807/1560-7917.ES2013.18.31.20549 [DOI] [PubMed] [Google Scholar]
- Potron A., Schrenzel J., Poirel L., Renzi G., Cherkaoui A., Nordmann P. (2012). Emergence of OXA-48-producing Enterobacteriaceae in Switzerland. Int. J. Antimicrob. Agents 40 563–564. 10.1016/j.ijantimicag.2012.07.003 [DOI] [PubMed] [Google Scholar]
- Pournaras S., Poulou A., Tsakris A. (2010a). Inhibitor-based methods for the detection of KPC carbapenemase-producing Enterobacteriaceae in clinical practice by using boronic acid compounds. J. Antimicrob. Chemother. 65 1319–1321. 10.1093/jac/dkq124 [DOI] [PubMed] [Google Scholar]
- Pournaras S., Poulou A., Voulgari E., Vrioni G., Kristo I., Tsakris A. (2010b). Detection of the new metallo-β-lactamase VIM-19 along with KPC-2, CMY-2 and CTX-M-15 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 65 1604–1607. 10.1093/jac/dkq190 [DOI] [PubMed] [Google Scholar]
- Qi Y., Wei Z., Ji S., Du X., Shen P., Yu Y. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66 307–312. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- Qin S., Fu Y., Zhang Q., Qi H., Wen J. G., Xu H., et al. (2014). High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob. Agents Chemother. 58 4275–4282. 10.1128/AAC.02813-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles M. G., Rocchetti T. T., Fehlberg L. C., Kusano E. J., Chebabo A., Pereira R. M., et al. (2015). Unusual association of NDM-1 with KPC-2 and armA among Brazilian Enterobacteriaceae isolates. Braz. J. Med. Biol. Res. 48 174–177. 10.1590/1414-431X20144154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones D., Hart M., Espinosa F., Garcia S., Carmona Y., Ghosh S., et al. (2014). Emergence of Klebsiella pneumoniae clinical isolates producing KPC-2 carbapenemase in Cuba. New Microbes New Infect. 2 123–126. 10.1002/nmi2.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabnia R., Asgharpour F., Ferdosi Shahandashti E., Moulana Z. (2015). Nosocomial emerging of (VIM1) carbapenemase-producing isolates of in North of Iran. Iran J. Microbiol. 7 88–93. [PMC free article] [PubMed] [Google Scholar]
- Rasheed J. K., Kitchel B., Zhu W., Anderson K. F., Clark N. C., Ferraro M. J., et al. (2013). New Delhi metallo-β-lactamase-producing Enterobacteriaceae, United States. Emerg. Infect. Dis. 19 870–878. 10.3201/eid1906.121515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. K., Balne P. K., Reddy R. K., Mathai A., Kaur I. (2010). Development and evaluation of loop-mediated isothermal amplification assay for rapid and inexpensive detection of cytomegalovirus DNA in vitreous specimens from suspected cases of viral retinitis. J. Clin. Microbiol. 48 2050–2052. 10.1128/JCM.02248-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S. M., de la Fuente-Nunez C., Baquir B., Faria-Junior C., Franco O. L., Hancock R. E. (2015). Antibiofilm peptides increase the susceptibility of carbapenemase-producing Klebsiella pneumoniae clinical isolates to b-lactam antibiotics. Antimicrob. Agents Chemother. 59 3906–3912. 10.1128/AAC.00092-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S. N., Frasson I., Bergo C., Parisi S., Cavallaro A., Palù G. (2011). Transfer of KPC-2 Carbapenemase from Klebsiella pneumoniae to Escherichia coli in a patient: first case in Europe. J. Clin. Microbiol. 49 2040–2042. 10.1128/JCM.00133-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S. N., Frasson I., Franchin E., Bergo C., Lavezzo E., Barzon L., et al. (2012). KPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009-December 2011: massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care units. Gut Pathog. 4:7 10.1186/1757-4749-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimrang B., Chanawong A., Lulitanond A., Wilailuckana C., Charoensri N., Sribenjalux P., et al. (2012). Emergence of NDM-1- and IMP-14a-producing Enterobacteriaceae in Thailand. J. Antimicrob. Chemother. 67 2626–2630. 10.1093/jac/dks267 [DOI] [PubMed] [Google Scholar]
- Robert J., Pantel A., Merens A., Lavigne J. P., Nicolas-Chanoine M. H., Group O. N. S. C. R. S. (2014). Incidence rates of carbapenemase-producing Enterobacteriaceae clinical isolates in France: a prospective nationwide study in 2011-12. J. Antimicrob. Chemother. 69 2706–2712. 10.1093/jac/dku208 [DOI] [PubMed] [Google Scholar]
- Robustillo Rodela A., Diaz-Agero Perez C., Sanchez Sagrado T., Ruiz-Garbajosa P., Pita Lopez M. J., Monge V. (2012). Emergence and outbreak of carbapenemase-producing KPC-3 Klebsiella pneumoniae in Spain, September 2009 to February 2010: control measures. Euro. Surveill. 17:20086. [PubMed] [Google Scholar]
- Roche C., Cotter M., O Connell N., Crowley B. (2009). First identification of class A carbapenemase-producing Klebsiella pneumoniae in the Republic of Ireland. Euro. Surveill. 14:19163. [PubMed] [Google Scholar]
- Rock C., Thom K. A., Masnick M., Johnson J. K., Harris A. D., Morgan D. J. (2014). Frequency of Klebsiella pneumoniae carbapenemase (KPC)-producing and non-KPC-producing Klebsiella species contamination of healthcare workers and the environment. Infect. Control Hosp. Epidemiol. 35 426–429. 10.1086/675598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Avial I., Pena I., Picazo J. J., Rodriguez-Avial C., Culebras E. (2015). In vitro activity of the next-generation aminoglycoside plazomicin alone and in combination with colistin, meropenem, fosfomycin or tigecycline against carbapenemase-producing Enterobacteriaceae strains. Int. J. Antimicrob. Agents 46 616–621. 10.1016/j.ijantimicag.2015.07.021 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martinez J. M., Nordmann P., Fortineau N., Poirel L. (2010). VIM-19, a metallo-β-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54 471–476. 10.1128/AAC.00458-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas L. J., Mojica M. F., Blanco V. M., Correa A., Montealegre M. C., De La Cadena E., et al. (2013). Emergence of Klebsiella pneumoniae Coharboring KPC and VIM Carbapenemases in Colombia. Antimicrob. Agents Chemother. 57 1101–1102. 10.1128/AAC.01666-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose W. E., Rybak M. J. (2006). Tigecycline: first of a new class of antimicrobial agents. Pharmacotherapy 26 1099–1110. 10.1592/phco.26.8.1099 [DOI] [PubMed] [Google Scholar]
- Rosenblum R., Khan E., Gonzalez G., Hasan R., Schneiders T. (2011). Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 38 39–45. 10.1016/j.ijantimicag.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Garbajosa P., Curiao T., Tato M., Gijon D., Pintado V., Valverde A., et al. (2013). Multiclonal dispersal of KPC genes following the emergence of non-ST258 KPC-producing Klebsiella pneumoniae clones in Madrid, Spain. J. Antimicrob. Chemother. 68 2487–2492. 10.1093/jac/dkt237 [DOI] [PubMed] [Google Scholar]
- Ruzin A., Visalli M. A., Keeney D., Bradford P. A. (2005). Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49 1017–1022. 10.1128/AAC.49.3.1017-1022.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R., Takahashi R., Sawabe E., Koyano S., Takahashi Y., Shima M., et al. (2014). First report of KPC-2 Carbapenemase-producing Klebsiella pneumoniae in Japan. Antimicrob. Agents Chemother. 58 2961–2963. 10.1128/AAC.02072-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsen O., Naseer U., Tofteland S., Skutlaberg D. H., Onken A., Hjetland R., et al. (2009). Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J. Antimicrob. Chemother. 63 654–658. 10.1093/jac/dkp018 [DOI] [PubMed] [Google Scholar]
- Sattar H., Toleman M., Nahid F., Zahra R. (2014). Co-existence of blaNDM-1 and blaKPC-2 in clinical isolates of Klebsiella pneumoniae from Pakistan. J. Chemother. 10.1179/1973947814Y.0000000223 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Sautrey G., Duval R. E., Chevalley A., Fontanay S., Clarot I. (2015). Capillary electrophoresis for fast detection of heterogeneous population in colistin-resistant Gram-negative bacteria. Electrophoresis 36 2630–2633. 10.1002/elps.201500064 [DOI] [PubMed] [Google Scholar]
- Seara N., Oteo J., Carrillo R., Perez-Blanco V., Mingorance J., Gomez-Gil R., et al. (2015). Interhospital spread of NDM-7-producing Klebsiella pneumoniae belonging to ST437 in Spain. Int. J. Antimicrob. Agents 46 169–173. 10.1016/j.ijantimicag.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Seiffert S. N., Marschall J., Perreten V., Carattoli A., Furrer H., Endimiani A. (2014). Emergence of Klebsiella pneumoniae co-producing NDM-1, OXA-48, CTX-M-15, CMY-16, QnrA and ArmA in Switzerland. Int. J. Antimicrob. Agents 44 260–262. 10.1016/j.ijantimicag.2014.05.008 [DOI] [PubMed] [Google Scholar]
- Semin-Pelletier B., Cazet L., Bourigault C., Juvin M. E., Boutoille D., Raffi F., et al. (2015). Challenges of controlling a large outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in a French university hospital. J. Hosp. Infect. 89 248–253. 10.1016/j.jhin.2014.11.018 [DOI] [PubMed] [Google Scholar]
- Shahcheraghi F., Nobari S., Rahmati Ghezelgeh F., Nasiri S., Owlia P., Nikbin V. S., et al. (2013). First report of New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in Iran. Microb. Drug Resist. 19 30–36. 10.1089/mdr.2012.0078 [DOI] [PubMed] [Google Scholar]
- Shanmugam P., Meenakshisundaram J., Jayaraman P. (2013). blaKPC gene detection in clinical isolates of carbapenem resistant enterobacteriaceae in a tertiary care hospital. J. Clin. Diagn. Res. 7 2736–2738. 10.7860/JCDR/2013/7759.3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Wei Z., Jiang Y., Du X., Ji S., Yu Y., et al. (2009). Novel genetic environment of the carbapenem-hydrolyzing β-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53 4333–4338. 10.1128/AAC.00260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibl A., Al-Agamy M., Memish Z., Senok A., Khader S. A., Assiri A. (2013). The emergence of OXA-48- and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 17:e1130–3 10.1016/j.ijid.2013.06.016 [DOI] [PubMed] [Google Scholar]
- Shin S. Y., Bae I. K., Kim J., Jeong S. H., Yong D., Kim J. M., et al. (2012). Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J. Med. Microbiol. 61 239–245. 10.1099/jmm.0.037036-0 [DOI] [PubMed] [Google Scholar]
- Shoma S., Kamruzzaman M., Ginn A. N., Iredell J. R., Partridge S. R. (2014). Characterization of multidrug-resistant Klebsiella pneumoniae from Australia carrying blaNDM-1. Diagn. Microbiol. Infect. Dis. 78 93–97. 10.1016/j.diagmicrobio.2013.08.001 [DOI] [PubMed] [Google Scholar]
- Sidjabat H. E., Townell N., Nimmo G. R., George N. M., Robson J., Vohra R., et al. (2015). Dominance of IMP-4-producing enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob. Agents Chemother. 59 4059–4066. 10.1128/AAC.04378-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanki R., Vanjari L., Ede N., Gungi A., Soory A., Vemu L. (2013). Evaluation of LAMP assay using phenotypic tests and conventional PCR for detection of blaNDM-1 and blaKPC genes among carbapenem-resistant clinical Gram-negative isolates. J. Med. Microbiol. 62 1540–1544. 10.1099/jmm.0.059907-0 [DOI] [PubMed] [Google Scholar]
- Solanki R., Vanjari L., Subramanian S., B A., E N., Lakshmi V. (2014). Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of carbapenemases among gram negative bacilli. J. Clin. Diagn. Res 8 DC23–DC26 10.7860/JCDR/2014/10794.5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnevend A., Al Baloushi A., Ghazawi A., Hashmey R., Girgis S., Hamadeh M. B., et al. (2013). Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J. Med. Microbiol. 62 1044–1050. 10.1099/jmm.0.059014-0 [DOI] [PubMed] [Google Scholar]
- Sonnevend A., Ghazawi A., Darwish D., AlDeesi Z., Kadhum A. F., Pal T. (2015a). Characterization of KPC-type carbapenemase-producing Klebsiella pneumoniae strains isolated in the Arabian Peninsula. J. Antimicrob. Chemother. 70 1592–1593. 10.1093/jac/dku576 [DOI] [PubMed] [Google Scholar]
- Sonnevend A., Ghazawi A. A., Hashmey R., Jamal W., Rotimi V. O., Shibl A. M., et al. (2015b). Characterization of carbapenem-resistant enterobacteriaceae with high rate of autochthonous transmission in the Arabian Peninsula. PLoS ONE 10:e0131372 10.1371/journal.pone.0131372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu T., Fiori B., D’Inzeo T., Canu G., Campoli S., Giani T., et al. (2012). Evaluation of the new NucliSENS EasyQ KPC test for rapid detection of Klebsiella pneumoniae carbapenemase genes (blaKPC). J. Clin. Microbiol. 50 2783–2785. 10.1128/JCM.00284-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulou A., Bartzavali C., Vamvakopoulou S., Marangos M., Anastassiou E. D., Spiliopoulou I., et al. (2016). The first NDM Metallo-β-lactamase producing Klebsiella pneumoniae isolate in a University Hospital of Southwestern Greece. J. Chemother. 10.1179/1973947815Y.0000000003 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Steinmann J., Kaase M., Gatermann S., Popp W., Steinmann E., Damman M., et al. (2011). Outbreak due to a Klebsiella pneumoniae strain harbouring KPC-2 and VIM-1 in a German university hospital, July 2010 to January 2011. Euro. Surveill. 16:19944. [PubMed] [Google Scholar]
- Stoesser N., Giess A., Batty E. M., Sheppard A. E., Walker A. S., Wilson D. J., et al. (2014). Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob. Agents Chemother. 58 7347–7357. 10.1128/AAC.03900-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studentova V., Dobiasova H., Hedlova D., Dolejska M., Papagiannitsis C. C., Hrabak J. (2015). Complete nucleotide sequences of two NDM-1-encoding plasmids from the same sequence type 11 Klebsiella pneumoniae strain. Antimicrob. Agents Chemother. 59 1325–1328. 10.1128/AAC.04095-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Cai Y., Liu X., Bai N., Liang B., Wang R. (2013). The emergence of clinical resistance to tigecycline. Int. J. Antimicrob. Agents 41 110–116. 10.1016/j.ijantimicag.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Swayne R., Ellington M. J., Curran M. D., Woodford N., Aliyu S. H. (2013). Utility of a novel multiplex TaqMan PCR assay for metallo-β-lactamase genes plus other TaqMan assays in detecting genes encoding serine carbapenemases and clinically significant extended-spectrum β-lactamases. Int. J. Antimicrob. Agents 42 352–356. 10.1016/j.ijantimicag.2013.06.018 [DOI] [PubMed] [Google Scholar]
- Székely E., Damjanova I., Jánvári L., Vas K. E., Molnár S., Bilca D. V., et al. (2013). First description of blaNDM-1, blaOXA-48, blaOXA-181 producing Enterobacteriaceae strains in Romania. Int. J. Med. Microbiol. 303 697–700. 10.1016/j.ijmm.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Tada T., Miyoshi-Akiyama T., Dahal R. K., Mishra S. K., Ohara H., Shimada K., et al. (2013). Dissemination of multidrug-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (NDM-1 and OXA-72) and 16S rRNA methylases (ArmA, RmtC and RmtF) in Nepal. Int. J. Antimicrob. Agents 42 372–374. 10.1016/j.ijantimicag.2013.06.014 [DOI] [PubMed] [Google Scholar]
- Tamayo M., Santiso R., Otero F., Bou G., Lepe J. A., McConnell M. J., et al. (2013). Rapid determination of colistin resistance in clinical strains of Acinetobacter baumannii by use of the micromax assay. J. Clin. Microbiol. 51 3675–3682. 10.1128/JCM.01787-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangden T., Giske C. G. (2015). Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J. Intern. Med. 277 501–512. 10.1111/joim.12342 [DOI] [PubMed] [Google Scholar]
- Tangden T., Hickman R. A., Forsberg P., Lagerback P., Giske C. G., Cars O. (2014). Evaluation of double- and triple-antibiotic combinations for VIM- and NDM-producing Klebsiella pneumoniae by in vitro time-kill experiments. Antimicrob. Agents Chemother. 58 1757–1762. 10.1128/AAC.00741-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tascini C., Tagliaferri E., Giani T., Leonildi A., Flammini S., Casini B., et al. (2013). Synergistic activity of colistin plus rifampin against colistin-resistant KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57 3990–3993. 10.1128/AAC.00179-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin E., Adler A., Lerner A., Carmeli Y. (2014). Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann. N. Y. Acad. Sci. 1323 22–42. 10.1111/nyas.12537 [DOI] [PubMed] [Google Scholar]
- Teo J. W. P., Kurup A., Lin R. T. P., Hsien K. T. (2013). Emergence of clinical Klebsiella pneumoniae producing OXA-232 carbapenemase in Singapore. New Microbes New Infect. 1 13–15. 10.1002/2052-2975.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. P., Moore L. S., Elamin N., Doumith M., Zhang J., Maharjan S., et al. (2013). Early (2008–2010) hospital outbreak of Klebsiella pneumoniae producing OXA-48 carbapenemase in the UK. Int. J. Antimicrob. Agents 42 531–536. 10.1016/j.ijantimicag.2013.08.020 [DOI] [PubMed] [Google Scholar]
- Tijet N., Boyd D., Patel S. N., Mulvey M. R., Melano R. G. (2013). Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57 4578–4580. 10.1128/AAC.00878-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofteland S., Naseer U., Lislevand J. H., Sundsfjord A., Samuelsen O. (2013). A long-term low-frequency hospital outbreak of KPC-producing Klebsiella pneumoniae involving Intergenus plasmid diffusion and a persisting environmental reservoir. PLoS ONE 8:e59015 10.1371/journal.pone.0059015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo P. V., Aranha Junior A. A., Arend L. N., Ribeiro V., Zavascki A. P., Tuon F. F. (2015). Activity of antimicrobial combinations against KPC-2-producing Klebsiella pneumoniae in a rat model and time-kill assay. Antimicrob. Agents Chemother. 59 4301–4304. 10.1128/AAC.00323-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman M. A., Spencer J., Jones L., Walsh T. R. (2012). blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56 2773–2776. 10.1128/AAC.06297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollentino F. M., Polotto M., Nogueira M. L., Lincopan N., Neves P., Mamizuka E. M., et al. (2011). High prevalence of blaCTX-M extended spectrum β-lactamase genes in Klebsiella pneumoniae isolates from a tertiary care hospital: first report of blaSHV-12, blaSHV-31 blaSHV-38 and blaCTX-M-15 in Brazil. Microb. Drug Resist. 17 7–16. 10.1089/mdr.2010.0055 [DOI] [PubMed] [Google Scholar]
- Toth A., Damjanova I., Puskas E., Janvari L., Farkas M., Dobak A., et al. (2010). Emergence of a colistin-resistant KPC-2-producing Klebsiella pneumoniae ST258 clone in Hungary. Eur. J. Clin. Microbiol. Infect. Dis. 29 765–769. 10.1007/s10096-010-0921-3 [DOI] [PubMed] [Google Scholar]
- Tsai Y. K., Liou C. H., Fung C. P., Lin J. C., Siu L. K. (2013). Single or in combination antimicrobial resistance mechanisms of Klebsiella pneumoniae contribute to varied susceptibility to different carbapenems. PLoS ONE 8:e79640 10.1371/journal.pone.0079640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakris A., Kristo I., Poulou A., Markou F., Ikonomidis A., Pournaras S. (2008). First occurrence of KPC-2-possessing Klebsiella pneumoniae in a Greek hospital and recommendation for detection with boronic acid disc tests. J. Antimicrob. Chemother. 62 1257–1260. 10.1093/jac/dkn364 [DOI] [PubMed] [Google Scholar]
- Tsakris A., Poulou A., Bogaerts P., Dimitroulia E., Pournaras S., Glupczynski Y. (2015). Evaluation of a new phenotypic OXA-48 disk test for differentiation of OXA-48 carbapenemase-producing Enterobacteriaceae clinical isolates. J. Clin. Microbiol. 53 1245–1251. 10.1128/JCM.03318-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakris A., Poulou A., Pournaras S., Voulgari E., Vrioni G., Themeli-Digalaki K., et al. (2010). A simple phenotypic method for the differentiation of metallo-β-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J. Antimicrob. Chemother. 65 1664–1671. 10.1093/jac/dkq210 [DOI] [PubMed] [Google Scholar]
- Tseng I. L., Liu Y. M., Wang S. J., Yeh H. Y., Hsieh C. L., Lu H. L., et al. (2015). Emergence of carbapenemase producing Klebsiella pneumoniae and spread of KPC-2 and KPC-17 in Taiwan: a nationwide study from 2011 to 2013. PLoS ONE 10:e0138471 10.1371/journal.pone.0138471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello M., Trecarichi E. M., De Rosa F. G., Giannella M., Giacobbe D. R., Bassetti M., et al. (2015). Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J. Antimicrob. Chemother. 70 2133–2143. 10.1093/jac/dkv086 [DOI] [PubMed] [Google Scholar]
- Tumbarello M., Viale P., Viscoli C., Trecarichi E. M., Tumietto F., Marchese A., et al. (2012). Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin. Infect. Dis. 55 943–950. 10.1093/cid/cis588 [DOI] [PubMed] [Google Scholar]
- Tzouvelekis L. S., Markogiannakis A., Piperaki E., Souli M., Daikos G. L. (2014). Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Infect. 20 862–872. 10.1111/1469-0691.12697 [DOI] [PubMed] [Google Scholar]
- Tzouvelekis L. S., Miriagou V., Kotsakis S. D., Spyridopoulou K., Athanasiou E., Karagouni E., et al. (2013). KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob. Agents Chemother. 57 5144–5146. 10.1128/AAC.01052-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M. (2010). Polymyxins and their novel derivatives. Curr. Opin. Microbiol. 13 574–581. 10.1016/j.mib.2010.09.002 [DOI] [PubMed] [Google Scholar]
- van der Zwaluw K., de Haan A., Pluister G. N., Bootsma H. J., de Neeling A. J., Schouls L. M. (2015). The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE 10:e0123690 10.1371/journal.pone.0123690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasoo S., Cunningham S. A., Kohner P. C., Simner P. J., Mandrekar J. N., Lolans K., et al. (2013). Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J. Clin. Microbiol. 51 3097–3101. 10.1128/JCM.00965-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L., Poirel L., Nordmann P., Carta C., Carattoli A. (2012). Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J. Antimicrob. Chemother. 67 1645–1650. 10.1093/jac/dks114 [DOI] [PubMed] [Google Scholar]
- Villegas M. V., Lolans K., Correa A., Suarez C. J., Lopez J. A., Vallejo M., et al. (2006). First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 50 2880–2882. 10.1128/AAC.00186-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgincar N., Iyer S., Stacey A., Maharjan S., Pike R., Perry C., et al. (2011). Klebsiella pneumoniae producing KPC carbapenemase in a district general hospital in the UK. J. Hosp. Infect. 78 293–296. 10.1016/j.jhin.2011.03.016 [DOI] [PubMed] [Google Scholar]
- Voulgari E., Gartzonika C., Vrioni G., Politi L., Priavali E., Levidiotou-Stefanou S., et al. (2014). The balkan region: NDM-1-producing Klebsiella pneumoniae ST11 clonal strain causing outbreaks in Greece. J. Antimicrob. Chemother. 69 2091–2097. 10.1093/jac/dku105 [DOI] [PubMed] [Google Scholar]
- Voulgari E., Zarkotou O., Ranellou K., Karageorgopoulos D. E., Vrioni G., Mamali V., et al. (2013). Outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in Greece involving an ST11 clone. J. Antimicrob. Chemother. 68 84–88. 10.1093/jac/dks356 [DOI] [PubMed] [Google Scholar]
- Vourli S., Giakkoupi P., Miriagou V., Tzelepi E., Vatopoulos A. C., Tzouvelekis L. S. (2004). Novel GES/IBC extended-spectrum β-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol. Lett. 234 209–213. 10.1016/j.femsle.2004.03.028 [DOI] [PubMed] [Google Scholar]
- Vrioni G., Daniil I., Voulgari E., Ranellou K., Koumaki V., Ghirardi S., et al. (2012). Comparative evaluation of a prototype chromogenic medium (ChromID CARBA) for detecting carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J. Clin. Microbiol. 50 1841–1846. 10.1128/JCM.06848-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J., Yamane K., Suzuki S., Kimura K., Arakawa Y. (2010). Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob. Agents Chemother. 54 3061–3064. 10.1128/AAC.01834-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wailan A. M., Paterson D. L., Kennedy K., Ingram P. R., Bursle E., Sidjabat H. E. (2015). Genomic characteristics of NDM-producing Enterobacteriaceae in Australia and their blaNDM genetic contexts. Antimicrob. Agents Chemother. 60 136–141. 10.1128/AAC.01243-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Chen J., Yang L., Mou Y., Yang Y. (2014a). Phenotypic and enzymatic comparative analysis of the KPC variants, KPC-2 and its recently discovered variant KPC-15. PLoS ONE 9:e111491 10.1371/journal.pone.0111491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hou W., Chen J., Mou Y., Yang L., Yang L., et al. (2014b). Characterization of the blaKPC-2 and blaKPC-3 genes and the novel blaKPC-15 gene in Klebsiella pneumoniae. J. Med. Microbiol. 63 981–987. 10.1099/jmm.0.073841-0 [DOI] [PubMed] [Google Scholar]
- Wang L., Gu H., Lu X. (2012). A rapid low-cost real-time PCR for the detection of Klebsiella pneumoniae carbapenemase genes. Ann. Clin. Microbiol. Antimicrob. 11:9 10.1186/1476-0711-11-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen S., Guo Y., Xiong Z., Hu F., Zhu D., et al. (2011). Occurrence of false positive results for the detection of carbapenemases in carbapenemase-negative Escherichia coli and Klebsiella pneumoniae isolates. PLoS ONE 6:e26356 10.1371/journal.pone.0026356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. D., Cai J. C., Zhou H. W., Zhang R., Chen G. X. (2009). Reduced susceptibility to carbapenems in Klebsiella pneumoniae clinical isolates associated with plasmid-mediated β-lactamase production and OmpK36 porin deficiency. J. Med. Microbiol. 58(Pt 9), 1196–1202. 10.1099/jmm.0.008094-0 [DOI] [PubMed] [Google Scholar]
- Wang X., Chen G., Wu X., Wang L., Cai J., Chan E. W., et al. (2015). Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the blaNDM-1 element and clonal spread of progenitor resistant strains. Front. Microbiol. 6:595 10.3389/fmicb.2015.00595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li H., Zhao C., Chen H., Liu J., Wang Z., et al. (2014c). Novel NDM-9 metallo-β-lactamase identified from a ST107 Klebsiella pneumoniae strain isolated in China. Int. J. Antimicrob. Agents 44 90–91. 10.1016/j.ijantimicag.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Wang X., Xu X., Li Z., Chen H., Wang Q., Yang P., et al. (2014d). An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb. Drug Resist. 20 144–149. 10.1089/mdr.2013.0100 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang F., Zhao C., Wang Z., Nichols W. W., Testa R., et al. (2014e). In vitro activities of ceftazidime-avibactam and aztreonam-avibactam against 372 Gram-negative bacilli collected in 2011 and 2012 from 11 teaching hospitals in China. Antimicrob. Agents Chemother. 58 1774–1778. 10.1128/AAC.02123-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg G., Hidalgo-Grass C., Partridge S. R., Tolmasky M. E., Temper V., Moses A. E., et al. (2012). A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6’)-Ib. J. Antimicrob. Chemother. 67 898–901. 10.1093/jac/dkr552 [DOI] [PubMed] [Google Scholar]
- Wendt C., Schutt S., Dalpke A. H., Konrad M., Mieth M., Trierweiler-Hauke B., et al. (2010). First outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 29 563–570. 10.1007/s10096-010-0896-0 [DOI] [PubMed] [Google Scholar]
- Weterings V., Zhou K., Rossen J. W., van Stenis D., Thewessen E., Kluytmans J., et al. (2015). An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur. J. Clin. Microbiol. Infect. Dis. 34 1647–1655. 10.1007/s10096-015-2401-2 [DOI] [PubMed] [Google Scholar]
- Willemse-Erix D., Bakker-Schut T., Slagboom-Bax F., Jachtenberg J. W., Lemmens-den Toom N., Papagiannitsis C. C., et al. (2012). Rapid typing of extended-spectrum β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolates by use of SpectraCell RA. J. Clin. Microbiol. 50 1370–1375. 10.1128/JCM.05423-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D. A., Heffernan H., Sidjabat H., Roberts S. A., Paterson D., Smith L. M., et al. (2011). Intercontinental transfer of OXA-181-producing Klebsiella pneumoniae into New Zealand. J. Antimicrob. Chemother. 66 2888–2890. 10.1093/jac/dkr396 [DOI] [PubMed] [Google Scholar]
- Williamson D. A., Sidjabat H. E., Freeman J. T., Roberts S. A., Silvey A., Woodhouse R., et al. (2012). Identification and molecular characterisation of New Delhi metallo-β-lactamase-1 (NDM-1)- and NDM-6-producing Enterobacteriaceae from New Zealand hospitals. Int. J. Antimicrob. Agents 39 529–533. 10.1016/j.ijantimicag.2012.02.017 [DOI] [PubMed] [Google Scholar]
- Wiskirchen D. E., Nordmann P., Crandon J. L., Nicolau D. P. (2013). Efficacy of humanized carbapenem exposures against New Delhi metallo-β-lactamase (NDM-1)-producing enterobacteriaceae in a murine infection model. Antimicrob. Agents Chemother. 57 3936–3940. 10.1128/AAC.00708-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiskirchen D. E., Nordmann P., Crandon J. L., Nicolau D. P. (2014a). Efficacy of humanized carbapenem and ceftazidime regimens against Enterobacteriaceae producing OXA-48 carbapenemase in a murine infection model. Antimicrob. Agents Chemother. 58 1678–1683. 10.1128/AAC.01947-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiskirchen D. E., Nordmann P., Crandon J. L., Nicolau D. P. (2014b). In vivo efficacy of human simulated regimens of carbapenems and comparator agents against NDM-1-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 58 1671–1677. 10.1128/AAC.01946-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter D. J., Kurpiel P. M., Woodford N., Palepou M. F., Goering R. V., Hanson N. D. (2009). Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 53 557–562. 10.1128/AAC.00734-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Tierno P. M., Jr., Young K., Tysall L., Palepou M. F. I., Ward E., et al. (2004). Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class a β-Lactamase, KPC-3, in a New York medical center. Antimicrob. Agents Chemother. 48 4793–4799. 10.1128/AAC.48.12.4793-4799.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Zhang J., Warner M., Kaufmann M. E., Matos J., Macdonald A., et al. (2008). Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J. Antimicrob. Chemother. 62 1261–1264. 10.1093/jac/dkn396 [DOI] [PubMed] [Google Scholar]
- Wrenn C., O’Brien D., Keating D., Roche C., Rose L., Ronayne A., et al. (2014). Investigation of the first outbreak of OXA-48-producing Klebsiella pneumoniae in Ireland. J. Hosp. Infect. 87 41–46. 10.1016/j.jhin.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Xiang D. R., Li J. J., Sheng Z. K., Yu H. Y., Deng M., Bi S., et al. (2015). Complete sequence of a novel IncR-F33:A-:B- Plasmid, pKP1034, Harboring fosA3 blaKPC-2 blaCTX-M-65 blaSHV-12 and rmtB from an epidemic Klebsiella pneumoniae Sequence Type 11 Strain in China. Antimicrob. Agents Chemother. 60 1343–1348. 10.1128/AAC.01488-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Takano T., Fusegawa T., Shibuya T., Hung W. C., Higuchi W., et al. (2013). Electron microscopic structures, serum resistance, and plasmid restructuring of New Delhi metallo-β-lactamase-1 (NDM-1)-producing ST42 Klebsiella pneumoniae emerging in Japan. J. Infect. Chemother. 19 118–127. 10.1007/s10156-012-0470-z [DOI] [PubMed] [Google Scholar]
- Yan Y., Yang H., Pan L., Sun K., Fan H., Lu Y., et al. (2014). Improving the efficiency of the modified Hodge test in KPC-producing Klebsiella pneumoniae isolates by incorporating an EDTA disk. Curr. Microbiol. 69 47–52. 10.1007/s00284-014-0552-5 [DOI] [PubMed] [Google Scholar]
- Yang J., Ye L., Guo L., Zhao Q., Chen R., Luo Y., et al. (2013). A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin. Microbiol. Infect. 19 E509–E515. 10.1111/1469-0691.12275 [DOI] [PubMed] [Google Scholar]
- Yigit H., Queenan A. M., Anderson G. J., Domenech-Sanchez A., Biddle J. W., Steward C. D., et al. (2001). Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45 1151–1161. 10.1128/AAC.45.4.1151-1161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J. S., Kim H. M., Yoo J. I., Yang J. W., Kim H. S., Chung G. T., et al. (2013). Detection of clonal KPC-2-producing Klebsiella pneumoniae ST258 in Korea during nationwide surveillance in 2011. J. Med. Microbiol. 62 1338–1342. 10.1099/jmm.0.059428-0 [DOI] [PubMed] [Google Scholar]
- Yu W. L., Lee M. F., Tang H. J., Chang M. C., Walther-Rasmussen J., Chuang Y. C. (2015). Emergence of KPC new variants (KPC-16 and KPC-17) and ongoing outbreak in southern Taiwan. Clin. Microbiol. Infect. 21 375.e5–375.e8 10.1016/j.cmi.2014.11.030 [DOI] [PubMed] [Google Scholar]
- Yusuf E., Van Der Meeren S., Schallier A., Pierard D. (2014). Comparison of the Carba NP test with the Rapid CARB screen Kit for the detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 33 2237–2240. 10.1007/s10096-014-2199-3 [DOI] [PubMed] [Google Scholar]
- Zagorianou A., Sianou E., Iosifidis E., Dimou V., Protonotariou E., Miyakis S., et al. (2012). Microbiological and molecular characteristics of carbapenemase-producing Klebsiella pneumoniae endemic in a tertiary Greek hospital during 2004-2010. Euro. Surveill. 17:20088. [PubMed] [Google Scholar]
- Zaidi A. K., Huskins W. C., Thaver D., Bhutta Z. A., Abbas Z., Goldmann D. A. (2005). Hospital-acquired neonatal infections in developing countries. Lancet 365 1175–1188. 10.1016/S0140-6736(05)71881-X [DOI] [PubMed] [Google Scholar]
- Zarfel G., Hoenigl M., Wurstl B., Leitner E., Salzer H. J., Valentin T., et al. (2011). Emergence of carbapenem-resistant Enterobacteriaceae in Austria, 2001-2010. Clin. Microbiol. Infect. 17 E5–E8. 10.1111/j.1469-0691.2011.03659.x [DOI] [PubMed] [Google Scholar]
- Zhang X., Li X., Wang M., Yue H., Li P., Liu Y., et al. (2015). Outbreak of NDM-1-producing Klebsiella pneumoniae causing neonatal infection in a teaching hospital in mainland China. Antimicrob. Agents Chemother. 59 4349–4351. 10.1128/AAC.03868-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jiang X., Wang Y., Li G., Tian Y., Liu H., et al. (2014). Contribution of β-lactamases and porin proteins OmpK35 and OmpK36 to carbapenem resistance in clinical isolates of KPC-2-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 58 1214–1217. 10.1128/AAC.02045-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Xu H., Chen D., Zhou H., Hu X., Cheng G. (2014). First emergence of acrAB and oqxAB mediated tigecycline resistance in clinical isolates of Klebsiella pneumoniae pre-dating the use of tigecycline in a Chinese hospital. PLoS ONE 9:e115185 10.1371/journal.pone.0115185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zowawi H. M., Forde B. M., Alfaresi M., Alzarouni A., Farahat Y., Chong T. M., et al. (2015). Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci. Rep. 5:15082 10.1038/srep15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zowawi H. M., Sartor A. L., Balkhy H. H., Walsh T. R., Al Johani S. M., AlJindan R. Y., et al. (2014). Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob. Agents Chemother. 58 3085–3090. 10.1128/AAC.02050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zujic Atalic V., Bedenic B., Kocsis E., Mazzariol A., Sardelic S., Barisic M., et al. (2014). Diversity of carbapenemases in clinical isolates of Enterobacteriaceae in Croatia–the results of a multicentre study. Clin. Microbiol. Infect. 20 O894–O903 10.1111/1469-0691.12635 [DOI] [PubMed] [Google Scholar]