Abstract
We infected mice with the 2009 influenza A pandemic virus (H1N1pdm09), boosted with an inactivated vaccine, and cloned immunoglobulins (Igs) from HA-specific B cells. Based on the redundancy in germline gene utilization, we inferred that between 72–130 unique IgH VDJ and 35 different IgL VJ combinations comprised the anti-HA recall response. The IgH VH1 and IgL VK14 variable gene families were employed most frequently. A representative panel of antibodies were cloned and expressed to confirm reactivity with H1N1pdm09 HA. The majority of the recombinant antibodies were of high avidity and capable of inhibiting H1N1pdm09 hemagglutination. Three of these antibodies were subtype-specific cross-reactive, binding to the HA of A/South Carolina/1/1918(H1N1), and one further reacted with A/swine/Iowa/15/1930(H1N1). These results help define the genetic diversity of the influenza anti-HA antibody repertoire profile induced following infection and vaccination, which may facilitate the development of influenza vaccines that are more protective and broadly neutralizing.
Importance
Protection against influenza viruses is mediated mainly by antibodies, and in most cases this antibody response is narrow, only providing protection against closely-related viruses. In spite of this limited range of protection, recent findings indicate individuals immune to one influenza virus may contain antibodies (generally a minority of the overall response) that are more broadly reactive. These findings have raised the possibility that influenza vaccines could induce a more broadly protective response, reducing the need for frequent vaccine strain changes. However, interpretation of these observations is hampered by the lack of quantitative characterization of the antibody repertoire. In this study, we used single-cell cloning of influenza HA-specific B cells to assess the diversity and nature of the antibody response to influenza hemagglutinin in mice. Our findings help put bounds on the diversity of the anti-hemagglutinin antibody response, as well as characterizing the cross-reactivity, affinity, and molecular nature of the antibody response.
Introduction
Influenza viruses are common pathogens of humans and animals. In humans, influenza virus infections cause substantial morbidity and mortality through seasonal epidemics and occasional pandemics (1, 2). Vaccination remains the key component of public health protection against influenza virus infection. However, durable protection is limited by the ability of influenza virus to undergo rapid genetic and antigenic change, allowing it to escape from pre-existing immunity. As a result, influenza vaccines require updating on a regular basis (3).
Antibodies that target the principal viral surface protein, hemagglutinin (HA), play a major role in protection against influenza virus infection and provide the basis for current vaccine design and the tests by which vaccine efficacy is assessed (4). HA is a trimeric glycoprotein consisting of the membrane-distal globular head and the stalk region. The head is responsible for receptor binding, while the HA2 subunit is required for viral fusion with cellular membranes. Antibodies that bind to the globular head of HA and block receptor binding can be detected by the hemagglutination inhibition (HI) assay, and HI antibody threshold titers ≥32 are associated with a reduction in the risk of influenza infection (5). There are four or five antigenic sites in the globular head region of the HA (6, 7) and antibodies binding to these antigenic sites generally have virus-neutralizing activity, but non-neutralizing globular-head binding antibodies also have been described (8–12). Antibodies binding outside these antigenic sites, including to the stalk region of HA, have also been identified. Some of these antibodies confer protection against infection, either by directly blocking virus infectivity or by playing a role in other functions of the immune system, such as antibody-dependent cellular cytotoxicity (ADCC)-mediated activation of NK cells (13–15) or complement (16, 17).
Influenza virus can tolerate significant sequence variation in antigenic sites, and sequence changes in these regions (“antigenic drift”) often reduce binding of the existing antibody repertoire. Some anti-HA antibodies, however, are broadly cross-reactive, and can confer protection to a range of viruses within a particular HA subtype or even across subtypes. These antibodies have been reported to bind to conserved regions within the globular head of HA (18–22) or the stalk region (18, 19, 23–28).
Clearly the goal of influenza vaccination is to increase the proportion of antibodies that are protective, and ideally to increase the proportion that are cross-protective against multiple strains. Generating antibodies after vaccination that are cross-protective against antigenically drifted strains of the same subtype may reduce the need for frequent updates of vaccine strains. Furthermore, a vaccine that could elicit broadly cross-reactive antibodies that protect against multiple subtypes of HA would be an important public health tool in the event of a newly emerged virus with pandemic potential. However, without understanding the relative frequency of strain-specific versus cross-reactive, it is difficult to preferentially stimulate an optimal antibody response.
Evaluating the anti-influenza repertoire in humans is complicated by the fact that, with increasing age, individuals are repeatedly exposed to antigenically diverse influenza viruses, including both different subtypes and drifted strains within a subtype. Exposure to any particular HA is likely to preferentially stimulate and expand previously primed B cells, eclipsing the stimulation of naive B cells and thereby potentially impairing the generation of strain-specific, high-affinity antibodies that provide a more protective response.
At the molecular level, mouse antibody responses are generally similar to that of humans, but the exposure of laboratory mice to influenza viruses can be controlled. To understand the antibody repertoire arising from infection with influenza virus, we infected naïve C57BL/6 mice with the 2009 pandemic influenza A virus (H1N1pdm09) and boosted them with an inactivated influenza vaccine. B cells specific to HA were purified, and the immunoglobulin (Ig) heavy and light chain variable regions were cloned from single B cells to characterize the diversity and specificity of the anti-HA antibody repertoire.
Materials and Methods
Cells, virus, and recombinant hemagglutinin
Human embryonic kidney 293T cells (293T) were obtained from ATCC (Manassas, VA) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (Thermo Fisher Scientific, Pittsburg, PA) and penicillin streptomycin (Invitrogen, Carlsbad, CA).
Influenza viruses were grown in embryonated chicken eggs and titers were determined on MDCK cells by plaque assay. Table 1 summarizes the viruses and recHAs used, including the abbreviations used in this report.
Table 1.
Viruses and recombinant proteins used in this study. recHA, recombinant hemagglutinin. TIV, trivalent inactivated vaccine. MIV, monovalent inactivated vaccine.
| Strain | Abbreviation | Subtype | Group | HA Accession # | HA (HA1) % AA identity to OH/07 | Comment |
|---|---|---|---|---|---|---|
| A/Ohio/07/2009 | OH/07(H1pdm) | H1N1pdm09 | 1 | ACQ76386.2 | 100 | Wild-type virus for mouse infection |
| A/California/04/2009 | CA/04(H1pdm) | H1N1pdm09 | 1 | ACP41105.1 | 99 (98) | recHA/FACS bait/ELISA antigen |
| A/California/04/2009 | CA/04(HA1pdm) | H1N1pdm09 | 1 | (98) | recHA/ELISA antigen TIV | |
| A/California/07/2009 | CA/07(H1pdm) | H1N1pdm09 | 1 | AFM72832.1 | 99 (98) | component/ELISA antigen/HI, recHA/ForteBio |
| A/Shorebird/Delaware/68/2004 | DE/68(H13) | H13N9 | 1 | ABB87334.1 | 50 (45) | recHA/FACS bait/ELISA antigen TIV |
| A/Victoria/210/2009(H3N2) | VIC/210(H3) | H3N2 | 2 | AFM72894.1 | 43 (36) | component/ELISA antigen/HI TIV |
| B/Brisbane/60/2008 | Bris/60(B) | B (Victoria lineage) | - | ADN32817.1 | 28 (22) | component/ELISA antigen/HI |
| A/South Carolina/1/1918 | SC/1918(H1) | H1N1 | 1 | EPI15571 | 86 (83) | recHA/ELISA antigen |
| A/swine/Iowa/15/1930 | IA/1930(H1) | H1N1 | 1 | AF091308.1 | 86 (83) | recHA/ELISA antigen |
| A/Brisbane/59/2007 | Bris/59(H1) | H1N1 | 1 | EPI162496 | 80 (73) | recHA/ELISA antigen |
The full-length recombinant his-tagged HA (recHA) from CA/04(H1pdm) and DE/68(H13) used for fluorescence-activated cell sorting (FACS) analysis were obtained from the Influenza Reagent Resource (IRR) (Manassas, VA). For ELISA studies, full-length recHA from CA/04(H1pdm), SC/1918(H1), IA/1930(H1) and Bris/59(H1) were also obtained from IRR (Manassas, VA). For kinetic studies, the CA/07(H1pdm) recHA protein was produced in a baculovirus expression system as previously described (29).
Inoculation of mice and isolation of leukocytes
All animal experiments were performed in accordance with the guidelines of the CDC Institutional Animal Care and Use Committee in an Association for the Assessment and Accreditation of Laboratory Animal Care accredited facility. Female 8–12 week old C57BL/6 mice were anesthetized by isoflurane inhalation and inoculated intranasally with 50 μL containing 104–105 plaque-forming units of OH/07(H1pdm) diluted in sterile PBS, pH 7.4. Three to four weeks post-infection, mice were boosted by IP injection with 200 μL of the 2011/12 trivalent inactivated influenza vaccine (TIV) (Fluarix: GSK, Research Triangle Park, NC), which contained the HA from CA/07(H1pdm), VIC/210(H3), and BR/60(B) viruses (6 μg of each HA). Three days post-boost, four mice per group were euthanized by isoflurane overdose followed by cervical dislocation, and spleens were harvested and pooled in order to remove inter-individual bias. In brief, the spleens were ground between two frosted glass slides, and the cells were dissociated through a 70-μm mesh strainer. Cells were then centrifuged at 550 x g for 20 min over an 18.23% histodenz (Sigma-Aldrich, St Louis, MO) cushion at room temperature with the brake off. The low-density cells were collected, washed with FACS buffer and incubated in red blood cell lysing media (Sigma-Aldrich) for 5 min at room temperature. The cells were again washed and resuspended in FACS buffer for further analysis.
Biotinylation of recHA
CA/04(H1pdm) and DE/68(H13) recHA (2 mg of each) were buffer exchanged into 0.1 M sodium acetate, pH 5.5 (oxidation buffer), by centrifugation in Amicon Ultra-4 desalting centrifugal filter units (Millipore, Billerica, MA) and diluted to a final volume of 1 ml (2 mg/ml). The glycans on the proteins were oxidized by adding 1 ml of 20 mM sodium meta-periodate (Sigma-Aldrich) and incubating for 30 min on ice, in the dark. The buffer was then exchanged to PBS, pH 7.2, in a total volume of 3.6 ml, using a desalting column. Four hundred μl of 50 mM Pierce Biotin-PEG4-Hydrazide in DMSO (Thermo Fisher Scientific) was added to the protein, which was then incubated for 2 h at room temperature with gentle rocking, protected from light. The protein was again buffer exchanged into 50 mM Tris-Cl, pH 7.4 containing 100 mM NaCl and 15 % w/v glycerol. The concentration of the protein was determined using the Coomassie Plus Protein Assay Reagent (Thermo Fisher Scientific), and the molar ratio of biotin to protein was quantified using the Pierce Biotin Quantitation Kit (Thermo Fisher Scientific). The protein was stored at −80 °C in single-use aliquots.
B cell labeling and flow cytometry
The isolated leukocytes were labeled with biotinylated HA, followed by streptavidin-APC (BD Biosciences). In addition, the following surface markers were used: CD3e-V450 (500A2), CD4-V450 (RM4-5), CD8a-V450 (53-6.7), CD11b-V450 (M1/70), IgD-V450 (11-26c.2a), Gr1-V450 (RB6-8C5), CD19-PE-Cy7 (1D3), CD138-PE (281-2) and IgM-FITC (R6-60.2) (BD Biosciences, San Jose, CA). All staining was performed in the following antibody dilution buffer: Hank’s Balanced Salt Solution (HBSS) containing 5 % FBS (Hyclone), 10 mM of EDTA, 10mM of HEPES, 1% Fc Block (BD Biosciences, San Jose, CA) and 5% each of normal hamster serum, normal mouse serum and normal rat serum (Jackson ImmunoResearch). The optimal staining conditions identified were 5 μg/ml of biotinylated-HA and 1.25 μg/ml of streptavidin-APC.
Cells were analyzed using a BD FACS CANTO II, and single cell sorting was performed using a BD FACS Aria II. The cells were gated on FSC-A/FSC-H to remove doublets and FSC-A/SSC-A to remove debris. Single antigen-positive B cells were sorted based on IgM-/CD19+/HA+ surface staining onto the 48 reaction sites of an AmpliGrid AG480F slide (Beckman Coulter, Houston, TX). A visual analysis of cell deposition on the AmpliGrid AG480F slide using a microscope was used to monitor the accuracy of the sort.
Molecular cloning of Ig genes
A Multiplex One-step RT-PCR (Qiagen, Valencia, CA) was performed directly on the slide in a 1 μL reaction containing 1x Qiagen RT-PCR buffer, 0.6 pmol of primer mix, 0.4 mM of each dNTP, 0.8 U of RNAsin and 0.04 μL of Qiagen enzyme mix. The primer mix was adopted from Tiller et al. with slight modifications (Supplemental Table 1) (30, 31). The reactions were overlaid with 5 μL of sealing solution (Advalytix AG, Munich, Germany), and amplification was performed on an AmpliSpeed slide cycler (Beckman Coulter, Houston, TX) as follows: 58 °C for 30 min, 93 °C for 10 min, 40 cycles of 93 °C for 45 sec/54 °C for 1 min/72 °C for 90 sec and a final incubation at 72 °C for 10 min. This initial reaction was transferred to a 96-well PCR plate and diluted to final volume of 10 μL with RNase free water to serve as template for subsequent reactions. In total, 1 μL of the 1st round reaction was used as a template for a semi-nested (IgH) and nested (IgK) reactions. The PCR was performed in 25 μL reactions as follows: 94 °C for 5 min, 35 cycles of 93 °C for 45 sec/55 °C (IgH) or 45 °C (IgK) for 1 min/72 °C for 1.5 min and a final incubation at 72 °C for 10 min. Amplification products were screened by agarose gel electrophoresis, and positive amplicons were sequenced for identification of germline variable and joining gene usage by the International ImMunoGeneTics information system (IMGT)(32). Second-round reactions were then repeated using 1 μL of the 1st round template and targeted, gene-specific, V and J primers incorporating restriction sites for directional cloning into pFUSE2ss-CLIg-hG1 and pFUSE2ss-CLIg-hk expression vectors (InvivoGen, San Diego, CA). EcoRI and BsiWI were used for IgH and EcoRI and NheI for IgK ligations, respectively. TOPO cloning was used as an alternative strategy for Ig’s with internal restriction sites.
Recombinant monoclonal antibody production and quantification
293T cells in T160 flasks at 80% confluency were washed to remove serum IgG, and covered with OPTIMEM (Invitrogen, Carlsbad, CA) without serum. IgH and IgK constructs were co-transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The cell culture supernatants were harvested up to 10 days post transfection and concentrated using Amicon Ultra-30 Centrifugal Filter Unit (Millipore, Billerica, MA). The concentration of each recombinant monoclonal antibody (rmAb) was determined by ELISA, and all of the samples were normalized to a 25 μg/mL working stock.
ELISA
Costar Hi Bind plates (Corning Inc., Tewksbury, MA) were coated overnight with TIV or the individual monovalent inactivated virus constituents at 4 °C. For 2011/12 TIV, 3 μg/ml of total HA was adsorbed, and 1 ug/ml total HA was adsorbed for the individual vaccine constituents. For recHA, 1 ug/ml of respective protein was absorbed. The plates were blocked for 1 h with PBS/0.1 % Tween20 (PBST) containing 1.5 % BSA (blocking buffer) for 1 h at room temperature. All rmAb were serially titrated in blocking buffer and allowed to incubate for 1 h at room temperature. After three PBST washes, the wells were probed with goat anti-human (H&L)-HRP (Thermo Scientific) for 1 h at room temperature. The plates were washed three times with PBST, and the signal was developed with 1 step Turbo TMB-ELISA reagent (Thermo Scientific). The reactions were stopped with 1 N sulfuric acid, and the absorbance was read at 450 nm.
Hemagglutination inhibition assay
The indicated rmAb were tested for reactivity with the following BPL-inactivated virus antigens: H1N1 A/California/07/2009 NYMC X-179A (FR-187), H3N2 A/Victoria/210/2009 X-187 (FR-644) and B/Brisbane/60/2008 (FR-47) (IRR), by hemagglutination (HI) assay as previously described (33) using 4 hemagglutinating units (HAU) of antigen and 0.5 % turkey red blood cells.
BioLayer interferometry
The binding of each rmAb to CA/07 was analyzed by BioLayer interferometry (BLI) on an Octet Red instrument (Fortebio, Inc., Menlo Park, CA) according to the manufacturer’s instructions. The CA/07(H1pdm) recHA protein (34) was diluted to 75 μg/ml in kinetics buffer (KB; PBS containing 0.02 % Tween 20, 0.005 % sodium azide and 100 μg/ml bovine serum albumin, purchased from Fortebio as 10 x stock) and were coupled to offline (prior to running assay) anti-Penta-His biosensors using a Sidekick instrument (Fortebio). A twofold dilution series (25–0.391 μg/ml) for each rmAb in KB was prepared and analyzed, including a commercial anti-A/California/06/2009 (H1N1pdm09) mAb (Immune Technology Corp., CA) as a control. The data were analyzed using the system software and exported as a Microsoft Excel file for analysis and presentation in other software packages.
Alignments and phylogenetic trees
CDR3 regions were aligned using MUSCLE (35) and a maximum likelihood tree was generated using MEGA5 (36).
Estimates of germline diversity
To estimate the diversity of the germline population from which our samples of anti-HA heavy and light chains were drawn, we used the methods of Burnham and Overton (37, 38), Chao (39), Chao and Lee (40), Chao and Bunge (41), Wang and Lindsay (42), and Wang (43), as implemented in the R module SPECIES (44).
Results
Anti-HA B cell identification and purification
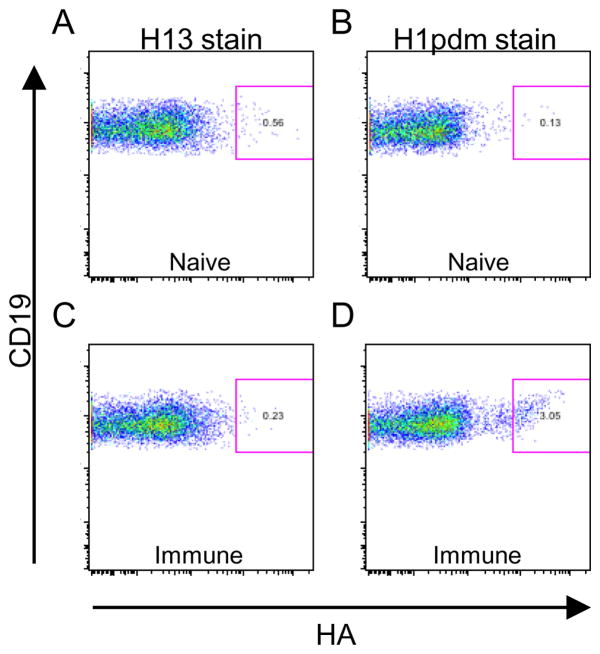
To induce an anti-HA response, C57BL/6 mice were administered a sub-lethal infection with OH/07(H1pdm), and three weeks later, the mice were boosted with 2011/2012 TIV. Three days post-boost, spleens from four mice were harvested and pooled. Antigen positive B cells (1.5–8 % of the CD19/IgM- population: (Figure 1D vs. B) were single cell-sorted onto glass slides based on their ability to bind CA/04(H1pdm) recHA. As a control, lymphocytes were stained with the distantly-related recHA of DE/68(H13) (Figure 1A, C).
Figure 1. Flow cytometry of splenocytes from mice immunized with H1pdm.
C57BL/6 mice were immunized with H1N1pdm09 virus and boosted with TIV as described. Splenocytes from naïve mice (A, B) or immunized mice (C,D) were stained with a cocktail of antibodies including CD19, and with H13 as a control (A,C) or H1pdm (B,D). Splenocytes were gated as described to show the percent of HA-specific B cells.
Anti-HA IgH and IgL gene segment usage
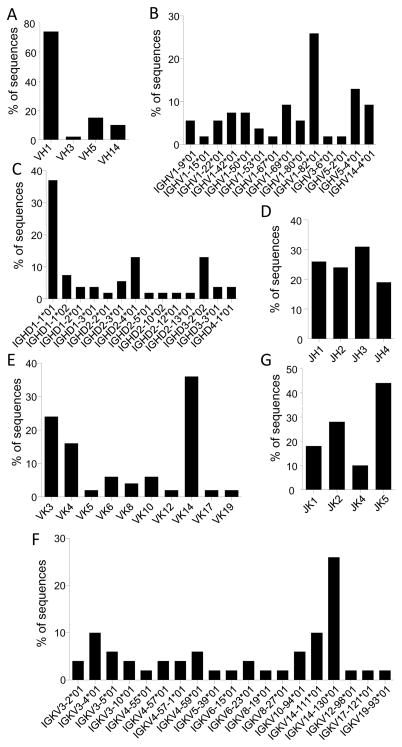
The amplification of the IgH gene from the single cells yielded 54 heavy chains for which respective VDJH germline genes were identified according to the international ImMunoGeneTics database (IMGT) (32). In total, genes representing 4 of the 15 murine VH families were identified (Figure 2A). The VH chains displayed a strong bias toward the use of the VH1 family (74%), which represents approximately 50% of the VH gene repertoire available and is located most distal from the diversity genes (45–47). Within the four VH families, the VH genes included 14 of the possible 92 functional C57BL/6 VH genes (46), of which IGHV1-82*01 was the most common (26%), followed by IGHV5-4*01 (13%) (Figure 2B). The DH segment of IgH most frequently employed was the DH1-1*01 gene (37%) (Figure 2C). For the JH gene segment, genes 1–4 were used with similar frequencies, ranging from 20–30% each (Figure 2D).
Figure 2. IgH and IgL germline usage.
HA-specific B cells were sorted and IgH and IgL variable regions were amplified from individual B cells and sequenced. Germline usage was determined using the international ImMunoGeneTics database of mouse immunoglobulin sequences. Usage of VH gene families (A), VH genes (B), DH genes (C), JH genes (D), VK gene families (E), VK genes (F) and JK genes (G) are shown as a percentage of total heavy or light chain sequences respectively.
Because C57BL/6 mice predominately use the kappa light chain (45), and preliminary experiments confirmed that the anti-HA response in our experiments was comprised of 85–90% kappa light chain usage (data not shown), we only amplified these VL chains. Of the 18 known VK families, 10 (55.5%) were identified in our screen with a bias toward use of the VK14 family (36%) (Figure 2E). VK3 and 4 were also well represented, with 24% and 16% of germline sequences used, respectively. Of the possible 93 murine-expressed VK genes, 19 were recovered from the HA-specific B cells (20%) (Figure 2F), and of these, segment IGKV14-130*01 was obtained most frequently (26%), followed by IGKV3-4*01 and 14-11*01 (10% each). All four possible IgL J genes were used, with JK5 represented most often (44%) (Figure 2G).
Germ-line gene combinations and CDR3 sequence analysis of the IgH and IgL chain anti-HA repertoire
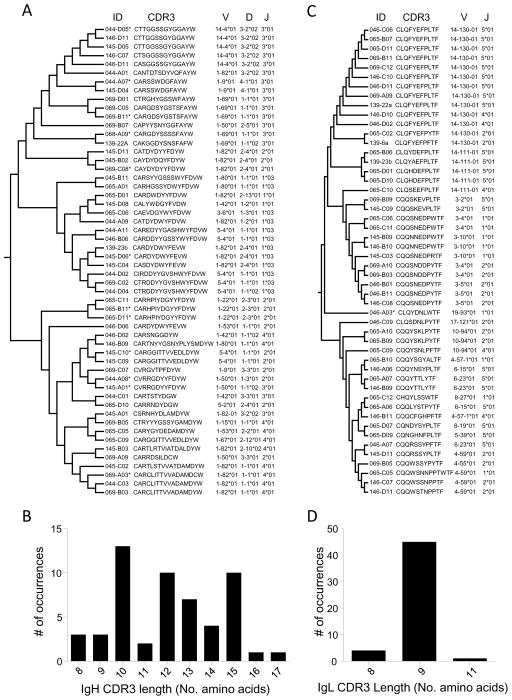
The 54 individual anti-HA IgH chains included 32 distinct VDJH combinations, with 11 being employed multiple times (from 2–6 times) (Figure 3A). The length of the CDR3 region (produced through joining the 3’ end of VH, a D segment, and the 5’ end of JH) ranged from 8 to 17 amino acids (Figure 3B). As well as a number of identical CDR3 sequences, clusters of similar CDR3 sequences were observed, mainly but not solely driven by similar VDJ usage (Figure 3A). For example, the sequence C[T/A][T/S]GGSSGYGGAYW was seen 5 times, and was generated by the use of the same V, D, and J gene segments (14-4*01, 3-2*02, and 3*01 respectively), while the sequence CVRRGDYYFDYW, identified twice, was generated from either 1-50*01/1-1*02/3*01 or 1-50*01/1-3*01/2*01 V/D/J regions.
Figure 3. IgH V, D and J IgL V and J gene combinations and CDR3 sequences.
CDR3 sequences were aligned using MUSCLE [1] and a maximum likelihood dendrogram was generated using MEGA5 for IgH (A) and IgL (C). Columns indicate the IgH or IgL identifier (“ID”), the CDR3 sequence (CDR3), and the identity of the V, D, and J (IgH) or V and J (IgL) gene segments used. “*” indicates the segment was sequenced but not cloned. The overall distribution of IgH (B) and IgL (D) CDR3 length.
The 50 IgL chains encode 25 unique VJL combinations, 12 of which were represented multiple times (from 2–9 instances) (Figure 3C). For IgL, the CDR3 region is formed by the 3’ end of VL and the 5’ end of JL. The length of the IgL CDR3 ranged from 8 to 11 amino acids, with 90% encoding 9 residues (Figure 3D). The VL CDR3 sequences were less diverse than those of VH, with three major clusters identified, including 9 that had identical sequences (Figure 3C). As with IgH, some evidence for convergence on sets of CDR3 sequences was observed. Similar CDR3 sequences were usually generated by identical V/J germline usage (e.g., 9 instances of CLQFYEFPLTF generated by 14-130-01/5*01), but in some cases, similar CDR3 sequences were generated by different V and J germlines (CQQS[K/N]E[V/D]P[L/W]TF generated by 3-2*01/5*01 or 3-4*01/1*01).
In 18 cases we amplified both the IgH and IgL chain from the same B cell, allowing analysis of the complete antibody gene segment utilization (Table 2), as well as the cloning of an rmAb for functional analysis. In some cases (e.g., 146-D11 and 146-C07), the use of a similar IgH sequence (CDR3: CT[T/S]GGSSGYGGAYW, V/D/J : 14-4*01/3-2*02/3*01) and an identical light chain (CDR3: CQQWSSNPPTF,V/J: 4-59*01/2*01) suggests that these samples may represent clonal expansion and somatic mutation of a single naïve B cell. However, rmAb 046-D11, with the same IgH CDR3 and V/D/J usage as 146-D11 and 146-C07, was paired with a different light chain (CDR3 sequence CLQFYEFPLTF, 14-130-01/5*01 V/J). Similarly, within IgL sequences, the IGKV14-130*01/IgJ5*01 combination was seen most often (18% of sequences: Figure 3C), but of the three rmAb with this IgL sequence (046-D11, 069-A09, and 139-22a: Figure 3C and Table2), each was paired with a different IgH, showing that these were not produced by clonal expansion and again supporting the notion of convergence upon a common paratope.
Table 2. rmAb germline gene usage and number of mutations.
For eighteen individual B cells, both heavy and light chain variable sequences were sequenced. The germline usage for VDJ and VJ (VH/VL) genes was defined using the IMGT database. The number of amino acid mutations (“ ”) was identified by comparison of the germline sequence to the amino acid sequence of the respective genes.
| rmAb | VH | DH | JH | Δ | VL | JL | Δ |
|---|---|---|---|---|---|---|---|
| 046-D02 | 1-42*01 | 1-1*01 | 4*01 | 3 | 14-130*01 | 4*01 | 3 |
| 046-D11 | 14-4*01 | 3-2*02 | 3*01 | 9 | 14-130*01 | 5*01 | 4 |
| 065-C05 | 1-53*01 | 2-2*01 | 4*01 | 4 | 4-59*01 | 1*01 | 2 |
| 065-C06 | 3-6*01 | 2-3*01 | 1*03 | 0 | 3-4*01 | 1*01 | 0 |
| 065-C09 | 1-67*01 | 2-12*01 | 4*01 | 11 | 10-94*01 | 4*01 | 6 |
| 065-C11 | 1-22*01 | 2-3*01 | 2*01 | 7 | 3-4*01 | 1*01 | 3 |
| 065-D01 | 1-82*01 | 2-4*01 | 1*01 | 6 | 14-111*01 | 5*01 | 3 |
| 065-D10 | 5-2*01 | 2-4*01 | 2*01 | 7 | 14-111*01 | 5*01 | 8 |
| 069-A09 | 1-50*01 | 2-10*02 | 2*01 | 7 | 14-130*01 | 5*01 | 1 |
| 069-B03 | 1-82*01 | 1-1*01 | 4*01 | 4 | 3-4*01 | 2*01 | 4 |
| 069-B05 | 1-15*01 | 1-1*01 | 4*01 | 8 | 4-55*01 | 2*01 | 1 |
| 139-22a | 1-69*01 | 2-5*01 | 3*01 | 15 | 14-130*01 | 5*01 | 1 |
| 139-23b | 1-82*01 | 2-4*01 | 1*03 | 10 | 14-111*01 | 5*01 | 3 |
| 145-C09 | 5-4*01 | 1-1*01 | 2*01 | 5 | 3-2*01 | 5*01 | 0 |
| 145-D11 | 1-82*01 | 2-4*01 | 2*01 | 2 | 4-57*01 | 3*01 | 4 |
| 146-B09 | 1-80*01 | 1-1*01 | 4*01 | 7 | 6-23*01 | 5*01 | 5 |
| 146-C07 | 14-4*01 | 3-2*02 | 3*01 | 6 | 4-59*01 | 2*01 | 3 |
| 146-D11 | 14-4*01 | 3-2*02 | 3*01 | 4 | 4-59*01 | 2*01 | 4 |
Diversity of the anti-HA antibody response
Our sample contained 32 different VDJ sequences among 54 total heavy chain sequences, and 25 different VJ sequences among 50 total light chain sequences. We used several approaches (37–44) to predict the overall germline sequence diversity within the anti-HA immunoglobulin response, based on the numbers of repeated (“recaptured”) sequences in our sample. The algorithms consistently inferred that our sample drew from a pool of 72-130 different heavy chain VDJ combinations, and 35 light chain VJ combinations (Table 3).
Table 3. IgH and IgL diversity estimates.
The “SPECIES” module for R was used to estimate the diversity of the combinations of germline V, D, and J segments (IgH chain) or V and J segments (light chain) in the population of antibodies specific for HA, based on sampled IgH and IgL sequences. Estimates and 95% confidence intervals (CI) are shown for each method.
| VDJ (H)
|
VJ (L)
|
|||
|---|---|---|---|---|
| Estimate | 95% CI | Estimate | 95% CI | Reference |
|
|
|
|||
| 113 | 55–321 | 35 | 29–60 | Chao (1984) |
| 75 | 47–148 | 38 | 30–60 | Chao & Lee (1992) (ACE method) |
| 92 | 51–223 | 38 | 29–63 | Chao & Lee (1992) (ACE-1 method) |
| 72 | 40–171 | 35 | 28–60 | Chao & Bunge (2002) |
| 111 | 63–159 | 39 | 29–49 | Burnham & Overton (1978), Burnham & Overton (1979) |
| 129 | 47–251 | 38 | 28–80 | Wang & Lindsay (2005) |
| 97 | 63–114 | 37 | 32–71 | Wang (2010) |
Generation of mouse-human chimeric antibodies
In eighteen cases, we cloned both an IgH and an IgL variable region from the same individual B cell (Table 2). All except two (146-C07 and 146-D11) were unique in terms of germline IgH and IgL combinations. When sorting single B cells by flow cytometry, we selected IgG-expressing rather than IgM-expressing cells. Since this population has undergone class switching, they should be dominated by memory cells and likely contain some level of somatic mutation in the IgH and IgL regions. Varying levels of somatic mutation, at the amino acid level, were present in these genes (0–15 mutations for H and 0–8 for L) (Table 2, “Δ”). Only one rmAb (065-C06) retained germline sequence in both IgH and IgL, indicating that as expected most antibodies had undergone some amount of affinity maturation during the infection and vaccine boost.
The matched IgH and IgL chains were cloned into a vector supplying an IgG1 human constant region (heavy chain) or human kappa constant region (light chain), as well as the IL2 signal sequence and promoter elements to allow expression in mammalian cells. For 16 of the pairs, co-expression of the IgH and IgL chains in 293T cells resulted in secretion of recombinant monoclonal antibodies (rmAb), which migrated at approximately 150 kDa when subjected to SDS-PAGE, the same size as a control Ig (data not shown). Two of the antibody IgH/IgL pairs (065-C09 and 065-D10) did not efficiently assemble into full-sized mature Ig, and predominately showed bands of an apparent molecular mass of 50 and 25 kDa (data not shown); therefore, these pairs were not analyzed further. The concentration of each rmAb was determined by ELISA and normalized by concentrating the tissue-culture supernatant to approximately 25 μg/mL.
Most cloned Igs recognize influenza hemagglutinin
The 16 rmAb were screened for reactivity by ELISA using the 2011/12 TIV as antigen. In total, 15 of the rmAb reacted with TIV (defined as a signal greater than twice background level, at ≤25 μg/mL), and 10 of these bound at a concentration of <1 μg/mL (Table 4), indicating that the sorting process isolated B cells that do in fact target HA.
Table 4. Binding of rmAb to inactivated vaccines and hemagglutination inhibition assays.
rmAb produced as described were screened by ELISA for reactivity with TIV (containing CA/07(H1pdm), VIC/210(H3), and BR/60(B)) and with the individual monovalent inactivated vaccine (MIV) constituents: CA/07(H1pdm), VIC/210(H3), and BR/60(B). Data are shown as minimum rmAb concentration (ng/ml) required to produce a 2-fold signal above negative control. “-“, no reactivity seen at concentrations ≤25 μg/mL. “NT”, not tested. rmAb were also tested in HI assays against CA/07(H1pdm), VIC/210(H3), and B/BR/60. Values are shown as minimum concentration (ng/mL) rmAb required to inhibit 4 HA units of antigen. “*“, no HI activity seen at concentrations ≤12.5 μg/mL.
| ELISA titer
|
HI titer
|
||||||
|---|---|---|---|---|---|---|---|
| rmAb | TIV | CA/07 (H1pdm) | VIC/210 (H3) | BR/60 (B) | CA/07 (H1pdm) | VIC/210 (H3) | BR/60 (B) |
|
|
|
||||||
| 046-D02 | - | - | NT | NT | * | * | * |
| 046-D11 | 1390 | - | - | - | * | * | * |
| 065-C05 | 1.9 | 1.14 | - | - | 780 | * | * |
| 065-C06 | 25000 | - | NT | NT | * | * | * |
| 065-C11 | 50 | 3.43 | 5.7 | - | * | * | * |
| 065-D01 | 5.7 | 3.43 | - | - | 98 | * | * |
| 069-A09 | 930 | 30.86 | - | - | 98 | * | * |
| 069-B03 | 25000 | 25000 | - | - | * | * | * |
| 069-B05 | 25000 | - | NT | NT | * | * | * |
| 139-22a | 1.9 | 1.14 | - | - | 98 | * | * |
| 139-23b | 25000 | - | NT | NT | * | * | * |
| 145-C09 | 150 | 3.43 | - | - | 780 | * | * |
| 145-D11 | 1.9 | 3.43 | 25000 | - | 98 | 12500 | * |
| 146-B09 | 5.7 | 1.14 | 25000 | - | 49 | * | * |
| 146-C07 | 1.9 | 1.14 | - | - | 98 | * | * |
| 146-D11 | 1.9 | 1.14 | 25000 | - | 98 | * | * |
The rmAb were further characterized by ELISA for reactivity to the individual viruses that were components of the TIV. Eleven of the rmAb were reactive with the monovalent vaccine containing the HA from CA/07(H1pdm), while none reacted with inactivated BR/60(B) (Table 4). Four rmAb were also reactive with inactivated VIC/210(H3), although for three rmAb (145-D11, 146-B09 and 146-D11) this reactivity was only seen at the highest concentration tested. The fourth rmAb (065-C11) was strongly reactive with VIC/210(H3) as well as CA/07 (H1pdm) (Table 4), suggesting that it is a subtype cross-reactive antibody.
Cloned Igs recognize H1 with high affinity
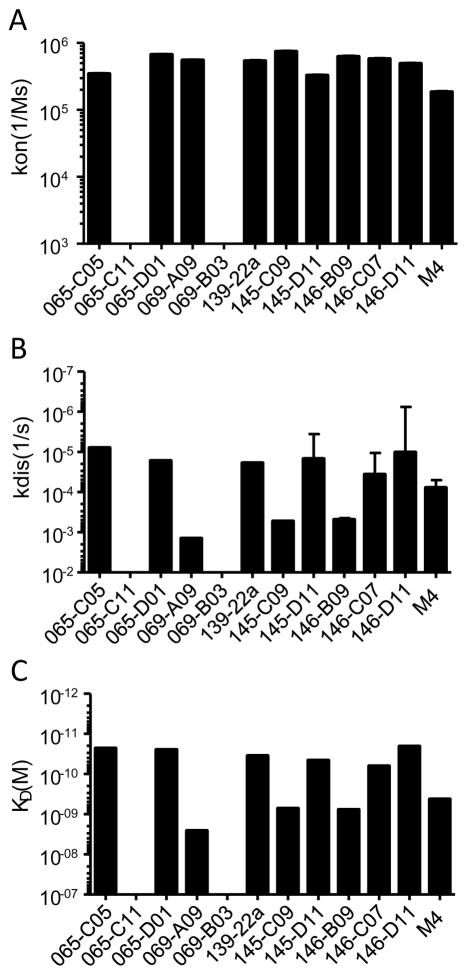
BioLayer interferometry (BLI) was performed on the 11 rmAb with the highest binding activity toward H1N1pdm by ELISA analysis, to determine the relative binding rate constants to recHA of CA/07(H1pdm). Two rmAb (065-C11 and 069-B03) did not bind detectably bind recHA in this assay, while the nine remaining rmAb bound recHA with similar association rates, surpassing that of the control Ab (Figure 4A). These nine rmAb can be divided into two categories based on dissociation rates: those ranging from 8 x 10−6(1/s) to 3.7 x 10−5(1/s) (065-C05, 065-D01, 139-22a, 154-C09, 145-D11, 146-C07 and 164-D11) and a second with faster dissociation rates ranging from 4.9 x 10−5(1/s) to 1.4 x 10−4(1/s) (069-A09, 145-C09 and 146-B09) (Figure 4B). As the rmAb association rates were similar, each group also has similar steady state affinities, of about 2 x 10−11 to 6.4 x 10−11 KD for the first group and 7.8 x 10−10 to 2.6 x 10−9 KD for the second (Figure 4C). Interestingly, 065-C11, an antibody that displayed strong reactivity with the TIV as well as with the individual H1N1pdm and H3N2 virus components of the TIV in ELISA (Table 4), failed to detectably bind to recombinant H1pdm in this assay.
Figure 4. Biolayer interferometry analysis of binding kinetics.
BioLayer Interferometry (BLI) using an Octet Red instrument (Fortebio) was performed as described. The association rate (Kon) (A), dissociation rate (Kdis) (B), and the steady state affinity (KD) (C) were calculated for each rmAb. Error bars represent the standard deviation generated by running multiple rmAb concentrations.
Cloned Igs are able to inhibit hemagglutination
We also tested the ability of the 16 rmAb to block hemagglutination of CA/07(H1pdm) viruses. The HI assay is widely used to measure influenza post-infection and post-vaccination antibody responses that are biologically meaningful, since HI titers ≥32 or 40 are correlated with protection against influenza infection (5). All 9 rmAb that were strongly reactive in the recHA BLI assay also showed HI activity against the CA/07(H1pdm) virus at concentrations ranging from 0.098 to 0.78 μg/mL (Table 4). Only 145-D11, a rmAb that was weakly reactive with VIC/210(H3) by ELISA, showed HI activity against VIC/210(H3) virus, and then only at 12.5 μg/ml. 065-C11, which showed binding activity toward TIV in ELISA assays (Table 4) but no reactivity to the recHA in BLI (Figure 4), also lacked HI reactivity. All rmAb failed to inhibit hemagglutination of BR/60(B) (Table 4).
ELISA activity against H1N1 recHA’s
Having established that the panel of rmAb display only limited cross-clade reactivity, we further tested the ability of the eleven rmAb reactive to the H1pdm vaccine component (Table 4) to bind a set of recHA within the H1 lineage. The amino acid identities of each HA, compared to OH/07, are shown in Table 1, and range from 80% (BR/59) to 86% (SC/1918 and IA/1930). As a control we also included HA from the H13 lineage (DE/68).
All but 065-C11 bound to the full length recHA from CA/04(H1pdm). Consistent with the HI activity of these rmAb, the same rmAb also bound to the HA1 domain of CA/04. Three rmAb (065-D01, 069-A09 and 139-22a) also showed reactivity with the recHA of SC/1918, at levels comparable to that of CA/04 (Table 5). 065-D01 further reacted with the recHA of IA/1930, although at >200 fold increased concentration. None of the rmAb bound detectably to recHA from Bris/59. The rmAb 065-C11, which efficiently bound to both H1N1pdm09 and H3N2 vaccine antigen but failed to bind recHA in BLI analysis, also failed to bind recHA in the more sensitive ELISA assay. This suggests the 065-C11 rmAb may recognize a cross-reactive conformational epitope present in virus-associated HA that is not preserved in the recHA used for ELISA assays.
Table 5. Binding of rmAb to H1N1 recHA.
rmAb were screened by ELISA for reactivity with the respective recHA. Data are shown as minimum rmAb concentration (ng/ml) required to produce a 2-fold signal above negative control. “-“, no reactivity seen at concentrations ≤12.5 μg/mL.
| recHA
|
||||||
|---|---|---|---|---|---|---|
| rmAb | CA/04 (H1pdm) | CA/04 (HA1pdm) | SC/1918 (H1) | IA/1930 (H1) | Bris/59 (H1) | DE/68 (H13) |
| 065-C05 | 1.9 | 1.9 | - | - | - | - |
| 065-C11 | - | - | - | - | - | - |
| 065-D01 | 1.9 | 1.9 | 5.7 | 463 | - | - |
| 069-A09 | 1.9 | 1.9 | 51 | - | - | - |
| 069-B03 | 12500 | 12500 | - | - | - | - |
| 139-22a | 1.9 | 0.635 | 3 | - | - | - |
| 145-C09 | 1.9 | 0.57 | - | - | - | - |
| 145-D11 | 1.9 | 1.9 | - | - | - | - |
| 146-B09 | 460 | 150 | - | - | - | - |
| 146-C07 | 0.635 | 1.9 | - | - | - | - |
| 146-D11 | 0.635 | 1.9 | - | - | - | - |
Discussion
The antibody response to HA following influenza virus infection or vaccination includes a wide range of specificities. Some antibodies bind to highly strain-specific regions and confer protection, others bind to conserved regions that can lead to varying degrees of cross-reactive protection, and some may bind to HA without neutralizing the virus in vitro, although in some cases (though not all (8, 48)) such antibodies may confer some protection in vivo (49). Although antibodies with these different specificities and functions can be measured using different assays (e.g. ELISA, microneutralization (MN), and HI), the relative proportion of antibodies with each type of specificity and function that is stimulated by a given exposure event is unclear. In this study, we isolated mouse memory B cells specific to influenza H1pdm HA, analyzed individual IgH and IgL genes to characterize the genetic diversity of the response, and produced recombinant antibodies to analyze specificity and binding kinetics. Although there are some molecular differences between mouse and human antibodies, in general the B cell response is similar in the two species. Because humans are typically repeatedly exposed to multiple strains of influenza throughout their lifetime, leading to unpredictable expansion of particular B cell clones depending on the precise history of virus exposure, the mouse is a simpler model that can be used to assess antibody responses to the virus.
Overall, the anti-HA antibody response generated in these mice was relatively constrained. The 54 heavy chain variable genes analyzed in our sample included 14 different VH genes (representing 4 different families), while the 50 light chain variable regions included 19 different VK genes from 10 different families. We infer that our samples drew from a population of approximately 72–130 different heavy chain VDJ combinations, and approximately 35 different light chain VJ combinations. This suggests that there must be considerable redundancy in the germline usage for the anti-HA response, as previously proposed based on studies of anti-HA monoclonal antibody repertoire (50, 51). This inference assumes that the purification of the HA-specific B cells and cloning of variable regions was unbiased, and that relevant anti-HA antibodies raised against virion-associated HA will bind to recombinant HA. However, while it is difficult to validate this without already knowing the global anti-HA repertoire, sorting was performed using the full HA ectodomain and PCR primers covered the full range of heavy and kappa light chain sequences, so there is no a priori reason to expect bias. Considering the further diversity contributed by heavy/light chain pairing, junctional diversity, and somatic hypermutation, the actual pool of H1pdm anti-HA antibody repertoire must be significantly larger than these estimates, roughly consistent with the BALB/c PR8 anti-HA paratypic repertoire of approximately 1500 specificities estimated by Staudt and Gerhard in 1983 (51).
CDR3 regions are generally a principal determinant in antigen molecule binding specificity (52, 53). IgH chain CDR3 sequences showed some redundancy, while the IgL CDR3 repertoire was less diverse, suggesting that a limited number of CDR3 sequences (especially in IgL) are preferred for binding to H1pdm HA. The fact that very similar CDR3 sequences were generated from different IgH V/D/J IgL V/J combinations also suggests convergence on a specific set of CDR3 sequences. The lower diversity seen in the IgL germline usage and CDR3 sequences compared to IgH (Figure 3A vs. 3C) may imply that the IgL region is more important than the IgH region in determining binding specificity, and is therefore more constrained.
The analysis of 16 paired IgH/IgL chains, each derived from a single B cell (i.e. variable regions from the natural antibody), indicated that the majority of these Igs (15 out of 16) were indeed specific to the boosting antigen, in that they were capable of binding TIV in ELISA assays. Of these, 10 rmAb bound to inactivated CA/07 at low concentrations (<31 ng/mL), and 9 bound recHA with high affinity (>10−9 K). However, these assays may underestimate HA binding to B cells because previous studies have shown that B cell Igs that are capable of binding HA when expressed on the surface of a B cell (as membrane-bound molecules) may have low affinity or fail to bind to HA when expressed as secreted antibodies in the germline form (54). In particular, this may explain the poor binding activity of 065-C06, a rmAb that maintained strictly germline identity. This rmAb, as a soluble human/mouse chimeric molecule, bound TIV only at the highest concentration tested by ELISA. In addition, it is possible that the use of human constant regions for these mouse variable regions may have led to some changes in activity. For example, 2 of the cloned IgH/IgL pairs failed to assemble when expressed in the context of the human constant regions (Table 2 and data not shown), even though they were derived from a single B cell and presumably capable of effective assembly in vivo.
As well as binding to inactivated virus, 9 of 16 rmAb bound recHA1 (the globular head of HA) and were active in HI assays. Thus, >50% of the cloned antibodies have the ability to inhibit virus receptor binding, a function of neutralizing and protective antibodies (55).
Three rmAb further showed subtype-specific cross-reactivity with the antigenically related recHA of SC/1918, and one also reacted weakly with IA/30; none reacted with the recent seasonal H1N1 virus BR/59. These viruses share just 73–83% identity to OH/07 in the HA1 region; however, specific antigenic sites are more similar. For example, the Sa region of SC/1918 is 92% identical to OH/07, and previous studies have identified cross-reactive antibodies isolated from humans that target this region (56). Four of the antibodies showed some cross-reactivity with H3N2, although three of these reacted only weakly. One of these also had HI activity against H3N2, although again, this activity was only detected at high concentrations. This is not surprising as the B cells were sorted based on the ability to bind recHA of H1pdm, which shares only 43% AA identity with HA of VIC/210. Further, a lack of H3 HA cross-reactivity is likely due to the timing of splenocyte harvest (day 3 post TIV boost). Interestingly, the rmAb with the most cross-reactivity in ELISA using inactivated vaccine (Table 4, 065-C11) did not show detectable HI activity against any virus, nor did it react with recHA in BLI or ELISA assays. This rmAb failed to react with the distantly-related influenza B virus, suggesting that the broad influenza A cross-reactivity is specific. These findings suggest that 065-C11 may react with a conserved, conformationally sensitive epitope on HA and future experiments will aim to identify this interaction. Thus, although some cross-subtype reactivity was identified in our characterization, the cross-reactivity was weak and poorly associated with HI activity.
In conclusion, these findings suggest that the antibody recall response to HA following H1N1pdm infection and vaccination of naïve animals is diverse, but based on a relatively limited number of germline sequences. The majority of cloned antibodies are directed against the globular head of HA and exhibited HI activity and are therefore likely capable of protecting against influenza disease in vivo. A subset of these antibodies further showed subtype-specific cross-reactivity with the antigenically related recHA of SC/1918 while minimal subtype cross-reactivity was identified. Further studies with this approach will help delimit the diversity of the antibody repertoire profile in response to novel vaccination strategies, with the goal of identifying approaches that induce a high yield of broadly reactive and neutralizing antibodies.
Supplementary Material
Acknowledgments
We thank Marina Khristova for her help sequencing amplicons for germline gene identification and subsequent direct cloning
Funding. This work was supported by the Centers for Disease Control and Prevention (CDC). JRW received financial support for this work from the Oak Ridge Institute for Science and Education, Oak Ridge, TN.
Footnotes
Conflicts of Interest. All authors: no reported commercial or other association that may pose a conflict of interest.
References
- 1.Thompson M, Shay D, Zhou H, Bridges C, Cheng P, Burns M, Bresee J, Cox N. Estimates of Deaths Associated with Seasonal Influenza –United States, 1976 –2007. MMWR. 2010:59. [PubMed] [Google Scholar]
- 2.Molinari N-AM, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine. 2007;25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 3.Grohskopf L, Uyecki T, Bresee J, Cox N, Shimabukuro T. Prevention and Control of Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices (ACIP)—United States, 2012–13 Influenza Season. MMWR. 2012;61:613–618. [PubMed] [Google Scholar]
- 4.Ellebedy AH, Webby RJ. Influenza vaccines. Vaccine. 2009;27(Supplement 4):D65–D68. doi: 10.1016/j.vaccine.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katz JM, Hancock K, Xu X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert review of anti-infective therapy. 2011;9:669–683. doi: 10.1586/eri.11.51. [DOI] [PubMed] [Google Scholar]
- 6.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 7.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 8.Khurana S, Loving CL, Manischewitz J, King LR, Gauger PC, Henningson J, Vincent AL, Golding H. Vaccine-Induced Anti-HA2 Antibodies Promote Virus Fusion and Enhance Influenza Virus Respiratory Disease. Science translational medicine. 5:200ra114. doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- 9.Daniels RS, Jeffries S, Yates P, Schild GC, Rogers GN, Paulson JC, Wharton SA, Douglas AR, Skehel JJ, Wiley DC. THE RECEPTOR-BINDING AND MEMBRANE-FUSION PROPERTIES OF INFLUENZA-VIRUS VARIANTS SELECTED USING ANTI-HEMAGGLUTININ MONOCLONAL-ANTIBODIES. Embo Journal. 1987;6:1459–1465. doi: 10.1002/j.1460-2075.1987.tb02387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laeeq S, Smith CA, Wagner SD, Thomas DB. Preferential selection of receptor-binding variants of influenza virus hemagglutinin by the neutralizing antibody repertoire of transgenic mice expressing a human immunoglobulin mu minigene. J Virol. 1997;71:2600–2605. doi: 10.1128/jvi.71.4.2600-2605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temoltzin-Palacios F, Thomas DB. Modulation of immunodominant sites in influenza hemagglutinin compromise antigenic variation and select receptor-binding variant viruses. The Journal of experimental medicine. 1994;179:1719–1724. doi: 10.1084/jem.179.5.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yewdell JW, Caton AJ, Gerhard W. Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J Virol. 1986;57:623–628. doi: 10.1128/jvi.57.2.623-628.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, Kent SJ. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol. 2013;190:1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- 14.Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ. Antibody-Dependent Cellular Cytotoxicity Is Associated with Control of Pandemic H1N1 Influenza Virus Infection of Macaques. J Virol. 2013;87:5512–5522. doi: 10.1128/JVI.03030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava V, Yang Z, Hung IF, Xu J, Zheng B, Zhang MY. Identification of dominant antibody-dependent cell-mediated cytotoxicity epitopes on the hemagglutinin antigen of pandemic H1N1 influenza virus. J Virol. 2013;87:5831–5840. doi: 10.1128/JVI.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Co MD, Cruz J, Takeda A, Ennis FA, Terajima M. Comparison of complement dependent lytic, hemagglutination inhibition and microneutralization antibody responses in influenza vaccinated individuals. Human vaccines & immunotherapeutics. 2012;8:1218–1222. doi: 10.4161/hv.21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terajima M, Cruz J, Co MD, Lee JH, Kaur K, Wrammert J, Wilson PC, Ennis FA. Complement-dependent lysis of influenza a virus-infected cells by broadly cross-reactive human monoclonal antibodies. J Virol. 2011;85:13463–13467. doi: 10.1128/JVI.05193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corti D, Suguitan AL, Jr, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, Weiss RA, Sallusto F, Subbarao K, Lanzavecchia A. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. The Journal of clinical investigation. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O'Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of experimental medicine. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS pathogens. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 28.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H, Carney P, Stevens J. Structure and Receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS currents. 2010;2:RRN1152. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiller T, Busse CE, Wardemann H. Cloning and expression of murine Ig genes from single B cells. Journal of Immunological Methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Seidl KJ, MacKenzie JD, Wang D, Kantor AB, Kabat EA, Herzenberg LA, Herzenberg LA. Frequent occurrence of identical heavy and light chain Ig rearrangements. International immunology. 1997;9:689–702. doi: 10.1093/intimm/9.5.689. [DOI] [PubMed] [Google Scholar]
- 32.Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Research. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. Manual for the Laboratory diagnosis and virological surveillance of influenza. 2011 www.who.int/csr/disease/influenza/manual_diagnosis_surveillance_influenza/en/index.html.
- 34.Chen LM, Rivailler P, Hossain J, Carney P, Balish A, Perry I, Davis CT, Garten R, Shu B, Xu X, Klimov A, Paulson JC, Cox NJ, Swenson S, Stevens J, Vincent A, Gramer M, Donis RO. Receptor specificity of subtype H1 influenza A viruses isolated from swine and humans in the United States. Virology. 2011;412:401–410. doi: 10.1016/j.virol.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burnham KP, Overton WS. Estimation of the Size of a Closed Population when Capture Probabilities vary Among Animals. Biometrika. 1978;65:625–633. [Google Scholar]
- 38.Burnham KP, Overton WS. Robust Estimation of Population Size When Capture Probabilities Vary among Animals. Ecology. 1979;60:927–936. [Google Scholar]
- 39.Chao A. Nonparametric Estimation of the Number of Classes in a Population. Scandinavian Journal of Statistics. 1984;11:265–270. [Google Scholar]
- 40.Chao A, Lee S-M. Estimating the Number of Classes via Sample Coverage. Journal of the American Statistical Association. 1992;87:210–217. [Google Scholar]
- 41.Chao A, Bunge J. Estimating the Number of Species in a Stochastic Abundance Model. Biometrics. 2002;58:531–539. doi: 10.1111/j.0006-341x.2002.00531.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang JPZ, Lindsay BG. Penalized Nonparametric Maximum Likelihood Approach to Species Richness Estimation. Journal of American Statistical Association. 2005;100:942–959. [Google Scholar]
- 43.Wang JP. Estimating species richness by a Poisson-compound gamma model. Biometrika. 2010;97:727–740. doi: 10.1093/biomet/asq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang JP. SPECIES: An R Package for Species Richness Estimation. J Stat Softw. 2011;40:1–15. [Google Scholar]
- 45.Schroeder HW., Jr Similarity and divergence in the development and expression of the mouse and human antibody repertoires. Developmental and comparative immunology. 2006;30:119–135. doi: 10.1016/j.dci.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 46.de Bono B, Madera M, Chothia C. VH gene segments in the mouse and human genomes. Journal of molecular biology. 2004;342:131–143. doi: 10.1016/j.jmb.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 47.Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 48.To KK, Zhang AJ, Hung IF, Xu T, Ip WC, Wong RT, Ng JC, Chan JF, Chan KH, Yuen KY. High titer and avidity of nonneutralizing antibodies against influenza vaccine antigen are associated with severe influenza. Clinical and vaccine immunology : CVI. 2012;19:1012–1018. doi: 10.1128/CVI.00081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLain L, Dimmock NJ. Protection of mice from lethal influenza by adoptive transfer of non-neutralizing haemagglutination-inhibiting IgG obtained from the lungs of infected animals treated with defective interfering virus. The Journal of general virology. 1989;70( Pt 10):2615–2624. doi: 10.1099/0022-1317-70-10-2615. [DOI] [PubMed] [Google Scholar]
- 50.Caton AJ, Stark SE, Kavaler J, Staudt LM, Schwartz D, Gerhard W. Many variable region genes are utilized in the antibody response of BALB/c mice to the influenza virus A/PR/8/34 hemagglutinin. The Journal of Immunology. 1991;147:1675–1686. [PubMed] [Google Scholar]
- 51.Staudt LM, Gerhard W. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. I. Significant variation in repertoire expression between individual mice. The Journal of experimental medicine. 1983;157:687–704. doi: 10.1084/jem.157.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis MM, Lyons DS, Altman JD, McHeyzer-Williams M, Hampl J, Boniface JJ, Chien Y. T cell receptor biochemistry, repertoire selection and general features of TCR and Ig structure. Ciba Foundation symposium. 1997;204:94–100. doi: 10.1002/9780470515280.ch7. discussion 100–104. [DOI] [PubMed] [Google Scholar]
- 53.Xu JL, Davis MM. Diversity in the CDR3 Region of VH Is Sufficient for Most Antibody Specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 54.Lingwood D, McTamney PM, Yassine HM, Whittle JRR, Guo X, Boyington JC, Wei C-J, Nabel GJ. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katz JM, Hancock K, Xu X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert review of anti-infective therapy. 2011;9:669–683. doi: 10.1586/eri.11.51. [DOI] [PubMed] [Google Scholar]
- 56.Krause JC, Tumpey TM, Huffman CJ, McGraw PA, Pearce MB, Tsibane T, Hai R, Basler CF, Crowe JE., Jr Naturally occurring human monoclonal antibodies neutralize both 1918 and 2009 pandemic influenza A (H1N1) viruses. J Virol. 2010;84:3127–3130. doi: 10.1128/JVI.02184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.