Abstract
We previously showed that the organic extract of a blue-green alga, Spirulina platensis (SPE), had potent anti-inflammatory effects in macrophages. As the interplay between macrophages and adipocytes is critical for adipocyte functions, we investigated the contribution of the anti-inflammatory effects of SPE in macrophages to adipogenesis/lipogenesis in 3T3-L1 adipocytes. 3T3-L1 preadipocytes were treated with 10% conditioned medium from lipopolysaccharide (LPS)-stimulated RAW 264.7 macrophages (CMC) or LPS-stimulated, but SPE-pretreated, macrophages (CMS) at different stages of adipocyte differentiation. The expression of adipocyte differentiation markers, such as CCAAT/enhancer-binding protein α, peroxisome proliferator-activated receptor γ, and perilipin, was significantly repressed by CMC when added on day 3, while the repression was attenuated by CMS. Oil Red O staining confirmed that adipocyte maturation in CMS-treated cells, but not in CMC-treated cells, was equivalent to that of control cells. Nuclear translocation of nuclear factor κB (NF-κB) p65 was decreased by CMS compared to CMC. In lipid-laden adipocytes, CMC promoted the loss of lipid droplets, while CMS had minimal effects. Histone deacetylase 9 mRNA and protein levels were increased during adipocyte maturation, which were decreased by CMC. In conclusion, by cross-talking with adipocytes, the anti-inflammatory effects of SPE in macrophages promoted adipocyte differentiation/maturation, at least in part, by repressing the activation of NF-κB inflammatory pathways, which otherwise can be compromised in inflammatory conditions.
Key Words: : adipocyte differentiation, blue-green algae, inflammation, macrophages, Spirulina platensis
Introduction
Obesity currently affects 34.6% of U.S. adults.1,2 Chronic inflammation triggered primarily by recruited macrophages in adipose tissue has been widely recognized as a causative factor in the development of obesity-induced insulin resistance and type 2 diabetes.3–5 Macrophage infiltration into adipose tissue can result in a state of chronic inflammation by releasing proinflammatory cytokines, such as tumor necrosis factor α (TNFα), interleukin (IL)-1β, and IL-6.2,3 Lipid-laden adipocytes can also secrete these cytokines, although less than macrophages.2,6 The proinflammatory cytokines can disturb metabolic events in adipocytes in autocrine and paracrine manner.7 In particular, TNFα has been demonstrated to inhibit insulin signaling, decrease fatty acid esterification, enhance lipolysis, and activate nuclear factor κB (NF-κB) in adipocytes.8 The activation of NF-κB negatively regulates peroxisome proliferator-activated receptor γ (PPARγ), the master transcriptional regulator of adipogenesis.9 Furthermore, it has been shown that the NF-κB activity is obligatory for the effect of TNFα on metabolic dysfunctions of adipocytes.10 Therefore, cross talk between adipocytes and macrophages play a critical role in the development of systemic insulin resistance and other metabolic diseases in obesity.
Histone acetylation controls gene transcription through the regulation of chromatin states.11 The dynamics of histone acetylation is controlled by the opposing action of histone acetyltransferases (HATs) and histone deacetylases (HDACs), which add the acetyl moiety to and remove it from histones, respectively.12 Recently, HDAC inhibitors have been shown to increase energy expenditure and insulin sensitivity, while decreasing macrophage infiltration to the adipose tissue in obese mice.13,14 NF-κB is directly associated with HDAC1 and the inhibition of HDAC activity increases NF-κB activation in macrophages.15 In contrast, HDAC3 has been shown to be required for inflammatory gene expression in macrophages.16 In the case of adipocytes, HDAC3 in association with retinoblastoma protein has been shown to repress the PPARγ transcriptional activity, whereas the inhibition of HDAC activity allows for the PPARγ transcriptional activity.17 Furthermore, it has been demonstrated that HDAC9 is a negative regulator of adipogenic differentiation by repressing upstream stimulatory factor-1 (USF1), which is responsible for the transcription of CCAAT/enhancer-binding protein α (C/EBPα), a transcription factor critical for adipogenesis.18 Therefore, as epigenic regulators, HDACs are closely involved in adipocyte biology.
We previously reported that the organic extract of Spirulina platensis (SPE), a blue-green alga, inhibited the expression and secretion of proinflammatory cytokines in macrophages by inhibiting NF-κB translocation into the nucleus.19 We further demonstrated that the repression of proinflammatory gene expression by SPE is, at least in part, due to the modulation of HDACs and histone acetylation.19 In this study, we showed that the potent anti-inflammatory effect of SPE in macrophages can enhance adipocyte differentiation/maturation in inflammatory conditions by attenuating NF-κB activation in 3T3-L1 adipocytes.
Materials and Methods
SPE preparation
SPE powder (Earthrise® Natural Spirulina) was kindly provided by Earthrise Nutritionals (Irvine, CA, USA). SPE was prepared by the extraction of SPE powder with chloroform/methanol (1:2, v/v) and stored under N2 gas at −20°C for short term and −80°C for long term as we previously described.20,21 To deliver SPE into the cell culture medium, it was dried down under N2 to remove solvents and then dissolved in dimethyl sulfoxide (DMSO). The final DMSO concentration in the cell medium was <0.5%.
Cell culture, adipocyte differentiation, and treatments
RAW 264.7 macrophages (RAW macrophages) and 3T3-L1 preadipocytes were purchased from ATCC (Manassas, VA, USA). RAW macrophages were maintained in RPMI-1640 containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, 1× vitamins, and 2 mmol/L l-glutamine. 3T3-L1 preadipocytes were maintained in high-glucose DMEM containing 10% calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mmol/L l-glutamine. Two days postconfluence (Day 0), 3T3-L1 preadipocytes were induced to differentiate in high-glucose DMEM supplemented with 10% FBS, 1 μg/mL insulin, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mmol/L l-glutamine in the presence of differentiation cocktail (1 μg/mL insulin, 1 μmol/L dexamethasone, 0.5 mmol/L methyl-isobutylxanthine). After 3 days of differentiation, cells were maintained in high-glucose DMEM containing 10% FBS and 1 μg/mL insulin until the indicated day of each experiment. All cells were kept in a humidified chamber at 37°C with 5% CO2. All cell culture supplies were purchased from Hyclone (Logan, UT, USA).
For the generation of conditioned medium, RAW macrophages were plated in 12-well plates at a density of 0.5 × 106/well. At ∼90% confluence, macrophages were pretreated with vehicle (DMSO) only or 100 μg/mL of SPE for 12 h in serum-free media, after which they were stimulated with 100 ng/mL of Salmonella enterica serotype typhimurium lipopolysaccharide (LPS; Sigma-Aldrich, St. Louis, MO, USA) with or without 100 μg/mL of SPE for an additional 18 h. The conditioned medium was then collected, filter sterilized, and stored at −80°C until use. The conditioned medium from the cells stimulated with LPS only is called conditioned medium control (CMC), while the conditioned medium from LPS-stimulated, SPE-pretreated cells is called conditioned medium spirulina (CMS) hereafter. Experimental scheme is shown in Figure 1A.
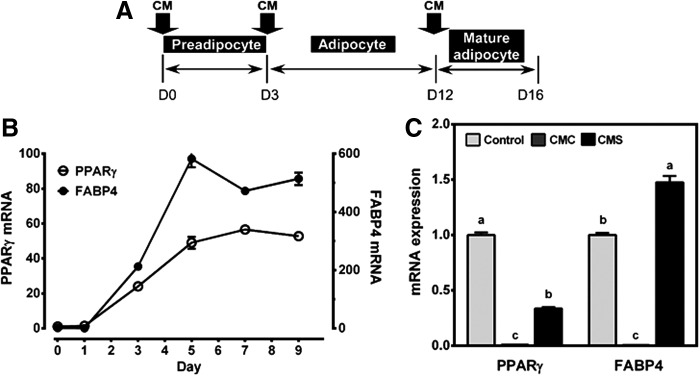
FIG. 1.
Role of macrophage CMs on the differentiation of 3T3-L1 preadipocyte. (A) Schematic of experimental design. Arrows point to days during adipocyte differentiation when CMs were first introduced. When CM is introduced on day 0, the cells were differentiated and kept until day 8, with change in media with CM every 2 days. When CM is introduced on day 3, the cells were grown in media with CM until day 8 with media changed every 2 days. When CM is introduced on day 12, the cells were grown in media with CM for 4 days (until day 16) with media change every 2 days. (B) mRNA levels of PPARγ and FABP4 in adipocytes from preadipocyte on day 0 to mature adipocytes on day 9. Mean ± SEM. n = 3 (C) mRNA expression of PPARγ and FABP4 in the adipocytes treated with 10% CMC or CMS on day 0 until day 8. Mean ± SEM, n = 3. Bars with different letters are significantly different (P < .05). CM, conditioned medium; CMC, conditioned medium control; CMS, conditioned medium spirulina; FABP4, fatty acid-binding protein 4; PPARγ, peroxisome proliferator-activated receptor γ.
Total RNA isolation and quantitative real-time PCR
Total RNA was extracted from cells using TRIzol® reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's protocol. Reverse transcription for cDNA synthesis and quantitative real-time PCR (qRT-PCR) analysis were performed using a Bio-Rad CFX96 Real-Time system (Bio-Rad, Hercules, CA, USA) as previously described.19 Ribosomal protein p0 (RPLP0) was used as an internal control for normalization. Primers were designed according to GenBank database using the Beacon Designer 7 software (PREMIER Biosoft International, Palo Alto, CA, USA). Primer sequences will be made available upon request.
Western blot analysis
Whole cell lysates were prepared and Western blot analysis was performed as we described.21 HDAC9 antibody was purchased from Santa Cruz Biotechnology (Dallas, TX, USA) and β-actin loading control from Sigma-Aldrich. The blots were developed using a SuperSignal™ West Pico chemiluminescent substrate (Thermo Fisher Scientific) and imaged using a Chemidoc XRS+ system (Bio-Rad) and Image Lab software (Bio-Rad).
NF-κB p65 nuclear translocation
Cytoplasmic and nuclear fractions were collected from 3T3-L1 adipocytes treated with 10% CMC or CMS from day 3 of differentiation for 2 and 5 days (day 5 and 7, respectively) using a Nuclear Extraction Kit (Cayman Chemicals, Ann Arbor, MI, USA) according to the manufacturer's protocol. Antibodies against p65 and a cytoplasmic fractionation control glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). TATA box-binding protein was used as a loading control and a marker of nuclear fraction purity, and its antibody was purchased from Abcam (Cambridge, MA, USA). Western blot was conducted as described above.
Oil Red O staining
Oil Red O stock solution was prepared by dissolving 500 mg Oil Red O in 100 mL of isopropanol overnight at room temperature and subsequently filtered with a Whatman 40 filter paper. For a working solution, the stock was diluted in distilled water (3:2, v/v). After completion of treatments, cells were washed twice with 1× PBS and fixed with 3.7% formaldehyde in 1× PBS for 20 min. Subsequently, the cells were washed once with 1× PBS and then twice with 60% isopropanol. When cells were completely dried, they were incubated with Oil Red O working solution at room temperature for 30 min, after which they were washed thrice with 1× PBS and imaged using an Axio Vert.A1 microscope (Carl Zeiss, Jena, Germany).
Statistical analysis
One-way analysis of variance and Newman–Keuls post hoc analysis or unpaired t-test were performed with Graphpad Prism 6.0 (GraphPad Software, La Jolla, CA, USA) to detect differences between group means. P-values <.05 were considered significant. Values were expressed as mean ± SEM.
Results
CMC inhibited adipocyte differentiation and maturation
The conditioned medium from macrophages incubated with LPS only (i.e., CMC) or LPS together with SPE (i.e., CMS) was added to 3T3-L1 preadipocytes to determine their effect on the differentiation of preadipocytes to adipocytes. When 3T3-L1 preadipocytes (day 0) became adipocytes (day 3), there was an ∼50-fold increase in PPARγ expression, the master regulator of adipocyte differentiation, with a concomitant increase in mRNA of fatty acid-binding protein 4 (FABP4), a PPARγ target gene (Fig. 1B). PPARγ mRNA peaked on day 7 and remained elevated. The addition of CMC, but not CMS, at the start of the differentiation program (day 0 until day 8) significantly repressed the expression of PPARγ and FABP4 to near undetectable levels (Fig. 1C).
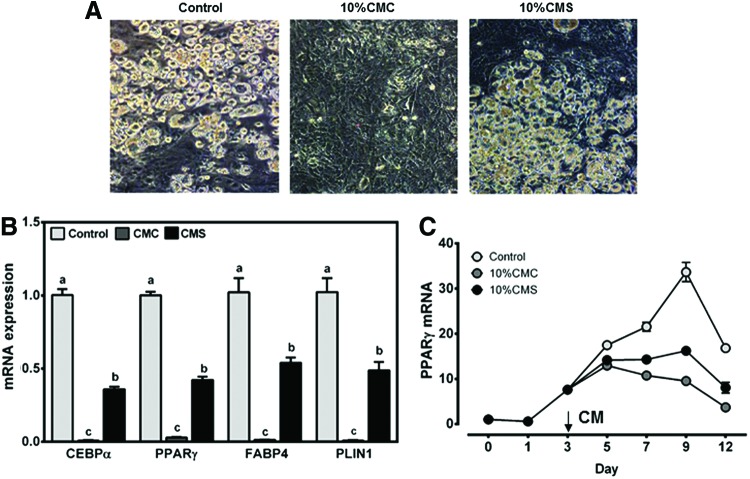
We next evaluated the effect of CMC and CMS on adipocyte maturation. When 10% CMC or CMS was added to adipocytes on day 3 until day 8, cells treated with CMC stayed fibroblastic with virtual absence of lipid droplets, while control and CMS-treated cells had abundant lipid droplets (Fig. 2A). The expression of adipocyte differentiation markers, such as C/EBPα, PPARγ, FABP4, and perilipin1 (PLIN1), was significantly repressed in the presence of CMC, while the repression was much less in the cells treated with CMS (Fig. 2B). Consistently, the inhibitory effect of CMC, but not CMS, on adipocyte maturation, as judged by PPARγ expression, was observed when the conditioned media were added on day 3 (Fig. 2C).
FIG. 2.
Effects of macrophage CMs on adipocyte maturation/lipid accumulation. 3T3-L1 adipocytes were treated with 10% CMC or CMS on day 3 until day 8 or 12. (A) Representative images of 3T3-L1 adipocytes treated with CMC or CMS on day 3 until day 8. (B) mRNA expression of adipocyte differentiation markers in 3T3-L1 adipocytes treated with CMC or CMS on day 3 until day 8. Mean ± SEM. n = 6. Bars with different letters are significantly different (P < .05). (C) PPARγ mRNA expression of cells treated with CMC or CMS on day 3 until day 12. Mean ± SEM. n = 3. Color images available online at www.liebertpub.com/jmf
CMC facilitated intracellular lipid loss in lipid-laden adipocytes
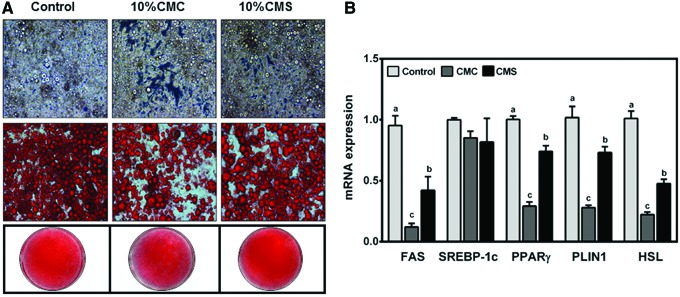
The role of CMC and CMS in maintaining phenotypes of mature adipocytes was explored. Differentiated 3T3-L1 adipocytes maintained for at least 12 days were fully engorged with lipids (Fig. 3A). The addition of CMC for 4 days to mature adipocytes facilitated the loss of lipid droplets as evidenced by visible loss of intracellular lipid droplets and decreased Oil Red O staining, whereas lipid loss was limited in cells treated with CMS. To gain insight into the facilitated loss of lipid droplets by CMC, the expression of genes involved in lipogenesis, for example, fatty acid synthase (FAS) and sterol regulatory element binding protein 1C (SREBP-1C), and lipolysis, for example, hormone-sensitive lipase (HSL), was measured. The loss of lipid droplets by CMC was associated with significant repression of FAS, but not SREBP-1C (Fig. 3B). The mRNA levels of PPARγ, PLIN1, and HSL were also significantly repressed by CMC compared to control and CMS.
FIG. 3.
Effects of macrophage CMs on lipid droplets in mature lipid-engorged adipocytes. 3T3-L1 adipocytes were treated with 10% CMC or CMS on day 12 for 4 days. (A) Micropictographs of 3T3-L1 adipocytes. From top to bottom, representative images of unstained cells (top panel), cells stained with Oil Red O (middle panel), and entire wells stained with Oil Red O (bottom). (B) mRNA expression of lipogenic and lipolytic genes. Mean ± SEM. n = 6. Bars with different letters for each gene are significantly different (P < .05). No letters in a gene indicates that there is no significant difference between groups. Color images available online at www.liebertpub.com/jmf
CMC increased nuclear translocation of NF-κB p65 and proinflammatory gene expression in adipocytes
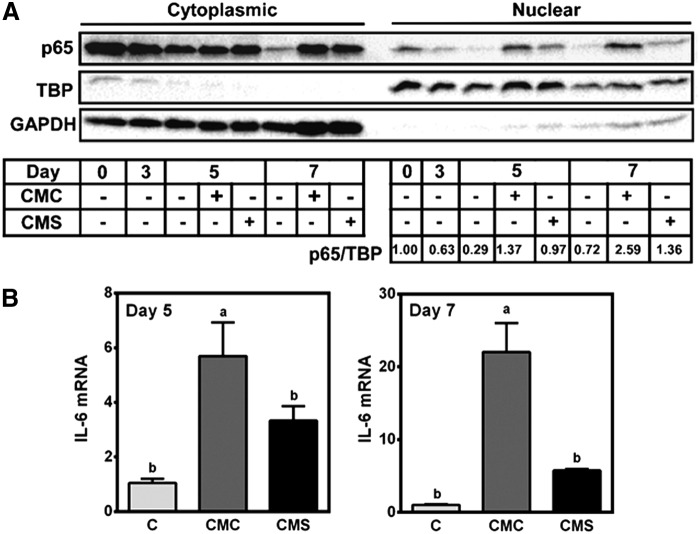
It has been demonstrated that PPARγ antagonizes p65.22 Therefore, the activation of p65 by CMC may antagonize PPARγ, inhibiting adipocyte differentiation. The differentiation of preadipocytes to adipocytes was accompanied by decreased p65 protein levels both in the cytoplasmic and nuclear fractions (Fig. 4A). The addition of CMC after 3 days of differentiation markedly increased p65 translocation into the nucleus, which was attenuated in CMS-treated cells, although slightly higher than control. The increase in p65 nuclear translocation was not acute, rather persisted for several days after the addition of CMC and CMS. The marked increase in p65 translocation into the nucleus on day 5 and 7 by CMC occurred concomitantly with significant increases in IL-6 expression (Fig. 4B).
FIG. 4.
Effect of macrophage CM on NF-κB and IL-6 expression. 3T3-L1 adipocytes were incubated with 10% CMC or CMS on day 3 until day 5 or 7. (A) A representative blot of Western blot analysis of p65 in cytoplasmic and nuclear fractions. GAPDH and TBP were used as a control for cytoplasmic and nuclear fraction, respectively. Densitometry quantification of p65/TBP ratio relative to day 0 is shown. (B) IL-6 mRNA expression. Mean ± SEM. n = 6. Bars with different letters are significantly different (P < .05). GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IL, interleukin; NF-κB, nuclear factor κB; TBP, TATA-binding protein.
HDAC9 expression was induced during adipocyte maturation
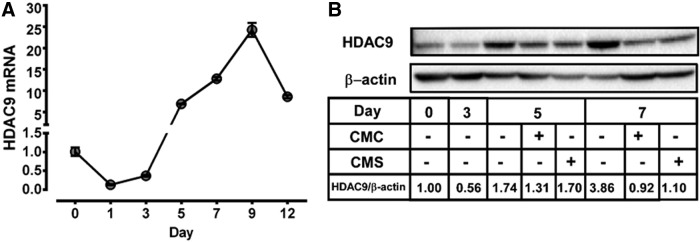
As HDAC9 has been implicated in adipocyte differentiation,18 we explored if HDAC9 may be involved in the regulation of SPE conditioned medium on adipocyte differentiation. On day 1 of 3T3-L1 preadipocyte differentiation, there was an ∼80% reduction in the expression of HDAC9 compared to undifferentiated preadipocytes on day 0, which was maintained until day 3 (Fig. 5A). By day 5, the expression of HDAC9 increased by ∼7 fold compared to undifferentiated preadipocytes and peaked on day 9. Similar to changes in its mRNA levels during adipocyte differentiation and maturation, MEF2-interacting transcription repressor (MITR), an HDAC9 isoform resulting from alternative splicing of the HDAC9 transcripts,23 was reduced on day 3, but increased by day 5 and 7 (Fig. 5B). The addition of CMC and CMS on day 3 noticeably decreased HDAC9/MITR protein levels by day 5. On day 7, there were noticeable differences in the HDAC9/MITR protein between CMC- and CMS-treated cells, with more HDAC9/MITR present in CMS-treated cells.
FIG. 5.
Effect of macrophage conditioned medium on HDAC9 expression during adipocyte differentiation and maturation. (A) HDAC9 mRNA levels of 3T3-L1 preadipocytes and adipocytes from day 0 to day 12. n = 3 Mean ± SEM. (B) A representative blot image of Western blot analysis of HDAC9 protein on indicated days of adipocyte differentiation in whole cell lysate. CMs were added on day 3 until day 5 or day 7. Densitometry quantification of HDAC9/β-actin ratios relative to day 0 is shown. HDAC, histone deacetylase.
Discussion
Adipocyte differentiation and adipose tissue expansion in response to excess caloric intake are important to avoid the ectopic deposition of lipids in nonadipose tissues. Evidence from mice and humans suggests that ectopic lipid deposition can lead to the development of metabolic diseases, such as hyperlipidemia and insulin resistance.8,24,25 In contrast, factors, such as thiazolidinediones, PPARγ agonists, which facilitate adipose tissue expansion and lipid storage can prevent insulin resistance despite increasing fat mass in the body.26,27 Increased numbers of adipose tissue macrophages are associated with insulin resistance in obese humans by disturbing adipocyte functions to store fats.28–30 We previously reported that SPE possesses potent anti-inflammatory properties in macrophages by inhibiting proinflammatory cytokine expression and secretion through inhibition of NF-κB translocation into the nucleus.19 As inflammatory properties of macrophages have a large impact on adipocyte functions, SPE may elicit positive metabolic influence in adipocytes as to adipogenesis/lipogenesis due to its anti-inflammatory properties. In this study, we demonstrated that the anti-inflammatory effect of SPE in macrophages can promote adipocyte differentiation/maturation in inflammatory conditions, which otherwise can be compromised in metabolic disorders associated with obesity.
Macrophages surrounding lipid-engorged adipocytes have been characterized as classically activated “M1” macrophages, while resident adipose tissue macrophages are alternatively activated anti-inflammatory “M2” macrophages.31 As macrophages can be polarized to an M1 phenotype by LPS stimulation,31 in our experimental setting, the incubation of adipocytes with the conditioned medium collected from macrophages activated by LPS, that is, CMC, should approximate the interaction of inflammatory macrophages with adipocytes. Based on our previous study using cytokine arrays, we confirmed that CMC had marked increases in IL-6, TNFα, granulocyte macrophage colony-stimulating factor, and chemokine (C-C motif) ligand 5 compared to CMS.19
In this study, when preadipocytes were incubated with CMC or CMS at different stages of differentiation, we observed that preadipocytes exposed to an inflammatory milieu, that is, CMC, on day 0 did not undergo any differentiation and stayed fibroblastic. In contrast, preadipocytes exposed to CMS resulted in noticeable differentiation and lipid accumulation. Also, in fully differentiated lipid-laden adipocytes on day 12, CMC facilitated the loss of intracellular lipid droplets with concomitant decreases in the expression of PPARγ and its downstream genes. PPARγ controls the expression of lipoprotein lipase (LPL), glucose transporter 4 (GLUT4), and FAS, which can promote the uptake of glucose and fatty acids for triglyceride synthesis and storage in lipid droplets, respectively.32 Therefore, the inhibition of PPARγ by CMC may be responsible for the loss of lipid droplets in mature adipocytes.
Furthermore, the reduction in the expression of PLIN1, which has been demonstrated to increase lipolysis33 by CMC may also contribute to the progressive loss of lipid droplets in lipid-laden adipocytes. Overall, the addition of CMC on day 0, 3, or 12 of adipocyte differentiation/maturation elicited a negative effect on adipocyte differentiation and lipid storage, which was significantly attenuated in CMS-treated adipocytes. As adipocytes have been estimated to have a turnover rate of ∼10% annually in adult humans,34 the exposure of adipocytes to inflammatory cytokines in inflamed adipose tissue may reduce the ability of preadipocytes to differentiate, diminishing the pool of adipocytes. Decreased number of adipocytes due to disturbed adipocyte differentiation and increased lipid loss from adipocytes could facilitate ectopic deposition of lipids in nonadipose tissue, notably skeletal muscle, triggering the onset of local and ultimately systemic insulin resistance.3,8 In this aspect, it appears that the anti-inflammatory effect of SPE on macrophages may prevent metabolic dysfunctions in adipose tissue induced by inflammation.
TNFα alters many aspects of adipocyte biology. TNFα decreases GLUT4, LPL, and PPARγ protein, resulting in decreased glucose transport, fatty acid uptake, and lipogenesis in adipocytes.8 TNFα also has stimulatory effects on lipolysis by increasing the levels of cAMP and decreasing PLIN1.8,33 In addition, studies have shown that the binding of TNFα to its receptor on adipocytes inhibits adipocyte differentiation,35 which is largely attributed to its inhibitory action on PPARγ, while activating NF-κB.9,10,36
In this study, we observed that the incubation of adipocytes with CMC chronically increased nuclear translocation of NF-κB p65, which was not evident with CMS treatment. Upon translocation into the nucleus, p65 induces the transcription of inflammatory genes, such as TNFα, IL-6, and IL-1β.37 In agreement with this study, there was elevated IL-6 expression in the adipocytes treated with CMC, while IL-6 induction by CMS was much less. Interestingly, p65 nuclear translocation and transcriptional activity remained elevated for several days. Typically, the translocation of NF-κB p65 into the nucleus is a rapid event, taking only minutes with remaining effective for several hours.38 In addition to the induction of cytokine expression, NF-κB also transcribes inhibitor of kappa B (IκB), as a part of an autoregulatory loop, which can bind p65 in the nucleus for exiting p65 back into the cytoplasm.39 It is unknown whether adipocytes possess this autoregulatory loop for the resolution of inflammatory responses. It is tempting to speculate that adipocytes may not have a self-resolving capacity, leading to extended residency of NF-κB in the nucleus for ultimate repression of adipocyte differentiation. This aspect warrants further investigation.
It has been suggested that HDAC9 is a negative regulator of adipocyte differentiation by repressing the expression of USF1, a transcription factor responsible for C/EBPα expression.18 The study also demonstrated that during adipocyte differentiation, HDAC9 is downregulated and correlated negatively with lipid accumulation. We observed that after the addition of the differentiation cocktail, there was an ∼80% reduction in HDAC9 mRNA abundance, supporting the idea that HDAC9 is a negative regulator of adipocyte differentiation. Surprisingly, the expression of HDAC9 increased by ∼24 fold during adipocyte maturation. Furthermore, the increase in HDAC9 expression translated into increases in HDAC9/MITR protein levels. The discrepancy between these two studies as to whether HDAC9 decreases or increases during lipid accumulation in adipocytes may be due to cell-type differences, that is, primary human preadipocytes vs. mouse 3T3-L1. In addition, it is not immediately clear what HDAC9 isoform was measured in the study by Chatterjee et al.,18 as there are multiple HDAC9 isoforms due to alternative splicing.23 When adipocytes are removed from the differentiation cocktail, they actively synthesize lipid droplets and become mature. Therefore, there was a strong correlation between lipid accumulation in the adipocytes and HDAC9/MITR expression in this study. As CMC reduced lipid accumulation within differentiated adipocytes, as well as HDAC9 protein levels, our data suggest that HDAC9 may be associated with adipocyte lipogenesis.
In obesity, there is a need for adipose tissue expansion to facilitate triglyceride storage, which is typically met by adipocyte hypertrophy (increase cell size) or hyperplasia (increased cell number). Hyperplasia through increased adipocyte differentiation from preadipocytes increases triglyceride storage capacity without stress associated with adipocyte hypertrophy, such as endoplasmic reticulum stress.40 In obesity, inflammation due to increased macrophage infiltration into the adipose tissue creates an environment that is not conducive to adipocyte differentiation. Therefore, dietary products with anti-inflammatory properties may be able to prevent obesity-associated metabolic diseases by inhibiting metabolic disturbances in the adipocytes triggered by inflammation. In this study, we demonstrated that the anti-inflammatory effect of SPE in macrophages can prevent perturbation of adipocyte differentiation/maturation in inflammatory conditions, supporting a potential use of SPE as a dietary agent for the prevention of obesity-associated metabolic and inflammatory disorders.
Acknowledgments
This work was supported by National Institute Health grant R21AT005152 and U.S. Department of Agriculture Multi-state Hatch CONS 00916 to J.-Y.L.; and by U.S. Department of Agriculture AFRI NIFA Predoctoral Fellowship (2014-01870) to T.X.P.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Go AS, Mozaffarian D, Roger VL, et al. : Executive summary: Heart disease and stroke statistics—2013 update: A report from the American Heart Association. Circulation 2013;127:143–152 [DOI] [PubMed] [Google Scholar]
- 2.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW, Jr.: Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 2003;112:1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Barnes GT, Yang Q, et al. : Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003;112:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McArdle MA, Finucane OM, Connaughton RM, McMorrow AM, Roche HM: Mechanisms of obesity-induced inflammation and insulin resistance: Insights into the emerging role of nutritional strategies. Front Endocrinol (Lausanne) 2013;4:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surmi BK, Hasty AH: Macrophage infiltration into adipose tissue: Initiation, propagation and remodeling. Future Lipidol 2008;3:545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotamisligil GS, Shargill NS, Spiegelman BM: Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science 1993;259:87–91 [DOI] [PubMed] [Google Scholar]
- 7.Curat CA, Miranville A, Sengenes C, et al. : From blood monocytes to adipose tissue-resident macrophages: Induction of diapedesis by human mature adipocytes. Diabetes 2004;53:1285–1292 [DOI] [PubMed] [Google Scholar]
- 8.Guilherme A, Virbasius JV, Puri V, Czech MP: Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol 2008;9:367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B, Berger J, Hu E, et al. : Negative regulation of peroxisome proliferator-activated receptor-gamma gene expression contributes to the antiadipogenic effects of tumor necrosis factor-alpha. Mol Endocrinol 1996;10:1457–1466 [DOI] [PubMed] [Google Scholar]
- 10.Ruan H, Hacohen N, Golub TR, Van Parijs L, Lodish HF: Tumor necrosis factor-alpha suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: Nuclear factor-kappaB activation by TNF-alpha is obligatory. Diabetes 2002;51:1319–1336 [DOI] [PubMed] [Google Scholar]
- 11.Bannister AJ, Kouzarides T: Regulation of chromatin by histone modifications. Cell Res 2011;21:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang XJ, Seto E: HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007;26:5310–5318 [DOI] [PubMed] [Google Scholar]
- 13.Galmozzi A, Mitro N, Ferrari A, et al. : Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes 2013;62:732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Z, Yin J, Zhang J, et al. : Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009;58:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner BP, Westerheide SD, Baldwin AS, Jr.: The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol 2001;21:7065–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Barozzi I, Termanini A, et al. : Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci USA 2012;109:E2865–E2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fajas L, Egler V, Reiter R, et al. : The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell 2002;3:903–910 [DOI] [PubMed] [Google Scholar]
- 18.Chatterjee TK, Idelman G, Blanco V, et al. : Histone deacetylase 9 is a negative regulator of adipogenic differentiation. J Biol Chem 2011;286:27836–27847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ku CS, Pham TX, Park Y, et al. : Edible blue-green algae reduce the production of pro-inflammatory cytokines by inhibiting NF-kappaB pathway in macrophages and splenocytes. Biochim Biophys Acta 2013;1830:2981–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park YK, Rasmussen HE, Ehlers SJ, et al. : Repression of proinflammatory gene expression by lipid extract of Nostoc commune var sphaeroides Kutzing, a blue-green alga, via inhibition of nuclear factor-kappaB in RAW 264.7 macrophages. Nutr Res 2008;28:83–91 [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen HE, Blobaum KR, Park YK, Ehlers SJ, Lu F, Lee JY: Lipid extract of Nostoc commune var. sphaeroides Kutzing, a blue-green alga, inhibits the activation of sterol regulatory element binding proteins in HepG2 cells. J Nutr 2008;138:476–481 [DOI] [PubMed] [Google Scholar]
- 22.Pascual G, Fong AL, Ogawa S, et al. : A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 2005;437:759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrie K, Guidez F, Howell L, et al. : The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem 2003;278:16059–16072 [DOI] [PubMed] [Google Scholar]
- 24.Bindlish S, Presswala LS, Schwartz F: Lipodystrophy: Syndrome of severe insulin resistance. Postgrad Med 2015;127:511–516 [DOI] [PubMed] [Google Scholar]
- 25.Brochu M, Tchernof A, Dionne IJ, et al. : What are the physical characteristics associated with a normal metabolic profile despite a high level of obesity in postmenopausal women? J Clin Endocrinol Metab 2001;86:1020–1025 [DOI] [PubMed] [Google Scholar]
- 26.Larsen TM, Toubro S, Astrup A: PPARgamma agonists in the treatment of type II diabetes: Is increased fatness commensurate with long-term efficacy? Int J Obes Relat Metab Disord 2003;27:147–161 [DOI] [PubMed] [Google Scholar]
- 27.Lu Q, Li M, Zou Y, Cao T: Induction of adipocyte hyperplasia in subcutaneous fat depot alleviated type 2 diabetes symptoms in obese mice. Obesity 2014;22:1623–1631 [DOI] [PubMed] [Google Scholar]
- 28.Wentworth JM, Naselli G, Brown WA, et al. : Pro-inflammatory CD11c+CD206+ adipose tissue macrophages are associated with insulin resistance in human obesity. Diabetes 2010;59:1648–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorisky A, Molgat AS, Gagnon A: Macrophage-induced adipose tissue dysfunction and the preadipocyte: Should I stay (and differentiate) or should I go? Adv Nutr 2013;4:67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amano SU, Cohen JL, Vangala P, et al. : Local proliferation of macrophages contributes to obesity-associated adipose tissue inflammation. Cell Metab 2014;19:162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lumeng CN, Bodzin JL, Saltiel AR: Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest 2007;117:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tontonoz P, Spiegelman BM: Fat and beyond: The diverse biology of PPARgamma. Annu Rev Biochem 2008;77:289–312 [DOI] [PubMed] [Google Scholar]
- 33.Ryden M, Arvidsson E, Blomqvist L, Perbeck L, Dicker A, Arner P: Targets for TNF-alpha-induced lipolysis in human adipocytes. Biochem Biophys Res Commun 2004;318:168–175 [DOI] [PubMed] [Google Scholar]
- 34.Spalding KL, Arner E, Westermark PO, et al. : Dynamics of fat cell turnover in humans. Nature 2008;453:783–787 [DOI] [PubMed] [Google Scholar]
- 35.Xing H, Northrop JP, Grove JR, Kilpatrick KE, Su JL, Ringold GM: TNF alpha-mediated inhibition and reversal of adipocyte differentiation is accompanied by suppressed expression of PPARgamma without effects on Pref-1 expression. Endocrinology 1997;138:2776–2783 [DOI] [PubMed] [Google Scholar]
- 36.Medina-Gomez G, Gray S, Vidal-Puig A: Adipogenesis and lipotoxicity: Role of peroxisome proliferator-activated receptor gamma (PPARgamma) and PPARgammacoactivator-1 (PGC1). Public Health Nutr 2007;10:1132–1137 [DOI] [PubMed] [Google Scholar]
- 37.Oeckinghaus A, Ghosh S: The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 2009;1:a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao CY, Kim C, Huang F, Wu R: Requirements for two proximal NF-kappaB binding sites and IkappaB-zeta in IL-17A-induced human beta-defensin 2 expression by conducting airway epithelium. J Biol Chem 2008;283:15309–15318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D: The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev 1993;7:1266–1276 [DOI] [PubMed] [Google Scholar]
- 40.Rutkowski JM, Stern JH, Scherer PE: The cell biology of fat expansion. J Cell Biol 2015;208:501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]