Abstract
Background and Aims Several unrelated lineages such as plastids, viruses and plasmids, have converged on quadripartite genomes of similar size with large and small single copy regions and a large inverted repeat (IR). Except for Erodium (Geraniaceae), saguaro cactus and some legumes, the plastomes of all photosynthetic angiosperms display this structure. The functional significance of the IR is not understood and Erodium provides a system to examine the role of the IR in the long-term stability of these genomes. We compared the degree of genomic rearrangement in plastomes of Erodium that differ in the presence and absence of the IR.
Methods We sequenced 17 new Erodium plastomes. Using 454, Illumina, PacBio and Sanger sequences, 16 genomes were assembled and categorized along with one incomplete and two previously published Erodium plastomes. We conducted phylogenetic analyses among these species using a dataset of 19 protein-coding genes and determined if significantly higher evolutionary rates had caused the long branch seen previously in phylogenetic reconstructions within the genus. Bioinformatic comparisons were also performed to evaluate plastome evolution across the genus.
Key Results Erodium plastomes fell into four types (Type 1–4) that differ in their substitution rates, short dispersed repeat content and degree of genomic rearrangement, gene and intron content and GC content. Type 4 plastomes had significantly higher rates of synonymous substitutions (dS) for all genes and for 14 of the 19 genes non-synonymous substitutions (dN) were significantly accelerated. We evaluated the evidence for a single IR loss in Erodium and in doing so discovered that Type 4 plastomes contain a novel IR.
Conclusions The presence or absence of the IR does not affect plastome stability in Erodium. Rather, the overall repeat content shows a negative correlation with genome stability, a pattern in agreement with other angiosperm groups and recent findings on genome stability in bacterial endosymbionts.
Keywords: chloroplast, plastome, inverted repeat, repeated sequences, inversions, genome evolution, Geraniaceae, Erodium
INTRODUCTION
The majority of flowering plants harbour plastid genomes (plastomes) with a nearly identical gene complement, gene order and quadripartite structure (Bock, 2007; Ruhlman and Jansen, 2014). This structure includes a large [usually ∼25-kilobase (kb)] recombinogenic inverted repeat (IR) that gives rise to isomers differing in the relative orientation of their single copy regions, termed the large and small single copy regions (LSC and SSC, respectively) (Palmer, 1983). Expansion and contraction of the IR presumably occur through illegitimate recombination between opposite junctions of the IR and single copy regions (Goulding et al., 1996), resulting in some variation in IR gene content in particular lineages. With the exception of Monsonia speciosa (Geraniaceae), the only known example among autotrophic angiosperms to lack a portion of the ribosomal operon (Guisinger et al., 2011), the IR always contains this feature in its entirety. In some algal plastomes, the IR comprises the ribosomal operon exclusively (Yamada, 1991).
Although most photosynthetic angiosperms contain the IR, early restriction fragment mapping studies identified two independent lineages in which one repeat copy had been lost (Downie and Palmer, 1992): the genus Erodium (Geraniaceae) and a large clade within legumes (Fabaceae) (Palmer et al., 1987; Downie and Palmer, 1992) subsequently termed the IR Lacking Clade (IRLC; Wojciechowski et al., 2004). The IR losses in Erodium and legumes left no clear indicators of illegitimate recombination such as homology on both sides of the tract that underwent gene conversion, or at least none remains, as neither loss is recent. The increasing availability of plastome sequences from diverse lineages may reveal, as it has for the saguaro cactus (Carnegiea gigantea), more cases of IR loss (Sanderson et al., 2015).
The geranium family (Geraniaceae) is an interesting study system for IR evolution because it contains not only one of the three autotrophic angiosperm lineages lacking the IR (Erodium) but also the lineage with the largest known IR (Pelargonium) as well as the only autotrophic group in which the IR has contracted such that it does not contain the entire ribosomal operon (Monsonia) (Blazier et al., 2011; Guisinger et al., 2011). As the inferred ancestral plastome organization for Geraniaceae includes a relatively normal IR in terms of size and content, these large and lineage-specific fluctuations likely occurred independently in each genus (Weng et al., 2014). Given the rarity of plastome rearrangement across angiosperms it is tempting to speculate that the rearrangements in the major genera of Geraniaceae, although independent and different in outcome, share an underlying mechanism (Guisinger et al., 2008, 2011) such as reduced efficacy of plastid DNA repair or of cellular machinery maintaining the structural integrity of these genomes (Marechal and Brisson, 2010; Tremblay-Belzile et al., 2015; Zhang et al., 2016).
Conservation of the IR across angiosperms and its presence in many other land plant and algal lineages suggest it has some functional importance. Several functions for the IR have been suggested, including replication initiation (Heinhorst and Cannon, 1993), stabilization of the plastome (Palmer and Thompson, 1982; Hirao et al., 2008) and conservation of genes encoding the translational machinery (Palmer and Thompson, 1982), as genes in the IR have been shown to evolve approximately three-fold more slowly than those in the single-copy regions (Wolfe et al., 1987; Perry and Wolfe, 2002; Zhu et al., 2016). The early observation that legumes lacking the IR have undergone more frequent genomic rearrangement than those retaining the IR led to the hypothesis that the IR functioned to stabilize the plastome (Palmer and Thompson, 1982). Indeed, sequencing of the highly rearranged plastome of Erodium texanum seemed to lend further support for this hypothesis (Guisinger et al., 2011).
Phylogenetic reconstructions (Fiz et al., 2006) parsed the Erodium lineage into two highly supported major clades, Clade I and Clade II. Among the unusual features of the E. texanum (Clade I) plastome was the loss and/or divergences of some canonical plastid genes (Guisinger et al., 2011). While the genes encoding the NADH dehydrogenase (NDHA–K; NDH) were found intact, PCR surveys indicated that nested within Erodium Clade I was a lineage that lacked the entire suite of NDH genes: the so-called long branch clade (LBC; Blazier et al., 2011). Indeed it appeared that these IR-lacking, dispersed repeat-rich plastomes had been destabilized relative to other photosynthetic angiosperms. However, sequencing of plastomes from Erodium carvifolium (Blazier et al., 2011), a representative of Clade II, and California (Weng et al., 2014), the monotypic genus sister to Erodium, did not support the notion of IR mediated genome stabilization. The California macrophylla plastome (IR ∼ 22·3 kb) shows a single unique inversion relative to the inferred ancestral gene order for Geraniaceae (Weng et al., 2014). Erodium carvifolium, despite having lost one IR copy, differs from C. macrophylla by just two inversions. The high level of synteny between these two plastomes demonstrates that the extensive rearrangement in all major genera within Geraniaceae has occurred separately and that the presence of the IR appears neither necessary nor sufficient to stabilize gene order. In fact, next to C. macrophylla, the IR-lacking Erodium plastomes from Clade II (including E. carvifolium) are among the least rearranged in the family. Conversely, the great expansion of the IR in Pelargonium × hortorum may have mediated the severe genomic rearrangements seen in this species (Chumley et al., 2006; Guisinger et al., 2011; Weng et al., 2014).
An alternative hypothesis is that the IR may play an important role in replication initiation. The primary origins of replication lie within the IR of Nicotiana tabacum, Oenothera, Zea mays and several algal plastomes (Kunnimalaiyaan and Nielsen, 1997; Krishnan and Rao, 2009). Similarly, the quadripartite structure of an IR separated by two single copy regions has been found in herpes simplex virus 1 (Lehman and Boehmer, 1999; Okamoto et al., 2011) and in the 2-micron circular plasmids of Saccharomyces cerevisiae (Broach and Volkert, 1991). This structure is inferred to have evolved independently several times in herpes viruses (Davison, 1998; Davison et al., 2005). These genomes are all present as multiple isomers differing in the relative orientation of the single copy regions to the IRs, a signature of double rolling circle replication initiated by recombination between inverted repeats (Okamoto et al., 2011). The functional significance of double rolling circle amplification is unknown, as replication is not perceptibly impaired in IRLC legumes and Erodium plastids, and herpes virus genomes with IRs disrupted through insertional mutagenesis were capable of independent replication (Jenkins and Roizman, 1986). Although the patterns of IR expansion, loss and genomic rearrangement in Geraniaceae plastomes do not support the genome stability hypothesis for IR retention, convergence on this genome architecture in a virus, a plasmid and in plastomes suggests functional significance.
In this study we focused on plastome evolution in Erodium. We categorized 17 complete Erodium plastomes into four types and describe distinct evolutionary trajectories in the two major clades of the genus. We explore evidence of a single IR loss and describe LBC (Blazier et al., 2011) plastomes that contain a large IR. We analysed the branch leading to the LBC to determine whether it shows significantly higher nucleotide substitution rates than other branches in the genus. Finally, we discuss the functional significance of the IR in plastids and other genomes with a similar architecture and relate repeat content to genomic rearrangement with comparisons to recent studies of bacterial endosymbiont genomes.
MATERIALS AND METHODS
Taxon sampling
The dataset consisted of 19 taxa (18 Erodium plus California macrophylla), 18 complete plastomes and one draft genome from which protein-coding genes were extracted for phylogenetic and evolutionary rate analyses (Supplementary Data Table S1). Of the 18 complete plastomes, three were previously published (Blazier et al., 2011; Guisinger et al., 2011; Weng et al., 2014). The 16 new taxa were chosen based on a molecular phylogeny of the genus to ensure taxon sampling across the genus (Fiz et al., 2006) (Supplementary Data Fig. S1). Plants were obtained from commercial sources (Geraniaceae.com and B and T World Seeds) or grown from seed provided by J. J. Aldasoro (Real Jardín Botánico de Madrid, Spain), and voucher specimens are deposited at TEX.
DNA isolation and sequencing
Three different sequencing technologies were used. Supplementary Data Table S1 itemizes the sequencing platforms used for each taxon. For pyrosequenced genomes, plastids were isolated and plastid DNA was amplified as previously described (Jansen et al., 2005; Blazier et al., 2011). Sequencing was conducted on the 454 FLX platform at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign. For Illumina-sequenced genomes, total genomic DNA was isolated from fresh leaf tissue using a modified version of the hexadecyltrimethyl-ammonium bromide procedure of Doyle and Doyle (1987). Total genomic DNA was sequenced using Illumina HiSeq 2000 system at Beijing Genomics Institute Corporation or the Genome Sequence and Analysis Facility at the University of Texas at Austin. For each species, approx. 60 million 100-bp paired-end reads were generated from a sequencing library with ∼750-bp inserts. For genomes with Pacific Biosciences (PacBio) single molecule, real-time (SMRT) sequencing data, DNA was extracted using the same protocol used for Illumina sequencing. One SMRT cell of data per species was obtained from 10-kb insert libraries at the University of Florida’s Genomics Core Laboratory.
Genome assembly and annotation
Pyrosequenced genomes were assembled de novo in the native 454 assembler (Newbler package) under default settings as well as in MIRA v.3.4 using the ‘accurate’ setting (Chevreux et al., 1999). Illumina data were assembled de novo with Velvet v.1.2.07 (Zerbino and Birney, 2008) using a range of kmer sizes from 71 to 93, with and without scaffolding enabled. Plastid contigs were identified by BLAST searches against a database of Geraniaceae plastid protein-coding genes using custom Python scripts. Nuclear and mitochondrial contigs containing plastid DNA insertions were excluded using 1000× coverage cutoff. Assembly and filtering were performed on the Lonestar Linux Cluster at the Texas Advanced Computing Center (TACC). Correction of PacBio data was done using the LSC program (Au et al., 2012) and all ∼60 million 100-bp paired-end Illumina reads on the Lonestar Linux Cluster at TACC.
For all data types, contigs were assembled and edited in Geneious 7.0.4 (www.biomatters.com) and annotated in DOGMA (Wyman et al., 2004). Whole genome alignments were created using MAUVE v.2.3.1 (Darling et al., 2010) as implemented in Geneious 7.0.4 under default settings.
Prediction of tRNA genes
Genes encoding tRNAs were predicted in DOGMA under default settings. For two tRNA genes found to be missing from some species, trnG-GCC and trnV-GAC, tRNAscan-SE (Schattner et al., 2005) was used under ‘cove-only’ and ‘organellar’ parameters to verify the absence of these genes.
Rates analyses
For the 19-gene dataset, genes were extracted from DOGMA and inspected in Geneious. Genes were aligned in MAFFT (Katoh et al., 2009) as implemented in Geneious, and a concatenated alignment of all 19 genes (26 985 bp; Supplementary Data Table S2) was used to generate a constraint tree in Garli (Zwickl, 2008) under default settings as implemented in Geneious. For rates analyses, codon alignments were generated using MAFFT and the translation align function in Geneious.
Using the constraint tree (Fig. 1), plastid genes were analysed with codon-based models to quantify the rates of synonymous (dS) and non-synonymous (dN) substitution. Analyses were conducted in PAML 4.7 (Yang, 2007) using custom Python scripts on the Lonestar Linux Cluster at TACC. The F3 × 4 model was used to calculate codon frequencies, and the free-ratio model was used to compute dN/dS values. Transition/transversion and dN/dS ratios were estimated with the initial values of 2 and 0·4, respectively, consistent with other studies examining evolutionary rate heterogeneity in angiosperm organellar genomes (Sloan et al., 2009; Weng et al., 2012).
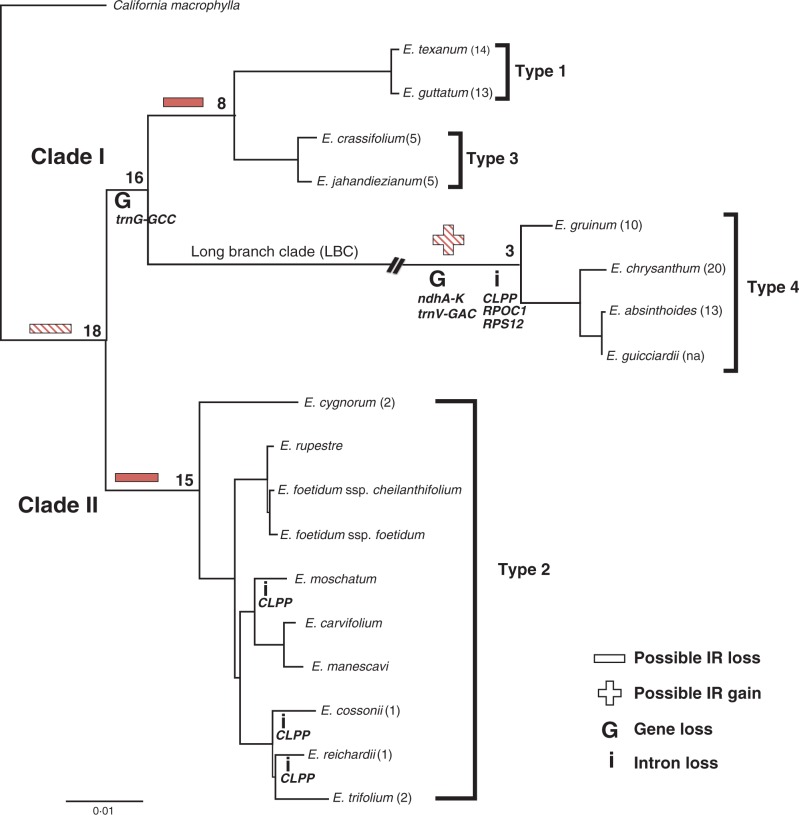
Fig. 1.
Phylogram depicting relationships among selected Erodium species. The maximum likelihood tree (score 70^914·4940 lnL) was generated from a concatenated alignment of sequences of 19 protein-coding genes (26 985 bp) for 18 Erodium species and the outgroup California macrophylla and represents the constraint topology for rates analyses. The two major clades (sensu Fiz et al., 2006) within the genus are labelled along with the long branch clade (LBC) and the four types of plastome characterized. The branch leading to LBC has been interrupted for concision. The number of inversions relative to California is given in parentheses after each species. Numerals at the nodes indicate divergence time estimates (Fiz et al., 2008). The two hatched and two solid symbols (+/–) indicating IR status represent paired events that are alternatives of each other as discussed in the text. Gene and intron losses are indicated on the relevant branches. The scale bar indicates the number of substitutions per site.
Likelihood ratio tests (LRTs) were conducted in HyPhy v.2.1.1 Beta for Mac (Kosakovsky Pond and Muse, 2005) to detect whether the branch leading to the clade that includes E. absinthoides, E. chrysanthum, E. gruinum and E. guicciardii (i.e. LBC) was significantly different from the other branches. The LRTs were conducted between two models, the null model with globally constrained dS shared by all branches and the alternative model with the branch leading to the LBC free from this constraint. The same setting was applied to the LRTs for dN. Because the constraint tree used for estimating rates has 34 branches, the P-value was multiplied by a Bonferroni correction factor of 34 to account for multiple comparisons.
Repeat analyses and genomic rearrangement estimates
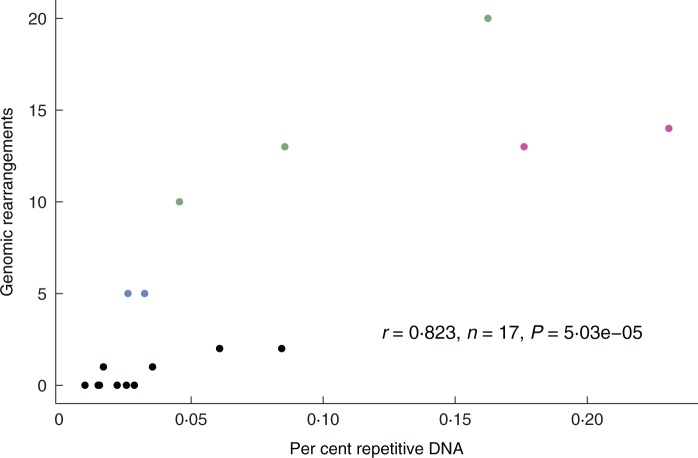
Repetitive DNA was identified by BLAST search of each genome against itself using blastn under default parameters and an e-value of 1e−10. One copy of the IR was removed from genomes with an IR (C. macrophylla and three Erodium species). The number of genomic rearrangements was determined by enumeration of rearranged co-linear blocks of genes in MAUVE alignments of each genome against the unrearranged Type 2 genome of E. carvifolium. For highly rearranged genomes and those containing a novel IR the most conservative estimate possible was used. Thus for E. guttatum it was assumed that the one inversion separating it from E. texanum had occurred in E. texanum such that E. texanum had an estimated 14 gene order changes and E. guttatum an estimated 13 gene order changes. For the three Type 4 genomes with novel IRs, the IR was not counted as a genomic rearrangement. For repeat content and genomic rearrangements a Pearson product-moment correlation coefficient was computed using the ‘Hmisc’ library in the R statistical package.
Verification of IR boundaries
Presence of the IR was verified independently for each genome. For E. gruinum, four sets of primers were designed to span the putative LSC/IR and SSC/IR junctions. Primer sequences are given in Supplementary Data Table S3. Sanger sequencing of PCR products was performed on an ABI 3730 platform at the core facility of the Institute of Cellular and Molecular Biology at The University of Texas at Austin. For E. absinthoides and E. chrysanthum, long, corrected PacBio reads >5 kb spanning the IR/single-copy region boundaries were used to verify the assemblies.
Divergence time estimates
Divergence time estimates were derived from a previous study of Geraniaceae (Fiz et al., 2008).
RESULTS
The dataset contained 17 completed plastomes representing the major clades in Erodium and outgroup California macrophylla (Supplementary Data Fig. S1, Table S1). Erodium plastomes fell into four types (hereafter Types 1–4) that differ in their short dispersed repeat (SDR) content and degree of genomic rearrangement (Fig. 2), gene and intron content, GC content (Fig. 1, Table 1), and substitution rates. The four plastome types corresponded to their respective clades in the phylogram (Fig. 1). Among the four (Table 1), Type 1 plastomes have the highest repeat content and 13–14 gene order changes. Type 3 species were sister to Type 1 in the phylogram, although these plastomes have much reduced repeat content and many fewer gene order changes. Type 4 plastomes represent the LBC, contain IRs, many gene order changes and lowest gene content among all types. Finally, Type 2 plastomes are characterized by the fewest gene order changes and the lowest repeat content.
Fig. 2.
Genomic rearrangement correlates with repetitive DNA content in Erodium plastomes. Repetitive DNA was identified by BLAST search of each genome against itself using blastn under default parameters and an e-value of 1e−10. One copy of the IR was removed from genomes with an IR (C. macrophylla and three Erodium species). The number of genomic rearrangements was determined by counting rearranged co-linear blocks of genes in MAUVE alignments of each genome against the unrearranged Type 2 genome of E. carvifolium. Coloured points represent Type 1 (pink), Type 2 (black), Type 3 (blue) and Type 4 (green) plastomes.
Table 1.
Plastid genome statistics for 18 Erodium species
| Clade I | Type 1 |
Type 3 |
Type 4 (LBC) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. texanum | E. guttatum | E. jahandiezianum | E. crassifolium | E. gruinum | E. absinthoides | E. chrysanthum | E. guicciardii | |||
| Size (bp) | 130 812 | 128 510 | 121 692 | 121 393 | 142 208 | 162 618 | 168 946 | genes only | ||
| Protein-coding genes | 75 | 75 | 75 | 75 | 64 | 64 | 64 | 64 | ||
| tRNA genes | 28 | 28 | 28 | 28 | 28 | 28 | 28 | 28 | ||
| Introns | 14 | 14 | 14 | 14 | 11 | 11 | 11 | 11 | ||
| GC content (%) | 39·5 | 39·4 | 39·1 | 39·1 | 43·0 | 42·9 | 42·9 | 42·3 | ||
| GC (%) 19-gene dataset | 41·9 | 41·8 | 41·7 | 41·6 | 44·1 | 44·0 | 44·1 | 44·0 | ||
| SDR (%) | 23·09 | 17·61 | 2·61 | 3·244 | 4·56 | 8·55 | 16·24 | unknown | ||
| Estimated gene order changes† | 14 | 13 | 5 | 5 | 10 | 13 | 20 | unknown | ||
| IR size (bp) | np | np | np | np | 25 508 | 45 490 | 47 428 | unknown | ||
| Clade II Type 2 | E. carvifolium | E. cossonii | E. cygnorum | E. foetidum ssp. cheil. | E. foetidum ssp. foet. | E. manescavi | E. moschatum | E. reichardii | E. rupestre | E. trifolium |
| Size (bp) | 116 935 | 121 465 | 121 905 | 116 340 | 115 794 | 116 810 | 119 078 | 117 753 | 116 810 | 123 865 |
| Protein-coding genes | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 |
| tRNAs | 28 | 28 | 28 | 28 | 28 | 28 | 28 | 28 | 28 | 28 |
| Introns | 18 | 17 | 18 | 18 | 18 | 18 | 17 | 17 | 18 | 18 |
| GC content (%) | 39·0 | 39·2 | 39·2 | 38·9 | 38·9 | 39·1 | 39·1 | 39·2 | 38·9 | 39·3 |
| GC (%) 19-gene dataset | 41·9 | 41·8 | 41·8 | 41·8 | 41·8 | 41·9 | 41·8 | 41·8 | 41·8 | 41·9 |
| SDR (%) | 2·85 | 3·54 | 8·48 | 1·53 | 1·48 | 0·98 | 2·20 | 1·68 | 2·55 | 6·08 |
| Estimated gene order changes† | 0 | 1* | 2 | 0 | 0 | 0 | 0 | 1* | 0 | 2 |
| IR size (bp) | np | np | np | np | np | np | np | np | np | np |
*Same inversion.
†Gene order changes were calculated relative to C. macrophylla.
SDR, short dispersed repeats; np, not present.
Gene and intron loss
Erodium plastomes lack several canonical tRNA genes, protein-coding genes and introns. Some of these losses show homoplasy (e.g. CLPP intron 1) or show no discernible pattern. For example, it was unclear whether all Type 2 genomes encode TRNK-UUU, as the first exon was not detected in E. moschatum; however, this gene is divergent in all four genome types with no clear pattern to the loss or divergence of this gene. Overall the pattern of gene and intron loss in Erodium parallels that of genomic rearrangement, with Clade I showing greater variation than Clade II and the greatest number of losses in the LBC (Fig. 1). The uncertain status of ribosomal protein gene RPL23 in the LBC could be resolved by RNA editing. We identified three CAA codons upstream of RPL23 or in the 5′ end of the first exon that could undergo C-to-U editing to produce TAA stop codons. If the CAA codon downstream of RPL23 is edited to serve as its stop codon, the resulting reading frame would be approx. 339 bp, plausible considering the 288-bp RPL23 gene found in other Clade I species.
Increased GC, elevated substitution rates in Type 4 plastomes
While GC content in Types 1–3 was within the expected range for vascular plants (36–40 %; Ruhlman and Jansen, 2014), the Erodium Type 4 (LBC) plastomes were the highest among all sampled from across angiosperms, as high as 43 % GC (Table 1). When calculated for the set of 19 protein-coding genes used in the evolutionary rates analysis (below), GC content remained elevated (44·0–44·1 %, Table 1). The 19-gene dataset (Supplementary Data Table S2) did not include NDH sequences, suggesting the elevation of GC content in LBC species is unrelated to the status of these GC-poor plastid genes.
The GC content of the three complete Type 4 plastomes (plus an estimate based on a concatenation of non-redundant contigs from a fourth species, E. guiccardii) was compared with that of 472 plastomes representing the major lineages of Viridiplantae. Only four taxa, a lycophyte and three algae, had GC content equal to or higher than the Type 4 species (Supplementary Data Table S4). A t-test determined whether the GC content of Type 4 plastomes was significantly different from the other 472 Viridiplantae. As the variances of the two groups were unequal, a two-sample test assuming unequal variances was conducted, and the difference between the mean GC content of the two groups was statistically significant (P = 5·634e−166).
The clade comprising Type 4 plastomes has been called the LBC because the branch leading to this lineage was distinctly long in phylogenetic reconstructions based on nucleotide sequence data (Blazier et al., 2011). We conducted a more rigorous analysis of evolutionary rates in the expanded Erodium dataset to test whether the branch leading to the LBC has a significantly higher nucleotide substitution rate considering codon positions. The dataset contained 19 genes (Supplementary Data Table S2), with two representatives of each functional class, all four RPO genes, and MATK, CLPP and RBCL. The branch leading to the LBC showed significant acceleration (P < 0·05) in the rate of synonymous substitutions (dS) for all genes and for non-synonymous substitutions (dN) for 14 of the 19 genes (Supplementary Data Table S2). Of the five genes for which dN was not significantly different, two were significant before a conservative (Bonferroni) correction for multiple comparisons. The other three genes, PETB, PSBC and RBCL, have a very low rate of non-synonymous substitution such that many branches in the analysis had dN values of zero or close to zero, which may have confounded the likelihood ratio test (Supplementary Data Table S2). The concatenated dataset with 19 genes showed that the branch leading to the LBC was significantly higher in both dS and dN. This branch experienced rate acceleration with respect to dS for all genes, and dN for all but the slowest evolving genes.
Inverted repeat loss in Erodium plastomes
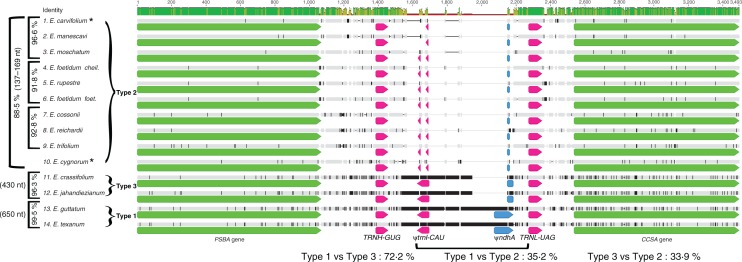
Loss of the IR was previously documented in two Erodium species: E. texanum (Clade I, Type 1; Guisinger et al., 2011) and E. carvifolium (Clade II, Type 2; Weng et al., 2014). Additional Erodium plastomes were compared to establish if a single loss of the IR had occurred in the genus. Among Clade I species, Type 1 and Type 3 plastomes, represented by E. texanum and E. guttatum, and E. crassifolium and E. jahandiezianum, respectively, were lacking the large repeat. Genomic features including a pseudogene of ndhA (exon 1) and a full copy of TRNI-CAU in Types 1 and 3 were detected in the alignment (Fig. 3). These genes are located at distal ends within the IR of the inferred ancestral genome for all Geraniaceae genera (Fig. 3; Weng et al., 2014). In the Type 2 genomes only small fragments of ndhA and trnI-CAU were detectable (Fig. 3). The upstream (PSBA, TRNH-GUG) and downstream (TRNL-UAG, CCSA) sequences represent single copy genes situated outside the ancestral IRa region, supporting the loss of the same IR copy in all cases.
Fig. 3.
Erodium species lacking the IR have all lost IRa. Annotated nucleotide alignment of the region formerly flanking the copy of the IR lost on the branch leading to Erodium. Pseudogenes are indicated by the (ψ) symbol. Upper histogram indicates nucleotide identities across the alignment. Nucleotide identities were calculated within and between plastome types and are reported to the left (within) and below (between) the alignment. For within-type comparisons (left) square brackets indicate which species were included in each comparison with the length of sequences compared (nt) and per cent identity. For Clade II (Type 2), species comparisons within subclades are also given (left). The bracket below indicates the extent of the region compared for identity values, and the length of these regions for each type is reported (nt) to the left of each group. For between-type comparisons, two Type 2 species (asterisks) were selected representing the two major clades within Clade II. Identity values were based on alignments of two species from each type, for a total of four species included in each between-clade comparison.
Nucleotide identities within and between clades were calculated from the alignment in Fig. 3. Erodium cygnorum and E. carvifolium were selected to represent Type 2 plastomes for this analysis. The region comprising the trnI-CAU pseudogene through the beginning of TRNL-UAG was compared and values are reported in Fig. 3.
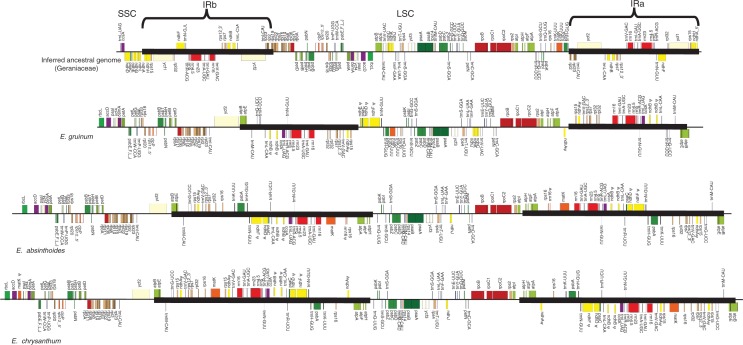
Identification of a novel IR in Type 4 plastomes
Assembly of those plastomes classified as Type 4 revealed the presence of a large inverted repeat (∼25·5 to ∼47·5 kb) that includes all four genes encoding the RNAs of the plastid ribosome. Depending on the species, the large repeat comprised a number of other genes, among them both canonical IR sequences as well as others encoded in the LSC and SSC regions in the ancestor of Geraniaceae as inferred by Weng et al. (2014; Fig. 4). Assembled contigs from the draft plastome of E. guiccardii included the IR boundaries, one of which is shown in alignment with extractions from the completed LBC plastomes (Supplementary Data Fig. S2) suggesting the presence of the novel IR in a fourth species of Erodium.
Fig. 4.
Schematic linear representation of Type 4 Erodium plastomes. The inferred ancestral plastome with gene annotations was adapted from Weng et al. (2014). Type 4 plastome maps were adapted from maps generated using OrganellarGenomeDRAW (Lohse et al., 2013). Pseudogenes are indicated by the (ψ) symbol. Thick, black lines indicate IR extent.
The presence of the IR was validated independently for the completed assemblies. For E. gruinum, four sets of primers (Supplementary Data Table S3) were designed to span the putative LSC/IR and SSC/IR junctions. Amplified products were Sanger sequenced and verified the predicted junctions. For E. absinthoides and E. chrysanthum, genomic DNA was submitted for PacBio sequencing. Corrected PacBio reads >5 kb spanning all the assembled IR/single-copy region boundaries confirmed the assemblies for E. absinthoides and E. chrysanthum.
DISCUSSION
With more available sequences for comparative genomics comes the opportunity to explore variable evolutionary trajectories across taxa. Despite a high degree of overall conservation in structure and content, plastomes in a few unrelated lineages have evolved in unique ways. The molecular signatures of divergence and convergence on genome architecture may reveal common factors that have shaped the contemporary genomic landscape. We have demonstrated that in Erodium, as in the family, studies of plastome evolution are best conducted at the species level.
Elevated GC content and rate acceleration in Type 4 plastomes
Geraniaceae plastomes display elevated GC content (Guisinger et al., 2011), but in LBC taxa it is elevated even further. In fact, LBC Erodium species have the highest GC content of any angiosperm plastome examined, and are surpassed by a single lycophyte and a few algal taxa (Supplementary Data Table S4). The genome-wide acceleration in both dN and dS, together with the increase in GC content seen in the four LBC species, suggests that an aberration in mutation rate and/or DNA repair may be responsible for this long branch as there is no obvious difference in habit or life history that would explain the elevated GC content, acceleration in rates of nucleotide substitution, gene and intron loss, or genomic rearrangement in this clade.
On the differential evolution of the IR in Erodium
In an effort to illuminate the IR loss thought to have occurred in the lineage leading to Erodium, we were surprised to identify a large IR containing the entire plastid ribosomal operon in the three completed plastomes from the LBC. The repeats range in size from ∼25 to ∼47 kb in length and largely contribute to size variation between plastomes in this group. Especially evident in the larger plastomes of E. absinthioides and E. chrysanthum, the so-called LSC and SSC regions are of very similar size due to the incorporation of non-canonical IR genes into the repeats in these species (Fig. 4).
With this finding we must consider whether the IR loss in Erodium occurred independently in two lineages or once on the branch leading to the genus, with subsequent reappearance of the IR in the LBC. The data presented in Fig. 3 clearly demonstrate that IRa was lost in Types 1–3. Furthermore the remaining sequences adjacent to the loss site indicate that, were there two losses, the SSC was in the same orientation at the time of loss in each case. Either scenario could be considered equally likely according to Occam’s razor, under which the most plausible path is the one that requires the least number of assumptions. Either path would require two steps: one loss and one gain or, alternatively, two losses. IR loss presents a special case, however, as this structural change may not adhere to weighting criteria used in parsimony analyses as applied to individual genes. If no fitness advantage (or disadvantage) were conferred by retention of the canonical quadripartite structure of the plastome then loss or gain would be equally likely to occur. While all of the genes in IRs are present in duplicate, one copy may be sufficient to support overall function of the plastid. Nonetheless, given the rarity of IR loss, it seems unlikely that this structure is neutral with regard to fitness, although the lineages where the IR has been lost do not display impaired phenotypes.
If the state of the IR was indeed neutral, then loss or retention should be stochastic. Under this assumption, the likelihood of two independent losses where the same IR copy was lost with the SSC in the same orientation would be 25 %. Among land plants, IR loss has been documented five times. Among those instances only Pinaceae has lost IRb (Wu et al., 2011). The cupressophytes (Wu et al., 2011), IRLC (Wojciechowski et al., 2004), Carnegiea gigantea (saguaro; Sanderson et al., 2015) and Erodium species have all retained IRb. This observation suggests IRa may be more dispensible than IRb, perhaps related to preservation of transcriptional units (i.e. the RPL23 operon). In Erodium and IRLC species the 5′ end of the operon, including RPL23 and RPL2, is situated within IRb. The losses in Pinaceae and cupressophytes may indeed be random as these lineages have retained different copies of the IR. It is worth noting that the leading genes of the RPL23 operon are not duplicated in the IR where present among gymnosperms (Wu et al., 2011).
Comparison of the region containing the former IR-SSC and IR-LSC boundaries (CCSA/TRNL-UAG and TRNH-GUG/PSBA, respectively) in plastome Types 1, 2 and 3 could be offered as support for a single loss of the IR in Erodium, with greater reduction of the resulting intergenic region in Type 2. Types 1, 2 and 3 each contain pseudogenes of ndhA and trnI, although these sequences are greatly reduced in Type 2 genomes, where a region of 8 bp for ndhA and two adjacent regions of 7 and 5 bp for trnI remain (Fig. 3). However, the argument could also be made that the retention pattern in this region suggests two losses as Types 1 and 3 show much greater similarity to each other (72·7 % pairwise nucleotide identity) than to Type 2 (35·2 and 33·9 %, respectively). In an alignment of the analogous region extracted from IRLC plastomes, congeners were 91·1 % identical, which dropped to 70·0 % when more distant relatives were considered. The IRLC loss is widely accepted to have resulted from a single event at the base of the clade (Wojciechowski et al., 2004) and comparisons across different genera robustly support a single IR loss over more than 25 million years (Wojciechowski, 2003)
Loss of the IR has not destabilized the plastome in Erodium, nor has IR presence in the LBC stabilized those genomes. Although they share the loss of the IR, Clade II contains species with few if any unique genomic rearrangements whereas the three types of Clade I plastomes all show considerable rearrangement. Furthermore, the most highly rearranged plastomes have retained the IR [e.g. Pelargonium, Geranium, Trachelium (Campanulaceae), Jasminum (Oleaceae), Vaccinium and Arbutus (Ericaceae), and lobelioids (Campanulaceae)] (Chumley et al., 2006; Lee et al., 2007; Haberle et al., 2008; Fajardo et al., 2013; Martínez-Alberola et al., 2013; Knox, 2014).
Destabilization of bacterial endosymbiont and Erodium plastid genomes
It is possible that the IR plays some role in stabilizing the plastome, but only in genomes with low repeat content. In rearranged plastomes evidence of illegitimate recombination has been detected (Ogihara et al., 2002; Maréchal et al., 2009), which is thought to underlie gene order changes and movement of the IR boundaries. When the repeat content of a plastome is very low, illegitimate recombination should be minimized. Under such conditions, intramolecular recombination could be largely confined to the IRs and genome stability would be maintained. However, once the repeat content of a plastome increases, illegitimate recombination within single copy regions or between regions flanking the IRs become more likely, causing inversions and expansion or contraction of the IR, respectively. Thus, the presence of the IR may be less important to plastome stability than the absence of other large repeats that serve as substrates for rearrangements.
The positive correlation between repeat content and genomic rearrangement (Fig. 2) is comparable with trends in genome evolution in bacterial endosymbionts. Plastid genomes resemble those of bacterial endosymbionts in some respects, which is unsurprising given that plastids descended from ancient bacterial symbionts (Sagan, 1967). The two types of genomes generally share a massive loss of genes, relatively low GC content, low repeat content and little genomic rearrangement. Proteomic constraint has been proposed to explain the paucity of DNA repair genes, and the lack of genomic rearrangement, in bacterial endosymbionts (Garcia-Gonzalez et al., 2013). In short, the larger the proteome the more DNA repair genes a bacterial genome maintains, a correlation that holds in both free-living and intracellular bacteria. The loss of DNA repair genes involved in recombination has been hypothesized to underlie the conservation of gene order among related endosymbionts over great time scales (Garcia-Gonzalez et al., 2013), although recent evidence calls this hypothesis into question (Sloan and Moran, 2013).
A key difference between plastome evolution and that of insect endosymbiont genomes is the role of recombination. As previously noted, the reduction in recombination, along with low repeat content, has been implicated in long-term genome stability in endosymbionts such as Buchnera (Tamas et al., 2002). The plastomes of Erodium and a few other unusual lineages aside, plastomes of flowering plants have remained co-linear with nearly identical gene content for well over 100 million years. However, this co-linearity has not been maintained through lack of recombination; in fact, just the opposite. The majority of angiosperm plastomes have large inverted repeats that recombine to produce distinct isomers differing in the relative orientation of the single-copy regions, and recombination is integral to both replication and repair of plastid DNA (Day and Madesis, 2007). A recent study of closely related Portiera whitefly endosymbiont biotypes with high-frequency genomic rearrangement but also lacking genes for recombination challenges the hypothesis that genomic stability in endosymbionts is maintained through elimination of recombination (Sloan and Moran, 2013). In both endosymbiont and plastid genomes, high repeat content may contribute to genomic instability, suggesting that limiting recombinogenic substrates, rather than eliminating recombination per se, may favour stability. Taken together, the repeat-rich Portiera genomes that undergo rearrangement in the absence of recombination genes and the positive correlation between non-IR repeat content and rearrangement in Erodium and other plastomes support the notion of repeat-mediated genomic instability regardless of the state of recombination genes.
Although the comparison between Erodium plastomes and endosymbiont genomes provides the insight that repetitive DNA mediates genomic rearrangement irrespective of the available pathways governing recombination, it raises the obvious question as to the origin of the repeats. How do some genomes accumulate a high proportion of repetitive DNA while others do not remains an open question. Illegitimate recombination between repeats has been implicated in many plastome rearrangements. However, it is increasingly likely that illegitimate recombination is a proximate cause, and the mechanism generating the repeats is the ultimate cause of genome rearrangement.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: a maximum-likelihood tree for Erodium based on the trnL-F spacer for 72 species, adapted from Fiz et al. (2006) and Blazier et al. (2011). Figure S2: nucleotide alignments of IR/SSC boundary in the 5′ end of ATPB in four LBC species. Table S1: taxon sampling and accession numbers. Table S2: nineteen-gene data set used in evolutionary analyses. Table S3: PCR primers used to confirm IR boundaries in E. gruinum. Table S4: viridiplantae plastomes with GC content > 42 %.
ACKNOWLEDGEMENTS
This paper represents a portion of J.C.B.’s PhD thesis in the Plant Biology Program at UT-Austin. We thank TACC at the University of Texas at Austin for access to supercomputers. We also thank Juan José Aldasoro for providing seeds of selected Erodium species, Robin Parer from Geranicaeae.com for providing live plants of selected species and Plant Resources Center at the University of Texas at Austin for storage of voucher specimens. This work was supported by an NSF GRF predoctoral fellowship to J.C.B. and a National Science Foundation grant (IOS-1027259) to R.K.J., T.A.R. and J.P.M., and the S. F. Blake Centennial Professorship to R.K.J. J.C.B. designed the research, assembled and analysed genomic data. J.C.B. and T.A.R. wrote the manuscript, designed and composed figures and tables. T.A.R. isolated DNA samples for Illumina and PacBio sequencing. J.P.M. assisted in the assembly of the E. chrysanthum and E. absinthoides genomes. M.G. annotated and submitted genomes to GenBank. J.Z. provided custom Python scripts for genome assembly and manipulation of Illumina data. M.W. conducted PAML analyses. R.K.J. assisted in designing the research and editing the manuscript. All authors read and approved the final paper.
LITERATURE CITED
- Au KF, Underwood JG, Lee L, Wong WH. 2012. Improving PacBio long read accuracy by short read alignment. PLoS One 7: e46679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier JC, Guisinger MM, Jansen RK. 2011. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Molecular Biology 76: 263–272. [DOI] [PubMed] [Google Scholar]
- Bock R. 2007. Structure, function, and inheritance of plastomes In: Bock R, ed. Cell and molecular biology of plastids. Topics in current genetics. Berlin: Springer, 29–63. [Google Scholar]
- Broach JR, Volkert FC. 1991. Circular DNA plasmids of yeasts In: Broach JR, Pringle JR, Jones EW, eds. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics, Vol. I New York: Cold Spring Harbor Laboratory Press, 297–331. [Google Scholar]
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. In: Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB), 45–56. doi:10.1.1.23.7465.
- Chumley TW, Palmer JD, Mower JP. et al. 2006. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Molecular Biology and Evolution 23: 2175–2190. [DOI] [PubMed] [Google Scholar]
- Darling AD, Mau B, Perna NP. 2010. progressiveMauve: Multiple genome alignment with gene gain, loss, and rearrangement. PLoS One 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ. 1998. The genome of salmonid herpesvirus 1. Journal of Virology 72: 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Trus BL, Cheng N. et al. 2005. A novel class of herpesvirus with bivalve hosts. Journal of General Virology 86: 41–53. [DOI] [PubMed] [Google Scholar]
- Day A, Madesis P. 2007. DNA replication, recombination, and repair in plastids In: Bock R, ed. Cell and molecular biology of plastids, Vol. 19 Berlin: Springer, 65–119. [Google Scholar]
- Downie SR, Palmer JD. 1992. Use of chloroplast DNA rearrangements in reconstructing plant phylogeny In: Soltis PS, Soltis DE, Doyle JJ, eds. Molecular systematics of plants. New York: Springer, 14–35. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue . Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Fajardo D, Senalik D, Ames M, et al. 2013. Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genetics and Genomes 9: 489–498. [Google Scholar]
- Fiz O, Vargas P, Alarcon ML, Aldasoro JJ. 2006. Phylogenetic relationships and evolution in Erodium (Geraniaceae) based on trnL-trnF sequences. Systematic Botany 31: 739–763. [Google Scholar]
- Fiz O, Vargas P, Alarcón M, Aedo C, García JL, Aldasoro JJ. 2008. Phylogeny and historical biogeography of Geraniaceae in relation to climate changes and pollination ecology. Systematic Botany 33: 326–342. [Google Scholar]
- Garcia-Gonzalez A, Vicens L, Alicea M, Massey SE. 2013. The distribution of recombination repair genes is linked to information content in bacteria. Gene 528: 295–303. [DOI] [PubMed] [Google Scholar]
- Goulding SE, Wolfe KH, Olmstead RG, Morden CW. 1996. Ebb and flow of the chloroplast inverted repeat. Molecular and General Genetics 252: 195–206. [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2008. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proceedings of the National Academy of Sciences USA 105: 18424–18429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2011. Extreme reconfiguration of plastomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Molecular Biology and Evolution 28: 583–600. [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. Journal of Molecular Evolution 66: 350–361. [DOI] [PubMed] [Google Scholar]
- Heinhorst S, Cannon GC. 1993. DNA replication in chloroplasts. Journal of Cell Science 104: 1–9. [Google Scholar]
- Hirao T, Watanabe A, Kurita M, Kondo T, Takata K. 2008. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biology 8: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Raubeson LA, Boore JL. 2005. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods in Enzymology 395: 348–384. [DOI] [PubMed] [Google Scholar]
- Jenkins FJ, Roizman B. 1986. Herpes simplex virus 1 recombinants with noninverting genomes frozen in different isomeric arrangements are capable of independent replication. Journal of Virology 59: 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT In: Posada D, ed. Bioinformatics for DNA sequence analysis, Vol. 537 Totowa, NJ: Humana Press, 39–64. [DOI] [PubMed] [Google Scholar]
- Knox EB. 2014. The dynamic history of plastomes in the Campanulaceae sensu lato is unique among angiosperms. Proceedings of the National Academy of Sciences USA 111: 11097–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies In: Nielsen R, ed. Statistical methods in molecular evolution. New York: Springer, 125–181. [Google Scholar]
- Krishnan NM, Rao BJ. 2009. A comparative approach to elucidate chloroplast genome replication. BMC Genomics 10: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL. 1997. Fine mapping of replication origins (oriA and oriB) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Research 25: 3681–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-L, Jansen RK, Chumley TW, Kim K-J. 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Molecular Biology and Evolution 24: 1161–1180. [DOI] [PubMed] [Google Scholar]
- Lehman IR, Boehmer PE. 1999. Replication of herpes simplex virus DNA. Journal of Biological Chemistry 274: 28059–28062. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research 41: W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Brisson N. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytologist 186: 299–317. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Véronneau-Lafortune F, Joyeux A, Lang BF, Brisson N. 2009. Whirly proteins maintain plastid genome stability in Arabidopsis. Proceedings of the National Academy of Sciences USA 106: 14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Alberola F, Del Campo EM, Lazaro-Gimeno D. et al. 2013. Balanced gene losses, duplications and intensive rearrangements led to an unusual regularly sized genome in Arbutus unedo chloroplasts. PLoS One 8: e79685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y, Isono K, Kojima T. et al. 2002. Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Molecular Genetics and Genomics 266: 740–746. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Horiuchi T, Watanabe T. 2011. Double rolling circle replication (DRCR) is recombinogenic. Genes to Cells 16: 503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. 1983. Chloroplast DNA exists in two orientations. Nature 301: 92–93. [Google Scholar]
- Palmer JD, Thompson WF. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29: 537–550. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Osorio B, Aldrich J, Thompson WF. 1987. Chloroplast DNA evolution among legumes: loss of a large inverted repeat occurred prior to other sequence rearrangements. Current Genetics 11: 275–286. [Google Scholar]
- Perry AS, Wolfe KH. 2002. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. Journal of Molecular Evolution 55: 501–508. [DOI] [PubMed] [Google Scholar]
- Ruhlman TA, Jansen RK. 2014. The plastomes of flowering plants In: Maliga P, ed. Chloroplast biotechnology. Methods in molecular biology. New York: Humana Press, 3–38. [Google Scholar]
- Sagan L. 1967. On the origin of mitosing cells. Journal of Theoretical Biology 14: 255–274. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Copetti D, Búrquez AI. et al. 2015. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. American Journal of Botany 102: 1115–1127. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Research 33: W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. 2013. The evolution of genomic instability in the obligate endosymbionts of whiteflies. Genome Biology and Evolution 5: 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Oxelman B, Rautenberg A, Taylor DR. 2009. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evolutionary Biolology 9: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas I, Klasson L, Canback B. et al. 2002. 50 Million years of genomic stasis in endosymbiotic bacteria. Science 296: 2376–2379. [DOI] [PubMed] [Google Scholar]
- Tremblay-Belzile A, Lepage E, Zampini E, Brisson N. 2015. Short-range inversions: rethinking organelle genome stability: template switching events during DNA replication destabilize organelle genomes. Bioessays 37: 1086–1094. [DOI] [PubMed] [Google Scholar]
- Weng M-L, Ruhlman TA, Gibby M, Jansen RK. 2012. Phylogeny, rate variation, and genome size evolution of Pelargonium (Geraniaceae). Molecular Phylogenetics and Evolution 64: 654–670. [DOI] [PubMed] [Google Scholar]
- Weng M-L, Blazier JC, Govindu M, Jansen RK. 2014. Reconstruction of the ancestral plastome in Geraniaceae reveals a correlation between genome rearrangements, repeats and nucleotide substitution rates. Molecular Biology and Evolution 31: 645–659. [DOI] [PubMed] [Google Scholar]
- Wojciechowski MF. 2003. Reconstructing the phylogeny of legumes (Leguminosae): an early 21st century perspective In: Klitgaard BB, Bruneau A, eds. Advances in legume systematics, part 10, higher level systematics. Kew: Royal Botanic Gardens, 5–35. [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. American Journal of Botany 91: 1846–1862. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences USA 84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. 2011. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biology and Evolution 3: 1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255. [DOI] [PubMed] [Google Scholar]
- Yamada T. 1991. Repetitive sequence-mediated rearragements in Chlorella ellipsoidea chloroplast DNA: Completion of nucleotide sequence of the large inverted repeat. Current Genetics 19: 139–147. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ruhlman TA, Sabir JSM. et al. 2016. Coevolution between nuclear encoded DNA replication, recombination and repair genes and plastid genome complexity. Genome Biology and Evolution 8: 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytologist 209: 1747–1756. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. 2008. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD Thesis, University of Texas at Austin, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.