Abstract
Gene co-option, usually after gene duplication, in the evolution of development is found to contribute to vertebrate morphological innovations, including the endothelium-based vascular system. Recently, a zebrafish kank gene was found expressed in the vascular vessel primordium, suggesting KANK genes are a component of the developmental tool kit for the vertebrate vascular system. However, how the KANK gene family is involved in vascular vessel development during evolution remains largely unknown. First, we analyzed the molecular evolution of the KANK genes in metazoan, and found that KANK1, KANK2, KANK3 and KANK4 emerged in the lineage of vertebrate, consistent with the two rounds of vertebrate whole-genome duplications (WGD). Moreover, KANK genes were further duplicated in teleosts through the bony-fish specific WGD, while only kank1 and kank4 duplicates were retained in some of the examined fish species. We also found all zebrafish kank genes, except kank1b, are primarily expressed during embryonic vascular development. Compared to invertebrate KANK gene expression in the central nervous system, the vascular expression of zebrafish kank genes suggested KANK genes were co-opted for vertebrate vascular development. Given the cellular roles of KANK genes, our results suggest that this co-option may facilitate the evolutionary origin of vertebrate vascular vessels.
One of the central goals of evolution is to understand the morphological complexity of living organisms such as vertebrates. This subphylum is defined by the presence of key morphological characteristics including neural crest cells and their derivatives such as craniofacial structures, neurogenic placodes, elaborate segmented brain, endoskeleton (bone and cartilage), and endothelium-based vasculature1,2,3. Decades of developmental genetic studies have revealed that such morphological structures are controlled by the genetic tool kit: developmental regulatory genes and tissue-specific genes4. Thus, the molecular phylogeny of such genes in the tool kit could provide us key information about the evolutionary origin of novel morphological structures. Whole genome duplications (WGD) were found as a way for increasing the diversity of genetic tool kits, and the vertebrate innovations were associated with the early WGD, around the origin of the subphylum5,6,7,8. WGD does not only increase the size of gene families, but also create novel functions of the duplicates (sub- or neo-functionalization) by co-option9,10,11,12. For example, we previously found WGD to contribute to the origin of vertebrate novelties such as cartilage by increasing the number of clade A collagen genes13,14,15.
The KANK protein family is characterized by an N-terminal KN motif, coiled-coil motifs, and four to five ankyrin repeat domains located in the C-terminus16. In C. elegans, VAB-19, the orthologous gene of KANK, is localized at the basement membrane and in trans-epidermal attachments17,18. VAB-19 is also required for later localization of attachment structures to muscle-adjacent regions of epidermis due to its role in actin remodeling mediated by Ras to Rac, key regulators of cell migration19. Fruit fly, also have a KANK gene, CG10249, which was found to be expressed in the midline of the central nervous system20. CG10249 is also expressed at the attachment region of muscle and epidermal cells in fruit fly embryos, and it interacted with EB1 at the microtubule plus end20,21. In human, there are four KANK genes: KANK1, KANK2, KANK3 and KANK416. KANK1 was originally identified as a candidate tumor suppressor gene on chromosome 9p in renal cell carcinoma patients, and when overexpressed in renal cell lines it was found to inhibit cell growth22. In addition, KANK1 functions as a regulator of actin polymerization, actin stress fiber formation, and cell migration through RhoA signaling23,24. Similar to KANK1, the other three human KANK genes were also found to be able to regulate actin polymerization and cell mobility16. An actin stress fiber is a bundle of approximately 10–30 actin filaments, and it plays important roles in cell migration and contractility of non-muscle vasculature cells. The stress fibers in these cells are important in dealing with mechanical stresses such as hydrostatic pressure, blood flow shear, and cyclic stretch25. Moreover, during vasculogenesis, endothelial cells undergo dramatic polarization, migration, rearrangements, and cell shape alterations26,27. All of the changes rely on cyctoskeleton and actin polymers which are critical for making the vascular capillary structures26,27. Recently, kank3 was found expressed in the blood vessel primordium of zebrafish embryos28, suggesting KANK genes may play important roles in vertebrate blood vessel development. However, how the KANK gene family is involved in vascular vessel development as a component of the genetic tool kit during evolution remains largely unknown.
To explore this direction, we took advantage of the zebrafish model system, which has a complex closed endothelial circulatory system. Zebrafish vasculature anatomy, the vessel assembling process and the molecular regulatory mechanisms were found to be similar to humans29,30. We first analyzed the evolutionary history of the KANK gene family in major taxa of metazoan, and then examined expression patterns of zebrafish kank genes during early development. We found that the KANK gene family was expanded through WGD and the zebrafish kank genes were primarily expressed during vascular development. As invertebrate KANK genes are not expressed in vascular system during development, the vascular expression domains of zebrafish kank genes suggested that the KANK gene family is co-opted for vertebrate endothelial vascular development after vertebrate WGD around the separation of vertebrate subphylum.
Results
Vertebrates have four groups of KANK genes
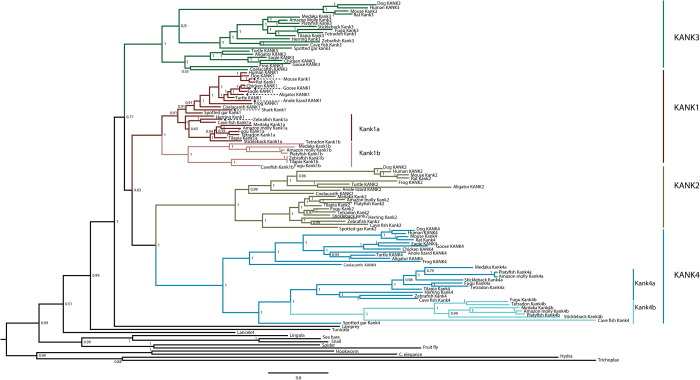
To better understand how novel morphological characters can arise at the cellular and developmental level, it is important to learn the evolutionary history of the key genes of the related developmental tool kit. Because we were interested in a large span of evolutionary time, and DNA sequence most likely underwent multiple substitutions through this time, we chose KANK protein sequences from major metazoan taxa for phylogenetic analysis. We first retrieved KANK protein sequences from Ensembl and other genome databases by BLASTp search. Then multiple protein sequences were aligned using MUSCLE program31. We found that all examined KANK proteins possess both KN motifs at the N-terminus and ankyrin repeat domains at the C-terminus. Molecular phylogeny analyses were performed using Bayesian analysis (BP) and maximum likelihood (ML) methods32. We were able to trace back KANK to the basal metazoans such as Trichoplax and Hydra (Fig. 1 and Fig. S1). The invertebrate KANK proteins form a distinct group from the vertebrate KANKs, and the tunicate KANK forms the closest outgroup of vertebrate KANK proteins. The overall phylogenetic relationship of KANKs is consistent with our current knowledge on the phylogeny of metazoan taxa33,34.
Figure 1. Extended majority-rule consensus tree for the Bayesian phylogenetic analysis of KANK proteins.
Numbers at each node indicate posterior probability (pp) values based on twenty million runs. Branch lengths are proportional to means of the pp densities for their expected replacements per site. The ML phylogenetic tree (Fig. S1.) was generally in agreement with the BP phylogeny: all the invertebrates have one KANK, while there are four KANKs in vertebrates (KANK1-KANK4). There are extra Kank1 and Kank4 proteins in teleosts. The tunicate (urochordate) and lancelet (cephalochordate) form the closest outgroups of vertebrates. The tree was rooted with Tricoplax and Hydra.
In vertebrates, there are four distinct clades of KANK proteins, KANK1, KANK2, KANK3 and KANK4 (Fig. 1 and Fig. S1). This is consistent with the current knowledge that humans and mice have four KANK genes. The appearance of these four clades arose around the origin of the vertebrates, suggesting that the birth of the four KANK clades was one of the consequences of the consecutive WGD events7,35. Within each vertebrate KANK clade, sarcopterigii (lobe-finned fish) and actinoperigi (ray-finned fish) generally formed distinct groups. Interestingly, the lamprey KANK formed an outgroup of the gnathostomes in the BP phylogenetic tree (Fig. 1), while it was clustered within gnathostome KANK groups with low supporting bootstrap value (233/1000) in the ML phylogenetic tree (Fig. S1). This may be due to the fact that the lamprey genome might have undergone unusual independent genomic events, as evidenced by other lamprey genes such as collagens and HOX genes36,37.
Loss of KANK genes in certain vertebrate taxa
Since it is known that some duplicated genes may be lost during evolution38, we examined the presence of the four KANK genes in the major taxa, and found that the KANK2 gene is missing from the birds’ genomes (chicken, turkey etc.). To confirm this loss, we did a Blast search for KANK2 orthologous genes in the bird genomes, however, no KANK2 ortholog was found. In addition, we found that KANK3 was missing from the current anole lizard genome, yet both KANK2 and KANK3 genes were identified in their relatives (turtle and crocodile) (Fig. 1 and Fig. S1). These data suggested the losses of KANK2 and KANK3 happened independently in the bird and lizard lineages, respectively. To further confirm these findings, we examined the evolutionarily conserved syntenies that contain these genes in the Synteny Database39 and Genomicus browser40. We found that the KANK2 and DOX6 genes are linked together in other jawed vertebrates, but these two genes are missing from current bird genomes. Similarly, KANK3 is linked with ANGPTL4, RPS28 and NDUFA7A in basal and higher vertebrates, but the synteny is missing from anole lizard genome.
Duplicated kank genes in teleosts
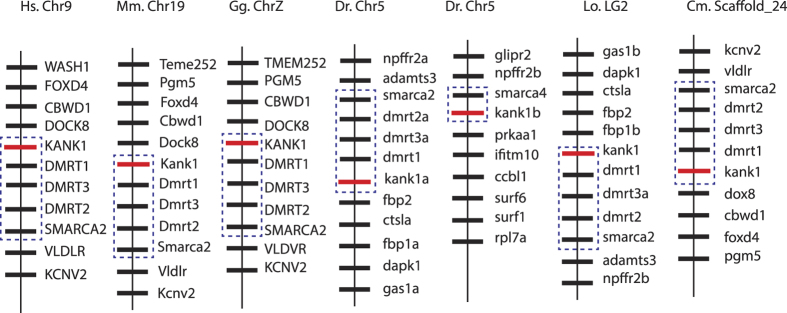
Bony fish are the most diverse group of species in the vertebrate subphylum. We found that of the four kank genes, kank1 and kank4 genes were duplicated in teleosts (Fig. 1 and Fig. S1). For the KANK1 genes, kank1a is grouped with KANK1 genes of tetrapod, spotted gar and elephant shark, rather than the teleost kank1b; suggesting that kank1b diverged from its ancestral state. Although widely used, phylogenetic analysis alone may not be sufficient for determining the gene orthologous relationships, especially in the unique situation of gene duplications followed by gene losses or rapid lineage-specific gene expansions38. To confirm the closer relationship between tetrapod KANK1 and teleost kank1a, we did a syntenic analysis on the represented vertebrate KANK1 genes. Interestingly, we found the kank1a in teleost are still in an evolutionarily conserved synteny, while kank1b has lost its neighboring genes and is only linked with the smarca4 gene in zebrafish (Fig. 2). To further examine the relationships between the duplicated kank1 genes in teleosts, we performed protein domain analysis using SMART program41. We found that both zebrafish Kank1a and Kank1b have similar protein domains when compared to human KANK1, except that Kank1b is 300aa shorter than Kank1a in the non-conserved middle region (Fig. S2). Furthermore, we analyzed the genomic intron-exon structural differences between zebrafish kank1a and kank1b. Kank1a has a similar intron-exon structure with human and spotted gar, while kank1b has a distinct structure with regard to the size and arrangement of exons (Fig. S3). Within the Kank4 clade, Kank4a and Kank4b clustered more closely compared to tetrapod KANK4 genes and both clustered with spotted gar, indicating that the two groups of genes appeared at the same time within this lineage (Fig. 1 and Fig. S1). Interestingly, zebrafish and herring have only a single kank4 gene, and they are located within the kank4a lineage suggesting that kank4b genes were lost in these two species.
Figure 2. KANK1 gene is located in an evolutionary conserved synteny in vertebrates.
Seven representative vertebrate species were analyzed. The illustration of the genes and their sizes are not proportional to the length of the bars. KANK1 is highlighted in red, and the synteny is highlighted in blue dashed-line boxes. The zebrafish kank1a is within the evolutionary conserved synteny. The zebrafish kank1b is distinct from the rest as it is only linked with smarca4. Hs, human; Mm, mouse; Gg, chicken; Dr, zebrafish; Lo, Spotted gar; Cm, elephant shark. Chr, chromosome. LG2, linkage group 2.
Zebrafish kank genes have similar yet distinct expression patterns
WGD usually leads to gene co-option, the functional specialization of paralogous genes, and contributes to vertebrate morphological innovations. Recently, kank3 was found expressed in the blood vessel primordium of zebrafish embryos28, suggesting KANK genes may play important roles in vertebrate vascular development. As zebrafish are a widely used vertebrate model system in developmental biology, we reasoned that examining the gene expression patterns of the five zebrafish kank genes might shed light on the evolutionary origin of the vertebrate vascular system.
Expression of kank1a and kank1b
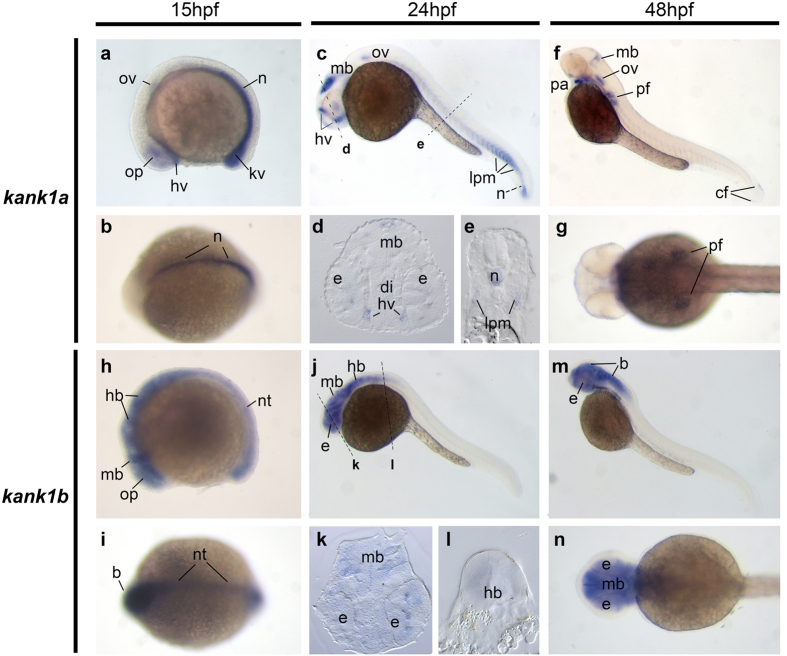
Zebrafish have two kank1 genes, kank1a and kank1b. At the developmental stage 15 hpf (hours post fertilization), kank1a is expressed in the optic vesicles, otic vesicles, head blood vessels, notochord and Kupffer’s vesicle (Fig. 3a,b), while kank1b is expressed throughout the central nervous system (Fig. 3h,i). By 24 hpf, kank1a is expressed within distinct patches of the midbrain, head blood vessels, otic vesicles, notochord, and lateral plate mesoderm, which gives rise to trunk intersegmental vessels (Fig. 3c–e). In contrast to kank1a’s distinct expression patterns, kank1b is expressed broadly throughout the midbrain, hindbrain and eyes at 24 hpf (Fig. 3j–l), with this expression pattern continued to 48 hpf (Fig. 3m,n). By 48 hpf, kank1a expression in the notochord and somites has decreased anteriorly to posteriorly. In addition, while kank1a is still expressed in the midbrain and otic vesicle, at 48 hpf, there is additional expression in the pharyngeal arch, pectoral and caudal fin buds (Fig. 3f,g).
Figure 3. Gene expression patterns of kank1a and kank1b in zebrafish embryos.
Whole-mount in situ hybridization of zebrafish embryos at stages 15 hpf (a,b,h and i), 24 hpf (c–e,j–l) and 48 hpf (f,g,m,n). Anterior is to the left in all the whole-mount images, and dorsal is to the top in all transverse sections. (a,b). Lateral and dorsal view of the expression of kank1a at 15 hpf. (c,f). Lateral view of kank1a expression at 24 hpf and 48 hpf, respectively. (h,i). Lateral and dorsal view of the expression of kank1b at 15 hpf. (j,m). Lateral view of kank1b expression at 24 hpf and 48 hpf, respectively. The dashed lines indicate the positions of sections. The letters below the dashed lines correspond to the panels. b, brain; cf, caudal fin bud; di, diencephalon; e, eye; hb, hindbrain; hv, head blood vessels; kv, Kupffer’s vesicle; lpm, lateral plate mesoderm; mb, midbrain; n, notochord; nt, neural tube; op, optic vesicle; ov, otic vesicle; pa, pharyngeal arch; pf, pectoral fin.
Expression of kank2, kank3, and kank4
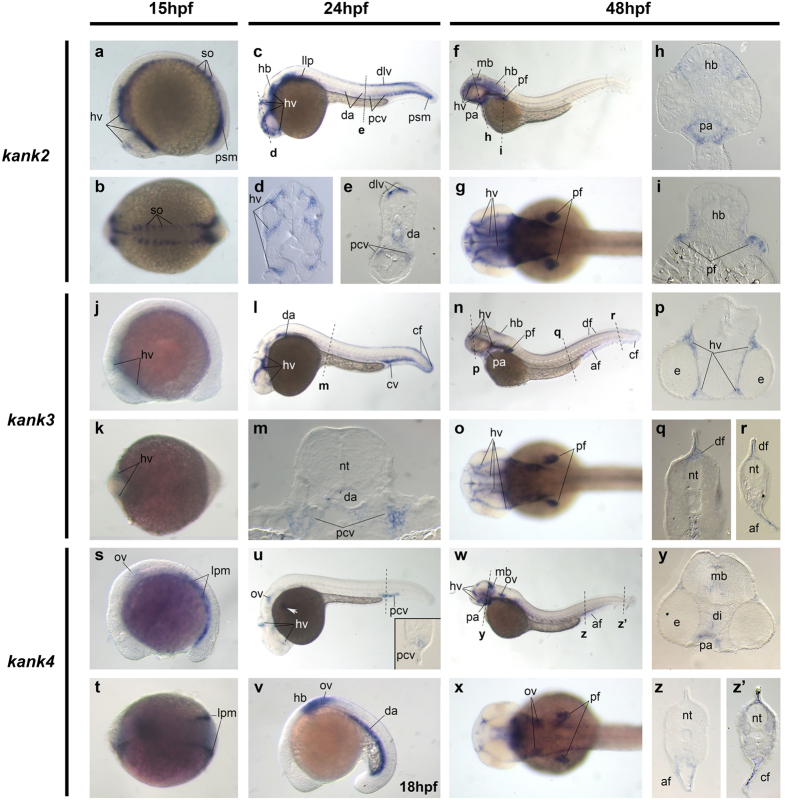
Next, we examined kank2 gene expression, which is strongly expressed in the head blood vessels, somites and presomitic mesoderm of the tail bud at 15 hpf (Fig. 4a,b). At 24 hpf, it is expressed in the head blood vessels, midbrain, and lateral line primordia, with expanding expression from the presomatic mesoderm to the trunk blood vessels (Fig. 4c–e). By 48 hpf, kank2 expression is restricted to the head region with retained expression in the head blood vessels, midbrain and hindbrain. New expression was detected in the pharyngeal arches, as well as the pectoral and annual fin buds (Fig. 4f–i). Kank3 has limited expression in the head blood vessel at 15 hpf (Fig. 4j,k). By 24 hpf, kank3 expression in the head blood vessels remains, with additional expression in the dorsal aorta, caudal vein, and caudal fin bud (Fig. 4l,m). Kank3 retains this expression pattern at 48 hpf (Fig. 4n–r). Our kank3 expression results are consistent with previous reports that this gene is expressed in the head blood vessels during zebrafish development28. Lastly, at 15 hpf, kank4 is expressed in the otic vesicles and lateral plate mesoderm, which gives rise to angioblasts in zebrafish (Fig. 4s,t). When the embryo is at 18 hpf, kank4 expression is also found in the hindbrain and dorsal aorta (Fig. 4v). The dorsal aorta expression of kank4 is greatly reduced anteriorly to posteriorly by 24 hpf, where its expression is also seen in the head blood vessels, otic vesicle and in the posterior cardinal veins (Fig. 4u). At 48 hpf, kank4 is still expressed in the otic vesicle and head blood vessels, with additional expression in the midbrain, pharyngeal arch, anal and caudal fin (Fig. 4w–z’).
Figure 4. Kank2, kank3 and kank4 gene expression during zebrafish development.
Whole mount in situ hybridization of zebrafish embryos at stages 15 hpf (a,b,j,k,s,t), 18 hpf (v), 24 hpf (c–e,l,m,u) and 48 hpf (f–i,n–q,w–z’). Anterior is to the left in all whole-mount images, and dorsal is to the top in all transverse sections. (a–i). Gene expression of kank2. (j–r). Gene expression of kank3. (s–z’). Gene expression of kank4. The dashed lines indicate the positions of section. The letters below the dashed lines correspond to the panels. The inset within panel (u) is the transverse section of the dashed line above. The white arrow points to the kank4 expression on the vascular vessels on the surface of the yolk. af, anal fin bud; cf, caudal fin bud; cv, caudal vein; da, dorsal aorta; df, dorsal fin bud; dlv, dorsal longitudinal vein; e, eye; hb, hindbrain; hv, head vessels; llp, lateral line premordia; psm, presomatic mesoderm; lpm, lateral plate mesoderm; mb, midbrain; nt, neural tube; ov, otic vesicle; pa, pharyngeal arch; pcv, posterior cardinal vein; pf, pectoral fin bud; so, somite.
Discussion
Understanding the morphological complexity of living organisms such as vertebrates is one of the central questions for evolutionary biology. The separation of the vertebrate lineage was accompanied with key morphological characteristics, one of which was the endothelium-based vascular system. Such morphological innovations are usually generated through tinkering or modification of the corresponding developmental genetic tool kits. Thus, the evolution and gene expression of key genes in such genetic tool kits provide us important information for understanding the evolutionary origin of morphological novelties. Here, we first analyzed the evolution of the KANK gene family and then examined the zebrafish kank gene expression patterns during early development. Our results suggested KANK genes were co-opted for vertebrate vascular development after vertebrate WGD, and KANK gene duplication and diversification may facilitate the evolutionary origin of vertebrate vascular vessels.
Evolutionary history of KANK genes
The KANK genes form an evolutionary conserved gene family. We were able to track their evolution as far back as basal metazoans such as placozoa and cnidarian. In the invertebrate species we analyzed, there is only one KANK gene in each species, while there are four KANK genes (KANK1-KANK4) in vertebrates. The expansion of the four KANK gene clades coincides with the origin of vertebrates. As there are two consecutive WGD (2R) in the vertebrate common ancestors, the four KANK genes are most likely the result of these whole genome duplications5,6,7,8. In addition, we found duplicates of kank1 and kank4 in teleosts. This is consistent with the bony fish specific WGD (3R)42,43. The duplicated paralogous genes through WGD (ohnolog) can be lost distinctively in different species, and only about one third of duplicated genes are retained in the zebrafish genome38,44. For example, zebrafish have only one kank4, while other fish such as fugu, amazon molly, platyfish, etc. have two kank4 genes (kank4a and kank4b).
We did not find any duplicates of kank2 or kank3 in the bony fish species we analyzed, suggesting that one of the ohnologs of kank2 and kank3 were lost after the teleost WGD. To our surprise, we did not find any KANK2 genes in the currently sequenced representative bird genomes. However, this gene was found in lizard, turtles and crocodiles, indicating that the KANK2 gene was lost specifically in the lineage of birds. Currently, KANK2 is known for its functions as a steroid receptor coactivator, inhibitor of apoptosis and regulator of actin dynamics in kidney podocyte foot processes45,46,47. The biological consequences of KANK2 gene loss in birds are currently unknown.
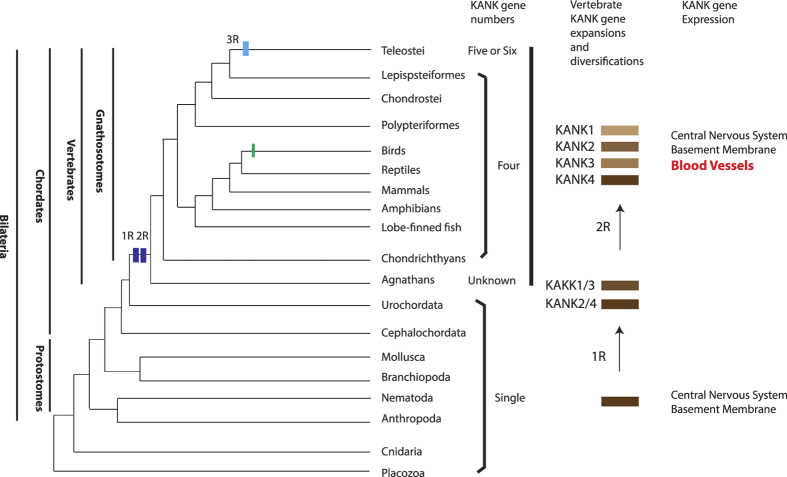
Based on our analysis on KANK genes’ evolution and our current knowledge of the WGD, we propose a model for KANK gene evolution, as shown in Fig. 5. Briefly, an invertebrate KANK gene was duplicated twice through the 2 consecutive WGD (2R) and created the vertebrate KANK1-KANK4. In teleost, all the kank1-kank4 were further duplicated through bony fish specific WGD (3R), and one of the duplicates of kank2 and kank3 were lost, and only one copy of each of these two genes retained in examined current teleost species. Similarly, in birds, KANK2 may have been lost independently after the separation of the bird lineage.
Figure 5. The molecular evolution of KANK genes and the origin of vertebrate blood vessels.
The phylogenetic relationships of bilateria and vertebrata are based on the recent phylogenomic analyses and our past collagen gene phylogenetic analyses. The branch lengths are not proportional to the time of diversification. The two vertical dark blue bars on the tree indicate the WGD events (R1 and R2) at the origin of the vertebrates. The other light blue vertical bar represents the teleost specific WGD. The green bar on the bird lineage indicates the loss of KANK2. The process of KANK gene duplication and diversification through WGD is illustrated by the brown bars. The co-option of the vertebrate four KANK genes’ expression coincides with the vertebrate vascular vessels.
Overlapping and distinct expression patterns of zebrafish kank genes
Gene expression usually corresponds to gene’s functions during embryonic development. We have found that zebrafish kank1a, kank1b and kank4 are expressed in the central nerve system, while kank2, kank3, and kank4 are mainly expressed in the blood vessel primordium. Additionally, kank1a, kank2, kank3, and kank4 are also expressed in the paired and median fin folds. These expression domains suggest a developmental function for kank genes in these tissue and organs. These zebrafish kank genes’ overlapping and distinct expression domains most likely resulted from WGD and subsequent gene-coption: sub-functionalization and neo-functionalization9. After gene duplication, the two duplicates may simply split the original functions of ancestral gene, leading to sub-functionalization. Or, one gene duplicate may retain the original function(s) of the ancestral gene, while the other duplicated gene may gain new expression domain(s) and evolve new functions, leading to neo-functionalization. Usually gene co-options were caused by changes in the gene regulatory regions after gene duplication, as coding regions are generally subject to more functional constrains11. Consistent to this, we found all the human and zebrafish KANK genes share very similar protein structures (Fig. S2), and all four human KANK genes have been reported to have the same functions in regulation of cytoskeletons16.
KANK genes were co-opted for vascular vessel development in vertebrates
The sub-functionalization and neo-functionalization of developmental genes usually lead to gene co-options for new developmental expression domains that are related to morphological novelties9,10,11,12. One of the vertebrate innovations is the blood vascular system that is essential for transportation of oxygen, nutrition, and waste products3,30. Compared to invertebrates, the major difference is the endothelial lining which makes the blood circulation more efficient3,48,49. Because blood vessels do not fossilize as do skeletons, it is unrealistic to trace the evolution of the circulatory system using fossils. Thus, molecular evolution and developmental studies are important to understand the evolutionary origin of this system.
Recently, the expression of zebrafish kank3 was reported in the blood vessel primordium28, suggesting the KANK genes are a component of the genetic tool kit for vertebrate blood vessel development. The fruit fly KANK gene was found expressed in the central nervous system20, where we also found zebrafish kank1a, kank1b and kank4 to be expressed. This suggests that one of the primitive functions of KANK genes is regulating development of the central nervous system. The new expression domain of kank genes (kank1a, kank2, kank3 and kank4) in zebrafish blood vessel primordium revealed that the KANK genes were co-opted for vascular development after the two rounds of vertebrate WGDs (Fig. 5). Given the regulatory functions of KANK genes in cytoskeleton and cell migration16,23,24, and how vasculogenesis requires extensive cell shape change and movement, the co-option of KANK genes from neural development to vascular development might facilitate the evolutionary origin of the vertebrate vascular system. Interestingly, the similarities of neural and vascular development have long been noticed since both processes utilize very similar molecular regulatory networks, such as VEGF and Ephrin–Eph signaling49,50. Alternatively, the zebrafish kank genes only express specifically in blood vessels of fish. This is unlikely since most orthologous genes in zebrafish express in similar locations during development compared with that in tetrapod51,52,53. These include vegf and other vascular regulatory genes such as vecdn, estrp and fli129,30. It is even more unlikely that all five kank ohnologs are fish specific. Future studies should define KANKs’ role in the evolution of blood vessels in vertebrates by determining the expression and functions of KANKs in urochordates, cephalochordates, agnathans, chondrichthyans, and tetrapods.
Methods
Zebrafish strains and husbandry
Zebrafish were raised and maintained following the procedures described in the zebrafish book54. The Purdue animal housing facility is an AAALAC-approved animal facility and all experiments were carried out according to the protocols approved by the Purdue Animal Care and Use Committee (PACUC) (Protocol # 1210000750). The wild type line used in this study is of the TAB background. The zebrafish embryos were sorted and staged using the Kimmel’s staging guide55.
KANK protein sequence retrieval and analysis
KANK protein sequences were identified by a BLASTp search using human KANK1 sequences as a query. The lamprey KANK sequence was retrieved from the Japanese lamprey genome website (http://jlampreygenome.imcb.a-star.edu.sg), and the lancelet KANK sequence was retrieved from the Chinese lancelet genome website (http://mosas.sysu.edu.cn/genome/index.php). The rest of the KANK protein sequences of the representative’s metazoan taxa were retrieved from either Ensembl or NCBI56,57 (Table S1). The longest sequence was preferentially chosen when there were multiple sequences. Multiple protein sequences were aligned using MUSLE program31, and the FASTA format alignment can be found in the supplementary File 1. To identify the best evolutionary model for phylogenetic analysis, we carried out a best model test using maximum likelihood and default parameters in MEGA658. The models with lowest BIC (Bayesian Information Criterion) scores were considered to describe the substitution pattern the best, and JTT+G was chosen. Then, we constructed phylogenetic trees using Bayesian analysis (BP) and maximum likelihood (ML) methods that are currently the most reliable for inferring phylogeny32. The ML and BP analyses were conducted as described using PhyML 3.1 and MrBayes 3.2.6, respectively59,60. For BP phylogenetic analysis, 20 million generations were run using the following parameters: nruns = 2, nchains = 4, aamodel = fixed(Jones), rates = gamma ngammacat = 8, samplefreq = 500, burninfrac = 0.25. ML phylogenetic analysis was performed using JTT + G with 1000 bootstrap replicates. The final phylogenetic trees were viewed and generated with FigTree V1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). Gene intron-exon structures were analyzed using the longest transcripts in Ensembl. The synteny of KANK genes were analyzed using Ensembl, UCSC genome browser, Synteny Database39 and Genomicus browser40. Protein domain analysis was carried out using the SMART online tool41.
Gene cloning, in situ hybridization and imaging
Full-length coding regions of zebrafish kank genes were amplified by RT-PCR using gene specific primers designed according to the current DNA sequence in Ensembl. One-three day old zebrafish embryos were mixed for total RNA isolation using TRIzol reagent (Thermo Fisher) and reverse transcription was performed using SuperScript® III First-Strand Synthesis System (Thermo Fisher) following the manual’s instructions. Phusion® High-Fidelity DNA Polymerase master mix (New England Biolabs) was used for PCR amplification. PCR primers used here are: kank1a: 5′ATGGCCCAAACCATGCACATTAACG 3′ and 5′GACAGAACCTGAAGGAATAGCAGCTC 3′; kank1b: 5′ATGACTCAAGAAACGCACTTCCACC 3′, 5′CCATATTCGTCGTGTGATTCAACACC3′; kank2: 5′ATGACAATCATGGCTCAGGTGCT 3′, 5′CTTCAGTTCAGAGGACGAAGGAGGA 3′; kank3: 5′ATGACCCAATCTGTGCACATAACCT 3′, 5′TTTCTCCGAGGGCCATGTTTTTCTA 3′; kank4: 5′ATGGACAAGAAAAGTGCAAATGGCT 3′, 5′GAGCGGGGCCGCTGAATCT 3′. The PCR products were purified using GeneJET Gel Extraction Kit (Thermo Scientific) before they were cloned into pJet1.2 vectors using the CloneJET PCR Cloning Kit (Thermo Scientific). Orientations of gene inserts were verified by Sanger sequencing. Riboprobes were synthesized through in vitro transcription using T7 DNA polymerase (New England Biolabs) and DIG RNA Labeling Mix (Roche, 11277073910). Then the riboprobes were purified by SigmaSpinTM post-reaction clean-up columns (Sigma, S5059). Whole mount in situ hybridization was carried out according to previously published method61 with the following modification for proteinase K treatment: 8–18 hours embryos, no treatment; 24 hours embryos, 5 minutes; 48 hours embryos for 30 minutes at room temperature. The riboprobe hybridization was carried out at 65 °C. For histological analysis, post-hybridization embryos were equilibrated in 15% sucrose then 30% sucrose in 20% gelatin, after which they were embedded in 20% gelatin for cryosectioning (6–12 μm). Images were acquired using AxioCam MRc camera on Zeiss SteREO Discovery.V12 and Axio Imager 2 compound microscope.
Additional Information
How to cite this article: Hensley, M. R. et al. Evolutionary and developmental analysis reveals KANK genes were co-opted for vertebrate vascular development. Sci. Rep. 6, 27816; doi: 10.1038/srep27816 (2016).
Supplementary Material
Acknowledgments
We graciously thank the Heyward Foundation and Purdue University College of Veterinary Medicine for supporting our research. We also thank Jyothi Thimmapuram and Ketaki Bhide for help on the phylogenetic analysis, and David A. Chasey and Lev Gorenstein for IT help on using computer clusters. The authors thank the support from the Purdue University Center for Cancer Research, NIH grant P30 CA023168.
Footnotes
Author Contributions G.J.Z. designed and coordinated research; M.R.H., Z.B.C., R.F.M.C., S.S., N.L.S. and G.J.Z. performed research; J.Y.Y. and Y.F.L. contributed discussions of results and interpretations. G.J.Z. analyzed data and wrote the paper. All authors read and approved the final manuscript. All authors have no competing interests.
References
- Shimeld S. M. & Holland P. W. Vertebrate innovations. Proc Natl Acad Sci USA 97, 4449–4452 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. W., Garcia-Fernandez J., Williams N. A. & Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl, 125–133 (1994). [PubMed] [Google Scholar]
- Monahan-Earley R., Dvorak A. M. & Aird W. C. Evolutionary origins of the blood vascular system and endothelium. J Thromb Haemost 11 Suppl 1, 46–66, doi: 10.1111/jth.12253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B. Endless forms: the evolution of gene regulation and morphological diversity. Cell 101, 577–580 (2000). [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. (Springer Science+Business Media, LLC, 1970). [Google Scholar]
- Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin Cell Dev Biol 10, 517–522, doi: 10.1006/scdb.1999.0332 (1999). [DOI] [PubMed] [Google Scholar]
- Dehal P. & Boore J. L. Two rounds of whole genome duplication in the ancestral vertebrate. PLos Biol 3, e314, doi: 10.1371/journal.pbio.0030314 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong R. F. & Holland P. W. Were vertebrates octoploid? Philos Trans R Soc Lond B Biol Sci 357, 531–544, doi: 10.1098/rstb.2001.1035 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A. et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G. C. & Wolfe K. H. Turning a hobby into a job: How duplicated genes find new functions. Nature Reviews Genetics 9, 938–950, doi: 10.1038/nrg2482 (2008). [DOI] [PubMed] [Google Scholar]
- True J. R. & Carroll S. B. Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol 18, 53–80, doi: 10.1146/annurev.cellbio.18.020402.140619 (2002). [DOI] [PubMed] [Google Scholar]
- Sanetra M., Begemann G., Becker M. B. & Meyer A. Conservation and co-option in developmental programmes: the importance of homology relationships. Front Zool 2, 15, doi: 10.1186/1742-9994-2-15 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R., Zhang G. & Cohn M. J. Evidence that mechanisms of fin development evolved in the midline of early vertebrates. Nature 442, 1033–1037, doi: 10.1038/nature04984 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang G. & Cohn M. J. Hagfish and lancelet fibrillar collagens reveal that type II collagen-based cartilage evolved in stem vertebrates. Proc Natl Acad Sci USA 103, 16829–16833, doi: 10.1073/pnas.0605630103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. & Cohn M. J. Genome duplication and the origin of the vertebrate skeleton. Curr Opin Genet Dev 18, 387–393, doi: 10.1016/j.gde.2008.07.009 (2008). [DOI] [PubMed] [Google Scholar]
- Kakinuma N., Zhu Y., Wang Y., Roy B. C. & Kiyama R. Kank proteins: structure, functions and diseases. Cell Mol Life Sci 66, 2651–2659, doi: 10.1007/s00018-009-0038-y (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M., Goncharov A., Jin Y. & Chisholm A. D. C. elegans ankyrin repeat protein VAB-19 is a component of epidermal attachment structures and is essential for epidermal morphogenesis. Development 130, 5791–5801, doi: 10.1242/dev.00791 (2003). [DOI] [PubMed] [Google Scholar]
- Ihara S. et al. Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nat Cell Biol 13, 641–651, doi: 10.1038/ncb2233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M. et al. The Cell Signaling Adaptor Protein EPS-8 Is Essential for C-elegans Epidermal Elongation and Interacts with the Ankyrin Repeat Protein VAB-19. PLos One 3, doi: ARTN e334610.1371/journal.pone.0003346 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. W., Park K. W. & Levine M. S. Temporal regulation of single-minded target genes in the ventral midline of the Drosophila central nervous system. Dev Biol 380, 335–343, doi: 10.1016/j.ydbio.2013.05.015 (2013). [DOI] [PubMed] [Google Scholar]
- Clohisey S. M., Dzhindzhev N. S. & Ohkura H. Kank Is an EB1 interacting protein that localises to muscle-tendon attachment sites in Drosophila. PLos One 9, e106112, doi: 10.1371/journal.pone.0106112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S. et al. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J Biol Chem 277, 36585–36591, doi: 10.1074/jbc.M204244200 (2002). [DOI] [PubMed] [Google Scholar]
- Kakinuma N., Roy B. C., Zhu Y., Wang Y. & Kiyama R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J Cell Biol 181, 537–549, doi: 10.1083/jcb.200707022 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B. C., Kakinuma N. & Kiyama R. Kank attenuates actin remodeling by preventing interaction between IRSp53 and Rac1. J Cell Biol 184, 253–267, doi: 10.1083/jcb.200805147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S. & Mellor H. Actin stress fibres. J Cell Sci 120, 3491–3499, doi: 10.1242/jcs.018473 (2007). [DOI] [PubMed] [Google Scholar]
- Lammert E. & Axnick J. Vascular lumen formation. Cold Spring Harbor perspectives in medicine 2, a006619, doi: 10.1101/cshperspect.a006619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M., Gerhardt H. & Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887, doi: 10.1016/j.cell.2011.08.039 (2011). [DOI] [PubMed] [Google Scholar]
- Boggetti B. et al. NBP, a zebrafish homolog of human Kank3, is a novel Numb interactor essential for epidermal integrity and neurulation. Dev Biol 365, 164–174, doi: 10.1016/j.ydbio.2012.02.021 (2012). [DOI] [PubMed] [Google Scholar]
- Isogai S., Horiguchi M. & Weinstein B. M. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev Biol 230, 278–301, doi: 10.1006/dbio.2000.9995 (2001). [DOI] [PubMed] [Google Scholar]
- Gore A. V., Monzo K., Cha Y. R., Pan W. & Weinstein B. M. Vascular development in the zebrafish. Cold Spring Harbor perspectives in medicine 2, a006684, doi: 10.1101/cshperspect.a006684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797, doi: 10.1093/nar/gkh340 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Inferring Phylogenies. (2004). [Google Scholar]
- Nosenko T. et al. Deep metazoan phylogeny: when different genes tell different stories. Mol Phylogenet Evol 67, 223–233, doi: 10.1016/j.ympev.2013.01.010 (2013). [DOI] [PubMed] [Google Scholar]
- Maddison D. R. & Schulz K.-S. The Tree of Life Web Project, < http://tolweb.org/Animals/2374/2002.01.01> (2007).
- Meyer A. & Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27, 937–945 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang G., Miyamoto M. M. & Cohn M. J. Lamprey type II collagen and Sox9 reveal an ancient origin of the vertebrate collagenous skeleton. Proc Natl Acad Sci USA 103, 3180–3185, doi: 10.1073/pnas.0508313103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta T. K. et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). P Natl Acad Sci USA 110, 16044–16049, doi: 10.1073/pnas.1315760110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait J. H. The zebrafish genome in context: ohnologs gone missing. J Exp Zool B Mol Dev Evol 308, 563–577, doi: 10.1002/jez.b.21137 (2007). [DOI] [PubMed] [Google Scholar]
- Catchen J. M., Conery J. S. & Postlethwait J. H. Automated identification of conserved synteny after whole-genome duplication. Genome Res 19, 1497–1505, doi: 10.1101/gr.090480.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis A., Muffato M. & Roest Crollius H. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res 41, D700–705, doi: 10.1093/nar/gks1156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T. & Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43, D257–260, doi: 10.1093/nar/gku949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O. et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957, doi: 10.1038/nature03025 (2004). [DOI] [PubMed] [Google Scholar]
- Angel Amores A. F. & Yan Yi-Lin, Lucille Joly, Chris Amemiya, Andreas Fritz, Robert K. Ho, James Langeland, Victoria Prince, Yan-Ling Wang, Monte Wester?eld, Marc Ekker, John H. Postlethwait. Zebrafish hox Clusters and Vertebrate Genome Evolution. Science 282, 1711–1714 (1998). [DOI] [PubMed] [Google Scholar]
- Inoue J., Sato Y., Sinclair R., Tsukamoto K. & Nishida M. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci USA 112, 14918–14923, doi: 10.1073/pnas.1507669112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. et al. Steroid receptor coactivator-interacting protein (SIP) inhibits caspase-independent apoptosis by preventing apoptosis-inducing factor (AIF) from being released from mitochondria. J Biol Chem 287, 12612–12621, doi: 10.1074/jbc.M111.334151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang H., Liang J., Yu W. & Shang Y. SIP, a novel ankyrin repeat containing protein, sequesters steroid receptor coactivators in the cytoplasm. EMBO J 26, 2645–2657, doi: 10.1038/sj.emboj.7601710 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee H. Y. et al. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest, doi: 10.1172/JCI79504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Chapuli R. Evolution of angiogenesis. Int J Dev Biol 55, 345–351, doi: 10.1387/ijdb.103212rm (2011). [DOI] [PubMed] [Google Scholar]
- Munoz-Chapuli R., Carmona R., Guadix J. A., Macias D. & Perez-Pomares J. M. The origin of the endothelial cells: an evo-devo approach for the invertebrate/vertebrate transition of the circulatory system. Evol Dev 7, 351–358, doi: 10.1111/j.1525-142X.2005.05040.x (2005). [DOI] [PubMed] [Google Scholar]
- Carmeliet P. Blood vessels and nerves: common signals, pathways and diseases. Nat Rev Genet 4, 710–720, doi: 10.1038/nrg1158 (2003). [DOI] [PubMed] [Google Scholar]
- Sprague J. et al. The Zebrafish Information Network (ZFIN): the zebrafish model organism database. Nucleic Acids Res 31, 241–243 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh T. et al. A gene expression screen in zebrafish embryogenesis. Genome Res 11, 1979–1987, doi: 10.1101/gr.209601 (2001). [DOI] [PubMed] [Google Scholar]
- Amsterdam A. et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA 101, 12792–12797, doi: 10.1073/pnas.0403929101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th ed edn, (Univ. of Oregon Press, 2000). [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. & Schilling T. F. Stages of embryonic development of the zebrafish. Dev Dyn 203, 253–310, doi: 10.1002/aja.1002030302 (1995). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
- Cunningham F. et al. Ensembl 2015. Nucleic Acids Res 43, D662–669, doi: 10.1093/nar/gku1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–2729, doi: 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59, 307–321, doi: 10.1093/sysbio/syq010 (2010). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Nieto M. A., Patel K. & Wilkinson D. G. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol 51, 219–235 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.