Abstract
Elucidation of structural changes involved in protein misfolding and amyloid formation is crucial for unraveling the molecular basis of amyloid formation. Here we report structural analyses of the amyloidogenic intermediate and insoluble aggregates of transthyretin (TTR) using solution and solid-state NMR spectroscopy. Our solution NMR results show that one of the two main β-sheet structures (CBEF β-sheet) is maintained in the aggregation-competent intermediate, while the other DAGH β-sheet is more flexible on ms time scales. Magic-angle-spinning solid-state NMR revealed that AB loop regions interacting with strand A in DAGH β-sheet undergo conformational changes, leading to the destabilized DAGH β-sheet.
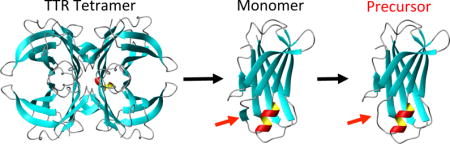
Transthyretin (TTR) is a 127-residue homo-tetrameric protein (55 kDa) that acts a carrier of the thyroid hormone thyroxine and retinol-binding protein. Rate-limiting tetramer dissociation, followed by monomer misfolding and aggregation of wild type (WT) TTR, is known to cause senile systemic amyloidosis (cardiomyopathy).1 More than 100 single-point mutations accelerate the dissociation and amyloid formation, leading to earlier onset of amyloidoses such as familial amyloidotic polyneuropathy (FAP) and familial amyloidotic cardiomyopathy (FAC).2
The native TTR monomer adopts a primarily β-sheet tertiary structure, in which eight β-strands are arranged into a β-sandwich consisting of two β-sheets comprising strands CBEF and DAGH (Figure 1a).3 The amyloid formation process of the globular protein is initiated by local unfolding to form aggregation-prone intermediates, which self-assemble into amyloid.4 It was proposed that aggregation-prone states generated by local unfolding of natively folded proteins retain extensive native-like structures.5 Investigating whether there is a native-like structure in amyloidogenic intermediates as well as in amyloid is, therefore, of great importance in understanding the molecular mechanism of misfolding and aggregation.
Figure 1.
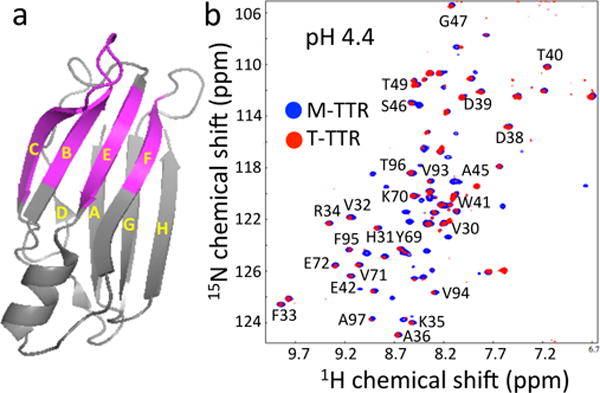
(a) A ribbon diagram representation of TTR monomer showing residues (pink) observed in the HSQC spectra. (b) Overlay of 1H/15N HSQC NMR spectra of WT TTR (T-TTR) and a monomeric variant (F87M/L110M, M-TTR) acquired at pH 4.4 (15 °C) at a 1H frequency of 500 MHz. In order to slow down aggregation kinetics, a very low protein concentration of 0.15 mg/mL (12 μM) was used to probe the monomeric aggregation-competent state at pH 4.4, where the aggregation rate reaches a maximum. At higher protein concentrations, no NMR signals were observed at pH 4.4 due to aggregation.
Previous structural studies of TTR suggested that amyloidogenic intermediates and the end product of aggregation, amyloid, contain extensive native-like β-sheet structures. However, the extent of native structure has been controversial.1,6–9 Previous solid-state NMR structural studies of amyloid formed by a short TTR peptide fragment (105–115) provided valuable structural information on its amyloid state.10 However, full-length TTR has different amyloidogenic properties and likely forms a distinct amyloid state, requiring structural analyses of the full-length TTR amyloid. In this study, we employed solution and solid-state NMR to scrutinize native-like structural features in amyloidogenic intermediates and to identify critical regions undergoing structural changes during amyloid formation.
TTR dissociation, misfolding and amyloid formation is greatly accelerated under mildly acidic conditions (pH 4 – 5).1 In order to probe the soluble amyloidogenic intermediate state, a two dimensional 1H/15N heteronuclear single-quantum coherence (HSQC) NMR spectrum of WT TTR was acquired at low concentration under the most amyloidogenic pH of 4.4 (Figure 1b). Given that TTR efficiently forms amyloid at higher protein concentrations (> 0.2 mg/ml) under these experimental conditions, the NMR parameters have substantial contributions from amyloidogenic precursor states. It is notable that about one third of the NMR resonances are observable, while the rest of the resonances are undetectable due to broadening from conformational exchange on ms time scales. The remaining NMR resonances are well-dispersed and their chemical shifts are also similar to those of the native tetrameric TTR (BMRB 5507), indicating that the aggregation competent state contains native-like conformations.
The WT tetramer has been shown to substantially dissociate to monomers at acidic pH.1 Thus, the HSQC spectrum of the WT TTR tetramer (T-TTR) was compared to that of a monomeric TTR variant (F87M/L110M, M-TTR) that exists predominantly as monomer (> 99 % at a protein concentration of 0.1 mM).11 Most of the observable NMR resonances of WT TTR appeared at identical positions with similar linewidth to the cross peaks in the spectrum of M-TTR, suggesting that TTR is predominantly in the monomeric state at the amyloidogenic pH. The observable NMR resonances were assigned using pH-titration experiments of M-TTR12 and the available backbone assignment of WT tetramer (BMRB 5507; the chemical shifts of the residues in CBEF β-sheet were not affected by the two mutations (F87M/L110M))7. Interestingly, the observed NMR resonances in WT TTR are mapped to residues in strands C, B, E and F and the BC loop (Figure 1a), while other resonances are undetectable due to the extensive line-broadening caused by ms time scale dynamics, consistent with our previous relaxation dispersion NMR experiments on M-TTR.12 The solution NMR results described above indicate that the aggregation-competent state of WT TTR contains native-like structure in the CBEF β-sheet, while the DAGH β-sheet structure appears to be perturbed. In order to investigate structural changes in the DAGH β-sheet, magic-angle-spinning (MAS) solid-state NMR was used for the structural studies of TTR amyloid.
Magic-angle-spinning (MAS) solid-state NMR has provided valuable structural information on various amyloids, such as the location of β-strands and unstructured regions within the amyloid.13–20 Despite the great progress in solid-state NMR methodology, full structural characterization of the amyloid derived from this 127-residue protein remains an enormous challenge. The current structural study is, therefore, focused on specific regions that may undergo structural changes during amyloid formation (Figure 2a). Previous relaxation dispersion NMR experiments on monomeric variant12 and tetrameric TTR21 demonstrated that AB loop regions undergo extensive conformational exchanges on ms time scales, which might be associated with TTR misfolding and amyloid formation. Thus, six amino acids (A, D, L, R, S, V) were uniformly 13C/15N labeled to achieve sequential labeling for the AB loop regions (Materials and Methods in Supplemental Information).
Figure 2.
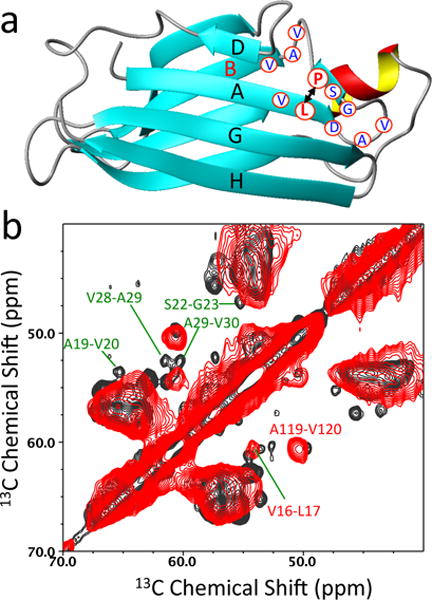
(a) A ribbon diagram representation of TTR monomer showing labeled residues (blue) in the AB loop regions. Pro-24 and Leu-17 are labeled in red. (b) Cross-polarization (CP) based two-dimensional 13C-13C correlation spectra of native (black) and amyloid (red) states of TTR obtained using a mixing time of 50 ms at a 1H frequency of 800 MHz. 64 FIDs were accumulated for the 2D spectra with a spinning frequency of 11.3 kHz.
Two-dimensional proton-driven spin-diffusion (PDSD)22 experiments were carried out to investigate structural features of the AB loop regions in the native (black) and amyloid (red) states of TTR (Figure 2b and Supplemental Figure S2). The cross-peaks from the AB loop regions are well resolved and unambiguously assigned for the native state based on the selective labeling of the six amino acids and the available backbone assignment (BMRB 5507). The strong cross-peaks in the CP based 13C-13C correlation spectrum indicate that the AB loop is well structured in the native state. In contrast to the native state, the cross-peaks from the AB loop region (19 – 30) are not observed in amyloid state (Figure 2b).
The solid-state NMR results described above suggest that the loop region may adopt distinct non-native conformations or become disordered in amyloid state. In particular, hydrogen-bonding interactions between the short strand in the AB loop and strand A might be disrupted in amyloid state. In order to examine the structural change, we utilized specific labeling schemes, which generate isolated 13C-13C dipolar-coupled spin pairs in the native structure (Figure 2a). For example, 13CO-Pro and 13Cα-Leu labeled protein has two dipolar-coupled 13CO-13Cα spin pairs at distances of 2–5 Å (P11-L12, 2.4 Å; P24-L17, 4.9 Å). The dipolar-coupled spin pairs in the native tetramer can then be probed and compared to those in amyloid state using solid-state NMR experiments that can detect spin-pairs with a separation of up to 6 Å
2D PDSD22 experiments were carried out to probe the dipolar-coupled spin pairs in the native, and also in the amyloid state(s) (Figure 3a). A longer mixing time of 500 ms was used to probe the weakly-coupled spin pair (P24-L17, 4.9 Å) and the spinning speed was set slightly below the Δωiso = 2ωr rotational resonance condition23 for more efficient polarization transfer. The cross-peaks from the two spin pairs (P11-L12 and P24-L17) were clearly observed in the 2D spectrum of the native tetrameric TTR (the assignment of the cross-peaks were made from the previous assignment, BMRB 5507). On the other hand, only one cross-peak was observed in amyloid state of TTR. In order to assign the cross-peak in the TTR amyloid, 13Cα-Val and 13Cα-Leu labeled TTR was prepared, resulting in only one V16-L17 (3.3 Å) spin pair with a separation of less than 5 Å. The 2D PDSD experiment on the 13Cα-Val and 13Cα-Leu labeled TTR confirms the assignment of the cross-peak in the amyloid state (P11-L12, red). The disappearance of the cross-peak from the P24-L17 spin-pair in TTR amyloid supports the occurrence of structural changes in the AB loop regions (Figure 2a). The cross-peak between the two residues may be broadened by conformational disorder of the loop regions and/or the short β-strand in the AB loop and strand A may not be in close proximity in amyloid state.
Figure 3.
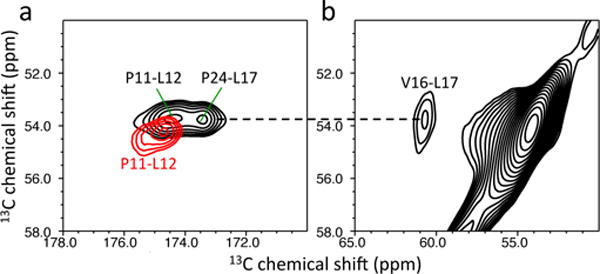
(a) 2D PDSD spectra of native (black) and amyloid (red) states of the 13CO-Pro and 13Cα-Leu labeled TTR. (b) A 2D PDSD spectrum of the amyloid state for the 13Cα-Leu and 13Cα-Val labeled TTR. The NMR spectra were obtained using a mixing time of 500 ms at a 1H frequency of 830 MHz.
In order to obtain additional evidence for the conformational disorder of the AB loop regions, the 2D 13C-13C correlation experiment was carried out on 13CO-Pro, 13Cα-Ala, and 13Cα-Leu labeled TTR amyloid that contains two strongly dipolar-coupled 13CO-13Cα spin pairs (P11-L12 and P24-A25; 2.4 Å, Supplemental Figure S4). In the 2D PDSD spectrum, the cross-peak for P24-A25 was not detected in amyloid state of TTR, while the cross-peak for P11-L12 was clearly observed in the amyloid state spectra. These NMR results clearly indicate that the AB loop regions are disordered in amyloid state.
Previous solid-state NMR studies of various amyloid states formed by small peptide fragments and full-length proteins (90 – 150 residues) showed that NMR signals are observed mainly from residues in rigid, structured amyloid core regions in the cross-polarization (CP) based solid-state NMR spectra, which utilize substantial dipole-dipole couplings for efficient magnetization transfer from 1H to 13C.19,20,24,25 NMR peaks are usually not detected from flexible and/or disordered regions due to severe line-broadening and/or inefficient cross-polarization caused by motional averaging of dipolar interactions.19,20,24,25 The strong cross-peaks observed in TTR amyloid spectra (Figure 2 and 3) are comparable to those of its native tetrameric state, indicating that the amyloid state contains a rigid, structured amyloid core. The presence of a β-structured amyloid core is further evidenced in two-dimensional 13C-13C correlation MAS spectra (Supplemental Figure S2 and S3). On the other hand, NMR peaks from the AB loop regions in TTR amyloid state were not detectable in the solid-state NMR spectra, suggesting that the loop regions become disordered during the misfolding and amyloid formation.
Recent relaxation dispersion NMR experiments on monomeric variant (F87M/L110M)12 and tetrameric forms of TTR21 revealed that the residues in the DAGH β-sheet and AB loop regions undergo conformational exchange on ms time scales, while residues in the CBEF β-sheet remain stable. The dynamics experiments are in good agreement with our solution and solid-state NMR results. The tetrameric native state of TTR is stabilized by dimerization of dimers through the interactions between the AB and GH loops. The structural perturbations of the AB loop may facilitate dissociations of the native tetramers into monomers, which is the critical step for TTR misfolding and amyloid formation.
In summary, the solution HSQC NMR results described here show that the soluble amyloidogenic precursor state of TTR contains a native-like CBEF β-structure. The stable CBEF β-sheet native structure in the amyloidogenic intermediate state is somewhat contracted to the previous hydrogen/deuterium (H/D) exchange6 and protease digestion studies9 of TTR amyloids that suggested the CBEF β-structure is partly disrupted in amyloid state. A very recent study, however, showed that TTR amyloid is in a dynamic equilibrium with soluble intermediate states in solution,26 rendering analyses of the H/D exchange and protease digestion studies of TTR amyloid more complicated.
In contrast to the CBEF β-structure, NMR signals from the DAGH β-sheet were not observed in solution due to extensive line-broadening. Our solid-state NMR experiments indicate that the well-structured AB loop regions in the native state become disordered during amyloid formation. The structural perturbations of the AB loop regions including the disruption of the interactions between the short β-strand in the AB loop and strand A may affect the stability of the whole DAGH β-sheet, rendering the β-sheet more dynamic on ms time scales and unobservable in the HSQC NMR spectrum.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants NS084138 (K.H.L), AG10770 (D.E.W.), DK46335 (J.W.K.) and the Skaggs Institute of Chemical Biology (J.W.K.). The high-field solid-state NMR spectra were acquired at the National High Magnetic Field Laboratory, which is supported by NSF Cooperative Agreement No. DMR-1157490 and the State of Florida. We’d also like to thank Prof. Peter Wright for reading the manuscript and for helpful discussions.
ABBREVIATIONS
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single-quantum coherence
- MAS
magic-angle spinning
- CP
cross-polarization
- WT
wild-type
- TTR
transthyretin
- PDSD
proton-driven spin-diffusion
- TEM
transmission electron microscopy
- ThT
thioflavin T
- Leu
leucine
- Val
Valine
- Pro
proline
- H/D
hydrogen/deuterium
Footnotes
Supporting Information. Materials and methods. TEM image of TTR amyloid. 2D PDSD spectrum obtained at 800 MHz 1H frequency. One-dimensional slices of the 2D PDSD spectra. 2D PDSD spectrum for 13CO-Pro/13Cα-Leu/13Cα-Ala, and 13CO-Ile/13Cα-Ala labeled TTR. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Funding Sources
No competing financial interests have been declared.
References
- 1.Lai ZH, Colon W, Kelly JW. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 2.Saraiva MJ. Hum Mutat. 1995;5:191–6. doi: 10.1002/humu.1380050302. [DOI] [PubMed] [Google Scholar]
- 3.Blake CC, Geisow MJ, Oatley SJ, Rerat B, Rerat C. J Mol Biol. 1978;121:339–56. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- 4.Colon W, Kelly JW. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F, Dobson CM. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 6.Olofsson A, Ippel JH, Wijmenga SS, Lundgren E, Ohman A. J Biol Chem. 2004;279:5699–707. doi: 10.1074/jbc.M310605200. [DOI] [PubMed] [Google Scholar]
- 7.Liu K, Cho HS, Lashuel HA, Kelly JW, Wemmer DE. Nat Struct Biol. 2000;7:754–757. doi: 10.1038/78980. [DOI] [PubMed] [Google Scholar]
- 8.Serag AA, Altenbach C, Gingery M, Hubbell WL, Yeates TO. Nat Struct Biol. 2002;9:734–9. doi: 10.1038/nsb838. [DOI] [PubMed] [Google Scholar]
- 9.Bateman DA, Tycko R, Wickner RB. Biophys J. 2011;101:2485–2492. doi: 10.1016/j.bpj.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. Proc Natl Acad Sci USA. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X, Smith CS, Petrassi HM, Hammarstrom P, White JT, Sacchettini JC, Kelly JW. Biochemistry. 2001;40:11442–11452. doi: 10.1021/bi011194d. [DOI] [PubMed] [Google Scholar]
- 12.Lim KH, Dyson HJ, Kelly JW, Wright PE. J Mol Biol. 2013;425:977–988. doi: 10.1016/j.jmb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang M, Comellas G, Rienstra CM. Acc Chem Res. 2013;46:2080–2088. doi: 10.1021/ar4000168. [DOI] [PubMed] [Google Scholar]
- 14.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debelouchina GT, Bayro MJ, Fitzpatrick AW, Ladizhansky V, Colvin MT, Caporini MA, Jaroniec CP, Bajaj VS, Rosay M, Macphee CE, Vendruscolo M, Maas WE, Dobson CM, Griffin RG. J Am Chem Soc. 2013;135:19237–19247. doi: 10.1021/ja409050a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 17.Helmus JJ, Surewicz K, Apostol MI, Surewicz WK, Jaroniec CP. J Am Chem Soc. 2011;133:13934–13937. doi: 10.1021/ja206469q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linser R, Dasari M, Hiller M, Higman V, Fink U, Lopez del Amo JM, Markovic S, Handel L, Kessler B, Schmieder P, Oesterhelt D, Oschkinat H, Reif B. Angew Chem Int Ed Engl. 2011;50:4508–4512. doi: 10.1002/anie.201008244. [DOI] [PubMed] [Google Scholar]
- 19.Barbet-Massin E, Ricagno S, Lewandowski JR, Giorgetti S, Bellotti V, Bolognesi M, Emsley L, Pintacuda G. J Am Chem Soc. 2010;132:5556–5557. doi: 10.1021/ja1002839. [DOI] [PubMed] [Google Scholar]
- 20.Daebel V, Chinnathambi S, Biernat J, Schwalbe M, Habenstein B, Loquet A, Akoury E, Tepper K, Muller H, Baldus M, Griesinger C, Zweckstetter M, Mandelkow E, Vijayan V, Lange A. J Am Chem Soc. 2012;134:13982–13989. doi: 10.1021/ja305470p. [DOI] [PubMed] [Google Scholar]
- 21.Das JK, Mall SS, Bej A, Mukherjee S. Angew Chem Int Ed Engl. 2014;53:12781–12784. doi: 10.1002/anie.201407323. [DOI] [PubMed] [Google Scholar]
- 22.Szeverenyi NM, Sullivan MJ, Maciel GE. J Magn Reson. 1982;47:462. [Google Scholar]
- 23.Raleigh DP, Levitt MH, Griffin RG. Chem Phys Lett. 1988;147:71. [Google Scholar]
- 24.Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. J Am Chem Soc. 2010;132:2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. J Am Chem Soc. 2010;132:10414–10423. doi: 10.1021/ja102775u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groenning M, Campos RI, Hirschberg D, Hammarstrom P, Vestergaard B. Sci Rep. 2015;5:11443. doi: 10.1038/srep11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


