Abstract
The methylenetetrahydrofolate reductase (MTHFR) gene is one of the most investigated of the genes associated with chronic human diseases because of its associations with hyperhomocysteinemia and toxicity. It has been proposed as a prototype gene for the prevention of colorectal cancer (CRC). The major objectives of this meta-analysis were to examine the polymorphism-mutation patterns of MTHFR and their associations with risk for CRC as well as potential contributing factors for mutations and disease risks. This analysis included 33,626 CRC cases and 48,688 controls across 92 studies for MTHFR 677 and 16,367 cases and 24,874 controls across 54 studies for MTHFR 1298, comprising data for various racial and ethnic groups, both genders, and multiple cancer sites. MTHFR 677 homozygous TT genotype was protective (p < .05) for CRC for all included populations; however, with heterogeneity across various racial–ethnic groups and opposing findings, it was a risk genotype for the subgroup of Hispanics (p < .01). Additional countries for which subgroup analyses resulted in 677 TT as a risk genotype included Turkey, Romania, Croatia, Hungary, Portugal, Mexico, Brazil, U.S. Hawai’i, Taiwan, India, and Egypt. Countries with the highest mutation rates and risks for both MTHFR 677 and 1298 genotypes are presented using global maps to visualize the grouping patterns. Meta-predictive analyses revealed that air pollution levels were associated with gene polymorphisms for both genotypes. Future nursing research should be conducted to develop proactive measures to protect populations in cities where air pollution causes more deaths.
Keywords: meta-analysis, meta-prediction, methylenetetrahydrofolate reductase gene, colorectal cancer
The methylenetetrahydrofolate reductase gene (MTHFR) is one of the most investigated of the genes associated with various chronic human diseases through the mechanism of epigenetics, as identified through the genome application framework (McBride, 2012; Wade, McBride, Kardia, & Brody, 2010). Of particular interest in the context of the present meta-analysis, the National Human Genome Research Institute lists MTHFR as a prototype gene for the application of prevention studies for colorectal cancer (CRC). The two most common loci polymorphism-mutation variations involved with MTHFR are C677T (rs 1801133) and A1298C (rs 1801131). These two single nucleotide polymorphisms are about 2,000 base pairs of deoxyribonucleic acid (DNA) apart in the complete human genome sequence (http://ghr.nlm.nih.gov/gene/MTHFR). As the MTHFR enzyme is encoded by the MTHFR gene for homocysteine remethylation to methionine, polymorphism mutations in MTHFR are associated with MTHFR enzyme deficiency, which can lead to hyperhomocysteinemia and toxicity and health issues including neurologic symptoms and occlusive vascular symptoms such as thrombosis (Balion & Kapur, 2011; Refsum et al., 2006; Sibani et al., 2003). With an MTHFR 677 TT homozygous mutation, up to 70% of the enzyme function is lost; and with a 677 CT heterozygous mutation, there is a 35% loss of enzymatic function. In contrast, with an MTHFR 1298CC homozygous mutation, there is approximately a 30% loss of enzyme function, while a 1298 AC heterozygous mutation leads to a 15% loss of function. Individuals with compound polymorphisms or mutations of both the MTHFR 677 and 1298 loci are rare and are at increased risk of developing severe health conditions with neurologic and vascular symptoms.
In laboratory testing, with 5 min of heat treatment at 46°C followed by a return to 37°C, MTHFR enzyme deficiency was associated with marked hyperhomocysteinemia and homocystinuria (Kang, Zhou, Wong, Kowalisyn, & Strokosch, 1998; Leclerc, Sibani, & Rozen, 2000). The degree of MTHFR enzyme thermolability (defined as a reduced level of activity in response to the action of moderate heat) is much greater with the MTHFR 677 homozygous mutation TT genotype as compared to the 677 CT heterozygous mutation, and polymorphism mutations in MTHFR 1298 and other loci are not associated with MTHFR enzyme thermolability (Kang et al., 1998; Leclerc et al., 2000; Sibani et al., 2003). Investigators have discovered that additional MTHFR loci polymorphisms and mutations, other than those at 677 and 1298, are associated with reduced enzyme activity; however, these associations have not yet been fully investigated (Leclerc et al., 2000; Sibani et al., 2003).
The mechanism of low folate levels, regulated by MTHFR, is well documented regarding its various subtypes and their associations with cancer as well as a plethora of major cardiovascular and neurodevelopmental diseases associated with high homocysteine levels (Chen et al., 2010; Crider, Maneval, et al., 2012; Crider, Yang, Berry, & Bailey, 2012). Through the one-carbon metabolism enzyme pathways, the folate, MTHFR gene, and methylation pathways are critical to basic biological processes involving DNA and protein methylation as well as DNA replication and mutation (Crider, Maneval, et al., 2012; Inoue-Choi et al., 2012, 2013; Jamaluddin, Yang, & Wang, 2007). Methylation pathways and MTHFR gene variations affect the development of cancer and disease prognoses; thus, their effects need to be monitored closely with the cancer treatment. Furthermore, the effects of genome variations in the methylation pathways on health outcomes, including variations on the MTHFR gene, need to be validated with effective triangulation methods and disseminated for cancer prevention.
Individuals with gene mutations in the methylation pathways and dysfunctional regulation of DNA methylation have particular trouble processing environmental toxicants. Air pollution, so common in modern societies, causes damage to the methylation pathways similar to that caused by cigarette smoking, and the damage is worse in the individuals with gene mutations (Guo et al., 2014; Kloog, Ridgway, Koutrakis, Coull, & Schwartz, 2013; Mallone et al., 2011). Air pollution, like cigarette smoking, is a well-known carcinogen that causes DNA damage. The toxicants are inhaled and pass through the respiratory system into other organs (Pope et al., 2011). Pollution particles, particulate matter smaller than 2.5 μm (PM 2.5), can pass through the lungs, leading to plaque deposits in the cardiovascular system that cause systemic inflammation (Myers et al., 2013; Wellenius et al., 2012) and increased homocysteine levels (Ren et al., 2010). The result is a progressive impairment in the conversion of key enzymes in the regulatory pathways, producing free radicals, such as peroxynitrite and superoxides and inflammatory substances that cause a range of health problems.
While various studies and meta-analyses have reported the polymorphism-mutation patterns of MTHFR loci for various racial and ethnic groups, the risks for CRC associated with these polymorphisms have been inconsistent across studies. Additionally, while there have been16 studies published to date on polymorphism mutations in MTHFR using the conventional meta-analysis method, there has been little consistency across these studies in recognizing the risk genotypes or the variation of risk across different populations. Thus, a gap exists in the literature regarding a reliable meta-analysis that identifies the risk genotypes of MTHFR across different populations. Deeks, Higgins, Altman and the Cochrane Statistical Methods Group (2008) have suggested that meta-predictive analysis is an appropriate method to use in cases of heterogeneity in meta-analysis findings. Therefore, the next level of innovative meta-analytic approaches beyond the standardized ratios can be applied to provide further insights into the identification of the polymorphism mutations of the MTHFR genotypes. In particular, big-data analytics can be applied to identify potential factors contributing to the heterogeneity of findings, including the use of data visualization techniques to identify distribution patterns on a global map of polymorphism mutations of MTHFR genotypes and of potentially associated health risks. Accordingly, the major objective of the present meta-analysis, extending findings of previous meta-analyses on this topic, is to examine the polymorphism-mutation patterns and risk subtypes of the MTHFR gene for CRC for the world population. The secondary objective is to use meta-predictive analysis to identify the potential contributing factors for MTHFR gene polymorphisms and associated risks for CRC.
Method
Following the guidelines for reporting meta-analyses of observational studies (Stroup et al., 2000), we searched the online databases of PubMed, PubMed Central, and Airiti Library from all available studies, from 1996 (year in which the first related study was published) to February 2015. We used the search terms colon cancer, colorectal cancer, MTHFR gene, MTHFR in colorectal cancer, environment, diet prevention, folate, lifestyle, behavior, case–control design, and meta-analysis, limiting the results to human studies. Additionally, we used previous meta-analysis and review papers to cross check and trace back to all original studies. We searched the various databases thoroughly at three different times at least 3 months apart until we could identify no additional articles. We entered the resulting articles into a database organized by key words. Two raters, one who was familiar with the literature search process and organization and one who was familiar with meta-analytic methods, conducted the literature search to identify all possible case–control studies.
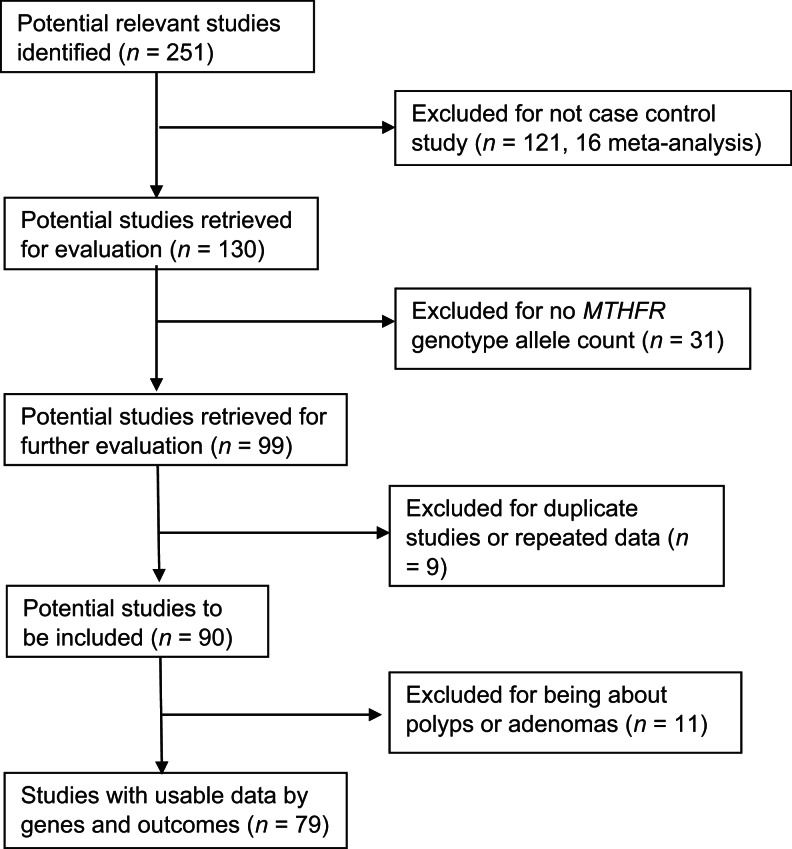
We excluded articles that did not report case–control studies or have the appropriate genotype allele counts per case and control groups. We did not restrict our search based on language of publication or country of study as long as the articles included an abstract in English and tables that clearly listed the genotype allele counts. Figure 1 presents our study selection process. Of the 251 articles we identified involving MTHFR and CRC, we excluded 105 from the analysis because they were not case–control studies as well as 16 previous meta-analyses. From the remaining 130 articles, we excluded 31 for not having MTHFR genotype allele counts or for having only one case cohort group. Of the remaining 99 studies, 9 involved subsidiary or redundant use of data contained in other included studies that had more current and/or complete information, thus we excluded them as well. After further review, we excluded 11 more articles that were about polyps or adenomas rather than cancer. Finally, we had 79 articles with usable genotype data for pooled analysis (Figure 1). We checked data extractions and entry for accuracy and ran the preliminary analyses to make sure the ranges of entries and pooled results were accurate for all studies.
Figure 1.
Progression on the selection of studies for the meta-analysis.
We evaluated each study for quality using a set of indicators appropriate for the current state of science for the field, integrated from multiple sources on the assessment of studies. The sources for these criteria included the U.S. QUOROM consensus process on the quality of meta-analysis (Moher et al., 1999), guidelines on quality reporting for observational studies (Downs & Black, 1998; Stroup et al., 2000), and the quality evidence from a previous meta-analysis of MTHFR and CRC (Kennedy et al., 2012). The details of the quality indicators that we used to assess the studies included in the meta-analysis are presented in Supplementary Table S1. The range for the total quality score is 0–29. We combined three subscores to obtain the total score: (1) external validity, with 10 items on demographic data (score range of 0–11); (2) internal validity, with 12 items on research methods and procedures (score range of 0–12); and (3) Data and study results reporting (score range of 0–6).
Characteristics of Original Studies
We separated studies with genotype allele counts for additional subgroups of racial or ethnic groups, gender, and cancer types from the 79 studies included and listed them per case and control groups, yielding an additional 13 studies for a total of 92 study groups (Table S2). Specifically, two studies had data more than one racial or ethnic group for both case and control groups (Keku et al., 2002; Le Marchand et al., 2002), yielding three additional study groups, while 10 studies included data by gender groups and/or by cancer locations yielding a total of 92 study groups (Table S2). The studies in this meta-analysis were published from 1996 to 2014 and comprised 92 studies, or study groups, with data for the MTHFR 677 genotype and 54 that included MTHFR 1298 genotype counts. Study populations were drawn from countries and continents across the globe (Australia, Europe, North America, South America, and Asia and Africa). We checked the racial and ethnic compositions of each study to be sure we properly accounted for data from distinct groups versus mixed racial or ethnic groups. The most investigated racial or ethnic populations in these studies were White (45 studies) followed by Asian (28 studies, including 7 Southeastern Asians and Pacific Islanders), then mixed race (10 studies), Middle-Eastern Asian (5 studies), Hispanics (2 studies), and African (2 studies). The list of studies included in this analysis, as well as key studies and meta-analyses excluded, follows Supplemental Table S2.
Along with the geographical location of each study, we also entered the population sizes and checked the air quality data for that location. Specifically, we verified from various sources of the most current air pollution data, including the death rates from air pollution (death rates per million, Level 2 = 50–100, Level 3 = 101–250, Level 4 = 251–400, and greater; Kenworthy & Laube, 2002; World Health Organization [WHO], 2004, 2009, 2012, 2015) and further verified the scales on air pollution status with current scales on air pollution data (http://www.airnow.gov/index.cfm?action=aqibasics.aqi) before entering the data into the analyses.
The range of total quality scores for all studies was 15–25 (out of a possible range of 0–29). The reviewed studies thus all scored above 50% of the total possible score, suggesting that their findings are trustworthy (Moher et al., 1999). For all of the studies, we checked the Hardy-Weinberg Equilibrium (HWE) analysis, which was developed to assess the distribution equilibrium of the evolutionary mechanisms in population genetics (Sha & Zhang, 2011; Wittke-Thompson, Pluzhnikov, & Cox, 2005). Departure from the HWE with a p value < .05 may be associated with factors such as population migration or stratification and disease association. As presented in Table S2, we checked HWE status in the reports of original studies and verified the reported findings with additional calculations. Where there were discrepancies between the HWE reported in the original articles and our subsequent calculations, we have reported the results of our calculations in the table, with a notation added. As there were a number of studies for which original results or our subsequent calculations indicated a departure from HWE, we performed subgroup analyses with these studies. The results of these subgroup analyses, however, did not yield significant differences for pooled analyses. Therefore, we included all studies, regardless of HWE status, in the meta-analysis. This approach is consistent with the approaches used in recent meta-analyses involving MTHFR and CRC (Kennedy et al., 2012; Teng et al., 2013).
All studies in this meta-analysis included the use of biological samples of blood, tissue, buccal saliva, or a combination of two of these for DNA analysis. Researchers collected these biological samples prospectively with 100% accuracy for testing from all those studies that reported accuracy; based on the current literature, we would thus expect 100% accuracy for the genotyping analyses. We checked the distributions for all three genotypes in each study to be sure that they were within the limit of expected ranges. In addition, we pooled the distributions for each of the wild type, heterozygous mutation or homozygous mutation types within each country to check for the total number of samples accrued per country (see Supplemental Figures S1 for MTHFR 677 polymorphism-mutation distributions and Figure S2 for MTHFR 1298 polymorphism-mutation distributions for control groups and CRC cases.
Data Synthesis and Analysis
We entered data into Excel spreadsheets (Microsoft Corp, Redmond, WA) and used StatsDirect, version 2.4.7 (Cheshire, UK), to pool data analyses. We calculated pooled risk ratios (RRs) and odds ratios (ORs) for the MTHFR polymorphism subtypes between cases and controls and 95% confidence intervals (CIs) for the associations of MTHFR polymorphism genotypes with CRC. For standardized risk ratios, a pooled RR is preferred and has been used in most recent consensus reports (Deeks, Higgins, & Altman, 2008); however, as most of the previous meta-analysis reports on this topic presented ORs, we also pooled ORs to check the differences between the results with RRs. An RR of 1 represents “no effect,” < 1 indicates a protective effect (favoring the case or the CRC group), and > 1 indicates increased risks for CRC. We defined significant findings as those with p values < .05.
Because the data presented heterogeneity with regional differences on mutation rates for polymorphisms and risks, we used geographic information system (GIS) maps to visualize the distributions of polymorphisms or mutations and risks on the global maps. These GIS maps were helpful for visually identifying geographic patterns (Albrechit, 2007; ESRI, 2015). In addition, we used recursive partition trees in the JMP 11 program (SAS Institute) to examine how an independent variable (air pollution, total quality score and quality score subcategories, gender, cancer site, and type of controls) can make a decisive split of the data by partitioning the original group into pairs of subgroups with reference to the dependent variable (polymorphism-mutation rates and risks; Breiman, Friedman, Olshen, & Stone, 1984; Speybroeck, 2012; Vanitha & Niraimathi, 2013; Yu, 2010). The goodness of the partition can be judged using Akaike’s information criterion correction (AICc). A smaller AIC suggests a better model (Akaike, 1973; Bozdogan, 2000; Burnham & Anderson, 2002; Symonds & Moussalli, 2011). Both GIS maps and recursive trees are common big-data analytical techniques for handling large-scale and multidimensional data sets. Unlike conventional hypothesis testing, data mining or big-data analytics does not start with a predetermined hypothesis. Rather, data-driven pattern recognition plays the central role.
For triangulation purpose, we employed a conventional multiple comparison procedure (Tukey test) to examine whether partition trees and Tukey tests concurred with each other. Additionally, we used nonlinear fit to examine the associations between contributing factors such as air quality and the outcome variables (mutation rates and risks). Further insights can be unveiled when the scatterplot of bivariate distributions is converted into a heat map, in which the color spectrum is utilized to represent the frequency counts (Deng, Wang, Liu, Cheng, & Xue, 2014; Deu-Pons, Schroeder, & Lopez-Bigas, 2014; Yu, 2014). We employed the preceding techniques for meta-prediction, which aims to generate more precise predictions by integrating data from diverse sources. One of the main differences between conventional statistical methods and data mining (e.g., recursive partition trees) is that big-data analytical techniques can be used to verify the results by cross validation (Mao, 2014; Yu, 2010), yielding more accurate meta-prediction.
Results
For pooled analysis of MTHFR 677, we included a total of 33,626 CRC cases and 48,688 controls in 92 study groups, including the tests of heterogeneity and association (Table 1). As presented in Table 1, using the control groups to represent the proportion of the general healthy population with polymorphism mutations, the rank order of populations for percentage of polymorphisms on homozygous TT for MTHFR 677 is Hispanics (20%), East Asian (18.24%), Mideast Asian (13.77%), mixed populations (12.02%), White (11.13%), African (2.52%), and finally South Asian (1.86%). Consistent with most previous reports, MTHFR 677 TT homozygous mutation type was protective for all populations pooled together (RR = 0.92, 95% CI [0.87, 0.99], p = .0172). The TT homozygous mutation genotype was also significant as a protective type for mixed populations in 10 studies, according to subgroup analysis (RR = 0.85, 95% CI [0.76, 0.96], p = .0057). Contrary to these results, the TT genotype was a risk genotype for Hispanics in two studies (RR = 1.51, 95% CI [1.13, 2.02], p = .0054). While not statistically significant, pooled TT genotype showed a tendency toward being a risk genotype for CRC among the African population in two studies and the South Asian population in seven studies. By contrast, pooled TT genotype also showed a tendency to be a protective genotype from CRC for White, East Asian, and Mideast Asian populations, although, again, without statistical significance.
Table 1.
Pooled Meta-Analysis: MTHFR 677 Genotypes and Risk of CRC.
| Genotype by Race or Ethnicity (Number of Studies) | CRC Cases (N = 33,626) n (%) | Controls (N = 48,688) n (%) | Test of Heterogeneity | Statistical Model | Test of Association | |||
|---|---|---|---|---|---|---|---|---|
| Q | p | I2 (%) | Risk Ratio (95% Cl) | p | ||||
| TT (92) | 3,886 (11.56) | 6,016 (12.36) | 183.18 | <.0001 | 50.3 | Random | 0.93 [0.87, 0.99] | .0172 |
| White (45) | 2,146 (10.51) | 3,277 (11.13) | 79.56 | .0008 | 44.7 | Random | 0.94 [0.87, 1.01] | .1097 |
| East Asian (21) | 1,096 (15.82) | 1,643 (18.24) | 57.00 | <.0001 | 64.9 | Random | 0.89 [0.78, 1.01] | .0801 |
| South Asian (7)a | 19 (1.98) | 24 (1.86) | 5.77 | .4495 | 0 | Fixed | 1.16 [0.66, 2.04] | .6032 |
| Mixed (10) | 413 (10.25) | 903 (12.02) | 12.72 | .1757 | 29.2 | Fixed | 0.85 [0.76, 0.95] | .0057 |
| Mideast (5)a | 67 (11.86) | 103 (13.77) | 6.42 | .1696 | 37.7 | Fixed | 0.81 [0.62, 1.07] | .1440 |
| Hispanic (2)a | 136 (30.70) | 56 (20.00) | 2.18 | .1395 | — | Fixed | 1.51 [1.13, 2.02] | .0054 |
| African (2)a | 9 (3.22) | 10 (2.52) | 2.49 | .1143 | — | Fixed | 1.45 [0.62, 3.40] | .3895 |
| CT (92) | 14,802 (44.02) | 20,922 (42.97) | 179.61 | <.0001 | 49.3 | Random | 1.02 [1.00, 1.05] | .0952 |
| White (45) | 9,062 (44.37) | 12,813 (43.50) | 83.61 | .0003 | 47.4 | Random | 1.02 [0.98, 1.05] | .3149 |
| East Asian (21) | 3,363 (48.53) | 4,337 (48.15) | 12.39 | .9019 | 0 | Fixed | 1.01 [0.97, 1.04] | .6548 |
| South Asian (7) | 188 (19.58) | 188 (14.57) | 13.91 | .0307 | 56.9 | Random | 1.26 [0.96, 1.68] | .1012 |
| Mixed (10) | 1,735 (43.07) | 3,078 (40.98) | 4.92 | .8413 | 0 | Fixed | 1.04 [0.99, 1.09] | .0855 |
| Mideast (5) | 220 (38.94) | 296 (39.57) | 36.45 | <.0001 | 89.0 | Random | 1.16 [0.76, 1.77] | .5023 |
| Hispanic (2) | 168 (37.92) | 131 (46.79) | 7.48 | .0062 | — | Random | 0.93 [0.57, 1.51] | .7724 |
| African (2) | 66 (23.66) | 79 (19.90) | 8.48 | .0036 | — | Random | 1.47 [0.64, 3.34] | .3614 |
| CC (92) | 14,938 (44.42) | 21,750 (44.67) | 202.80 | <.0001 | 55.1 | Random | 1.00 [0.98, 1.03] | .7775 |
| White (45) | 9,214 (45.12) | 13,365 (45.37) | 79.56 | .0008 | 44.7 | Random | 0.94 [0.87, 1.01] | .1097 |
| East Asian (21) | 2,470 (35.65) | 3,027 (33.61) | 29.81 | .073 | 32.9 | Fixed | 1.06 [1.02, 1.11] | .006 |
| South Asian (7) | 753 (78.44) | 1,078 (83.57) | 10.05 | .1226 | 40.3 | Fixed | 0.95 [0.91, 0.99] | .0124 |
| Mixed (10) | 1,880 (46.67) | 3,530 (47.00) | 9.59 | .3846 | 6.2 | Fixed | 1.00 [0.96, 1.04] | .9379 |
| Mideast (5) | 278 (49.20) | 349 (46.66) | 40.61 | <.0001 | 90.2 | Random | 0.94 [0.63, 1.40] | .7588 |
| Hispanic (2) | 139 (31.38) | 93 (33.21) | 1.70 | .1918 | — | Fixed | 0.89 [0.71, 1.12] | .3114 |
| African (2) | 204 (73.12) | 308 (77.58) | 15.50 | <.0001 | — | Random | 0.54 [0.12, 2.37] | .4146 |
| CC + CT (92) | 29,740 (88.44) | 42,672 (87.64) | 197.38 | <.0001 | 53.9 | Random | 1.01 [1.00, 1.02] | .0017 |
| TT + CT (92) | 18,688 (55.58) | 26,938 (55.33) | 223.49 | <.0001 | 59.3 | Random | 1.00 [0.98, 1.02] | .9745 |
| Subgroups | ||||||||
| TT risk > 1 | 3,422 | 4,689 | ||||||
| TT (19) | 425 (12.42) | 397 (8.47) | 16.82 | .5356 | 0 | Fixed | 1.32 [1.15, 1.52] | <.0001 |
| CT (19) | 1,293 (37.78) | 1,626 (34.68) | 63.96 | <.0001 | 71.9 | Random | 1.14 [1.01, 1.28] | .0366 |
| CC (19) | 1,704 (49.80) | 2,666 (56.85) | 34.13 | .0121 | 47.3 | Random | 0.90 [0.85, 0.95] | .0003 |
| CC + CT (19) | 2,997 (87.58) | 4,292 (91.53) | 73.97 | <.0001 | 75.7 | Random | 0.97 [0.95, 0.99] | .0108 |
| TT + CT (19) | 1,718 (50.20) | 2,023 (43.15) | 47.43 | .0002 | 62.0 | Random | 1.17 [1.08, 1.27] | .0001 |
| TT risk < 1 | 16,075 | 23,336 | ||||||
| TT (38) | 1,761 (10.96) | 3,132 (13.42) | 41.95 | .2649 | 11.8 | Fixed | 0.79 [0.75, 0.83] | <.0001 |
| CT (38) | 7,263 (45.18) | 10,110 (43.32) | 36.42 | .4958 | 0 | Fixed | 1.03 [1.01, 1.06] | .0061 |
| CC (38) | 7,051 (43.86) | 10,094 (43.26) | 51.08 | .0616 | 27.6 | Fixed | 1.03 [1.01, 1.06] | .0054 |
| CC + CT (38) | 14,314 (89.04) | 20,204 (86.58) | 43.47 | .2151 | 14.9 | Fixed | 1.03 [1.03, 1.04] | <.0001 |
| TT + CT (38) | 9,024 (56.14) | 13,242 (56.74) | 50.95 | .0631 | 27.4 | Fixed | 0.98 [0.96, 0.99] | .0056 |
| TT risk varied | 14,129 | 20,663 | ||||||
| TT (35) | 1,700 (12.03) | 2,487 (12.03) | 68.00 | .0003 | 51.5 | Random | 1.00 [0.91, 1.11] | .9638 |
| CT (35) | 6,246 (44.21) | 9,186 (44.46) | 69.39 | .0003 | 51.0% | Random | 0.99 [0.95, 1.03] | .6434 |
| CC (35) | 6,183 (43.76) | 8,990 (43.51) | 87.22 | <.0001 | 61.0 | Random | 1.01 [0.97, 1.06] | .5969 |
| CC + CT (35) | 12,429 (87.97) | 18,176 (87.97) | 67.96 | .0005 | 50.0 | Random | 1.00 [0.99, 1.02] | .5869 |
| TT + CT (35) | 7,946 (56.24) | 11,673 (56.49) | 90.66 | <.0001 | 62.5 | Random | 0.99 [0.96, 1.03] | .6791 |
Note. Data included from 92 studies. Q = Cochran’s Q; CRC = colorectal cancer; CI = confidence interval.
aTotal sample size (combining all three genotypes) did not reach 1,000 cases or 1,000 controls.
Pooled analyses for all populations did not reach statistical significance for the other two genotypes on MTHFR 677 and heterozygous mutation CT genotype or the CC wild type. However, for the subgroup analysis, MTHFR 677 wild type CC was a protective genotype from CRC in South Asian populations (RR = 0.95, 95% CI [0.91, 0.99], p = .0124) but a risk genotype in East Asian populations (RR = 1.06, 95% CI [1.01, 1.11], p = .006). For visual examination of the geographic distributions of polymorphism mutations and associated CRC risks for MTHFR 677 to detect regional patterns for further investigation, we generated GIS maps using the JMP program (see Supplemental Figures S3 for combined TT and CT polymorphism-mutation genotypes, Supplemental Figure S4 for TT homozygous mutation genotype, and Supplemental Figure S5 for CT heterozygous mutation genotype).
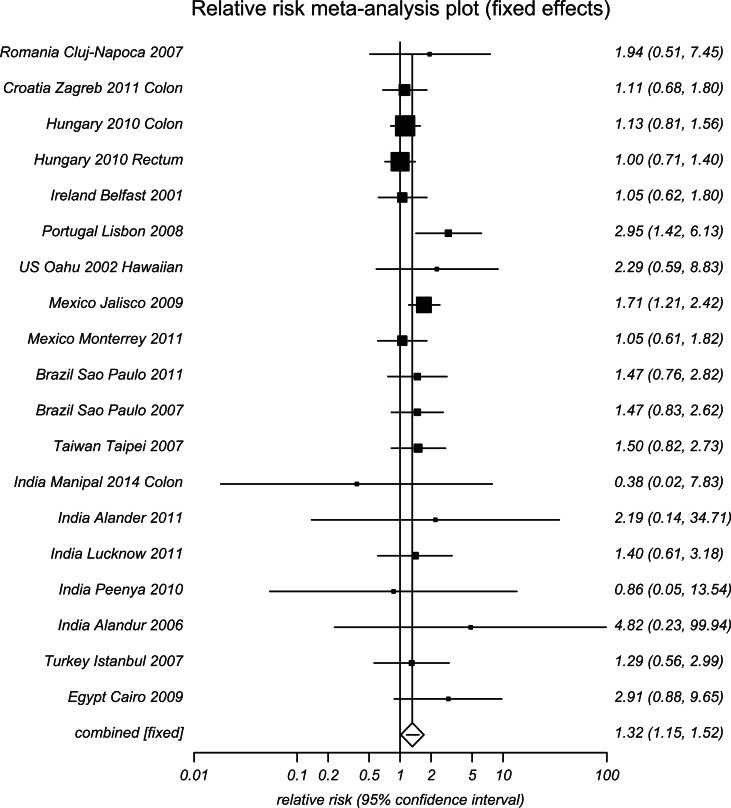
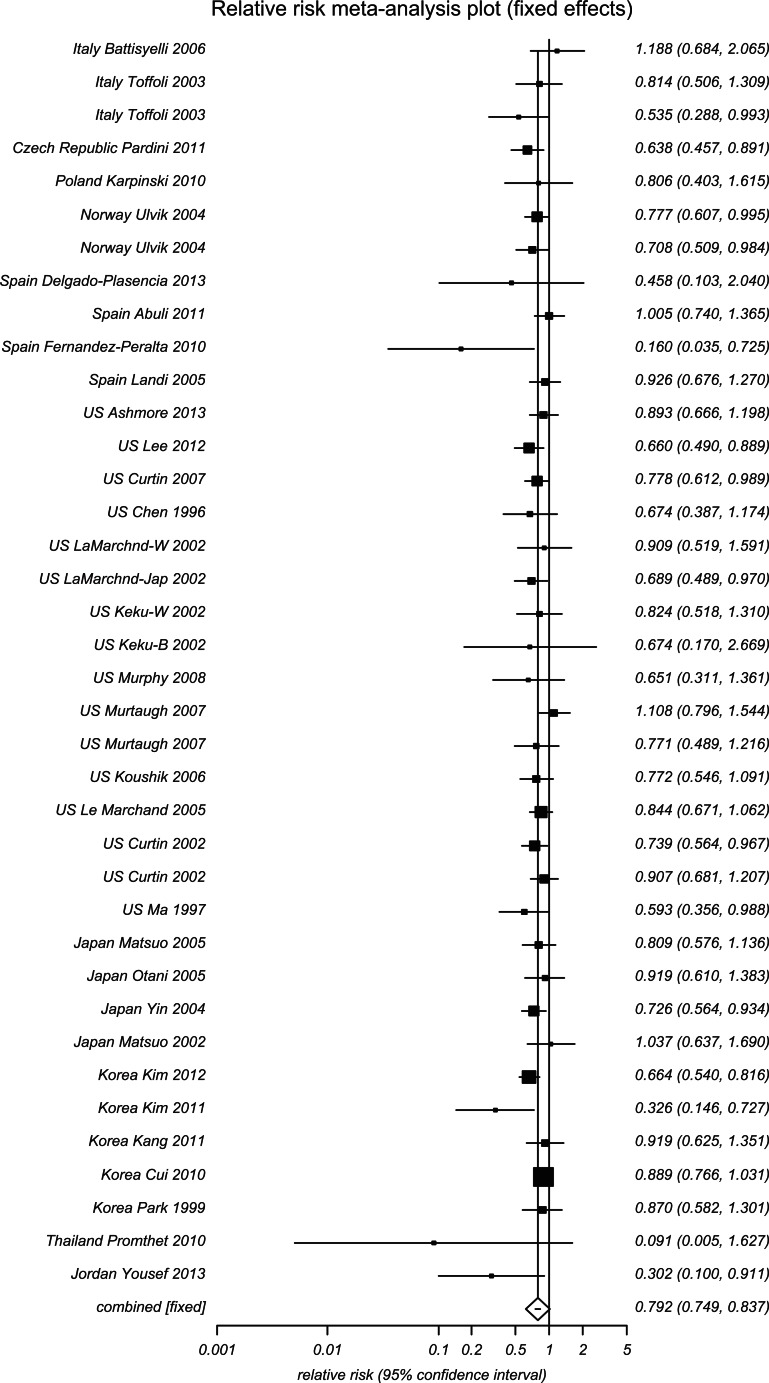
In Table 1, for the heterogeneity of the pooled results within each racial or ethnic subgroup, we also pooled additional analyses for the countries that had TT as a risk genotype (RR > 1) versus those that had it as a protective genotype (RR < 1) and others (RR varied around 1). This way of subgroup distinctions also allowed clear data presentation for various countries within limited space (Figures 2 and 3 and Supplemental Figure S6, forest plots). The RRs for all genotypes were statistically significant for one of two opposing subgroups: MTHFR 677 TT as a risk genotype or as a protective genotype for development of CRC. MTHFR 677 homozygous mutation TT genotype was a risk type for Eastern European countries (Turkey, Romania, Croatia, and Hungary) as well as Portugal, Mexico (Hispanic), Brazil (reported as European descendent of White), U.S. Hawai’i, Taiwan, India, and Egypt (Figure 2). In contrast, MTHFR 677 TT genotype was a protective type for many European countries (Italy, Czech Republic, Poland, Norway, and Spain) as well as the United States, Japan, South Korea, Thailand, and Jordan. Other countries presented diversity for the distribution of MTHFR 677 TT genotype, including Australia, Germany, France, the Netherlands, Sweden, the United Kingdom, China, and Iran (Figure S6).
Figure 2.
Forest plot for meta-analysis of MTHFR 677 TT, countries with risks > 1.
Figure 3.
Forest plot for meta-analysis of MTHFR 677 TT, countries with risks < 1.
As most previous meta-analyses have reported ORs (with the base denominator of the ORs being inconsistently used in previous reports, with either CC wild genotype or TT polymorphism genotype used as the denominator), we also pooled the standardized ORs for the present analyses using the total counts as the base of ratios for MTHFR 677 genotypes (the total of three genotype counts as the base denominator of the ratios). We found consistent results between the ORs and RRs for all genotypes for the tests of heterogeneity and association (see Supplemental Table S3a for MTHFR 677). However, as expected, for tests of association, RRs presented more conservative results than ORs for MTHFR 677 genotypes.
For pooled analyses on MTHFR 1298 genotypes, we included a total of 16,367 CRC cases and 24,874 controls from 54 study groups for the tests of heterogeneity and association (Table 2). Consistent with most previous reports, pooled results for MTHFR 1298 for all three genotypes with all populations combined did not reach statistical significance. As presented in Table 2, using the control group as the basis to check the mutation rates in the general populations, the rank order for populations in terms of polymorphism mutations on homozygous CC for MTHFR 1298 is Mideast Asian (11.96%), White (11.09%), mixed populations (10.38%), South Asian (9.01%), African (4.53%), and finally East Asian (3.37%). For visual examination of the geographical distributions of polymorphism mutations and CRC risks for MTHFR 1298 genotypes, we generated GIS maps using the JMP program (see Supplemental Figure S7 for combined mutations of CC and AC types, Figure S8 for CC homozygous mutation genotype, and Figure S9 for AC heterozygous mutation genotype). GIS maps enabled visual detection of regional patterns, leading to further investigation. Using MTHFR 1298 risk (RR) for CRC as an example (Figure S7, third map), the highest risk areas (in red) are nations in South Asia and South East Asia. MTHFR 1298 heterozygous mutation AC genotype distribution among the CRC case groups is another good example (Figure S9, the second map); in this map, the highest proportion of polymorphism mutations falls into South Asia, South East Asia, and North Africa.
Table 2.
Pooled Meta-Analysis: MTHFR 1298 Genotypes and Risk of CRC.
| Genotype (Number of Studies) | CRC Cases (N = 16,367) n (%) | Controls (N = 24,874) n (%) | Test of Heterogeneity | Statistical Model | Test of Association | |||
|---|---|---|---|---|---|---|---|---|
| Q | p | I2 (%) | Risk Ratio (95% Cl) | p | ||||
| CC (54) | 1,439 (8.79) | 2,332 (9.38) | 87.10 | .0022 | 39.2 | Random | 0.95 [0.87, 1.05] | .3434 |
| White (23) | 973 (10.54) | 1,603 (11.09) | 36.10 | .0296 | 39.1 | Random | 0.99 [0.86, 1.10] | .8282 |
| East Asian (14) | 91 (3.09) | 155 (3.37) | 11.56 | .5638 | 0 | Fixed | 0.86 [0.66, 1.11] | .2488 |
| South Asian (5)a | 58 (7.60) | 80 (9.01) | 15.37 | .004 | 74.0 | Random | 0.88 [0.40, 1.95] | .7491 |
| Mixed (7) | 267 (9.16) | 440 (10.38) | 6.26 | .3943 | 4.2 | Fixed | 0.86 [0.75, 1.00] | .0466 |
| Mideast (3)a | 33 (14.16) | 36 (11.96) | 4.28 | .1174 | 53.3 | Fixed | 1.18 [0.76, 1.84] | .456 |
| African (2)a | 17 (6.12) | 18 (4.53) | 4.46 | .0346 | — | Random | 1.66 [0.41, 6.80] | .4781 |
| AC (54) | 6,792 (41.50) | 10,059 (40.44) | 71.65 | .0448 | 26.0 | Random | 1.03 [1.00, 1.06] | .0646 |
| White (23) | 3,985 (43.15) | 6,167 (42.66) | 18.83 | .6556 | 0 | Fixed | 1.01 [0.98, 1.04] | .3995 |
| East Asian (14) | 929 (31.56) | 1,468 (31.94) | 10.55 | .6481 | 0 | Fixed | 0.98 [0.92, 1.06] | .6677 |
| South Asian (5) | 425 (55.70) | 419 (47.18) | 19.32 | .0007 | 79.3 | Random | 1.19 [0.97, 1.47] | .0994 |
| Mixed (7) | 1,243 (42.66) | 1,726 (40.74) | 5.64 | .4642 | 0 | Fixed | 1.04 [0.98, 1.10] | .2161 |
| Mideast (3) | 111 (47.64) | 143 (47.51) | 2.35 | .3081 | 15.1 | Fixed | 0.99 [0.83, 1.19] | .955 |
| African (2) | 99 (35.61) | 136 (34.26) | 0.02 | .8758 | — | Fixed | 1.07 [0.88, 1.32] | .49 |
| AA (54) | 8,136 (49.71) | 12,483 (50.18) | 93.13 | .0005 | 43.1 | Random | 1.00 [0.97, 1.02] | .7702 |
| White (23) | 4,277 (46.31) | 6,685 (46.25) | 23.45 | .3766 | 6.2 | Fixed | 0.99 [0.96, 1.02] | .4861 |
| East Asian (14) | 1,924 (65.35) | 2,973 (64.69) | 13.02 | .4459 | 0.2 | Fixed | 1.02 [0.98, 1.05] | .3912 |
| South Asian (5) | 280 (36.70) | 389 (43.81) | 40.38 | <0.0001 | 90.1 | Random | 0.81 [0.53, 1.23] | .3204 |
| Mixed (7) | 1,404 (48.18) | 2,071 (48.88) | 7.53 | .2746 | 20.3 | Fixed | 1.00 [0.95, 1.05] | .9848 |
| Mideast (3) | 89 (38.20) | 122 (40.53) | 0.83 | .6604 | 0 | Fixed | 0.95 [0.73, 1.19] | .6590 |
| African (2) | 162 (58.27) | 243 (61.21) | 5.04 | .0248 | — | Random | 0.66 [0.25, 1.74] | .4034 |
| AA + AC (54) | 14,928 (91.21) | 22,542 (90.62) | 89.63 | .0012 | 40.9 | Random | 1.01 [1.00, 1.01] | .0828 |
| CC + AC (54) | 8,231 (50.29) | 12,391 (49.82) | 110.64 | <.0001 | 52.1 | Random | 1.02 [0.99, 1.05] | .3034 |
| Subgroups | ||||||||
| CC risk > 1 | 2,880 | 4,460 | ||||||
| CC (11) | 328 (11.39) | 481 (10.79) | 7.70 | .6585 | 0 | Fixed | 1.18 [1.03, 1.35] | .0152 |
| AC (11) | 1,187 (41.21) | 1,823 (40.87) | 11.76 | .3018 | 14.9 | Fixed | 1.00 [0.95, 1.06] | .8879 |
| AA (11) | 1,365 (47.40) | 2,156 (48.34) | 16.73 | .0806 | 40.2 | Fixed | 0.96 [0.91, 1.01] | .1002 |
| AA + AC (11) | 2,552 (88.61) | 3,979 (89.21) | 7.25 | .7019 | 0 | Fixed | 0.98 [0.96, 1.00] | .0174 |
| CC + AC (11) | 1,515 (52.60) | 2,304 (51.66) | 18.78 | .0432 | 46.7 | Random | 1.05 [0.98, 1.13] | .1534 |
| CC risk < 1 | 7,610 | 11,454 | ||||||
| CC (25) | 560 (7.36) | 1,042 (9.10) | 16.94 | .851 | 0 | Fixed | 0.81 [0.73, 0.89] | <.0001 |
| AC (25) | 3,106 (40.81) | 4,545 (39.68) | 22.37 | .5574 | 0 | Fixed | 1.03 [0.99, 1.06] | .1379 |
| AA (25) | 3,944 (51.83) | 5,867 (51.22) | 24.72 | .4212 | 2.9 | Fixed | 1.01 [0.99, 1.04] | .3415 |
| AA + AC (25) | 3,666 (48.17) | 5,587 (48.78) | 21.84 | .5887 | 0 | Fixed | 1.02 [1.01, 1.03] | <.0001 |
| CC + AC (25) | 7,050 (92.64) | 10,412 (90.90) | 24.80 | .4169 | 3.2 | Fixed | 0.99 [0.96, 1.02] | .3429 |
| CC risk varied | 5,877 | 8,960 | ||||||
| CC (18) | 551 (9.38) | 809 (9.03) | 41.49 | .0008 | 59.0 | Random | 1.04 [0.85, 1.28] | .6735 |
| AC (18) | 2,499 (42.52) | 3,691 (41.19) | 36.85 | .0035 | 53.9 | Random | 1.05 [0.98, 1.12] | .1441 |
| AA (18) | 2,827 (48.10) | 4,460 (49.78) | 46.86 | .0001 | 63.7 | Random | 0.97 [0.91, 1.03] | .3248 |
| AA + AC (18) | 5,326 (90.62) | 8,151 (90.97) | 42.17 | .0006 | 59.7 | Random | 1.00 [0.98, 1.02] | .915 |
| CC + AC (18) | 3,050 (51.90) | 4,500 (50.22) | 61.91 | <.0001 | 72.5 | Random | 1.04 [0.98, 1.12] | .2107 |
Note. Data included from 54 studies. Q = Cochran’s Q; CRC = colorectal cancer; CI = confidence interval.
aTotal sample size (combining all three genotypes) did not reach 1,000 cases or 1,000 controls.
We also pooled additional subgroup analyses for MTHFR 1298 with the countries that had homozygous mutation genotype CC as a risk type (RR > 1) versus those that had it as a protective type (RR < 1) and others (RR varied around 1; Supplemental Figures S10–S12 ). MTHFR 1298 homozygous mutation CC genotype was a risk type for Romania, France, the Netherlands, Sweden, Spain, Brazil, Jordan, and Egypt (Figure S10), whereas it was a protective type for Australia, Croatia, Germany, United States, South Korea, Japan, Taiwan, China, and Thailand (Figure S11). Other countries presented diversity for MTHFR 1298 CC genotype distributions, including Italy, the Czech Republic, the United Kingdom, Japan, India, and Iran (Figure S12). We also pooled the standardized ORs for MTHFR 1298 genotypes, using the total count of all three genotypes as the base denominator (vs. paired ratios using one of the genotypes as the base denominator as in most previous reports; see Supplemental Table S3b for MTHFR 1298). Results were consistent between ORs and RRs for MTHFR 1298 genotypes for the tests of heterogeneity and association. As expected, for the tests of association, RRs presented more conservative results than ORs for MTHFR 1298 genotypes, as with those for MTHFR 677 genotypes.
For the big-data analytics approach, the classification tree yielded the most crucial variable that could make a decisive split of the mutation and risk data. Death from air pollution trumped all other potential contributing variables (i.e., population size, source of controls, cancer site, quality score, and gender) for the polymorphism mutation and risk data in all partition tree analyses. Hence, we based subsequent analyses on the one-split partition trees (see Supplemental Figure S13 as an example). The partition tree (split groups) and Tukey’s test (with p values) results are presented side by side for MTHFR 677 genotype rates and risks in Table 3. Looking at the percentage of TT for MTHFR 677 in the control group (TT%ct) as an example, the partition tree splits the data into two groups by death from air pollution (AP [death rates from air pollution] death) with Level 2 distinguished from Levels 3 and 4. In the Tukey’s test, there were significant differences between Levels 2 and 3 (p = .015) and between Levels 2 and 4 (p = .025) for percentage of TT on MTHFR 677. Obviously, the results from the partition tree and the Tukey’s test reached a consensus for percentage of TT genotype for the control group. However, we did not find significant results in the partition trees for the RRs on the various genotypes (RRCC, RRCT, and RRTT), and the Tukey’s test did not show statistical significance despite its small AICc (smaller is better).
Table 3.
Meta-Prediction: Death From Air Pollution (AP Death) on MTHFR 677 Genotypes for Ct and CRC Cases and CRC Risks.
| Variable | AICc | Partition Tree | Tukey’s Test | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AP death | Count | Mean | SD | Levels compared | Difference | SE difference | Lower CI | Upper CI | p | ||
| TT%ct | 586.248 | 2 | 22 | 8.989 | 5.56 | 3/2 | 4.426 | 1.549 | 0.734 | 8.119 | .015 |
| 3 and 4 | 70 | 13.329 | 5.769 | 4/2 | 4.294 | 1.612 | 0.451 | 8.136 | .025 | ||
| 3/4 | 0.133 | 1.399 | −3.203 | 3.468 | .995 | ||||||
| TT%ca | 589.758 | 2 and 3 | 61 | 10.313 | 5.988 | 4/2 | 5.021 | 1.640 | 1.112 | 8.930 | .008 |
| 4 | 31 | 14.688 | 5.502 | 4/3 | 4.034 | 1.424 | 0.641 | 7.428 | .016 | ||
| 3/2 | 0.986 | 1.576 | −2.770 | 4.743 | .806 | ||||||
| CT%ct | 661.195 | 2 | 22 | 34.975 | 13.339 | 4/2 | 9.805 | 2.406 | 4.068 | 15.542 | .000 |
| 3 and 4 | 70 | 43.539 | 6.499 | 3/2 | 7.502 | 2.313 | 1.989 | 13.016 | .005 | ||
| 4/3 | 2.303 | 2.089 | −2.678 | 7.284 | .515 | ||||||
| CT%ca | 64.711 | 2 | 22 | 36.816 | 12.323 | 3/2 | 7.898 | 2.373 | 2.240 | 13.556 | .004 |
| 3 and 4 | 70 | 44.548 | 7.344 | 4/2 | 7.490 | 2.470 | 1.602 | 13.378 | .009 | ||
| 3/4 | 0.408 | 2.144 | −4.703 | 5.520 | .980 | ||||||
| CC%ct | 733.734 | 2 | 22 | 56.035 | 18.243 | 2/4 | 14.099 | 3.585 | 5.552 | 22.646 | .001 |
| 3 and 4 | 70 | 43.131 | 10.526 | 2/3 | 11.929 | 3.445 | 3.715 | 20.142 | .002 | ||
| 3/4 | 2.170 | 3.112 | −5.250 | 9.590 | .766 | ||||||
| CC%ca | 664.711 | 2 | 22 | 53.516 | 17.746 | 2/4 | 12.511 | 3.415 | 4.368 | 20.653 | .001 |
| 3 and 4 | 70 | 42.997 | 9.942 | 2/3 | 8.884 | 3.282 | 1.059 | 16.709 | .022 | ||
| 3/4 | 3.626 | 2.965 | −3.443 | 10.695 | .443 | ||||||
| RRTT | 150.235 | 2 and 3 | 60 | 0.92 | 0.492 | 4/3 | 0.389 | 0.131 | 0.077 | 0.702 | .011 |
| 4 | 31 | 1.249 | 0.623 | 4/2 | 0.225 | 0.153 | −0.140 | 0.590 | .311 | ||
| 2/3 | 0.165 | 0.147 | −0.187 | 0.516 | .507 | ||||||
| RRCT | 32.106 | 2 and 3 | 60 | 1.107 | 0.331 | 2/4 | 0.153 | 0.079 | −0.035 | 0.341 | .134 |
| 4 | 32 | 0.998 | 0.149 | 3/4 | 0.084 | 0.069 | −0.079 | 0.248 | .438 | ||
| 2/3 | 0.069 | 0.076 | −0.112 | 0.250 | .639 | ||||||
| RRCC | −35.557 | 2 and 4 | 54 | 0.979 | 0.148 | 3/2 | 0.091 | 0.053 | −0.035 | 0.216 | .203 |
| 3 | 38 | 1.048 | 0.247 | 3/4 | 0.054 | 0.048 | −0.059 | 0.168 | .490 | ||
| 4/2 | 0.036 | 0.055 | −0.094 | 0.167 | .786 | ||||||
Note. AICc = Akaike’s information criterion correction; AP death = death rates from air pollution levels per million (2 = 50–100, 3 = 100–250, 4 = 250–400 and greater); RR = risk ratio; ct = controls; MTHFR = methylenetetrahydrofolate reductase; CI = confidence interval; CRC = colorectal cancer.
We also analyzed the partition tree and Tukey’s tests for MTHFR 1298 (Supplementary Table S4). Consider the example of percentage of heterozygous mutation AC genotype on MTHFR 1298 for CRC group (AC%ca): The partition tree suggests that Levels 2 and 3 were separated from Level 4, and the Tukey’s tests show significant differences between Levels 2and 4 (p = .002) and Levels 3 and 4 (p = .026). Fewer of the Tukey’s tests presented with statistical significance for MTHFR 1298 than for MTHFR 677 analyses. It is important to point out that the α cutoff (p < .05) of hypothesis testing is a convention, whereas pattern recognition may unveil certain hidden insights into the data.
We also used nonlinear associations to explore the association of the potential contributing factor, death from air pollution (AP death), with MTHFR 677 homozygous mutation percentage of TT genotype of the control group (TT%7ct; Figure S14a). With a change in air pollution from Level 2 to Level 3, there was a substantial increase in the percentage of homozygous mutation of TT genotype. However, the line decreases slightly as air pollution increased from Level 3 to Level 4. The results on the heat map were more revealing for data density (see Supplementary Figure S15), with the red blocks being the areas of high data concentration and the nonlinear fit line following the dense data (the red cells). For the nonlinear association between the levels of death from air pollution (AP death) and the percentage of MTHFR 1298 CC homozygous mutation in the control group (see Supplemental Figure S14b, CC%8ct), the nonlinear fit line was virtually flat as air pollution changed from Level 2 to Level 3. However, there was a sharp drop from Level 3 to Level 4 on the fit line and also on the heat map (see Supplemental Figure S16. the red and purple cells).
Discussion
The present study presents the most comprehensive report on the largest number of studies of any of the previous meta-analyses on this topic. In addition, we corrected errors coded and used in the previous study results and identified repeated use of data in subsidiary reports (Teng et al., 2013). Consistent with the reports from previous meta-analyses, in the present pooled analyses for all populations, MTHFR 677 homozygous mutation TT genotype presented as a protective genotype for CRC with statistical significance (p < .05); however, there were heterogeneity and opposing findings for the subgroup of Hispanics (p < .01). Previous meta-analyses consistently indicated heterogeneity or discrepancies for MTHFR and CRC risk for various populations, perhaps due in part to the dominant population stratification from Australia, the United States, and Brazil (Haerian & Haerian, 2015). In this meta-analysis study, the subgroup analysis with those countries that had findings contrary to those majority countries with the MTHFR 677 TT homozygous mutation genotype as a protective type was revealing to delineate the impacts of population stratification. Subgroup analyses for countries that presented MTHFR 677 TT as a risk genotype included Turkey, Romania, Croatia, Hungary, Portugal, Mexico, Brazil, U.S. Hawai'i, Taiwan, India, and Egypt. These countries tend to be located in the southern regions with warmer climates relative to the countries that had MTHFR 677 TT homozygous mutation genotype as a protective type for CRC. The global pollution levels adding to the global warming effects for countries with warmer climates played an important role in the MTHFR polymorphisms (677 vs. 1298 loci) and associated risks for CRC.
We presented countries with the highest mutation rates and CRC risk on both MTHFR 677 and 1298 genotypes using global maps to visualize grouping patterns. Additionally, we presented the polymorphism-mutation rates per country (Figures S1–S5 and S7–S9) to visualize the potential sources of heterogeneity for the regional patterns on the global maps. As the risk ratios were based on the polymorphism-mutation rates of cases versus controls, the polymorphism-mutation rates need to be identified for the grouping patterns in the global context. Meta-predictive analyses revealed that air pollution levels were associated with gene polymorphisms on both MTHFR 677 and 1298. Additionally, we used standardized ratios for RRs and ORs using the total count as the denominator for all three genotypes (homozygous mutation, heterozygous mutation, and wild types) to depict the standardized RRs (vs. use of only one of the genotypes as denominator) to further understand the sources of heterogeneity of the findings. Furthermore, we examined the heterogeneity of results for the pooled analyses by RR of 1 as well as the sources of heterogeneity, including geographic regions and additional potential contributing factors such as air pollution levels and other identified nonsignificant factors. When we compared the RRs and ORs, the results on the tests of heterogeneity and association were similar, but RRs presented more conservative results than ORs for pooled analyses on both MTHFR 677 and MTHFR 1298 genotype analyses.
When interpreting the source of heterogeneity of analytical findings, an understanding of the underlying physiological mechanisms can be revealing. In a previous meta-analyses of the associations between MTHFR 677 and various cancer types and chronic diseases, which was the largest such analysis prior to this one, authors speculated that the protective effects of MTHFR 677 homozygous mutation TT genotype in CRC were perhaps related to the phenomenon of global hypomethylation in all cancers and chronic diseases (Zacho, Yazdanyar, Bojesen, Tybjærg-Hansen, & Nordestgaard, 2011). That is, global hypomethylation in association with the MTHFR polymorphism mutations could limit DNA strand breakage in the tissues of colorectal epithelium or bone marrow that require high DNA synthesis, thus playing a protective role in CRC and leukemia. However, the source of heterogeneity and opposing findings regarding MTHFR 677 TT genotype in various regions in the global context is still not clear, especially when considering historic global human migrations along with global climate changes and environmental pollution.
To further explore potential explanations for this heterogeneity, we can consider the observation that MTHFR 677 polymorphism mutations present with thermolability (reduced enzyme activity in response to the action of moderate heat) when compared to MTHFR 1298 mutations (Kang et al., 1998; Leclerc et al., 2000). The populations in southern regions, including South Asia, had fewer polymorphism mutations on MTHFR 677 but greater polymorphism mutations on MTHFR 1298 than did those in northern locations. It is also well known that heat can worsen the health effects of air pollution. Additionally, global pollution combined with warmer climate may further diminish the function of folate enzymes (for MTHFR 677 function), compromising methylation pathways and affecting health status negatively for populations with various chronic diseases. Therefore, polymorphism-mutation patterns of the MTHFR gene in various geographic locations need to be investigated further in association with the enzyme activities of various loci polymorphism mutations and environmental toxicants from air pollutions. Along with a better understanding of methylation pathways and the environmental exposures to toxicants, such investigation could lead to potential prevention strategies through the methylation or detox pathways, which could play critical roles for human health. Thus, these heterogeneous findings warrant further multidisciplinary investigations for potential proactive strategies to improve the health of various populations.
We applied big-data analytical techniques in this study, in addition to the conventional pooled-analysis technique, to visualize the heterogeneity using additional advanced meta-predictive analyses. While meta-regression is used commonly for advanced meta-analysis for meta-prediction (Deeks et al., 2008), it is important to point out that regression analysis, as a linear model, is unable to detect nonlinear patterns. In this study, we performed meta-prediction using recursive partition tree, nonlinear fit, and heat maps for data visualization to reveal nonlinear patterns. Further, it is well known that regression based on R2 tends to yield a complex and overfitted model because R2 always goes up with additional predictors. On the other hand, AIC or AICc does not necessarily change with the addition of variables. Rather, it varies based upon the composition of the predictors; thus, it is more likely to yield an optimal model (Faraway, 2005).
The results that the classification trees yielded in the present study were astonishingly parsimonious. Out of many potential predictors, only air pollution could decisively demarcate the polymorphism mutation and risk outcomes. Although in some analysis population size is the second-level splitter of the tree, this variable did not input additional information into the model because big cities are highly correlated with poor air quality. Indeed, including population size into the explanatory model would be misleading because population size per se could not increase the risk of polymorphism mutations. At most, it is a proxy measure of other risk factors such as air pollution. The nonlinear association between air pollution and polymorphism mutations or risks, as indicated by the classification trees, Tukey’s tests, and heat maps, was revealing. As air pollution increased, polymorphism-mutation rates for MTHFR 677 (TT genotype) increased, while the same rates decreased for MTHFR 1298 (CC genotype).
In conclusion, our analyses revealed differences in the rate of MTHFR gene polymorphism mutations and the associated risks with CRC across different global populations. Geographical location (i.e., southern vs. northern) played an important role in the rate of MTHFR (677 vs. 1209 loci) gene polymorphisms. Coupled with increased air pollution levels, global warming may impact health negatively in populations with various chronic diseases. Specific levels of air pollution may have significant implications for policy makers for environmental health; thus, proactive measures could be implemented in cities where air pollution causes more deaths than in other regions, according to the World Health Organization’s classification. Nursing and health-care research focused on developing proactive measures for population health through increasing our understanding of the methylation pathways might play a major role in improving the health of various populations.
Supplementary Material
Supplemental Material, BRN628054_Supplementary_FigsS1and2 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplementary Material
Supplemental Material, BRN628054_Supplementary_FigsS3ThruS5 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplementary Material
Supplemental Material, BRN628054_Supplementary_FigS6 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplementary Material
Supplemental Material, BRN628054_Supplementary_FigS7thru9 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplementary Material
Supplemental Material, BRN628054_Supplementary_FigS10thru12 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplementary Material
Supplemental Material, BRN628054_Supplementary_FigS13 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplementary Material
Supplemental Material, Shiao_15060052_toSage_FigS14 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplementary Material
Supplemental Material, BRN628054_Supplementary_FigS15thru16 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplementary Material
Supplemental Material, BRN628054supplementary_tablesl for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Acknowledgements
The authors acknowledge assistance from Amanda Lie, BSN, who retrieved the literature and coded the data, and Veronica Nunez, DNP, FNP, who coded and double checked the data of studies in Spanish with abstracts and tables in English.
Footnotes
Author Contribution: S. P. K. Shiao contributed to conception, design, acquisition, analysis, and interpretation; drafted and critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy. C. H. Yu contributed to conception, design, analysis, and interpretation; drafted and critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported in part by funding from the Center for the Study of Health Disparities and the general research fund of the School of Nursing, Azusa Pacific University.
Supplementary Material: The online appendices are available at http://brn.sagepub.com/supplemental
References
- Akaike H. (1973). Information theory and an extension of the maximum likelihood principle In Petrov B. N., Csaki F. (Eds.), International symposium on information theory (pp. 267–281). Budapest, Hungary: Akademia Kiado. [Google Scholar]
- Albrecht J. (2007). Key concepts and techniques in GIS. London, England: Sage. [Google Scholar]
- Balion C., Kapur B. M. (2011). Folate. Clinical utility of serum and red blood cell analysis. American Association for Clinical Chemistry Clinical Laboratory News, 37, 8–10. [Google Scholar]
- Bozdogan H. (2000). Akaike’s information criterion and recent developments in information complexity. Journal of Mathematical Psychology, 44, 62–91. doi:10.1006jmps.1999.1277 [DOI] [PubMed] [Google Scholar]
- Breiman L., Friedman J. H., Olshen R. A., Stone C. J. (1984). Classification and regression trees. Monterey, CA: Wadsworth International Group. [Google Scholar]
- Burnham K. P., Anderson D. R. (2002). Model selection and multimodel inference: A practical information-theoretic approach (2nd ed). New York, NY: Springer-Verlag. [Google Scholar]
- Chen N. C., Yang F., Capecci L. M., Gu Z. Y., Schafer A. I., Durante W.…Wang H. (2010). Regulation of homocysteine metabolism and methylation in human and mouse tissues. FASEB Journal, 24, 2804–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider K. S., Maneval D. R., Dowling N. F., Bailey L. B., Kaudwell G., Hao L.…Berry R. J. (2012). Variants in one-carbon metabolism and blood folate, homocysteine and B12 deficiency in a population-based study. Proceedings of the American Society of Human Genetics 62nd Annual Meeting (1304W), San Francisco, CA. [Google Scholar]
- Crider K. S., Yang T. P., Berry R. J., Bailey L. B. (2012). Folate and DNA methylation: A review of molecular mechanisms and the evidence for folate’s role. Advanced Nutrition, 3, 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks J. J., Higgins J. P. T., Altman D. G. on behalf of the Cochrane Statistical Methods Group. (Eds.). (2008). Analysing data and undertaking meta-analyses In Higgins J. P. T., Green S. (Eds.), Cochrane handbook for systematic reviews of interventions (version 5.0.1, Chapter 9). The Cochrane Collaboration: Retrieved from www.cochrane-handbook.org [Google Scholar]
- Deng W., Wang Y. B., Liu Z., Cheng H., Xue Y. (2014). HemI: A toolkit for illustrating heatmaps. PLOS One, 9, e111988 Retrieved from http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deu-Pons J., Schroeder M. P., Lopez-Bigas N. (2014). jHeatmap: An interactive heatmap viewer for the Web. Bioinformatics, 30, 1757–1758. [DOI] [PubMed] [Google Scholar]
- Downs S. H., Black N. (1998). The feasibility of creating a checklist for the assessment of the methodological quality both of randomised and non-randomised studies of health care interventions. Journal of Epidemiology and Community Health, 52, 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Environmental Systems Research Institute (ESRI). (2015). What is GIS? Retrieved from http://www.esri.com/what-is-gis
- Faraway J. (2005). Extending the linear model with R: Generalized linear, mixed effects and nonparametric regression models. London, England: Chapman and Hall. [Google Scholar]
- Guo L., Byun H. M., Zhong J., Motta V., Barupal J., Zheng Y.…Baccarelli A. A. (2014). Effects of short-term exposure to inhalable particulate matter on DNA methylation of tandem repeats. Environmental and Molecular Mutagenesis, 55, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerian B. S., Haerian M. S. (2015). Evaluation of association studies and meta-analyses of MTHFR gene polymorphisms in colorectal cancer. Pharmacogenomics, 16, 413–425. [DOI] [PubMed] [Google Scholar]
- Inoue-Choi M., Nelson H. H., Robien K., Arning E., Bottiglieri T., Koh W., Yuan J.-M. (2012). One-carbon metabolism nutrient status and plasma S-adenosylmethionine concentrations in middle-aged and older Chinese in Singapore. International Journal of Molecular Epidemiology and Genetics, 3, 160–173. [PMC free article] [PubMed] [Google Scholar]
- Inoue-Choi M., Nelson H. H., Robien K., Arning E., Bottiglieri T., Koh W., Yuan J.-M. (2013). Plasma S-adenosylmethionine, DNMT polymorphisms, and peripheral blood LINE-1 methylation among healthy Chinese adults in Singapore. BMC Cancer, 13, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaluddin M. S., Yang X., Wang H. (2007). Hyperhomocysteinemia, DNA methylation and vascular disease. Clinical Chemistry and Laboratory Medicine, 45, 1660–1666. [DOI] [PubMed] [Google Scholar]
- Kang S. S., Zhou J., Wong P. W., Kowalisyn J., Strokosch G. (1998). Intermediate homocysteinemia: A thermolabile variant of methylenetetrahydrofolate reductase. American Journal of Human Genetics, 43, 414–421. [PMC free article] [PubMed] [Google Scholar]
- Keku T., Millikan R., Worley K., Winkel S., Eaton A., Biscocho L.…Sandler R. (2002). 5,10-methylenetetrahydrofolate reductase codon 677 and 1298 polymorphisms and colon cancer in African Americans and Whites. Cancer Epidemiology, Biomarkers & Prevention, 11, 1611–1621. [PubMed] [Google Scholar]
- Kennedy D. A., Stern S. J., Matok I., Moretti M. E., Sarkar M., Adams-Webber T., Koren G. (2012). Folate intake, MTHFR polymorphisms, and the risk of colorectal cancer: A systematic review and meta-analysis. Journal of Cancer Epidemiology, 952508 doi:10.1155/2012/952508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy J., Laube F. (2002). Urban transport patterns in a global sample of cities and their linkages to transport infrastructures, land use, economics and environment. World Transport Policy and Practice, 8, 5–20. [Google Scholar]
- Kloog I., Ridgway B., Koutrakis P., Coull B. A., Schwartz J. D. (2013). Long- and short-term exposure to PM2.5 and mortality: Using novel exposure models. Epidemiology, 24, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc D., Sibani S., Rozen R. (2000). Molecular biology of methylenetetrahydrofolate reductase (MTHFR) and overview of mutations/polymorphisms In Madame Curie bioscience database. Austin, TX: Landes Bioscience; Retrieved from http://www.ncbi.nlm.nih.gov/books/NBK6561/ [Google Scholar]
- Le Marchand L., Donlon T., Hankin J. H., Kolonel L. N., Wilkens L. R., Seifried A. (2002). B-vitamin intake, metabolic genes, and colorectal cancer risk (United States). Cancer Causes and Control, 13, 239–248. [DOI] [PubMed] [Google Scholar]
- Mallone S., Stafoggia M., Faustini A., Gobbi G. P., Marconi A., Forastiere F. (2011). Saharan dust and associations between particulate matter and daily mortality in Rome, Italy. Environmental Health Perspective, 119, 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W. (2014). Leave-one-out cross-validation-based model selection for multi-input multi-output support vector machine. Neural Computing & Applications, 24, 441–451. [Google Scholar]
- McBride C. (2012, April 18). Applications of genomics to improve public health [Lecture 12]. National Human Genome Research Institute’s Current Topics in Genome Analysis 2012. Retrieved from http://www.genome.gov/Course2012/
- Moher D., Cook D. J., Eastwood S., Olkin I., Rennie D., Stroup D. F. (1999). Improving the quality of reports of meta-analyses of randomized controlled trials: The QUOROM statement. Lancet, 354, 1896–1900. [DOI] [PubMed] [Google Scholar]
- Myers V., Broday D. M., Steinberg D. M., Yuval D. Y., Gerber Y. (2013). Exposure to particulate air pollution and long-term incidence of frailty after myocardial infarction. Annals of Epidemiology, 23, 395–400. [DOI] [PubMed] [Google Scholar]
- Pope C. A.,, 3rd, Burnett R. T., Turner M. C., Cohen A., Krewski D., Jerrett M.…Thun M. J. (2011). Lung cancer and cardiovascular disease mortality associated with ambient air pollution and cigarette smoke: Shape of the exposure-response relationships. Environmental Health Perspective, 119, 1616–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsum H., Nurk E., Smith A. D., Ueland P. M., Gjesdal C. G., Bjelland I.…Vollset S. E. (2006). The Hordaland Homocysteine study: A community-based study of homocysteine, its determinants, and associations with disease. Journal of Nutrition, 136, 1731S–1740S. [DOI] [PubMed] [Google Scholar]
- Ren C., Park S. K., Vokonas P. S., Sparrow D., Wilker E., Baccarelli A.…Schwartz J. (2010). Air pollution and homocysteine: More evidence that oxidative stress-related genes modify effects of particulate air pollution. Epidemiology, 21, 198–206. doi:10.1097/EDE.0b013e3181cc8bfc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Q., Zhang S. (2011). A test of Hardy-Weinberg equilibrium in structured populations. Genetic Epidemiology, 35, 671–678. doi:10.1002/gepi.20617 [DOI] [PubMed] [Google Scholar]
- Sibani S., Leclerc D., Weisberg I. S., O’Ferrall E., Watkins D., Artigas C.…Rozen R. (2003). Characterization of mutations in severe methylenetetrahydrofolate reductase deficiency reveals an FAD-responsive mutation. Human Mutation, 21, 509–520. [DOI] [PubMed] [Google Scholar]
- Speybroeck N. (2012). Classification and regression trees. International Journal of Public Health, 57, 243–246. [DOI] [PubMed] [Google Scholar]
- Stroup D. F., Berlin J. A., Morton S. C., Olkin I., Williamson G. D., Rennie D.…Thacker S. B. (2000). Meta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis of Observational Studies in Epidemiology (MOOSE) group. Journal of American Medical Association, 283, 2008–2012. [DOI] [PubMed] [Google Scholar]
- Symonds M., Moussalli A. (2011). A brief guide to model selection, multimodel inference and model averaging in behavioral ecology using Akaike’s information criterion. Behavioral Ecology and Sociobiology, 65, 13–21. doi:10.1007/s00265-010-1037-6 [Google Scholar]
- Teng Z., Wang L., Cai S., Yu P., Wang J., Gong J., Liu Y. (2013). The 677C>T (rs1801133) polymorphism in the MTHFR gene contributes to colorectal cancer risk: A meta-analysis based on 71 research studies. PLoS One, 8, e55332 doi:10.1371/journal.pone.0055332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitha A., Niraimathi S. (2013). Study on decision tree: Competent data classification. International Journal of Computer Science and Mobile Computing, 2, 365–370. [Google Scholar]
- Wade D. H., McBride C. M., Kardia S. L. R., Brody L. C. (2010). Considerations for designing a prototype genetic test for use in translational research. Public Health Genomics, 13, 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenius G. A., Burger M. R., Coull B. A., Schwartz J., Suh H. H., Koutrakis P.…Mittleman M. A. (2012). Ambient air pollution and the risk of acute ischemic stroke. Archives of Internal Medicine, 172, 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke-Thompson J. K., Pluzhnikov A., Cox N. J. (2005). Rational inferences about departures from Hardy-Weinberg equilibrium. American Journal of Human Genetics, 76, 967–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2004). Deaths attributable to urban air pollution. Retrieved from http://www.who.int/heli/risks/urban/en/uapmap.1.pdf?ua=1
- World Health Organization. (2009). Global health risks. Retrieved from http://www.who.int/healthinfo/global_burden_disease/GlobalHealthRisks_report_full.pdf
- World Health Organization. (2012). Global health risks. Retrieved from https://commons.wikimedia.org/wiki/File:Deaths_from_air_pollution.png
- World Health Organization. (2015). The urban environment. Retrieved from http://www.who.int/heli/risks/urban/urbanenv/en/
- Yu C. H. (2010). Exploratory data analysis in the context of data mining and resampling. International Journal of Psychological Research, 3, 9–22. Retrieved from http://revistas.usb.edu.co/index.php/IJPR/article/view/819 [Google Scholar]
- Yu C. H. (2014). Dancing with the data: The art and science of data visualization. Saarbrucken, Germany: Lambert Academic. [Google Scholar]
- Zacho J., Yazdanyar S., Bojesen S. E., Tybjærg-Hansen A., Nordestgaard B. G. (2011). Hyperhomocysteinemia, methylenetetrahydrofolate reductase c.677C>T polymorphism and risk of cancer: Cross-sectional and prospective studies and meta-analyses of 75,000 cases and 93,000 controls. International Journal of Cancer, 128, 644–652. doi:10.1002/ijc.25375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, BRN628054_Supplementary_FigsS1and2 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplemental Material, BRN628054_Supplementary_FigsS3ThruS5 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplemental Material, BRN628054_Supplementary_FigS6 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplemental Material, BRN628054_Supplementary_FigS7thru9 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplemental Material, BRN628054_Supplementary_FigS10thru12 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplemental Material, BRN628054_Supplementary_FigS13 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplemental Material, Shiao_15060052_toSage_FigS14 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplemental Material, BRN628054_Supplementary_FigS15thru16 for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing
Supplemental Material, BRN628054supplementary_tablesl for Meta-Prediction of MTHFR Gene Polymorphism Mutations and Associated Risk for Colorectal Cancer by S. P. K. Shiao, and C. H. Yu in Biological Research For Nursing