Abstract
The stomatal apparatus consists of a pair of guard cells and regulates gas exchange between the leaf and atmosphere. In guard cells, blue light (BL) activates H+-ATPase in the plasma membrane through the phosphorylation of its penultimate threonine, mediating stomatal opening. Although this regulation is thought to be widely adopted among kidney-shaped guard cells in dicots, the molecular basis underlying that of dumbbell-shaped guard cells in monocots remains unclear. Here, we show that H+-ATPases are involved in the regulation of dumbbell-shaped guard cells. Stomatal opening of rice was promoted by the H+-ATPase activator fusicoccin and by BL, and the latter was suppressed by the H+-ATPase inhibitor vanadate. Using H+-ATPase antibodies, we showed the presence of phosphoregulation of the penultimate threonine in Oryza sativa H+-ATPases (OSAs) and localization of OSAs in the plasma membrane of guard cells. Interestingly, we identified one H+-ATPase isoform, OSA7, that is preferentially expressed among the OSA genes in guard cells, and found that loss of function of OSA7 resulted in partial insensitivity to BL. We conclude that H+-ATPase is involved in BL-induced stomatal opening of dumbbell-shaped guard cells in monocotyledon species.
Keywords: Dumbbell-type stomata, H+-ATPase, Light-induced stomatal opening, Rice
Introduction
The stomatal apparatus is responsible for gas exchange between the leaf and the atmosphere. The stomata open in response to light to take up CO2 required for photosynthesis, while they close under water-limited conditions to minimize transpiration. This plastic regulation is one of the most fundamental characteristics of sessile land plants to adapt to the fluctuating environment (Shimazaki et al. 2007). Dicots possess a stomatal complex with a pair of kidney-shaped guard cells, while those of monocots typically consist of dumbbell-shaped guard cells with adjacently arranged subsidiary cells. Subsidiary cells function as an ion reservoir for guard cells such as K+ and Cl–, which are utilized during stomatal regulation (Raschke and Fellows 1971, Büchsenschütz et al. 2005). Despite the morphological differences between kidney-shaped and dumbbell-shaped guard cells, stomatal opening depends on swelling of the guard cells in both types. In kidney-shaped cells, the H+ pump, later identified as H+-ATPase in the plasma membrane, plays a central role in stomatal opening (Assmann et al. 1985, Shimazaki et al. 1986, Shimazaki et al. 1999). H+-ATPases create an electrochemical gradient across the plasma membrane and trigger K+ uptake through the voltage-gated K+ channel, resulting in an increase in osmotic pressure and subsequent inflow of water (Assmann and Shimazaki 1999, Schroeder et al. 2001). Genetic analyses of Arabidopsis mutants have suggested that the blue light (BL) receptor phototropin (phot1 and phot2) regulates stomatal opening through the activation of H+-ATPases in the plasma membrane (Kinoshita et al. 2001).
H+-ATPase in the plasma membrane is self-inhibited by the C-terminal domain binding to the catalytic domain (CAT domain) (Palmgren et al. 1990, Morsomme et al. 1996, Palmgren et al. 2001). Activation of H+-ATPase, such as in response to BL, occurs by phosphorylation of the C-terminal penultimate threonine followed by the binding of 14-3-3 proteins, which causes a conformational change and results in de-repression of the CAT domain (Kinoshita and Shimazaki 1999, Kinoshita and Shimazaki 2002). To date, neither the protein kinase nor the protein phosphatase directly regulating phosphorylation of the penultimate threonine in guard cells has been identified, although the candidate phosphatase is within the protein phosphatase type 2C (PP2C) family proteins based on biochemical approaches (Hayashi et al. 2010). It is noteworthy that PP2C D-subfamily proteins were recently shown to regulate H+-ATPase activity negatively to control hypocotyl elongation (Spartz et al. 2014).
Genetic analyses have suggested the involvement of H+-ATPases in stomatal regulation of kidney-type stomata. In Arabidopsis, dominant mutations of H+-ATPase (ost2-1D/ost2-2D), which lead to the expression of its constitutively active form, showed defects in stomatal closure (Merlot et al. 2007). In tobacco, suppression of H+-ATPase (PMA4) showed the stomatal closure phenotype and displayed insensitivity to fusicoccin (FC), a fungal toxin that activates H+-ATPase (Zhao et al. 2000). Moreover, overexpression of H+-ATPase in guard cells or increasing the localization of H+-ATPase in the plasma membrane of guard cells resulted in the promotion of light-induced stomatal opening (Hashimoto-Sugimoto et al. 2013, Y. Wang et al. 2014a). However, to the best of our knowledge, H+-ATPase loss-of-function mutants that affect stomatal opening have not been reported.
Compared with the extensive studies that have been performed on kidney-shaped guard cells, limited studies have characterized the physiological responses of dumbbell-shaped guard cells. Increases in stomatal conductance in rice and the transpiration rate in wheat and sugarcane were observed after BL irradiation (Karlsson et al. 1986, Assmann and Grantz, 1990, Shimazaki et al. 2007). Using a voltage clamp against intact guard cells of barley, changes in plasma membrane potential in response to light have been observed (Kollist et al. 2014). These results suggest that the regulation of dumbbell-shaped guard cells is at least in part similar to that of kidney-shaped guard cells; however, the molecular mechanisms underlying the regulation of dumbbell-shaped guard cells have remained unclear. In this study, we characterized stomata of rice, which harbors dumbbell-shaped guard cells. The stomata of rice were opened by FC and BL, and the BL-induced stomatal opening was suppressed by vanadate, an H+-ATPase inhibitor. Further molecular analysis showed that phosphorylation of the penultimate threonine in Oryza sativa H+-ATPases (OSAs) was actively regulated. Moreover, immunohistochemical detection using H+-ATPase antibodies showed localization of OSAs in the plasma membrane of guard cells. Interestingly, we found that one isoform, OSA7, was abundantly expressed in guard cells, and dysfunction of OSA7 resulted in the impairment of BL-induced stomatal opening. Our results not only show the involvement of H+-ATPase in the regulation of dumbbell-shaped guard cells but also provide the first report of an H+-ATPase loss-of-function mutant that affects BL-dependent stomatal opening in higher plants.
Results
H+-ATPase is involved in the regulation of dumbbell-type stomatal openings
To explore whether H+-ATPases are involved in the regulation of dumbbell-shaped guard cells, we examined the response of stomata in rice. To visualize the stomatal pores, epidermal fractions were detached from mesophyll tissues using a waring blender (Fig. 1A). In rice, when epidermal fractions were incubated in the dark, only a small proportion of the stomata were open (Fig. 1B). However, application of the fungal toxin FC, which activates H+-ATPases via phosphorylation of the penultimate threonine of H+-ATPases (Olsson et al. 1998, Svennelid et al. 1999, Kinoshita and Shimazaki 2001), increased the proportion of open stomata. Similar effects were observed by light irradiation. BL combined with red light (RL) enhanced stomatal opening, whereas RL alone had a limited effect. The BL-induced stomatal opening was over-ridden in the presence of vanadate, an inhibitor of H+-ATPase (Gepstein et al. 1982, Amodeo et al. 1992). Moreover, the addition of the phytohormone ABA further decreased the proportion of open stomata, even under RL and BL treatment. It is noteworthy that stomata of oat, which also harbors dumbbell-shaped guard cells, showed similar responses to rice (Supplementary Fig. S1). These results suggest that dumbbell-shaped guard cells are responsive to BL, and that H+-ATPase is involved in this regulation.
Fig. 1.
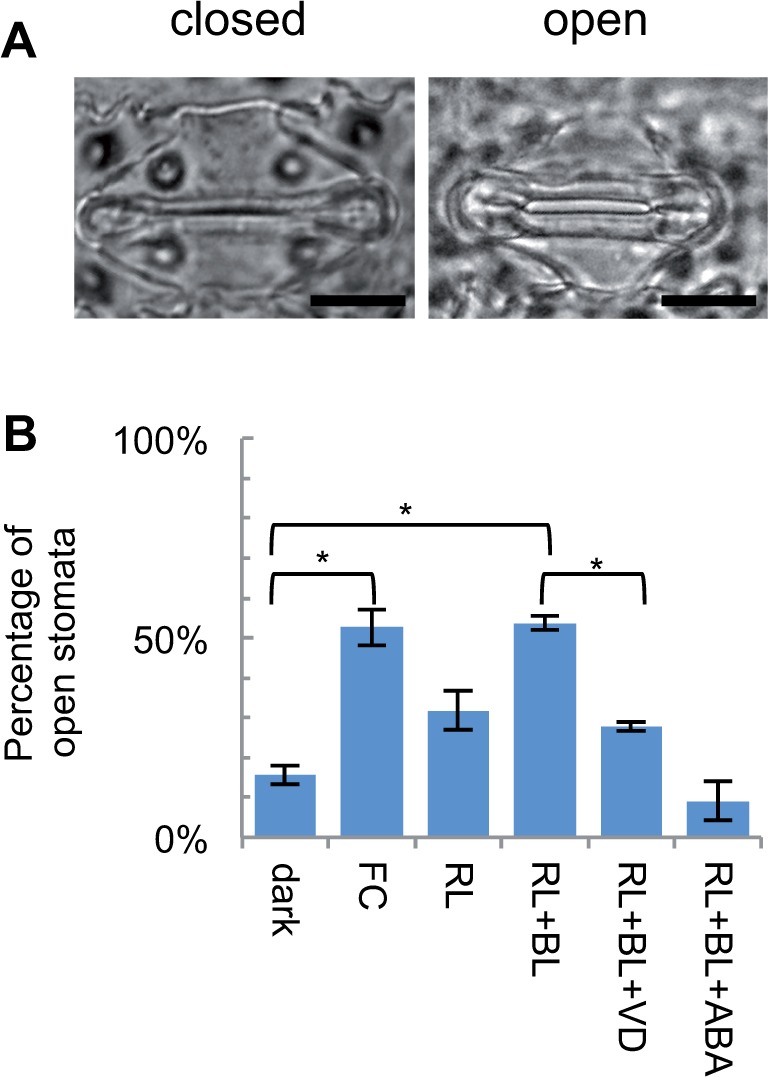
H+-ATPase is involved in the regulation of rice dumbbell-type stomata. (A) Representative images of open and closed stomata of 5-day-old rice seedlings. Scale bars = 5 μm. (B) The percentage of opened stomata observed under various conditions. Mean ± SD (n = 3; at least 50 stomata were observed for each replicate). FC, 10 μM fusicoccin for 3 h; RL+BL, 150 μmol m−2 s−1 red light and 50 μmol m−2 s−1 blue light for 4 h; RL+BL+VD, RL+BL treatment with 1 mM vanadate; RL+BL+ABA, RL+BL treatment with 20 μM ABA. Asterisks indicate statistical differences (P < 0.05) based on the Student’s t-test.
To increase our understanding of the morphology of stomata with dumbbell-shaped guard cells, we performed non-destructive observations of rice stomata using environmental scanning electron microscopy (ESEM). Supplementary Fig. S2A shows the typical appearance of stomata of leaf blades incubated under light conditions. When leaves were treated with FC, not only the enlargement of stoma but also the withering of subsidiary cells was observed (Supplementary Fig. S2B). In contrast, treatment with ABA resulted in the expansion of subsidiary cells along with stomatal closure (Supplementary Fig. S2C). These observations support the results in Fig. 1 and suggest that subsidiary cells play an important role in stomatal regulation. To observe the dynamics of guard cells during stomatal opening, we performed bright field time-lapse imaging against FC-treated oat stoma (Supplementary Movie S1). Bulbous ends of guard cells pushed each other apart, resulting in the formation of the stomatal pore. To the best of our knowledge, this is the first live imaging of stomatal opening of dumbbell-shaped guard cells.
Sequential and phylogenetic analysis of OSAs
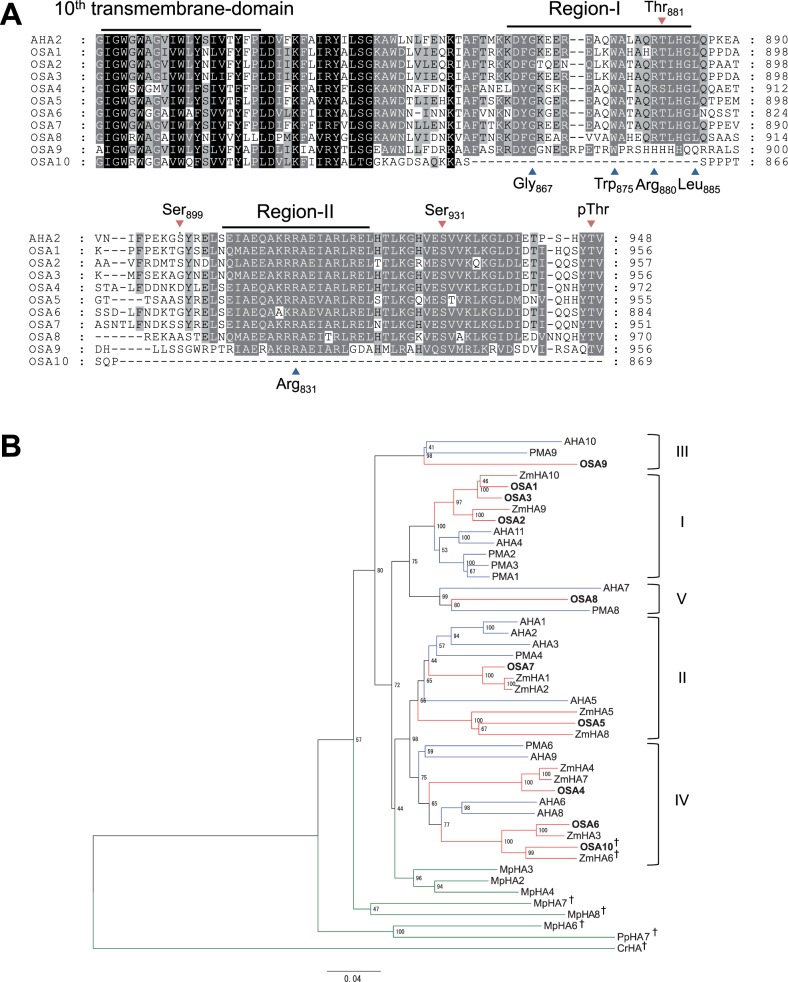
We performed further molecular analysis in rice; its genome has been reported to encode 10 OSAs (Arrango et al. 2003, Baxter et al. 2003). Comparing the C-terminal amino acid sequence of H+-ATPases, excluding OSA10, other OSAs harbored the inhibitory domain (Fig. 2A). Gly867, Trp875, Arg880, Leu885 and Arg931 in Region-I and Region-II, which were considered critical for the self-inhibiting function of H+-ATPase (Axelsen et al. 1999), were also conserved in OSA1–OSA8, but only partially in OSA9 (Fig. 2a, blue arrowheads). This suggested that OSA1–OSA9 possess a functional self-inhibitory domain, but the mode of the self-inhibition of OSA9 probably differs from that of the other isoforms.
Fig. 2.
Sequential and phylogenetic analysis of H+-ATPases in rice. (A) Sequence alignment of the C-terminal inhibitory domain of Oryza sativa H+-ATPases (OSAs) and Arabidopsis AHA2. The 10th transmembrane domain and the inhibitory motif (Region-I and Region-II) within the C-terminal inhibitory domain are shown. Blue arrowheads below the sequence indicate amino acids that are critical for the function of the inhibitory domain of AHA2 (Axelsen et al. 1999). Red arrowheads over the sequence indicate phosphorylation target sites in AHA2 (Fugisang et al. 2007, Niittylä et al. 2007, Haruta et al. 2014). (B) Phylogenetic tree of H+-ATPases of rice, maize (ZmHAs), Arabidopsis (AHAs), tobacco (PMAs), M. polymorpha (MpHAs), P. patens (PpHA) and C. reinhardtii (CrHA). The phylogenetic tree was constructed using the full-length amino acid sequences of H+-ATPases. The scale bar indicates 0.04 amino acid substitutions per site. Blue, red and green nodes represent H+-ATPases of dicots, monocots and others, respectively. CrHA was used as an outgroup. Daggers indicate non-pT-type H+-ATPases. Roman numerals indicate subfamilies defined by Arango et al. (2003). PMA5, PMA7, MpHA1 and MpHA5 were not incorporated into this analysis because full-length sequences were not available.
The repressive function of the C-terminal inhibitory domain is controlled by the phosphorylation of specific Ser/Thr residues (Thr947, Ser899, Thr881 and Ser931; Fugisang et al. 2007, Niittylä et al. 2007, Hayashi et al. 2010, Haruta et al. 2014). The rate of preservation of these residues varied among isoforms (Fig. 2A, red arrowheads). Importantly, OSA1–OSA9 all possessed the penultimate threonine; its phosphorylation was reported to be critical for the activation of H+-ATPases via binding of 14-3-3 proteins (Kinoshita and Shimazaki 1999, Kinoshita and Shimazaki 2002). Here, we define OSA1–OSA9 as penultimate threonine-containing (pT-type) H+-ATPases and OSA10 as a non-pT-type H+-ATPase, following the classification of Okumura et al. (2012a).
Phylogenetic analyses of OSAs were performed with H+-ATPases of monocot maize (ZmHAs), dicot Arabidopsis (AHAs) and tobacco (PMAs), bryophytes Marchantia polymorpha (MpHAs) and Physcomitrella patens (PpHA7) (Okumura et al. 2012b), and Chlamydomonas reinhardtii (CrHA) (Fig. 2B). OSAs were classified into subfamilies I–V together with H+-ATPases of both monocots and dicots, but not with that of bryophytes and Chlamydomonas. In subfamily I, OSA1, OSA2 and OSA3 formed a clade together with maize (but not with dicots), supported by a bootstrap value of 100. Similarly, OSA6 and OSA10 in subfamily IV were separate from AHA6 and AHA8. This indicates that gene duplications that led to the establishment of at least five pT-type H+-ATPases occurred prior to monocot–dicot divergence; however, further duplications within the respective subfamilies may have occurred during the lineage of monocot, which is in agreement with the previous report (Arrango et al. 2003).
It is noteworthy that the non-pT H+-ATPases OSA10 and ZmHA6 (Supplementary Fig. S3) were categorized into subfamily IV, while non-pT-type H+-ATPases of bryophytes and Chlamydomonas (MpHA6, MpHA7, MpHA8, PpHA7 and CrHA) constituted independent clades (Fig. 2B). This suggests that monocots have reacquired non-pT-type H+-ATPase from the descent of subfamily IV.
Regulation of penultimate threonine phosphorylation in OSAs
To determine whether the regulation of penultimate threonine phosphorylation occurs in pT-type OSAs, we performed immunoblot analysis using polyclonal antibodies recognizing the CAT domain (anti-CAT) and the phosphorylated penultimate threonine of Arabidopsis H+-ATPase AHA2 (anti-pThr) (Hayashi et al. 2010). Using protein extracted from rice leaf blades of seedlings grown under light conditions, a band near 97 kDa was most strongly detected by anti-CAT, whereas a single band near 97 kDa was detected by anti-pThr (Fig. 3A). As the predicted sizes of OSAs were in the range of 95–105 kDa (Supplementary Fig. S4), the major band detected by anti-CAT is likely to be OSA1–OSA10, while a band detected by anti-pThr is likely to be pT-type OSA1–OSA9.
Fig. 3.
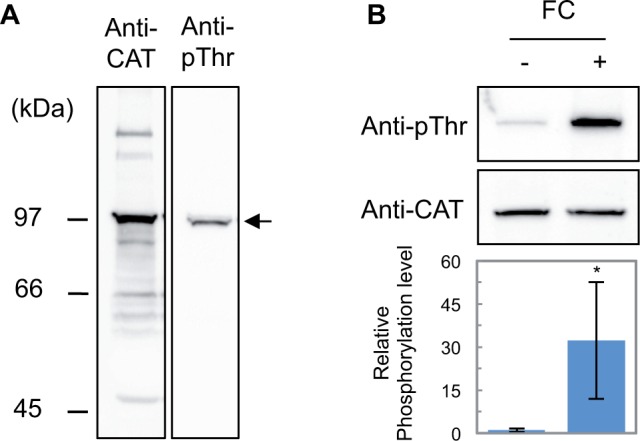
Presence of phosphoregulation of the penultimate threonine in pT-type OSAs. (A) Western blot showing the specificity of antibodies (anti-CAT and anti-pThr) against protein extracts of rice leaf blades harvested under light. Molecular weights are shown on the left. The positions of OSAs are indicated by arrows. (B) Penultimate threonine phosphorylation in H+-ATPase of rice. Rice seedlings at 5 d after germination were adapted to dark for >12 h prior to 10 μM FC (+) or mock (–) treatment. Top column, immunoblot using anti-CAT and anti-pTh. Bottom column, graphs quantifying the relative phosphorylation level (anti-pThr detection level normalized by the anti-CAT detection level). Mean ± SD (n = 3).
Next, we treated rice leaves with FC, which activates H+-ATPase through the inhibition of dephosphorylation of the penultimate threonine (Camoni et al. 2000, Maudoux et al. 2000, Kinoshita and Shimazaki 2001). FC treatment against dark-adapted leaves enhanced the level of phosphorylated penultimate threonine (Fig. 3B). Importantly, treatment with FC did not affect the detection level of anti-CAT, indicating that the total amount of H+-ATPase did not change by this treatment (Fig. 3B). Similar to Arabidopsis, penultimate threonine phosphorylation of H+-ATPase is actively regulated in rice.
pT-type H+-ATPases are expressed in dumbbell-shaped guard cells in rice
To increase our understanding of the expression profile of OSA genes, tissue-specific expression patterns were investigated in rice seedlings using reverse transcription–PCR (RT–PCR) (Fig. 4). In addition to roots and aerial tissues, the guard cell-enriched fraction (GCEF) and mesophyll cell protoplasts (MCPs) isolated from leaf blades were prepared. The enrichment of the guard cells in the GCEF was evaluated by quantifying the expression level of OsKAT2, which was reported to be expressed specifically in guard cells (Hwang et al. 2013). Compared with MCPs, the GCEF showed 10-fold higher expression of OsKAT2, while the difference in the expression of UBQ5, a housekeeping gene that displays stable expression in various tissues (Jain et al. 2001), was indistinguishable (Supplementary Fig. S5). In rice seedlings, OSA3, OSA5, OSA7 and OSA8 tended to show ubiquitous expression, while other isoforms displayed tissue specificity (Fig. 4). OSA4, OSA9 and OSA10 were weakly detected under this experimental condition. Importantly, in the GCEF, the expression of at least seven isoforms was detected (OSA1, OSA2, OSA3, OSA5, OSA6, OSA7, and OSA8), where OSA7 was the most abundant. These results suggest that multiple isoforms of pT-type OSAs were expressed in guard cells.
Fig. 4.
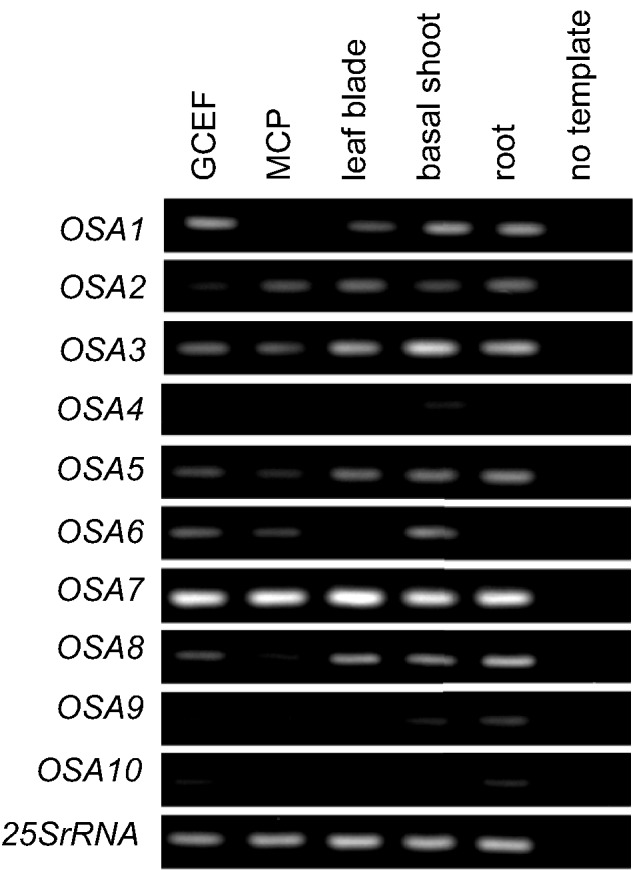
Expression pattern of OSAs in rice seedling. Tissue-specific expression pattern of OSAs in tissues obtained from 5-day-old seedlings were examined using RT–PCR analysis. Expression levels of 25SrRNA were used as a control. GCEF, guard cell-enriched fraction; MCP, mesophyll cell protoplasts.
Next, we performed immunohistochemical detection of H+-ATPase in rice guard cells to clarify the localization of OSAs in the stomatal complex. Due to the low specificity of anti-CAT (Fig. 3A), we used anti-pThr for this analysis. When anti-pThr was used against leaf blades harvested under light, a signal that inscribes the guard cell wall was detected (Fig. 5B, D). However, no signals were detected in guard cells when pre-immune serum was used (Fig. 5A, C). This suggests that pT-type H+-ATPases localize in the plasma membrane of rice guard cells, and that their penultimate threonine is phosphorylated under light conditions.
Fig. 5.
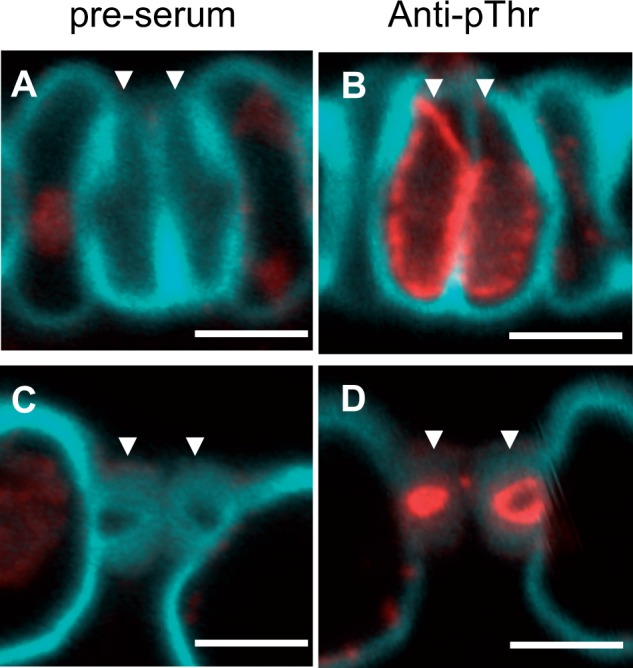
Detection of pT-type OSAs in rice guard cells. Subcellular localization of pT-type OSAs in guard cells was confirmed by immunostaining using (A, C) pre-immune serum or (B, D) anti-pThr against longitudinal sections of rice leaf blades. (A, B) The bulbous end and (C, D) central part of guard cells, respectively, were observed. Pre-immune serum was used as a negative control. Red and blue signals indicate phosphorylated penultimate threonine in OSAs and cell wall autofluorescence, respectively. A set of guard cells is represented by arrowheads in each panel. Scale bars = 5 μm.
Dysfunction of OSA7 results in impaired stomatal regulation
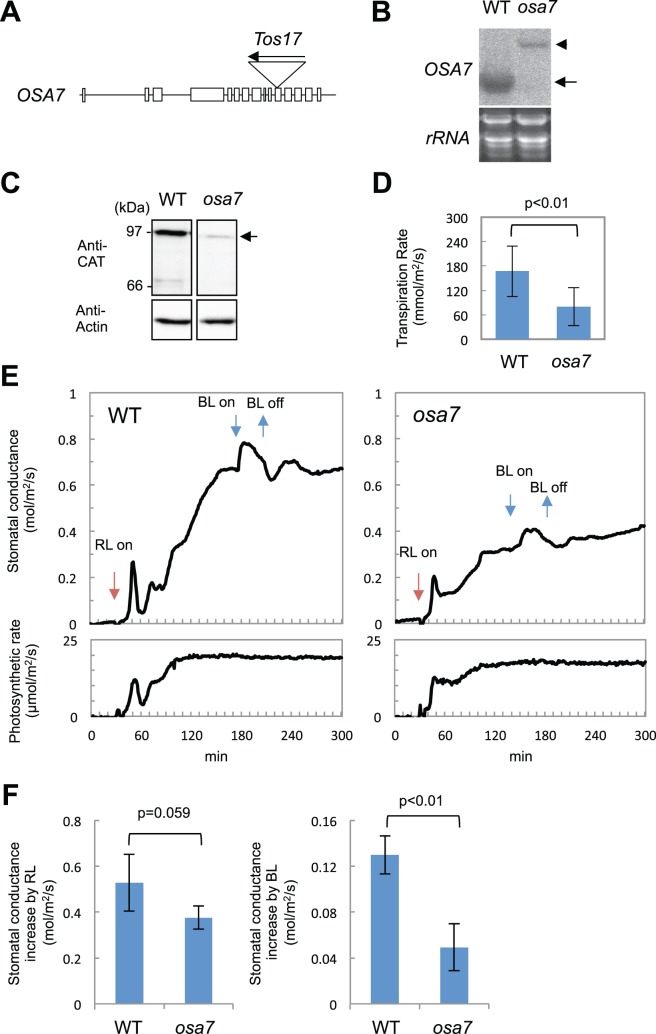
Based on the finding that OSA7 was preferentially expressed among the OSA genes that were detected in the GCEF (Fig. 4), we examined whether dysfunction of OSA7 affects stomatal movements. From the rice TOS17 retrotransposon mutant panel (Miyao et al. 2003), we obtained a rice mutant harboring a TOS17 insertion in the 11th exon of OSA7 (Fig. 6A). Northern blot analysis showed that the mRNA of OSA7 was undetectable in the osa7 homozygote, but expression of a longer truncated mRNA was confirmed (Fig. 6B). This transcript was predicted to harbor a premature stop codon and produce a protein of 686 amino acids (74 kDa) (Supplementary Fig. S6); however, this protein was not present in osa7 (Fig. 6C). Therefore, we concluded that osa7 is a null mutant. We note that the 97 kDa band, which was faintly detected in osa7, is possibly due to the presence of other isoforms of OSAs that have a molecular mass similar to that of OSA7.
Fig. 6.
osa7 displays defects in light-induced stomatal opening. (A) Schematic genome structure of OSA7. Lines indicate untranslated regions and introns. Exons are drawn as white boxes. Positions of the TOS17 insertion are shown. (B) Expression of OSA7 in leaf blades of the wild type and osa7 observed by Northern blot analysis. The arrow and arrowheads indicate the position of the native form and truncated form of OSA7 mRNA, respectively. rRNA confirms the equal loading of the total RNA. Data represent the results of two biological replicates. (C) Detection of OSA7 protein in leaf blades of the wild type and osa7. The arrow indicates the position of OSA7. Antibody recognizing actin protein (Anti-Actin) was used to confirm equal loading of total protein. Data represent the results of two biological replicates. (D) Transpiration rate of wild-type and osa7 leaf blades evaluated under daylight. Mean ± SD (n ≥ 7). The P-value indicates the Student’s t-test. (E) Light-induced increases in stomatal conductance in the wild type(left) and osa7 (right). Leaf blades of dark-adapted rice seedlings were subjected to measurement of stomatal conductance (top panels) and photosynthetic rate (bottom panels). Points of red light (RL) and blue light (BL) irradiation are indicated by arrows. Data represent the results of three biological replicates, each measured in the same day. RL, 700 μmol m−2 s−1 red light; BL, 3 μmol m−2 s−1 blue light. (F) Graphs quantifying the increase in the stomatal conductance by RL (left) and BL superimposed on RL (right), respectively. Stomatal conductance increase by RL was calculated by subtracting the stomatal conductance in dark conditions from stable stomatal conductance by RL. The increase in stomatal conductance induced by BL was calculated by subtracting the stable stomatal conductance by RL from the maximum stomatal conductance by BL in the background of RL. Mean ± SD (n = 3). P-values indicate the Student’s t-test.
When grown in a greenhouse, osa7 showed severe growth defects (Supplementary Fig. S7), and the majority of the osa7 blighted before reaching its reproductive stage. Therefore, we examined the phenotype of osa7 in seedlings that segregated from heterozygotes; the genotype of each segregant was confirmed by genomic PCR. We first found that the osa7 homozygote showed a low transpiration rate compared with the wild-type segregant under daylight in our growth conditions (Fig. 6D). To clarify whether the low transpiration rate in osa7 is the result of impairment of light-induced stomatal opening, we observed the response of stomatal conductance to short-term light irradiation. Under dark conditions, wild-type and osa7 seedlings both showed stomatal conductance close to zero, indicating that the majority of stomata in both genotypes are closed in such conditions (Fig. 6E). In the wild type, RL irradiation significantly enhanced stomatal conductance along with the increased photosynthetic rate (Fig. 6E, left). After stabilization of stomatal conductance under RL, BL was further superimposed. BL increased stomatal conductance without affecting the photosynthetic rate (Fig. 6E, left). In osa7, a similar increase in stomatal conductance was observed by both RL and BL; however, its amplitude was reduced (Fig. 6E, right). Reduction of the stomatal conductance increase by RL in osa7 was marginally significant (Fig. 6F, left), whereas that by BL in osa7 was significantly suppressed (Fig. 6F, right). These results suggest that OSA7 is involved in BL-induced stomatal opening of rice.
Discussion
We observed BL-induced stomatal opening in rice (Figs. 1B, 6E), consistent with a previous study (Shimazaki et al. 2007). Importantly, BL-dependent stomatal opening was inhibited by vanadate, an H+-ATPase inhibitor, but was promoted by FC, an H+-ATPase activator (Fig. 1B). Moreover, FC stimulated penultimate threonine phosphorylation of OSA (Fig. 3B), and phosphorylated penultimate threonine OSA was detected under light in guard cells (Fig. 5). We conclude that BL promotes stomatal opening via penultimate threonine phosphorylation of OSA in dumbbell-shaped guard cells.
Notably, the stomatal response of oat, which also harbors dumbbell-shaped guard cells, was similar to that of rice (Supplementary Fig. S1). Therefore, this mechanism is not specific to rice; it may occur in all monocot grasses with dumbbell-shaped guard cells. The presence of H+-ATPase-mediated stomatal opening in both monocots and dicots illustrates the evolutionary conservation of mechanisms that underlie stomatal regulation in the lineage of higher plants. Moreover, the mode of regulation in kidney-shaped and dumbbell-shaped guard cells may share the same signaling components. This includes upstream factors involved in the regulation of H+-ATPase identified in Arabidopsis, such as the BL receptor phototropin (phot1 and phot2) and its substrate kinase BLUS1 (Kinoshita et al. 2001, Takemiya et al. 2013). It has been reported that homologs of phototropin and BLUS1 are encoded in the rice genome (Kanegae et al. 2000, Takemiya et al. 2013), but their roles in stomatal regulation remain unclear. Further investigation is required to determine whether counterparts of these components in dumbbell-shaped guard cells also constitute the signaling pathway underlying BL-induced stomatal opening.
The pT-type H+-ATPase was first observed in the liverwort M. polymorpha, which is the oldest lineage of terrestrial plants (Okumura et al. 2012a). In such species, penultimate threonine phosphorylation is regulated in response to external signals, including light, sucrose and osmotic stress (Okumura et al., 2012a). In dicot Arabidopsis, on the other hand, penultimate threonine phosphorylation is involved in stomatal opening regulated by BL, flowering time factors, sucrose treatment and auxin-induced hypocotyl elongation (Kinoshita et al. 1999, Niittylä et al. 2007, Kinoshita et al. 2011, Takahashi et al. 2012). In this study, we showed that penultimate threonine phosphorylation is involved in BL-dependent stomatal regulation of monocot grass, exemplified by observations in rice and oat. Considering that penultimate threonine phosphorylation was observed in both dicots and monocots, it is suggested that this is one of the most widely adopted post-transcriptional regulation mechanisms that land plants have acquired.
In addition to penultimate threonine phosphorylation of H+-ATPase, other phosphorylation sites in the inhibitory domain of AHA2 have been shown to regulate its activity. Phosphorylation of Ser931 by a Ser/Thr kinase 5 (PKS5) is involved in repression of AHA2, associated with the adaptation to high external pH (Fuglsang et al. 2007), whereas Thr881 is involved in activation in response to sucrose treatment (Niittylä et al. 2007). More recently, Ser899 was reported to be controlled by a ligand–receptor pair, RALF/FERONIA, which mediates growth inhibition (Haruta et al. 2014). The existence of multiple phosphorylation sites illustrates the requirement for H+-ATPase activity to be precisely regulated. Because these sites are partially conserved in OSAs (Fig. 2A), similar regulation may also exist in rice. Indeed, phosphoproteomic analyses of rice root and shoot plasma membranes have identified threonine phosphorylation (corresponding to Thr881 of AHA2; Fig. 2A) in OSA3 (Whiteman et al. 2008). Moreover, OSAs are involved in broad physiological processes, such as adaptation to acid rain and low pH of the soil, adaptation to nitrogen availability, and phosphorus uptake and translocation (Chang et al. 2009, Zhu et al. 2009, Sperandio et al. 2011, E. Wang et al. 2014, Liang et al. 2015). Further investigations are required to clarify whether these regulations are also involved in the regulation of OSAs in stomatal opening or other physiological processes.
In this study, we found that disruption of OSA7 impaired BL-induced stomatal opening. However, the BL response was not completely lost (Fig. 6E, F). Therefore, it is suggested that other OSAs are also redundantly involved in the stomatal opening of rice. Indeed, expression of several pT-type OSA genes in addition to OSA7 was present in guard cells (OSA1, OSA2, OSA3, OSA5, OSA6 and OSA8 in Fig. 4). In Arabidopsis, expression of all AHA isoforms was confirmed in guard cell protoplasts (Ueno et al. 2005). The abundance of multiple isoforms of H+-ATPases in guard cells highlights the importance of H+-ATPase-dependent physiological processes in stomatal regulation.
In addition to the suppression of BL-induced stomatal opening, osa7 also shows severe growth defects (Supplementary Fig. S7). Because H+-ATPases are responsible for various physiological functions, such as cell elongation, nutrient uptake and translocation, and sucrose loading in phloem (reviewed in Sondergaard et al. 2004; Y. Wang et al. 2014b), dysfunction of such processes may account for the growth defects observed in osa7, as expression of OSA7 was ubiquitously observed (Fig. 4). Further analysis of osa7 will increase our understanding of the physiological processes involving rice H+-ATPases. We here note that technical issues such as a remarkable GC-rich region in the genomic sequence of OSA7, and poor callus formation in the osa7 homozygote, prevented us from carrying out the complementation assay of osa7. However, considering the fact that both transcript and protein of OSA7 were diminished in osa7, and that depression of BL-induced stomatal opening was linked to osa7 mutation, it is suggested that OSA7 is involved in the regulation of stomatal opening. Nonetheless, as OSA7 is considered a suitable material for genetic analysis to elucidate the diverse role of H+-ATPases in plants, continuous trials to reinforce the evidence of the involvement of OSA7 are underway.
In this research, we found that OSA7 shows predominant expression compared with other OSA genes in seedlings (Fig. 4). Reference to the public microarray database showed that OSA7 was ubiquitously and highly expressed in various tissues and stages compared with other isoforms (Supplementary Fig. S8). Thus, OSA7 is considered one of the major isoforms of rice H+-ATPases. Interestingly, Arabidopsis AHA1 and AHA2 and maize ZmHA1 and ZmHA2, which are the closest homologs of OSA7 (Fig. 2B), were strongly expressed in various tissues, resembling the expression profile of OSA7 (Supplementary Fig. S8). Expression analysis performed by Haruta et al. (2010) also showed that AHA1 and AHA2 are the most abundantly expressed isoforms in Arabidopsis. Moreover, co-suppression of PMA4, a tobacco homolog of OSA7, causes defects in plant growth, sugar transport, stomatal opening and photosynthesis, indicative of the involvement of PMA4 in pleiotropic physiological processes (Zhao et al. 2000). Our results suggest that within subfamily II, OSA7 and its homologs are the major H+-ATPases in higher plants.
Because CO2 uptake through the stomata is considered the limiting step for photosynthesis, manipulation of the stomata is an essential technique to enhance the photosynthetic activity of terrestrial plants. Recently, Y. Wang et al. (2014a) reported that guard cell-specific overexpression of AHA2 in Arabidopsis not only enhanced light-induced stomatal opening but also resulted in an increase in the photosynthetic rate and biomass. Given our observation that dumbbell-shaped guard cells are regulated in a similar manner to kidney-shaped guard cells, it is suggested that the pT-type H+-ATPase-targeted stomatal manipulation techniques developed in dicots are compatible in monocot crops. Further studies on the molecular mechanisms underlying the regulation of monocot stomata will allow us to develop new strategies to enhance plant biomass, which will contribute to a low carbon society.
Materials and Methods
Plant growth conditions
Rice wild-type (Oryza sativa cv. Nipponbare) and osa7 (TOS17 Line NE1507) seedlings were grown at 28 °C under a photoperiod of 14 h light/10 h dark or in a greenhouse at room temperature (25–32 °C). Oat (Avena sativa) seedlings were grown at 22 °C under a photoperiod of 16 h white light (50 μmol m−2 s−1)/8 h dark.
Classification of stomatal status
Using 5-day-old rice seedlings grown in a plant box with 1/2 MS agar medium and 7-day-old oat seedlings grown in vermiculite, leaf blades of dark-adapted seedlings were cut into small sections with scissors under dim red light and blended for 40 s in distilled water using a commercially available waring blender. Tissues were collected using a 58 μm nylon mesh and washed gently with water. Epidermal fractions were transferred to Petri dishes with observation buffer (10 mM KCl, 1 mM CaCl2, 0.4 M mannitol, 5 mM MES-KOH; pH 6.0). Samples were subjected to chemical treatments [FC, vanadate or dimethylsulfoxide (DMSO)] and/or light irradiation [RL (LED-R;EYELA) and/or BL (Stick-B-32)] for 3–4 h.
For the classification of rice and oat stomata, intact stomatal complexes in the epidermal layer with no mesophyll tissues attached under the stomata were selected for observation. Fully opened stomata with straight pore outlines of the long axis side as in Fig. 1A and Supplementary Fig. S1A were classified as Opened. Stomata in which the pore was not visible were classified as Closed. The third biological replicate of rice stomatal observation was performed in a blind test.
Phylogenetic analysis
Phylogenetic trees were constructed using MEGA5.2.2 and the maximum-likelihood method. Bootstrap values were obtained from 1,000 replications. Multiple sequence alignment was performed using the MUSCLE algorithm in MEGA5.2.2. Output data were visualized using GENEDOC (http://www.nrbsc.org/gfx/) and Figtree (http://tree.bio.ed.ac.uk/software/figtree). Maize H+-ATPases were obtained based on a Blastp search against the maize genome database (B73 RefGen_v3 sequence of maizeGDB, http://maizegdb.org) using the full-length sequence of AHA2.
Immunodetection of OSAs
Western blot analysis was performed as described previously (Hayashi et al. 2010). Immunohistochemical detection of OSAs was performed according to previous methods (Yamaji and Ma 2007) with minor modifications. Briefly, young leaf blades of 24-day-old rice seedlings grown in a greenhouse were harvested under light and placed in fixation buffer (4% paraformaldehyde, 60 mM sucrose and 50 mM cacodylic acid; pH 7.4) for 2 h at room temperature. Fixed samples were washed three times with phosphate-buffered saline (PBS) and embedded in 5% agar dissolved in PBS. Sections of 100 μm thickness were prepared using a vibratome (ZERO1; Dosaka EM) and placed on a glass slide. Samples were treated with enzyme solution (0.1% pectolyase and 0.3% Triton X-100 in PBS) for 2 h, washed three times with PBS, once with blocking solution (5% bovine serum albumin) for 10 min and further with primary antibody (anti-pThr) diluted to 1,000-fold in PBS overnight. On the second day, samples were washed three times with PBS, blocking solution for 10 min and secondary antibody (Alexa 545 diluted 1,000-fold in PBS) for 2 h. Finally, the samples were observed by confocal laser scanning microscopy (FV-10i; Olympus). Northern blot analysis was performed as described previously (Takahashi et al. 2007) with the probe amplified using the primers listed in Supplementary Table S1.
Tissue preparation and expression analysis
Preparation of the GCEF was performed as described previously (Huang et al. 2009) with minor modifications. Leaf blades of 5-day-old seedlings were cut into small sections with scissors and blended for 40 s in distilled water using a commercially available waring blender. Tissues were collected using a 58 μm nylon mesh and treated with 0.6 M mannitol for 15 min, and then with enzyme buffer (1.5% Cellulase RS, 0.75% Macerozyme R-10, 0.01% cordycepcin, 0.0033% actinomycin D, 10 mM CaCl2, 0.6 M mannitol, 10 mM MES-KOH; pH 5.7) for 5–8 h. Enzyme treatment was continued until the majority of mesophyll tissues detached from the epidermis. Finally, the GCEF was collected using the 58 μm nylon mesh. MCPs were prepared similarly; however, enzyme solution (1.5% Cellulase R-10, 0.75% Macerozyme R-10, 0.6 M mannitol, 10 mM MES-KOH; pH 5.7) was used and the flow-through filtered using a 58 μm nylon mesh was collected.
Total RNAs were extracted from tissues using an RNeasy Plant Mini kit (Qiagen). Reverse transcription followed by DNase treatment was performed using a Quantitect Rev transcription kit (Qiagen). First-strand cDNA was subjected to RT–PCR analysis with Primestar GXL (TAKARA) or to real-time RT–PCR with SYBR GREEN amplification master mix (Bio-rad) and StepOne (Applied Biosystems) using the primer sets listed in Supplementary Table S1. Specifically, primers for the expression analysis of OSA genes, 25SrRNA and UBQ5 were based on previous reports (Jain et al. 2006, Sperandio et al. 2011).
Measurement of transpiration rate and stomatal conductance
AP4 (Delta-T) was used to measure the transpiration rate of the abaxial side of the leaf blade. Calibration was performed at least once before the start of the measurement so that the measurement error was always kept below 5%. Evaluation of the photosynthetic rate and stomatal conductance using the LI-6400 system (Li-Cor) was performed as described previously (Ando et al. 2013). Briefly, the leaf blade of rice seedlings incubated in the dark overnight was clamped in a standard chamber and under dark treatment for 30 min. Then a strong RL (700 μmol m−2 s−1) was illuminated from the top of the chamber. After photosynthesis and stomatal conductance reached the steady state, a weak BL (3 μmol m−2 s−1) was added for 30 min. The leaf temperature and relative humidity in the chamber were kept constant at 24 °C and 65–70%, respectively.
Accession numbers
Sequence data from this article can be found in GenBank/EMBL, RAP-DB (http://rapdb.dna.affrc.go.jp) or the maizeGDB (http://maizegdb.org) under the accession numbers listed in Supplementary Table S2.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japan Science and Technology Agency [the Advanced Low Carbon Technology Research and Development Program]; Okayama University Institute of Plant Science and Resources [International Joint Usage/Research Center Project]; the Ministry of Education, Culture, Sports, Science and Technology of Japan [Scientific Research on Priority Areas (15H059555, 15H059556 and 22119005)]; the Japan Society for the Promotion of Science [KAKENHI 15H04386 to T.K.and 23780047 to A.T.].
Supplementary Material
Acknowledgments
We thank Y. Kamiya for the initial evaluation of anti-CAT and anti-pThr against rice, Dr. Y. Tsuchiya and E. Ando for helpful discussions, T. Kitaoka for technical assistance in general, Ms. S. Hatanaka and FEI for the assistance with ESEM observations, and M. Okumura for providing amino acid sequences of MpHAs, PpHA7, and CrHA.
Glossary
Abbreviations
- AHA
Arabidopsis plasma membrane H+-ATPase
- BL
blue light
- CAT domain
catalytic domain
- FC
fusicoccin
- GCEF
guard cell-enriched fraction
- MCP
mesophyll cell protoplast
- OSA
Oryza sativa plasma membrane H+-ATPase
- PBS
phosphate-buffered saline
- RL
red light
- RT–PCR
reverse transcription–PCR
Disclosures
The authors have no conflicts of interest to declare.
References
- Amodeo G., Srivastava A., Zeiger E. (1992) Vanadate inhibits blue light-stimulated swelling of Vicia guard cell protoplasts. Plant Physiol. 100: 1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando E., Ohnishi M., Wang Y., Matsushita T., Watanabe A., Hayashi Y., et al. (2013) TWIN SISTER OF FT, GIGANTEA, and CONSTANS have a positive but indirect effect on blue light-induced stomatal opening in Arabidopsis. Plant Physiol. 162: 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango M., Gévaudant F., Oufattole M., Boutry M. (2003) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216: 355–365. [DOI] [PubMed] [Google Scholar]
- Assmann S.M., Grantz D.A. (1990) Stomatal response to humidity in sugarcane and soybean: effect of vapour pressure difference on the kinetics of the blue light response. Plant Cell Environ. 13: 163–169. [Google Scholar]
- Assmann S.M., Shimazaki K. (1999) The multisensory guard cell. Stomatal responses to blue light and abscisic acid. Plant Physiol. 119: 809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann S.M., Simoncini L., Schroeder J.I. (1985) Blue light activates electrogenic ion pumping in guard cell protoplasts of Vicia faba. Nature 318: 285–287. [Google Scholar]
- Axelsen K.B., Venema K., Jahn T., Baunsgaard L., Palmgren M.G. (1999) Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38: 7227–7234. [DOI] [PubMed] [Google Scholar]
- Baxter I., Tchieu J., Sussman M.R., Boutry M., Palmgren M.G., Gribskov M., et al. (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 132: 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchsenschütz K., Marten I., Becker D., Philippar K., Ache P., Hedrich R. (2005) Differential expression of K+ channels between guard cells and subsidiary cells within the maize stomatal complex. Planta 222: 968–976. [DOI] [PubMed] [Google Scholar]
- Camoni L., Iori V., Marra M., Aducci P. (2000) Phosphorylation-dependent interaction between plant plasma membrane H(+)-ATPase and 14-3-3 proteins. J. Biol. Chem. 275: 9919–9923. [DOI] [PubMed] [Google Scholar]
- Chang C., Hu Y., Sun S., Zhu Y., Ma G., Xu G. (2009) Proton pump OsA8 is linked to phosphorus uptake and translocation in rice. J. Exp. Bot. 60: 557–565. [DOI] [PubMed] [Google Scholar]
- Fuglsang A.T., Guo Y., Cuin T.A., Qiu Q., Song C., Kristiansen K.A., et al. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S., Jacobs M., Taiz L. (1982) Inhibition of stomatal opening in Vicia faba epidermal tissue by vanadate and abscisic acid. Plant Sci. Lett. 28: 63–72. [Google Scholar]
- Haruta M., Burch H.L., Nelson R.B., Barrett-Wilt G., Kline K.G., Mohsin S.B., et al. (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 285: 17918–17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Sugimoto M., Higaki T., Yaeno T., Nagami A., Irie M., Fujimi M., et al. (2013) A Munc13-like protein in Arabidopsis mediates H+-ATPase translocation that is essential for stomatal responses. Nat. Commun. 4: 2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Nakamura S., Takemiya A., Takahashi Y., Shimazaki K., Kinoshita T. (2010) Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol. 51: 1186–1196. [DOI] [PubMed] [Google Scholar]
- Hwang H., Yoon J., Kim H.Y., Min M.K., Kim J.A., Choi E.H., et al. (2013) Unique features of two potassium channels, OsKAT2 and OsKAT3, expressed in rice guard cells. PLoS One 8: e72541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Tyagi A.K., Khurana J.P. (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345: 646–651. [DOI] [PubMed] [Google Scholar]
- Kanegae H., Tahir M., Savazzini F., Yamamoto K., Yano M., Sasaki T., et al. (2000) Rice NPH1 homologs, OsNPH1a and OsNPH1b, are differently photoregulated. Plant Cell Physiol. 41: 415–423. [DOI] [PubMed] [Google Scholar]
- Karlsson P.E. (1986) Blue light regulation of stomata in wheat seedlings. I. Influence of red background illumination and initial conductance level. Physiol. Plant. 66: 202–206. [Google Scholar]
- Kinoshita T., Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. (2001) phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Ono N., Hayashi Y., Morimoto S., Nakamura S., Soda M., et al. (2011) FLOWERING LOCUS T regulates stomatal opening. Curr. Biol. 21: 1232–1238. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Shimazaki K. (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J. 18: 5548–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Shimazaki K. (2001) Analysis of the phosphorylation level in guard-cell plasma membrane H+-ATPase in response to fusicoccin. Plant Cell Physiol. 42: 424–432. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Shimazaki K. (2002) Biochemical evidence for the requirement of 14-3-3 protein binding in activation of the guard-cell plasma membrane H+-ATPase by blue light. Plant Cell Physiol. 43: 1359–1365. [DOI] [PubMed] [Google Scholar]
- Kollist H., Nuhkat M., Roelfsema M.R.G. (2014) Closing gaps: linking elements that control stomatal movement. New Phytol. 203: 44–62. [DOI] [PubMed] [Google Scholar]
- Liang C., Ge Y., Su L., Bu J. (2015) Response of plasma membrane H+-ATPase in rice (Oryza sativa) seedlings to simulated acid rain. Environ. Sci. Pollut. Res. Int. 22: 535–545. [DOI] [PubMed] [Google Scholar]
- Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., et al. (2000) A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275: 17762–17770. [DOI] [PubMed] [Google Scholar]
- Merlot S., Leonhardt N., Fenzi F., Valon C., Costa M., Piette L., et al. (2007) Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26: 3216–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A., Tanaka K., Murata K., Sawaki H., Takeda S., Abe K., et al. (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P., de Kerchove d’Exaerde A., De Meester S., Thinès D., Goffeau A., Boutry M. (1996) Single point mutations in various domains of a plant plasma membrane H+-ATPase expressed in Saccharomyces cerevisiae increase H+-pumping and permit yeast growth at low pH. EMBO J. 15: 5513–5526. [PMC free article] [PubMed] [Google Scholar]
- Niittyla T., Fuglsang A.T., Palmgren M.G., Frommer W.B., Schulze W.X. (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics 6: 1711–1726. [DOI] [PubMed] [Google Scholar]
- Okumura M., Inoue S., Takahashi K., Ishizaki K., Kohchi T., Kinoshita T. (2012a) Characterization of the plasma membrane H+-ATPase in the liverwort Marchantia polymorpha. Plant Physiol. 159: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M., Takahashi K., Inoue S., Kinoshita T. (2012b) Evolutionary appearance of the plasma membrane H+-ATPase containing a penultimate threonine in the bryophyte. Plant Signal. Behav. 7: 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A., Svennelid F., Ek B., Sommarin M., Larsson C. (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 118: 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren M.G. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 817–845. [DOI] [PubMed] [Google Scholar]
- Palmgren M.G., Larsson C., Sommarin M. (1990) Proteolytic activation of the plant plasma membrane H+-ATPase by removal of a terminal segment. J. Biol. Chem. 265: 13423–13426. [PubMed] [Google Scholar]
- Raschke K., Fellows M.P. (1971) Stomatal movement in Zea mays: shuttle of potassium and chloride between guard cells and subsidiary cells. Planta 101: 296–316. [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Allen G.J., Hugouvieux V., Kwak J.M., Waner D. (2001) Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 627–658. [DOI] [PubMed] [Google Scholar]
- Shimazaki K., Doi M, Assmann S.M., Kinoshita T. (2007) Light regulation of stomatal movement. Annu. Rev. Plant Biol. 58: 219–247. [DOI] [PubMed] [Google Scholar]
- Shimazaki K., Goh C., Kinoshita T. (1999) Involvement of intracellular Ca2+ in blue light-dependent proton pumping in guard cell protoplasts from Vicia faba. Physiol. Plant. 105: 554–561. [Google Scholar]
- Shimazaki K., Iino M., Zeiger E. (1986) Blue light-dependent proton extrusion by guard-cell protoplasts of Vicia faba. Nature 319: 324–326. [Google Scholar]
- Sondergaard T.E., Schulz A., Palmgren M.G. (2004) Energization of transport processes in plants. Roles of the plasma membrane H+-ATPase. Plant Physiol. 136: 2475–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A.K., Ren H., Park M.Y., Grandt K.N., Lee S.H., Murphy A.S., et al. (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio M.V.L., Santos L.A., Bucher C.A., Fernandes M.S., de Souza S.R. (2011) Isoforms of plasma membrane H+-ATPase in rice root and shoot are differentially induced by starvation and resupply of NO3– or NH4+. Plant Sci. 180: 251–258. [DOI] [PubMed] [Google Scholar]
- Svennelid F., Olsson A., Piotrowski M., Rosenquist M., Ottman C., Larsson C., et al. (1999) Phosphorylation of Thr-948 at the C-terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11: 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Agrawal G., Yamazaki M., Onosato K., Miyao A., Kawasaki T., et al. (2007) Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell 19: 2940–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Hayashi K., Kinoshita T. (2012) Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 159: 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A., Sugiyama N., Fujimoto H., Tsutsumi T., Yamauchi S., Hiyama A., et al. (2013) Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat. Commun. 4: 2094. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Sugano S.S., Shimada T., Hara-Nishimura I. (2013) Enhancement of leaf photosynthetic capacity through increased stomatal density in Arabidopsis. New Phytol. 198: 757–764. [DOI] [PubMed] [Google Scholar]
- Ueno K., Kinoshita T., Inoue S., Emi T., Shimazaki K. (2005) Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 46: 955–963. [DOI] [PubMed] [Google Scholar]
- Wang E., Yu N., Bano S., Liu C., Miller A.J., Cousins D., et al. (2014) A H+-ATPase that energizes nutrient uptake during mycorrhizal symbioses in rice and Medicago truncatula. Plant Cell 26: 1818–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Noguchi K., Ono N., Inoue S., Terashima I., Kinoshita T. (2014a) Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc. Natl. Acad. Sci. USA 111: 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shimazaki K., Kinoshita T. (2014b) Multiple roles of the plasma membrane H+-ATPase and its regulation. Enzymes 35: 191–211. [DOI] [PubMed] [Google Scholar]
- Whiteman S.A., Nühse T.S., Ashford D.A., Sanders D., Maathuis F.J.M. (2008) A proteomic and phosphoproteomic analysis of Oryza sativa plasma membrane and vacuolar membrane. Plant J. 56: 146–156. [DOI] [PubMed] [Google Scholar]
- Yamaji N., Ma J.F. (2007) Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 143: 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Dielen V., Kinet J.M., Boutry M. (2000) Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell 12: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Di T., Xu G., Chen X., Zeng H., Yan F., et al. (2009) Adaptation of plasma membrane H+-ATPase of rice roots to low pH as related to ammonium nutrition. Plant Cell Environ. 32: 1428–1440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.