Structure-function analysis reveals domains in LORELEI critical for its role in pollen tube reception at the synergid surface, which it performs in conjunction with the receptor-like kinase FERONIA.
Abstract
In angiosperms, pollen tube reception by the female gametophyte is required for sperm release and double fertilization. In Arabidopsis thaliana lorelei (lre) mutants, pollen tube reception fails in most female gametophytes, which thus remain unfertilized. LRE encodes a putative glycosylphosphatidylinositol (GPI)-anchored surface protein with a modified eight-cysteine motif (M8CM). LRE fused to citrine yellow fluorescent protein (LRE-cYFP) remains functional and localizes to the synergid plasma membrane-rich filiform apparatus, the first point of contact between the pollen tube and the female gametophyte. Structure-function analysis using LRE-cYFP showed that the role of LRE in pollen tube reception requires the M8CM, but not the domains required for GPI anchor addition. Consistently, LRE-cYFP-TM, where GPI anchor addition domains were replaced with a single-pass transmembrane domain, fully complemented the pollen tube reception defect in lre-7 female gametophytes. Ectopically expressed and delivered LRE-cYFP from pollen tubes could non-cell-autonomously complement the pollen tube reception defect in lre female gametophytes, only if they expressed FERONIA. Additionally, pollen tube-expressing LRE variants lacking domains critical for GPI anchor addition also rescued lre female gametophyte function. Therefore, LRE and FERONIA jointly function in pollen tube reception at the interface of the synergid cell and pollen tube.
INTRODUCTION
The success of plant reproduction depends on a series of cell-cell interactions between the male and female gametophytes (Kessler and Grossniklaus, 2011; Palanivelu and Tsukamoto, 2012; Beale and Johnson, 2013; Qu et al., 2015). In Arabidopsis thaliana, the mature male gametophyte (pollen) consists of two sperm cells and a vegetative cell (Twell, 2011), whereas the female gametophyte consists of five accessory cells (three antipodal cells and two synergids) and two gametic cells (one egg cell and one central cell) (Yadegari and Drews, 2004; Yang et al., 2010; Song et al., 2014). Synergids facilitate the fusion of one sperm cell to the egg cell and one sperm cell to the central cell to complete double fertilization, forming an embryo and endosperm, respectively (Russell, 1992). Synergids are specialized secretory cells with a unique structure called the filiform apparatus (FA) at the micropylar pole. In the FA, the plasma membrane surface area is vastly increased by many finger-like projections, which extend into the synergid cytoplasm (Mansfield et al., 1991; Huang and Russell, 1992; Kasahara et al., 2005). Thus, the unique structure of the FA increases the area of contact between the pollen tube and the synergid cell and facilitates secretion of small peptides to influence pollen tube attraction and pollen tube reception, which is defined as pollen tube burst and release of sperm cells for double fertilization (Russell, 1992; Huck et al., 2003; Okuda et al., 2009; Amien et al., 2010; Takeuchi and Higashiyama, 2012).
In Arabidopsis, genes expressed in female and male gametophytes regulate pollen tube reception. For example, the female gametophyte-expressed gene FERONIA (FER) encodes a receptor-like kinase. The fer female gametophyte fails to induce pollen tube reception and consequently remains unfertilized (Huck et al., 2003; Escobar-Restrepo et al., 2007; Haruta et al., 2014). Many mutants cause a similar phenotype due to defects in the female gametophyte: these include scylla (syl), lorelei (lre), nortia (nta), evan, and turan (Capron et al., 2008; Rotman et al., 2008; Kessler et al., 2010; Tsukamoto et al., 2010; Lindner et al., 2015). Of these, the identity of SYL remains unknown. LRE encodes a putative glycosylphosphatidylinositol (GPI)-anchored surface protein (Capron et al., 2008; Tsukamoto et al., 2010), and NTA encodes a transmembrane protein, a member of the Mildew Resistance Locus O family (Kessler et al., 2010). LRE and NTA function with FER in pollen tube reception (Kessler et al., 2010; Duan et al., 2014; Li et al., 2015; also see below). TURAN and EVAN may N-glycosylate synergid-expressed proteins that are involved in pollen tube reception (Lindner et al., 2015). Also, maize (Zea mays) EMBRYO SAC4 induces pollen tube burst via opening of the potassium channel KZM1 in the pollen tube (Amien et al., 2010).
In the male gametophyte, transcription factor genes MYB97, MYB101, and MYB120 are preferentially induced in pollen tubes growing through pistils (Qin et al., 2009; Leydon et al., 2013). In the triple myb mutant, the pollen tube coils in the female gametophyte and fails to discharge sperm cells (Leydon et al., 2013; Liang et al., 2013), demonstrating an active role for the pollen tube in pollen tube reception. Loss of AUTO-INHIBITED Ca2+ ATPASE9 (ACA9), which encodes a calcium efflux pump, results in some pollen tubes failing to discharge the sperm, even though they reach the synergid and arrest growth in the synergid cell; thus, aca9 tubes differ from triple myb mutant tubes in that they fail to coil and burst (Schiøtt et al., 2004). Interestingly, abstinence by mutual consent (amc) mutants show pollen tube reception defects only when an amc pollen tube encounters an amc female gametophyte, indicating that pollen tube reception requires interactions between the male and the female gametophyte and that both gametophytes share common signaling components (Boisson-Dernier et al., 2008).
Among the proteins involved in pollen tube reception, FER and LRE function together in the FER-RAC/ROP signaling complex to regulate reactive oxygen species (ROS) production in the micropylar region of female gametophytes (Duan et al., 2014). Additionally, ROS produced in the female gametophyte are important for pollen tube reception, and ROS-mediated pollen tube rupture requires calcium (Duan et al., 2014). Indeed, changes in cytoplasmic calcium ([Ca2+]cyto) observed during pollen tube-synergid interactions (Iwano et al., 2012; Denninger et al., 2014; Hamamura et al., 2014) showed that the initiation of calcium oscillations and the increase in [Ca2+]cyto are affected in fer and lre mutant synergids (Ngo et al., 2014). In addition to affecting ROS and calcium production in ovules, LRE also has an intracellular role in chaperoning FER from the endoplasmic reticulum (ER) to the FA of the synergids (Li et al., 2015). FER and NTA also function together in pollen tube reception: The preferential relocalization of NTA from puncta in synergids to the FA upon pollen tube arrival requires FER (Kessler et al., 2010), indicating that NTA functions downstream of FER. Calcium may also play an important role in mediating the function of NTA. Unlike FER and LRE, NTA is required to modulate the magnitude of the calcium signatures in the synergids and therefore likely functions downstream of FER and LRE (Ngo et al., 2014).
LRE is expressed in ovules but not in the pollen or pollen tubes (Tsukamoto et al., 2010). Consistent with this, lre mutants only showed defects in the female gametophyte (Capron et al., 2008; Tsukamoto et al., 2010). LRE encodes a putative GPI-anchored surface protein. In eukaryotes, GPI-anchored pre-pro-proteins (nascent full-length protein) usually contain an N-terminal signal peptide (SP) for ER translocation, a proline-rich unstructured ω-11 region, and two domains that are critical for addition of the GPI anchor: (1) the ω-site region containing four tiny residues, one of which will serve as the ω site, and (2) a hydrophobic tail at the C terminus of the protein (Eisenhaber et al., 1998, 2003). In the ER lumen, the transamidase cleaves the pro-protein (from which the SP has been removed) at the ω site, removes the hydrophobic tail, and covalently links the remainder of the pro-protein to a GPI anchor, which is synthesized in the ER. Subsequently, the mature protein with the GPI-anchor traffics through the endomembrane system to the cell surface, where it associates with the plasma membrane using the GPI anchor (Varma and Mayor, 1998; Mayor and Riezman, 2004).
Consistent with the prediction that the GPI anchors LRE to the membrane, transiently expressed GFP-LRE localized to the surface of Arabidopsis mesophyll protoplasts (Capron et al., 2008) and citrine yellow fluorescent protein (cYFP)-LRE localized to the FA (Lindner et al., 2015). However, whether the predicted GPI anchor addition domains in LRE are important for its function in pollen tube reception has not been experimentally tested. LRE interacts with an extracellular domain of FER in yeast two-hybrid and pull-down assays, and FER-GFP localization in the FA is affected in lre mutants (Li et al., 2015). These results and characterization of LLG1, a putative paralog of LRE, indicate that LRE/LLG1 interacts with FER in the ER lumen and functions as a chaperone to bring FER to the FA (Li et al., 2015). Once localized to the FA, LRE could function as a coreceptor with FER to perceive signals from the pollen tube to trigger calcium profile changes and ROS production (Denninger et al., 2014; Duan et al., 2014; Ngo et al., 2014). However, the amino acid residues in LRE that are important for it to function in pollen tube reception are not yet clear. Additionally, whether LRE has a separate function at the synergid cell surface, which is independent of its intracellular function in the synergids, remains to be established.
We hypothesized that LRE localizes to the synergid cell surface using a GPI anchor and regulates pollen tube reception. Our analysis showed that the predicted GPI anchor addition domains in LRE are critical for its localization in the FA but not its function. We discovered that a unique 12-amino acid domain between the 5th and the 6th cysteines, including the highly conserved Asn-Asp dipeptide, is critical for LRE function in pollen tube reception. Finally, non-cell-autonomous and extracellular complementation of lre synergids with pollen tube-expressed LRE demonstrated that LRE and FER have a synergid cell surface-specific function in pollen tube reception.
RESULTS
LRE Encodes a Cysteine-Rich Protein with Domains Implicated in GPI Anchor Addition
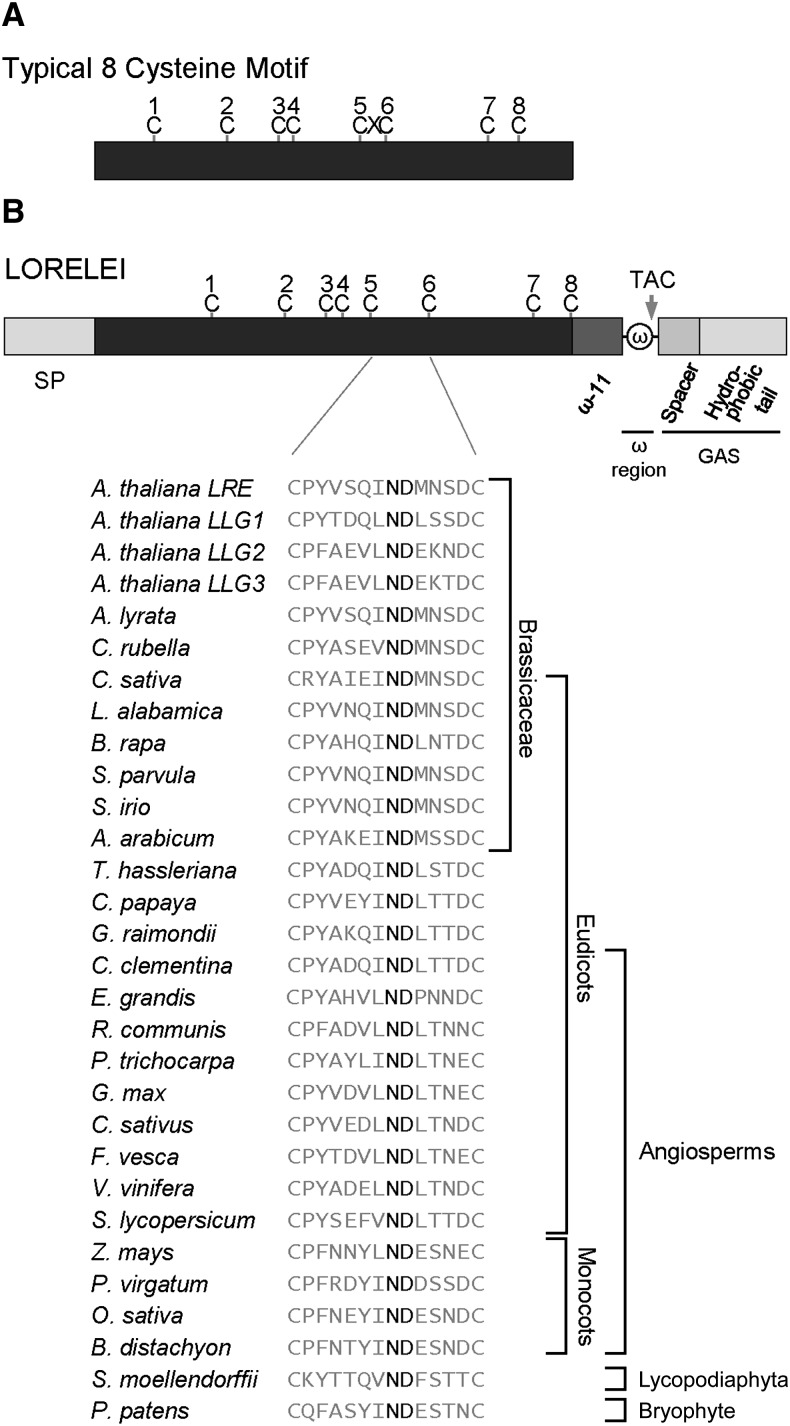
LRE encodes a putative GPI-anchored surface protein (Capron et al., 2008; Tsukamoto et al., 2010), as it has the domains predicted to be required for GPI anchor addition (ω-site region and GAS domain; Figure 1; Supplemental Figure 1). Additional protein sequence analysis revealed that LRE encodes a cysteine-rich protein (CRP) with eight cysteines in the ectodomain, similar to proteins containing an eight-cysteine motif (8CM; José-Estanyol et al., 2004). The cysteines in an 8CM protein form disulfide bridges to maintain the tertiary structure of a scaffold containing conserved helical regions connected by variable loops, which confer functional specificity to the protein (José-Estanyol et al., 2004). In a typical 8CM, one amino acid separates the 5th and the 6th cysteines; by contrast, in LRE, a unique 12-amino acid domain separates them (Figure 1; Supplemental Figure 1). Sequence alignment showed conservation of spacing of the eight cysteines and the 12-amino acid domain between the 5th and the 6th cysteines (Supplemental Figure 1). Interestingly, this 12-amino acid domain is relatively variable with a highly conserved Asn-Asp dipeptide (Figure 1; Supplemental Figure 1), suggesting that the Asn-Asp dipeptide is critical for LRE function. Thus, LRE encodes a putative GPI-anchored surface protein with a modified 8CM (M8CM).
Figure 1.
LRE Encodes a Putative GPI-Anchored Surface Protein with a M8CM.
(A) Diagram of a typical 8CM, in which the 3rd and 4th cysteines are adjacent to each other and the 5th and 6th cysteines are separated by one amino acid. C, cysteine; X, any other amino acid.
(B) Diagram of the LRE pre-pro protein. Predicted domains critical for GPI anchor addition to the pro-protein and positions of eight cysteines in LRE are indicated. A unique domain of 12-amino acids between the 5th and the 6th cysteines in LRE from Arabidopsis and in proteins from indicated plants that share the highest sequence similarity with full-length Arabidopsis LRE are shown. The highly conserved Asn-Asp dipeptide (ND) in this 12-amino acid domain is shown in bold. Multiple sequence alignment of full-length protein sequences shown here is presented in Supplemental Figure 1. ω, omega site; GAS, GPI attachment signal with a spacer region (darker gray) and a hydrophobic tail (lighter gray). Arrow points to the omega site cleaved by the transamidase complex (TAC).
Polar Localization of LRE-cYFP in the Filiform Apparatus of Synergids
To determine if the predicted GPI anchor addition domains and the M8CM in LRE are necessary for its function, we generated LRE-cYFP fusions for structure-function analysis. We used cYFP as the reporter because it is pH insensitive (Griesbeck et al., 2001) and can fluoresce even in the acidic apoplast (Gjetting et al., 2012), where the ectodomain of GPI-anchored surface proteins is expected to localize (Schultz et al., 1998). Additionally, cYFP was used to determine plasma membrane localization of other Arabidopsis GPI-anchored surface proteins (Simpson et al., 2009).
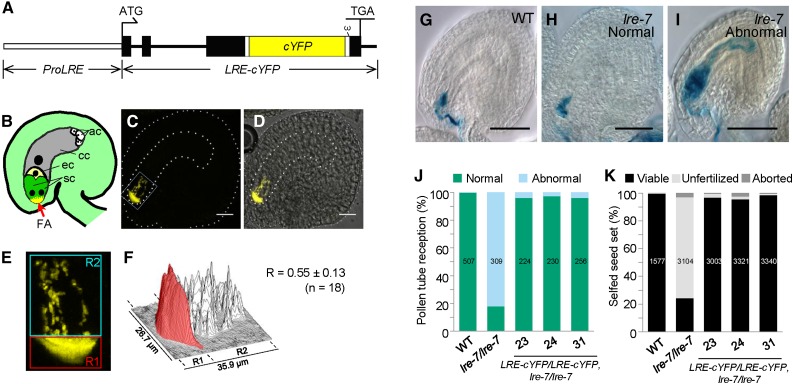
We generated N-terminal (ProLRE:cYFP-LRE) and C-terminal (ProLRE:LRE-cYFP) constructs (Figure 2A; Supplemental Figure 2A), determined the subcellular localization of LRE-reporter fusion proteins in the synergids, and verified that they retained LRE function. Although ProLRE:cYFP-LRE (N-terminal fusion construct) complemented the reduced seed set defect in lre-7/lre-7 plants (Supplemental Figure 2D), cYFP expression was not detected in unfertilized ovules of all 14 primary transformants (T1) analyzed, and these were not characterized further. However, T1 plants carrying ProLRE:LRE-cYFP (C-terminal fusion construct) expressed cYFP in synergids (Supplemental Figures 3A, 3B, and 3I; also see below) and complemented the reduced seed set defect in lre-7/lre-7 plants (Supplemental Figure 2D). We isolated three single-insertion lines in the T2 generation (Supplemental Table 1), identified plants that are homozygous for the transgene, and used these to analyze the subcellular localization and function of LRE-cYFP in greater detail; this approach was also undertaken for other constructs reported here (see below).
Figure 2.
LRE-cYFP Shows a Polarized Localization in the Filiform Apparatus and Is Fully Functional in Pollen Tube Reception.
(A) Diagram of ProLRE:LRE-cYFP in which the cYFP is placed toward the C-terminal portion of the LRE protein and the LRE-cYFP is expressed from the LRE promoter (ProLRE, open rectangle). Filled black rectangles and black lines refer to exons and introns, respectively, in the LRE gene. ω, omega site; ATG and TGA, the start and stop codons, respectively, in LRE. The small white rectangle that appears before and after cYFP points to 9-amino acid and 11-amino acid linkers, respectively.
(B) Diagram of an unfertilized Arabidopsis ovule containing a mature female gametophyte. sc, synergid cell; ec, egg cell; cc, central cell; ac, antipodal cell.
(C) A representative fluorescent image showing LRE-cYFP localization in an ovule. The female gametophyte and the ovule are outlined with thick and thin white dashed lines, respectively. Bar = 20 µm.
(D) A merged image of fluorescent image shown in (C) and a bright-field image (data not shown) of the same ovule. Bar = 20 µm.
(E) A close-up view of the portion of the ovule within the white rectangle in (C). cYFP signal in the region of the filiform apparatus (red box) and the remainder of the synergids (cyan box) were quantified. R1 and R2, regions of interest 1 and 2, respectively.
(F) Surface plot of quantification of cYFP signal intensity within the boxed areas in (E). cYFP signal intensity in the filiform apparatus region is highlighted in red. R (mean ± sd) indicates the proportion of cYFP signal intensity in the filiform apparatus relative to the total cYFP signal intensity in the synergids [R = R1/(R1 + R2)]. n, number of images, from three independent transformants ([J] and [K]), used to calculate R.
(G) to (I) Representative images showing pollen tube reception in ovules of indicated genotype when crossed to pLAT52:GUS pollen. This GUS staining-based assay was used to score pollen tube reception (Figures 2J, 3F, 4F, and 5D). Bar = 50 µm.
(G) Normal pollen tube reception in a wild-type ovule.
(H) Normal pollen tube reception in an lre-7 ovule.
(I) Abnormal pollen tube reception (pollen tube coiling due to overgrowth) in an lre-7 ovule.
(J) and (K) The ProLRE:LRE-cYFP construct fully complements the pollen tube reception defect (J) and the reduced seed set defect (K) in lre-7/lre-7 plants. Total number of ovules (J) or seeds (K) analyzed are in the center of each column. Number below each column, three lines used that are single insertion and homozygous for the ProLRE:LRE-cYFP transgene in the lre-7/lre-7 background.
Confocal laser scanning microscopy showed that LRE-cYFP localized at the micropylar end of synergids (Figures 2B to 2F), where the FA is located (Huang and Russell, 1992; Kasahara et al., 2005). Indeed, the FA region contained nearly 56% of the total cYFP signal in synergids (Figures 2E and 2F). This is in contrast to EGFP-ROP6C, a membrane protein that localizes both in the FA and outside the FA, throughout the synergid plasma membrane (Escobar-Restrepo et al., 2007). Polar localization of LRE-cYFP in the FA is consistent with LRE being a putative GPI-anchored surface protein that functions in pollen tube reception (Capron et al., 2008; Tsukamoto et al., 2010). Besides its polarized localization in the FA, we consistently detected LRE-cYFP fluorescence in puncta in the synergid cell cytoplasm (Figures 2B to 2F). To explore the intracellular localization of LRE-cYFP, we performed colocalization experiments with an organelle marker (Golgi, ER, or peroxisome; Nelson et al., 2007) expressed under the control of the LRE promoter (Supplemental Figure 4A). None of these organelle markers colocalized with LRE-cYFP puncta in the synergids (Supplemental Figures 4B to 4M), and the identity of intracellular LRE-cYFP puncta remains unknown.
Upon pollen tube arrival in the female gametophyte, NTA-GFP relocalizes from the fluorescent puncta in the cytoplasm to the FA of synergids (Kessler et al., 2010). To test if LRE-cYFP localization also changes upon pollen tube arrival, ProLRE:LRE-cYFP pistils were pollinated with pollen carrying ProLAT52:DsRed. The localization of LRE-cYFP did not change in the FA after pollen tube reception (Supplemental Figures 3C to 3H and 3J).
LRE-cYFP Functions in Pollen Tube Reception
To test if LRE-cYFP is functional, we performed four experiments. Using a GUS staining-based assay (Tsukamoto et al., 2010), we showed that LRE-cYFP fully complemented the pollen tube reception defect in the lre-7 female gametophytes (Figures 2G to 2J). Additionally, the seed set in lre-7/lre-7 was also fully restored to wild-type levels (Figure 2K), indicating that LRE-cYFP is functional.
If LRE-cYFP is functional, it should also rescue reduced transmission of the lre mutation through the female gametophyte (Capron et al., 2008; Tsukamoto et al., 2010). Indeed, in lines containing ProLRE:LRE-cYFP, the transmission efficiency of the lre-7 mutation increased from 0.11 (11%) to nearly 1 (∼100% transmission; Table 1). Additionally, when the transgenic plant was the female parent, the progeny of the cross revealed an increased transmission of ProLRE:LRE-cYFP, showing that LRE-cYFP complemented the defects in the lre-7 female gametophyte (Supplemental Table 1); no such increase in transmission was observed when the transgenic plant was the male parent (Supplemental Table 1). Taken together, our results demonstrated that LRE-cYFP is functional.
Table 1. ProLRE:LRE-cYFP Restored the Reduction in Transmission of the lre-7 Mutation through the Female Gametophyte.
| Female Parenta | Male Parenta | Observed No. of Progeny |
TE (R/S)b | χ2c | P Value | |
|---|---|---|---|---|---|---|
| BastaRd | BastaSd | |||||
| Wild type | lre-7/+ | 180 | 164 | 1.10 | 0.740 | 0.388 |
| lre-7/+ | Wild type | 22 | 194 | 0.11 | 137.000 | <0.001 |
| Line 23 | Wild type | 141 | 140 | 1.01e | 0.004 | 0.952 |
| Wild type | Line 23 | 138 | 131 | 1.05 | 0.180 | 0.670 |
Line numbers refer to a ProLRE:LRE-cYFP transformant in the lre-7/lre-7 background containing a single insertion of the ProLRE:LRE-cYFP transgene. Genotype of the transgenic line used is homozygous for the transgene (ProLRE:LRE-cYFP/ProLRE:LRE-cYFP) and heterozygous for the lre-7 mutation (lre-7/+).
Transmission efficiency (TE) was calculated as the ratio of Basta resistance (R) to susceptibility (S) in the progeny of the indicated cross.
χ2 was calculated based on the expectation of a 1:1 segregation of Basta resistance to susceptibility in the progeny of a cross between the wild type and lre-7/+.
Basta-resistant (BastaR) and Basta-susceptible (BastaS) progeny. Basta resistance gene is linked with the T-DNA that is inserted into the LRE gene in lre-7 mutant.
When segregation ratio in row three was compared to that in row two as the expected segregation ratio, the χ2 = 491.31 and P value < 0.001.
The Predicted Signal Peptide Is Essential for LRE-cYFP Expression in the Synergids
To test the importance of the predicted SP in LRE, we generated 12 T1 transformants with ProLRE:LREΔSP-cYFP (Supplemental Figure 2B); none showed cYFP signal in unfertilized ovules. RT-PCR on five randomly chosen lines showed that ProLRE:LREΔSP-cYFP was transcribed in unpollinated pistils (Supplemental Figure 2C). The lack of an LREΔSP-cYFP signal may be due to degradation of mislocalized LREΔSP-cYFP pro-protein, containing the C-terminal hydrophobic tail, in the cytoplasm (Hessa et al., 2011). Consistent with the lack of expression, ProLRE:LREΔSP-cYFP did not complement the seed set defect in lre-7/lre-7 plants (Supplemental Figure 2D), showing that the predicted SP in LRE is essential for its expression in the synergids.
Loss of the GPI Anchor Addition Domains in LRE Disrupts LRE-cYFP Localization in the Filiform Apparatus
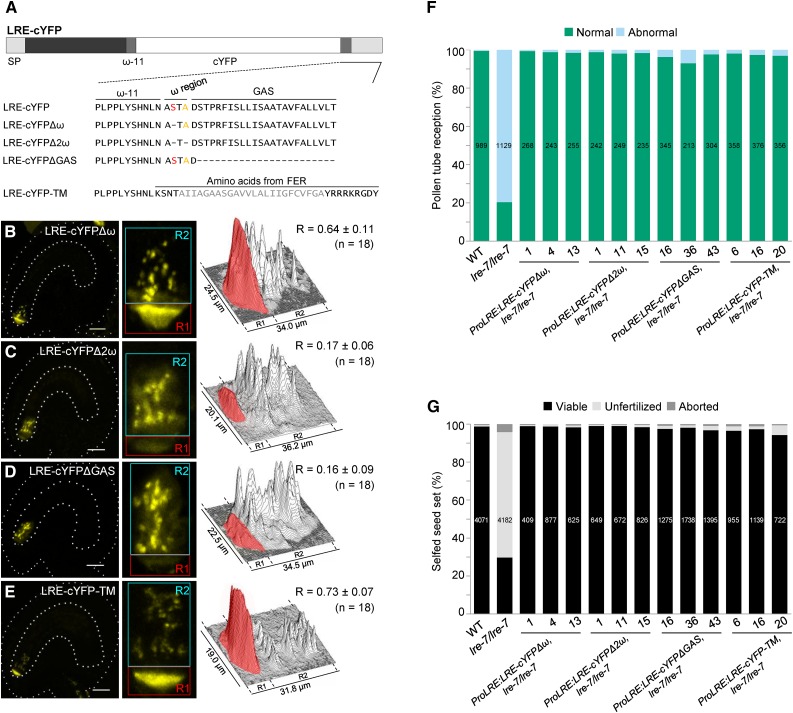
A prediction algorithm for plant GPI-anchored surface proteins, Big-PI plant predictor (Eisenhaber et al., 2003), identified Ser-139 as the ω site in LRE. Loss of the ω site and the resulting failure to receive a GPI anchor could disrupt LRE-cYFP localization in the plasma membrane-rich FA. However, synergid cell localization of LRE-cYFPΔω (Figure 3A), in which Ser-139 was deleted, was indistinguishable from LRE-cYFP localization (compare Figure 3B with Figures 2C to 2F). One possible interpretation of these results is that a cryptic ω site in LRE may function in the absence of Ser-139. Indeed, in an LRE sequence lacking Ser-139, Big-PI plant predictor identified Ala-141 as the ω site, and it identified no other ω site in an LRE sequence that lacked both Ser-139 and Ala-141. When both Ser-139 and Ala-141 were deleted (Figure 3A), LRE-cYFPΔ2ω localization in the FA was dramatically lower compared with LRE-cYFP (cYFP signal intensity decreased from ∼56 to ∼17%; Figure 3C). Additionally, there was a notable increase in the diffuse cYFP signal in the synergid cell cytoplasm. These results raise the possibility that removal of the predicted ω sites led to defects in GPI anchor addition and disrupted the trafficking of LRE-cYFPΔ2ω to the FA, consistent with studies showing that GPI attachment is required for transport of GPI-anchored proteins from the ER to the cell surface (Doering and Schekman, 1996; Mao et al., 2003).
Figure 3.
Predicted GPI Anchor Addition Domains Are Necessary for LRE Localization in the Filiform Apparatus but Not Its Function in Pollen Tube Reception.
(A) Diagram of the LRE-cYFP protein. Wild-type or altered amino acid sequence on either side of the predicted ω amino acid in each construct is indicated below the diagram. In LRE, the best predicted ω and a cryptic ω site are labeled in red and orange, respectively. Each dash in the protein sequence represents a deletion of the corresponding amino acid in wild-type LRE protein sequence. GAS, GPI attachment signal (lighter gray rectangle). In LRE-cYFP-TM, predicted transmembrane region of FER is in gray.
(B) to (E) Localization of LRE-cYFPΔω (B), LRE-cYFPΔ2ω (C), LRE-cYFPΔGAS (D), and LRE-cYFP-TM (E) in the synergids of the female gametophyte. Left panels, representative image showing the localization of each fusion protein; middle panels, close-up view of the micropylar region in corresponding left panel images; right panels, surface plots showing quantification of cYFP signal in the region of filiform apparatus (red box) and the remainder of the synergids (cyan box) as in Figure 2F. Bar = 20 µm.
(F) and (G) Constructs with altered GPI addition domains fully complement pollen tube reception defect (F) and the reduced seed set defect (G) in lre-7/lre-7 plants. Total number of ovules (F) and seeds (G) analyzed are in the center of each column. Number below each column, the three lines used that are single insertion and homozygous for the indicated transgenes in the lre-7/lre-7 background.
If loss of ω sites affects LRE-cYFP localization in the FA, then deletion of the GAS domain (Ser-143–Thr-165) should also produce LRE-cYFP without a GPI anchor (Mao et al., 2003) that fails to accumulate in the FA. Indeed, we observed a dramatic reduction in FA localization of LRE-cYFPΔGAS (cYFP signal intensity decreased from ∼56 to ∼16%; Figure 3D). Additionally, the absence of the GPI anchor addition domains was associated with an enhanced diffuse signal in the cytoplasm (Figure 3D), which may be due to inefficient protein sorting and/or enhanced retention of these proteins in the ER (Doering and Schekman, 1996). These results were similar to LRE-cYFPΔ2ω localization and consistent with the model that LRE localization in the FA requires a GPI anchor. Unlike in LRE-cYFP and LRE-cYFPΔ2ω, in 4/4 lines analyzed, LRE-cYFPΔGAS was seen in the intercellular spaces of the integument cells surrounding the micropyle, and the number of ovules showing this extracellular accumulation increased over time (Supplemental Figure 5), indicating that LRE-cYFPΔGAS is released outside the female gametophyte. These results are consistent with other GPI-anchored surface proteins lacking the GAS domain, which were released from the cell into the extracellular space at enhanced levels (Mao et al., 2003).
LRE-cYFP Localization in the Filiform Apparatus Is Disrupted in a Mutant Defective in GPI8, a Putative Subunit of the Transamidase Involved in GPI Anchor Addition
To further test if the GPI anchor addition domains in LRE are important for its localization in the FA, we examined LRE-cYFP localization in a mutant defective in GPI8, a critical subunit of the transamidase, which catalyzes the endoproteolysis reaction that removes the GAS from the pro-protein before covalently attaching the GPI anchor to the ω site. We reasoned that if GPI anchor addition domains were important for localization of LRE-cYFP in the FA, then the transamidase that uses these domains to catalyze GPI anchor addition would also be critical for the FA localization of LRE-cYFP. Based on this, it could be expected that the FA localization of LRE-cYFP would be affected in a gpi8 mutant female gametophyte, as the GPI anchor will not be added to LRE-cYFP.
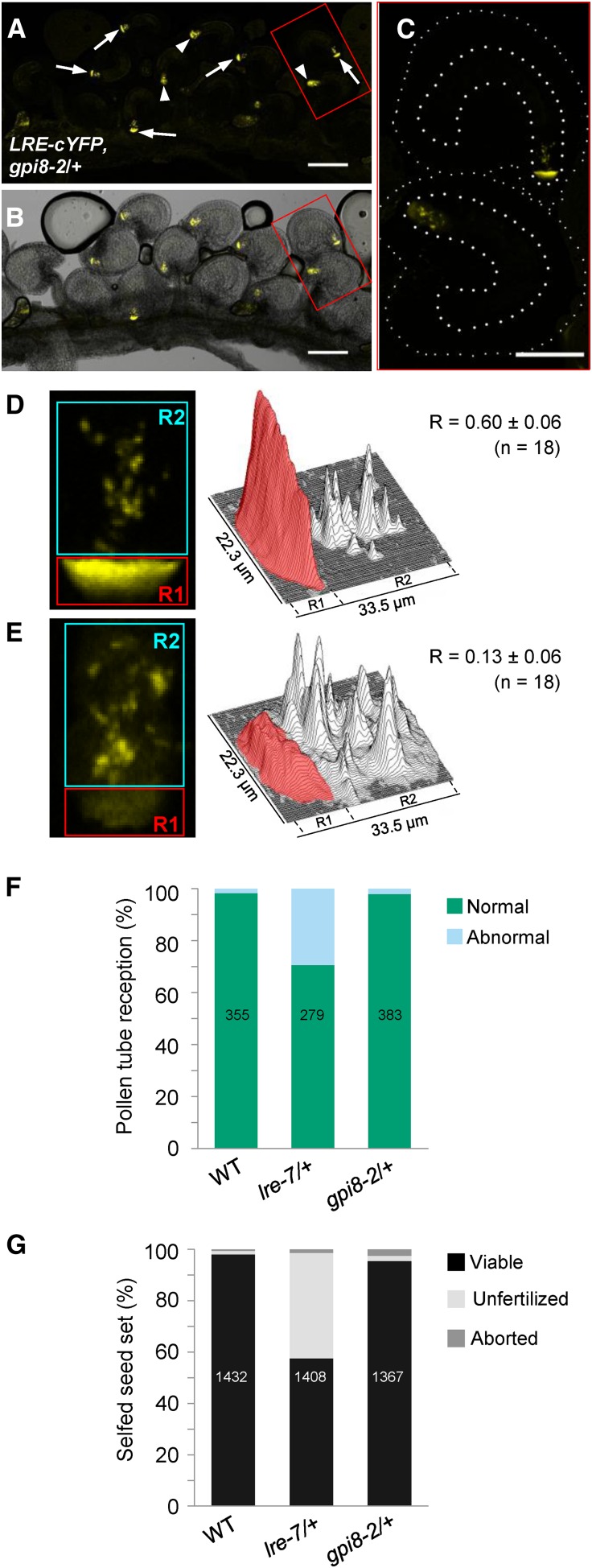
In yeast (Schizosaccharomyces pombe) and human cells, gpi8 mutants are defective in GPI attachment and cause a decrease in cell surface display of GPI-anchored proteins (Benghezal et al., 1996; Chen et al., 1996; Yu et al., 1997). The Arabidopsis genome contains a single gene (GPI8) that encodes a protein with high sequence similarity to yeast (43.8% identity) and human (45.7% identity) GPI8 and contains the postulated protease catalytic site that is conserved in C13 clade of cysteine proteases (Supplemental Figures 6A and 6B) (Zacks and Garg, 2006). The expression of SKU5, a known GPI-anchored surface protein (Sedbrook et al., 2002), is affected in gpi8-1, a weaker point mutant allele of GPI8 that was identified as an enhancer of stomatal clustering in an erecta-like1 erecta-like2 double mutant; this decrease in expression is likely due to the failure in removing the GAS from SKU5 (Bundy et al., 2016). Many GPI-anchored proteins play essential roles in the male gametophyte (Lalanne et al., 2004). Consistent with this, transmission of gpi8-2, a stronger T-DNA allele of GPI8 (Supplemental Figure 6C), through the male gametophyte is abolished (Supplemental Table 2); however, gpi8-2 is transmitted through the female gametophyte (Supplemental Table 2). This result, combined with the finding that GPI8 is expressed in the synergids (Wuest et al., 2010), makes it feasible to test the effect of gpi8-2 on LRE-cYFP localization in the FA.
We crossed ProLRE:LRE-cYFP homozygous plants with the gpi8-2/+ mutants and analyzed LRE-cYFP localization in the F1 progeny, which were either +/+ or gpi8-2/+ (Supplemental Table 3 and Supplemental Figure 6D). In all the progeny that were wild type for GPI8, cYFP-positive ovules showed only one pattern of localization: in fluorescent puncta and a polarized localization in the FA, as expected for LRE-cYFP. However, in every gpi8-2/+ progeny analyzed, the cYFP-positive ovules showed two localization patterns: about half of the cYFP-positive ovules had polarized LRE-cYFP localization and in the other half, the cYFP signal was diffuse and dramatically reduced in the FA (from ∼60 to ∼13%; Figures 4A to 4E; Supplemental Table 3), a localization pattern similar to LRE-cYFPΔ2ω and LRE-cYFPΔGAS (Figures 3C and 3D).
Figure 4.
LRE-cYFP Localization in the Filiform Apparatus Is Affected by a Mutation in GPI8, a Critical Component of the Transamidase.
(A) and (B) Localization of the LRE-cYFP fusion protein in a gpi8-2/+ pistil. White arrows, female gametophytes with a polarized cYFP localization in the filiform apparatus; white arrowheads, sibling female gametophytes with a diffuse cYFP localization throughout the synergids. Bar = 100 µm.
(C) Enlarged view of two ovules within the red box in (A) and (B). Ovules and female gametophytes are outlined in thin and thick white dashed lines, respectively. Bar = 50 µm.
(D) and (E) cYFP signal intensity quantification in ovules within a gpi8-2/+ pistil. Left panels, representative images of the micropylar region of ovules with polarized (D) and diffuse (E) cYFP localization in the synergids of ovules within a gpi8-2/+ pistil. cYFP signal in the region of filiform apparatus (red) and the remainder of the synergids (cyan) were quantified as in Figure 2F.
(F) and (G) gpi8-2 mutation does not cause pollen tube reception (F) or seed set defect (G).
To test if the effect of gpi8-2 is specific to proteins with GPI anchor addition domains, such as LRE-cYFP, we generated gpi8-2/+ plants expressing a variant of LRE-cYFP, in which a single-pass transmembrane (TM) domain of FER, an integral membrane protein, replaced the GPI anchor addition domains (Figure 3A). LRE-cYFP-TM is expected to be unaffected in gpi8-2 mutants, as TM-containing proteins do not require the transamidase for their localization to the cell surface. Indeed, all cYFP-positive ovules in gpi8-2/+ pistils carrying ProLRE:LRE-cYFP-TM showed only one localization pattern: a preferential accumulation of cYFP in the FA (Supplemental Figure 7 and Supplemental Table 4), compared with the two localization patterns of ProLRE:LRE-cYFP in gpi8-2/+ pistils (Figures 4A to 4E; Supplemental Table 3). These results are consistent with our finding that GPI anchor addition domains in LRE play a role in localizing LRE-cYFP to the FA (Figure 3).
The GPI Anchor Addition Domains in LRE Are Not Necessary for Its Function in Pollen Tube Reception
If LRE localization in the FA is affected by the loss of transamidase, gpi8-2 and lre female gametophytes may show the same defects in pollen tube reception and seed set. Contrary to this expectation, gpi8-2 mutants showed wild-type pollen tube reception and seed set (Figures 4F and 4G). Transmission of the gpi8-2 mutation through female gametophytes was also not affected (Supplemental Table 2), suggesting that LRE function does not require the GPI anchor addition domains.
The lack of pollen tube reception and seed set phenotypes in gpi8-2/+ plants prompted us to perform complementation tests with LRE-cYFPΔ2ω and LRE-cYFPΔGAS to investigate if LRE function requires the GPI anchor addition domains. Despite dramatically reduced localization in the FA (Figures 3C and 3D), LRE-cYFPΔ2ω and LRE-cYFPΔGAS almost fully complemented the pollen tube reception and seed set defects in lre-7/lre-7 plants (Figures 3F and 3G). Transmission of either ProLRE:LRE-cYFPΔ2ω or ProLRE:LRE-cYFPΔGAS through the lre-7 female gametophyte (Supplemental Table 5) was also comparable to the transmission of ProLRE:LRE-cYFP through the lre-7 female gametophyte (Supplemental Table 1), indicating that LRE can induce pollen tube reception despite lacking the potential ω sites or GAS domain and suggesting that LRE function does not require GPI anchor addition domains.
An LRE Variant Lacking the GPI Anchor Addition Domains but Attached to a Transmembrane Domain Can Function in Pollen Tube Reception
To further test if the GPI anchor addition domains in LRE are necessary for its function in pollen tube reception, we tested if LRE-cYFP-TM is functional in pollen tube reception. We chose LRE-cYFP-TM because in this transgenic protein, both the ω site and the GAS domain of LRE have been replaced with a single-pass TM domain (Figure 3A). Additionally, it localized to the FA of the female gametophyte (Figure 3E) and is expected to produce an LRE variant that remains tethered to the plasma membrane via the TM.
LRE-cYFP-TM almost fully rescued the pollen tube reception and seed set defects in lre-7/lre-7 plants (Figures 3F and 3G; Supplemental Figure 8). Transmission of ProLRE:LRE-cYFP-TM through the lre-7 female gametophyte was also enhanced, confirming that LRE-cYFP-TM is functional (Supplemental Table 5). These results indicate that even if it lacks GPI anchor addition domains but is attached to a TM domain, LRE can still function in pollen tube reception. Complementation by LRE-cYFP-TM and by LRE variants that lack the GPI anchor addition domains (LRE-cYFPΔ2ω and LRE-cYFPΔGAS) indicates that a domain shared by these three constructs, such as the M8CM in the ectodomain of LRE, might be important for its function in pollen tube reception.
LRE Function in Pollen Tube Reception Requires a Unique 12-Amino Acid Domain in the M8CM
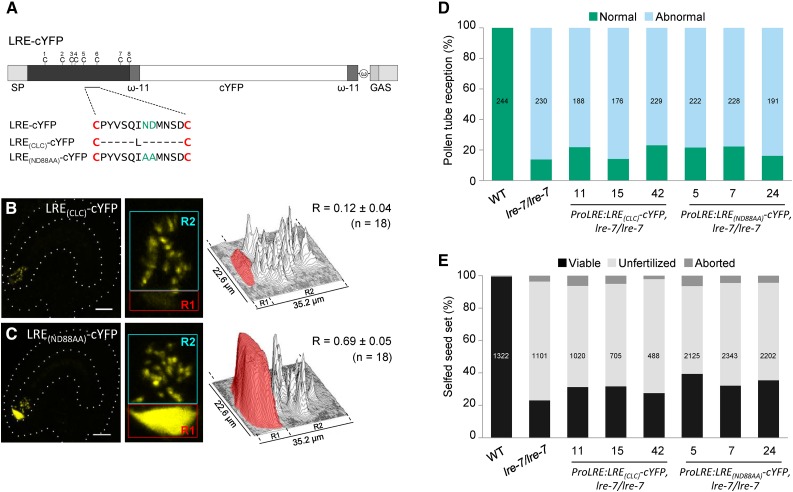
To identify which domain within the LRE ectodomain is critical for its function in pollen tube reception, we tested if loss of the 12-amino acid domain in the M8CM of the ectodomain, uniquely found in LRE family proteins (Figure 1; Supplemental Figure 1), affects LRE function. We produced an LRE with a typical 8CM by replacing the 12-amino acid domain between the 5th and the 6th cysteines in LRE with a single Leu (Figure 5A), the most prevalent amino acid in Arabidopsis proteins containing a typical 8CM (61 out of 98 proteins; José-Estanyol et al., 2004). LRE(CLC)-cYFP signal intensity in the FA was ∼4.6 times lower than that of LRE-cYFP (from ∼56 to ∼12%; compare Figure 5B with Figures 2C to 2F). LRE(CLC)-cYFP did not rescue pollen tube reception, seed set, or transmission defects in lre-7/lre-7 plants (Figures 5D and 5E; Supplemental Figure 8 and Supplemental Table 6). These results demonstrated that the 12-amino acid domain between the 5th and the 6th cysteines in LRE is necessary for its localization in the FA and its function in pollen tube reception.
Figure 5.
The 12-Amino Acid Residues between the 5th and 6th Cysteines of LRE and the Conserved Asn-Asp Dipeptide Are Necessary for LRE Function in Pollen Tube Reception.
(A) Diagram of the LRE-cYFP fusion protein. Amino acid sequence between the 5th and the 6th cysteines of LRE in each construct are indicated below the diagram. Each dash in the protein sequence represents a deletion of the corresponding amino acid in the wild-type LRE protein sequence. ω, omega site; GAS, GPI attachment signal containing a spacer region (darker gray rectangle) and a hydrophobic tail (lighter gray rectangle).
(B) and (C) LRE(CLC)-cYFP (B) and LRE(ND88AA)-cYFP (C) fusion proteins in the synergids of the female gametophyte. Left panels, representative image showing the localization of indicated fusion protein; middle panels, close-up view of the micropylar region in corresponding left panel images; right panels, surface plots showing quantification of cYFP signal in the region of filiform apparatus (red box) and the remainder of the synergids (cyan box) as in Figure 2F. Bar = 20 µm.
(D) and (E) None of these three constructs complemented the pollen tube reception defect (D) or the reduced seed set (E) defect in lre-7/lre-7 plants. Total number of ovules (D) and seeds (E) analyzed are in the center of each column. Number below each column, three lines used that are that are single insertion and homozygous for the indicated transgenes in the lre-7/lre-7 background.
To investigate if the highly conserved Asn-Asp dipeptide in the 12-amino acid domain (Figure 1; Supplemental Figure 1) is necessary for LRE localization and function, we replaced the Asn-Asp with two alanines in LRE-cYFP (Figure 5A). Unlike LRE(CLC)-cYFP, LRE(ND88AA)-cYFP localized to the FA normally in unfertilized ovules (Figure 5C), indicating that loss of Asn-Asp does not affect subcellular localization of LRE. However, LRE(ND88AA)-cYFP is not functional, as evident from pollen tube reception and seed count assays and transmission analysis (Figures 5D and 5E; Supplemental Figure 8 and Supplemental Table 6). These results showed that loss of the Asn-Asp dipeptide abolished LRE function in pollen tube reception. Taken together, these results show that the unique 12-amino acid domain in the M8CM of LRE is important for LRE function in pollen tube reception.
Pollen Tube-Expressed LRE Complements the Pollen Tube Reception Defect in the lre Female Gametophyte
Intracellular and synergid cell surface (FA) localization of LRE-cYFP raises the question of where LRE functions in pollen tube reception. As the first point of contact between the pollen tube and the synergid cell, the FA may be the critical location for LRE function. Alternatively, intracellular LRE may be critical for pollen tube reception for the following reasons: First, in our study, the levels of the two fusion proteins that successfully complemented lre-7 (LRE-cYFPΔ2ω and LRE-cYFPΔGAS) were dramatically reduced in the FA, but they did localize intracellularly (Figures 3D to 3F and 3G; Supplemental Table 5); second, a recent report showed that LLG1/LRE functions intracellularly as a chaperone to facilitate cell surface localization of the FER receptor kinase (Li et al., 2015). However, since the intracellular function of LLG1/LRE precedes its function in the cell surface, it remains unclear whether LRE has a separate function in the synergid cell surface.
To test these possibilities, we supplied LRE-cYFP extracellularly from the pollen tube, a cell where LRE is not normally expressed, and determined if it can complement the pollen tube reception defect in lre synergids. To avoid potential complications from ectopically expressed LRE-cYFP interfering with pollen development and germination, we expressed LRE-cYFP from the PECTIN LYASE-LIKE SUPERFAMILY PROTEIN3 (PLL3) promoter (Sun and van Nocker, 2010), which is preferentially induced in pollen tubes after their growth in the pistil (Qin et al., 2009) (Supplemental Figures 9A and 9B).
LRE may inhibit pollen tube growth by initiating a signal cascade in the pollen tube when it reaches the synergid cell, probably after being released from the GPI anchor (Capron et al., 2008) upon cleavage by enzymes such as phospholipase C or D (Wang, 2001; Sharom and Lehto, 2002). Therefore, ectopically expressed LRE-cYFP may affect pollen tube growth. However, we observed normal transmission of ProPLL3:LRE-cYFP through the wild-type male gametophyte, when in competition with nontransgenic pollen, indicating that LRE-cYFP expression did not compromise in vivo pollen tube functions (Supplemental Table 7). We therefore used ProPLL3:LRE-cYFP to test if pollen tube-expressed LRE can induce pollen tube reception in lre synergids.
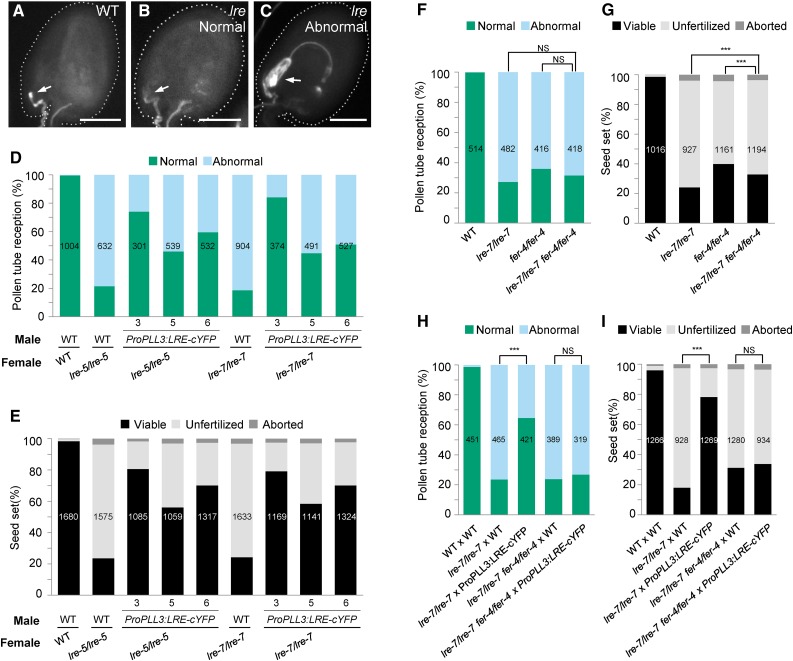
Indeed, we observed increased seed set in lre-5 pistils pollinated with pollen from ProPLL3:LRE-cYFP T1 plants (Supplemental Figure 9C), suggesting that pollen tube-expressed LRE-cYFP can function in pollen tube reception. To further characterize this non-cell-autonomous complementation, we isolated three single insertion lines (Supplemental Table 7) that were homozygous for ProPLL3:LRE-cYFP. An aniline blue staining-based pollen tube reception assay showed that substantially more lre female gametophytes undergo pollen tube reception with ProPLL3:LRE-cYFP pollen compared with wild-type pollen (Figures 6A to 6D). Consistent with this, lre pistils also showed improved seed set when pollen tubes expressing LRE-cYFP were used (Figure 6E). To further test whether pollen tubes expressing LRE-cYFP can rescue the lre female gametophyte defect, we examined the transmission of the lre-5 and lre-7 mutations through the female gametophyte, using homozygous ProPLL3:LRE-cYFP plants as the male parent. As expected, ProPLL3:LRE-cYFP pollen enhanced the transmission of lre-5 and lre-7 through the female gametophyte from 18% to 62 to 72% and 12% to 60 to 72%, respectively (Table 2). Consistent with these results, ProPLL3:LRE-cYFP is also transmitted at an enhanced rate to the progeny if crossed to lre-7 females compared with wild-type females (Supplemental Table 7). These results suggested that at the interface of the pollen tube and the synergid cell, LRE has a function that is independent of its intracellular function in the synergids.
Figure 6.
LRE-cYFP from Pollen Tubes Rescues Female Gametophyte Defects Caused by the lre-7 Mutation.
(A) to (C) Representative images from pollen tube reception assays using aniline blue staining. Ovules are outlined in white dashed line. Normal pollen tube reception in wild-type (A) and lre-7/lre-7 ovules (B); abnormal pollen tube reception in lre-7/lre-7 ovule (C). Bar = 50 µm.
(D) and (E) Pollen tube-expressed LRE-cYFP rescued pollen tube reception (D) and seed set defects (E) of the lre-7 female gametophyte, respectively. Total number of ovules (D) and seeds (E) analyzed, respectively, is in the center of each column.
(F) and (G) lre-7/lre-7 fer-4/fer-4 double mutant showed similar level of pollen tube reception as lre-7/lre-7 and fer-4/fer-4 single mutant and a small but significant difference (χ2 test) in seed set defect when compared with either lre-7/lre-7 or fer-4/fer-4 single mutant (G). Total number of ovules (F) and seeds (G) analyzed is in the center of each column. NS, not significant. P values = 0.192 (column 2 versus column 4 in [F]) and 0.1957 (column 3 versus column 4 in [F]), respectively. ***P value < 0.001 (P values = 6.34 × 10−5 for column 2 versus column 4 and 0.0007 for column 3 versus column 4 in [G]).
(H) and (I) FER is required in synergids to rescue the pollen tube reception (H) and seed set (I) defects in the lre-7 female gametophyte by the pollen tube-expressed LRE. Total number of ovules (H) and seeds (I) analyzed is in the center of each column. NS, not significant. P value = 0.3994 (column 4 versus column 5 in [H]) and 0.4094 (column 4 versus column 5 in [I]) respectively. ***P value < 0.001; P value < 2.2 × 10−16 for column 2 versus column 3 in both (H) and (I).
Table 2. ProPLL3:LRE-cYFP in the Male Gametophyte Restored the Reduction in Transmission of the lre-7 Mutation through the Female Gametophyte.
| Female Parent |
Male Parenta |
Observed No. of Progeny |
TE (R/S)b |
χ2c |
P Value |
|
|---|---|---|---|---|---|---|
| Resistantd | Susceptibled | |||||
| Wild type | lre-7/+ | 405 | 397 | 1.02 | – | – |
| lre-7/+ | Wild type | 100 | 542 | 0.18 | – | – |
| lre-7/+ | Line 3 | 150 | 215 | 0.70 | 180.77 | <0.001 |
| lre-7/+ | Line 5 | 176 | 285 | 0.62 | 179.09 | <0.001 |
| lre-7/+ | Line 6 | 226 | 316 | 0.72 | 281.24 | <0.001 |
| Wild type | lre-5/+ | 435 | 414 | 1.05 | – | – |
| lre-5/+ | Wild type | 53 | 448 | 0.12 | – | – |
| lre-5/+ | Line 3 | 94 | 130 | 0.72 | 233.25 | <0.001 |
| lre-5/+ | Line 5 | 203 | 328 | 0.62 | 429.17 | <0.001 |
| lre-5/+ | Line 6 | 158 | 264 | 0.60 | 321.88 | <0.001 |
Line numbers refer to three independent transformants in the wild-type background containing a single insertion of the ProPLL3:LRE-cYFP transgene; genotype of each transgenic line used is homozygous for the transgene (ProPLL3:LRE-cYFP / ProPLL3:LRE-cYFP).
Transmission efficiency (TE) was calculated as the ratio of Basta or kanamycin resistance (R) to susceptibility (S) in the progeny of the indicated cross.
χ2 was calculated based on Basta or kanamycin resistance to susceptibility segregation ratio (R:S) in the progeny of the crosses lre-7/+(♀) × wild type(♂) or lre-5/+(♀) × wild type(♂), respectively, as the expected segregation ratio.
Resistant and susceptible progeny to Basta or kanamycin. Basta and kanamycin resistance genes are linked to the T-DNA inserted in the LRE gene in lre-7 and lre-5, respectively.
In the synergid cells, complementing lre-7 female gametophyte with LRE variants LRE-cYFPΔ2ω or LRE-cYFPΔGAS revealed that the GPI anchor addition domains in LRE are not necessary for its function in pollen tube reception. To investigate if this is the case with pollen tube-expressed LRE, we ectopically expressed LRE-cYFPΔ2ω or LRE-cYFPΔGAS in the pollen tube and examined if they are functional in pollen tube reception. Our results showed that pollen carrying ProPLL3:LRE-cYFPΔ2ω or ProPLL3:LRE-cYFPΔGAS transgenes rescued the pollen tube reception defect in the lre female gametophyte to a similar extent as did ProPLL3:LRE-cYFP (Supplemental Figure 9C). These results showed that GPI anchor addition domains in LRE are not necessary for pollen tube-expressed LRE at the interface of the pollen tube and the synergid cell to induce pollen tube reception.
Alternatively, the pollen tube-expressed LRE-cYFP or LRE-cYFPΔ2ω or LRE-cYFPΔGAS might be released from the pollen tubes and endocytosed into synergids and then carry out an intracellular function. To test this possibility, we used the PLL3 promoter to express LRE-cYFP-TM (Supplemental Figures 9A and 9B), as it was almost fully functional when expressed in synergids (Figures 3A, 3F, and 3G) and is expected to remain tethered to the pollen tube plasma membrane. Although the YFP signal was primarily in the cytoplasm and there was only a weak plasma membrane localization in the pollen tubes, in all seven T1 transformants, seed set was restored when pollen tube-expressed LRE-cYFP-TM was pollinated onto lre/lre pistils (Supplemental Figure 9C). ProPLL3:LRE-cYFP-TM also was transmitted at an enhanced rate to the progeny if crossed to an lre-7 female compared with the wild type (Supplemental Table 8), confirming that LRE-cYFP-TM is functional when expressed in pollen tubes. These results are consistent with the model that LRE has a function at the interface of the pollen tube and the synergid cell, independent of its intracellular function in the synergids.
LRE and FER Require Each Other at the Interface of the Pollen Tube and the Synergid Cell to Induce Pollen Tube Reception
LRE acts as a chaperone for FER in the synergid cell and promotes its localization in the FA (Li et al., 2015). If FER localization in the FA is entirely dependent on LRE, then complementation of pollen tube reception defect in the lre female gametophyte by pollen tube-expressed LRE must depend on other protein(s) in the FA, besides FER, to induce pollen tube reception. We tested this possibility by examining if pollen tube-expressed LRE can complement the pollen tube reception defect in a female gametophyte that lacks both LRE and FER.
For this, we first established an lre-7/lre-7 fer-4/fer-4 double homozygous mutant and assessed the seed set and pollen tube reception phenotypes in the double mutant pistils. There was no statistically significant difference in the pollen tube reception defect in the double mutant compared with either the lre-7/lre-7 or fer-4/fer-4 single mutant (Figure 6F). However, there was a small but statistically significant difference in seed set between each of the single mutants when compared with the lre/lre fer/fer double mutant (Figure 6G). Nevertheless, similar levels of pollen tube reception defect in single and double mutants provide genetic evidence in support of the proposal that, similar to its paralog LLG1, LRE functions with FER to induce pollen tube reception in the synergid cell (Li et al., 2015).
We next crossed ProPLL3:LRE-cYFP pollen onto the lre-7/lre-7 fer-4/fer-4 double mutant pistil and found that pollen tube-expressed LRE-cYFP did not rescue the pollen tube reception defect in the lre-7 fer-4 double mutant female gametophyte (Figure 6H) and consequently also did not rescue the seed set defect in the lre-7/lre-7 fer-4/fer-4 double mutant pistil (Figure 6I). Taken together, these results indicated that LRE-cYFP delivered by the pollen tube requires FER in the synergid cell to rescue the pollen tube reception defect in the lre female gametophyte and that LRE and FER require each other at the interface of the pollen tube and the synergid cell to induce pollen tube reception.
DISCUSSION
Non-Cell-Autonomous and Extracellular Complementation Analysis with Pollen Tube-Expressed LRE Clarified the Role of LRE in Pollen Tube Reception
Based on the excessive pollen tube growth phenotype in lre female gametophytes, we initially hypothesized that LRE participates in pollen tube reception by inhibiting/reducing pollen tube growth after it interacts with the synergids. This pause in pollen tube growth may then activate additional signaling between the pollen tube and the synergids to complete pollen tube reception. Near normal transmission efficiency of ProPLL3:LRE-cYFP in the wild-type background to the progeny through the male gametophyte, even in direct competition with nontransgenic pollen, showed that LRE-cYFP in pollen tubes did not affect any male gametophyte function (Supplemental Table 7; ProPLL3:LRE-cYFP/+ crossed to the wild type). These results argue against the possibility that LRE by itself functions as a growth arrest signal in the pollen tube. Consistent with this, pollen tubes did not show premature growth arrest when interacting with ovules showing extracellular accumulation LRE-cYFPΔGAS in the micropyle (Supplemental Figure 5).
Still, pollen tube-expressed LRE-cYFP complemented pollen tube reception defects in the lre-7 female gametophyte (Figures 6D and 6E), showing that a pollen tube interacting with the synergid cell is necessary for LRE function. One way synergids may regulate LRE function is by possessing a mechanism to release the LRE ectodomain from the GPI anchor and activate it, akin to what is known in other organisms (Müller and Bandlow, 1993; Wang, 2001; Sharom and Lehto, 2002; Capron et al., 2008; Haruta et al., 2014). However, non-cell-autonomous and extracellular complementation of the lre-7 female gametophyte defects by pollen tube-expressed LRE-cYFP-TM, which is expected to remain tethered to the pollen tube plasma membrane, suggests that pollen tube reception does not require release of the LRE ectodomain. Additionally, synergid-expressed LRE-cYFP-TM fully complemented pollen tube reception defects in the lre-7 female gametophytes, arguing against release of the LRE ectodomain as a mode of LRE function in pollen tube reception.
Another proposed function for LRE is in synergid maturation, when they acquire the competency to induce pollen tube reception (Capron et al., 2008; Rotman et al., 2008). In this model, LRE does not mediate interactions between the pollen tube and the synergids; instead, it is part of a signaling pathway important for synergid development prior to the arrival of the pollen tube. Non-cell-autonomous and extracellular complementation of the lre-7 female gametophyte defects by pollen tube-expressed LRE-cYFP argues against such a function for LRE, as it is highly unlikely that lre mutant synergids use pollen tube-expressed LRE-cYFP to rapidly mature and induce pollen tube reception. Indeed, a pollen tube reception assay did not show any overt abnormalities in the interactions between LRE-cYFP-expressing pollen tubes and lre female gametophytes (Figure 6D).
Separate, FER-Dependent Function for LRE at the Interface of the Pollen Tube and the Synergid Cell in Pollen Tube Reception
LRE is proposed to have two functions in the synergid cell: chaperoning FER in the ER en route to the FA and functioning as a coreceptor with FER in the FA (Li et al., 2015). However, since FER localization in the FA depends on intracellular LRE, it has not been experimentally demonstrated if LRE has any synergid surface-specific function, such as its proposed role as a coreceptor with FER (Li et al., 2015) to signal ROS production (Duan et al., 2014). That is, the lre phenotype related to FER signaling at the plasma membrane can also be interpreted as an indirect consequence of the failure of FER to reach the plasma membrane in the absence of LRE. To directly investigate if there is an independent function for LRE in the interface of the pollen tube and the synergids, we ectopically expressed LRE in the pollen tube and used it to rescue the pollen tube reception function of lre synergids. Non-cell-autonomous and extracellular complementation of lre revealed that indeed LRE has a function at the interface of the pollen tube and the synergid cell and that the two functions of LRE can be uncoupled.
Based on the current model of LRE function as a coreceptor with FER (Li et al., 2015), it would be expected that pollen tube-expressed LRE functioned with FER in the FA, when it rescued pollen tube reception defects in lre female gametophytes (Figures 6D and 6E; Supplemental Figure 9C). However, since optimal FER localization in the FA is dependent on LRE (Li et al., 2015), it is not clear what protein functioned with pollen tube-expressed LRE to rescue the pollen tube reception phenotype in the lre female gametophyte. We speculated that in lre female gametophytes, perhaps sufficient amounts of FER reached the FA and functioned with pollen tube-expressed LRE. We considered this possibility based on the finding that all lre ovules showed some FER-GFP in the FA region despite increased intracellular retention of FER-GFP in the lre synergids (Li et al., 2015). Consistent with this, in this study, we showed that pollen tube-expressed LRE-cYFP requires FER in the lre female gametophyte to rescue the pollen tube reception defect associated with that lre female gametophyte (Figures 6H and 6I), indicating that some amount of FER is present in the FA of lre synergid cell, at least sufficient enough to rescue the pollen tube reception defect.
Although pollen tube-expressed LRE-cYFP, LRE-cYFPΔ2ω, LRE-cYFPΔGAS, or LRE-cYFP-TM complemented the lre-7 phenotypes, restoration was not to wild-type levels (Figures 6D and 6E; Supplemental Figure 9C), pointing to additional roles for LRE in the synergid cell, such as intracellular LRE chaperoning FER to the synergid cell surface (Li et al., 2015). Alternatively, the incomplete complementation could be due to the possibility that pollen tube-expressed LRE variants, compared with their synergid cell-expressed counterparts, are not optimal for functioning in pollen tube reception. Nevertheless, non-cell-autonomous, extracellular complementation of the pollen tube reception defect in the lre female gametophyte by pollen tube-expressed LRE-cYFP, LRE-cYFPΔ2ω, LRE-cYFPΔGAS, or LRE-cYFP-TM establishes that LRE performs a synergid cell surface-specific function during interactions between the pollen tube and the synergid cell.
Both lre and fer are partially transmitted through the female gametophyte, and normal pollen tube reception is detected in a limited number of lre or fer female gametophytes (Figures 6F and 6G). Such an outcome is perhaps possible due to at least two scenarios: Either synergid-expressed proteins that function redundantly with LRE-FER complex to induce pollen tube reception or additional players, besides LRE-FER complex, might mediate pollen tube reception. Our observation that the lre-7 fer-4 double mutant had similar levels of pollen tube reception and seed set defects compared with either lre-7 or fer-4 single mutants argues against the first possibility. Instead, our results demonstrate that LRE and FER require each other to function in pollen tube reception and predict the existence of another minor signaling pathway to induce pollen tube reception.
The GPI Anchor Addition Domains Are Not Necessary for LRE Function in Pollen Tube Reception
Prediction programs identified GPI anchor addition domains in the LRE sequence with high confidence, and these domains are also highly conserved in LRE family members. Deleting these domains in LRE (LRE-cYFPΔ2ω and LRE-cYFPΔGAS) led to a dramatic decrease in protein localization in the FA. Localization of LRE-cYFP in the FA was also affected in gpi8-2 female gametophytes, which are expected to lack a core component of the transamidase. These results point to the need for a GPI anchor in localizing LRE in the FA. Surprisingly, mutant proteins lacking GPI anchor addition domains fully complemented pollen tube reception and seed set defects. Our complementation results indicate that the GPI anchor addition domains are not necessary for LRE function in pollen tube reception.
An alternative interpretation of the complementation results with LRE-cYFPΔ2ω and LRE-cYFPΔGAS is that sufficient amounts of these proteins may be available in the FA or pollen tube surface for function in the transgenic lines. In both synergid cells and pollen tubes, for example, sufficient amounts of LRE-cYFPΔ2ω may have reached the cell surface and associated with the plasma membrane using the lingering C-terminal hydrophobic tail (Galian et al., 2012), which is expected to be removed by the transamidase from the wild-type LRE pro-protein. Or, in LRE-cYFPΔGAS, although much of the mutant protein is retained in the cytoplasm (Figure 3D; Supplemental Figure 9B), detectable levels of the protein were released from the female gametophyte to the surrounding integument cells lining the micropyle (Supplemental Figure 5). During this release, perhaps sufficient amounts of mutant proteins were in the FA or in the pollen tube surface and induced pollen tube reception.
LRE functions as a chaperone of FER from the ER to the synergid cell periphery and is required for optimal localization of FER in the FA (Li et al., 2015). It remains to be experimentally demonstrated if this intracellular function was also rescued by LRE-cYFPΔ2ω and LRE-cYFPΔGAS expressed in the synergids. Our results showing that pollen tube-expressed LRE complemented lre female gametophytes in a FER-dependent manner indicated that some amount of FER is present in the FA of lre female gametophytes. Even if LRE-cYFPΔ2ω or LRE-cYFPΔGAS were unable to rescue the chaperone function of LRE in the synergid cells, they most likely functioned with the available FER in FA of lre female gametophytes to induce pollen tube reception.
Based on the full complementation of pollen tube reception and seed set defects in the lre-7 female gametophyte by synergid-expressed LRE-cYFP-TM, which lacks GPI anchor addition domains, we propose that the GPI anchor is not necessary for LRE function in pollen tube reception. Consistent with this proposal, LRE homologs in some plants do not contain the GAS domain or the ω site region, even though they all contain the portion of the protein that shares sequence similarity to the LRE ectodomain (Supplemental Figure 1). Still, most putative LRE homologs have highly conserved GPI anchor addition domains. One possibility that may reconcile these observations is that the GPI anchor may enhance the efficiency of LRE function in pollen tube reception and hence remain fixed in the population. For example, the GPI anchor may afford flexibility to LRE in setting up signaling microdomains, similar to mammalian GPI-anchored surface proteins (Varma and Mayor, 1998), at the site of the pollen tube-synergid cell interaction, as the exact site of this interaction is determined stochastically.
A Novel 12-Amino Acid Domain in the M8CM of LRE Is Essential for LRE Function in Pollen Tube Reception
Results with synergid-expressed LRE-cYFP-TM also showed that the LRE ectodomain without the GPI anchor addition domains is sufficient to induce pollen tube reception. For some GPI-anchored proteins, proper structure of the ectodomain requires a GPI anchor (Bütikofer et al., 2001). However, LRE does not appear to require the GPI anchor, as LRE lacking the GPI anchor addition domains fully complements the pollen tube reception defects in lre female gametophytes. Lack of dominant-negative phenotypes in lre synergids and pollen tubes expressing LRE-cYFP-TM indicates it did not acquire novel functions, as in some GPI-anchored surface proteins in which a TM was swapped for a GPI anchor (Shenoy-Scaria et al., 1992, 1993).
We identified LRE as a CRP, as it contains eight cysteines that are arranged in a pattern reminiscent of cysteines in an 8CM protein. This adds LRE to the list of CRPs that play diverse roles in plant reproduction (Marshall et al., 2011; Beale and Johnson, 2013). However, LRE is notable in that it contains an M8CM, with a unique 12-amino acid domain separating the 5th and the 6th cysteines. Additionally, unlike other CRPs, most of which are released from the cell, LRE belongs to a group of CRPs that contain GPI-anchor addition domains (Fliegmann et al., 2011). The novel 12-amino acid domain in LRE is ancient, as it is present even in the putative LRE homolog in the moss (Physcomitrella patens) (Figure 1; Supplemental Figure 1). Identification of the M8CM in LRE supports the hypothesis that the 8CM serves as a sequence scaffold to evolve new proteins with different functions (José-Estanyol et al., 2004).
Our study showed that a 12-amino acid domain between the 5th and the 6th cysteines, especially the highly conserved Asn-Asp dipeptide domain, in LRE is critical for its function and thus provided insights into the mechanism of LRE function in the female gametophyte. M8CM in LRE awaits additional characterization, including biochemical approaches to determine its disulfide bond linkages (Tang and Speicher, 2004) and structural studies to solve its three-dimensional structure. Preliminary three-dimensional structural prediction of LRE in the Phyre2 structure prediction website (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) showed that this 12-amino acid domain may form a loop between two alpha helices and may mediate interactions between LRE and other proteins.
Although LRE(CLC)-cYFP accumulated to normal levels in the unknown organelle within synergid cells, it was barely detectable in the FA, indicating that this 12-amino acid domain in the M8CM may mediate interactions between LRE and protein sorting machinery of the endomembrane system that is critical for its trafficking to the FA or between LRE and other proteins that are necessary for retention of LRE in the FA. It is also possible that the 12-amino acid domain in M8CM is critical for chaperoning FER from the ER to the FA (Duan et al., 2014; Li et al., 2015). Alternatively, this domain may mediate functional interactions between LRE and FER during pollen tube reception. For example, the interaction may facilitate binding of the LRE/FER complex with their putative ligand from the pollen tube or synergids or activate FER either by aiding it in reaching the correct confirmation or attaining maturation to initiate signaling in the synergid cell. Failure to mediate functional interactions with FER might be the basis for LRE (ND88AA)-cYFP not complementing the defects in the lre female gametophyte, despite localizing normally in the synergid cell.
An Integrated Approach to Study Subcellular Localization and Function of LRE in Pollen Tube Reception in the Synergids
We analyzed the subcellular localization of LRE and its function in pollen tube reception in the synergids. For localization studies, the FA, an easily identifiable structure of a highly secretory cell, allowed quick monitoring of localization of LRE. Whether it is due to preferential secretion of LRE into the FA or concentration of LRE in the plasma membrane-rich structure of the FA, polarized localization of LRE in the FA allowed us to rapidly test the role of the GPI-anchor addition domains in subcellular localization of LRE in the synergids.
For functional analysis, the synergids also presented an important opportunity to perform genetic studies. Synergids are haploid and essential for sperm release from the pollen tube for fertilization. Loss of LRE function affects the function of the synergids in pollen tube reception, reducing lre transmission through the female gametophyte (Capron et al., 2008; Tsukamoto et al., 2010). Conversely, complementation of the pollen tube reception defect by an LRE transgene will increase the transmission of both the lre mutation and the transgene. With LRE-cYFP, both types of transmission analysis produced the same conclusions (Table 1; Supplemental Table 1). Therefore, for functional analysis of all the variants of LRE, we used enhanced transmission of the transgene through the lre female gametophyte to assay complementation, as it was easier to establish the plants of the required genotype (LRE transgene/+ lre-7/lre-7) for this analysis.
Utilizing these two important opportunities, in this study, we fused the cYFP reporter to the LRE variants and used stable transformants to perform subcellular localization, pollen tube reception and seed set assays, transmission analysis, and non-cell-autonomous complementation studies. This integrated analysis allowed us to test the relevance of subcellular localization of LRE to its function, which led to the surprising finding that even when lacking domains critical for GPI anchor addition, LRE can still function in pollen tube reception by the synergid cell.
METHODS
LRE Sequence Analysis
Proteins with highest sequence similarity to Arabidopsis thaliana LRE in the indicated plants were identified, assembled, aligned, and box-shaded to identify the M8CM. Genes encoding LRE-like proteins in the indicated plants were identified by comparing the LRE coding sequence against genomic sequence of corresponding species using tBLASTx on the Accelerating Comparative Genomics (CoGe) Platform (https://genomevolution.org/CoGe/CoGeBlast.pl), with default settings (E-value cutoff, 0.001; word size, 3; gap existence penalty,11; gap extending penalty, 1). Top 100 hits were retrieved from BLAST and assembled in Geneious software (Geneious R6.1.8) using default settings; ∼3 kb of the genomic sequence on each side of the assembly was retrieved from the CoGe Organism View tool (https://genomevolution.org/CoGe/OrganismView.pl), and the coding region was identified by aligning it with the LRE coding sequence. For Physcomitrella patens, the coding sequences of LRE-like genes were identified using RNaseq reads (Chang et al., 2014) in Integrated Genome Browser (http://bioviz.org/igb/index.html). In plants where multiple LRE-like genes were found, the one that shared highest identity with Arabidopsis LRE was chosen for further analysis. Protein alignment was performed on ClustalW Web server (http://www.ch.embnet.org/software/ClustalW.html), with the default settings (scoring matrix, Blosum; opening gap penalty, 10; extending gap penalty, 0.05; end gap penalty, 10; separation gap penalty, 0.05), and the box shade was generated by Mobyle box shade tool (http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::boxshade). In each protein sequence, the signal peptide was predicted by SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011), and the ω site was predicted by BIG-PI plant predictor (http://mendel.imp.ac.at/gpi/plant_server.html) (Eisenhaber et al., 2003).
For GPI8 sequence analysis, Homo sapiens GPI8 (hGPI8) and Schizosaccharomyces pombe GPI8 (yGPI8) protein sequence were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/) and aligned with Arabidopsis GPI8 using ClustalW. Pairwise protein identity and similarity were calculated using the SIAS tool (http://imed.med.ucm.es/Tools/sias.html).
Plant Materials and Growth Conditions
Arabidopsis seeds were surface sterilized and plated on 0.5× Murashige and Skoog plates containing the corresponding antibiotics. Columbia (Col-0) is the ecotype of all Arabidopsis seeds used in this study. Seeds on plates were stratified at 4°C for 2 to 3 d before moving them to the growth chamber maintained at 20°C and continuous light (Philips F17T8/TL741 fluorescent tube light bulb, 100 to 150 µmol·m−2·s−1). The 7- to 10-d-old seedlings were transferred to the soil and were grown as described (Kessler et al., 2010). lre-5, lre-7 (CS66104), and fer-4 were reported previously (Tsukamoto et al., 2010; Li et al., 2015). The gpi8-2/+ (CS853564) seeds were from ABRC.
Cloning Transgenic Constructs
The cYFP (E1403; Tian et al., 2004) reporter gene was fused to wild-type and mutated LRE and expressed from the LRE promoter. To fuse the cYFP (E1403; Tian et al., 2004) reporter gene either toward the N terminus or C terminus of LRE, fragments were PCR amplified using primers and templates listed in Supplemental Table 9. The fragments were then fused by overlap PCR and the LRE-cYFP fusion under the control of LRE promoter was cloned into pENTR/D-TOPO (Invitrogen; K2400-20). For the N-terminal fusion construct (ProLRE:cYFP-LRE), the cYFP coding sequence (Tian et al., 2004) was cloned immediately downstream of the predicted secretion signal sequence (Supplemental Figure 2A). The C-terminal fusion construct (ProLRE:LRE-cYFP) was generated by placing the cYFP coding sequence slightly upstream of the predicted ω site (Figure 2A). Cloned transgenes in pENTR/D-TOPO were swapped into the Gateway destination vector pH7WG using Clonase II enzyme mix (Life Technologies; 11791020). The resultant plasmids containing the N-terminal and C-terminal fusions of cYFP to LRE were designated ProLRE:cYFP-LRE and ProLRE:LRE-cYFP, respectively. Swapping also resulted in these plasmids inheriting unique AscI restriction site from the entry vector, which facilitated cloning of modified LRE-cYFP constructs (see below).
For all other mutated, truncated, or ectopically expressed LRE-YFP constructs used in this study, fragments with desired changes in LRE coding regions were introduced by PCR using primers and templates listed in Supplemental Table 9. Full-length constructs were assembled by overlap PCR and then cloned into the ProLRE:LRE-cYFP plasmid that was linearized with SpeI/AscI and using In-Fusion HD Cloning Plus (Clontech; 638909). Linearizing the plasmid followed by in-fusion cloning essentially resulted in swapping wild-type LRE-cYFP with the modified LRE-cYFP transgene.
For constructs with organelle markers expressed from the LRE promoter, vectors carrying the organelle marker (G-rb, Px-rb, and ER-rb plasmids; Nelson et al., 2007) were used to PCR amplify the organelle marker. The LRE promoter and the mCherry coding sequence along with NOS or 35S terminator were PCR amplified using the primers and templates listed in Supplemental Table 9. These fragments were then assembled by overlap PCR to obtain the desired transgene that was cloned into EcoRI linearized pFGC19 using In-Fusion HD Cloning Plus.
Plant Transformation
Transformation solution containing Agrobacterium tumefaciens (GV3101 strain) harboring the desired plasmid were sprayed onto Arabidopsis inflorescences (Chung et al., 2000). Hygromycin-resistant transformants were selected as described (Harrison et al., 2006).
Isolation of Single Insertion Lines
For each construct, multiple T1 hygromycin-resistant transformants were selected and moved to soil. T1 plants are expected to be heterozygous for the transgene at the insertion locus. Upon bolting, stage 12c buds (Smyth et al., 1990) were emasculated and 24 h after emasculation cYFP expression in synergid cells was scored. For each construct, the number of T1 lines scored for cYFP expression and seed set are provided in Supplemental Figures 2 and 8. Among these lines, candidate single insertion lines were identified based on ∼50% ovules containing the cYFP signal. Pollen from candidate single insertion lines were then crossed to wild-type female and progeny were analyzed for segregation of hygromycin resistance. Those lines that showed a 1:1 segregation of hygromycin resistance:susceptibility were considered single insertion lines.
The above approach was used for all constructs except ProLRE:LRE-cYFPΔGAS. When the progeny of crosses were checked for hygromycin resistance, we obtained slightly higher than expected transmission of the ProLRE:LRE-cYFPΔGAS transgene through the male gametophyte (we do not yet understand why), for the construct carrying this transgene (Supplemental Table 5). Hence, for this construct, single insertion lines were merely identified using 1:1 segregation of cYFP-positive ovules in T1 hygromycin-resistant transformants.
RNA Isolation and RT-PCR
RNA was isolated according to Qin et al. (2009). Briefly, ∼20 pistils were collected 24 h after emasculation for each sample and were frozen in −80°C until RNA extraction. RNA was isolated using RNeasy Plant Mini Kit (Qiagen; catalog #74904) according to manufacturer’s instruction. RNA samples were then subjected to RNase-free DNase I (Life Technologies; catalog #AM2222) treatment to remove potential DNA contamination, cleaned up using the RNeasy MinElute Cleanup Kit (Qiagen; catalog #74204), and tested for RNA integrity on Agilent Bioanalyzer 2100 (Agilent Technologies).
For RT-PCR, cDNA was synthesized from 5 µg total RNA using the ThermoScript RT-PCR System (Life Technologies; catalog #11146-024), and genes of interest (LRE-cYFP transgene and ACTIN11) were amplified using TaKaRa Ex Taq DNA polymerase (Fisher Scientific; catalog #TAK_RR01BM), with primers listed in Supplemental Table 9.
Confocal Imaging
Fluorescent images were taken using a Leica SP5 confocal laser scanning microscope system. For cYFP imaging, samples were excited with a 488-nm laser line, and emission spectra between 510 and 550 nm were collected. For colocalization analysis, cYFP and mCherry signals were detected in sequential scan mode, with the excitation at 488 and 543 nm, and emission spectra at 510 to 550 nm and 580 to 660 nm, respectively, were collected. YFP images were processed and quantified with ImageJ software (http://imagej.nih.gov/ij/). Single confocal sections were used for colocalization analyzed on Leica Application Suite (LAS 2.6.0.7200).
Pollen Tube Reception Assays
In vivo pollen tube reception assays were done as previously described (Tsukamoto et al., 2010). Briefly, ProLAT52:GUS pollen was crossed to emasculated stage 14 pistils of the indicated plants. Crossed pistils were then collected 24 h after pollination, stained for GUS activity, and imaged using differential interference contrast optics in a Zeiss Axiovert 100 microscope. Aniline blue staining was performed according to Mori et al. (2006). Stained pistils were mounted with 15% glycerol, and pollen tube behavior was scored under a Zeiss Axiovert 100 fluorescent microscope.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers GPI8 (AT1G08750), human GPI8 (NP_005473), yeast GPI8 (CAC13970), LRE (At4g26466), and FER (At3g51550).
Supplemental Data
Supplemental Figure 1. Multiple sequence alignment of LRE-like proteins.
Supplemental Figure 2. cYFP-LRE and LRE-cYFP, but not LREΔSP-cYFP, complemented the lre-7 seed set defect.
Supplemental Figure 3. FA localization of LRE-cYFP remains unchanged upon pollen tube arrival or burst.
Supplemental Figure 4. Puncta of LRE-cYFP in synergids does not colocalize with the ER, Golgi, or peroxisome markers.
Supplemental Figure 5. LRE-cYFPΔGAS is released from the synergids.
Supplemental Figure 6. Genotyping of F1 progeny from a cross between gpi8-2/+(♀) and ProLRE:LRE-cYFP (♂).
Supplemental Figure 7. The gpi8-2 mutation does not affect LRE-cYFP-TM localization in the FA.
Supplemental Figure 8. Seed set of T1 plants carrying synergid cell-expressed constructs.
Supplemental Figure 9. Pollen tube-expressed LRE-cYFP, LRE-cYFPΔ2ω, LRE-cYFPΔGAS, and LRE-cYFP-TM complemented lre seed set defect.
Supplemental Table 1. Enhanced transmission of the ProLRE:LRE-cYFP transgene through the lre-7 female gametophyte.
Supplemental Table 2. Transmission of the gpi8-2 mutation through the male gametophyte, but not through the female gametophyte, is abolished.
Supplemental Table 3. Abnormal LRE-cYFP localization cosegregates with the gpi8-2 mutation.
Supplemental Table 4. LRE-cYFP-TM localization in the FA of synergids is not affected by the gpi8-2 mutation.
Supplemental Table 5. Enhanced transmission of transgenes with mutated GPI anchor addition domains through the lre-7 female gametophyte.
Supplemental Table 6. Normal transmission of mutated M8CM transgenes through the lre-7 female gametophyte.
Supplemental Table 7. ProPLL3:LRE-cYFP is transmitted at an enhanced rate to the progeny when crossed to a lre-5 female but not wild-type female.
Supplemental Table 8. ProPLL3:LRE-cYFP-TM is transmitted at an enhanced rate to the progeny when crossed to a lre-7 female but not wild-type female.
Supplemental Table 9. Primers used in this study.
Supplementary Material
Acknowledgments
We thank B. Fane (University of Arizona) for discussions on structural analysis of 8CM in LRE; A. Cheung (University of Massachusetts, Amherst) for fer-4 seeds; A. Nebenführ (University of Tennessee) for organelle markers; C. Seffren, K. Clark, and D. Byrne for technical assistance; J. Mach for language editing; and M. Johnson for critical reading of the manuscript. J.N. was supported by National Science Foundation Graduate Research Fellowship Grant DGE-1143953. This work was supported by a grant from the National Science Foundation to R.P. (IOS-1146090).
AUTHOR CONTRIBUTIONS
X.L., E.S., and R.P. designed the experiments. X.L., C.C., N.P., Y.W., J.N., M.B., and C.H. performed the experiments. R.P. and X.L. wrote the article.
Glossary
- GPI
glycosylphosphatidylinositol
- ROS
reactive oxygen species
- FA
filiform apparatus
- ER
endoplasmic reticulum
- SP
signal peptide
- 8CM
eight-cysteine motif
- M8CM
modified 8CM
- CRP
cysteine-rich protein
References
- Amien S., Kliwer I., Márton M.L., Debener T., Geiger D., Becker D., Dresselhaus T. (2010). Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol. 8: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale K.M., Johnson M.A. (2013). Speed dating, rejection, and finding the perfect mate: advice from flowering plants. Curr. Opin. Plant Biol. 16: 590–597. [DOI] [PubMed] [Google Scholar]
- Benghezal M., Benachour A., Rusconi S., Aebi M., Conzelmann A. (1996). Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 15: 6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Frietsch S., Kim T.H., Dizon M.B., Schroeder J.I. (2008). The peroxin loss-of-function mutation abstinence by mutual consent disrupts male-female gametophyte recognition. Curr. Biol. 18: 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy M.G.R., Kosentka P.Z., Willet A.H., Zhang L., Miller E.J., Shpak E.D. (2016). A mutation in the catalytic subunit of the glycosylphosphatidylinositol transamidase disrupts growth, fertility and stomata formation in Arabidopsis. Plant Physiol. http://dx.doi.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bütikofer P., Malherbe T., Boschung M., Roditi I. (2001). GPI-anchored proteins: now you see 'em, now you don’t. FASEB J. 15: 545–548. [DOI] [PubMed] [Google Scholar]
- Capron A., Gourgues M., Neiva L.S., Faure J.E., Berger F., Pagnussat G., Krishnan A., Alvarez-Mejia C., Vielle-Calzada J.P., Lee Y.R., Liu B., Sundaresan V. (2008). Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell 20: 3038–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.Y., Lin W.D., Tu S.L. (2014). Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 165: 826–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Udenfriend S., Prince G.M., Maxwell S.E., Ramalingam S., Gerber L.D., Knez J., Medof M.E. (1996). A defect in glycosylphosphatidylinositol (GPI) transamidase activity in mutant K cells is responsible for their inability to display GPI surface proteins. Proc. Natl. Acad. Sci. USA 93: 2280–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.H., Chen M.K., Pan S.M. (2000). Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res. 9: 471–476. [DOI] [PubMed] [Google Scholar]
- Denninger P., Bleckmann A., Lausser A., Vogler F., Ott T., Ehrhardt D.W., Frommer W.B., Sprunck S., Dresselhaus T., Grossmann G. (2014). Male-female communication triggers calcium signatures during fertilization in Arabidopsis. Nat. Commun. 5: 4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering T.L., Schekman R. (1996). GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J. 15: 182–191. [PMC free article] [PubMed] [Google Scholar]
- Duan Q., Kita D., Johnson E.A., Aggarwal M., Gates L., Wu H.M., Cheung A.Y. (2014). Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 5: 3129. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Bork P., Eisenhaber F. (1998). Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 11: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Wildpaner M., Schultz C.J., Borner G.H., Dupree P., Eisenhaber F. (2003). Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol. 133: 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo J.M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W.C., Grossniklaus U. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660. [DOI] [PubMed] [Google Scholar]
- Fliegmann J., Uhlenbroich S., Shinya T., Martinez Y., Lefebvre B., Shibuya N., Bono J.J. (2011). Biochemical and phylogenetic analysis of CEBiP-like LysM domain-containing extracellular proteins in higher plants. Plant Physiol. Biochem. 49: 709–720. [DOI] [PubMed] [Google Scholar]
- Galian C., Björkholm P., Bulleid N., von Heijne G. (2012). Efficient glycosylphosphatidylinositol (GPI) modification of membrane proteins requires a C-terminal anchoring signal of marginal hydrophobicity. J. Biol. Chem. 287: 16399–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjetting K.S., Ytting C.K., Schulz A., Fuglsang A.T. (2012). Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot. 63: 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O., Baird G.S., Campbell R.E., Zacharias D.A., Tsien R.Y. (2001). Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276: 29188–29194. [DOI] [PubMed] [Google Scholar]
- Hamamura Y., Nishimaki M., Takeuchi H., Geitmann A., Kurihara D., Higashiyama T. (2014). Live imaging of calcium spikes during double fertilization in Arabidopsis. Nat. Commun. 5: 4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.J., Mott E.K., Parsley K., Aspinall S., Gray J.C., Cottage A. (2006). A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T., Sharma A., Mariappan M., Eshleman H.D., Gutierrez E., Hegde R.S. (2011). Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 475: 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.Q., Russell S.D. (1992). Female germ unit - Organization, isolation, and function. Int. Rev. Cytol. 140: 233–293. [Google Scholar]
- Huck N., Moore J.M., Federer M., Grossniklaus U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159. [DOI] [PubMed] [Google Scholar]
- Iwano M., Ngo Q.A., Entani T., Shiba H., Nagai T., Miyawaki A., Isogai A., Grossniklaus U., Takayama S. (2012). Cytoplasmic Ca2+ changes dynamically during the interaction of the pollen tube with synergid cells. Development 139: 4202–4209. [DOI] [PubMed] [Google Scholar]
- José-Estanyol M., Gomis-Rüth F.X., Puigdomènech P. (2004). The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 42: 355–365. [DOI] [PubMed] [Google Scholar]
- Kasahara R.D., Portereiko M.F., Sandaklie-Nikolova L., Rabiger D.S., Drews G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S.A., Grossniklaus U. (2011). She’s the boss: signaling in pollen tube reception. Curr. Opin. Plant Biol. 14: 622–627. [DOI] [PubMed] [Google Scholar]
- Kessler S.A., Shimosato-Asano H., Keinath N.F., Wuest S.E., Ingram G., Panstruga R., Grossniklaus U. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971. [DOI] [PubMed] [Google Scholar]
- Lalanne E., Honys D., Johnson A., Borner G.H., Lilley K.S., Dupree P., Grossniklaus U., Twell D. (2004). SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leydon A.R., Beale K.M., Woroniecka K., Castner E., Chen J., Horgan C., Palanivelu R., Johnson M.A. (2013). Three MYB transcription factors control pollen tube differentiation required for sperm release. Curr. Biol. 23: 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., et al. (2015). Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Tan Z.M., Zhu L., Niu Q.K., Zhou J.J., Li M., Chen L.Q., Zhang X.Q., Ye D. (2013). MYB97, MYB101 and MYB120 function as male factors that control pollen tube-synergid interaction in Arabidopsis thaliana fertilization. PLoS Genet. 9: e1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H., Kessler S.A., Müller L.M., Shimosato-Asano H., Boisson-Dernier A., Grossniklaus U. (2015). TURAN and EVAN mediate pollen tube reception in Arabidopsis synergids through protein glycosylation. PLoS Biol. 13: e1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S.G., Briarty L.G., Erni S. (1991). Early embryogenesis in Arabidopsis thaliana. 1. The mature embryo sac. Can. J. Bot. 69: 447–460. [Google Scholar]
- Mao Y., Zhang Z., Wong B. (2003). Use of green fluorescent protein fusions to analyse the N- and C-terminal signal peptides of GPI-anchored cell wall proteins in Candida albicans. Mol. Microbiol. 50: 1617–1628. [DOI] [PubMed] [Google Scholar]
- Marshall E., Costa L.M., Gutierrez-Marcos J. (2011). Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J. Exp. Bot. 62: 1677–1686. [DOI] [PubMed] [Google Scholar]
- Mayor S., Riezman H. (2004). Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5: 110–120. [DOI] [PubMed] [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8: 64–71. [DOI] [PubMed] [Google Scholar]
- Müller G., Bandlow W. (1993). Glucose induces lipolytic cleavage of a glycolipidic plasma membrane anchor in yeast. J. Cell Biol. 122: 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ngo Q.A., Vogler H., Lituiev D.S., Nestorova A., Grossniklaus U. (2014). A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev. Cell 29: 491–500. [DOI] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361. [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Tsukamoto T. (2012). Pathfinding in angiosperm reproduction: pollen tube guidance by pistils ensures successful double fertilization. Wiley Interdiscip. Rev. Dev. Biol. 1: 96–113. [DOI] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Qin Y., Leydon A.R., Manziello A., Pandey R., Mount D., Denic S., Vasic B., Johnson M.A., Palanivelu R. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L.J., Li L., Lan Z., Dresselhaus T. (2015). Peptide signalling during the pollen tube journey and double fertilization. J. Exp. Bot. 66: 5139–5150. [DOI] [PubMed] [Google Scholar]
- Rotman N., Gourgues M., Guitton A.E., Faure J.E., Berger F. (2008). A dialogue between the SIRENE pathway in synergids and the fertilization independent seed pathway in the central cell controls male gamete release during double fertilization in Arabidopsis. Mol. Plant 1: 659–666. [DOI] [PubMed] [Google Scholar]
- Russell S.D. (1992). Double fertilization. Int. Rev. Cytol. 140: 357–387. [Google Scholar]
- Schiøtt M., Romanowsky S.M., Baekgaard L., Jakobsen M.K., Palmgren M.G., Harper J.F. (2004). A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc. Natl. Acad. Sci. USA 101: 9502–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C., Gilson P., Oxley D., Youl J., Bacic A. (1998). GPI-anchors on arabinogalactan-proteins: implications for signalling in plants. Trends Plant Sci. 3: 426–431. [Google Scholar]
- Sedbrook J.C., Carroll K.L., Hung K.F., Masson P.H., Somerville C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharom F.J., Lehto M.T. (2002). Glycosylphosphatidylinositol-anchored proteins: structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochem. Cell Biol. 80: 535–549. [DOI] [PubMed] [Google Scholar]
- Shenoy-Scaria A.M., Gauen L.K., Kwong J., Shaw A.S., Lublin D.M. (1993). Palmitylation of an amino-terminal cysteine motif of protein tyrosine kinases p56lck and p59fyn mediates interaction with glycosyl-phosphatidylinositol-anchored proteins. Mol. Cell. Biol. 13: 6385–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy-Scaria A.M., Kwong J., Fujita T., Olszowy M.W., Shaw A.S., Lublin D.M. (1992). Signal transduction through decay-accelerating factor. Interaction of glycosyl-phosphatidylinositol anchor and protein tyrosine kinases p56lck and p59fyn 1. J. Immunol. 149: 3535–3541. [PubMed] [Google Scholar]
- Simpson C., Thomas C., Findlay K., Bayer E., Maule A.J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Yuan L., Sundaresan V. (2014). Antipodal cells persist through fertilization in the female gametophyte of Arabidopsis. Plant Reprod. 27: 197–203. [DOI] [PubMed] [Google Scholar]
- Sun L., van Nocker S. (2010). Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol. 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H., Higashiyama T. (2012). A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 10: e1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.Y., Speicher D.W. (2004). Determination of disulfide-bond linkages in proteins. Curr. Protoc. Protein Sci. 11: 11. [DOI] [PubMed] [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T., Qin Y., Huang Y., Dunatunga D., Palanivelu R. (2010). A role for LORELEI, a putative glycosylphosphatidylinositol-anchored protein, in Arabidopsis thaliana double fertilization and early seed development. Plant J. 62: 571–588. [DOI] [PubMed] [Google Scholar]
- Twell D. (2011). Male gametogenesis and germline specification in flowering plants. Sex. Plant Reprod. 24: 149–160. [DOI] [PubMed] [Google Scholar]
- Varma R., Mayor S. (1998). GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394: 798–801. [DOI] [PubMed] [Google Scholar]
- Wang X. (2001). Plant phospholipases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 211–231. [DOI] [PubMed] [Google Scholar]
- Wuest S.E., Vijverberg K., Schmidt A., Weiss M., Gheyselinck J., Lohr M., Wellmer F., Rahnenführer J., von Mering C., Grossniklaus U. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20: 506–512. [DOI] [PubMed] [Google Scholar]
- Yadegari R., Drews G.N. (2004). Female gametophyte development. Plant Cell 16 (suppl.): S133–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.C., Shi D.Q., Chen Y.H. (2010). Female gametophyte development in flowering plants. Annu. Rev. Plant Biol. 61: 89–108. [DOI] [PubMed] [Google Scholar]
- Yu J., Nagarajan S., Knez J.J., Udenfriend S., Chen R., Medof M.E. (1997). The affected gene underlying the class K glycosylphosphatidylinositol (GPI) surface protein defect codes for the GPI transamidase. Proc. Natl. Acad. Sci. USA 94: 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacks M.A., Garg N. (2006). Recent developments in the molecular, biochemical and functional characterization of GPI8 and the GPI-anchoring mechanism [review]. Mol. Membr. Biol. 23: 209–225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.