NIP4;1 and NIP4;2, two pollen-specific aquaporins, are paralogous genes displayed in tandem that are critical for appropriate pollen development and pollen tube growth.
Abstract
In flowers with dry stigmas, pollen development, pollination, and pollen tube growth require spatial and temporal regulation of water and nutrient transport. To better understand the molecular mechanisms involved in reproductive processes, we characterized NIP4;1 and NIP4;2, two pollen-specific aquaporins of Arabidopsis thaliana. NIP4;1 and NIP4;2 are paralogs found exclusively in the angiosperm lineage. Although they have 84% amino acid identity, they displayed different expression patterns. NIP4;1 has low expression levels in mature pollen, while NIP4;2 expression peaks during pollen tube growth. Additionally, NIP4;1pro:GUS flowers showed GUS activity in mature pollen and pollen tubes, whereas NIP4;2pro:GUS flowers only in pollen tubes. Single T-DNA mutants and double artificial microRNA knockdowns had fewer seeds per silique and reduced pollen germination and pollen tube length. Transport assays in oocytes showed NIP4;1 and NIP4;2 function as water and nonionic channels. We also found that NIP4;1 and NIP4;2 C termini are phosphorylated by a pollen-specific CPK that modifies their water permeability. Survival assays in yeast indicated that NIP4;1 also transports ammonia, urea, boric acid, and H2O2. Thus, we propose that aquaporins NIP4;1 and NIP4;2 are exclusive components of the reproductive apparatus of angiosperms with partially redundant roles in pollen development and pollination.
INTRODUCTION
In angiosperms, water and nutrient status regulation is relevant for pollen development, germination, and pollen tube growth (Samuel et al., 2009; Pacini et al., 2011; Firon et al., 2012). During its developmental program and functioning, pollen grains experience changes in water and nutrient content in relation to the surrounding environment (reviewed in Firon et al., 2012). While pollen develops inside the anther immersed in the locular fluid, pollen volume increases through hydration and vacuolation. During the dehydration phase, pollen water content decreases, reaching a minimum at maturity. Upon landing on a compatible stigma, pollen grains rapidly hydrate and germinate to produce pollen tubes (Preuss et al., 1993). The rate of pollen hydration depends on both the capacity of the stigma to support the flow of water and the degree of hydration of the pollen grains at the time of the capture (Heslop-Harrison, 1979). In plants with dry stigmas, pollen-stigma adhesion is a highly species-specific process in which the stigma has the ability to recognize pollen from compatible species while rejecting pollen from unrelated species or self-incompatible pollen. In Arabidopsis thaliana, adhesion first relies on the pollen exine wall (Zinkl et al., 1999) and then the lipid- and protein-rich coat flows out to form an interface between the two cell surfaces, promoting the transfer of water, nutrients, and other small molecules (Murphy, 2006). Regardless of the still unknown transfer mechanisms, pollen hydration is regulated both temporally and spatially. It has been reported that inappropriate hydration inhibits fertilization due to premature germination within the anther (Johnson and McCormick, 2001) or to germination on an unsuitable surface (Lolle et al., 1998). Finally, during pollen tube growth, additional water entry through the plasma membrane is necessary to adjust turgor pressure and cytosolic concentration of ions (such as Ca2+, K+, H+, and Cl−) and other nutrients (Sommer et al., 2008) relevant for pollen tube growth.
Changes in pollen hydration and pollen tube growth require molecular mechanisms that control water and solute levels. Aquaporins (AQPs), also named membrane intrinsic proteins, are highly diversified channel proteins, which facilitate the bidirectional transport of water, small solutes such as H2O2, urea, glycerol, metalloids (boric, silicic, and arsenious acid), lactic acid, and/or dissolved gas molecules (CO2 and NH3) across cell membranes in all living organisms (Soto et al., 2012; Perez Di Giorgio et al., 2014; Abascal et al., 2014). Plant AQPs are classified into seven subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), X intrinsic proteins (XIPs), hybrid intrinsic proteins, and GlpF-like intrinsic proteins (Abascal et al., 2014). AQPs have been identified in most tissues of the anthers (Ruiter et al., 1997; O’Brien et al., 2002; Alexandersson et al., 2005; Bots et al., 2005a; Fujita et al., 2010) and some are pollen specific (Bock et al., 2006; Maeshima and Ishikawa, 2008; Soto et al., 2008; this study). Pollen AQPs transport water, small solutes, and/or gases during pollen development, germination, and pollen tube growth (Bock et al., 2006; Soto et al., 2008; Wudick et al., 2014). It has been reported that transient expression of the Arabidopsis PIP2 group members in lily (Lilium longiflorum) pollen enhanced water channel activity, suggesting that they play a role in pollen water transport (Sommer et al., 2008). AQPs of the PIP2 class were also suggested to be required for anther dehydration in tobacco (Nicotiana tabacum; Bots et al., 2005b). Recently, simulations using an osmotic model of pollen tube growth predicted the involvement of mercury-sensitive water channels as osmosensors, suggesting the presence of AQP-like molecules at the plasma membrane of pollen tubes (Shachar-Hill et al., 2013). However, the function of AQPs during pollen development and pollen tube growth has not yet been clearly reported.
In Arabidopsis, only four AQP genes (out of 35 loci) are specifically expressed in mature pollen and/or pollen tubes: TIP1;3, TIP5;1, NIP4;1, and NIP4;2. Of these four pollen-specific genes, TIP1;3 and TIP5;1 have been previously characterized (Soto et al., 2008, 2010; Wudick et al., 2014). TIP1;3 is expressed in vesicles and vacuoles of vegetative cells. TIP5;1 is expressed in vacuoles of sperm cells, when expressed under its own promoter (Wudick et al., 2014), or in mitochondria of vegetative cells, when heterologously expressed under the control of the LAT52 promoter (Soto et al., 2010). Single tip1;3 and tip5;1 mutant plants showed no apparent growth defect phenotype and no significant distortion in the male transmission rate. However, double tip1;3 tip5;1 mutant plants showed an abnormal incidence of sterile pods, especially under water- or nutrient-deficient conditions (Wudick et al., 2014). Soto et al. (2008) established that TIP1;3 and TIP5;1 are AQPs with intermediate levels of permeability to water and high permeability to urea. In addition, it was shown that single tip1;3 and tip5;1 and double tip1;3 tip5;1 mutant plants showed reduced pollen tube elongation only when germinated in vitro in medium lacking nitrogen. These results suggested that TIP5;1 and TIP1;3 are involved in the nitrogen metabolic pathway during pollen tube growth (Soto et al., 2008, 2010).
In this study, we showed that NIP4;1 and NIP4;2, the two previously uncharacterized Arabidopsis pollen-specific AQP members, are important for proper pollen development, pollen germination, and pollen tube growth. Furthermore, we showed that while NIP4;1 functions during pollen development and germination, NIP4;2, functions exclusively during pollen tube growth. This observation, along with the fact that NIP4;1 and NIP4;2 belong to the NIP1 group, which is exclusive to angiosperms (Anderberg et al., 2012; Abascal et al., 2014), suggests that NIP4;1 and NIP4;2 contribute specifically to pollen germination and pollen tube growth in angiosperms.
RESULTS
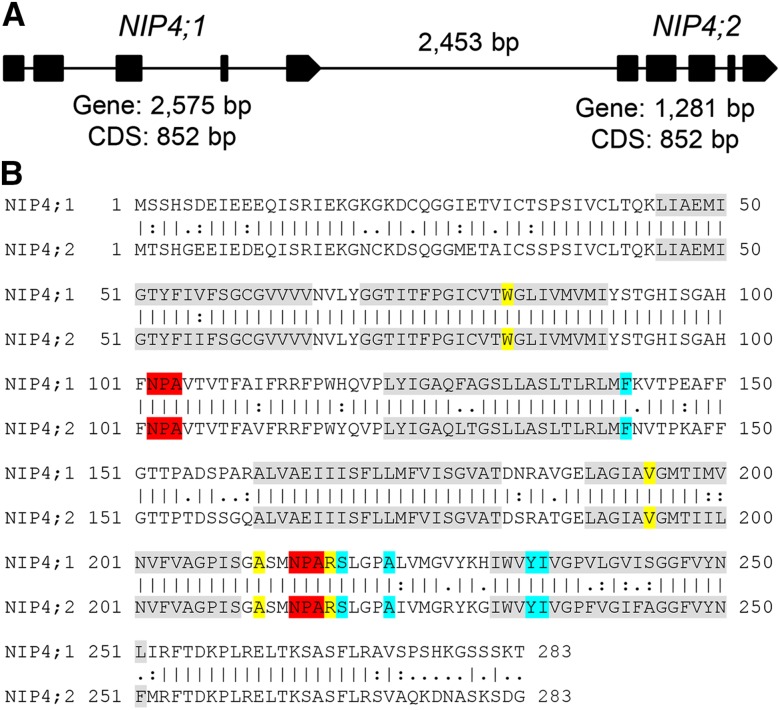
NIP4;1 and NIP4;2 are 88% identical at the nucleotide level and encode proteins with 84% amino acid identity. These genes are arranged in tandem, separated only by 2453 bp (Figure 1). AQP solute selectivity and permeability is mainly applied by the ar/R (aromatic/arginine) constriction, the narrowest constriction located close to the extracellular vestibule (Hove and Bhave, 2011). There is also evidence that the ar/R region creates a repulsive barrier against protons at the channel entrance (Kreida and Törnroth-Horsefield, 2015). According to the ar/R filter, NIPs can be further divided into three groups (Rougé and Barre, 2008). Group 1 is characterized by residues W V/I A R with permeability to water, glycerol, lactic acid, ammonia, arsenite, and hydrogen peroxide (Wallace and Roberts, 2004; Rougé and Barre, 2008; Perez Di Giorgio et al., 2014). NIP4;1 and NIP4;2 have the same conserved residues, also for other specificity motifs (Figure 1B), suggesting similar substrate transport specificity and, thus, possibly, functional redundancy.
Figure 1.
Gene Structure and Protein Sequence Alignment of Arabidopsis NIP4;1 and NIP4;2.
(A) Schematic structures of genomic DNA of NIP4;1 and NIP4;2. Exons are shown as black boxes. The length of each gene, coding DNA sequence (CDS), and distance between genes are indicated.
(B) Pairwise sequence alignment of NIP4;1 (Q9FIZ9) and NIP4;2 (Q9FIZ8) proteins using the EMBOSS Stretcher tool (www.ebi.ac.uk/Tools/psa/emboss_stretcher). Identity “|” = 238/283 (84.1%), similarity (“:” very similar and “.” moderately similar residues) = 258/283 (91.2%), and gaps = 0/283 (0.0%). Transmembrane domains (TM 1 to 6) are highlighted in gray, filter NPA residues in red, filter ar/R residues in yellow, and P1-P5 residues in light blue, as determined by DASTY3 (http://www.ncbi.nlm.nih.gov/pubmed/21798964).
In addition, syntenic analysis showed that NIP4;1 and NIP4;2 probably arose from a tandem gene duplication event in the Brassicaceae, suggesting that they are paralogous genes (Supplemental Figure 1 and Supplemental Data Set 1). Their fixation, long-term persistence, and conservation of amino acid sequences within different species of Brassicaceae suggest that these putative paralogs confer a selective advantage in this lineage, possibly due to the subdivision of their function and/or expression patterns.
Expression of NIP4;1 and NIP4;2
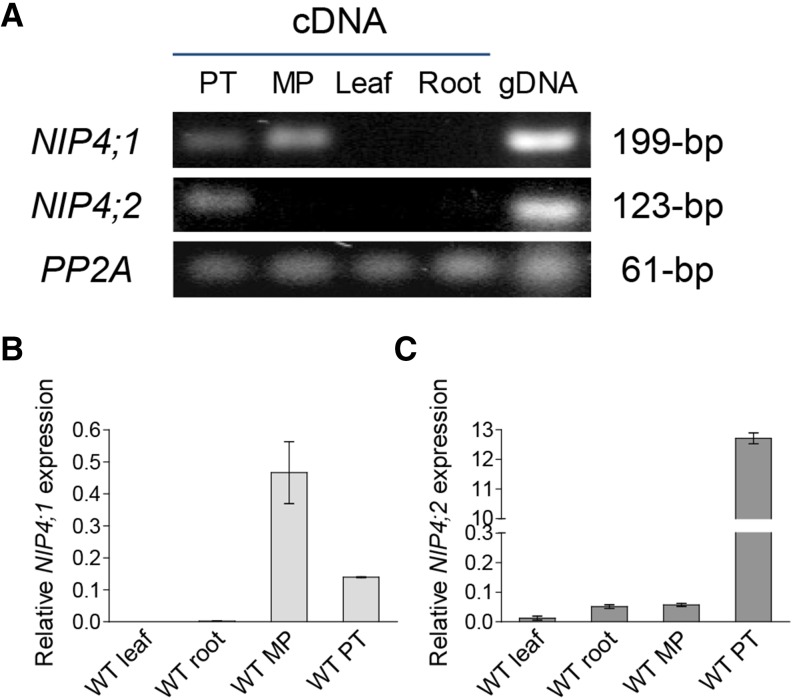
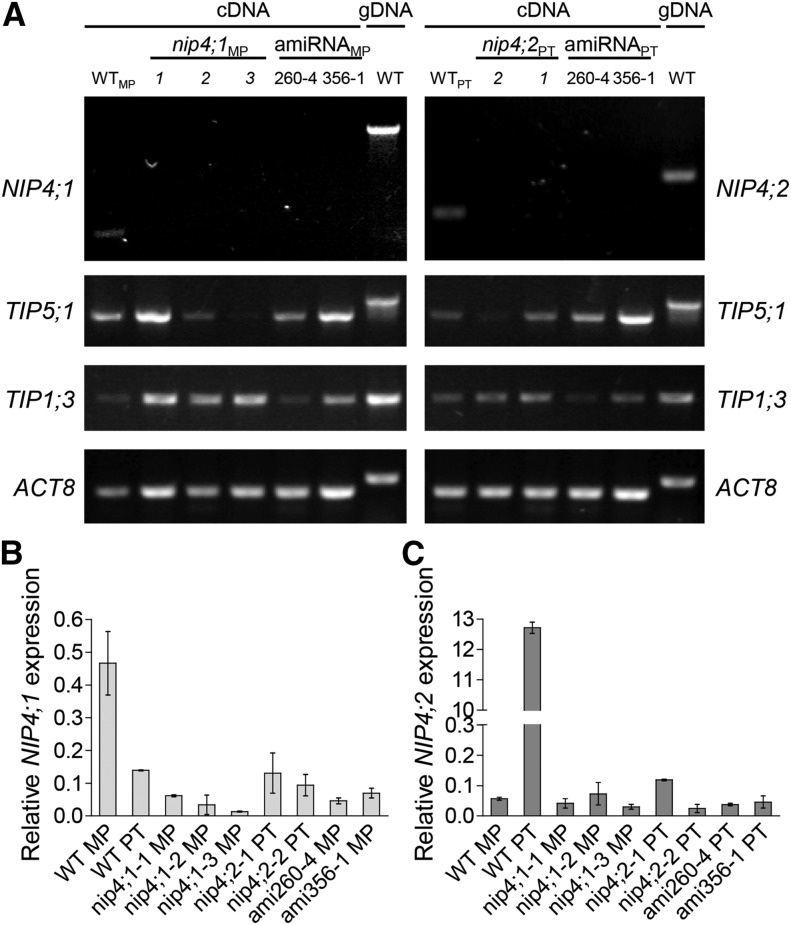
Based on comparative pollen transcriptomic studies, NIP4;1/NIP4;2 are expressed during pollen development, with moderate expression in the tricellular pollen stage. Expression decreases in mature pollen but peaks upon pollen germination (Honys and Twell, 2004; Wang et al., 2008; Qin et al., 2009; Boavida et al., 2011) (Table 1). Higher expression values were observed in pollen tubes grown in vitro than in mature pollen (Wang et al., 2008) and also in pollen grown semi-in vivo with respect to those grown in vitro (Qin et al., 2009), suggesting that the pistil influences NIP4;1/NIP4;2 expression. As ATH1 chip probes do not distinguish between these two genes, we performed RT-PCR and real-time quantitative-PCR (RT-qPCR) assays. Figures 2A to 2C show that both NIP4;1 and NIP4;2 transcripts were expressed in pollen. However, while NIP4;1 showed low expression at mature pollen when compared with other pollen-specific AQPs, such as TIP5;1 and TIP1;3 (Honys and Twell, 2004), NIP4;2 was strongly expressed in pollen tubes, with its expression being ∼224 times that in mature pollen (Figure 2C).
Table 1. Expression Levels of Arabidopsis Pollen Aquaporin Genes TIP5;1, TIP1;3, NIP4;1, and NIP4;2 during Pollen Development and Pollen Tube Growth.
| Microarray | Aquaporin | UCP | BCP | TCP | MP | HP | In Vitro PT | Semi-in Vivo PT | Sperm Cells |
|---|---|---|---|---|---|---|---|---|---|
| Honys and Twell (2004) | TIP5;1 | 1,018 | 1,263 | 1,981 | 3,101 | ||||
| TIP1;3 | 852 | 1,899 | 4,792 | 1,563 | |||||
| NIP4;1 NIP4;2 | 917 | 1,143 | 1,754 | 560 | |||||
| Wang et al. (2008) | TIP5;1 | 6,117 | 6,200 | 4,612 | |||||
| TIP1;3 | 555 | 490 | 1,606 | ||||||
| NIP4;1 NIP4;2 | 42 | 55 | 5,198 | ||||||
| Qin et al. (2009) | TIP5;1 | 29,889 | 29,625 | 25,765 | 25,816 | ||||
| TIP1;3 | 3,020 | 3,194 | 3,244 | 5,306 | |||||
| NIP4;1 NIP4;2 | 361 | 510 | 6,166 | 18,426 | |||||
| Borges et al. (2008) | TIP5;1 | 6,119 | 23,230 (P) | ||||||
| TIP1;3 | 5,397 | 69 (A) | |||||||
| NIP4;1 NIP4;2 | 201 | 63 (P) |
Expression values from unicellular pollen (UCP), bicellular pollen (BCP), tricellular pollen (TCP), and mature pollen (MP) were taken from Honys and Twell (2004). Hydrated pollen grains (HP), in vitro pollen tubes (PT), and semi-in vivo pollen tubes (PT) from Wang et al. (2008) and Qin et al. (2009). Sperm cells were from Borges et al. (2008). (A), absent; (P), present. Data correspond to the mean absolute values obtained in each microarray.
Figure 2.
NIP4;1 and NIP4;2 Expression in Arabidopsis.
Total RNA was isolated from the following tissues: pollen tubes (PT), mature pollen (MP), leaves, and roots of wild type Col-0 plants.
(A) RT-PCR analysis of NIP4;1 and NIP4;2 transcripts. Protein phosphatase 2 subunit A3 (PP2A) gene and genomic DNA (gDNA) were used as controls.
(B) and (C) RT-qPCR analysis of NIP4;1 and NIP4;2 transcripts. The expression of each gene was normalized to the expression of PP2A. Data are mean relative expression values ± se of two (leaf and root) and three (MP and PT) independent experiments in triplicate.
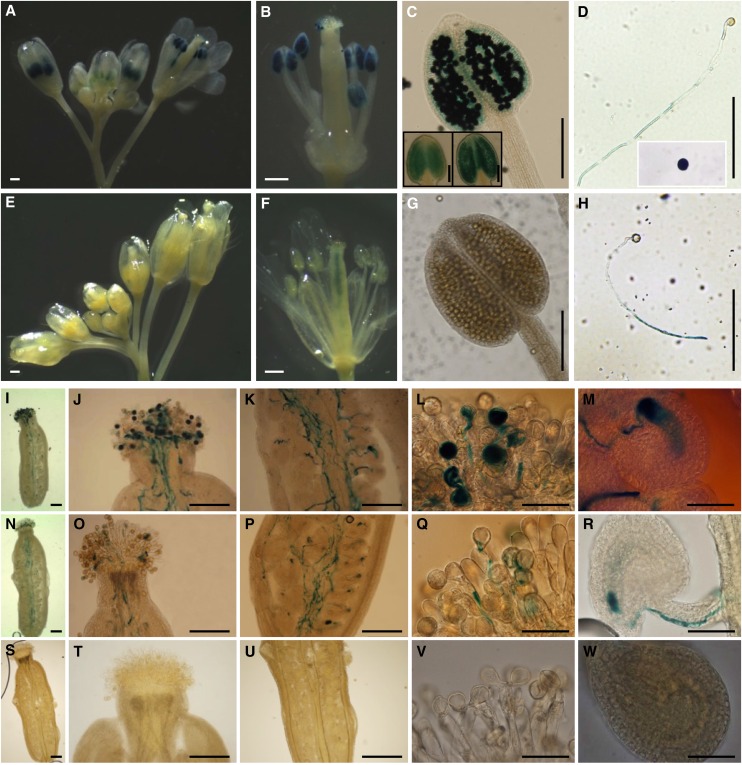
To confirm these results, we fused the putative promoter regions encompassing 2966 and 1959 bp for NIP4;1 and NIP4;2, respectively, to a GUS reporter gene and introduced these fusions into wild-type Arabidopsis plants. Two independent reporter lines for NIP4;1 and three for NIP4;2 were generated (Figure 3). In the F2 generation, GUS activity of NIP4;1pro:GUS flowers was observed in pollen grains inside the anthers (Figures 3A to 3C) and adhered to stigmatic cells (Figure 3J) and in pollen tubes (Figures 3I to 3M; Supplemental Figure 3A). In anthers, GUS activity was relatively weak during the early stages of pollen development (Figure 3C, inset) but increased in mature pollen (Figure 3C). By contrast, NIP4;2pro:GUS activity was only detected in self-pollinated pistils, but not in pollen grains (Figures 3E to 3G). Additionally, when pollen was germinated in vitro, GUS signal was observed in pollen tubes for both genes (Figures 3D and 3H). To distinguish whether NIP4;2pro:GUS activity corresponded with expression in pollen tubes or in the style transmitting tract, wild-type pistils were pollinated with NIP4;2pro:GUS pollen. Figures 3N to 3R and Supplemental Figure 3C demonstrate that NIP4;2 is indeed expressed in pollen tubes. As negative controls, the wild type (Figures 3S to 3W), NIP4;1pro:GUS pistils (Supplemental Figure 3B), and NIP4;2pro:GUS pistils (Supplemental Figure 3D) pollinated with wild-type pollen were used. No GUS activity was detected in NIP4;1pro:GUS and NIP4;2pro:GUS 5- and 10-d-old seedlings (Supplemental Figures 3E to 3N).
Figure 3.
Differential Expression of NIP4;1 and NIP4;2 in Pollen.
GUS staining of NIP4;1pro:GUS and NIP4;2pro:GUS transgenic Arabidopsis plants. Results were consistent among the lines. Bars = 500 µm in (A), (B), (E), and (F), 200 µm in (C), (D), (G) to (K), (N) to (P), and (S) to (U), and 50 µm in (L), (M), (Q), (R), (V), and (W).
(A) to (C) NIP4;1pro:GUS flowers showing GUS activity in developing anthers and mature pollen grains. Insets in (C) show GUS activity in two different stages of pollen development.
(D) NIP4;1pro:GUS pollen tube germinated in vitro and mature pollen grain (inset) showing GUS activity.
(E) to (G) NIP4;2pro:GUS flowers showing GUS activity in the transmitting tract of the pistils (F) but not in the developing anthers or mature pollen grains.
(H) NIP4;2pro:GUS pollen tube germinated in vitro for 4 h showing strong GUS activity in the tube.
(I) to (M) Wild-type pistils hand-pollinated with heterozygous NIP4;1pro:GUS pollen showing GUS activity in pollen grains and tubes.
(N) to (R) Wild-type pistils hand-pollinated with homozygote NIP4;2pro:GUS pollen showing GUS activity in pollen tubes.
(S) to (W) Wild-type pistils hand-pollinated with wild-type pollen used as a negative control.
(I), (N), and (S) Pollen grains and tubes in a pistil.
(J), (O), and (T) Pollen tubes entering the style.
(K), (P), and (U) Pollen tubes growing through the style.
(L), (Q), and (V) Pollen grains germinating at the stigma surface.
(M), (R), and (W) A pollen tube discharging in an ovule.
In summary, all these results indicate that NIP4;2 is only expressed in pollen tubes, while NIP4;1 is expressed in pollen grains and possibly also in pollen tubes. However, considering the RT-qPCR results, the GUS activity observed for NIP4;1pro:GUS in pollen tubes could be a consequence of its expression in mature pollen.
Subcellular Localization of GFP-Tagged NIP4;1 and NIP4;2
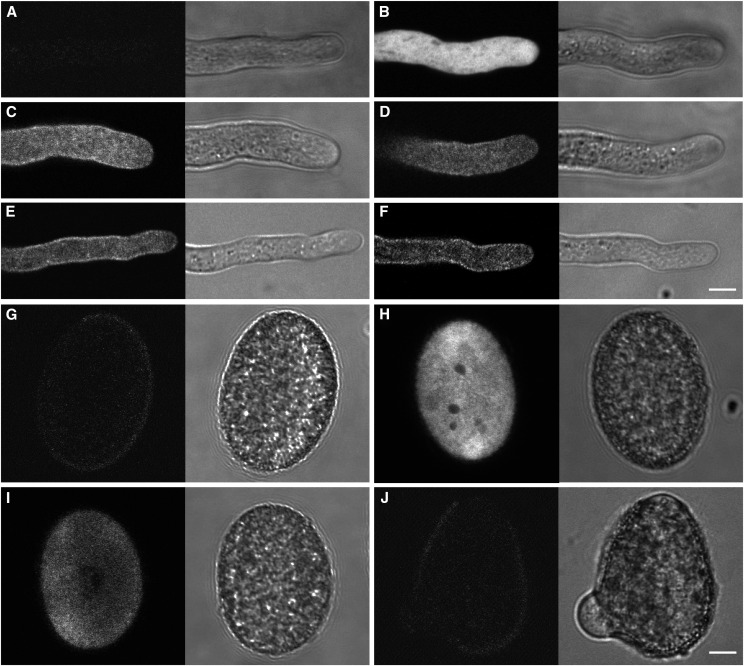
NIP4;1-enhanced GFP (EGFP) and NIP4;2-EGFP fusions were expressed in transgenic wild type and their respective nip4;1 and nip4;2 mutant plants under the control of their native promoters. NIP4;1 is localized in the plasma membrane and intracellular vesicles of pollen tubes (Figures 4C and 4D) and pollen grains (Figure 4I), while NIP4;2 is localized in the plasma membrane and intracellular vesicles only in pollen tubes (Figures 4E and 4F) but not in pollen grains (Figure 4J). Wild-type (Figures 4A and 4G) and LAT52pro:GFP (Figures 4B and 4H) pollen were used as negative and positive controls, respectively.
Figure 4.
Subcellular Localization of EGFP-NIP4;1 and EGFP-NIP4;2 in Pollen.
EGFP-NIP4;1 and EGFP-NIP4;2 were expressed in transgenic Arabidopsis plants under the control of their respective native promoters. Representative fluorescence confocal images (left panels) and differential interference contrast (right panels) of pollen tubes ([A] to [F]) and mature pollen ([G] to [J]). Bars = 10 µm.
(A) and (G) Wild-type pollen showing autofluorescence background.
(B) and (H) GFP signal in the LAT52pro:EGFP pollen tube and mature pollen, as controls.
(C) and (I) GFP signal in the PM and internal vesicles of NIP4;1pro:EGFP-NIP4;1 pollen tube and mature pollen (line NIP4;1-WT-15).
(D) GFP signal in the PM and internal vesicles of a NIP4;1pro:EGFP-NIP4;1 pollen tube (line NIP4;1-WT-20).
(E) GFP signal in the PM and internal vesicles of a NIP4;2pro:EGFP-NIP4;2 pollen tube (line NIP4;2-835-7).
(F) and (J) GFP signal in the PM and internal vesicles of a NIP4;2pro:EGFP-NIP4;2 pollen tube, but not in mature pollen (line NIP4;2-142-9). A total of 13 independent germinations analyzing at least 20 grains and pollen tubes for each genotype and condition were performed.
To determine whether these NIPs would cycle between plasma membrane and intracellular vesicles, germinating pollen was treated with Brefeldin A (BFA), a drug that inhibits the endoplasmic reticulum to Golgi transport and, therefore, exocytosis. Supplemental Figures 5B to 5E and 5J show a slight accumulation of EGFP-NIP4;1 in intracellular vesicles, especially at the tip of pollen tubes; then, after 60 min, a bubble tip and a large vacuole in the subapical region appeared, suggesting the arrest of pollen tube growth (Supplemental Figure 5E). In mature pollen, we also observed accumulation of EGFP-NIP4;1 vesicles near the germination zone (Supplemental Figure 5G), but not for EGFP-NIP4;2 lines (Supplemental Figure 5H). When germinating pollen was treated with tyrphostin A23, a drug that blocks endocytosis, we observed a stronger signal of EGFP-NIP4;1 at the membrane of the pollen tube tip (Supplemental Figure 5K). These results suggest that under our conditions of pollen tube growth, EGFP-NIP4;1 would experience cycling between the plasma membrane and intracellular vesicles.
Single T-DNA nip4;1 and nip4;2 Mutants and Double Artificial MicroRNA Knockdowns Showed Abnormal Pollen Transmission
To characterize the function of NIP4;1 and NIP4;2 in pollen development and pollination, three independent T-DNA insertion mutant alleles for NIP4;1 (SALK_038278.54.75, SALK_013924.51.75, and SALK_007730.42.45) and two for NIP4;2 (SALK_142789.54.50 and SAIL_799_E09) were obtained and named nip4;1-1, nip4;1-2, and nip4;1-3, and nip4;2-1 and nip4;2-2, respectively. As confirmed by PCR genotyping and sequencing, T-DNAs were inserted in exon 3, intron 2, and intron 3 for nip4;1-1, nip4;1-2, and nip4;1-3, respectively, and in the 5′ untranslated region and exon 5 for nip4;2-1 and nip4;2-2, respectively (Supplemental Figure 6).
To obtain the double mutant for NIP4;1 and NIP4;2, we had to employ artificial microRNA (amiRNA) technology because both genes were separated by only 2453 bp. Thus, we generated two amiRNA constructs that targeted 21-bp fragments at positions 260 and 356 in the coding region of both genes under the control of the LAT52 promoter (LAT52pro) (Supplemental Figure 7). Three independent lines, designated as ami260-4, ami356-1, and ami356-4 (in the wild-type background), and one qrt ami260-1 line (in the qrt background) were chosen (Supplemental Figure 8). We confirmed that all T-DNA insertion lines were knockout mutants, while ami260-4 and ami356-1 lines showed significantly reduced levels of NIP4;1 and NIP4;2 in mature pollen (Figure 5). Specific pollen aquaporin genes TIP5;1 and TIP1;3 were normally expressed in all of the amiRNA lines (Figure 5), suggesting that the amiRNAs were specific to both NIP4;1 and NIP4;2.
Figure 5.
Validation of Single T-DNA Mutant and Double amiRNA Knockdown Lines.
Total RNA was isolated from mature pollen (MP) and pollen tubes (PT) of wild-type Col-0 plants, single T-DNA mutant lines nip4;1-1, -2, and -3 and nip4;2-1 and -2, and double amiRNA knockdown lines 260-4 and 356-1.
(A) RT-PCR analysis of pollen-specific transcripts NIP4;1, NIP4;2, TIP5;1, and TIP1;3. To distinguish the cDNA from the gDNA (gDNA) band, the Actin 8 (ACT8) gene and genomic DNA were used as controls.
(B) and (C) RT-qPCR analysis of NIP4;1 and NIP4;2 transcripts. The expression of each gene is normalized to that of the protein phosphatase 2 subunit A3 (PP2A). Data are mean relative expression values ± se of three (wild-type, nip4;1, and nip4;2 lines) or two (ami260-4 and ami356-1 lines) independent experiments in triplicate.
When we analyzed the segregation of self-crosses of heterozygous plants for single mutants nip4;1-1, nip4;1-2, nip4;1-3, nip4;2-1, and nip4;2-2 (Table 2, single mutants, self-crosses), we observed a statistically significant distortion of the expected 1:2:1 segregation ratio, with a marked deficiency in the number of obtained homozygous nip4;1 and nip4;2 mutants. Next, we observed that the progeny of all the double knockdown lines displayed a significant deviation from the expected 3:1 ratio, indicating a gametophytic defect (Table 2, double knockdowns resulting from self-crosses). To analyze whether these abnormal segregations were due to a defect in the male or female gametophyte, we conducted reciprocal crosses (Table 2, single mutants and double knockdowns resulting from reciprocal crosses, respectively). While an expected Mendelian 1:1 transmission was observed through the female gametophyte, a reduced transmission was found when mutant pollen was used. All these results suggest that T-DNA nip4;1 and nip4;2 mutants and double amiRNA knockdowns result only in defective pollen.
Table 2. Segregation Analysis of Single T-DNA nip4;1 and nip4;2 Mutants and Double amiRNA Knockdowns.
| Cross | Parent Female × Male | F1 Segregation Genotype or Phenotype | Expected Ratio | Observed Ratio | Observed Ratio | χ2 P Value |
|---|---|---|---|---|---|---|
| Single mutants (self-crosses) | nip4;1-1+/− | NIP4;1+/+:+/−:−/− | 1:2:1 | 3.0:2.8:1 | 86:81:29 | P < 0.0001 |
| nip4;1-3+/− | NIP4;1+/+:+/−:−/− | 1:2:1 | 2.0:2.1:1 | 107:111:53 | P < 0.0001 | |
| nip4;1-2+/− | NIP4;1+/+:+/−:−/− | 1:2:1 | 1.9:2.3:1 | 46:54:24 | P < 0.01 | |
| nip4;2-2+/− | NIP4;2+/+:+/−:−/− | 1:2:1 | 2.0:1.9:1 | 122:110:58 | P < 0.0001 | |
| nip4;2-1+/− | NIP4;2+/+:+/−:−/− | 1:2:1 | 2.4:2.1:1 | 79:68:33 | P < 0.0001 | |
| Double knockdowns (self-crosses) | ami356-1+/− | KanR:KanS | 3:1 | 1.7:1 | 1037:600 | P < 0.0001 |
| ami356-4+/− | KanR:KanS | 3:1 | 1.4:1 | 112:80 | P < 0.0001 | |
| ami260-4+/− | KanR:KanS | 3:1 | 1.6:1 | 546:342 | P < 0.0001 | |
| qrt ami260-1+/− | KanR:KanS | 3:1 | 1.2:1 | 234:201 | P < 0.0001 | |
| Single mutants (reciprocal crosses) | wt × nip4;1-1+/− | KanR:KanS | 1:1 | 0.8:1 | 162:213 | P < 0.01 |
| nip4;1-1+/− × wt | KanR:KanS | 1:1 | 1.0:1 | 50:48 | P = 0.84 | |
| wt × nip4;1-3+/− | KanR:KanS | 1:1 | 0.7:1 | 95:140 | P < 0.01 | |
| nip4;1-3+/− × wt | KanR:KanS | 1:1 | 1.0:1 | 120:123 | P = 0.85 | |
| wt × nip4;1-2+/− | KanR:KanS | 1:1 | 0.7:1 | 145:200 | P < 0.01 | |
| nip4;1-2+/− × wt | KanR:KanS | 1:1 | 1.0:1 | 97:97 | P = 1.00 | |
| wt × nip4;2-2+/− | BastaR:BastaS | 1:1 | 0.8:1 | 284:367 | P < 0.01 | |
| nip4;2-2+/− × wt | BastaR:BastaS | 1:1 | 1.0:1 | 63:62 | P = 0.93 | |
| Double knockdowns (reciprocal crosses) | wt × ami356-1+/− | KanR:KanS | 1:1 | 0.7:1 | 82:125 | P < 0.01 |
| ami356-1+/− × wt | KanR:KanS | 1:1 | 1.0:1 | 94:98 | P = 0.77 | |
| Complementation lines (self-crosses) | nip4;1-2−/− NIP4;1-9+/− | BastaR:BastaS | 3:1 | 3.7:1 | 246:67 | P = 0.14 |
| nip4;2-1−/− NIP4;2-9+/− | BastaR:BastaS | 3:1 | 3.9:1 | 324:83 | P < 0.05 |
Disruption of NIP4;1 and NIP4;2 genes caused defective male-specific gene transmission. Segregation analysis of complementation lines showed that EGFP-NIP4;1 and EGFP-NIP4;2 rescued the nip4;1 and nip4;2 pollen transmission defect. wt, the wild type.
To confirm that the male transmission phenotype was caused by a loss of function of NIP4;1 and NIP4;2, we generated complementation transgenic lines expressing EGFP-NIP4;1 and EGFP-NIP4;2 under endogenous NIP4;1 and NIP4;2 promoters, respectively, in the nip4;1-2 and nip4;2-1 backgrounds. We found that nip4;1-2(−/−)/EGFP-NIP4;1-9(+/−) and nip4;2-1(−/−)/EGFP-NIP4;2-9(+/−) lines rescued the defective pollen mutant (Table 2, complementation lines).
Single T-DNA nip4;1 and nip4;2 Mutants and Double amiRNA Knockdowns Have Reduced Fertility
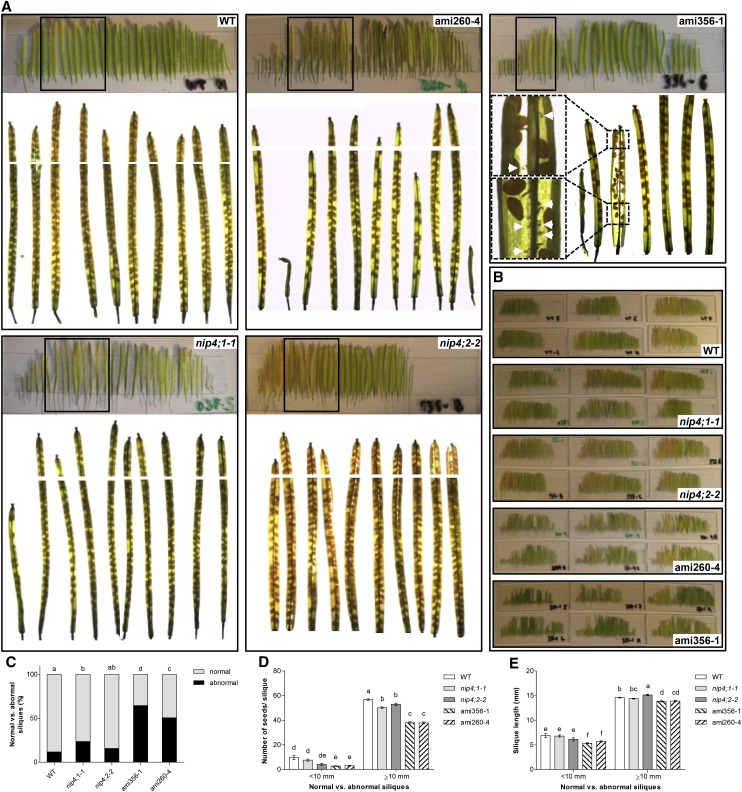
To analyze in more detail the impairment found in mutant pollen, we measured the length of the siliques and counted the number of seeds in all mature siliques of the primary shoot of self-crosses of single mutants nip4;1-1 and nip4;2-2 and double knockdowns ami356-1 and ami260-4 (Figures 6A and 6B). Data were analyzed in two categories: abnormal stunted siliques (length <10 mm) and normal siliques (≥10 mm).
Figure 6.
Fertility Assays: Number of Seeds per Silique and Silique Length.
(A) Siliques of the primary shoot of wild-type, single T-DNA nip4;1-1 and nip4;2-2 mutants, and double knockdown ami260-4 and ami356-1 plants. The boxed areas correspond to the enlarged images of cleared siliques shown underneath. Images were captured to cover the entire length of the siliques and used for seed counts and silique length measurements. White lines show the junction between the images. Insets in ami356-1 show an enlargement of the indicated silique section, and white arrows indicate unfertilized or aborted ovules.
(B) Siliques from wild-type, nip4;1-1, nip4;2-2, ami260-4, and ami356-1 mutant plants placed on slides. Each slide corresponds to the siliques from left to right in order of appearance in the primary shoot of a plant.
(C) to (E) Percentage of abnormal (<10 mm) and normal (≥10 mm) siliques (C), number of seeds per silique (D), and silique length (E) of all mature siliques in the primary shoot of wild-type, single mutant nip4;1-1 and nip4;2-2, and double knockdown ami260-4 and ami356-1 plants (n = 22, 27, 11, 31, and 24 plants per genotype, respectively). Data represent the mean values ± se. Different letters indicate significant difference (P < 0.05, one-way ANOVA, Tukey’s test).
We found that the percentage of abnormal siliques was higher only in nip4;1-1 and in double knockdown plants (Figure 6C). In contrast to wild-type plants, in which the first two flowers of the inflorescence were only occasionally infertile, in double knockdown lines, the percentage of abnormal siliques with undeveloped ovules was significantly higher (Figure 6A; see white arrows in the inset of ami356-1). After the onset of flowering, and approximately for the first 2 weeks, double knockdown plants developed stunted siliques that were mostly seedless. Then, the plants produced siliques of normal size, but still with fewer seeds than the wild type, alternating with cycles of abnormal, stunted siliques (Figure 6A). Regarding the number of seeds and silique length, nip4;1-1, nip4;2-2, and double knockdown lines had fewer seeds (Figure 6D), while only the double knockdown had shorter siliques (P < 0.0001) compared with wild-type plants (Figure 6E). Finally, the average ratio of seeds per silique length was 3.5 (P < 0.0001) for both single mutants nip4;1-1 and nip4;2-2, and 2.7 (P < 0.0001) for double knockdowns, compared with 3.9 for wild-type plants. All these results suggest that fertility is compromised in the single mutants and double knockdowns.
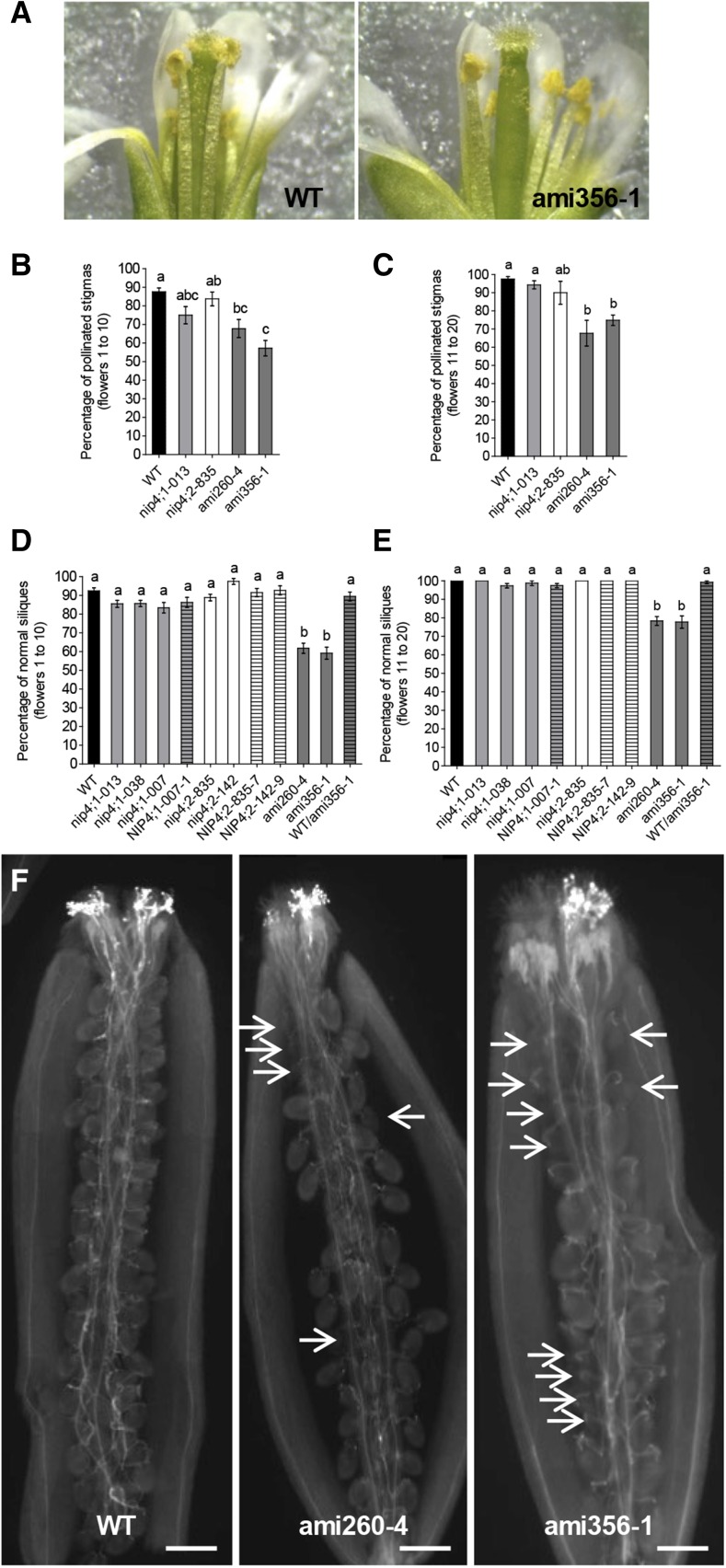
We then examined the pollen content on the stigma surface of opened flowers (stages late 13 and 14) of the primary shoot. During stage 13, anthesis and pollination take place while in stage 14, long anthers extend above the stigma surface, and fertilization occurs (Ferrándiz et al., 1999). Stigmas were classified as pollinated and nonpollinated with no pollen grains adhered to the stigma (Figure 7A). We compared the first 10 flowers (flowers 1 to 10, corresponding to the first 2 weeks after flowering onset) to flowers 11 to 20 (from the third and fourth week). In both cases, only the double knockdowns showed significantly lower percentages of pollinated stigmas compared with the wild type (Figures 7B and 7C). We also evaluated the percentage of normal and abnormal, stunted siliques in the primary shoot of the mutants. Figures 7D and 7E show that the percentages of normal siliques in double knockdown lines were significantly lower than in wild-type plants. Interestingly, in double knockdown lines, the percentage of abnormal siliques was similar to that of nonpollinated pistils.
Figure 7.
Pollination Defects in Double Knockdown Lines.
(A) Stigmas were classified into two categories: pollinated (wild type) or nonpollinated (double knockdown ami356-1).
(B) and (C) Percentage of pollinated stigmas of the flowers in positions 1 to 10 (B) and 11 to 20 (C) in the primary shoot of wild-type (n = 17), single mutant nip4;1-2 (n = 8) and nip4;2-2 (n = 8), and double knockdown ami260-4 (n = 16) and ami356-1 (n = 17) plants. Data represent the mean values ± se. Different letters indicate significant difference (P < 0.05, one-way ANOVA, Tukey’s test).
(D) and (E) Percentage of normal siliques (≥10 mm) in positions 1 to 10 (D) and 11 to 20 (E) in the primary shoot of wild-type (n = 33), single mutant nip4;1-2 (n = 29), nip4;1-1 (n = 49), nip4;1-2 (n = 26), nip4;2-2 (n = 27), and nip4;2-1 (n = 8), homozygous ami260-4 (n = 36) and ami356-1 (n = 36), and heterozygous wild-type/ami356-1+/− (n = 14) plants. Data represent the mean values ± se. Different letters indicate significant difference (P < 0.05, one-way ANOVA, Tukey’s test).
(F) Aniline blue staining of pistils corresponding to the first 10 self-crossed flowers from stage 15 in the primary shoot of wild-type and double knockdown ami260-4 and ami356-1 plants (n = 6 for each genotype). White arrows indicate nonfertilized ovules (only funiculus staining is observed). Bar = 100 µm.
In summary, double knockdown plants showed a defect in pollination and a reduced number of normal siliques. They either failed to mature in a coordinated way with the female organs and/or to adhere to the stigma. However, considering that pollen is always in excess, it is feasible to expect complete fertilization despite reduced pollination.
To analyze whether fertilization is also compromised in the knockdown lines, we stained pollen tubes with aniline blue in self-pollinated pistils from stage 15 postfertilization (Ferrándiz et al., 1999). While wild-type pollen reached the ovules (Figure 7F), ami260-4 and ami356-1 pollen led to a high number of unfertilized ovules and a lack of pollen tubes in the vicinity (Figure 7F, arrows; Supplemental Figures 9E and 9F). Therefore, these results suggest that double knockdown pollen would have not only deficient pollination but also defective fertilization due to reduced pollen germination and/or pollen tube growth.
When pollen morphology was evaluated, we found that double knockdown pollen grains showed a collapsed shape on the stigma surface (Supplemental Figures 9A to 9D), suggesting defects in development and/or rehydration.
Pollen Development Is Compromised in Single T-DNA nip4;1 Mutant and Double Knockdown amiRNA Plants
To establish whether the reduced fertility of mutants nip4;1 and nip4;2 and double knockdown plants was caused by a defect in pollen development, we stained nuclei of mature pollen grains with Hoechst 33,342 (Supplemental Figure 10). We found a slight increase of immature (uni- and bicellular) pollen grains only for nip4;1-2 and ami356-1 and ami260-4, when compared with wild-type flowers (Supplemental Table 1). Then, we evaluated the viability of mature pollen after fluorescein diacetate treatment (FDA) (Supplemental Figure 11). We found a minor increased percentage of dead and/or collapsed pollen and a reduced relative volume only in single nip4;1 mutants and double knockdowns, compared with wild-type flowers (Supplemental Table 2).
All these results suggest that in the double knockdown and single mutant nip4;1, but not in nip4;2 plants, pollen development is affected with a delay at the uni- and bicellular stages losing viability and collapsing in some cases. Thus, defects in pollen maturation would be associated with the absence of NIP4;1, but not NIP4;2, in good agreement with the expression pattern of NIP4;1 during pollen development and the restricted expression of NIP4;2 in pollen tubes.
Pollen Tube Growth Is Affected in Single T-DNA nip4;2 Mutant and Double Knockdown amiRNA Plants
To assess whether the reduction in the transmission of the male mutant allele was caused by a deficiency in pollen germination and/or pollen tube growth, we performed in vivo and in vitro pollen tube growth assays.
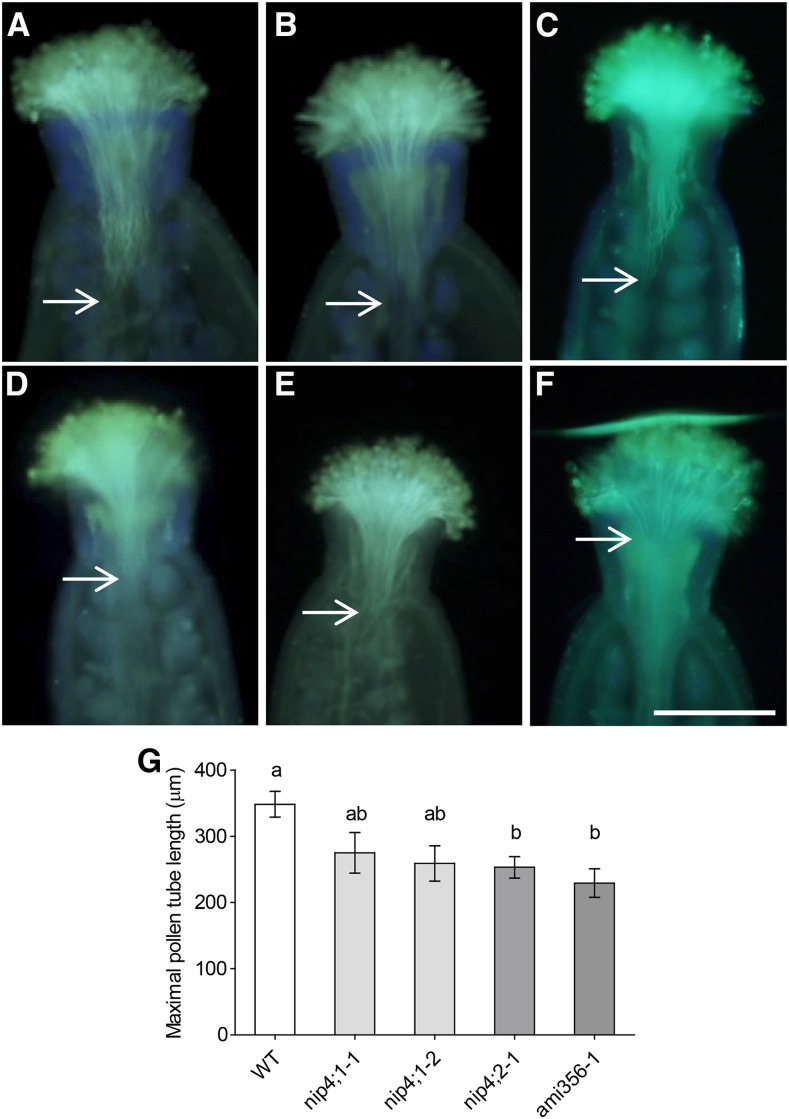
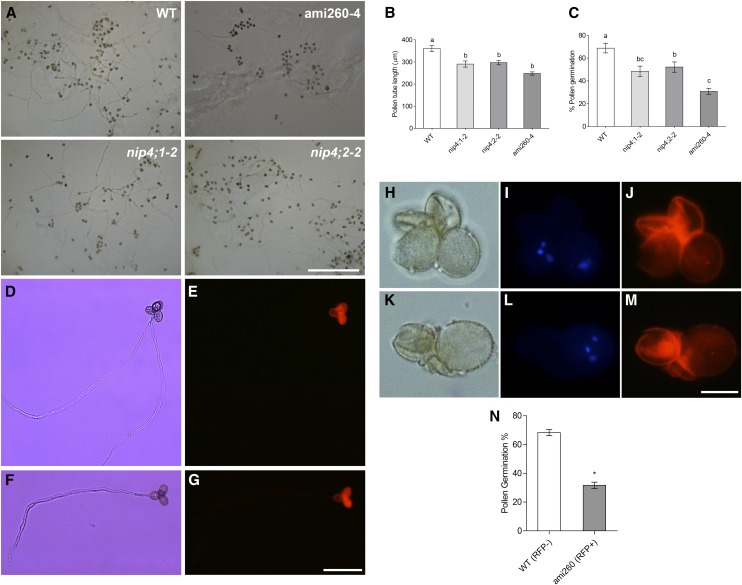
First, we examined in vivo pollen tube growth with aniline blue staining. Figure 8G shows that the pollen tubes of nip4;2-1 and double knockdown plants, but not nip4;1 mutants, were shorter than those of the wild type, suggesting a role for NIP4;2 in pollen tube growth. Additionally, aniline blue staining of self-crossed pistils of double knockdown and single mutant flowers with fully pollinized stigmas showed nonfertilized ovules at stage 15 postfertilization, suggesting a pollen growth defect associated with the absence of both NIP4;1 and NIP4;2 (Supplemental Figure 12). Then, we performed in vitro germination assays in solid pollen germination medium (PGM) for 5 h. Figures 9A to 9C show that nip4;1-2, nip4;2-2, and ami260-4 pollen had a reduced germination percentage and shorter pollen tubes when compared with wild-type pollen.
Figure 8.
Aniline Blue Staining of Pollen Tubes in Vivo.
(A) to (F) Wild-type pistils were cross-pollinated with wild-type (A), single mutant nip4;1-1 (B), nip4;1-3 (C), nip4;2-1 (D), and double knockdown ami356-1 ([E] and [F]) pollen. Aniline blue staining of pollen tubes was performed 1 h after hand-pollination. Bar = 200 µm.
(G) Maximal pollen tube length achieved 1 h after pollination. Data represent the mean values ± se of 12 to 20 independent pollinations per pollen genotype. Different letters indicate significant difference (P < 0.05, one-way ANOVA, Tukey’s test).
Figure 9.
In Vitro Pollen Germination and Pollen Tube Growth Assays.
(A) Germination of wild-type, single mutants nip4;1-2 and nip4;2-2, and double knockdown ami260-4 pollen in solid PGM after 5 h. Bar = 500 µm.
(B) and (C) Pollen tube length (B) and percentage of germinated pollen (C) after 5 h. Data represent mean values ± se of four to six replicates (i.e., plants) for each genotype repeated four times. We analyzed only repetitions for which we could measure ≥100 pollen tubes and counted ≥200 pollen grains. Different letters indicate significant difference (P < 0.05, one-way ANOVA, Tukey’s test).
(D) to (N) Germination of heterozygous qrt ami260-1/wild type (RFP+/RFP−) pollen in liquid PGM after 5 h. Bright-field ([D], [F], [H], and [K]) and RFP ([E], [G], [J], and [M]) or Hoechst 33342 ([I] and [L]) images.
(D) to (G) Tetrads showing germination deficiency of double knockdown ami260-4 pollen. Bar = 100 µm.
(H) to (M) Pollen grain tetrads showing double knockdown grains with a collapsed shape. Nuclei were stained with Hoechst 33342 ([I] and [L]). Bar = 20 µm.
(N) Percentage of germinated pollen after 5 h ± se of three replicates (i.e., plants) (*P < 0.05, paired t test).
To confirm that the pollen disorder in the double knockdown plants was due to the expression of the amiRNA, we generated transgenic ami260-1 plants heterozygous for the LAT52pro:RFP-LAT52pro:ami260 construct in the quartet mutant background (Johnson-Brousseau and McCormick, 2004). In vitro germination assays showed that 5.9% of the double knockdown pollen was abnormal, with a collapsed shape (Figures 9H to 9M), in accordance with our scanning electron microscopy images (Supplemental Figure 9) and FDA assays (Supplemental Figure 11). When pollen germination was analyzed, only 31.7% of the pollen tubes was RFP positive (Figures 9D to 9G and 9N). Despite the fact that we only analyzed one of the amiRNA transgenic lines in the quartet background, these results support our previous findings that double knockdown plants have defects in pollen development and germination.
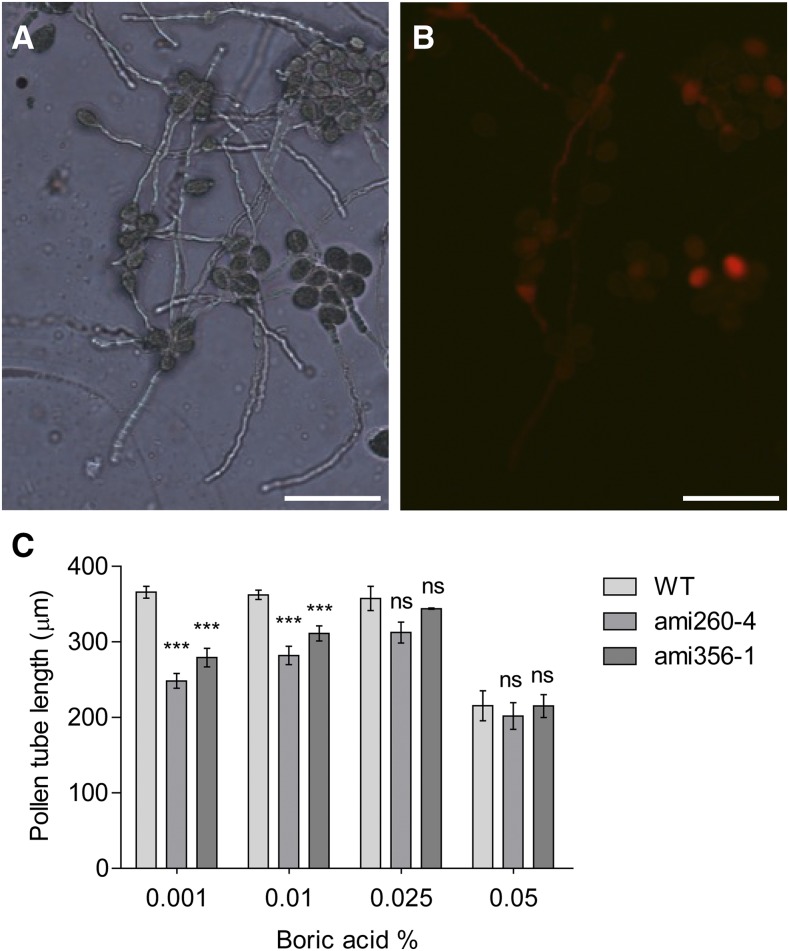
Finally, we investigated whether pollen tube length is affected when double knockdown pollen was germinated under different stress conditions. We used heterozygous ami260-4+/− and ami356-1+/− plants in which double knockdown pollen is expressed in association with the RFP marker, allowing measurement of the length of double knockdown and wild type pollen tubes in the same germination assay (Figures 10A to 10C). We first evaluated four different boric acid concentrations: 0.001%, 0.01% (standard concentration in Arabidopsis PGM), 0.025%, and 0.05%. We observed a significant decrease in the mean length of double knockdown pollen tubes in the presence of 0.01% boric acid, being more significant with 0.001% (Figure 10C). However, no differences were found with 0.025%, suggesting that increasing boric acid concentration could compensate for the germination defects of the double knockdowns. In the presence of 0.05% boric acid, pollen tube length decreased for all genotypes, possibly due to a toxic effect of boron. These results suggest that boron limitation is an important aspect on double knockdown pollen fitness.
Figure 10.
Effect of Boric Acid on Pollen Tube Growth.
(A) and (B) In vitro pollen germination assay of pollen isolated from heterozygous ami356-1/wild type (RFP+/RFP−) plants in solid PGM with 0.001% boric acid under white light (A) and using a red filter to differentiate double knockdown ami356-1 pollen (RFP+) from wild-type pollen (RFP−) (B). Bars = 100 µm.
(C) Mean pollen tube length ± se of heterozygous ami260-4/wild type and ami356-1/wild type plants germinated in solid PGM with 0.001, 0.01, 0.025, or 0.05% boric acid. Assays were performed in four to five replicates (i.e., plants) and ≥100 pollen tubes were measured for each replicate. One-way ANOVA, Tukey’s test, indicates differences between amiRNA and wild-type pollen tube length for the 0.01 and 0.001% (**P < 0.01 and ***P < 0.001), but not for 0.025 and 0.05% (ns, not significant) boric acid treatment.
When we analyzed salinity, temperature, and osmotic stresses, we observed a general reduction of pollen tube length for wild-type and double knockdown pollen (Supplemental Figure 13), suggesting that all of these stress conditions affect both backgrounds to the same extent. In contrast to standard conditions, we only found a tendency toward decreased pollen tube length in double knockdowns grown in the presence of 0.08 M glycerol. Higher concentrations (0.16, 0.24, and 0.32 M) of glycerol resulted in pollen tube bursting. These results suggest that under mild osmotic stress, the pollen tube defects of the double knockdowns are visible.
Phosphorylation of C Termini of NIP4;1 and NIP4;2
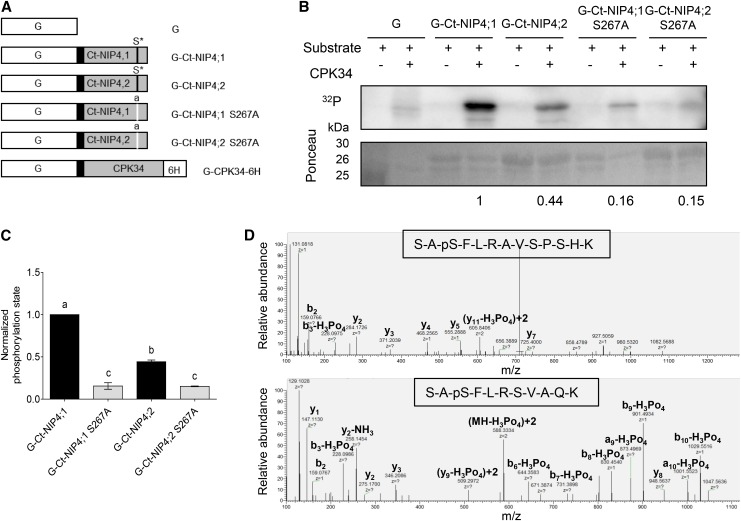
Proteome-wide mapping of in vivo phosphorylation sites in Arabidopsis showed that NIP4;1 and NIP4;2 are phosphorylated at Ser-267 (Sugiyama et al., 2008), as seen in soybean (Glycine max) NOD26 and in other NIP proteins (Wallace et al., 2006). Curran et al. (2011) demonstrated that a synthetic peptide based on the NIP4;1 sequence (jfh-752: LRELTKSAS*FLRAVS) was phosphorylated in vitro by calcium-dependent protein kinases (CPKs) 1, 10, and 34, with substrate specificity differences among the CPKs (relative phosphorylation levels were 28, 7, and 35, respectively). Using mutant peptides where selected serines or threonines (underlined) were replaced with alanines, the phospho-site was mapped to Ser-267 (indicated by an asterisk).
To analyze whether Ser-267 phosphorylation is involved in the regulation of the transport activity, we first performed in vitro phosphorylation of NIP4;1 and NIP4;2 by CPK, using their complete C-terminal domain (Figure 11A). We chose CPK34 because it is essential for normal polarized pollen tube growth in Arabidopsis (Myers et al., 2009). Both NIP4;1 and NIP4;2 wild-type C-terminal domains are phosphorylated (Figures 11B and 11C), and phosphorylation levels of NIP4;1 S267A and NIP4;2 S267A markedly decreased. Tandem mass spectrometry analysis revealed that NIP4;1 and NIP4;2 C-terminal domains are indeed specifically in vitro phosphorylated at Ser-267 by CPK34 (Figure 11D).
Figure 11.
Phosphorylation of NIP4;1 and NIP4;2 C-Terminal Domains.
(A) Diagram of the GST fusion proteins used for analysis of CPK phosphorylation sites of the NIP4;1 and NIP4;2 C-terminal domains. pGEX-6P-1 vectors expressing GST (G, white boxes) or GST fused to wild-type NIP4 or mutated NIP4-S267A C-terminal domains (Ct-NIP, gray boxes), linked by a PreScission protease site (black boxes). Asterisk labeled S* residue indicates wild-type phosphorylation site; lowercase “a” indicates an S-to-A substitution at position 267 (S267A) by site-directed mutagenesis of nucleotide T at position 799 to G (t799g). pGEX-4T vector expressing CDPK34 located between N-terminal GST and a C-terminal 6 His tag (6H; kindly provided by J.F. Harper).
(B) Kinase reactions showing in vitro phosphorylation of GST only (G), G-Ct-NIP4;1, G-Ct-NIP4;2, G-Ct-NIP4;1-S267A, and G-Ct-NIP4;2-S267A C-terminal fusion proteins. Autoradiogram of stained gel showing proteins labeled by 32P-phosphorylation. Ponceau-stained gel showing relative loading amounts of substrates for each kinase reaction. Results shown are representative of three independent analyses. Note that the purified G-Ct-NIP4s and G-Ct-NIP4s S267A fusion proteins run as two bands corresponding to full-length fusion protein (30 kD) and GST (26 kD), respectively. The 25-kD band in the first row corresponds to the protein marker.
(C) Relative phosphorylation levels were normalized to the amount of Ponceau-stained protein for each substrate calculated by densitometry, and the background phosphorylation level of GST was subtracted from each sample. Results shown are the average of three independent analyses for each variant. Error bars show se. Different letters indicate statistically significant differences (P < 0.05, one-way ANOVA, Tukey’s test).
(D) Mass spectra of NIP4;1 and NIP4;2 S267-phosphorylated peptides. A loss of 98 D (H3PO4) is observed only for the Ser-267 residue.
NIP4;1 and NIP4;2 Transport Water and Glycerol in Xenopus laevis Oocytes
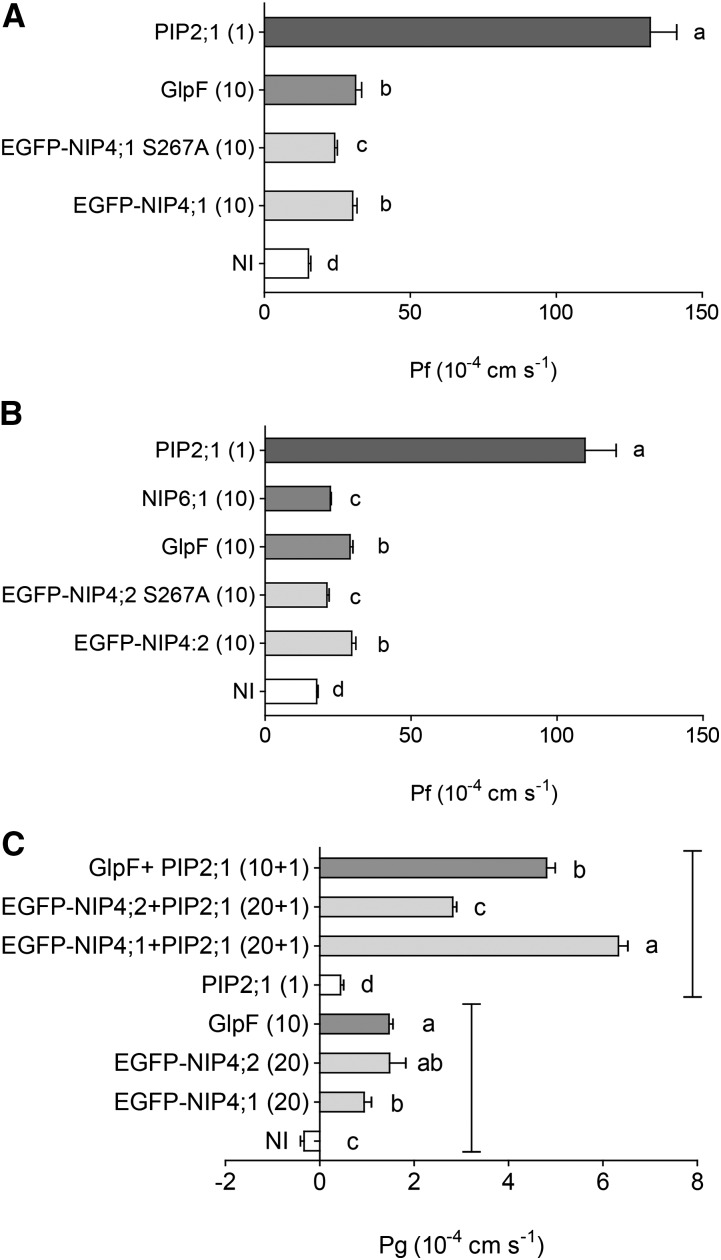
To functionally characterize NIP4;1 and NIP4;2 transport activity, we used X. laevis oocytes. Figures 12A and 12B show that EGFP-NIP4;1 and EGFP-NIP4;2 displayed low plasma membrane (PM) osmotic water permeability values (Pf: 30.33 × 10−4 cm/s and 29.68 × 10−4 cm/s, respectively) relative to the levels of the positive control Fragaria ananassa PIP2;1 (Pf: 132.19 − 109.60 × 10−4 cm/s). For EGFP-NIP4;1 and EGFP-NIP4;2, the values were ∼2-fold higher than Pf of noninjected oocytes. These results are consistent with AQPs that transport not only water but also nonionic solutes such as Arabidopsis NIP6;1 and Escherichia coli GlpF (Pf: 31.33 × 10−4 and 29.15 × 10−4 cm/s, respectively) (Figures 12A and 12B). Glycerol transport assays showed that when EGFP-NIP4;1 and EGFP-NIP4;2 were coinjected with F. ananassa PIP2;1, glycerol permeability increased ∼14 and 6 times (Pg: 6.33 × 10−4 and 2.82 × 10−4 cm/s, respectively) compared with oocytes injected only with PIP2;1 (Pg: 0.44 × 10−4 cm/s) (Figure 12C). Considering that NIP4;1 and NIP4;2 reached the PM in X. laevis oocytes (Supplemental Figure 14), this result suggests that NIP4;1 and NIP4;2 transport glycerol with levels comparable to E. coli GlpF (Pg: 4.81 × 10−4 cm/s).
Figure 12.
NIP4;1 and NIP4;2 Transport Water and Glycerol in X. laevis Oocytes.
Water ([A] and [B]) and glycerol (C) transport assays in X. laevis oocytes. Permeability measurements of oocytes injected with EGFP-NIP4;1, EGFP-NIP4;1-S267A, EGFP-NIP4;2, and EGFP-NIP4;2-S267A cRNA and coinjections. The amount of cRNA (ng) is shown in parentheses. F. ananassa PIP2;1, Arabidopsis NIP6;1, and E. coli GlpF were used as positive controls for water and glycerol transport. Noninjected (NI) oocytes were used as negative controls. Representative result of three independent experiments is shown. Shown are the mean permeability values for water (Pf) and glycerol (Pg) ± se, n = 6 to 12. Different letters indicate significant differences among treatments (P < 0.05, one-way ANOVA, Tukey test).
Regarding a potential role for S267 phosphorylation in the regulation of transport activity, EGFP-NIP4;1 S267A and EGFP-NIP4;2 S267A expressing oocytes showed lower levels of water permeability than EGFP-NIP4;1 and EGFP-NIP4;2 (Pf: 24.14 × 10−4 and 21.17 × 10−4 cm/s, respectively) (Figures 12A and 12B). Considering that both mutants reached the PM in X. laevis oocytes (Supplemental Figure 14), our results suggest a potential role for Ser-267 phosphorylation in the regulation of water transport, in accordance with previous studies in which phosphorylation of soybean NOD26 at Ser-262 enhanced water transport (Guenther et al., 2003).
Survival Assays of NIP4;1 and Mutant NIP4;1-S267A in Saccharomyces cerevisiae
We further characterized NIP4;1 transport using yeast. Yeast spheroplasts transformed with NIP4;1, NIP4;1-S267A, or human AQP8 as a positive control were used. While hAQP8 showed its typical high water permeability, expression of NIP4;1 resulted in a moderate increased swelling rate compared with the negative control (Supplemental Figure 15A). This is in accordance with the water permeability found in X. laevis oocytes, and of other NIPs for which a moderate water permeability was shown (Choi and Roberts, 2007; Tanaka et al., 2008; Katsuhara et al., 2014). When NIP4;1 was compared with NIP4;1-S267A, there was no significant reduction in water permeability, suggesting that phosphorylation site S267A is not active in yeast (Figure 12A; Supplemental Figure 15A). Thus, taking into account that the oocytes’ Pf differences found for S267A mutants are statistically significant but minor at the physiological level, our results regarding water transport of NIP mutants should be interpreted cautiously.
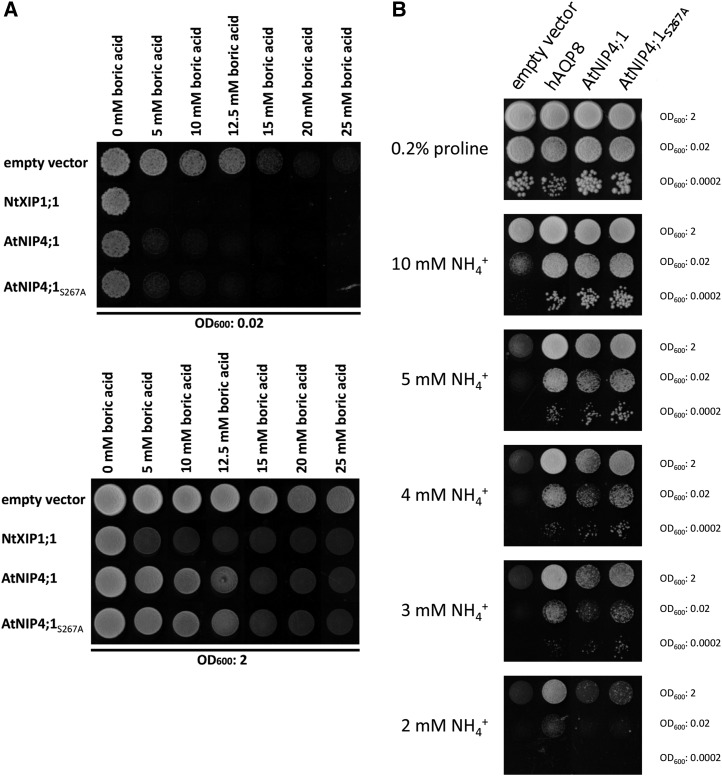
Then, we used the yeast mutant YNVW1 (∆dur3), which carries a deletion in the DUR3 transporter gene that influences boron transport (Nozawa et al., 2006). Figure 13A shows that NIP4;1 and NIP4;1-S267A displayed a growth inhibition phenotype with increasing boric acid concentrations, suggesting that NIP4;1 is weakly permeable to boric acid. These data suggest that the shorter pollen tubes for the knockdown lines in boron-deficient medium might indicate a role for NIP4 in boron transport (Figure 10).
Figure 13.
Functional Assays of NIP4;1 and Mutant NIP4;1-S267A in Yeast.
(A) Yeast growth and survival on synthetic medium with different concentrations of boric acid. Yeast strain ∆dur3 transformed with the empty vector pYeDP60u or pYeDP60u containing the indicated AQP homologs were spotted at an OD600 of 0.02 and 2 on medium containing various concentrations of boric acid (0, 5, 10, 12.5, 15, 20, and 25 mM). Growth was recorded after 6 d at 30°C. All data were repeated in three independent experiments.
(B) Yeast growth and survival on synthetic medium with different concentrations of ammonia. The ∆mep1-3 yeast mutant transformed with the empty vector pYeDP60u or pYeDP60u containing the indicated AQP homologs was spotted at an OD600 of 2, 0.02, or 0.0002 on medium containing various concentrations of ammonia (2, 3, 4, 5, and 10 mM) or 0.2% proline as sole nitrogen source and growth was recorded after 9 d at 30°C. All data were repeated in three independent experiments.
We found also that expression of NIP4;1 and NIP4;1-S267A isoforms complemented the growth inhibition of the 31019b (∆mep1-3) yeast mutant, which carries deletions in the MEP1-3 ammonium transporters, on medium with low concentrations of ammonium as a sole nitrogen source (Figure 13B). It has been shown that H2O2 crosses biological membranes via AQPs (Bienert et al., 2007; Dynowski et al., 2008). Then, YNVW1 (∆dur3) and ∆mep1-3 mutant yeast strains were transformed with vectors expressing NIP4;1, NIP4;1-S267A, or human AQP8 as a positive control (Bienert et al., 2011). Expression of NIP4;1 and NIP4;1-S267A resulted in an increased sensitivity of ∆dur3 and ∆mep1-3 yeast toward externally supplied H2O2 compared with the negative control (Supplemental Figure 15B). Finally, we used the yeast mutant YNVW1 (∆dur3), which carries a deletion in the DUR3 urea transporter gene and is unable to grow on medium containing 5 mM or lower concentrations of urea as a sole nitrogen source (Liu et al., 2003). Supplemental Figure 15C shows that yeast expressing NIP4;1 and NIP4;1-S267A were able to grow on medium containing 3, 4, or 5 mM urea as a sole nitrogen source. However, their complementation efficiency was lower compared with the positive control XIP1;1α.
DISCUSSION
In this study, we showed that the poor fertility parameters found in the single mutants and double knockdown of NIP4;1 and NIP4;2 resulted from partial male sterility, caused by a defect in pollen development and in pollen germination and pollen tube growth.
NIP4;1 and NIP4;2 Are Angiosperm-Exclusive Paralogs
The abrupt occurrence of angiosperms and their fast rise to dominance can be in part explained by their rapid reproduction, due in large part to accelerated pollen germination and pollen tube growth rates (Darwin and Seward, 1903; Williams, 2008). Arabidopsis NIP4;1 and NIP4;2 encode proteins belonging to the NIP1 group, only present in angiosperms (Anderberg et al., 2012; Abascal et al., 2014). In agreement with the general pattern of angiosperm genome evolution where recurrent gene duplication and retention of paralogous genes play a critical role in the adaptation of species to different environments (Fischer et al., 2014), our syntenic analysis showed that Arabidopsis NIP4;1 and NIP4;2 are paralogs arising from a recent gene duplication event in the common ancestor of the Brassicaceae lineage (Supplemental Figure 1). To explain the maintenance of both paralogs, models that assume either neofunctionalization or subfunctionalization have been proposed (Conant and Wolfe, 2008). The partitioning of NIP4;1 and NIP4;2 expression between different pollen stages and their common function in reproduction are empirical evidence of subfunctionalization in which each paralog retains a subset of its original ancient function. Contrary to the common approaches comparing paralogous genes from closely related species (Kim et al., 2014; Yang et al., 2014), we suggest that subfunctionalization processes within a single species occurred through modification in the temporal expression pattern.
NIP4;1 and NIP4;2 Are Pollen-Specific Genes with Different Expression Patterns
According to the microarray data, the signal for both NIP4;1 and NIP4;2 remains low throughout pollen development (Honys and Twell, 2004) and a major expression peak appears after pollen germination (Wang et al., 2008; Qin et al., 2009) (Table 1). High-throughput sequencing of Arabidopsis mature dry pollen detected NIP4;1, but not NIP4;2, expression (Loraine et al., 2013). Here, we showed that NIP4;1 is modestly expressed in mature pollen (Figure 2B), while NIP4;2 is not expressed in mature pollen but is strongly expressed in pollen tubes (Figure 2C). Therefore, our results suggest that the mature pollen values obtained from the ATH1 chip (Honys and Twell, 2004; Wang et al., 2008; Qin et al., 2009) would correspond to NIP4;1, while the high expression found in pollen tubes (Wang et al., 2008; Qin et al., 2009) is due mainly to NIP4;2. Consistent with these results, analysis of NIP4;1pro:GUS plants showed that NIP4;1 is expressed in pollen grains (Figures 3A to 3C) and also in pollen tubes (Figures 3I to 3M). In contrast, NIP4;2 is only expressed in pollen tubes (Figures 3N to 3R). Altogether, these findings suggest that although NIP4;1 and NIP4;2 may have similar functions (based on protein sequence identity), their roles may not be redundant (based on their expression pattern).
NIP4;1 and NIP4;2 Are Localized in the Plasma Membrane and in Internal Vesicles
We showed that EGFP-NIP4;1 and EGFP-NIP4;2 are localized in the plasma membrane and also in intracellular vesicles of pollen tubes (Figure 4). Plant AQPs have been localized in mainly all subcellular compartments, including plasma membrane, tonoplast, endoplasmic reticulum, Golgi apparatus, and chloroplast, and in multiple localizations for some of them (Li et al., 2014). In EGFP-NIP4;1 mature pollen grains and tubes treated with BFA, there was an increase in the intracellular localization, suggesting an active endocytosis and recycling of NIP4;1 between the plasma membrane and intracellular vesicles (Supplemental Figure 5). In pollen tubes, a tight regulation of vesicle trafficking, cytoskeleton dynamics, and ions fluxes is essential for proper polarized cell growth (Steinhorst and Kudla, 2013). It has also been reported that PIP1;2 and PIP2;1 increased their dynamic cycling under salt stress conditions in Arabidopsis roots (Li et al., 2011b; Luu et al., 2012).
Single nip4;1 and nip4;2 Mutants and Double Knockdown nip4;1 nip4;2 Lines Affected Fertility Parameters
Our results showed a defect in single mutant nip4;1 and nip4;2 and double knockdown lines specifically associated with the male gamete (Table 2). The single nip4;1 mutant and double knockdown plants showed a significantly higher frequency of abnormal, stunted siliques and fewer seeds when compared with the wild type (Figure 6). A similar phenotype was observed for Arabidopsis mutants of class XI myosins XI-1, XI-2, and XI-K, essential for organelle transport and cell expansion. Ojangu et al. (2012) showed that after the onset of flowering, up to 60% of flowers of the primary shoot of xi-1/xi-2/xi-k plants remained mostly seedless because the pistils were unpollinated or poorly pollinated. About 3 weeks after bolting, plants started to produce siliques only slightly underdeveloped or normal in size with up to 28% of unfertilized ovules (Ojangu et al., 2012). Here, we observed that the defective development of mature pollen would be the main reason for the reduced fertility of nip4;1 mutant plants.
Double knockdown plants had a lower percentage of pistils with pollen adhered to the stigma surface, compared with wild-type plants (Figures 7A and 7B), suggesting that reduced fertility in double knockdown plants is produced, at least in part, by a defect in adhesion of the pollen to the stigma. This could be due to a failure in development, possibly preventing a coordinated maturation with the female organs, mainly in the early flowers. Considering that pollen is always in excess with respect to the number of ovules, we would expect complete fertilization despite the fact that pistils were subpollinated. However, when pollen tube growth was analyzed in subpollinated pistils, we observed that wild-type pistils were fully fertilized, whereas double knockdown pistils showed nonfertilized ovules (Figure 7F), indicating an additional defect in double knockdown lines related to pollen germination and/or pollen tube growth.
NIP4;1 and NIP4;2 Transport Water and Glycerol in Oocytes
Our results showed that in X. laevis oocytes NIP4;1 and NIP4;2 have relative low permeability to water, but transport solutes as glycerol at similar levels to E. coli glycerol uptake facilitator GlpF (Figure 12). Glycerol is a common substrate for NIPs, but there is no physiological evidence for a membrane intrinsic protein-mediated glycerol transport in plants. Hu et al. (2014) demonstrated that exogenous glycerol inhibited primary root growth. In bacteria, it has been postulated that glycerol modulates water permeation in the micromolar range, whereas at millimolar concentrations, E. coli GlpF is glycerol saturated, occluding the conducting pore (Chen, 2013). In yeast, S. cerevisiae Fps1 plays critical roles in osmoregulation by modulating the accumulation of glycerol (reviewed in Ahmadpour et al., 2014). Here, we showed that under mild osmotic stress, double knockdowns displayed shorter pollen tubes, suggesting a role for NIP4s in osmoregulation. In Arabidopsis, triacylglycerol (TAG) is the major form of carbon storage in developing seeds, flower petals, tapetal cells of the anther, and pollen (Footitt et al., 2007). In pollen, it has been proposed that TAG provides energy and carbon skeletons to support germination and pollen tube growth for fertilization (Footitt et al., 2007). Interestingly, complete disruption of TAG synthesis results in embryo and pollen lethality in Arabidopsis (Zhang et al., 2009).
Additionally, functional survival yeast analysis indicated that NIP4;1 could be involved in the transport of hydrogen peroxide, urea, and boric acid with relative low efficiency and ammonia with high efficiency (Figure 13; Supplemental Figure 15), suggesting that NIP4;1 plays a role in the response to different kinds of biotic and abiotic factors. It has been suggested that water and H2O2 use the same pathway through the channel as their transport is inhibited by the same mutations (reviewed in Bienert and Chaumont, 2014).
Boron is an essential element for cell wall structure and, thus, for plant growth and reproduction (Blevins and Lukaszewski, 1998). One of the primary functions of boron in plants is to serve in the cross-linking of rhamnogalacturonan-II, a component of cell wall pectic polysaccharides. Interestingly, a large number of NIPs, which transport metalloids, are simultaneously impermeable or only poorly permeable to water (reviewed in Pommerrenig et al., 2015). In Arabidopsis, NIP5;1, NIP6;1, and NIP7;1 are boron transporters with organ-specific roles in boron nutrition (Takano et al., 2006; Tanaka et al., 2008; Li et al., 2011a). NIP5;1 exhibits root-specific expression that can be induced by boric acid limitation (Takano et al., 2006), while NIP6;1 is expressed in a shoot-specific manner (Tanaka et al., 2008). NIP5;1 and NIP6;1 T-DNA insertional mutants exhibit an enhanced sensitivity to B deficiency that results in reductions in cell elongation and tissue expansion (O’Neill et al., 2004). It has been also reported that boron plays a role in plant reproduction, including inflorescence formation, pollen development and germination, pollen tube growth, and seed and fruit set (Dell and Huang, 1997; Pommerrenig et al., 2015). Arabidopsis NIP7;1, which is selectively expressed in developing pollen grains of stage 9-11 anthers, was also identified as boric acid channel (Li et al., 2011a). The NIP7;1 T-DNA insertional mutant showed defects in pollen tube growth in the absence of boric acid, suggesting that NIP7;1 might be involved in boric acid uptake (Li et al., 2011a). Conversely, our results suggest that NIP4;1 could play a role in boric acid uptake during pollen development, germination, and pollen tube growth under boron-deficient conditions (Figures 10 and 13).
Besides the physiological interpretation of the function of each solute during pollen development and/or pollination, these results could also suggest a role for a solute-solvent interplay during hydration. Thus, pollen-specific aquaporins could be part of a biochemical machinery involved in the temporal and spatial regulation of pollen hydration.
NIP4;1 and NIP4;2 Are in Vitro Phosphorylated at Ser-267
Our results show that NIP4;1 and NIP4;2 C termini are phosphorylated in Ser-267 in vitro by a pollen CPK (Figure 11). Both NIP4s possess a conserved CPK phosphorylation sequence (LTKSAS) similar to that of soybean NOD26 (ITKSAS) and other NIPs (hydrophobic-X-basic-X-X-Ser/Thr) (Guenther et al., 2003; Wallace et al., 2006), suggesting that phosphorylation by CPK may be a common mechanism to regulate their activity or/and targeting. Considering the low water permeability of NIP4;1 and NIP4;2, regulation by phosphorylation could be an important feature. In the case of NOD26, phosphorylation is regulated during nodule development and maintained at steady state levels throughout the N2-fixing portion of the nodule lifespan, but also by osmotic stress signals (Guenther et al., 2003; Masalkar et al., 2010). In particular, CPKs are important regulators of pollen tube growth (Estruch et al., 1994; Yoon et al., 2006). In Arabidopsis, CPK17 and CPK34 are pollen specific (Honys and Twell, 2004) and essential for pollen fitness and pollen tube growth (Myers et al., 2009), while CPK11 and CPK24 together mediate the Ca2+-dependent inhibition of the inward K+ channels and participate in the regulation of pollen tube growth (Zhao et al., 2013).
NIP4;1 Plays a Role in Pollen Development
We observed a higher percentage of immature (uni- and bicellular) (Supplemental Figure 10 and Supplemental Table 1) and dead pollen grains (Supplemental Figure 11 and Supplemental Table 2) only in the single mutant nip4;1 and double knockdown lines. Therefore, these results suggest that the single nip4;1, but not nip4;2, mutant and double knockdown lines have a defect in pollen development, often losing the cytoplasm and collapsing. These phenotypes, observed only for NIP4;1, correlate well with its expression pattern.
NIP4;1 and NIP4;2 Have a Role in Pollen Germination and Pollen Tube Growth
Aniline blue experiments showed that in vivo pollen tubes of nip4;2, but not nip4;1, and double knockdown plants were shorter than wild-type tubes (Figure 8), suggesting that NIP4;2 plays a role in pollen tube growth. In vitro germination assays showed that nip4;1 and nip4;2 mutants and double knockdown lines had lower pollen germination percentages and pollen tube lengths when compared with the wild type (Figures 9B and 9C). The fact that for nip4;1 we have observed differences in vitro but not in vivo may be explained based on the parameters evaluated in each assay. For the in vivo analysis, we measured the maximum length reached within the pistil; thus, the average length reduction of pollen tubes could have been masked by a normal elongation of some tubes. Considering that NIP4;1 is predominantly expressed during pollen development, and pollen tube growth depends on the rehydration and germination capacity of mature pollen, poor tube growth of nip4;1 may be explained by deficiencies during pollen development. Nonetheless, EGFP-NIP4;1 is present in pollen tubes, suggesting that NIP4;1 could also function together with NIP4;2 to regulate pollen tube growth.
The defects found in pollen development, germination, and pollen tube growth are statistically significant but mild at the physiological level. This is probably due to the expression of other pollen AQPs (TIP5;1, TIP1;3, SIP1;1, SIP2;1, PIP2;8, TIP1;1, NIP2;1, and NIP7;1; Supplemental Figure 2) apart from NIP4;1 and NIP4;2, which may have some functional redundancy masking more pronounced phenotypes. TIP5;1 and TIP1;3 were shown to transport water and urea; SIP1;1, water; TIP1;1, water, urea, arsenite, and H2O2; NIP2;1, water, glycerol, and H2O2; and NIP7;1, water, glycerol, urea, boron, arsenite, and H2O2 (revised in Perez Di Giorgio et al., 2014). It has been shown that aquaporin mutant-associated phenotypes were mostly observed under nutrient limiting growth conditions (Takano et al., 2006; Schnurbusch et al., 2010; Li et al., 2011a; Hanaoka et al., 2014; Wudick et al., 2014; Xu et al., 2015), while the nip4;1 and nip4;2 phenotypes described here are under normal growth conditions.
We propose that NIP4;1 and NIP4;2 activity, which is possibly regulated by modifying their subcellular localization or their phosphorylation status in response to either pollination stage, pistil guidance, and/or stress cues could influence pollen nutrient levels and that this is critical for achieving efficient pollen development and pollination.
METHODS
Bioinformatic Analysis of NIP4;1 and NIP4;2
Whole-genome syntenic analysis was performed for nine dicot species using Phytozome v9.1 (www.phytozome.net).
Plant Material and Growth Conditions
Seeds from Arabidopsis thaliana (ecotype Columbia-0) wild type, single and double mutants, and transgenic plants were sown on soil, sterified for 3 d at 4°C, and grown at 22°C on a 16-h-day/8-h-dark cycle.
Single nip4;1 and nip4;2 Mutants
Arabidopsis (ecotype Columbia-0) T-DNA insertion mutants for NIP4;1: SALK_038278.54.75.x (designated as nip4;1-1), SALK_013924.51.75.x (nip4;1-2), and SALK_007730.42.45.x (nip4;1-3); and NIP4;2: SALK_142789.54.50.x (nip4;2-1), and SAIL_799_E09 (nip4;2-2), were obtained from the ABRC. Homozygous plants were identified by two PCR reactions using (1) gene-specific left and right primers (LP + RP) designed to anneal on either side of the T-DNA insertion, and (2) the T-DNA border-specific primer LBa1 (for SALK lines) or LB1 (for SAIL line) + the RP primer. To confirm the location of the T-DNA insertion in each line, PCR products using LB + RP primers containing regions of NIP4;1 or NIP4;2 and the transposon sequences were sequenced. T-DNA mutants were selected by screening for kanamycin resistance (KanR; SALK lines, except for nip4;2-1) and Glufosinate Ammonium (BASTA) resistance (BastaR; SAIL line) and confirmed by PCR genotyping. The sequences of the used primers are shown in Supplemental Data Set 2.
amiRNA Double Knockdown (nip4;1 nip4;2)
Two 21-bp amiRNAs that both simultaneously silence NIP4;1 and NIP4;2 (named ami260 and ami356 according to their matching position in the coding region of both genes) were identified with the WMD Web Micro-RNA Designer (www.weigelworld.org). The amiRNAs were constructed according to Schwab et al. (2006) by overlapping PCRs using four specific primers for each amiRNA (I to IV) and two common primers (pRS300-A and -B). The resulting miR precursors were PCR amplified with primers Pf NheI-amiRNA and pRS300-B and initially transferred into a pZD05 vector (AmpR) under the pollen-specific promoter LAT52 (LAT52pro) via digestion with NheI and BamHI. The LAT52pro:amiRNA sequences were PCR amplified with primers Pf SpeI-LAT52pro:amiRNA and Pr SpeI-LAT52pro:amiRNA and cloned into a pK7WG2D:LAT52pro:mRFP vector (SepR in bacteria and KanR in plant) via digestion with SpeI. The LAT52pro:mRFP sequence was amplified from the pZD05 vector, cloned into a pENTR1A vector (KanR), and subsequently introduced into the pK7WG2D Gateway vector (Sm/SpR for bacteria, KanR for plants) by LR recombination. Both resulting constructs, LAT52pro:mRFP-LAT52pro:amiRNA 356 and 260, were confirmed by sequencing and transferred into wild-type Arabidopsis plants (Col-0) via Agrobacterium tumefaciens (EHA105) transformation by the floral dip method (Zhang et al., 2006). Transformants were identified by screening for KanR and confirmed by PCR. The sequences of primers are shown in Supplemental Data Set 2.
Complementation Lines
To complement nip4;1 and nip4;2 single mutants, we generated pollen-specific EGFP-NIP4;1 and EGFP-NIP4;2 expression constructs under their own promoters. Briefly, NIP4;1 and NIP4;2 coding sequences were amplified from mature pollen grain and pollen tube RNA with Pf BamHI-NIP4;1-Pr XhoI-NIP4;1 and Pf BamHI-NIP4;2-Pr XhoI-NIP4;2, respectively. The resulting PCR products were cloned into the pENTR1A Gateway vector (KanR) via digestion with BamHI and XhoI and subsequently inserted into the pZY03 Gateway binary vector (Sm/SpR for bacteria, BastaR for plants) by LR recombination. The resulting vectors expressing EGFP-NIP4;1 and EGFP-NIP4;2 under the control of LAT52pro were monitored in tobacco (Nicotiana tabacum) pollen assays as described (Zhang et al., 2008) (Supplemental Figure 5). Thereafter, NIP4;1 and NIP4;2 promoter fragments (containing 2984 and 1937 bp upstream of the ATG start codon, respectively) were amplified using primers Pf SacI-NIP4;1pro-Pr SpeI-NIP4;1pro and Pf SacI-NIP4;2pro-Pr SpeI-NIP4;2pro, respectively. The fragments were then cloned into pZY03 LAT52pro:EGFP-NIP4 vectors, replacing LAT52pro via digestion with SacI and SpeI. The resulting expression vectors pZY03 NIP4;1pro:EGFP-NIP4;1 and pZY03 NIP4;2pro:EGFP-NIP4;2 were confirmed by sequencing and transferred into nip4;1-1, nip4;1-2, and nip4;1-3, and nip4;2-1 and nip4;2-2 backgrounds via Agrobacterium (EHA105) transformation by the floral dip method (Zhang et al., 2006). Transformants were identified by screening for BastaR and confirmed by PCR genotyping with primers Pf-Pr EGFP and Pf5-Pr 3 pZY03. The sequences of primers are shown in Supplemental Data Set 2. Two independent LAT52pro:EGFP-NIP4;1 lines in the wild-type background (named WT/NIP4;1-15 and WT/NIP4;1-20) and three independent NIP4;1pro:EGFP-NIP4;1 lines in the nip4;1-1, nip4;1-2, and nip4;1-3, backgrounds (named nip4;1-1/EGFP-NIP4;1-1, nip4;1-2/EGFP-NIP4;1-9, and nip4;1-3/EGFP-NIP4;1-1, respectively) and two independent NIP4;2pro:EGFP-NIP4;2 lines in the nip4;2-1 and nip4;2-2 backgrounds (named nip4;2-1/EGFP-NIP4;2-9 and nip4;2-2/EGFP-NIP4;2-7, respectively) were analyzed.
GUS Expression Lines
The Gateway entry vector pENTR1A GUS (KanR) was recombined into the pZY03 destination vector. The resulting vector expressed GUS fused to EGFP in the N terminus under the control of LAT52pro. NIP4;1 and NIP4;2 promoter fragments were amplified using the primers Pf NIP4;1pro-SacI - Pr NIP4;1pro-NcoI and Pf NIP4;2prom-SacI - Pr NIP4;2prom-NcoI and cloned into the binary vector pZY03 LAT52pro:EGFP-GUS (Sm/SpR for bacteria, BastaR for plants), replacing LAT52pro and EGFP via digestion with SacI and NcoI. The resulting expression vectors pZY03 NIP4;1pro:GUS and pZY03 NIP4;2pro:GUS were confirmed by sequencing and transferred into a wild-type (Col-0) background via Agrobacterium (EHA105) transformation by the floral dip method (Zhang et al., 2006). Transformants were selected by screening for BastaR and confirmed by PCR genotyping with primers Pf 4 pZY03-Pr GUS (for NIP4;1pro:GUS) and Pf 7 pZY03-Pr GUS (for NIP4;2pro:GUS). Primer sequences are shown in Supplemental Data Set 2. Two NIP4;1pro:GUS (1 and 16) and three NIP4;2pro:GUS (11, 12, and 15) independent lines were analyzed.
Growth Conditions
Sterilized seeds were plated on 0.5× Murashige and Skoog (1962) medium with 1% sucrose, 0.7% agar, and the corresponding selective agent (50 mg/L kanamycin or 12.5 mg/L glufosinate ammonium, for KanR and BastaR lines, respectively) and cold stratified 4 d in the dark at 4°C. Seeds were germinated and grown with continuous light at 22°C for 7 d. Seedlings were then transferred to soil or peat, mixed with vermiculite and perlite (2:1:1), and grown in chamber at 22°C under long-day (16/8 h light/dark) photoperiod and 60% relative humidity.
RT-PCR and RT-qPCR
Total RNA (100 mg) was extracted from leaves, roots, mature pollen, and pollen tubes with an RNeasy Plant Mini Kit (Qiagen). At least two independent RNA extractions were made for each tissue and genotype. cDNAs were synthesized using MMLV reverse transcriptase according to the manufacturer’s instructions (Promega) using oligo(dT) primers (Invitrogen). For RT-PCR analysis, the cDNAs (20 ng) from mature pollen and pollen tubes were used for amplification of Arabidopsis NIP4;1, NIP4;2, TIP5;1, and TIP1;3, using gene-specific primers (Pf-Pr NIP4;1, Pf-Pr NIP4;2, Pf-Pr TIP5;1, and Pf-Pr TIP1;3). As an internal control, Actin-8, a constitutively expressed gene, was amplified with primers Pf-Pr ACT8. All RT-PCRs were performed in duplicate and the PCR products were analyzed by agarose gel electrophoresis. For qPCR analysis, cDNA samples from leaves, roots, mature pollen, and pollen tubes were amplified with NIP4; 1- and NIP4;2-specific primers (Pf-Pr qPCR NIP4;1 and Pf-Pr qPCR NIP4;2) using HOT FIREPol EvaGreen qPCR Mix Plus (no ROX; Solis BioDyne) in a Rotor-Gene 6000 qPCR system (Corbett Life Science). Expression levels of NIP4s were calculated by the absolute quantification method using plasmid reference curves (Whelan et al., 2003) and normalized to Protein Phosphatase 2 subunit A3 (PP2A) (Czechowski et al., 2005). All qPCRs were performed in triplicate. Primer sequences are shown in Supplemental Data Set 2.
Assessment of Fluorescently Tagged NIP Proteins in Oocyte Plasma Membranes
Confocal fluorescence microscopy was used to localize the respective NIP4;1 and NIP4;2 isoforms tagged with EGFP in Xenopus laevis oocytes. As a marker of the interior of the oocyte, tetrametylrhodamine (TMR) dextran (10,000 MW; Invitrogen-Molecular Probes), an unconjugated nonspecific fluorochrome marker that distinguishes plasma membrane from cytosol (Brooks and Wessel, 2003), was used. Briefly, 3 d after cRNA injection and 40 min prior to imaging, oocytes were microinjected with 50 nL of a 33 µM aqueous solution of TMR-dextran. Fluorescence images of EGFP distribution, together with TMR, were obtained with a FluoView1000 spectral confocal scanning microscope (Spectral FV1000; Olympus), using a 60× UPLSAPO oil immersion objective lens (numerical aperture 1.35). To avoid crosstalk, images were recorded line by line in a sequential order. EGFP and TMR were excited using the 488-nm line of the argon laser and the 543-nm He-Ne laser, respectively, and the emitted fluorescence was detected in the 500 to 540 nm and 570 to 670 nm range. Autofluorescence (monitored in control oocytes) was negligible in comparison with cells expressing fluorescent NIPs. Three to five oocytes from each of at least two donor frogs were analyzed.
GUS Staining
GUS staining and whole-mount clearing preparations of flowers and seedlings from NIP4pro:GUS-expressing lines were performed as described (Johnson et al., 2004). In addition, wild-type pistils hand-crossed with NIP4pro:GUS or wild-type pollen were examined 12 h after pollination. Pistils were excised, mounted on double-sided tape, and cut longitudinally along the septum with a 27.5-gauge needle (Becton Dickinson). Plant organs were placed in 100 μL 80% acetone for 30 min to fix cells and remove chlorophyll, incubated overnight at 37°C in X-Gluc solution (0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid in DMSO, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 50 mM buffer sodium phosphate, pH 7.0), and washed three times in 70% ethanol for 1 h until complete discoloration. Cleared preparations were mounted on microscope slides in 50% glycerol and observed using a Leica EZ4 D stereoscope or an Olympus BX41 microscope with differential interference contrast.
Pollen Development
Pollen grain viability was assessed after 20 µM FDA treatment for 10 min at room temperature in the dark (Heslop-Harrison and Heslop-Harrison, 1970) using an Olympus BX41microscope with a GFP filter. The number of dead and abnormal pollen grains (shrunken-shaped) was scored. To determine their hydration capability of pollen grains, the pollen grain volume was calculated as V = (π × A × B2)/6, where A is the length of the major axis and B the minor axis, as measured with ImageJ software (Williams, 2012). Pollen nuclei were visualized by staining with Hoechst 33342 (NucBlue Live Ready Probes Reagent; Molecular Probes). Pollen was observed with an epifluorescence Olympus BX41 microscope using UV light emission. For each assay, at least four biological replicates were scored for each pollen genotype.
In Vitro Pollen Germination Assays
In vitro Arabidopsis pollen germination experiments were conducted as described (Boavida and McCormick, 2007). Briefly, pollen from 15 flowers of the same plant was germinated in 100 μL of liquid PGM (0.01% boric acid, 5 mM CaCl2, 5 mM KCl, 1 mM MgSO4, and 10% sucrose, pH 7.6) at 22°C for 0.5, 1, 1.5, 3, or 5 h. Additionally, pollen tube growth assays were performed in 80 μL of solid PGM (0.01% boric acid, 5 mM CaCl2, 5 mM KCl, 1 mM MgSO4, 10% sucrose, and 1 or 2% low melting agarose, pH 7.6) at 22°C for 3 or 5 h. For boron transports assays, the boric acid content of the solid PGM was adjusted to 0.001, 0.1, 0.025, and 0.05%. For temperature, salinity, and osmolality stress conditions, pollen was germinated at 28°C, in modified PGM with 5× or 10× KCl and with 7.5, 5, 2.5, or 0% sucrose plus 0.08, 0.16, 0.24, or 0.32 M glycerol, respectively. A pollen grain was classified as germinated if the pollen tube length was equal to or greater than the pollen grain diameter. At least four biological replicates (i.e., sibling plants for each genotype) were used for each assay, and the assays were repeated three or four times. Two hundred pollen grains were scored for pollen germination rates and 100 pollen tubes were measured in each replicate.
In Vivo Pollination and Aniline Blue Staining of Pollen Tubes
For in vivo Arabidopsis pollen germination experiments, pistils of wild-type emasculated flowers were hand-pollinated with pollen from single mutants and double knockdown plants. Pollen exclusively from the first 10 flowers on the main stem of the plants was used. One hour after saturated pollination, pistils were collected, fixed, cleared, and stained with aniline blue. Independent pollinations (n = 12 to 20) were performed for each pollen genotype. Additionally, in vivo pollination and pollen tube growth of self-pollinated pistils (stages 13/14 and 15) were visualized by aniline blue staining as described (Mori et al., 2006). Briefly, self-pollinated or cross-pollinated pistils were fixed for 2 h at room temperature (RT) in acetic acid/ethanol (1:3) fixative, rehydrated at RT through 70, 50, and 30% ethanol and ddH2O, for 10 min each, softened with 8 M NaOH overnight at RT, washed with ddH2O, and stained with 0.1% decolorized aniline blue solution (in 108 mM K2HPO4 buffer, pH 11) with 2% (v/v) glycerol for 2 h in darkness at RT. Finally, pistils were mounted on a microscope slide using a drop of decolorized aniline blue solution with 5% (v/v) glycerol and carefully pressed with a cover slip to open the pistil longitudinally. Aniline blue fluorescence was observed in an Olympus BX41 or a Zeiss Axio Imager Z1 inverted microscope using UV light emission. Pollen germination and pollen tube lengths were measured with ImageJ software (public domain; http://imagej.nih.gov/ij/).
Fertility Assays
Fertility of wild-type, single nip4;1-1 and nip4;2-2 mutants, and double knockdown ami356-1 and 260-4 plants was evaluated by measuring the length of the siliques and counting the number of seeds in mature siliques produced in the main stem 5 weeks after the onset of flowering. All siliques were harvested and stuck on a slide with double-sided tape, from left to right in order of appearance on the main stem of each plant. Images of cleared siliques were captured using a Leica EZ4 D stereomicroscope, and measurements were made using ImageJ software. Pollen content on the stigma of wild-type, single nip4;1-2 and nip4;2-2 mutants, and double knockdown ami356-1 and 260-4 flowers of stages late 13 and 14 on the main stem of the plants was evaluated using a Leica EZ4 D stereomicroscope.
Scanning Electron and Confocal Microscopy
To visualize pollen hydration and adhesion to the stigma, newly opened flowers were fixed and prepared as previously described (Chebli et al., 2012). Samples were observed with a FEI Quanta 200 3D microscope operating at 20 kV.
In Arabidopsis complementation lines, subcellular localization of NIP4;1 and NIP4;2 tagged with EGFP in mature pollen and pollen tubes was determined by confocal microscopy. Images were obtained with a FluoView1000 spectral confocal scanning microscope (Spectral FV1000; Olympus), using a 60× water immersion objective lens (numerical aperture 1.42). For cycling experiments, pollen was germinated in 100 μL of standard germination medium for 30 min and then BFA (3 to 6 µg/mL final concentration) or tyrphostin A23 (100 µM final concentration), in combination with FM4-64 (1 µM final concentration), was added to the medium. Excitation/emission parameters were as follows: ECFP (excitation 488 nm/emission 500 to 540 nm), FM4-64 (excitation 514 nm/emission 580 to 630 nm). Autofluorescence (monitored in wild-type mature pollen and pollen tubes) was negligible in comparison with pollen expressing fluorescent NIPs.
Segregation Analysis
Three self-crosses between wild-type (Col-0) females and heterozygous males from single T-DNA nip4;1 or nip4;2 mutants or double amiRNA knockdown plants were performed. Segregation of F2 progeny was analyzed by PCR genotyping. Five reciprocal crosses between wild-type (Col-0) females and single mutants or double knockdown heterozygous males, and their reciprocal crosses, were performed. The genotype of individual F1 seeds was determined using antibiotic selection.
Plasmid Constructs for Kinase Assays
Plasmids pGEX-6P-1 (GE Healthcare Life Sciences) expressing the native C termini of NIP4;1 and NIP4;2 fused to GST (G-Ct-NIP4) or their respective mutant variants S267A(G-Ct-NIP4-S267A) were constructed as follows. Standard PCRs performed with DNA polymerase Pfx Platinum (Invitrogen) were used to amplify the last 31 amino acids at the C termini of NIP4;1 and NIP4;2 and the mutants NIP4;1-S267A and NIP4;2-S267A using pSGEM T7pro:EGFP-NIP4 and pSGEM T7pro:EGFP-NIP4-S267A vectors as templates (see Plasmid Constructs for Oocyte Assays) and the following specific primers (Supplemental Data Set 2): Pf BamHI-NIPCtPr NIP41Ct-XhoIPr NIP42Ct-XhoI. The forward primer contained a BamHI restriction site (underlined), an ATG (methionine) initial codon (in bold), and the rare codon AGA (arginine) substituted by CGC (in italics), and the reverse primers contained a XhoI restriction site (underlined) and an extra TTA stop codon (in bold). The coding sequences for the C termini of wild-type and mutant NIP4;1 and NIP4;2 were cloned into the BamHI and XhoI restriction sites of a pGEX-6P-1 Escherichia coli expression vector (GE Healthcare) with an N-terminal GST affinity tag to produce recombinant C-terminal domains that could later be purified using a GST affinity tag. Substrate expression clones were confirmed by sequencing.
pGEX-4T G-CPK34-6H (ps897), a kinase expression clone encoding CDPK34 between an N-terminal GST tag and a C-terminal 6His tag, was kindly provided by J.F. Harper (University of Nevada, Reno, NV).
Expression and Affinity Purification of Substrate and Kinase Expression Vectors
GST-tagged (G) substrate expression vectors G-Ct-NIP4;1, G-Ct-NIP4;2, G-Ct-NIP4;1-S267A, and G-Ct-NIP4;2-S267A and kinase expression vector G-CPK34-6H, modified to include a C-terminal 6 His tag (6H), were used to transform E. coli strain BL21(DE3) pLysE (Merck) by standard procedures. Batch purification of fusion proteins using Glutathione Sepharose 4B media (GE Healthcare) was performed as described (GST Gene Fusion System Handbook 18-1157-58, edition AA; GE Healthcare). Briefly, a single colony of E. coli cells (BL21 PlysE) harboring each recombinant plasmid was grown under ampicillin selection in nutrient broth 2XYT. Overnight cultures grown at 37°C with vigorous shaking were diluted 100-fold and grown for 2 h at 37°C (for G-Ct-NIP4s) or 10-fold and grown for 3 h at 30°C (for G-CPK34-6H) before IPTG was added to a final concentration of 0.5 mM and growth was continued for an additional 2 h at 30°C. All subsequent purification steps were performed at 4°C. Each culture was centrifuged for 10 min at 8000 rpm, and the pellet was resuspended in 50 μL of ice-cold binding buffer (1× PBS: 140 mM NaCl, 27 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2HPO4, pH 7.3) per milliliter of culture. Suspended cells were disrupted by sonication on ice and 20% Triton X-100 (final concentration 1%) was added and mixed gently for 30 min at RT. Cellular debris and unlysed cells were removed by centrifugation at 10,000 rpm for 10 min. The cleared supernatant was incubated for 30 min with 2 mL of 50% slurry Glutathione Sepharose 4B equilibrated with 1× PBS (1 mL of bed volume) per 100 mL of sonicate. The beads were sedimented by centrifugation for 5 min at 500g and washed three times with 1× PBS. Protein was eluted with 1 mL elution buffer (50 mM Tris-HCl and 10 mM reduced glutathione, pH 8.0) per milliliter of bed volume 10 min at RT. Fusion proteins were stored at −20°C.
Kinase Activity and Mass Spectrometry Assays
In vitro kinase assays were performed as previously described (Rodriguez Milla et al., 2006). Briefly, 50 µM ATP (with 2.5 µCi gamma-P32) was added to start the kinase reaction (final volume 10 or 15 µL), which consisted of 1 ng/μL purified CPK, 100 ng/μL fusion protein substrate, and standard kinase reaction buffer (20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM EGTA, and 1.1 mM CaCl2) The reactions were incubated for 15 min at 30°C and stopped by adding 1 μL ATP 100 mM, 1 μL EGTA 50 mM, and 3 μL 5× SDS sample buffer. Proteins were separated by 12% SDS-PAGE and semi-dry transferred to nitrocellulose membrane. The dried membrane was processed using either a phosphor imager (Storm 820; GE Healthcare) or exposure to x-ray film (Amersham Hyperfilm; GE Healthcare). Images were processed using ImageJ software to determine the relative intensity of labeling. Relative phosphorylation levels were normalized to the Ponceau-stained protein for each substrate (calculated by densitometry) and the background phosphorylation level of GST was subtracted from each sample. Three independent kinase activity assays were performed.
For mass spectrometry (MS) analysis, in vitro kinase assays were performed with G-Ct-NIP4;1 and G-Ct-NIP4;2, cold ATP, and CPK34. The target bands were excised from SDS-PAGE gels stained with Coomassie Brilliant Blue G 250. Protein digestion and MS analysis were performed at the Proteomics Core Facility CEQUIBIEM, at the University of Buenos Aires/CONICET as follows: Excised protein bands were sequentially washed with 50 mM ammonium bicarbonate, 25 mM ammonium bicarbonate 50% acetonitrile, and 100% acetonitrile; reduced and alkylated with 10 mM DTT and 20 mM iodoacetamide; and in-gel digested with 100 ng of Lys-C in 25 mM ammonium bicarbonate overnight at 37°C. Peptides were recovered by elution with 50% acetonitrile-0.5% trifluoroacetic acid, including brief sonication, and further concentrated by speed-vacuum drying. Samples were resuspended in 15 μL of water containing 0.1% formic acid, desalted using C18 zip tips (Merck Millipore), and eluted in 10 μL of water:acetonitrile:formic acid 40:60:0.1%. Digests were analyzed by nanoLC-MS/MS in a Thermo Scientific QExactive mass spectrometer. A 75-min gradient of water:acetonitrile at a flow of 33 nL/min was used with a C18 2-mm Easy Spray column × 150 mm. The data-dependent MS/MS method was used to fragment the top 12 peaks in each cycle. Data analysis was performed using Proteome Discoverer 1.4.
Plasmid Constructs for Oocyte Assays
To construct pSGEM T7pro:EGFP-NIP4, the coding regions of NIP4;1 and NIP4;2 fused to EGFP at the N-terminal region were obtained from pZY03 NIP4;1pro:EGFP-NIP4;1 and pZY03 NIP4;1pro:EGFP-NIP4;2 (see Complementation Lines) and cloned into the SpeI and XhoI sites of a pSGEM vector derived from pGEM containing T7 RNA polymerase promoter and carrying the 5′- and 3′-translated region of the X. laevis β-globin gene for enhanced expression. Constructs were confirmed by DNA sequencing.
To generate pSGEM T7pro:EGFP-NIP4-S267A, mutated cDNAs encoding NIP4;1-S267A and NIP4;2-S267A were obtained by site-directed mutagenesis (Quickchange II site-directed mutagenesis kit; Stratagene) according to the manufacturer’s protocol. To mutagenize the serine (UCU) at position 267 to alanine (GCU) (S267A), pSGEM T7pro:EGFP-NIP4 vectors as templates and primers NIP4;1 t799g S2-AS2 and NIP4;2 t799g S-AS were used. Mutations were confirmed by DNA sequencing.
X. laevis expression constructs of Fragaria ananassa PIP2;1 prepared in the pT7 vector (Alleva et al., 2010) and Arabidopsis NIP6;1 and E. coli GlpF in the pXbG-ev-1 vector (Wallace and Roberts, 2005; Maurel et al., 1994) were used as positive controls for water and glycerol transport.
In Vitro RNA Synthesis for Oocyte Assays
Capped cRNA encoding EGFP-NIP4;1, EGFP-NIP4;2, EGFP-NIP4;1-S267A, EGFP-NIP4;2-S267A, F. ananassa PIP2;1, Arabidopsis NIP6;1, and E. coli GlpF were synthesized in vitro using the mMESSAGE mMACHINE kit (Ambion) after linearization of the pSGEM-, pXβG-ev-1-, and pT7T-derived vectors with NheI, XbaI, and EcoRI, respectively. The synthesized products were suspended at a final concentration of 0.1 µg µL−1 in RNase-free water supplemented with Recombinant RNasin (ribonuclease inhibitor; Promega) and stored at −20°C until used (Preston et al., 1992). The cRNA was quantified by fluorescence using a Quant-iT RNA assay kit (Invitrogen). Agarose gel electrophoresis and GelRed (BioAmerica Biotech) staining were used to verify the absence of unincorporated nucleotides in the cRNA.
Oocyte Transport Assays
Defolliculated X. laevis oocytes were injected with cRNA of EGFP-NIP4;1, EGFP-NIP4;2, EGFP-NIP4;1-S267A, EGFP-NIP4;2-S267A, Arabidopsis NIP6;1, E. coli GlpF (10 ng), or F. ananassa PIP2;1 (1 ng) in a final volume of 50 nL, using an automatic injector (Drummond Scientific). Injected oocytes were incubated for 72 h at 18°C in ND96 medium supplemented with 1 μg·mL−1 gentamycin sulfate (Bellati et al., 2010). Osmotic water permeability (Pf) was determined by measuring the rate of oocyte swelling induced by a hypo-osmotic shock of 160 mOsm·kg−1. Changes in cell volume were video monitored and Pf was calculated according to previous reports (Zhang and Verkman, 1991; Agre et al., 1999). Glycerol permeability (Pg) was tested by transferring the oocytes into iso-osmotic ND96 medium where iso-osmolarity was achieved by adding glycerol to a 5-fold diluted ND96 medium (Hansen et al., 2002; Beitz et al., 2004). A combination of EGFP-NIP4;1, EGFP-NIP4;2, NIP6;1, or GlpF cRNA alone or together with PIP2;1 cRNA was injected into individual oocytes. Noninjected oocytes and cRNA from PIP2;1, NIP6;1, and GlpF were used as controls. Solute permeabilities were compared analyzing the initial swelling rates (d(V/V0)/dt). All osmolarities were determined using a vapor pressure osmometer (5520C; Wescor). Three independent experiments with different oocyte batches were assayed. Results were reported in the form of means ± se. Significant differences between treatments were calculated using ANOVA and Tukey’s test.
Plasmid Constructs for Yeast Assays
Vectors pSGEM T7pro:EGFP-NIP4s and pSGEM T7pro:EGFP-NIP4s t799g were used to amplify NIP4;1 cDNA and its respective S267A mutant. PCR products were directionally subcloned into the yeast expression vectors pYeDP60u (Hamann and Møller, 2007) or pRS426-pTPI-N-ter-GFPu and verified by DNA sequencing. pRS426-pTPI-N-ter-GFPu was generated from pRS426-pTPIu (Bienert et al., 2014).
Spheroplast Swelling Assay
Wild-type Saccharomyces cerevisiae strain BY4741 was transformed with pYeDP60u empty vector or pYeDP60u containing NIP4;1, NIP4;1-S267A, or hAQP8. Transformants were selected on synthetic medium (2% agar, 2% glucose, 50 mM succinic acid/Tris base, pH 5.5, and 0.7% YNB [yeast nitrogen base] without amino acids [Difco]) supplemented according to auxotrophic requirements with histidine, methionine, and leucine. Transformants were grown in 4 mL of the above-described synthetic medium for 12 h at 30°C and then transferred to 25 mL of synthetic medium (2% glucose replaced by 2% galactose) for 36 h at 30°C. After centrifugation, cells were resuspended in 3 mL of 50 mM KH2PO4 (pH 7.2) plus 6 μL of 2-mercaptoethanol and incubated for 15 min at 30°C. Six milliliters of spheroblasting buffer (2.4 M sorbitol, 50 mM KH2PO4, pH 7.2, 200 mg BSA, and 10 mg of Zymolase 20T [Amsbio]) was added to the cell suspension. The cells were incubated for 60 min at 30°C. Following centrifugation at 1500g for 15 min at 4°C, spheroplasts were washed once and finally resuspended in 10 mM Tris/MES, pH 8.0, 5 mM CaCl2, 50 mM NaCl, and 1.8 M sorbitol, at an OD600 of 1.5. Kinetics of spheroplast swelling were measured essentially as described previously (Laize et al., 1999). Cell osmotic water permeability was measured after exposing spheroplasts to hypoosmotic conditions (transfer from 1.8 to 1.2 mM sorbitol buffer). Volume changes were recorded at 25°C as light scattering at an angle of 90° and 450 nm using a fast kinetics instrument (SFM-3000; BioLogic). All data presented are averages of 15 to 30 trace recordings. The rate constant of the decrease of scattered light intensity is proportional to the water permeability coefficient (Kozono et al., 2003; Calamita et al., 2005). Two independent experiments were performed and gave consistent results.
Yeast Strains and Growth Assays
For the boric acid growth assay, S. cerevisiae mutant strain YNVW1 (∆dur3) was transformed with the vectors described above. In addition, pYeDP60u empty vector and pYeDP60u carrying tobacco XIP1;1α (Bienert et al., 2011) were used as controls. Transformants were selected on synthetic medium containing 2% agar, 2% glucose, 0.7% YNB without amino acids (Difco), and 50 mM succinic acid/Tris base, pH 5.5. For the toxicity growth assay, transformants were spotted on similar medium supplemented with different concentrations of boric acid and galactose instead of glucose as a carbon source.
For the ammonia complementation assay, S. cerevisiae mutant strain 31019b (∆mep1-3) (Marini et al., 1997) was transformed with the vectors described above. In addition, pYeDP60u empty vector and pYeDP60u carrying hAQP8 were used as controls. Growth of the transformants was tested as described by Jahn et al. (2004).
After 6 to 11 d of incubation at 30°C, differences in growth and survival in the different assays were recorded. All yeast growth assays were repeated in three independent experiments with consistent results.
For H2O2 growth assays, S. cerevisiae mutant strains YNVW1 (∆dur3) and 31019b (∆mep1-3) were transformed with pYeDP60u empty vector or pYeDP60u carrying NIP4;1, NIP4;1-S267A, or tobacco XIP1;1 (Bienert et al., 2011). Transformants were selected on synthetic medium (2% agar, 2% glucose, 50 mM succinic acid/Tris base, pH 5.5, and 0.7% YNB without amino acids [Difco]). The toxicity growth assay was performed as described by Bienert et al. (2007). Transformants were spotted on synthetic medium supplemented with different concentrations of H2O2 and galactose instead of glucose as a carbon source necessary for the induction of the galactose-inducible promoter.
For the urea complementation assay, the S. cerevisiae mutant strain YNVW1 (∆dur3) was transformed with pYeDP60u empty vector or pYeDP60u carrying NIP4;1, NIP4;1-S267A, or tobacco XIP1;1α. Transformants were selected on synthetic medium (2% agar, 2% glucose, 50 mM succinic acid/Tris base, pH 5.5, and 0.17% YNB without amino acids and ammonium [Difco]). For the complementation growth assay, transformants were spotted on similar medium supplemented with 1 mM arginine or different concentrations of urea as the sole nitrogen source and galactose instead of glucose as a carbon source. After 6 to 9 d of incubation at 30°C, differences in growth and survival in the different assays were recorded. All yeast growth assays were performed in three independent experiments with consistent results.
Statistical Analysis
Results are expressed as means ± se; probability values of <0.05 were considered statistically significant. Statistical analysis of the data and further processing were performed using GraphPad Prism version 6.00 for Windows (GraphPad Software).
Accession Numbers
Sequence data from this article can be found in the GenBank/Phytozome/Plaza data libraries under the following accession numbers: Arabidopsis thaliana CDPK34 (AT5G19360), NIP1;1 (AT4G19030), NIP1;2 (AT4G18910), NIP2;1 (AT2G34390), NIP3;1 (AT1G31885), NIP4;1 (AT5G37810), NIP4;2 (AT5G37820), NIP5;1 (AT4G10380), NIP6;1 (AT1G80760), NIP7;1 (AT3G06100), PIP1;1 (AT3G61430), TIP1;1 (AT2G36830), TIP5;1 (AT3G47440), TIP1;3 (AT4G01470), and SIP1;1 (AT3G04090); Arabidopsis lyrata (Loc330314, Loc891407); Capsella rubella (Carubv10006501m and Carubv10007677m); Brassica rapa (rapaBra025437, rapaBra025436, and rapaBra02543); Thellungiella halophila (Thhalv10028080m, Thhalv10027876m, and Thhalv10028303m); Carica papaya (evm.model.supercontig_65.137); Theobroma cacao (Thecc1EG019213t1); Vitis vinifera (GSVIVT01030857001); and Solanum tuberosum (PGSC0003DMP400053063).Other accession numbers in the GenBank/EMBL databases for the protein sequences in transport assays are as follows: E. coli GlpF (P18156), F. ananassa PIP2;1 (GQ390799), Glycine max NOD26 (P08995), N. tabacum XIP1;1α (HM475295), and Homo sapiens AQP8 (O94778).
Supplemental Data
Supplemental Figure 1. Synteny analysis of NIP4;1 and NIP4;2.
Supplemental Figure 2. Expression patterns of Arabidopsis thaliana aquaporin genes.
Supplemental Figure 3. GUS staining of pistils and seedlings.
Supplemental Figure 4. Pollen bombardment assays.
Supplemental Figure 5. Disruption of EGFP-NIP4;1 cycling.
Supplemental Figure 6. Single T-DNA nip4;1 and nip4;2 mutant lines.
Supplemental Figure 7. Double knockdown amiRNA lines.
Supplemental Figure 8. Double knockdown amiRNA lines expressing RFP reporter gene.
Supplemental Figure 9. Scanning electron microscopy images of pollinated pistils and developing seeds.
Supplemental Figure 10. Pollen microgametogenesis.
Supplemental Figure 11. Viability of mature pollen grains.
Supplemental Figure 12. Pollen tube growth in self-crossed pistils.
Supplemental Figure 13. Effect of temperature, salt, and osmotic stress on pollen tube growth.
Supplemental Figure 14. Subcellular localization of EGFP-NIP4;1 and EGFP-NIP4;2 in X. laevis oocytes.
Supplemental Figure 15. Functional assays of NIP4;1 and mutant NIP4;1-S267A in yeast
Supplemental Table 1. Microgametogenesis of single mutant nip4;1 and nip4;2 and double knockdown pollen grains.
Supplemental Table 2. Viability and hydration capacity of mature pollen grains.
Supplemental Data Set 1. Alignment used to generate the phylogeny in Supplemental Figure 1.
Supplemental Data Set 2. List of primers used for PCR and sequencing.
Supplementary Material
Acknowledgments
We thank Silvia Moreno and Pia Valacco of the CEQUIBIEM Proteomic Facility (www.qb.fcen.uba.ar/cequibiem) for their expertise in the MS phospho-site identification. This work was supported by Grants PICT 2011-1698, PICT 2012-0007, and PICT2014-0423 to J.P.M. G.P.B. was supported by Emmy Noether Grant 1668/1-1 from the Deutsche Forschungsgemeinschaft.
AUTHOR CONTRIBUTIONS
J.A.P.D.G., A.Y., G.P.B., N.D.A., G.C.S., G.A., and J.P.M. designed the research. J.A.P.D.G. generated the transgenic lines and performed all the experiments in planta and oocyte transport assays. G.C.S. and N.D.A. performed the phylogenetic analyses. G.P.B. performed the yeast experiments. A.Y. and G.A. contributed to the oocyte assays. M.L.B. and M.A.M contributed to the confocal microscopy localizations. J.A.P., G.P.B., G.C.S., N.D.A., G.A., and J.P.M. analyzed and interpreted the data and wrote the manuscript.
Glossary
- AQP
aquaporin
- ar/R
aromatic/arginine
- BFA
Brefeldin A
- amiRNA
artificial microRNA
- FDA
fluorescein diacetate treatment
- PGM
pollen germination medium
- PM
plasma membrane
- TAG
triacylglycerol
- TMR
tetrametylrhodamine
- MS
mass spectrometry
References
- Abascal F., Irisarri I., Zardoya R. (2014). Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta 1840: 1468–1481. [DOI] [PubMed] [Google Scholar]
- Agre P., Mathai J.C., Smith B.L., Preston G.M. (1999). Functional analyses of aquaporin water channel proteins. Methods Enzymol. 294: 550–572. [DOI] [PubMed] [Google Scholar]
- Ahmadpour D., Geijer C., Tamás M.J., Lindkvist-Petersson K., Hohmann S. (2014). Yeast reveals unexpected roles and regulatory features of aquaporins and aquaglyceroporins. Biochim. Biophys. Acta 1840: 1482–1491. [DOI] [PubMed] [Google Scholar]
- Alexandersson E., Fraysse L., Sjövall-Larsen S., Gustavsson S., Fellert M., Karlsson M., Johanson U., Kjellbom P. (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59: 469–484. [DOI] [PubMed] [Google Scholar]
- Alleva K., Marquez M., Villarreal N., Mut P., Bustamante C., Bellati J., Martínez G., Civello M., Amodeo G. (2010). Cloning, functional characterization, and co-expression studies of a novel aquaporin (FaPIP2;1) of strawberry fruit. J. Exp. Bot. 61: 3935–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderberg H.I., Kjellbom P., Johanson U. (2012). Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front. Plant Sci. 3: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitz E., Pavlovic-djuranovic S., Yasui M., Agre P., Schultz J.E. (2004). Molecular dissection of water and glycerol permeability of the aquaglyceroporin from Plasmodium falciparum by mutational analysis. Proc. Natl. Acad. Sci. USA 101: 1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellati J., Alleva K., Soto G., Vitali V., Jozefkowicz C., Amodeo G. (2010). Intracellular pH sensing is altered by plasma membrane PIP aquaporin co-expression. Plant Mol. Biol. 74: 105–118. [DOI] [PubMed] [Google Scholar]
- Bienert G.P., Bienert M.D., Jahn T.P., Boutry M., Chaumont F. (2011). Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 66: 306–317. [DOI] [PubMed] [Google Scholar]
- Bienert G.P., Chaumont F. (2014). Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta 1840: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Bienert G.P., Heinen R.B., Berny M.C., Chaumont F. (2014). Maize plasma membrane aquaporin ZmPIP2;5, but not ZmPIP1;2, facilitates transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta 1838: 216–222. [DOI] [PubMed] [Google Scholar]
- Bienert G.P., Møller A.L.B., Kristiansen K.A., Schulz A., Møller I.M., Schjoerring J.K., Jahn T.P. (2007). Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 282: 1183–1192. [DOI] [PubMed] [Google Scholar]
- Blevins D.G., Lukaszewski K.M. (1998). Boron in plant structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 481–500. [DOI] [PubMed] [Google Scholar]
- Boavida L.C., Borges F., Becker J.D., Feijó J.A. (2011). Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 155: 2066–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida L.C., McCormick S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582. [DOI] [PubMed] [Google Scholar]
- Bock K.W., Honys D., Ward J.M., Padmanaban S., Nawrocki E.P., Hirschi K.D., Twell D., Sze H. (2006). Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 140: 1151–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Gomes G., Gardner R., Moreno N., McCormick S., Feijó J.A., Becker J.D. (2008). Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 148: 1168–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bots M., Feron R., Uehlein N., Weterings K., Kaldenhoff R., Mariani T. (2005a). PIP1 and PIP2 aquaporins are differentially expressed during tobacco anther and stigma development. J. Exp. Bot. 56: 113–121. [DOI] [PubMed] [Google Scholar]
- Bots M., Vergeldt F., Wolters-Arts M., Weterings K., van As H., Mariani C. (2005b). Aquaporins of the PIP2 class are required for efficient anther dehiscence in tobacco. Plant Physiol. 137: 1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J.M., Wessel G.M. (2003). Selective transport and packaging of the major yolk protein in the sea urchin. Dev. Biol. 261: 353–370. [DOI] [PubMed] [Google Scholar]
- Calamita G., Ferri D., Gena P., Liquori G.E., Cavalier A., Thomas D., Svelto M. (2005). The inner mitochondrial membrane has aquaporin-8 water channels and is highly permeable to water. J. Biol. Chem. 280: 17149–17153. [DOI] [PubMed] [Google Scholar]
- Chebli Y., Kaneda M., Zerzour R., Geitmann A. (2012). The cell wall of the Arabidopsis pollen tube--spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol. 160: 1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. (2013). Glycerol modulates water permeation through Escherichia coli aquaglyceroporin GlpF. Biochim. Biophys. Acta 1828: 1786–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.-G., Roberts D.M. (2007). Arabidopsis NIP2;1, a major intrinsic protein transporter of lactic acid induced by anoxic stress. J. Biol. Chem. 282: 24209–24218. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9: 938–950. [DOI] [PubMed] [Google Scholar]
- Curran A., Chang I.-F., Chang C.-L., Garg S., Miguel R.M., Barron Y.D., Li Y., Romanowsky S., Cushman J.C., Gribskov M., Harmon A.C., Harper J.F. (2011). Calcium-dependent protein kinases from Arabidopsis show substrate specificity differences in an analysis of 103 substrates. Front. Plant Sci. 2: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin F., Seward A.C., eds (1903). More Letters of Charles Darwin: A Record of His Work in a Series of Hitherto Unpublished Letters, Vol. 2. (London: J. Murray). [Google Scholar]
- Dell B., Huang L. (1997). Physiological response of plants to low boron. Plant Soil 193: 103–120. [Google Scholar]
- Dynowski M., Schaaf G., Loque D., Moran O., Ludewig U. (2008). Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem. J. 414: 53–61. [DOI] [PubMed] [Google Scholar]
- Estruch J.J., Kadwell S., Merlin E., Crossland L. (1994). Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc. Natl. Acad. Sci. USA 91: 8837–8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C., Pelaz S., Yanofsky M.F. (1999). Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 68: 321–354. [DOI] [PubMed] [Google Scholar]
- Firon N., Nepi M., Pacini E. (2012). Water status and associated processes mark critical stages in pollen development and functioning. Ann. Bot. (Lond.) 109: 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer I., Dainat J., Ranwez V., Glémin S., Dufayard J.-F., Chantret N. (2014). Impact of recurrent gene duplication on adaptation of plant genomes. BMC Plant Biol. 14: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S., Dietrich D., Fait A., Fernie A.R., Holdsworth M.J., Baker A., Theodoulou F.L. (2007). The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. 144: 1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., et al. (2010). Rice expression atlas in reproductive development. Plant Cell Physiol. 51: 2060–2081. [DOI] [PubMed] [Google Scholar]
- Guenther J.F., Chanmanivone N., Galetovic M.P., Wallace I.S., Cobb J.A., Roberts D.M. (2003). Phosphorylation of soybean nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell 15: 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T., Møller B.L. (2007). Improved cloning and expression of cytochrome P450s and cytochrome P450 reductase in yeast. Protein Expr. Purif. 56: 121–127. [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Uraguchi S., Takano J., Tanaka M., Fujiwara T. (2014). OsNIP3;1, a rice boric acid channel, regulates boron distribution and is essential for growth under boron-deficient conditions. Plant J. 78: 890–902. [DOI] [PubMed] [Google Scholar]
- Hansen M., Kun J.F.J., Schultz J.E., Beitz E. (2002). A single, bi-functional aquaglyceroporin in blood-stage Plasmodium falciparum malaria parasites. J. Biol. Chem. 277: 4874–4882. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1979). An interpretation of the hydrodynamics of pollen. Am. J. Bot. 66: 737–743. [Google Scholar]
- Heslop-Harrison J., Heslop-Harrison Y. (1970). Evaluation of pollen viability by enzymatically induced fluorescence; intracellular hydrolysis of fluorescein diacetate. Stain Technol. 45: 115–20. [DOI] [PubMed] [Google Scholar]
- Honys D., Twell D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hove R.M., Bhave M. (2011). Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Mol. Biol. 75: 413–430. [DOI] [PubMed] [Google Scholar]
- Hu J., Zhang Y., Wang J., Zhou Y. (2014). Glycerol affects root development through regulation of multiple pathways in Arabidopsis. PLoS One 9: e86269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T.P., Møller A.L.B., Zeuthen T., Holm L.M., Klaerke D.A., Mohsin B., Kühlbrandt W., Schjoerring J.K. (2004). Aquaporin homologues in plants and mammals transport ammonia. FEBS Lett. 574: 31–36. [DOI] [PubMed] [Google Scholar]
- Johnson M.A., von Besser K., Zhou Q., Smith E., Aux G., Patton D., Levin J.Z., Preuss D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.A., McCormick S. (2001). Pollen germinates precociously in the anthers of raring-to-go, an Arabidopsis gametophytic mutant. Plant Physiol. 126: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau S.A., McCormick S. (2004). A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 39: 761–775. [DOI] [PubMed] [Google Scholar]
- Katsuhara M., Sasano S., Horie T., Matsumoto T., Rhee J., Shibasaka M. (2014). Functional and molecular characteristics of rice and barley NIP aquaporins transporting water, hydrogen peroxide and arsenite. Plant Biotechnol. 219: 213–219. [Google Scholar]
- Kim S., Lieberman T.D., Kishony R. (2014). Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl. Acad. Sci. USA 111: 14494–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozono D., Ding X., Kwasaki I., Meng X., Kamagata Y., Agre P., Kitagawa Y. (2003). Functional expression and characterization of an archaeal aquaporin. J. Biol. Chem. 278: 10649–10656. [DOI] [PubMed] [Google Scholar]
- Kreida S., Törnroth-Horsefield S. (2015). Structural insights into aquaporin selectivity and regulation. Curr. Opin. Struct. Biol. 33: 126–134. [DOI] [PubMed] [Google Scholar]
- Laize V., Rousselet G., Hohmann S., Ripoche P. (1999). Molecular and functional study of AQY1 from Saccharomyces cerevisiae: Role of the C-terminal domain. Biochem. Biophys. Res. Commun. 144: 139–144. [DOI] [PubMed] [Google Scholar]
- Li G., Santoni V., Maurel C. (2014). Plant aquaporins: roles in plant physiology. Biochim. Biophys. Acta 1840: 1574–1582. [DOI] [PubMed] [Google Scholar]
- Li T., Choi W., Wallace I.S., Baudry J., Roberts D.M. (2011a). Arabidopsis thaliana NIP7;1: an anther-specific boric acid transporter of the aquaporin superfamily regulated by an unusual tyrosine in helix 2 of the transport pore. Biochemistry 50: 6633–6641. [DOI] [PubMed] [Google Scholar]
- Li X., Wang X., Yang Y., Li R., He Q., Fang X., Luu D.-T., Maurel C., Lin J. (2011b). Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23: 3780–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-H., Ludewig U., Gassert B., Frommer W.B., von Wirén N. (2003). Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol. 133: 1220–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolle S.J., Hsu W., Pruitt R.E. (1998). Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine A.E., Mccormick S., Estrada A., Patel K., Qin P. (2013). RNA-Seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 162: 1092–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu D.-T., Martinière A., Sorieul M., Runions J., Maurel C. (2012). Fluorescence recovery after photobleaching reveals high cycling dynamics of plasma membrane aquaporins in Arabidopsis roots under salt stress. Plant J. 69: 894–905. [DOI] [PubMed] [Google Scholar]
- Maeshima M., Ishikawa F. (2008). ER membrane aquaporins in plants. Pflugers Arch. 456: 709–716. [DOI] [PubMed] [Google Scholar]
- Marini A.-M., Soussi-Boudekou S., Vissers S., Andre B. (1997). A family of ammonium transporters in Saccharomyces cerevisiae Mol. Cell. Biol. 17: 4282–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masalkar P., Wallace I.S., Hwang J.H., Roberts D.M. (2010). Interaction of cytosolic glutamine synthetase of soybean root nodules with the C-terminal domain of the symbiosome membrane nodulin 26 aquaglyceroporin. J. Biol. Chem. 285: 23880–23888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C., Reizer J., Schroeder J.I., Chrispeels M.J., Saier M.H. Jr (1994). Functional characterization of the Escherichia coli glycerol facilitator, GlpF, in Xenopus oocytes. J. Biol. Chem. 269: 11869–11872. [PubMed] [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8: 64–71. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Murphy D.J. (2006). The extracellular pollen coat in members of the Brassicaceae: composition, biosynthesis, and functions in pollination. Protoplasma 228: 31–39. [DOI] [PubMed] [Google Scholar]
- Myers C., Romanowsky S.M., Barron Y.D., Garg S., Azuse C.L., Curran A., Davis R.M., Hatton J., Harmon A.C., Harper J.F. (2009). Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 59: 528–539. [DOI] [PubMed] [Google Scholar]
- Nozawa A., Takano J., Kobayashi M., von Wirén N., Fujiwara T. (2006). Roles of BOR1, DUR3, and FPS1 in boron transport and tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 262: 216–222. [DOI] [PubMed] [Google Scholar]
- O’Brien M., Bertrand C., Matton D.P. (2002). Characterization of a fertilization-induced and developmentally regulated plasma-membrane aquaporin expressed in reproductive tissues, in the wild potato Solanum chacoense Bitt. Planta 215: 485–493. [DOI] [PubMed] [Google Scholar]
- O’Neill M.A., Ishii T., Albersheim P., Darvill A.G. (2004). Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu. Rev. Plant Biol. 55: 109–139. [DOI] [PubMed] [Google Scholar]
- Ojangu E.-L., Tanner K., Pata P., Järve K., Holweg C.L., Truve E., Paves H. (2012). Myosins XI-K, XI-1, and XI-2 are required for development of pavement cells, trichomes, and stigmatic papillae in Arabidopsis. BMC Plant Biol. 12: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini E., Jacquard C., Clément C. (2011). Pollen vacuoles and their significance. Planta 234: 217–227. [DOI] [PubMed] [Google Scholar]
- Perez Di Giorgio J., Soto G., Alleva K., Jozefkowicz C., Amodeo G., Muschietti J.P., Ayub N.D. (2014). Prediction of aquaporin function by integrating evolutionary and functional analyses. J. Membr. Biol. 247: 107–125. [DOI] [PubMed] [Google Scholar]
- Pommerrenig B., Diehn T.A., Bienert G.P. (2015). Metalloido-porins: Essentiality of Nodulin 26-like intrinsic proteins in metalloid transport. Plant Sci. 238: 212–227. [DOI] [PubMed] [Google Scholar]
- Preston G.M., Carroll T.P., Guggino W.B., Agre P. (1992). Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 256: 385–387. [DOI] [PubMed] [Google Scholar]
- Preuss D., Lemieux B., Yen G., Davis R.W. (1993). A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 7: 974–985. [DOI] [PubMed] [Google Scholar]
- Qin Y., Leydon A.R., Manziello A., Pandey R., Mount D., Denic S., Vasic B., Johnson M.A., Palanivelu R. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Milla M.A., Uno Y., Chang I.-F., Townsend J., Maher E.A., Quilici D., Cushman J.C. (2006). A novel yeast two-hybrid approach to identify CDPK substrates: characterization of the interaction between AtCPK11 and AtDi19, a nuclear zinc finger protein. FEBS Lett. 580: 904–911. [DOI] [PubMed] [Google Scholar]
- Rougé P., Barre A. (2008). A molecular modeling approach defines a new group of Nodulin 26-like aquaporins in plants. Biochem. Biophys. Res. Commun. 367: 60–66. [DOI] [PubMed] [Google Scholar]
- Ruiter R.K., van Eldik G.J., van Herpen M.M., Schrauwen J.A., Wullems G.J. (1997). Expression in anthers of two genes encoding Brassica oleracea transmembrane channel proteins. Plant Mol. Biol. 34: 163–168. [DOI] [PubMed] [Google Scholar]
- Samuel M.A., Chong Y.T., Haasen K.E., Aldea-Brydges M.G., Stone S.L., Goring D.R. (2009). Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurbusch T., Hayes J., Hrmova M., Baumann U., Ramesh S.A., Tyerman S.D., Langridge P., Sutton T. (2010). Boron toxicity tolerance in barley through reduced expression of the multifunctional aquaporin HvNIP2;1. Plant Physiol. 153: 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar-Hill B., Hill A.E., Powell J., Skepper J.N., Shachar-Hill Y. (2013). Mercury-sensitive water channels as possible sensors of water potentials in pollen. J. Exp. Bot. 64: 5195–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer A., Geist B., Da Ines O., Gehwolf R., Schäffner A.R., Obermeyer G. (2008). Ectopic expression of Arabidopsis thaliana plasma membrane intrinsic protein 2 aquaporins in lily pollen increases the plasma membrane water permeability of grain but not of tube protoplasts. New Phytol. 180: 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G., Alleva K., Amodeo G., Muschietti J., Ayub N.D. (2012). New insight into the evolution of aquaporins from flowering plants and vertebrates: orthologous identification and functional transfer is possible. Gene 503: 165–176. [DOI] [PubMed] [Google Scholar]
- Soto G., Alleva K., Mazzella M.A., Amodeo G., Muschietti J.P. (2008). AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 582: 4077–4082. [DOI] [PubMed] [Google Scholar]
- Soto G., Fox R., Ayub N., Alleva K., Guaimas F., Erijman E.J., Mazzella A., Amodeo G., Muschietti J. (2010). TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. Plant J. 64: 1038–1047. [DOI] [PubMed] [Google Scholar]
- Steinhorst L., Kudla J. (2013). Calcium - a central regulator of pollen germination and tube growth. Biochim. Biophys. Acta 1833: 1573–1581. [DOI] [PubMed] [Google Scholar]
- Sugiyama N., Nakagami H., Mochida K., Daudi A., Tomita M., Shirasu K., Ishihama Y. (2008). Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Wada M., Ludewig U., Schaaf G., von Wirén N., Fujiwara T. (2006). The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18: 1498–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Wallace I.S., Takano J., Roberts D.M., Fujiwara T. (2008). NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20: 2860–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace I.S., Choi W.-G., Roberts D.M. (2006). The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochim. Biophys. Acta 1758: 1165–1175. [DOI] [PubMed] [Google Scholar]
- Wallace I.S., Roberts D.M. (2005). Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry 44: 16826–16834. [DOI] [PubMed] [Google Scholar]
- Wallace I.S., Roberts D.M. (2004). Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol. 135: 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W.-Z., Song L.-F., Zou J.-J., Su Z., Wu W.-H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148: 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J.A., Russell N.B., Whelan M.A. (2003). A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods 278: 261–269. [DOI] [PubMed] [Google Scholar]
- Williams J.H. (2008). Novelties of the flowering plant pollen tube underlie diversification of a key life history stage. Proc. Natl. Acad. Sci. USA 105: 11259–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.H. (2012). The evolution of pollen germination timing in flowering plants: Austrobaileya scandens (austrobaileyaceae). AoB Plants 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick M.M., Luu D.-T., Tournaire-Roux C., Sakamoto W., Maurel C. (2014). Vegetative and sperm cell-specific aquaporins of Arabidopsis highlight the vacuolar equipment of pollen and contribute to plant reproduction. Plant Physiol. 164: 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Dai W., Yan H., Li S., Shen H., Chen Y., Xu H., Sun Y., He Z., Ma M. (2015). Arabidopsis NIP3;1 plays an important role in arsenic uptake and root-to-shoot translocation under arsenite stress conditions. Mol. Plant 8: 722–733. [DOI] [PubMed] [Google Scholar]
- Yang Z., Wang Y., Gao Y., Zhou Y., Zhang E., Hu Y., Yuan Y., Liang G., Xu C. (2014). Adaptive evolution and divergent expression of heat stress transcription factors in grasses. BMC Evol. Biol. 14: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon G.M., Dowd P.E., Gilroy S., McCubbin A.G. (2006). Calcium-dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. Plant Cell 18: 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Wengier D., Shuai B., Gui C.-P., Muschietti J., McCormick S., Tang W.-H. (2008). The pollen receptor kinase LePRK2 mediates growth-promoting signals and positively regulates pollen germination and tube growth. Plant Physiol. 148: 1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Fan J., Taylor D.C., Ohlrogge J.B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.B., Verkman A.S. (1991). Water and urea permeability properties of Xenopus oocytes: expression of mRNA from toad urinary bladder. Am. J. Physiol. 260: C26–C34. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zhao L.-N., Shen L.-K., Zhang W.-Z., Zhang W., Wang Y., Wu W.-H. (2013). Ca2+-dependent protein kinase11 and 24 modulate the activity of the inward rectifying K+ channels in Arabidopsis pollen tubes. Plant Cell 25: 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl G.M., Zwiebel B.I., Grier D.G., Preuss D. (1999). Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.