Plant xanthine dehydrogenase contributes to H2O2 accumulation for pathogen defense in epidermal cells and also generates uric acid to remove H2O2 in chloroplasts, protecting mesophyll cells from oxidative stress.
Abstract
While plants produce reactive oxygen species (ROS) for stress signaling and pathogen defense, they need to remove excessive ROS induced during stress responses in order to minimize oxidative damage. How can plants fine-tune this balance and meet such conflicting needs? Here, we show that XANTHINE DEHYDROGENASE1 (XDH1) in Arabidopsis thaliana appears to play spatially opposite roles to serve this purpose. Through a large-scale genetic screen, we identified three missense mutations in XDH1 that impair XDH1’s enzymatic functions and consequently affect the powdery mildew resistance mediated by RESISTANCE TO POWDERY MILDEW8 (RPW8) in epidermal cells and formation of xanthine-enriched autofluorescent objects in mesophyll cells. Further analyses revealed that in leaf epidermal cells, XDH1 likely functions as an oxidase, along with the NADPH oxidases RbohD and RbohF, to generate superoxide, which is dismutated into H2O2. The resulting enrichment of H2O2 in the fungal haustorial complex within infected epidermal cells helps to constrain the haustorium, thereby contributing to RPW8-dependent and RPW8-independent powdery mildew resistance. By contrast, in leaf mesophyll cells, XDH1 carries out xanthine dehydrogenase activity to produce uric acid in local and systemic tissues to scavenge H2O2 from stressed chloroplasts, thereby protecting plants from stress-induced oxidative damage. Thus, XDH1 plays spatially specified dual and opposing roles in modulation of ROS metabolism during defense responses in Arabidopsis.
Plants produce reactive oxygen species (ROS) such as superoxide (O2⋅–) and hydrogen peroxide (H2O2) under biotic or abiotic stress conditions. While ROS at low levels act as signaling molecules to activate cellular programs to cope with such stresses, high levels of ROS can cause oxidative damage to cells unless they are locally deployed for fighting against pathogens. Thus, plants must have evolved efficient systems for both ROS production and ROS scavenging to adapt to environmental changes. In Arabidopsis thaliana, plasma membrane-localized NADPH oxidases RbohD (and RbohF in some cases) have been reported to be primarily responsible for ROS production during pathogen-associated molecular pattern-triggered immunity (Kadota et al., 2014; Li et al., 2014b) and effector-triggered immunity (Torres et al., 2002). However, in some plant-pathogen interactions, the source of H2O2 is unclear. For example, massive amounts of H2O2 are produced in powdery mildew-invaded epidermal cells expressing classic nucleotide binding leucine-rich repeat resistance (R) proteins in barley (Hordeum vulgare; Huckelhoven et al., 1999), atypical R proteins RPW8.1 and RPW8.2 in Arabidopsis (Xiao et al., 2001, 2003), or the Ol-1 R protein in tomato (Solanum lycopersicum; Li et al., 2012). Yet, the enzyme(s) that contributes to H2O2 production in powdery mildew-invaded epidermal cells remains to be identified.
Xanthine dehydrogenase (XDH; EC 1.17.1.4.) is a highly conserved housekeeping enzyme that has been extensively studied, mostly in animal systems, for over 80 years (Booth, 1935; Vorbach et al., 2003). During hydroxylation of hypoxanthine to xanthine and of xanthine to uric acid, both plant and animal XDHs are capable of producing ROS when O2 is used as the electron acceptor. In mammals, XDH can be converted into xanthine oxidase (XO) via posttranslational modification (Nishino et al., 2008); thus, it is often called xanthine oxidoreductase (XOR). While XOR is capable of producing both superoxide (O2⋅–) and H2O2, which may be utilized to kill pathogens in infected cells (Vorbach et al., 2003), its enzymatic product uric acid may function as a ROS scavenger to maintain redox homeostasis (Kim et al., 2001; Valko et al., 2007; Brychkova et al., 2008b). By contrast, plant XDHs are known only to exist as xanthine dehydrogenase capable of producing O2⋅– but not H2O2, though O2⋅– is rapidly dismutated to H2O2 (Yesbergenova et al., 2005). The complexity of XOR’s in vivo physiological functions in animals is still becoming clear (Khambata et al., 2015; Madigan et al., 2015; Suzuki et al., 2015); XOR has been shown or suggested to play a role in immune responses (Vorbach et al., 2003; Martin et al., 2004; Ives et al., 2015), in maintaining redox homeostasis, and in milk droplet secretion in mammals (Vorbach et al., 2002; Jeong et al., 2009; Jeong et al., 2013). Whether plant XDHs play a role in defense against pathogens while also protecting other cells from oxidative damage has not been determined.
In this study, we demonstrate that while Arabidopsis XANTHINE DEHYDROGENASE1 (XDH1), along with RbohD, contributes to RPW8-dependent and RPW8-independent H2O2 accumulation in powdery mildew-invaded leaf epidermal cells and basal resistance, XDH1-derived uric acid is probably essential for removing H2O2 from stressed chloroplasts in leaf mesophyll cells. Thus, our results support dual and opposing roles of Arabidopsis XDH1 in ROS metabolism and provide insights into how plants resolve conflicting needs in harnessing ROS during biotic stress.
RESULTS
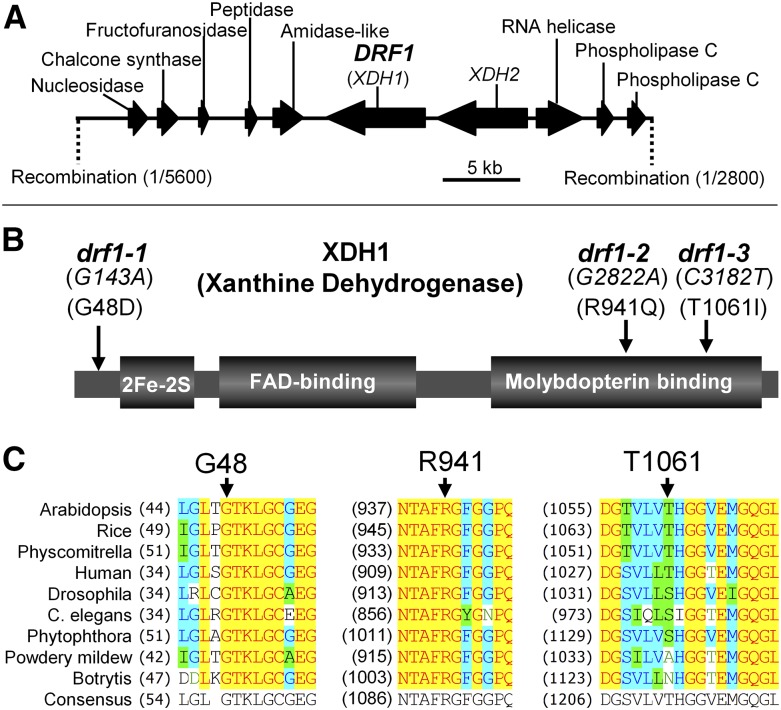
drf1 Mutants Contain Missense Mutations in XDH1
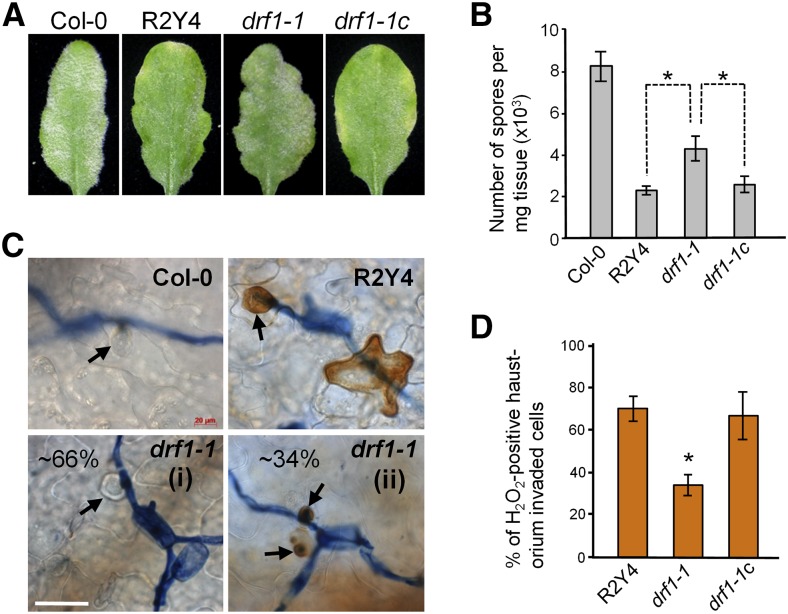
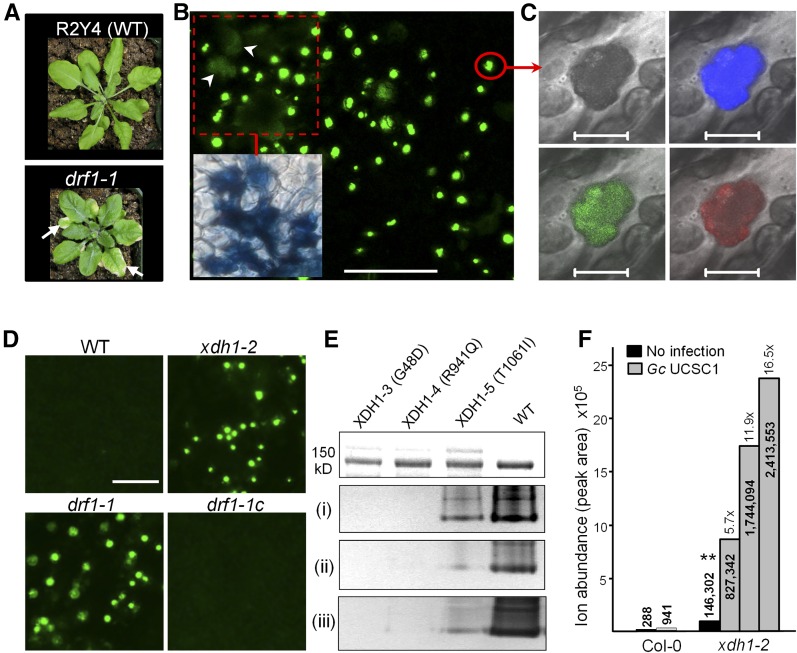
The atypical disease R genes RPW8.1 and RPW8.2 from Arabidopsis accession MS-0 confer broad-spectrum resistance against powdery mildew fungal pathogens (Xiao et al., 2001, 2003, 2005). Accession Col-0 lacks these two R genes and thus is susceptible to powdery mildew (Xiao et al., 2001, 2004). RPW8.2 is specifically targeted to and functions at the extrahaustorial membrane (EHM) that encases the fungal feeding structure named the haustorium (Wang et al., 2009, 2013). To understand how RPW8.2 activates haustorium-targeted defense including H2O2 accumulation in the host-pathogen interface and the haustorial complex (Wang et al., 2009), we chemically mutagenized R2Y4, a Col-0 transgenic line expressing RPW8.2-YFP from the RPW8.2 promoter (Wang et al., 2009), and screened for mutants that were defective in RPW8.2 function (drf) using a well-adapted powdery mildew isolate Golovinomyces cichoracearum UCSC1. We identified 15 potential drf mutants, three of which fell into the same genetic complementation group based on characterization and rescue of the mutant phenotypes (see below) and thus were named drf1-1, drf1-2, and drf1-3. We derived an F2 segregating population by crossing drf1-1 with Landsberg erecta (Ler) and found that the mutant phenotypes cosegregated with a single missense mutation G143A (resulting in the Gly48-to-Asp substitution) in At4g34890, a gene previously designated XDH1 (Hesberg et al., 2004) (Figures 1A and 1B). Targeted sequencing of XDH1 revealed missense mutations G2822A (resulting in R941Q) and C3182T (resulting in T1061I) in the remaining two mutants, respectively (Figure 1B). The other homologous XDH gene XDH2 (At4g34900) is located adjacent to XDH1 and contained no mutations in the three mutants. These three drf1 mutants showed significant reduction in RPW8.2-mediated resistance to Gc UCSC1 (Figures 2A and 2B), despite the fact that RPW8.2-YFP’s EHM localization (Wang et al., 2009) was not grossly affected. Examination of subcellular defense responses revealed that while there was no apparent difference in the formation of the callosic haustorial encasement (Wang et al., 2009), RPW8.2-YFP-triggered whole-cell and haustorial complex-confined H2O2 was significantly reduced in the mutants compared with the parental line (Figures 2C and 2D). Interestingly, bright autofluorescent objects (AFOs) of varying sizes (0.5 to 15 μm) were found in leaf tissues of 7-week‐old drf1 mutant plants (Figure 3; Supplemental Figure 1A). These plants also showed sign of cell death (Figure 3B) and early senescence (Figure 3A). To further demonstrate that DRF1 is XDH1, we obtained a T-DNA line in which XDH1 is knocked out in Col-0 (GK-049D04) (Supplemental Figure 2). AFOs also occurred in leaves of this mutant lacking RPW8.2 expression around 7 weeks old (Figure 3D), indicating that AFO formation is not RPW8.2 dependent. We expressed the genomic sequence of XDH1 from the native promoter in drf1-1. All 24 transgenic lines showed disappearance of AFOs (Figure 3D) and restoration of RPW8.2-mediated resistance (Figures 2A, 2B, and 2D). In addition, RNAi silencing of XDH1 in Col-0 also resulted in AFO formation (Supplemental Figure 1B) and infiltration of allopurinol, an inhibitor of XDH (Klinenberg et al., 1965), induced AFOs in both Arabidopsis and Nicotiana tabacum (Supplemental Figure 1C). Thus, based on the compelling genetic evidence, we conclude that impairment of XDH1 results in reduction in RPW8.2-mediated, haustorium complex-enriched H2O2 and powdery mildew resistance as well as the formation of age-dependent AFOs. To standardize the nomenclature, we designated the xdh1 mutant alleles in the GK-049D04 knockout line as xdh1-2 and those in drf1-1, drf1-2, and drf1-3 mutant lines as xdh1-3, xdh1-4, and xdh1-5, respectively, with xdh1-1 denoting the knockdown allele in SALK_148364 where a T-DNA is inserted in the 11th intron of XDH1 described earlier (Yesbergenova et al., 2005).
Figure 1.
Cloning of DRF1.
(A) Map-based cloning of DRF1 using an F2 population derived from drf1-1 × Ler.
(B) The nature and position of the three drf1 mutations in XDH1 that encodes a xanthine dehydrogenase with three functional domains.
(C) Protein sequence alignment showing conservation of the three mutated residues (indicated by arrows) among XDH homologs from rice (AAT81740), Physcomitrella patens (EDQ74505), human (NP_000370), Drosophila melanogaster (NP_524337), Caenorhabditis elegans (NP_500531), Phytophthora infestans (EEY63796), powdery mildew (CCU77189), and Botrytis cinerea (CCD52002).
Figure 2.
Characterization of the Defense Phenotypes of the drf1-1 Mutant.
(A) Representative leaves of indicated genotypes showing whitish fungal mass. Six-week-old plants were inoculated with Gc UCSC1 and pictures were taken at 10 dpi. R2Y4 is a homozygous Col-0 line expressing RPW8.2-YFP from the RPW8.2 promoter. drf1-1c is a representative line of drf1-1 genetically complemented with XDH1 expressed from the XDH1 promoter.
(B) Quantification of disease susceptibility of plants in (A). Data are means ± se from four replicated experiments. Asterisks indicate significant difference (P < 0.05, n = 4) for the paired comparisons using Tukey’s HSD test following one-way ANOVA.
(C) Representative microscopic images of invaded epidermal cells of indicated genotypes showing whole-cell or haustorial complex-confined H2O2 stained by DAB at 52 hpi. Note (i) and (ii) denote two types of reactions of drf1-1 mutant plants. Arrows indicate haustoria. Bar = 50 μm.
(D) Frequencies of H2O2-positive epidermal cells (whole-cell H2O2 + haustorium complex-confined H2O2 of indicated genotypes used in [C]). At least 100 haustorium-invaded cells were assessed for each genotype. Data are means ± sd from three replicated experiments. Asterisk indicates significant difference when compared with other two genotypes (P < 0.01; n = 3, Student’s t test).
Figure 3.
Characterization of AFOs.
(A) Compared with the parental R2Y4 line (WT) expressing RPW8.2-YFP, drf1-1 showed slightly reduced stature with shorter petioles and displayed early leaf senescence (arrows).
(B) Mature leaves of 7-week-old (or more) drf1-1 plants developed AFOs of various sizes that were subsequently associated with cell death shown by diffuse autofluorescence, indicated by arrowheads and trypan blue staining (inset). Bars = 100 μm.
(C) Confocal images of an AFO from (B) showing autofluorescence under 405-, 514-, and 561-nm laser excitation. Bars = 5 μm.
(D) Development of AFOs in drf1-1 and an XDH1 knockout line (GK-049D04; xdh1-2) and lack of AFOs in drf1-1 genetically complemented by expressing XDH1 from its native promoter (drf1-1c). Bars = 50 μm.
(E) Assay for the enzymatic activities of three mutant proteins, XDH1-3 (G48D), XDH1-4 (R941Q), and XDH1-5 (T1061I), in comparison with XDH1. Recombinant proteins (∼2 μg) purified after heterologous expression in P. pastoris (visualized in gel by Coomassie blue staining in the top panel) were used for measuring (i) the basic XDH activity using hypoxanthine and NAD+ as substrate, (ii) superoxide production using hypoxanthine and O2 as substrate, or (iii) superoxide production using NADH and O2 as substrate (for details, see Methods).
(F) Xanthine levels in AFOs from uninfected or Gc UCSC1-infected leaves of 12-week-old xdh1-2 and the equivalent extracts from leaves of 12-week-old Col-0 grown under the same conditions. Leaves were collected at 7 dpi. AFOs were enriched by sucrose gradient centrifugation and subjected to liquid chromatography-mass spectrometry analysis (see Methods). The numbers within or on top of the columns are exact peak areas reporting the total ion abundance. The numbers followed by “x” are fold increase of xanthine levels compared with that indicated by asterisks.
To link the enzymatic properties of XDH1 variants with their biological functions, we expressed the coding sequence of the three new XDH1 mutant alleles in the methylotrophic yeast Pichia pastoris. Purified XDH1-3 (G48D), XDH1-4 (R941Q), and XDH1-4 (T1061I) variants were then subjected to assays for (1) basic dehydrogenase activities where the oxidation of hypoxanthine was followed in the presence of O2 and phenazine methosulfate as electron acceptor, (2) superoxide (O2⋅–) production where hypoxanthine served as substrate and O2 as electron acceptor, and (3) O2⋅– production where NADH was used as substrate in the presence of O2 as electron acceptor. In addition, the basic dehydrogenase activity with hypoxanthine as substrate and NAD+ as the naturally preferred electron acceptor was monitored (in the presence of molecular oxygen). While XDH1-3 showed no or barely detectable activities, XDH1-4 and XDH1-5 displayed 90 to 95% and 60 to 80% reduction in these activities, respectively (Figure 3E; Supplemental Figure 3). The lower the levels of the in vitro enzymatic activities of these recombinant proteins encoded by the three mutant alleles, the higher the levels of AFO formation found in these three drf1 mutants (i.e., drf1-1 showed the highest while drf1-3 the lowest levels; Figure 3E; Supplemental Figure 1A), implying similar reductions in in vivo enzymatic activities of the corresponding mutant proteins. Interestingly, both G48 and R941 are absolutely conserved in all XDH/XOR homologs across kingdoms, while the T1061 site is less conserved (Figure 1C). These results suggest that G48 is essential, R941 is important, and T1061 is less important for the biochemical functions of XDHs in plants and perhaps in animals.
Xanthine Accumulation in xdh1 Reports Higher Purine Catabolic Activity during Defense Responses
In our efforts to characterize AFOs formed as a consequence of genetic impairment of XDH1, we found that AFOs were resistant to proteases, lipases, and various detergents (see Methods) but could be dissolved in multiple buffers with a pH value of 8.0 or higher in situ or in vitro (Supplemental Figure 4). Because pure xanthine becomes water insoluble and crystallizes in pH 6 to 7 aqueous solution (Supplemental Figure 5B), we suspected that xanthine crystallines may be part of AFOs in leaf cells (with a cytosolic pH around 7.0) of xdh1 mutant plants where xanthine accumulates due to loss of XDH activity. Relevantly, there was one report that described xanthine crystalline deposition in unfixed sections of skeletal muscle biopsies from two human patients (Chalmers et al., 1969). We thus purified AFOs by sucrose gradient centrifugation from mature leaves of xdh1-2 plants and found that the AFOs could indeed dissolve in pH 8.0 Tris-HCl buffer (Supplemental Figure 5A). We then collected leaves of 12-week-old xdh1-2 plants infected with powdery mildew or uninoculated xdh1-2 plants at 7 d postinoculation (dpi) for AFO isolation and purification. We also prepared the AFO-equivalent fraction from powdery mildew-infected or uninoculated Col-0 plants as control. Mass spectrometry analysis showed that the AFO-enriched fraction from the xdh1-2 samples contained over 500 times more xanthine than the corresponding fraction of the Col-0 sample following the same extraction and centrifugation procedures (Figure 3F; Supplemental Figure 6B), while there was only an ∼6-fold increase of xanthine content in whole-leaf tissue of xdh1-2 compared with that of the wild type based on our HPLC analysis (Supplemental Figure 6A). Thus, it appeared that xanthine was highly enriched in AFOs formed in xdh1 mutant plants. Moreover, xanthine levels in the AFO samples from powdery mildew-infected xdh1-2 plants were at least 5-fold higher than that in the AFO sample from uninfected xdh1-2 plants (Figure 3F), suggesting that powdery mildew infection could further induce xanthine accumulation, likely due to upregulation of purine catabolism in infected host cells.
Since XDH also converts hypoxanthine to xanthine, we measured the hypoxanthine levels in the above samples and found that hypoxanthine levels in xdh1-2 were at least two orders of magnitude lower compared with xanthine levels, despite a slight increase (2 to 4×) in powdery mildew-infected xdh1-2 compared with uninfected xdh1-2 (Supplemental Figure 7). This result is consistent with previous reports that hypoxanthine may be reverted within a salvage pathway to inosine, inosine monophosphate, xanthosine monophosphate, xanthosine, and finally to xanthine (Brychkova et al., 2008a).
To further confirm that AFO formation was due to xanthine accumulation, we infiltrated 10 mM xanthine (dissolved in 100 mM Tris-HCl, pH8.0) in Col-0 or N. tabacum leaves and detected AFOs similar to those in drf1 mutants (Supplemental Figure 5C). Using the lambda scan function of Zeiss LSM710, we determined the emission spectra of AFOs purified from xdh1-3 and pure xanthine crystallines under different laser excitation wavelengths. We found that the former had much broader emission spectra that also largely covered the peaks at ∼580 nm from the latter (Supplemental Figure 5D), suggesting that AFOs in plant cells may contain other, unknown autofluorescent compounds in addition to xanthine crystallines. Regardless, it appears that AFO formation could provide a convenient and reliable visual marker for xanthine accumulation in cells and tissues in xdh1 mutant plants, which can further reflect purine catabolic activity at a high spatiotemporal resolution in Arabidopsis and perhaps in other plant species (Supplemental Figure 1C). As detailed in Supplemental Figures 8 to 11, our observations surrounding AFOs in xdh1 plants suggest that purine catabolic activity (1) is higher in mesophyll cells compared with epidermal cells, as AFOs are mostly found in the former but rarely, if ever, found in the latter; (2) increases as plants/organs mature and age, (3) increases in response to fungal infections, and (4) increases in plants expressing RPW8.1 and RPW8.2.
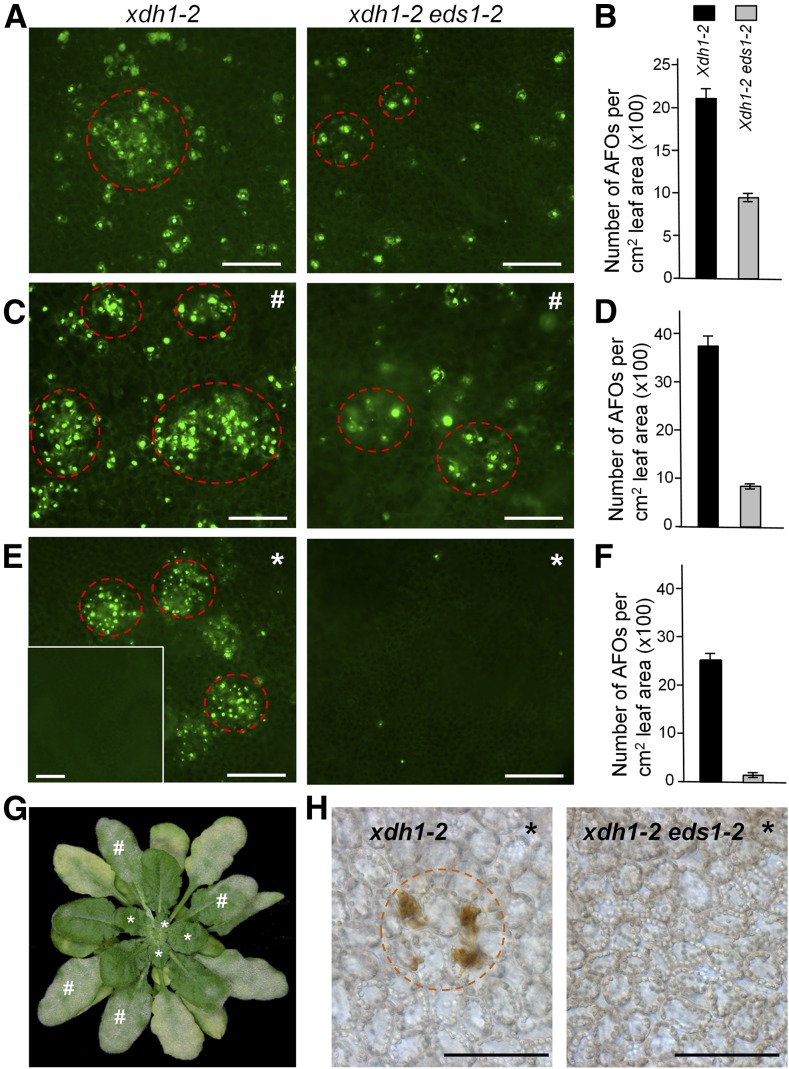
Interestingly, there appeared to be a local induction and amplification of AFOs in leaves of XDH1-defective plants, especially when they were challenged by G. cichoracearum UMSG1, a sow thistle powdery mildew poorly adapted to Arabidopsis (Wen et al., 2011) (Supplemental Figure 10), or overexpressing RPW8.1 and RPW8.2 from their native promoters (Xiao et al., 2003) (Supplemental Figure 11). These observations suggest that defense signals may enhance purine catabolic activity in local cells adjacent to the site of infection or cells undergoing defense-associated cell death. To see if purine catabolic activity is systemically enhanced by defense signals, we challenged fully expanded leaves without age-dependent AFOs in 8-week-old xdh1-2 plants with Gc UCSC1, a well-adapted powdery mildew isolate. Not surprisingly, Gc UCSC1 infection could induce local AFO formation (Figure 4C). More interestingly, newly emerged leaves free from powdery mildew infection also developed numerous clustered AFOs from 5 dpi on (Figures 4E and 4G), whereas no AFOs were found in similar leaves of uninfected xdh1-2 plants (inset in Figure 4E).
Figure 4.
EDS1 Is Required for Local and Systemic Amplification of AFO Formation.
Eight-week-old plants of xdh1-2 or xdh1-2 eds1-2 were examined for AFO formation. All bars = 100 μm.
(A) and (B) AFO formation in mature leaves of uninfected plants from both genotypes as control for (C,D). Note that AFOs in xdh1-2 eds1-2 were in lower density with fewer clusters as seen in xdh1-2 (circled by dashed red lines).
(C) and (D) AFO induction by powdery mildew (Gc UCSC1) infection in AFO-free leaves of plants from both genotypes. Images were taken at 14 dpi.
(E) and (F) Systemic AFOs in uninfected young leaves (indicated by an asterisk, as seen in [G]) induced by powdery mildew infection on older leaves (indicated by a pound sign, as seen in [G]) of the indicated genotypes from 5 dpi on. No AFOs were found in similar young leaves of uninfected xdh1-2 (inset in [E]). Images were taken at 14 dpi. Data in (B), (D), and (F) represent means ± se, calculated from six similarly aged leaves (two from one plant) for each genotype.
(G) One representative 8-week-old plant infected with Gc UCSC1 at 10 dpi.
(H) H2O2 accumulation in chloroplasts of uninfected younger leaves (asterisks) revealed by DAB staining.
This “systemic” AFO formation to some extent resembled the micro-oxidative burst that is induced by local immune response in mature leaves and required for establishment of systemic acquired resistance (SAR) (Alvarez et al., 1998). To examine if pathogen-induced local and systemic AFO formation requires defense signaling, we introduced a null allele of EDS1 (i.e., eds1-2) (Bartsch et al., 2006) into xdh1-2 by crossing with eds1-2 in the Col-0 background. EDS1 is an essential component for local resistance triggered by TIR-NB-LRRs (Parker et al., 1996; Aarts et al., 1998), or RPW8.1 and RPW8.2 (Xiao et al., 2001, 2005), as well as SAR (Breitenbach et al., 2014; Wittek et al., 2014). We found that although xdh1-2 eds1-2 plants still developed age-dependent AFOs, the AFO density was only ∼50% of that in xdh1-2, and AFOs were rarer in clusters (Figures 4A and 4B). We then inoculated fully expanded leaves of xdh1-2 eds1-2 that had not developed age-dependent AFOs with Gc UCSC1 and found that Gc UCSC1-induced local AFO formation was reduced to 20% of the level seen in infected leaves of xdh1-2 (Figures 4C and 4D). Strikingly, systemic AFO formation was almost completely abolished in xdh1-2 eds1-2 (Figures 4E and 4F). These results suggest that purine catabolic activity is potentially geared up with both local and systemic defense responses, presumably for delimiting local and systemic ROS-dependent hypersensitive response (HR) (Alvarez et al., 1998). Consistent with this, XDH1 was found to be induced to higher levels (∼2 to 3×) by powdery mildew infection, and this transcriptional upregulation seemed to require its own protein function as the xdh1-3 allele did not show powdery mildew-induced elevation (Supplemental Figures 12A and 12B). In addition, XDH1 was induced to higher levels after spray with 5% H2O2 solution both at the mRNA and the protein level, which was also associated with increased xanthine dehydrogenase activity (Supplemental Figures 12C and 12D). Examination of the 5′ promoter region of XDH1 identified three W boxes (TTGACC/T) upstream of the ATG start codon (i.e., at −74, −187, and −576 bp; Supplemental Figure 12E). Since W boxes (TTGACC/T) are known to be binding sites for defense-related WRKY transcription factors (Eulgem and Somssich, 2007), it is likely that the transcription of XDH1 may be intrinsically connected to defense responses.
XDH1 Is Required for Scavenging Age-Dependent and Pathogen-Induced H2O2 in Chloroplasts
To understand the mechanistic connection between elevation of purine catabolic activity (reported by AFO formation) and age-dependent (Nakagawa et al., 2007; Brychkova et al., 2008b) or pathogen-induced early leaf senescence in xdh1 mutants (Supplemental Figure 13), we used 3,3′-diaminobenzidine (DAB) staining to investigate whether there is H2O2 accumulation before occurrence of senescence-associated cell death in xdh1-2. Interestingly, we found that AFO formation was often accompanied by high-level H2O2 accumulation in chloroplasts before collapse of the affected mesophyll cells (Figure 5A). Parallel to a local “amplification” of AFOs in xdh1-2 (Figure 4), H2O2 accumulation in chloroplasts also showed apparent “amplification,” as it started in individual chloroplasts, then spread to all chloroplasts in the same cell, and then spread to neighboring mesophyll cells but not epidermal cells (Figure 5A). Detailed examination showed that H2O2-positive chloroplasts lost their normal positioning along the anticlinal cell wall, aggregated, and then disintegrated, which was followed by the collapse of the affected individual and clustered mesophyll cells as revealed by trypan blue staining (Figures 5A and 5B). Macroscopically, sporadic chlorotic lesions corresponding to localized mesophyll cell death became visible to the naked eye and then further enlarged and coalesced and eventually spread to the entire leaf, exhibiting leaf senescence-like phenotypes (Supplemental Figure 9A). Importantly, DAB staining showed that powdery mildew infection-induced systemic AFOs in uninfected leaves also associated with H2O2 accumulation in chloroplasts of affected mesophyll cells in xdh1-2, whereas no H2O2 was detectable in similar leaves of xdh1-2 eds1-2 double mutant (Figure 4H), suggesting that XDH1 may function to limit SAR-associated micro-oxidative burst (Alvarez et al., 1998).
Figure 5.
Chloroplast-H2O2 Accumulation in Mesophyll Cells Due to Loss of XDH1 and Its Suppression by Exogenous Uric Acid.
(A) Five (1 to 5) serial images showing gradual amplification of chloroplast-H2O2 accumulation revealed by DAB staining in mesophyll cells of mature leaves of 7-week-old xdh1-2 plants and lack of H2O2 accumulation in epidermal cells (5′). Note that initial H2O2 accumulation occurred in one or few chloroplasts (arrows) and H2O2-positive chloroplasts were often disoriented, aggregated, and subsequently degraded (arrowheads). Bars = 50 μm.
(B) Three (1 to 3) serial images showing collapse of mesophyll cells whose chloroplasts accumulated H2O2 (inset in 2) 4 to 6 d after AFO formation in mature leaves of xdh1-2 plants stained by trypan blue. Bars = 200 μm.
(C) Images showing suppression of chloroplast-H2O2 accumulation in mature leaves of 6-week-old xdh1-2 plants by exogenous uric acid. Fully expanded leaves of xdh1-2 were inserted (with their petioles) into MS-agar medium supplemented with uric acid and cultured for 4 weeks with one transfer per week to fresh MS-agar plates containing the same levels of uric acid. Note that (i) MS-agar conditions, though good for supplying uric acid, attenuated chloroplast-H2O2 production in xdh1-2; and (ii) these images showed the highest local density of H2O2-positive mesophyll cells for each treatment. This experiment was repeated three times with similar results. Bars = 200 μm.
(D) Quantification of DAB-positive mesophyll cells in (C). Data are means ± se, calculated from four duplicated leaf samples of one experiment in (C).
H2O2 accumulation in chloroplasts appeared to be an early event of leaf senescence in both Col-0 and xdh1-2. However, the sporadic occurrence of a high level of H2O2 accumulation and aggregation of chloroplasts in clustered mesophyll cells of xdh1-2 was distinct from the relatively synchronized H2O2 accumulation in less aggregated chloroplasts of the majority of mesophyll cells in naturally senescing leaves of Col-0 (which occurs ∼2 to 3 weeks later than the former under the same experimental conditions) (Supplemental Figure 9B). Intriguingly, when we managed to visualize both AFOs and H2O2 in mesophyll cells of xdh1-2, we found that less than one-third of H2O2-positive cells contained very large AFOs (∼20 µm in diameter) and there was no strict co-occurrence of AFOs and H2O2 accumulation (Supplemental Figure 14A), possibly because small AFOs may dissolve or lose autofluorescence during chlorophyll clearing as part of DAB staining. This observation implies that xanthine accumulation per se may not be the direct cause for chloroplast-H2O2 accumulation in mesophyll cells. It has been reported that uric acid and/or its catabolic products can scavenge ROS and therefore could be important antioxidants in both plants and animals (Kim et al., 2001; Valko et al., 2007; Brychkova et al., 2008b).
To test if uric acid deficiency as a corollary of xanthine accumulation due to loss of XDH1 is responsible for chloroplast-H2O2 accumulation and subsequent cell death, we supplied uric acid to detached, fully expanded but AFO-free, xdh1-2 leaves by incubating them on Murashige and Skoog (MS)-agar medium containing uric acid for 4 weeks. We found that chloroplast-H2O2 accumulation was completely suppressed by addition of 200 µM uric acid to the MS-agar medium (Figures 5C and 5D), suggesting that endogenous uric acid may indeed protect chloroplasts from oxidative damage. Interestingly, although AFOs still developed sporadically in leaves incubated in uric acid-containing MS-agar medium for 4 weeks, amplification of AFOs in those leaves was significantly reduced (Supplemental Figure 14B), suggesting that chloroplast-localized oxidative stress can stimulate purine catabolism in local adjacent mesophyll cells. These results are consistent with the earlier findings that reduced expression of XDH1 results in accelerated senescence of mature leaves (Nakagawa et al., 2007) and that exogenous uric acid or its catabolic products can suppress natural or dark-induced early senescence of XDH1-impaired plants (Nakagawa et al., 2007; Brychkova et al., 2008b). Thus, it is likely that uric acid (and/or potentially its downstream catabolic products) derived from xanthine by XDH1 is essential for protecting chloroplasts in mesophyll cells from age-dependent and/or stress-induced oxidative damage in Arabidopsis.
XDH1 Plays Opposing Roles in H2O2 Metabolism in Haustorium-Affected Epidermal and Mesophyll Cells
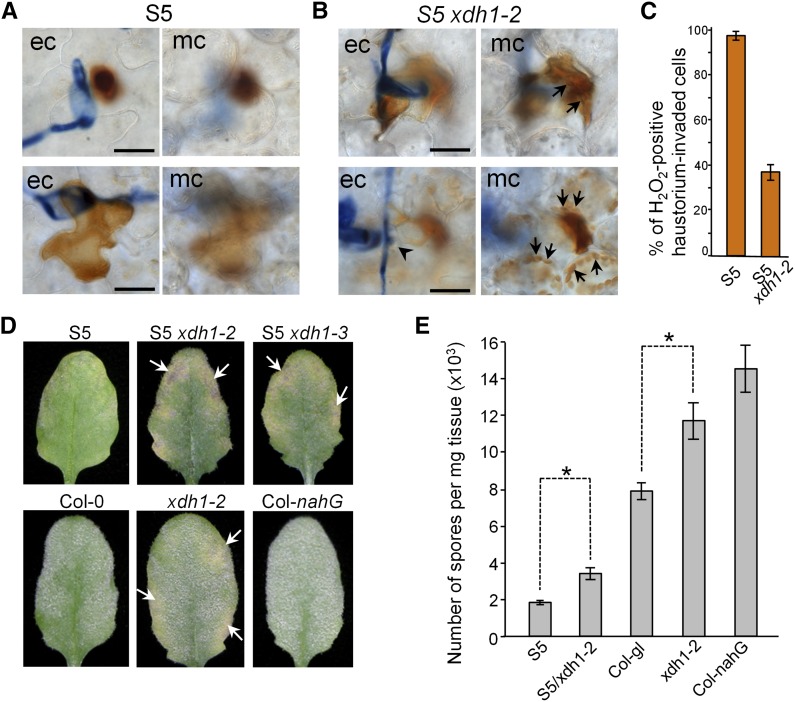
Our above genetic evidence supports seemingly opposite roles of XDH1 in H2O2 metabolism: While XDH1 contributes to RPW8.2-YFP-mediated H2O2 production in haustorium-invaded epidermal cells, it is required for H2O2 removal in the chloroplasts of stressed mesophyll cells. To evaluate these two opposing functional aspects of XDH1 under the same cellular context while reconfirming the requirement of XDH1 in RPW8.2-mediated resistance, we introduced the xdh1-2 knockout allele into S5, a Col-gl line transgenic for a single copy of a genomic fragment that contains RPW8.1 and RPW8.2 and their native promoters (referred to as RPW8 for simplicity unless otherwise indicated) (Xiao et al., 2003) rendering stable powdery mildew resistance for >20 generations. We inoculated 6-week-old plants of S5 and S5/xdh1-2 with the adapted pathogen Gc UCSC1 and found no apparent differences between S5 and S5/xdh1-2 in terms of callose deposition around haustoria. By contrast, DAB staining revealed that whole-cell H2O2 and haustorial complex-confined (HC) H2O2 (Figure 6A) together were reduced in S5/xdh1-2 to ∼38%, which is less than half of that (98%) in S5 (Figure 6C), confirming that XDH1 contributes to haustorium-induced, RPW8-mediated H2O2 in epidermal cells.
Figure 6.
XDH1 Is Required for RPW8-Mediated and Basal Resistance against Powdery Mildew.
(A) and (B) Loss of XDH1 compromised RPW8-dependent H2O2 production in haustorium-invaded epidermal cells (ec) but resulted in chloroplast H2O2 accumulation in underneath mesophyll cells (mc). Leaves from 6-week-old S5 and S5/xdh1-2 were evenly inoculated with Gc UCSC1, subjected to DAB staining at 60 hpi, and imaged with a Zeiss Axio microscope. H2O2 accumulation (reported by brownish DAB staining) was either confined in the haustorial complex or spread throughout the entire invaded epidermal cell (ec) in S5 (A). By contrast, while H2O2 accumulation in haustorium-invaded epidermal cells (arrowhead) was reduced, in ∼10% of penetration sites chloroplast H2O2 accumulation (indicated by arrowhead) occurred in the mesophyll cells underneath the infection site in S5/xdh1-2 at 60 hpi (B). Such induced H2O2 accumulation in affected mesophyll cells increased to >60% after 8 dpi. Bars = 50 µm.
(C) Quantitative assessment of H2O2 accumulation in haustorium-invaded epidermal cells. Data represent means ± se of four duplicate experiments in which >200 haustorium-invaded epidermal cells per genotype were examined.
(D) Representative leaves from indicated genotypes infected with Gc UCSC1 at 10 dpi. Note chlorotic or necrotic lesions (white arrows) induced by powdery mildew infection in plants impaired for XDH1.
(E) Quantification of disease susceptibility of the indicated genotypes. Infected leaves were assayed at 7 dpi before extensive chlorosis occurred in xdh1-2 plants. Data are means ± se, calculated from four duplicate samples per genotype in one of three independent experiments in which similar results were obtained. Asterisks indicate significant difference (P < 0.01; n = 4, Student’s t test).
Because AFOs were never detected in epidermal cells in xdh1 mutant plants (Supplemental Figure 8A), one may speculate that either XDH1 is not expressed or XDH1 may not catalyze conversion of xanthine to uric acid in epidermal cells. To examine if XDH1 is expressed in epidermal cells, we conducted XDH1 promoter-GUS and XDH1 promoter-YFP-XDH1 analyses and found that XDH1 is expressed in epidermal cells albeit at relatively lower levels compared with mesophyll cells based on GUS staining intensity (Supplemental Figures 8B and 8C). Next, we examined the cells surrounding the Gc UCSC1 haustorium-invaded cells in S5 and S5/xdh1-2. Haustorium-induced H2O2 accumulation in S5 was confined to the invaded epidermal cells and we rarely observed DAB-detectable H2O2 in neighboring epidermal or underneath mesophyll cells under our current experimental conditions (Figure 6A). However, in ∼10% of the fungal infection sites examined in mature leaves of 6-week-old plants, there was high-level chloroplast-H2O2 accumulation in the mesophyll cells underneath the haustorium-invaded epidermal cells of S5/xdh1-2 at 60 h postinoculation (hpi), despite much reduced H2O2 accumulation in the invaded epidermal cells (Figure 6B). The frequency of chloroplast-H2O2 accumulation sharply increased to >60% from 8 dpi on and/or as plants got older. Consequently, there were many more powdery mildew-induced necrotic/chlorotic lesions in S5/xdh1-2 compared with S5 (Figure 6D; Supplemental Figure 13).
Taken together, our observations suggest that XDH1 may utilize a substrate other than xanthine to generate O2⋅– and/or H2O2 in epidermal cells for fighting against powdery mildew infection, whereas it performs its housekeeping enzymatic function to convert xanthine to uric acid for scavenging biotic stress-induced and age-dependent H2O2 accumulation in the chloroplasts of affected mesophyll cells.
XDH1-Derived H2O2 Is Also Required for RPW8-Dependent and -Independent Basal Resistance
To confirm that XDH1-derived H2O2 in the haustorial complex or throughout the epidermal cells contributes to RPW8-mediated resistance, we quantified the disease phenotypes of S5 and S5/xdh1-2 plants upon Gc UCSC1 infection. We found that S5/xdh1-2 plants supported a small but significant increase of fungal sporulation compared with S5 at 7 dpi (Figure 6E). However, at later infection stages (10 to 14 dpi), we detected no significant differences between S5 and S5/xdh1-2 because massive powdery mildew-induced mesophyll cell death in the latter reduced fungal infection, probably passively, thus offsetting the effect caused by H2O2 decreases in epidermal cells. These observations suggest that in plants expressing RPW8, H2O2 derived from XDH1 and other sources (see below) in the epidermal cells is utilized to constrain haustoria, whereas uric acid derived from XDH1 may protect adjacent mesophyll cells from powdery mildew infection-induced oxidative stress.
To test if XDH1 also plays a role in basal resistance independent of RPW8, we compared Col-0 and xdh1-2 plants for their disease phenotypes upon infection with Gc UCSC1. We found that xdh1-2 supported significantly more fungal sporulation at 7 dpi (Figure 6E; Supplemental Figure 15A). As expected, xdh1-2 also displayed powdery mildew-induced chlorosis at 10 to 14 dpi (Figure 6D). This suggests that XDH1 may also contribute to H2O2 production during basal resistance, even though H2O2 accumulation in infected epidermal cells is largely suppressed by the well-adapted Gc UCSC1 isolate. Consistent with this, while weak DAB staining was detected in ∼5% of fungal penetration sites in Col-0, it was rarely (1 to 2%) observed in fungal penetration sites in xdh1-2 (Supplemental Figure 15B).
XDH1 and NADPH Oxidases Coordinate to Produce H2O2 at the Host-Pathogen Interface
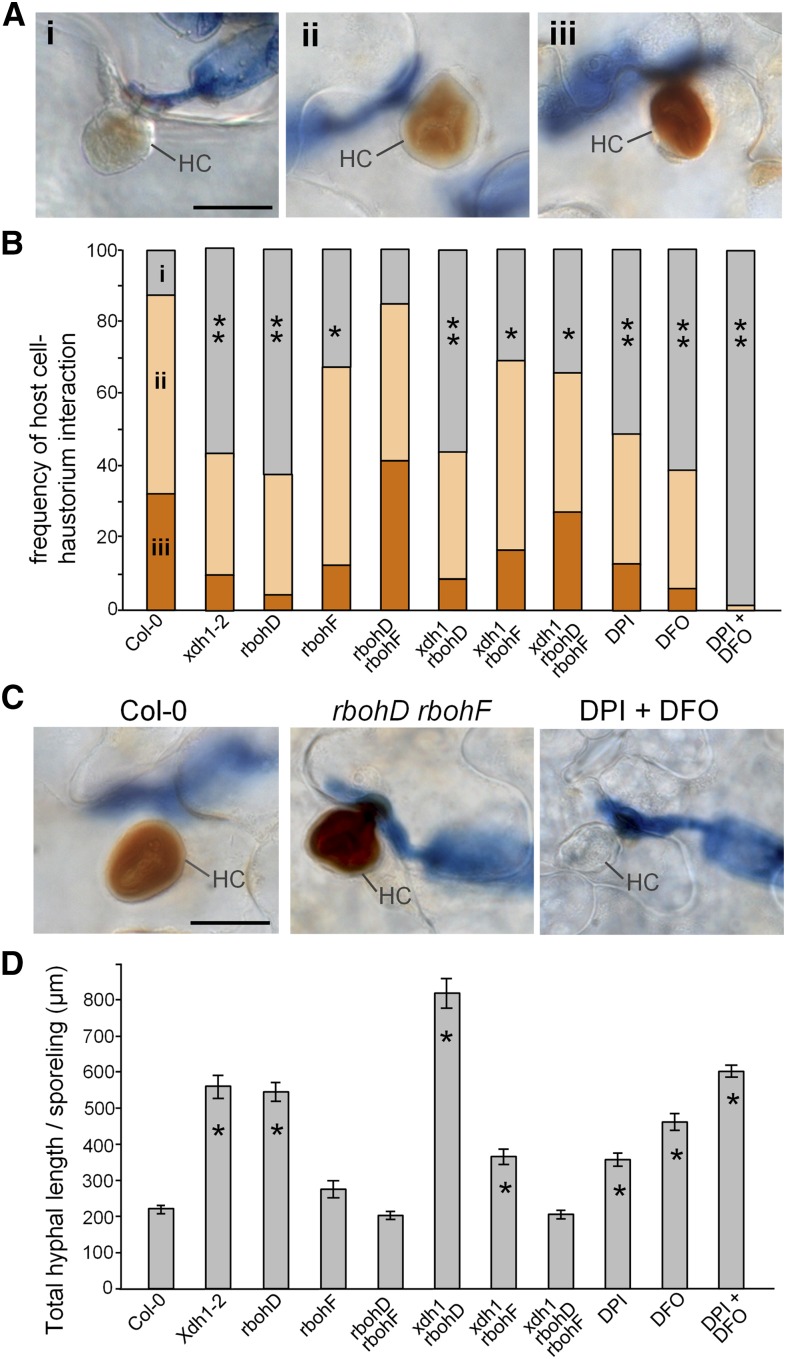
To further evaluate if XDH1 contributes to H2O2 in Col-0 independent of RPW8, we challenged Col-0 and xdh1-2 plants with Gc UMSG1, a powdery mildew isolate infectious on sow thistle (Wen et al., 2011). Gc UMSG1 overcomes penetration resistance but is invariably arrested at the postpenetration stage in >30 Arabidopsis accessions tested including Col-0 (Wen et al., 2011). DAB staining revealed that nearly 90% of invaded cells of Col-0 showed detectable H2O2 accumulation in HCs, while H2O2 was also occasionally found throughout the invaded epidermal cells. To assess the amount of powdery mildew-induced H2O2 in situ, we focused on HC-H2O2 (which dominates under this context) and divided haustorial complexes into three types: (1) those without H2O2, (2) those with medium levels of H2O2, and (3) those with high levels of H2O2 based on the intensity of DAB staining (Figure 7A). We found that the frequency of H2O2-positive haustorial complexes in xdh1 (∼43%) was significantly lower compared with that of Col-0 (∼87%) (P < 0.01, ANOVA in R) (Figure 7B). This result indicates that XDH1 indeed contributes to RPW8-independent H2O2 production during basal defense against poorly adapted pathogens and also implies that H2O2 from other sources must orchestrate haustorium-targeted defense.
Figure 7.
XDH1, RbohD, and RbohF Orchestrate H2O2 Generation in Haustorial Complexes.
Single, double, and triple mutants concerning XDH1 and NADPH oxidase-encoding RbohD and RbohF were inoculated with Gc UMSG1, a poorly adapted powdery mildew isolate. H2O2 accumulation in invaded epidermal cells was revealed by DAB staining.
(A) Three typical levels of H2O2 accumulation in HCs, i.e., no or very little (i), moderate (ii), and strong (iii) that were observed in most genotypes. Bars = 20 µm.
(B) Frequencies of the three levels of HC-H2O2 accumulation in eight indicated genotypes treated with 100 μM DPI and/or 2 mM DFO. Fully expanded leaves of 6-week-old plants were inoculated evenly with Gc UMSG1 and then subjected to DAB staining at 60 hpi to assess HC-H2O2 accumulation. At least 200 HCs were assayed for each genotype. Three different color bars represent the three levels of H2O2 accumulation in (A). Statistical analysis was conducted by comparing all genotypes or treatments to Col-0 wild type using ANOVA in R (www.r-project.org). One asterisk indicates significant difference at P < 0.05; two asterisks indicate significant difference at P < 0.01. This experiment was repeated three times with similar results.
(C) Three representative HC-H2O2 of the indicated genotypes. Bars = 20 µm.
(D) Basal resistance reflected by the total hyphal length per sporeling in eight indicated genotypes treated with DPI and/or DFO. Data are means ± se, calculated from at least 20 sporelings per genotype in one of three duplicate experiments. The asterisk indicates significant difference when compared with Col-0 wild type (P < 0.01; Student t test).
The Arabidopsis respiratory burst oxidase homologs RbohD and RbohF have been shown to be largely responsible for bacterial and oomycete pathogen-induced H2O2 production (Torres et al., 2002; Xu et al., 2014), and RbohD is responsible for pathogen elicitor-induced oxidative burst (Kadota et al., 2014; Li et al., 2014b). However, whether RbohD or RbohF contribute to HC-H2O2 has not been determined. We thus first tested if rbohD and rbohF single and rbohD/F double mutants show any altered HC-H2O2 phenotypes in comparison with Col-0 and xdh1. As shown in Figure 7B, HC-H2O2 frequency in rbohD was reduced to ∼37%, which is an even slightly greater reduction relative to that in xdh1-2 (∼43%). HC-H2O2 frequency in rbohF also showed a slight but significant reduction to ∼67% compared with Col-0 (P < 0.05). These results are consistent with a previous report that RbohD makes a major, while RbohF a minor, contribution to bacterium-induced H2O2 in Arabidopsis (Torres et al., 2002). Unexpectedly, we found that the rbohD rbohF double mutant showed a HC-H2O2 frequency (85%) close to that of Col-0, with the “high HC-H2O2” type being even higher (41%) than that of Col-0 (32%) (Figure 7B). In fact, the highest intensity of HC-H2O2 was most commonly observed in rbohD rbohF (Figure 7C). This striking observation suggests that there may be a compensatory mechanism for ROS generation in epidermal cells of Arabidopsis for haustorium-targeted defense. To further test this idea, we made xdh1 rbohD and xdh1 rbohF double and xdh1 rbohD rbohF triple mutants by genetic crossing. Soil-grown xdh1 rbohD rbohF seedlings developed massive necrosis within 3 weeks after seed germination and died at 4 to 5 weeks old. However, plants of the triple mutant grown in MS-agar plates for 4 weeks and then grown in sterile soil under >90% relative humidity could reach maturity, despite their smaller stature, thus enabling tests for H2O2 and disease phenotypes. Interestingly, while the xdh1 rbohD and xdh1 rbohF double mutants developed HC-H2O2 as frequently as or more frequently than their respective single mutants, the triple mutant exhibited significantly more HC-H2O2 of the high HC-H2O2 type than any of the single or double mutant except rbohD rbohF (Figure 7B). These results indicate that these three genes individually contributes to haustorium-induced H2O2 in epidermal cells with an order of RbohD ≥ XDH1 > RbohF. These results also suggest that plants might have evolved a compensatory mechanism at least in leaf epidermal cells to ensure adequate ROS production to ward off nonadapted or poorly adapted powdery mildew and perhaps other similar pathogens. Interestingly, loss of RbohF in particular seems to strongly activate the compensatory mechanism when the ROS generation capacity is severely compromised due to genetic lesions in (or possibly pathogen suppression of) other oxidases particularly RbohD. In this regard, it is worth noting that the rbohF single mutant was reported to have enhanced DAB staining and HR induced by an oomycete pathogen (Torres et al., 2002), implying a potential role for RbohF in regulating other ROS-producing enzymes in plants under attack by powdery mildew and oomycete pathogens.
Both Diphenylene Iodonium-Sensitive and -Insensitive Enzymes Contribute to H2O2 Production in Haustorium-Invaded Cells
Diphenylene iodonium (DPI) is often used to inhibit ROS production mediated by flavoenzymes, including XDH (Yesbergenova et al., 2005; Zarepour et al., 2010) and NAD(P)H oxidases (Jabs et al., 1996). The free iron chelator, deferoxamine (DFO), inhibits DAB-detectable H2O2 accumulation in powdery mildew-invaded wheat (Triticum aestivum) epidermal cells (Liu et al., 2007a). This observation implies that accumulation of reactive iron in the fungal penetrate site is required for local powdery mildew-induced H2O2 production/accumulation, although the underlying mechanism is unclear (Liu et al., 2007a). It has been suggested that DFO can affect oxidative reactions independent of its ability as an iron chelator; namely, it can act as a substrate for peroxidases and a scavenger of radicals (Reeder et al., 2008). When treated with either DPI (100 µM) or DFO (2 mM) at maximum concentrations that do not inhibit powdery mildew spore germination based on our study, Col-0 plants still produced H2O2 upon invasion by Gc UMSG1 albeit at significantly lower levels compared with untreated Col-0 plants (P < 0.01) (Figure 7B). However, DPI+DFO treatment almost completely abolished H2O2 production in Col-0 (Figures 7B and 7C), indicating that both DPI- and DFO-sensitive mechanisms are involved in HC-H2O2 generation in the epidermal cells of Arabidopsis. This result seems to differ from an early study in wheat where only DFO- but not DPI-sensitive mechanisms for H2O2 generation are detectable upon invasion by a nonadapted barley powdery mildew isolate (Liu et al., 2007a).
Haustorium Complex-Confined H2O2 Contributes to Basal Postpenetration Resistance
To directly assess the role of H2O2 confined in haustorial complexes in restricting fungal development, we measured the total hyphal length per germinated sporeling of Gc UMSG1 at 60 hpi. We found that compared with wild-type Col-0 plants, xdh1, rbohD, xdh1 rbohD, and xdh1 rbohF mutants supported significantly more hyphal growth, as did Col-0 plants treated with DPI or DFO and DPI+DFO in particular (Figure 7D; Supplemental Figure 16). Hence, there appeared to be an inverse correlation between HC-H2O2 and fungal growth, supporting an important role for HC-H2O2 in basal, haustorium-targeted resistance. Notably, despite not being the least in HC-H2O2, xdh1 rbohD plants supported the greatest amount of hyphal growth (i.e., ∼4× of Col-0) (Figure 7D), suggesting that an immune process in addition to H2O2 generation may also be defective in the xdh1 rbohD double mutant.
To further validate the above results, we inoculated plants of the same set of genotypes with the well-adapted isolate Gc UCSC1 and observed a similar tendency, i.e., those mutant plants with reduced HC-H2O2 in response to Gc UMSG1 were slightly but significantly more susceptible to Gc UCSC1 compared with the wild type (Supplemental Figure 17). This suggests that H2O2 derived from XDH1, RbohD, or RbohF in Col-0, although barely detectable by DAB staining except in the penetration site (Supplemental Figure 15B), still contributes to basal resistance.
Taken together, our data demonstrate that both DPI-sensitive (including XDH1 and NADPH oxidase based) and DPI-insensitive ROS generation mechanisms are engaged in leaf epidermal cells of Arabidopsis for HC-H2O2 accumulation, thereby contributing to basal haustorium-targeted resistance against powdery mildew fungi.
XDH1 Is Membrane Associated and Localized to the Tonoplast
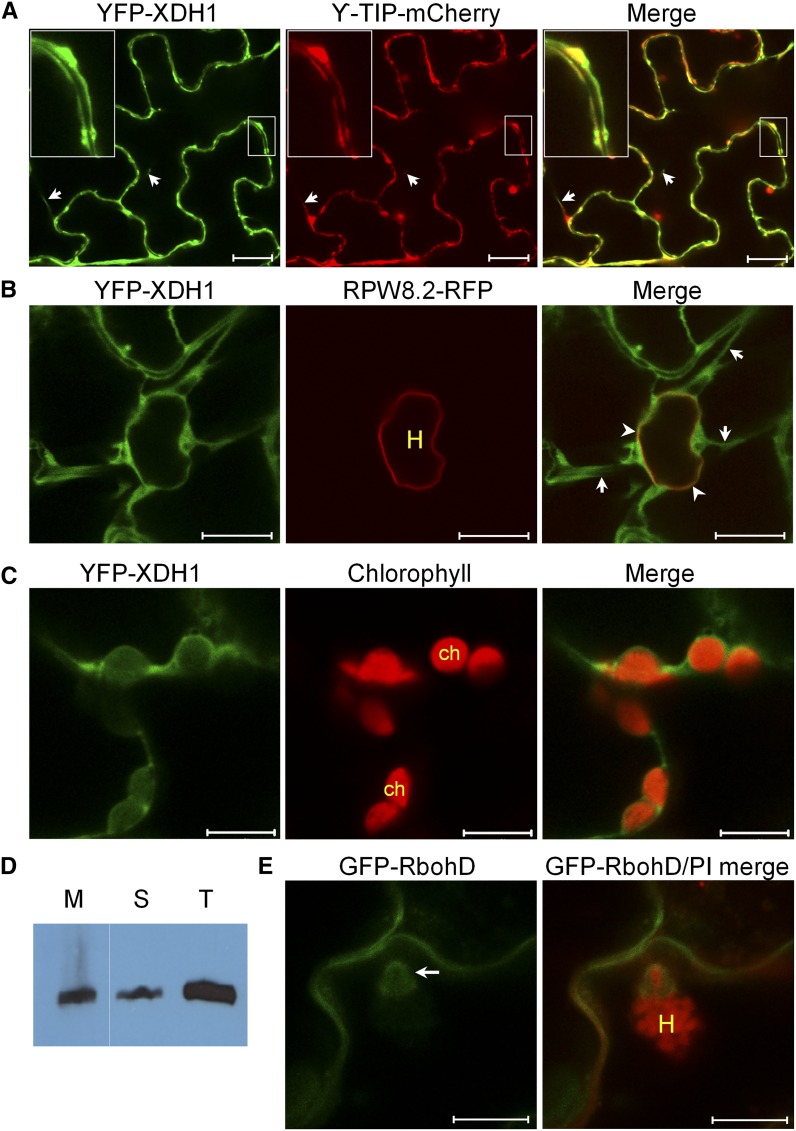
Accumulation of host-derived H2O2 in the haustorial complex raises a question as to whether H2O2 is generated in the extrahaustorial matrix by host oxidases at the EHM or it is transported there. To address this question, we made the YFP-tagged XDH1 fusion construct and found that YFP-XDH1 expressed from its native promoter or the 35S promoter eliminated AFO formation in xdh1-2 (Supplemental Figure 18A), indicating that YFP-XDH1 is functional. We then used xdh1-2 lines transgenic for 35S:YFP-XDH1 to determine the subcellular localization of YFP-XDH1 because YFP-XDH1 expressed from the native promoter was often difficult to detect by confocal microscopy. Since YFP-XDH1 in leaf epidermal cells appeared to be in the tonoplast, i.e., the vacuolar membrane (Supplemental Figure 18A), we transiently coexpressed YFP-XDH1 with the tonoplast marker gamma-TIP-mCherry, an aquaporin in the vacuolar membrane (Nelson et al., 2007) in Nicotiana benthamiana and found that YFP-XDH1 and γ-TIP-mCherry were precisely colocalized (Figure 8A). To confirm that XDH1 is membrane associated, we tagged XDH1 with the hemagglutinin (HA) epitope and stably expressed HA-XDH1 from the 35S promoter in Arabidopsis. A gel blot analysis showed that HA-XDH1 was partitioned between the soluble fraction (∼65%) and the membrane fraction (∼35%) (Figure 8D). We then examined epidermal cells invaded by haustoria from Gc UCSC1 and found that a portion of YFP-XDH1-labeled membrane tightly wrapped around the haustorium (Supplemental Figure 18C) and seemed to have partial colocalization with RPW8.2-RFP-labeled EHM (Figure 8B). Interestingly, dynamic transvacuolar strands were often seen to connect the perihaustorial membrane labeled by YFP-XDH1 with more distal portion of the tonoplast (Figure 8B). To assess if YFP-XDH1 is incorporated into the EHM, we subjected the powdery mildew-infected leaves to plasmolysis and found that YFP-XDH1-labeled perihaustorial membrane could be completely separated from RPW8.2-RFP-labeled EHM (Supplemental Figure 18D), indicating that even though the tonoplast tightly wraps around the EHM, it is unlikely fused into the EHM. We also examined if plasma membrane-localized GFP-RbohD (Hao et al., 2014) is recruited to the EHM and found that although GFP-RbohD appeared to be enriched in the plasma membrane around the penetration site and likely the haustorial neck region, it was absent from the EHM (Figure 8E). These observations suggest that XDH1 (and RbohD)-produced O2⋅– or its dismutation product H2O2 may have to be transported or diffuse across the EHM to contribute to HC-H2O2. Supporting this inference, H2O2-positive granules were found gathered around the haustorium with H2O2 highly enriched around the EHM and in the extrahaustorial matrix (Supplemental Figure 19). Similar H2O2-positive vesicles were also observed around the fungal perpetration site in barley epidermal cells (Huckelhoven et al., 1999; Collins et al., 2003).
Figure 8.
XDH1 Is Membrane Associated and Localized to the Tonoplast.
(A) Precise colocalization of YFP-XDH1 with a tonoplast marker γ-TIP-mCherry (Nelson et al., 2007) in leaf epidermal cells of N. benthamiana (transient expression) or Arabidopsis Col-0 (stable expression). YFP-XDH1 was expressed from the 35S promoter and γ-TIP-mCherry was expressed from the maize (Zea mays) ubiquitin promoter. Arrows indicate transvacuolar strands. Bars = 20 µm.
(B) Dynamic and intimate association of YFP-XDH1-labeled tonoplast with the EHM. A Col-0 line transgenic for both 35S:YFP-XDH1 and pRPW8.2:RPW8.2-RFP was inoculated with Gc UCSC1. Infected leaves were subjected to confocal imaging at 2 dpi. Note the seemingly partial co-localization between YFP-XDH1 and RPW8.2-RFP at the EHM (arrowheads) and extensive transvacuolar strands (arrows) connecting the distal portion of the tonoplast to the part wrapping the haustorium (H). Bars = 10 µm.
(C) Close association of YFP-XDH1-labeled tonoplast with the chloroplasts (ch) visualized by autofluorescence of chlorophyll. Bars = 10 µm.
(D) A gel blot assay showing that HA-XDH1 exists in both soluble (S) and membrane (M) fractions. Total protein (T) was extracted from leaves of Arabidopsis plants expressing HA-XDH1 from the 35S promoter, fractionated by ultracentrifugation, gel blotted, and analyzed using an anti-HA antibody.
(E) Localization of YFP-RbohD expressed from the native promoter (Hao et al., 2014) to the plasma membrane and possibly the haustorial neck or papilla (arrow). Bars = 10 µm.
In mesophyll cells, YFP-XDH1 was found in the tonoplast closely associated with chloroplasts (Figure 8C; Supplemental Figure 18B). This localization of XDH1 may allow production of uric acid in the vicinity of chloroplasts to protect them from oxidative damage via scavenging stress-induced ROS. How XDH1-derived uric acid might get across the chloroplast membrane remains to be resolved.
DISCUSSION
H2O2 as a Chemical Weapon for Constraining Haustoria of Powdery Mildew in Epidermal Cells
ROS including O2⋅– and H2O2 at low levels serve as signaling molecules in many plant physiological processes, including stomatal closure (Zhang et al., 2001; Desikan et al., 2006), root differentiation (Lee et al., 2013), and defense signaling (Lamb and Dixon, 1997; Alvarez et al., 1998). ROS at higher levels are thought to play a role in restricting pathogens in plants (Wojtaszek, 1997; Hückelhoven and Kogel, 2003), similar to their role in pathogen killing by animal phagocytotic cells (Nathan et al., 1979; Rada and Leto, 2008). However, unequivocal genetic evidence in support of this claim is sparse (Kadota et al., 2015). Moreover, there is controversy over the precise role of ROS in the activation of HR and R gene-mediated disease resistance in plants (Torres et al., 2002, 2005; Hückelhoven and Kogel, 2003; Trujillo et al., 2006). Previously, we demonstrated that RPW8-mediated haustorium-targeted resistance against powdery mildew correlates with H2O2 accumulation in the haustorial complexes and formation of callosic haustorial encasements (Xiao et al., 2001; Wang et al., 2009).
Here, we showed that XDH1 and RbohD are two major enzymes responsible for H2O2 accumulation in haustorial complexes (Figures 2, 6, and 7). We further found that reduction of this subcellular H2O2 due to genetic mutations or chemical inhibition of these two enzymes compromised both RPW8-dependent and RPW8-independent basal resistance against well-adapted or poorly adapted powdery mildew isolates (Figures 2, 6, and 7). Thus, our results provide direct genetic evidence for a positive role of host H2O2 in restricting fungal growth. Interestingly, our results also suggest that there exists a robust compensatory mechanism(s) for H2O2 generation in epidermal cells invaded by haustoria and that both DPI-sensitive (XDH1 and RbohD, etc.) and DPI-insensitive enzymes are engaged for H2O2 production under this cellular context (Figure 7). Conceivably, the enrichment and confinement of H2O2 in haustorial complexes must entail onsite production and/or targeted transport of H2O2. Based on our imaging analyses, neither XDH1 nor RbohD is exactly localized to the EHM (Figure 8), suggesting that at least a major proportion of H2O2 may be produced in the cytoplasm, the tonoplast, or other compartments and then mobilized via transport vesicles across the host-pathogen interface into the haustorial complexes (Supplemental Figure 19). Given the close proximity of the XDH1-labeled tonoplast to the EHM, XDH1-derived H2O2 may also reach the haustorium via simple diffusion. Future studies will be directed to identifying other enzymes contributing to HC-H2O2 and mechanisms underlying the likely targeted transport of H2O2 to and across the host-pathogen interface.
Purine Catabolism Protects the Chloroplast from Oxidative Damage
Chloroplasts produce a large amount of ROS from photosynthesis and photorespiration especially under abiotic and biotic stress conditions (Foyer et al., 1994; Asada, 2006; Pintó-Marijuan and Munné-Bosch, 2014). Plants have thus evolved an intrinsic and efficient antioxidative defense system that can scavenge excessive chloroplast-generated ROS to minimize unnecessary oxidative damage (Foyer et al., 1994; Davletova et al., 2005; Galvez-Valdivieso and Mullineaux, 2010). Our results suggest that XDH1-catalyzed uric acid production in mesophyll cells may be part of this complex antioxidative defense system, as evidenced by massive H2O2 accumulation in chloroplasts of aging and/or defense-active xdh1 mutant plants and prevention of age-dependent H2O2 accumulation in leaves of xdh1 mutant plants treated with 200 μM uric acid (Figures 5 and 6). Based on these observations, an important role of endogenous uric acid in reducing oxidative stresses of chloroplasts can be envisaged. However, despite repeated attempts, we did not observe any obvious differences in leaf uric acid levels between Col-0 and xdh1-2, presumably because uric acid accumulation is toxic to plants (Hauck et al., 2014); therefore, it is probably rapidly catabolized. In this context, AFO formation in mesophyll cells of xdh1 mutant plants provides a convenient visual marker to report spatiotemporal purine catabolic flux, therefore indirectly reflecting the level of uric acid production in mesophyll cells of wild-type plants under the same conditions.
The fact that ROS rapidly accumulates in infected plant cells during incompatible interaction with pathogens indicates a deliberate imbalance of the cellular redox system such that a large amount of ROS can be exploited for pathogen killing. Upon perception of invading pathogens, in addition to a rapid oxidative burst in the apoplast of infected cells following the activation of RbohD (Kadota et al., 2014; Li et al., 2014b), activation of mitogen-activated protein kinase cascades also leads to massive chloroplast-originated ROS production, resulting in HR cell death (Liu et al., 2007b). This implies that mitogen-activated protein kinase signaling may perturb chloroplast physiology and biochemistry for more ROS production and/or attenuates the ROS scavenging system, likely including downregulation of purine catabolism to allow ROS accumulation.
Consistent with this notion, several recent studies have revealed the chloroplast to be a major virulence (effector) target of various pathogens (Jelenska et al., 2007; Li et al., 2014a; Petre et al., 2016). It is particularly worth noting that ROS production in chloroplasts plays an early and important role in PTI signaling and is therefore a target for suppression by a subset of effectors of the virulent bacterial pathogen Pseudomonas syringae pv tomato (Pst) strain DC3000 (de Torres Zabala et al., 2015). It is also possible that virulent bacteria may hijack the purine catabolic pathway to enhance production of antioxidant uric acid, thereby suppressing chloroplast-based ROS production. Future testing of xdh1 and wild-type Col-0 plants with bacterial, fungal, or oomycete pathogens that invade mesophyll cells will be revealing.
The Dual and Opposing Roles of XDH1 in ROS Metabolism
Our results demonstrate that XDH1 contributes to H2O2 production in epidermal cells to fight haustoria, whereas it produces uric acid to scavenge chloroplast H2O2 in mesophyll cells to minimize oxidative damage. Thus, XDH1 is a yin-yang protein with dual and opposing roles in ROS metabolism in Arabidopsis and likely other plants, given the high level of sequence conservation in XDHs across kingdoms. A critical question is how the opposing roles of XDHs are realized in plants. In mammals, the posttranslational XDH-XO conversion is thought to provide such a functional switch (Vorbach et al., 2003). However, for organisms whose XDHs lack such a posttranslational modification mechanism, the above question has remained to be answered. Based on the results from this study, we propose that spatially distinct substrate availability in epidermal cells (NADH/O2) and mesophyll cells (xanthine/NAD+) specifies the two opposing roles of XDH1 in ROS metabolism (Figure 9). Our speculation that NADH may be the main substrate of XDH1 for generation of O2⋅– and then H2O2 in epidermal cells is based on the following circumstantial evidence. First, recombinant XDH1 exhibits exceptionally high NADH oxidase activity (the highest among several native and recombinant XDHs from different organisms) in generating O2⋅– (Zarepour et al., 2010), and not only Arabidopsis XDH but also tomato XDH possesses NADH oxidase activity (Yesbergenova et al., 2005). Second, O2⋅– is produced in epidermal cells of barley invaded by powdery mildew (Huckelhoven and Kogel, 1998), and O2⋅– can be converted to H2O2 by superoxide dismutase or spontaneously in plant cells (Lamb and Dixon, 1997). This inference is also compatible with the notion that early salicylic acid-dependent defense signaling results in cellular redox changes from an initial more oxidizing environment to a more reducing environment due to the accumulation of antioxidants (likely including NADH) (Vanacker et al., 2000; Mou et al., 2003). Finally, XDH1 is expressed in epidermal cells, although its expression seemed to be lower than that in mesophyll cells (Supplemental Figures 8B and 8C). Thus, it appears that the intrinsic capacity of XDH1 in producing O2⋅– via its NADH oxidase activity (Yesbergenova et al., 2005; Zarepour et al., 2010) and generating antioxidative uric acid via its basic xanthine dehydrogenase activity (Nakagawa et al., 2007; Brychkova et al., 2008b) underscores the biochemical basis of XDH1’s yin and yang roles in mesophyll and epidermal cells, respectively.
Figure 9.
A Working Model for the Opposing Roles of XDH1 in Epidermal and Mesophyll Cells.
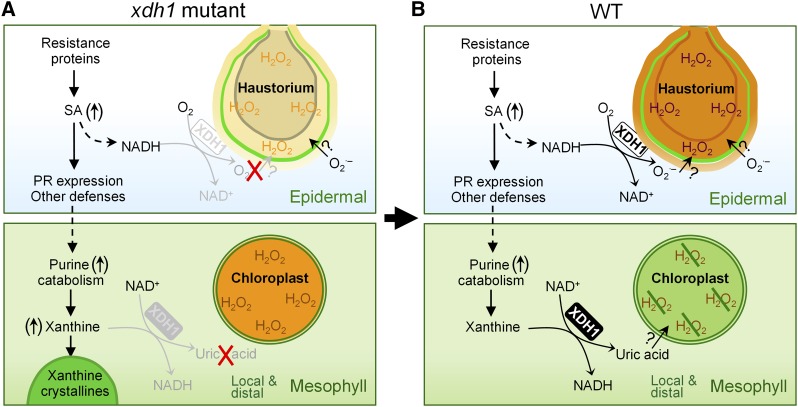
Based on our data from xdh1 mutants (A), we propose the following working model (B) to illustrate the dual functions of XDH1. In powdery mildew-infected epidermal cells, following activation of SA-dependent defense signaling (which leads to NADH generation), XDH1 mainly functions as NADH oxidase to produce O2⋅– that is dismutated to H2O2, thereby contributing to H2O2 enrichment in haustorial complexes as part of a basal defense response. By contrast, XDH1 in mesophyll cells carries out its basic enzymatic activity to convert (hypo)xanthine to uric acid. Uric acid is transported via an unknown mechanism to chloroplasts where it protects chloroplasts from stress-induced oxidative damage by scavenging excessive ROS, thereby dampening oxidative bursts in local as well as systemic tissues. Our data also suggest that purine catabolism may be intrinsically geared up (↑) with plant defense responses such that while adequate H2O2 is produced by XDH1 and other oxidases in the epidermis for fighting against pathogen invasion, more uric acid is produced by XDH1 in affected mesophyll cells that scavenge stress-induced H2O2, thereby confining local and systemic defense response.
Why such a functional specialization for XDH1? It is conceivable that epidermal cells constitute the first and major battleground for fighting against pathogen invasions. Thus, it is essential that epidermal cells possess the capacity to produce adequate H2O2 to kill invading pathogens. By contrast, the major function of mesophyll cells is to conduct photosynthesis, hence having a capacity to remove excessive ROS derived from photosynthesis and/or induced by various abiotic/biotic stresses is of paramount importance for mesophyll cells. Thus, it is likely that purine catabolism—a ubiquitous process for nutrient recycling by default wherein XDH performs its basic enzymatic function in hydroxylation of (hypo)xanthine—may be co-opted and geared up to produce more uric acid as a ROS scavenger in mesophyll cells especially during stress states. Our observations in this study support this hypothesis. First, there were more AFOs (reporting xanthine accumulation, which in turn reflects higher purine catabolic activity) in mature leaves/plants, which are more active in photosynthesis compared with young leaves/plants, indicating a positive correlation between purine catabolism and photosynthesis. Second, xdh1 mutant plants exhibit powdery mildew-inducible and age-dependent AFO formation and chloroplast-H2O2 accumulation (Figures 5A and 6) presumably due to insufficient uric acid production in mesophyll cells. Third, exogenous uric acid could prevent or significantly reduce chloroplast-H2O2 accumulation in leaves of xdh1-2 (Figures 5C and 5D), supporting a role of endogenous uric acid in scavenging chloroplast-generated H2O2.
Apart from a spatial correlation between purine catabolic activities and photosynthesis, we also obtained several lines of evidence to suggest likely crosstalk between defense signaling and purine catabolism as reported by AFO formation in xdh1-2. First, we observed local induction and amplification of xanthine accumulation in leaves by powdery mildew infection and/or enhanced RPW8 expression (Figure 4; Supplemental Figures 10 and 11), suggesting that purine catabolism is likely upregulated in the neighboring mesophyll cells of defense-active cells in both xdh1 mutant and wild-type plants. Consistent with this, both exogenous H2O2 and powdery mildew infection could elevate XDH1 expression in Col-0 (Supplemental Figure 12). Second, local powdery mildew infection was able to induce xanthine accumulation as reported by AFO formation and chloroplast H2O2 accumulation in distal uninfected tissues of xdh1-2 (Figure 4), mirroring the SAR-associated low-frequency micro-oxidative burst in mature wild-type plants (Alvarez et al., 1998). Last and more important, systemic upregulation of purine catabolism as reported by AFO formation in xdh1-2, just like SAR, requires the key salicylic acid signaling component EDS1 (Figure 4). Thus, a logical inference is that concomitant with the activation of local and systemic defense, purine catabolic activity is also upregulated through an unknown mechanism, leading to elevated production of uric acid, which may be utilized to remove excessive ROS generated in the respective affected mesophyll cells to delimit defense responses. Currently, the chemical composition of AFOs from xdh1 mutants is not known, except that they contain xanthine crystallines, nor is it clear whether there is any compositional difference between AFOs formed in different genetic backgrounds or under different stress conditions. More detailed mass spectrometry analysis is needed in the future to resolve these questions. More importantly, given the opposing roles of XDH1 in epidermal cells versus mesophyll cells, future challenges are to understand how Arabidopsis XDH1 (and its functional counterparts in other plants species) might modulate plant defense against pathogens that also invade mesophyll cells and how purine catabolism in general is mechanistically connected with defense regulation.
METHODS
Plant Materials, Growth Conditions, and Transformation
Seeds were sown on autoclaved Metro-Mix 360 soil and were cold treated (4°C for 2 d) before seed germination and seedling growth for ∼2 to 3 weeks under 75% relative humidity in short-day conditions (8 h of light ∼125 µmol·m−2·s−1 from Sylvania fluorescent tubes and 16 h of dark, 22°C). Seedlings were then transplanted individually and maintained in short-day conditions for 4 to 6 additional weeks before treatment, unless indicated otherwise. Plants of Arabidopsis thaliana accession Col-0 (or Col-gl derived from Col-0) or indicated mutant lines were used for Agrobacterium tumefaciens-mediated transformation of DNA constructs (Clough and Bent, 1998). The loss-of-function single and double mutant lines for atrobhD and atrbohF (Kwak et al., 2003) and the eds1-2 null allele in the Col-0 background were crossed with xdh1-2 to make xdh1 rbohD and xdh1 rbohF, xdh1 eds1 double, and xdh1 rbohD rbohF triple mutant lines. The primers used for genotyping xdh1-2 are listed in Supplemental Table 1. Because the double (except xdh1 eds1) and triple mutants show poor growth performance and some die before maturation in soil, we used the following growth scheme for preparation of big close-to-wild-type-size plants. Seeds of mutant lines and Col-0 were sterilized and sown on 0.5× MS-agar plates. Seedlings were grown in 0.5× MS-agar plates for 4 weeks and then transplanted into sterile soil and maintained under short-day conditions with a high relative humidity (∼90%) until treatment.
Isolation and Cloning of drf1-1
The three drf1 mutants were isolated from an EMS-mutagenized M2 population (∼50,000 M2 plants derived from 10,000 M1 plants) derived from R2Y4, a Col-0 transgenic line homozygous for the PRPW8.2:RPW8.2-YFP transgene (Wang et al., 2009) according to an established EMS mutagenesis protocol (Kim et al., 2006). The drf1-1 mutant was crossed with Ler to derive a segregating F2 population for mapping of drf1-1. About 3000 F2 individuals were used to fine-map the drf1-1 mutation in an ∼40-kb region in chromosome 4 by using various PCR-based markers (Supplemental Table 1). All genes located in the drf1-1 region were sequenced to reveal that drf1-1 occurred in XDH1 (At4g34890). Genetic complementation of drf1-1 with the wild-type XDH1 and targeted silencing of XDH1 using artificial microRNA (amiRNA) genes further confirmed that DRF1 is XDH1. One XDH1 knockdown allele in the SALK_148364 line had been reported earlier (Yesbergenova et al., 2005) and was designated xdh1-1 in this study. The xdh1 alleles in the GK-049D04, drf1-1, drf1-2, and drf1-3 mutant lines were named xdh1-2, xdh1-3, xdh1-4, and xdh1-5, respectively. To make the XDH1 constructs, we first used primers XDH1-tpF and XDH1-R to amplify the genomic sequence of XDH1, cloned it into the Gateway-compatible pENTR/D-TOPO vector, and then shuttled it to the binary vector pEG100 under control of the 35S promoter (Earley et al., 2006). The amiRNA fragments targeting XDH1 were amplified and cloned into binary vector pBTEX. All constructs generated by PCR were verified by sequencing. Primers for making the XDH1 constructs and amiRNA constructs are listed in Supplemental Table 1.
Pathogens, Inoculation, and Quantification of Disease Susceptibility
Powdery mildew isolates Golovinomyces cichoracearum UCSC1 (Gc UCSC1) and Gc UMSG1 were maintained on live eds1-2 plants and sow thistle plants, respectively, in separate growth chambers. Inoculation, visual scoring, and quantification of disease susceptibility in number of conidiophores per colony or total number of spores per milligram of leaf tissue or total hyphal length were performed as previously described (Xiao et al., 2005; Wen et al., 2011). Induction of AFOs was examined at 3 dpi and other times under a Zeiss Axio microscope coupled with an HBO 100 microscope illumination system.
In Situ Detection of H2O2 Accumulation and Cell Death
DAB staining was used to assess H2O2 production and accumulation in the haustorium-invaded cells and mesophyll cells, aniline blue staining was used to reveal callose deposition in the fungal penetration sites and around the haustorium, and trypan blue staining was used to visualize host cell death and/or fungal structures as previously described (Wang et al., 2009).
Gene and Protein Expression and Subcellular Localization and Imaging Analyses
Total RNA was extracted from leaf tissues using Trizol reagent (Invitrogen). cDNA was synthesized using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). Quantitative real-time-RT-PCR was performed using Taqman technology as described (Xiao et al., 2005). Conventional RT-PCR was used to assess if XDH1 is expressed in the xdh1-2 knockout mutant. For XDH1 promoter-GUS analysis, a 1.6-kb fragment upstream of the ATG start codon of XDH1 was amplified by primers EcoXDH1-npF and EcoXDH1-npR, and T/A cloned into pCX-GUS-P (Chen et al., 2009). Col-0 transgenic lines expressing the resulting promoter-GUS construct were subjected to GUS activity assays. To determine where XDH1 is localized, the XDH1 genomic DNA was amplified by primers XDH1-tpF and XDH1-nstopR, or XDH1-tpF and XDH1-R, and cloned into the Gateway-compatible pENTR/D-TOPO vector (Life Technologies) and then shuttled to binary vector pEG101 where XDH1 was in-frame fused to add YFP at the C terminus of XDH1, or to binary vector pEG104 (Earley et al., 2006), where XDH1 is in-frame fused to add YFP at the N terminus of XDH1. Both constructs were under control of the 35S promoter. To express YFP-XDH1 from the native promoter of XDH1, the YFP-XDH1 chimeric gene was amplified from the YFP-XDH1 fusion construct cloned in pEG104 with appropriate primers and T/A cloned into the binary vector pCX-DG (Chen et al., 2009) downstream of the 1.6-kb 5′ regulatory sequence of XDH1 that was inserted into the EcoRI site of pCX-DG. To determine if XDH1 is partitioned between soluble and membrane fractions, XDH1 was cloned into pEG201 containing the HA epitope (Earley et al., 2006) and leaf tissues of a Col-0 line transgenic for 35S:HA-XDH1 were used for extraction of total proteins, which were then separated into soluble and membrane fraction by ultracentrifugation (30,000g for 2 h), followed by immunoblot analysis using anti-HA antibody. All constructs generated by PCR were verified by sequencing and introduced into drf1-1, xdh1-2, and Col-0 by stable transformation. The expression and localization of the fusion proteins were examined by confocal microscopy using a Zeiss LSM710 microscope (Wang et al., 2013). Subcellular localization of RbohD in haustorium-invaded cells was determined using the Col-0 line expressing GFP-RbohD from the native promoter (Hao et al., 2014). Confocal image files were processed using the ZEN software (2009 edition) from Carl Zeiss and Adobe Photoshop CS4.
Pharmacological Treatments
For inhibition of xanthine dehydrogenase activity, Arabidopsis Col-0 seedlings were grown on 0.5× MS-agar plates containing 0.25 mM allopurinol for 3 weeks before examination for AFO formation under a Zeiss Axio microscope. In addition, 0.25 mM allopurinol was injected into leaves of 4-week-old Nicotiana tabacum plants; AFO formation was examined 2 weeks after treatment. For uric acid treatment, leaves were detached from 6-week-old Col-0 plants and inserted (with their petioles) into 0.5× MS-agar medium containing 50, 100, or 200 µM uric acid. After incubation for 4 weeks (with one transfer per week to fresh media containing the same concentration of uric acid), the leaves were subjected to DAB staining for visualizing H2O2 production and accumulation. To assess the enzymatic sources of H2O2 produced in the haustorium-invaded epidermal cells, 100 µM DPI and/or 2 mM DFO or buffer (sterile H2O) was infiltrated using a needleless syringe into detached, fully expanded leaves of 8-week-old Col-0 plants via the base of the leaves. The infiltrated leaves were inserted (with their petioles) into 0.5× MS-agar plates and incubated for 4 to 5 h before inoculation with powdery mildew Gc UCSC1 or Gc UMSG1. The leaves were subjected to DAB staining at ∼50 hpi.
Isolation, Purification, and Characterization of AFOs
Mature leaves of 12-week-old leaves of xdh1 mutant or Col-0 wild-type plants were ground into powder in liquid nitrogen. The leaf powder was resuspended in sterile water (0.1 g powder/mL water) and subjected to sucrose gradient (100, 80, 60, and 40%) centrifugation for AFO purification. The corresponding leaf extract from Col-0 was used as control. The AFO-containing sample was used for testing different reagents and conditions for dissolving AFOs. These include 0.5 units/μg protease (P6911; Sigma-Aldrich), 20 units/μg proteinase K (25530015; Invitrogen), 10% SDS (L6026; Sigma-Aldrich), 2% Triton X-100 (Sigma-Aldrich), and 1% Nonidet P-40 (L9080650; US Biological). For measuring xanthine content in leaf powder or purified AFOs, the samples were dissolved in Tris-HCl buffer (100 mM, pH 8.0) and subjected to HPLC-MS analysis using xanthine (CAS 69896; Sigma-Aldrich) as standard.
Detection of Xanthine by HPLC or LC-MS
Xanthine was extracted from leaf tissues according to the method of Gilmore and Bjorkman (1994) with minor modifications. Xanthine samples were analyzed on an Elite Lachrom HPLC system and detected with a DAD L-2450 detector. Xanthine was detected based on its retention time, absorption spectrum, and coelution with standard xanthine added in 1:1 ratio to the samples to be analyzed. The concentration of xanthine was estimated based on the HPLC calibration curve obtained with pure xanthine.
For identification and quantification of xanthine in AFO-related samples, the atmospheric pressure ionization time-of-flight mass spectrometer (AccuTOF; JEOL) equipped with an APCI ion source at a resolving power of 6000 (FWHM) and coupled with an Agilent 1100 HPLC system was used. The AccuTOF MS settings were as follows: needle voltage = 4000 V, desolvating chamber temperature = 400°C, orifice 1 temperature = 100°C, orifice 1 V= 30 V, orifice 2 = 5 V, ring V = 10 V.
For the liquid chromatography elution, the mobile phase composed of 0.1% formic acid solution and methanol was used in the positive mode. The elution was performed as follows: the concentration of solvent (i.e., methanol) was kept at 5% for 20 min. An Agilent TC-C18 column (4.6 × 250 mm, particle size 5 μm) was used in the liquid chromatography experiments, and the flow rate was set to 0.50 mL/min. The UV wavelength range was from 190 to 400 nm. The injection volume was 10 μL. Xanthine or hypoxanthine in the tested samples was identified by its molecular mass matching to the molecular weight of the standard xanthine (CAS 69896; Sigma-Aldrich) or hypoxanthine (CAS 68940; Sigma-Aldrich), respectively. Adonitol (A5502; Sigma-Aldrich) was used as an internal standard to normalize all measurements.
Expression and Enzymatic Assays of XDH1 Variants
For generation of XDH1 variants in a vector for protein expression in Pichia pastoris, PCR-based site-directed mutagenesis was performed using the pPICZA/AtXDH1 construct (Hesberg et al., 2004) as template and specific primers that contained the desired mutation (Supplemental Table 1). The resulting PCR fragments were cloned into KpnI and ApaI restriction sites of pPICZA (Invitrogen) to generate the vector constructs pPICZA/XDH1-G48D, pPICZA/XDH1-R941Q, and pPICZA/XDH1-T1061I. Correctness of the respective XDH1-cDNAs was confirmed by sequence analysis at MWG-Biotech. Transformation of XDH1 constructs into P. pastoris KM71 muts was performed by electroporation according to the EasySelect Pichia expression kit manual (Invitrogen). Confirmation of the respective XDH1-cDNAs in Zeocin-resistant colonies was performed by PCR and DNA gel blotting. Recombinant expression of His6-tagged XDH1 and its variants in P. pastoris and standard purification were performed as described previously (Zarepour et al., 2010). Concentration of the total soluble protein was determined by use of Roti Quant solution (Roth) according to Bradford (1976). XDH activity of recombinant XDH1 and its variants was visualized after electrophoresis of proteins in native 7.5% polyacrylamide gels according to Hesberg et al. (2004). The staining solution for the basic XDH activity contained 1 mM hypoxanthine as substrate, 1 mM 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and 0.1 mM phenazine methosulfate (PMS) in 250 mM Tris-HCl, pH 8.5. To demonstrate the production of superoxides, the staining was done likewise but in the absence of PMS. The staining solution for detection of NADH oxidase activity contained 0.25 mM NADH as substrate and 1 mM MTT in 250 mM Tris-HCl, pH 8.5. For both, the basic XDH activity as well as the production of superoxides, each lane of the native PAGE gel was loaded with 2 µg recombinant protein. Due to generally lower staining intensities, NADH oxidase activity was analyzed after electrophoresis of 10 µg recombinant protein per lane. The relative densities of the resulting activity bands were determined by using ImageJ software version 1.48 from the National Institutes of Health (http://imagej.nih.gov/ij). The specific activity of the xanthine/NAD+-dependent activity of XDH1 was determined by measuring the rate of oxidation of xanthine to uric acid at 295 nm using an absorption coefficient of 9.6 mM−1 cm−1 as described previously (Zarepour et al., 2010).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AF273059 (RPW8.2), AT4G34890 (Arabidopsis XDH1), AT4G34900 (Arabidopsis XDH2), NP_000370 (human XDH), NP_524337 (Drosophila melanogaster XDH), NP_500531 (C. elegans XDH), EDQ74505 (Physcomitrella patens XDH), EEY63796 (Phytophthora infestans XDH), CCD52002 (Botrytis cinerea XHD), CCU77189 (Blumeria graminis f. sp hordei XDH), AAT81740 (Oryza sativa XDH), AT5G47910 (Arabidopsis Respiratory Burst Oxidase Homolog D, RbohD), and AT1G64060 (Arabidopsis RbohF),
Supplemental Data
Supplemental Figure 1. Formation of Autofluorescent Objects in Plants Impaired for XANTHINE DEHYDROGENASE1 (XDH1).
Supplemental Figure 2. Molecular Characterization of the Arabidopsis XDH1 Knockout Line xdh1-2.
Supplemental Figure 3. Spectroscopic Determination of Uric Acid Formation as a Product of XDH1.
Supplemental Figure 4. AFOs Are Dissolvable in Aqueous Buffer with a pH Value Higher Than 8.
Supplemental Figure 5. AFOs Contain Xanthine Crystallines in Mesophyll Cells.
Supplemental Figure 6. Xanthine Is Highly Enriched in Autofluorescent Objects.
Supplemental Figure 7. Hypoxanthine Is Only Slightly Enriched in Autofluorescent Objects.
Supplemental Figure 8. AFOs Are Found in Mesophyll and Guard Cells but Not in Epidermal Cells.
Supplemental Figure 9. H2O2 Accumulation in and Disintegration of Chloroplasts in Mesophyll Cells Underscore xdh1-2-Triggered and Natural Leaf Senescence.
Supplemental Figure 10. Induction of AFOs in Leaves of xdh1-2 Plants by Pathogen Infection.
Supplemental Figure 11. RPW8 Overexpression Accelerates Amplification of AFOs in XDH1-Silenced Plants.
Supplemental Figure 12. Upregulation of XDH1 upon Powdery Infection or H2O2 Treatment.
Supplemental Figure 13. Accelerated Leaf Senescence of xdh1-2 Plants upon Activation of RPW8-Mediated or Basal Defenses.
Supplemental Figure 14. AFO Formation Is Not Always Associated with H2O2 Accumulation in Chloroplasts in xdh1-2 but Removal of H2O2 Reduces AFO Formation.
Supplemental Figure 15. XDH1-Derived H2O2 Contributes to Basal Resistance.
Supplemental Figure 16. XDH1- and NADPH Oxidase-Derived H2O2 Contributes to Basal Resistance against Poorly Adapted Powdery Mildew.
Supplemental Figure 17. Both XDH1 and RbohD Contribute to Basal Resistance against Adapted Powdery Mildew.
Supplemental Figure 18. XDH1 Is Localized to the Tonoplast.
Supplemental Figure 19. H2O2 May Be Transported Across the EHM into the Haustorial Matrix.
Supplemental Table 1. Sequences and Purposes of Primers Used.
Supplementary Material
Acknowledgments
We thank Ryan Cooper for maintaining the plant growth facility, Jane Parker for the eds1-2 mutant in the Col-0 background, June Kwak for seeds of rbohD, rbohF, and rbohD rbohF mutants, Jin-Xing Lin for the GFP-RbohD DNA construct, Xinnan Wang and Ryan Kwak for mutant screening, and particularly Ralf Mendel for guidance in biochemical characterization of XDH1 variants. This project was supported by a National Science Foundation grant (IOS-1146589 and IOS-1457033) to S.X., a collaborative grant from Hunan Agricultural University, China to X.M., and Deutsche Forschungsgemeinschaft grants (Bi 1075/4-1 and Bi 1075/5-1) to F.B.
AUTHOR CONTRIBUTIONS
X.M. performed most of the experiments with support from S.X., W.W., F.B., N.S., R.B., L.Z., H.K., Y.Z., Y.L., J.F., Y.W., L.T., and Q.Z. X.X., Z.D., and S.X. conceived the project. X.M., F.B., and S.X. designed the experiments. X.M., X.X., F.B., and S.X. analyzed the data and wrote the manuscript.
Glossary
- ROS
reactive oxygen species
- XDH
xanthine dehydrogenase
- XOR
xanthine oxidoreductase
- EHM
extrahaustorial membrane
- Ler
Landsberg erecta
- AFO
autofluorescent object
- dpi
days postinoculation
- SAR
systemic acquired resistance
- DAB
3,3′-diaminobenzidine
- HC
haustorial complex
- hpi
hours postinoculation
- DPI
diphenylene iodonium
- DFO
deferoxamine
- amiRNA
artificial microRNA
References
- Aarts N., Metz M., Holub E., Staskawicz B.J., Daniels M.J., Parker J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M.E., Pennell R.I., Meijer P.J., Ishikawa A., Dixon R.A., Lamb C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784. [DOI] [PubMed] [Google Scholar]
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., Parker J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth V.H. (1935). The identity of xanthine oxidase and the Schardinger enzyme. Biochem. J. 29: 1732–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Breitenbach H.H., et al. (2014). Contrasting roles of the apoplastic aspartyl protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT1 and LEGUME LECTIN-LIKE PROTEIN1 in Arabidopsis systemic acquired resistance. Plant Physiol. 165: 791–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brychkova G., Fluhr R., Sagi M. (2008a). Formation of xanthine and the use of purine metabolites as a nitrogen source in Arabidopsis plants. Plant Signal. Behav. 3: 999–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brychkova G., Alikulov Z., Fluhr R., Sagi M. (2008b). A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 54: 496–509. [DOI] [PubMed] [Google Scholar]
- Chalmers R.A., Watts R.W., Bitensky L., Chayen J. (1969). Microscopic studies on crystals in skeletal muscle from two cases of xanthinuria. J. Pathol. 99: 45–56. [DOI] [PubMed] [Google Scholar]
- Chen S., Songkumarn P., Liu J., Wang G.L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977. [DOI] [PubMed] [Google Scholar]
- Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D.J., Coutu J., Shulaev V., Schlauch K., Mittler R. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., Last K., Harrett-Williams R., Tagliavia C., Harter K., Hooley R., Hancock J.T., Neill S.J. (2006). Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 47: 907–916. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M., et al. (2015). Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nature Plants 1: 15074. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371. [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Lelandais M., Kunert K.J. (1994). PHOTOOXIDATIVE STRESS IN PLANTS. Physiol. Plant. 92: 696–717. [Google Scholar]
- Galvez-Valdivieso G., Mullineaux P.M. (2010). The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol. Plant. 138: 430–439. [DOI] [PubMed] [Google Scholar]
- Gilmore A.M., Bjorkman O. (1994). Adenine-nucleotides and the xanthophyll cycle in leaves. 1. effects of co2- and temperature-limited photosynthesis on adenylate energy charge and violaxanthin de-epoxidation. Planta 192: 526–536. [Google Scholar]
- Hao H., Fan L., Chen T., Li R., Li X., He Q., Botella M.A., Lin J. (2014). Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26: 1729–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck O.K., Scharnberg J., Escobar N.M., Wanner G., Giavalisco P., Witte C.P. (2014). Uric acid accumulation in an Arabidopsis urate oxidase mutant impairs seedling establishment by blocking peroxisome maintenance. Plant Cell 26: 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesberg C., Hänsch R., Mendel R.R., Bittner F. (2004). Tandem orientation of duplicated xanthine dehydrogenase genes from Arabidopsis thaliana: differential gene expression and enzyme activities. J. Biol. Chem. 279: 13547–13554. [DOI] [PubMed] [Google Scholar]
- Huckelhoven R., Kogel K.H. (1998). Tissue-specific superoxide generation at interaction sites in resistant and susceptible near-isogenic barley lines attacked by the powdery mildew fungus (Erysiphe graminis f. sp. hordei). Mol. Plant Microbe Interact. 11: 292–300. [Google Scholar]
- Hückelhoven R., Kogel K.H. (2003). Reactive oxygen intermediates in plant-microbe interactions: who is who in powdery mildew resistance? Planta 216: 891–902. [DOI] [PubMed] [Google Scholar]
- Huckelhoven R., Fodor J., Preis C., Kogel K.H. (1999). Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 119: 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives A., Nomura J., Martinon F., Roger T., LeRoy D., Miner J.N., Simon G., Busso N., So A. (2015). Xanthine oxidoreductase regulates macrophage IL1β secretion upon NLRP3 inflammasome activation. Nat. Commun. 6: 6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs T., Dietrich R.A., Dangl J.L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856. [DOI] [PubMed] [Google Scholar]
- Jelenska J., Yao N., Vinatzer B.A., Wright C.M., Brodsky J.L., Greenberg J.T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Rao A.U., Xu J., Ogg S.L., Hathout Y., Fenselau C., Mather I.H. (2009). The PRY/SPRY/B30.2 domain of butyrophilin 1A1 (BTN1A1) binds to xanthine oxidoreductase: implications for the function of BTN1A1 in the mammary gland and other tissues. J. Biol. Chem. 284: 22444–22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Lisinski I., Kadegowda A.K., Shin H., Wooding F.B., Daniels B.R., Schaack J., Mather I.H. (2013). A test of current models for the mechanism of milk-lipid droplet secretion. Traffic 14: 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C. (2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56: 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., Zipfel C. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Khambata R.S., Ghosh S.M., Ahluwalia A. (2015). “Repurposing” of xanthine oxidoreductase as a nitrite reductase: a new paradigm for therapeutic targeting in hypertension. Antioxid. Redox Signal. 23: 340–353. [DOI] [PubMed] [Google Scholar]
- Kim Y., Schumaker K.S., Zhu J.K. (2006). EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323: 101–103. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Nam H.J., Chung H.Y., Kim N.D., Ryu J.H., Lee W.J., Arking R., Yoo M.A. (2001). Role of xanthine dehydrogenase and aging on the innate immune response of Drosophila. J. Am. Aging Assoc. 24: 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinenberg J.R., Goldfinger S.E., Seegmiller J.E. (1965). The effectiveness of the xanthine oxidase inhibitor allopurinol in the treatment of gout. Ann. Intern. Med. 62: 639–647. [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C., Dixon R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 251–275. [DOI] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412. [DOI] [PubMed] [Google Scholar]
- Li C., et al. (2012). Characterization of polygenic resistance to powdery mildew in tomato at cytological, biochemical and gene expression level. Mol. Plant Pathol. 13: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Froehlich J.E., Elowsky C., Msanne J., Ostosh A.C., Zhang C., Awada T., Alfano J.R. (2014a). Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 77: 310–321. [DOI] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M. (2014b). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Liu G., Greenshields D.L., Sammynaiken R., Hirji R.N., Selvaraj G., Wei Y. (2007a). Targeted alterations in iron homeostasis underlie plant defense responses. J. Cell Sci. 120: 596–605. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ren D., Pike S., Pallardy S., Gassmann W., Zhang S. (2007b). Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 51: 941–954. [DOI] [PubMed] [Google Scholar]
- Madigan M.C., McEnaney R.M., Shukla A.J., Hong G., Kelley E.E., Tarpey M.M., Gladwin M., Zuckerbraun B.S., Tzeng E. (2015). Xanthine oxidoreductase function contributes to normal wound healing. Mol. Med. 21: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H.M., Hancock J.T., Salisbury V., Harrison R. (2004). Role of xanthine oxidoreductase as an antimicrobial agent. Infect. Immun. 72: 4933–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944. [DOI] [PubMed] [Google Scholar]
- Nakagawa A., Sakamoto S., Takahashi M., Morikawa H., Sakamoto A. (2007). The RNAi-mediated silencing of xanthine dehydrogenase impairs growth and fertility and accelerates leaf senescence in transgenic Arabidopsis plants. Plant Cell Physiol. 48: 1484–1495. [DOI] [PubMed] [Google Scholar]
- Nathan C., Nogueira N., Juangbhanich C., Ellis J., Cohn Z. (1979). Activation of macrophages in vivo and in vitro. Correlation between hydrogen peroxide release and killing of Trypanosoma cruzi. J. Exp. Med. 149: 1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nishino T., Okamoto K., Eger B.T., Pai E.F., Nishino T. (2008). Mammalian xanthine oxidoreductase - mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J. 275: 3278–3289. [DOI] [PubMed] [Google Scholar]
- Parker J.E., Holub E.B., Frost L.N., Falk A., Gunn N.D., Daniels M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre B., Lorrain C., Saunders D.G., Win J., Sklenar J., Duplessis S., Kamoun S. (2016). Rust fungal effectors mimic host transit peptides to translocate into chloroplasts. Cell. Microbiol. 18: 453–465. [DOI] [PubMed] [Google Scholar]
- Pintó-Marijuan M., Munné-Bosch S. (2016). Photo-oxidative stress markers as a measure of abiotic stress-induced leaf senescence: advantages and limitations. J. Exp. Bot. 65: 3845–3857. [DOI] [PubMed] [Google Scholar]
- Rada B., Leto T.L. (2008). Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib. Microbiol. 15: 164–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder B.J., Hider R.C., Wilson M.T. (2008). Iron chelators can protect against oxidative stress through ferryl heme reduction. Free Radic. Biol. Med. 44: 264–273. [DOI] [PubMed] [Google Scholar]
- Suzuki G., Okamoto K., Kusano T., Matsuda Y., Fuse A., Yokota H. (2015). Evaluation of neuronal protective effects of xanthine oxidoreductase inhibitors on severe whole-brain ischemia in mouse model and analysis of xanthine oxidoreductase activity in the mouse brain. Neurol. Med. Chir. (Tokyo) 55: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37: 1130–1134. [DOI] [PubMed] [Google Scholar]
- Trujillo M., Altschmied L., Schweizer P., Kogel K.H., Hückelhoven R. (2006). Respiratory burst oxidase homologue A of barley contributes to penetration by the powdery mildew fungus Blumeria graminis f. sp. hordei. J. Exp. Bot. 57: 3781–3791. [DOI] [PubMed] [Google Scholar]
- Valko M., Leibfritz D., Moncol J., Cronin M.T., Mazur M., Telser J. (2007). Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 39: 44–84. [DOI] [PubMed] [Google Scholar]
- Vanacker H., Carver T.L., Foyer C.H. (2000). Early H(2)O(2) accumulation in mesophyll cells leads to induction of glutathione during the hyper-sensitive response in the barley-powdery mildew interaction. Plant Physiol. 123: 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorbach C., Scriven A., Capecchi M.R. (2002). The housekeeping gene xanthine oxidoreductase is necessary for milk fat droplet enveloping and secretion: gene sharing in the lactating mammary gland. Genes Dev. 16: 3223–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorbach C., Harrison R., Capecchi M.R. (2003). Xanthine oxidoreductase is central to the evolution and function of the innate immune system. Trends Immunol. 24: 512–517. [DOI] [PubMed] [Google Scholar]
- Wang W., Wen Y., Berkey R., Xiao S. (2009). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21: 2898–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhang Y., Wen Y., Berkey R., Ma X., Pan Z., Bendigeri D., King H., Zhang Q., Xiao S. (2013). A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting. Plant Cell 25: 4242–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Wang W., Feng J., Luo M.C., Tsuda K., Katagiri F., Bauchan G., Xiao S. (2011). Identification and utilization of a sow thistle powdery mildew as a poorly adapted pathogen to dissect post-invasion non-host resistance mechanisms in Arabidopsis. J. Exp. Bot. 62: 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek F., Hoffmann T., Kanawati B., Bichlmeier M., Knappe C., Wenig M., Schmitt-Kopplin P., Parker J.E., Schwab W., Vlot A.C. (2014). Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY1 promotes systemic acquired resistance via azelaic acid and its precursor 9-oxo nonanoic acid. J. Exp. Bot. 65: 5919–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P. (1997). Oxidative burst: an early plant response to pathogen infection. Biochem. J. 322: 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Brown S., Patrick E., Brearley C., Turner J.G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Ellwood S., Calis O., Patrick E., Li T., Coleman M., Turner J.G. (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120. [DOI] [PubMed] [Google Scholar]
- Xiao S., Emerson B., Ratanasut K., Patrick E., O’Neill C., Bancroft I., Turner J.G. (2004). Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol. Biol. Evol. 21: 1661–1672. [DOI] [PubMed] [Google Scholar]
- Xiao S., Calis O., Patrick E., Zhang G., Charoenwattana P., Muskett P., Parker J.E., Turner J.G. (2005). The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 42: 95–110. [DOI] [PubMed] [Google Scholar]
- Xu J., Xie J., Yan C., Zou X., Ren D., Zhang S. (2014). A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77: 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesbergenova Z., Yang G., Oron E., Soffer D., Fluhr R., Sagi M. (2005). The plant Mo-hydroxylases aldehyde oxidase and xanthine dehydrogenase have distinct reactive oxygen species signatures and are induced by drought and abscisic acid. Plant J. 42: 862–876. [DOI] [PubMed] [Google Scholar]
- Zarepour M., Kaspari K., Stagge S., Rethmeier R., Mendel R.R., Bittner F. (2010). Xanthine dehydrogenase AtXDH1 from Arabidopsis thaliana is a potent producer of superoxide anions via its NADH oxidase activity. Plant Mol. Biol. 72: 301–310. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang L., Dong F., Gao J., Galbraith D.W., Song C.P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126: 1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.