Two Arabidopsis SUMO E3 ligase-like proteins interact with MOM1 to form a complex that mediates transcriptional silencing independently of changes in DNA methylation.
Abstract
The mechanism by which MORPHEUS’ MOLECULE1 (MOM1) contributes to transcriptional gene silencing has remained elusive since the gene was first identified and characterized. Here, we report that two Arabidopsis thaliana PIAS (PROTEIN INHIBITOR OF ACTIVATED STAT)-type SUMO E3 ligase-like proteins, PIAL1 and PIAL2, function redundantly to mediate transcriptional silencing at MOM1 target loci. PIAL1 and PIAL2 physically interact with each other and with MOM1 to form a high molecular mass complex. In the absence of either PIAL2 or MOM1, the formation of the high molecular mass complex is disrupted. We identified a previously uncharacterized IND (interacting domain) in PIAL1 and PIAL2 and demonstrated that IND directly interacts with MOM1. The CMM2 (conserved MOM1 motif 2) domain of MOM1 was previously shown to be required for the dimerization of MOM1. We demonstrated that the CMM2 domain is also required for the interaction of MOM1 with PIAL1 and PIAL2. We found that although PIAL2 has SUMO E3 ligase activity, the activity is dispensable for PIAL2’s function in transcriptional silencing. This study suggests that PIAL1 and PIAl2 act as components of the MOM1-containing complex to mediate transcriptional silencing at heterochromatin regions.
INTRODUCTION
In eukaryotic genomes, transposable elements, DNA repeats, and exogenous transgenes are usually subjected to transcriptional silencing (Slotkin and Martienssen, 2007). In Arabidopsis thaliana, transcriptional silencing is accompanied by DNA methylation and repressive histone marks (Law and Jacobsen, 2010; Matzke and Mosher, 2014; Du et al., 2015). Components involved in DNA methylation and repressive histone modification were previously identified and demonstrated to be required for transcriptional silencing (Law and Jacobsen, 2010; Matzke and Mosher, 2014; Du et al., 2015). The DNMT1-like DNA methyltransferase MET1 maintains DNA methylation at symmetric CG sites during DNA replication (Ronemus et al., 1996; Jones et al., 2001). DDM1, a SNF2-type chromatin-remodeling protein, allows DNA methyltransferases to access H1-containing heterochromatin (Zemach et al., 2013). The plant-specific DNA methyltransferase CMT3 specifically contributes to DNA methylation at CHG sites (Lindroth et al., 2001). The histone methyltransferases SUVH4/KYP, SUVH5, and SUVH6 mediate H3K9 dimethylation and act together with CMT3 to facilitate a self-reinforcing loop between CHG methylation and H3K9 methylation (Jackson et al., 2002; Ebbs and Bender, 2006; Du et al., 2015). DNA methylation at CHH sites is established and maintained by DRM2, a DNMT3-like DNA methyltransferase, and CMT2, a plant-specific DNA methyltransferase (Cao and Jacobsen, 2002; Stroud et al., 2014; Zemach et al., 2013).
To mediate de novo DNA methylation, DRM2 is guided by an RNA-directed DNA methylation (RdDM) pathway (Law and Jacobsen, 2010; Matzke and Mosher, 2014). In the RdDM pathway, 24-nucleotide small interfering RNA (siRNA) mediates de novo DNA methylation (Hamilton et al., 2002; Aufsatz et al., 2002; Matzke and Mosher, 2014). DNA-dependent RNA polymerases IV and V (Pol IV and Pol V) are two multisubunit DNA-dependent RNA polymerases that are unique to plants (Ream et al., 2009; Huang et al., 2009; Law and Jacobsen, 2010). NRPD1 and NRPE1 are the largest subunits of Pol IV and Pol V, respectively (Herr et al., 2005; Kanno et al., 2005; Pontier et al., 2005; Ream et al., 2009; Huang et al., 2009). The generation of 24-nucleotide siRNA is dependent on Pol IV, RNA-dependent RNA polymerase 2 (RDR2), and Dicer-like 3 (Xie et al., 2004; Haag and Pikaard, 2011). The 24-nucleotide siRNA is loaded into Argonaute 4 (AGO4) and forms an AGO4-siRNA complex that is thought to guide DRM2 to homologous genomic loci (Pontes et al., 2006; Zhong et al., 2014). Long noncoding RNA produced by Pol V is believed to base-pair with the AGO4-bound siRNA and is required for the occupancy of AGO4-siRNA on chromatin (Wierzbicki et al., 2008; Haag and Pikaard, 2011).
Alteration of DNA methylation is not always required for transcriptional silencing in Arabidopsis (Amedeo et al., 2000; Vaillant et al., 2006). The plant-specific CHD3-like protein MOM1 was identified by a forward genetic screen as a unique component in transcriptional silencing (Amedeo et al., 2000). In the mom1 mutant, transcriptional silencing is released even when DNA methylation is not affected (Amedeo et al., 2000; Vaillant et al., 2006), suggesting that MOM1 mediates transcriptional silencing through a mechanism that is different from DNA methylation. Although MOM1 mediates H3K9 dimethylation at a few target loci shared by RdDM and MOM1 (Numa et al., 2010), the involvement of MOM1 in transcriptional silencing at the whole-genome level is independent of changes not only in DNA methylation but also in histone H3K9 dimethylation (Probst et al., 2003; Vaillant et al., 2006). To understand how MOM1 contributes to transcriptional silencing, a series of truncated MOM1 sequences were transformed into the mom1 mutant for complementation assays (Caikovski et al., 2008). The results indicated that the conserved C-terminal CMM2 domain is necessary and sufficient for transcriptional silencing at some MOM1 target loci and that the putative SNF2 and DNA helicase domains in MOM1 are dispensable for transcriptional silencing. The CMM2 domain exists in an antiparallel coiled-coil structure and forms a homodimer required for transcriptional silencing (Nishimura et al., 2012). These results suggest that MOM1 most likely acts as an adaptor component in a multisubunit complex rather than as an ATP-dependent chromatin-remodeling enzyme. The molecular mechanism by which MOM1 contributes to transcriptional silencing remains to be elucidated.
Small ubiquitin-related modifier (SUMO) is a conserved posttranscriptional modification in eukaryotes. SUMO proteins are covalently attached to substrate proteins through the activities of an enzyme cascade composed of E1 (SUMO activating enzyme [SAE]), E2 (SUMO conjugating enzyme [SCE]), and E3 (SUMO ligase) (Gareau and Lima, 2010). In yeast and animals, SUMO modification is involved in various biological processes including chromatin organization and transcriptional regulation (Shin et al., 2005; Shiio and Eisenman, 2003; Nathan et al., 2006; Cubeñas-Potts and Matunis, 2013). Noncovalent interaction of SUMO proteins with chromatin-associated proteins is also required for SUMO-dependent transcriptional regulation (Stielow et al., 2008; Ouyang et al., 2009; Cubeñas-Potts and Matunis, 2013). In Arabidopsis, mass spectrometric analysis of purified sumoylated proteins demonstrated that many SUMO substrates are involved in chromatin structure regulation, transcription, and RNA metabolism (Budhiraja et al., 2009; Miller et al., 2010). Moreover, several chromatin-associated proteins were identified by a yeast two-hybrid screen as SUMO-interacting proteins (Elrouby et al., 2013). However, it is unknown whether and how SUMO proteins and SUMO-conjugating enzymes contribute to transcriptional silencing in plants.
Arabidopsis has three SUMO E1 activating enzymes (SAE1a, SAE1b, and SAE2), two SUMO E2 conjugating enzymes (SCE1a and SCE1b), and four SUMO E3 ligases (SIZ1, HPY2, PIAL1, and PIAL2) (Kurepa et al., 2003; Ishida et al., 2009; Tomanov et al., 2014). Analyses of mutants defective in these components suggest that they are required not only for growth and development but also for responses to a variety of stresses (Saracco et al., 2007; Miura and Hasegawa, 2010; Tomanov et al., 2014). SUMO E3 ligases contain a SP(SIZ-PAS)-RING zinc finger domain that is responsible for recruiting E2 to substrates (Gareau and Lima, 2010; Miura and Hasegawa, 2010). In this study, we performed a forward genetic screen and identified a SUMO E3 ligase-like protein PIAL2 as a regulator of transcriptional silencing. Our results demonstrate that PIAL2 and its homolog PIAL1 interact with each other and with MOM1 and form a novel complex that mediates transcriptional silencing independently of changes in DNA methylation.
RESULTS
Identification of PIAL2 as a Regulator of Transcriptional Silencing
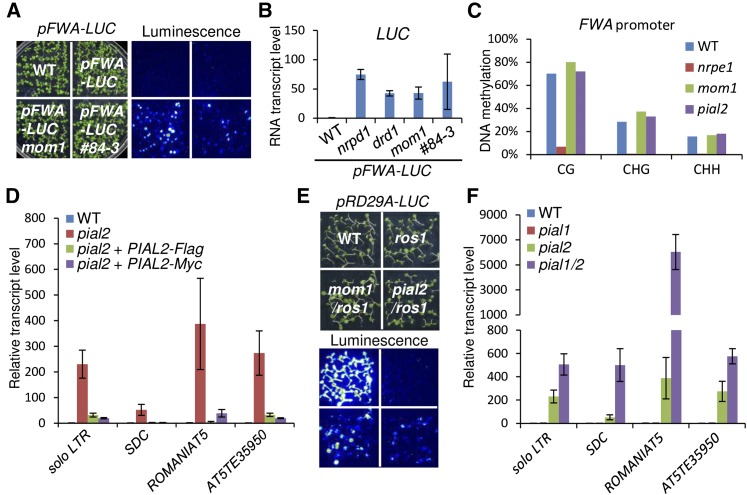
FWA (FLOWERING WAGENINGEN), a flowering repressor gene, is silenced by DNA methylation at the SINE-type repeats in the FWA transcription start region (Soppeet al., 2000; Kinoshita et al., 2007). Previous studies demonstrated that the full-length FWA transgene is an efficient target of DNA methylation and transcriptional silencing (Cao and Jacobsen, 2002; Chan et al., 2004). Here, we generated a luciferase reporter system under the control of the FWA promoter (pFWA-LUC) to screen for novel regulators of transcriptional silencing (Supplemental Figures 1 and 2). The pFWA-LUC reporter gene was initially transformed into the nrpe1-11 mutant and the reporter gene was expressed at high levels in the transgenic lines. A pFWA-LUC transgenic line was selected and then crossed to the wild-type Col-0. In the F2 generation, the release of silencing in pFWA-LUC cosegregated with the nrpe1-1 mutation (Supplemental Figures 1 and 2), suggesting that the silencing of the pFWA-LUC reporter gene requires NPRE1, a component of the RNA-directed DNA methylation pathway. A silenced pFWA-LUC transgenic line in the wild-type background was selected from the F2 generation, and its offspring were subjected to EMS mutagenesis to create a mutant library (Supplemental Figures 1 and 2). We screened for mutants that released the silencing of the pFWA-LUC transgene by luminescence imaging and identified the mutations by map-based cloning. Based on the screening, we identified several known RdDM components, including AGO4, NRPD1, NRPD2, NRPD4, DRD1, RDR2, and RDM4/DMS4, and various other silencing regulators, including MORC6/DMS11, MOM1, MET1, FPGS1, FAS1, and BRU1 (Supplemental Table 1). We also identified a silencing mutant, #84-3, in which the silencing of the pFWA-LUC transgene was released (Figure 1A).
Figure 1.
PIAL1 and PIAL2 Function Redundantly in the Silencing of Transgenes and Endogenous Genomic Loci.
(A) The silencing of the pFWA-LUC transgene in the mom1 and #84-3 mutants as determined by luminescence imaging. Seedlings were grown on MS medium for 10 d and then treated with luciferin for luminescence imaging.
(B) The transcript levels of pFWA-LUC in the wild type and the mutants as determined by RT-qPCR. The mutants including nrpd1, drd1, mom1, and #84-3 were identified from the mutant library in the wild-type background harboring the pFWA-LUC transgene. Error bars are sd of three biological replicates.
(C) The DNA methylation level of the FWA promoter region in the pFWA-LUC transgenic lines in the wild-type, nrpe1, mom1, and #84-3 backgrounds. The DNA methylation level was determined by whole-genome bisulfite sequencing analysis.
(D) Silencing of PIAL2 target loci is affected in the pial2 mutant SALK_043892 and is restored by either the Flag- and Myc-tagged PIAL2 transgenes. The transcript levels of PIAL2 target loci were assessed by RT-qPCR. Error bars are sd of three biological replicates.
(E) The pial2 mutation releases the silencing of the RD29A-LUC transgene in the ros1 mutant background. The pial2 mutation was introduced into the RD29A-LUC reporter system by crossing. The ros1/mom1 mutant identified in the RD29A-LUC system is shown as a control.
(F) The transcript levels of PIAL2 target loci are up-regulated synergistically in the pial1 pial2 double mutant relative to either of the pial1 and pial2 single mutants. The transcript levels were determined by RT-qPCR. Error bars are sd of three biological replicates.
We performed RT-qPCR analysis to determine the transcript level of pFWA-LUC and demonstrated that the silencing of pFWA-LUC was released in #84-3 as well as in nrpd1, drd1, and mom1 (Figure 1B). Whole-genome bisulfite sequencing analysis was performed to determine the DNA methylation status of the FWA promoter in the pFWA-LUC transgenic lines in the wild-type, nrpe1, mom1, and #84-3 backgrounds. We found that the DNA methylation level of the FWA promoter was markedly reduced in nrpe1 but not in mom1 and #84-3 (Figure 1C), suggesting that #84-3 as well as mom1 released the silencing of pFWA-LUC independently of changes in DNA methylation. By using map-based cloning in combination with whole-genome sequencing, we identified a G-to-A mutation at a splice site of AT5G41580 in the #84-3 mutant (Supplemental Figures 3A to 3C). AT5G41580 encodes PROTEIN INHIBITOR OF ACTIVATED STAT LIKE2 (PIAL2), which is distantly related to PIAS-type SUMO E3 ligases in animals (Supplemental Figure 4).
To demonstrate that the mutation in PIAL2 is responsible for the defect in transcriptional silencing, we obtained a homozygous T-DNA pial2 mutant (SALK_043892C, pial2-1) from the ABRC. Using RT-qPCR analysis, we found that the silencing of the endogenous silencing target loci, including solo LTR, SDC, ROMANIAT5, and AT5TE35950, was released in the pial2-1 mutant (Figure 1D). Furthermore, we generated constructs harboring the full-length PIAL2 genomic fragment fused with the C-terminal Flag or Myc tag (PIAL2-Flag and PIAL2-Myc) and transformed the constructs into the pial2-1 mutant for complementation assays. The results indicated that the PIAL2-Flag and PIAL2-Myc transgenes restored the silencing of these loci in the pial2 mutant (Figure 1D), demonstrating that PIAL2 is required for transcriptional silencing.
PIAL2 Is Involved in the Silencing of the RD29A-LUC Transgene in the ros1 Mutant
We previously identified silencing regulators using a pRD29A-LUC reporter system in the DNA demethylation mutant ros1 (He et al., 2009). The pRD29A-LUC reporter gene was silenced when the DNA demethylation component ROS1 was disrupted (Gong et al., 2002). By a genetic screen for suppressors of ros1, we identified many RdDM components that mediate the silencing of the pRD29A-LUC in the ros1 mutant background (He et al., 2009). Here, our further genetic screen identified mom1 as a suppressor of ros1 in the pRD29A-LUC reporter system (Figure 1E). To determine whether PIAL2 contributes to the silencing of the RD29A-LUC reporter gene in the ros1 mutant background, we crossed pial2-1 to ros1 to obtain a ros1/pial2 double mutant harboring the pRD29A-LUC reporter gene. We found that silencing of the pRD29A-LUC reporter gene was released not only in ros1/mom1 but also in ros1/pial2 (Figure 1E). The result demonstrates that PIAL2 as well as MOM1 are involved in the silencing of the pRD29A-LUC transgene in the ros1 mutant background.
PIAL1 and PIAL2 Function Redundantly in Transcriptional Silencing
PIAL2 has a homolog, PIAL1, in Arabidopsis and is highly conserved, especially at the N-terminal regions, in plants (Supplemental Figure 5). It is possible that PIAL1 functions redundantly with PIAL2 to mediate transcriptional silencing. We obtained a homozygous T-DNA pial1 mutant (CS358389) from ABRC and crossed the pial1 mutant with the T-DNA pial2 mutant to create a pial1 pial2 (pial1/2) double mutant. As determined by RT-qPCR analysis, the transcript levels of solo LTR, SDC, ROMANIAT5, and AT5TE35950 were markedly increased in pial2 and were either not increased or weakly increased in pial1. Moreover, the transcript levels of these loci were significantly higher in the pial1/2 double mutant than in either of the single mutants (Figure 1F), suggesting that the function of PIAL1 and PIAL2 in transcriptional silencing is partially redundant. We thereafter analyzed the function of PIAL1 and PIAL2 together in the pial1/2 double mutant.
PIAL1 and PIAl2 Share Common Targets with MOM1
Several well-known genomic loci were commonly used as MOM1 and/or RdDM targets in previous reports (Steimer et al., 2000; He et al., 2009; Yokthongwattana et al., 2010; Numa et al., 2010; Blevins et al., 2014). We examined the transcript levels of these loci in mom1, nrpe1, and pial1/2 by RT-qPCR analysis. Based on the expression patterns of these loci, we divided these loci into three classes (Figure 2A). Class I loci are markedly upregulated in mom1 and pial1/2 but are either not upregulated or are weakly upregulated in nrpe1; Class II loci are upregulated in all three mutants, and Class III loci are markedly upregulated in nrpe1 but are either not upregulated or are weakly upregulated in mom1 and pial1/2. The RT-qPCR results indicated that the expression patterns in mom1 and pial1/2 are almost the same but are different from that in nrpe1 (Figure 2A). Previous studies reported that the transcript level of ROS1 is decreased in mutants defective in the RdDM pathway (Huettel et al., 2006; He et al. 2009). Our RT-qPCR result indicated that although the ROS1 transcript level is decreased in the RdDM mutant nrpe1, it is not decreased in pial1/2 or mom1 (Figure 2A). These results suggest that PIAL1/2 and MOM1 are not components of the RdDM pathway.
Figure 2.
PIAL1 and PIAL2 Contribute to the Transcriptional Silencing of MOM1 Target Loci as Determined by Transcriptome Analysis.
(A) The transcript levels of the indicated loci were determined by RT-qPCR in the wild type, nrpe1, mom1, and pial1 pial2. The transcript level of ACT7 is shown as a control. I, target loci shared by MOM1 and PIAL1/2 but not by NRPE1; II, target loci common to NRPE1, MOM1, and PIAL1/2; III, loci targeted by NRPE1 but not by MOM1 or PIAL1/2. Error bars are sd of three biological replicates.
(B) Venn diagrams of overlaps between upregulated TEs or genes in mom1, pial1/2, and nrpe1 relative to the wild type. I, II, and III represent three classes of loci targeted by MOM1, PIAL1/2, and NRPE1 as explained above.
(C) Heat maps of differentially expressed TEs and genes in mom1, pial1/2, and nrpe1. Red and blue lines indicate up- and downregulated TEs or genes, respectively.
(D) The transcript levels of common target TEs of MOM1 and PIAL1/2 as determined by RNA deep sequencing analysis. FPKM, fragments per kilobase per million fragments mapped.
(E) Distribution of upregulated TEs throughout the five Arabidopsis chromosomes. Red, blue, and green bars represent upregulated TEs in nrpe1, pial1/2, and mom1, respectively.
(F) Box plots showing the sizes of upregulated TEs in nrpe1, mom1, and pial1/2 relative to the wild type.
To compare the targets of NRPE1, MOM1, and PIAL1/2 at the whole-genome level, we performed RNA deep sequencing to determine the transcriptomes of the wild type, nrpe1, mom1, and pial1/2. A total of 72, 153, and 105 transposable elements (TEs) are significantly upregulated in nrpe1, mom1, and pial1/2, respectively (Figure 2B; Supplemental Data Sets 1 to 3). In these mutants, upregulated TEs are much more abundant than downregulated TEs (Supplemental Data Sets 1 to 3), which is consistent with the notion that NRPE1, MOM1, and PIAL1/2 are involved in TE silencing. Among the 105 TEs upregulated in pial1/2, 89.5% (94/105; Classes I and II) are upregulated in mom1 but only 21.9% (23/105) are upregulated in nrpe1 (Figure 2B). A total of 230, 293, and 205 genes are upregulated in nrpe1, mom1, and pial1/2, respectively (Figure 2B; Supplemental Data Sets 4 to 6). Among the 205 genes upregulated in pial1/2, 77.1% (158/205; Classes I and II) are upregulated in mom1, but only 30.7% (63/205) are upregulated in nrpe1 (Figure 2B). Ten identified upregulated loci were randomly selected for validation by RT-PCR analysis, and the results demonstrated that the RNA deep sequencing data are reliable (Supplemental Figure 6). Heat maps indicated that the effects of pial1/2 and mom1 on transcriptomes are highly similar but are clearly different from that of nrpe1 (Figure 2C). These results demonstrate that PIAL1 and PIAL2 contribute to transcriptional silencing at most MOM1 target loci.
To determine the genetic relationship between MOM1 and PIAL2, we performed RNA deep sequencing to analyze the transcript levels of MOM1 and PIAL1/2 common target TEs in the wild type, mom1, pial2, and mom1 pial2. We found that the silencing of MOM1 and PIAL1/2 common target TEs was markedly released in mom1 and is slightly released in pial2 (Figure 2D). The weak effect of pial2 supports the notion that PIAL2 functions redundantly with PIAL1 in transcriptional silencing. The release of silencing in MOM1 and PIAL1/2 common target TEs was not significantly enhanced in the mom1 pial2 double mutant relative to the mom1 single mutant (Figure 2D), suggesting that MOM1 and PIAL2 function in the same pathway.
RdDM components tend to target the euchromatic regions of chromosome arms (Huettel et al., 2006; Cokus et al., 2008; Zemach et al., 2013), whereas MOM1 preferentially targets pericentromeric heterochromatin regions (Habu et al., 2006; Yokthongwattana et al., 2010). We localized upregulated TEs identified in nrpe1, mom1, and pial1/2 on five Arabidopsis chromosomes. The results suggest that PIAL1/2 and MOM1 tend to target TEs in pericentromeric heterochromatin regions, whereas NRPE1 tends to target TEs in euchromatic regions (Figure 2E). Long TEs are preferentially enriched in pericentromeric heterochromatin regions, whereas short TEs are mostly present at the intergenic regions of chromosome arms (Ahmed et al., 2011; Zemach et al., 2013). We found that TEs targeted by PIAL1/2 and MOM1 are markedly longer than those targeted by NRPE1 (Figure 2F). These results suggest that PIAL1/2 and MOM1 function together to mediate transcriptional silencing in heterochromatin regions.
The Involvement of PIAL1 and PIAL2 in Transcriptional Silencing Is Independent of Changes in DNA Methylation
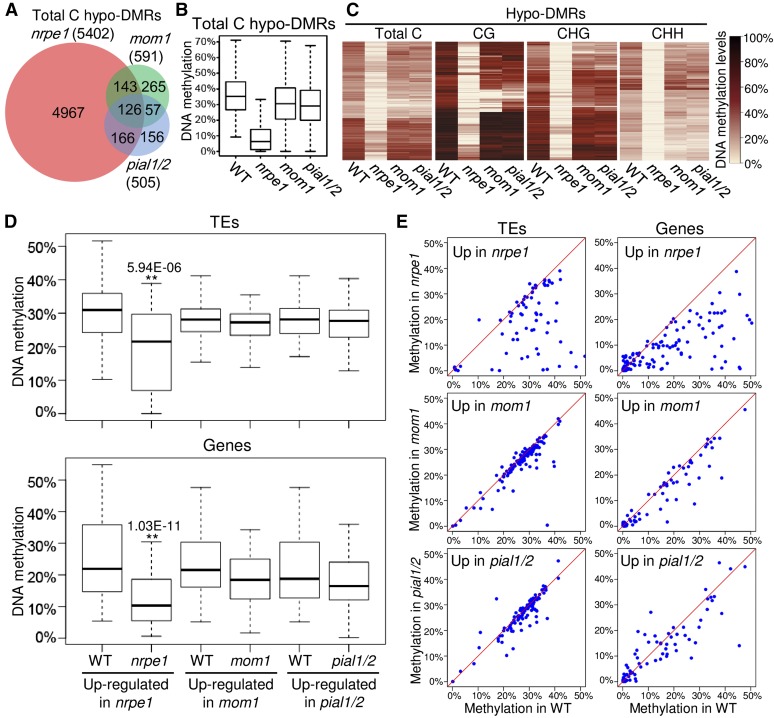
We performed whole-genome bisulfite sequencing analysis in the wild type, nrpe1, mom1, and pial1/2. DNA methylation was separately analyzed at total C, CG, CHG, and CHH sites for differentially methylated regions (DMRs). When DNA methylation of total C was analyzed, 5402 hypomethylated-DMRs (hypo-DMRs) were identified in nrpe1 (Figure 3A). When CG, CHG, and CHH sites were separately analyzed for DMRs, the number of CHH hypo-DMRs (5261) identified in nrpe1 is much higher than the numbers of CG hypo-DMRs (313) and CHG hypo-DMRs (874) (Supplemental Figure 7A), which supports the notion that the RdDM pathway affects DNA methylation especially at CHH sites. MOM1 was believed to mediate transcriptional silencing without changes in DNA methylation (Amedeo et al., 2000; Vaillant et al., 2006). When DNA methylation of total C was analyzed, 591 and 509 hypo-DMRs were identified in mom1 and pial1/2, respectively, which are much lower than number of the hypo-DMRs identified in npre1 (Figure 3A). When the three different cytosine contexts CG, CHG, and CHH were separately analyzed, the numbers of hypo-DMRs are comparable in mom1 and pial1/2 and are much lower than in nrpe1 (Supplemental Figure 7A). In the hypo-DMRs identified in nrpe1, DNA methylation is only slightly reduced in mom1 and pial1/2 (Figure 3B; Supplemental Figure 7B). Heat maps indicated that the DNA methylation patterns of mom1 and pial1/2 are similar and are comparable to that of the wild type but are different from that of the RdDM mutant nrpe1 (Figure 3C).
Figure 3.
The Involvement of PIAL1 and PIAL2 in Transcriptional Silencing Is Independent of Changes in DNA Methylation.
(A) Venn diagram showing overlaps of hypo-DMRs in nrpe1, mom1, and pial1/2 relative to the wild type. Bins were recognized as DMRs when their DNA methylation change is more than 10% in the mutants relative to the wild type.
(B) Box plots showing DNA methylation levels of hypo-DMRs in the wild type, nrpe1, mom1, and pial1/2. The hypo-DMRs in nrpe1 were analyzed.
(C) Heat maps showing DNA methylation of total C, CG, CHG, and CHH hypo-DMRs in the wild type, nrpe1, mom1, and pial1/2. The total C, CG, CHG, and CHH hypo-DMRs identified in nrpe1 are separately shown. Black and light yellow indicate high methylation and low methylation, respectively.
(D) and (E) DNA methylation levels of transcriptionally up-regulated TEs or genes are shown by box plots (D) and scatterplots (E) in nrpe1, mom1, and pial1/2 relative to the wild type. *P < 0.05 or **P < 0.01 was determined by t test. In the box plots, gene promoters were included for DNA methylation analysis only when their DNA methylation levels in the wild type were higher than 5%.
We next determined whether the upregulation of TEs and genes is correlated with the weak reduction of DNA methylation in mom1 and pial1/2. Our whole-genome bisulfite sequencing data suggest that DNA methylation of transcriptionally upregulated TEs in nrpe1 is preferentially reduced (Figures 3D and 3E; Supplemental Figures 8 and 9 and Supplemental Data Set 1), which is consistent with the role of NRPE1 in RdDM. For most of the transcriptionally upregulated TEs in mom1 and pial1/2, however, DNA methylation is not significantly affected (Figures 3D and 3E; Supplemental Figures 8 and 9 and Supplemental Data Sets 2 and 3), demonstrating that release of transcriptional silencing in pial1/2 and mom1 is independent of changes in DNA methylation. Promoter DNA methylation levels of transcriptionally upregulated genes were determined in nrpe1, mom1, and pial1/2 based on the whole-genome bisulfite sequencing data. When upregulated genes are methylated (methylated C/total C > 5%), promoter DNA methylation levels are markedly reduced in nrpe1 but are not reduced or are slightly reduced in mom1 and pial1/2 (Figures 3D and 3E; Supplemental Figures 8 and 9 and Supplemental Data Sets 4 to 6). These results suggest that both PAL1/2 and MOM1 contribute to transcriptional silencing independently of changes in DNA methylation.
PIAL1, PIAL2, and MOM1 Interact with Each Other and Form a High Molecular Mass Complex
To determine how PIAL2 functions in transcriptional silencing, we purified PIAL2-interacting proteins from Flag-PIAL2 transgenic plants using anti-Flag antibody. MOM1 peptides were identified by mass spectrometric analysis in PIAL2-copurified proteins (Figure 4A; Supplemental Data Set 7). Moreover, we purified MOM1-interacting proteins from MOM1-Flag transgenic plants and identified PIAL2 peptides in MOM1-copurified proteins (Figure 4A; Supplemental Data Set 7). These results suggest that PIAL2 physically interacts with MOM1 to form a complex in vivo, which is consistent with the coexpression between PIAL2 and MOM1 (Obayashi et al., 2009). Interestingly, two previously uncharacterized proteins were also identified by mass spectrometric analysis of MOM1-copurified proteins (Figure 4A). One (AT4G11560) was named BDT1 (Bromo-adjacent homology [BAH] domain-containing transcriptional regulator 1); the other (AT1G43770) was named PHD1 (PHD domain-containing protein 1). BDT1 was identified not only in MOM1-copurified proteins but also in PIAL2-copurified proteins (Figure 4A). Both the BAH domain and the PHD domain typically interact with histones or other chromatin-related proteins. It is possible that BDT1 and PHD1 act as components of the MOM1-containing complex on chromatin.
Figure 4.
PIAL1, PIAL2, and MOM1 Form a High Molecular Mass Complex in Vivo.
(A) PIAL2- and MOM1-copurified proteins identified by mass spectrometric analysis. Proteins were extracted from Flag-PIAL2 or MOM1-Flag transgenic plants and were subjected to affinity purification with anti-Flag antibody.
(B) The interaction between PIAL1, PIAL2, and MOM1. By crossing PIAL1-Myc, PIAL2-Myc, and MOM1-Myc, transgenes were separately introduced into MOM1-Flag transgenic plants, and PIAL1-Myc and PIAL2-Myc transgenes were separately introduced into Flag-PIAL2 transgenic plants. F1 generation plants were subjected to protein extraction and co-IP.
(C) Gel filtration analyses of PIAL1, PIAL2, and MOM1. Proteins were extracted from PIAL1-Myc, PIAL2-Myc, or MOM1-Myc transgenic plants in the wild-type background, from PIAL2-Myc transgenic plants in the mom1 mutant background, and from MOM1-Myc transgenic plants in the pial1/2 double mutant background. The proteins were eluted on a Superose 6 (10/300 GL) column. The epitope-tagged proteins in different fractions were detected by antibodies against the Myc tag. Arrows indicate the fractions that correspond to the standard proteins of 67, 220, and 669 kD.
Although PIAL1 and PIAL2 function redundantly in transcriptional silencing, the mass spectrometric analysis identified PIAL2 but not PIAL1 in MOM1-copurified proteins. We performed coimmunoprecipitation (co-IP) analysis to determine whether MOM1 interacts with PIAL1 as well as PIAL2. Transgenic plants harboring the PIAL1 or PIAL2 genomic sequence fused to a Myc tag were generated and crossed to the transgenic plants harboring the MOM1-Flag fusion transgene. Plants from the F1 generation were used to determine the interaction between MOM1 and PIAL1 or PAL2 by co-IP. The results indicated that MOM1-Flag copurified with PIAL1-Myc as well as PIAL2-Myc (Figure 4B), demonstrating that both PIAL1 and PIAL2 interact with MOM1 in vivo. MOM1 was previously demonstrated to contain a conserved MOM1 motif (CMM2), which is sufficient for the function of MOM1 in transcriptional silencing at a subset of MOM1 target loci (Caikovski et al., 2008; Nishimura et al., 2012). The bacterially expressed CMM2 domain folds into an unusual hendecad-based coiled-coil structure and forms a homodimer as determined by x-ray crystallography (Nishimura et al., 2012). However, it is unknown whether the full-length MOM1 forms a homodimer in Arabidopsis. Transgenic lines carrying the MOM1-Myc fusion transgene were generated and crossed to the MOM1-Flag transgenic line. F1 plants harboring both the MOM1-Myc and MOM1-Flag transgenes were used to assess dimerization of MOM1 by co-IP. We found that MOM1-Myc coprecipitated with MOM1-Flag (Figure 4B), demonstrating that MOM1 forms a homodimer in vivo. To determine whether PIAL2 interacts with PIAL2 and PIAL1, we generated Flag-PIAL2 transgenic plants and crossed them to PIAL2-Myc or PIAL1-Myc transgenic plants to obtain F1 plants carrying both Flag-PIAL2 and PIAL2-Myc or PIAL1-Myc. Co-IP assay indicated that both PIAL2-Myc and PIAL1-Myc coprecipitated with Flag-PIAL2 (Figure 4B), demonstrating that PIAL2 interacts not only with PIAL2 but also with PIAL1 in vivo.
The interaction between PIAL1, PIAL2, and MOM1 suggests that these proteins form a complex in vivo. We examined whether PIAL1, PIAL2, and MOM1 exist in a complex by gel filtration. Proteins were extracted from epitope-tagged transgenic plants and were separated on a Superose 6 (10/300 GL) column; the eluted fractions were detected by immunoblotting. We found that PIAL1-Myc and PIAL2-Myc were predominantly eluted in fractions of ∼220 kD, whereas MOM1 was absent in these fractions (Figure 4C). The ∼220-kD faction is much higher than the PIAL1 or PIAL2 monomer and corresponds to the PIAL1 and/or PIAL2 dimer. Given the interaction of PIAl2 with PIAL1 and PIAL2 as determined by co-IP (Figure 4B), PIAL1 and/or PIAl2 may form a homodimer or a heterodimer in vivo. Moreover, the elution of PIAL1 and PIAL2 was extended to fractions of >669 kD, whereas MOM1 was present in the fractions (Figure 4C), suggesting that PIAL1/2 and MOM1 form a high molecular mass complex in the fractions. To determine whether MOM1 is required for PIAL2 to form a complex, we expressed the PIAL2-Myc transgene in the mom1 mutant. The gel filtration result indicated that the elution of PIAL2 disappeared in the high molecular mass fractions in the mom1 mutant (Figure 4C), suggesting that MOM1 is required for the formation of the high molecular mass PIAL2-containing complex. Furthermore, we expressed the MOM1-Myc transgene in the pial1/2 double mutant to determine whether the pial1/2 mutation affects the MOM1 elution pattern in the gel filtration assay. Our result indicated that although MOM1 forms a complex in the pial1/2 mutant, the complex is clearly smaller than that in the wild type (Figure 4C), suggesting that PIAL1 and PIAL2 are required for the formation of the high molecular mass MOM1-containing complex. Our co-IP result demonstrated that MOM1 forms a homodimer (Figure 4B). The remaining MOM1-containing complex in the pial1/2 mutant may at least contain a MOM1 homodimer. Together, these results suggest that PIAL1, PIAL2, and MOM1 interact with each other and form a high molecular mass complex in vivo.
The IND Domains of PIAL1 and PIAL2 Interact with Each Other and with the CMM2 Domain of MOM1
The interaction of MOM1 with PIAL1 or PIAl2 was confirmed by yeast two-hybrid analysis (Figures 5A and 5B). To determine the interaction domain of MOM1, we performed yeast two-hybrid analysis with truncated versions of MOM1. MOM1 contains a domain that is distantly related to a part of the catalytic SNF2 domain in chromatin remodeling proteins (Amedeo et al., 2000). Additionally, MOM1 contains three conserved MOM1 motifs (CMM1, CMM2, and CMM3), which are specifically present in homologs of MOM1 in plants (Caikovski et al., 2008). A series of truncated versions of MOM1 were used in the yeast two-hybrid analysis. The results indicated that both PIAL1 and PIAL2 interact with MOM1-P2, MOM1-P4, and MOM1-P5 but not with MOM1-P1 or MOM1-P3 (Figures 5A and 5B), suggesting that the conserved CMM2 is necessary and sufficient for the interaction of MOM1 with PIAL1 and PIAL2.
Figure 5.
The IND Domain of PIAL1 and PIAL2 Interacts with the CMM2 Domain of MOM1.
(A) Truncated versions of MOM1 used in yeast two-hybrid analysis. P1, 1 to 832 amino acids; P2, 798 to 2001 amino acids; P3, 798 to 1662 amino acids; P4, 1660 to 2001 amino acids; P5, 1660 to 1860 amino acids.
(B) The interaction of a series of truncated forms of MOM1 with PIAL1 or PIAL2 as determined by yeast two-hybrid assays. The full-length MOM1 and truncated forms of MOM1 were fused with GAL4-BD; full-length PIAL1 and PIAL2 were fused with GAL4-AD. Yeast strains carrying the indicated combinations of fusion proteins were grown for yeast two-hybrid analysis. “Vec” represents the empty GAL4-AD or GAL4-BD vector.
(C) Truncated versions of PIAL2 used in yeast two-hybrid assays. P1, 1 to 301 amino acids; P2, 276 to 386 amino acids; P3, 358 to 760 amino acids; P4, 1 to 142 amino acids; P5, 143 to 301 amino acids.
(D) The interaction of the CMM2 domain with truncated forms of PIAL2 as determined by yeast two-hybrid assays. The full-length PIAL2 and truncated forms of PIAL2 were fused with GAL4-AD, and the CMM2 domain of MOM1 was fused with GAL4-BD.
(E) The IND domains of PIAL1 and PIAL2 interact with each other and with the CMM2 domain of MOM1 as determined by GST pull-down assays. PIAL1-IND, 143 to 301 amino acids; PIAL2-IND, 113 to 271 amino acids; MOM1-CMM2, 1700 to 1824 amino acids. GST- and HIS-tagged proteins were coexpressed in E. coli and purified with anti-GST antibody.
We further performed yeast two-hybrid analysis to identify the domain of PIAL2 that is required for interaction with the CMM2 domain of MOM1. A series of truncated versions of PIAL2 were analyzed in the yeast two-hybrid analysis, and the results indicated that the CMM2 domain of MOM1 interacts with PIAL2-P1 and PIAL2-P5 but not with PIAL2-P2, PIAL2-P3, or PIAL2-P4 (Figures 5C and 5D). The results suggest that a previously uncharacterized domain in PIAL2-P5 (143 to 301 amino acids) is necessary and sufficient for the interaction with the CMM2 domain of MOM1. This previously uncharacterized domain of PIAL2 was thereafter named IND (interacting domain).
To determine whether the CMM2 domain of MOM1 directly interacts with the IND domain of PIAL2, we examined the interaction by GST pull-down analysis. The CMM2 and IND domains were fused to the GST and HIS tags, respectively, and were coexpressed in Escherichia coli. GST pull-down analysis indicated that HIS-IND coprecipitated with GST-CMM2 (Figure 5E), demonstrating that the CMM2 domain of MOM1 directly interacts with the IND domain of PIAL2. Moreover, as determined by GST pull-down analysis, HIS-IND also coprecipitated with GST-IND (Figure 5E), suggesting that the IND domain is required for the dimerization of PIAL2. Because of the sequence similarity between PIAL1 and PIAL2, we performed GST pull-down to determine whether the PIAL1 fragment (PIAL1-IND, 113 to 271 amino acids) corresponding to the IND domain of PIAL2 is also required for dimerization and MOM1 interaction. The results indicated that PIAL1-IND interacts with PIAL1-IND, PIAL2-IND, and MOM1-CMM2 (Figure 5E), suggesting that the IND domains of PIAL1 and PIAL2 interact with each other and with the CMM2 domain of MOM1.
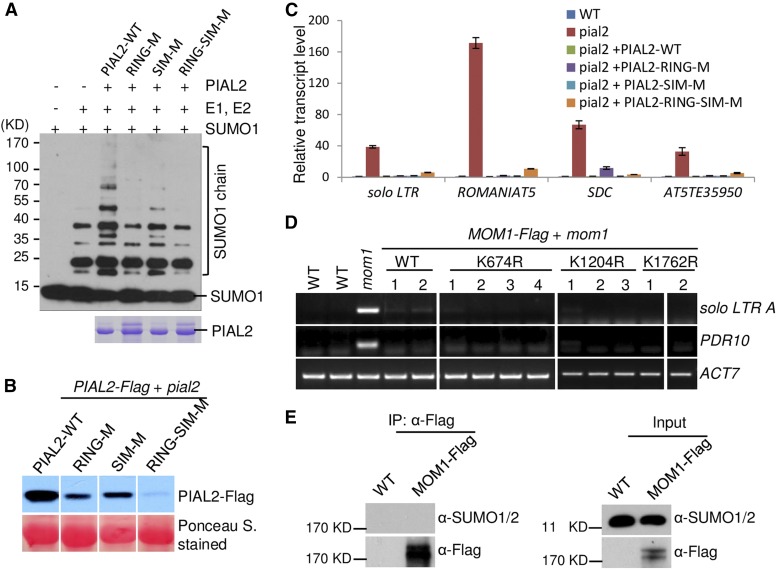
SUMO Ligase Activity Is Not Required for the Function of PIAL2 in Transcriptional Silencing
PIAL2 contains a RING zinc finger motif (RING) and a SUMO interaction motif (SIM), which are necessary for the SUMO ligase activity of PIAL2; mutations in RING and SIM impair the SUMO ligase activity of PIAL2 (Tomanov et al., 2014). We introduced mutations into the RING and SIM domains (RING-M, C329A/H331A; SIM-M, VFDL425AAAA) and performed a sumoylation assay to determine the effect of the RING and SIM mutations on the SUMO ligase activity of PIAL2 (Figure 6A). The result indicated that the SUMO ligase activity of PIAL2 was abolished by the mutation in the RING domain (C329A/H331A) and was partially affected by the mutation in the SIM domain (VFDL425AAAA) (Figure 6A). To determine whether the SUMO ligase activity is required for transcriptional silencing, we introduced the RING and SIM mutations into the full-length PIAL2 construct and performed complementation assays in the pial2 mutant. The expression levels of the wild-type and mutated PIAL2 transgenes were determined by immunoblot analysis in randomly selected transgenic lines. We found that the expression levels of the mutated transgenes PIAL2-RING-M and PIAL2-RING-SIM-M were markedly lower than that of the wild-type PIAL2 transgene, whereas the expression level of the mutated transgene PIAL2-SIM-M was only slightly lower than that of the wild-type PIAL2 transgene (Figure 6B; Supplemental Figure 10). RT-qPCR indicated that the wild-type PIAL2 transgene restored the silencing of the PIAL2 target loci (solo LTR, ROMANIAT5, SDC, and AT5TE35950) in the pial2 mutant (Figure 6C). The mutated transgene PIAL2-RING-M restored the silencing of solo LTR, ROMANIAT5, and AT5TE35950 to the same level of silencing as observed in the wild-type PIAL2 transgene, whereas it restored the silencing of SDC to a lesser degree (Figure 6C). The mutated transgene PIAL2-SIM-M restored the silencing of all of the PIAL2 target loci to an equal degree as the wild-type PIAL2 transgene (Figure 6C). The mutated transgene PIAL2-RING-SIM-M also restored the silencing of all the PIAL2 target loci even though the efficiency was slightly lower than that of the wild-type PIAL2 transgene (Figure 6C). The reduced expression levels of the mutated PIAL2 transgenes (PIAL2-RING-M and PIAL2-RING-SIM-M) may explain the reduced efficiency in the complementation assays. These results indicate that the mutations in RING and SIM do not markedly affect the silencing of PIAL2 target loci, suggesting that the SUMO ligase activity is not required for the function of PIAL2 in transcriptional silencing.
Figure 6.
The SUMO E3 Ligase Activity of PIAL2 Is Not Required for Transcriptional Silencing.
(A) The mutations in the RING and SIM domains of PIAL2 affect the SUMO E3 ligase activity. The SUMO activating enzyme E1, the SUMO conjugating enzyme E2, and SUMO1 were used for the SUMO conjugation assay in the presence of either the full-length wild-type PIAL2 (PIAL2-WT) or the full-length mutated PIAL2 (RING-M, SIM-M, and RING-SIM-M). The SUMO chain signals were visualized by immunoblotting. The wild-type and mutated PIAL2 proteins in the reaction are shown by Coomassie blue staining.
(B) The expression levels of the wild-type (PIAL2-WT) and mutated PIAL2-Flag transgenes (RING-M, SIM-M, and RING-SIM-M) harboring the mutations in the RING and SIM domains in the pial2 mutant background. The transgenic lines were selected from the lines shown in Supplemental Figure 10 and used for RT-qPCR analysis.
(C) Effect of the mutations in the RING and SIM domains on transcriptional silencing. The wild-type and mutated PIAL2-Flag transgenes were separately expressed in the pial2 mutant for complementation assays. The transcript levels of the indicated PIAL2 target loci were determined by RT-qPCR. ACT7 was used as an internal control. Two biological replicates were performed and similar results were obtained. The results of a set of representative experiments are shown. Error bars indicate sd of three technical replicates.
(D) Mutation of putative sumoylated sites in MOM1 does not affect MOM1-mediated transcriptional silencing. The wild-type MOM1-Flag or the mutated MOM1-Flag carrying the K674R, K1204R, or K1762R mutation was expressed in the mom1 mutant for complementation assays. The transcript levels of the MOM1 target loci solo LTR and PDR10 were determined by RT-PCR. ACT7 was used as a control. For the RT-PCR experiments, at least two biological replicates were performed.
(E) MOM1 is not sumoylated in vivo. The MOM1-Flag fusion protein was precipitated by anti-Flag antibody from the proteins of MOM1-Flag transgenic plants. Anti-SUMO1/2 antibody was used to detect possible sumoylated MOM1 signals. Free SUMO1 and SUMO2 signals were detected by anti-SUMO1/2 antibody in input proteins.
The interaction between PIAL2 and MOM1 suggests that MOM1 is probably a substrate of sumoylation. Prediction of sumoylated lysine sites using the SUMOylation prediction website (http://sumosp.biocuckoo.org/) indicated that the lysine sites in MOM1 most likely to be sumoylated are K674, K1204, and K1762. To determine whether the sumoylation of MOM1 is required for transcriptional silencing, we mutated the three lysine sites to arginine to block the possible sumoylation of these sites, and we then transformed each of the mutated MOM1 sequences into the mom1 mutant for complementation testing. RT-PCR analysis indicated that the silencing of the MOM1 target loci solo LTR and PDR10 was restored by the mutated MOM1 sequences as well as by the wild-type MOM1 sequence (Figure 6D), suggesting that even if MOM1 is sumoylated at these lysine sites, sumoylation is not required for the function of MOM1 in transcriptional silencing. To determine whether MOM1 is sumoylated, we precipitated the MOM1-Flag protein from the MOM1-Flag transgenic plants with anti-Flag antibody and determined whether a sumoylated MOM1 signal was present. No sumoylated MOM1 signal was detected by immunoblot analysis (Figure 6E). These results suggest that MOM1 is not a substrate of sumoylation even though MOM1 interacts with the SUMO E3 ligase-like proteins PIAL1 and PIAL2.
DISCUSSION
PIAS proteins were initially identified as protein inhibitors of activated STAT in animals (Chung et al., 1997; Liu et al., 1998). Later, these proteins were found to form a SUMO E3 ligase family defined by a RING zinc finger motif (Johnson and Gupta, 2001). PIAS proteins function in transcriptional regulation through various mechanisms in animals (Cubeñas-Potts and Matunis, 2013). In plants, it is unknown whether and how PIAS homologs are involved in transcriptional regulation. In this study, we found that the Arabidopsis SUMO E3 ligase-like proteins PIAL1 and PIAL2 interact with the plant-specific protein MOM1 and thus mediate transcriptional silencing, suggesting that the molecular mechanism underlying the function of PIAL1 and PIAL2 in plants is different from that of PIAS proteins in animals. This study revealed a novel mechanism by which SUMO E3 ligase-like proteins contribute to transcriptional silencing in plants.
SUMO E3 ligases interact with substrates and SUMO proteins, thereby facilitating sumoylation of substrates in animals (Gareau and Lima, 2010). PIAL1 and PIAL2 were shown to have SUMO ligase activity, which is required for SUMO chain formation (Tomanov et al., 2014). Although the RING and SIM domains are necessary for the SUMO ligase activity of PIAL2, they are dispensable for transcriptional silencing (Figures 6A to 6C). Thus, the SUMO ligase activity of PIAL2 is not required for the function of PIAL2 in transcriptional silencing. This result is consistent with the finding that MOM1 is not sumoylated in vivo (Figure 6E). Together, these results suggest that PIAL2 serves as an interaction partner of MOM1 rather than as a SUMO ligase for MOM1.
MOM1 contains a domain that is distantly related to the catalytic SNF2 domain in chromatin-remodeling proteins (Amedeo et al., 2000). The full SNF2 domain is composed of seven conserved DNA helicase motifs and is required for the function of chromatin-remodeling proteins (Thomä et al., 2005). However, the SNF2-related domain in MOM1 is dispensable for the function of MOM1 in transcriptional silencing (Caikovski et al., 2008). Instead, CMM2, a conserved MOM1 motif, is not only necessary but is also sufficient for transcriptional silencing at a subset of MOM1 target loci (Caikovski et al., 2008; Nishimura et al., 2012). The CMM2 domain forms a homodimer as determined by x-ray crystallography (Nishimura et al., 2012). Our results demonstrate that the CMM2 domain is required not only for forming a homodimer but also for interacting with PIAL1 and PIAL2 (Figures 5A to 5E). The dual functions of the CMM2 domain are consistent with the finding that this domain plays a critical role in transcriptional silencing (Caikovski et al., 2008; Nishimura et al., 2012). Our results suggest that PIAL1 and PIAL2 interact with each other and with MOM1 to form a novel PIAL1/2- and MOM1-containing complex in vivo. Considering that PIAL1 and PIAL2 are necessary for transcriptional silencing at most MOM1 target loci, we predict that the integrity of the complex is required for transcriptional silencing.
The involvement of MOM1 in transcriptional silencing is independent of changes in DNA methylation in heterochromatin regions and remains a poorly understood silencing mechanism in plants (Habu et al., 2006; Vaillant et al., 2006; Yokthongwattana et al., 2010). Previous reports showed that SUMO proteins and SUMO-conjugating enzymes mediate transcriptional repression at heterochromatin regions in yeast and animals (Shiio and Eisenman, 2003; Shin et al., 2005; Nathan et al., 2006), but it is unknown whether and how these proteins function at heterochromatin regions in plants. Our results demonstrate that the SUMO ligase-like proteins PIAL1 and PIAL2 interact with each other and with MOM1 to form a high molecular mass complex (Figures 4A to 4C). Although we demonstrate that the SUMO ligase activity is not required for the function of PIAL2 in transcriptional silencing (Figures 6A and 6C), we cannot exclude the possibility that the function of the PIAL1/2-MOM1 complex in transcriptional silencing is related to sumoylation. A yeast two-hybrid screen previously demonstrated that MOM1 physically interacts with SUMO1 (Elrouby et al., 2013). Several silencing regulators, including the RdDM components SUVR2, KTF1, and IDN2, have been demonstrated to be covalently sumoylated in vivo (Budhiraja et al., 2009; Miller et al., 2010, 2013). Sumoylation is probably necessary for these silencing regulators to function in transcriptional silencing. MOM1 was previously shown to be functionally linked to the RdDM pathway (Numa et al., 2010; Yokthongwattana et al., 2010). It is possible that the PIAL1/2-MOM1 complex cooperates with these sumoylated silencing regulators and thereby facilitates transcriptional silencing on chromatin.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seedlings were grown on MS (Murashige and Skoog) medium plates under standard long-day conditions (16 h/light, 8 h/dark, 22°C). The T-DNA insertion mutants pial2 (SALK_043892), pial1 (CS358389), and mom1-3 (SALK_141293) were in the Col-0 background and obtained from the ABRC. The ros1 mutant in the C24 background harboring both pRD29A-LUC and p35S-NPTII transgenes was previously reported (Gong et al., 2002). The ros1/pial2 mutant was generated by crossing the ros1 and pial2 single mutants. For luminescence imaging, seedlings were grown on MS medium for 10 d and then kept at 4°C for 2 d to activate the stress-responsive pRD29A-LUC transgene. Following the cold treatment, the seedlings were sprayed with luciferin and subjected to luminescence imaging.
The full length of PIAL1, PIAL2, or MOM1 driven by the corresponding native promoter was introduced into the modified pCAMBIA1305 or pRI909 vector to express the C-terminal 5xMyc- or 3xFlag-tagged proteins. The constructs were introduced into the Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis by the flower-dipping method (Clough and Bent, 1998). Site-directed mutagenesis was performed to introduce point mutations into the epitope-tagged PIAL2 and MOM1 constructs, and the function of the mutated PIAL2 and MOM1 proteins was then determined by complementation testing. In the N-terminal Flag-tagged PIAL2 construct, the PIAL2 native promoter was placed in front of the Flag-tag sequence in the pRI909 vector. All constructs were sequenced for verification, and the primers used for the construction are listed in Supplemental Data Set 8.
RT-PCR, RT-qPCR, and RNA Deep Sequencing
Total RNA was extracted with Trizol reagent (Invitrogen) from 10-d-old Arabidopsis seedlings or from rosette leaves. The cDNA was obtained using a reverse transcription kit (TaKaRa; RR012A). RT-qPCR was performed with SYBR Green Master Mix (TaKaRa) according to the manufacturer’s instruction. The actin gene ACT7 was used as an internal control. The primers used for RT-PCR are listed in Supplemental Data Set 8. Total RNA was extracted from 2-week-old Arabidopsis seedlings and then used to produce RNA libraries for deep sequencing (HiSeq 2000; Illumina). To analyze the data, the Arabidopsis genome sequences and annotated gene models were downloaded from TAIR10 (http://www.arabidopsis.org/). After the adaptor sequences were removed, 49-bp reads were mapped to the TAIR10 Arabidopsis genome using TopHat (v2.0.12, http://ccb.jhu.edu/software/tophat/), allowing up to two mismatches. Cufflinks (v2.2.1, http://cole-trapnell-lab.github.io/cufflinks/) was performed to assemble transcripts and calculate transcript abundances. Gene expression differences were evaluated using a combination of Fisher’s exact test (P < 0.01) and the fold change of the normalized reads (log2(fold change) > 1). The gplots package in R was used to draw the heat maps of the differentially expressed genes and TEs. The quantsmooth package in Bioconductor was modified to draw the distribution of upregulated TEs throughout the five Arabidopsis chromosomes. The raw RNA deep sequencing data have been deposited in the Gene Expression Omnibus database (accession number GSE80303).
Whole-Genome DNA Methylation Analysis
For whole-genome DNA methylation analysis, genomic DNA was extracted from the 2-week-old Arabidopsis seedlings with hexadecyltrimethyl ammonium bromide and subjected to bisulfite treatment that converted unmethylated cytosine to uracil. The converted genomic DNA was amplified and then used for whole-genome bisulfite sequencing with HiSeq 2000 (Illumina). Bisulfite sequencing reads were mapped to the TAIR10 reference genome using Bismark, allowing two mismatches (Krueger and Andrews, 2011). Reads that were mapped to more than one position were removed to retain only reads that were uniquely mapped. The methylation level of each cytosine site was represented by the percentage of the number of reads reporting a C relative to the total number of reads reporting a C or T. Only sites with at least 5-fold coverage were included in the results. Gene and TE annotations were downloaded from TAIR. The methylation levels of genes and TEs were estimated by pooling the read counts that show at least 5-fold coverage. The method for identifying DMRs was previously described (Stroud et al., 2013). Bins of 200 bp were analyzed, and the DNA methylation levels of total cytosine, CG, CHG, and CHH sites were separately evaluated. Bins were considered DMRs when the absolute DNA methylation change was more than 10, 40, 20, and 10% for total C, CG, CHG, and CHH, respectively. We compared the DNA methylation levels between each mutant and three different wild-type controls to obtain three batches of DMRs. Only those DMRs identified in the mutant relative to all three controls were selected for further analysis. Whole-genome DNA methylation of Col-0 was determined by bisulfite sequencing analysis two times, and the data were used for two wild-type controls; another wild-type control was from a previous report (Stroud et al., 2013). To evaluate the correlation between expression and DNA methylation, we determined the DNA methylation levels of different classes of differentially expressed loci. The raw bisulfite sequencing data have been deposited in the Gene Expression Omnibus database (accession number GSE80303).
Affinity Purification, Mass Spectrometric Analysis, Co-IP, and Gel Filtration
For affinity purification, 6 g of flower tissue was ground to a fine powder and then suspended in lysis buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40, 0.5 mM DTT, 1 mM PMSF, and 1 protease inhibitor cocktail tablet/50 mL [Roche]). The supernatant obtained by centrifugation was incubated with anti-Flag antibody-conjugated beads (Sigma-Aldrich; A2220) for 2.5 h at 4°C. After the beads were vigorously washed several times, the proteins were eluted with 3xFLAG peptides (Sigma-Aldrich; F4799) and separated on a 7.5% SDS-PAGE gel. The gel was stained with the ProteoSilver Silver Stain Kit (Sigma-Aldrich PROT-SIL1), and the bands were then subjected to mass spectrometric analysis. For co-IP, the protein extracted from flower tissue or from seedlings was incubated with anti-Flag M1 agarose (Sigma-Aldrich; A2220) for affinity purification. After the sample was washed, it was boiled and subjected to SDS-PAGE and then to immunoblotting. For gel filtration, 0.5 g of seedlings was ground to a powder and suspended in 2 mL of lysis buffer. After centrifugation, the supernatant was passed through a 0.22-micron filter, and 500 μL of the filtrate was loaded onto a Superose 6 (10/300 GL) column (GE Healthcare; 17-5172-01). The eluate was collected in a series of fractions (500 μL/fraction) and run on SDS-PAGE gel for immunoblotting.
Yeast Two-Hybrid Assay
The cDNA sequences and the different truncated forms of MOM1, PIAL1, and PIAL2 were cloned into pGADT7 and pGBKT7 vectors in frame to the 3′-termini of GAL4-AD and GAL4-BD using the One-Step Cloning Kit (Vazyme Biotech), respectively. The DNA primers used for cloning are listed in Supplemental Data Set 8. The yeast strains AH109 and Y187 were transformed with pGADT7 and pGBKT7 constructs and grown on synthetic dropout medium lacking Leu and Trp, respectively. The positive clones from the synthetic dropout medium minus Leu were mated with the positive clones from the synthetic dropout medium minus Trp in YPD medium for 16 to 20 h. The mixture was spotted on synthetic dropout medium minus Leu and Trp. The positive yeast colonies were spotted on both the synthetic dropout medium minus Trp and Leu, and the synthetic dropout medium minus Trp, Leu, and His. Growth of transformed, positive yeast strains on SD-TLH indicates the interaction between the GAL-AD fusion protein and the GAL4-BD fusion protein in corresponding yeast strains. A 5 mM solution of 3-amino-1,2,4-triazole was used to inhibit the background growth of transformed strains on the synthetic dropout medium minus Trp, Leu, and His.
Protein Induction and Pull-Down Assays in Bacteria
PIAL1-IND, PIAL2-IND, and MOM1-CMM2 were fused with 5′-terminal GST or 6xHIS in the constructs pGEX-6P-1 and pET28a+, respectively. The primers used for the construction are listed in Supplemental Data Set 8. The constructs expressing both GST- and HIS-tagged proteins were cotransformed into the Escherichia coli expression strain Transetta (DE3). After selection on solid Luria-Bertani (LB) medium containing both kanamycin and ampicillin (Amresco; both at 50 mg/L), a single colony was transferred to 5 mL of LB liquid medium containing antibiotics (50 mg/L kanamycin, 50 mg/L ampicillin, and 34 mg/L chloromycetin) and grown at 37°C with rapid shaking (>200 rpm) overnight. A 1-mL volume of the LB was then added to 100 mL of LB for growth under the same conditions. When the OD600 of the culture increased to 0.4 to 0.5, 1 mM IPTG was added and then the culture was shaken continuously at 18°C for another 24 h for protein induction.
The bacteria in the culture were collected by centrifugation, and 4 mL of the bacterial culture was suspended in 1.5 mL of protein extraction buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10% glycerol, 0.5% Tween 20, 15 mM β-mercaptoethanol, 1 mM PMSF, and 1 protease inhibitor cocktail tablet/50 mL [Roche]). The sample was sonicated four times (10 s on and 59 s off) and then centrifuged at 13,000 rpm for 10 min at 4°C. A 1.4-mL volume of the supernatant was transferred to a 1.5-mL Eppendorf tube. Of this supernatant, 200 μL was used as input, and 1.2 mL was incubated with 50 μL of GST-beads for a pull-down assay. After incubation for 1.5 h with gentle rotation at 4°C, the sample was centrifuged at 2000 rpm for 1 min at 4°C. The supernatant was discarded and the GST beads were washed six times (5 min per time) with 1.5 mL of protein extraction buffer at 4°C. After the last wash, 200 μL of protein extraction buffer and 50 μL of 5× SDS sample buffer were added to the tube. The samples were boiled for 6 min at 100°C and separated on 15% SDS-PAGE gels for immunoblotting with GST antibody (Abmart; 12G8) and HIS antibody (Abmart; 10E2).
In Vitro Sumoylation Assay
All proteins used in the in vitro sumoylation assay were expressed and purified in E. coli using Rosetta (DE3). SUMO1 was expressed in pET28a vector with a C-terminal His tag and was purified with Ni2+ beads, followed by removal of His tag with Ulp1 enzyme. SAE1b was expressed in pETDuet-1 vector with no tags, while SAE2 was expressed in a modified pET28a vector with an N-terminal His tag followed by a PreScission site. The two subunits of E1 enzyme (SAE1b and SAE2) were coexpressed in Rosetta (DE3) and purified with Ni2+ beads followed by His tag removal using PreScission protease. The E2 enzyme SCE1 was expressed in a modified pET28a vector with a His tag followed by PreScission site in the N terminus and was purified with Ni2+ beads followed by His tag removal. PIAL2 and its mutants were tagged with hexa-His in the N terminus and were purified with Ni2+ beads.
The in vitro sumoylation reaction buffer contains 50 mM Tris-HCl (pH 7.6), 100 mM NaCl, 15% glycerol, 5 mM ATP, and 5 mM MgCl2. For the assay, 8 µg SUMO1 was incubated with 0.5 µg E1 enzyme, 4 µg E2 enzyme, and 1 µg PIAL2 or its mutant for 2 h at 37°C. The total volume of the reaction was 50 µL. The samples were then separated by SDS-PAGE and analyzed by immunoblotting for detection of sumoylated bands.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PIAL1 (AT1G08910), PIAL2 (AT5G41580), MOM1 (AT1G08060), NRPD1 (AT1G63020), NRPE1 (AT2G40030), DRD1 (AT2G16390), BDT1 (AT4G11560), PHD1 (AT1G43770), FWA (AT4G25530), SDC (AT2G17690), ROS1 (AT2G36490), MORC6 (AT1G19100), AGO4 (AT2G27040), RDR2 (AT4G11130), NRPD4 (AT4G15950), RDM4 (AT2G30280), NRPD2 (AT3G23780), MET1 (AT5G49160), FPGS1 (AT5G05980), FAS1 (AT1G65470), and BRU1 (AT3G18730).
Supplemental Data
Supplemental Figure 1. The FWA-LUC reporter system.
Supplemental Figure 2. Luminescence imaging of pFWA-LUC transgenic plants.
Supplemental Figure 3. Map-based cloning and characterization of PIAL2.
Supplemental Figure 4. Alignment of PIAL1 and PIAL2 in Arabidopsis, PIAS2 in human, and SU(VAR)2-10 in Drosophila.
Supplemental Figure 5. Alignment of Arabidopsis PIAL1 and PIAL2 and their homologs in other plants including Glycine max, Brassica napus, Vitis vinifera, Populus trichocarpa, Oryza sativa, and Sorghum bicolor.
Supplemental Figure 6. Validation of the transcript levels of the MOM1, PIAL1/2, and/or NPRE1 target loci in the wild type, nrpe1, mom1, and pial1/2.
Supplemental Figure 7. Analyses of hypo-DMRs in nrpe1, mom1, and pial1/2.
Supplemental Figure 8. Box plots showing CG, CHG, and CHH methylation of transcriptionally upregulated TEs and genes in nrpe1, mom1, and pial1/2 relative to the wild type.
Supplemental Figure 9. Scatterplots showing CG, CHG, and CHH methylation of transcriptionally upregulated TEs or genes in nrpe1, mom1, and pial1/2 relative to the wild type.
Supplemental Figure 10. The expression levels of the wild-type and mutated PIAL2 transgenes in the pial2 mutant.
Supplemental Table 1. Full list of mutants identified by map-based cloning in this study.
Supplemental Data Set 1. RNA transcript and DNA methylation levels of differentially expressed TEs in nrpe1.
Supplemental Data Set 2. RNA transcript and DNA methylation levels of differentially expressed TEs in mom1.
Supplemental Data Set 3. RNA transcript and DNA methylation levels of differentially expressed TEs in pial1/2.
Supplemental Data Set 4. RNA transcript and DNA methylation levels of differentially expressed genes in nrpe1.
Supplemental Data Set 5. RNA transcript and DNA methylation levels of differentially expressed genes in mom1.
Supplemental Data Set 6. RNA transcript and DNA methylation levels of differentially expressed genes in pial1/2.
Supplemental Data Set 7. Full list of proteins identified by mass spectrometric analyses in affinity purification of MOM1 and PIAL2.
Supplemental Data Set 8. DNA oligonucleotides used in this study.
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program; 2012CB910900) and the 973 Program (2011CB812600) from the Chinese Ministry of Science and Technology.
AUTHOR CONTRIBUTIONS
Y.-F.H., Q.-Y.Z., L.-L.D., and X.-J.H. conceived and designed the experiments. Y.-F.H., Q.-Y.Z., L.-L.D., Y.-X.L., S.-S.C., C.-R.S., L.L., and S.C. performed the experiments. Y.-F.H., Q.-Y.Z., L.-L.D., H.-W.H., Y.-Q.L., T.C., and X.-J.H. analyzed the data. Y.-F.H., Q.-Y.Z., and X.-J.H. wrote the manuscript.
Glossary
- siRNA
small interfering RNA
- RdDM
RNA-directed DNA methylation
- TE
transposable element
- DMR
differentially methylated region
- co-IP
coimmunoprecipitation
- MS
Murashige and Skoog
- LB
Luria-Bertani
References
- Ahmed I., Sarazin A., Bowler C., Colot V., Quesneville H. (2011). Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 39: 6919–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedeo P., Habu Y., Afsar K., Mittelsten Scheid O., Paszkowski J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405: 203–206. [DOI] [PubMed] [Google Scholar]
- Aufsatz W., Mette M.F., van der Winden J., Matzke A.J., Matzke M. (2002). RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 (suppl 4.): 16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T., Pontvianne F., Cocklin R., Podicheti R., Chandrasekhara C., Yerneni S., Braun C., Lee B., Rusch D., Mockaitis K., Tang H., Pikaard C.S. (2014). A two-step process for epigenetic inheritance in Arabidopsis. Mol. Cell 54: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhiraja R., Hermkes R., Müller S., Schmidt J., Colby T., Panigrahi K., Coupland G., Bachmair A. (2009). Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. 149: 1529–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caikovski M., Yokthongwattana C., Habu Y., Nishimura T., Mathieu O., Paszkowski J. (2008). Divergent evolution of CHD3 proteins resulted in MOM1 refining epigenetic control in vascular plants. PLoS Genet. 4: e1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. (2002). Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12: 1138–1144. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Zilberman D., Xie Z., Johansen L.K., Carrington J.C., Jacobsen S.E. (2004). RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chung C.D., Liao J., Liu B., Rao X., Jay P., Berta P., Shuai K. (1997). Specific inhibition of Stat3 signal transduction by PIAS3. Science 278: 1803–1805. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C., Matunis M.J. (2013). SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell 24: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Johnson L.M., Jacobsen S.E., Patel D.J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs M.L., Bender J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N., Bonequi M.V., Porri A., Coupland G. (2013). Identification of Arabidopsis SUMO-interacting proteins that regulate chromatin activity and developmental transitions. Proc. Natl. Acad. Sci. USA 110: 19956–19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau J.R., Lima C.D. (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., David L., Zhu J.K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814. [DOI] [PubMed] [Google Scholar]
- Haag J.R., Pikaard C.S. (2011). Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12: 483–492. [DOI] [PubMed] [Google Scholar]
- Habu Y., Mathieu O., Tariq M., Probst A.V., Smathajitt C., Zhu T., Paszkowski J. (2006). Epigenetic regulation of transcription in intermediate heterochromatin. EMBO Rep. 7: 1279–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Pontes O., Zhu J., Lu J., Bressan R.A., Pikaard C., Wang C.S., Zhu J.K. (2009). NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 23: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120. [DOI] [PubMed] [Google Scholar]
- Huang L., Jones A.M., Searle I., Patel K., Vogler H., Hubner N.C., Baulcombe D.C. (2009). An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat. Struct. Mol. Biol. 16: 91–93. [DOI] [PubMed] [Google Scholar]
- Huettel B., Kanno T., Daxinger L., Aufsatz W., Matzke A.J., Matzke M. (2006). Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 25: 2828–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Fujiwara S., Miura K., Stacey N., Yoshimura M., Schneider K., Adachi S., Minamisawa K., Umeda M., Sugimoto K. (2009). SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560. [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Gupta A.A. (2001). An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744. [DOI] [PubMed] [Google Scholar]
- Jones L., Ratcliff F., Baulcombe D.C. (2001). RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11: 747–757. [DOI] [PubMed] [Google Scholar]
- Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J. (2005). Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 37: 761–765. [DOI] [PubMed] [Google Scholar]
- Kinoshita Y., Saze H., Kinoshita T., Miura A., Soppe W.J., Koornneef M., Kakutani T. (2007). Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 49: 38–45. [DOI] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Walker J.M., Smalle J., Gosink M.M., Davis S.J., Durham T.L., Sung D.Y., Vierstra R.D. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278: 6862–6872. [DOI] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Liu B., Liao J., Rao X., Kushner S.A., Chung C.D., Chang D.D., Shuai K. (1998). Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95: 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Mosher R.A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Miller M.J., Barrett-Wilt G.A., Hua Z., Vierstra R.D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.J., Scalf M., Rytz T.C., Hubler S.L., Smith L.M., Vierstra R.D. (2013). Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol. Cell. Proteomics 12: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Hasegawa P.M. (2010). Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 20: 223–232. [DOI] [PubMed] [Google Scholar]
- Nathan D., Ingvarsdottir K., Sterner D.E., Bylebyl G.R., Dokmanovic M., Dorsey J.A., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B., Johnson E.S., Berger S.L. (2006). Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 20: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Molinard G., Petty T.J., Broger L., Gabus C., Halazonetis T.D., Thore S., Paszkowski J. (2012). Structural basis of transcriptional gene silencing mediated by Arabidopsis MOM1. PLoS Genet. 8: e1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numa H., et al. (2010). Transduction of RNA-directed DNA methylation signals to repressive histone marks in Arabidopsis thaliana. EMBO J. 29: 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37: D987–D991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J., Shi Y., Valin A., Xuan Y., Gill G. (2009). Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell 34: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Li C.F., Costa Nunes P., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. (2006). The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92. [DOI] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. (2005). Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19: 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Fransz P.F., Paszkowski J., Mittelsten Scheid O. (2003). Two means of transcriptional reactivation within heterochromatin. Plant J. 33: 743–749. [DOI] [PubMed] [Google Scholar]
- Ream T.S., Haag J.R., Wierzbicki A.T., Nicora C.D., Norbeck A.D., Zhu J.K., Hagen G., Guilfoyle T.J., Pasa-Tolić L., Pikaard C.S. (2009). Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell 33: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M.J., Galbiati M., Ticknor C., Chen J., Dellaporta S.L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657. [DOI] [PubMed] [Google Scholar]
- Saracco S.A., Miller M.J., Kurepa J., Vierstra R.D. (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y., Eisenman R.N. (2003). Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100: 13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.A., Choi E.S., Kim H.S., Ho J.C., Watts F.Z., Park S.D., Jang Y.K. (2005). SUMO modification is involved in the maintenance of heterochromatin stability in fission yeast. Mol. Cell 19: 817–828. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285. [DOI] [PubMed] [Google Scholar]
- Soppe W.J., Jacobsen S.E., Alonso-Blanco C., Jackson J.P., Kakutani T., Koornneef M., Peeters A.J. (2000). The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6: 791–802. [DOI] [PubMed] [Google Scholar]
- Steimer A., Amedeo P., Afsar K., Fransz P., Mittelsten Scheid O., Paszkowski J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12: 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stielow B., Sapetschnig A., Krüger I., Kunert N., Brehm A., Boutros M., Suske G. (2008). Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol. Cell 29: 742–754. [DOI] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomä N.H., Czyzewski B.K., Alexeev A.A., Mazin A.V., Kowalczykowski S.C., Pavletich N.P. (2005). Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 12: 350–356. [DOI] [PubMed] [Google Scholar]
- Tomanov K., Zeschmann A., Hermkes R., Eifler K., Ziba I., Grieco M., Novatchkova M., Hofmann K., Hesse H., Bachmair A. (2014). Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26: 4547–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I., Schubert I., Tourmente S., Mathieu O. (2006). MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep. 7: 1273–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokthongwattana C., Bucher E., Caikovski M., Vaillant I., Nicolet J., Mittelsten Scheid O., Paszkowski J. (2010). MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 29: 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Du J., Hale C.J., Gallego-Bartolome J., Feng S., Vashisht A.A., Chory J., Wohlschlegel J.A., Patel D.J., Jacobsen S.E. (2014). Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.